- Molecular and Cellular Immunoregulation Section, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, United States
CD4 T helper (Th) cell subsets, including Th1, Th2 and Th17 cells, and their innate counterparts innate lymphoid cell (ILC) subsets consisting of ILC1s, ILC2s and ILC3s, display similar effector cytokine-producing capabilities during pro-inflammatory immune responses. These lymphoid cell subsets utilize the same set of lineage-determining transcription factors (LDTFs) for their differentiation, development and functions. The distinct ontogeny and developmental niches between Th cells and ILCs indicate that they may adopt different external signals for the induction of LDTF during lineage commitment. Increasing evidence demonstrates that many conserved cis-regulatory elements at the gene loci of LDTFs are often preferentially utilized for the induction of LDTF expression during Th cell differentiation and ILC development at different stages. In this review, we discuss the functions of lineage-related cis-regulatory elements in inducing T-bet, GATA3 or RORγt expression based on the genetic evidence provided in recent publications. We also review and compare the upstream signals involved in LDTF induction in Th cells and ILCs both in vitro and in vivo. Finally, we discuss the possible mechanisms and physiological importance of regulating LDTF dynamic expression during ILC development and activation.
Introduction
Innate lymphoid cells (ILCs) and adaptive CD4 T helper (Th) cells provide protection coordinately against pathogens and regulate tissue homeostasis, regeneration, and morphogenesis. Conventional effector Th cells are classified into multiple lineages, including interferon-gamma (IFN-γ)-producing type 1 Th (Th1) cells, interleukin-4 (IL-4)-, IL-5- and IL-13-producing Th2 cells, IL-17-producing Th17 cells, IL-21-producing follicular T helper (Tfh) cells, IL-9-producing Th9 cells, and IL-22-producing Th22 cells (1, 2). Lineage-determining transcription factors (LDTFs) T-bet (T-box expressed in T cells), GATA3 (GATA binding protein 3) and RORγt (retinoic acid-related orphan receptor gamma t) are critical for Th1, Th2 and Th17 cell differentiation and lineage maintenance, respectively (3–9). IFN-γ-producing type 1 ILCs (ILC1s) and natural killer (NK) cells belong to group 1 ILCs. ILC1s require T-bet for their development, whereas NK cells require both Eomes (Eomesodermin) and T-bet for their development and maturation (10, 11). IL-5- and IL-13-producing ILC2s depend on high levels of GATA3 for the lineage determination and development (12–14). RORγt is indispensable for the development of IL-17- and IL-22-producing ILC3s (15, 16).
Although ILC and Th cell subsets share some characteristics, they also exhibit different properties, including differences in ontologies, location, specificity and epigenetic modifications. Th cell differentiation occurs upon T cell receptor (TCR) engagement in pro-inflammatory cytokine environment. On the other hand, ILCs are pre-developed under homeostatic conditions and IL-7 signaling is required for their homeostasis. In addition, ILCs are not antigen-specific and thus develop independently of DNA recombination mediated by recombination-activating genes (RAGs) (17). Furthermore, ILCs are largely tissue-resident and rarely detected in peripheral secondary lymphoid organs or vascular system in steady state. Interestingly, most of the circulating NK cells do not express IL-7 receptor (IL-7R) but express inhibitory receptors, activating receptors and costimulatory receptors, which allow them to have certain levels of antigen-specific response in some conditions (18, 19). Genome-wide analyses of methylation and chromatin accessibility clearly show different epigenetic modification patterns between Th cell and ILC subsets (20–25). Although some cytokines that are involved in the induction of LDTF expression during Th cell differentiation can also upregulate LDTF expression in mature ILCs during their activation, these signals are usually dispensable for LDTF induction during ILC development (21, 26–28). Therefore, it is reasonable to assume that ILCs and Th cells could utilize different mechanisms for the initial induction of LDTF during their development and lineage commitment.
Lineage-specific landscapes at the LDTF gene loci
To which extent the mechanisms for LDTF induction during Th cell differentiation and ILC development are mirrored or specified has not been well studied in the past. Epigenetic modifications are highly correlated with gene expression. For example, high levels of demethylation at the Tbx21 and Gata3 gene locus are detected in NK/ILC1s and ILC2s, respectively (20). Gene accessibility at the Tbx21, Gata3 and Rorc loci in NK cells, ILC2s and ILC3s is also associated with their demethylation status (24). Recently, several studies have highlighted the lineage-specific landscapes at LDTF gene loci in Th cell and ILC subsets (21, 22, 29). In these studies, through CRISPR/Cas9-mediated genetic modification mouse models and in vivo disease models, genetic evidence has been provided to demonstrate the lineage-specific requirement of individual cis-regulatory element in response to external stimuli, including cytokines and cell-cell interactions, for the induction of LDTF in Th cells and ILCs.
Cis-regulatory elements at the Tbx21 locus in Th1 cells, NK cells and ILC1s
T-bet, a member of the T-box family of transcription factors, is encoded by the Tbx21 gene and plays critical roles in Th1 cell lineage determination (5–7), ILC1 development and NK cell maturation (10, 11). Through DNase I hypersensitive site sequencing (DHS-Seq) and comparison of chromatin accessibility at the Tbx21 gene locus in different ILC and Th cell subsets, several Th1-, NK- and ILC1-related DHSs have been identified (21) (Figure 1). While the chromatin accessibility at the promoter of Tbx21 is comparable in Th1 cells, NK cells and ILC1s, the conserved non-coding sequence (CNS) 12 kb upstream of the Tbx21 transcriptional start site (Tbx21-CNS-12) is highly accessible in Th1 cells, NK cells and ILC1s and contains STAT (signal transducer and activator of transcription) binding motifs. Tbx21-CNS-12 is also accessible in naïve CD4 T cells indicating the readiness of this element to respond to cytokines even at the naïve cell stage. Tbx21-CNS-12 is essential for T-bet induction during Th1 cell differentiation both in vitro and in vivo, but it is dispensable for the T-bet induction during NK cell and ILC1 development. By contrast, the element Tbx21-CNS-3 is an NK-specific DHS with partial accessibility in ILC1s, but it is inaccessible in all Th cell subsets, ILC2s and ILC3s (21, 30). Accordingly, Tbx21-CNS-3 is critical for optimal T-bet expression during NK cell development and thus for their maturation, but it has a minimal if any effect on T-bet expression in ILC1s and Th1 cells. Interestingly, while Tbx21-CNS-8.5 is also an NK- and ILC1-specific DHS, this element is dispensable for T-bet expression neither in NK cells nor in ILC1s. Furthermore, the protein levels of T-bet in ILC1s from the Tbx21-CNS-3 and Tbx21-CNS-8.5 double knockout mice are still relatively normal indicating that a functional cis-regulatory element used by ILC1s for T-bet induction during their development is yet to be discovered, if it exists.
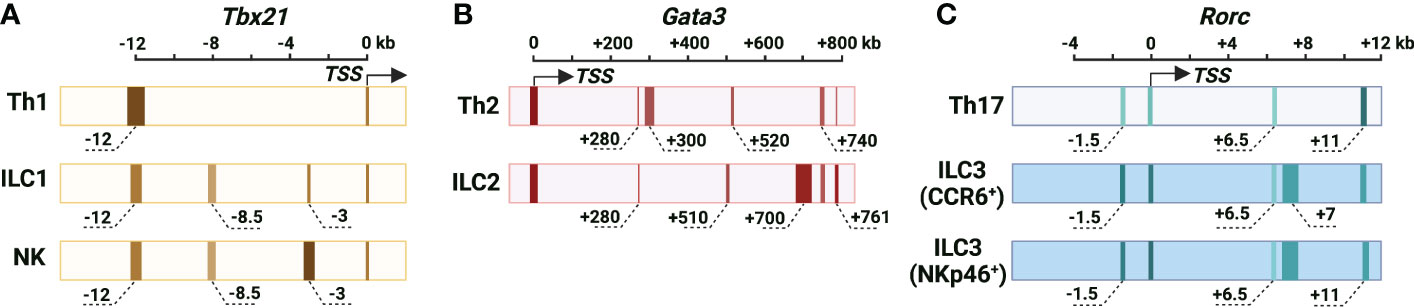
Figure 1 Cis-regulatory elements at the murine Tbx21, Gata3 and Rorc gene loci. (A) The chromatin accessibility of the Tbx21 gene locus in Th1 cells, NK cells and ILC1s. Tbx21 promoter region is partially accessible in Th1 cells, NK cells and ILC1s. Tbx21-CNS-12 is the major highly accessible site at the Tbx21 gene locus in Th1 cells, and it contains STAT binding motifs and is essential for IL-12/STAT4- and IFN-γ/STAT1-mediated T-bet expression during Th1 cell differentiation. Tbx21-CNS-12 is also a responsive element for T-bet expression during NK cell and ILC1 activation, but it is dispensable for T-bet induction during NK cell and ILC1 development. Tbx21-CNS-3 is an NK-specific DHS, and it is critical for T-bet induction during NK cell development. Tbx21-CNS-8.5 is an NK cell- and ILC1-specific DHS, but this region is dispensable for T-bet induction in NK cells and ILC1s. (B) The chromatin accessibility of the Gata3 gene locus in Th2 cells and ILC2s. Long-range elements and promoter region are critical for the Gata3 transcriptional activity. Gata3-CNS+280 is highly accessible and important for GATA3 expression during thymic T cell and ILCP development, but it is only partially accessible and dispensable for the high levels of GATA3 expression in Th2 cells and ILC2s. The chromatin accessibility around Gata3-CNS+700 increases as cells developing from ILCPs into ILC2Ps and ILC2s. The accessibility of Gata3-CNS+700 is much higher in ILC2s than in Th2 cells, and this region is important for GATA3 expression in ILC2s and has a minimal effect on GATA3 expression in Th2 cells. Gata3-CNS+761/762 has the Gata3 enhancer activity in Th2 cells and ILC2s. Gata3-CNS+740 is a Th2- and ILC2-specific DHS. Gata3-CNS+300 and Gata3-CNS+520 are Th2-specific DHSs. Gata3-CNS+510 is an ILC2-specific DHS. (C) Chromatin accessibility of the Rorc gene locus in Th17 cells, CCR6+ ILCs and NKp46+ ILC3s. Overall chromatin accessibility at the Rorc gene locus in ILC3s is much higher than that in Th17 cells. Rorc-CNS-1.5 and Rorc-CNS+6 are important for epigenetic modifications of the whole Rorc gene locus associated with active chromatin and the induction of RORγt during Th17 cell differentiation. These two elements are accessible in CCR6+ ILC3s and NKp46+ ILC3s, but they have limited effect on ILC3 population in steady state. Whether they are responsive elements for RORγt expression upon ILC3 activation is unknown. Rorc-CNS-11 is highly accessible in Th17 cells and ILC3s, and it is important for RORγt maintenance in Th17 cells. Rorc-CNS-7 is an ILC3-specific element. The color from light to dark corresponds to chromatin accessibility from low to high. The width of elements stands for the width of peaks according to DHS-Seq or ATAC-Seq results. The approximate distance of the middle of peaks from TSS is labeled, and the “-” is for upstream and “+” is for downstream of TSS.
