- 1Institute of Dermatology and Department of Dermatology, The First Affiliated Hospital, Anhui Medical University, Hefei, China
- 2Key Laboratory of Dermatology, Anhui Medical University, Ministry of Education, Hefei, China
Antineutrophil cytoplasmic antibody (ANCA)-associated vasculitis (AAV) is characterized by the inflammation of small and medium vessels and presence of proteinase 3-ANCA or myeloperoxidase-ANCA in the circulation. AAV comprises three clinical subtypes: granulomatosis with polyangiitis (GPA), microscopic polyangiitis (MPA), and eosinophilic GPA (EGPA). Although the pathogenesis of AAV is still unclear, genetic and environmental factors and the immune system are thought to be involved. Genetic factors have been confirmed to play an important role in AAV. Genome-wide association studies have identified numerous genetic variants in MHC and non-MHC regions associated with AAV. The strongest evidence of MHC association in AAV is human leukocyte antigen (HLA)-DP. A significant association between AAV and genetic variations in non-MHC regions, such as CTLA-4, FCGR2A, PTPN22, SERPINA1, and TLR9 has also been found. Moreover, different clinical subtypes of AAV have distinct genetic backgrounds. GPA is associated with HLA-DP1, MPA with HLA-DQ, and EGPA with HLA-DRB4. These findings could help elucidate the etiology of AAV and develop new biomarkers for diagnosis and targeted therapy. Herein, we briefly summarize the updates on the genetic pathogenesis and biomarkers of AAV.
Introduction
Antineutrophil cytoplasmic antibody (ANCA)-associated vasculitis (AAV) is a complex systemic autoimmune disease presenting with the inflammation of small and medium vessels that results in vascular destruction and tissue necrosis (1, 2). AAV is divided into three groups according to clinical features: granulomatosis with polyangiitis (GPA, formerly called Wegener’s granulomatosis), microscopic polyangiitis (MPA), and eosinophilic GPA (EGPA, formerly called Churg-Strauss syndrome) (3). The disease is characterized by the presence of proteinase 3 (PR3)-ANCA or myeloperoxidase (MPO)-ANCA in the serum. GPA is predominantly associated with PR3-ANCA, while MPA and EGPA are predominantly associated with MPO-ANCA, but are also occasionally ANCA-negative (4). EGPA is often divided into two subtypes, MPO-ANCA+ EGPA and ANCA– EGPA. Vasculitis can occur in any organ or tissue, commonly affecting the respiratory tract and kidneys, causing life-threatening kidney failure or pulmonary hemorrhage (3).
AAV has an estimated prevalence of 48–184/1,000,000 individuals worldwide, while the incidence and prevalence in Europe are higher than those in other regions (2). The annual incidence in Europe is 4.9–10.6/1,000,000 individuals for GPA, 2.7–11.6/1,000,000 individuals for MPA, and 0.5–3.1/1,000,000 individuals for EGPA. The epidemiological manifestations of AAV differ among geographical regions; GPA and PR3-ANCA AAV are more common in Europeans, while MPA and MPO-ANCA AAV are more common in Asians (5).
Although the pathogenesis of AAV remains elusive, it is believed that both genetic and environmental components are involved. Environmental factors, including silica exposure, bacterial or viral infections, and drugs suggest an association with the occurrence and relapse of AAV (6). Familial studies and genetic association studies have demonstrated that AAV has a background of genetic susceptibility (7–9). It has been estimated that 20% of AAV risk is due to genetic factors (10).
In this review, we mainly discuss the genetic studies on AAV, focusing on the identified susceptibility genes or loci to enrich our understanding of the disease.
Genetic Approaches for AAV
Different genetic approaches have revealed that a large number of genes are related to AAV. First, several candidate gene association studies have been used to identify susceptibility genes associated with AAV. Although these approaches are easy to carry out, they are prone to false-positive results. Human leukocyte antigen (HLA), PTPN22, CTLA-4, IL-10, and TLR9 have been found to be associated with AAV (11, 12). The emergence of genome-wide association studies (GWAS) has led to their wide use in exploring the genetic factors involved in various diseases. GWAS is a powerful approach to identify the genetic architecture of complex diseases, and it has made major contributions towards a better understanding of AAV genetics. Four GWAS have been performed in patients with AAV since 2012 (two for MPO and GPA (13, 14), one for GPA (15), and one for EGPA (16)). The latest GWAS was on EGPA in 676 cases and 6809 controls with the identification of 11 loci associated with EGPA. The study also revealed that EGPA comprises genetically distinct subgroups, that MPO-ANCA+ subgroup was strongly associated with HLA-DQ and MPO-ANCA- subgroup was associated with non-HLA regions, such as GPA33 and IL5/IRF1 (16). All these GWAS studies were conducted in populations with European ancestry and have identified more than 20 genes associated with AAV. The findings of GWAS are listed in Tables 1 and 2. In addition, meta-analysis and fine mapping have also been used to identify new gene variations and analyze the effect of the variants (17–21).
