- Department of Pathology, UMass Medical School, Worcester, MA, United States
Major histocompatibility class I (MHC I) molecules bind peptides derived from a cell's expressed genes and then transport and display this antigenic information on the cell surface. This allows CD8 T cells to identify pathological cells that are synthesizing abnormal proteins, such as cancers that are expressing mutated proteins. In order for many cancers to arise and progress, they need to evolve mechanisms to avoid elimination by CD8 T cells. MHC I molecules are not essential for cell survival and therefore one mechanism by which cancers can evade immune control is by losing MHC I antigen presentation machinery (APM). Not only will this impair the ability of natural immune responses to control cancers, but also frustrate immunotherapies that work by re-invigorating anti-tumor CD8 T cells, such as checkpoint blockade. Here we review the evidence that loss of MHC I antigen presentation is a frequent occurrence in many cancers. We discuss new insights into some common underlying mechanisms through which some cancers inactivate the MHC I pathway and consider some possible strategies to overcome this limitation in ways that could restore immune control of tumors and improve immunotherapy.
Introduction
Highly immunodeficient mice, which completely lack adaptive immunity, develop high rates of spontaneous and carcinogen-induced cancers (1, 2). Similarly, immunodeficient humans suffer from higher rates of malignancy (3–5). Therefore, the immune system is capable of recognizing and eliminating many cancers before they become clinically evident. Moreover, cancers that are infiltrated with activated T cells often have better prognosis, indicating that the immune system can exert some control on cancers, even after they have become clinically evident (6–15). Further evidence that the immune system has the potential to control and/or eliminate cancers has come from the success of immunotherapies, such as checkpoint blockade. In checkpoint blockade immunotherapy, patients are treated with antibodies that block negative regulatory molecules, such as PD-1/PD-L1 or CTLA4, which normally restrain T cell responses. This kind of therapy can reinvigorate a patient's anti-tumor T cell responses, which then can cause tumors to shrink and even lead to cures in some patients (16, 17). While all these observations show that the immune system has the capacity to fight cancer, the unfortunate fact is that once the majority of cancers have become clinically evident, untreated they almost always continue to progress and a majority fail to respond and/or be eliminated by checkpoint blockade immunotherapy. Therefore, understanding how cancers evade immune control is important for understanding tumor pathogenesis and for devising ways to improve immunotherapy.
While there are several immune effector mechanisms that can damage tumors, the most important ones are carried out by CD8 T cells. This has been shown [e.g., in experiments where tumor rejection was inhibited in mice that were depleted of CD8 T cells (18, 19)]. Similar principles are thought to apply in humans as shown [e.g., by the observations that the presence of activated CD8 T cells in cancers are associated with improved survival (20) and adoptive immunotherapy with T cells engineered to express TCRs from tumor-reactive CD8 T cells can lead to cancer regression (21)]. Tumor-reactive CD8 T cells identify cancers by recognizing peptide-MHC I complexes that are generated through the MHC I antigen presentation pathway (Figure 1 and below). Upon recognizing a cancer, CD8 T cells go on to kill these cells via perforin or FAS-dependent pathways and also can injure tumors by inciting inflammation. Such mechanisms are important in controlling cancer as shown [e.g., by the finding that higher frequencies of cancers develop in perforin-null or FAS-deficient mice (22, 23) compared to their wild type counterparts and potentially also in perforin-deficient humans (24)].
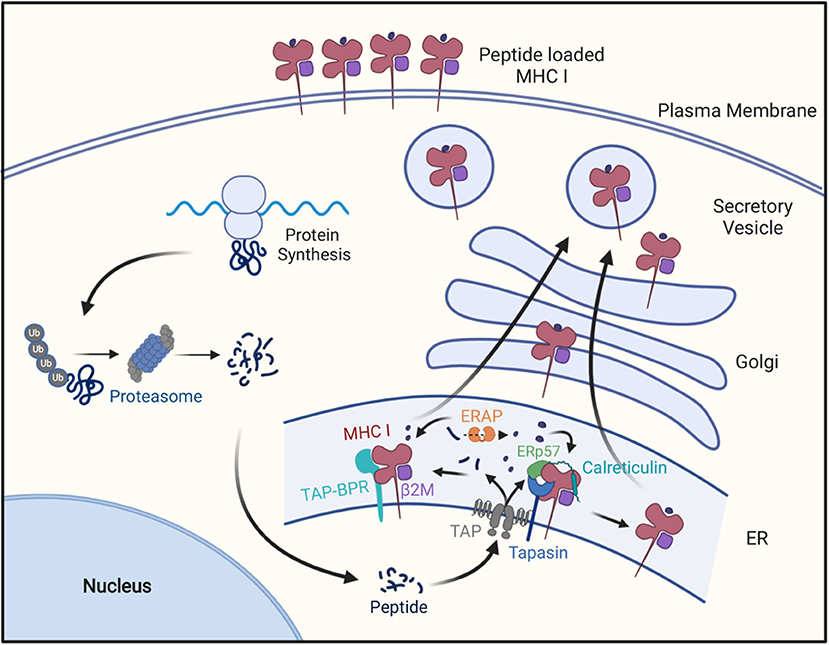
Figure 1. The MHC class I antigen presentation pathway. Cellular proteins are hydrolyzed by the ubiquitin-proteosome pathway into oligopeptides, which are subsequently transported into endoplasmic reticulum through the TAP transporter. In the ER these peptides may be further trimmed by ERAP1 and then peptides of the right length and sequence bind to MHC I molecules with the help of Tapasin in a peptide-loading complex containing Tapasin, TAP, calreticulim, and ERP57, or the with the help of TAPBPR. After MHCI molecules bind peptide, they are transported to the cell surface for display to CD8+ T cells.
In order to progress, cancers need to circumvent immune control. This was nicely illustrated by a study of carcinogen-induced cancers that arose in immunodeficient vs. immunosufficient mice. Cancers from immunodeficient mice grew when transplanted into other immunodeficient mice. However, these same cancers were generally rejected in wild type mice, showing that they were inherently immunogenic (1). In contrast, tumors that arose in wild type mice would often grow when transplanted into other wild type mice (1). These findings indicated that tumors that arose in the presence of the intact immune system in wild type mice evolved in ways that allowed them to evade immune elimination (2). This evolution of cancers under selection pressure from CD8 T cells has been referred to as “immunoediting” (2, 25).
Cancers are often genetically unstable and can lose expression of non-essential molecules through gene loss or epigenetic silencing. MHC I molecules and most of the other molecules of the MHC I antigen presentation pathway are not essential for cell viability or growth (see below). Consequently, cancers can down-regulate or lose MHC I antigen presentation, and thereby become less stimulatory or even invisible to CD8 T cells, without impairing their ability to grow and metastasize. In this article we will review the incidence, underlying mechanism, and therapeutic implications of loss of MHC I in cancers. Except where noted, this review primarily focuses on human cancers, because of their clinical importance. It should be noted that cancers can also evade immune elimination by expressing “non-classical” MHC class Ib molecules, HLA-E and HLA-G (26–28). However, since this immune evasion mechanism is not due to a loss of antigen presentation by “classical” MHC class Ia molecules, but rather through engagement of inhibitory receptors on T lymphocytes and other immune cells (26–28), this subject is not covered in this review, except as it relates to how MHC I low cancers may evade NK cell recognition. Similarly, MHC II molecules can play a role in cancer immunity, however, since MHC I and MHC II antigen presentation are separate and non-intersecting pathways, this review does not cover the MHC II pathway in cancer.
The MHC Class I Pathway of Antigen Presentation
To understand some of the mechanisms by which many cancers evade immune surveillance, it is necessary to first understand the MHC I pathway of antigen presentation (Figure 1). This pathway is the mechanism that allows CD8 T cells to identify cells producing “foreign” proteins, such as ones from viruses in infected cells or mutant genes in cancers. In this pathway, MHC I-presented peptides are generated as part of the normal catabolism of cellular proteins. All endogenously synthesized proteins are continuously degraded into oligopeptides by the ubiquitin-proteasome pathway (29). This catabolic pathway is responsible for making the initial cleavages, and particularly the proper C-terminal cut, needed for the generation of a majority of MHC I-presented peptides (29–32).
There are several forms of proteasomes, known as proteasomes, immunoproteasomes and thymoproteasomes (33). Immunoproteasomes are formed when three alternate versions of proteasome active site subunits are expressed in cells and preferentially incorporate into newly assembling proteasomes in place of the standard active site subunits. Since these alternate active sites have different catalytic properties, immunoproteasomes generate many different (as well as some of the same) peptides as proteasomes and it seems that the ones produced by immunoproteasomes are often better for presentation on MHC I molecules (34, 35). Cells and animals that genetically lack the three immunoproteasome subunits are viable (35).
A fraction of the peptides produced by proteasomes and immunoproteasomes are transferred into the lumen of the endoplasmic reticulum (ER) by a peptide transporter called TAP (36). TAP can transport most, but not all, peptides that are between 9 and 13 residues in length (37–39). TAP is composed of two different subunits (TAP1 and TAP2) and both are needed for transporting peptides (40–43). Upon transport into the lumen of the ER, peptides are in the vicinity of newly assembling MHC I molecules.
The heavy and light [ß2-microglobulin (ß2M)] chains of MHC I molecules are co-translationally transported into the ER where they fold into the MHC I heterodimer. Before binding these complexes are inherently unstable and are stabilized through interactions with chaperones such as calreticulin within a multi-protein complex, called the peptide-loading complex (44, 45). Other components of this complex include the peptide transporter TAP, the oxidoreductase ERP57 and the peptide “editor” Tapasin. Tapasin helps retain peptide-empty MHC I molecules in the ER and also promotes their loading with high affinity peptides (46, 47). There is another peptide-editor called TAPBPR, which is not part of the peptide-loading complex, that also promotes peptide-loading of MHC I molecules (48). Cells and animals that lack Tapasin, ERP57, or TAP are viable (49, 50).
The empty MHC I molecule contains a groove that binds peptides (51, 52). Peptides are bound via molecular interactions typically with two of their side chains, some of their main chain atoms and their free N- and C-termini (47). Because the interactions with the peptide's two ends contributes substantially to the affinity with which peptides are bound, most MHC I-bound peptides are of a uniform length, which depending on the particular MHC I molecule is typically 8, 9, or 10 residues (53). Proteasomes and immunoproteasomes make some peptides in this size range, but also many more that are too short or too long for stable binding to MHC I molecules (54). However, the long peptides can be trimmed to the proper size for presentation. Much of the trimming of long peptides occurs in the endoplasmic reticulum by an aminopeptidase called ERAP1 (ERAAP) (55, 56) and in humans also a second related peptidase called ERAP2 (57). ERAP1 is specialized in trimming long peptides to the optimal length for binding MHC I molecules, as it slows or stops trimming most peptide substrates when they are 8–9 residues in length (55). Long peptides can also be trimmed by aminopeptidases in the cytosol and the resulting shorter peptides can be transported by TAP into the ER (58). Similar to TAP and MHC I mentioned above, cells and animals that lack ERAP1 are also viable (59–62).
The peptides produced by these various mechanisms that have the right length and sequences can then bind to the empty MHC I molecules in the ER, often assisted by the peptide-editors Tapasin and TAPBPR. Upon binding peptides MHC I complexes are both stabilized and released from the ER, whereupon they follow the default exocytic pathway to the plasma membrane for display to CD8 T cells. In cells that have defects in making, transporting or MHC I-loading of peptides, most of their MHC I molecules are retained in ER and ultimately degraded, resulting in a paucity of MHC I molecules on the cell surface (29, 32, 63). As will be discussed further below, such defects underlie the MHC I low phenotype in many cancers.
Regulation of the MHC I Pathway of Antigen Presentation
Regulation of MHC I antigen presentation is also relevant to tumor immune evasion. The expression of most components of the MHC I antigen presentation pathway, including MHC I heavy chains, ß2M, immunoproteasome subunits, TAP, Tapasin and ERAP1, are coordinately regulated. This is because these antigen presentation components all have similar gene control elements in their promoters/enhancers (64, 65) (Figure 2). These elements include sequences that bind the transcription factors NLRC5-enhanceosome, NF-κB, and IRF1/IRF2 (66). Gene silencing or editing experiments have shown that the NLRC5, IRF1, and IRF2 transcription factors are essential for basal and/or interferon (IFN)-induced MHC I expression, but not for cell viability or growth (67–70). Since these are non-essential genes (71, 72), their expression can be lost in cancer cells, as will be discussed below.
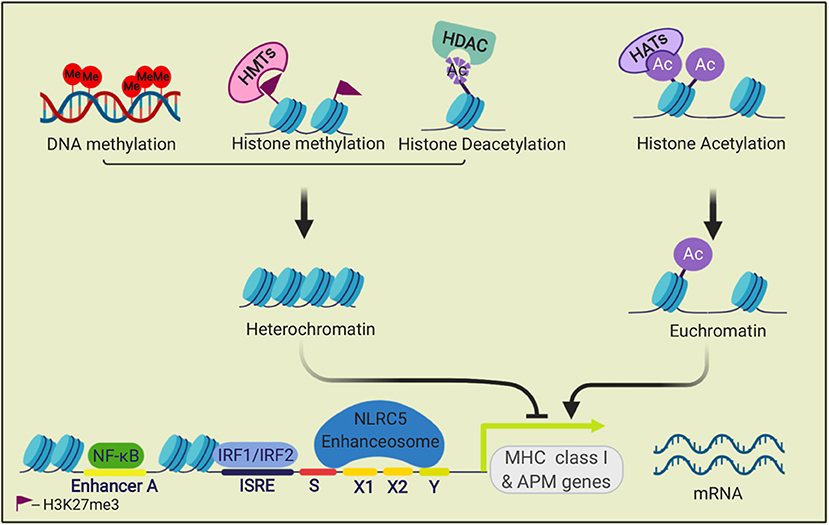
Figure 2. Transcriptional regulation of MHC class I genes. The transcription factors NF-κB, IRF1, and IRF2, and the NLRC5 enhanceosome bind to promoter and enhancer elements in the 5' upstream sequences of MHC I APM genes and drive their transcription. This process is regulated by epigenetic modifications. Methylation of histone (H3K27me3) by histone methyltransferases (HMTs) and DNA methylation repress transcription. Histone acetyltransferases (HATs) acetylate histones, which can open the chromatin for transcription. Histone deacetylases (HDACs) can remove histone acetylation marks and silence transcription.
Many cells of hematopoietic origin, such as dendritic cells and lymphocytes, constitutively express relatively high amounts of all of the MHC I antigen presentation components and consequently without any stimulation have high levels of MHC I molecules on their cell surface (73, 74). In contrast, under basal conditions, most other cells have lower expression of these components and have less MHC I on the cell surface (74). However, in all cells, the expression of MHC I pathway components and surface MHC I levels are increased upon stimulation with interferons, especially type 2 IFN (IFNγ) (75). In such responses, IFNs upregulate expression of IRF1, STAT1, and NLRC5 (76), which then binds to the promoters of the antigen presenting gene and drives their expression (Figure 3). IFNs are induced in response to infections and T cell responses, this upregulated expression is thought to enhance detection of pathological cells. That this mechanism is important in cancers is suggested from studies that have documented an increased incidence of cancer in mice that have defects in the IFN pathway (77, 78), and that in humans IFNγ-signaling signatures in cancers correlate with response to immunotherapy (79).
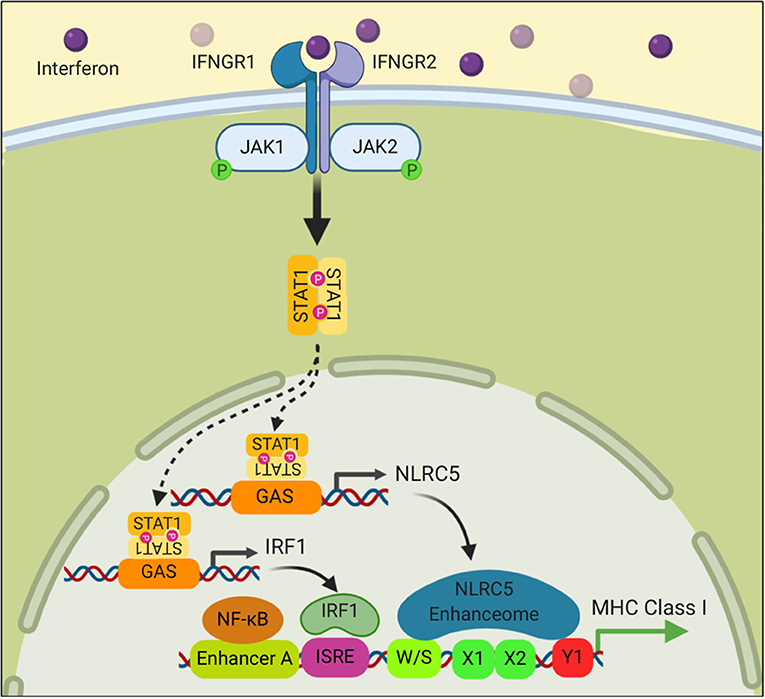
Figure 3. Interferon signaling stimulates the transcription of MHC class I genes. Binding of Interferon to its receptor stimulates phosphorylation of the Janus kinases, JAK1 and JAK2, which in turn phosphorylate STAT1. Phosphorylated STAT1 translocates into the nucleus where it binds to promoter elements of NLRC5 and IRF1 and drives their transcription. NLRC5 and IRF1 then stimulate MHC I gene transcription as described in Figure 2.
Cancers Antigens and Their Visibility to the Immune System
Through the mechanisms describe above, all cells display on their surface peptides from a majority of the proteins that they are making. This process allows CD8 T cells to identify and eliminate cells that are synthesizing “foreign” or other immunogenic proteins. Foreign (non-self) sequences in cancer may come from endogenous genes harboring mutations, which are often referred to as neoantigens, or in some cases from viral sequences in cancers (e.g., human papilloma viral proteins in human cervical carcinomas) (80).
Mutational burdens vary substantially among cancers. Tumors with higher mutational burdens are theoretically more immunogenic, and there is some evidence to support this concept. Melanomas and non-small cell lung carcinoma (NSCLC) often have high mutational burdens (81) and are considered more immunogenic tumors. There is also a correlation between the number of mutations in cancers and their responses to checkpoint blockade or adoptive T cell immunotherapy (82–84). This has been interpreted to suggest that cancers, which display many immunogenic peptides, will be much more likely to be attacked by CD8 T cells that have been reinvigorated by immunotherapy. In addition to mutated peptides, there are other kinds of immunogenic tumor antigens. For example, anti-cancer CD8 T cells can recognize de-repressed oncofetal antigens, cancer germline antigens, and even normal (non-mutated) cellular antigens, such as tyrosinase in melanomas and melanocytes (85, 86). In this latter case, the responding T cells are autoreactive ones that have escaped normal tolerance mechanisms and are present in the T cell repertoire. However, cancers that lack any immunogenic antigens are ones that can't be controlled by CD8 T cells.
Cancers that are initially immunogenic can lose visibility to CD8 T cells in two general ways. If the immunogenic antigens are non-essential for cell survival, and this is probably true for a majority of tumor antigens, then genetically unstable cancer cells can lose expression of the cancer antigens (87–90). After this occurs, CD8 T cells will be ineffective in controlling the cancer because despite the tumors having plenty of MHC I molecules, the cancer cells have lost all antigenic peptides that CD8 T cells can recognize. This route of immune evasion will be less likely in cancers that express many immunogenic cancer antigens because it would require simultaneous loss of expression of many independent gene products. This may be another reason as to why tumors with high mutational burdens are more susceptible to T cell immunotherapy. The other general way that cancers can lose visibility to CD8 T cells is by down regulating the MHC I antigen presentation pathway. The evidence that this occurs, and its underlying mechanisms and clinical importance are considered in the following sections.
Cancers Often Lose Expression of MHC I Molecules
A large number of many different types of human cancers have been reported to lose expression of MHC I molecules to varying degrees (Figure 4). An MHC-low phenotype has been observed in many of the most frequent human cancers including NSCLC, breast, prostate, colorectal, head and neck squamous cell carcinoma (HNSC), hepatocellular carcinoma, and melanoma. The number of cases that have lost MHC I expression varies for different types of these cancers and between different studies, and ranges from 0 to 93% (Figure 4). Cancers may not be homogeneous and can have variable expression of MHC I among its cells and/or in different regions. In addition, expression may change over time as a cancer progresses and may differ between the primary site and metastases (91–94).
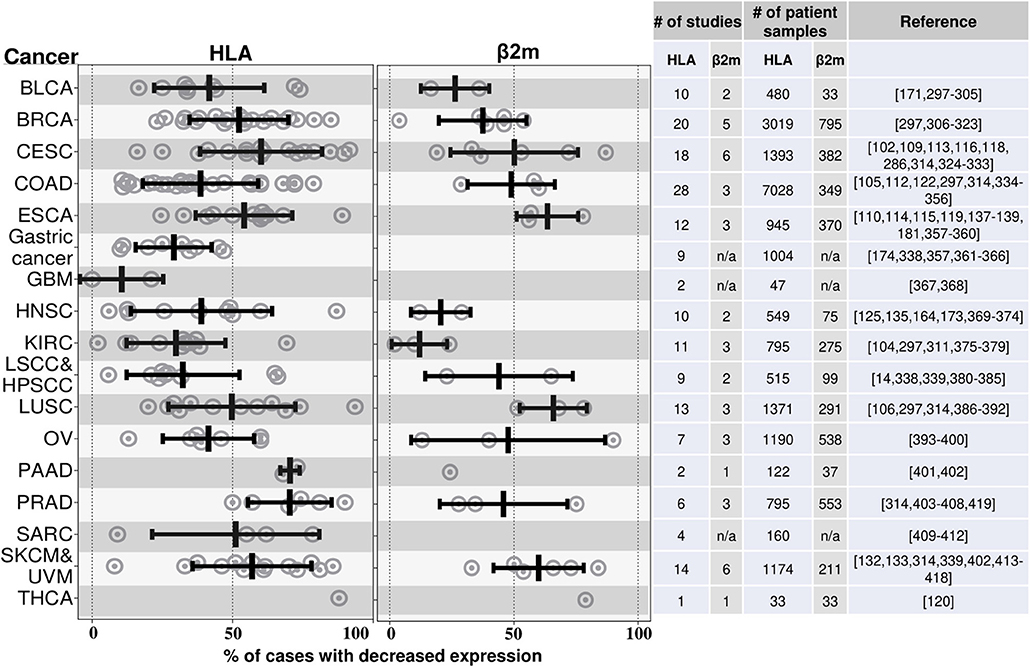
Figure 4. HLA and β2M are frequently downregulated in many different cancers. This graph illustrates the findings from a number of studies that have measured MHC I expression in various cancers by immunohistochemistry. Cancers are annotated by their TCGA abbreviations (see abbreviation list). Each dot represents the percent of cases with loss of MHC I expression in an individual study. The mean % reductions and standard deviations for all of the studies combined are shown by the black bars. The data to the right of the graph shows the number of studies and number of patients samples used to quantify the MHC class I. The studies that were included are shown in the references; this is not an exhaustive list of all such analyses. BLCA, Bladder urothelial carcinoma; BRCA, Breast invasive carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; COAD, Colon adenocarcinoma; ESCA, Esophageal carcinoma; GBM, Glioblastoma multiforme; HNSC, Head and neck squamous cell carcinoma; KIRC, Kidney renal clear cell carcinoma; LSCC, Lung squamous cell carcinoma; HPSCC, Hypopharyngeal squamous cell carcinoma; LUSC, Lung squamous cell carcinoma; OV, Ovarian serous cystadenocarcinoma; PAAD, Pancreatic adenocarcinoma; PRAD, Prostate adenocarcinoma; SARC, Sarcoma; SKCM, Skin cutaneous melanoma; UVM, Uveal melanoma; THCA, Thyroid carcinoma; IFN, Interferon; LIHC, Liver hepatocellular carcinoma; NSCLC, Nonsmall cell lung carcinoma.
