- 1Laboratory of Clinical Immunopathology, Research Institute for Fundamental and Clinical Immunology, Novosibirsk, Russia
- 2Laboratory of Molecular Immunology, Research Institute for Fundamental and Clinical Immunology, Novosibirsk, Russia
In this review, we described the structure and organization of antigen-recognizing repertoires of B and T cells from the standpoint of modern immunology. We summarized the latest advances in bioinformatics analysis of sequencing data from T and B cell repertoires and also presented contemporary ideas about the mechanisms of clonal diversity formation at different stages of organism development. At the same time, we focused on the importance of the allelic variants of the HLA genes and spectra of presented antigens for the formation of T-cell receptors (TCR) landscapes. The main idea of this review is that immune equilibrium and proper functioning of immunity are highly dependent on the interaction between the recognition and the presentation landscapes of antigens. Certain changes in these landscapes can occur during life, which can affect the protective function of adaptive immunity. We described some mechanisms associated with these changes, for example, the conversion of effector cells into regulatory cells and vice versa due to the trans-differentiation or bystander effect, changes in the clonal organization of the general TCR repertoire due to homeostatic proliferation or aging, and the background for the altered presentation of some antigens due to SNP mutations of MHC, or the alteration of the presenting antigens due to post-translational modifications. The authors suggest that such alterations can lead to an increase in the risk of the development of oncological and autoimmune diseases and influence the sensitivity of the organism to different infectious agents.
Introduction
The immune system is a complicated multilevel system of protection from different pathogens that contributes to the multicellularity and maintenance of genetic homeostasis (1–3). The development of adaptive immunity is associated with the appearance of RAG (recombination-activating gene) and two consecutive whole-genome duplications (4) that could be associated with the appearance of vertebrates and a transition from Agnatha to gnathostomes, which occurred around 500 million years ago (5, 6). The most important evolutionary advantage of adaptive immunity seems to be its specificity, which provides high precision and selectivity of the immune system activity. Another important advantage is the formation of immunological memory, which provides a quick and targeted reaction to the pathogen that the organism faced before (7). The so-called “price” that has to be paid for these advantages is the necessity to re-customize the adaptive immunity and form the immune memory individually in each generation.
The evolutionary development of the adaptive immune response is associated with the appearance of populations of T and B lymphocytes. Their precursors are found at the stage of early vertebrates (8). The main peculiarity of adaptive immunity is the formation of T and B lymphocytes with a high diversity of clones, wherein each clone has a unique antigen-recognizing receptor (TCR—T-cell receptor or BCR—B-cell receptor, respectively). For example, according to the recent data, the clonal diversity of only β chains of TCRs in the peripheral bloodstream is up to 108, which does not reflect the whole diversity that is comprised in the different organism compartments because the total number of T cells in the human organism is up to 1012 (9). At the same time, a recent evaluation indicates that the potential diversity of αβ TCRs varies from 1020 to 1061, which significantly exceeds the number of unique TCRs in the human organism (10, 11). In the case of B cells, the potential diversity of the BCR repertoire is also great and reaches 1020 (12–14). However, as for TCRs, the actual number is lower than the theoretical one and is approximately 108–109 of unique heavy BCR chains in the peripheral bloodstream (15, 16). These peculiarities lead to high personalization of repertoires, when the major part is private TCRs/BCRs and only a small part can be common in different individuals (public TCRs/BCRs). It is suggested that cross-reactivity plays an important role in the recognition of antigens because the approximate diversity of potential antigens reaches 209 and seems to exceed the summed actual diversity of T- and B-cell repertoires (11).
Such diversity is provided due to recombination of V(D)J gene segments of TCRs and BCRs caused by the activation of the RAG gene and due to the effect of terminal deoxynucleotidyl transferase (TdT) at the early stages of lymphocyte maturation (17, 18). The migration of T cells from the thymus begins at the end of the first trimester of the intrauterine development, while TdT, which randomly inserts nucleotides during V(D)J recombination, begins to express only in the middle of the second trimester (19). Thus, the majority of T cells in the fetus to the middle of gestation have zero nucleotide insertion in the region CDR3 (complementarity-determining region-3) (20). Still, their repertoire has a quite high diversity of TCRs due to V(D)J recombination. Unlike TCRs, the diversification of the BCR repertoire occurs earlier. For this reason, at the beginning of the second trimester, the BCR repertoire is characterized by a relatively high diversity, which gradually increases to the time of birth. After the birth, the diversity of the T-cell repertoire continues to increase to the involution of the thymus, while an increase in the diversity of the B-cell repertoire seems to be limited by the age-related degeneration of the bone marrow (20, 21). At the same time, at the early stages of development (less than 14 weeks), both repertoires of T and B cells are characterized by the oligoclonal organization that is replaced with polyclonal one by the 17th week of gestation, which is associated with a progressive increase in the number of sjTRECs (signal-joint T-cell receptor excision circles) and sjKRECs (signal-joint kappa-deleting recombination excision circles) (20). In early childhood, the diversity of T- and B-cell repertoires tends to its maximum (20, 22). Thus, in the fetal period and early childhood, the main diversity of the TCR and BCR repertoires is established that form the general landscape of recognition of antigens, which normally changes insignificantly within the life and tends to decrease with aging (22–24).
It is worth noting that MHC (major histocompatibility complex) molecules influence the formation of TCR repertoires of CD4+ and CD8+ lymphocytes. In other words, allele variants of MHCs limit the diversity of the represented antigens, which in turn, influences the formation of naïve and antigen-experienced TCR repertoires (25–27). At the same time, there are some differences in the formation of TCR repertoires of CD4+ and CD8+. This could be associated with different events observed in the thymus that determine the choice between the CD4+ or CD8+ cell differentiation. Double-positive CD4+CD8+ lymphocytes that receive a strong TCR-MHC-II signal, quickly stop the expression of CD8 and become single-positive CD4+ lymphocytes. In turn, CD4+CD8+ lymphocytes that do not receive a relatively strong TCR-MHC-II signal for a long time stop the expression of CD4+ and become single-positive CD8+ lymphocytes (28). Thus, CD8+ lymphocytes undergo a stricter selection in the thymus. Along with the possibility of recognizing epitopes presented by MHC-I, they lose the capability to recognize epitopes presented by MHC-II, which plays an important role in forming the naïve CD8+ TCR repertoire (29). This is confirmed by a small total amount of common TCRβ sequences in the populations of CD4+ and CD8+ lymphocytes, i.e. TCR repertoires of CD4+ and CD8+ overlap weakly, and there is a small amount of TCRs capable of reacting with both classes of MHC-I and MHC-II (30). Thus, the specificity of TCRs at the stage of CD4+CD8+ cells regulates the choice of CD4/CD8 differentiation. Further changes in the CD8+ repertoires could be associated with the allele variants of MHC-I, in particular, with their variants of fastidious or promiscuous binding that initiate oligoclonal or polyclonal variants of the immune response, respectively, by changing the number of certain clones (31–33). The difference between the CD4+ and CD8+ repertoires is in the formation of T-regulatory cells (Tregs) with a relatively high affinity of TCRs to self-antigens at the double-positive stage (34). Thus, the CD4+ repertoire contains cells with relatively high affinity to self-antigens, which is not observed in the CD8+ repertoire.
The formation of the naïve BCR repertoire of B cells is not so dependent on innate immunity. Similar to T cells, during maturation, B cells go through several stages of positive and negative selection. Each B cell can go through several cycles of rearrangement of V genes at different stages of maturation to increase the possibility of the formation of BCRs with the minimal capacity of reacting to self-antigens for the population of B-2 cells and a relatively higher affinity to self-antigens for the population of B-1a and B-1b cells (35, 36). Further formation of antigen-experienced BCR repertoire occurs in the process of somatic hypermutations (SHMs) during the maturation and activation of B cells on the periphery and under the mediated effect of Th cells, which suggests indirect involvement of the innate immunity (36).
Although the formation of the diversity of Ag-recognizing receptors occurs in a stochastic manner due to random V(D)J recombination and non-template nucleotide insertions (NIs), it is limited by a set of allele variants of MHC genes for a certain subject, at least, for CD4+ and CD8+ cells. Probably, this determines the individuality of the immune response and peculiarity of the homeostasis of the immune system in general in a certain organism. In this review, the authors will shortly describe the main peculiarities that can occur throughout life and affect the immune equilibrium increasing the risk of pathology.
Landscape of Recognition
As it was mentioned before, the general landscape of recognition is understood as a diversity of specific antigen-recognizing receptors that include TCRs of CD4+ and CD8+ cells, as well as BCRs of B cells. It is assumed that the higher the diversity of antigen-recognizing receptors, the wider the range of antigens that can potentially be recognized by the immune system, and, thus, the higher the effectiveness of the immune response against pathogens and altered self-antigens (37). It is suggested that the maintenance of auto-tolerance and the efficiency of anti-cancer immunity are also associated with the diversity of antigen-recognizing receptors of Tregs, which is critical in the context of antigen-specific action of these cells (38). Since the number of cells in the organism is limited, the formation of protective diversity should be based on the relation between the general diversity and the size of each antigen-specific clone. This idea is described in the Protecton Theory (39), wherein the protecton is the minimum number of cells of certain antigen specificity required for the timely provision of the sufficient number of effector cells per the unit of the body volume for the efficient protection against antigens (10, 39). Thus, knowledge of the clonal organization of T- and B-cell repertoires is important for the understanding of consistencies in the immune response in normal and pathological conditions, as well as for the identification of the peculiarities of the immune equilibrium maintenance in different conditions.
Organization of the Human T-Cell Repertoire
Recently, next-generation sequencing technologies and mathematic analysis have expanded the understanding of the clonal organization of the TCR repertoire in humans. It has been shown that the distribution of T-cell clones in the general TCR repertoire complies with the general consistency pattern within the human population and is not age-dependent (22, 40). The distribution of clones in the descending rank order (r) depending on the size of a clone (C) is subject to power-law distribution, i.e. the rank (r) of the largest clones correlates with their size (C) according to the power-law distribution r~C-α, wherein α is a scaling exponent (22, 41). The size of a clone is inversely proportional to its rank, i.e. the larger the clone, the lower is its rank, and vice versa. This dependence is described by a power-law function y = kC-α (40, 41). Empiric calculation of α is associated with a number of difficulties and depends on the used methods of sequencing and mathematical analysis. Thus, a recent study on the frequency distribution of T-cell clones of two independent cohorts showed a power-law relationship between the rank and size of the largest clones. At the same time, in both cohorts, α was almost similar and was equal to ~1.2 (22). The character of distribution of T-cell clones was similar in people of different ages. Thus, the general TCR repertoire in different people is characterized by a similar frequency distribution of T-cell clones regardless of age and is represented by a small number of dominant clones and a large variety of minor clones, which is consistent with the general type of Pareto distribution (Figure 1) (42).
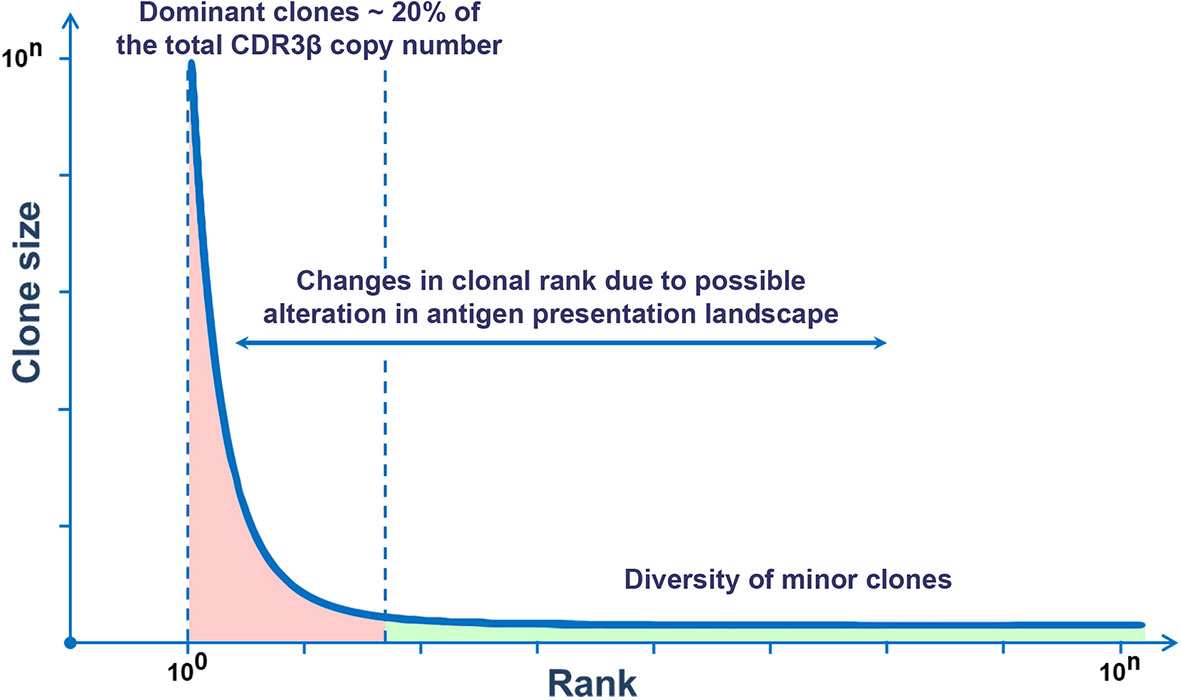
Figure 1 Rank–size frequency distribution follows a power-law distribution. This graph demonstrates the ranking of TCR clones by sizes. To the left, there are few dominant clones (red), and to the right, there is the long tail reflecting a multitude of minor clones (green).
