- 1Institute of Marine Sciences and Guangdong Provincial Key Laboratory of Marine Biotechnology, Shantou University, Shantou, China
- 2College of Fisheries, Henan Normal University, Xinxiang, China
- 3Guangxi Key Laboratory of Aquatic Genetic Breeding and Healthy Aquaculture, Guangxi Academy of Fishery Sciences, Nanning, China
- 4Southern Marine Science and Engineering Guangdong Laboratory, Guangzhou, China
Hemocyanin is an important non-specific innate immune defense molecule with phenoloxidase, antiviral, antibacterial, hemolytic, and antitumor activities. To better understand the mechanism of functional diversity, proteomics approach was applied to characterize hemocyanin (HMC) expression profiles from Litopenaeus vannamei. At first, hemocyanin was purified by Sephadex G-100 and DEAE-cellulose (DE-52) columns from shrimp serum, and 34 protein spots were identified as HMC on the 2-DE gels. Furthermore, we found that 9 HMC spots about 75 or 77 kDa were regulated by Streptococcus agalactiae and Vibrio parahaemolyticus infection at 6, 12, and 24 h. In addition, 6 different pathogen-binding HMC fractions, viz., HMC-Mix, HMC-Vp, HMC-Va, HMC-Vf, HMC-Ec, and HMC-Sa, showed different agglutinative and antibacterial activities. Moreover, lectin-blotting analysis showed significant differences in glycosylation level among HMC isomers and bacteria-binding HMC fractions. Particularly, the agglutinative activities of the HMC fractions were almost completely abolished when HMC was deglycosylated by O-glycosidase, which suggest that O-linked sugar chains of HMC played important roles in the innate immune recognition. Our findings demonstrated for the first time that L. vannamei HMC had molecular diversity in protein level, which is closely associated with its ability to recognize diverse pathogens, whereas glycan modification probably contributed to HMC’s diversity and multiple immune activities.
1 Introduction
Hemocyanin (HMC) is a large copper-containing respiratory protein found in the hemolymph of mollusks and arthropods. Initially, the immune significance of HMC was dismissed until the late 1990s when Decker and Rimke demonstrated unequivocally its phenoloxidase activity (1). Further research revealed that HMC may be a novel and important non-specific innate immune defense molecule (2, 3). It has been reported that HMC could be functionally converted into phenoloxidase (4) antimicrobial peptides (5–8), antiviral (9–12), antitumor agent (13–17), antifungal (18), agglutinin (19, 20), hemolysin (21), and regulatory proteins (22, 23). Moreover, our previous findings indicated that HMC from shrimp Litopenaeus vannamei reacted with human Ig as an antigen (5, 19, 24) and acted as an immune-enhancing protein (25, 26). These results show HMC’s roles in many immune activities and its importance in invertebrate innate immune system. However, so far, little is known about the molecular mechanism of HMC’s functional diversity in L. vannamei.
Despite the long belief that invertebrates lacked an adaptive system and possessed simple, innate immune systems, recent evidence suggests that diversification of immune-response proteins against pathogens and high immune specificities are also found in invertebrates (27–29). Interestingly, our recent observations and those of others indicated that L. vannamei HMC also displayed high molecular diversity, including single-nucleotide polymorphisms (SNPs) and different types of variants, both of which could be modulated by pathogenic infection (30–35), suggesting that HMC functional diversity may be associated with its molecular polymorphism. However, the diversity of HMC at the protein level has not been studied so far.
In this study, we demonstrated via proteomic approach the diversity of HMC at the protein level in L. vannamei. Furthermore, the association between polymorphism at the protein level and resistance to diverse pathogens, and the mechanisms were also characterized. These findings will be helpful for understanding the molecular basis of HMC multifunctionality and establishing the novel strategies for shrimp disease control.
2 Materials and Methods
2.1 Animal and Preparation of Shrimp Serum
Healthy penaeid shrimps (L. vannamei) with approximate size of 10–14 cm in length and 15–20 g in weight from Shantou Huaxun Aquatic Product Corporation were obtained and reared in 25-L seawater tanks at 25°C. Air was continuously supplied using an electric pump.
Hemolymph was taken directly from the pericardial sinus using a sterile tube and then allowed to clot overnight at 4°C. The serum was separated after centrifuging at 3,000 g for 20 min and kept at −20°C until analysis. All animal experiments were conducted in accordance with the recommendations set forth in the Animal Ethics Procedures and Guidelines of the People’s Republic of China.
2.2 Purification of HMC by Chromatographies
HMC purification was performed by gel-filtration chromatography and anion-exchange chromatography as previously described with some modifications (5). Briefly, 2 ml of L. vannamei serum was loaded onto a Sephadex G-100 column, and then, the column was washed with Tris–HCl buffer (0.05 M, pH 8.0) at a flowrate of 1 ml/min until absorbance at 280 nm reached baseline. The eluted proteins were concentrated by ultrafiltration centrifuge tube. Then, the concentrated proteins (8 ml) were loaded onto a DEAE-cellulose (DE-52) column, equilibrated with 0.01 M pH 7.5 phosphate-buffered saline (PBS) buffer. Elution was performed with 0.2 M NaCl at a flowrate of 1 ml/min. Eluted proteins from the main peak (potential HMC) was determined by a modified Bradford assay (Bio-Rad, USA) and stored at −20°C until analysis.
2.3 One-Dimensional Sodium Sulfate–Polyacrylamide Gel Electrophoresis and Its Immunoblotting
Identification of the potential HMC was carried out using one-dimensional sodium sulfate–polyacrylamide gel electrophoresis (1-DE) and its immunoblotting. In brief, 1-DE was performed using a 5% stacking gel (pH 6.8) and a 10% separating gel (pH 8.9) in Tris–glycine buffer (pH 8.3). Then, the gel was transferred to a polyvinylidene fluoride (PVDF) membrane with a semi-dry transfer apparatus according to the manufacturer’s instructions. The membrane was blocked for 1 h with 5% skim milk in Tris-buffered saline (TBS) (20 mM Tris, 0.15 M NaCl, pH 7.4) at room temperature, then incubated with rabbit anti-shrimp HMC antisera (1:1,000 dilution) and goat anti-rabbit immunoglobulin G (IgG)–horseradish peroxidase (HRP) (1:3,000 dilution) antibodies at room temperature for 40 min and 1 h, respectively. Finally, the membrane was washed and developed with substrate (3′3-diminobenzidine, DAB) until optimum color developed.
2.4 Two-Dimensional Polyacrylamide Gel Electrophoresis and its Immunoblotting
To investigate the diversity of HMC, two-dimensional polyacrylamide gel electrophoresis (2-DE) and its immunoblotting were performed as our previous descriptions with some modifications (36). Briefly, a total of 30 µg of HMC in rehydration buffer [containing 7 M urea, 2 M thiourea, 4% CHAPS, 65 mM dithiothreitol (DTT), 0.2% bio-lyte, and 0.001% bromophenol blue, pH 3–10] was used to rehydrate the IPG strip (7 cm, pH 4.7–5.9 Bio-Rad, Hercules, CA) for 12 h. The isoelectric focusing (IEF) was performed at a constant temperature of 20°C using a continuous increase in voltage (up to 4,000 V) until reaching 35,000 Vh. Prior to the second dimension, the focused IPG was incubated for 15 min in an equilibration buffer containing 20% w/v glycerol, 2% sodium dodecyl sulfate (SDS), 0.375 M Tris–HCl (pH 8.8), 2% DTT, then further equilibrated for 15 min in a similar buffer in which 2% DTT was replaced with 2.5% of iodoacetamide. The strip was placed onto the top of a 12% 1-DE gel. Low-melting point agarose was used to cover the IPG strip and filter paper. Separation of proteins was carried out in the same conditions as described above for 1-DE. Following 2-DE, immunoblotting analysis was carried out as the same descriptions in Section 2.3 for the identification of HMC. Gels were stained with colloidal Coomassie, and spot volumes were compared using the image analysis software PDQuest Analysis Software (Bio-Rad).
2.5 MALDI-TOF-TOF Mass Spectrometry
To identify the proteins reacted with rabbit anti-shrimp HMC antibodies in the 2-DE map, matrix-assisted laser desorption ionization–time of flight–time of flight (MALDI-TOF-TOF) mass spectrometry analysis was further performed as our previous descriptions (26). The spots were excised from 2-DE gels, and the gel plug was digested with trypsin; then, 0.5 µl of the peptide mixture was mixed with the matrix a-cyano-4-hydroxycinnamic acid (1:1) and spotted onto a stainless steel MALDI plate. MS spectra were obtained using the ABI 4700 Proteomics Analyzer MALDI-TOF-TOF mass spectrometer (Applied Biosystems, Foster City, CA) operating in a result-dependent acquisition mode. Peptide mass maps were acquired in reflection mode (1-keV accelerating voltage) with 1,000 laser shots per spectrum. Six external standards (mass standard kit for the 4700 Proteomics Analyzer calibration mixture, Part Number 4333604, Applied Biosystems, Foster City, CA) were used to calibrate each spectrum to a mass accuracy within 50 ppm. Selected peptide masses were submitted to Mascot (http://www.matrixscience.com/cgi/search_form.pl?FORMVER=2&SEARCH=PMF) for NCBInr databases search.
2.6 Pathogens Challenge Tests
For the pathogens challenge tests, two representative bacterial, namely, Streptococcus agalactiae (Gram-positive bacterium) and Vibrio parahaemolyticus (Gram-negative bacterium) were selected. The shrimps were inoculated intramuscularly by using 1-ml syringes in the second abdominal segment with 50 μl of bacterial inoculum (107–108 CFU/ml). The treated animals were then returned to the tanks at room temperature. Hemolymph was collected at 0, 6, 12, and 24 hpi. The following procedures for serum preparation and 2-DE analysis were performed as described in Sections 2.1 and 2.4.