The vast majority of these studies have analyzed MHC I expression in primary patient samples by immunohistochemistry (IHC) using antibodies specific for monomorphic determinants on the heavy chains of classical MHC I molecules (HLA-A, HLA-B, and HLA-C) or for ß2M. Therefore, many cancers have downregulated MHC I antigen presentation broadly. Loss of expression of a single MHC I molecule has also been reported (95, 96). Many studies have reported cancers that are MHC I negative, however because of the limits of sensitivity of IHC, it is possible that some of these cases may still express some MHC I molecules.
As described above, because peptide empty MHC I molecules are unstable without chaperone-binding and retained in the ER, defects almost anywhere in the MHC I pathway (e.g., loss of MHC I heavy chain, ß2-microglobulin, immunoproteasome subunits, TAP, Tapasin, and ERAP1) results in a loss of MHC I molecules from the cell surface. In mouse and human cells, genetic deletion of TAP reduces MHC I levels by 30–70% for most MHC I alleles (40, 97). Similarly, loss of Tapasin decreases MHC I expression by as much as 90% and deletion of immunoproteasome subunits reduces MHC I levels by ~50% (35, 98). MHC I levels are reduced in ERAP1 KO cells by 20–70% (59, 99, 100).
The expression of these antigen presentation pathway components in cancers has been studied (101) but much less extensively than for MHC I and ß2M. Of these components, TAP has been studied most extensively. Loss of TAP expression, ranging from 10 to 80.4%, has been documented in colorectal, renal cell cervical cancers, and melanomas (102–110) (Figure 5). There are more limited studies that have documented loss of expression of Tapasin (111–114), Immunoproteasomes (113, 115), and ERAP1 (116–119) (Figure 5). Individual cancers can lose expression of multiple of these antigen presenting components (see section on transcriptional regulation below) and the net effect of these multiple loses on MHC I expression should be compounded.
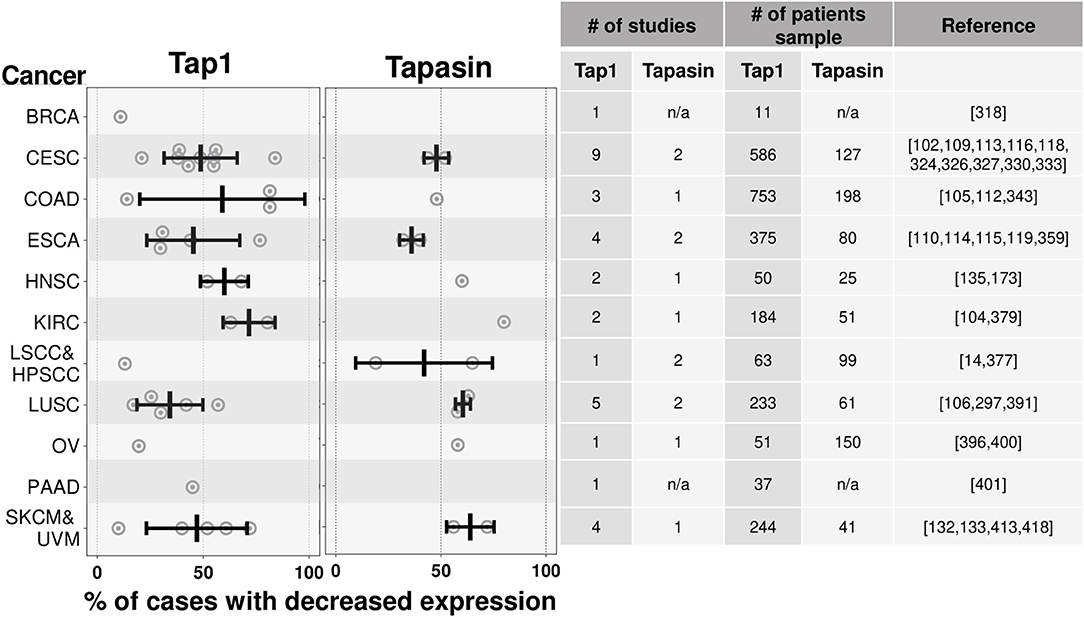
Figure 5. Tap1 and Tapasin are downregulated in many cancer types. Similar to Figure 4, except summarizing results from studies quantifying loss of Tap1 (one of the chains of the TAP transporter) and Tapasin, instead of MHC I. There are fewer studies of the expression of these proteins in cancers relative to the studies of MHC I expression in cancers. n/a-data is not available. Cancer abbreviation are as in Figure 4.
Clinical Importance of Loss of MHC I Expression In Cancers
As discussed above, the loss of MHC I antigen presentation will make cancers less visible to the immune system and this is predicted to impair control of such tumors by CD8 T cells. There are three lines of evidence that support this concept in human cancer patients. First, in some cancers the presence of tumor-infiltrating lymphocytes (TIL), which is often an indication of a host immune response, is positively correlated with MHC I molecule expression on tumor cells. For example, MHC I-low cancers (e.g., breast cancer) contain fewer TIL than their MHC I-high counterparts (10, 11, 120, 121). Since TILs are a positive prognostic feature in many cancers (7–10, 12, 13, 15, 122), the correlation of TIL with MHC I expression is consistent with a role of antigen presentation in immune control of cancers.
A second line of evidence for the clinical significance of MHC I-loss, comes from studies that have correlated MHC I expression with prognosis. In many cancers, including melanoma, glioblastoma, colorectal, bladder, uterine, cervical, head/neck, breast and other cancers, loss of MHC I is associated with worse clinical outcomes (14, 110, 111, 122–140). Since loss of the MHC I antigen presentation pathway does not alter intrinsic cell growth or viability, this correlation is also consistent with a role for antigen presentation in immune control of cancers (128).
The third line of evidence for the clinical importance of MHC I-loss comes from studies of immunotherapy. In several studies, loss of MHC I expression has been correlated with resistance to checkpoint blockade (124, 141–147) and adoptive immunotherapy (148, 149). Moreover, during immunotherapy it was observed that MHC I high metastases were the ones that regressed while MHC low metastases progressed (150). Similarly, defects in IFN-response pathways, which regulate MHC I levels, as described above, are also correlated with resistance to checkpoint immunotherapy (79, 122, 151–153).
These three lines of evidence point to the likely importance of MHC I-loss to clinical outcomes. However, there is a “chicken and egg issue” that should be considered. Since activated CD8 T cells and CD4 Th1 cells produce IFNγ, which can upregulate the MHC I pathway, this raises the question of which of the events came first: High MHC I or the T cell immune response. Similarly, if higher MHC I levels are a consequence of a preexisting T cell responses, then the presence of the responsive T cells could also be the reason that these cases are more responsive to checkpoint blockade immunotherapy. In other words, high MHC I could be an effect rather than a cause of TIL infiltration and the consequent improved clinical responses. While there is undoubtably interplay between MHC I stimulating T cells and T cells stimulating MHC I antigen presentation, the fact is that MHC I is needed to initiate this process (154). Therefore, loss of MHC I antigen presentation is likely causally related to clinical outcomes. In support of this concept, in an experimental human xenograft model, wherein the preexisting T cell repertoire is identical and the only variable is whether a tumor is MHC I high vs. low, loss of MHC I antigen presentation results in resistance to checkpoint blockade (141).
If MHC I expression is a key factor needed for immune control of cancers, as is expected from the underlying science and suggested by the above clinical data, then it is important to understand the underlying mechanisms for MHC I loss. This is of obvious importance for understanding pathogenesis and also for evaluating whether there are ways to potentially restore MHC I expression to improve therapy.
Loss OF MHC I Expression in Cancers Through Mutation or Deletion of Structural Genes
Many cancers are genetically unstable and can lose gene expression through deletions or mutation of chromosomal sequences (Figure 7). Many of the MHC I antigen presenting components (e.g., MHC I heavy chains, TAP, Tapasin, immunoproteasome subunits) are encoded in the MHC on Chromosome 6. Cells that sustain homozygous deletion of large regions of the MHC region are viable and proliferate (155–157) and therefore such chromosomal deletions are permissive in cancers, as are inactivating mutations in the antigen presenting components.
Loss of both copies of MHC I heavy chain genes or of ß2M will eliminate essentially all MHC I expression, and such loss does occur in cancers (96, 141, 158, 159). Loss of one copy of MHC I heavy chain or ß2M genes (loss of heterozygosity) also has been documented in many cancers (95, 143, 158, 160–165). In a survey of 59 cancer types, loss of MHC I heterozygosity was observed to occur in 17% of cancers (166). That this might be a consequence of immunoediting was suggested by the observation that this loss occurred more frequently in cancers with higher mutational burdens and therefore ones that were expressing potentially more immunogenic neoantigens (166). Because MHC I heavy chain genes are co-dominantly expressed from both chromosomes, loss of one copy of an MHC I heavy chain gene reduces MHC I expression by about 50% (35). In cells with MHC I loss of heterozygosity, a single inactivating mutation in a remaining MHC I gene will lead to a null phenotype and such mutations do occur in the coding regions of individual MHC I heavy chain genes (143, 167). Over time MHC I expression can decrease in patients, with e.g., primary lesions being MHC I positive but metastasis losing such expression, presumably the result of immunoediting (93, 168).
Mutations and deletions also occur in all of the other components of the MHC I antigen presentation and IFN pathways as shown in sequencing data of many human cancers (Figure 6) (171). Much of this data has not been analyzed to tell whether and how often these genetic alterations have led to a loss of function, nor how many of the various cancers are free of any mutation in an MHC I pathway component. However, there are a number of reports of inactivating mutations and deletions of several of these components (95).
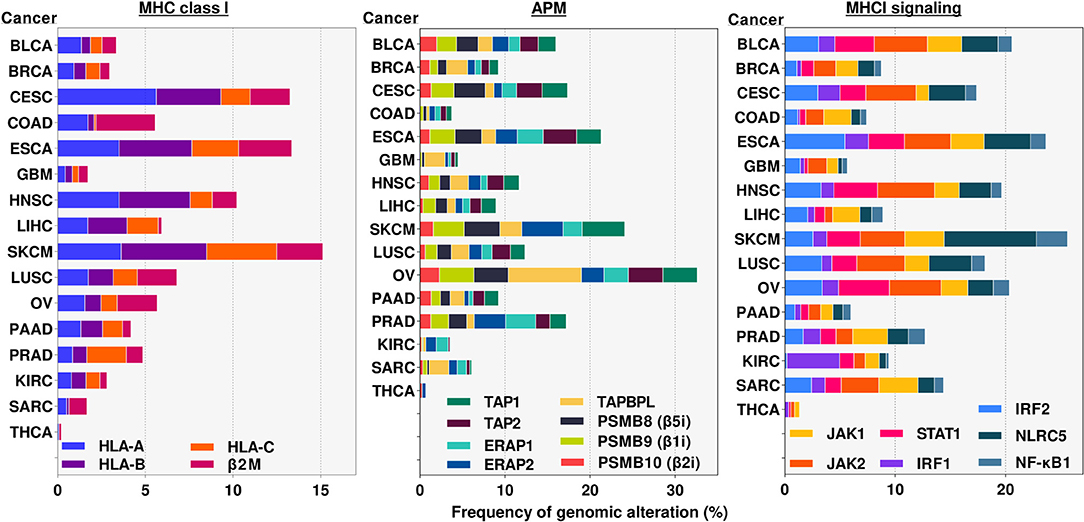
Figure 6. Frequency of alterations in genomic sequences of MHC class I pathway genes in various cancers. Genomic alterations include any mutation, deletion and/or amplification occurring in either intron or exon regions of the indicated genes. Results were obtained from cBioportal.org (169, 170). analyses of publicly available TCGA data sets for the indicated genes and cancers. APM, Antigen Presentation Machinery. Cancer abbreviation are as in Figure 4.
Loss OF the MHC I Antigen Presentation Pathway In Cancers Through Transcriptional Regulation
In many cancers with MHC I pathway defects, there is an underlying loss of transcription of MHC I pathway genes (172, 173). In an individual cancer, this process can affect the expression of multiple MHC I pathway genes at the same time, including MHC I heavy chains, ß2M, TAP, Tapasin, ERAP1, and immunoproteasome subunits (105, 173–176). The underlying mechanisms for such loss of MHC I pathway gene expression have been elucidated for some cancers.
One mechanism that affects transcription of MHC I pathway genes in cancers is a loss of key transcription factors. The NLRC5 transcription factor is reduced in multiple cancers including prostate, lung, uterine, melanoma, and thyroid cancers and this is correlated with a reduction in the expression of its target genes, including MHC I, ß2M, TAP, and immunoproteasome subunits (123) The loss of NLRC5 could arise from loss or mutation of the gene (Figure 6), or methylation of its promoter or associated histones (123, 177, 178). Loss of nuclear IRF1 in melanomas is associated with resistance to checkpoint blockade (179). Loss of expression of NF-κB and IRF1 in neuroblastomas results in a loss of MHC I expression (180). Loss of IRF2 caused a downregulation of many components of the MHC I antigen presentation pathway (MHC I heavy chains, immunoproteasomes, TAP, TAPBPR, and ERAP1) as well as an increase in CD274 (PDL-1) (69). Many human cancers (e.g., breast, NSCLC, prostate, colorectal, and uterine) downregulate IRF2 expression, which results in an immune evasion phenotype with MHC I low and PD-L1 high expression (69).
The expression of antigen presentation pathway genes can be downregulated through epigenetic silencing (Figures 2, 7). One such mechanism that has been observed in several cancer types is hypermethylation of the promoters or enhancers of these genes. This modification has been documented in the regulatory elements of MHC I (176, 181–183), TAP (128), Tapasin (184), IFNR pathway components (185–187). This DNA modification silences gene expression by recruiting repressive factors, such as methyl-CpG binding domain protein 1 (MBD1) and methyl-CpG binding protein 2 (MeCP2) and interfering with transcription. As cancer cells divide, these methylated sequences are passed onto daughter cells, thereby perpetuating the gene silencing. Treatment with agents that cause DNA demethylation has restored MHC I expression in some cancers, demonstrating the importance of this silencing mechanism (182, 188). Cancer neoantigen genes can also be subjected to DNA hypermethylation (189).
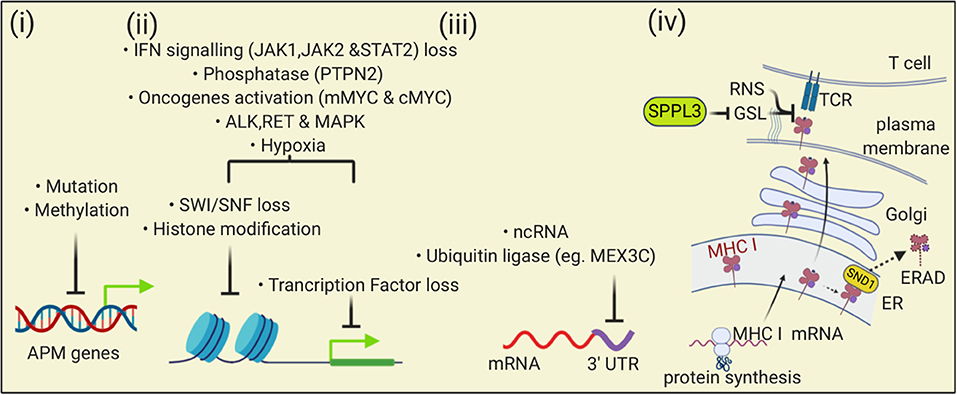
Figure 7. The MHC I antigen presentation pathway is down-regulated by multiple mechanisms in cancers. (i) At the level of DNA, mutation and methylation of nucleotides can reduce expression of APM genes; (ii) Transcription of APM can be reduced by changes in chromatin that impair gene accessibility or through loss of transcription factors. Multiple mechanisms can affect these processes including altered signaling pathways, oncogene activity, and the tumor microenvironment; (iii) At the level of transcription, binding of ncRNA or proteins to the 3′ UTR of APM mRNAs can reduce transcription; (iv) At the level of protein, Staphylococcal nuclease and tudor domain containing 1 (SND1) can bind to MHC I in the ER and trigger endoplasmic reticulum-associated degradation (ERAD). At the cell surface, loss of SPPL3 increases glycosphingolipids (GSL) that then sterically inhibit MHC I and TCR interactions. Reactive nitrogen species can nitrosylate peptide-MHC I complexes in ways that impair TCR interactions.
Another epigenetic silencing mechanism involves histone modifications, which are also heritable to daughter cells. Histone acetylation can alter chromatin in ways that increase DNA accessibility and thereby allowing entry and binding of transcription factors. Histone deacetylase (HDAC) inhibitors, which lead to increased acetylation levels, have restored expression of MHC I molecules and other antigen presentation components in some cancers, suggesting that histone deacetylation silences expression of MHC I pathway genes (190–194). Trimethylation of histones (e.g., H3K27me3) can also downregulate genes by affecting the state of heterochromatin. In some MHC I low cancers, H3K27me3 repressive marks are associated with the promoters of NLRC5, MHC I heavy chain genes, β2M, immunoproteasomes and TAP, and loss of this repressive modification results in an increase in MHC I pathway expression (177, 195).
The polycomb repressive complex 2 (PRC2) was found to be a repressor of MHC I expression in some cancer cells, such as neuroblastomas and small cell lung carcinomas (177). PRC2 silences the basal expression of NLRC2, MHC I, immunoproteasomes, and TAP and also inhibits IFNγ-induced MHC I upregulation (177). A subunit of the PRC2 complex binds and activates the lysine methyltransferase EZH2. Consistent with this mode of action, the repression caused by PRC2 was associated with increased H2K27me3 histone repressive marks associated with MHC I pathway genes, which when reversed, increased transcription factor binding and MHC I expression. Consistent with these results, deletion of EZH2 in leukemia cells increased MHC I expression (196) and activating mutations of EZH2 caused a loss of MHC I expression in these cancers (197).
SWI/SNF factors affect gene expression by regulating chromatin accessibility. The Polybromo-associated BAF (PBAF) SWI/SNF complexes were found to be a positive regulator of MHC I expression (198). The expression of PBAF in cancers is correlated with better prognosis and responsiveness to checkpoint blockade. Interestingly for PBAF, it particularly affects promoter accessibility of IRF2 and interferon-stimulated response elements (ISREs) (198).
loss of the MHC I Antigen Presentation Pathway In Cancers Through Post-Transcriptional/Pre-Translational Regulation
The expression of proteins can be regulated through post-transcriptional mechanisms (Figure 7) and one of these mechanisms is mediated by non-coding RNAs (ncRNA) (199). One class of ncRNAs are small (22 bp average length) microRNAs (miRNA). These sequences can bind to the 3′ untranslated regions (UTR) of mRNAs and inhibit their translation through repression or by targeting them for degradation.
There are a number of examples of miRNAs that regulate the expression of components of the MHC I antigen presentation pathway and can contribute to a loss of antigen presentation in cancers (66, 200). In some cancers, there is increased expression miRNAs and it has been shown that overexpression of these miRNAs leads to a reduction in MHC I pathway components. For example, in esophageal cancer, miR-148a-3p was found to bind to untranslated regions (UTR) of MHC I transcripts and miR-125a-5p bound to UTRs of TAP2 transcripts. Moreover, the overexpression of these miRNAs reduced the expression of these antigen presentation components (201). In melanomas, miR-26b-5p and miR-21-3P bind the UTR of TAP1 transcripts and downregulate TAP1 expression (202). In colorectal cancers, miR-27a expression is increased and causes reduced MHC I expression by suppressing expression of calreticulin (203). Mir-502-5P in gastric cancer and miR-23a in hepatocellular cancer were found to reduce IRF1 expression (204, 205). Thus, miRNAs, which can be highly expressed in cancers, can negatively regulate many of the components of the MHC I antigen presentation pathway. Investigations in this subject area have been relatively limited and therefore it is likely that many more examples of miRNA-mediated inhibition of MHC I antigen presentation in cancers are yet to be discovered.
Another class of ncRNAs are long (>200 bp) non-coding RNAs (lncRNA). These sequences can regulate gene expression in many ways, including epigenetically, transcriptionally and post-transcriptionally (199, 206). One interesting example of a lncRNA that regulates MHC I antigen presentation post-transcriptionally is LINK-A. In a breast cancer model, LINK-A inhibited antigen presentation by indirectly stimulating an E3 ubiquitin ligase which led to the degradation of the peptide-loading complex (207). Again, it is highly likely that additional lncRNAs will be found to negatively regulate components of the MHC I antigen presentation pathway.
ncRNAs can also be positive regulators of MHC I. For example, in head and neck squamous cell carcinomas, expression of the lncRNA, lnc02195, increases MHC I expression and is associated with a better prognosis (208). In nasopharyngeal carcinomas miR9 expression increases expression of MHC I molecules and TAP1 (209). Whether down-regulation of these ncRNAs in cancers is an important mechanism for immune evasion remains to be determined.
The UTR regions of mRNAs can be regulated not only by ncRNAs, but also by proteins binding to these sequences. An RNA-binding E3 ubiquitin ligase, MEX-3C, binds to the 3′ UTR of the transcript for MHC class I molecule HLA-A2 leading to its degradation (210); whether this mechanism is operative in and important to cancer immune evasion has not yet been examined.
Loss of the MHC I Antigen Presentation Pathway in Cancers Through Post-translational Mechanisms
There are post-translational mechanisms that can impair MHC I antigen presentation in cancers (Figure 7). One such post-translational mechanism is analogous to immune evasion mechanisms employed by some viral pathogens. Some viruses encode immune evasion molecules that cause MHC I complexes to be dislocated from the ER into the cytoplasm, where they are degraded through a process referred to as endoplasmic reticulum-associated degradation (ERAD). It turns out that the oncoprotein Staphylococcal nuclease and tudor domain containing 1 (SND1), which is highly expressed in a number of cancers (e.g., Prostate and Melanoma), binds MHC I molecules and causes them to undergo ERAD. Deletion of SND1 in some cancer cell lines increases MHC I expression (211).