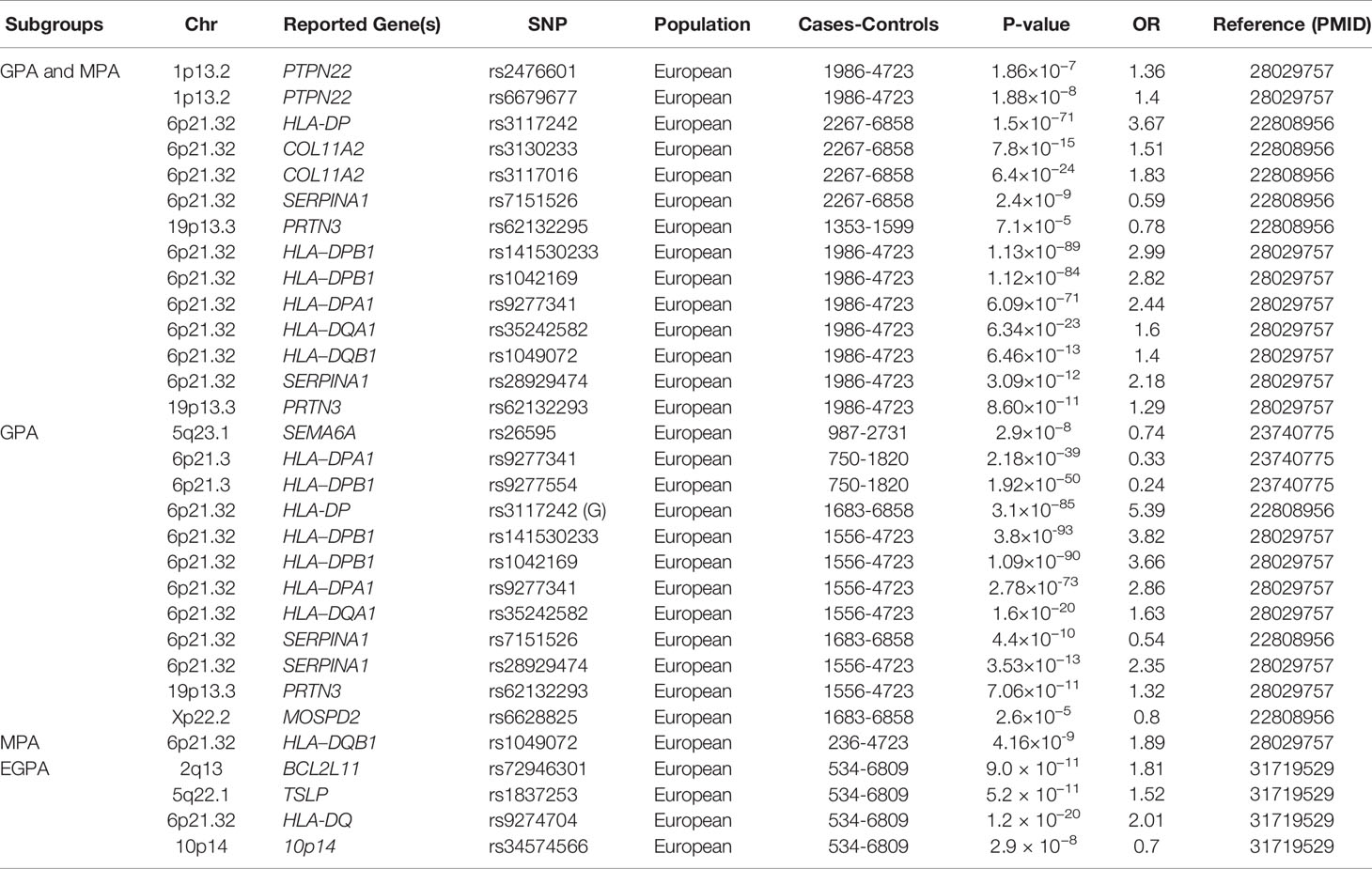
Table 1 A summary of main genetic associations with antineutrophil cytoplasmic antibody (ANCA)-associated vasculitis through genome-wide association studies, according to Clinical subgroups.
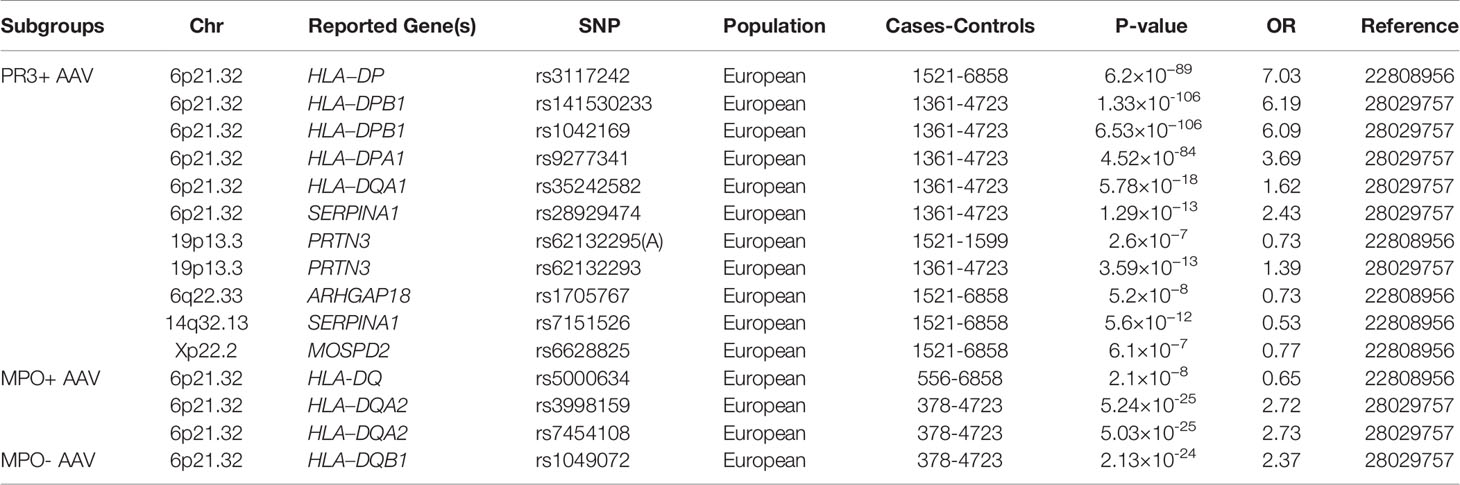
Table 2 Summary of main genetic associations with antineutrophil cytoplasmic antibody (ANCA)-associated vasculitis through genome-wide association studies, according to ANCA subgroups.
AAV susceptibility genes
HLA Region
HLA
HLA, located on chromosome 6p21, is the most gene-dense region of the human genome and contains diverse genes involved in key immune responses (22). Molecules encoded by HLA participate in a variety of immune and inflammatory pathways. HLA single-nucleotide polymorphisms (SNPs) are associated with numerous human diseases, especially autoimmune diseases, such as type 1 diabetes, rheumatoid arthritis (RA), and systemic lupus erythematosus (SLE) (23). Since AAV is considered an autoimmune disease, HLA may also be a potential predisposing factor for AAV. Candidate gene studies in HLA regions were performed in different populations, including the Swedish, Germans, and Italians in Europe and Japanese and Chinese in Asia. HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQB1, and HLA-DRB1 were found to be involved in AAV susceptibility.
(1). HLA and AAV
Two GWAS were performed to identify the genetic factors associated with AAV. The first GWAS in a European population in 2012 demonstrated that HLA-DP rs3117242 (G) was the strongest signal in the HLA region (P = 1.5×10−71, OR = 3.67) (13). Another GWAS in Canadian and American populations conducted in 2017 showed that SNPs rs141530233 and rs1042169 at HLA-DPB1 had the largest effect on the risk of developing AAV (P = 1.13×10−89, OR = 2.99; P = 1.12×10-84, OR = 2.82, respectively). HLA-DPA1, HLA-DQA1, and HLA-DQB1 are also risk alleles that may cause AAV (14). These studies not only indicated a highly significant association between AAV and HLA regions but also showed genetic distinctions between different clinical phenotypes and ANCA specificity. GPA and PR3-ANCA AAV are associated with HLA-DPB1 and HLA-DPA1, while MPA and MPO-ANCA AAV are associated with HLA-DQB1 and HLA-DQA2.