2.7 Bacterial Pull-Down Assay
To purify the HMC fractions binding with pathogens, bacterial pull-down assay was performed as described previously (37). In brief, five kinds of strains including S. agalactiae, V. parahaemolyticus, Vibrio alginolyticus, Vibrio fluvialis, and Escherichia coli K12 were used for incubation with L. vannamei serum in vitro. The bacterial cells were harvested at OD600 of 1.0 and washed three times with 0.9% (w/v) saline. Then, the supernatant was boiled for 10 min and harvested by centrifugation at 5,000 g for 10 min. Three hundred microliters of serum was mixed with 300 µl bacterial solution (1.0 × 109 CFU/ml), incubated at room temperature for 2 h, and the sample was centrifuged at 5,000 g for 10 min at 4°C. The pellet was washed three times with 0.9% saline and then resuspended in 1 M Tris–HCl (pH 8.0) containing 0.8 M NaCl at 37°C for 2 h. The supernatant incubated with S. agalactiae, V. parahaemolyticus, V. alginolyticus, V. fluvialis, and E. coli K12, and mixed five bacteria were collected and named as HMC-Sa, HMC-Vp, HMC-Va, HMC-Vf, HMC-Ec, and HMC-Mix, respectively. After determination with a modified Bradford assay (Bio-Rad, USA), these HMC isomers were identified by 1-DE, 2-DE, and their immunoblotting as described in Sections 2.3 and 2.4.
2.8 Agglutination Assays
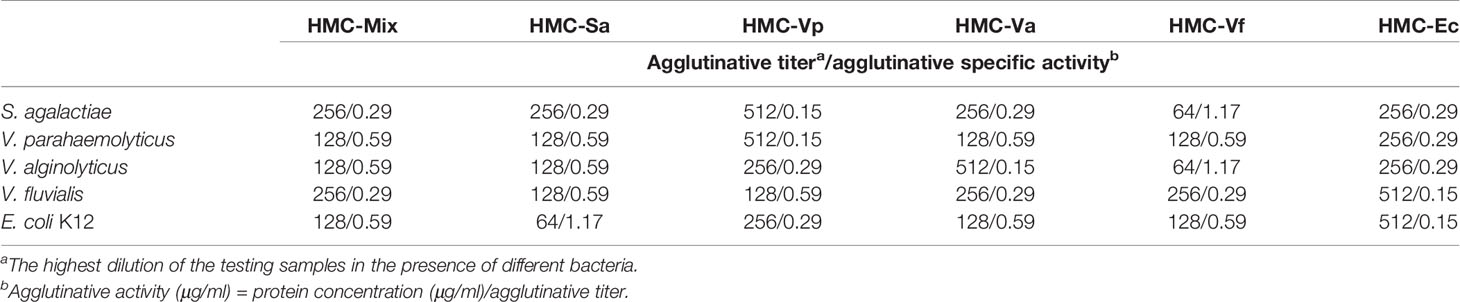
Agglutinative activities of six kinds of pathogens-binding HMC fractions, namely, HMC-Sa, HMC-Vp, HMC-Va, HMC-Vf, HMC-Ec, and HMC-Mix, were performed as our previous descriptions (38). Briefly, these bacteria were cultured in broth medium or Luria–Bertani medium overnight at 28 or 37°C (E. coli K12). The cells were harvested, washed, and diluted to 108 CFU/ml in TBS-Ca2+ (0.05 M Tris, 0.75% NaCl, 0.05 M CaCl2). Agglutination tests using the five bacteria and six specimens were performed at 37°C for 30 min. The proteins were twofold diluted in TBS-Ca2+ and added into 10 μl of each bacterium. Agglutination was observed in a light microscope and scored as positive (+) or negative (−) compared to a control placing the corresponding bacteria only in the TBS-Ca2+ buffer. Agglutinative titer was defined as the highest dilution of the test samples when the agglutination was appeared.
2.9 Antibacterial Activity Assays
Antibacterial activity was assessed by the numbers of bacterial colonies grown on the Petri dish according to our previous descriptions (5). V. alginolyticus and V. fluvialis were cultured in broth medium at 28°C for 12–24 h, then diluted to 103–104 CFU/ml with sterile PBS (0.01 M, pH 7.4). One hundred microliters of bacterial suspension was taken out and mixed with an equal volume of filtered HMC-Sa, HMC-Vp, HMC-Va, HMC-Vf, HMC-Ec, and HMC-Mix (experimental groups) or sterile 0.01 M pH 7.4 PBS (control group) at 37°C for 2 h. Fifty microliters of mixed solution was taken out and incubated on solid broth medium by the spread plate method at 37°C for 12 h, then counted the colony of the experiment group (A1) and control group (A0). The antibacterial activity was calculated as follows: antibacterial rate = (A0 − A1)/A0 × 100%. All samples were prepared in triplicate, and digital photomicrographs were taken with an Olympus BH-2 microscope.
2.10 Lectin-Blotting Analysis
Lectin-blotting analysis of HMC purified by gel-filtration chromatography and anion-exchange chromatography and the six kinds of pathogens-binding HMC fractions was carried out as previously described with some modifications (8). For 1D or 2D lectin blotting, 30 µg of HMC was separated by 1-DE or 2-DE and then transferred to a PVDF membrane as above descriptions in Sections 2.3 and 2.4. The membrane was blocked for 2.5 h with 5% bovine serum albumin (BSA) in TBS (20 mM Tris, 0.5 M NaCl, pH 7.4) at room temperature, then incubated with biotinylated lectin, viz., 1:1,000 dilution of concanavalin A (ConA), 1:100 dilution of peanut agglutinin (PNA), 1:500 dilution of ulex europaeus agglutinin (UEA), or 1:1,000 dilution dolichos biflorus agglutinin (DBA), for 1 h and avidin-peroxidase for 40 min at 37°C, respectively. Finally, the membrane was washed and developed with substrate (3′3-diminobenzidine, DAB) until optimum color developed. For Dot-lectin blotting, 1.5 μl of the six kinds of pathogens-binding HMC fractions (0.025, 0.05, or 0.5 mg/ml) was spotted onto a nitrocellulose (NC) membrane, which has been cut into desired size, soaked in TBS for 5 min, and then allowed to dry, using a regulatory pipe, respectively. After drying, the NC membrane was blocked with 2% polyvinylpyrrolidone (PVP, Sigma) for 1.5 h at room temperature. The following procedures for incubating with four biotinylated lectins and avidin-peroxidase and stained with DAB were performed as above described.
2.11 Total Glycan Measurement
The carbohydrate content measurement of HMC-Va, HMC-Vf, and HMC-Mix were determined by the colorimetric method as previously described with some modification (5). Briefly, 0, 100, 200, 300, and 400 μl of 0.01 M standard glucose solution were added to tubes; then, the duplicate distillated water was complemented to the final solution volume of 500 μl. Three hundred microliters of 6% (m/v) phenol and 1.5 ml sulfuric acid were added rapidly. After incubation at room temperature for 25 min, the absorbance was detected at 490 nm. Standard curve was constructed using sugar content as ordinate and the absorbance as abscissa. The total glycan content of samples were calculated according to the standard curve after detecting the absorbance as above using 200 μl of 250 μg/ml HMC-Va, HMC-Vf, and HMC-Mix, respectively.
2.12 Deglycosylation Assay
Deglycosylation of two HMC fractions was performed with O-glycosidase or N-glycosidase (New England Biolabs, USA) according to the manufacturer’s instructions and previous description (38). Briefly, HMC-Vp or HMC-Vf was deglycosylated under the following conditions: 5 μl of 10× G7 buffer, 3 μl of O-glycosidase or N-glycosidase, 5 μl of neuraminidase (only for O-glycosidase), then added HMC fractions (300 μg/ml for each) up to 50 μl; the reaction solution above was incubated 37°C for 4 h, respectively. The deglycosylated HMC fractions (HMC-Vp or HMC-Vf) by O-glycosidase and N-glycosidase were collected and named as dO-HMC-Vp and dO-HMC-Vf, and dN-HMC-Vp and dN-HMC-Vf, respectively. Deglycosylated HMC fractions (non-treated HMC fractions as control) were further processed by agglutinative activity assays as above described in Section 2.8.
2.13 Statistical Analysis
In this study, data are presented as mean ± standard deviation (SD). Statistical significance across groups was analyzed using one-way ANOVA analysis; differences were considered to be significant at p < 0.05 and extremely significant at p < 0.01.
