- 1Department of Pathology, Medical College of Georgia, Augusta University, Augusta, GA, United States
- 2Department of Neurosurgery, Augusta University, Augusta, GA, United States
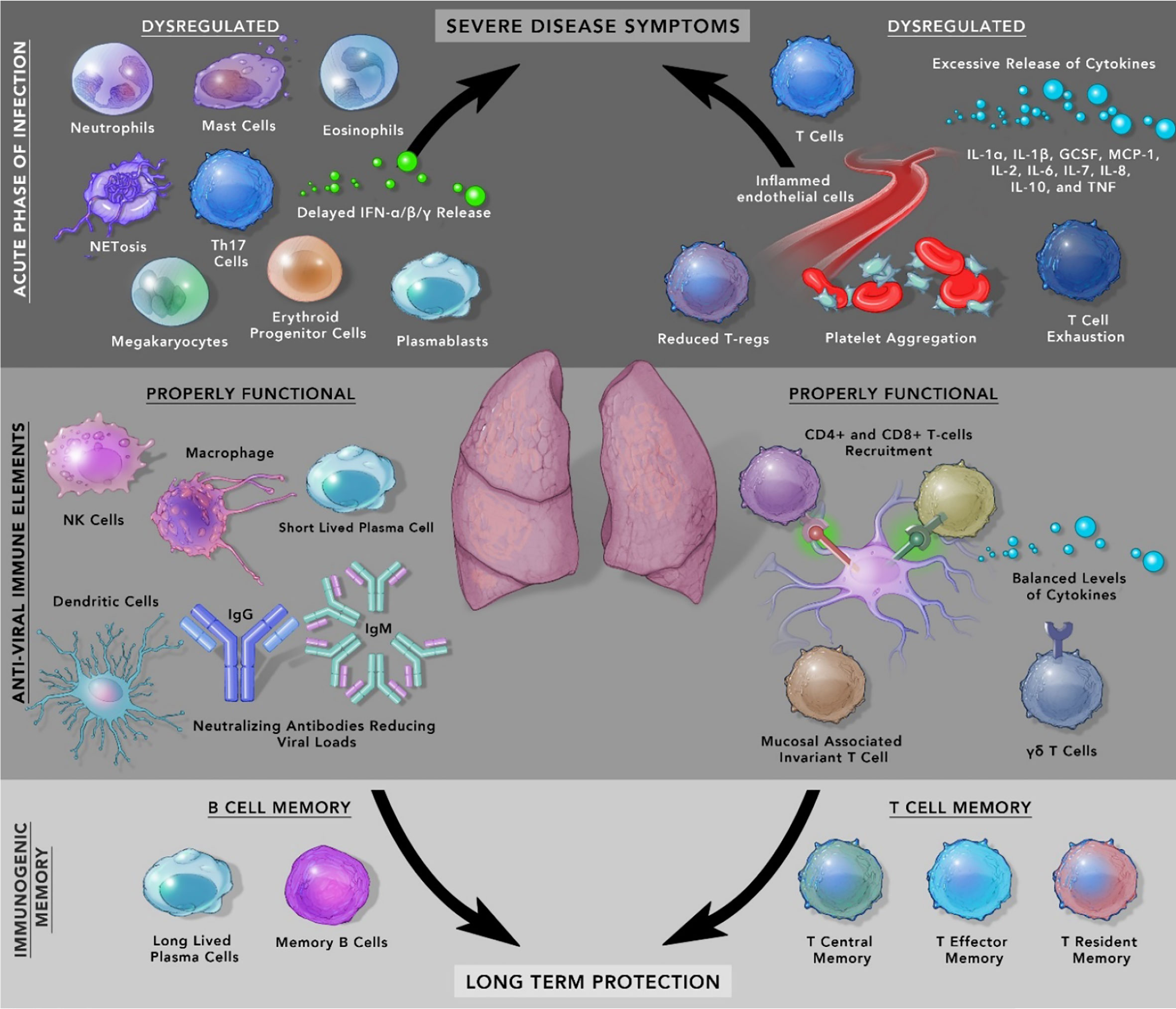
SARS-CoV-2 is the cause of a recent pandemic that has led to more than 3 million deaths worldwide. Most individuals are asymptomatic or display mild symptoms, which raises an inherent question as to how does the immune response differs from patients manifesting severe disease? During the initial phase of infection, dysregulated effector immune cells such as neutrophils, macrophages, monocytes, megakaryocytes, basophils, eosinophils, erythroid progenitor cells, and Th17 cells can alter the trajectory of an infected patient to severe disease. On the other hand, properly functioning CD4+, CD8+ cells, NK cells, and DCs reduce the disease severity. Detailed understanding of the immune response of convalescent individuals transitioning from the effector phase to the immunogenic memory phase can provide vital clues to understanding essential variables to assess vaccine-induced protection. Although neutralizing antibodies can wane over time, long-lasting B and T memory cells can persist in recovered individuals. The natural immunological memory captures the diverse repertoire of SARS-CoV-2 epitopes after natural infection whereas, currently approved vaccines are based on a single epitope, spike protein. It is essential to understand the nature of the immune response to natural infection to better identify ‘correlates of protection’ against this disease. This article discusses recent findings regarding immune response against natural infection to SARS-CoV-2 and the nature of immunogenic memory. More precise knowledge of the acute phase of immune response and its transition to immunological memory will contribute to the future design of vaccines and the identification of variables essential to maintain immune protection across diverse populations.
SARS-CoV-2 and the Acute Phase of Infection
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel strain of coronavirus responsible for the current pandemic that has infected more than 140 million and caused the death of more than 3 million individuals globally (https://coronavirus.jhu.edu/). Among infected individuals, 80% of patients display mild symptoms or are asymptomatic, 15% required oxygen (O2) and about 5% have critical pneumonia-like symptoms and require assisted ventilation (1). As more than a year has passed since the origin of this pathogen, there is tremendous interest in the long-term properties of immunological memory of recovered individuals as it can assist in design & improvement of next generation vaccines (2). As of March 2021, there are currently 8 vaccines approved for full use and 5 vaccines in early or limited use (3). Among these, the first two authorized vaccines were modified mRNA vaccines by Pfizer-BioNTech (Tozinameran) and Moderna (mRNA-1273). These two vaccines were predominantly rolled out in high-income countries and require ultra-cold chain infrastructure (-70°C for Pfizer vaccine). Other approved vaccines exhibit fewer logistic challenges and are suited for medium to low-income countries where they can be transported and stored in standard 2-8°C conditions (4). Among the most populous countries in the world, India and China have ramped up immunization efforts using their indigenous vaccines developed by Bharat Biotech and CanSino Biologics respectively. Further, there are 23 vaccine candidates in Phase 3 trials and its hoped that millions of individuals would be vaccinated in the next few months globally (3). Intensive research is underway to understand similarities and variations in the immune response in naturally recovered patients and vaccine-induced immunization. In this review, we will discuss the natural immune response to SARS-CoV-2 with particular emphasis on immunological memory.
The SARS-CoV-2 (+) RNA genome is known to encode 29 proteins (5, 6). The protein repertoire includes structural proteins: spike (S), membrane (M), envelope (E), nucleocapsid (N), and 16 nonstructural proteins (NSP 1-16) (7). Additionally, there are 9 accessory proteins (ORFs - 3a, 3b, 6, 7a, 7b, 8, 9b, 9c, 10) (8). Although main structural proteins and NSPs are studied in considerable detail, accessory proteins are emerging as important mediators of SARS-CoV-2 pathophysiology. In a recent study, accessory protein ORF9b was found to promote infection by binding to a mitochondrial chaperone protein named Tom70 (9).
The entry of the virus into the cell requires the presence of two host proteins, ACE2 and TMPRSS2. ACE2 acts as an entry receptor while TMPRSS2 acts as a cellular protease for priming of viral spike protein which is required for fusion with the host cell membrane (10). Recently, the expression of ACE2 and TMPRSS2 has been confirmed in salivary glands and oral mucosa epithelia which implicates the role of the oral cavity and saliva in the transmission of SARS-CoV-2 (11). In respiratory tissues, co-expression of ACE2 and TMPRSS2 is only restricted to type II pneumocytes and a subset of epithelial cells. Ziegler et al. reported that respiratory tissue shows a poor expression of ACE2 and TMPRSS2 with only 0.8% of type II pneumocytes with co-expression of both the proteins (12). Interestingly, SARS-CoV-2 has more than 10-20-fold higher affinity for ACE2 compared to other coronaviruses, and it is purposed as one of the several reasons for its harsh pathophysiology (13). Meanwhile, the expression of ACE2 has also been documented in two other cells: enterocytes of the small intestine and goblet secretory cells of the nasal mucosa (12). These findings hint at the presence of other receptors/pathways which can be used by the virus to infect the host cells. Recently, two studies have identified Neuropilin-1 as another novel cell surface receptor for the entry of SARS-CoV-2 (14, 15). In addition to these receptors, the role of other co-receptors such as sialic acids, heparan sulfate, CD147, or GRP78 needs to be investigated in greater detail as they can provide novel opportunities to design therapeutic interventions against COVID-19 (15, 16).