T-bet induction in Th1 cells, NK cells and ILC1s
Cytokines IL-12, IL-18, IL-21, IL-27 and IFN-γ are reported to be involved in T-bet expression during Th1 cell differentiation both in vitro and in vivo, however, it needs to be revisited whether the transcription factors activated by those cytokines directly induce T-bet expression or other secondary transcription factors that are induced by cytokines, in turn, regulate T-bet expression during Th1 cell differentiation. In addition, whether those cytokines are also involved in T-bet induction during NK cell and ILC1 development requires further investigation and discussion (Figure 2). IL-12 mediates the phosphorylation of STAT4 resulting in its binding to Tbx21-CNS-12 in T cells and NK cells to upregulate T-bet expression (21, 26, 31–34). Abolishing STAT binding motifs at Tbx21-CNS-12 or deleting STAT4 renders cells cultured under Th1 differentiation conditions incapable of expressing T-bet and effector cytokine IFN-γ (35, 36). However, mutating STAT binding motifs at Tbx21-CNS-12 has no effect on T-bet expression in NK cells and ILC1s (21). Indeed, IL-12/STAT4 signaling is dispensable for T-bet induction during NK cell and ILC1 development in steady state; neither T-bet protein nor Tbx21 mRNA are altered in Stat4-deficient NK cells and ILC1s, and neutralizing IL-12 doesn’t affect T-bet expression during NK cell development (21, 26).
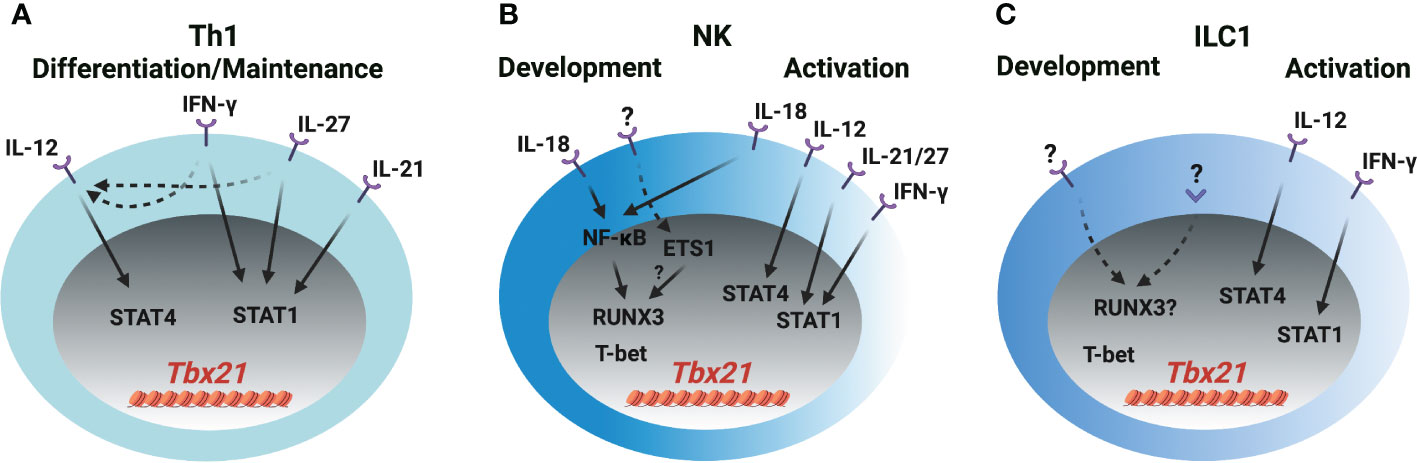
Figure 2 T-bet induction in Th1 cells, NK cells and ILC1s (A) IL-12/STAT4, IFN-γ/STAT1, IL-21/STAT1 and IL-27/STAT1 signaling induce T-bet expression through Tbx21-CNS-12 during Th1 cell differentiation and maintenance. IFN-γ and IL-27 also upregulate IL-12R expression in T cells. (B) IL-18/NF-κB signaling induces RUNX3 expression and plays a role in the initial T-bet induction through Tbx21-CNS-3 during NK cell development. ETS1 is critical for the T-bet expression in NKPs, pro-NK cells and mature NK cells. Whether ETS1 directly regulates T-bet induction or indirectly regulates T-bet induction through RUNX3 is not clear. T-bet may self-regulate its expression during NK cell development. IL-18/NF-κB/RUNX3 regulates T-bet expression through Tbx21-CNS-3, and IL-12/STAT4, IFN-γ/STAT1, IL-21/STAT1 and IL-27/STAT1 regulate T-bet expression through Tbx21-CNS-12 during NK cell activation. (C) The mechanism of T-bet induction during ILC1 development is not clear. IL-12/STAT4 and IFN-γ/STAT1 may regulate T-bet expression during ILC1 activation. Only a small proportion of ILC1s express IL-18R.
Th1 cells, NK cells and ILC1s are professional IFN-γ-producing cells. The IFN-γ/STAT1 pathway is a potent inducer for Tbx21 mRNA expression mainly through Tbx21-CNS-12, but such capability may be masked by the IL-12/STAT4 signaling as addition of exogenous IFN-γ in culture is unable to further upregulate T-bet expression at late stage of Th1 cell differentiation (21, 36–38). Nevertheless, Tbx21-CNS-12 is readily accessible in naïve CD4 T cells, and IFN-γ can quickly induce Tbx21 mRNA and subsequently Il12rb2 mRNA in those cells (21, 37). Altogether, IFN-γ would be a critical player for the induction of T-bet during Th1 cell differentiation both in vitro and in vivo. Since Tbx21-CNS-12 is highly accessible in NK cells and ILC1s, which also highly express IFN-γR (21, 39), it is plausible to anticipate an important role of IFN-γ in Tbx21 mRNA induction during NK cell and ILC1 development and/or activation. T-bet expression is substantially reduced in the NK cells from the Stat1-deficient mice, but it is likely due to altered environmental niche rather than a cell intrinsic defect (40). Furthermore, the expression of T-bet by Ifng-deficient or Tbx21-CNS-12-mutant NK/ILC1s is intact (21, 26). Therefore, the IFN-γ/STAT1 pathway is dispensable for the initial induction of T-bet during NK cell and ILC1 development, although it may be involved in dynamic regulation of T-bet expression in activated NK cells and ILC1s.
IL-27 activates downstream STAT1 and STAT3 (41, 42), and quickly induces T-bet expression in the human T cell line (Jurkat), NK cell line (NKL) and naïve T cells (41). CD4 T cells incubated with IL-27 for more than twenty-four hours in vitro express high levels of T-bet (42–45). STAT1 is important for the optimal T-bet induction in the IL-27 supplemented Th1 polarization conditions, but IL-27 can still further upregulate T-bet expression in the absence of STAT1 (42). This may be due to the induction of IL-12Rβ2 and IFN-γ (41–44, 46). Since mature NK cells express the ligand-specific component of the IL-27R, WSX-1 (47), and Tbx21-CNS-12 is accessible in NK cells and ILC1s, IL-27 may serve as an upstream signal for T-bet induction in these cells. Whether IL-27 signaling is involved in T-bet induction during early NK cell and ILC1 development needs further investigation.
IL-21 stimulation induces STAT1 binding to Tbx21-CNS-12 and upregulates T-bet expression in CD4 T cells, but IL-21-mediated STAT3 activation represses T-bet upregulation (48). IL-21 is still able to upregulate T-bet expression in the Tbx21-CNS-12 mutant mature NK cells, however, IL-21R is undetectable in the NK progenitors (NKPs) (21). A recent study shows that the effect of IL-18 on T-bet expression in T cells is minimal, and IL-18 alone doesn’t upregulate T-bet expression in CD4 T cells (21). However, IL-18 is capable of inducing IFN-γ expression in Th1 cells (21, 49) indicating that it may regulate T-bet expression through IFN-γ signaling pathway in CD4 T cells. On the other hand, IL-18 can upregulate T-bet expression in NK cells in a Tbx21-CNS-12 independent manner. Furthermore, IL-18R is expressed in NK progenitors as early as at the refined NKP (rNKP) stage, and lower amounts of T-bet protein are detected in the Il18r1-deficient NK progenitors compared to their wild type counterparts (21). Thus, IL-18 signaling is involved in T-bet expression during both early NK cell development and late activation. However, IL-18 signaling pathway doesn’t induce Tbx21 expression directly, instead RUNX3 induced by IL-18/NF-κB pathway promotes T-bet expression through Tbx21-CNS-3 (21).