3 Results
3.1 Protein Diversity of L. vannamei HMC
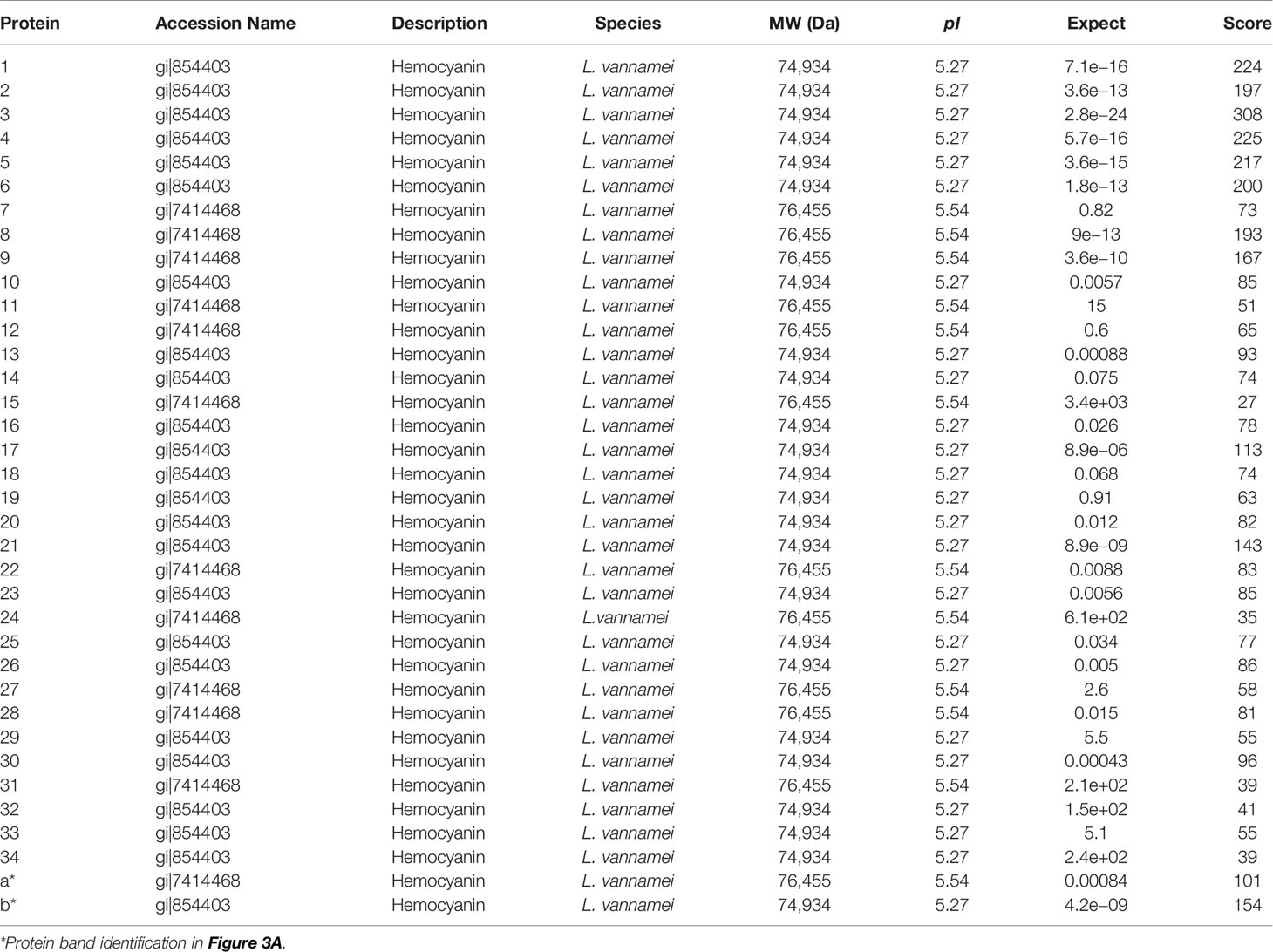
For a global assessment of HMC diversity in shrimp, HMC was first isolated from the hemolymph of L. vannamei by gel-filtration chromatography and anion-exchange chromatography and then identified with 1-DE and immunoblotting. As shown in Figure 1A, two bands at molecular weights approximately 75 and 77 kDa could react specifically with anti-shrimp HMC antibodies, suggesting that a good separation of the HMC has been achieved. Then, the purified HMC was analyzed by 2-DE (pH 4.7–5.9) and immunoblotting. The pattern of spots obtained from purified HMC is shown in Figure 1B. Over 40 distinct protein spots (mainly ranging from 25 to 77 kDa) were detected in 2-DE gels after Coomassie blue staining followed by analysis with the PDQuest software version 8.0. Immunoblotting showed a similar protein profile, indicating that all protein spots were HMC (Figure 1C). Furthermore, these spots were excised from gels and subjected to MALDI-TOF-TOF analysis. In total, 34 spots were successfully identified as L. vannamei HMC (Table 1), of which 6 spots (spots 1–6) and 3 spots (spots 7–9) were confirmed as HMC subunit with 75 and 77 kDa, respectively. Notably, 25 spots (spots 10–34), ranging from 25 to 66 kDa, also showed homology with HMC, suggesting that these may be HMC fragments or modified proteins. These results indicate that L. vannamei HMC might possess high molecular diversity at the protein level.
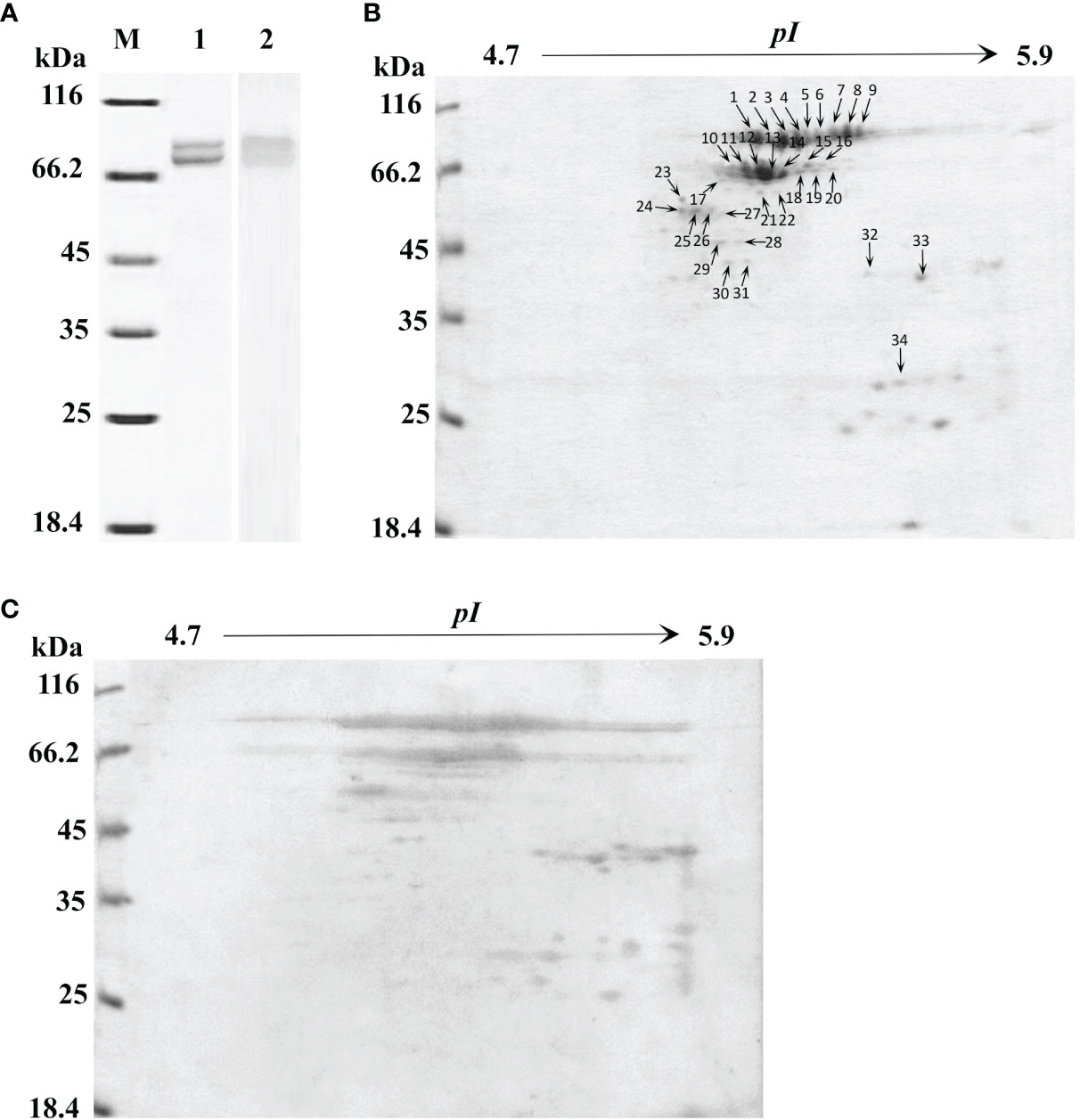
Figure 1 One-dimensional sodium sulfate-polyacrylamide gel electrophoresis (1-DE), two-dimensional polyacrylamide gel electrophoresis (2-DE) and immunoblotting analysis of L. hemocyanin. (A) 1-DE image of L. vannamei hemocyanin purified by gel-filtration chromatography and anion-exchange chromatography (lane 1) and its immunoblotting analysis (lane 2). M, molecular mass markers. Western blotting was performed using rabbit anti-shrimp HMC antisera (1:1,000 dilution) and goat anti-rabbit IgG-HRP (1: 3,000 dilution) antibodies. (B) 2-DE image of 30 µg hemocyanin from (A) with pH 4.7–5.9 IPG strips. The spots were excised from 2-DE gels, and submitted to protein identification by MALDI-TOF-TOF-MS analysis, the spots marked with number indicated that the HMC proteins identified. (C) Immunoblotting was carried out as the same descriptions with (A).
3.2 Responses of HMC Isomers to Pathogen Challenge In Vivo
In order to examine whether different HMC isomers responded to pathogen stimulation, 2-DE analysis was carried out to compare the variation in the nine protein spots in HMC of L. vannamei treated with S. agalactiae or V. parahaemolyticus. As shown in Figure 2A, the protein expression of HMC spots increased or decreased to some extent at different time points during the 24-h time period. After treatment with S. agalactiae for 6–24 h, spots 1–4 were upregulated, while spots 5 and 7 were downregulated compared with 0 h control group. However, spots 5–7 were upregulated after treatment with V. parahemolyticus for 6–24 h, while spots 4, 8, and 9 were downregulated at least at one time point post V. parahemolyticus treatment compared with the 0-h control group. Notably, a comparative analysis of the two gels indicated that spot 2 was upregulated about 2.5-, 2.5-, and 1.9-fold after stimulation with S. agalactiae at 6, 12, and 24 h, respectively, whereas no significant difference was observed in spot 2 during the entire 24-h period of infection by V. parahemolyticus (Figures 2B, C). Thus, these results indicated that L. vannamei HMC diversity might be related to shrimp’s resistance to specific bacteria in vivo.