A significant part of dominant clones in healthy people comprises zero insertion clones that are formed before birth and preserve in high abundance for several years with a tendency to a slight decrease throughout life (22, 24). At the same time, these clones provide the basis for public TCR repertoire in different individuals, which raises the issue of the presence of the inborn evolutionary determined set of T-cell clones within the adaptive immunity (11, 22, 24). If these clones exert certain functions or if they are a by-product of the formation of the TCR repertoire in the fetal period, it should be the subject of further studies.
Meier et al. (40) studied the frequency distribution of the TCRβ sequences at each level of the combination of gene segments (DJ, VDJ, and VDJ + NI) and revealed a fractal organization of the TCR repertoire and self-similarity of the frequency distribution of unique TCR clones (Figure 2) (40). An earlier study revealed the fractal organization of CD8+ TCR repertoires (43). At the same time, persons that are similar by human leukocyte antigens (HLAs) have a similar organization of TCR repertoires, which confirms the involvement of MHC genes in the formation of self-similarity pattern with a strict hierarchy of dominant and minor clones in the individual TCR repertoire (40).
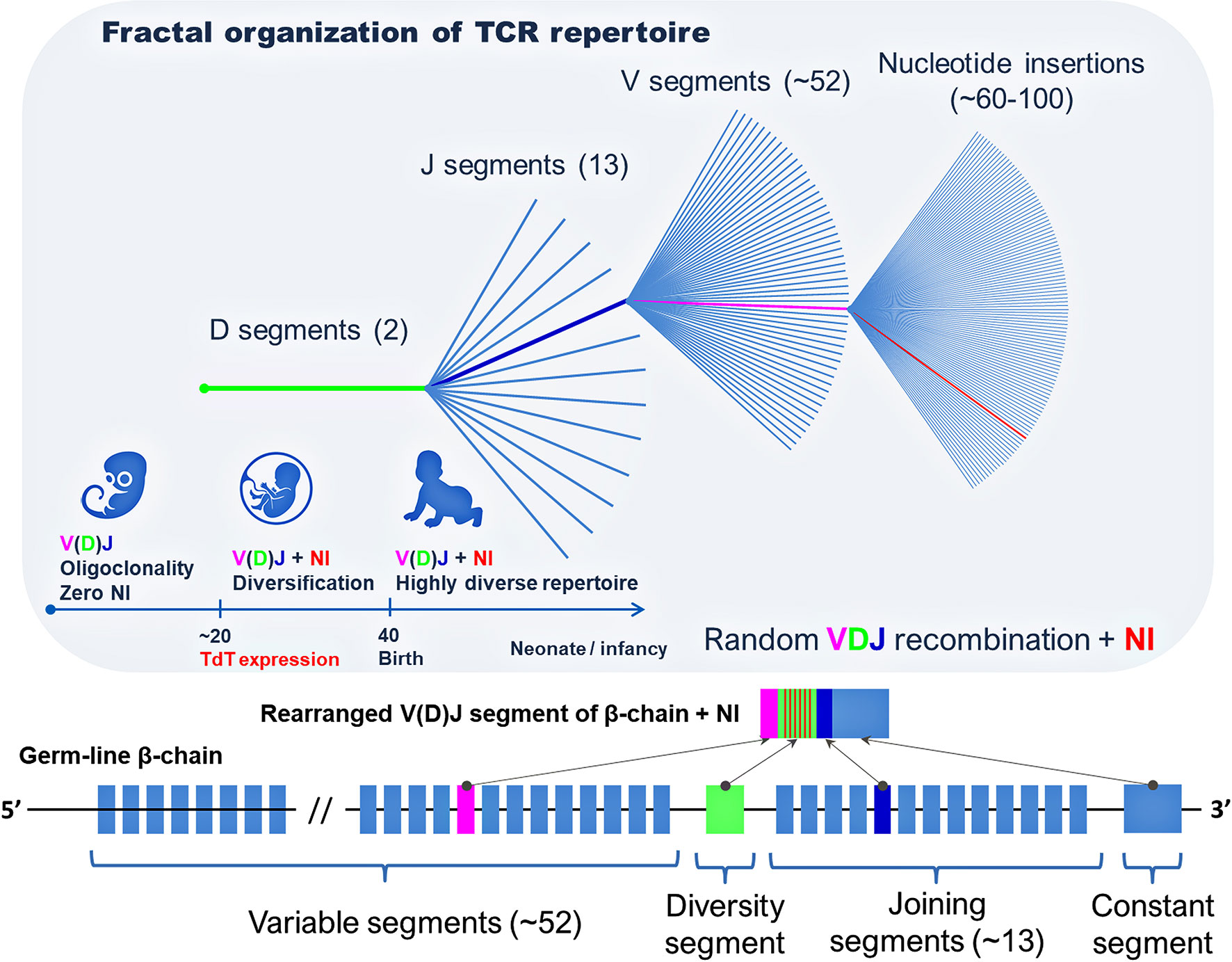
Figure 2 Fractal organization of T-cell repertoire. A fractal is a set with self-similarity (an object that exactly or approximately coincides with a part of itself, similar to itself on any magnification).
Besides, this study showed that the development of the “graft-versus-host disease” (GVHD) in recipients after transplantation of hematopoietic stem cells (HSCs) is associated with the changes in the clonal organization of the TCR repertoire and change of dominant clones within the first four ranks in comparison with the respective donors (40). Probably, such a shift in dominant clones is associated with incomplete identity by MHC genes between donors and recipients. GVHD is based on the incompliance between the landscapes of self-antigen presentation, which leads to the activation and expansion of minor self-reactive clones in recipients with GVHD (40, 44). This can be associated not only with quantitative changes in the clonal organization of the TCR repertoire but also with the plasticity of some subpopulations of T cells and the respective functional changes within these subpopulations (45, 46). In particular, the transition of some Tregs to some subpopulations of the effector cells or polarization of Th0 into Th17 cells in HSC recipients can lead to the development of GVHD (46, 47).
Transdifferentiation between the different T-cell subpopulations is well-known. However, not long ago, it was established that functionally different subpopulations of CD4+ cells expressed TCRs with different physicochemical properties and had different profiles of VDJ recombination, which affected their tendency to differentiate into each other (48). In their study, Kasatskaya et al. (48) focused on some characteristics of the CDR3 region in different subpopulations of T cells. The authors of that study evaluated different properties of amino acids in the CDR3 loop, the hydrophobicity of the loop (Kidera factor 4) (49), the length of the CDR3 loop, the predicted averaged binding energy of the TCR-pMHC (50, 51), and some other parameters that generally influence the affinity of Ag-specific TCR-pMHC interaction and the degree of TCR cross-reactivity (48). The study of these parameters revealed the differences in the physicochemical properties of the CDR3 TCR loop at the level of different subpopulations of T cells. It was shown that Treg cells have TCRs with high cross-reactivity, while follicular helpers Tfh have TCRs with minimal cross-reactivity (48). TCRs of Treg cells exert relatively higher affinity to self-antigens, bind cognate pMHC ligands less specifically and have lower averaged energy of TCR-pMHC binding than Tfh cells that bind cognate pMHC ligands with high affinity and have a higher energy of TCR-pMHC binding, which agrees with previous data (52–54). At the same time, such differences were also observed in other subpopulations: amino acidic characteristics of the CDR3 loop among the populations Th1/Th1-17/Th17 were similar to the characteristics of Tfh, while among populations Th22/Th2a/Th2, there was a similarity with Treg cells (48). Besides, different subpopulations of T cells were distinguished by a diversity of TCR repertoires. The highest diversity was observed in the subpopulation Tfh. A relatively high TCR diversity was observed in the subpopulations Th2, Th17, Th1, and Treg, while subpopulations Th22 and Th2a had signs of oligoclonal expansion, which indicated Ag-specific proliferation in these subpopulations (48). The physicochemical properties of TCRs are different in naïve TCR T cells and memory T cells, which was shown for populations of CD4+ and CD8+ lymphocytes. At the same time, for naïve nTreg and memory mTreg cells, such differences were not revealed (48, 55). Thus, the functional specialization of T cells depends on the structure of the CDR3 region and could be determined by the interaction of TCRs with the respective diversity of pMHC epitopes for each T-cell population (Figure 3). A paired analysis of the overlap of CDR3β diversity of different subpopulations of T cells revealed certain consistency in the transdifferentiation in healthy donors. A high plasticity was revealed between the functionally close populations Th17/Th22, Th17/Th2, Th22/Th2, Th2/Th2a. Lower plasticity was revealed between other populations Th17/Treg and Th1/Th17. At the same time, Tfh and Treg subpopulations were characterized by a high discreteness and had few CDR3β sequences common with other subpopulations of T cells (Figure 3) (48). Such data were first obtained for healthy donors and agreed with the data obtained in vitro or on animal models (38, 56, 57). Taking into account that the functional specialization of the subpopulation of T cells could be defined by specific interaction between TCR-pMHC and physicochemical properties of the CDR3 loop, the changes in the presentation landscape of antigens can lead to changes in the clonal structure of certain subpopulations of T cells. In some cases, such transitions of Ag-specific clones between subpopulations of T cells are involved in pathological processes. For example, an important role of a transition Treg ↔ Th17 was established in patients with different autoimmune conditions, graft rejection, and oncologic processes (58–63). Besides, plasticity Th17 → Th1 was revealed in patients with juvenile idiopathic arthritis and Crohn’s disease. In patients with allergic conditions and bronchial asthma, an enhanced transdifferentiation Th17 → Th2 was observed (64, 65). An imbalance between the subpopulations Th1/Th2 explains the pathogenesis of allergic and some oncologic diseases. However, in this case, an incorrect functional specialization of certain Ag-specific clones in the ontogenesis resulting from the changes in the conditions of presentation of the respective antigens is observed rather than a transdifferentiation (66–68).