(2). HLA and GPA
GWAS for GPA with 492 patients and 1,506 healthy individuals of European descent identified 32 SNPs across the HLA region; among them, HLA-DPB1 rs9277554 and HLA-DPA1 rs9277341 were significantly associated with GPA in the combined cohort (15). In another study with 150 GPA patients and 100 healthy controls conducted in northern Germany, HLA-DPB1*0401 was identified to be associated with GPA (P = 1.51 × 10-10, OR = 3.91), and DPB1*0401/RXRB03 haplotype frequency was significantly increased in patients with GPA (P = 7.13 × 10-17, OR = 6.41) (18). The result was replicated in an independent German cohort with 108 patients with GPA (P = 6.4 × 10-8) (20). HLA-DPB1 was also considered a genetic risk factor for GPA in a cohort of 176 Han Chinese patients with AAV (100 with GPA, 76 with MPA) and 485 healthy controls (P = 1.83 × 10-5, OR = 2.57). In this study, the result also showed that the HLA-DPB1 SNP rs3117242 variant T allele was significantly associated with GPA patients (P = 6.24 × 10-5, OR = 2.09), but not with MPA patients (24).
(3). HLA and MPA
There was no specific GWAS study for MPA, but the clinical subgroup analysis of AAV GWAS demonstrated that HLA-DQB1 was significantly associated with MPA (P = 4.16×10−9, OR = 1.89) (14). A significant association of HLA-DRB1*0901 with MPA (P = 0.0037, OR = 2.44) and MPO-AAV (P = 0.0014, OR = 2.44) was demonstrated in 50 MPA and 64 MPO-ANCA AAV cases compared with 265 controls in a Japanese cohort (25). The result was confirmed with new observations that DRB1*13:02 was associated with protection against MPA and MPO-ANCA AAV in 468 Japanese patients with AAV and 596 controls(P = 2.1 × 10-4, OR = 1.57) (26). In a small cohort of 50 patients with MPA and 77 Japanese controls, DQB1*0303 was significantly associated with MPA (P = 0.017, OR = 2.35) (27). A recent study genotyped HLA-DRB1, DQA1, DQB1, DPB1, and HLA-DP in 258 patients with MPO-AAV and 597 healthy controls in the Chinese population and found that HLA-DQA1*0302 (P = 3.45 × 10-9, OR = 2.34) and DQB1*0303 (P = 3.26 × 10-9, OR = 1.89) were risk alleles for MPO-ANCA AAV (28).
(4). HLA and EGPA
A recent GWAS for EGPA including 676 EGPA cases and 6,809 controls in European populations showed that HLA-DQ was strongly associated with EGPA at genome-wide significance (P = 1.2 × 10−20,OR = 2.01), especially MPO -ANCA EGPA (P = 1.1 × 10−28, OR = 5.68) (16). This result was consistent with a study comprising 102 German patients with EGPA (P = 2.00×10−5, OR = 1.87) (29) as well as another study with 48 Italian patients (P = 2.32 × 10−4, OR = 2.49) (30). The Italian study also revealed that HLA-DRB1*07 allele frequency was significantly higher in patients with EGPA than in controls (P = 0. 42 × 10−2, OR = 2.42) (30).
Furthermore, HLA alleles were associated with the severity, prognosis, and relapse of AAV. HLA-DRB1*04:05 was associated with poor renal prognosis; HLA-DRB1*04:02 was associated with high mortality (31); and DPB1*04:01 was significantly associated with an increased risk of relapse (32).
AAV is an autoimmune disease. The identification of HLA-alleles associated with AAV confirm the centrality of auto-reactivity in the development of AAV, help us make a better understanding about its autoimmune pathogenesis, and point to logical therapeutic strategies. The identification of HLA-alleles not only can help us to distinguish from cases of GPA, MPA and EGPA and their subtypes, but also to evaluate and predict the severity, prognosis, and relapse of AAV.
Retinoid X Receptor Beta
RXRB, located on 6p21.32, encodes a member of the retinoid X receptor family of nuclear receptors. Strong associations of RXRB with GPA were revealed through the markers located in the HLA region and RXRB loci. Because RXRB located in the HLA region near HLA-DPB1, HLA-DPB1*0401/RXRB03 haplotype was found to be strongly associated with GPA (P = 7.13 × 10-17, OR = 6.41) (18). RXRB SNP rs6531 (P = 5.20 × 10-5, OR = 1.88) was significantly increased among patients with GPA compared to controls (19). The result was confirmed in a later meta-analysis, which included 140 genetic variants associated with AAV (17).
Ring Finger Protein 1
RING is also located in the HLA region, near HLA-DPB1. RING is a member of the polycomb repressive complex 1 that mediates histone H2A polyubiquitination and monoubiquitination, regulating its gene expression. Three SNPs in the region near RING1 (rs213210, rs213209, and rs213208) were found to be associated with GPA in German patients (20). The HLA-DPB1/RING1 haplotype is strongly associated with GPA in ANCA-positive subjects.
Non-HLA Region
Genes Encoding ANCA Associated Proteins
Serpin family A Member 1
SERPINA1, located on 14q32.13, encodes α1-antitrypsin, which is the major inhibitor of PR3 and is thought to limit the damage to local tissues (33). In AAV, the genetic variants of SERPINA1 may lead to the decreased function of α1-antitrypsin, potentially resulting in ANCA generation as PR3 accumulates in tissues, causing the inflammation of blood vessels. The first AAV GWAS reported that the SERPINA1 Z allele along with the rs7151526 risk allele were significantly associated with AAV and were the most prominent non-HLA regions associated with both PR3-ANCA and GPA subgroups (13). Another GWAS confirmed the result with the identification of SERPINA1 rs28929474, which was associated with AAV (P = 3.09 × 10−12, OR = 2.18), especially associated with GPA (P = 3.53 × 10−13, OR = 2.35) (14). Several studies have investigated the role of the Z allele in AAV and showed that heterozygous patients for the Z variant of SERPINA1 have an increased risk of developing GPA than the general population (34, 35).
Proteinase 3 (PRTN3)
PRTN3, located on 19p13.3, encodes PR3. PR3 is a neutrophil intracellular protease that is the main antigen of ANCA autoantibodies. PR3 is located on the plasma membrane of a subset of neutrophils and stored in the neutrophil azurophilic granules. The membrane-bound form interacts directly with ANCA (33), decreasing neutrophil activation and endothelial adhesion.