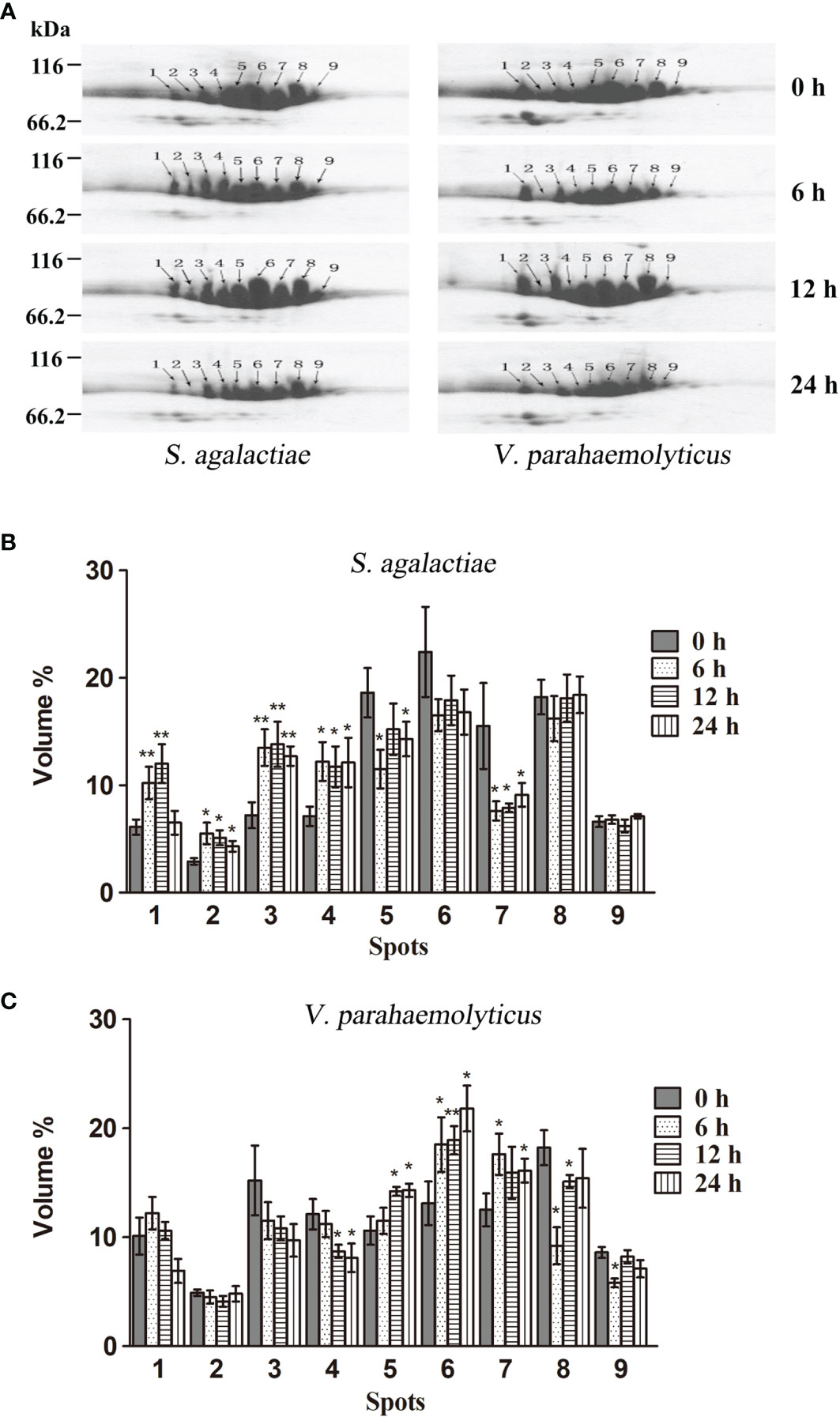
Figure 2 2-DE analysis of the hemocyanin from L. vannamei infected with pathogenic bacteria. (A) 2-DE analysis of 30 µg hemocyanin from L. vannamei treated with S. agalactiae (left) and V. parahaemolyticus (right) for 0–24 h; 0 h samples were used as control. (B, C) The expression level is normalized by densitometry analysis using the image analysis software PDQuest Analysis Software (Bio-Rad); 0 h samples were used as control. *p < 0.05, **p < 0.01.
3.3 Agglutinative and Antibacterial Activities of HMC Isomers Bound to Pathogens In Vitro
To further determine the relationship between HMC isomers and its resistance to different pathogens in vitro, six HMC fractions bound directly to mixed bacteria (HMC-Mix), S. agalactiae (HMC-Sa), V. parahaemolyticus (HMC-Vp), V. alginolyticus (HMC-Va), V. fluvialis (HMC-Vf), and E. coli K12 (HMC-Ec) were purified by pull-down. Results of the 1-DE and 1-D immunoblotting (1-D IB) showed two similar bands, which were identified as 77 and 75 kDa HMC (Figure 3A and Table 1). Comparative analysis of 2-DE and 2-D immunoblotting (2-D IB) profiles indicated that there were significant differences in HMC combined with different pathogenic bacteria. For example, the protein spots of HMC-Sa were mainly concentrated in the middle, while HMC-Vp and HMC-Vf were the opposite (Figure 3B). Thus, these results indicated that L. vannamei HMC diversity might be related to shrimp’s resistance to specific bacteria in vivo.
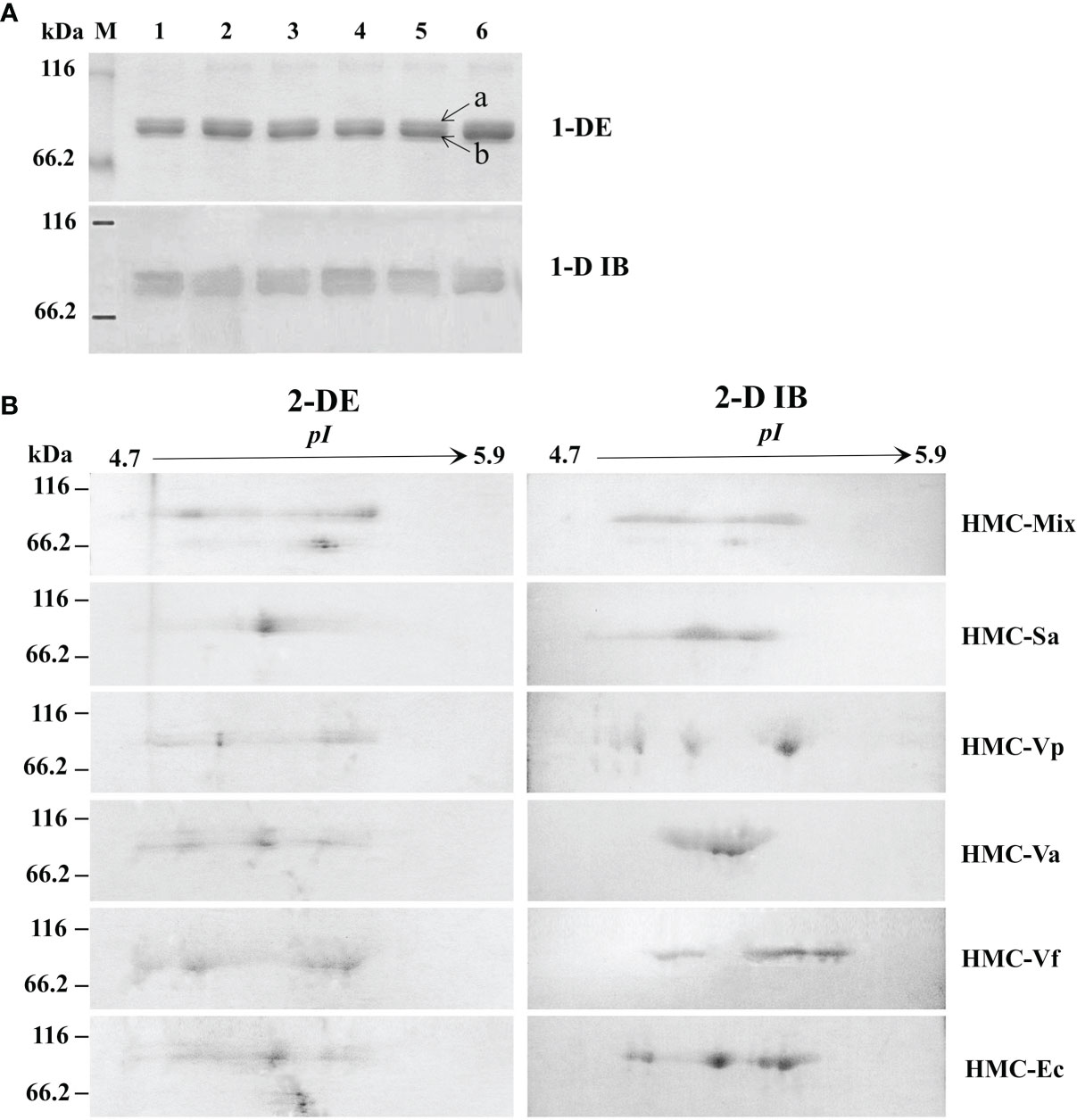
Figure 3 Proteins diversity analysis of six bacteria-binding hemocyanin fractions from L. vannamei. (A) 1-DE and 1-D immunoblotting analysis of six bacteria-binding hemocyanin fractions; 1–6, HMC-Mix, HMC-Sa, HMC-Vp, HMC-Va, HMC-Vf, and HMC-Ec, respectively. 1-D IB, 1-D immunoblotting. Bands a and b were identified by MALDI-TOF-TOF-MS. Western blotting was performed using rabbit anti-shrimp HMC antisera (1:1,000 dilution) and goat anti-rabbit IgG-HRP (1: 3,000 dilution) antibodies. (B) 2-DE and 2-D immunoblotting analysis of six bacteria-binding hemocyanin fractions, i.e., HMC-Mix, HMC-Sa, HMC-Vp, HMC-Va, HMC-Vf, and HMC-Ec. 2-D IB, 2-D immunoblotting. Western blotting was carried out as the same descriptions with (A).
Next, agglutinative and antibacterial activities were further performed to compare the immunological properties of the six HMC fractions. As shown in Table 2, agglutination could be observed when the six HMC fractions were separately bound with all of the five examined bacteria, showing agglutinative activities of 0.15–1.17 μg/ml. It is worth emphasizing that HMC-Sa, HMC-Vp, HMC-Va, HMC-Vf, and HMC-Ec possessed the strongest agglutinative activities against S. agalactiae, V. parahaemolyticus, V. alginolyticus, V. fluvialis, and E. coli K12, respectively, whose agglutinative titer was two- or fourfold as compared to the other four bacteria. Similarly, the six HMC fractions also showed a different degree of antibacterial activities against V. alginolyticus or V. fluvialis, respectively (Figure 4A). Higher antimicrobial activity was detected in HMC-Va against V. alginolyticus, and HMC-Vf and HMC-Ec against the two tested bacteria, showing inhibition of about 54.9–83.3%, but lower in HMC-Sa and HMC-Vp against V. alginolyticus. However, no activity was detected in HMC-Vp and HMC-Va to V. fluvialis and HMC-Mix against V. alginolyticus and V. fluvialis under the same conditions (Figure 4A). Similar results were found with counted colony method in Figure 4B. These results indicate that the specific bacterial resistance of HMC depends on co-effect of HMC isomers.