Another interesting mechanism that inhibits MHC I antigen presentation is a change in glycolipids on the plasma membrane that occurs in cancer cells that lose the signal peptide peptidase-like 3 (SPPL3) protease (212). SPPL3 cleaves and inactivates a glycosyltransferases B3GNT5, and loss of B3GNT5 reduces levels of negatively charged glycosphingolipids (GSL). Loss of SPPL3 results in an increase in these GSLs, which then associate with MHC I molecules in ways that appear to sterically inhibit their interaction with T cells. This process occurs in gliomas (and potentially some other cancers) and impairs T cell responses to these cells.
Yet another interesting post-translational mechanism that interferes with MHC I antigen presentation is modification of amino acid residues in the peptide-binding groove of MHC I molecules that alters peptide binding; this mechanism is described in more detail in the next section.
Loss of the MHC I Antigen Presentation Pathway In Cancers Due to Signaling Mechanisms and Extrinsic Stimuli From the Tumor Microenvironment
Alterations in signaling pathways can lead to MHC I downregulation in cancers (Figure 7). MAPKs, which are activated in some cancers, are negative regulators of MHC I (213, 214). Inhibiting or silencing of MAPKs increased levels of IRF1 and STAT1 (215) as well as MHC I expression (214). MAPK inhibitors increased mRNA expression of MHC I, TAP, and ß2M (214). Similarly, inhibition of the ALK and RET kinases, which are upstream activators of MAPK, also increase MHC I expression and interestingly also results in the presentation of a different repertoire of peptides (216). Another example is that signaling though the EGFR oncogene HER2/neu is associated with a loss of transcripts for immunoproteasome subunits and TAP, resulting in a loss of MHC I surface expression and antigen presentation (217, 218). Yet another example is that n-MYC and c-MYC overexpression caused loss of MHC I expression, potentially by affecting NF-κB (219–221).
IFNs that are present in the tumor microenvironment [e.g., IFNγ produced by activated T cells or type I interferons produced by a variety of cells, bind to interferon receptors (IFNR) on tumor cells]. Signaling through the IFNRs (Figure 2) leads to an increase in expression of many components of the MHC I antigen presentation pathway (e.g., MHC I, TAP, Tapasin, immunoproteasomes, and ERAP1). Components of the IFN pathway can also be lost (Figure 7) and this prevents IFN-induced upregulation of the MHC I pathway of antigen presentation. Such loss can also reduce basal levels of MHC I molecule expression (222, 223). IFN receptors signal through Janus kinases (Jak1 and Jak2) and STAT (STAT 1 and STAT2) proteins (224) (Figure 3). LOH and/or mutations in Jaks, and STATs are observed in cancers (143, 171). Loss of function mutations in Jak kinases with consequent loss of responsiveness to INFγ were found in Melanomas that became resistant to checkpoint blockade, pointing to the likely clinical significance of the inactivation of the IFN pathway (147, 151, 225). Loss of function in a receptor (APLNR) that interacts with Jak1, reduces IFNγ-stimulated Jak1 and STAT signaling and MHC I upregulation. Mutations in this receptor are found in melanoma patients that are resistant to checkpoint blockade and similarly such resistance is conferred upon knock out of this receptor from mouse melanoma cells (142). Loss of the tyrosine protein phosphatase Ptpn2 that represses IFNγ signaling by dephosphorylating both JAK1 and STAT1. Deletion of Ptpn2 in mouse tumors increases MHC I antigen presentation and improve immunotherapy (226); whether increased Ptpn2 activity in human tumors causes a loss of MHC I antigen presentation is not known. Finally, TGF-ß, which can be present in the tumor microenvironment, can cause a down-regulation of MHC I molecules in some cancers (e.g., ovarian, prostate, and ocular melanoma) (227–229).
Other events in the tumor microenvironment can lead to impaired MHC I antigen presentation in cancers (Figure 7). Tumor microenvironments can be hypoxic and hypoxia can impair MHC I antigen presentation in cancers, in part by inhibition of STAT1 (230). Tumor-infiltrating myeloid cells produce reactive nitrogen species in the tumor microenvironment, and this can impair MHC I antigen presentation in cancers. In this situation, the reactive nitrogen species cause nitrosylation of residues in the MHC I peptide binding site, which can inhibit the binding of peptides (231). In tumor-bearing mice, myeloid suppressor cells cause defective IFN responses in host cells (responses in tumors were not examined) likely due to a STAT1 defect potentially caused by nitrosylation (232).
Gene Disruptions That Affect MHC I Antigen Presentation: Evidence From Forward Genetic Screens In Cancers
Recently, a number of forward genetic screens have been performed in cancer cells subjected to selection for decreased or increased MHC I expression and have identified a large number of new gene candidates that are potentially involved in MHC class I antigen presentation (69, 142, 167, 174, 177, 226, 233–235). In fact, several of the genes described above (IRF2, PBAF, PRC2, and SPPL3) were discovered in such screens. It is important to note that many of the gene candidates that are initially identified may be artifacts. Therefore, all candidates require further validation and analyses to determine whether they are affecting the MHC I antigen presentation pathway and involved in cancer immune evasion.
A recent CRISPR-cas9 screen in B cell lymphoma cell lines did repeat gene disruptions for individual candidates and were able to reproduce a loss or increase in MHC I expression upon disruption of ~200 genes (196). Among these genes were ones that are thought to be involved in endocytosis and vesicular trafficking, ubiquitin conjugation, ER quality control, as well as other processes. Further work is needed on these and candidates from other screens to determine whether they are involved in cancer immune evasion. However, interestingly, 30 of these genes showed correlations with CD8 T cell infiltration in multiple cancers; 10 negative-regulatory genes were correlated with less tumor infiltrating CD8 T cells and 20 positive-regulatory were correlated with more infiltrating CD8 T cells. The field can look forward to much more information on the role of these genes and other validated ones in MHC I antigen presentation and cancer immune evasion.
Potential for Restoring MHC I Expression In Cancers
The fact that the loss of MHC I antigen presentation is common in cancers and allows these cells to evade immune surveillance, raises the question of whether the MHC I pathway defects could be reversed so as to reestablish immune control and responses to immunotherapy. For cancers with deletions or inactivating mutations in structural antigen presenting genes, this would require gene replacement or editing in most cases. In vitro, this has been successfully accomplished by transfection of MHC I pathway genes into cancer cell lines. Similarly, gene therapy with a ß2M-adeno-viral vector has been successful in restoring MHC I expression in vivo in a murine model (236–238). However, for gene transfer or editing to be a viable therapy, it will likely require that all cancer cells (in the primary site and metastasis) to be transduced and “repaired,” because otherwise MHC I-low clones would continue to grow. Achieving this level of gene expression or repair is probably not feasible with current gene therapy technology. An exception for overcoming the loss of structural genes is the situation where the function of the lost gene can be replaced by inducing another functionally redundant gene. One example of this is where MHC I expression in cancer cells was lost by deletion of the IRF2 transcription factor, but then restored by inducing IRF1 with IFN-stimulation (69). In this case IRF1 and IRF2 are both activating transcription factors that bind to the same promotor element (239).
In situations where the MHC I pathway structural genes are intact but their expression is downregulated, there is the potential to restore gene expression. In some MHC I low cancers, treatment with IFNs has increased MHC I levels (120, 240, 241). The mechanism as to how IFN is restoring MHC I expression has not been investigated in detail, except in one study, IFN was shown to cause increased histone acetylation, DNA demethylation of the promoters of TAP and immunoproteasome genes, and increased transcription of these and other antigen presenting genes (241). In addition, it is possible that IFN is also increasing MHC I levels in MHC I-low cancers through induction of IRF1, which then drives more transcription of the MHC I pathway genes (69), but this has not generally been examined. There are recombinant type I and II IFNs that work in vivo and are FDA-approved for other indications. In a small phase 2 trial in which two patients had MHC I negative melanomas, systemic IFNγ-administration induced MHC I expression (240). IFNγ has been shown to improve outcomes of checkpoint blockade in one clinical trial in melanoma (242), however whether and how much this had to do with MHC I expression is unknown.
For cancers that have lost MHC I expression due to epigenetic silencing mechanisms, there may be the potential to restore MHC I expression by reversing the repressive epigenetic marks. There are several examples where MHC I low cancer cell lines have been treated with drugs that inhibit DNA methyltransferases, the enzymes that are responsible for methylating DNA (181, 182, 243, 244), and thereby reverse gene silencing, presumably through demethylation of promoters. Such treatment has increased MHC I expression in several MHC I low cancers cell lines (181, 182, 188, 245). Where examined, this class of epigenetic modifying drugs was found to restore MHC I expression by upregulating expression of many IFN-responsive gene (246) including MHC I antigen presentation pathway genes in cell lines (95, 241, 245). These findings raised the possibility that this class of agents could augment T cell-based immunotherapy. Consistent with this idea, this class of agents has been shown to augment or give additive effects with checkpoint inhibitors in preclinical mouse models (182, 247, 248). There are several DNA methyltransferase inhibitors that are approved by the FDA for cancer treatment, although the exact basis for their efficacy (i.e., what are the key pathways that are affected to give the anti-cancer effects, isn't known). These drugs have been shown to increase expression of IFN and MHC I antigen presentation pathway genes in cancers in vivo (182, 245) and in limited clinical trials have improved responses to checkpoint blockade immunotherapy (249) and a tumor vaccine (250). Currently there are further ongoing trials of these agents in combination with immunotherapy (251). Inhibitors of the EZH2 methyltransferase, which as described above is a negative regulator of MHC I antigen presentation, can restore MHC I levels in lymphomas (196, 197).
Similarly, a number of MHC I low cancer cell lines have been treated with histone deacetylase (HDAC) inhibitors, which by increasing histone acetylation can restore promoter activity. HDAC inhibitors have also increased MHC I expression and MHC I pathway components in cancer cell lines (182, 190, 192, 193, 243, 252, 253). There are FDA-approved HDAC and DNA methyltransferase inhibitors that are used to treat cancers. Where studied in vivo, methyltransferase inhibitors increased expression of MHC I and MHC I pathway components in multiple types of cancers in patients (182, 245) and a xenograft model (192). In preclinical models, HDAC inhibitors and anti-PDL1 antibody (186, 193, 254–257) or with T cell therapy (258) gave additive effects. Combinations of HDAC inhibitors and checkpoint blockade have and continue to be tested in clinical trials (194, 259–261).
Based on the data just discussed, it is clear that epigenetic modifying agents can increase the MHC I antigen presentation pathway in some MHC I low cancers and that these drugs can improve responses to immunotherapy, however whether these two observations are causally related is not yet established. This is because epigenetic modifying drugs effect the promoter landscapes in potentially all cells. Therefore, these agents can affect not only the tumor, but also cells within the tumor microenvironment and immune system. Moreover, the drugs can affect the expression of many genes within these cells. In most studies, which of the key gene regulatory events that are responsible for therapeutic effects of these drugs is not known. Interestingly, in one animal study the therapeutic effect of an HDAC inhibitor was lost in immunodeficient mice (191), providing evidence that the drug was acting to improve immune control a cancer; however, whether this effect is via the restoration of the MHC I pathway or some other immune mechanism is not known.
The broad effects of the epigenetic modifying agents lead to multiple and sometimes opposing effects. For example, global DNA hypomethylation may increase MHC I expression, but also upregulate immunosuppressive mechanisms such as suppressive cytokines and checkpoint inhibitors (262, 263). Such complexity might be overcome, and outcomes improved if there were ways to more selectively modify epigenetic marks of particular genes. At present, drugs that inhibit individual HDAC enzymes are available, and perhaps even more selective agents will be developed. New gene editing approaches using modified Cas9 fusion proteins (e.g., Cas9-p300 acetyltransferase, Cas9-methyltransferases can Cas9-demethylases) have the ability to alter epigenetic marks and/or transcriptionally activate or repress expression of specific genes (264, 265). Whether such approaches could somehow be used in vivo to efficiently edit all cancer cells remains to be seen.
MicroRNAs that reduce MHC I antigen presentation are a potential therapeutic target. MiRNAs can be blocked in cells by treatment with complementary anti-sense RNAs (antimirRs/antagomiRs) and overexpression of miRNAs can be achieved using miRNA duplexes (miRNA mimetics) (199, 266). These approaches require chemical modifications to stabilize RNA oligonucleotides and methods to deliver these compounds into cells (e.g., lipid nanoparticles). Such agents have been used successfully in preclinical models and have and/or are being tested in phase 1 and 2 clinical trials, but not yet for affecting MHC I antigen presentation. LncRNAs can be targeted for degradation with anti-sense oligonucleotides.
Inhibitors of enzymes that lead to a loss of MHC I antigen presentation also have the potential to restore antigen presentation in some cancers. Inhibitors of ALK, RET, and MAPK kinases can reverse the downregulation of the MHC I antigen presentation caused by these enzymes (213, 214, 216). There are FDA approved inhibitors of glycosyltransferases, and these agents were shown to reverse the suppression of MHC I stimulatory activity caused by the increase in negatively charged glycosphingolipids from loss of SPPL3 (212). Inhibitors of thymidylate synthetase were found to increase MHC I levels in lymphoma cells in a targeted small molecule screen, via an as yet unknown mechanism (196).
Finally, in cells that have lost some MHC I antigen presentation pathway components, such as TAP (40, 267) and ERAP1 (60, 100), novel peptides peptides [aka “T cell epitopes associated with impaired peptide processing (TEIPP)] are presented that are not displayed in wild type cells and can be immunogenic. There is limited data that immunization with such sequences can elicit anti-tumor responses (268, 269). Therefore, an alternate approach to restoring the loss of MHC I antigen presentation components could be to exploit the presentation of unique antigen peptides that are displayed on such cancer cells (269).
The elucidation of the many mechanisms that lead to a loss of MHC I antigen presentation and the identification of tractable therapeutic targets to reverse this loss, brings the hope of restoring immune control and improving T cell-based immunotherapy. Given the differences between different cancer types, the heterogeneity within a single type of cancer, and the many different mechanisms that can disable the MHC class I pathway, it seems likely that such approaches will require precision medicine, where the cause of immune evasion in an individual patient is identified and then the appropriate therapy selected. The advances in identifying the underlying mechanisms that cripple MHC I antigen presentation are necessary steps in attempting to achieve this goal.
Recognition and Control OF MHC I Low Cancers by NK Cells
Cancers that have lost MHC I expression through the mechanisms discussed above, can avoid control and elimination by CD8 T lymphocytes. This is analogous to the situation where some viruses encode immune evasion molecules that inhibit MHC I antigen presentation and thereby allow virally infected cells to avoid being killed by CD8 T cells and help establish chronic infections. In these situations, there is a second line of defense that can kill these abnormal cells and this immune function is provided by natural killer (NK) cells. NK cells identify these cells in part by sensing the presence or absence of MHC I molecules.
NK cells are a lineage of lymphocyte that is distinct from B and T cells. These cells have similar effector functions (cytotoxicity and cytokine production), as CD8 T cells. However, NK cells are innate lymphoid cells (ILC) and the receptors they use to recognize their target cells are fundamentally different from the ones used by T (and B) lymphocytes. Instead of employing a mechanism that recombines gene segments to generate clonally unique and diverse receptors, NK cells use non-rearranging germ-line encoded receptors of several different types (270). Some of these NK receptors are activating ones and others provide inhibitory signals.
Human NK cells express several killer inhibitory receptors (KIR) that upon engagement of their ligands, impart inhibitory signals through ITIM motifs in the receptors' intracytoplasmic domains. HLA-A and HLA-B are ligands for KIR3D receptors and HLA-C is recognized by KIR2D receptors. In addition, NK cells express other types of inhibitory receptors that recognize MHC I molecules, including CD94-NKG2A, which recognizes HLA-E, and LILRB1, which recognizes all MHC I molecules. Moreover, NK cells express other inhibitory receptors that are not involved in MHC I recognition (270). Because of their inhibitory receptors that recognize MHC I molecules, NK cells ignore normal MHC I-sufficient cells but are disinhibited when they encounter abnormal-MHC I low cells. This loss of inhibition is a necessary but not a sufficient event to trigger the NK cell's effector mechanisms.
Activation of NK cells requires engagement of activating receptors, which in humans include NKG2D, NKp20, NKp44, and NKp46 (270). These receptors associate with and signal through ITAM-containing proteins (270). Other stimulatory receptors expressed by NK include 2B4 (CD244) and DNAM1 (CD226) (270).
The ligands of some of the activating receptors are ones whose expression is induced on cancers, virally infected and stressed cells. The best characterized examples are the MHC class I polypeptide–related sequence A and B (MICA and MICB) molecules, which are the ligands of the activating NKG2D receptor. MIC A and MIC B are structurally similar to HLA class heavy chains but are not associated with ß2M and do not bind peptides. Because of these properties, MICA's and MICB's expression is not affected by defects in the MHC I antigen presentation pathway and therefore can be expressed in MHC I negative cancer cells. The activation of NK cells depends on the balance of stimulatory and inhibitor signals they receive. Cancers or virally infected cells that both express activating ligands and lack inhibitory ligands can be killed by NK cells.
Mice that lack functional NKs cell due to antibody treatment or genetic knock outs develop a higher frequency of some cancers (271, 272). Similarly, humans that have NK cell defects have increases in some cancers, particularly ones that may be induced by viruses (273). A caveat in many of these studies is that the mouse models and NK deficient humans may have defects beyond just a loss of NK cells (273). Nevertheless, the data in aggregate suggest that NK cells play a role in immune surveillance. Whether the protection against carcinogenesis afforded by NK cells is primarily against MHC I low cancers is not clear (274).
NK cells may still exert some control after cancers have arisen. Depletion of NK cells in mouse models promotes the growth and metastasis of transplanted tumors (271, 275, 276). Moreover, adoptive transfer of NK cells into tumor-bearing mice can lead to tumor rejection. In humans, the level of cytolytic activity of circulating NK cells correlates with the risk of cancer (277) and infiltration of NK cells in some cancers is associated with a better prognosis (278, 279). Similarly, adoptive transfer of NK cells into human cancer patients has shown therapeutic effects in some patients, demonstrating that similar principles likely apply in humans (280). Such studies have led to considerable interest in exploiting NK cells for tumor therapy and there are many efforts underway to do so [e.g., developing CAR-NK cell therapy and antibodies that recruit activated NK cells to cancers (270, 281, 282)].
Despite NK cells being a potential second line of defense against tumors that have lost MHC I, once such tumors become clinically evident, they almost always progress. In fact, as noted above, loss of MHC I is often a negative prognostic indicator. Moreover, there is no evidence that MHC I negative cancers are infiltrated with more NK cells than MHC I sufficient cancers (283). Therefore, for many MHC I low cancers, either they were never targets of NK cells or such tumors have evolved ways to evade control by NK cells.
In fact, there are many mechanisms through which cancers can evade NK cells. For example, HLA A, B&C low cancers can express the non-classical MHC I molecules HLA-G and HLA-E, which can inhibit NK cells by engaging their inhibitory receptors (284–288). In addition, NK cells may not penetrate into solid tumors or once within the cancer can become anergic or exhausted, including in MHC I low tumors (289–291). Furthermore, tumors can shed their MIC molecules, thereby removing an activation signal and creating a soluble ligand that can block the NK cell's cognate receptor (292, 293). Moreover, cancers can also create an immunosuppressive environment [e.g., producing TGFß, which can lead to inhibition of NK cells (294)]. A fuller consideration of these mechanisms is beyond the scope of this article and readers are referred to recent reviews (270, 295, 296).
Future Directions
While there is abundant evidence that loss of MHC I antigen presentation is a frequent event in cancers that results in immune evasion, we still have much to learn. As reviewed above, new mechanisms for MHC I downregulation have recently been discovered and there will be more to be uncovered. Forward genetic screens are identifying new components that contribute to the MHC I pathway and it will be of importance to investigate how they contribute to MHC I phenotypes in cancers. Even among the known mechanisms for loss of antigen presentation, a majority of the analyses have been performed in limited cancer types and a more comprehensive understanding across more types of cancer is needed. Moreover, many of the underlying mechanisms for MHC I pathway loss need to be elucidated at higher resolution (e.g., causes and specific targets of epigenetic modifications).
Given the importance of MHC I antigen presentation for the immune control and immunotherapy of cancers, there is a need to develop therapeutic approaches to restore the MHC I pathway in cancers and this should be feasible in at least some cancers. This might itself be an immunotherapy by allowing a restoration of immune control. It might also provide adjunct therapy that could improve the percentage of patients that respond to immunotherapies and potentially extend the efficacy of immunotherapies to cancers that have been largely resistant to such therapy. The various mechanisms that cause MHC I pathway-loss might also serve as biomarkers to help identify patients that have the potential to respond, or not, to immunotherapy and/or have the potential for the loss of MHC I to be reversed. The hope for such biomarkers is that they could make immunotherapy more personalized (e.g., sparing those patients who won't respond to such therapy from the risks and enormous expense of the treatment).
Conclusions
A sizable percentage of many different types of cancers lose MHC I antigen presentation partially or completely. This is almost certainly the result of immunoediting where MHC I low variants emerge under selection pressure imposed by CD8 T cells. The result of this process is that CD8 T cells can no longer “see” these MHC I-deficient variants and are therefore unable to control or eliminate them. This process reflects the fact that the MHC I pathway is non-essential for viability and growth and therefore when lost does not compromise cancer progression. Where examined, this escape of immune control is generally associated with worse prognoses and resistance to immunotherapy. There are many mechanisms that underlie the loss of MHC I antigen presentation. Some mechanisms involve mutations and deletions of structural genes of one or more component(s) of the antigen presentation pathway; others effect transcription of pathway genes via loss of transcription factors or epigenetic silencing of gene regulatory elements; and yet others can affect the stability of mRNAs for MHC I pathway components or the molecules themselves, or signaling pathways that regulate MHC I expression. Some of these alterations are unique to an individual cancer and others are common in multiple patients and cancer types. It will be important to further understand the multiple mechanisms for loss of the MHC I pathway that are operative in all cancer types and their clinical significance. The hope is that in the future, characterizing MHC I pathway lesions in individual patient samples would lead to actionable information about what therapies will or will not be likely to work and prognosis. Moreover, some of the mechanisms that cause the loss of the MHC I pathway in cancers are reversible and may be amenable to the development of therapeutic interventions that could make T cell-based immunotherapies more efficacious in more patients and in more kinds of cancer.