The SARS-CoV-2 infection leads to broad activation of innate and adaptive limbs of the immune system in humans. In the adaptive immune system, T cells play a critical role in managing the immune response to the viral pathogens (17). For its activation, T cells depend on the interaction between TCR (T Cell Receptor) and peptide-MHC complex (Major Histocompatibility Complex also known as HLA - Human Leukocyte Antigen in humans). One of the important variables to be considered in this interaction is the effect of genetic polymorphism in the process of antigen presentation and its association with the risk of COVID-19 (18). Individuals with specific allele, particularly HLA-B*46:01 are found to be pre-disposed to severe form of COVID-19 disease. On the other hand, individuals with HLA-B*15:03 had higher capacity to present SARS-CoV-2 epitopes to T cells and were associated with mild COVID-19 symptoms (19, 20). Although further studies are required to establish this correlation in diverse populations, it has been proposed that individuals with high-risk HLA alleles could be prioritized for vaccination (20).
At the clinical level, COVID-19 patients show diverse symptoms, ranging from mild to life-threatening acute respiratory distress syndrome (ARDS) which were also present during MERS and SARS-CoV-1 infection (21). The accumulation of pro-inflammatory cytokines like IL-1α, IL-1β, IL-17A, IL-12 p70, IFN-α, IFN-γ, TNFα and other cytokines included in the ‘cytokine release syndrome’ increases proportionally with an increase in viral load (22). In some patients with comorbidities, symptoms can be severe from septic shock, multi-organ failure due to capillary leaks, to the formation of thromboemboli and organ dysfunction. Another important variable in the form of sex difference has emerged, which has resulted in males developing a higher risk of mortality and severe illness compared to females. It is likely due to inherent immunological differences between males and females. One of the reasons for this difference could be higher levels of baseline pro-inflammatory cytokine, chemokines, and the presence of improved T-cell activation in females (23). In circulation, lower numbers of blood lymphocytes characterized as Lymphopenia is a prognostic indicator in COVID-19 patients as its level drops drastically in patients with severe disease whereas, moderately ill patients showed less fluctuation (24) Besides, rise in neutrophils, low lymphocyte-to-CRP and low neutrophil-to-lymphocyte ratio (NLR) have emerged as indicators of severe COVID-19 progression (25, 26). Several other immune cells play critical roles in COVID-19 patients and are shown in Figure 1 and Table 1.
In addition to understanding the immune response from circulating blood, two other sources with much more information from the site of infection are Bronchoalveolar lavage fluid (BALF) and autopsy specimen. BALF analysis has identified increased infiltration of macrophages in patients with severe disease compared to moderately ill patients. CD8+ T cells in severe patients showed higher proliferation, energy production, and translation capacity whereas, patients with moderate symptoms had T-cells in the proliferative stage (76). In another study, single-cell analysis identified pro-inflammatory macrophages at higher levels in severe patients compared to clonally expanded CD8+ T cells (77). Lung autopsy of COVID-19 patients has shown 2 distinct categories of interferon-stimulated genes (ISGs), ISGhigh (high viral load and pro-inflammatory cytokines) with early mortality compared to ISGlow with low viral load. Interestingly, lung morphology was relatively intact in ISGhigh patients compared to ISGlow patients which showed significant lung damage. In this study, activated T cell signature was found in ISGlow patients with a low viral count which might be indicative of their importance in antiviral immunity (78).
SARS-CoV-2 and the Immunity Phase
When infected by respiratory pathogens, innate and adaptive immune cells eliminate the pathogen and lead to formation of memory immune cells for rapid immune response against future infection. There is evidence of protective immunity conferred by B-cells in SARS-CoV-2 infected patients. The spike and nucleocapsid proteins specific antibodies are detectable as early as 6 days after the confirmation of COVID-19 (79, 80). It has been shown that the presence of neutralizing titers of IgG antibodies against SARS-CoV-2 can offer protection against re-infection (81). There is emerging evidence of potent memory B cell response in COVID-19 recovered individuals (82). It has been additionally shown in longitudinal studies of mildly infected patients, that while the initial spike in IgA antibodies drops significantly, the levels of IgG antibodies remain elevated for duration of at least first 3 months (83). In a recent study, IgG antibodies against spike proteins were found to be stably produced for over 6+ months post-infection. SARS-CoV-2 CD4+ T cells and CD8+ T cells showed a half-life of 3-5 months (84). A recent comparative study between severe and mild patients showed better humoral immunity in severe patients because of a higher level of BCR clonal expansion and activation (85). In contrast to these observations, Gattinger et al. used inhibition assay and found that S- (Spike) specific and RBD- (Receptor-Binding Domain) specific antibodies did not always correlate with inhibition of RBD binding to ACE2. In this small study, 25 convalescent patients showed a range of inhibition levels, from complete lack of inhibition to even enhanced RBD binding in some of the subjects (86). It should be pointed out that natural SARS-CoV-2 infection does not lead to protective antibody response in all patients. These factors should be carefully assessed while quantifying the efficacy of vaccine-induced immune memory.
In addition to the humoral response, SARS-CoV-2 infection elicits protective immunity as shown by the persistence of BM (Memory B cells) and TM (Memory T cells) in recovered patients (87–89). The resolution phase after an infection is followed by the death of most of the immune cells except around 10% ‘long-lived memory cells’ (90). These memory cells can provide long-term robust protection against future infections. Interestingly, the transition of IgM+ and IgG+ BM generates a transitionary bi-phasic appearance during the post-infection period. IgM+ BM cells were found to be present during the initial 20 days of infection up to 150 days, which was followed by IgG+ antibodies (84). Further, there has been evidence of a decline in circulating virus-specific antibodies against spike protein, NCP, or non-structural proteins (91–93). Despite this drop, a numerically significant number of RBD- and NCP-specific BM cells remain in circulation as much as 240 days after onset of symptoms (94). However, most of the RBD- BM cells showed CD27+ phenotype compared to NCP- BM cells which suggest that RBD- cells have undergone more cell divisions with higher number of somatic hypermutations (94) (95). It has been shown that BM transitions through phases, as antigen-exposed BM cells demonstrate multiple replication cycles, class switching, and somatic hypermutation (95) (96). CD21low CD27+ activated BM cells are present for a short duration for about 2 weeks and are followed by resting BM expressing CD21+ CD27- (97, 98). The presence of TAC1, CD80, CD180 cells shows that they can activate quickly when exposed to infection again (95, 99). SARS-CoV-2 studies have shown robust B cell memory response as long as 8 months after the start of COVID-19 symptoms (84). Further, these BM cells have been found to correlate positively with T follicular helper cells (Tfh) cells which suggests increased germinal center activity (94). Tfh cells help B cells in affinity maturation and are essential for the generation of memory plasma cells (100). This study has shown that although antibody numbers are reduced over time, the memory B cells compensate for this drop and are ready for active deployment. Some researchers have proposed the identification of these BM cells to be better predictors of long-term immunity compared to antibodies (94).
When infected by respiratory pathogens, the innate, and cellular components of the adaptive immune system function in succession to eliminate and generate immunogenic memory against the pathogen (101). The humoral immune response to SARS-CoV-2 is short-lived which puts specific focus on T cell immune memory. In comparison, B cell response to SARS-CoV-1 was also short-lived but virus-specific CD8+ T cells have been shown to persist for 6-11 years (102, 103). In another study, memory T cells against SARS-CoV-1 were detected even 17 years after the previous pandemic, hinting at the long-term immune response against the pathogens (17). In a recent study, almost half of donor blood samples between 2015 and 2018 showed reactivity to SARS-CoV-2, well before the emergence of this pathogen in the human population. It is speculated that this might be due to the cross-reactivity of other common cold coronaviruses with SARS-CoV-2 (104). This type of ‘heterologous immunity’ is from peptides of diverse viruses and is considered a widespread phenomenon but its role in protection against SARS-CoV-2 needs to be investigated.