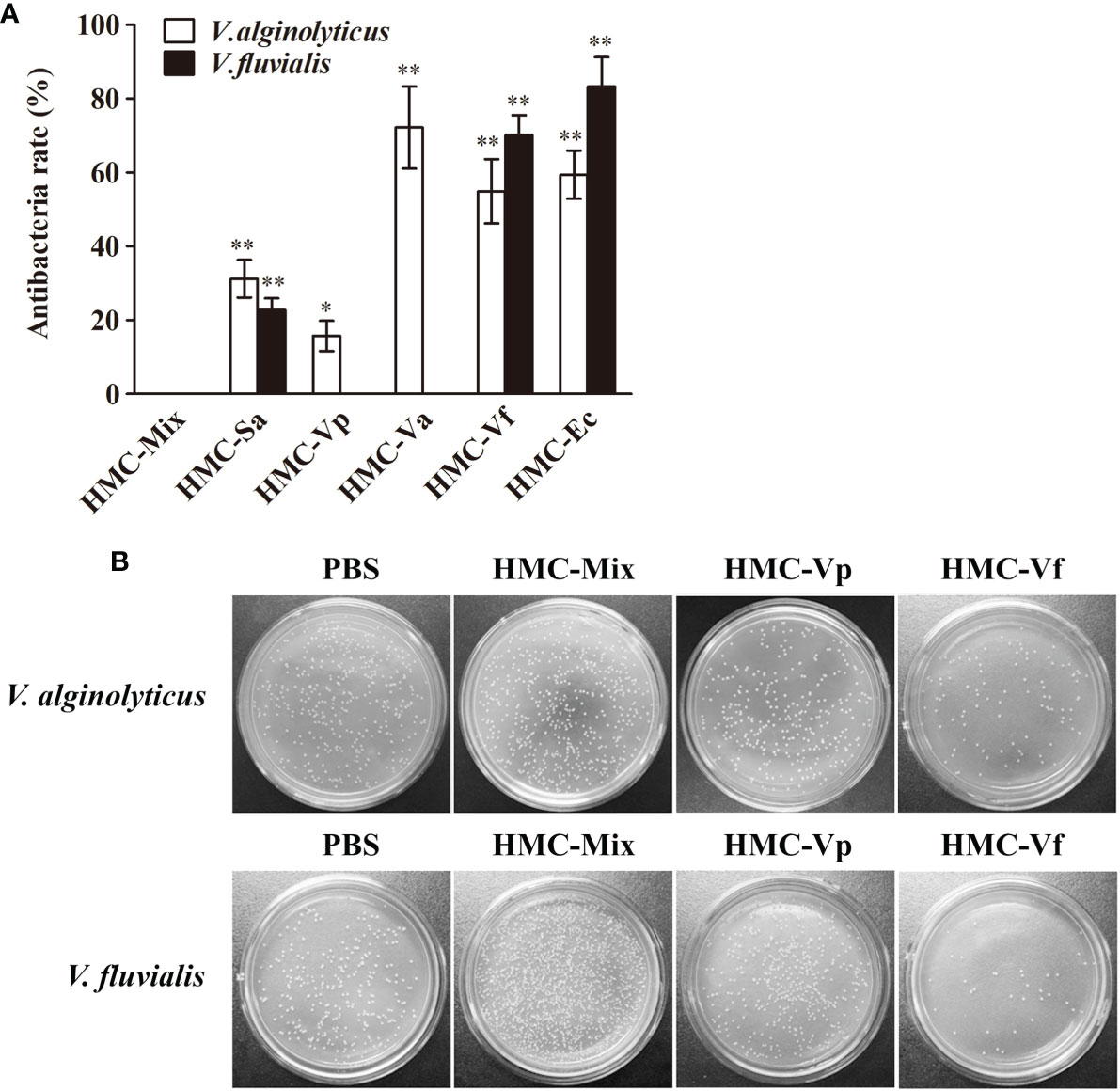
Figure 4 Antibacterial activities analysis of six bacteria-binding hemocyanin fractions from L. vannamei. (A) Comparison of antibacterial activities of 125 μg/ml HMC-Mix, HMC-Sa, HMC-Vp, HMC-Va, HMC-Vf, and HMC-Ec to V. alginolyticus and V. fluvialis, respectively. Data represent mean ± SD of at least three separate experiments. *p < 0.05, **p < 0.01. (B) Bacterial colonies of V. alginolyticus and V. fluvialis in Petri dishes treated with 50 μl HMC-Mix, HMC-Vp, HMC-Vf (125 μg/ml for each), respectively, 0.01 M pH 7.4 PBS as negative control.
3.4 Glycosylation Modification Might Be Responsible for HMC Diversity
Since glycosylation is one of the most important post-translational modifications for immune molecules to regulate their function (39), we hypothesize that glycosylation difference may be also present in HMC isomers. To confirm the hypothesis, 1-D and 2-D lectin blotting was first applied. Lectins including concanavalin A (ConA, recognizing α-D-mannose > α-D-glucose), peanut agglutinin [PNA, recognizing β-D-gal-(1,3)-D- acetyl galactosamine], ulex europaeus agglutinin 1 (UEA, recognizing α-L-fucose), and dolichos biflorus agglutinin (DBA, recognizing N-acetyl-D-galactosamine) were used to blot the type of glycans in L. vannamei HMC purified by gel-filtration chromatography and anion-exchange chromatography. As expected, two bands at molecular weights approximately 75 and 77 kDa were observed, which could react with four lectins to different degrees (Figure 5A). For 2-D immunoblotting analysis, nine spots were detected (Figure 5B), and for 2-D lectin-blotting analysis, five to nine spots of HMC could bind specifically to the four examined lectins. Specifically, PNA, UEA, and DBA can recognize nine, nine, and eight HMC spots, respectively. However, ConA only recognized five HMC spots (Figure 5B). This indicates that different spots of HMC had different glycosylation modification. Next, dot lectin blotting and total glycan measurement were further performed to characterize glycosylation diversity of the above bacteria-binding HMC fractions and HMC-Mix as a control. As shown in Figure 5C, the six HMC fractions could bind to the four lectins, but the degree of binding was different. Among these, HMC-Va, HMC-Vp, HMC-Sa, HMC-Vf, and HMC-Ec could react with four lectins (ConA, PNA, UEA, and DBA), three lectins (ConA, PNA, and UEA), two lectins (ConA and PNA), one lectin (UEA), and one lectin (ConA), respectively, whereas HMC-Mix showed the lowest reaction to the four lectins compared with the other five HMC fractions. Consistently, the total glycan content of HMC-Va and HMC-Vf exhibited very significant difference (p < 0.01) compared with that of HMC-Mix. The HMC-Va carbohydrate content was 6.38%, which was about 2.6- and 5.4-fold to HMC-Vf and HMC-Mix (Figure 5D). These results suggest that glycosylation diversity might exist in HMC and HMC fractions bound to different bacteria.
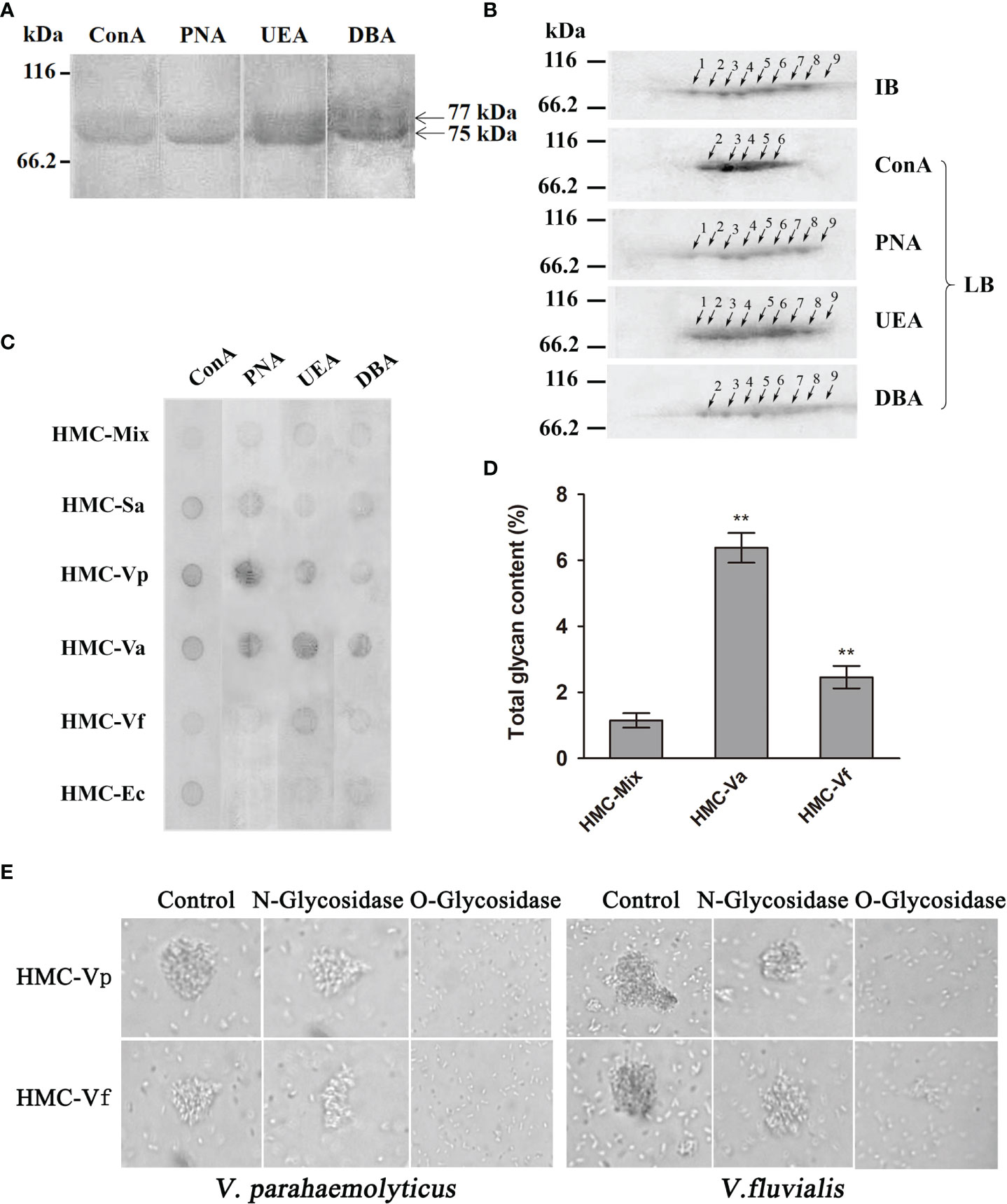
Figure 5 Analysis of glycosylation of hemocyanin from L. vannamei. (A) Lectin-blotting analysis of hemocyanin with ConA (1:1,000), PNA (1:100), UEA (1:500), and DBA (1:1000), respectively. (B) 2-D immunoblotting and 2-D lectin blotting with ConA, PNA, UEA, and DBA analysis of hemocyanin. IB, immunoblotting; LB, lectin blotting. Western blotting was performed using rabbit anti-shrimp HMC antisera (1:1,000 dilution) and goat anti-rabbit IgG-HRP (1:3,000 dilution) antibodies. Lectin-blotting analysis were carried out as the same descriptions with (A). (C) Comparative analysis of the sugar content among six bacteria-binding hemocyanin by dot lectin blotting. ConA (1:20), PNA (1:5), UEA (1:5), DBA (1:5), and 50 μg/ml of HMC-Mix, HMC-Sa, HMC-Vp, HMC-Va, HMC-Vf, and HMC-Ec were used. (D) Comparison of sugar content of 200 μl of 250 μg/ml HMC-Mix, HMC-Va, and HMC-Vf by the method of phenol-sulfate acid. (E) Comparison of agglutinative activities of 50 μl HMC-Vp and HMC-Vf (300 μg/ml) treated with O-glycosidase or N-glycosidase against V. parahaemolyticus (left) or V. fluvialis (right) (2,000×), respectively. Untreated HMC-Vp and HMC-Vf were used as a control. **p < 0.01.
