- Pathovet Labs, Puerto Montt, Chile
Piscirickettsiosis (SRS) has been the most important infectious disease in Chilean salmon farming since the 1980s. It was one of the first to be described, and to date, it continues to be the main infectious cause of mortality. How can we better understand the epidemiological situation of SRS? The catch-all answer is that the Chilean salmon farming industry must fight year after year against a multifactorial disease, and apparently only the environment in Chile seems to favor the presence and persistence of Piscirickettsia salmonis. This is a fastidious, facultative intracellular bacterium that replicates in the host’s own immune cells and antigen-presenting cells and evades the adaptive cell-mediated immune response, which is why the existing vaccines are not effective in controlling it. Therefore, the Chilean salmon farming industry uses a lot of antibiotics—to control SRS—because otherwise, fish health and welfare would be significantly impaired, and a significantly higher volume of biomass would be lost per year. How can the ever-present risk of negative consequences of antibiotic use in salmon farming be balanced with the productive and economic viability of an animal production industry, as well as with the care of the aquatic environment and public health and with the sustainability of the industry? The answer that is easy, but no less true, is that we must know the enemy and how it interacts with its host. Much knowledge has been generated using this line of inquiry, however it remains insufficient. Considering the state-of-the-art summarized in this review, it can be stated that, from the point of view of fish immunology and vaccinology, we are quite far from reaching an effective and long-term solution for the control of SRS. For this reason, the aim of this critical review is to comprehensively discuss the current knowledge on the interaction between the bacteria and the host to promote the generation of more and better measures for the prevention and control of SRS.
Piscirickettsiosis: The Usual Enemy
Piscirickettsiosis (SRS) is caused by the Gram-negative facultative intracellular bacterium Piscirickettsia salmonis, which belongs to the subdivision of gamma-Proteobacteria with Coxiella and Francisella genera (1). P. salmonis has been confirmed as the causative agent for SRS in coho salmon (Oncorhynchus kisutch), Atlantic salmon (Salmo salar), and rainbow trout (Oncorhynchus mykiss) in Norway, Canada, Scotland, Ireland, and Chile (1). However, SRS especially affects salmon farming in Chile. In 2020, SRS was responsible for 47.8% and 67.3% of mortality related to infectious diseases and 11.7% and 10.7% of total mortality in the production of Atlantic salmon and rainbow trout, respectively (2). The most important determinants of the prevalence of SRS are the number of infected farms in upstream waters, followed by seawater salinity and temperature (3). On downstream farms of infected farms, the prevalence of SRS at 25 weeks of the cycle was close to 100%; while on farms with little or no exposure to infected upstream- farms, the prevalence only reached ~10% at week 56. Similarly, the prevalence of SRS within a group of concessions or “neighbourhoods” reached 100% at an average of 46 weeks after fallowing (4).
At the same time, we observed an average reduction of 18.3% in the volume of antimicrobials used in the Chilean salmon industry between 2016 and 2019, although during 2020, there was a slight increase of 7.6% compared to 2019 (5). This is very important because a median of 93.5% of the total volume of antimicrobials used between 2017 and 2020 was used for SRS control. These results could be a consequence of the positive impact of the health strategy for SRS control based on a specific official epidemiological surveillance program, improved genetic resistance plans, regulatory changes, and good production practices, including animal welfare aspects, lower stocking densities, fallow periods, vaccination, among other practices. However, the slight increase observed in 2020 is an early warning indicator that has been observed, along with the detection of P. salmonis, the presentation of earlier clinical findings of SRS after transfer of smolts to seawater, and increased SRS-related mortality, all of which are indicators that support the relative efficacy of commercially available vaccines to control the disease.
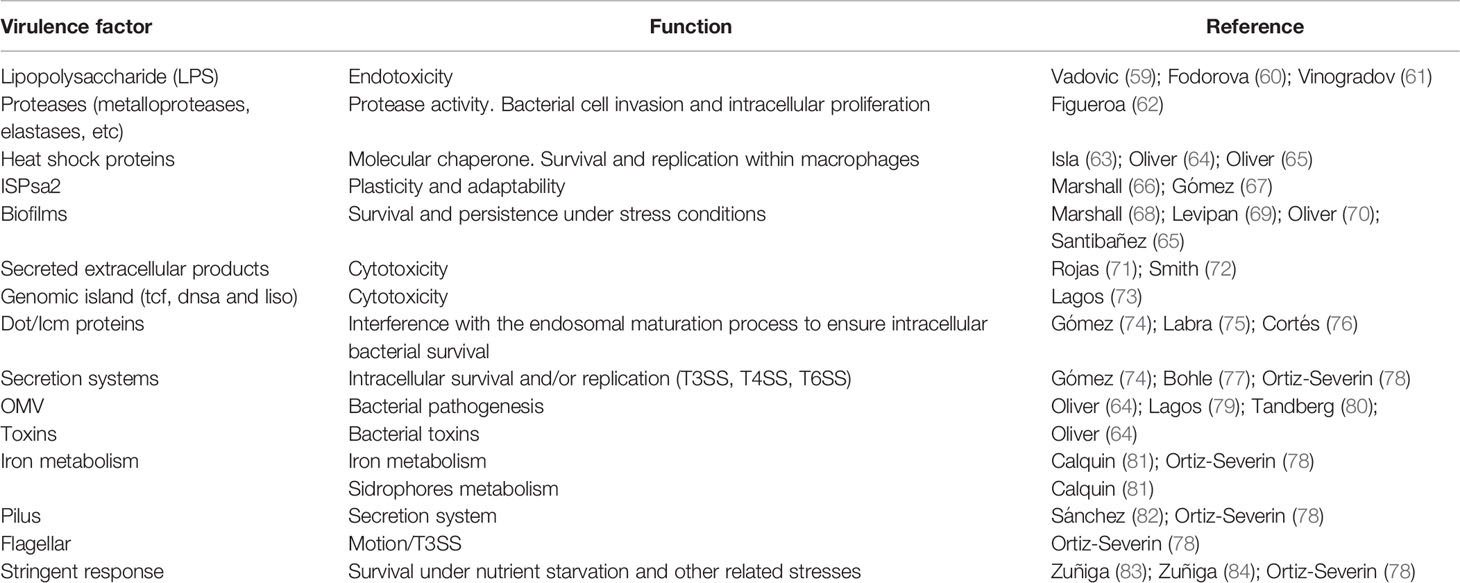
P. salmonis enters fish mainly through mucosal surfaces such as the skin and gills and, to a lesser extent, through the intestine (6–8). P. salmonis can even cross the intact skin in healthy fish (7), and although P. salmonis is not significantly negatively affected under exposure to the skin mucus of rainbow trout and Atlantic salmon over time, the cytotoxic effect of the bacterium pre-exposed to salmonid skin mucus is delayed (9). Rozas-Serri et al. (8) first reported a cohabitation challenge for P. salmonis, which was used to comparatively describe SRS pathogenesis using LF-89 and EM-90 isolates. This study concluded that the pathogenesis of postsmolt Atlantic salmon infected with LF-89-like and EM-90-like P. salmonis isolates was different. Fish infected with EM-90 showed higher cumulative mortality and a shorter time to mortality than fish infected with LF-89. EM-90 isolate produces an acute systemic and hemorrhagic disease characterized by lesions in all internal organs, whereas the most frequent internal lesions in fish infected with LF-89 show the more classical pathological picture of SRS with splenomegaly, renomegaly, large yellowish-white nodules in the kidney and liver, fibrinous pseudomembranes in the liver and heart, hydropericardium and ecchymosis in the liver (Figure 1). By histology, the yellowish-white nodules were found to be granulomas typically consisting of central necrosis along with the presence of bacteria and were surrounded by macrophages at different stages, neutrophils, putative dendritic cells (DCs), and natural killer cells; finally, all nodules are surrounded by putative T- and B-cells (Figure 1). However, a detailed molecular and cellular characterization of the granuloma arising during acute and chronic P. salmonis infection remains incomplete, mainly because antibodies to identify the different cell populations with confidence do not exist or are not accessible. P. salmonis induces flagellin-dependent tlr5 activation, which results in the up-regulation of tnfa, il1b, il8 and il16 (10–13), and promotes an intense proinflammatory response supported by leukocyte activation, and chemotaxis of neutrophils and macrophages towards the site of infection. At the same time, fish infected with P. salmonis showed up-regulation of relb and down-regulation of nfkbiz, which determine the activation of the NF- kB pathway (10, 12), promoting the up-regulation of genes encoding acute phase proteins (lect2, ltbr4, gpr84 and gpr43) and chemokines (cxcl7, cxcl9, cxcl10, cxcl12 and cxcr1) (10, 12, 13). Fish from both groups showed alterations in different blood biochemical parameters related to liver and kidney function, although the most severe alterations were present in fish infected with EM-90. In addition, the presence of P. salmonis was detected in the gills of coinhabiting fish of both groups at 21 days post-infection, confirming that the gills are the main point of entry of the bacterium into the host. Furthermore, the detection of the bacterium in the gills at 21 dpi and the occurrence of the first deaths at 36 dpi in fish with EM-90 and at 40 dpi in fish with LF-89 could indicate that these isolates have incubation periods of 15 and 20 days, respectively.