There has been accumulating evidence that SARS-CoV-2 memory T cells are produced and maintained in individuals recovered from illness. The response of T cells against a range of SARS-CoV-2 epitopes such as M, N, and 6 other ORFs along with spike protein has been identified. CD4+ T cell response against SARS-CoV-2 spike protein was found to correlate with levels of IgG and IgA titers (105). Further, in COVID-19 patients, a positive correlation of lymphopenia with disease severity was found. However, the qualitative response of CD4+ T cells was found to be impaired in critically ill patients as they had a lower number of virus-specific CD4+ T cells with decreased IL-21, IL-4, and IFN-γ production (106). In one study, polyfunctional Th1-specific response with the secretion of IFN-γ, TNF-α, IL-2 was seen in patients with mild symptoms whereas, it was found to shift toward Th2 in ICU patients (106). Additionally, a higher frequency of Th17 cells and release of IL-17 was found to be associated with severe cases of COVID-19 (75). In contrast, another study found a lower distribution of Th1, TH17 compared to a higher percentage of Th2 cells in COVID-19 patients. Interestingly, this study also found an increase in senescent Th2 cells in patients who died from COVID-19 (107). In a different post-mortem study on severe COVID patients, lymph and spleen analysis revealed a lack of germinal centres, an increase in Th1 cells and a decrease in Th2 cells (108). Additionally, allergic diseases and asthma with its characteristic type-2 inflammation might not be a risk factor for SARS-CoV-2 infection. Apart from IL-4 and IL-5, IL-13 has been shown to reduce the expression of ACE-2 (109, 110). Asthma patients have been broadly divided into Type-2 asthma and Non-Type 2 asthma with different inflammatory profiles and resulting risk susceptibility. While T2 asthma patients elicit a strong TH2 response and possess lower COVID risk, Non-T2 asthma patients express higher Th1 response and COVID disease severity with pre-dominant lung destruction (111). In another interesting study, ex vivo analysis of IL-13 reduced expression of both ACE2 and TMPRSS2 in airway epithelial cells. Further, this study also found higher expression of TMPRSS2 but lower expression of ACE2 in nasal epithelial cells of type 2 asthma and allergy patients (112). The influence of the Th1/Th2 paradigm and its relationship to SARS-CoV-2 infection and disease severity needs to be characterized in greater detail.
There are several other SARS-CoV-2 induced variations at the cellular and metabolic level that play an essential role in shaping the immune response of humans. Apart from the reduction of CD4+T cells, lower number of NK cells and an increase in inflammatory monocytes has been observed in COVID 19 patients (50, 113). In another study, similar findings in severe patients have been reported along with the downregulation of HLA class II, increase in inflammatory cytokines, and neutrophil count (114). At the cellular level, an epigenetic modification was observed with hypomethylation of genes corresponding to innate immune signaling, the release of IL-1β, and TNF-α. Comparatively, genes of ATP metabolism and T cell receptor signaling showed decreased expression (50). Metabolic exhaustion of immune cells has also shown to play a vital role in COVID-19 pathogenesis. The activation process of lymphocytes involves switching between glycolysis and mitochondrial respiration that require additional mitochondrial biogenesis, protein translocation, and glycosylation pathways (115). In COVID-19 patients, Plasmoblasts (PBs) are shown to be metabolically active with the excessive shuttling of glucose for antibody glycosylation that might lead to metabolic exhaustion of these cells (116, 117). Further, if exacerbated, alteration in antibody formation such as the formation of fucosylated-IgG antibodies might lead to critical illness in COVID 19 patients (118). In another study, increase in the frequency of PBs was found in severe patients that normalized after sickness (50, 119). Further, Megakaryocytes (MKs) were found to be significantly high characterized by high Interferon expression signature (50). In the autopsy analysis of COVID 19 patients, MKs and thrombosis were observed in several organs including a rare finding of platelet-derived microthrombi in the heart (120). Systematic multi-organ complications might be facilitated by these damaging interactions of immune cells and tissue. Critically ill patients with poor oxygen circulation have shown an increase in numbers of Erythroid progenitor cells which derive from bone marrow due to hypoxic stress. The upregulation of GATA1 through HIF1 regulation leads to an increase in circulation of these cells and has been linked to heightened immune response (50, 121). The emerging role of epigenetic regulation and metabolic exhaustion in the generation of immunogenic memory should increase our understanding of their participation in immune memory against SARS-CoV-2.
Upon activation, T cells can move along multiple trajectories for clearance of pathogen but the T cells with effector functions die after few days of the onslaught, leaving behind a pool of memory T cells. These T cells can be divided into three broad categories based on their trafficking pattern, anatomical location, and activity: T central memory (TCM), T effector memory (TEM), and T resident memory (TRM) (122). TEM displays rapid generation granzyme and IFN-γ but very low proliferative capacity. TCM on the other hand, with CD62L and CCR7 show homing capacity for secondary lymphoid organs where they can be deployed to prevent systemic infection, activate effector functions, and infiltrate peripheral tissues for rapid response against pathogens. This fits into the biphasic response whereby TEM initial onslaught against pathogen and later new range of T cells from TCM pool gets ready for the final elimination of the pathogen (123). Overall, the difference between effector T cell response and memory T cell response includes an increased pool of memory T cells reactive against the pathogen, pre-programmed specific effector response generates rapid response against a specific pathogen, and generation of resident memory T cells TRM in peripheral tissues (124). Additionally, other memory immune cells are also being identified with their role in SARS-CoV-2 pathogenesis. In a recent study, SARS-CoV-2 specific CD8+ T cells was found to consist of two prominent populations of TEMRA cells (terminally differentiated effector memory cells re‐expressing CD45RA) and Tscm (T stem cell memory) cells. TEMRA cells have been identified to play an important in protection against SARS-CoV-2. These SARS-CoV-2 antigen experienced tissue resident CD8+ T cells re-express CD45RA, a naïve cell marker known to offer protection in tissues such as spleen, blood, and lung (125). The role of TEMRA cells was explored in a different study using deep immune profiling analysis that identified three major immunotypes of COVID-19. Immunotype 1 was associated with disease severity, activated CD4+ T cells, low circulatory circulating follicular helper cells, and TEMRA cells. Immunotype 2 group did not show any association with disease severity and showed lower CD4 T cell activation and proliferation memory B cells. Immunotype 3 was characterized by lack of T and B cell response and negative association with disease severity (119). Further, the importance of other immune cells, particularly memory Tfh cells is critical against SARS-CoV-2 as these CD4+ T cells originating from GC-Tfh cells play a central role in the event of re-exposure to the antigen (126). Whether the SARS-CoV-2 long-term immunity can maintain this pool of memory Tfh cells remains to be determined. Cumulatively, these studies show that the complexity of immune memory in SARS-CoV-2 infection will form a critical component while analyzing the long-term efficacy of vaccine-induced memory response to SARS-CoV-2.
There is another critical host factor that plays a central role in shaping the detrimental effect of SARS-CoV-2 infection. “Immunosenescence”, characterized by decline and dysregulation of immune response due to aging plays a significant role in COVID-19 pathogenesis. It includes gradual deterioration in the form of reduced thymus function, chronic stimulation by antigens and corresponding increase in proinflammatory mediators, latent reactivation of pathogens, and poor response to vaccines. This “Inflammaging” also assists in alternative activation of M2-macrophages with a dysregulated phagocytic capacity which can lead to adverse outcomes in many infectious diseases (127) (128). The co-morbidity of heart and lung diseases puts the elderly population at mortality risk from viral infections (129). As the individual grows older, the pool of naïve T cells declines along with reduction of proper environment, a conducive milieu in the form of cytokines, cell-cell interaction, and chemokines, which are essential for the proper functioning of the immune response. The lack of a proper environment for their functionality can sometimes lead to an excessive life-threatening situation of a cytokine storm. On the other end, paucity of proper signaling, ineffective priming of T cells by antigen-presenting cells can lead to exhausted T cells with chronic poor effector response.
SARS-CoV-2 Vaccine and Mediators of Durable Immunity
The greater understanding of the immune response to SARS-CoV-2 is assisting the scientific community in the identification of key variables essential to study the long-term immune response to this novel pathogen. There are several important variables to be considered to understand the durability of vaccine-induced immune memory. To begin with, epigenetic programming of innate cells has shown protection against SARS-CoV-2. In a randomized controlled study, BCG administration showed significant delay in first infection compared to the placebo group (median: 16 months Vs, 11 months respectively) (130). BCG or other vaccines can trigger ‘pathogen-agnostic antimicrobial resistance’ due to increased baseline immunity (131). It has been proposed that these live vaccines (including oral polio and measles) can boost trained immunity through reprogramming of immune cells at epigenetic, transcriptional, and functional levels (132). It should be further explored as to how much ‘trained immunity’ can contribute to protection against infective agents of the pandemic.