Author Contributions
The text was written by KR, KD, and JC. Figures were made by KD and JC. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants R01CA247624, R01AI114495, and R01 AI145932 to KR.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. (2001) 410:1107–11. doi: 10.1038/35074122
2. Koebel CM, Vermi W, Swann JB, Zerafa N, Rodig SJ, Old LJ, et al. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. (2007) 450:903–7. doi: 10.1038/nature06309
3. Mayor PC, Eng KH, Singel KL, Abrams SI, Odunsi K, Moysich KB, et al. Cancer in primary immunodeficiency diseases: cancer incidence in the United States Immune Deficiency Network Registry. J Allergy Clin Immunol. (2018) 141:1028–35. doi: 10.1016/j.jaci.2017.05.024
4. Vajdic CM, van Leeuwen MT. Cancer incidence and risk factors after solid organ transplantation. Int J Cancer. (2009) 125:1747–54. doi: 10.1002/ijc.24439
5. Mortaz E, Tabarsi P, Mansouri D, Khosravi A, Garssen J, Velayati A, et al. Cancers related to immunodeficiencies: update and perspectives. Front Immunol. (2016) 7:365. doi: 10.3389/fimmu.2016.00365
6. Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. (2003) 348:203–13. doi: 10.1056/NEJMoa020177
7. Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci USA. (2005) 102:18538–43. doi: 10.1073/pnas.0509182102
8. Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. (2006) 313:1960–4. doi: 10.1126/science.1129139
9. Pages F, Kirilovsky A, Mlecnik B, Asslaber M, Tosolini M, Bindea G, et al. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J Clin Oncol. (2009) 27:5944–51. doi: 10.1200/JCO.2008.19.6147
10. Rusakiewicz S, Semeraro M, Sarabi M, Desbois M, Locher C, Mendez R, et al. Immune infiltrates are prognostic factors in localized gastrointestinal stromal tumors. Cancer Res. (2013) 73:3499–510. doi: 10.1158/0008-5472.CAN-13-0371
11. Ryschich E, Notzel T, Hinz U, Autschbach F, Ferguson J, Simon I, et al. Control of T-cell-mediated immune response by HLA class I in human pancreatic carcinoma. Clin Cancer Res. (2005) 11:498–504.
12. Sharma P, Shen Y, Wen S, Yamada S, Jungbluth AA, Gnjatic S, et al. CD8 tumor-infiltrating lymphocytes are predictive of survival in muscle-invasive urothelial carcinoma. Proc Natl Acad Sci USA. (2007) 104:3967–72. doi: 10.1073/pnas.0611618104
13. Matsumoto H, Thike AA, Li H, Yeong J, Koo SL, Dent RA, et al. Increased CD4 and CD8-positive T cell infiltrate signifies good prognosis in a subset of triple-negative breast cancer. Breast Cancer Res Treat. (2016) 156:237–47. doi: 10.1007/s10549-016-3743-x
14. Ogino T, Shigyo H, Ishii H, Katayama A, Miyokawa N, Harabuchi Y, et al. HLA class I antigen down-regulation in primary laryngeal squamous cell carcinoma lesions as a poor prognostic marker. Cancer Res. (2006) 66:9281–9. doi: 10.1158/0008-5472.CAN-06-0488
15. Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. (2012) 12:298–306. doi: 10.1038/nrc3245
16. Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. (2017) 168:707–23. doi: 10.1016/j.cell.2017.01.017
17. Patel SA, Minn AJ. Combination cancer therapy with immune checkpoint blockade: mechanisms and strategies. Immunity. (2018) 48:417–33. doi: 10.1016/j.immuni.2018.03.007
18. Boesen M, Svane IM, Engel AM, Rygaard J, Thomsen AR, Werdelin O. CD8+ T cells are crucial for the ability of congenic normal mice to reject highly immunogenic sarcomas induced in nude mice with 3-methylcholanthrene. Clin Exp Immunol. (2000) 121:210–5. doi: 10.1046/j.1365-2249.2000.01292.x
19. Walsh SR, Simovic B, Chen L, Bastin D, Nguyen A, Stephenson K, et al. Endogenous T cells prevent tumor immune escape following adoptive T cell therapy. J Clin Invest. (2019) 129:5400–10. doi: 10.1172/JCI126199
20. Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. (2005) 353:2654–66. doi: 10.1056/NEJMoa051424
21. Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. (2006) 314:126–9. doi: 10.1126/science.1129003
22. van den Broek ME, Kagi D, Ossendorp F, Toes R, Vamvakas S, Lutz WK, et al. Decreased tumor surveillance in perforin-deficient mice. J Exp Med. (1996) 184:1781–90. doi: 10.1084/jem.184.5.1781
23. Brennan AJ, Chia J, Trapani JA, Voskoboinik I. Perforin deficiency and susceptibility to cancer. Cell Death Differ. (2010) 17:607–15. doi: 10.1038/cdd.2009.212
24. Voskoboinik I, Trapani JA. Perforinopathy: a spectrum of human immune disease caused by defective perforin delivery or function. Front Immunol. (2013) 4: 441. doi: 10.3389/fimmu.2013.00441
25. Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. (2011) 331:1565–70. doi: 10.1126/science.1203486
26. Rizzo R, Fainardi E, Rouas-Freiss N, Morandi F. The role of HLA-class Ib molecules in immune-related diseases, tumors, and infections. J Immunol Res. (2017) 2017:2309574. doi: 10.1155/2017/2309574
27. da Silva IL, Montero-Montero L, Ferreira E, Quintanilla M. New insights into the role of Qa-2 and HLA-G non-classical MHC-I complexes in malignancy. Front Immunol. (2018) 9:2894. doi: 10.3389/fimmu.2018.02894
28. Bukur J, Jasinski S, Seliger B. The role of classical and non-classical HLA class I antigens in human tumors. Semin Cancer Biol. (2012) 22:350–8. doi: 10.1016/j.semcancer.2012.03.003
29. Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. (1994) 78:761–71. doi: 10.1016/S0092-8674(94)90462-6
30. Michalek MT, Grant EP, Gramm C, Goldberg AL, Rock KL. A role for the ubiquitin-dependent proteolytic pathway in MHC class I-restricted antigen presentation. Nature. (1993) 363:552–4. doi: 10.1038/363552a0
31. Shen L, Rock KL. Cellular protein is the source of cross-priming antigen in vivo. Proc Natl Acad Sci USA. (2004) 101:3035–40. doi: 10.1073/pnas.0308345101
32. Wei J, Zanker D, Di Carluccio AR, Smelkinson MG, Takeda K, Seedhom MO, et al. Varied role of ubiquitylation in generating MHC class I peptide ligands. J Immunol. (2017) 198:3835–45. doi: 10.4049/jimmunol.1602122
33. Murata S, Takahama Y, Kasahara M, Tanaka K. The immunoproteasome and thymoproteasome: functions, evolution and human disease. Nat Immunol. (2018) 19:923–31. doi: 10.1038/s41590-018-0186-z
34. Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. (1999) 17:739–79. doi: 10.1146/annurev.immunol.17.1.739
35. Kincaid EZ, Che JW, York I, Escobar H, Reyes-Vargas E, Delgado JC, et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat Immunol. (2011) 13:129–35. doi: 10.1038/ni.2203
36. Eggensperger S, Tampe R. The transporter associated with antigen processing: a key player in adaptive immunity. Biol Chem. (2015) 396:1059–72. doi: 10.1515/hsz-2014-0320
37. Roelse J, Gromme M, Momburg F, Hammerling G, Neefjes J. Trimming of TAP-translocated peptides in the endoplasmic reticulum and in the cytosol during recycling. J Exp Med. (1994) 180:1591–7. doi: 10.1084/jem.180.5.1591
38. Momburg F, Roelse J, Hammerling GJ, Neefjes JJ. Peptide size selection by the major histocompatibility complex-encoded peptide transporter. J Exp Med. (1994) 179:1613–23. doi: 10.1084/jem.179.5.1613
39. Schumacher TN, Kantesaria DV, Heemels MT, Ashton-Rickardt PG, Shepherd JC, Fruh K, et al. Peptide length and sequence specificity of the mouse TAP1/TAP2 translocator. J Exp Med. (1994) 179:533–40. doi: 10.1084/jem.179.2.533
40. Van Kaer L, Ashton-Rickardt PG, Ploegh HL, Tonegawa S. TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4-8+ T cells. Cell. (1992) 71:1205–14. doi: 10.1016/S0092-8674(05)80068-6
41. Sandberg JK, Chambers BJ, Van Kaer L, Karre K, Ljunggren HG. TAP1-deficient mice select a CD8+ T cell repertoire that displays both diversity and peptide specificity. Eur J Immunol. (1996) 26:288–93. doi: 10.1002/eji.1830260203
42. Gadola SD, Moins-Teisserenc HT, Trowsdale J, Gross WL, Cerundolo V. TAP deficiency syndrome. Clin Exp Immunol. (2000) 121:173–8. doi: 10.1046/j.1365-2249.2000.01264.x
43. Hanalioglu D, Ayvaz DC, Ozgur TT, van der Burg M, Sanal O, Tezcan I. A novel mutation in TAP1 gene leading to MHC class I deficiency: report of two cases and review of the literature. Clin Immunol. (2017) 178:74–8. doi: 10.1016/j.clim.2017.01.011
44. Cresswell P, Bangia N, Dick T, Diedrich G. The nature of the MHC class I peptide loading complex. Immunol Rev. (1999) 172:21–8. doi: 10.1111/j.1600-065X.1999.tb01353.x
45. Blees A, Januliene D, Hofmann T, Koller N, Schmidt C, Trowitzsch S, et al. Structure of the human MHC-I peptide-loading complex. Nature. (2017) 551:525–8. doi: 10.1038/nature24627
46. Schoenhals GJ, Krishna RM, Grandea AG 3rd, Spies T, Peterson PA, Yang Y, et al. Retention of empty MHC class I molecules by tapasin is essential to reconstitute antigen presentation in invertebrate cells. EMBO J. (1999) 18:743–53. doi: 10.1093/emboj/18.3.743
47. Hafstrand I, Sayitoglu EC, Apavaloaei A, Josey BJ, Sun R, Han X, et al. Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin. Proc Natl Acad Sci USA. (2019) 116:5055–60. doi: 10.1073/pnas.1807656116
48. Boyle LH, Hermann C, Boname JM, Porter KM, Patel PA, Burr ML, et al. Tapasin-related protein TAPBPR is an additional component of the MHC class I presentation pathway. Proc Natl Acad Sci USA. (2013) 110:3465–70. doi: 10.1073/pnas.1222342110
49. Grandea AG 3rd, Golovina TN, Hamilton SE, Sriram V, Spies T, Brutkiewicz RR, et al. Impaired assembly yet normal trafficking of MHC class I molecules in Tapasin mutant mice. Immunity. (2000) 13:213–22. doi: 10.1016/S1074-7613(00)00021-2
50. Coe H, Jung J, Groenendyk J, Prins D, Michalak M. ERp57 modulates STAT3 signaling from the lumen of the endoplasmic reticulum. J Biol Chem. (2010) 285:6725–38. doi: 10.1074/jbc.M109.054015
51. Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. (1987) 329:506–12. doi: 10.1038/329506a0
52. Bjorkman PJ, Parham P. Structure, function, and diversity of class I major histocompatibility complex molecules. Annu Rev Biochem. (1990) 59:253–88. doi: 10.1146/annurev.bi.59.070190.001345
53. Rammensee HG, Falk K, Rotzschke O. Peptides naturally presented by MHC class I molecules. Annu Rev Immunol. (1993) 11:213–44. doi: 10.1146/annurev.iy.11.040193.001241
54. Cascio P, Hilton C, Kisselev AF, Rock KL, Goldberg AL. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. (2001) 20:2357–66. doi: 10.1093/emboj/20.10.2357
55. York IA, Chang SC, Saric T, Keys JA, Favreau JM, Goldberg AL, et al. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8–9 residues. Nat Immunol. (2002) 3:1177–84. doi: 10.1038/ni860
56. Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. (2002) 419:480–3. doi: 10.1038/nature01074
57. Evnouchidou I, Weimershaus M, Saveanu L, van Endert P. ERAP1-ERAP2 dimerization increases peptide-trimming efficiency. J Immunol. (2014) 193:901–8. doi: 10.4049/jimmunol.1302855
58. Hearn A, York IA, Bishop C, Rock KL. Characterizing the specificity and cooperation of aminopeptidases in the cytosol and endoplasmic reticulum during MHC class I antigen presentation. J Immunol. (2010) 184:4725–32. doi: 10.4049/jimmunol.0903125
59. York IA, Brehm MA, Zendzian S, Towne CF, Rock KL. Endoplasmic reticulum aminopeptidase. (2006) 1 (ERAP1) trims MHC class I-presented peptides in vivo and plays an important role in immunodominance. Proc Natl Acad Sci USA 103:9202–7. doi: 10.1073/pnas.0603095103
60. Nagarajan NA, de Verteuil DA, Sriranganadane D, Yahyaoui W, Thibault P, Perreault C, et al. ERAAP shapes the peptidome associated with classical and nonclassical MHC class I molecules. J Immunol. (2016) 197:1035–43. doi: 10.4049/jimmunol.1500654
61. Kanaseki T, Blanchard N, Hammer GE, Gonzalez F, Shastri N. ERAAP synergizes with MHC class I molecules to make the final cut in the antigenic peptide precursors in the endoplasmic reticulum. Immunity. (2006) 25:795–806. doi: 10.1016/j.immuni.2006.09.012
62. Kanaseki T, Lind KC, Escobar H, Nagarajan N, Reyes-Vargas E, Rudd B, et al. ERAAP and tapasin independently edit the amino and carboxyl termini of MHC class I peptides. J Immunol. (2013) 191:1547–55. doi: 10.4049/jimmunol.1301043
63. Townsend A, Ohlen C, Bastin J, Ljunggren HG, Foster L, Karre K. Association of class I major histocompatibility heavy and light chains induced by viral peptides. Nature. (1989) 340:443–8. doi: 10.1038/340443a0
64. van den Elsen PJ, Holling TM, Kuipers HF, van der Stoep N. Transcriptional regulation of antigen presentation. Curr Opin Immunol. (2004) 16:67–75. doi: 10.1016/j.coi.2003.11.015
65. Kelly A, Trowsdale J. Genetics of antigen processing and presentation. Immunogenetics. (2019) 71:161–70. doi: 10.1007/s00251-018-1082-2
66. Jongsma MLM, Guarda G, Spaapen RM. The regulatory network behind MHC class I expression. Mol Immunol. (2019) 113:16–21. doi: 10.1016/j.molimm.2017.12.005
67. Meissner TB, Li A, Biswas A, Lee KH, Liu YJ, Bayir E, et al. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci USA. (2010) 107:13794–9. doi: 10.1073/pnas.1008684107
68. Hobart M, Ramassar V, Goes N, Urmson J, Halloran PF. IFN regulatory factor-1 plays a central role in the regulation of the expression of class I and II MHC genes in vivo. J Immunol. (1997) 158:4260–9
69. Kriegsman BA, Vangala P, Chen BJ, Meraner P, Brass AL, Garber M, et al. Frequent loss of IRF2 in cancers leads to immune evasion through decreased MHC class I antigen presentation and increased PD-L1 expression. J Immunol. (2019) 203:1999–2010. doi: 10.4049/jimmunol.1900475
70. Chang CH, Hammer J, Loh JE, Fodor WL, Flavell RA. The activation of major histocompatibility complex class I genes by interferon regulatory factor-1 (IRF-1). Immunogenetics. (1992) 35:378–84. doi: 10.1007/BF00179793
71. Biswas A, Meissner TB, Kawai T, Kobayashi KS. Cutting edge: impaired MHC class I expression in mice deficient for Nlrc5/class I transactivator. J Immunol. (2012) 189:516–20. doi: 10.4049/jimmunol.1200064
72. Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. (1993) 75:83–97. doi: 10.1016/S0092-8674(05)80086-8
73. Uhlen M, Karlsson MJ, Zhong W, Tebani A, Pou C, Mikes J, et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science. (2019) 366:aax9198. doi: 10.1126/science.aax9198
74. Boegel S, Lower M, Bukur T, Sorn P, Castle JC, Sahin U. HLA and proteasome expression body map. BMC Med Genom. (2018) 11:36. doi: 10.1186/s12920-018-0354-x
75. Zhou F. Molecular mechanisms of IFN-gamma to up-regulate MHC class I antigen processing and presentation. Int Rev Immunol. (2009) 28:239–60. doi: 10.1080/08830180902978120
76. Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. (1998) 95:15623–8. doi: 10.1073/pnas.95.26.15623
77. Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, et al. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci USA. (1998) 95:7556–61. doi: 10.1073/pnas.95.13.7556
78. Reyal F, Guyader C, Decraene C, Lucchesi C, Auger N, Assayag F, et al. Molecular profiling of patient-derived breast cancer xenografts. Breast Cancer Res. (2012) 14:R11. doi: 10.1186/bcr3095
79. Grasso CS, Tsoi J, Onyshchenko M, Abril-Rodriguez G, Ross-Macdonald P, Wind-Rotolo M, et al. Conserved interferon-gamma signaling drives clinical response to immune checkpoint blockade therapy in melanoma. Cancer Cell. (2020) 38:500–15 e3. doi: 10.1016/j.ccell.2020.08.005
80. Schumacher TN, Scheper W, Kvistborg P. Cancer neoantigens. Annu Rev Immunol. (2019) 37:173–200. doi: 10.1146/annurev-immunol-042617-053402
81. Galuppini F, Dal Pozzo CA, Deckert J, Loupakis F, Fassan M, Baffa R. Tumor mutation burden: from comprehensive mutational screening to the clinic. Cancer Cell Int. (2019) 19:209. doi: 10.1186/s12935-019-0929-4
82. Yarchoan M, Johnson BA 3rd, Lutz ER, Laheru DA, Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer. (2017) 17:209–22. doi: 10.1038/nrc.2016.154
83. Maleki Vareki S. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J Immunother Cancer. (2018) 6:157. doi: 10.1186/s40425-018-0479-7
84. Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. (2017) 377:2500–1. doi: 10.1056/NEJMc1713444
85. Ilyas S, Yang JC. Landscape of tumor antigens in T cell immunotherapy. J Immunol. (2015) 195:5117–22. doi: 10.4049/jimmunol.1501657
86. Disis ML, Bernhard H, Jaffee EM. Use of tumour-responsive T cells as cancer treatment. Lancet. (2009) 373:673–83. doi: 10.1016/S0140-6736(09)60404-9
87. Xiong D, Wang Y, You M. Tumor intrinsic immunity related proteins may be novel tumor suppressors in some types of cancer. Sci Rep. (2019) 9:10918. doi: 10.1038/s41598-019-47382-3
88. Safonov A, Jiang T, Bianchini G, Gyorffy B, Karn T, Hatzis C, et al. Immune gene expression is associated with genomic aberrations in breast cancer. Cancer Res. (2017) 77:3317–24. doi: 10.1158/0008-5472.CAN-16-3478
89. Kim R, Emi M, Tanabe K. Cancer immunoediting from immune surveillance to immune escape. Immunology. (2007) 121:1–14. doi: 10.1111/j.1365-2567.2007.02587.x
90. Teng MW, Swann JB, Koebel CM, Schreiber RD, Smyth MJ. Immune-mediated dormancy: an equilibrium with cancer. J Leukoc Biol. (2008) 84:988–93. doi: 10.1189/jlb.1107774
91. Schrors B, Lubcke S, Lennerz V, Fatho M, Bicker A, Wolfel C, et al. HLA class I loss in metachronous metastases prevents continuous T cell recognition of mutated neoantigens in a human melanoma model. Oncotarget. (2017) 8:28312–27. doi: 10.18632/oncotarget.16048
92. Romero I, Garrido C, Algarra I, Chamorro V, Collado A, Garrido F, et al. MHC intratumoral heterogeneity may predict cancer progression and response to immunotherapy. Front Immunol. (2018) 9:102. doi: 10.3389/fimmu.2018.00102
93. del Campo AB, Kyte JA, Carretero J, Zinchencko S, Mendez R, Gonzalez-Aseguinolaza G, et al. Immune escape of cancer cells with beta2-microglobulin loss over the course of metastatic melanoma. Int J Cancer. (2014) 134:102–13. doi: 10.1002/ijc.28338
94. Erdogdu IH. MHC class 1 and PDL-1 status of primary tumor and lymph node metastatic tumor tissue in gastric cancers. Gastroenterol Res Pract. (2019) 2019:4785098. doi: 10.1155/2019/4785098
95. Chang CC, Pirozzi G, Wen SH, Chung IH, Chiu BL, Errico S, et al. Multiple structural and epigenetic defects in the human leukocyte antigen class I antigen presentation pathway in a recurrent metastatic melanoma following immunotherapy. J Biol Chem. (2015) 290:26562–75. doi: 10.1074/jbc.M115.676130
96. Maeurer MJ, Gollin SM, Storkus WJ, Swaney W, Karbach J, Martin D, et al. Tumor escape from immune recognition: loss of HLA-A2 melanoma cell surface expression is associated with a complex rearrangement of the short arm of chromosome 6. Clin Cancer Res. (1996) 2:641–52.
97. Wei ML, Cresswell P. HLA-A2 molecules in an antigen-processing mutant cell contain signal sequence-derived peptides. Nature. (1992) 356:443–6. doi: 10.1038/356443a0
98. Williams AP, Peh CA, Purcell AW, McCluskey J, Elliott T. Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity. (2002) 16:509–20. doi: 10.1016/S1074-7613(02)00304-7
99. Yan J, Parekh VV, Mendez-Fernandez Y, Olivares-Villagomez D, Dragovic S, Hill T, et al. In vivo role of ER-associated peptidase activity in tailoring peptides for presentation by MHC class Ia and class Ib molecules. J Exp Med. (2006) 203:647–59. doi: 10.1084/jem.20052271
100. Hammer GE, Gonzalez F, James E, Nolla H, Shastri N. In the absence of aminopeptidase ERAAP, MHC class I molecules present many unstable and highly immunogenic peptides. Nat Immunol. (2007) 8:101–8. doi: 10.1038/ni1409
101. Cai L, Michelakos T, Yamada T, Fan S, Wang X, Schwab JH, et al. Defective HLA class I antigen processing machinery in cancer. Cancer Immunol Immunother. (2018) 67:999–1009. doi: 10.1007/s00262-018-2131-2
102. Cromme FV, Airey J, Heemels MT, Ploegh HL, Keating PJ, Stern PL, et al. Loss of transporter protein, encoded by the TAP-1 gene, is highly correlated with loss of HLA expression in cervical carcinomas. J Exp Med. (1994) 179:335–40. doi: 10.1084/jem.179.1.335
103. Seliger B, Hohne A, Knuth A, Bernhard H, Ehring B, Tampe R, et al. Reduced membrane major histocompatibility complex class I density and stability in a subset of human renal cell carcinomas with low TAP and LMP expression. Clin Cancer Res. (1996) 2:1427–33.
104. Seliger B, Atkins D, Bock M, Ritz U, Ferrone S, Huber C, et al. Characterization of human lymphocyte antigen class I antigen-processing machinery defects in renal cell carcinoma lesions with special emphasis on transporter-associated with antigen-processing down-regulation. Clin Cancer Res. (2003) 9:1721–7.