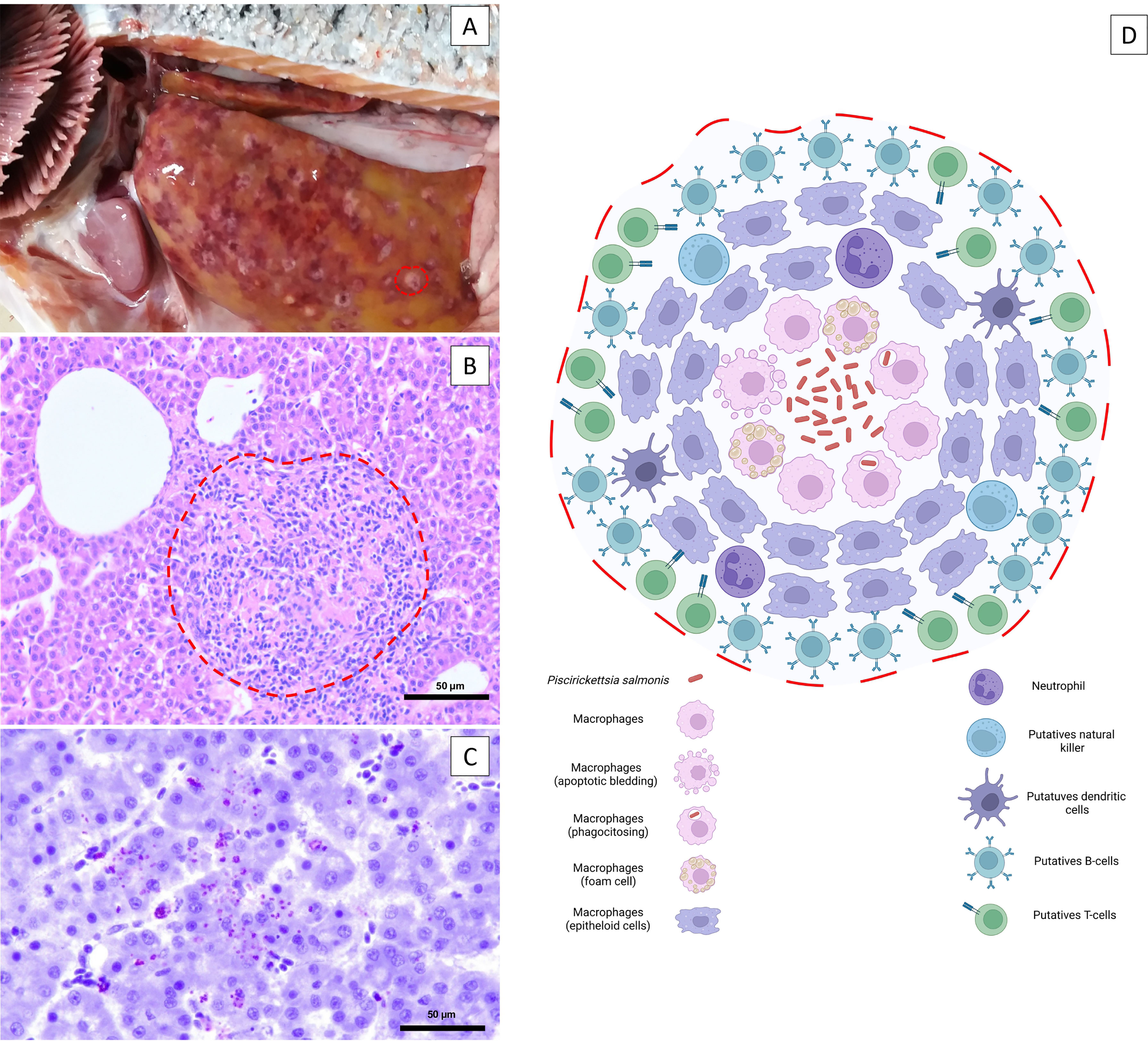
Figure 1 Macroscopic pathology and microscopic lesions in the liver associated with SRS infection. (A) Pale liver with subcapsular and circular reddish and gray–yellow mottled areas approximately 1-6 mm in diameter. The dashed red line delineates the borders of a gray–white discrete nodule characteristic of P. salmonis infection. (B) Multifocal necrosis of hepatocytes, diffuse infiltration of inflammatory cells and focal granuloma and/or multifocal coalescent granulomas. The dashed red line delineates the borders of a focal granuloma. Bar. 50 µm. (C) Immunolocalization of extracellular P. salmonis in the central zone of necrotic lesions and intracellular P. salmonis within macrophages using immunohistochemistry. Bar. 50 µm. (D) The yellowish-white nodules are granulomas typically consisting of a central necrosis with the presence of the bacteria, surrounded by macrophages at different stages, neutrophils, putative dendritic cells, and natural killer cells; all of these are surrounded by lymphocytes (putative T- and B-cells).
Cohabitation and immersion challenges are generally considered a better strategy compared to intraperitoneal injection challenges in terms of mimicking natural infection (8, 14, 15). There are only small differences between the intraperitoneal and cohabitation challenge models considering mortality and pathological changes (8, 14, 15). Meza et al. (14) reported that the incubation period was followed by an acute outbreak, with mortality reaching 100% in both challenged groups. Conversely, Rozas-Serri et al. (8) described a cumulative mortality of 84% and 100% in fish infected intraperitoneally with LF-89 and EM-90, respectively, but in fish infected using a cohabitation challenge model, the cumulative mortality reached 70 and 95%. In both studies, the pathological and histopathological changes were more visible during the clinical phase of the disease. Nevertheless, the death of fish in the cohabitant group was recorded earlier in the study from Meza et al. (14) than Rozas-Serri et al. (8) (28 dpc vs. 36 dpc, respectively), which could be due to the environmental variables of the experimental design; e.g., fish were kept in seawater at 32‰ and 15°C in the first case, while in the second case, the study was conducted in brackish water at 15‰ and 12°C. Finally, Long et al. (15) demonstrated that in tanks containing infected Atlantic salmon, using the immersion challenge model, the bacterium was first detected at 12 dpi, and levels peaked at 36 dpi, 3 days before the mean day of death. From 44 dpi onward, P. salmonis levels in the water column remained close to the detection limit and coincided with the cessation of acute mortality. Thus, the highest levels of P. salmonis are shed shortly before death, and Atlantic salmon experiencing an SRS outbreak are most infectious between 18 and 42 dpi.
Commercial vaccines are dominated by bacterin, recombinant (1) and attenuated live bacterial vaccines, administered primarily in water-in-oil emulsions (25) (Table 1). Regardless of the species of salmon farmed, these vaccines against SRS have not been fully protective in reducing total mortality or in delaying the time to onset of the first SRS outbreak under field conditions in Chile (1, 26, 27). Oral immunization is an attractive alternative, primarily because of its lower cost and ease of administration to farmed fish. It has been suggested that several oral immunizations for SRS are essential to maintain the humoral adaptive immune response in farmed fish, expressed in terms of SRS-specific antibodies, and to keep the fish protected throughout the entire production cycle (22, 28). This scenario might be biologically plausible in the case of diseases caused by viruses or extracellular bacteria, but the facultative intracellular nature of P. salmonis requires that a truly effective vaccine must activate the cell-mediated adaptive (CMI) response for a sufficiently long time to protect the salmon population throughout the production cycle in seawater. Unfortunately, P. salmonis precisely modulates and compromises the CMI response in infected fish as a mechanism to escape the host defense (10–13, 29, 30). Other intracellular bacteria of importance in aquaculture, such as Renibacterium salmoninarum and Edwardsiella tarda, modulate CMI in a similar way. Rozas-Serri et al. (31) demonstrated that load of R. salmoninarum had a significant positive correlation with the down-regulation of ifnγ, eomes, tbet, gata3, il2, il12, cd8 and mpeg1 (perforin) in the head-kidney of Atlantic salmon pre-smolts; consequently, R. salmoninarum did not trigger a CMI response in fish infected at 11 or 15°C. Yamasaki et al. (32) showed the important role of CMI rather than humoral immunity against E. tarda infection in ginbuna cross carp. Bacterial clearance in the kidney and spleen was observed after elevated cytotoxic activity and increased numbers of CTLs, but E. tarda-specific antibody titers did not increase until after bacterial clearance, suggesting that induction of humoral immunity is late to provide protection. The length of time that CD8+ cells remain active to “kill” infected cells following infection with P. salmonis and/or vaccination is unknown, so optimal levels of CD8+ cell activation capable of conferring protection in salmon are also unknown. Moreover, the extent to which mucosal vaccination can evoke protective immunity against intracellular replicating bacteria remains unclear, unlike in the case of viral and extracellular bacterial infections, in which protective mechanisms have been widely studied (33, 34). Hence, the practical conditions under which intracellular vaccination translates into protective CMI responses have not been determined for SRS vaccines.