Several studies have explored immune variations with protection against SARS-CoV-2 but currently, there is a lack of immune correlate of protection associated with the COVID-19 (133). Currently, nABs (neutralizing antibodies) are considered the benchmark to quantify protection but a decline in production of nABs can be observed in some of the SARS-CoV-2 recovered patients (134). In a recent study, a mechanistic link was found between SARS-CoV-2 infection and resulting B cell response with limited durability. In this study, severe SARS-CoV-2 infected patients showed blunt germinal centers, reduction in Bcl-6 expressing B cells and Tfh cells. Further, the presence of a high amount of TNF can also reduce GC response and block differentiation of Tfh cells necessary for B cell maturation (108). There are other mediators of immunity such as T-cells, that can complement nABs as a potential variable to quantify immune protection. In fact, heterogeneous immunological memory has shown discordance where recovered individuals displayed both ends of the spectrum: High nAB/low T cell and low nAB/high T cell (135). In another study, 93% exposed asymptomatic individuals mounted T cell while only 60% showed seropositive status (136) On the other hand, T cells have shown reactivity even in the absence of antibodies in asymptomatic individuals thus, highlighting the potent role of memory T cells in COVID-19 infection (88). Further, CD8+ T cells of recovered individuals showed higher expression of TCF1 and can quickly differentiate into diverse T cells such as Tfh in case of re-exposure to SARS-CoV-2 (88). It has been speculated that the presence of other means of protection such as TRM can lower the threshold of nAB required for protection against COVID-19. It can cause rapid deployment of effector T cells and other leukocytes to provide synergistic protection. TRM can also directly influence the production of antibodies (137, 138). Further, T cell activation and resulting reduction of Treg cells can also play a critical role in maintaining the immunity in T cell patients. T cells are known to express activation markers such as CD38, HLA-DR, and Ki67 (54). It has been observed that the exhaustion markers such as programmed cell death marker 1 (PD-1) and the receptor mucin domain-containing protein-3 (TIM-3) leads to the poor effector function of T cells (139). Interestingly, T cell hyperactivation fuels its exhaustion which leads to a reduction in the activity of T-reg cells. In fact, circulating levels of Treg cells were found to be reduced in severe patients compared to COVID-19 with mild symptoms (58, 60). It would be interesting to explore the relative contribution of reduction of T-reg cells in SARS-CoV-2 induced pathogenesis.
The activation of T cells requires interaction between T cell receptor and antigen-MHC complex I (Human leukocyte antigen – HLA in humans) to induce an immune response against infection. The spike protein is a major T cell epitope in other coronaviruses whereas in SARS-CoV-2, the epitopes are spread out from spike to nucleocapsid and matrix protein which results in multiple co-dominant CD4 T cell epitopes (138). Clinically, the magnitude and range of epitope coverage was found to be higher in severe cases compared to mild ones after SARS-CoV-2 infection (140). Further, a higher number of multi-cytokines producing T cells (M/NP) compared to anti-spike CD8+ T cells was found in recovered patients which indicates the importance of other viral proteins as a target for future vaccines (140). Among first approved vaccines, Pfizer, Moderna, and many other candidate vaccines are targeting spike protein (S) but other vaccine candidates are targeting other proteins such as viral protease (Mpro) and RNA-dependent polymerase (RdRp) among others in an effort to induce potent immune response and protection against COVID-19 (141). In another attempt to provide broad immunity by activating multifunctional CD4 T cells, vaccine developer Novavax have incorporated antigens such as M (Matrix protein) as an adjuvant along with standard trimeric SARS-CoV-2 S protein as a vaccine target (142). Apart from these antigens, a recent study has addressed the need for novel antigens by mapping the full landscape of exact antigens involved in SARS-CoV-2 infection. This study recognized a total of 122 immunogenic epitopes in SARS-CoV-2 infected patients among a total of 3141 epitopes. They also found that most immunodominant peptides are located in ORF1 and ORF3 of SARS-CoV-2 genome (143). These observations might help in understanding the quality of memory immune response especially considering that ORF1 is highly conserved among coronaviruses and there is baseline heterologous immunity against other coronaviruses in general population (143). Further, these antigens expand the horizon for vaccine design as it has been speculated that an increase in the number of epitopes may be beneficial in the generation of a more robust CD8+ T cell response (138). It would be interesting to compare and analyze the extent and durability of immune protection provided by vaccines based on single spike protein, other antigen(s) or the use of antigens in the form of adjuvant. In this light, any promising vaccine should define a correlate of protection in the form of durable B and/or T cell response. The unprecedented pace of discovery has led to the development of vaccines in a record time and the continuation of subsequent research is bound to decode the complexity of mediators required for protection in a wider population.
Author Contributions
PA, RK, KV, and MA drafted the manuscript. AM, NS, and AR provided intellectual inputs. RK supervised the project. All authors contributed to the article and approved the submitted version.
Funding
KV and RK acknowledges the support provided in the form of start-up grant by Augusta University. The funding bodies had no role in design, analysis, and interpretation pertaining to this study.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Colby Zahn for providing the illustration for this article.
References
1. de Candia P, Prattichizzo F, Garavelli S, Matarese G. T Cells: Warriors of SARS-Cov-2 Infection. Trends Immunol (2021) 42:18–30. doi: 10.1016/j.it.2020.11.002
2. Teijaro JR, Farber DL. COVID-19 Vaccines: Modes of Immune Activation and Future Challenges. Nat Rev Immunol (2021) 21:195–7. doi: 10.1038/s41577-021-00526-x
4. Murphy A, Mbau L, McKee M, Hanson K, Torreele E. Can We Do for Other Essential Medicines What We are Doing for the COVID-19 Vaccine? BMJ Glob Health (2021) 6. doi: 10.1136/bmjgh-2021-005158
5. Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, et al. Genomic Characterization of the 2019 Novel Human-Pathogenic Coronavirus Isolated From a Patient With Atypical Pneumonia After Visiting Wuhan. Emerg Microbes Infect (2020) 9:221–36. doi: 10.1080/22221751.2020.1719902
6. Yao H, Song Y, Chen Y, Wu N, Xu J, Sun C, et al. Molecular Architecture of the SARS-Cov-2 Virus. Cell (2020) 183:730–8.e13. doi: 10.1016/j.cell.2020.09.018
7. Yoshimoto FK. The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS Cov-2 or N-COV19), the Cause of COVID-19. Protein J (2020) 39:198–216. doi: 10.1007/s10930-020-09901-4