105. Kaklamanis L, Townsend A, Doussis-Anagnostopoulou IA, Mortensen N, Harris AL, Gatter KC. Loss of major histocompatibility complex-encoded transporter associated with antigen presentation (TAP) in colorectal cancer. Am J Pathol. (1994) 145:505–9.
106. Lou Y, Vitalis TZ, Basha G, Cai B, Chen SS, Choi KB, et al. Restoration of the expression of transporters associated with antigen processing in lung carcinoma increases tumor-specific immune responses and survival. Cancer Res. (2005) 65:7926–33. doi: 10.1158/0008-5472.CAN-04-3977
107. Bernal M, Garcia-Alcalde F, Concha A, Cano C, Blanco A, Garrido F, et al. Genome-wide differential genetic profiling characterizes colorectal cancers with genetic instability and specific routes to HLA class I loss and immune escape. Cancer Immunol Immunother. (2012) 61:803–16. doi: 10.1007/s00262-011-1147-7
108. Bernal M, Ruiz-Cabello F, Concha A, Paschen A, Garrido F. Implication of the beta2-microglobulin gene in the generation of tumor escape phenotypes. Cancer Immunol Immunother. (2012) 61:1359–71. doi: 10.1007/s00262-012-1321-6
109. Keating PJ, Cromme FV, Duggan-Keen M, Snijders PJ, Walboomers JM, Hunter RD, et al. Frequency of down-regulation of individual HLA-A and -B alleles in cervical carcinomas in relation to TAP-1 expression. Br J Cancer. (1995) 72:405–11. doi: 10.1038/bjc.1995.346
110. Tanaka K, Tsuchikawa T, Miyamoto M, Maki T, Ichinokawa M, Kubota KC, et al. Down-regulation of Human Leukocyte Antigen class I heavy chain in tumors is associated with a poor prognosis in advanced esophageal cancer patients. Int J Oncol. (2012) 40:965–74. doi: 10.3892/ijo.2011.1274
111. Shionoya Y, Kanaseki T, Miyamoto S, Tokita S, Hongo A, Kikuchi Y, et al. Loss of tapasin in human lung and colon cancer cells and escape from tumor-associated antigen-specific CTL recognition. Oncoimmunology. (2017) 6:e1274476. doi: 10.1080/2162402X.2016.1274476
112. Sokol L, Koelzer VH, Rau TT, Karamitopoulou E, Zlobec I, Lugli A. Loss of tapasin correlates with diminished CD8(+) T-cell immunity and prognosis in colorectal cancer. J Transl Med. (2015) 13:279. doi: 10.1186/s12967-015-0647-1
113. Hasim A, Abudula M, Aimiduo R, Ma JQ, Jiao Z, Akula G, et al. Post-transcriptional and epigenetic regulation of antigen processing machinery (APM) components and HLA-I in cervical cancers from Uighur women. PLoS ONE. (2012) 7:e44952. doi: 10.1371/journal.pone.0044952
114. Sheyhidin I, Hasim A, Zheng F, Ma H. Epigenetic changes within the promoter regions of antigen processing machinery family genes in Kazakh primary esophageal squamous cell carcinoma. Asian Pac J Cancer Prev. (2014) 15:10299–306. doi: 10.7314/APJCP.2014.15.23.10299
115. Liu Q, Hao C, Su P, Shi J. Down-regulation of HLA class I antigen-processing machinery components in esophageal squamous cell carcinomas: association with disease progression. Scand J Gastroenterol. (2009) 44:960–9. doi: 10.1080/00365520902998679
116. Mehta AM, Jordanova ES, Kenter GG, Ferrone S, Fleuren GJ. Association of antigen processing machinery and HLA class I defects with clinicopathological outcome in cervical carcinoma. Cancer Immunol Immunother. (2008) 57:197–206. doi: 10.1007/s00262-007-0362-8
117. Stratikos E, Stamogiannos A, Zervoudi E, Fruci D. A role for naturally occurring alleles of endoplasmic reticulum aminopeptidases in tumor immunity and cancer pre-disposition. Front Oncol. (2014) 4:363. doi: 10.3389/fonc.2014.00363
118. Lee D, Das Gupta J, Gaughan C, Steffen I, Tang N, Luk KC, et al. In-depth investigation of archival and prospectively collected samples reveals no evidence for XMRV infection in prostate cancer. PLoS ONE. (2012) 7:e44954. doi: 10.1371/journal.pone.0044954
119. Ayshamgul H, Ma H, Ilyar S, Zhang LW, Abulizi A. Association of defective HLA-I expression with antigen processing machinery and their association with clinicopathological characteristics in Kazak patients with esophageal cancer. Chin Med J. (2011) 124:341–6. doi: 10.3760/cma.j.issn.0366-6999.2011.03.004
120. Angell TE, Lechner MG, Jang JK, LoPresti JS, Epstein AL. MHC class I loss is a frequent mechanism of immune escape in papillary thyroid cancer that is reversed by interferon and selumetinib treatment in vitro. Clin Cancer Res. (2014) 20:6034–44. doi: 10.1158/1078-0432.CCR-14-0879
121. Jang M, Yew PY, Hasegawa K, Ikeda Y, Fujiwara K, Fleming GF, et al. Characterization of T cell repertoire of blood, tumor, and ascites in ovarian cancer patients using next generation sequencing. Oncoimmunology. (2015) 4:e1030561. doi: 10.1080/2162402X.2015.1030561
122. Simpson JA, Al-Attar A, Watson NF, Scholefield JH, Ilyas M, Durrant LG. Intratumoral T cell infiltration, MHC class I and STAT1 as biomarkers of good prognosis in colorectal cancer. Gut. (2010) 59:926–33. doi: 10.1136/gut.2009.194472
123. Yoshihama S, Roszik J, Downs I, Meissner TB, Vijayan S, Chapuy B, et al. NLRC5/MHC class I transactivator is a target for immune evasion in cancer. Proc Natl Acad Sci USA. (2016) 113:5999–6004. doi: 10.1073/pnas.1602069113
124. Ito S, Okano S, Morita M, Saeki H, Tsutsumi S, Tsukihara H, et al. Expression of PD-L1 and HLA class I in esophageal squamous cell carcinoma: prognostic factors for patient outcome. Ann Surg Oncol. (2016) 23:508–15. doi: 10.1245/s10434-016-5376-z
125. Yoo SH, Keam B, Ock CY, Kim S, Han B, Kim JW, et al. Prognostic value of the association between MHC class I downregulation and PD-L1 upregulation in head and neck squamous cell carcinoma patients. Sci Rep. (2019) 9:7680. doi: 10.1038/s41598-019-44206-2
126. Rodig SJ, Gusenleitner D, Jackson DG, Gjini E, Giobbie-Hurder A, Jin C, et al. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci Transl Med. (2018) 10. doi: 10.1126/scitranslmed.aar3342
127. Pedersen MH, Hood BL, Beck HC, Conrads TP, Ditzel HJ, Leth-Larsen R. Downregulation of antigen presentation-associated pathway proteins is linked to poor outcome in triple-negative breast cancer patient tumors. Oncoimmunology. (2017) 6:e1305531. doi: 10.1080/2162402X.2017.1305531
128. Ling A, Lofgren-Burstrom A, Larsson P, Li X, Wikberg ML, Oberg A, et al. TAP1 down-regulation elicits immune escape and poor prognosis in colorectal cancer. Oncoimmunology. (2017) 6:e1356143. doi: 10.1080/2162402X.2017.1356143
129. Basile MS, Mazzon E, Russo A, Mammana S, Longo A, Bonfiglio V, et al. Differential modulation and prognostic values of immune-escape genes in uveal melanoma. PLoS ONE. (2019) 14:e0210276. doi: 10.1371/journal.pone.0210276
130. Thuring C, Follin E, Geironson L, Freyhult E, Junghans V, Harndahl M, et al. HLA class I is most tightly linked to levels of tapasin compared with other antigen-processing proteins in glioblastoma. Br J Cancer. (2015) 113:952–62. doi: 10.1038/bjc.2015.297
131. Marincola FM, Jaffee EM, Hicklin DJ, Ferrone S. Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Adv Immunol. (2000) 74:181–273. doi: 10.1016/S0065-2776(08)60911-6
132. Kageshita T, Hirai S, Ono T, Hicklin DJ, Ferrone S. Down-regulation of HLA class I antigen-processing molecules in malignant melanoma: association with disease progression. Am J Pathol. (1999) 154:745–54. doi: 10.1016/S0002-9440(10)65321-7
133. Kamarashev J, Ferrone S, Seifert B, Boni R, Nestle FO, Burg G, et al. TAP1 down-regulation in primary melanoma lesions: an independent marker of poor prognosis. Int J Cancer. (2001) 95:23–83. doi: 10.1002/1097-0215(20010120)95:1<23::AID-IJC1004>3.0.CO
134. Ogino T, Bandoh N, Hayashi T, Miyokawa N, Harabuchi Y, Ferrone S. Association of tapasin and HLA class I antigen down-regulation in primary maxillary sinus squamous cell carcinoma lesions with reduced survival of patients. Clin Cancer Res. (2003) 9:4043–51.
135. Bandoh N, Ogino T, Katayama A, Takahara M, Katada A, Hayashi T, et al. HLA class I antigen and transporter associated with antigen processing downregulation in metastatic lesions of head and neck squamous cell carcinoma as a marker of poor prognosis. Oncol Rep. (2010) 23:933–9. doi: 10.3892/or_00000717
136. Hosch SB, Meyer AJ, Schneider C, Stoecklein N, Prenzel KL, Pantel K, et al. Expression and prognostic significance of HLA class I, ICAM-1, and tumor-infiltrating lymphocytes in esophageal cancer. J Gastrointest Surg. (1997) 1:316–23. doi: 10.1016/S1091-255X(97)80051-0
137. Hosch SB, Izbicki JR, Pichlmeier U, Stoecklein N, Niendorf A, Knoefel WT, et al. Expression and prognostic significance of immunoregulatory molecules in esophageal cancer. Int J Cancer. (1997) 74:582–73. doi: 10.1002/(SICI)1097-0215(19971219)74:6<582::AID-IJC4>3.0.CO
138. Mizukami Y, Kono K, Maruyama T, Watanabe M, Kawaguchi Y, Kamimura K, et al. Downregulation of HLA Class I molecules in the tumour is associated with a poor prognosis in patients with oesophageal squamous cell carcinoma. Br J Cancer. (2008) 99:1462–7. doi: 10.1038/sj.bjc.6604715
139. Zhang X, Lin A, Zhang JG, Bao WG, Xu DP, Ruan YY, et al. Alteration of HLA-F and HLA I antigen expression in the tumor is associated with survival in patients with esophageal squamous cell carcinoma. Int J Cancer. (2013) 132:82–9. doi: 10.1002/ijc.27621
140. Zhao J, Guo C, Xiong F, Yu J, Ge J, Wang H, et al. Single cell RNA-seq reveals the landscape of tumor and infiltrating immune cells in nasopharyngeal carcinoma. Cancer Lett. (2020) 477:131–43. doi: 10.1016/j.canlet.2020.02.010
141. Gettinger S, Choi J, Hastings K, Truini A, Datar I, Sowell R, et al. Impaired HLA class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer. Cancer Discov. (2017) 7:1420–35. doi: 10.1158/2159-8290.CD-17-0593
142. Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, et al. Identification of essential genes for cancer immunotherapy. Nature. (2017) 548:537–42. doi: 10.1038/nature23477
143. Sade-Feldman M, Jiao YJ, Chen JH, Rooney MS, Barzily-Rokni M, Eliane JP, et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun. (2017) 8:1136. doi: 10.1038/s41467-017-01062-w
144. Paulson KG, Voillet V, McAfee MS, Hunter DS, Wagener FD, Perdicchio M, et al. Acquired cancer resistance to combination immunotherapy from transcriptional loss of class I HLA. Nat Commun. (2018) 9:3868. doi: 10.1038/s41467-018-06300-3
145. Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. (2018) 359:582–7. doi: 10.1126/science.aao4572
146. Lee JH, Shklovskaya E, Lim SY, Carlino MS, Menzies AM, Stewart A, et al. Transcriptional downregulation of MHC class I and melanoma de- differentiation in resistance to PD-1 inhibition. Nat Commun. (2020) 11:1897. doi: 10.1038/s41467-020-15726-7
147. Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. (2016) 375:819–29. doi: 10.1056/NEJMoa1604958
148. Lauss M, Donia M, Harbst K, Andersen R, Mitra S, Rosengren F, et al. Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nat Commun. (2017) 8:1738. doi: 10.1038/s41467-017-01460-0
149. Khong HT, Wang QJ, Rosenberg SA. Identification of multiple antigens recognized by tumor-infiltrating lymphocytes from a single patient: tumor escape by antigen loss and loss of MHC expression. J Immunother. (2004) 27:184–90. doi: 10.1097/00002371-200405000-00002
150. Carretero R, Romero JM, Ruiz-Cabello F, Maleno I, Rodriguez F, Camacho FM, et al. Analysis of HLA class I expression in progressing and regressing metastatic melanoma lesions after immunotherapy. Immunogenetics. (2008) 60:439–47. doi: 10.1007/s00251-008-0303-5
151. Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, et al. Loss of IFN-gamma Pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. (2016) 167:397–404 e9. doi: 10.1016/j.cell.2016.08.069
152. Burrack AL, Spartz EJ, Raynor JF, Wang I, Olson M, Stromnes IM. Combination PD-1 and PD-L1 blockade promotes durable neoantigen-specific T cell-mediated immunity in pancreatic ductal adenocarcinoma. Cell Rep. (2019) 28:2140–55 e6. doi: 10.1016/j.celrep.2019.07.059
153. Chew GL, Campbell AE, De Neef E, Sutliff NA, Shadle SC, Tapscott SJ, et al. DUX4 suppresses MHC class I to promote cancer immune evasion and resistance to checkpoint blockade. Dev Cell. (2019) 50:658–71 e7. doi: 10.1016/j.devcel.2019.06.011
154. Buferne M, Chasson L, Grange M, Mas A, Arnoux F, Bertuzzi M, et al. IFNgamma producing CD8(+) T cells modified to resist major immune checkpoints induce regression of MHC class I-deficient melanomas. Oncoimmunology. (2015) 4:e974959. doi: 10.4161/2162402X.2014.974959
155. Kavathas P, Bach FH, DeMars R. Gamma ray-induced loss of expression of HLA and glyoxalase I alleles in lymphoblastoid cells. Proc Natl Acad Sci USA. (1980) 77:4251–5. doi: 10.1073/pnas.77.7.4251
156. Erlich H, Lee JS, Petersen JW, Bugawan T, DeMars R. Molecular analysis of HLA class I and class II antigen loss mutants reveals a homozygous deletion of the DR, DQ, and part of the DP region: implications for class II gene order. Hum Immunol. (1986) 16:205–19. doi: 10.1016/0198-8859(86)90049-2
157. Shimizu Y, Geraghty DE, Koller BH, Orr HT, DeMars R. Transfer and expression of three cloned human non-HLA-A,B,C class I major histocompatibility complex genes in mutant lymphoblastoid cells. Proc Natl Acad Sci USA. (1988) 85:227–31. doi: 10.1073/pnas.85.1.227
158. McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell. (2017) 171:1259–71 e11. doi: 10.1016/j.cell.2017.10.001
159. Benitez R, Godelaine D, Lopez-Nevot MA, Brasseur F, Jimenez P, Marchand M, et al. Mutations of the beta2-microglobulin gene result in a lack of HLA class I molecules on melanoma cells of two patients immunized with MAGE peptides. Tissue Antigens. (1998) 52:520–9. doi: 10.1111/j.1399-0039.1998.tb03082.x
160. Maleno I, Aptsiauri N, Cabrera T, Gallego A, Paschen A, Lopez-Nevot MA, et al. Frequent loss of heterozygosity in the beta2-microglobulin region of chromosome. (2011) 15 in primary human tumors. Immunogenetics. 63:65–71. doi: 10.1007/s00251-010-0494-4
161. Maleno I, Cabrera CM, Cabrera T, Paco L, Lopez-Nevot MA, Collado A, et al. Distribution of HLA class I altered phenotypes in colorectal carcinomas: high frequency of HLA haplotype loss associated with loss of heterozygosity in chromosome region 6p21. Immunogenetics. (2004) 56:244–53. doi: 10.1007/s00251-004-0692-z
162. Maleno I, Lopez-Nevot MA, Cabrera T, Salinero J, Garrido F. Multiple mechanisms generate HLA class I altered phenotypes in laryngeal carcinomas: high frequency of HLA haplotype loss associated with loss of heterozygosity in chromosome region 6p21. Cancer Immunol Immunother. (2002) 51:389–96. doi: 10.1007/s00262-002-0296-0
163. Maleno I, Romero JM, Cabrera T, Paco L, Aptsiauri N, Cozar JM, et al. LOH at 6p21.3 region and HLA class I altered phenotypes in bladder carcinomas. Immunogenetics. (2006) 58:503–10. doi: 10.1007/s00251-006-0111-8
164. Feenstra M, Veltkamp M, van Kuik J, Wiertsema S, Slootweg P, van den Tweel J, et al. HLA class I expression and chromosomal deletions at 6p and 15q in head and neck squamous cell carcinomas. Tissue Antigens. (1999) 54:235–45. doi: 10.1034/j.1399-0039.1999.540304.x
165. Seliger B, Ritz U, Abele R, Bock M, Tampe R, Sutter G, et al. Immune escape of melanoma: first evidence of structural alterations in two distinct components of the MHC class I antigen processing pathway. Cancer Res. (2001) 61:8647–50.