In a Swedish cohort of 79 patients with GPA and 129 controls, the coding and promoter sequences of PRTN3 were investigated. An association with GPA was demonstrated by PRTN3 A564G variation in the promoter region affecting a transcription factor-binding site (P < 0.00001, OR = 4.2) (36). The association of PRTN3 has been reported in two GWAS of AAV. Subgroup analysis revealed that PRTN3 SNP rs62132295 and rs62132293 were significantly associated with AAV, especially PR3-AAV or GPA (P = 2.6 × 10−5, OR = 0.78; P = 8.60 × 10−11, OR = 1.29) (13, 14). As observed for the above-mentioned HLA association, the strength of the PRTN3 SNP signal increased in the PR3-ANCA-positive subgroup, independent of the clinical diagnosis (P = 2.6 × 10−7, OR = 0.73). No association between PRTN3 and patients with MPA or MPO-AAV was found, which indicates that PRTN3 only plays a crucial role in the pathogenesis of anti-PR3-positive AAV.
Immunoregulatory Genes for AAV
Protein Tyrosine Phosphatase, Non-Receptor Type 22
PTPN22, located on 1p13.2, encodes lymphoid tyrosine phosphatase (Lyp), a member of the non-receptor class 4 subfamily of the protein tyrosine phosphatase family. PTPN22 620W variation causes the substitution of arginine with tryptophan at amino acid residue 620, disturbing the function of PTPN22, which is involved in B-cell receptor and T cell receptor signaling, causing autoimmune diseases. It is associated with type 1 diabetes, RA, SLE, and other autoimmune disorders (37, 38).
The 620W allele conferred susceptibility to AAV in a German cohort (P = 0.002, OR = 1.75), and the allele frequency was significantly increased in ANCA-positive GPA (P < 0.0002) (39). This result has been confirmed in a British cohort (P = 1.4 × 10-4, OR = 1.40) (40) and an Italian cohort (P = 0.005, OR = 1. 91) (41). A meta-analysis of four studies in 1,399 patients of European descent found the association between the PTPN22 620W allele and the occurrence and development of AAV (42).
Cytotoxic T Lymphocyte-Associated Antigen-4
CTLA4, located on 2q33.2, encodes an inhibitory glycoprotein expressed on activated T cells, which transmits an inhibitory signal to T cells. CTLA4 competes with CD28 to bind to CD80 and CD86, which function antagonistically. CTLA4 is a negative regulator of T cell activation and inhibits the immune response, while CD28 transmits a stimulatory signal. The expression of CTLA4 on CD4 T cells is increased in GPA (43). Mutations on this gene have been associated with Graves’ disease, celiac disease, SLE, and other autoimmune diseases.
Although the CTLA4 SNP did not show genome-wide association in the GWAS of AAV, the association with AAV was found in candidate gene studies in different populations. CTLA4 rs3087243 showed an association with AAV in 641 British AAV cases and 9,115 controls (P = 6.4 ×10-3, OR=0.84). CTLA4 exon 1 (+49) and 4 (CT60) polymorphisms showed an association with AAV in another independent British cohorts with 222 cases and 629 controls(P(+49) = 0.004, P (CT60) = 0.002, respectively) (40, 44). The similar result was confirmed in a Swedish cohort with 101 cases and 200 controls (P = 0.049) (45).
Toll-Like Receptors
TLRs, located on chromosome 3p21.2, encode a family of innate receptors whose specificities are predetermined in the germline. TLRs can recognize microbiological structures and activate immune responses that participate in the development of autoimmune diseases (46). Infection, particularly Staphylococcus aureus infection, is considered a potential trigger of AAV, and TLR9 signaling may be involved in disease pathology, favoring models of infectious agents triggering AAV development (47). SNPs in TLR9 were genotyped in a German cohort comprising 863 patients with AAV and 1,344 healthy controls to investigate the contribution of genetic polymorphisms of TLR9 to the susceptibility and clinical manifestation. Strong association of TLR9 genotypes and haplotypes with the subgroups of GPA and MPA was observed, but no association was observed with EGPA. In a cohort of 426 Dutch and British AAV cases, the findings were not replicated (21). TLR9 signaling may be involved in disease pathology, favoring models of infectious agents triggering AAV development.
Fc Gamma Receptors
FCGRs are a group of proteins expressed on the surface of different cell types with different affinities for the Fc portions of different IgG subclasses. The key role of Fc receptors is the regulation of inflammatory and immune responses. FCGR3B encodes FcgRIIIB, which is expressed by neutrophils and eosinophils and is important in the tethering of neutrophils to immune complexes and the clearance of immune complexes (48). The FCGR3B copy number may play a role in the development of SLE or other autoimmune diseases (49).
In a study consisted with two independent cohorts of individuals with GPA from Britain and France, strong association between FCGR3B copy numbers and risk of GPA was shown. In British patients, low FCGR3B copy number was found to be associated with GPA (P = 0.015, OR = 2.46), but in French GPA patients, the association with high FCGR3B copy number was identified (P = 0.002, OR = 0.28) (50). The study also explored the relationship between FCGR3B and MPA in a separate British cohort and observed a significant association between low FCGR3B copy number and MPA(P = 0.013, OR = 2.56). Later, the results were replicated in a large Chinese cohort with 139 AAV patients and 564 controls (P = 0.040, OR = 1.72) with the result that low FCGR3A CNVs were significantly associated with AAV susceptibility (P = 0.042, OR = 2.64) (51).
Possible associations of SNPs of FCGR2A, FCGR2B, FCGR3A and the FCGR3B haplotype NA1/NA2 and EGPA have been discussed in a recent study, which included 130 EGPA patients and 181 controls. FCGR3B haplotype NA1/NA2 was found to be associated with relapse-free survival of EGPA that relapse-free survival was significantly lower in carriers of FCGR3B haplotype NA1-NA1 than the others (P = 0.029). MPO-ANCA+ EGPA subgroup was also associated with relapse(P = 0.032) while no association was observed in MPO-ANCA- EGPA subgroup(P = 0.68) (52).
Interleukin-10
IL-10 is an anti-inflammatory cytokine produced by T helper cells, and its expression increases in patients with AAV (53). Two small studies in German and Caucasian cohorts proposed an association of IL-10 SNP (–1082) in GPA (54); interestingly, one study also demonstrated a signal for the IL-10 SNP in 36 patients with MPA (P < 1.00× 10−6) (55). In a large German cohort of 403 patients with GPA, 103 patients with EGPA and 507 controls, none of the IL-10 polymorphisms were associated with GPA, but IL-10 -3575/-1082/-592 TAC haplotype was highly significantly associated with ANCA-negative EGPA (P = 3.00 × 10−5, OR = 2.16) (56).