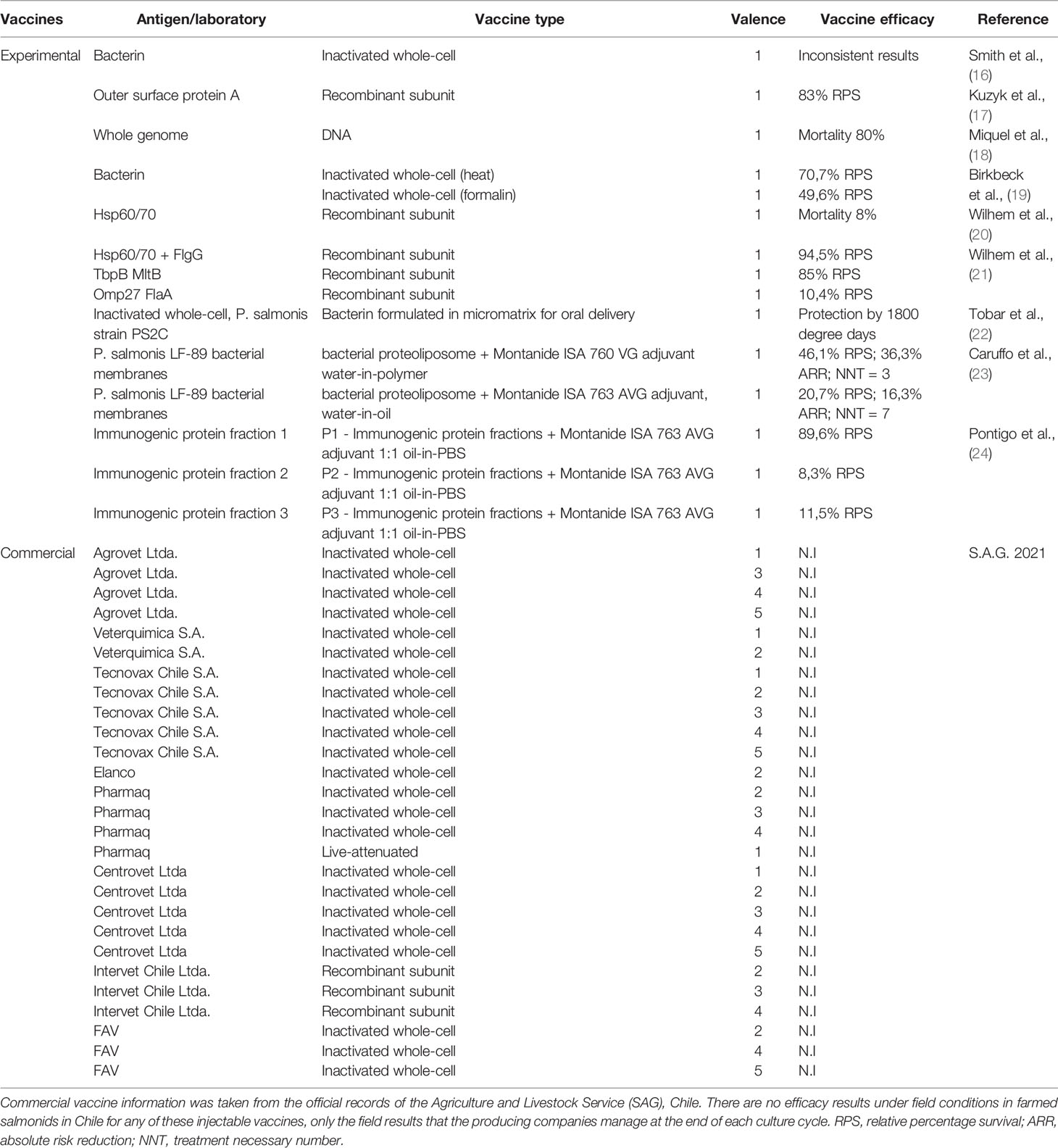
Table 1 Summary of experimental studies on efficacy evaluation of injectable and oral vaccines, types of antigens used and availability of commercial vaccines.
Yamasaki et al. (35) also showed the importance of CMI against E. tarda infection using vaccine trials comparing the effects of live vs. formalin-killed bacteria. Live cell-vaccinated fish showed high survival rates, high IFN-γ and T-bet gene expression levels, and increased CTLs. On contrary, all bacterin-vaccinated fish died following E. tarda infection and induced high IL4/13a and IL-10 expression levels, whereas Th1-like responses were suppressed. Rozas-Serri et al. (29) showed that a bacterin P. salmonis vaccinated-fish exhibited MHCI, MHCII, and CD4 overexpression but a significant downregulation of CD8b and IgM, suggesting that the formalin-killed bacteria promoted the CD4+ T-cell response but did not induce an immune response mediated by CD8+ T cells or a humoral response. The level of SRS-specific antibodies generated by vaccines and/or parenteral or oral boosters does not correlate with the level of protection or a reduction in the SRS-related mortality rate under field conditions. For this reason, no significant differences are observed between fish receiving a pentavalent injectable vaccination plus an oral booster and those receiving the same regimen but without the oral booster (27). Similarly, there is also no difference in overall mortality or time to first outbreak in fish receiving one or more saltwater booster vaccines and fish not receiving the booster (26).
In the last four years, our research group has evaluated the immune response of fish under field conditions against different vaccines, including the live attenuated P. salmonis vaccine. Overall, the activation of the innate immune response and the cell-mediated adaptive response modulated by overexpression of ifnγ, il2, il10, il12, il12, mhc1, cd4 and cd8 in the head kidney of Atlantic salmon smolts at 24 hours postvaccination and 150-, 300-, 460- and 600-degree days before transfer to seawater were observed. These findings are frequently noted in fish farmed in open-flow water hatcheries at different water temperatures (between 7 and 14°C) and in recirculation aquaculture systems (RAS), but the response is always significantly better in fish kept at higher temperatures (> 12°C). Acute and chronic exposure to suboptimal temperatures often has suppressive effects on the immune response of fish, especially on adaptive immunity (36). Sanhueza et al. (37) demonstrated that behavioural fever in ectotherms leads to a neuroimmune interaction that could modulate the systemic inflammatory response during pathogen infection. Results from the last time point of the monitoring plan conducted at 1,800-degree days postvaccination (1,200-degree days in seawater) showed a reduction in cd4 and cd8 gene expression, a time that usually coincides with the onset of the first SRS outbreaks in Atlantic salmon seawater farms. The downregulation of cd4 and cd8 at this point would indicate a reduced CMI response, probably associated with the gradual use of the different immune components previously induced by the vaccine and/or associated with a natural challenge.
These results are consistent with the transient stimulation of the CMI response described by Vargas et al. (38) in fish immunized with the same vaccine under field conditions, as they observed increased gene expression of ifna, ifnγ, cd4, cd8a, il10 and tgfb at 5 days postvaccination but a decreased response at 15 and 45 days postvaccination. However, the results of the latter study were obtained from fish vaccinated with an injectable booster vaccine in seawater when they were on average between 1 and 1.5 kg in weight. In Chile, 100% of Atlantic salmon smolts entering the sea are vaccinated with a vaccine containing the P. salmonis component, so these results could be influenced by pentavalent injectable primo-vaccination and natural challenge after entering seawater. Although we know that current vaccines do not sufficiently activate the CMI response to protect fish throughout the production cycle, current vaccines are important in the relative control of SRS. Nevertheless, a decrease in protection in terms of the time to the first detection of P. salmonis and SRS outbreaks, an increased number of therapies, and increased associated mortality, among other indicators, has been noted.
Piscirickettsia salmonis: A Fastidious Bacterium
P. salmonis is a facultative intracellular bacterium because although it infects lymphoid cells such as macrophages, monocytes and nonlymphoid cells in vitro and in vivo (39, 40), it can be cultured in vitro in solid and liquid media enriched with cysteine (41). Cortés et al. (42) identified the metabolic characteristics of P. salmonis that differentiate the two main clades of the species and demonstrated that P. salmonis could benefit from different experimentally tested carbon sources in newly defined media. The genome of P. salmonis contains one circular chromosome of 3,184,851 bp and three plasmids, pPSLF89-1 (180,124 bp), pPSLF89-2 (33,516 bp) and pPSLF89-3 (51,573 bp) (43). The in silico analysis of several strains of P. salmonis showed an open pangenome of 3463 genes and a coregenome of 1732 genes (44). Plasmids are extrachromosomal circular or linear double-stranded DNA molecules that vary in size, and some bacteria contain plasmids with no obvious functions that are classified as cryptic. Ortíz-Severin et al. (45) identified four large cryptic plasmids in the P. salmonis reference strain LF-89, in addition to twelve putative virulence factors and two global transcriptional regulators. These plasmids would be predicted to be critical in the bacterium’s ability to adapt to the environment, as they encode proteins related to nutrient mobilization, transport, and utilization.