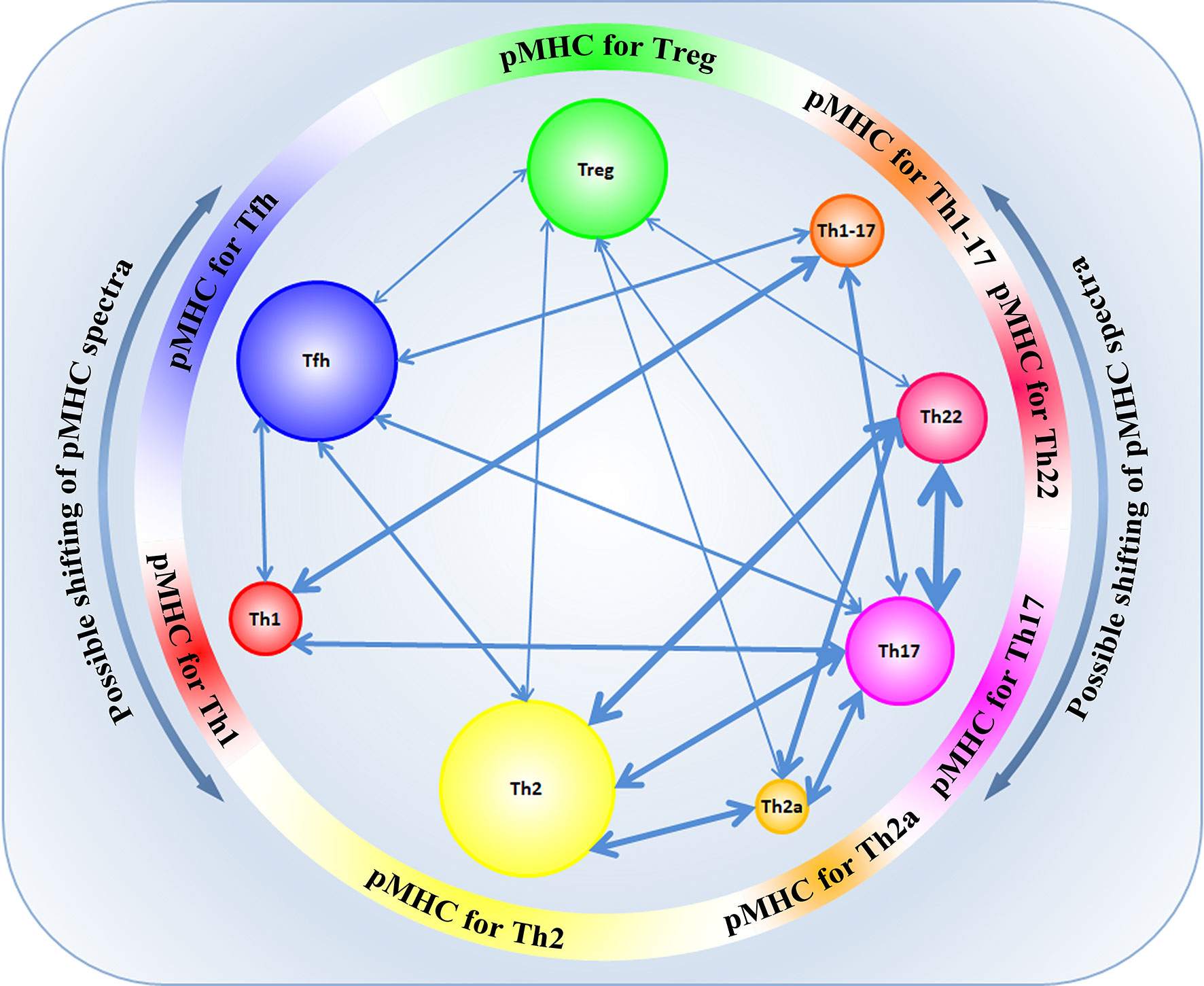
Figure 3 The landscape of presentation shapes different CD4+ subpopulations according to their CDR3 physicochemical properties and specificity. Possible shifts in pMHC spectra can contribute to transdifferentiation between some subpopulations of CD4+ T cells. The width of the arrows reflects the number of common TCR clonotypes between subpopulations.
Similar to CD4+ lymphocytes, an imbalance between effector CD8+ Teff and regulatory CD8+ Treg cells is significant for the maintenance of the immune equilibrium. Along with humoral factors of suppression, CD8+ Treg cells can exert Ag-specific suppressive activity mediated by the interaction with antigen-representing cells (69). Shifts in the represented antigen spectra and changes of conditions of their presentation can contribute to the irrelevant CD8+ Teff ↔ CD8+ Treg transdifferentiation (70). It was shown that such plasticity between subpopulations of CD8+ lymphocytes significantly contributed to the pathogenesis of different autoimmune and infectious diseases and oncological processes and took part in the graft rejections (69, 71–73). Still, despite the present achievements in the understanding of the organization of the T-cell repertoire, the identification of certain clones involved in the pathogenesis of different diseases attracts the attention of scientists in modern immunology and opens perspectives for personalized medicine.
Organization of the Human B-Cell Repertoire
The immunoglobulin gene rearrangement of B cell in the bone marrow results in the formation of a highly diverse repertoire of naïve (antigen-inexperienced) B cells that get into the peripheral circulation (74, 75). Similar to T cells, this process occurs under the influence of a complex of RAG, TdT, and a number of enzymes. Their activation induces V(D)J recombination and P- and N-insertions in the CDR3 loop of naive B cells (75–78). Further diversification of the BCR repertoire is associated with SHMs that occur in a cell under the influence of activation-induced deaminase (AID) in the peripheral lymph nodes with a cognate antigen (79, 80). This process underlies the affinity maturation of antibodies and targets an increase in the specificity of antigen recognition (75). Similar to T cells, the frequency distribution of clones in the general repertoire of B cells complies with power-law distribution (Figure 1) and agrees with the general biological type of Pareto distribution (81, 82). In other words, the BCR repertoire contains a relatively low number of dominant clones and an extremely high diversity of minor clones that form a long tail of distribution. Besides, as for T cells, the repertoires of naïve B cells are characterized by a tree-like fractal organization (Figure 4) (82, 83).
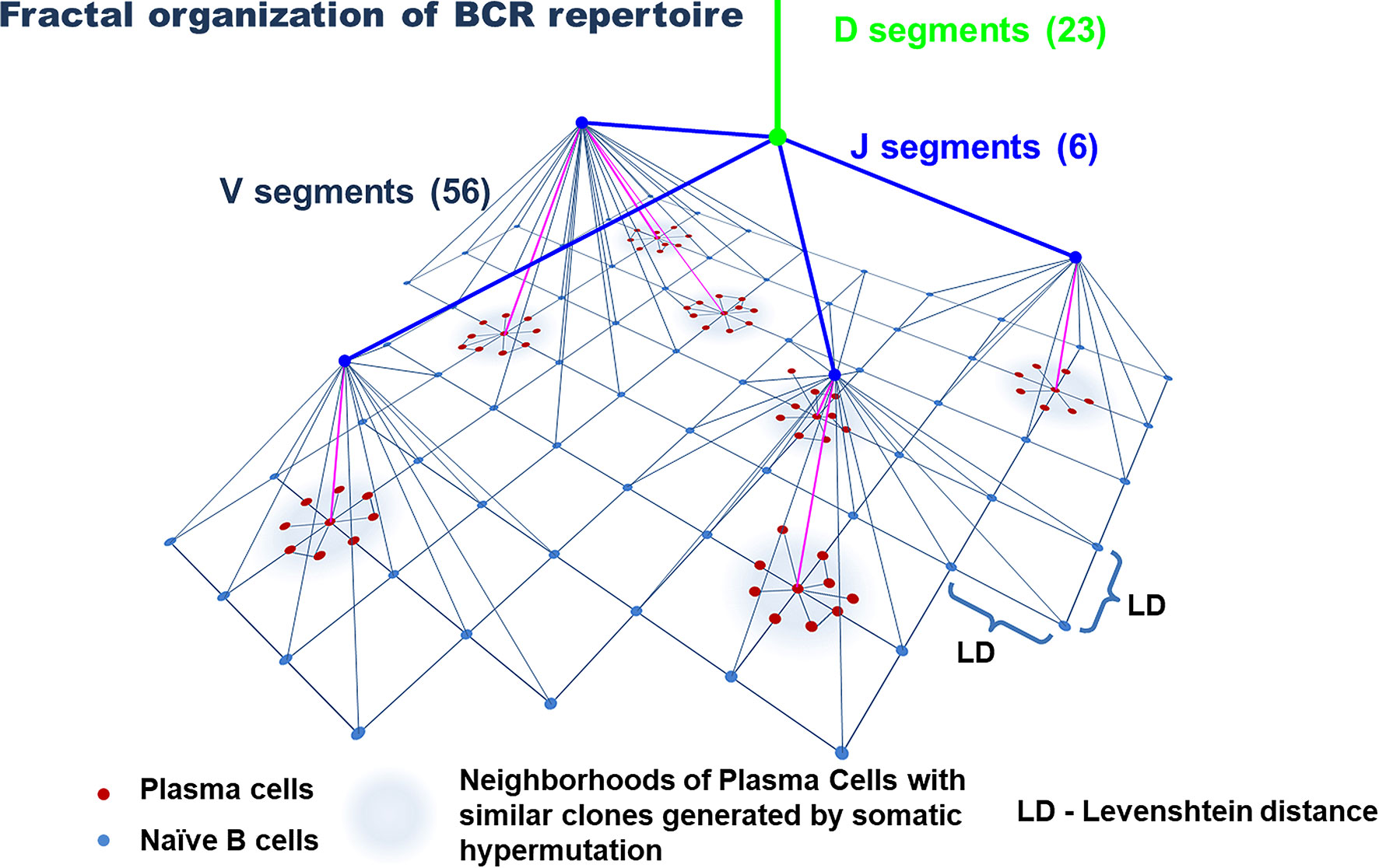
Figure 4 Model of the organization of the B-cell repertoire. Tree-like structures generated by VDJ recombination and nucleotide additions/deletions and a star-like structure for plasma cells likely generated by somatic hypermutation. The uniform distribution of naive B cells in the similarity layer schematically reflects a homogeneously interconnected network (by Levenshtein distances) of these cells, in contrast to plasma cells, that form highly interconnected subnetworks of similar clones. The number of V, D, and J segments is indicated for the IgH chain.
However, the architecture of the BCR repertoire has some peculiarities that are closely associated with the process of SHM and the formation of memory plasma cells. The diversity of the repertoire of these cells is significantly lower than the diversity of naïve B cells (84), which is associated with the history of antigen challenges that an organism faces throughout its life. At the same time, a tree-like structure of the repertoire of naïve B cells, which is generated due to VDJ recombination and nucleotide insertions, is replaced by a star-like structure for memory B cells and plasma cells, which is associated with the process of SHM (Figure 4) (82, 85). Such star-like structures reflect the process of activation of one or several B-cell clones closed by specificity. In the course of further expansion and SHMs, these B-cell clones form a set of antigen-experienced B-cell clone neighboring in the common space of CDR3 sequences. Part of these cells later becomes plasma cells. In this case, for the evaluation of similarity/closeness of the clones, Levenshtein distances were used (82, 86). Finally, the activity of AID is capable of mediating the shift of heavy chains from IgM/IgD to IgG, IgA, or IgE during SHM (74). It should be noted that despite the high personalization of BCR repertoires, different individuals have a similar organization of BCR and antigen repertoires (82, 87), which indicates general principles of the formation of BCRs and Ig diversity in different individuals. At the same time, the diversity of naïve B cells is affected by self-antigens and the repertoire of naïve B cells is limited by positive and negative selection (35, 36). The diversity of plasma cells directly depends on the diversity of antigen challenges within the life and depends on the functional activity of T cells, which is confirmed by a significant decrease in SHM in T cell-deficient mice (36, 87).
It is well-known that B cells play a central role in humoral immunity as antibody producers, can express some cytokines, and act as antigen-presenting cells (88–90). During the past years, many studies have been dedicated to the subpopulation of B cells with regulatory functions that were called B regulatory cells (Bregs) (91–94). Bregs exert their functions due to the production of anti-inflammatory cytokines, inhibit different populations of immune cells, and can induce the formation of Tregs from effector T cells acting as tolerogenic antigen-presenting cells, which do not exclude the Ag-specific effect of Bregs (91, 95, 96). Similar to T cells, irrelevant induction of Bregs and an imbalance between effector and regulatory B cells play a significant role in the pathogenesis of different autoimmune and oncologic processes, in patients with chronic infections and graft rejections (97–101). However, in this case, a transition Beff ↔ Breg could be primarily associated with the peculiarities of the microenvironment and only indirectly mediated by the shifts of spectra of the presented antigens and the conditions of their presentations via T cells (91, 102–104). Still, in some cases, the induction and functional activity of Bregs depend on the recognition of cognate antigens by Breg cells; and the suppression activity of Bregs can be mediated by direct B–T cellular interaction, which confirms the possibility of the Ag-specific effect of Bregs (105, 106). Thus, the conditions of the microenvironment and spectra of B-dependent antigens in the microenvironment of B cells influence their functional specialization; and an irrelevant transdifferentiation Beff ↔ Breg can underlie the pathogenesis of different pathologies.
An extremely high diversity of Ag-recognizing receptors of T and B cells provides the formation of qualitatively new properties that distinguish adaptive immunity from innate. The most important of them is the specificity of antigen recognition. Another important property is universality, i.e. the adaptive immune system can potentially specifically recognize any antigen of all possibilities. Redundancy—the same antigen can be recognized by different Ag-recognizing receptors due to cross-reactivity and the fact that one antigen can have different epitopes. Clones that are close by their specificity can duplicate and replace each other during the formation of the immune response. This underlies the robustness of adaptive immunity. In general, these properties provide the reliability of the immune system functioning and reflect the qualitative–quantitative transition, when a high diversity of Ag-recognizing receptors provides a qualitatively new level of the immune system functioning. However, changes in the conditions of antigen presentation can lead to situations when a disturbance of functional specialization of some subpopulations of T and B cells occurs, which can underlie the pathogenesis of different pathologies and be the main factor in the disturbances of the adaptive immunity.
Presentation Landscape
Rearrangement of a genome underlies a colossal diversity of Ag-recognizing receptors (17, 75, 107). However, the final formation of naïve repertoires of T and B cells is observed during the process of positive, and then, negative selection and depends on the diversity of the antigens presented in the thymus (108, 109) and the diversity of self-antigens in bone marrow (110, 111). In the past years, more data have been accumulated that confirm the influence of allele variants of MHC on the formation of individual TCR landscape (25–27, 112, 113).