Anti-CD3 treatment quickly induces Tbx21 mRNA in Th1 cell clones (5). Tbx21 can be induced in the CD8-depleted Stat4-deficient lymph node cells under IL-12 and IL-18 neutralized T cell activation conditions after two days (50). However, TCR signaling alone is not able to induce T-bet expression in the Tbx21-CNS-12 mutant CD4 T cells under Th1 conditions (21). Thus, the effect of auto-secreted IFN-γ on T-bet expression in the in vitro T cell activation system needs to be considered. Nevertheless, the T-bet expression in the Th1 cells from the early stage of Influenza- and Salmonella-infected mice is independent of IL-12, IFN-γ or IL-18 signaling (51), suggesting that the redundancy of these signaling pathways or other signals (e.g. TCR) may contribute to the initial T-bet induction.
Over-expression of RUNX3 promotes IFN-γ production, but RUNX3 is unable to upregulate T-bet expression in wild type Th1, Tbx21-CNS-12 mutant “Th1”, or Stat4 and Ifngr1 double knockout “Th1” cells (21). Therefore, RUNX3 may not directly induce T-bet expression in CD4 T cells. T-bet expressing-ILC1s, NK cells and NKp46+ ILC3s are reduced in the NKp46-Cre- and PLZF-Cre-mediated Cbfb-deficient mice (52, 53). Yet, the protein levels of T-bet in the residual ILC1s, NK cells and NKp46+ ILC3s are relatively normal in the NKp46-Cre-mediated Cbfb-deficient cells (53). It is noticed that the expression of RUNX3 is earlier than T-bet induction during NK cell development (21, 53). According to the above observations, the functions of RUNXs/CBF-β on T-bet expression during innate cell development and maintenance require further investigation. ETS1 functions at early stages of NK cell development presumably through its binding to the Tbx21 gene locus, and decreased levels of Tbx21 mRNA is observed in the Ets1-deficient NKPs, pro-NK cells and mature NK (mNK) cells (54). Therefore, ETS1 may work as an important regulator for T-bet expression during NK cell development. However, ETS1 expression is reduced in the activated NK cells and NK cell lines compared with unstimulated mNK cells (54). In addition, ETS1 promotes RUNX3 expression in DP thymocytes (55), but whether ETS1 also regulates RUNX3 expression during ILC development is not known. Therefore, it will be interesting to further explore the upstream signals for the induction of RUNX family proteins and ETS1 during ILC development.
T-bet only binds to Tbx21-CNS-12 in Th1 cells (21, 56), however, it binds to multiple elements, including Tbx21-CNS-3, Tbx21-CNS-8.5 and Tbx21-CNS-12, in NK cells (21). Ectopic expression of T-bet promotes the endogenous expression of Tbx21 under Th2 polarization conditions (50), but this autoactivation of T-bet may mainly be due to the secreted IFN-γ as such an effect is abrogated in the Stat1-deficient cells (38). Furthermore, the self-regulation of T-bet expression is only observed in the NK cells, but not in the Th1 cells by using the bacterial artificial chromosome (BAC)-transgenic T-bet reporter system (21, 57). Therefore, the binding of T-bet to Tbx21-CNS-12 is not involved in regulating Tbx21 expression in Th1 cells. Similarly, T-bet binds to Tbx21-CNS-8.5 in NK cells, nevertheless, this element is dispensable for T-bet expression in NK cells at steady state (21). Finally, although NK cells and ILC1s are classified as two different lineages, it is difficult to define early ILC1 and NK progenitors, and thus it remains challenging to compare the initial mechanism of T-bet induction in ILC1 or NK progenitors.
Cis-regulatory elements at the Gata3 locus in Th2 cells and ILC2s
GATA3, encoded by gene Gata3, is required for the early T cell and ILC development, and the high levels of GATA3 are essential for Th2 cell differentiation and ILC2 lineage determination and development (3, 4, 12–14, 58–61). Different from the fact that the transcriptional activity of Tbx21 and Rorc gene is regulated by the cis-regulatory elements near their promoter region, the expression of Gata3 is largely controlled by the long-range elements in T cells and ILCs (22, 62, 63) (Figure 1). An element located at 280 kb downstream of the Gata3 transcriptional start site (Gata3-CNS+280) is important for the GATA3 expression in thymic T cells and peripheral CD4 T cells by using BAC-transgenic strategy and CRISPR/Cas9-mediated knockout approach (22, 62, 63). Gata3-CNS+280 deficiency also has a pan effect on ILC progenitors (ILCPs) and ILC subsets (22). However, this element seems dispensable for the high levels of GATA3 expression in Th2 cells and ILC2s (22, 62, 63). It is consistent with the observation that chromatin accessibility of Gata3-CNS+280 is reduced in the mature Th2 cells and ILC2s comparing to naïve CD4 T cells and ILCPs by using assay for transposase-accessible chromatin using sequencing (ATAC-Seq) and DHS-Seq (21, 22). Together, Gata3-CNS+280 is critical for the GATA3 expression at early stage of T cell and ILCP development, but it is not required for the high levels of GATA3 induction during Th2 cell differentiation and ILC2 lineage determination.
No ILC2- vs. Th2-specific chromatin accessible site at the large region around Gata3-CNS+700 is found from ATAC-Seq data (22, 30). However, DHS-Seq result shows that the accessibility at Gata3-CNS+700 region is much higher in ILC2s than that in Th2 cells and naïve CD4 T cells (21). Consistently, the deficiency of Gata3-CNS+700 and nearby region results in a severer defect in GATA3 expression in ILC2s than in Th2 cells (22). The chromatin accessibility of Gata3-CNS+700 region is increased as cells developing from ILCPs into ILC2Ps and then ILC2s, but this region remains closed in the other ILC subsets. The functions of ILC2s, but not of Group 1 ILCs and ILC3s, are greatly disturbed in the absence of Gata3-CNS+700 and nearby region, indicating that this region has no effect on ILCP development (22). Gata3-CNS+761/762 is considered as a type 2-specific enhancer through H11 Gata3-CNS+761/762 EGPF reporter assay (22), but this site is only partially accessible in Th2 cells (21).
There are two Th2-specific DHSs located around Gata3-CNS+300 and Gata3-CNS+520, one ILC2-specific DHS located around Gata3-CNS+510, and one Th2- and ILC2-specific DHS located around Gata3-CNS+740 comparing to naïve CD4 T cells, other Th cell and ILC subsets (21). So far, the functions of those elements on GATA3 induction and Th2 cell differentiation and ILC2 development are still unknown.
GATA3 induction in Th2 cells and ILC2s
IL-4 activates downstream STAT6 and vigorously induces GATA3 expression through distal and proximal promoters during naïve CD4 T cells differentiating into Th2 cells (64–67) (Figure 3). The binding of STAT6 to the responsive Gata3 promoters is essential for the induction of GATA3 in T cells. STAT6 is critical for the reduction of repressive H3K27me3 modification and the increase of active H3K4me3 modification at the promoter region to induce Gata3 expression (68, 69). IL-4 can upregulate GATA3 expression in the T cells which are not previously exposed to IL-4, but it fails to further upregulate GATA3 in already differentiated Th2 cells (66). Interestingly, Stat6-deficient IL-4-producing Th2 cells express intermediate levels of GATA3 (70). Therefore, an IL-4/STAT6-independent pathway may play a role in GATA3 induction in CD4 T cells. Although STAT5A and STAT5B can bind to the Gata3 promoter, IL-2/STAT5 signaling likely works through IL-4 production to regulate GATA3 expression (71–74). The ILC2 population is intact in Il4/Il13-deficient mice (75), which indicates that IL-4 and IL-13 signaling is dispensable for the high levels of GATA3 expression in ILC2s. Similarly, GATA3 expression is also normal in the Stat6-deficient ILC2s (76), and IL-4/STAT6 only has a subtle effect on the activated GATA3hi ILC2s from the parasite infected mice (77).
