- 1Department of Immunology, Leiden University Medical Center, Leiden, Netherlands
- 2Leiden Computational Biology Center, Leiden University Medical Center, Leiden, Netherlands
By Khatri I, Staal FJT, van Dongen JJM. Front Immunol 2020;11:2258. doi: 10.3389/fimmu.2020.570018
In the original article, there was a mistake in Figure 1 as published. In the structure of the CoV2 Spike protein, at amino-acid position 501, G (Gly) was erroneously indicated and should be N (Asn). In the CoV1 Spike protein, Y (Tyr) position was erroneously mentioned at 477 which is position 475. The corrected Figure 1 appears below.
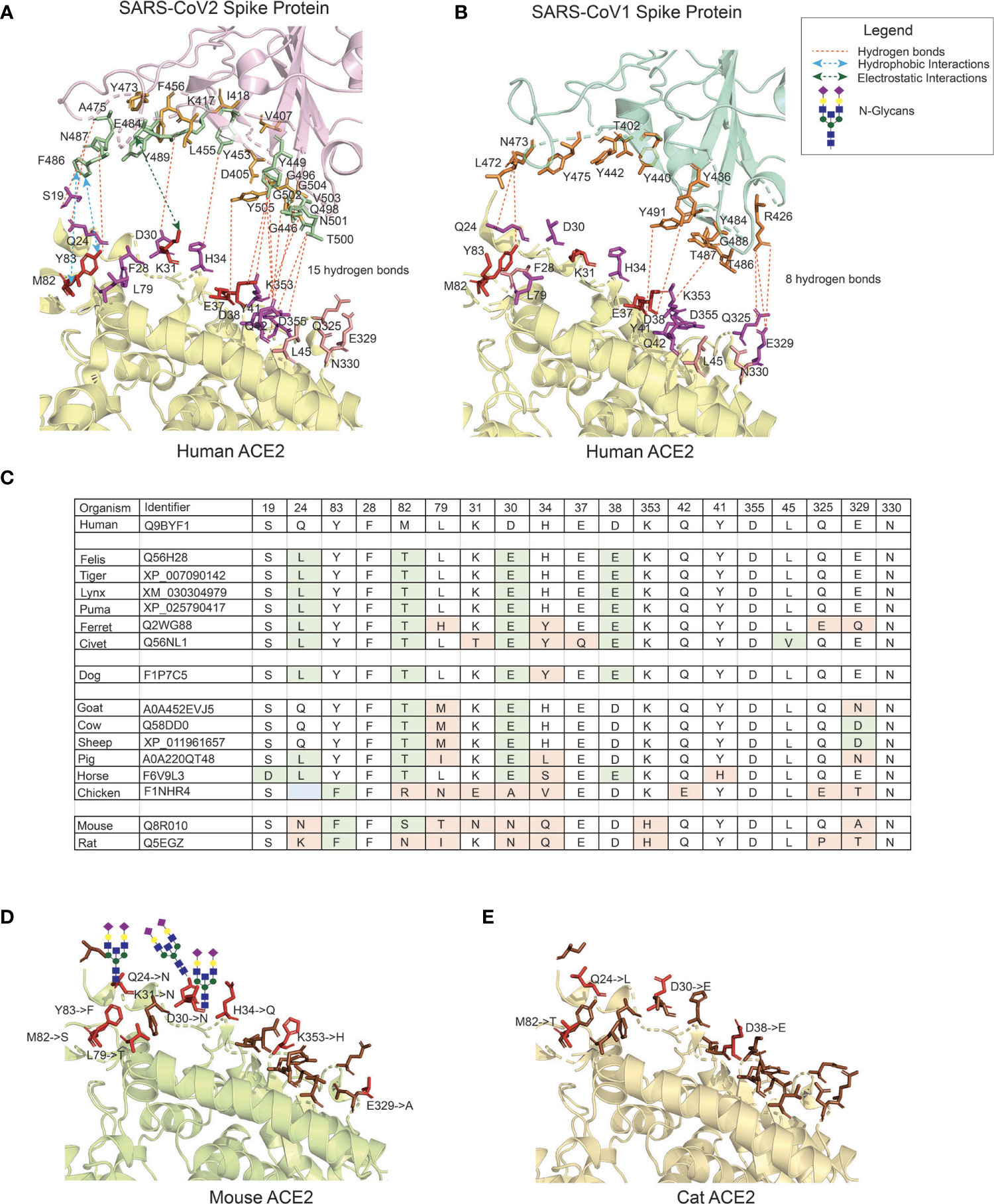
Figure 1 The interface of ACE2 protein in different organisms with important amino-acid residues for interacting with the Spike (S) protein of SARS-CoV viruses. (A) The interacting interface between hACE2 protein and CoV2S (PDB ID: 6LZG). The residues in hACE2 proteins are colored in red, magenta and pink. Red residues are the most important and pink the least. The interacting residues in CoV2S are colored in green and orange colors where green residues are the important interacting residues. The distance between CoV2Spike and hACE2 proteins is increased to better visualize the residues and the interactions. (B) The interacting interface between hACE2 protein and CoV1S (PDB ID: 2AJF). Red residues on hACE2 protein are important residues for maintaining the interaction between hACE2 and CoV1S proteins. All the residues in CoV1S interacting with hACE2 are colored in orange. The distance between CoV1Spike and hACE2 proteins is increased to better visualize the residues and the interactions. Hydrogen bonds (1A and 1B) as described in the structures of these interaction (PDB ID: 6LZG and 2AJF) and electrostatic and hydrophobic bonds in CoV1S-hACE2 interaction are depicted from Brielle et al. (4). (C) The positions mutated in ACE2 proteins in selected vertebrates that are either pets, domesticated or live in vicinity of humans. The mutated residues are shaded with green or orange background. The green background represents mutations resulting in similar property residue i.e. polar -> polar or non-polar -> non-polar. The orange background represents mutations resulting in changes in the residue property i.e. polar -> non-polar. (D) The interface of mouse’s ACE2 protein. Red color residues represent the mutated residues. Three mutations on the left interacting region has resulted in an Arginine, introducing a glycan site. The basic structure of glycan is shown on the sites. (E) The interface of cat’s ACE2 protein. The red color residues represent the mutated residues. The mouse’s and cat’s ACE2 structure are modelled with modeller-9.24 (65) using 6LZG as a template.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Keywords: ACE2, SARS-CoV, interaction-synapse, antibody, binding-affinity, felines, interface
Citation: Khatri I, Staal FJT and van Dongen JJM (2021) Corrigendum: Blocking of the High-Affinity Interaction-Synapse Between SARS-CoV-2 Spike and Human ACE2 Proteins Likely Requires Multiple High-Affinity Antibodies: An Immune Perspective. Front. Immunol. 12:659375. doi: 10.3389/fimmu.2021.659375
Received: 27 January 2021; Accepted: 26 March 2021;
Published: 14 April 2021.
Edited by:
Anke Huckriede, University Medical Center Groningen, NetherlandsReviewed by:
Jagadeesh Bayry, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceCopyright © 2021 Khatri, Staal and van Dongen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Indu Khatri, aS5raGF0cmlAbHVtYy5ubA==; Jacques J. M. van Dongen, Si5KLk0udmFuX0RvbmdlbkBsdW1jLm5s
 Indu Khatri
Indu Khatri Frank J. T. Staal
Frank J. T. Staal Jacques J. M. van Dongen
Jacques J. M. van Dongen