T-Cell Presentation Landscape
The significance of MHC restriction for the development of T cells is well-known and can be illustrated by a recent study. It showed that during positive and negative selection, a selection of T cells with certain properties of the CDR3 loop occurred (114). Thus, during the process of positive selection, MHC restriction provides the selection of TCRs with the length of CDR3 (8–13 amino acid residues) and limits the selection of TCRs with positively charged and hydrophobic amine acid residues in the CDR3 loop. During the process of negative selection, it prevents the selection of TCRs with the residues of cysteine in the Ag-binding regions of the CDR3 loop (114). Probably, in this case, the selection of TCRs is primarily influenced by the physicochemical properties of MHC molecules and not certain epitopes in their composition. It should be noted that MHC restriction does not lead to the selection of TCRs with certain sequences of amino acids in the CDR3 loop and preserves high randomness of amino acid sequences in the CDR3 loop and sufficient diversity of TCRs for the recognition of the variety of potential antigens (9, 114, 115).
Apart from the common physicochemical properties of MHC molecules, an important role in the formation of the naïve repertoire of TCRs is played by epitopes presented in the thymus as a part of MHC molecules. The formation of the central auto-tolerance occurs due to the independent activity of transcriptional factors AIRE and Fezf2 that induce the expression of different tissue-restricted antigens by medullary thymic epithelial cells (mTECs) and thymic B cells (AIRE), which provide the elimination of self-reactive T cells during negative selection (109, 116–119). At the same time, some T cells, that exert a relatively high affinity to self-antigens, become Treg cells (38, 117). The affinity of TCR Treg cells to self-antigens is 100-fold lower than in the self-reactive T cells that undergo negative selection (38, 120). It was established that the transcriptional factors AIRE and Fezf2 provided the expression of nearly 60% of tissue-restricted antigens presented in the thymus (116). It is suggested that other antigens are presented in the thymus by different subpopulations of dendritic cells (DCs) (121). Presently, at least three DC subpopulations are known to provide the presentation of antigens in the thymus: CD8α+, Sirpα+, and B220+ plasmacytoid dendritic cell (pDCs). CD8α+ DCs occupy ~50% of the pool of thymic DCs (121), develop from the precursors in the thymus (122), and present tissue-restricted antigens, obtained from mTECs during trogocytosis or uptake of extracellular matrix, to T cells (123). Sirpα+ DCs occupy ~20% of the pool of thymic DCs, are found primarily in the corticomedullary perivascular spaces (124), and present antigens taken up from the bloodstream or acquired in the peripheral tissues before the migration to the thymus (121). The remaining ~30% of the pool of thymic DCs represent B220+ pDCs and present antigens obtained primarily on the periphery before the migration to the thymus (121). The presentation of self-antigens by the thymic DCs provides a negative selection of self-reactive CD4+ and CD8+ cells and probably contributes to the formation of Treg cells. However, precise mechanisms of this process are understudied (121, 125, 126). It is suggested that the migration of DCs from the periphery and the pathologic migration of B cells to the thymus (127) create the risks of inadequate formation of the central tolerance, for example, to the tumor or infectious antigens. However, this assumption requires experimental confirmation. Besides, it should be noted that the mechanisms of central tolerance do not exert absolute efficiency, and some self-reactive T cells can get to the peripheral bloodstream, which should be suppressed by Treg cells in normal conditions (54). This creates additional risks for the development of autoimmune diseases.
Thus, as a result of the described process, two varieties of T cells are formed that leave the thymus: T cells with relatively high affinity to self-antigens (Tregs) and T cells with low affinity to self-antigens (future anti-pathogen T cells) (54). After leaving the thymus, recent thymic emigrants (RTEs) migrate to the peripheral lymph nodes. During the maturation under the influence of the microenvironmental factors, they form a variety of mature naïve T cells (128). It is suggested that on the periphery, RTEs are subject to additional selection that is provided by MHC molecules in the peripheral lymph nodes. Only some RTEs with certain physicochemical properties of the CDR3 loop become mature naïve T cells (128). Probably, this stage of peripheral selection is necessary for the culling of potentially self-reactive T cells that manage to avoid negative selection in the thymus and deletion of T cells that cannot recognize effectively MHC molecules (48, 55, 128). At the same time, it is suggested that Treg cells do not undergo this stage of additional peripheral selection, which is confirmed by the lack of differences in physicochemical properties of the CDR3 loop between immature and mature subpopulations of Treg cells (48). Thus, the presentation landscape on the periphery provides an additional stage of the selection of CD4+ and CD8+ cells. As a result, a formation of the repertoire of mature naïve T cells occurs that will further go through a functional specialization according to their physicochemical properties and the specificity of their TCRs and form the main subpopulations of Th lymphocytes (48, 55, 129–131). It is evident that the changes in the landscape of presentation of antigens associated with different antigen challenges in the ontogenesis will influence the further formation of the TCR repertoire and clonal organization of different subpopulations of T cells in a certain individual. At the same time, it should be noted that the personalization of TCR repertoires of CD4+ and CD8+ is significantly provided by the individual set of allele variants of MHC genes (26, 31–33, 112).
Genetic Variants of MHC Molecules Influence the Landscape of Recognition
During the past decade, a lot of attention has been paid to the study of the influence of MHC allele variants on the repertoire of TCRs in different individuals. Associations were revealed between MHC genetic variability and the profiles of expression of TCR V genes (132). It was established that such associations were provided not only by the contact of TCRs with a peptide in the MHC complex but also by a physical contact between V-regions of the TCR β-chain and complementary regions of MHC molecules (132, 133), which indicated a direct influence of MHC genotypes on the formation of individual TCR repertoires. This agrees with the assumption of Niels Jerne on the co-evolution of MHC and TCR genes for a better predisposition to interact with each other (134). Still, the influence of the MHC genotype is primarily observed on the CD8+ lymphocytes. This is explained by a closer contact between TCRs of CD8+ cells and MHC-I molecules, while the regions CDR1 and CDR2 in TCRs of CD4+ cells have a weaker contact with complementary regions of the MHC-II molecules, and the region CDR3 primarily contacts with a peptide in the MHC-II complex (133, 135). This could provide the lack of influence of individual polymorphism of MHC-II genes on the diversity of the CD4+ repertoire, while a higher polymorphism of the MHC-I gene in heterozygotes is associated with a higher diversity of CD8+ cells (27). Still, a recent study on animal models showed a direct influence of MHC-II allele variants on the diversity and clonal organization of the CD4+ repertoire, including Treg cells (112). This confirms the hypothesis that allele variants of MHC-I and MHC-II play an important role in the formation of TCR repertoires of CD4+ and CD8+, respectively. Besides, it was shown that mutations in the conservative regions of MHC-I and MHC-II that contact with the complementary regions of TCRs influence the profiles of expression of TRAV and TRBV in the CD4+ and CD8+ repertoires and change their clonal organization (136, 137).
Thus, it is evident that the individual HLA phenotype defines epitope spectra that could be presented with the highest possibility, i.e. HLA phenotype is responsible for the formation of immunopeptidomes of MHC-I and MHC-II-associated antigens (138, 139). In turn, this affects the selection of T cells in the thymus, plays an important role in the formation of individual TCR repertoires on the periphery, and determines the individualization of the immune response. Such association between HLA genes and TCR repertoires reflects a close functional and phylogenetic association between innate and adaptive immunity.
Potential Risks of Immune Disequilibrium
The equilibrium in the immune system is achieved due to fine coordination between the innate and adaptive branches of immunity. Potential risks of the immune disequilibrium can be associated with different genetic factors, all possible antigenic challenges, and the influence of unfavorable factors of the environment.
It is well-known that there is a genetic predisposition to auto-immune diseases (ADs). It is hypothesized that the influence of different HLA variants and other genes associated with ADs in combination with epigenetic factors and unfavorable exogenous conditions contributes to the development of ADs (140). However, the presence of genetic predisposition does not always lead to the realization of the risk of ADs. Probably, an additional trigger is required (long-term lymphopenia or some immune regulatory disturbance) (141, 142). During the past two decades, the role of homeostatic proliferation in the development of ADs has been widely discussed (54, 143–147). This is a physiological process of the quantitative restoration of the peripheral pool of T cells after lymphopenia of any etiology by means of the antigen-specific proliferation of lymphocytes under the influence of IL-7 and IL-15 that could acquire pathological traits depending on the depth of lymphopenia (54, 148, 149). It was shown that this process could result in the selection of potentially self-reactive clones of T cells due to the competition for the contact of TCRs with self-pMHC, in a decrease in the diversity of the general TCR repertoire, and in a decrease of functional activity of Treg cells because of the deficiency of IL-2 in the conditions of lymphopenia (54, 150–153). Besides, a disturbance of the functional specialization of Treg cells and their conversion into pathogenic Th lymphocytes can occur (154). Some studies showed that Treg cells could not suppress the proliferation of T cells that received a strong TCR signal under the influence of IL-7 and IL-15, which was important in the context of homeostatic proliferation when a strong TCR signal gives advantages to T cells in the competition for the factors of survival (155, 156). This fact is interesting taking into account that AD-associated variants of MHCs contribute to a better presentation of antigens associated with the disease, and thus, homeostatic proliferation can contribute to the expansion of self-reactive T-clones in people with genetic predisposition (157, 158). Thus, a shift of spectra of the presented antigens towards self-antigens that are provided by AD-associated HLA variants and mediated via homeostatic proliferation of the changes in the clonal organization of TCR repertoires can underlie the disturbances in the immune equilibrium in patients with ADs.
Homeostatic proliferation can lead to a favorable antitumor immune response (159, 160). This response is formed as a result of polyclonal homeostatic expansion in the lymph nodes and is characterized by CD8+-cell cytotoxicity, an increase in the concentration of IFNγ, and the formation of memory cells (159). Besides, some data indicate that the shift in focus of homeostatic proliferation from CD8+ to CD4+ cells can be one of the causes of the development of ADs (159, 161). At the same time, the homeostatic proliferation of B cells does not lead to negative effects because it is exerted via an Ag-independent pathway and does not influence the diversity and clonal organization of the BCR repertoire (162, 163).
Probably, homeostatic proliferation can also contribute to a decrease in the general diversity of TCRs and the TCR diversity of naïve T cells with age (23), which negatively affects the protective function of the immune system against infections or other antigenic challenges in senior age (23, 164). It was shown that increased sensitivity to viral and oncological disease was associated with a decrease in the diversity of TCRs and connected with the formation of holes in the TCR repertoires (164–167).
The conditions of the microenvironment can significantly affect the functional activity of T and B cells causing their activation or leading to anergy and inducing the formation of Treg and Breg cells. An inflammatory microenvironment and co-stimulating signals that are transmitted during contact with the neighboring cells can lead to non-specific activation of different lymphocyte clones due to a so-called bystander effect (168). Since self-antigens can be present in the site of inflammation or immune response to an infection and any other antigen, the bystander effect can potentially cause unfavorable activation of self-reactive clones and increase the risk of ADs (168–171). Probably, a functional modulation of the TCR activation threshold due to the factors of co-stimulation and inflammatory microenvironment can contribute to the non-specific activation of T cells (172, 173). It was established that as a result of the bystander effect, a disturbance in the functional specialization of different subpopulations of lymphocytes could occur, for example, a transition of Treg cells into pathogenic Th lymphocytes, which could be also associated with the risk of the development of ADs (174). In other words, the bystander effect can lead to irrelevant Ag-independent activation of self-reactive lymphocytes and their expansion changing the structural organization of TCR and BCR repertoires at the level of separate clones and can contribute to the development of ADs.
Apart from the activation of lymphocytes, the factors of the microenvironment can cause anergy or even induce lymphocytes with regulatory functions by the Ag-independent bystander suppression. Similar effects are observed in the microenvironment of tumors that express some suppressor factors forming a tolerogenic medium and avoiding the immune surveillance (175, 176). At the same time, in the microenvironment of the tumor, a population of tolerogenic dendrite cells is formed. These cells are responsible for the formation of tumor-specific tolerance that is provided by T and B lymphocytes with regulatory functions (177–180). Besides, there is a possibility of the formation of central tolerance due to a migration of some dendrite cells loaded with tumor antigens to the thymus, wherein they can potentially be involved in the process of negative selection of T cells (121, 124–126). However, this assumption is hypothetical, and this issue requires additional research. Thus, the changes in the conditions of presentation of tumor antigens can shift the immune response from the immunogenic to tolerogenic and result in the respective changes in the clonal organization of T- and B-cell repertoires (181, 182). Besides, it changes the functional specialization of different lymphocyte populations providing the progression of the tumor growth.