Mauel et al. (46) showed that P. salmonis isolated in different parts of the world, including the reference strain LF-89, are closely related to each other (over >99% similarity in their 16S, ITS and 23S genes), but the Chilean isolate EM-90 is unique and genetically divergent from the others. Later, these findings were confirmed with more evidence from evaluations of their genomic structures and phylogenetic relationships to support the existence of two genogroups of P. salmonis, LF-89 and EM-90 (44, 47–52). P. salmonis pangenome LF-89 and EM-90 show 148 and 273 unique proteins, respectively (44), and different geographical distribution patterns and susceptibilities of salmon species have been described (53). This notion has also been experimentally supported by studies based on multilocus sequence typing (MLST) and PCR amplification of 16S rRNA genes followed by restriction fragment length polymorphism (PCR-RFLP) typing (51, 54). Although different real-time TaqMan® PCR assays have been used for several years by private diagnostic laboratories to identify and discriminate between LF-89 and EM-90 isolates in the Chilean salmon industry, a multiplex PCR protocol to differentiate both genogroups has recently been reported (52). P. salmonis can survive ≥120 h and replicate in cell cultures enriched with Atlantic salmon macrophages (55). This strategy would be induced by a limited lysosomal response that could be associated with P. salmonis host immune evasion mechanisms. Later, Pérez-Stuardo et al. (56) showed that IgM bead treatment promotes lysosomal activity in Atlantic salmon macrophage-enriched cell cultures infected with P. salmonis by reducing the lysosomal pH and increasing the proteolytic activity within the lysosome, reducing the bacterial load and the cytotoxicity induced by P. salmonis. Artificially induced iron deprivation during P. salmonis infection in vitro and in vivo using iron chelators generates a protective response of infected cells coincident with a reduction in bacterial load and cell damage (57, 58).
Although numerous virulence-related genes have been identified in P. salmonis, only a few encoded virulence factors have been characterized (Table 2). Proteolytic enzymes are important virulence factors because they are involved in cell invasion and intracellular proliferation. P. salmonis can synthesize and secrete siderophores (81), confirming its ability to utilize different sources of iron. In addition, P. salmonis has protease activity that increases significantly when the bacterium infects cells (62). At the same time, genomic islands have been described in the genomes of different P. salmonis strains (73), which could explain the different virulence levels of P. salmonis genogroups, pathogenesis (8, 14) and immune response observed in both fish experimentally challenged with different P. salmonis genogroups and vaccinated fish (11, 12, 14, 29). Intracellular bacterial pathogens use different strategies that allow them to adhere to, invade, and replicate in host cells and modulate intracellular processes such as membrane trafficking, signaling pathways, metabolism, and cell death and survival (85). One of the strategies used by these bacteria is secretion systems (SSs), which are complex multiprotein transmembrane nanomachines that form a channel that allows for the exportation of different molecules, including virulence effectors (86). P. salmonis is deficient in organelle trafficking/intracellular multiplication (Dot/Icm) secretion system genes and is classified as a type IVB secretion system (T4SS) (74, 75), which has been described as the main virulence mechanism of Legionella pneumophila and Coxiella burnetii and is responsible for their intracellular survival and replication (87). Temporal acidification of cell-free media results in overexpression of P. salmonis genes that inhibit phagosome-lysozyme fusion to prevent phagolysosome killing (74). Complementarily, the Sec-dependent pathway and Type 4B secretion system are biologically active during in vitro P. salmonis infection (76). The type VI (T6SS) and type III (T3SS) secretion system is also present in the P. salmonis genome, but no studies have demonstrated its functional mechanism.
Putative plasmid-encoded toxins are secreted in P. salmonis extracellular vesicles (EVs) (64, 79, 80, 88), which suggests a possible role in bacterial virulence of this important mobile genetic element. Leiva et al. (89) identified 35 unique proteins in serum EVs from P. salmonis-infected fish, including proteasome subunits, granulins and major histocompatibility classes I (MHC-I) and II (MHC-II). Therefore, these results suggest that the release of EVs could be part of a mechanism in which host stimulatory molecules are released from infected cells to promote an immune response. Additionally, other mechanisms of pathogenicity have been described in P. salmonis, such as outer membrane vesicles (OMVs) containing different proteins related to critical survival functions, plasmid-encoded toxins (64, 79, 80, 88) and type IV pili, which are filamentous structures on the bacterial surface important for adherence to host cell surfaces (82) (Table 3). Biofilms play an important role in bacterial pathogenicity, as their physical and spatial arrangement impedes access to antimicrobials and increases their resistance to phagocytosis. P. salmonis forms viable, stable, and fish skin-mucus tolerant biofilms on abiotic surfaces and under conditions of severe nutrient starvation (69). The cytotoxic response of the salmon head kidney cell line to P. salmonis showed interisolate differences because LF-89 isolate biofilms were sensitive to Atlantic salmon skin mucus during early formation, whereas EM-90 isolate biofilms were more tolerant. Complementarily, Santibañez et al. (90) showed that the planktonic isolate EM-90 and the sessile LF-89 would generate the highest levels of virulence in vitro by the modulation of the proinflammatory response (il1b, il8, nfkb, and ikba). NaCl and Fe significantly increase biofilm production. Oliver et al. (65) suggested that the presence of Hsp60 (GroEL) in P. salmonis OMVs would insinuate that they may be important in interacting with host proteins and/or modulating biofilm formation.
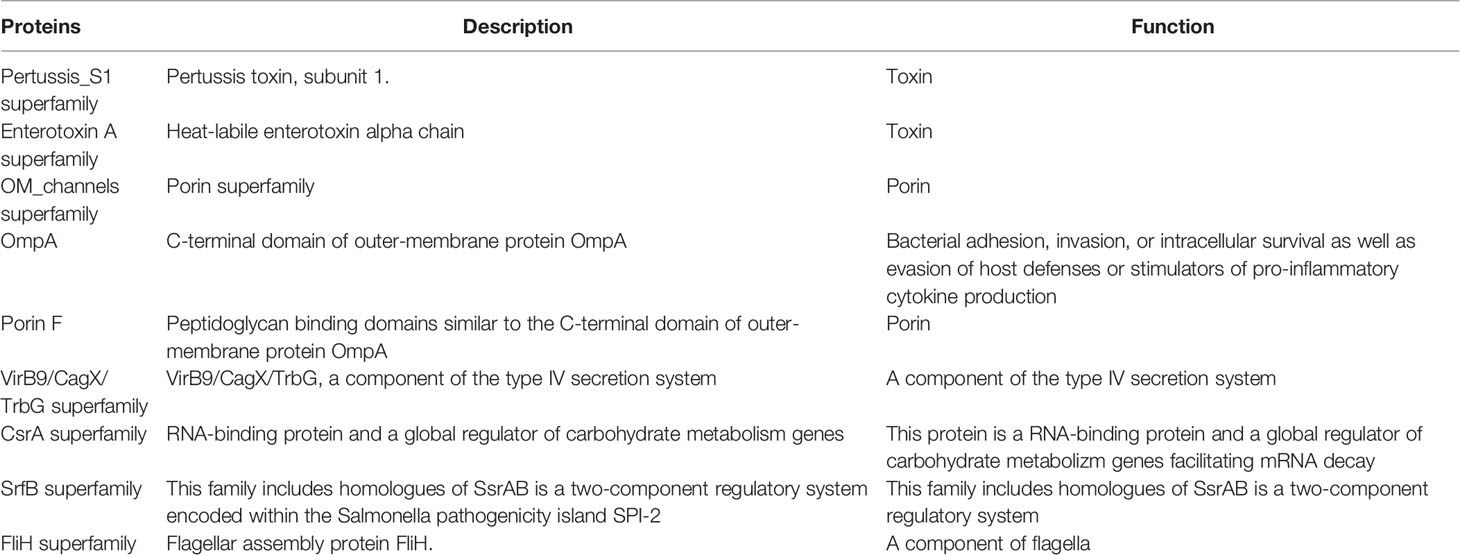
Table 3 Major virulence-related proteins described in membrane vesicles of P. salmonis strain type LF-89.