166. Montesion M, Murugesan K, Jin DX, Sharaf R, Sanchez N, Guria A, et al. Somatic HLA class I loss is a widespread mechanism of immune evasion which refines the use of tumor mutational burden as a biomarker of checkpoint inhibitor response. Cancer Discov. (2020) 11:282–92. doi: 10.1158/2159-8290.CD-20-0672
167. Chang CC, Campoli M, Restifo NP, Wang X, Ferrone S. Immune selection of hot-spot beta. (2005) 2-microglobulin gene mutations, HLA-A2 allospecificity loss, and antigen-processing machinery component down-regulation in melanoma cells derived from recurrent metastases following immunotherapy. J Immunol 174:1462–71. doi: 10.4049/jimmunol.174.3.1462
168. Garrido F, Aptsiauri N. Cancer immune escape: MHC expression in primary tumours versus metastases. Immunology. (2019) 158:255–66. doi: 10.1111/imm.13114
169. Carretero R, Cabrera T, Gil H, Saenz-Lopez P, Maleno I, Aptsiauri N, et al. Bacillus Calmette-Guerin immunotherapy of bladder cancer induces selection of human leukocyte antigen class I-deficient tumor cells. Int J Cancer. (2011) 129:839–46. doi: 10.1002/ijc.25733
170. Cabrera T, Pedrajas G, Cozar JM, Garrido A, Vicente J, Tallada M, et al. HLA class I expression in bladder carcinomas. Tissue Antigens. (2003) 62:324–7. doi: 10.1034/j.1399-0039.2003.00104.x
171. Shen T, Chen Z, Zhao ZJ, Wu J. Genetic defects of the IRF1-mediated major histocompatibility complex class I antigen presentation pathway occur prevalently in the JAK2 gene in non-small cell lung cancer. Oncotarget. (2017) 8:60975–86. doi: 10.18632/oncotarget.17689
172. Georgopoulos NT, Proffitt JL, Blair GE. Transcriptional regulation of the major histocompatibility complex (MHC) class I heavy chain, TAP1 and LMP2 genes by the human papillomavirus (HPV) type 6b, 16 and 18 E7 oncoproteins. Oncogene. (2000) 19:4930–5. doi: 10.1038/sj.onc.1203860
173. Romero JM, Jimenez P, Cabrera T, Cozar JM, Pedrinaci S, Tallada M, et al. Coordinated downregulation of the antigen presentation machinery and HLA class I/beta2-microglobulin complex is responsible for HLA-ABC loss in bladder cancer. Int J Cancer. (2005) 113:605–10. doi: 10.1002/ijc.20499
174. Restifo NP, Esquivel F, Kawakami Y, Yewdell JW, Mule JJ, Rosenberg SA, et al. Identification of human cancers deficient in antigen processing. J Exp Med. (1993) 177:265–72. doi: 10.1084/jem.177.2.265
175. Meissner M, Reichert TE, Kunkel M, Gooding W, Whiteside TL, Ferrone S, et al. Defects in the human leukocyte antigen class I antigen processing machinery in head and neck squamous cell carcinoma: association with clinical outcome. Clin Cancer Res. (2005) 11:2552–60. doi: 10.1158/1078-0432.CCR-04-2146
176. Ye Q, Shen Y, Wang X, Yang J, Miao F, Shen C, et al. Hypermethylation of HLA class I gene is associated with HLA class I down-regulation in human gastric cancer. Tissue Antigens. (2010) 75:30–9. doi: 10.1111/j.1399-0039.2009.01390.x
177. Burr ML, Sparbier CE, Chan KL, Chan YC, Kersbergen A, Lam EYN, et al. An evolutionarily conserved function of polycomb silences the MHC class I antigen presentation pathway and enables immune evasion in cancer. Cancer Cell. (2019) 36:385–401 e8. doi: 10.1016/j.ccell.2019.08.008
178. Ozcan M, Janikovits J, von Knebel Doeberitz M, Kloor M. Complex pattern of immune evasion in MSI colorectal cancer. Oncoimmunology. (2018) 7:e1445453. doi: 10.1080/2162402X.2018.1445453
179. Smithy JW, Moore LM, Pelekanou V, Rehman J, Gaule P, Wong PF, et al. Nuclear IRF-1 expression as a mechanism to assess “Capability” to express PD-L1 and response to PD-1 therapy in metastatic melanoma. J Immunother Cancer. (2017) 5:25. doi: 10.1186/s40425-017-0229-2
180. Lorenzi S, Forloni M, Cifaldi L, Antonucci C, Citti A, Boldrini R, et al. IRF1 and NF-kB restore MHC class I-restricted tumor antigen processing and presentation to cytotoxic T cells in aggressive neuroblastoma. PLoS ONE. (2012) 7:e46928. doi: 10.1371/journal.pone.0046928
181. Serrano A, Tanzarella S, Lionello I, Mendez R, Traversari C, Ruiz-Cabello F, et al. Rexpression of HLA class I antigens and restoration of antigen-specific CTL response in melanoma cells following 5-aza-2'-deoxycytidine treatment. Int J Cancer. (2001) 94:243–51. doi: 10.1002/ijc.1452
182. Luo N, Nixon MJ, Gonzalez-Ericsson PI, Sanchez V, Opalenik SR, Li H, et al. DNA methyltransferase inhibition upregulates MHC-I to potentiate cytotoxic T lymphocyte responses in breast cancer. Nat Commun. (2018) 9:248. doi: 10.1038/s41467-017-02630-w
183. Nie Y, Yang G, Song Y, Zhao X, So C, Liao J, et al. DNA hypermethylation is a mechanism for loss of expression of the HLA class I genes in human esophageal squamous cell carcinomas. Carcinogenesis. (2001) 22:1615–23. doi: 10.1093/carcin/22.10.1615
184. Jiang Q, Pan HY, Ye DX, Zhang P, Zhong LP, Zhang ZY. Downregulation of tapasin expression in primary human oral squamous cell carcinoma: association with clinical outcome. Tumour Biol. (2010) 31:451–9. doi: 10.1007/s13277-010-0054-4
185. Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. (2015) 162:974–86. doi: 10.1016/j.cell.2015.07.011
186. Stone ML, Chiappinelli KB, Li H, Murphy LM, Travers ME, Topper MJ, et al. Epigenetic therapy activates type I interferon signaling in murine ovarian cancer to reduce immunosuppression and tumor burden. Proc Natl Acad Sci USA. (2017) 114: E10981–90. doi: 10.1073/pnas.1712514114
187. Kim JY, Choi JK, Jung H. Genome-wide methylation patterns predict clinical benefit of immunotherapy in lung cancer. Clin Epigenetics. (2020) 12:119. doi: 10.1186/s13148-020-00907-4
188. Fonsatti E, Nicolay HJ, Sigalotti L, Calabro L, Pezzani L, Colizzi F, et al. Functional up-regulation of human leukocyte antigen class I antigens expression by 5-aza-2'-deoxycytidine in cutaneous melanoma: immunotherapeutic implications. Clin Cancer Res. (2007) 13:3333–8. doi: 10.1158/1078-0432.CCR-06-3091
189. Rosenthal R, Cadieux EL, Salgado R, Bakir MA, Moore DA, Hiley CT, et al. Neoantigen-directed immune escape in lung cancer evolution. Nature. (2019) 567:479–85. doi: 10.1038/s41586-019-1032-7
190. Magner WJ, Kazim AL, Stewart C, Romano MA, Catalano G, Grande C, et al. Activation of MHC class I, II, and CD40 gene expression by histone deacetylase inhibitors. J Immunol. (2000) 165:7017–24. doi: 10.4049/jimmunol.165.12.7017
191. Setiadi AF, Omilusik K, David MD, Seipp RP, Hartikainen J, Gopaul R, et al. Epigenetic enhancement of antigen processing and presentation promotes immune recognition of tumors. Cancer Res. (2008) 68:9601–7. doi: 10.1158/0008-5472.CAN-07-5270
192. Ritter C, Fan K, Paschen A, Reker Hardrup S, Ferrone S, Nghiem P, et al. Epigenetic priming restores the HLA class-I antigen processing machinery expression in Merkel cell carcinoma. Sci Rep. (2017) 7:2290. doi: 10.1038/s41598-017-02608-0
193. Briere D, Sudhakar N, Woods DM, Hallin J, Engstrom LD, Aranda R, et al. The class I/IV HDAC inhibitor mocetinostat increases tumor antigen presentation, decreases immune suppressive cell types and augments checkpoint inhibitor therapy. Cancer Immunol Immunother. (2018) 67:381–92. doi: 10.1007/s00262-017-2091-y
194. Ugurel S, Spassova I, Wohlfarth J, Drusio C, Cherouny A, Melior A, et al. MHC class-I downregulation in PD-1/PD-L1 inhibitor refractory Merkel cell carcinoma and its potential reversal by histone deacetylase inhibition: a case series. Cancer Immunol Immunother. (2019) 68:983–90. doi: 10.1007/s00262-019-02341-9
195. Zhou L, Mudianto T, Ma X, Riley R, Uppaluri R. Targeting EZH2 enhances antigen presentation, antitumor immunity, and circumvents anti-PD-1 resistance in head and neck cancer. Clin Cancer Res. (2020) 26:290–300. doi: 10.1158/1078-0432.CCR-19-1351
196. Almeida L, Dhillon-LaBrooy A, Castro CN, Adossa N, Carriche GM, Guderian M, et al. Ribosome-targeting antibiotics impair T cell effector function and ameliorate autoimmunity by blocking mitochondrial protein synthesis. Immunity. (2020) 54:68–83.e6. doi: 10.1101/832956
197. Ennishi D, Takata K, Beguelin W, Duns G, Mottok A, Farinha P, et al. Molecular and genetic characterization of MHC deficiency identifies EZH2 as therapeutic target for enhancing immune recognition. Cancer Discov. (2019) 9:546–63. doi: 10.1158/2159-8290.CD-18-1090
198. Pan D, Kobayashi A, Jiang P, Ferrari de Andrade L, Tay RE, Luoma AM, et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science. (2018) 359:770–5. doi: 10.1126/science.aao1710
199. Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. (2019) 179:1033–55. doi: 10.1016/j.cell.2019.10.017
200. Liu L, Wang Q, Qiu Z, Kang Y, Liu J, Ning S, et al. Non-coding RNAs: the shot callers in tumor immune escape. Signal Transduct Target Ther. (2020) 5:102. doi: 10.1038/s41392-020-0194-y
201. Mari L, Hoefnagel SJM, Zito D, van de Meent M, van Endert P, Calpe S, et al. microRNA 125a regulates MHC-I expression on esophageal adenocarcinoma cells, associated with suppression of antitumor immune response and poor outcomes of patients. Gastroenterology. (2018) 155:784–98. doi: 10.1053/j.gastro.2018.06.030
202. Lazaridou MF, Massa C, Handke D, Mueller A, Friedrich M, Subbarayan K, et al. Identification of microRNAs targeting the transporter associated with antigen processing TAP1 in Melanoma. J Clin Med. (2020) 9:92690. doi: 10.3390/jcm9092690
203. Colangelo T, Polcaro G, Ziccardi P, Pucci B, Muccillo L, Galgani M, et al. Proteomic screening identifies calreticulin as a miR-27a direct target repressing MHC class I cell surface exposure in colorectal cancer. Cell Death Dis. (2016) 7:e2120. doi: 10.1038/cddis.2016.28
204. Yan Y, Liang Z, Du Q, Yang M, Geller DA. MicroRNA-23a downregulates the expression of interferon regulatory factor-1 in hepatocellular carcinoma cells. Oncol Rep. (2016) 36:633–40. doi: 10.3892/or.2016.4864
205. Wang B, Yang H, Shen L, Wang J, Pu W, Chen Z, et al. Rs56288038 (C/G) in 3′UTR of IRF-1 regulated by MiR-502-5p promotes gastric cancer development. Cell Physiol Biochem. (2016) 40:391–9. doi: 10.1159/000452554
206. Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. (2019) 21:542–51. doi: 10.1038/s41556-019-0311-8
207. Hu Q, Ye Y, Chan LC, Li Y, Liang K, Lin A, et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol. (2019) 20:835–51. doi: 10.1038/s41590-019-0400-7
208. Li H, Xiong HG, Xiao Y, Yang QC, Yang SC, Tang HC, et al. Long non-coding RNA LINC02195 as a regulator of MHC I molecules and favorable prognostic marker for head and neck squamous cell carcinoma. Front Oncol. (2020) 10:615. doi: 10.3389/fonc.2020.00615
209. Gao F, Zhao ZL, Zhao WT, Fan QR, Wang SC, Li J, et al. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem Biophys Res Commun. (2013) 431:610–6. doi: 10.1016/j.bbrc.2012.12.097
210. Cano F, Bye H, Duncan LM, Buchet-Poyau K, Billaud M, Wills MR, et al. The RNA-binding E3 ubiquitin ligase MEX-3C links ubiquitination with MHC-I mRNA degradation. EMBO J. (2012) 31:3596–606. doi: 10.1038/emboj.2012.218
211. Wang Y, Wang X, Cui X, Zhuo Y, Li H, Ha C, et al. Oncoprotein SND1 hijacks nascent MHC-I heavy chain to ER-associated degradation, leading to impaired CD8(+) T cell response in tumor. Sci Adv. (2020) 6:aba5412. doi: 10.1126/sciadv.aba5412
212. Jongsma MLM, de Waard AA, Raaben M, Zhang T, Cabukusta B, Platzer R, et al. The SPPL3-defined glycosphingolipid repertoire orchestrates HLA class I-mediated immune responses. Immunity. (2020) 54:132–50.e9. doi: 10.1016/j.immuni.2021.01.016
213. Mimura K, Shiraishi K, Mueller A, Izawa S, Kua LF, So J, et al. The MAPK pathway is a predominant regulator of HLA-A expression in esophageal and gastric cancer. J Immunol. (2013) 191:6261–72. doi: 10.4049/jimmunol.1301597
214. Brea EJ, Oh CY, Manchado E, Budhu S, Gejman RS, Mo G, et al. Kinase regulation of human MHC class I molecule expression on cancer cells. Cancer Immunol Res. (2016) 4:936–47. doi: 10.1158/2326-6066.CIR-16-0177
215. Lulli D, Carbone ML, Pastore S. The MEK inhibitors trametinib and cobimetinib induce a type I interferon response in human keratinocytes. Int J Mol Sci. (2017) 18:102227. doi: 10.3390/ijms18102227
216. Oh CY, Klatt MG, Bourne C, Dao T, Dacek MM, Brea EJ, et al. ALK and RET inhibitors promote HLA class I antigen presentation and unmask new antigens within the tumor immunopeptidome. Cancer Immunol Res. (2019) 7:1984–97. doi: 10.1158/2326-6066.CIR-19-0056
217. Mimura K, Ando T, Poschke I, Mougiakakos D, Johansson CC, Ichikawa J, et al. T cell recognition of HLA-A2 restricted tumor antigens is impaired by the oncogene HER2. Int J Cancer. (2011) 128:390–401. doi: 10.1002/ijc.25613
218. Maruyama T, Mimura K, Sato E, Watanabe M, Mizukami Y, Kawaguchi Y, et al. Inverse correlation of HER2 with MHC class I expression on oesophageal squamous cell carcinoma. Br J Cancer. (2010) 103:552–9. doi: 10.1038/sj.bjc.6605772
219. Bernards R, Dessain SK, Weinberg RA. N-myc amplification causes down-modulation of MHC class I antigen expression in neuroblastoma. Cell. (1986) 47:667–74. doi: 10.1016/0092-8674(86)90509-X
220. Versteeg R, Noordermeer IA, Kruse-Wolters M, Ruiter DJ, Schrier PI. c-myc down-regulates class I HLA expression in human melanomas. EMBO J. (1988) 7:1023–9. doi: 10.1002/j.1460-2075.1988.tb02909.x
221. van 't Veer LJ, Beijersbergen RL, Bernards R. N-myc suppresses major histocompatibility complex class I gene expression through down-regulation of the p50 subunit of NF-kappa B. EMBO J. (1993) 12:195–200. doi: 10.1002/j.1460-2075.1993.tb05645.x
222. Goes N, Sims T, Urmson J, Vincent D, Ramassar V, Halloran PF. Disturbed MHC regulation in the IFN-gamma knockout mouse. Evidence for three states of MHC expression with distinct roles for IFN-gamma. J Immunol. (1995) 155:4559–66.
223. Lee CK, Gimeno R, Levy DE. Differential regulation of constitutive major histocompatibility complex class I expression in T and B lymphocytes. J Exp Med. (1999) 190:1451–64. doi: 10.1084/jem.190.10.1451
224. Majoros A, Platanitis E, Kernbauer-Holzl E, Rosebrock F, Muller M, Decker T. Canonical and non-canonical aspects of JAK-STAT signaling: lessons from interferons for cytokine responses. Front Immunol. (2017) 8:29. doi: 10.3389/fimmu.2017.00029
225. Sucker A, Zhao F, Pieper N, Heeke C, Maltaner R, Stadtler N, et al. Acquired IFNgamma resistance impairs anti-tumor immunity and gives rise to T-cell-resistant melanoma lesions. Nat Commun. (2017) 8:15440. doi: 10.1038/ncomms15440
226. Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. (2017) 547:413–8. doi: 10.1038/nature23270
227. Marijt KA, Sluijter M, Blijleven L, Tolmeijer SH, Scheeren FA, van der Burg SH, et al. Metabolic stress in cancer cells induces immune escape through a PI3K-dependent blockade of IFNgamma receptor signaling. J Immunother Cancer. (2019) 7:152. doi: 10.1186/s40425-019-0627-8
228. Lu T, Ramakrishnan R, Altiok S, Youn JI, Cheng P, Celis E, et al. Tumor-infiltrating myeloid cells induce tumor cell resistance to cytotoxic T cells in mice. J Clin Invest. (2011) 121:4015–29. doi: 10.1172/JCI45862
229. Mundy-Bosse BL, Lesinski GB, Jaime-Ramirez AC, Benninger K, Khan M, Kuppusamy P, et al. Myeloid-derived suppressor cell inhibition of the IFN response in tumor-bearing mice. Cancer Res. (2011) 71:5101–10. doi: 10.1158/0008-5472.CAN-10-2670
230. Dong MB, Wang G, Chow RD, Ye L, Zhu L, Dai X, et al. Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells. Cell. (2019) 178:1189–204 e23. doi: 10.1016/j.cell.2019.07.044
231. Cruz FM, Colbert JD, Merino E, Kriegsman BA, Rock KL. The biology and underlying mechanisms of cross-presentation of exogenous antigens on MHC-I molecules. Annu Rev Immunol. (2017) 35:149–76. doi: 10.1146/annurev-immunol-041015-055254
232. Dersh D, Phelan JD, Gumina ME, Wang B, Arbuckle JH, Holly J, et al. Genome-wide screens identify lineage- and tumor-specific genes modulating MHC-I- and MHC-II-restricted immunosurveillance of human lymphomas. Immunity. (2020) 54:116–31.e10. doi: 10.2139/ssrn.3584739
233. del Campo AB, Aptsiauri N, Mendez R, Zinchenko S, Vales A, Paschen A, et al. Efficient recovery of HLA class I expression in human tumor cells after beta2-microglobulin gene transfer using adenoviral vector: implications for cancer immunotherapy. Scand J Immunol. (2009) 70:125–35. doi: 10.1111/j.1365-3083.2009.02276.x
234. del Campo AB, Carretero J, Aptsiauri N, Garrido F. Targeting HLA class I expression to increase tumor immunogenicity. Tissue Antigens. (2012) 79:147–54. doi: 10.1111/j.1399-0039.2011.01831.x
235. Del Campo AB, Carretero J, Munoz JA, Zinchenko S, Ruiz-Cabello F, Gonzalez-Aseguinolaza G, et al. Adenovirus expressing beta2-microglobulin recovers HLA class I expression and antitumor immunity by increasing T-cell recognition. Cancer Gene Ther. (2014) 21:317–32. doi: 10.1038/cgt.2014.32
236. Oshima S, Nakamura T, Namiki S, Okada E, Tsuchiya K, Okamoto R, et al. Interferon regulatory factor. (2004) 1 (IRF-1) and IRF-2 distinctively up-regulate gene expression and production of interleukin-7 in human intestinal epithelial cells. Mol Cell Biol 24:6298–310. doi: 10.1128/MCB.24.14.6298-6310.2004
237. Propper DJ, Chao D, Braybrooke JP, Bahl P, Thavasu P, Balkwill F, et al. Low-dose IFN-gamma induces tumor MHC expression in metastatic malignant melanoma. Clin Cancer Res. (2003) 9:84–92.
238. Vlkova V, Stepanek I, Hruskova V, Senigl F, Mayerova V, Sramek M, et al. Epigenetic regulations in the IFNgamma signalling pathway: IFNgamma-mediated MHC class I upregulation on tumour cells is associated with DNA demethylation of antigen-presenting machinery genes. Oncotarget. (2014) 5:6923–35. doi: 10.18632/oncotarget.2222
239. Davar D, Wang H, Chauvin JM, Pagliano O, Fourcade JJ, Ka M, et al. Phase Ib/II study of pembrolizumab and pegylated-interferon alfa-2b in advanced melanoma. J Clin Oncol. (2018) 36:JCO1800632. doi: 10.1200/JCO.18.00632
240. Mora-Garcia Mde L, Duenas-Gonzalez A, Hernandez-Montes J, De la Cruz-Hernandez E, Perez-Cardenas E, Weiss-Steider B, et al. Up-regulation of HLA class-I antigen expression and antigen-specific CTL response in cervical cancer cells by the demethylating agent hydralazine and the histone deacetylase inhibitor valproic acid. J Transl Med. (2006) 4:55. doi: 10.1186/1479-5876-4-55
241. van den Elsen PJ, Holling TM, van der Stoep N, Boss JM. DNA methylation and expression of major histocompatibility complex class I and class II transactivator genes in human developmental tumor cells and in T cell malignancies. Clin Immunol. (2003) 109:46–52. doi: 10.1016/S1521-6616(03)00200-6
242. Li H, Chiappinelli KB, Guzzetta AA, Easwaran H, Yen RW, Vatapalli R, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget. (2014) 5:587–98. doi: 10.18632/oncotarget.1782
243. Chiossone L, Dumas PY, Vienne M, Vivier E. Natural killer cells and other innate lymphoid cells in cancer. Nat Rev Immunol. (2018) 18:671–88. doi: 10.1038/s41577-018-0061-z
244. Covre A, Coral S, Nicolay H, Parisi G, Fazio C, Colizzi F, et al. Antitumor activity of epigenetic immunomodulation combined with CTLA-4 blockade in syngeneic mouse models. Oncoimmunology. (2015) 4:e1019978. doi: 10.1080/2162402X.2015.1019978
245. Wang L, Amoozgar Z, Huang J, Saleh MH, Xing D, Orsulic S, et al. Decitabine enhances lymphocyte migration and function and synergizes with CTLA-4 blockade in a murine ovarian cancer model. Cancer Immunol Res. (2015) 3:1030–41. doi: 10.1158/2326-6066.CIR-15-0073
246. Daver N, Garcia-Manero G, Basu S, Boddu PC, Alfayez M, Cortes JE, et al. Efficacy, safety, and biomarkers of response to azacitidine and nivolumab in relapsed/refractory acute myeloid leukemia: a nonrandomized, open-label, phase II study. Cancer Discov. (2019) 9:370–83. doi: 10.1158/2159-8290.CD-18-0774
247. Odunsi K, Matsuzaki J, James SR, Mhawech-Fauceglia P, Tsuji T, Miller A, et al. Epigenetic potentiation of NY-ESO-1 vaccine therapy in human ovarian cancer. Cancer Immunol Res. (2014) 2:37–49. doi: 10.1158/2326-6066.CIR-13-0126
248. Emran AA, Chatterjee A, Rodger EJ, Tiffen JC, Gallagher SJ, Eccles MR, et al. Targeting DNA methylation and EZH2 activity to overcome melanoma resistance to immunotherapy. Trends Immunol. (2019) 40:328–44. doi: 10.1016/j.it.2019.02.004
249. Khan AN, Gregorie CJ, Tomasi TB. Histone deacetylase inhibitors induce TAP, LMP, Tapasin genes and MHC class I antigen presentation by melanoma cells. Cancer Immunol Immunother. (2008) 57:647–54. doi: 10.1007/s00262-007-0402-4
250. Sun T, Li Y, Yang W, Wu H, Li X, Huang Y, et al. Histone deacetylase inhibition up-regulates MHC class I to facilitate cytotoxic T lymphocyte-mediated tumor cell killing in glioma cells. J Cancer. (2019) 10:5638–45. doi: 10.7150/jca.34471
251. Terranova-Barberio M, Thomas S, Ali N, Pawlowska N, Park J, Krings G, et al. HDAC inhibition potentiates immunotherapy in triple negative breast cancer. Oncotarget. (2017) 8:114156–72. doi: 10.18632/oncotarget.23169
252. Woods DM, Sodre AL, Villagra A, Sarnaik A, Sotomayor EM, Weber J. HDAC inhibition upregulates PD-1 ligands in melanoma and augments immunotherapy with PD-1 blockade. Cancer Immunol Res. (2015) 3:1375–85. doi: 10.1158/2326-6066.CIR-15-0077-T
253. Knox T, Sahakian E, Banik D, Hadley M, Palmer E, Noonepalle S, et al. Selective HDAC6 inhibitors improve anti-PD-1 immune checkpoint blockade therapy by decreasing the anti-inflammatory phenotype of macrophages and down-regulation of immunosuppressive proteins in tumor cells. Sci Rep. (2019) 9:6136. doi: 10.1038/s41598-019-42237-3
254. Khan AN, Magner WJ, Tomasi TB. An epigenetic vaccine model active in the prevention and treatment of melanoma. J Transl Med. (2007) 5:64. doi: 10.1186/1479-5876-5-64
255. Vo DD, Prins RM, Begley JL, Donahue TR, Morris LF, Bruhn KW, et al. Enhanced antitumor activity induced by adoptive T-cell transfer and adjunctive use of the histone deacetylase inhibitor LAQ824. Cancer Res. (2009) 69:8693–9. doi: 10.1158/0008-5472.CAN-09-1456