Conclusion
The maintenance of the equilibrium in the immune system is an intricate dynamic process associated with constant changes in the landscapes of presentation and recognition, wherein genetic HLA variants play an important role, influence the formation of TCR repertoires, and determine the individualization of the immune response. In general, the disturbance of the immune equilibrium (autoimmune, infectious, or oncogenic process) is associated with the changes in the conditions of presentation and the spectra of the presented antigens, as well as with the transformation of T- and B-cell repertoires and a shift in the functional specialization of some T and B cells. In this case, the most important role is played by the genetic background and the influence of external environmental factors.
It is suggested that the study of genetic HLA variants and immunopeptidomes associated with a disease in a certain individual and the identification of certain clones of T and B cells involved in the pathogenesis of the disease will allow using personalized approaches to the therapy of different pathologies based on a targeted, specific effect on certain pathology mechanisms.
Author Contributions
DS contributed to the conception, drafting of the manuscript, and design. VT contributed to the conception and revision. VK contributed to the revision and final approval of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The research was supported by the grant of the Russian Science Foundation project no. 21-65-00004, https://rscf.ru/project/21-65-00004/.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Chaplin DD. Overview of the Immune Response. J Allergy Clin Immunol (2010) 125(2 Suppl 2):S3–23. doi: 10.1016/j.jaci.2009.12.980
2. Bich L, Pradeu T, Moreau JF. Understanding Multicellularity: The Functional Organization of the Intercellular Space. Front Physiol (2019) 10:1170. doi: 10.3389/fphys.2019.01170
3. Medina KL. Overview of the Immune System. Handb Clin Neurol (2016) 133:61–76. doi: 10.1016/B978-0-444-63432-0.00004-9
4. Flajnik MF, Kasahara M. Origin and Evolution of the Adaptive Immune System: Genetic Events and Selective Pressures. Nat Rev Genet (2010) 11(1):47–59. doi: 10.1038/nrg2703
5. Dehal P, Boore JL. Two Rounds of Whole Genome Duplication in the Ancestral Vertebrate. PloS Biol (2005) 3(10):e314. doi: 10.1371/journal.pbio.0030314
6. Holland LZ, Ocampo Daza D. A New Look at an Old Question: When did the Second Whole Genome Duplication Occur in Vertebrate Evolution? Genome Biol (2018) 19(1):209. doi: 10.1186/s13059-018-1592-0
7. Bonilla FA, Oettgen HC. Adaptive Immunity. J Allergy Clin Immunol (2010) 125(2 Suppl 2):S33–40. doi: 10.1016/j.jaci.2009.09.017
8. Cooper MD, Alder MN. The Evolution of Adaptive Immune Systems. Cell (2006) 124(4):815–22. doi: 10.1016/j.cell.2006.02.001
9. Qi Q, Liu Y, Cheng Y, Glanville J, Zhang D, Lee JY, et al. Diversity and Clonal Selection in the Human T-Cell Repertoire. Proc Natl Acad Sci USA (2014) 111(36):13139–44. doi: 10.1073/pnas.1409155111
10. Zarnitsyna VI, Evavold BD, Schoettle LN, Blattman JN, Antia R. Estimating the Diversity, Completeness, and Cross-Reactivity of the T Cell Repertoire. Front Immunol (2013) 4:485. doi: 10.3389/fimmu.2013.00485
11. de Greef PC, Oakes T, Gerritsen B, Ismail M, Heather JM, Hermsen R, et al. The Naive T-Cell Receptor Repertoire has an Extremely Broad Distribution of Clone Sizes. Elife (2020) 9:e49900. doi: 10.7554/eLife.49900
12. Elhanati Y, Sethna Z, Marcou Q, Callan CG Jr, Mora T, Walczak AM. Inferring Processes Underlying B-Cell Repertoire Diversity. Philos Trans R Soc Lond B Biol Sci (2015) 370(1676):20140243. doi: 10.1098/rstb.2014.0243
13. Imkeller K, Wardemann H. Assessing Human B Cell Repertoire Diversity and Convergence. Immunol Rev (2018) 284(1):51–66. doi: 10.1111/imr.12670
14. Chaudhary N, Wesemann DR. Analyzing Immunoglobulin Repertoires. Front Immunol (2018) 9:462. doi: 10.3389/fimmu.2018.00462
15. Briney B, Inderbitzin A, Joyce C, Burton DR. Commonality Despite Exceptional Diversity in the Baseline Human Antibody Repertoire. Nature (2019) 566(7744):393–7. doi: 10.1038/s41586-019-0879-y
16. Soto C, Bombardi RG, Branchizio A, Kose N, Matta P, Sevy AM, et al. High Frequency of Shared Clonotypes in Human B Cell Receptor Repertoires. Nature (2019) 566(7744):398–402. doi: 10.1038/s41586-019-0934-8
17. Krangel MS. Mechanics of T Cell Receptor Gene Rearrangement. Curr Opin Immunol (2009) 21(2):133–9. doi: 10.1016/j.coi.2009.03.009
18. Ralph DK, Matsen FA4. Consistency of VDJ Rearrangement and Substitution Parameters Enables Accurate B Cell Receptor Sequence Annotation. PloS Comput Biol (2016) 12(1):e1004409. doi: 10.1371/journal.pcbi.1004409
19. Park JE, Jardine L, Gottgens B, Teichmann SA, Haniffa M. Prenatal Development of Human Immunity. Science (2020) 368(6491):600–3. doi: 10.1126/science.aaz9330
20. Rechavi E, Lev A, Lee YN, Simon AJ, Yinon Y, Lipitz S, et al. Timely and Spatially Regulated Maturation of B and T Cell Repertoire During Human Fetal Development. Sci Transl Med (2015) 7(276):276ra25. doi: 10.1126/scitranslmed.aaa0072
21. Stras SF, Werner L, Toothaker JM, Olaloye OO, Oldham AL, McCourt CC, et al. Maturation of the Human Intestinal Immune System Occurs Early in Fetal Development. Dev Cell (2019) 51(3):357–73.e5. doi: 10.1016/j.devcel.2019.09.008
22. Gaimann MU, Nguyen M, Desponds J, Mayer A. Early Life Imprints the Hierarchy of T Cell Clone Sizes. Elife (2020) 9:e61639. doi: 10.7554/eLife.61639
23. Britanova OV, Putintseva EV, Shugay M, Merzlyak EM, Turchaninova MA, Staroverov DB, et al. Age-Related Decrease in TCR Repertoire Diversity Measured With Deep and Normalized Sequence Profiling. J Immunol (2014) 192(6):2689–98. doi: 10.4049/jimmunol.1302064
24. Pogorelyy MV, Elhanati Y, Marcou Q, Sycheva AL, Komech EA, Nazarov VI, et al. Persisting Fetal Clonotypes Influence the Structure and Overlap of Adult Human T Cell Receptor Repertoires. PloS Comput Biol (2017) 13(7):e1005572. doi: 10.1371/journal.pcbi.1005572
25. Hesnard L, Legoux F, Gautreau L, Moyon M, Baron O, Devilder MC, et al. Role of the MHC Restriction During Maturation of Antigen-Specific Human T Cells in the Thymus. Eur J Immunol (2016) 46(3):560–9. doi: 10.1002/eji.201545951
26. Migalska M, Sebastian A, Radwan J. Major Histocompatibility Complex Class I Diversity Limits the Repertoire of T Cell Receptors. Proc Natl Acad Sci USA (2019) 116(11):5021–6. doi: 10.1073/pnas.1807864116
27. Krishna C, Chowell D, Gönen M, Elhanati Y, Chan TA. Genetic and Environmental Determinants of Human TCR Repertoire Diversity. Immun Ageing (2020) 17:26. doi: 10.1186/s12979-020-00195-9
28. Karimi MM, Guo Y, Cui X, Pallikonda HA, Horková V, Wang YF, et al. The Order and Logic of CD4 Versus CD8 Lineage Choice and Differentiation in Mouse Thymus. Nat Commun (2021) 12(1):99. doi: 10.1038/s41467-020-20306-w
29. Koning D, Costa AI, Hoof I, Miles JJ, Nanlohy NM, Ladell K, et al. CD8+ TCR Repertoire Formation is Guided Primarily by the Peptide Component of the Antigenic Complex. J Immunol (2013) 190(3):931–9. doi: 10.4049/jimmunol.1202466
30. Li HM, Hiroi T, Zhang Y, Shi A, Chen G, De S, et al. Tcrβ Repertoire of CD4+ and CD8+ T Cells is Distinct in Richness, Distribution, and CDR3 Amino Acid Composition. J Leukoc Biol (2016) 99(3):505–13. doi: 10.1189/jlb.6A0215-071RR
31. Kaufman J. Generalists and Specialists: A New View of How MHC Class I Molecules Fight Infectious Pathogens. Trends Immunol (2018) 39(5):367–79. doi: 10.1016/j.it.2018.01.001
32. Rao X, Hoof I, Costa AI, van Baarle D, Keşmir C. HLA Class I Allele Promiscuity Revisited. Immunogenetics (2011) 63(11):691–701. doi: 10.1007/s00251-011-0552-6
33. Chappell P, Meziane el K, Harrison M, Magiera Ł, Hermann C, Mears L, et al. Expression Levels of MHC Class I Molecules are Inversely Correlated With Promiscuity of Peptide Binding. Elife (2015) 4:e05345. doi: 10.7554/eLife.05345
34. Caramalho Í, Nunes-Cabaço H, Foxall RB, Sousa AE. Regulatory T-Cell Development in the Human Thymus. Front Immunol (2015) 6:395. doi: 10.3389/fimmu.2015.00395
35. Nemazee D. Mechanisms of Central Tolerance for B Cells. Nat Rev Immunol (2017) 17(5):281–94. doi: 10.1038/nri.2017.19
36. Wang Y, Liu J, Burrows PD, Wang JY. B Cell Development and Maturation. Adv Exp Med Biol (2020) 1254:1–22. doi: 10.1007/978-981-15-3532-1_1
37. Wang GC, Dash P, McCullers JA, Doherty PC, Thomas PG. T Cell Receptor αβ Diversity Inversely Correlates With Pathogen-Specific Antibody Levels in Human Cytomegalovirus Infection. Sci Transl Med (2012) 4(128):128ra42. doi: 10.1126/scitranslmed.3003647
38. Shevyrev D, Tereshchenko V. Treg Heterogeneity, Function, and Homeostasis. Front Immunol (2020) 10:3100. doi: 10.3389/fimmu.2019.03100
39. Vrisekoop N, Monteiro JP, Mandl JN, Germain RN. Revisiting Thymic Positive Selection and the Mature T Cell Repertoire for Antigen. Immunity (2014) 41(2):181–90. doi: 10.1016/j.immuni.2014.07.007
40. Meier J, Roberts C, Avent K, Hazlett A, Berrie J, Payne K, et al. Fractal Organization of the Human T Cell Repertoire in Health and After Stem Cell Transplantation. Biol Blood Marrow Transpl (2013) 19(3):366–77. doi: 10.1016/j.bbmt.2012.12.004
41. Oakes T, Heather JM, Best K, Byng-Maddick R, Husovsky C, Ismail M, et al. “Quantitative Characterization of the T Cell Receptor Repertoire of Naïve and Memory Subsets Using an Integrated Experimental and Computational Pipeline Which Is Robust, Economical, and Versatile.” Front Immunol (2017) 8:1267. doi: 10.3389/fimmu.2017.01267