Interleukin 2 Receptor Subunit Alpha
IL2RA encodes the high-affinity IL-2 receptor, which is represented not only on the surface of T cells, but also on activated B cells, NK cells, and monocytes (57). The IL2-IL2RA pathway is critical for the physiological function of the immune system; survival, proliferation, and activation of T cells and development of a normal Treg repertoire (58). An association has been reported in a UK cohort of 670 patients with AAV for the IL2RA SNP rs41295061 (P = 1.22× 10−2) (59), but no correlation between IL12RA and AAV was found during the replication stage of the first GWAS validation.
Other Susceptible Genes
Defensin Beta 4A
DEFB4 is an antibiotic peptide. The copy number variation of DEFB4 reportedly causes susceptibility to inflammatory disorders (60). Higher DEFB4 copy number was demonstrated to be significantly higher in a small cohort of 112 Chinese patients with AAV than in controls, which supported the role of the defensin system in autoimmunity (61).
Rho GTPase-Activating Protein 18
ARHGAP18 belongs to a family of Rho GTPase-activating proteins that modulate cell signaling. The subgroup analysis of the first GWAS for AAV revealed that ARHGAP18 was associated with GPA (P = 3.3 × 10-7, OR = 0.84) and PR3-ANCA AAV (P = 5.2 ×10-8, OR = 0.87) (13).
BCL2-Like11 and MIR4435-2HG(MORRBID)
BCL2L11 encodes BIM, which belongs to the BCL-2 protein family and is crucial for controlling apoptosis, immune homeostasis, and autoimmune diseases (62, 63). MORRBID encodes a long non-coding RNA that regulates BIM transcription, controls eosinophil apoptosis, and is dysregulated in hypereosinophilic syndrome (64). In the GWAS of EGPA, SNPs near BCL2L11 and MORRBID were identified to be associated with EGPA (P = 9 × 10-11, OR = 1.66) (16).
Thymic Stromal Lymphopoietin
TSLP is located on 5q22.1. Its product, TSLP, is released by stromal and epithelial cells in response to inflammatory stimuli, which drives eosinophilia and enhances TH2 responses that are associated with immunity in various inflammatory diseases, including asthma, allergic inflammation, and chronic obstructive pulmonary disease (65). TSLP SNP rs1837253 which lies immediately upstream of TSLP was observed to be EGPA susceptibility variant (P = 5.2 × 10-11, OR = 1.42) (16).
ETS Proto-Oncogene 1
ETS1, located on chromosome 11q24.3, encodes a member of the ETS family of transcription factors. ETS1 inhibits the differentiation of Th17 cells and induces the development of Tregs. Genetic variation may cause an increase in Th17 in patients with GPA, causing inflammation of small vessels. A GWAS in a Chinese population reported that ETS1 SNP rs1128334 was significantly associated with SLE (66). In order to observe whether this gene was associated with other autoimmune diseases, a Japanese study which contained 466 patients with AAV and 1099 healthy controls was carried out. ETS1 SNP rs1128334 was genotyped and found to be associated with GPA (P = 0.030, OR = 1.54) and PR3-ANCA AAV (P = 0.021, OR = 1.72) (67).
Telomerase Reverse Transcriptase and Desmoplakin
TERT is the catalytic subunit of telomerase and contributes to the maintenance of telomere length (68). TERT rs2736100A is associated with shorter leukocyte telomere length (69). TERT also has an anti-apoptotic effect, and a decreased expression of TERT associated with the risk allele may result in the enhancement of apoptosis (70). With the evidence that a proportion of T cells show short telomeres in patients with GPA and an enhancement in neutrophil apoptosis in patients with AAV, TERT has been proposed as a susceptibility gene for AAV (71). In a Japanese cohort comprising 544 patients with AAV and 5558 controls, the frequency of TERT rs2736100A alleles was significantly increased in MPA (P = 2.3 × 10−2, OR= 1.38) and MPO-AAV (P = 0.15× 10−2, OR = 1.33) (72). DSP rs2076295G showed similar results to TERT rs2736100A (P = 0.69× 10-2, OR = 1.32) in MPO-AAV (P = 0.011, OR = 1.26) (72). DSP reportedly modulates Wnt/β-catenin signaling, which is involved in cell proliferation, differentiation, immune response, and carcinogenesis. Wnt signaling plays a role in autoimmune diseases and may also be associated with the onset of AAV.
All the genes associated with AAV were summarized in Table 3.

Table 3 Summary of genetic associations with antineutrophil cytoplasmic antibody (ANCA)-associated vasculitis excluding those derived from genome-wide association studies.
Conclusion
In the past 20 years, genetic studies, including GWAS and candidate gene studies, have identified numerous key loci that are associated with the risk of AAV. These genes have not only provided novel etiological clues to deepen our understanding of AAV but also novel therapeutic and prophylactic biomarkers for new approaches to treat and prevent AVV. Biomarker-targeting therapies and preventing recurrence in the future may be more effective. With the increasing number of GWAS being conducted, it is desirable to combine these findings and improve the statistical power. To date, GWAS for AAV have been performed in the European population; in the future, more GWAS with large samples in different populations and different subgroups of AAV are required to reveal the pathogenesis of the disease and identify reliable biomarkers for precision medicine.
Author Contributions
YS is the investigator for the review article and has contributed to the concept and planning of the article, collection of data, and reporting of the work described. WL, HH, MC, and TY contributed to the collection of data, and reporting of the work described. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank all the study participants in this study.