To examine the relationship between immunological function and glycosylation modification of HMC, agglutination of V. parahaemolyticus and V. fluvialis was performed. We found that deglycosylated HMC fractions, dO-HMC-Vp and dO-HMC-Vf, by O-glycosidase, subsequently led to a general significant decrease in agglutinative activities to five kinds of pathogens, even completely abolished, whereas no effect was observed for those of deglycosylated HMC fractions, dN-HMC-Vp and dN-HMC-Vf, by N-glycosidase (Table 3 and Figure 5E). These results suggest that O-linked carbohydrates play a crucial role in agglutinative activities to five kinds of pathogens.
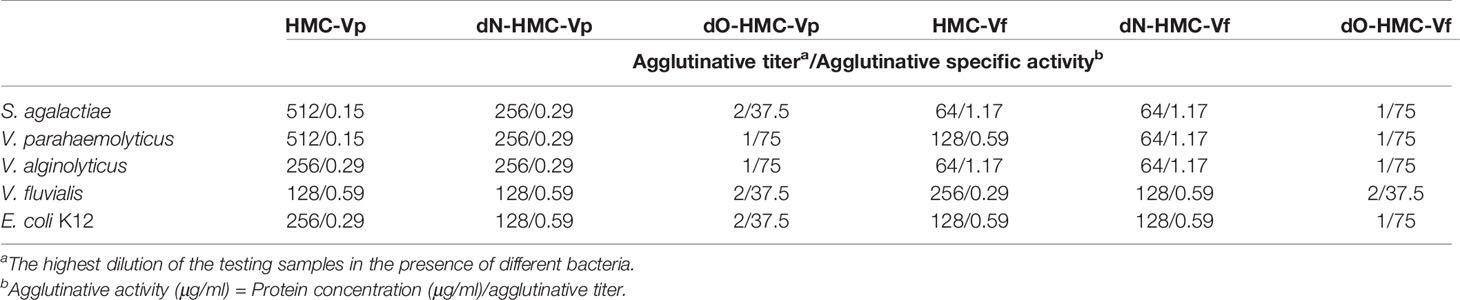
Table 3 Comparison of agglutinative activity of two bacteria-binding hemocyanin and deglycosylation hemocyanin with 75 μg/ml.
4 Discussion
Some non-specific immune molecules in invertebrates have been found to possess diversity at the protein level. Down syndrome cell adhesion molecule (Dscam) of Drosophila have the potential to express more than 18,000 isoforms of the immunoglobulin (Ig)-superfamily receptor (40). Similarly, the C-type lectin-like domain (CTLD) of C. elegans and fibrinogen-related proteins (FREPs) in the Pacific oyster Crassostrea gigas have diversity at nucleotide and protein levels (41, 42). Hemocyanin (HMC) is a large copper-containing respiratory protein found in the hemolymph of mollusks and arthropods. Recently, research revealed that HMC may be a novel and important non-specific innate immune defense molecule (2, 3). In crustaceans, HMC is thought to be composed of three distinct classes (α-type, β-type, or γ-type), although a single subunit is able to aggregate into hexameric structures (43). Within penaeid shrimp, β- and γ-type HMCs have been identified in Litopenaeus vannamei (33, 44–46). In this study, we identified at least nine protein spots as HMC small subunit (75 kDa) or large subunit (77 kDa). It is consistent with our previous research about small subunit HMC gene (47–49) and large subunit HMC gene (33, 34, 50), suggesting that HMC identified in this study belongs to γ-type and possesses significant HMC diversity at the protein level. Interestingly, other spots ranging from 20 to 60 kDa were also identified as HMC, which should be HMC fragments or modification isoforms (Figure 1). In combination with previous reports that HMC possessed polymorphisms including SNPs (30, 31) and alternative splicing variant (32) and could generate various antimicrobial peptides (7, 12, 17, 18, 51, 52) with resistance to pathogens infection, all these results indicated that HMC varied at nucleotide and protein levels.
Recently, many studies have indicated that HMC has multiple immune functions and could be involved in immune defense in invertebrate (2–26). Our previous publication displayed obvious molecular diversity including SNPs and variants at genomic and complementary DNA (cDNA) levels, which might contribute to the functional diversity of hemocyanin (30–34, 50). Some studies by other research groups also show that various hemocyanin isoforms were identified in L. vannamei (44–46). To further understand the responses of HMC isomers to pathogen challenge in vivo, the expression profile of the nine protein spots were examined; the detailed criteria are as follows: (1) the nine protein spots were detected repeatedly in different 2-DE gels; (2) the nine protein spots were identified as HMC subunit 75 or 77 kDa by MALDI-TOF-TOF-MS; (3) the molecular weights of the nine protein spot in the 2-DE gels were approximately 75 and 77 kDa, which is consistent with the two bands in 1-DE. After 6, 12, and 24 h of S. agalactiae and V. parahaemolyticus infection, spots 1–9 had different expressions (Figure 2A), indicating that HMC isoforms have different expression profile, which could be modulated by different bacteria. Similar results indicated that the expression of two hemocyanin subunits genes (PjHcL and PjHcY) from the shrimp P. japonicus could be strongly induced by WSSV infection, and PjHcL is more sensitive to WSSV infection than PjHcY (53). In black tiger shrimp P. monodon, 2D gels have identified several truncated HMC isoforms that were not only upregulated in response to bacterial infection but also showed in vitro antibacterial, antiviral, or agglutination activities (12, 51). Havanapan et al. reported that C-terminal HMC fragments were upregulated, whereas the N-terminal fragments were downregulated during Taura syndrome virus (TSV) infection in hemocytes of P. vannamei (54). This strongly suggested that the diversity of HMC is closely associated with its ability to recognize diverse pathogens.
Generally considered, the main mechanism of non-specific immune molecules recognizing pathogens was regarded as pathogen-associated molecular patterns (PAMPs), which are shared by these pathogens, representing conserved molecular patterns and absolutely essential for their physiology (29, 55). Interestingly, there is increasing evidence to support that several non-specific immune molecules can bind different pathogens and had the inhibitory property against bacteria (5, 56–59). C-type lectin, an important immune factor, might serve as LPS-specific pattern recognition receptor (PRR) to specifically recognize opportunistic bacterial and viral pathogens and thus play a role in the immune defense of aquatic shrimp via the binding and agglutination (56–58). Chitin-binding protein (CBP) from the kuruma shrimp Marsupenaeus japonicus could specifically recognize lipoteichoic acid, lipopolysaccharides, and peptidoglycans in the surface of several Gram-positive and Gram-negative bacteria, and facilitate the clearance of V. anguillarum (59). Notably, HMC directly bound to bacterial PAMPs, i.e., OmpT, OmpW, OmpX, OmpC, OmpA, and FadL, instead of intracellular and bacterial serine proteases and/or antimicrobial peptides. This binding can result in diverse biological actions including bacterial agglutination and growth inhibition, and human erythrocytes hemagglutination and hemolysis (5). However, the recognition mechanism of HMC to bacteria is not clear. In this study, immunological functions-based investigation showed that six kinds of pathogens-binding HMC fractions, namely, HMC-Sa, HMC-Vp, HMC-Va, HMC-Vf, HMC-Ec, and HMC-Mix, with diverse 2-DE profiles (Figure 3B), appeared to have different agglutinative and antibacterial activities against different bacteria. It is interesting to note that HMC-Sa showed higher agglutinative activities against Gram-positive bacteria S. agalactiae compared with Gram-negative bacteria, V. alginolyticus, and V. fluvialis. Similarly, high agglutinative and antimicrobial activities were detected in HMC-Va against V. alginolyticus, respectively (Table 2 and Figure 4). These cumulative evidence for shrimp hemocyanin suggests that various HMC fractions involved in recognition of a specific microorganism may be a novel PRRs molecule to recognize pathogenic microorganisms in invertebrate.
We previously reported that HMC had an Ig-like conserved domain and could react with goat anti-human IgG, IgM, and IgA (5, 19, 24). Furthermore, we also found that the diversity of human IgG constituents and glycosylation levels may have functional significance (5, 36, 38). Therefore, to determine whether polymorphism of HMC glycosylation is associated with its functional diversity, glycan content and glycosylation level were investigated. The results showed that lectins binding varied markedly among the experimental groups (Figure 5C), and the total glycan content of HMC-Va and HMC-Vf exhibited significant difference (p < 0.01) compared with control (Figure 5D). Interestingly, deglycosylation of HMCs by O-glycosidase led to complete abolished agglutinative activities (Table 3 and Figure 5E). In molluscan HMCs, the oligosaccharide structures of the structural subunits RvH2 from Rapana venosa hemocyanin (RvH), βc-Helix lucorum hemocyanin (βc-HlH), and Megatura crenulata keyhole limpet (KLH) reveal a complex N-glycan pattern combining typical structural features of different higher organisms; these glycosylation plays a crucial physiological role in the structural stability, immunostimulatory, and therapeutic effect of HMC (60, 61). Our previous research also found that L. vannamei HMC was deglycosylated using O-glycosidase; its agglutinative activity reduced about four- to eightfold (8, 38). The results from this study indicated that binding of different pathogens with HMC fractions showed diverse optimum immunological activities, and the differences in their immunological activities may be related to their glycosylation diversity. The remarkable diversity of hemocyanin glycan content is an important feature of their immune function and provides a new concept in pathogen–host interaction and is involved in a large number of biological recognition events in the host immune system. However, besides glycosylation diversity of HMC, other forms of diversity in protein level, such as degradation fragments and other post-translational modification of HMC, need further research.