8. Michel CJ, Mayer C, Poch O, Thompson JD. Characterization of Accessory Genes in Coronavirus Genomes. Virol J (2020) 17:131. doi: 10.1186/s12985-020-01402-1
9. Gordon DE, Hiatt J, Bouhaddou M, Rezelj VV, Ulferts S, Braberg H, et al. Comparative Host-Coronavirus Protein Interaction Networks Reveal Pan-Viral Disease Mechanisms. Science (2020) 370. doi: 10.1126/science.abe9403
10. Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-Cov-2 Cell Entry Depends on ACE2 and TMPRSS2 and is Blocked by a Clinically Proven Protease Inhibitor. Cell (2020) 181:271–80 e8. doi: 10.1016/j.cell.2020.02.052
11. Huang N, Perez P, Kato T, Mikami Y, Okuda K, Gilmore RC, et al. SARS-Cov-2 Infection of the Oral Cavity and Saliva. Nat Med (2021) 1–12. doi: 10.1038/s41591-021-01296-8
12. Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. SARS-Cov-2 Receptor ACE2 is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and is Detected in Specific Cell Subsets Across Tissues. Cell (2020) 181:1016–35.e19. doi: 10.1016/j.cell.2020.04.035
13. Davidson AM, Wysocki J, Batlle D. Interaction of SARS-Cov-2 and Other Coronavirus With ACE (Angiotensin-Converting Enzyme)-2 as Their Main Receptor: Therapeutic Implications. Hypertension (2020) 76:1339–49. doi: 10.1161/HYPERTENSIONAHA.120.15256
14. Daly JL, Simonetti B, Klein K, Chen KE, Williamson MK, Anton-Plagaro C, et al. Neuropilin-1 is a Host Factor for SARS-Cov-2 Infection. Science (2020) 370:861–5. doi: 10.1126/science.abd3072
15. Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, et al. Neuropilin-1 Facilitates SARS-Cov-2 Cell Entry and Infectivity. Science (2020) 370:856–60. doi: 10.1126/science.abd2985
16. Zamorano Cuervo N, Grandvaux N. ACE2: Evidence of Role as Entry Receptor for SARS-Cov-2 and Implications in Comorbidities. Elife (2020) 9. doi: 10.7554/eLife.61390
17. Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-Cov-2-Specific T Cell Immunity in Cases of COVID-19 and SARS, and Uninfected Controls. Nature (2020) 584:457–62. doi: 10.1038/s41586-020-2550-z
18. Shah VK, Firmal P, Alam A, Ganguly D, Chattopadhyay S. Overview of Immune Response During SARS-Cov-2 Infection: Lessons From the Past. Front Immunol (2020) 11:1949. doi: 10.3389/fimmu.2020.01949
19. Shkurnikov M, Nersisyan S, Jankevic T, Galatenko A, Gordeev I, Vechorko V, et al. Association of HLA Class I Genotypes With Severity of Coronavirus Disease-19. Front Immunol (2021) 12:641900. doi: 10.3389/fimmu.2021.641900
20. Nguyen A, David JK, Maden SK, Wood MA, Weeder BR, Nellore A, et al. Human Leukocyte Antigen Susceptibility Map for Severe Acute Respiratory Syndrome Coronavirus 2. J Virol (2020) 94. doi: 10.1128/JVI.00510-20
21. Carsana L, Sonzogni A, Nasr A, Rossi RS, Pellegrinelli A, Zerbi P, et al. Pulmonary Post-Mortem Findings in a Series of COVID-19 Cases From Northern Italy: A Two-Centre Descriptive Study. Lancet Infect Dis (2020) 20:1135–40. doi: 10.1016/S1473-3099(20)30434-5
22. Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, et al. Longitudinal Analyses Reveal Immunological Misfiring in Severe COVID-19. Nature (2020) 584:463–9. doi: 10.1038/s41586-020-2588-y
23. Takahashi T, Ellingson MK, Wong P, Israelow B, Lucas C, Klein J, et al. Sex Differences in Immune Responses That Underlie COVID-19 Disease Outcomes. Nature (2020) 588:315–20. doi: 10.1038/s41586-020-2700-3
24. Tan L, Wang Q, Zhang D, Ding J, Huang Q, Tang YQ, et al. Lymphopenia Predicts Disease Severity of COVID-19: A Descriptive and Predictive Study. Signal Transduct Target Ther (2020) 5:33. doi: 10.1038/s41392-020-0159-1
25. Li X, Liu C, Mao Z, Xiao M, Wang L, Qi S, et al. Predictive Values of Neutrophil-to-Lymphocyte Ratio on Disease Severity and Mortality in COVID-19 Patients: A Systematic Review and Meta-Analysis. Crit Care (2020) 24:647. doi: 10.1186/s13054-020-03374-8
26. Lagunas-Rangel FA. Neutrophil-to-Lymphocyte Ratio and Lymphocyte-to-C-Reactive Protein Ratio in Patients With Severe Coronavirus Disease 2019 (COVID-19): A Meta-Analysis. J Med Virol (2020) 92:1733–4. doi: 10.1002/jmv.25819
27. Camp JV, Jonsson CB. A Role for Neutrophils in Viral Respiratory Disease. Front Immunol (2017) 8:550. doi: 10.3389/fimmu.2017.00550
28. Liu J, Pang Z, Wang G, Guan X, Fang K, Wang Z, et al. Advanced Role of Neutrophils in Common Respiratory Diseases. J Immunol Res (2017) 2017:6710278. doi: 10.1155/2017/6710278
29. Arcanjo A, Logullo J, Menezes CCB, de Souza Carvalho Giangiarulo TC, Dos Reis MC, de Castro GMM, et al. The Emerging Role of Neutrophil Extracellular Traps in Severe Acute Respiratory Syndrome Coronavirus 2 (COVID-19). Sci Rep (2020) 10:19630. doi: 10.21203/rs.3.rs-40461/v1
30. Didangelos A. COVID-19 Hyperinflammation: What About Neutrophils? mSphere (2020) 5. doi: 10.1128/mSphere.00367-20
31. Rosa BA, Ahmed M, Singh DK, Choreno-Parra JA, Cole J, Jimenez-Alvarez LA, et al. IFN Signaling and Neutrophil Degranulation Transcriptional Signatures are Induced During SARS-Cov-2 Infection. Commun Biol (2021) 4(1):1–14. doi: 10.1038/s42003-021-01829-4
32. Liu J, Li S, Liu J, Liang B, Wang X, Wang H, et al. Longitudinal Characteristics of Lymphocyte Responses and Cytokine Profiles in the Peripheral Blood of SARS-Cov-2 Infected Patients. EBioMedicine (2020) 55:102763. doi: 10.1016/j.ebiom.2020.102763
33. Erjefalt JS. Mast Cells in Human Airways: The Culprit? Eur Respir Rev (2014) 23:299–307. doi: 10.1183/09059180.00005014
34. Campillo-Navarro M, Chavez-Blanco AD, Wong-Baeza I, Serafin-Lopez J, Flores-Mejia R, Estrada-Parra S, et al. Mast Cells in Lung Homeostasis: Beyond Type I Hypersensitivity. Curr Respir Med Rev (2014) 10:115–23. doi: 10.2174/1573398X10666141024220151
35. Afrin LB, Weinstock LB, Molderings GJ. Covid-19 Hyperinflammation and Post-Covid-19 Illness May Be Rooted in Mast Cell Activation Syndrome. Int J Infect Dis (2020) 100:327–32. doi: 10.1016/j.ijid.2020.09.016
36. Conti P, Caraffa A, Tete G, Gallenga CE, Ross R, Kritas SK, et al. Mast Cells Activated by SARS-Cov-2 Release Histamine Which Increases IL-1 Levels Causing Cytokine Storm and Inflammatory Reaction in COVID-19. J Biol Regul Homeost Agents (2020) 34:1629–32. doi: 10.23812/20-2EDIT
37. Schwartz C, Eberle JU, Voehringer D. Basophils in Inflammation. Eur J Pharmacol (2016) 778:90–5. doi: 10.1016/j.ejphar.2015.04.049
38. Stone KD, Prussin C, Metcalfe DD. Ige, Mast Cells, Basophils, and Eosinophils. J Allergy Clin Immunol (2010) 125:S73–80. doi: 10.1016/j.jaci.2009.11.017
39. Cohen M, Giladi A, Gorki AD, Solodkin DG, Zada M, Hladik A, et al. Lung Single-Cell Signaling Interaction Map Reveals Basophil Role in Macrophage Imprinting. Cell (2018) 175:1031–44.e18. doi: 10.1016/j.cell.2018.09.009
40. Rodriguez L, Pekkarinen PT, Lakshmikanth T, Tan Z, Consiglio CR, Pou C, et al. Systems-Level Immunomonitoring From Acute to Recovery Phase of Severe COVID-19. Cell Rep Med (2020) 1:100078. doi: 10.1016/j.xcrm.2020.100078
41. Marichal T, Mesnil C, Bureau F. Homeostatic Eosinophils: Characteristics and Functions. Front Med (Lausanne) (2017) 4:101. doi: 10.3389/fmed.2017.00101
42. Kopf M, Schneider C, Nobs SP. The Development and Function of Lung-Resident Macrophages and Dendritic Cells. Nat Immunol (2015) 16:36–44. doi: 10.1038/ni.3052