References
1. Hunter RW, Welsh N, Farrah TE, Gallacher PJ, Dhaun N. ANCA associated vasculitis. BMJ (2020) 369:m1070. doi: 10.1136/bmj.m1070
2. Kitching AR, Anders HJ, Basu N, Brouwer E, Gordon J, Jayne DR, et al. ANCA-associated vasculitis. Nat Rev Dis Primers (2020) 6:71. doi: 10.1038/s41572-020-0204-y
3. Berden A, Goceroglu A, Jayne D, Luqmani R, Rasmussen N, Bruijn JA, et al. Diagnosis and management of ANCA associated vasculitis. BMJ (2012) 344:e26. doi: 10.1136/bmj.e26
4. Ugarte-Gil MF, Espinoza LR. Genetics of ANCA-associated Vasculitides. Curr Rheumatol Rep (2014) 16:428. doi: 10.1007/s11926-014-0428-5
5. Berti A, Cornec D, Crowson CS, Specks U, Matteson EL. The Epidemiology of Antineutrophil Cytoplasmic Autoantibody-Associated Vasculitis in Olmsted County, Minnesota: A Twenty-Year US Population-Based Study. Arthritis Rheumatol (2017) 69:2338–50. doi: 10.1002/art.40313
6. Chen M, Kallenberg CG. The environment, geoepidemiology and ANCA-associated vasculitides. Autoimmun Rev (2010) 9:A293–8. doi: 10.1016/j.autrev.2009.10.008
7. Prendecki M, Cairns T, Pusey CD. Familial vasculitides: granulomatosis with polyangitis and microscopic polyangitis in two brothers with differing anti-neutrophil cytoplasm antibody specificity. Clin Kidney J (2016) 9:429–31. doi: 10.1093/ckj/sfw016
8. Papiha SS, Murty GE, Ad’Hia A, Mains BT, Venning M. Association of Wegener’s granulomatosis with HLA antigens and other genetic markers. Ann Rheum Dis (1992) 51:246–8. doi: 10.1136/ard.51.2.246
9. Bonatti F, Reina M, Neri TM, Martorana D. Genetic Susceptibility to ANCA-Associated Vasculitis: State of the Art. Front Immunol (2014) 5:577. doi: 10.3389/fimmu.2014.00577
10. Wallace ZS, Stone JH. Personalized Medicine in ANCA-Associated Vasculitis ANCA Specificity as the Guide? Front Immunol (2019) 10:2855. doi: 10.3389/fimmu.2019.02855
11. Salama AD. Genetics and pathogenesis of small-vessel vasculitis. Best Pract Res Clin Rheumatol (2018) 32:21–30. doi: 10.1016/j.berh.2018.10.002
12. Alberici F, Martorana D, Vaglio A. Genetic aspects of anti-neutrophil cytoplasmic antibody-associated vasculitis. Nephrol Dial Transplant (2015) 30 Suppl 1:i37–45. doi: 10.1093/ndt/gfu386
13. Lyons PA, Rayner TF, Trivedi S, Holle JU, Watts RA, Jayne DR, et al. Genetically distinct subsets within ANCA-associated vasculitis. N Engl J Med (2012) 367:214–23. doi: 10.1056/NEJMoa1108735
14. Merkel PA, Xie G, Monach PA, Ji X, Ciavatta DJ, Byun J, et al. Identification of Functional and Expression Polymorphisms Associated With Risk for Antineutrophil Cytoplasmic Autoantibody-Associated Vasculitis. Arthritis Rheumatol (2017) 69:1054–66. doi: 10.1002/art.40034
15. Xie G, Roshandel D, Sherva R, Monach PA, Lu EY, Kung T, et al. Association of granulomatosis with polyangiitis (Wegener’s) with HLA-DPB1*04 and SEMA6A gene variants: evidence from genome-wide analysis. Arthritis Rheum (2013) 65:2457–68. doi: 10.1002/art.38036
16. Lyons PA, Peters JE, Alberici F, Liley J, Coulson RMR, Astle W, et al. Genome-wide association study of eosinophilic granulomatosis with polyangiitis reveals genomic loci stratified by ANCA status. Nat Commun (2019) 10:5120. doi: 10.1038/s41467-019-12515-9
17. Rahmattulla C, Mooyaart AL, van Hooven D, Schoones JW, Bruijn JA, Dekkers OM, et al. Genetic variants in ANCA-associated vasculitis: a meta-analysis. Ann Rheum Dis (2016) 75:1687–92. doi: 10.1136/annrheumdis-2015-207601
18. Jagiello P, Gencik M, Arning L, Wieczorek S, Kunstmann E, Csernok E, et al. New genomic region for Wegener’s granulomatosis as revealed by an extended association screen with 202 apoptosis-related genes. Hum Genet (2004) 114:468–77. doi: 10.1007/s00439-004-1092-z
19. Szyld P, Jagiello P, Csernok E, Gross WL, Epplen JT. On the Wegener granulomatosis associated region on chromosome 6p21.3. BMC Med Genet (2006) 7:21. doi: 10.1186/1471-2350-7-21
20. Heckmann M, Holle JU, Arning L, Knaup S, Hellmich B, Nothnagel M, et al. The Wegener’s granulomatosis quantitative trait locus on chromosome 6p21.3 as characterised by tagSNP genotyping. Ann Rheum Dis (2008) 67:972–9. doi: 10.1136/ard.2007.077693
21. Husmann CA, Holle JU, Moosig F, Mueller S, Wilde B, Cohen Tervaert JW, et al. Genetics of toll like receptor 9 in ANCA associated vasculitides. Ann Rheum Dis (2014) 73:890–6. doi: 10.1136/annrheumdis-2012-202803
22. Ghodke Y, Joshi K, Chopra A, Patwardhan B. HLA and disease. Eur J Epidemiol (2005) 20:475–88. doi: 10.1007/s10654-005-5081-x
23. Matzaraki V, Kumar V, Wijmenga C, Zhernakova A. The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biol (2017) 18:76. doi: 10.1186/s13059-017-1207-1
24. Wu Z, Wu Q, Xu J, Chen S, Sun F, Li P, et al. HLA-DPB1 variant rs3117242 is associated with anti-neutrophil cytoplasmic antibody-associated vasculitides in a Han Chinese population. Int J Rheum Dis (2017) 20:1009–15. doi: 10.1111/1756-185X.12561
25. Tsuchiya N, Kobayashi S, Kawasaki A, Kyogoku C, Arimura Y, Yoshida M, et al. Genetic background of Japanese patients with antineutrophil cytoplasmic antibody-associated vasculitis: association of HLA-DRB1*0901 with microscopic polyangiitis. J Rheumatol (2003) 30:1534–40.