In summary, the present study showed that HMC of L. vannamei possessed protein diversity. Furthermore, the polymorphism appeared to be closely associated with HMC’s recognition and resistance to diverse pathogens and interpretation of HMC with multiple immune activities. These results will contribute to an understanding of HMC’s diversity and its multifunctional mechanisms. Our findings may also be helpful to enrich and develop the knowledge of invertebrate’s immune system and immune prevention.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author Contributions
XZ, JA, and YLZ wrote the manuscript. XZ, JQ, and YLZ conceptualized and designed the project. XZ, JA, XC, and YZZ performed data analysis. XZ, JQ, PZ, and ZZ performed experiments and collected samples. All authors contributed to the article and approved the submitted version.
Funding
This work was sponsored by the National Natural Science Foundation of China (Nos. 31072237, 31872596, and 31502204), 2020 Li Ka Shing Foundation Cross-Disciplinary Research Grant (No. 2020LKSFG01E), and Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (No. GML2019ZD0606).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Decker H, Rimke T. Tarantula Hemocyanin Shows Phenoloxidase Activity. J Biol Chem (1998) 273:25889–92. doi: 10.1074/jbc.273.40.25889
2. Coates CJ, Decker H. Immunological Properties of Oxygen-Transport Proteins: Hemoglobin, Hemocyanin and Hemerythrin. Cell Mol Life Sci (2017) 74:293–317. doi: 10.1007/s00018-016-2326-7
3. Aweya JJ, Zheng ZH, Zheng XY, Yao DF, Zhang YL. The Expanding Repertoire of Immune-Related Molecules With Antimicrobial Activity in Penaeid Shrimps: A Review. Rev Aquacult (2021) 13:1907–1937. doi: 10.1111/raq.12551
4. Nagai T, Osaki T, Kawabata SI. Functional Conversion of Hemocyanin to Phenoloxidase by Horseshoe Crab Antimicrobial Peptides. J Biol Chem (2001) 276:27166–70. doi: 10.1074/jbc.M102596200
5. Zhang YL, Peng B, Li H, Yan F, Wu HK, Zhao XL, et al. C-Terminal Domain of Hemocyanin, a Major Antimicrobial Protein From Litopenaeus Vannamei: Structural Homology With Immunoglobulins and Molecular Diversity. Front Immunol (2017) 8:611. doi: 10.3389/fimmu.2017.00611
6. Jiang N, Tan NS, Ho B, Ding JL. Respiratory Protein-Generated Reactive Oxygen Species as an Antimicrobial Strategy. Nat Immunol (2007) 8:1114–22. doi: 10.1038/ni1501
7. Yang S, Huang H, Aweya JJ, Zheng ZH, Liu GM, Zhang YL. PvHS9 Is a Novel In Silico Predicted Antimicrobial Peptide Derived From Hemocyanin of Penaeus Vannamei. Aquaculture (2021) 530:735926. doi: 10.1016/j.aquaculture.2020.735926
8. Zhang ZH, Wang F, Chen CD, Zheng Z, Aweya JJ, Zhang YL. Glycosylation of Hemocyanin in Litopenaeus Vannamei Is an Antibacterial Response Feature. Immunol Lett (2017) 192:42–7. doi: 10.1016/j.imlet.2017.10.008
9. Zhang X, Huang C, Qin Q. Antiviral Properties of Hemocyanin Isolated From Shrimp Penaeus Monodon. Antiviral Res (2004) 61:93–9. doi: 10.1016/j.antiviral.2003.08.019
10. Bao SY, Zheng ZH, Aweya JJ, Yao DF, Li SK, Sun CH, et al. microRNA-589-5p Modulates the Expression of Hemocyanin as Part of the Anti-WSSV Immune Response in Litopenaeus Vannamei. Dev Comp Immunol (2020) 107:103642. doi: 10.1016/j.dci.2020.103642
11. Zagorodnya SD, Dolashka P, Baranova GV, Golovan AV, Nesterova NV. Anti-EBV Activity of Hemocyanin Isolated From Helix Lucorum. Antiviral Res (2011) 90:A66. doi: 10.1016/j.antiviral.2011.03.134
12. Zhan SX, Aweya JJ, Wang F, Yao DF, Zhong MQ, Chen JH, et al. Litopenaeus Vannamei Attenuates White Spot Syndrome Virus Replication by Specific Antiviral Peptides Generated From Hemocyanin. Dev Comp Immunol (2019) 91:50–61. doi: 10.1016/j.dci.2018.10.005
13. Stoyanova E, Mihaylova N, Manoylov I, Bradyanova S, Raynova Y, Idakieva K, et al. Intensive Therapy With Gastropodan Hemocyanins Increases Their Antitumor Properties in Murine Model of Colon Carcinoma. Int Immunopharmacol (2020) 84:106566. doi: 10.1016/j.intimp.2020.106566
14. Palacios M, Tampe R, Del Campo M, Zhong TY, López MN, Salazar-Onfray F, et al. Antitumor Activity and Carrier Properties of Novel Hemocyanins Coupled to a Mimotope of GD2 Ganglioside. Eur J Med Chem (2018) 150:74–86. doi: 10.1016/j.ejmech.2018.02.082
15. Zheng LY, Zhao XL, Zhang P, Chen CD, Liu SJ, Huang RQ, et al. Hemocyanin From Shrimp Litopenaeus Vannamei has Antiproliferative Effect Against HeLa Cell In Vitro. PloS One (2016) 11:e0151801. doi: 10.1371/journal.pone.0151801
16. Liu SJ, Zheng LY, Aweya JJ, Zheng Z, Zhong MQ, Chen JH, et al. Litopenaeus Vannamei Hemocyanin Exhibits Antitumor Activity in S180 Mouse Model In Vivo. PloS One (2017) 12:e0183783. doi: 10.1371/journal.pone.0183783
17. Liu SJ, Aweya JJ, Zheng LY, Wang F, Zheng Z, Zhong MQ, et al. ALitopenaeus Vannamei Hemocyanin-Derived Antimicrobial Peptide (Peptide B11) Attenuates Cancer Cells' Proliferation. Molecules (2018) 23:3202. doi: 10.3390/molecules23123202
18. Monteiro ML, Lima DB, Menezes RRPPB, Sampaio TL, Silva BP, Serra Nunes JV, et al. Antichagasic Effect of Hemocyanin Derived From Antimicrobial Peptides of Penaeus Monodon Shrimp. Exp Parasitol (2020) 215:107930. doi: 10.1016/j.exppara.2020.107930
19. Zhang YL, Wang SY, Xu AL, Chen J, Lin BK, Peng XX. Affinity Proteomic Approach for Identification of an IgA-Like Protein in Litopenaeus Vannamei and Study on Its Agglutination Characterization. J Proteome Res (2006) 5:815–21. doi: 10.1021/pr0503984
20. Yan F, Zhang YL, Jiang RP, Zhong MQ, Hu Z, Du H, et al. Identification and Agglutination Properties of Hemocyanin From the Mud Crab (Scylla Serrata). Fish Shellfish Immunol (2011) 30:354–60. doi: 10.1016/j.fsi.2010.11.004
21. Zhang YL, Yan F, Hu Z, Zhao XL, Min SY, Du ZH, et al. Hemocyanin From Shrimp Litopenaeus Vannamei Shows Hemolytic Activity. Fish Shellfish Immunol (2009) 27:330–5. doi: 10.1016/j.fsi.2009.05.017
22. Xu J, Wu S, Zhang X. Novel Function of QM Protein of Shrimp (Penaeus Japonicus) in Regulation of Phenol Oxidase Activity by Interaction With Hemocyanin. Cell Physiol Biochem (2008) 21:473–80. doi: 10.1159/000129640
23. Yao DF, Wang ZH, Wei MH, Zhao XL, Aweya JJ, Zhong MQ, et al. Analysis of Litopenaeus Vannamei Hemocyanin Interacting Proteins Reveals Its Role in Hemolymph Clotting. J Proteomics (2019) 201:57–64. doi: 10.1016/j.jprot.2019.04.013
24. Zhang YL, Wang SY, Peng XX. Identification of a Type of Human IgG-Like Protein in Shrimp Penaeus Vannamei by Mass Spectrometry. J Exp Mar Biol Ecol (2004) 301:39–54. doi: 10.1016/j.jembe.2003.09.011
25. Qiao J, Du ZH, Zhang YL, Du H, Guo LL, Zhong MQ, et al. Proteomic Identification of the Related Immune-Enhancing Proteins in Shrimp Litopenaeus Vannamei Stimulated With Vitamin C and Chinese Herbs. Fish Shellfish Immunol (2011) 31:736–45. doi: 10.1016/j.fsi.2011.07.005
26. Cao JS, Wang ZH, Zhang YL, Qu FL, Guo LL, Zhong MQ, et al. Identification and Characterization of the Related Immune-Enhancing Proteins in Crab Scylla Paramamosain Stimulated With Rhubarb Polysaccharides. Mol Immunol (2014) 57:263–73. doi: 10.1016/j.molimm.2013.10.003
27. Netea MG, Schlitzer A, Placek K, Joosten LAB, Schultze JL. Innate and Adaptive Immune Memory: An Evolutionary Continuum in the Host's Response to Pathogens. Cell Host Microbe (2019) 25:13–26. doi: 10.1016/j.chom.2018.12.006
28. Pees B, Yang W, Zárate-Potes A, Schulenburg H, Dierking K. High Innate Immune Specificity Through Diversified C-Type Lectin-Like Domain Proteins in Invertebrates. J Innate Immunol (2016) 8:129–42. doi: 10.1159/000441475
29. Tetreau G, Pinaud S, Portet A, Galinier R, Gourbal B, Duval D. Specific Pathogen Recognition by Multiple Innate Immune Sensors in an Invertebrate. Front Immunol (2017) 8:1249. doi: 10.3389/fimmu.2017.01249
30. Zhao XL, Guo LL, Zhang YL, Liu Y, Zhang XY, Lun JS, et al. SNPs of Hemocyanin C-Terminal Fragment in Shrimp Litopenaeus Vannamei. FEBS Lett (2012) 586:403–10. doi: 10.1016/j.febslet.2011.12.038