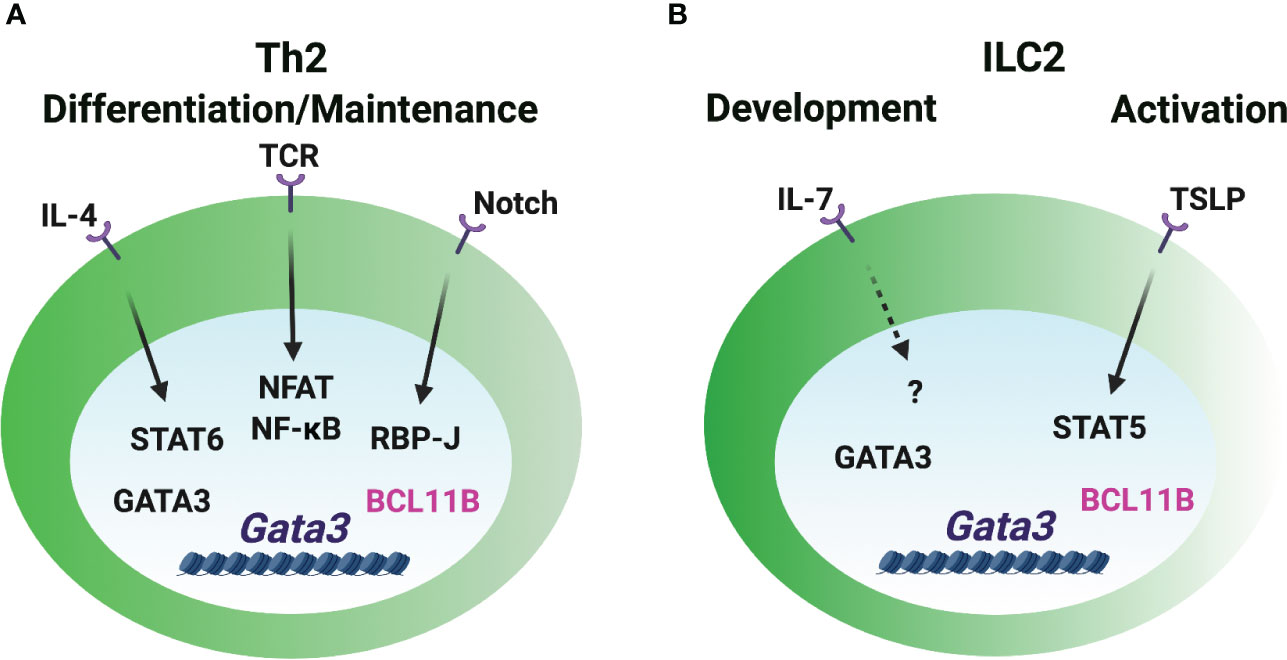
Figure 3 GATA3 induction in Th2 cells and ILC2s. (A) The Gata3 promoters are critical for the GATA3 induction during Th2 cell differentiation. IL-4/STAT6 directly and vigorously induces GATA3 expression. Notch signal regulates Gata3 promoter activity in CD4 T cells. TCR/NFAT/NF-κB signal is involved in GATA3 induction. GATA3 may self-regulate its expression in Th2 cells under certain circumstances. BCL11B works as a repressive regulator of GATA3 expression in already differentiated Th2 cells. (B) The booster of GATA3 induction during ILC2 lineage determination is unclear. IL-7 signaling may play a role in GATA3 expression in ILC2s. The release of repressive factor BCL11B may be important for the high levels of GATA3 expression. TSLP/STAT5 regulates GATA3 expression in the mature ILC2s, but it is dispensable for GATA3hi ILC2 development.
Certain strength of TCR signal induces early GATA3 expression (72). NFAT1, a TCR-inducible TF, binds to the Gata3 promoters and several CNSs close to promoters, and it regulates GATA3 expression in a calcineurin-dependent manner (65). Furthermore, NF-κB plays a critical role in GATA3 induction during Th2 cell differentiation (78). RBP-J, a Notch effector molecule, binds to and induces the activity of the Gata3 promoter in CD4 T cells (79). IL-2 receptor common γ chain (γc)-cytokine-mediated signaling seems to be dispensable for GATA3 expression in early ILC progenitors and their development (59), but IL-7 signal may be partially involved in the GATA3 expression during ILC2 development and activation in vivo (80). TSLP (thymic stromal lymphopoietin) activates STAT5 phosphorylation and induces GATA3 expression in the short-term cultured ILC2s (13). However, TSLP-TSLPR signal is dispensable for GATA3hi ILC2 development and maintenance as it doesn’t affect GATA3 levels in ILC2s in steady state (14, 80).
Besides the importance of transcriptional activity of the Gata3 promoters, long-distant cis-regulatory elements are also regulated by multiple TFs to induce GATA3 expression. Transcription factors TCF-1, HEB and CSL/RBP-J bind to Gata3-CNS+280. Transcription factors GATA3, CBF-β, RUNX1, RUNX3, GFI1 and BCL11B bind to Gata3-CNS+761/762 in Th2 cells and ILC2s (22, 52, 69, 81–84), but they do not bind to Gata3-CNS+280. Whether those TFs are involved in GATA3 induction in Th2 cells and ILC2s is unclear. Repressive regulator, such as BCL11B, may play an essential role in controlling the levels of GATA3 during ILC2 and Th2 cell lineage determination. Deleting BCL11B upregulates GATA3 expression in ILC2s and Th2 cells in vitro (81, 85). Deletion of ETS1 results in a reduction of ILC2s in the bone marrow and mesenteric lymph nodes, but GATA3 expression is normal in the residual Ets1-deficient ILC2s (86). Since ETS1 is critical for the TF ID2 expression in the common helper-liker innate lymphoid progenitors (CHILPs) and ILC2s (86), ETS1 may not directly regulate GATA3 expression during ILC2 development. Ectopic expression of GATA3 promotes the endogenous Gata3 induction under Th1- and Th2-polarization conditions in the absence of IL-4/STAT6 signaling (70). GATA3 binds to multiple Gata3 cis-regulatory elements in Th2 cells and ILC2s (22, 81). Gata3-CNS+761/762 bound by GATA3 in both Th2 cells and ILC2s exhibits Gata3 enhancer activity (22). Th2-specific DHSs around Gata3-CNS+300 are strongly bound by GATA3 (21, 81), yet, the activity of this element in Th2 cells has not been assessed. Taken together, the initial expression of GATA3 may be induced by TCR signaling in T cells and IL-7R signaling in ILCs through the promoter regions, and intracellular and extracellular signals triggered by these stimuli can further upregulate GATA3 expression. It is also possible that the release of repressive signal plays an important role in modulating the levels of GATA3, and GATA3 self-regulation may play a role in the maintenance of GATA3 levels under certain circumstances.
Cis-regulatory elements at the Rorc locus in Th17 cells and ILC3s
The truncated form of the nuclear hormone receptor RORγ (RORγt), encoded by gene Rorc, is critical for Th17 cell differentiation and ILC3 lineage determination (8, 9, 15, 16). Strikingly, the overall chromatin accessibility at the Rorc gene locus in ILC3s is much higher than that in Th17 cells, and there are multiple DHSs at the Rorc gene locus in ILC3s (21) (Figure 1). A recent study shows that two cis-regulatory elements, Rorc-CNS-1.5 (so called “CNS6” in the paper) and Rorc-CNS+6.5 (so called “CNS9” in the paper) are important for RORγt induction during Th17 cell differentiation both in vitro and in vivo (29). Those two elements synergistically affect the optimal expression of RORγt in Th17 cells. Surprisingly, these elements do not function as enhancers, since neither Rorc-CNS-1.5 nor Rorc-CNS+6.5 promotes Rorc promoter activity in a dual luciferase reporter assay. Instead, they inhibit Rorc promoter activity. Nevertheless, they may play a critical role in the formation of DNA loops that facilitates enhancer-promoter interactions. Rorc-CNS-1.5 and Rorc-CNS+6.5 are important for active epigenetic modifications, such as H3Ac, H3K27Ac, H3K4me3 and RNA polymerase II (RNA Pol II) binding, at the whole Rorc gene locus in Th17 cells (29). These observations indicate that Rorc-CNS-1.5 and Rorc-CNS+6.5 are critical for the permissive structure of the Rorc gene locus in Th17 cells. Rorc-CNS-11 is also highly accessible in Th17 cells and important for the maintenance of RORγt expression in Th17 cells (21, 29, 87).
Interestingly, although Rorc-CNS-1.5 and Rorc-CNS+6.5 are highly accessible in ILC3s, deleting Rorc-CNS-1.5 or Rorc-CNS+6.5 has a limited effect on ILC3 and thymic double-positive T cell population in steady state (21, 29). However, it is possible that these two elements will be utilized to regulate RORγt expression during ILC3 activation and thus their plasticity. While Rorc-CNS-11 is accessible in both Th17 cells and ILC3s, Rorc-CNS-7 is an ILC3-specific element (21). The functions of Rorc-CNS-7 and Rorc-CNS-11 on RORγt induction during ILC3 lineage determination require further investigation.
RORγt induction in Th17 cells and ILC3s
It is well accepted that cytokines IL-6 and TGF-β are essential for the initial induction of Rorc during Th17 cell differentiation (8, 9) (Figure 4). IL-6 stimulation induces the binding of STAT3 to Rorc-CNS-1.5, Rorc-CNS+6.5 and Rorc promoter and changes the chromatin structure of the Rorc gene locus (23, 29, 88). Rorc-CNS+6.5 is more important than Rorc-CNS-1.5 in regulating chromatin changes of the Rorc gene locus and RORγt expression upon IL-6 engagement in CD4 T cells (29). The function of TGF-β on Th17 cell differentiation depends on other pro-inflammatory cytokines and the dose of TGF-β (89, 90). Rorc-CNS-1.5 works as the main responsive element for TGF-β-induced RORγt expression and chromatin changes. TGF-β downstream transcription factors SMAD2/3, SMAD4 and c-MAF bind to Rorc-CNS-1.5, but not Rorc-CNS+6.5. Interestingly, the presence of Rorc-CNS+6.5 is essential for the binding of TFs to Rorc-CNS-1.5, and Rorc-CNS-1.5 affects STAT3 binding to Rorc-CNS+6.5 (29). The synergistic effects of cis-regulatory elements Rorc-CNS-1.5 and Rorc-CNS+6.5 optimize the IL-6- and TGF-β-mediated RORγt expression and Th17 cell function.