26. Kawasaki A, Hasebe N, Hidaka M, Hirano F, Sada KE, Kobayashi S, et al. Protective Role of HLA-DRB1*13:02 against Microscopic Polyangiitis and MPO-ANCA-Positive Vasculitides in a Japanese Population: A Case-Control Study. PloS One (2016) 11:e0154393. doi: 10.1371/journal.pone.0154393
27. Tsuchiya N, Kobayashi S, Hashimoto H, Ozaki S, Tokunaga K. Association of HLA-DRB1*0901-DQB1*0303 haplotype with microscopic polyangiitis in Japanese. Genes Immun (2006) 7:81–4. doi: 10.1038/sj.gene.6364262
28. Wang HY, Cui Z, Pei ZY, Fang SB, Chen SF, Zhu L, et al. Risk HLA class II alleles and amino acid residues in myeloperoxidase-ANCA-associated vasculitis. Kidney Int (2019) 96:1010–9. doi: 10.1016/j.kint.2019.06.015
29. Wieczorek S, Hellmich B, Gross WL, Epplen JT. Associations of Churg-Strauss syndrome with the HLA-DRB1 locus, and relationship to the genetics of antineutrophil cytoplasmic antibody-associated vasculitides: comment on the article by Vaglio et al. Arthritis Rheum (2008) 58:329–30. doi: 10.1002/art.23209
30. Vaglio A, Martorana D, Maggiore U, Grasselli C, Zanetti A, Pesci A, et al. HLA-DRB4 as a genetic risk factor for Churg-Strauss syndrome. Arthritis Rheum (2007) 56:3159–66. doi: 10.1002/art.22834
31. Chang DY, Luo H, Zhou XJ, Chen M, Zhao MH. Association of HLA genes with clinical outcomes of ANCA-associated vasculitis. Clin J Am Soc Nephrol (2012) 7:1293–9. doi: 10.2215/CJN.13071211
32. Hilhorst M, Arndt F, Joseph Kemna M, Wieczorek S, Donner Y, Wilde B, et al. HLA-DPB1 as a Risk Factor for Relapse in Antineutrophil Cytoplasmic Antibody-Associated Vasculitis: A Cohort Study. Arthritis Rheumatol (2016) 68:1721–30. doi: 10.1002/art.39620
33. Campbell EJ, Campbell MA, Owen CA. Bioactive proteinase 3 on the cell surface of human neutrophils: quantification, catalytic activity, and susceptibility to inhibition. J Immunol (2000) 165:3366–74. doi: 10.4049/jimmunol.165.6.3366
34. Elzouki AN, Segelmark M, Wieslander J, Eriksson S. Strong link between the alpha 1-antitrypsin PiZ allele and Wegener’s granulomatosis. J Intern Med (1994) 236:543–8. doi: 10.1111/j.1365-2796.1994.tb00842.x
35. Baslund B, Szpirt W, Eriksson S, Elzouki AN, Wiik A, Wieslander J, et al. Complexes between proteinase 3, alpha 1-antitrypsin and proteinase 3 anti-neutrophil cytoplasm autoantibodies: a comparison between alpha 1-antitrypsin PiZ allele carriers and non-carriers with Wegener’s granulomatosis. Eur J Clin Invest (1996) 26:786–92. doi: 10.1046/j.1365-2362.1996.2070553.x
36. Gencik M, Meller S, Borgmann S, Fricke H. Proteinase 3 gene polymorphisms and Wegener’s granulomatosis. Kidney Int (2000) 58:2473–7. doi: 10.1046/j.1523-1755.2000.00430.x
37. Lamsyah H, Rueda B, Baassi L, Elaouad R, Bottini N, Sadki K, et al. Association of PTPN22 gene functional variants with development of pulmonary tuberculosis in Moroccan population. Tissue Antigens (2009) 74:228–32. doi: 10.1111/j.1399-0039.2009.01304.x
38. Vang T, Congia M, Macis MD, Musumeci L, Orru V, Zavattari P, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet (2005) 37:1317–9. doi: 10.1038/ng1673
39. Jagiello P, Aries P, Arning L, Wagenleiter SE, Csernok E, Hellmich B, et al. The PTPN22 620W allele is a risk factor for Wegener’s granulomatosis. Arthritis Rheum (2005) 52:4039–43. doi: 10.1002/art.21487
40. Carr EJ, Niederer HA, Williams J, Harper L, Watts RA, Lyons PA, et al. Confirmation of the genetic association of CTLA4 and PTPN22 with ANCA-associated vasculitis. BMC Med Genet (2009) 10:121. doi: 10.1186/1471-2350-10-121
41. Martorana D, Maritati F, Malerba G, Bonatti F, Alberici F, Oliva E, et al. PTPN22 R620W polymorphism in the ANCA-associated vasculitides. Rheumatology (Oxford) (2012) 51:805–12. doi: 10.1093/rheumatology/ker446
42. Cao Y, Liu K, Tian Z, Hogan SL, Yang J, Poulton CJ, et al. PTPN22 R620W polymorphism and ANCA disease risk in white populations: a metaanalysis. J Rheumatol (2015) 42:292–9. doi: 10.3899/jrheum.131430
43. Steiner K, Moosig F, Csernok E, Selleng K, Gross WL, Fleischer B, et al. Increased expression of CTLA-4 (CD152) by T and B lymphocytes in Wegener’s granulomatosis. Clin Exp Immunol (2001) 126:143–50. doi: 10.1046/j.1365-2249.2001.01575.x
44. Kamesh L, Heward JM, Williams JM, Gough SC, Chavele KM, Salama A, et al. CT60 and +49 polymorphisms of CTLA 4 are associated with ANCA-positive small vessel vasculitis. Rheumatology (Oxford) (2009) 48:1502–5. doi: 10.1093/rheumatology/kep280
45. Persson U, Gullstrand B, Pettersson A, Sturfelt G, Truedsson L, Segelmark M. A candidate gene approach to ANCA-associated vasculitis reveals links to the C3 and CTLA-4 genes but not to the IL1-Ra and Fcgamma-RIIa genes. Kidney Blood Press Res (2013) 37:641–8. doi: 10.1159/000355744
46. Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol (2006) 6:823–35. doi: 10.1038/nri1957
47. Popa ER, Stegeman CA, Abdulahad WH, van der Meer B, Arends J, Manson WM, et al. Staphylococcal toxic-shock-syndrome-toxin-1 as a risk factor for disease relapse in Wegener’s granulomatosis. Rheumatology (Oxford) (2007) 46:1029–33. doi: 10.1093/rheumatology/kem022
48. Smith KG, Clatworthy MR. FcgammaRIIB in autoimmunity and infection: evolutionary and therapeutic implications. Nat Rev Immunol (2010) 10:328–43. doi: 10.1038/nri2762
49. Mueller M, Barros P, Witherden AS, Roberts AL, Zhang Z, Schaschl H, et al. Genomic pathology of SLE-associated copy-number variation at the FCGR2C/FCGR3B/FCGR2B locus. Am J Hum Genet (2013) 92:28–40. doi: 10.1016/j.ajhg.2012.11.013