43. Zhou R, To KK, Wong YC, Liu L, Zhou B, Li X, et al. Acute SARS-Cov-2 Infection Impairs Dendritic Cell and T Cell Responses. Immunity (2020) 53:864–77.e5. doi: 10.1016/j.immuni.2020.07.026
44. Groeneweg L, Hidalgo A, A-Gonzalez N. Emerging Roles of Infiltrating Granulocytes and Monocytes in Homeostasis. Cell Mol Life Sci (2020) 77:3823–30. doi: 10.1007/s00018-020-03509-8
45. Dunbar PR, Cartwright EK, Wein AN, Tsukamoto T, Tiger Li ZR, Kumar N, et al. Pulmonary Monocytes Interact With Effector T Cells in the Lung Tissue to Drive TRM Differentiation Following Viral Infection. Mucosal Immunol (2020) 13:161–71. doi: 10.1038/s41385-019-0224-7
46. Matic S, Popovic S, Djurdjevic P, Todorovic D, Djordjevic N, Mijailovic Z, et al. SARS-Cov-2 Infection Induces Mixed M1/M2 Phenotype in Circulating Monocytes and Alterations in Both Dendritic Cell and Monocyte Subsets. PloS One (2020) 15:e0241097. doi: 10.1371/journal.pone.0241097
47. Desai N, Neyaz A, Szabolcs A, Shih AR, Chen JH, Thapar V, et al. Temporal and Spatial Heterogeneity of Host Response to SARS-Cov-2 Pulmonary Infection. Nat Commun (2020) 11:6319. doi: 10.1038/s41467-020-20139-7
48. Kato A, Hulse KE, Tan BK, Schleimer RP. B-Lymphocyte Lineage Cells and the Respiratory System. J Allergy Clin Immunol (2013) 131:933–57; quiz 958. doi: 10.1016/j.jaci.2013.02.023
49. Laing AG, Lorenc A, Del Molino Del Barrio I, Das A, Fish M, Monin L, et al. A Dynamic COVID-19 Immune Signature Includes Associations With Poor Prognosis. Nat Med (2020) 26:1623–35. doi: 10.1038/s41591-020-01186-5
50. Bernardes JP, Mishra N, Tran F, Bahmer T, Best L, Blase JI, et al. Longitudinal Multi-Omics Analyses Identify Responses of Megakaryocytes, Erythroid Cells, and Plasmablasts as Hallmarks of Severe COVID-19. Immunity (2020) 53:1296–314 e9. doi: 10.1016/j.immuni.2020.11.017
51. Chen K, Kolls JK. T Cell-Mediated Host Immune Defenses in the Lung. Annu Rev Immunol (2013) 31:605–33. doi: 10.1146/annurev-immunol-032712-100019
52. Chen Z, John Wherry E. T Cell Responses in Patients With COVID-19. Nat Rev Immunol (2020) 20:529–36. doi: 10.1038/s41577-020-0402-6
53. Uddback I, Cartwright EK, Scholler AS, Wein AN, Hayward SL, Lobby J, et al. Long-Term Maintenance of Lung Resident Memory T Cells is Mediated by Persistent Antigen. Mucosal Immunol (2020) 14(1):92–9. doi: 10.1038/s41385-020-0309-3
54. Altmann DM, Boyton RJ. SARS-Cov-2 T Cell Immunity: Specificity, Function, Durability, and Role in Protection. Sci Immunol (2020) 5. doi: 10.1126/sciimmunol.abd6160
55. Allie SR, Bradley JE, Mudunuru U, Schultz MD, Graf BA, Lund FE, et al. The Establishment of Resident Memory B Cells in the Lung Requires Local Antigen Encounter. Nat Immunol (2019) 20:97–108. doi: 10.1038/s41590-018-0260-6
56. Ogega CO, Skinner NE, Blair PW, Park HS, Littlefield K, Ganesan A, et al. Durable SARS-Cov-2 B Cell Immunity After Mild or Severe Disease. J Clin Invest (2021) 131(7). doi: 10.1172/JCI145516
57. Duan W, Croft M. Control of Regulatory T Cells and Airway Tolerance by Lung Macrophages and Dendritic Cells. Ann Am Thorac Soc (2014) 11 Suppl 5:S306–13. doi: 10.1513/AnnalsATS.201401-028AW
58. Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and Immunological Features of Severe and Moderate Coronavirus Disease 2019. J Clin Invest (2020) 130:2620–9. doi: 10.1172/JCI137244
59. Wang F, Hou H, Luo Y, Tang G, Wu S, Huang M, et al. The Laboratory Tests and Host Immunity of COVID-19 Patients With Different Severity of Illness. JCI Insight (2020) 5. doi: 10.1172/jci.insight.137799
60. Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis (2020) 71:762–8. doi: 10.1093/cid/ciaa248
61. Dorhoi A, Du Plessis N. Monocytic Myeloid-Derived Suppressor Cells in Chronic Infections. Front Immunol (2017) 8:1895. doi: 10.3389/fimmu.2017.01895
62. Sacchi A, Grassi G, Bordoni V, Lorenzini P, Cimini E, Casetti R, et al. Early Expansion of Myeloid-Derived Suppressor Cells Inhibits SARS-Cov-2 Specific T-Cell Response and May Predict Fatal COVID-19 Outcome. Cell Death Dis (2020) 11:921. doi: 10.1038/s41419-020-03125-1
63. Cong J, Wei H. Natural Killer Cells in the Lungs. Front Immunol (2019) 10:1416. doi: 10.3389/fimmu.2019.01416
64. van Eeden C, Khan L, Osman MS, Cohen Tervaert JW. Natural Killer Cell Dysfunction and Its Role in COVID-19. Int J Mol Sci (2020) 21. doi: 10.3390/ijms21176351
65. Pahl JHW, Cerwenka A, Ni J. Memory-Like NK Cells: Remembering a Previous Activation by Cytokines and NK Cell Receptors. Front Immunol (2018) 9:2796. doi: 10.3389/fimmu.2018.02796
66. Varchetta S, Mele D, Oliviero B, Mantovani S, Ludovisi S, Cerino A, et al. Unique Immunological Profile in Patients With COVID-19. Cell Mol Immunol (2020) 18(3):604–12. doi: 10.21203/rs.3.rs-23953/v1
67. Stehle C, Hernandez DC, Romagnani C. Innate Lymphoid Cells in Lung Infection and Immunity. Immunol Rev (2018) 286:102–19. doi: 10.1111/imr.12712
68. Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, et al. Innate Lymphoid Cells Promote Lung-Tissue Homeostasis After Infection With Influenza Virus. Nat Immunol (2011) 12:1045–54. doi: 10.1038/ni.2131
69. Kuri-Cervantes L, Pampena MB, Meng W, Rosenfeld AM, Ittner CAG, Weisman AR, et al. Comprehensive Mapping of Immune Perturbations Associated With Severe COVID-19. Sci Immunol (2020) 5. doi: 10.1126/sciimmunol.abd7114
70. Zheng J, Liu Y, Lau YL, Tu W. Gammadelta-T Cells: An Unpolished Sword in Human Anti-Infection Immunity. Cell Mol Immunol (2013) 10:50–7. doi: 10.1038/cmi.2012.43
71. Toubal A, Nel I, Lotersztajn S, Lehuen A. Mucosal-Associated Invariant T Cells and Disease. Nat Rev Immunol (2019) 19:643–57. doi: 10.1038/s41577-019-0191-y
72. Parrot T, Gorin JB, Ponzetta A, Maleki KT, Kammann T, Emgard J, et al. MAIT Cell Activation and Dynamics Associated With COVID-19 Disease Severity. Sci Immunol (2020) 5. doi: 10.1101/2020.08.27.20182550
73. Martinez NE, Sato F, Kawai E, Omura S, Chervenak RP, Tsunoda I. Regulatory T Cells and Th17 Cells in Viral Infections: Implications for Multiple Sclerosis and Myocarditis. Future Virol (2012) 7:593–608. doi: 10.2217/fvl.12.44
74. Gurczynski SJ, Moore BB. IL-17 in the Lung: The Good, the Bad, and the Ugly. Am J Physiol Lung Cell Mol Physiol (2018) 314:L6–16. doi: 10.1152/ajplung.00344.2017
75. De Biasi S, Meschiari M, Gibellini L, Bellinazzi C, Borella R, Fidanza L, et al. Marked T Cell Activation, Senescence, Exhaustion and Skewing Towards TH17 in Patients With COVID-19 Pneumonia. Nat Commun (2020) 11:3434. doi: 10.1038/s41467-020-17292-4
76. Garcia LF. Immune Response, Inflammation, and the Clinical Spectrum of COVID-19. Front Immunol (2020) 11:1441. doi: 10.3389/fimmu.2020.01441
77. Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. Single-Cell Landscape of Bronchoalveolar Immune Cells in Patients With COVID-19. Nat Med (2020) 26:842–4. doi: 10.1038/s41591-020-0901-9
78. Nienhold R, Ciani Y, Koelzer VH, Tzankov A, Haslbauer JD, Menter T, et al. Two Distinct Immunopathological Profiles in Autopsy Lungs of COVID-19. Nat Commun (2020) 11:5086. doi: 10.1038/s41467-020-18854-2
79. Leung DT, Tam FC, Ma CH, Chan PK, Cheung JL, Niu H, et al. Antibody Response of Patients With Severe Acute Respiratory Syndrome (SARS) Targets the Viral Nucleocapsid. J Infect Dis (2004) 190:379–86. doi: 10.1086/422040