Legionella pneumophila is an aquatic organism that interacts with amoebae and ciliated protozoa as natural hosts, and this interaction plays a central role in bacterial ecology and infectivity (70). Declerck et al. (91) demonstrated that while L. pneumophila was present in 100% of floating biofilms in anthropogenic aquatic systems, Naegleria spp. and Acanthamoeba spp. were present in 50-92% and 67-72% of floating biofilm samples, respectively. Recently, Labra et al. (92) demonstrated that P. salmonis is detected only in adults of the crustacean ectoparasite Caligus rogercresseyi and that it is present only transiently after removal of P. salmonis-infected hosts, but the bacterium is not detected in the chalimus stages or in planktonic larvae of the parasites. However, the relationship of P. salmonis with Neoparamoeba perurans, a cosmopolitan marine amoeba and causative agent of AGD, remains unclear.
The Intracellular Invasion and Survival Strategy of P. salmonis
More information is now available to understand the process of invasion and survival of P. salmonis at the intracellular level (78, 83, 84). Although this knowledge remains to be validated in vivo using fish ideally challenged by cohabitation or immersion and under field conditions, it is an important step forward in the systematization and understanding of bacterial biology in host cells. However, like other intracellular bacteria in mammals and fish, we usually see that this battle between P. salmonis and fish defenses is won by the bacteria, probably because it has a wide arsenal of highly efficient virulence factors that allow it to evade the adaptive cell-mediated immune responses and modify the cell-autonomous immunity of the fish cells to its benefit, promoting a favorable environment for its replication and chronic maintenance in animals (8).
P. salmonis can infect, survive, and replicate primarily within the cytoplasmic vacuoles of macrophages and polymorphonuclear leukocytes without inducing a characteristic cytopathic effect (40) (Figure 2), although its replication in other dedicated phagocytic cells, such as DCs and B cells, remains to be elucidated. While macrophage biology has been characterized in mammals, macrophage differentiation and activation in teleost remain to be adequately described. Macrophages have a high degree of plasticity and can be activated by the classical (M1 or pro-inflammatory) or alternative (M2 or anti-inflammatory) pathway (108). M1 macrophages are activated by IFN-γ and TNF-α and produce proinflammatory cytokines and ROS to protect against pathogens (109). M2 macrophages are activated by il4/il13, il10, tgfb and are characterized by inducing lower microbicidal activity, immunosuppression and promoting cell growth and wound healing (110, 111). In teleost monocytes/macrophages, inducible nitric oxide synthase (iNOS) is an M1-type marker and arginase 2 is an M2-type marker (112). Smith et al. (113) observed a change in the morphology, phagocytic ability, and miRNA profile of Atlantic salmon head-kidney leukocytes (HKLs) in vitro, showing that the cells differentiate from “monocyte-like” (Day 1) to “macrophage-like” (Day 5). At the same time, the abundance of some miRNAs in EVs was significantly different from the abundance of miRNAs in HKLs, suggesting that these miRNAs are involved in the immune response and/or macrophage activation (114). Smith et al. (115) revealed major changes in the transcriptome of HDLs at day 1 and 5, including changes in the expression of macrophage and immune-related transcripts (csf1r, arg1, tnfa, mx2), lipid metabolism (fasn, dhcr7, fabp6) and transcription factors related to macrophage function and differentiation (klf2, klf9, irf7, irf8, stat1). Thus, the HKLs population differentiates in vitro to become macrophages without the addition of exogenous factors.
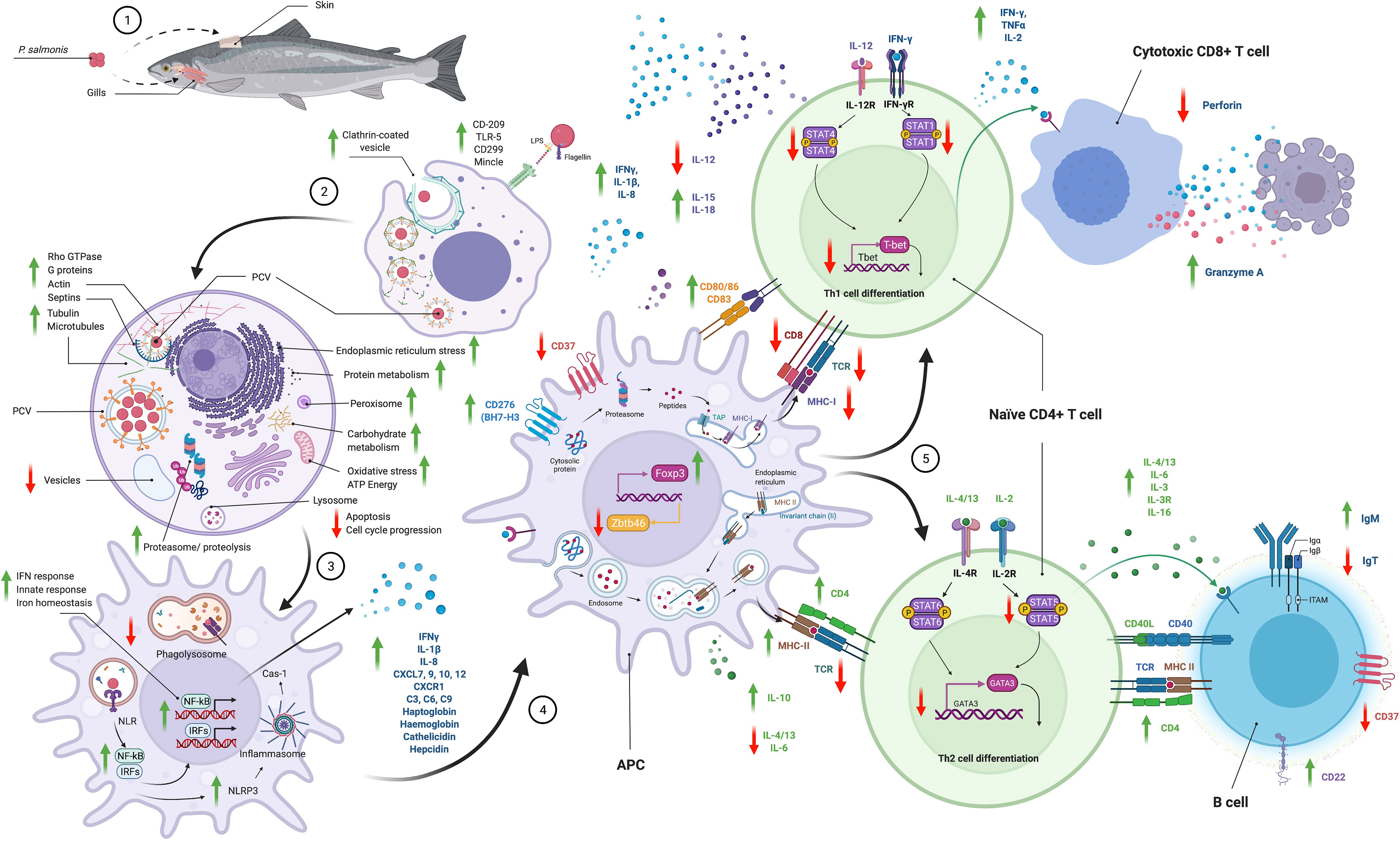
Figure 2 Schematic summary of the pathways used by P. salmonis to enter to the cell and the immune responses modulated. This information was consolidated with results descrbed in different in vitro and in vivo transcriptomic and proteomic studies. Phagocytosis, key in the life cycle of P. salmonis, is also its primary mode of pathogenesis. The interaction of P. salmonis with host cells has been described by different transcriptomic and proteomic studies in vitro and in vivo (10–13, 29, 30, 78, 89, 93–107). P. salmonis infection consists of an early or vacuolization stage and a late or spreading stage. (1) Several studies have reported that the main route of entry of the bacterium into the fish is through the gills and skin and, to a much lesser extent, the oral route. (2) P. salmonis is internalized by clathrin-dependent endocytosis into phagocytic cells. Pilus proteins, beta-hemolysin, and the T6SS are a characteristic finding of the vacuolization stage. The transport of carbohydrates, amino acids, peptides, iron, and other nutrients is increased inside the PCVs. (3) Once inside, a major reorganization of the cytoskeleton occurs by altering actin filaments, tubulins, myosins, and septins, and P. salmonis simultaneously promotes an inactive GTPase state. P. salmonis induces a significant inhibition of the antioxidant response that would promote the generation of an oxidative niche. Overall, P. salmonis alters cytoskeletal remodeling, intracellular transport, organelle organization, vesicle and endosome trafficking and early endosomal components. The antigen recognition (LPS, flagellin, etc.) is conducted by macrophage and dendritic cell (DC) PRRs, such as TLR5, DC-SIGN or CD209, C-type lectin (CD299, Mincle), and NLR. DCs play an important role in the response to P. salmonis by modulating NF-kB activation, pathogen recognition, phagocytosis and the production of cytokines and chemokines led by the IFN-mediated response that promotes Th1 polarization of T cells. At the same time, to enhance its survival in infected cells, P. salmonis upregulates IL-10 but downregulates IL-12, which promotes Th1 polarization. P. salmonis induces flagellin-dependent activation of TLR5, resulting in TNFα, IL-1β and IL-8 production. P. salmonis promotes the expression of antimicrobial peptides, such as hepcidin and cathelicidin, and acute-phase components such as haptoglobin, hemoglobin, collectrins, mannose-bound protein C, complement components (C3, C6, C9) and CD163. All these findings confirm that P. salmonis creates a specific environment that promotes their survival and replication in macrophages. During the propagation stage, the size and number of P. salmonis cells and vesicles increase, and nutrient availability is restricted, initiating a stringent response. Biosynthetic processes are increased in both P. salmonis and host cells, as are iron acquisition, iron transporter proteins and acute-phase responses. Expression of Dot/Icm T4SS genes, toxins, effector proteins, mobilome, transposons, and phage-related proteins is increased, probably in preparation for exiting the cell. Many virulence factors upregulated in both infection stages correspond to plasmid-encoded proteins, which supports the hypothesis of the importance of P. salmonis plasmids in the infective process. P. salmonis modulates the immune response to intracellular pathogens by promoting cell cycle proliferation and suppressing apoptosis and by altering vesicle trafficking and paracellular permeability. P. salmonis alters peroxisome activity as part of its infection strategy, thereby inducing an altered cellular redox balance, inflammation, and immune response. Moreover, P. salmonis