256. Banik D, Moufarrij S, Villagra A. Immunoepigenetics combination therapies: an overview of the role of HDACs in cancer immunotherapy. Int J Mol Sci. (2019) 20:92241. doi: 10.3390/ijms20092241
257. Gray JE, Saltos A, Tanvetyanon T, Haura EB, Creelan B, Antonia SJ, et al. Phase I/Ib study of pembrolizumab plus vorinostat in advanced/metastatic non-small cell lung cancer. Clin Cancer Res. (2019) 25:6623–32. doi: 10.1158/1078-0432.CCR-19-1305
258. Wang X, Waschke BC, Woolaver RA, Chen Z, Zhang G, Piscopio AD, et al. Histone deacetylase inhibition sensitizes PD1 blockade-resistant B-cell lymphomas. Cancer Immunol Res. (2019) 7:1318–31. doi: 10.1158/2326-6066.CIR-18-0875
259. Elashi AA, Sasidharan Nair V, Taha RZ, Shaath H, Elkord E. DNA methylation of immune checkpoints in the peripheral blood of breast and colorectal cancer patients. Oncoimmunology. (2019) 8:e1542918. doi: 10.1080/2162402X.2018.1542918
260. Sasidharan Nair V, El Salhat H, Taha RZ, John A, Ali BR, Elkord E. DNA methylation and repressive H3K9 and H3K27 trimethylation in the promoter regions of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, and PD-L1 genes in human primary breast cancer. Clin Epigenetics. (2018) 10:78. doi: 10.1186/s13148-018-0512-1
261. Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. (2015) 33:510–7. doi: 10.1038/nbt.3199
262. Haldeman JM, Conway AE, Arlotto ME, Slentz DH, Muoio DM, Becker TC, et al. Creation of versatile cloning platforms for transgene expression and dCas9-based epigenome editing. Nucleic Acids Res. (2019) 47:e23. doi: 10.1093/nar/gky1286
263. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. (2017) 16:203–22. doi: 10.1038/nrd.2016.246
264. El Hage F, Durgeau A, Mami-Chouaib F. TAP expression level in tumor cells defines the nature and processing of MHC class I peptides for recognition by tumor-specific cytotoxic T lymphocytes. Ann N Y Acad Sci. (2013) 1283:75–80. doi: 10.1111/j.1749-6632.2012.06777.x
265. Durgeau A, Virk Y, Gros G, Voilin E, Corgnac S, Djenidi F, et al. Human preprocalcitonin self-antigen generates TAP-dependent and -independent epitopes triggering optimised T-cell responses toward immune-escaped tumours. Nat Commun. (2018) 9:5097. doi: 10.1038/s41467-018-07603-1
266. Marijt KA, Van Der Burg SH, van Hall T. TEIPP peptides: exploration of unTAPped cancer antigens. Oncoimmunology. (2019) 8:1599639. doi: 10.1080/2162402X.2019.1599639
267. Miller JS, Lanier LL. Natural killer cells in cancer immunotherapy. Ann Rev Cancer Biol. (2019) 3:77–103. doi: 10.1146/annurev-cancerbio-030518-055653
268. Kim S, Iizuka K, Aguila HL, Weissman IL, Yokoyama WM. In vivo natural killer cell activities revealed by natural killer cell-deficient mice. Proc Natl Acad Sci USA. (2000) 97:2731–6. doi: 10.1073/pnas.050588297
269. Smyth MJ, Swann J, Cretney E, Zerafa N, Yokoyama WM, Hayakawa Y. NKG2D function protects the host from tumor initiation. J Exp Med. (2005) 202:583–8. doi: 10.1084/jem.20050994
270. Moon WY, Powis SJ. Does natural killer cell deficiency (NKD) increase the risk of cancer? NKD may increase the risk of some virus induced cancer. Front Immunol. (2019) 10:1703. doi: 10.3389/fimmu.2019.01703
271. Marcus A, Gowen BG, Thompson TW, Iannello A, Ardolino M, Deng W, et al. Recognition of tumors by the innate immune system and natural killer cells. Adv Immunol. (2014) 122:91–128. doi: 10.1016/B978-0-12-800267-4.00003-1
272. Wu J, Lanier LL. Natural killer cells and cancer. Adv Cancer Res. (2003) 90:127–56. doi: 10.1016/S0065-230X(03)90004-2
273. Hayakawa Y, Smyth MJ. Innate immune recognition and suppression of tumors. Adv Cancer Res. (2006) 95:293–322. doi: 10.1016/S0065-230X(06)95008-8
274. Imai K, Matsuyama S, Miyake S, Suga K, Nakachi K. Natural cytotoxic activity of peripheral-blood lymphocytes and cancer incidence: an 11-year follow-up study of a general population. Lancet. (2000) 356:1795–9. doi: 10.1016/S0140-6736(00)03231-1
275. Coca S, Perez-Piqueras J, Martinez D, Colmenarejo A, Saez MA, Vallejo C, et al. The prognostic significance of intratumoral natural killer cells in patients with colorectal carcinoma. Cancer. (1997) 79:2320–83. doi: 10.1002/(SICI)1097-0142(19970615)79:12<2320::AID-CNCR5>3.0.CO;2-P
276. Ishigami S, Natsugoe S, Tokuda K, Nakajo A, Che X, Iwashige H, et al. Prognostic value of intratumoral natural killer cells in gastric carcinoma. Cancer. (2000) 88:577–833. doi: 10.1002/(SICI)1097-0142(20000201)88:3<577::AID-CNCR13>3.0.CO;2-V
277. Hsu KC, Keever-Taylor CA, Wilton A, Pinto C, Heller G, Arkun K, et al. Improved outcome in HLA-identical sibling hematopoietic stem-cell transplantation for acute myelogenous leukemia predicted by KIR and HLA genotypes. Blood. (2005) 105:4878–84. doi: 10.1182/blood-2004-12-4825
278. Xie G, Dong H, Liang Y, Ham JD, Rizwan R, Chen J. CAR-NK cells: a promising cellular immunotherapy for cancer. EBioMedicine. (2020) 59:102975. doi: 10.1016/j.ebiom.2020.102975
279. Liu E, Marin D, Banerjee P, Macapinlac HA, Thompson P, Basar R, et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N Engl J Med. (2020) 382:545–53. doi: 10.1056/NEJMoa1910607
280. Garrido F, Aptsiauri N, Doorduijn EM, Garcia Lora AM, van Hall T. The urgent need to recover MHC class I in cancers for effective immunotherapy. Curr Opin Immunol. (2016) 39:44–51. doi: 10.1016/j.coi.2015.12.007
281. de Kruijf EM, Sajet A, van Nes JG, Natanov R, Putter H, Smit VT, et al. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J Immunol. (2010) 185:7452–9. doi: 10.4049/jimmunol.1002629
282. Urosevic M, Dummer R. Human leukocyte antigen-G and cancer immunoediting. Cancer Res. (2008) 68:627–30. doi: 10.1158/0008-5472.CAN-07-2704
283. Lin A, Yan WH. Heterogeneity of HLA-G expression in cancers: facing the challenges. Front Immunol. (2018) 9:2164. doi: 10.3389/fimmu.2018.02164
284. Wagner SN, Rebmann V, Willers CP, Grosse-Wilde H, Goos M. Expression analysis of classic and non-classic HLA molecules before interferon alfa-2b treatment of melanoma. Lancet. (2000) 356:220–1. doi: 10.1016/S0140-6736(00)02486-7
285. Rodriguez JA, Galeano L, Palacios DM, Gomez C, Serrano ML, Bravo MM, et al. Altered HLA class I and HLA-G expression is associated with IL-10 expression in patients with cervical cancer. Pathobiology. (2012) 79:72–83. doi: 10.1159/000334089
286. Ardolino M, Azimi CS, Iannello A, Trevino TN, Horan L, Zhang L, et al. Cytokine therapy reverses NK cell anergy in MHC-deficient tumors. J Clin Invest. (2014) 124:4781–94. doi: 10.1172/JCI74337
288. Platonova S, Cherfils-Vicini J, Damotte D, Crozet L, Vieillard V, Validire P, et al. Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res. (2011) 71:5412–22. doi: 10.1158/0008-5472.CAN-10-4179
289. Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. (2002) 419:734–8. doi: 10.1038/nature01112
290. Salih HR, Rammensee HG, Steinle A. Cutting edge: down-regulation of MICA on human tumors by proteolytic shedding. J Immunol. (2002) 169:4098–102. doi: 10.4049/jimmunol.169.8.4098
291. Castriconi R, Cantoni C, Della Chiesa M, Vitale M, Marcenaro E, Conte R, et al. Transforming growth factor beta 1 inhibits expression of NKp30 and NKG2D receptors: consequences for the NK-mediated killing of dendritic cells. Proc Natl Acad Sci USA. (2003) 100:4120–5. doi: 10.1073/pnas.0730640100
292. Souza-Fonseca-Guimaraes F, Cursons J, Huntington ND. The emergence of natural killer cells as a major target in cancer immunotherapy. Trends Immunol. (2019) 40:142–58. doi: 10.1016/j.it.2018.12.003
293. Wu Y, Li J, Jabbarzadeh Kaboli P, Shen J, Wu X, Zhao Y, et al. Natural killer cells as a double-edged sword in cancer immunotherapy: a comprehensive review from cytokine therapy to adoptive cell immunotherapy. Pharmacol Res. (2020) 155:104691. doi: 10.1016/j.phrs.2020.104691
294. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. (2012) 2:401–4. doi: 10.1158/2159-8290.CD-12-0095
295. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. (2013) 6:pl1. doi: 10.1126/scisignal.2004088
296. Torigoe T, Asanuma H, Nakazawa E, Tamura Y, Hirohashi Y, Yamamoto E, et al. Establishment of a monoclonal anti-pan HLA class I antibody suitable for immunostaining of formalin-fixed tissue: unusually high frequency of down-regulation in breast cancer tissues. Pathol Int. (2012) 62:303–8. doi: 10.1111/j.1440-1827.2012.02789.x
297. Mariya T, Hirohashi Y, Torigoe T, Asano T, Kuroda T, Yasuda K, et al. Prognostic impact of human leukocyte antigen class I expression and association of platinum resistance with immunologic profiles in epithelial ovarian cancer. Cancer Immunol Res. (2014) 2:1220–9. doi: 10.1158/2326-6066.CIR-14-0101
298. Rolland P, Deen S, Scott I, Durrant L, Spendlove I. Human leukocyte antigen class I antigen expression is an independent prognostic factor in ovarian cancer. Clin Cancer Res. (2007) 13:3591–6. doi: 10.1158/1078-0432.CCR-06-2087
299. Krishnakumar S, Abhyankar D, Sundaram AL, Pushparaj V, Shanmugam MP, Biswas J. Major histocompatibility antigens and antigen-processing molecules in uveal melanoma. Clin Cancer Res. (2003) 9:4159–64.
300. Lee HW, Min SK, Ju YS, Sung J, Lim MS, Yang DH, et al. Prognostic significance of HLA class I expressing in gastric carcinoma defined by monoclonal anti-pan HLA class I antibody, EMR8-5. J Gastrointest Surg. (2011) 15:1336–43. doi: 10.1007/s11605-011-1545-3
301. Krishnakumar S, Lakshmi S, Abhyankar D, Biswas J. Expression of HLA class I, beta(2)-microglobulin and HLA class II antigens in primary orbital melanoma. Orbit. (2003) 22:257–63. doi: 10.1076/orbi.22.4.257.17242
302. Saenz-Lopez P, Gouttefangeas C, Hennenlotter J, Concha A, Maleno I, Ruiz-Cabello F, et al. Higher HLA class I expression in renal cell carcinoma than in autologous normal tissue. Tissue Antigens. (2010) 75:110–8. doi: 10.1111/j.1399-0039.2009.01409.x
303. Morabito A, Dozin B, Salvi S, Pasciucco G, Balbi G, Laurent S, et al. Analysis and clinical relevance of human leukocyte antigen class I, heavy chain, and beta2-microglobulin downregulation in breast cancer. Hum Immunol. (2009) 70:492–5. doi: 10.1016/j.humimm.2009.04.029
304. Sanders H, McCue P, Graham SD Jr. ABO(H) antigens and beta-2 microglobulin in transitional cell carcinoma. Predictors of response to intravesical bacillus Calmette-Guerin. Cancer. (1991) 67:3024–83. doi: 10.1002/1097-0142(19910615)67:12<3024::AID-CNCR2820671216>3.0.CO;2-Q
305. Urosevic M, Kurrer MO, Kamarashev J, Mueller B, Weder W, Burg G, et al. Human leukocyte antigen G up-regulation in lung cancer associates with high-grade histology, human leukocyte antigen class I loss and interleukin-10 production. Am J Pathol. (2001) 159:817–24. doi: 10.1016/S0002-9440(10)61756-7
306. Koopman LA, Mulder A, Corver WE, Anholts JD, Giphart MJ, Claas FH, et al. HLA class I phenotype and genotype alterations in cervical carcinomas and derivative cell lines. Tissue Antigens. (1998) 51:623–36. doi: 10.1111/j.1399-0039.1998.tb03005.x
307. Vitale M, Rezzani R, Rodella L, Zauli G, Grigolato P, Cadei M, et al. HLA class I antigen and transporter associated with antigen processing (TAP1 and TAP2) down-regulation in high-grade primary breast carcinoma lesions. Cancer Res. (1998) 58:737–42.
308. Gudmundsdottir I, Gunnlaugur Jonasson J, Sigurdsson H, Olafsdottir K, Tryggvadottir L, Ogmundsdottir HM. Altered expression of HLA class I antigens in breast cancer: association with prognosis. Int J Cancer. (2000) 89:500–53. doi: 10.1002/1097-0215(20001120)89:6<500::AID-IJC6>3.0.CO
309. Atkins D, Breuckmann A, Schmahl GE, Binner P, Ferrone S, Krummenauer F, et al. MHC class I antigen processing pathway defects, ras mutations and disease stage in colorectal carcinoma. Int J Cancer. (2004) 109:265–73. doi: 10.1002/ijc.11681
310. Seliger B, Stoehr R, Handke D, Mueller A, Ferrone S, Wullich B, et al. Association of HLA class I antigen abnormalities with disease progression and early recurrence in prostate cancer. Cancer Immunol Immunother. (2010) 59:529–40. doi: 10.1007/s00262-009-0769-5
311. Iwayama Y, Tsuruma T, Mizuguchi T, Furuhata T, Toyota N, Matsumura M, et al. Prognostic value of HLA class I expression in patients with colorectal cancer. World J Surg Oncol. (2015) 13:36. doi: 10.1186/s12957-015-0456-2
312. Esteban F, Concha A, Delgado M, Perez-Ayala M, Ruiz-Cabello F, Garrido F. Lack of MHC class I antigens and tumour aggressiveness of the squamous cell carcinoma of the larynx. Br J Cancer. (1990) 62:1047–51. doi: 10.1038/bjc.1990.437
313. Sandel MH, Speetjens FM, Menon AG, Albertsson PA, Basse PH, Hokland M, et al. Natural killer cells infiltrating colorectal cancer and MHC class I expression. Mol Immunol. (2005) 42:541–6. doi: 10.1016/j.molimm.2004.07.039
314. Levin I, Klein T, Kuperman O, Segal S, Shapira J, Gal R, et al. The expression of HLA class I antigen in prostate cancer in relation to tumor differentiation and patient survival. Cancer Detect Prev. (1994) 18:443–5.
315. Tsuda H, Hirohashi S, Higuchi K, Shimosato Y. Beta-2-microglobulin expression in relation to amplification of oncogenes and prognosis in breast carcinoma. Histopathology. (1990) 16:500–2. doi: 10.1111/j.1365-2559.1990.tb01552.x
316. Kitamura H, Torigoe T, Honma I, Asanuma H, Nakazawa E, Shimozawa K, et al. Expression and antigenicity of survivin, an inhibitor of apoptosis family member, in bladder cancer: implications for specific immunotherapy. Urology. (2006) 67:955–9. doi: 10.1016/j.urology.2005.11.052
317. Na HY, Park Y, Nam SK, Lee KS, Oh HK, Kim DW, et al. Expression of human leukocyte antigen class I and beta2-microglobulin in colorectal cancer and its prognostic impact. Cancer Sci. (2021) 112:91–100. doi: 10.1111/cas.14723
318. Ferris RL, Hunt JL, Ferrone S. Human leukocyte antigen (HLA) class I defects in head and neck cancer: molecular mechanisms and clinical significance. Immunol Res. (2005) 33:113–33. doi: 10.1385/IR:33:2:113
319. Grandis JR, Falkner DM, Melhem MF, Gooding WE, Drenning SD, Morel PA. Human leukocyte antigen class I allelic and haplotype loss in squamous cell carcinoma of the head and neck: clinical and immunogenetic consequences. Clin Cancer Res. (2000) 6:2794–802.
320. Mattijssen V, De Mulder PH, Schalkwijk L, Manni JJ, Van 't Hof-Grootenboer B, Ruiter DJ. HLA antigen expression in routinely processed head and neck squamous cell carcinoma primary lesions of different sites. Int J Cancer Suppl. (1991) 6:95–100. doi: 10.1002/ijc.2910470719
321. Kabawat SE, Bast RC Jr, Welch WR, Knapp RC, Bhan AK. Expression of major histocompatibility antigens and nature of inflammatory cellular infiltrate in ovarian neoplasms. Int J Cancer. (1983) 32:547–54. doi: 10.1002/ijc.2910320505
322. Hilders CG, Houbiers JG, Krul EJ, Fleuren GJ. The expression of histocompatibility-related leukocyte antigens in the pathway to cervical carcinoma. Am J Clin Pathol. (1994) 101:5–12. doi: 10.1093/ajcp/101.1.5
323. Yuan J, Liu S, Yu Q, Lin Y, Bi Y, Wang Y, et al. Down-regulation of human leukocyte antigen class I (HLA-I) is associated with poor prognosis in patients with clear cell renal cell carcinoma. Acta Histochem. (2013) 115:470–4. doi: 10.1016/j.acthis.2012.11.002
324. Kaklamanis L, Leek R, Koukourakis M, Gatter KC, Harris AL. Loss of transporter in antigen processing 1 transport protein and major histocompatibility complex class I molecules in metastatic versus primary breast cancer. Cancer Res. (1995) 55:5191–4.
325. Shen YQ, Zhang JQ, Miao FQ, Zhang JM, Jiang Q, Chen H, et al. Relationship between the downregulation of HLA class I antigen and clinicopathological significance in gastric cancer. World J Gastroenterol. (2005) 11:3628–31. doi: 10.3748/wjg.v11.i23.3628
326. Cordon-Cardo C, Fuks Z, Drobnjak M, Moreno C, Eisenbach L, Feldman M. Expression of HLA-A,B,C antigens on primary and metastatic tumor cell populations of human carcinomas. Cancer Res. (1991) 51:6372–80.
327. Yang Y, Zhang J, Miao F, Wei J, Shen C, Shen Y, et al. Loss of heterozygosity at 6p21 underlying [corrected] HLA class I downregulation in Chinese primary esophageal squamous cell carcinomas. Tissue Antigens. (2008) 72:105–14. doi: 10.1111/j.1399-0039.2008.01078.x
328. Vermeulen CF, Jordanova ES, ter Haar NT, Kolkman-Uljee SM, de Miranda NF, Ferrone S, et al. Expression and genetic analysis of transporter associated with antigen processing in cervical carcinoma. Gynecol Oncol. (2007) 105:593–9. doi: 10.1016/j.ygyno.2007.02.016
329. Ishigami S, Arigami T, Uenosono Y, Matsumoto M, Okumura H, Uchikado Y, et al. Cancerous HLA class I expression and regulatory T cell infiltration in gastric cancer. Cancer Immunol Immunother. (2012) 61:1663–9. doi: 10.1007/s00262-012-1225-5
330. Kaklamanis L, Gatter KC, Hill AB, Mortensen N, Harris AL, Krausa P, et al. Loss of HLA class-I alleles, heavy chains and beta. (1992) 2-microglobulin in colorectal cancer. Int J Cancer 51:379–85. doi: 10.1002/ijc.2910510308
331. Hanak L, Slaby O, Lauerova L, Kren L, Nenutil R, Michalek J. Expression pattern of HLA class I antigens in renal cell carcinoma and primary cell line cultures: methodological implications for immunotherapy. Med Sci Monit. (2009) 15:CR638–43.
332. Kloth JN, Gorter A, Fleuren GJ, Oosting J, Uljee S, ter Haar N, et al. Elevated expression of SerpinA1 and SerpinA3 in HLA-positive cervical carcinoma. J Pathol. (2008) 215:222–30. doi: 10.1002/path.2347
333. Redondo M, Concha A, Oldiviela R, Cueto A, Gonzalez A, Garrido F, et al. Expression of HLA class I and II antigens in bronchogenic carcinomas: its relationship to cellular DNA content and clinical-pathological parameters. Cancer Res. (1991) 51:4948–54.
334. Ishigami S, Natsugoe S, Nakajo A, Arigami T, Kitazono M, Okumura H, et al. HLA-class I expression in gastric cancer. J Surg Oncol. (2020) 97:605–8. doi: 10.1002/jso.21029
335. Klein B, Klein T, Nyska A, Shapira J, Figer A, Schwartz A, et al. Expression of HLA class I and class II in gastric carcinoma in relation to pathologic stage. Tumour Biol. (1991) 12:68–74. doi: 10.1159/000217690
336. Tsuchikawa T, Ikeda H, Cho Y, Miyamoto M, Shichinohe T, Hirano S, et al. Association of CD8+ T cell infiltration in oesophageal carcinoma lesions with human leucocyte antigen (HLA) class I antigen expression and survival. Clin Exp Immunol. (2011) 164:50–6. doi: 10.1111/j.1365-2249.2010.04311.x
337. Kaneko K, Ishigami S, Kijima Y, Funasako Y, Hirata M, Okumura H, et al. Clinical implication of HLA class I expression in breast cancer. BMC Cancer. (2011) 11:454. doi: 10.1186/1471-2407-11-454
338. Dierssen JW, de Miranda NF, Mulder A, van Puijenbroek M, Verduyn W, Claas FH, et al. High-resolution analysis of HLA class I alterations in colorectal cancer. BMC Cancer. (2006) 6:233. doi: 10.1186/1471-2407-6-233
339. Wintzer HO, Benzing M, von Kleist S. Lacking prognostic significance of beta 2-microglobulin, MHC class I and class II antigen expression in breast carcinomas. Br J Cancer. (1990) 62:289–95. doi: 10.1038/bjc.1990.280
340. Redondo M, Garcia J, Villar E, Rodrigo I, Perea-Milla E, Serrano A, et al. Major histocompatibility complex status in breast carcinogenesis and relationship to apoptosis. Hum Pathol. (2003) 34:1283–9. doi: 10.1016/j.humpath.2003.06.001
341. Maiorana A, Cesinaro AM, Fano RA, Collina G. Expression of MHC class I and class II antigens in primary breast carcinomas and synchronous nodal metastases. Clin Exp Metastasis. (1995) 13:43–8. doi: 10.1007/BF00144017
342. Lopez-Nevot MA, Esteban F, Ferron A, Gutierrez J, Oliva MR, Romero C, et al. HLA class I gene expression on human primary tumours and autologous metastases: demonstration of selective losses of HLA antigens on colorectal, gastric and laryngeal carcinomas. Br J Cancer. (1989) 59:221–6. doi: 10.1038/bjc.1989.45
343. Baba T, Shiota H, Kuroda K, Shigematsu Y, Ichiki Y, Uramoto H, et al. Clinical significance of human leukocyte antigen loss and melanoma-associated antigen 4 expression in smokers of non-small cell lung cancer patients. Int J Clin Oncol. (2013) 18:997–1004. doi: 10.1007/s10147-012-0491-8
344. Ueda Y, Ishikawa K, Shiraishi N, Yokoyama S, Kitano S. Clinical significance of HLA class I heavy chain expression in patients with gastric cancer. J Surg Oncol. (2008) 97:451–5. doi: 10.1002/jso.20985
345. Saito T, Tanaka R, Yoshida S, Washiyama K, Kumanishi T. Immunohistochemical analysis of tumor-infiltrating lymphocytes and major histocompatibility antigens in human gliomas and metastatic brain tumors. Surg Neurol. (1988) 29:435–42. doi: 10.1016/0090-3019(88)90137-1
346. Mink SR, Hodge A, Agus DB, Jain A, Gross ME. Beta-2-microglobulin expression correlates with high-grade prostate cancer and specific defects in androgen signaling. Prostate. (2010) 70:1201–10. doi: 10.1002/pros.21155
347. Zeestraten EC, Van Hoesel AQ, Speetjens FM, Menon AG, Putter H, van de Velde CJ, et al. FoxP3- and CD8-positive infiltrating immune cells together determine clinical outcome in colorectal cancer. Cancer Microenviron. (2013) 6:31–9. doi: 10.1007/s12307-011-0071-x
348. Reimers MS, Bastiaannet E, Langley RE, van Eijk R, van Vlierberghe RL, Lemmens VE, et al. Expression of HLA class I antigen, aspirin use, and survival after a diagnosis of colon cancer. JAMA Intern Med. (2014) 174:732–9. doi: 10.1001/jamainternmed.2014.511
349. Ferron A, Perez-Ayala M, Concha A, Cabrera T, Redondo M, Oliva MR, et al. MHC class I and II antigens on gastric carcinomas and autologous mucosa. J Immunogenet. (1989) 16:413–23. doi: 10.1111/j.1744-313X.1989.tb00489.x
350. Zeestraten EC, Reimers MS, Saadatmand S, Goossens-Beumer IJ, Dekker JW, Liefers GJ, et al. Combined analysis of HLA class I, HLA-E and HLA-G predicts prognosis in colon cancer patients. Br J Cancer. (2014) 110:459–68. doi: 10.1038/bjc.2013.696
351. Redondo M, Ruiz-Cabello F, Concha A, Cabrera T, Perez-Ayala M, Oliva MR, Garrido F. Altered HLA class I expression in non-small cell lung cancer is independent of c-myc activation. Cancer Res. (1991) 51:2463–8.