42. Newman MEJ. Power Laws, Pareto Distributions and Zipf’s Law. Contemp Phys (2005) 46(5):323–51. doi: 10.1080/00107510500052444
43. Naumov YN, Naumova EN, Hogan KT, Selin LK, Gorski J. A Fractal Clonotype Distribution in the CD8+ Memory T Cell Repertoire Could Optimize Potential for Immune Responses. J Immunol (2003) 170(8):3994–4001. doi: 10.4049/jimmunol.170.8.3994
44. Berrie JL, Kmieciak M, Sabo RT, Roberts CH, Idowu MO, Mallory K, et al. Distinct Oligoclonal T Cells are Associated With Graft Versus Host Disease After Stem-Cell Transplantation. Transplantation (2012) 93(9):949–57. doi: 10.1097/TP.0b013e3182497561
45. Malard F, Bossard C, Brissot E, Chevallier P, Guillaume T, Delaunay J, et al. Increased Th17/Treg Ratio in Chronic Liver GVHD. Bone Marrow Transpl (2014) 49(4):539–44. doi: 10.1038/bmt.2013.215
46. Forcade E, Paz K, Flynn R, Griesenauer B, Amet T, Li W, et al. An Activated Th17-Prone T Cell Subset Involved in Chronic Graft-Versus-Host Disease Sensitive to Pharmacological Inhibition. JCI Insight (2017) 2(12):e92111. doi: 10.1172/jci.insight.92111
47. Malard F, Gaugler B, Lamarthee B, Mohty M. Translational Opportunities for Targeting the Th17 Axis in Acute Graft-vs.-Host Disease. Mucosal Immunol (2016) 9(2):299–308. doi: 10.1038/mi.2015.143
48. Kasatskaya SA, Ladell K, Egorov ES, Miners KL, Davydov AN, Metsger M, et al. Functionally Specialized Human CD4+ T-Cell Subsets Express Physicochemically Distinct TCRs. Elife (2020) 9:e57063. doi: 10.7554/eLife.57063
49. Kidera A, Konishi Y, Oka M, Scheraga H. Statistical Analysis of the Physical Properties of the 20 Naturally Occurring Amino Acids. J Protein Chem (1985) 4:23–55. doi: 10.1007/BF01025492
50. Miyazawa S, Jernigan RL. Residue-Residue Potentials With a Favorable Contact Pair Term and an Unfavorable High Packing Density Term, for Simulation and Threading. J Mol Biol (1996) 256(3):623–44. doi: 10.1006/jmbi.1996.0114
51. Kosmrlj A, Read EL, Qi Y, Allen TM, Altfeld M, Deeks SG, et al. Effects of Thymic Selection of the T-Cell Repertoire on HLA Class I-Associated Control of HIV Infection. Nature (2010) 465(7296):350–4. doi: 10.1038/nature08997
52. Bolotin DA, Poslavsky S, Davydov AN, Frenkel FE, Fanchi L, Zolotareva OI, et al. Antigen Receptor Repertoire Profiling From RNA-Seq Data. Nat Biotechnol (2017) 35(10):908–11. doi: 10.1038/nbt.3979
53. Wyss L, Stadinski BD, King CG, Schallenberg S, McCarthy NI, Lee JY, et al. Affinity for Self Antigen Selects Treg Cells With Distinct Functional Properties. Nat Immunol (2016) 17(9):1093–101. doi: 10.1038/ni.3522
54. Daniil S, Valeriy T, Olesya M, Vladimir K. Homeostatic Proliferation as a Physiological Process and a Risk Factor for Autoimmune Pathology. AIMS Allergy Immunol (2021) 5(1):18–32. doi: 10.3934/Allergy.2021002
55. De Simone G, Mazza EMC, Cassotta A, Davydov AN, Kuka M, Zanon V, et al. CXCR3 Identifies Human Naive CD8+ T Cells With Enhanced Effector Differentiation Potential. J Immunol (2019) 203(12):3179–89. doi: 10.4049/jimmunol.1901072
56. Hegazy AN, Peine M, Helmstetter C, Panse I, Fröhlich A, Bergthaler A, et al. Interferons Direct Th2 Cell Reprogramming to Generate a Stable GATA-3(+)T-Bet(+) Cell Subset With Combined Th2 and Th1 Cell Functions. Immunity (2010) 32(1):116–28. doi: 10.1016/j.immuni.2009.12.004
57. Peck A, Mellins ED. Plasticity of T-Cell Phenotype and Function: The T Helper Type 17 Example. Immunology (2010) 129(2):147–53. doi: 10.1111/j.1365-2567.2009.03189.x
58. DuPage M, Bluestone JA. Harnessing the Plasticity of CD4(+) T Cells to Treat Immune-Mediated Disease. Nat Rev Immunol (2016) 16(3):149–63. doi: 10.1038/nri.2015.18
59. Hua J, Inomata T, Chen Y, Foulsham W, Stevenson W, Shiang T, et al. Pathological Conversion of Regulatory T Cells Is Associated With Loss of Allotolerance. Sci Rep (2018) 8(1):7059. doi: 10.1038/s41598-018-25384-x
60. Knochelmann HM, Dwyer CJ, Bailey SR, Amaya SM, Elston DM, Mazza-McCrann JM, et al. When Worlds Collide: Th17 and Treg Cells in Cancer and Autoimmunity. Cell Mol Immunol (2018) 15(5):458–69. doi: 10.1038/s41423-018-0004-4
61. Downs-Canner S, Berkey S, Delgoffe GM, Edwards RP, Curiel T, Odunsi K, et al. Suppressive IL-17a+Foxp3+ and Ex-Th17 IL-17anegfoxp3+ Treg Cells Are a Source of Tumour-Associated Treg Cells. Nat Commun (2017) 8:14649. doi: 10.1038/ncomms14649
62. Shevyrev D, Tereshchenko V, Kozlov V, Sizikov A, Chumasova O, Koksharova V. T-Regulatory Cells From Patients With Rheumatoid Arthritis Retain Suppressor Functions In Vitro. Exp Ther Med (2021) 21(3):209. doi: 10.3892/etm.2021.9641
63. Qiu R, Zhou L, Ma Y, Zhou L, Liang T, Shi L, et al. Regulatory T Cell Plasticity and Stability and Autoimmune Diseases. Clin Rev Allergy Immunol (2020) 58(1):52–70. doi: 10.1007/s12016-018-8721-0
64. Cosmi L, Maggi L, Santarlasci V, Liotta F, Annunziato F. T Helper Cells Plasticity in Inflammation. Cytometry A (2014) 85(1):36–42. doi: 10.1002/cyto.a.22348
65. Mirlekar B. Co-Expression of Master Transcription Factors Determines CD4+ T Cell Plasticity and Functions in Auto-Inflammatory Diseases. Immunol Lett (2020) 222:58–66. doi: 10.1016/j.imlet.2020.03.007
66. Maggi E. The TH1/TH2 Paradigm in Allergy. Immunotechnology (1998) 3(4):233–44. doi: 10.1016/s1380-2933(97)10005-7
67. Lee HL, Jang JW, Lee SW, Yoo SH, Kwon JH, Nam SW, et al. Inflammatory Cytokines and Change of Th1/Th2 Balance as Prognostic Indicators for Hepatocellular Carcinoma in Patients Treated With Transarterial Chemoembolization. Sci Rep (2019) 9(1):3260. doi: 10.1038/s41598-019-40078-8
68. Lin W, Zhang HL, Niu ZY, Wang Z, Kong Y, Yang XS, et al. The Disease Stage-Associated Imbalance of Th1/Th2 and Th17/Treg in Uterine Cervical Cancer Patients and Their Recovery With the Reduction of Tumor Burden. BMC Womens Health (2020) 20(1):126. doi: 10.1186/s12905-020-00972-0
69. Vieyra-Lobato MR, Vela-Ojeda J, Montiel-Cervantes L, López-Santiago R, Moreno-Lafont MC. Description of CD8+ Regulatory T Lymphocytes and Their Specific Intervention in Graft-Versus-Host and Infectious Diseases, Autoimmunity, and Cancer. J Immunol Res (2018) 2018:3758713. doi: 10.1155/2018/3758713
70. Yu Y, Ma X, Gong R, Zhu J, Wei L, Yao J. Recent Advances in CD8+ Regulatory T Cell Research. Oncol Lett (2018) 15(6):8187–94. doi: 10.3892/ol.2018.8378
71. Zhang S, Wu M, Wang F. Immune Regulation by CD8+ Treg Cells: Novel Possibilities for Anticancer Immunotherapy. Cell Mol Immunol (2018) 15(9):805–7. doi: 10.1038/cmi.2018.170
72. Shimokawa C, Kato T, Takeuchi T, Ohshima N, Furuki T, Ohtsu Y, et al. CD8+ Regulatory T Cells Are Critical in Prevention of Autoimmune-Mediated Diabetes. Nat Commun (2020) 11(1):1922. doi: 10.1038/s41467-020-15857-x
73. Heinrichs J, Li J, Nguyen H, Wu Y, Bastian D, Daethanasanmak A, et al. CD8(+) Tregs Promote GVHD Prevention and Overcome the Impaired GVL Effect Mediated by CD4(+) Tregs in Mice. Oncoimmunology (2016) 5(6):e1146842. doi: 10.1080/2162402X.2016.1146842
74. Kreer C, Gruell H, Mora T, Walczak AM, Klein F. Exploiting B Cell Receptor Analyses to Inform on HIV-1 Vaccination Strategies. Vaccines (Basel) (2020) 8(1):13. doi: 10.3390/vaccines8010013
75. Chi X, Li Y, Qiu X. V(D)J Recombination, Somatic Hypermutation and Class Switch Recombination of Immunoglobulins: Mechanism and Regulation. Immunology (2020) 160(3):233–47. doi: 10.1111/imm.13176
76. Landau NR, Schatz DG, Rosa M, Baltimore D. Increased Frequency of N-Region Insertion in a Murine Pre-B-Cell Line Infected With a Terminal Deoxynucleotidyl Transferase Retroviral Expression Vector. Mol Cell Biol (1987) 7(9):3237–43. doi: 10.1128/mcb.7.9.3237
77. Komori T, Okada A, Stewart V, Alt FW. Lack of N Regions in Antigen Receptor Variable Region Genes of TdT-Deficient Lymphocytes. Science (1993) 261(5125):1171–5. doi: 10.1126/science.8356451
78. Lewis S, Gifford A, Baltimore D. DNA Elements are Asymmetrically Joined During the Site-Specific Recombination of Kappa Immunoglobulin Genes. Science (1985) 228(4700):677–85. doi: 10.1126/science.3158075
79. Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, et al. Specific Expression of Activation-Induced Cytidine Deaminase (AID), A Novel Member of the RNA-Editing Deaminase Family in Germinal Center B Cells. J Biol Chem (1999) 274(26):18470–6. doi: 10.1074/jbc.274.26.18470
80. Maul RW, Gearhart PJ. AID and Somatic Hypermutation. Adv Immunol (2010) 105:159–91. doi: 10.1016/S0065-2776(10)05006-6
81. Mora T, Walczak. How Many Different Clonotypes do Immune Repertoires Contain? Curr Opin Syst Biol (2019) 18:104–10. doi: 10.1016/j.coisb.2019.10.001
82. Miho E, Roškar R, Greiff V, Reddy ST. Large-Scale Network Analysis Reveals the Sequence Space Architecture of Antibody Repertoires. Nat Commun (2019) 10(1):1321. doi: 10.1038/s41467-019-09278-8
83. Burgos JD. Fractal Representation of the Immune B Cell Repertoire. Biosystems (1996) 39(1):19–24. doi: 10.1016/0303-2647(95)01574-4
84. Kovaltsuk A, Raybould MIJ, Wong WK, Marks C, Kelm S, Snowden J, et al. Structural Diversity of B-Cell Receptor Repertoires Along the B-Cell Differentiation Axis in Humans and Mice. PloS Comput Biol (2020) 16(2):e1007636. doi: 10.1371/journal.pcbi.1007636
85. Prechl J. Network Organization of Antibody Interactions in Sequence and Structure Space: The RADARS Model. Antibodies (Basel) (2020) 9(2):13. doi: 10.3390/antib9020013
86. Levenshtein VI. Binary Codes Capable of Correcting Deletions, Insertions, and Reversals. Sov Phys Dokl (1966) 10(8):707–10.