reduces the rate of protein degradation by the ubiquitin proteasome system related to the response to cellular and endoplasmic reticulum stress-associated unfolded proteins as a mechanism to increase its survival within host cells. (4) Processing and presentation of antigens. T cells recognize only antigen fragments that are bound to MHC-I or MHC-II on APCs. Antigens presented by MHC-I are processed through the proteasome and transferred to the endoplasmic reticulum by a transporter associated with antigen processing (TAP) where they associate with MHC-I and are finally transported to the cell membrane. MHC-II-presenting antigens are incorporated into cells by endocytosis, digested in lysosomes and loaded onto MHC-II molecules prior to migration to the cell surface. However, P. salmonis inhibits the MHC-I pathway (mhc1, cd8, tcra, tcrb gene underexpression) but activates the MHC-II pathway (mhc2, cd4), so its strategy is to evade the CD8+ T cell-mediated immune response. P. salmonis increases the expression of important co-stimulatory molecules on the macrophage surfaces (cd80/86, cd83) and reduces the expression of zbtb46, a transcriptional factor that inhibits APCs maturation (5) T cell activation and differentiation. CD4+ T helper cells can differentiate into Th1, Th2, Th17 and Treg populations, which play different roles in the immune response. IL-12 promotes the differentiation of CD4 cells into Th1 cells to eliminate intracellular pathogens, while IL-2, IL-4, IL-13A, and IL-10 promote the differentiation of Th2 cells in response to bacteria and extracellular parasites. However, P. salmonis promotes IFN-γ production but reduces IL-12 production, reduces the expression of Th1 polarization-specific transcription factors (tbet, eomes) and, while promoting granzyme A expression (gzma), reduces perforin (mpeg1) production. In addition, P. salmonis increases the expression of Treg polarization-specific transcription factor (foxp3) suggesting the activation of an immune tolerance response.
Macrophages infected with P. salmonis induce an anti-inflammatory milieu, probably involved in the development of its bacterial virulence mechanism to ensure replication and survival (11, 12, 93). The impact of an infection depends on the balance between the ability of macrophages to recognize and destroy bacterial pathogens and the ability of pathogens to disrupt the functions of these macrophages (112). Intracellular pathogens share several mechanisms of subversion of host immune responses (116), such as: (a) evasion of host immune recognition such as modulation of microbial surfaces, secretion of immunomodulators, antigenic variation, and concealment in target cells or tissues; and (b) modulation and suppression of host immune responses such as evasion of phagocytosis, innate immune receptors, the complement system, cytokines or chemokines, inhibition of apoptosis, resistance to host effector mechanisms, and induction of inappropriate immune responses such as immunosuppression and induction of Tregs. One of the most important strategies used by P. salmonis is the evasion of phago-lysosomal degradation (55, 56, 74, 76), but it is also capable of activating a mechanism to subvert host defense by inhibiting fusion with the host lysosomal compartment and altering lysosomal pH (74). Moreover, P. salmonis can escapes into the cytosol and replicate in the host cell cytoplasm (39). P. salmonis can resist host effector mechanisms because it can persist in macrophages (63), inhibiting apoptosis (40), inhibiting oxidative stress processes, and promoting cell cycle (10, 12, 13, 78, 83). Finally, P. salmonis induces an inappropriate immune response, immunosuppression (10–13, 29), and differentiation of Tregs (94).
Upon entering macrophages through phagocytosis, P. salmonis orchestrates the formation of vacuoles called P. salmonis-containing vacuoles (PCVs), and it evades the lysosomal degradation pathway by escaping into the cytoplasm (39, 40, 74). Clathrin and the actin cytoskeleton play pivotal roles in P. salmonis internalization and multiplication, respectively (12, 95, 96). PCVs are generated because P. salmonis fully exploits disorganized and de novo synthesized actin in its favor, generating tridimensional vacuoles, apparently made exclusively of actin during the later stages of infection (95). Clathrin-mediated endocytosis is a well-documented portal of entry for many intracellular bacteria and viruses (117). Phagocytosis, the key to the life cycle of L. pneumophila and P. salmonis, is also involved in their pathogenesis. In both cases, a key part of the pathogenesis process are the genes and gene complexes that they use to secrete morphological structures (e.g., pili) or reactive proteins. Several proteins produced by L. pneumophila allow it to evade the cellular immune response and replicate inside macrophages. Some of these proteins are the virulence factor BipA/TypA and the heat shock protein ClpB, both of which have been extensively characterized (118). Isla et al. (63) showed a significant increase in the expression of ClpB and BipA proteins during infection of SHK-1 cells by P. salmonis, whereby these virulence factors would be used by the bacterium to evade cell degradation mechanisms and promote its replication inside macrophages.
Inside the cells, P. salmonis alternates between a replicative and a stationary phase in which a strict response is activated (78, 83, 84). P. salmonis induces an interruption of translation during its intracellular replication, and the Dot/Icm secretion system type IVB plays an important role during this process (83); however, there are still many unexplored genes that are active during intracellular infection of the bacterium. Global proteomic profiling to identify differentially expressed proteins in macrophage-like cells of Atlantic salmon challenged with P. salmonis at different stages of infection has been described (78, 84). These results confirm previous findings at the transcriptomic level, and they are valuable to understand the biological pathways of P. salmonis infection in vitro at the protein level, but the need to obtain this knowledge from experimental and naturally infected fish in vivo remains. A model for the infective process of P. salmonis based on a vacuolization stage and a propagation stage can be proposed from the transcriptomic and proteomic results (10, 12, 13, 30, 78, 83, 84, 93, 95–97) (Figure 2).
The stringent response is one such adaptive mechanism through which P. salmonis can survive under nutrient starvation and other related stresses (119). It has been suggested that amino acid and fatty acid starvation triggers RelA and SpoT to produce the alarmones guanosine tetra- and pentaphosphate ((p)ppGpp) in different bacteria (120). Zúñiga et al. (84) described the upregulation of these key genes during the P. salmonis stationary phase and intracellular growth, including genes encoding the two-component sensor kinase LetS, the stationary phase sigma factor RpoS, and the (p)ppGpp synthetase and/or hydrolase RelA and SpoT, which have all been shown to play an important role in regulating virulence in L. pneumophila and C. burnetii (121). During the late stages of infection, the LCV becomes disrupted, leading to bacterial egress into the cytosol. Upon nutrient depletion, RelA and SpoT are triggered, leading to increased levels of ppGpp, which triggers phenotypic transition into stationary phase (122).
Piscirickettsia salmonis Evades the Cell-Mediated Adaptive Immune Response: Is It Checkmate?