352. Ruiter DJ, Bergman W, Welvaart K, Scheffer E, van Vloten WA, Russo C, et al. Immunohistochemical analysis of malignant melanomas and nevocellular nevi with monoclonal antibodies to distinct monomorphic determinants of HLA antigens. Cancer Res. (1984) 44:3930–5.
353. Vitale M, Pelusi G, Taroni B, Gobbi G, Micheloni C, Rezzani R, et al. HLA class I antigen down-regulation in primary ovary carcinoma lesions: association with disease stage. Clin Cancer Res. (2005) 11:67–72.
354. Menon AG, Morreau H, Tollenaar RA, Alphenaar E, Van Puijenbroek M, Putter H, et al. Down-regulation of HLA-A expression correlates with a better prognosis in colorectal cancer patients. Lab Invest. (2002) 82:1725–33. doi: 10.1097/01.LAB.0000043124.75633.ED
355. Ichinokawa K, Nakanishi Y, Hida Y, Tsuchikawa T, Kato T, Itoh T, et al. Downregulated expression of human leukocyte antigen class I heavy chain is associated with poor prognosis in non-small-cell lung cancer. Oncol Lett. (2019) 18:117–26. doi: 10.3892/ol.2019.10293
356. Kasajima A, Sers C, Sasano H, Johrens K, Stenzinger A, Noske A, et al. Down-regulation of the antigen processing machinery is linked to a loss of inflammatory response in colorectal cancer. Hum Pathol. (2010) 41:1758–69. doi: 10.1016/j.humpath.2010.05.014
357. Ericsson C, Seregard S, Bartolazzi A, Levitskaya E, Ferrone S, Kiessling R, et al. Association of HLA class I and class II antigen expression and mortality in uveal melanoma. Invest Ophthalmol Vis Sci. (2001) 42:2153–6.
358. Kooi S, Zhang HZ, Patenia R, Edwards CL, Platsoucas CD, Freedman RS. HLA class I expression on human ovarian carcinoma cells correlates with T-cell infiltration in vivo and T-cell expansion in vitro in low concentrations of recombinant interleukin-2. Cell Immunol. (1996) 174:116–28. doi: 10.1006/cimm.1996.0301
359. Shehata M, Mukherjee A, Deen S, Al-Attar A, Durrant LG, Chan S. Human leukocyte antigen class I expression is an independent prognostic factor in advanced ovarian cancer resistant to first-line platinum chemotherapy. Br J Cancer. (2009) 101:1321–8. doi: 10.1038/sj.bjc.6605315
360. Yabe H, Tsukahara T, Kawaguchi S, Wada T, Torigoe T, Sato N, et al. Prognostic significance of HLA class I expression in Ewing's sarcoma family of tumors. J Surg Oncol. (2011) 103:380–5. doi: 10.1002/jso.21829
361. Nouri AM, Smith ME, Crosby D, Oliver RT. Selective and non-selective loss of immunoregulatory molecules (HLA-A,B,C antigens and LFA-3) in transitional cell carcinoma. Br J Cancer. (1990) 62:603–6. doi: 10.1038/bjc.1990.338
362. Kikuchi E, Yamazaki K, Torigoe T, Cho Y, Miyamoto M, Oizumi S, et al. HLA class I antigen expression is associated with a favorable prognosis in early stage non-small cell lung cancer. Cancer Sci. (2007) 98:1424–30. doi: 10.1111/j.1349-7006.2007.00558.x
363. Koelzer VH, Dawson H, Andersson E, Karamitopoulou E, Masucci GV, Lugli A, et al. Active immunosurveillance in the tumor microenvironment of colorectal cancer is associated with low frequency tumor budding and improved outcome. Transl Res. (2015) 166:207–17. doi: 10.1016/j.trsl.2015.02.008
364. Haimiti A, Hailiman Y, Gulina A, Du J, Hao Z, Rong XL, et al. Reduced expression of members of the mhc-i antigen processing machinery in ethnic Uighur women with cervical cancer in the Xinjiang region of China. Curr Oncol. (2014) 21:e67–74. doi: 10.3747/co.21.1595
365. Dierssen JW, de Miranda NF, Ferrone S, van Puijenbroek M, Cornelisse CJ, Fleuren GJ, et al. HNPCC versus sporadic microsatellite-unstable colon cancers follow different routes toward loss of HLA class I expression. BMC Cancer. (2007) 7:33. doi: 10.1186/1471-2407-7-33
366. Houck JR, Sexton FM, Zajdel G. HLA class I and class II antigen expression on squamous cell carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg. (1990) 116:1181–5. doi: 10.1001/archotol.1990.01870100075016
367. Cabrera T, Angustias Fernandez M, Sierra A, Garrido A, Herruzo A, Escobedo A, et al. High frequency of altered HLA class I phenotypes in invasive breast carcinomas. Hum Immunol. (1996) 50:127–34. doi: 10.1016/0198-8859(96)00145-0
368. Carretero FJ, Del Campo AB, Flores-Martin JF, Mendez R, Garcia-Lopez C, Cozar JM, et al. Frequent HLA class I alterations in human prostate cancer: molecular mechanisms and clinical relevance. Cancer Immunol Immunother. (2016) 65:47–59. doi: 10.1007/s00262-015-1774-5
369. Vermeulen CF, Jordanova ES, Zomerdijk-Nooijen YA, ter Haar NT, Peters AA, Fleuren GJ. Frequent HLA class I loss is an early event in cervical carcinogenesis. Hum Immunol. (2005) 66:1167–73. doi: 10.1016/j.humimm.2005.10.011
370. Levin I, Klein T, Goldstein J, Kuperman O, Kanetti J, Klein B. Expression of class I histocompatibility antigens in transitional cell carcinoma of the urinary bladder in relation to survival. Cancer. (1991) 68:2591–43. doi: 10.1002/1097-0142(19911215)68:12<2591::AID-CNCR2820681212>3.0.CO;2-L
371. Miyagi K, Ingram M, Techy GB, Jacques DB, Freshwater DB, Sheldon H. Immunohistochemical detection and correlation between MHC antigen and cell-mediated immune system in recurrent glioma by APAAP method. Neurol Med Chir. (1990) 30:649–55. doi: 10.2176/nmc.30.649
372. Krishnakumar S, Abhyankar D, Lakshmi SA, Pushparaj V, Shanmugam MP, Biswas J. HLA expression in choroidal melanomas: correlation with clinicopathological features. Curr Eye Res. (2004) 28:409–16. doi: 10.1080/02713680490503741
373. Liu S, Qi L, Yu Q, Song Y, Han W, Zu X, et al. Survivin and HLA-I expression predicts survival of patients with clear cell renal cell carcinoma. Tumour Biol. (2014) 35:8281–8. doi: 10.1007/s13277-014-2058-y
374. Toquet C, Jarry A, Bou-Hanna C, Bach K, Denis MG, Mosnier JF, et al. Altered Calreticulin expression in human colon cancer: maintenance of Calreticulin expression is associated with mucinous differentiation. Oncol Rep. (2007) 17:1101–7. doi: 10.3892/or.17.5.1101
375. Garrido MA, Rodriguez T, Zinchenko S, Maleno I, Ruiz-Cabello F, Concha A, et al. HLA class I alterations in breast carcinoma are associated with a high frequency of the loss of heterozygosity at chromosomes 6 and 15. Immunogenetics. (2018) 70:647–59. doi: 10.1007/s00251-018-1074-2
376. Speetjens FM, de Bruin EC, Morreau H, Zeestraten EC, Putter H, van Krieken JH, et al. Clinical impact of HLA class I expression in rectal cancer. Cancer Immunol Immunother. (2008) 57:601–9. doi: 10.1007/s00262-007-0396-y
377. Ramnath N, Tan D, Li Q, Hylander BL, Bogner P, Ryes L, et al. Is downregulation of MHC class I antigen expression in human non-small cell lung cancer associated with prolonged survival? Cancer Immunol Immunother. (2006) 55:891–9. doi: 10.1007/s00262-005-0085-7
378. Tao J, Li Y, Liu YQ, Li L, Liu J, Shen X, et al. Expression of transporters associated with antigen processing and human leucocyte antigen class I in malignant melanoma and its association with prognostic factors. Br J Dermatol. (2008) 158:88–94. doi: 10.1111/j.1365-2133.2007.08294.x
379. Tsukahara T, Kawaguchi S, Torigoe T, Asanuma H, Nakazawa E, Shimozawa K, et al. Prognostic significance of HLA class I expression in osteosarcoma defined by anti-pan HLA class I monoclonal antibody, EMR8-5. Cancer Sci. (2006) 97:1374–80. doi: 10.1111/j.1349-7006.2006.00317.x
380. Kostine M, Cleven AH, de Miranda NF, Italiano A, Cleton-Jansen AM, Bovee JV. Analysis of PD-L1, T-cell infiltrate and HLA expression in chondrosarcoma indicates potential for response to immunotherapy specifically in the dedifferentiated subtype. Mod Pathol. (2016) 29:1028–37. doi: 10.1038/modpathol.2016.108
381. Cabrera CM, Jimenez P, Cabrera T, Esparza C, Ruiz-Cabello F, Garrido F. Total loss of MHC class I in colorectal tumors can be explained by two molecular pathways: beta2-microglobulin inactivation in MSI-positive tumors and LMP7/TAP2 downregulation in MSI-negative tumors. Tissue Antigens. (2003) 61:211–9. doi: 10.1034/j.1399-0039.2003.00020.x
382. Watson NF, Ramage JM, Madjd Z, Spendlove I, Ellis IO, Scholefield JH, et al. Immunosurveillance is active in colorectal cancer as downregulation but not complete loss of MHC class I expression correlates with a poor prognosis. Int J Cancer. (2006) 118:6–10. doi: 10.1002/ijc.21303
383. Reimers MS, Engels CC, Putter H, Morreau H, Liefers GJ, van de Velde CJ, et al. Prognostic value of HLA class I, HLA-E, HLA-G and Tregs in rectal cancer: a retrospective cohort study. BMC Cancer. (2014) 14:486. doi: 10.1186/1471-2407-14-486
384. Zhao X, Sun Q, Tian H, Cong B, Jiang X, Peng C. Loss of heterozygosity at 6p21 and HLA class I expression in esophageal squamous cell carcinomas in China. Asian Pac J Cancer Prev. (2011) 12:2741–5.
385. Han LY, Fletcher MS, Urbauer DL, Mueller P, Landen CN, Kamat AA, et al. HLA class I antigen processing machinery component expression and intratumoral T-Cell infiltrate as independent prognostic markers in ovarian carcinoma. Clin Cancer Res. (2008) 14:3372–9. doi: 10.1158/1078-0432.CCR-07-4433
386. Ogino T, Moriai S, Ishida Y, Ishii H, Katayama A, Miyokawa N, et al. Association of immunoescape mechanisms with Epstein-Barr virus infection in nasopharyngeal carcinoma. Int J Cancer. (2007) 120:2401–10. doi: 10.1002/ijc.22334
387. Kloor M, Becker C, Benner A, Woerner SM, Gebert J, Ferrone S, et al. Immunoselective pressure and human leukocyte antigen class I antigen machinery defects in microsatellite unstable colorectal cancers. Cancer Res. (2005) 65:6418–24. doi: 10.1158/0008-5472.CAN-05-0044
388. Atkins D, Ferrone S, Schmahl GE, Storkel S, Seliger B. Down-regulation of HLA class I antigen processing molecules: an immune escape mechanism of renal cell carcinoma? J Urol. (2004) 171:885–9. doi: 10.1097/01.ju.0000094807.95420.fe
389. Vora AR, Rodgers S, Parker AJ, Start R, Rees RC, Murray AK. An immunohistochemical study of altered immunomodulatory molecule expression in head and neck squamous cell carcinoma. Br J Cancer. (1997) 76:836–44. doi: 10.1038/bjc.1997.472
390. Zhang H, Melamed J, Wei P, Cox K, Frankel W, Bahnson RR, et al. Concordant down-regulation of proto-oncogene PML and major histocompatibility antigen HLA class I expression in high-grade prostate cancer. Cancer Immun. (2003) 3:2. doi: 10.1111/j.1399-0039.2007.00892.x
391. de Miranda NF, Nielsen M, Pereira D, van Puijenbroek M, Vasen HF, Hes FJ, et al. MUTYH-associated polyposis carcinomas frequently lose HLA class I expression—a common event amongst DNA-repair-deficient colorectal cancers. J Pathol. (2009) 219:69–76. doi: 10.1002/path.2569
392. Kitamura H, Torigoe T, Asanuma H, Honma I, Sato N, Tsukamoto T. Down-regulation of HLA class I antigens in prostate cancer tissues and up-regulation by histone deacetylase inhibition. J Urol. (2007) 178:692–6. doi: 10.1016/j.juro.2007.03.109
393. Saio M, Teicher M, Campbell G, Feiner H, Delgado Y, Frey AB. Immunocytochemical demonstration of down regulation of HLA class-I molecule expression in human metastatic breast carcinoma. Clin Exp Metastasis. (2004) 21:243–9. doi: 10.1023/B:CLIN.0000037707.07428.ff
394. Sato H, Suzuki Y, Ide M, Katoh T, Noda SE, Ando K, et al. HLA class I expression and its alteration by preoperative hyperthermo-chemoradiotherapy in patients with rectal cancer. PLoS ONE. (2014) 9:e108122. doi: 10.1371/journal.pone.0108122
395. de Boer MA, Jordanova ES, van Poelgeest MI, van den Akker BE, van der Burg SH, Kenter GG, et al. Circulating human papillomavirus type. (2007) 16 specific T-cells are associated with HLA Class I expression on tumor cells, but not related to the amount of viral oncogene transcripts. Int J Cancer 121:2711–5. doi: 10.1002/ijc.23035
396. Momburg F, Degener T, Bacchus E, Moldenhauer G, Hammerling GJ, Moller P. Loss of HLA-A,B,C and de novo expression of HLA-D in colorectal cancer. Int J Cancer. (1986) 37:179–84. doi: 10.1002/ijc.2910370203
397. Cabrera T, Salinero J, Fernandez MA, Garrido A, Esquivias J, Garrido F. High frequency of altered HLA class I phenotypes in laryngeal carcinomas. Hum Immunol. (2000) 61:499–506. doi: 10.1016/S0198-8859(00)00097-5
398. de Kruijf EM, van Nes JG, Sajet A, Tummers QR, Putter H, Osanto S, et al. The predictive value of HLA class I tumor cell expression and presence of intratumoral Tregs for chemotherapy in patients with early breast cancer. Clin Cancer Res. (2010) 16:1272–80. doi: 10.1158/1078-0432.CCR-09-1844
399. van Duinen SG, Ruiter DJ, Broecker EB, van der Velde EA, Sorg C, Welvaart K, et al. Level of HLA antigens in locoregional metastases and clinical course of the disease in patients with melanoma. Cancer Res. (1988) 48:1019–25.
400. Hanagiri T, Shigematsu Y, Shinohara S, Takenaka M, Oka S, Chikaishi Y, et al. Clinical significance of expression of cancer/testis antigen and down-regulation of HLA class-I in patients with stage I non-small cell lung cancer. Anticancer Res. (2013) 33:2123–8.
401. Li K, Du H, Lian X, Yang S, Chai D, Wang C, et al. Characterization of beta2-microglobulin expression in different types of breast cancer. BMC Cancer. (2014) 14:750. doi: 10.1186/1471-2407-14-750
402. Cromme FV, van Bommel PF, Walboomers JM, Gallee MP, Stern PL, Kenemans P, et al. Differences in MHC and TAP-1 expression in cervical cancer lymph node metastases as compared with the primary tumours. Br J Cancer. (1994) 69:1176–81. doi: 10.1038/bjc.1994.231
403. van Driel WJ, Tjiong MY, Hilders CG, Trimbos BJ, Fleuren GJ. Association of allele-specific HLA expression and histopathologic progression of cervical carcinoma. Gynecol Oncol. (1996) 62:33–41. doi: 10.1006/gyno.1996.0186
404. Pandha H, Rigg A, John J, Lemoine N. Loss of expression of antigen-presenting molecules in human pancreatic cancer and pancreatic cancer cell lines. Clin Exp Immunol. (2007) 148:127–35. doi: 10.1111/j.1365-2249.2006.03289.x
405. Koene G, Mulder A, van der Ven K, Eijsink C, Franke M, Slootweg P, et al. Human monoclonal antibodies as a tool for the detection of HLA class I allele-specific expression loss in head-and-neck squamous cell carcinoma and corresponding lymph node metastases. Hum Immunol. (2006) 67:692–9. doi: 10.1016/j.humimm.2006.06.001
406. Homma I, Kitamura H, Torigoe T, Tanaka T, Sato E, Hirohashi Y, et al. Human leukocyte antigen class I down-regulation in muscle-invasive bladder cancer: its association with clinical characteristics and survival after cystectomy. Cancer Sci. (2009) 100:2331–4. doi: 10.1111/j.1349-7006.2009.01329.x
407. Korkolopoulou P, Kaklamanis L, Pezzella F, Harris AL, Gatter KC. Loss of antigen-presenting molecules (MHC class I and TAP-1) in lung cancer. Br J Cancer. (1996) 73:148–53. doi: 10.1038/bjc.1996.28
408. Lu QL, Abel P, Mitchell S, Foster C, Lalani EN. Decreased HLA-A expression in prostate cancer is associated with normal allele dosage in the majority of cases. J Pathol. (2000) 190:169–763. doi: 10.1002/(SICI)1096-9896(200002)190:2169::AID-PATH517>3.0.CO;2-#
409. Tomita Y, Matsumoto Y, Nishiyama T, Fujiwara M. Reduction of major histocompatibility complex class I antigens on invasive and high-grade transitional cell carcinoma. J Pathol. (1990) 162:157–64. doi: 10.1002/path.1711620209
410. Connor ME, Stern PL. Loss of MHC class-I expression in cervical carcinomas. Int J Cancer. (1990) 46:1029–34. doi: 10.1002/ijc.2910460614
411. Leffers N, Lambeck AJ, de Graeff P, Bijlsma AY, Daemen T, van der Zee AG, et al. Survival of ovarian cancer patients overexpressing the tumour antigen p53 is diminished in case of MHC class I down-regulation. Gynecol Oncol. (2008) 110:365–73. doi: 10.1016/j.ygyno.2008.04.043
412. Jimenez P, Canton J, Concha A, Cabrera T, Fernandez M, Real LM, et al. Microsatellite instability analysis in tumors with different mechanisms for total loss of HLA expression. Cancer Immunol Immunother. (2000) 48:684–90. doi: 10.1007/s002620050017
413. Nada OH, Ahmed NS, Abou Gabal HH. Prognostic significance of HLA EMR8-5 immunohistochemically analyzed expression in osteosarcoma. Diagn Pathol. (2014) 9:72. doi: 10.1186/1746-1596-9-72
414. Madjd Z, Spendlove I, Pinder SE, Ellis IO, Durrant LG. Total loss of MHC class I is an independent indicator of good prognosis in breast cancer. Int J Cancer. (2005) 117:248–55. doi: 10.1002/ijc.21163
415. Esteban F, Concha A, Huelin C, Perez-Ayala M, Pedrinaci S, Ruiz-Cabello F, et al. Histocompatibility antigens in primary and metastatic squamous cell carcinoma of the larynx. Int J Cancer. (1989) 43:436–42. doi: 10.1002/ijc.2910430316
416. Pistillo MP, Nicolo G, Salvi S, Capanni P, Perdelli L, Pasciucco G, et al. Biochemical analysis of HLA class I subunits expression in breast cancer tissues. Hum Immunol. (2000) 61:397–407. doi: 10.1016/S0198-8859(99)00179-2
Glossary
APM, Antigen presentation machinery; ß2M, Beta2 microglobulin; CTLA-4, Cytotoxic T-lymphocyte-associated protein; ER, Endoplasmic reticulum; ERAD, Endoplasmic-reticulum-associated protein degradation; ERAP1, Endoplasmic reticulum aminopeptidase 1; EZH2, Enhancer of zeste 2 polycomb repressive complex 2; GSL, Glycosphingolipid; HDAC, Histone deacyetylase; HLA, Human leukocyte antigen; ILC, Innate lymphoid cells; JAK, Janus kinase; KIR, Killer inhibitory receptor; lnkRNA, Long noncoding RNA; NK, Natural killer; NLRC5, NLR Family CARD domain Containing 5; MHC, Major histocompatibility complex; miRNA, microRNA; PD1/PD-L1, Programmed cell death protein 1/ Programmed death-ligand 1; TAP, Transporter of antigen presentation; TIL, Tumor infiltrating lymphocytes; UTR, Untranslated region.
Keywords: antigen presentation, cancer immune evasion, MHC I antigen presentation, interferon, TAP1, Tapasin, epigenetic regulation
Citation: Dhatchinamoorthy K, Colbert JD and Rock KL (2021) Cancer Immune Evasion Through Loss of MHC Class I Antigen Presentation. Front. Immunol. 12:636568. doi: 10.3389/fimmu.2021.636568
Received: 01 December 2020; Accepted: 05 February 2021;
Published: 09 March 2021.
Edited by:
Luis C. Anton, Consejo Superior de Investigaciones Científicas (CSIC), SpainReviewed by:
Barbara Seliger, Martin Luther University of Halle-Wittenberg, GermanyDavid A. Scheinberg, Memorial Sloan Kettering Cancer Center, United States
Copyright © 2021 Dhatchinamoorthy, Colbert and Rock. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kenneth L. Rock, S2VubmV0aC5Sb2NrQHVtYXNzbWVkLmVkdQ==
 Karthik Dhatchinamoorthy
Karthik Dhatchinamoorthy Jeff D. Colbert
Jeff D. Colbert