80. Grzelak L, Temmam S, Planchais C, Demeret C, Tondeur L, Huon C, et al. A Comparison of Four Serological Assays for Detecting Anti-SARS-Cov-2 Antibodies in Human Serum Samples From Different Populations. Sci Transl Med (2020) 12. doi: 10.1126/scitranslmed.abc3103
81. Suthar MS, Zimmerman MG, Kauffman RC, Mantus G, Linderman SL, Hudson WH, et al. Rapid Generation of Neutralizing Antibody Responses in COVID-19 Patients. Cell Rep Med (2020) 1:100040. doi: 10.1101/2020.05.03.20084442
82. Rodda LB, Netland J, Shehata L, Pruner KB, Morawski PA, Thouvenel C, et al. Functional SARS-Cov-2-Specific Immune Memory Persists After Mild COVID-19. Res Sq (2020). doi: 10.1101/2020.08.11.20171843
83. Rodda LB, Netland J, Shehata L, Pruner KB, Morawski PA, Thouvenel CD, et al. Functional SARS-Cov-2-Specific Immune Memory Persists After Mild COVID-19. Cell (2020) 184(1):169–83. doi: 10.1101/2020.08.11.20171843
84. Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, et al. Immunological Memory to SARS-Cov-2 Assessed for Up to 8 Months After Infection. Science (2021) 371(6529). doi: 10.1101/2020.11.15.383323
85. Zhang F, Gan R, Zhen Z, Hu X, Li X, Zhou F, et al. Adaptive Immune Responses to SARS-Cov-2 Infection in Severe Versus Mild Individuals. Signal Transduct Target Ther (2020) 5:156. doi: 10.1038/s41392-020-00263-y
86. Gattinger P, Borochova K, Dorofeeva Y, Henning R, Kiss R, Kratzer B, et al. Antibodies in Serum of Convalescent Patients Following Mild COVID-19 Do Not Always Prevent Virus-Receptor Binding. Allergy (2021) 76:878–83. doi: 10.1111/all.14523
87. Juno JA, Tan HX, Lee WS, Reynaldi A, Kelly HG, Wragg K, et al. Humoral and Circulating Follicular Helper T Cell Responses in Recovered Patients With COVID-19. Nat Med (2020) 26:1428–34. doi: 10.1038/s41591-020-0995-0
88. Sekine T, Perez-Potti A, Rivera-Ballesteros O, Stralin K, Gorin JB, Olsson A, et al. Robust T Cell Immunity in Convalescent Individuals With Asymptomatic or Mild COVID-19. Cell (2020) 183:158–68.e14. doi: 10.1101/2020.06.29.174888
89. Lineburg KE, Srihari S, Altaf M, Swaminathan S, Panikkar A, Raju J, et al. Rapid Detection of SARS-Cov-2-Specific Memory T-Cell Immunity in Recovered COVID-19 Cases. Clin Transl Immunol (2020) 9:e1219. doi: 10.1002/cti2.1219
90. Ruterbusch M, Pruner KB, Shehata L, Pepper M. In Vivo CD4(+) T Cell Differentiation and Function: Revisiting the Th1/Th2 Paradigm. Annu Rev Immunol (2020) 38:705–25. doi: 10.1146/annurev-immunol-103019-085803
91. Gudbjartsson DF, Norddahl GL, Melsted P, Gunnarsdottir K, Holm H, Eythorsson E, et al. Humoral Immune Response to SARS-Cov-2 in Iceland. N Engl J Med (2020) 383:1724–34. doi: 10.1056/NEJMoa2026116
92. Long QX, Tang XJ, Shi QL, Li Q, Deng HJ, Yuan J, et al. Clinical and Immunological Assessment of Asymptomatic SARS-Cov-2 Infections. Nat Med (2020) 26:1200–4. doi: 10.1038/s41591-020-0965-6
93. Hachim A, Kavian N, Cohen CA, Chin AWH, Chu DKW, Mok CKP, et al. ORF8 and ORF3b Antibodies are Accurate Serological Markers of Early and Late SARS-Cov-2 Infection. Nat Immunol (2020) 21:1293–301. doi: 10.1038/s41590-020-0773-7
94. Nguyen-Contant P, Embong AK, Kanagaiah P, Chaves FA, Yang H, Branche AR, et al. S Protein-Reactive Igg and Memory B Cell Production After Human SARS-Cov-2 Infection Includes Broad Reactivity to the S2 Subunit. mBio (2020) 11. doi: 10.1128/mBio.01991-20
95. Berkowska MA, Driessen GJ, Bikos V, Grosserichter-Wagener C, Stamatopoulos K, Cerutti A, et al. Human Memory B Cells Originate From Three Distinct Germinal Center-Dependent and -Independent Maturation Pathways. Blood (2011) 118:2150–8. doi: 10.1182/blood-2011-04-345579
96. de Jong BG, IJspeert H, Marques L, van der Burg M, van Dongen JJ, Loos BG, et al. Human Igg2- and Igg4-Expressing Memory B Cells Display Enhanced Molecular and Phenotypic Signs of Maturity and Accumulate With Age. Immunol Cell Biol (2017) 95:744–52. doi: 10.1038/icb.2017.43
97. Ellebedy AH, Jackson KJ, Kissick HT, Nakaya HI, Davis CW, Roskin KM, et al. Defining Antigen-Specific Plasmablast and Memory B Cell Subsets in Human Blood After Viral Infection or Vaccination. Nat Immunol (2016) 17:1226–34. doi: 10.1038/ni.3533
98. Andrews SF, Chambers MJ, Schramm CA, Plyler J, Raab JE, Kanekiyo M, et al. Activation Dynamics and Immunoglobulin Evolution of Pre-Existing and Newly Generated Human Memory B Cell Responses to Influenza Hemagglutinin. Immunity (2019) 51:398–410.e5. doi: 10.1016/j.immuni.2019.06.024
99. Berkowska MA, Schickel JN, Grosserichter-Wagener C, de Ridder D, Ng YS, van Dongen JJ, et al. Circulating Human CD27-Iga+ Memory B Cells Recognize Bacteria With Polyreactive Igs. J Immunol (2015) 195:1417–26. doi: 10.4049/jimmunol.1402708
100. Crotty S. T Follicular Helper Cell Differentiation, Function, and Roles in Disease. Immunity (2014) 41:529–42. doi: 10.1016/j.immuni.2014.10.004
101. Ahluwalia P, Ahluwalia M, Vaibhav K, Mondal A, Sahajpal N, Islam S, et al. Infections of the Lung: A Predictive, Preventive and Personalized Perspective Through the Lens of Evolution, the Emergence of SARS-Cov-2 and Its Pathogenesis. EPMA J (2020) 1–21. doi: 10.1007/s13167-020-00230-1
102. Ng OW, Chia A, Tan AT, Jadi RS, Leong HN, Bertoletti A, et al. Memory T Cell Responses Targeting the SARS Coronavirus Persist Up to 11 Years Post-Infection. Vaccine (2016) 34:2008–14. doi: 10.1016/j.vaccine.2016.02.063
103. Welsh RM, Selin LK. No One is Naive: The Significance of Heterologous T-Cell Immunity. Nat Rev Immunol (2002) 2:417–26. doi: 10.1038/nri820
104. Sette A, Crotty S. Pre-Existing Immunity to SARS-Cov-2: The Knowns and Unknowns. Nat Rev Immunol (2020) 20:457–8. doi: 10.1038/s41577-020-0389-z
105. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T Cell Responses to SARS-Cov-2 Coronavirus in Humans With COVID-19 Disease and Unexposed Individuals. Cell (2020) 181:1489–501.e15. doi: 10.1016/j.cell.2020.05.015
106. Oja AE, Saris A, Ghandour CA, Kragten NAM, Hogema BM, Nossent EJ, et al. Divergent SARS-Cov-2-Specific T- and B-Cell Responses in Severe But Not Mild COVID-19 Patients. Eur J Immunol (2020) 50:1998–2012. doi: 10.1002/eji.202048908
107. Gil-Etayo FJ, Suarez-Fernandez P, Cabrera-Marante O, Arroyo D, Garcinuno S, Naranjo L, et al. T-Helper Cell Subset Response is a Determining Factor in COVID-19 Progression. Front Cell Infect Microbiol (2021) 11:624483. doi: 10.3389/fcimb.2021.624483
108. Kaneko N, Kuo HH, Boucau J, Farmer JR, Allard-Chamard H, Mahajan VS, et al. And GMassachusetts Consortium on Pathogen Readiness Specimen Working, Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19. Cell (2020) 183:143–57.e13. doi: 10.1016/j.cell.2020.08.025
109. Jackson DJ, Busse WW, Bacharier LB, Kattan M, O’Connor GT, Wood RA, et al. Association of Respiratory Allergy, Asthma, and Expression of the SARS-Cov-2 Receptor ACE2. J Allergy Clin Immunol (2020) 146:203–6.e3. doi: 10.1016/j.jaci.2020.04.009
110. Skevaki C, Karsonova A, Karaulov A, Fomina D, Xie M, Chinthrajah S, et al. SARS-Cov-2 Infection and COVID-19 in Asthmatics: A Complex Relationship. Nat Rev Immunol (2021) 21:202–3. doi: 10.1038/s41577-021-00516-z
111. Skevaki C, Karsonova A, Karaulov A, Xie M, Renz H. Asthma-Associated Risk for COVID-19 Development. J Allergy Clin Immunol (2020) 146:1295–301. doi: 10.1016/j.jaci.2020.09.017