Microbial infections are characterized by a constant interplay between pathogens and hosts, with pathogens exploiting various host functions during infection and hosts reacting with appropriate defense responses. Therefore, understanding host–pathogen interactions is crucial for the development of effective vaccines and therapies. Antigen-presenting cells (APCs) such as macrophages and dendritic cells (DC) sense bacteria through pathogen recognition receptors (PRRs) activated by pathogen-associated molecular patterns (PAMPs). Among the main PRRs and PAMPs are Toll-like receptors (TLRs) and flagellin, respectively (Figure 2). P. salmonis has a complete and organized set of flagellar genes, although no structural flagellum has ever been reported for this bacterium (123). Flagellin binding to TLR5 activates the MyD88-dependent pathway, leading to the activation of IRAK-1/4 and TRAF-6 and resulting in the activation of NF-κB, which induces the expression of proinflammatory cytokines (124). Enrichment analyses of the differentially expressed genes revealed several central signatures following infection, including positive regulation of TLR5 signaling, which converged at the NF-kB level to modulate the proinflammatory cytokine response (12). Flagellin from different bacteria induces the upregulation of cathelicidin (camp) in vitro, and TLR5 is involved in the signaling pathway (125). Rozas-Serri et al. (12) observed upregulation of tlr5 and camp in fish infected with P. salmonis. Lately, Muñoz-Flores et al. (126) showed that MyD88 is an essential adaptor protein in the activation of the P. salmonis flagellin-mediated TLR5M/TLR5S signaling pathway during in vitro infection. Several in vitro and in vivo assays have suggested a possible use for flagellin as an immunostimulant or vaccine adjuvant (127). The flagellar protein FlgG of P. salmonis achieved the highest level of protection, with a relative percent survival (RPS) of 95% (21). Recently, González-Stegmaier et al. (128) demonstrated that the full recombinant flagellin B from Vibrio anguillarum (rFLA) and its recombinant D1 domain (rND1) induced the expression of genes involved in an inflammatory response quickly and for a short time and that these effects can occur when the molecules are used alone or in combination with a commercial vaccine against P. salmonis.
P. salmonis induces significant cytoskeletal reorganization but decreases lysosomal protease activity and causes the degradation of proteins associated with cellular stress (12, 95) (Figure 2). Infection with P. salmonis also delays protein transport, antigen processing, vesicle trafficking and autophagy but promotes cell survival and proliferation and inhibits apoptosis (12). As P. salmonis has been shown to undergo both replication and degradation within rainbow trout head kidney macrophages, bacterial antigens could potentially be presented by the MHC-II system (39). Alternatively, as the bacterium has been shown to inhibit the fusion of phagosomes and lysosomes, P. salmonis could remain within phagosomes for replication followed by subsequent release or escape (74).
It has been reported that IFN would promote increased expression of APC-related markers (MHC-I and MHC-II) and down-regulation of ZBTB46 in rainbow trout, which is a transcriptional factor that inhibits APC maturation (129, 130). Recently, IFN-γ has been shown to promote the expression of cell surface markers (CD80/86, CD83 and MHC-II) and decrease the expression of zbtb46 in mononuclear splenocyte subpopulations of Atlantic salmon (98). Taken together, these results would confirm that IFN-γ promoted by P. salmonis modulates the interaction between APCs and T cell polarization, and that an optimal antigen presentation process is essential for the activation of a protective CMI response. Nevertheless, it is necessary to complement these findings with the expression of the counterpart molecular components of these markers on T cells, such as CD28 and CTLA4. Furthermore, an up-regulation of the transcription factor foxp3 has been described in rainbow trout splenocytes co-cultured with both IFN-stimulated cells and cells stimulated with P. salmonis proteins (94), demonstrating an intercommunication between APCs and lymphocytes that would promote a polarization towards a Treg phenotype. However, different transcriptomics results have shown that P. salmonis effectively modulates the upregulation of MHC-II and CD4 T cells and antibody responses, but these indicators are not correlated with the protection of vaccines or better survival rates in field conditions. Moreover, P. salmonis induces a downregulation of the CMI response led by cytotoxic CD8 T cells.
Transcriptome analysis has revealed a global translation shutdown during intracellular growth of P. salmonis, and it has been proposed that intracellular P. salmonis alternates between a replicative phase and a stationary phase during which a stringent response is activated (83). In vitro and in vivo functional genomic studies of P. salmonis infection in salmon have focused on changes in coding gene expression (10, 12, 13, 30, 83, 97–99), small/microcoding RNA expression (100), and long noncoding RNA expression (96) (Table 4). In addition, Leiva et al. (89) showed a significant DNA methylation alteration in P. salmonis-infected Coho salmon with a temporal pattern during infection. The number of differentially methylated regions and the associated metabolic pathways would support the hypothesis that epigenetic changes in the genome of infected Coho salmon could be modulated by P. salmonis. Trout skeletal muscle is an immunologically active organ that can implement an early immune response against P. salmonis (101). In fact, this response could be differentially regulated by cortisol, which could lead to bacterial outbreaks in muscle under stress conditions (102, 103). A common transcriptional response associated with clathrin-mediated endocytosis and iron homeostasis has been shown in different tissues (12, 96, 97, 104, 105). Dual global transcriptomic analysis revealed a bacterial dependency on host metabolism and nutrient accessibility (30). Interestingly, genome-wide comparisons of P. salmonis revealed the absence of the biosynthetic pathway for valine, leucine, and isoleucine. When these amino acids are restricted, bacterial growth dynamics are significantly impaired, but this condition is reversed when amino acids are supplemented again. All these transcriptomic analyses have provided evidence of host biological processes, cellular components, and molecular functions during P. salmonis infection to evade the immune response, induce significant cytoskeletal reorganization, and promote intracellular survival and replication. Nucleotide-binding oligomerization domain-like receptors, or NOD-like receptors (NLRs), are intracellular receptors responsible for recognizing pathogens. Pontigo et al. (106) described two different isoforms of SsNLRC3 in P. salmonis-infected Atlantic salmon, whereby the isoform might have a different function in the recognition of bacterial ligands during infection. However, these results suggest that, similar to L. pneumophila (131), P. salmonis promotes pyroptosis, a type of programmed cell death associated with intracellular pathogen infection characterized by inflammasome formation, caspase-1 activation and proinflammatory cytokine production.
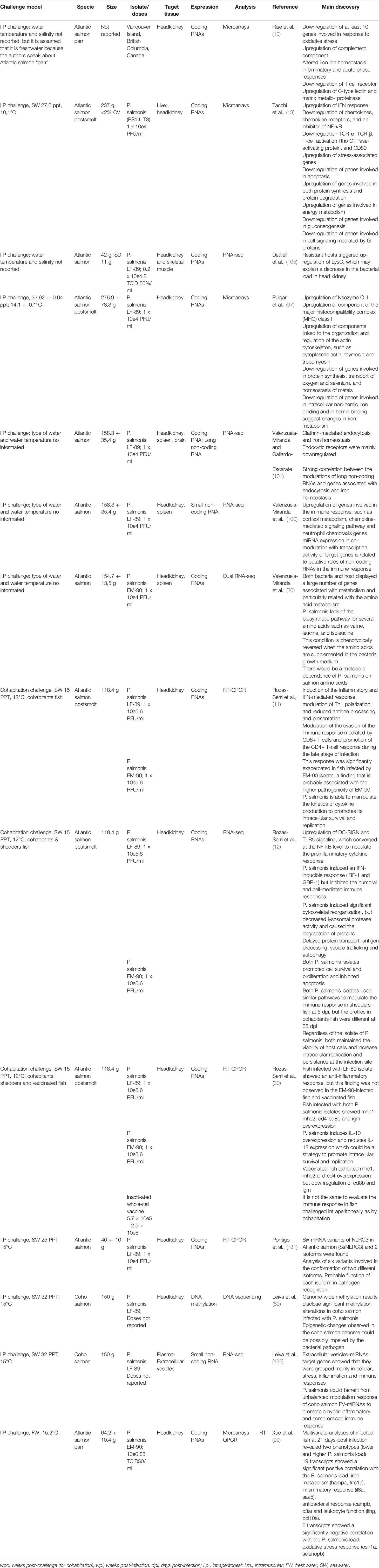
Table 4 In vivo transcriptomic studies describing innate and adaptive immune responses in Atlantic salmon and Coho salmon infected with P. salmonis.
In salmonids, the adaptive immune system consists of CMI, whose mechanism of action is to “kill” and eliminate pathogen-infected cells, and humoral immunity, which relies on antibodies to neutralize pathogens in fluids and tissues. As P. salmonis is a bacterium that replicates within host cells, it is inaccessible to neutralizing antibodies in the extracellular matrix, so cell-mediated immunity plays a key role. All T cells possess a T cell receptor (TCR) by which they recognize peptides presented by MHC, along with CD3 and costimulatory (CD28) and coinhibitory (CTLA-4) surface molecules (133). T cell-associated genes and their encoded proteins with T cell activity in fish have been well documented (134). The presence of cytotoxic T cells (CTLs) and Th cells in fish have been identified as CD8+ and CD4+ T cells, respectively (135, 136). In the case of CMI against P. salmonis, CD8 T cells should recognize infected cells by binding to MHC-I molecules expressing peptides processed from intracellular pathogens. MHC-I ligands bind to their respective TCRs on the surface of CD8+ T cells (137). In addition, CD28 costimulatory or CTLA-4 negative regulatory markers must mediate the interaction between CD8+ cells and MHC molecules (138). Upon binding to MHC-I molecules, naïve CD8 cells are activated into effector CTLs that secrete cytotoxic granules containing perforins and granzymes. Perforins form spores in target cell membranes, enabling granzymes, which are serine protease enzymes, to enter the target cells and cleave to host proteins to induce apoptosis. To execute their effector functions, CD8+ T cells are helped by CD4+ T cells.