31. Guo LL, Zhao XL, Zhang YL, Wang ZH, Zhong MQ, Li SK, et al. Evidences of SNPs in the Variable Region of Hemocyanin Ig-Like Domain in Shrimp Litopenaeus Vannamei. Fish Shellfish Immunol (2013) 35:1532–8. doi: 10.1016/j.fsi.2013.08.024
32. Zhao S, Lu X, Zhang YL, Zhao XL, Zhong MQ, Li SK, et al. Identification of a Novel Alternative Splicing Variant of Hemocyanin From Shrimp Litopenaeus Vannamei. Immunol Lett (2013) 154:1–6. doi: 10.1016/j.imlet.2013.08.003
33. Xu JX, Ruan LW, Li Z, Yu XM, Li SD, Shi H, et al. Characterization of Four Hemocyanin Isoforms in Litopenaeus Vannamei. Acta Oceanol Sin (2015) 34:36–44. doi: 10.1007/s13131-015-0588-9
34. Zhao XL, Guo LL, Lu X, Lu H, Wang F, Zhong MQ, et al. Evidences of Abundant Hemocyanin Variants in Shrimp Litopenaeus Vannamei. Mol Immunol (2016) 77:103–12. doi: 10.1016/j.molimm.2016.07.017
35. Fan JH, Li XM, Lu H, Lin RH, Aweya JJ, Zhang YL. N-Terminal Diversity of Litopenaeus Vannamei Hemocyanin and Immunity. Mol Immunol (2019) 112:360–8. doi: 10.1016/j.molimm.2019.06.019
36. Min SY, Yan F, Zhang YL, Ye XQ, Zhong MQ, Cao JS, et al. Characterization of a Novel Hemolytic Activity of Human IgG Fractions Arising From Diversity in Protein and Oligosaccharide Components. PloS One (2014) 9:e85711. doi: 10.1371/journal.pone.0085711
37. Liu X, She XT, Zhu QF, Li H, Peng XX. Heterogeneous Interactome Between Litopenaeus Vannamei Plasma Proteins and Vibrio Parahaemolyticus Outer Membrane Proteins. Fish Shellfish Immunol (2013) 34:192–8. doi: 10.1016/j.fsi.2012.10.023
38. Zhang ZH, Li RW, Aweya JJ, Wang F, Zhong MQ, Zhang YL. Identification and Characterization of Glycosylation Sites on Litopenaeus Vannamei Hemocyanin. FEBS Lett (2019) 593:820–30. doi: 10.1002/1873-3468.13367
39. Macedo-da-Silva J, Santiago VF, Rosa-Fernandes L, Marinho CRF, Palmisano G. Protein Glycosylation in Extracellular Vesicles: Structural Characterization and Biological Functions. Mol Immunol (2021) 135:226–46. doi: 10.1016/j.molimm.2021.04.017
40. Watson FL, Püttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, et al. Extensive Diversity of Ig-Superfamily Proteins in the Immune System of Insects. Science (2005) 309:1874–8. doi: 10.1126/science.1116887
41. Pees B, Yang W, Kloock A, Petersen C, Peters L, Fan L, et al. Effector and Regulator: Diverse Functions of C. Elegans C-Type Lectin-Like Domain Proteins. PloS Pathog (2021) 17:e1009454. doi: 10.1371/journal.ppat.1009454
42. Huang B, Zhang L, Li L, Tang X, Zhang G. Highly Diverse Fibrinogen-Related Proteins in the Pacific Oyster Crassostrea Gigas. Fish Shellfish Immunol (2015) 43:485–90. doi: 10.1016/j.fsi.2015.01.021
43. Stoeva S, Idakieva K, Georgieva DN, Voelter W, Genov N. Penaeus Monodon (Tiger Shrimp) Hemocyanin: Subunit Composition and Thermostability. Z Naturforsch C (2001) 56:416–22. doi: 10.1515/znc-2001-5-616
44. Johnson JG, Burnett LE, Burnett KG. Uncovering Hemocyanin Subunit Heterogeneity in Penaeid Shrimp Using RNA-Seq. Integr Comp Biol (2016) 56:1080–91. doi: 10.1093/icb/icw088
45. Wang JS, Janech MG, Burnett KG. Protein-Level Evidence of Novel Beta-Type Hemocyanin and Heterogeneous Subunit Usage in the Pacific Whiteleg Shrimp, Litopenaeus Vannamei. Front Mar Sci (2019) 6:687. doi: 10.3389/fmars.2019.00687
46. Mendoza-Porras O, Kamath S, Harris JO, Colgrave ML, Huerlimann R, Lopata AL, et al. Resolving Hemocyanin Isoform Complexity in Haemolymph of Black Tiger Shrimp Penaeus Monodon - Implications in Aquaculture, Medicine and Food Safety. J Proteomics (2020) 218:103689. doi: 10.1016/j.jprot.2020.103689
47. Yang PK, Aweya JJ, Yao DF, Wang F, Lun JS, Hong YJ, et al. The Kruppel-Like Factor of Penaeus Vannamei Negatively Regulates Transcription of the Small Subunit Hemocyanin Gene as Part of Shrimp Immune Response. Fish Shellfish Immunol (2020) 100:397–406. doi: 10.1016/j.fsi.2020.03.035
48. Yang PK, Yao DF, Aweya JJ, Wang F, Ning P, Li SK, et al. C-Jun Regulates the Promoter of Small Subunit Hemocyanin Gene of Litopenaeus Vannamei. Fish Shellfish Immunol (2019) 84:639–47. doi: 10.1016/j.fsi.2018.10.054
49. Zhang ZX, Yao DF, Yang PK, Zheng ZH, Aweya JJ, Lun JS, et al. Nuclear Receptor E75 Is a Transcription Suppressor of the Litopenaeus Vannamei Small Subunit Hemocyanin Gene. Dev Comp Immunol (2020) 107:103662. doi: 10.1016/j.dci.2020.103662
50. Lu X, Lu H, Guo L, Zhang Z, Zhao X, Zhong M, et al. Cloning and Characterization of a Novel Hemocyanin Variant LvHMCV4 From Shrimp Litopenaeus Vannamei. Fish Shellfish Immunol (2015) 46:398–405. doi: 10.1016/j.fsi.2015.06.022
51. Wen Y, Zhan SX, Huang H, Zhong MQ, Chen JH, You CH, et al. Identification and Characterization of an 18.4kda Antimicrobial Truncation From Shrimp Litopenaeus Vannamei Hemocyanin Upon Vibrio Parahaemolyticus Infection. Fish Shellfish Immunol (2016) 56:450–8. doi: 10.1016/j.fsi.2016.08.002
52. Yang S, Huang H, Wang F, Aweya JJ, Zheng ZH, Zhang YL. Prediction and Characterization of a Novel Hemocyanin-Derived Antimicrobial Peptide From Shrimp Litopenaeus Vannamei. Amino Acids (2018) 50:995–1005. doi: 10.1007/s00726-018-2575-x
53. Lei KY, Li F, Zhang MC, Yang HJ, Luo T, Xu X. Difference Between Hemocyanin Subunits From Shrimp Penaeus Japonicus in Anti-WSSV Defense. Dev Comp Immunol (2008) 32:808–13. doi: 10.1016/j.dci.2007.11.010
54. Havanapan PO, Kanlaya R, Bourchookarn A, Krittanai C, Thongboonkerd V. C-Terminal Hemocyanin From Hemocytes of Penaeus Vannamei Interacts With ERK1/2 and Undergoes Serine Phosphorylation. J Proteome Res (2009) 8:2476–83. doi: 10.1021/pr801067e
55. Amarante-Mendes GP, Adjemian S, Branco LM, Zanetti LC, Weinlich R, Bortoluci KR. Pattern Recognition Receptors and the Host Cell Death Molecular Machinery. Front Immunol (2018) 9:2379. doi: 10.3389/fimmu.2018.02379
56. Wongpanya R, Sengprasert P, Amparyup P, Tassanakajon A. A Novel C-Type Lectin in the Black Tiger Shrimp Penaeus Monodon Functions as a Pattern Recognition Receptor by Binding and Causing Bacterial Agglutination. Fish Shellfish Immunol (2017) 60:103–13. doi: 10.1016/j.fsi.2016.11.042
57. Alenton RR, Koiwai K, Miyaguchi K, Kondo H, Hirono I. Pathogen Recognition of a Novel C-Type Lectin From Marsupenaeus Japonicus Reveals the Divergent Sugar-Binding Specificity of QAP Motif. Sci Rep (2017) 7:45818. doi: 10.1038/srep45818
58. Wang XW, Vasta GR, Wang JX. The Functional Relevance of Shrimp C-Type Lectins in Host-Pathogen Interactions. Dev Comp Immunol (2020) 109:103708. doi: 10.1016/j.dci.2020.103708
59. Xu S, Jing M, Kong DM, Wang YR, Zhou Q, Liu WY, et al. Chitin Binding Protein From the Kuruma Shrimp Marsupenaeus Japonicus Facilitates the Clearance of Vibrio Anguillarum. Dev Comp Immunol (2021) 117:103981. doi: 10.1016/j.dci.2020.103981
60. Salazar ML, Jimenez JM, Villar J, Rivera M, Baez M, Manubens A, et al. N-Glycosylation of Mollusk Hemocyanins Contributes to Their Structural Stability and Immunomodulatory Properties in Mammals. J Biol Chem (2019) 294:19546–64. doi: 10.1074/jbc.RA119.009525
Keywords: Litopenaseus vannamei, hemocyanin, proteomics approach, diversity, pathogen recognition, glycosylation
Citation: Zhao X, Qiao J, Zhang P, Zhang Z, Aweya JJ, Chen X, Zhao Y and Zhang Y (2021) Protein Diversity and Immune Specificity of Hemocyanin From Shrimp Litopenaeus vannamei. Front. Immunol. 12:772091. doi: 10.3389/fimmu.2021.772091
Received: 07 September 2021; Accepted: 15 November 2021;
Published: 07 December 2021.
Edited by:
Jin-Xing Wang, Shandong University (Qingdao), ChinaCopyright © 2021 Zhao, Qiao, Zhang, Zhang, Aweya, Chen, Zhao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yueling Zhang, emhhbmd5bEBzdHUuZWR1LmNu
†These authors have contributed equally to this work
 Xianliang Zhao
Xianliang Zhao Jie Qiao1†
Jie Qiao1† Jude Juventus Aweya
Jude Juventus Aweya Yueling Zhang
Yueling Zhang