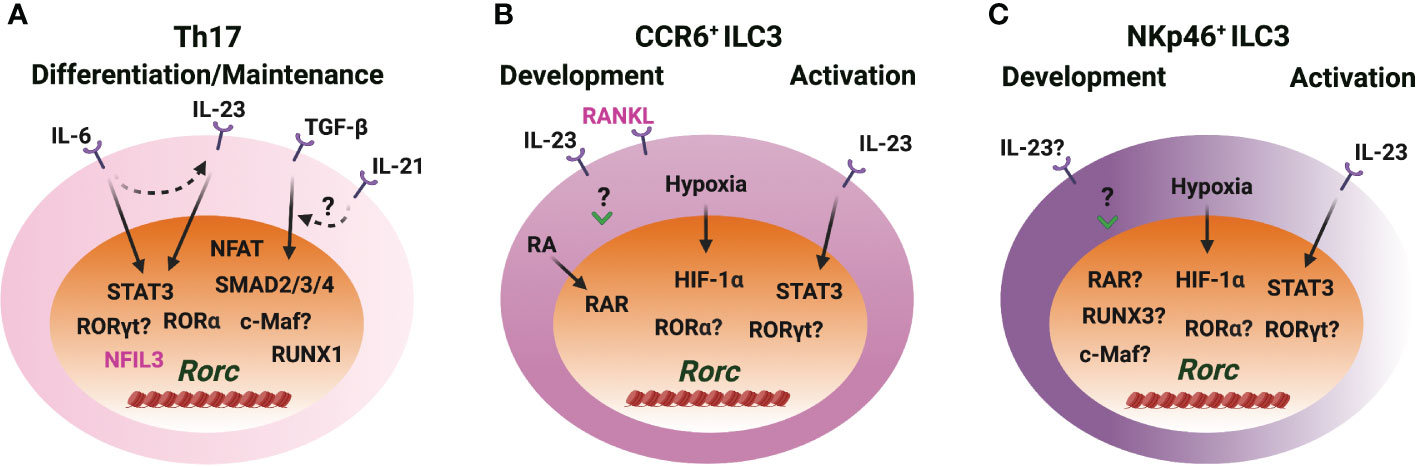
Figure 4 RORγt induction in Th17 cells and ILC3s. (A) IL-6/STAT3 and TGF-β/SMAD signaling induce RORγt expression through Rorc-CNS-1.5 and Rorc-CNS+6 during Th17 cell differentiation. IL-23/STAT3 plays a role in RORγt expression when IL-23R is upregulated upon naïve CD4 T cell activation. IL-21 synergizes with TGF-β to upregulate RORγt. RORα and RORγt bind to Rorc-CNS-11 and play important roles in maintaining RORγt levels in Th17 cells. NFIL3 represses RORγt expression in CD4 T cells. (B, C) IL-23 upregulates RORγt expression during CCR6+ ILC3 development in vitro, but STAT3 is dispensable for RORγt expression during ILC3 development in vivo. IL-23/STAT3 signaling regulates RORγt expression in mature ILC3s. Hypoxia induced HIF-1α regulates RORγt expression in ILC3s. RA receptor RAR binds to the Rorc gene locus and regulates RORγt expression during CCR6+ ILC3 development and ILC3 activation. RUNX3 is critical for the RORγt expression in NKp46+ ILC3s. The role of RORα and RORγt in regulating RORγt expression in ILC3s is unclear. c-Maf benefits RORγt levels, but whether this is a direct effect is unknown. RANKL represses RORγt expression in CCR6+ ILC3s.
IL-21 can synergize with TGF-β, but not with IL-6, to further upregulate RORγt expression during naïve CD4 T cell activation (91, 92). STAT3 directly binds to the Il21 promoter and is critical for the production of IL-21 by Th17 cells (92). Thus, IL-21 works as an autocrine factor to promote RORγt expression and Th17 cell differentiation through STAT3 (91–94). Moreover, IL-6-induced IL-21 is independent of RORγt; IL-6 and IL-21 induce IL-23R expression in an RORγt-dependent manner; IL-23 upregulates Rorc expression in the IL-23R enforced-expressing CD4 T cells; TGF-β represses IL-6- and IL-21-mediated IL-23R upregulation (91). Altogether, the network and the feedback loops formed by IL-6, TGF-β, IL-21 and IL-23 signaling pathways control RORγt expression and Th17 cell differentiation and function.
Increasing evidence indicates that Th17 cells and ILC3s may also utilize distinct mechanisms for RORγt expression. Indeed, STAT3 is dispensable for RORγt expression during ILC3 development (95), although IL-23 stimulation in vitro significantly upregulates Rorc expression in the CD45+Lin− leukocytes from the embryonic intestine on day E18.5 (96). Whether IL-6 or TGF-β plays a role in controlling high levels of RORγt upon ILC3 lineage activation is unknown. RUNX3, rather than RUNX1, is critical for RORγt expression in ILC3s (52, 53). Retrovirus-mediated over expression of RUNX3 fails to upregulate Rorc expression in Th17 cells (97). Over-expression of RUNX1 promotes Rorc expression and knockdown of RUNX1 reduces Rorc expression in Th17 cells (98). Luciferase activity assay also shows that RUNX1 promotes Rorc promoter activity while T-bet represses RUNX1-mediated Rorc promoter activity in Th17 cells (99). These studies indicate that RUNX1 is a positive regulator of Rorc during Th17 cell differentiation.
It is shown that c-Maf plays an important role in the balance between NKp46+ and CCR6+ ILC3s, and c-Maf harnesses T-bet expression and has a moderate effect on RORγt expression in NKp46-CCR6- ILC3s (100, 101). c-Maf binds to Rorc-CNS-1.5 in Th17 cells and in NKp46+ ILC3-like cell line MNK3 (29, 101). NFIL3, an important TF for early ILC development (102–105), directly binds to the Rorc promoter and negatively regulates RORγt expression in CD4 T cells (106). NFAT binds to the Rorc promoter and promotes RORγt expression in the activated CD4 T cells (107). RORγt expression is upregulated in ILC3s exposed to hypoxia ex vivo three hours in the presence of IL-1β and IL-23 and in MNK3 cells exposed to hypoxia without additional cytokines (108). Hypoxia induces HIF-1α binding to the Rorc gene locus in ILC3s (108). During Th17 cell differentiation, HIF-1α is upregulated in a STAT3-dependent manner, and HIF-1α binds to and activates Rorc luciferase reporter (109). Retinoic acid (RA), a metabolite of vitamin A, upregulates RORγt expression. RA receptors RAR and RXR can bind to the Rorc gene locus in CCR6+ ILC3s (110). Tumor necrosis factor (TNF) superfamily member receptor activator of nuclear factor κB ligand (RANKL) represses RORγt expression in CCR6+ ILC3s, but it has no effect in NKp46+ ILC3s and Th17 cells (111). RORγt and RORα bind to Rorc-CNS-11 and maintain RORγt expression in Th17 cells (21, 29, 87). So far, many cues show that cytokines, physiological factors and stress are involved in RORγt expression during Th17 cell differentiation and ILC3 development.
Conclusion and perspectives
Our previous understanding of LDTF induction during Th cell differentiation and ILC development is mainly based on the observations from in vitro studies. On one hand, it provides direct evidence to reveal the mechanisms of LDTF expression. On the other hand, regulatory network, feedback and space-time of multiple signals in vivo may be concealed. Seminal works on the differential requirements of Th cell subset- and ILC subset-specific cis-regulatory elements for LDTF induction in Th cells and ILCs provide a perspective to investigate the LDTF induction both in vitro and in vivo, as well as the cell development and functions (21, 22, 29).
Signals for LDTF induction during ILC development are less described in the past decade. Since ILCs that are not antigen-specific and may develop under sterile conditions, physiological factors or environmental stress may be involved in LDTF induction during ILC development. Endogenous factors, including mitochondrial reactive oxygen species (mROS), hypoxia, metabolites, products of lipid metabolism and complement, regulate sterile NLRP3 inflammasome priming and IL-1β and IL-18 production (112). IL-18 significantly induces RUNX3 expression, and RUNX3 plays multiple roles in controlling the development of several ILC subsets (21, 53). In addition, the factors mentioned above may be directly involved in LDTF expression through their downstream transcription factors. For example, hypoxia induces HIF-1α binding to the Rorc gene locus to upregulate RORγt expression in an ILC3 cell line (108). Notch signaling, a conserved developmental pathway, is also involved in ILC subset determination (113).
The induction of LDTF in vivo may differ from that in vitro depending on different properties of microbes and non-microbial substances. Although LDTFs antagonize each other (114), co-expression of two LDTFs are found in T cells and ILCs in vivo. T-bet-expressing pathogenic Th17 cells (115, 116), T-bet-, GATA3- or RORγt-expressing regulatory T (Treg) cells (89, 90, 117–119), and T-bet+RORγt+ ILC3s have been reported (120–122). Interestingly, during Treg cell development in the thymus (tTreg) and in the periphery (pTreg) as well as their maintenance in different tissues, distinct CNS elements are sequentially utilized for the induction and/or maintenance of Treg LDTF forkhead box P3 (Foxp3) expression (123–127). Therefore, it is likely that different CNS elements at the other LDTF loci may be preferentially used for fine-tuning the expression of these genes at different stages of T cell/ILC differentiation/development and activation. Furthermore, some cytokines, such as IFN-γ and IL-4, function as both LDTF drivers and the Th effector cytokines, which can result in a positive feedback loop for LDTF expression. In this review we focus on the positive regulators of LDTF expression, and it is possible that the release of repressive signals also plays critical roles in controlling LDTF expression. Therefore, the composition, concentration and time-space of cytokine environment together define the induction of LDTFs and cell differentiation/development.
Identifying Th cell subset- or ILC subset-specific cis-regulatory elements, evaluating the function of the elements on LDTF expression, finding TF binding motifs in the elements, and predicting upstream signals would be important strategies to reveal the mechanisms of LDTF induction during Th cell differentiation/maintenance and ILC development/activation. It should be noticed that some CNS cis-regulatory elements may code long noncoding RNAs (lncRNAs), which may regulate LDTF expression and cell development. Similarly, it will be important to analyze CNS elements at effector cytokine loci to determine cell type-specific regulation of these critical genes in T cell and ILC subsets. Finally, dissecting the developmental difference between ILCs and Th cells would help understand the space-time necessaries of innate ILCs and adaptive Th cells during immune responses, find new lineage-specific non-coding targets to engineer lymphocyte function, and discover novel interventions in treating immune diseases.