112. Kimura H, Francisco D, Conway M, Martinez FD, Vercelli D, Polverino F, et al. Type 2 Inflammation Modulates ACE2 and TMPRSS2 in Airway Epithelial Cells. J Allergy Clin Immunol (2020) 146:80–8.e8. doi: 10.1016/j.jaci.2020.05.004
113. Schulte-Schrepping J, Reusch N, Paclik D, Bassler K, Schlickeiser S, Zhang B, et al. Severe COVID-19 is Marked by a Dysregulated Myeloid Cell Compartment. Cell (2020) 182:1419–40.e23. doi: 10.1016/j.cell.2020.08.001
114. Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martinez-Colon GJ, McKechnie JL, et al. A Single-Cell Atlas of the Peripheral Immune Response in Patients With Severe COVID-19. Nat Med (2020) 26:1070–6. doi: 10.1038/s41591-020-0944-y
115. Kurupati RK, Haut LH, Schmader KE, Ertl HCJ. Age-Related Changes in B Cell Metabolism. Aging-Us (2019) 11:4367–81z. doi: 10.18632/aging.102058
116. Joshi CJ, Schinn SM, Richelle A, Shamie I, O’Rourke EJ, Lewis NE. Standep: Capturing Transcriptomic Variability Improves Context-Specific Metabolic Models. PloS Comput Biol (2020) 16:e1007764. doi: 10.1371/journal.pcbi.1007764
117. Corcoran LM, Nutt SL. Long-Lived Plasma Cells Have a Sweet Tooth. Immunity (2016) 45:3–5. doi: 10.1016/j.immuni.2016.07.003
118. Lee WS, Wheatley AK, Kent SJ, DeKosky BJ. Antibody-Dependent Enhancement and SARS-Cov-2 Vaccines and Therapies. Nat Microbiol (2020) 5:1185–91. doi: 10.1038/s41564-020-00789-5
119. Mathew D, Giles JR, Baxter AE, Oldridge DA, Greenplate AR, Wu JE, et al. Deep Immune Profiling of COVID-19 Patients Reveals Distinct Immunotypes With Therapeutic Implications. Science (2020) 369. doi: 10.1126/science.abc8511
120. Rapkiewicz AV, Mai X, Carsons SE, Pittaluga S, Kleiner DE, Berger JS, et al. Megakaryocytes and Platelet-Fibrin Thrombi Characterize Multi-Organ Thrombosis At Autopsy in COVID-19: A Case Series. EClinicalMedicine (2020) 24:100434. doi: 10.1016/j.eclinm.2020.100434
121. Zhang FL, Shen GM, Liu XL, Wang F, Zhao YZ, Zhang JW. Hypoxia-Inducible Factor 1-Mediated Human GATA1 Induction Promotes Erythroid Differentiation Under Hypoxic Conditions. J Cell Mol Med (2012) 16:1889–99. doi: 10.1111/j.1582-4934.2011.01484.x
122. Chang JT, Wherry EJ, Goldrath AW. Molecular Regulation of Effector and Memory T Cell Differentiation. Nat Immunol (2014) 15:1104–15. doi: 10.1038/ni.3031
123. Pennock ND, White JT, Cross EW, Cheney EE, Tamburini BA, Kedl RM. T Cell Responses: Naive to Memory and Everything in Between. Adv Physiol Educ (2013) 37:273–83. doi: 10.1152/advan.00066.2013
124. Jarjour NN, Masopust D, Jameson SC. T Cell Memory: Understanding Covid-19. Immunity (2021) 54:14–8. doi: 10.1016/j.immuni.2020.12.009
125. Neidleman J, Luo X, Frouard J, Xie G, Gill G, Stein ES, et al. SARS-Cov-2-Specific T Cells Exhibit Phenotypic Features of Helper Function, Lack of Terminal Differentiation, and High Proliferation Potential. Cell Rep Med (2020) 1:100081. doi: 10.1016/j.xcrm.2020.100081
126. Crotty S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity (2019) 50:1132–48. doi: 10.1016/j.immuni.2019.04.011
127. Oh SJ, Lee JK, Shin OS. Aging and the Immune System: The Impact of Immunosenescence on Viral Infection, Immunity and Vaccine Immunogenicity. Immune Netw (2019) 19:e37. doi: 10.4110/in.2019.19.e37
128. Ahluwalia PK, Pandey RK, Sehajpal PK, Prajapati VK. Perturbed Microrna Expression by Mycobacterium Tuberculosis Promotes Macrophage Polarization Leading to Pro-Survival Foam Cell. Front Immunol (2017) 8:107. doi: 10.3389/fimmu.2017.00107
129. Gruver AL, Hudson LL, Sempowski GD. Immunosenescence of Ageing. J Pathol (2007) 211:144–56. doi: 10.1002/path.2104
130. Giamarellos-Bourboulis EJ, Tsilika M, Moorlag S, Antonakos N, Kotsaki A, Dominguez-Andres J, et al. Activate: Randomized Clinical Trial of BCG Vaccination Against Infection in the Elderly. Cell (2020) 183:315–323 e9. doi: 10.1016/j.cell.2020.08.051
131. Mantovani A, Netea MG. Trained Innate Immunity, Epigenetics, and Covid-19. N Engl J Med (2020) 383:1078–80. doi: 10.1056/NEJMcibr2011679
132. Netea MG, Giamarellos-Bourboulis EJ, Dominguez-Andres J, Curtis N, van Crevel R, van de Veerdonk FL, et al. Trained Immunity: A Tool for Reducing Susceptibility to and the Severity of SARS-Cov-2 Infection. Cell (2020) 181:969–77. doi: 10.1016/j.cell.2020.04.042
133. Poland GA, Ovsyannikova IG, Kennedy RB. SARS-Cov-2 Immunity: Review and Applications to Phase 3 Vaccine Candidates. Lancet (2020) 396:1595–606. doi: 10.1016/S0140-6736(20)32137-1
134. Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA, et al. Longitudinal Observation and Decline of Neutralizing Antibody Responses in the Three Months Following SARS-Cov-2 Infection in Humans. Nat Microbiol (2020) 5:1598–607. doi: 10.1038/s41564-020-00813-8
135. Reynolds CJ, Swadling L, Gibbons JM, Pade C, Jensen MP, Diniz MO, et al. Discordant Neutralizing Antibody and T Cell Responses in Asymptomatic and Mild SARS-Cov-2 Infection. Sci Immunol (2020) 5. doi: 10.1126/sciimmunol.abf3698
136. Canete PF, Vinuesa CG. COVID-19 Makes B Cells Forget, But T Cells Remember. Cell (2020) 183:13–5. doi: 10.1016/j.cell.2020.09.013
137. Arunachalam PS, Charles TP, Joag V, Bollimpelli VS, Scott MKD, Wimmers F, et al. T Cell-Inducing Vaccine Durably Prevents Mucosal SHIV Infection Even With Lower Neutralizing Antibody Titers. Nat Med (2020) 26:932–40. doi: 10.1038/s41591-020-0858-8
138. Grigoryan L, Pulendran B. The Immunology of SARS-Cov-2 Infections and Vaccines. Semin Immunol (2020) 50:101422. doi: 10.1016/j.smim.2020.101422
139. Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front Immunol (2020) 11:827. doi: 10.3389/fimmu.2020.00827
140. Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, et al. Broad and Strong Memory CD4(+) and CD8(+) T Cells Induced by SARS-Cov-2 in UK Convalescent Individuals Following COVID-19. Nat Immunol (2020) 21:1336–45. doi: 10.1038/s41590-020-0782-6
141. Dos Santos WG. Impact of Virus Genetic Variability and Host Immunity for the Success of COVID-19 Vaccines. BioMed Pharmacother (2021) 136:111272. doi: 10.1016/j.biopha.2021.111272
142. Keech C, Albert G, Cho I, Robertson A, Reed P, Neal S, et al. Phase 1-2 Trial of a SARS-Cov-2 Recombinant Spike Protein Nanoparticle Vaccine. N Engl J Med (2020) 383:2320–32. doi: 10.1056/NEJMoa2026920
Keywords: SARS–CoV-2, Pathogen, Immune system, Immune memory, Vaccine, Vaccine design, Immunological memory, T cells
Citation: Ahluwalia P, Vaibhav K, Ahluwalia M, Mondal AK, Sahajpal N, Rojiani AM and Kolhe R (2021) Infection and Immune Memory: Variables in Robust Protection by Vaccines Against SARS-CoV-2. Front. Immunol. 12:660019. doi: 10.3389/fimmu.2021.660019
Received: 28 January 2021; Accepted: 27 April 2021;
Published: 11 May 2021.
Edited by:
Gennady Bocharov, Institute of Numerical Mathematics (RAS), RussiaReviewed by:
Filippo Castiglione, National Research Council (CNR), ItalyAntonina Karsonova, I.M. Sechenov First Moscow State Medical University, Russia
Copyright © 2021 Ahluwalia, Vaibhav, Ahluwalia, Mondal, Sahajpal, Rojiani and Kolhe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ravindra Kolhe, cmtvbGhlQGF1Z3VzdGEuZWR1
 Pankaj Ahluwalia
Pankaj Ahluwalia Kumar Vaibhav
Kumar Vaibhav Meenakshi Ahluwalia2
Meenakshi Ahluwalia2 Ashis K. Mondal
Ashis K. Mondal Amyn M. Rojiani
Amyn M. Rojiani Ravindra Kolhe
Ravindra Kolhe