87. Lindner C, Wahl B, Föhse L, Suerbaum S, Macpherson AJ, Prinz I, et al. Age, Microbiota, and T Cells Shape Diverse Individual IgA Repertoires in the Intestine. J Exp Med (2012) 209(2):365–77. doi: 10.1084/jem.20111980
88. Tarlinton D. B Cells Still Front and Centre in Immunology. Nat Rev Immunol (2019) 19(2):85–6. doi: 10.1038/s41577-018-0107-2
89. Hua Z, Hou B. The Role of B Cell Antigen Presentation in the Initiation of CD4+ T Cell Response. Immunol Rev (2020) 296(1):24–35. doi: 10.1111/imr.12859
90. Vazquez MI, Catalan-Dibene J, Zlotnik A. B Cells Responses and Cytokine Production Are Regulated by Their Immune Microenvironment. Cytokine (2015) 74(2):318–26. doi: 10.1016/j.cyto.2015.02.007
91. Wortel CM, Heidt S. Regulatory B Cells: Phenotype, Function and Role in Transplantation. Transpl Immunol (2017) 41:1–9. doi: 10.1016/j.trim.2017.02.004
92. Mauri C, Menon M. Human Regulatory B Cells in Health and Disease: Therapeutic Potential. J Clin Invest (2017) 127(3):772–9. doi: 10.1172/JCI85113
93. Dubois F, Limou S, Chesneau M, Degauque N, Brouard S, Danger R. Transcriptional Meta-Analysis of Regulatory B Cells. Eur J Immunol (2020) 50(11):1757–69. doi: 10.1002/eji.201948489
94. Chekol Abebe E, Asmamaw Dejenie T, Mengie Ayele T, Dagnew Baye N, Agegnehu Teshome A, Tilahun Muche Z. The Role of Regulatory B Cells in Health and Diseases: A Systemic Review. J Inflammation Res (2021) 14:75–84. doi: 10.2147/JIR.S286426
95. Zhang AH, Li X, Onabajo OO, Su Y, Skupsky J, Thomas JW, et al. B-Cell Delivered Gene Therapy for Tolerance Induction: Role of Autoantigen-Specific B Cells [Published Correction Appears in J Autoimmun. J Autoimmun (2010) 35(2):107–13. doi: 10.1016/j.jaut.2010.05.002
96. Matsushita T, Tedder TF. Identifying Regulatory B Cells (B10 Cells) That Produce IL-10 in Mice. Methods Mol Biol (2011) 677:99–111. doi: 10.1007/978-1-60761-869-0_7
97. Horii M, Matsushita T. Regulatory B Cells and T Cell Regulation in Cancer. J Mol Biol (2021) 433(1):166685. doi: 10.1016/j.jmb.2020.10.019
98. Shang J, Zha H, Sun Y. Phenotypes, Functions, and Clinical Relevance of Regulatory B Cells in Cancer. Front Immunol (2020) 11:582657. doi: 10.3389/fimmu.2020.582657
99. Cherukuri A, Mohib K, Rothstein DM. Regulatory B Cells: TIM-1, Transplant Tolerance, and Rejection. Immunol Rev (2021) 299(1):31–44. doi: 10.1111/imr.12933
100. Horst AK, Kumashie KG, Neumann K, Diehl L, Tiegs G. Antigen Presentation, Autoantibody Production, and Therapeutic Targets in Autoimmune Liver Disease. Cell Mol Immunol (2021) 18(1):92–111. doi: 10.1038/s41423-020-00568-6
101. Dasgupta S, Dasgupta S, Bandyopadhyay M. Regulatory B Cells in Infection, Inflammation, and Autoimmunity. Cell Immunol (2020) 352:104076. doi: 10.1016/j.cellimm.2020.104076
102. Baba Y, Matsumoto M, Kurosaki T. Signals Controlling the Development and Activity of Regulatory B-Lineage Cells. Int Immunol (2015) 27(10):487–93. doi: 10.1093/intimm/dxv027
103. Wang X, Wei Y, Xiao H, Liu X, Zhang Y, Han G, et al. Pre-Existing CD19-Independent GL7(-) Breg Cells are Expanded During Inflammation and in Mice With Lupus-Like Disease. Mol Immunol (2016) 71:54–63. doi: 10.1016/j.molimm.2016.01.011
104. Gallego-Valle J, Pérez-Fernández VA, Correa-Rocha R, Pion M. Generation of Human Breg-Like Phenotype With Regulatory Function In Vitro With Bacteria-Derived Oligodeoxynucleotides. Int J Mol Sci (2018) 19(6):1737. doi: 10.3390/ijms19061737
105. Kimura S, Rickert CG, Kojima L, Aburawi M, Tanimine N, Fontan F, et al. Regulatory B Cells Require Antigen Recognition for Effective Allograft Tolerance Induction. Am J Transpl (2020) 20(4):977–87. doi: 10.1111/ajt.15739
106. Mohib K, Cherukuri A, Zhou Y, Ding Q, Watkins SC, Rothstein DM. Antigen-Dependent Interactions Between Regulatory B Cells and T Cells at the T:B Border Inhibit Subsequent T Cell Interactions With DCs. Am J Transpl (2020) 20(1):52–63. doi: 10.1111/ajt.15546
107. Tonegawa S. Somatic Generation of Antibody Diversity. Nature (1983) 302(5909):575–81. doi: 10.1038/302575a0
108. Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and Negative Selection of the T Cell Repertoire: What Thymocytes See (and Don’t See). Nat Rev Immunol (2014) 14(6):377–91. doi: 10.1038/nri3667
109. Takaba H, Takayanagi H. The Mechanisms of T Cell Selection in the Thymus. Trends Immunol (2017) 38(11):805–16. doi: 10.1016/j.it.2017.07.010
110. Xu X, Deobagkar-Lele M, Bull KR, Crockford TL, Mead AJ, Cribbs AP, et al. An Ontogenetic Switch Drives the Positive and Negative Selection of B Cells. Proc Natl Acad Sci USA (2020) 117(7):3718–27. doi: 10.1073/pnas.1915247117
111. Eschbach C, Bach MP, Fidler I, Pelanda R, Köhler F, Rajewsky K, et al. Efficient Generation of B Lymphocytes by Recognition of Self-Antigens. Eur J Immunol (2011) 41(8):2397–403. doi: 10.1002/eji.201041344
112. Logunova NN, Kriukova VV, Shelyakin PV, Egorov ES, Pereverzeva A, Bozhanova NG, et al. MHC-II Alleles Shape the CDR3 Repertoires of Conventional and Regulatory Naïve CD4+ T Cells. Proc Natl Acad Sci USA (2020) 117(24):13659–69. doi: 10.1073/pnas.2003170117
113. DeWitt WS 3rd, Smith A, Schoch G, Hansen JA, Matsen FA4, Bradley P. Human T Cell Receptor Occurrence Patterns Encode Immune History, Genetic Background, and Receptor Specificity. Elife (2018) 7:e38358. doi: 10.7554/eLife.38358
114. Lu J, Van Laethem F, Bhattacharya A, Craveiro M, Saba I, Chu J, et al. Molecular Constraints on CDR3 for Thymic Selection of MHC-Restricted TCRs From a Random Pre-Selection Repertoire. Nat Commun (2019) 10(1):1019. doi: 10.1038/s41467-019-08906-7
115. Goldrath AW, Bevan MJ. Selecting and Maintaining a Diverse T-Cell Repertoire. Nature (1999) 402(6759):255–62. doi: 10.1038/46218
116. Takaba H, Morishita Y, Tomofuji Y, Danks L, Nitta T, Komatsu N, et al. Fezf2 Orchestrates a Thymic Program of Self-Antigen Expression for Immune Tolerance. Cell (2015) 163(4):975–87. doi: 10.1016/j.cell.2015.10.013
118. St-Pierre C, Trofimov A, Brochu S, Lemieux S, Perreault C. Differential Features of AIRE-Induced and AIRE-Independent Promiscuous Gene Expression in Thymic Epithelial Cells. J Immunol (2015) 195(2):498–506. doi: 10.4049/jimmunol.1500558
119. Cepeda S, Cantu C, Orozco S, Xiao Y, Brown Z, Semwal MK, et al. Age-Associated Decline in Thymic B Cell Expression of Aire and Aire-Dependent Self-Antigens. Cell Rep (2018) 22(5):1276–87. doi: 10.1016/j.celrep.2018.01.015
120. Lee HM, Bautista JL, Scott-Browne J, Mohan JF, Hsieh CS. A Broad Range of Self-Reactivity Drives Thymic Regulatory T Cell Selection to Limit Responses to Self. Immunity (2012) 37(3):475–86. doi: 10.1016/j.immuni.2012.07.009
121. Oh J, Shin JS. The Role of Dendritic Cells in Central Tolerance. Immune Netw (2015) 15(3):111–20. doi: 10.4110/in.2015.15.3.111
122. Luche H, Ardouin L, Teo P, See P, Henri S, Merad M, et al. The Earliest Intrathymic Precursors of CD8α(+) Thymic Dendritic Cells Correspond to Myeloid-Type Double-Negative 1c Cells. Eur J Immunol (2011) 41(8):2165–75. doi: 10.1002/eji.201141728
123. Perry JSA, Lio CJ, Kau AL, Nutsch K, Yang Z, Gordon JI, et al. Distinct Contributions of Aire and Antigen-Presenting-Cell Subsets to the Generation of Self-Tolerance in the Thymus. Immunity (2014) 41(3):414–26. doi: 10.1016/j.immuni.2014.08.007
124. Baba T, Nakamoto Y, Mukaida N. Crucial Contribution of Thymic Sirp Alpha+ Conventional Dendritic Cells to Central Tolerance Against Blood-Borne Antigens in a CCR2-Dependent Manner. J Immunol (2009) 183(5):3053–63. doi: 10.4049/jimmunol.0900438
125. Herbin O, Bonito AJ, Jeong S, Weinstein EG, Rahman AH, Xiong H, et al. Medullary Thymic Epithelial Cells and CD8α+ Dendritic Cells Coordinately Regulate Central Tolerance But CD8α+ Cells are Dispensable for Thymic Regulatory T Cell Production. J Autoimmun (2016) 75:141–9. doi: 10.1016/j.jaut.2016.08.002
126. Hasegawa H, Matsumoto T. Mechanisms of Tolerance Induction by Dendritic Cells In Vivo. Front Immunol (2018) 9:350. doi: 10.3389/fimmu.2018.00350
127. Hodge DL, Reynolds D, Cerbán FM, Correa SG, Baez NS, Young HA, et al. MCP-1/CCR2 Interactions Direct Migration of Peripheral B and T Lymphocytes to the Thymus During Acute Infectious/Inflammatory Processes. Eur J Immunol (2012) 42(10):2644–54. doi: 10.1002/eji.201242408
128. Houston EG Jr, Fink PJ. MHC Drives TCR Repertoire Shaping, But Not Maturation, in Recent Thymic Emigrants. J Immunol (2009) 183(11):7244–9. doi: 10.4049/jimmunol.0902313
129. Campbell DJ, Koch MA. Phenotypical and Functional Specialization of FOXP3+ Regulatory T Cells. Nat Rev Immunol (2011) 11(2):119–30. doi: 10.1038/nri2916
130. Luckheeram RV, Zhou R, Verma AD, Xia B. Cd4⁺T Cells: Differentiation and Functions. Clin Dev Immunol (2012) 2012:925135. doi: 10.1155/2012/925135
131. Mittrücker HW, Visekruna A, Huber M. Heterogeneity in the Differentiation and Function of CD8⁺ T Cells. Arch Immunol Ther Exp (Warsz) (2014) 62(6):449–58. doi: 10.1007/s00005-014-0293-y
132. Sharon E, Sibener LV, Battle A, Fraser HB, Garcia KC, Pritchard JK. Genetic Variation in MHC Proteins is Associated With T Cell Receptor Expression Biases. Nat Genet (2016) 48(9):995–1002. doi: 10.1038/ng.3625
133. Gao K, Chen L, Zhang Y, Zhao Y, Wan Z, Wu J, et al. Germline-Encoded TCR-MHC Contacts Promote TCR V Gene Bias in Umbilical Cord Blood T Cell Repertoire. Front Immunol (2019) 10:2064. doi: 10.3389/fimmu.2019.02064
134. Jerne NK. The Somatic Generation of Immune Recognition. Eur J Immunol (1971) 1(1):1–9. doi: 10.1002/eji.1830010102
135. Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW. Unconventional Topology of Self Peptide-Major Histocompatibility Complex Binding by a Human Autoimmune T Cell Receptor. Nat Immunol (2005) 6(5):490–6. doi: 10.1038/ni1187
136. Block MS, Hansen MJ, Van Keulen VP, Pease LR. MHC Class I Gene Conversion Mutations Alter the CD8 T Cell Repertoire. J Immunol (2003) 171(8):4006–10. doi: 10.4049/jimmunol.171.8.4006
137. Silberman D, Krovi SH, Tuttle KD, Crooks J, Reisdorph R, White J, et al. Class II Major Histocompatibility Complex Mutant Mice to Study the Germ-Line Bias of T-Cell Antigen Receptors. Proc Natl Acad Sci USA (2016) 113(38):E5608–17. doi: 10.1073/pnas.1609717113
138. Sofron A, Ritz D, Neri D, Fugmann T. High-Resolution Analysis of the Murine MHC Class II Immunopeptidome. Eur J Immunol (2016) 46(2):319–28. doi: 10.1002/eji.201545930