Author contributions
DF prepared the first draft. AH provided critical comments and edited the manuscript. DF and JZ finalized the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
This work is supported by the Division of Intramural Research of the NIAID (grant 1ZIA-AI-001169).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. O'Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science (2010) 327:1098–102. doi: 10.1126/science.1178334
2. Sun B. Advances in experimental medicine and biology. Dordrecht: Springer Netherlands : Imprint: Springer (2014). p. 1.
3. Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: Essential functions before and after T-helper-2-cell differentiation. Nat Rev Immunol (2009) 9:125–35. doi: 10.1038/nri2476
4. Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature (1996) 384:474–8. doi: 10.1038/384474a0
5. Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell (2000) 100:655–69. doi: 10.1016/S0092-8674(00)80702-3
6. Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science (2002) 295:338–42. doi: 10.1126/science.1065543
7. Finotto S, Neurath MF, Glickman JN, Qin S, Lehr HA, Green FH, et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science (2002) 295:336–8. doi: 10.1126/science.1065544
8. Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell (2006) 126:1121–33. doi: 10.1016/j.cell.2006.07.035
9. Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, et al. T Helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity (2008) 28:29–39. doi: 10.1016/j.immuni.2007.11.016
10. Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, et al. The transcription factors T-bet and eomes control key checkpoints of natural killer cell maturation. Immunity (2012) 36:55–67. doi: 10.1016/j.immuni.2011.11.016
11. Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife (2014) 3:e01659. doi: 10.7554/eLife.01659
12. Klein Wolterink RG, Serafini N, van Nimwegen M, Vosshenrich CA, de Bruijn MJ, Fonseca Pereira D, et al. Essential, dose-dependent role for the transcription factor Gata3 in the development of IL-5+ and IL-13+ type 2 innate lymphoid cells. Proc Natl Acad Sci USA (2013) 110:10240–5. doi: 10.1073/pnas.1217158110
13. Mjosberg J, Bernink J, Golebski K, Karrich JJ, Peters CP, Blom B, et al. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity (2012) 37:649–59. doi: 10.1016/j.immuni.2012.08.015
14. Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity (2012) 37:634–48. doi: 10.1016/j.immuni.2012.06.020
15. Buonocore S, Ahern PP, Uhlig HH, Ivanov II, Littman DR, Maloy KJ, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature (2010) 464:1371–5. doi: 10.1038/nature08949
16. Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littman DR. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol (2004) 5:64–73. doi: 10.1038/ni1022
17. Colonna M. Innate lymphoid cells: Diversity, plasticity, and unique functions in immunity. Immunity (2018) 48:1104–17. doi: 10.1016/j.immuni.2018.05.013
18. Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells: 10 years on. Cell (2018) 174:1054–66. doi: 10.1016/j.cell.2018.07.017
19. Mujal AM, Delconte RB, Sun JC. Natural killer cells: From innate to adaptive features. Annu Rev Immunol (2021) 39:417–47. doi: 10.1146/annurev-immunol-101819-074948
20. Beckstette M, Lu CW, Herppich S, Diem EC, Ntalli A, Ochel A, et al. Profiling of epigenetic marker regions in murine ILCs under homeostatic and inflammatory conditions. J Exp Med (2022) 219:e20210663. doi: 10.1084/jem.20210663
21. Fang D, Cui K, Cao Y, Zheng M, Kawabe T, Hu G, et al. Differential regulation of transcription factor T-bet induction during NK cell development and T helper-1 cell differentiation. Immunity (2022) 55:639–655 e637. doi: 10.1016/j.immuni.2022.03.005
22. Kasal DN, Liang Z, Hollinger MK, O'Leary CY, Lisicka W, Sperling AI, et al. A Gata3 enhancer necessary for ILC2 development and function. Proc Natl Acad Sci USA (2021) 118:e2106311118. doi: 10.1073/pnas.2106311118
23. Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, et al. A validated regulatory network for Th17 cell specification. Cell (2012) 151:289–303. doi: 10.1016/j.cell.2012.09.016
24. Peng V, Xing X, Bando JK, Trsan T, Di Luccia B, Collins PL, et al. Whole-genome profiling of DNA methylation and hydroxymethylation identifies distinct regulatory programs among innate lymphocytes. Nat Immunol (2022) 23:619–31. doi: 10.1038/s41590-022-01164-8
25. Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, et al. The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell (2016) 166:1231–1246 e1213. doi: 10.1016/j.cell.2016.07.043
26. Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, et al. T-Bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity (2004) 20:477–94. doi: 10.1016/S1074-7613(04)00076-7
27. Chaix J, Tessmer MS, Hoebe K, Fuseri N, Ryffel B, Dalod M, et al. Cutting edge: Priming of NK cells by IL-18. J Immunol (2008) 181:1627–31. doi: 10.4049/jimmunol.181.3.1627
28. Takeda K, Tsutsui H, Yoshimoto T, Adachi O, Yoshida N, Kishimoto T, et al. Defective NK cell activity and Th1 response in IL-18-deficient mice. Immunity (1998) 8:383–90. doi: 10.1016/S1074-7613(00)80543-9
29. Chang D, Xing Q, Su Y, Zhao X, Xu W, Wang X, et al. The conserved non-coding sequences CNS6 and CNS9 control cytokine-induced rorc transcription during T helper 17 cell differentiation. Immunity (2020) 53:614–626 e614. doi: 10.1016/j.immuni.2020.07.012
30. Shih HY, Sciume G, Mikami Y, Guo L, Sun HW, Brooks SR, et al. Developmental acquisition of regulomes underlies innate lymphoid cell functionality. Cell (2016) 165:1120–33. doi: 10.1016/j.cell.2016.04.029
31. Madera S, Geary CD, Lau CM, Pikovskaya O, Reiner SL, Sun JC. Cutting edge: Divergent requirement of T-box transcription factors in effector and memory NK cells. J Immunol (2018) 200:1977–81. doi: 10.4049/jimmunol.1700416
32. Rapp M, Lau CM, Adams NM, Weizman OE, O'Sullivan TE, Geary CD, et al. Core-binding factor beta and runx transcription factors promote adaptive natural killer cell responses. Sci Immunol (2017) 2:eaan3796. doi: 10.1126/sciimmunol.aan3796
33. Yang Y, Ochando JC, Bromberg JS, Ding Y. Identification of a distant T-bet enhancer responsive to IL-12/Stat4 and IFNgamma/Stat1 signals. Blood (2007) 110:2494–500. doi: 10.1182/blood-2006-11-058271
34. Morinobu A, Gadina M, Strober W, Visconti R, Fornace A, Montagna C, et al. STAT4 serine phosphorylation is critical for IL-12-induced IFN-gamma production but not for cell proliferation. Proc Natl Acad Sci USA (2002) 99:12281–6. doi: 10.1073/pnas.182618999
35. Hoey T, Zhang S, Schmidt N, Yu Q, Ramchandani S, Xu X, et al. Distinct requirements for the naturally occurring splice forms Stat4alpha and Stat4beta in IL-12 responses. EMBO J (2003) 22:4237–48. doi: 10.1093/emboj/cdg393
36. Thieu VT, Yu Q, Chang HC, Yeh N, Nguyen ET, Sehra S, et al. Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity (2008) 29:679–90. doi: 10.1016/j.immuni.2008.08.017
37. Schulz EG, Mariani L, Radbruch A, Hofer T. Sequential polarization and imprinting of type 1 T helper lymphocytes by interferon-gamma and interleukin-12. Immunity (2009) 30:673–83. doi: 10.1016/j.immuni.2009.03.013
38. Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, et al. T-Bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol (2002) 3:549–57. doi: 10.1038/ni794
39. Cautivo KM, Matatia PR, Lizama CO, Mroz NM, Dahlgren MW, Yu X, et al. Interferon gamma constrains type 2 lymphocyte niche boundaries during mixed inflammation. Immunity (2022) 55:254–271e257. doi: 10.1016/j.immuni.2021.12.014
40. Meissl K, Simonovic N, Amenitsch L, Witalisz-Siepracka A, Klein K, Lassnig C, et al. STAT1 isoforms differentially regulate NK cell maturation and anti-tumor activity. Front Immunol (2020) 11:2189. doi: 10.3389/fimmu.2020.02189
41. Hibbert L, Pflanz S, Malefyt RD, Kastelein RA. IL-27 and IFN-alpha signal via Stat1 and Stat3 and induce T-bet and IL-12R beta 2 in naive T cells. J Interf Cytok Res (2003) 23:513–22. doi: 10.1089/10799900360708632
42. Lucas S, Ghilardi N, Li J, de Sauvage FJ. IL-27 regulates IL-12 responsiveness of naive CD4+ T cells through Stat1-dependent and -independent mechanisms. Proc Natl Acad Sci USA (2003) 100:15047–52. doi: 10.1073/pnas.2536517100
43. Takeda A, Hamano S, Yamanaka A, Hanada T, Ishibashi T, Mak TW, et al. Cutting edge: role of IL-27/WSX-1 signaling for induction of T-bet through activation of STAT1 during initial Th1 commitment. J Immunol (2003) 170:4886–90. doi: 10.4049/jimmunol.170.10.4886