50. Fanciulli M, Norsworthy PJ, Petretto E, Dong R, Harper L, Kamesh L, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet (2007) 39:721–3. doi: 10.1038/ng2046
51. Qi Y, Zhou X, Bu D, Hou P, Lv J, Zhang H. Low copy numbers of FCGR3A and FCGR3B associated with Chinese patients with SLE and AASV. Lupus (2017) 26:1383–9. doi: 10.1177/0961203317700485
52. Alberici F, Bonatti F, Adorni A, Daminelli G, Sinico RA, Gregorini G, et al. FCGR3B polymorphism predicts relapse risk in eosinophilic granulomatosis with polyangiitis. Rheumatology (Oxford) (2020) 59:3563–6. doi: 10.1093/rheumatology/keaa134
53. Wang Y, Zhang S, Zhang N, Feng M, Liang Z, Zhao X, et al. Reduced activated regulatory T cells and imbalance of Th17/activated Treg cells marks renal involvement in ANCA-associated vasculitis. Mol Immunol (2020) 118:19–29. doi: 10.1016/j.molimm.2019.11.010
54. Murakozy G, Gaede KI, Ruprecht B, Gutzeit O, Schurmann M, Schnabel A, et al. Gene polymorphisms of immunoregulatory cytokines and angiotensin-converting enzyme in Wegener’s granulomatosis. J Mol Med (Berl) (2001) 79:665–70. doi: 10.1007/s001090100263
55. Bartfai Z, Gaede KI, Russell KA, Murakozy G, Muller-Quernheim J, Specks U. Different gender-associated genotype risks of Wegener’s granulomatosis and microscopic polyangiitis. Clin Immunol (2003) 109:330–7. doi: 10.1016/s1521-6616(03)00211-0
56. Wieczorek S, Hellmich B, Arning L, Moosig F, Lamprecht P, Gross WL, et al. Functionally relevant variations of the interleukin-10 gene associated with antineutrophil cytoplasmic antibody-negative Churg-Strauss syndrome, but not with Wegener’s granulomatosis. Arthritis Rheum (2008) 58:1839–48. doi: 10.1002/art.23496
57. Caruso C, Candore G, Cigna D, Colucci AT, Modica MA. Biological significance of soluble IL-2 receptor. Mediators Inflamm (1993) 2:3–21. doi: 10.1155/S0962935193000018
58. Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat Immunol (2005) 6:1142–51. doi: 10.1038/ni1263
59. Carr EJ, Clatworthy MR, Lowe CE, Todd JA, Wong A, Vyse TJ, et al. Contrasting genetic association of IL2RA with SLE and ANCA-associated vasculitis. BMC Med Genet (2009) 10:22. doi: 10.1186/1471-2350-10-22
60. Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, Rodijk-Olthuis D, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet (2008) 40:23–5. doi: 10.1038/ng.2007.48
61. Zhou XJ, Cheng FJ, Lv JC, Luo H, Yu F, Chen M, et al. Higher DEFB4 genomic copy number in SLE and ANCA-associated small vasculitis. Rheumatology (Oxford) (2012) 51:992–5. doi: 10.1093/rheumatology/ker419
62. Bouillet P, Purton JF, Godfrey DI, Zhang LC, Coultas L, Puthalakath H, et al. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature (2002) 415:922–6. doi: 10.1038/415922a
63. Enders A, Bouillet P, Puthalakath H, Xu Y, Tarlinton DM, Strasser A. Loss of the pro-apoptotic BH3-only Bcl-2 family member Bim inhibits BCR stimulation-induced apoptosis and deletion of autoreactive B cells. J Exp Med (2003) 198:1119–26. doi: 10.1084/jem.20030411
64. Kotzin JJ, Spencer SP, McCright SJ, Kumar DBU, Collet MA, Mowel WK, et al. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature (2016) 537:239–43. doi: 10.1038/nature19346
65. Ziegler SF, Roan F, Bell BD, Stoklasek TA, Kitajima M, Han H. The biology of thymic stromal lymphopoietin (TSLP). Adv Pharmacol (2013) 66:129–55. doi: 10.1016/B978-0-12-404717-4.00004-4
66. Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PloS Genet (2010) 6:e1000841. doi: 10.1371/journal.pgen.1000841
67. Kawasaki A, Yamashita K, Hirano F, Sada KE, Tsukui D, Kondo Y, et al. Association of ETS1 polymorphism with granulomatosis with polyangiitis and proteinase 3-anti-neutrophil cytoplasmic antibody positive vasculitis in a Japanese population. J Hum Genet (2018) 63:55–62. doi: 10.1038/s10038-017-0362-2
68. Nagpal N, Agarwal S. Telomerase RNA processing: Implications for human health and disease. Stem Cells (2020). doi: 10.1002/stem.3270
69. Codd V, Nelson CP, Albrecht E, Mangino M, Deelen J, Buxton JL, et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet (2013) 45:422–7, 427e1-2. doi: 10.1038/ng.2528
70. Sung YH, Choi YS, Cheong C, Lee HW. The pleiotropy of telomerase against cell death. Mol Cells (2005) 19:303–9.
71. Vogt S, Iking-Konert C, Hug F, Andrassy K, Hansch GM. Shortening of telomeres: Evidence for replicative senescence of T cells derived from patients with Wegener’s granulomatosis. Kidney Int (2003) 63:2144–51. doi: 10.1046/j.1523-1755.2003.00037.x
72. Kawasaki A, Namba N, Sada KE, Hirano F, Kobayashi S, Nagasaka K, et al. Association of TERT and DSP variants with microscopic polyangiitis and myeloperoxidase-ANCA positive vasculitis in a Japanese population: a genetic association study. Arthritis Res Ther (2020) 22:246. doi: 10.1186/s13075-020-02347-0
Keywords: vasculitis, antineutrophil cytoplasmic antibody, genetic, genome-wide association studies, variation
Citation: Li W, Huang H, Cai M, Yuan T and Sheng Y (2021) Antineutrophil Cytoplasmic Antibody-Associated Vasculitis Update: Genetic Pathogenesis. Front. Immunol. 12:624848. doi: 10.3389/fimmu.2021.624848
Received: 01 November 2020; Accepted: 11 March 2021;
Published: 26 March 2021.
Edited by:
Alexandre Wagner Silva De Souza, Federal University of São Paulo, BrazilReviewed by:
Jan Stephan Sanders, University Medical Center Groningen, NetherlandsRenato Alberto Sinico, University of Milano-Bicocca, Italy
Copyright © 2021 Li, Huang, Cai, Yuan and Sheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yujun Sheng, YWhtdXN5akAxNjMuY29t
 Weiran Li
Weiran Li He Huang
He Huang Minglong Cai
Minglong Cai Tao Yuan
Tao Yuan Yujun Sheng
Yujun Sheng