139. Vizcaíno JA, Kubiniok P, Kovalchik KA, Ma Q, Duquette JD, Mongrain I, et al. The Human Immunopeptidome Project: A Roadmap to Predict and Treat Immune Diseases. Mol Cell Proteomics (2020) 19(1):31–49. doi: 10.1074/mcp.R119.001743
140. Ramos PS, Shedlock AM, Langefeld CD. Genetics of Autoimmune Diseases: Insights From Population Genetics. J Hum Genet (2015) 60(11):657–64. doi: 10.1038/jhg.2015.94
141. Krupica T Jr, Fry TJ, Mackall CL. Autoimmunity During Lymphopenia: A Two-Hit Model. Clin Immunol (2006) 120(2):121–8. doi: 10.1016/j.clim.2006.04.569
142. L’Huillier A, Ren G, Shi Y, Zhang J. A Two-Hit Model of Autoimmunity: Lymphopenia and Unresponsiveness to TGF-β Signaling. Cell Mol Immunol (2012) 9(5):369–70. doi: 10.1038/cmi.2012.25
143. Jones JL, Thompson SA, Loh P, Davies JL, Tuohy OC, Curry AJ, et al. Human Autoimmunity After Lymphocyte Depletion Is Caused by Homeostatic T-Cell Proliferation. Proc Natl Acad Sci USA (2013) 110(50):20200–5. doi: 10.1073/pnas.1313654110
144. Monti P, Piemonti L. Homeostatic T Cell Proliferation After Islet Transplantation. Clin Dev Immunol (2013) 2013:217934. doi: 10.1155/2013/217934
145. Le Campion A, Gagnerault MC, Auffray C, Bécourt C, Poitrasson-Rivière M, Lallemand E, et al. Lymphopenia-Induced Spontaneous T-Cell Proliferation as a Cofactor for Autoimmune Disease Development. Blood (2009) 114(9):1784–93. doi: 10.1182/blood-2008-12-192120
146. Ellestad KK, Anderson CC. Two Strikes and You’re Out? The Pathogenic Interplay of Coinhibitor Deficiency and Lymphopenia-Induced Proliferation. J Immunol (2017) 198(7):2534–41. doi: 10.4049/jimmunol.1601884
147. Knipper JA, Wright D, Cope AP, Malissen B, Zamoyska R. PTPN22 Acts in a Cell Intrinsic Manner to Restrict the Proliferation and Differentiation of T Cells Following Antibody Lymphodepletion. Front Immunol (2020) 11:52. doi: 10.3389/fimmu.2020.00052
148. Moxham VF, Karegli J, Phillips RE, Brown KL, Tapmeier TT, Hangartner R, et al. Homeostatic Proliferation of Lymphocytes Results in Augmented Memory-Like Function and Accelerated Allograft Rejection. J Immunol (2008) 180(6):3910–8. doi: 10.4049/jimmunol.180.6.3910
149. Min B. Spontaneous T Cell Proliferation: A Physiologic Process to Create and Maintain Homeostatic Balance and Diversity of the Immune System. Front Immunol (2018) 9:547. doi: 10.3389/fimmu.2018.00547
150. Ge Q, Rao VP, Cho BK, Eisen HN, Chen J. Dependence of Lymphopenia-Induced T Cell Proliferation on the Abundance of Peptide/MHC Epitopes and Strength of Their Interaction With T Cell Receptors. Proc Natl Acad Sci USA (2001) 98(4):1728–33. doi: 10.1073/pnas.98.4.1728
151. Kassiotis G, Zamoyska R, Stockinger B. Involvement of Avidity for Major Histocompatibility Complex in Homeostasis of Naive and Memory T Cells. J Exp Med (2003) 197(8):1007–16. doi: 10.1084/jem.20021812
152. Kieper WC, Burghardt JT, Surh CD. A Role for TCR Affinity in Regulating Naive T Cell Homeostasis. J Immunol (2004) 172(1):40–4. doi: 10.4049/jimmunol.172.1.40
153. Zorn E, Nelson EA, Mohseni M, Porcheray F, Kim H, Litsa D, et al. IL-2 Regulates FOXP3 Expression in Human CD4+CD25+ Regulatory T Cells Through a STAT-Dependent Mechanism and Induces the Expansion of These Cells In Vivo. Blood (2006) 108(5):1571–9. doi: 10.1182/blood-2006-02-004747
154. Duarte JH, Zelenay S, Bergman ML, Martins AC, Demengeot J. Natural Treg Cells Spontaneously Differentiate Into Pathogenic Helper Cells in Lymphopenic Conditions. Eur J Immunol (2009) 39(4):948–55. doi: 10.1002/eji.200839196
155. Heninger AK, Theil A, Wilhelm C, Petzold C, Huebel N, Kretschmer K, et al. IL-7 Abrogates Suppressive Activity of Human CD4+CD25+FOXP3+ Regulatory T Cells and Allows Expansion of Alloreactive and Autoreactive T Cells. J Immunol (2012) 189(12):5649–58. doi: 10.4049/jimmunol.1201286
156. Shevyrev D, Tereshchenko V, Blinova E, Knauer N, Pashkina E, Sizikov A, et al. Regulatory T Cells Fail to Suppress Fast Homeostatic Proliferation In Vitro. Life (Basel) (2021) 11(3):245. doi: 10.3390/life11030245
157. Matzaraki V, Kumar V, Wijmenga C, Zhernakova A. The MHC Locus and Genetic Susceptibility to Autoimmune and Infectious Diseases. Genome Biol (2017) 18(1):76. doi: 10.1186/s13059-017-1207-1
158. Bodis G, Toth V, Schwarting A. Role of Human Leukocyte Antigens (HLA) in Autoimmune Diseases. Rheumatol Ther (2018) 5(1):5–20. doi: 10.1007/s40744-018-0100-z
159. Dummer W, Niethammer AG, Baccala R, Lawson BR, Wagner N, Reisfeld RA, et al. T Cell Homeostatic Proliferation Elicits Effective Antitumor Autoimmunity. J Clin Invest (2002) 110(2):185–92. doi: 10.1172/JCI15175
160. Datta S, Sarvetnick N. Lymphocyte Proliferation in Immune-Mediated Diseases. Trends Immunol (2009) 30(9):430–8. doi: 10.1016/j.it.2009.06.002
161. Theofilopoulos AN, Dummer W, Kono DH. T Cell Homeostasis and Systemic Autoimmunity. J Clin Invest (2001) 108(3):335–40. doi: 10.1172/JCI12173
162. Woodland RT, Schmidt MR. Homeostatic Proliferation of B Cells. Semin Immunol (2005) 17(3):209–17. doi: 10.1016/j.smim.2005.02.006
163. Jasiulewicz A, Lisowska KA, Pietruczuk K, Frąckowiak J, Fulop T, Witkowski JM. Homeostatic ‘Bystander’ Proliferation of Human Peripheral Blood B Cells in Response to Polyclonal T-Cell Stimulation In Vitro. Int Immunol (2015) 27(11):579–88. doi: 10.1093/intimm/dxv032
164. Yager EJ, Ahmed M, Lanzer K, Randall TD, Woodland DL, Blackman MA. Age-Associated Decline in T Cell Repertoire Diversity Leads to Holes in the Repertoire and Impaired Immunity to Influenza Virus. J Exp Med (2008) 205(3):711–23. doi: 10.1084/jem.20071140
165. Xu Y, Xu L, Chen C, Zhang Y, Zeng C, Jin Z, et al. Age-Related Immune Profile of the T Cell Receptor Repertoire, Thymic Recent Output Function, and miRNAs. BioMed Res Int (2020) 2020:5910823. doi: 10.1155/2020/5910823
166. Simnica D, Akyüz N, Schliffke S, Mohme M, V Wenserski L, Mährle T, et al. T Cell Receptor Next-Generation Sequencing Reveals Cancer-Associated Repertoire Metrics and Reconstitution After Chemotherapy in Patients With Hematological and Solid Tumors. Oncoimmunology (2019) 8(11):e1644110. doi: 10.1080/2162402X.2019.1644110
167. Charles J, Mouret S, Challende I, Leccia MT, De Fraipont F, Perez S, et al. T-Cell Receptor Diversity as a Prognostic Biomarker in Melanoma Patients. Pigment Cell Melanoma Res (2020) 33(4):612–24. doi: 10.1111/pcmr.12866
168. Pacheco Y, Acosta-Ampudia Y, Monsalve DM, Chang C, Gershwin ME, Anaya JM. Bystander Activation and Autoimmunity. J Autoimmun (2019) 103:102301. doi: 10.1016/j.jaut.2019.06.012
169. Fujinami RS, von Herrath MG, Christen U, Whitton JL. Molecular Mimicry, Bystander Activation, or Viral Persistence: Infections and Autoimmune Disease. Clin Microbiol Rev (2006) 19(1):80–94. doi: 10.1128/CMR.19.1.80-94.2006
170. Kim TS, Shin EC. The Activation of Bystander CD8+ T Cells and Their Roles in Viral Infection. Exp Mol Med (2019) 51(12):1–9. doi: 10.1038/s12276-019-0316-1
171. Burroughs NJ, Ferreira M, Oliveira BMPM, Pinto AA. Autoimmunity Arising From Bystander Proliferation of T Cells in an Immune Response Model. Math Comput Model (2011) 53, 7–8(April, 2011):1389–93. doi: 10.1016/j.mcm.2010.01.020
172. Slichter CK, McDavid A, Miller HW, Finak G, Seymour BJ, McNevin JP, et al. Distinct Activation Thresholds of Human Conventional and Innate-Like Memory T Cells. JCI Insight (2016) 1(8):e86292. doi: 10.1172/jci.insight.86292
173. Au-Yeung BB, Smith GA, Mueller JL, Heyn CS, Jaszczak RG, Weiss A, et al. IL-2 Modulates the TCR Signaling Threshold for CD8 But Not CD4 T Cell Proliferation on a Single-Cell Level. J Immunol (2017) 198(6):2445–56. doi: 10.4049/jimmunol.1601453
174. Komatsu N, Okamoto K, Sawa S, Nakashima T, Oh-hora M, Kodama T, et al. Pathogenic Conversion of Foxp3+ T Cells Into TH17 Cells in Autoimmune Arthritis. Nat Med (2014) 20(1):62–8. doi: 10.1038/nm.3432
175. Pitt JM, Marabelle A, Eggermont A, Soria JC, Kroemer G, Zitvogel L. Targeting the Tumor Microenvironment: Removing Obstruction to Anticancer Immune Responses and Immunotherapy. Ann Oncol (2016) 27(8):1482–92. doi: 10.1093/annonc/mdw168
176. Labani-Motlagh A, Ashja-Mahdavi M, Loskog A. The Tumor Microenvironment: A Milieu Hindering and Obstructing Antitumor Immune Responses. Front Immunol (2020) 11:940. doi: 10.3389/fimmu.2020.00940
177. DeVito NC, Plebanek MP, Theivanthiran B, Hanks BA. Role of Tumor-Mediated Dendritic Cell Tolerization in Immune Evasion. Front Immunol (2019) 10:2876. doi: 10.3389/fimmu.2019.02876
178. Demoulin S, Herfs M, Delvenne P, Hubert P. Tumor Microenvironment Converts Plasmacytoid Dendritic Cells Into Immunosuppressive/Tolerogenic Cells: Insight Into the Molecular Mechanisms. J Leukoc Biol (2013) 93(3):343–52. doi: 10.1189/jlb.0812397
179. Alonso R, Flament H, Lemoine S, Sedlik C, Bottasso E, Péguillet I, et al. Induction of Anergic or Regulatory Tumor-Specific CD4+ T Cells in the Tumor-Draining Lymph Node. Nat Commun (2018) 9(1):2113. doi: 10.1038/s41467-018-04524-x
180. Sharonov GV, Serebrovskaya EO, Yuzhakova DV, Britanova OV, Chudakov DM. B Cells, Plasma Cells and Antibody Repertoires in the Tumour Microenvironment. Nat Rev Immunol (2020) 20(5):294–307. doi: 10.1038/s41577-019-0257-x
181. Li N, Yuan J, Tian W, Meng L, Liu Y. T-Cell Receptor Repertoire Analysis for the Diagnosis and Treatment of Solid Tumor: A Methodology and Clinical Applications. Cancer Commun (Lond) (2020) 40(10):473–83. doi: 10.1002/cac2.12074
Keywords: adaptive immunity, immune equilibrium, T-cell receptor repertoire, B-cell receptor repertoire, antigen presentation/recognition, homeostatic proliferation, a rank-size frequency distribution of T- and B-cell receptors, immunopeptidome
Citation: Shevyrev D, Tereshchenko V and Kozlov V (2021) Immune Equilibrium Depends on the Interaction Between Recognition and Presentation Landscapes. Front. Immunol. 12:706136. doi: 10.3389/fimmu.2021.706136
Received: 06 May 2021; Accepted: 12 July 2021;
Published: 30 July 2021.
Edited by:
Nirupama Darshan Verma, University of New South Wales, AustraliaReviewed by:
Andrey Shaw, Genentech, Inc., United StatesHenrique Borges da Silva, Mayo Clinic Arizona, United States
Copyright © 2021 Shevyrev, Tereshchenko and Kozlov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniil Shevyrev, ZHIuZGFuaWlsMjVAbWFpbC5ydQ==
 Daniil Shevyrev
Daniil Shevyrev Valeriy Tereshchenko
Valeriy Tereshchenko Vladimir Kozlov
Vladimir Kozlov