44. Kamiya S, Owaki T, Morishima N, Fukai F, Mizuguchi J, Yoshimoto T. An indispensable role for STAT1 in IL-27-induced T-bet expression but not proliferation of naive CD4+ T cells. J Immunol (2004) 173:3871–7. doi: 10.4049/jimmunol.173.6.3871
45. Owaki T, Asakawa M, Morishima N, Hata K, Fukai F, Matsui M, et al. A role for IL-27 in early regulation of Th1 differentiation. J Immunol (2005) 175:2191–200. doi: 10.4049/jimmunol.175.4.2191
46. Pflanz S, Timans JC, Cheung J, Rosales R, Kanzler H, Gilbert J, et al. IL-27, a heterodimeric cytokine composed of EBI3 and p28 protein, induces proliferation of naive CD4+ T cells. Immunity (2002) 16:779–90. doi: 10.1016/S1074-7613(02)00324-2
47. Villarino AV, Larkin J 3rd, Saris CJ, Caton AJ, Lucas S, Wong T, et al. Positive and negative regulation of the IL-27 receptor during lymphoid cell activation. J Immunol (2005) 174:7684–91. doi: 10.4049/jimmunol.174.12.7684
48. Wan CK, Andraski AB, Spolski R, Li P, Kazemian M, Oh J, et al. Opposing roles of STAT1 and STAT3 in IL-21 function in CD4+ T cells. Proc Natl Acad Sci USA (2015) 112:9394–9. doi: 10.1073/pnas.1511711112
49. Tominaga K, Yoshimoto T, Torigoe K, Kurimoto M, Matsui K, Hada T, et al. IL-12 synergizes with IL-18 or IL-1beta for IFN-gamma production from human T cells. Int Immunol (2000) 12:151–60. doi: 10.1093/intimm/12.2.151
50. Mullen AC, High FA, Hutchins AS, Lee HW, Villarino AV, Livingston DM, et al. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science (2001) 292:1907–10. doi: 10.1126/science.1059835
51. Krueger PD, Goldberg MF, Hong SW, Osum KC, Langlois RA, Kotov DI, et al. Two sequential activation modules control the differentiation of protective T helper-1 (Th1) cells. Immunity (2021) 54:687–701 e684. doi: 10.1016/j.immuni.2021.03.006
52. Miyamoto C, Kojo S, Yamashita M, Moro K, Lacaud G, Shiroguchi K, et al. Runx/Cbfbeta complexes protect group 2 innate lymphoid cells from exhausted-like hyporesponsiveness during allergic airway inflammation. Nat Commun (2019) 10:447. doi: 10.1038/s41467-019-08365-0
53. Ebihara T, Song C, Ryu SH, Plougastel-Douglas B, Yang L, Levanon D, et al. Runx3 specifies lineage commitment of innate lymphoid cells. Nat Immunol (2015) 16:1124–33. doi: 10.1038/ni.3272
54. Ramirez K, Chandler KJ, Spaulding C, Zandi S, Sigvardsson M, Graves BJ, et al. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity (2012) 36:921–32. doi: 10.1016/j.immuni.2012.04.006
55. Zamisch M, Tian L, Grenningloh R, Xiong Y, Wildt KF, Ehlers M, et al. The transcription factor Ets1 is important for CD4 repression and Runx3 up-regulation during CD8 T cell differentiation in the thymus. J Exp Med (2009) 206:2685–99. doi: 10.1084/jem.20092024
56. Kanhere A, Hertweck A, Bhatia U, Gokmen MR, Perucha E, Jackson I, et al. T-Bet and GATA3 orchestrate Th1 and Th2 differentiation through lineage-specific targeting of distal regulatory elements. Nat Commun (2012) 3:1268. doi: 10.1038/ncomms2260
57. Zhu J, Jankovic D, Oler AJ, Wei G, Sharma S, Hu G, et al. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity (2012) 37:660–73. doi: 10.1016/j.immuni.2012.09.007
58. Zhu J. GATA3 regulates the development and functions of innate lymphoid cell subsets at multiple stages. Front Immunol (2017) 8:1571. doi: 10.3389/fimmu.2017.01571
59. Zhong C, Zheng M, Cui K, Martins AJ, Hu G, Li D, et al. Differential expression of the transcription factor GATA3 specifies lineage and functions of innate lymphoid cells. Immunity (2020) 52:83–95 e84. doi: 10.1016/j.immuni.2019.12.001
60. George KM, Leonard MW, Roth ME, Lieuw KH, Kioussis D, Grosveld F, et al. Embryonic expression and cloning of the murine GATA-3 gene. Development (1994) 120:2673–86. doi: 10.1242/dev.120.9.2673
61. Yagi R, Zhong C, Northrup DL, Yu F, Bouladoux N, Spencer S, et al. The transcription factor GATA3 is critical for the development of all IL-7Ralpha-expressing innate lymphoid cells. Immunity (2014) 40:378–88. doi: 10.1016/j.immuni.2014.01.012
62. Hosoya-Ohmura S, Lin YH, Herrmann M, Kuroha T, Rao A, Moriguchi T, et al. An NK and T cell enhancer lies 280 kilobase pairs 3' to the gata3 structural gene. Mol Cell Biol (2011) 31:1894–904. doi: 10.1128/MCB.05065-11
63. Ohmura S, Mizuno S, Oishi H, Ku CJ, Hermann M, Hosoya T, et al. Lineage-affiliated transcription factors bind the Gata3 Tce1 enhancer to mediate lineage-specific programs. J Clin Invest (2016) 126:865–78. doi: 10.1172/JCI83894
64. Ouyang W, Ranganath SH, Weindel K, Bhattacharya D, Murphy TL, Sha WC, et al. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity (1998) 9:745–55. doi: 10.1016/S1074-7613(00)80671-8
65. Scheinman EJ, Avni O. Transcriptional regulation of GATA3 in T helper cells by the integrated activities of transcription factors downstream of the interleukin-4 receptor and T cell receptor. J Biol Chem (2009) 284:3037–48. doi: 10.1074/jbc.M807302200
66. Asnagli H, Afkarian M, Murphy KM. Cutting edge: Identification of an alternative GATA-3 promoter directing tissue-specific gene expression in mouse and human. J Immunol (2002) 168:4268–71. doi: 10.4049/jimmunol.168.9.4268
67. Kurata H, Lee HJ, O'Garra A, Arai N. Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity (1999) 11:677–88. doi: 10.1016/S1074-7613(00)80142-9
68. Onodera A, Yamashita M, Endo Y, Kuwahara M, Tofukuji S, Hosokawa H, et al. STAT6-mediated displacement of polycomb by trithorax complex establishes long-term maintenance of GATA3 expression in T helper type 2 cells. J Exp Med (2010) 207:2493–506. doi: 10.1084/jem.20100760
69. Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity (2010) 32:840–51. doi: 10.1016/j.immuni.2010.06.003
70. Ouyang W, Lohning M, Gao Z, Assenmacher M, Ranganath S, Radbruch A, et al. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity (2000) 12:27–37. doi: 10.1016/S1074-7613(00)80156-9
71. Zhu J, Cote-Sierra J, Guo L, Paul WE. Stat5 activation plays a critical role in Th2 differentiation. Immunity (2003) 19:739–48. doi: 10.1016/S1074-7613(03)00292-9
72. Yamane H, Zhu J, Paul WE. Independent roles for IL-2 and GATA-3 in stimulating naive CD4+ T cells to generate a Th2-inducing cytokine environment. J Exp Med (2005) 202:793–804. doi: 10.1084/jem.20051304
73. Cote-Sierra J, Foucras G, Guo L, Chiodetti L, Young HA, Hu-Li J, et al. Interleukin 2 plays a central role in Th2 differentiation. Proc Natl Acad Sci USA (2004) 101:3880–5. doi: 10.1073/pnas.0400339101
74. Guo L, Wei G, Zhu J, Liao W, Leonard WJ, Zhao K, et al. IL-1 family members and STAT activators induce cytokine production by Th2, Th17, and Th1 cells. Proc Natl Acad Sci USA (2009) 106:13463–8. doi: 10.1073/pnas.0906988106
75. Symowski C, Voehringer D. Th2 cell-derived IL-4/IL-13 promote ILC2 accumulation in the lung by ILC2-intrinsic STAT6 signaling in mice. Eur J Immunol (2019) 49:1421–32. doi: 10.1002/eji.201948161
76. Doherty TA, Khorram N, Chang JE, Kim HK, Rosenthal P, Croft M, et al. STAT6 regulates natural helper cell proliferation during lung inflammation initiated by alternaria. Am J Physiol Lung Cell Mol Physiol (2012) 303:L577–88. doi: 10.1152/ajplung.00174.2012
77. Liang HE, Reinhardt RL, Bando JK, Sullivan BM, Ho IC, Locksley RM. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat Immunol (2011) 13:58–66. doi: 10.1038/ni.2182
78. Das J, Chen CH, Yang L, Cohn L, Ray P, Ray A. A critical role for NF-kappa b in GATA3 expression and TH2 differentiation in allergic airway inflammation. Nat Immunol (2001) 2:45–50. doi: 10.1038/83158
79. Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, et al. Direct regulation of Gata3 expression determines the T helper differentiation potential of notch. Immunity (2007) 27:89–99. doi: 10.1016/j.immuni.2007.05.021
80. Sheikh A, Lu J, Melese E, Seo JH, Abraham N. IL-7 induces type 2 cytokine response in lung ILC2s and regulates GATA3 and CD25 expression. J Leukoc Biol (2022) 112:1105–13. doi: 10.1002/JLB.3AB1220-819RRR
81. Fang D, Cui K, Hu G, Gurram RK, Zhong C, Oler AJ, et al. Bcl11b, a novel GATA3-interacting protein, suppresses Th1 while limiting Th2 cell differentiation. J Exp Med (2018) 215:1449–62. doi: 10.1084/jem.20171127
82. Zhong C, Cui K, Wilhelm C, Hu G, Mao K, Belkaid Y, et al. Group 3 innate lymphoid cells continuously require the transcription factor GATA-3 after commitment. Nat Immunol (2016) 17:169–78. doi: 10.1038/ni.3318
83. Hosokawa H, Romero-Wolf M, Yang Q, Motomura Y, Levanon D, Groner Y, et al. Cell type-specific actions of Bcl11b in early T-lineage and group 2 innate lymphoid cells. J Exp Med (2020) 217:e20190972. doi: 10.1084/jem.20190972
84. Spooner CJ, Lesch J, Yan D, Khan AA, Abbas A, Ramirez-Carrozzi V, et al. Specification of type 2 innate lymphocytes by the transcriptional determinant Gfi1. Nat Immunol (2013) 14:1229–36. doi: 10.1038/ni.2743
85. Yu Y, Wang C, Clare S, Wang J, Lee SC, Brandt C, et al. The transcription factor Bcl11b is specifically expressed in group 2 innate lymphoid cells and is essential for their development. J Exp Med (2015) 212:865–74. doi: 10.1084/jem.20142318
86. Zook EC, Ramirez K, Guo X, van der Voort G, Sigvardsson M, Svensson EC, et al. The ETS1 transcription factor is required for the development and cytokine-induced expansion of ILC2. J Exp Med (2016) 213:687–96. doi: 10.1084/jem.20150851
87. Hall JA, Pokrovskii M, Kroehling L, Kim BR, Kim SY, Wu L, et al. Transcription factor RORalpha enforces stability of the Th17 cell effector program by binding to a rorc cis-regulatory element. Immunity (2022) 55:2027–43. doi: 10.1016/j.immuni.2022.09.013
88. Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity (2010) 32:605–15. doi: 10.1016/j.immuni.2010.05.003
89. Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature (2008) 453:236–40. doi: 10.1038/nature06878
90. Lochner M, Peduto L, Cherrier M, Sawa S, Langa F, Varona R, et al. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J Exp Med (2008) 205:1381–93. doi: 10.1084/jem.20080034
91. Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol (2007) 8:967–74. doi: 10.1038/ni1488
92. Wei L, Laurence A, Elias KM, O'Shea JJ. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J Biol Chem (2007) 282:34605–10. doi: 10.1074/jbc.M705100200
93. Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem (2007) 282:9358–63. doi: 10.1074/jbc.C600321200
94. Mathur AN, Chang HC, Zisoulis DG, Stritesky GL, Yu Q, O'Malley JT, et al. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J Immunol (2007) 178:4901–7. doi: 10.4049/jimmunol.178.8.4901
95. Guo X, Qiu J, Tu T, Yang X, Deng L, Anders RA, et al. Induction of innate lymphoid cell-derived interleukin-22 by the transcription factor STAT3 mediates protection against intestinal infection. Immunity (2014) 40:25–39. doi: 10.1016/j.immuni.2013.10.021
96. Chen L, He Z, Slinger E, Bongers G, Lapenda TLS, Pacer ME, et al. IL-23 activates innate lymphoid cells to promote neonatal intestinal pathology. Mucosal Immunol (2015) 8:390–402. doi: 10.1038/mi.2014.77
97. Wang Y, Godec J, Ben-Aissa K, Cui K, Zhao K, Pucsek Alexandra B, et al. The transcription factors T-bet and runx are required for the ontogeny of pathogenic interferon-γ-Producing T helper 17 cells. Immunity (2014) 40:355–66. doi: 10.1016/j.immuni.2014.01.002
98. Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol (2008) 9:1297–306. doi: 10.1038/ni.1663
99. Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, et al. T-Bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol (2011) 12:96–104. doi: 10.1038/ni.1969
100. Tizian C, Lahmann A, Holsken O, Cosovanu C, Kofoed-Branzk M, Heinrich F, et al. C-maf restrains T-bet-driven programming of CCR6-negative group 3 innate lymphoid cells. Elife (2020) 9:e52549. doi: 10.7554/eLife.52549
101. Parker ME, Barrera A, Wheaton JD, Zuberbuehler MK, Allan DSJ, Carlyle JR, et al. C-maf regulates the plasticity of group 3 innate lymphoid cells by restraining the type 1 program. J Exp Med (2020) 217(1):e20191030. doi: 10.1084/jem.20191030
102. Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol (2009) 10:1118–24. doi: 10.1038/ni.1787
103. Geiger TL, Abt MC, Gasteiger G, Firth MA, O'Connor MH, Geary CD, et al. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J Exp Med (2014) 211:1723–31. doi: 10.1084/jem.20140212
104. Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, Chopin M, et al. Nfil3 is required for the development of all innate lymphoid cell subsets. J Exp Med (2014) 211:1733–40. doi: 10.1084/jem.20140145
105. Xu W, Domingues RG, Fonseca-Pereira D, Ferreira M, Ribeiro H, Lopez-Lastra S, et al. NFIL3 orchestrates the emergence of common helper innate lymphoid cell precursors. Cell Rep (2015) 10:2043–54. doi: 10.1016/j.celrep.2015.02.057
106. Yu X, Rollins D, Ruhn KA, Stubblefield JJ, Green CB, Kashiwada M, et al. TH17 cell differentiation is regulated by the circadian clock. Science (2013) 342:727–30. doi: 10.1126/science.1243884
107. Yahia-Cherbal H, Rybczynska M, Lovecchio D, Stephen T, Lescale C, Placek K, et al. NFAT primes the human RORC locus for RORgammat expression in CD4(+) T cells. Nat Commun (2019) 10:4698. doi: 10.1038/s41467-019-12680-x
108. Fachi JL, Pral LP, Dos Santos JAC, Codo AC, de Oliveira S, Felipe JS, et al. Hypoxia enhances ILC3 responses through HIF-1alpha-dependent mechanism. Mucosal Immunol (2021) 14:828–41. doi: 10.1038/s41385-020-00371-6
109. Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell (2011) 146:772–84. doi: 10.1016/j.cell.2011.07.033
110. van de Pavert SA, Ferreira M, Domingues RG, Ribeiro H, Molenaar R, Moreira-Santos L, et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature (2014) 508:123–7. doi: 10.1038/nature13158
111. Bando JK, Gilfillan S, Song C, McDonald KG, Huang SC, Newberry RD, et al. The tumor necrosis factor superfamily member RANKL suppresses effector cytokine production in group 3 innate lymphoid cells. Immunity (2018) 48:1208–1219 e1204. doi: 10.1016/j.immuni.2018.04.012
112. Patel MN, Carroll RG, Galvan-Pena S, Mills EL, Olden R, Triantafilou M, et al. Inflammasome priming in sterile inflammatory disease. Trends Mol Med (2017) 23:165–80. doi: 10.1016/j.molmed.2016.12.007
113. Golub R. The notch signaling pathway involvement in innate lymphoid cell biology. BioMed J (2021) 44:133–43. doi: 10.1016/j.bj.2020.12.004
114. Fang D, Zhu J. Dynamic balance between master transcription factors determines the fates and functions of CD4 T cell and innate lymphoid cell subsets. J Exp Med (2017) 214:1861–76. doi: 10.1084/jem.20170494
115. Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol (2012) 13:991–9. doi: 10.1038/ni.2416
116. Yang Y, Weiner J, Liu Y, Smith AJ, Huss DJ, Winger R, et al. T-Bet is essential for encephalitogenicity of both Th1 and Th17 cells. J Exp Med (2009) 206:1549–64. doi: 10.1084/jem.20082584
117. Yu F, Sharma S, Edwards J, Feigenbaum L, Zhu J. Dynamic expression of transcription factors T-bet and GATA-3 by regulatory T cells maintains immunotolerance. Nat Immunol (2015) 16:197–206. doi: 10.1038/ni.3053
118. Dominguez-Villar M, Baecher-Allan CM, Hafler DA. Identification of T helper type 1–like, Foxp3+ regulatory T cells in human autoimmune disease. Nat Med (2011) 17:673–5. doi: 10.1038/nm.2389
119. Koch MA, Thomas KR, Perdue NR, Smigiel KS, Srivastava S, Campbell DJ. T-Bet(+) treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor beta2. Immunity (2012) 37:501–10. doi: 10.1016/j.immuni.2012.05.031
120. Sciume G, Hirahara K, Takahashi H, Laurence A, Villarino AV, Singleton KL, et al. Distinct requirements for T-bet in gut innate lymphoid cells. J Exp Med (2012) 209:2331–8. doi: 10.1084/jem.20122097
121. Rankin LC, Groom JR, Chopin M, Herold MJ, Walker JA, Mielke LA, et al. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the notch pathway. Nat Immunol (2013) 14:389–95. doi: 10.1038/ni.2545
122. Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d'Hargues Y, et al. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature (2013) 494:261–5. doi: 10.1038/nature11813
123. Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature (2010) 463:808–12. doi: 10.1038/nature08750
124. Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature (2012) 482:395–9. doi: 10.1038/nature10772
125. Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell (2012) 151:153–66. doi: 10.1016/j.cell.2012.06.053
126. Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol (2008) 9:194–202. doi: 10.1038/ni1549
Keywords: lineage-determining transcription factor, epigenetic modification, CD4 T helper cells, innate lymphoid cells, Th cell differentiation, ILC development
Citation: Fang D, Healy A and Zhu J (2023) Differential regulation of lineage-determining transcription factor expression in innate lymphoid cell and adaptive T helper cell subsets. Front. Immunol. 13:1081153. doi: 10.3389/fimmu.2022.1081153
Received: 26 October 2022; Accepted: 14 December 2022;
Published: 04 January 2023.
Edited by:
Cyril Seillet, The University of Melbourne, AustraliaReviewed by:
Han-Yu Shih, National Eye Institute (NIH), United StatesJu Qiu, Shanghai Institute of Nutrition and Health (CAS), China
Copyright © 2023 Fang, Healy and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Difeng Fang, ZGlmZW5nLmZhbmdAbmloLmdvdg==; Jinfang Zhu, amZ6aHVAbmlhaWQubmloLmdvdg==
 Difeng Fang
Difeng Fang Ayanna Healy
Ayanna Healy Jinfang Zhu
Jinfang Zhu