The differentiation of naïve CD4+ T cells into Th1 cells is mediated by IL-12 and IFN-γ (139), which play crucial roles in the paracrine and autocrine modulation of APCs in the activation of CD8+ T cells against intracellular pathogens. Therefore, a better understanding of cytokine signaling that promotes CD4+ to CD8+ T-cell differentiation, as well as co-stimulatory signals between APCs and T-cells, is needed to understand the immunomodulators capable of activating a protective CMI response against P. salmonis. Antibody responses are normally observed in fish vaccinated against SRS under field conditions, but these humoral immune responses show no correlation with mortality rates and therefore fail to confer protection throughout the production cycle (1, 26, 27). However, although antibodies could react with P. salmonis before entering host cells or during cell-to-cell transmission, the real challenge is to develop protective vaccines based on CMI. Rozas-Serri et al. (11, 12) demonstrated that P. salmonis induces an IFN-inducible response (IRF-1, GBP-1, IFN), but CD4 overexpression and CD8b underexpression were observed, suggesting that P. salmonis modulates CD8+ T cell-driven evasion of CMI and promotes the CD4+ T cell response during the late phase of infection as a mechanism to escape host defenses (Figure 2). This mechanism could explain the high bacterial loads, severe pathological lesions, high cumulative mortality, and low survival in P. salmonis-infected fish (8). In general, the mechanisms used by intracellular bacteria to trigger CD8+ cell activation have not been fully elucidated in fish, as demonstrated in viral infections (140).
Viruses with economic importance in salmon farming, e.g., ISAV, IPNV, and SAV, are relatively well controlled by vaccines, including conventional vaccines such as virins; these viruses are integrated into a more global strategy that considers the genetic management of resistance, control of production conditions, epidemiological surveillance, and specific regulations for control. ISAV activates a rapid and long-lasting induction of MHC-I pathway genes in Atlantic salmon kidney cells, an effect mediated by virally induced type I IFN (141). These observations suggest that salmon type I IFN has important immunomodulatory functions in activating the MHC-I machinery in response to ISAV infection and that, unlike influenza and many other viruses, ISAV does not seem to interfere with MHC and IFN expression. This could support the thesis that salmon can mount an efficient CMI response, since the viruses are phagocytized, incorporated into antigen-presenting cells, and processed in the cytoplasm of vacuoles and their immunogenic structures are submitted by MHC-I to T cells to finally differentiate into CD8+ T cells. So, what happens to P. salmonis if it also replicates within vacuoles in the cell’s cytoplasm? When the bacterium is alive and infects fish, it is plausible that the bacterium’s virulence factors support its strategy to escape antigen processing and/or that MHC-I does not present the antigens efficiently, and the bacterium finally succeeds in evading the fish’s response mechanisms. The next question would then be whether this is the reason why fish are also unable to activate a response against a bacterin-like vaccine or even a live attenuated vaccine. Hence, elucidating which P. salmonis proteins are involved in altering the processing and/or presentation of its antigens in host cells and how they act could provide information to confirm the hypothesis that the bacterium is able to alter this cellular mechanism to evade CMI.
This could presumably be because of the selective pressure exerted by the immune system, many viruses have evolved proteins that interfere with antigen presentation by MHC-I molecules (142). Viral proteins have been characterized to exploit bottlenecks in the MHC-I pathway, such as peptide translocation by transporters associated with antigen processing (143). Alternatively, viral proteins can cause the degradation or mislocalization of MHC-I molecules. This is often achieved by the subversion of the host cell’s own protein degradation and trafficking pathways. Antigen processing and presentation by MHC molecules is a cornerstone in vertebrate immunity. Six different MHC-I lineages have been described in teleosts: U, Z, S, L, P, and H (144, 145). Although structurally similar to classical MHC-I molecules, all belong to the U lineage (Sasa-uba), and many nonclassical/MHC-I-like molecules (L lineage genes Sasa-lda, Sasa-lia, Sasa-lca, Sasa-lfa, Sasa-lga, and Sasa-lha) have functions other than peptide presentation, ranging from host homeostasis to immune regulation (146). Using two separate in vivo challenge models in Atlantic salmon with different kinetics and immune pathologies combined with in vitro stimulation using viral and bacterial TLR ligands, Svenning et al. (147) showed that de novo synthesis of different L lineage genes is distinctly regulated in response to different types of immune challenges. In this way, while salmonid alpha virus 3 (SAV3) strongly induced the expression of lia, lga and, to a lesser extent, lha, but not lda, lca or lfa, infection by P. salmonis resulted predominantly in positive regulation from lga. The induction of lca, which is predominantly expressed in primary and secondary lymphoid tissues, was marginal except for a transient upregulation in the pancreas following SAV3 challenge.
Vaccination Against P. salmonis: Mission Possible?
Biosecurity, vaccination, and selective breeding for disease resistance are the main tools for controlling infectious diseases in aquaculture. However, host genetics and vaccines provide only partial protection, raising concerns about their effectiveness in the field. Pathogens can be divided into extracellular, facultative intracellular, and obligate intracellular pathogens. Intracellular pathogens pose special challenges to the immune system because regardless of whether they are exposed extracellularly, they cannot be contacted directly by immune cells or by humoral factors such as antibodies. The aim of vaccination is to train the adaptive immune system of fish by exposure to pathogenic antigens so that upon subsequent exposure, the immune system mounts a rapid and long-term protective response against the same pathogen. The success of vaccination against bacterial diseases in fish is mainly attributed to vaccines targeting extracellular pathogens, as they activate antibody-mediated humoral adaptive immunity (148). However, circulating antibodies do not protect against P. salmonis infection or disease development, because P. salmonis is a facultative intracellular bacterium. Stimulation of the antigen presentation and processing process of P. salmonis is therefore the initial target of vaccines, which then aim to modulate immunological memory; but it is precisely these processes that constitute one of the current limitations in fish. Although IFN-γ has been reported to promote the expression of APC surface markers that are key in co-stimulating the signal for T-cell activation and Th1 polarization, it would also promote polarization towards Treg (94), indicating an immune tolerance profile that needs further investigation because it could be the cause of inhibition of the inflammatory response to promote an anti-inflammatory response that determines persistent infection and chronic disease course in fish. It is not known whether an increased inflammatory response driven by highly destructive proinflammatory macrophages, although they are an important part of Th1 polarization, could drive a CMI capable of controlling P. salmonis evasion mechanisms. On contrary, exacerbated activation of the IFN-γ-mediated inflammatory response (11, 12) appears to increase the susceptibility of infected fish, culminating in progression of the response from an acute to a chronic stage and resulting in increased intracellular replication of P. salmonis, high bacterial loads, serious pathological lesions and low survival (8). In addition, Iliev et al. (149) showed that the potent immunostimulatory properties of a TLR ligand would not necessarily translate into enhanced APC functions and highlight the complexity of the activation of fish immune cells by TLR ligands.
That said, there is a need to modulate and activate the CMI response, but available vaccines do not trigger effective antigen presentation through MHC-I, and there is a lack of adequate activation and expansion of T cells, especially CD8+ cytotoxic T cells (29, 38). The efficacy of vaccines reported from experimental challenges has always been acceptable (1, 25), but current vaccination strategies under field conditions, whether based on replicating or nonreplicating vaccines, have shown only a transient activation of the humoral immune response and specifically of the CMI response, which are not strong or long-lasting enough to achieve effective control of SRS (1, 26, 27, 29, 38). Based on current understanding of the limitations of vaccines for SRS control and the intracellular nature of P. salmonis, other vaccines may be advantageous. A new immunoinformatics-based strategy to design vaccines using epitopes of antigenic proteins from bacteria and viruses that directly promote T- and B-cell activation and differentiation has been used in humans. Although it is not yet a popular strategy in fish, the design and development of multi-epitope vaccines against Flavobacterium columnare (150), E. tarda y F. columnare (151), Streptococcus agalactiae (152), and E. ictaluri (153) have been reported. This in silico strategy was used by a consortium of Chilean entities to design a multi-epitope chimeric vaccine targeting different epitopes of P. salmonis.
The variable and relative protection of current vaccines against SRS under field conditions could be a consequence of different environmental variables (1), the choice of the vaccination strategy (26, 27, 137), coinfection with other pathogens such as sea lice (154) or the variation in genetic resistance to SRS among families (155). In this way, different factors related to fish production can lead to increased susceptibility, coinfection, and mortality. Everson et al. (156) showed the importance of environmental factors, host genetics, and vaccination in relation to infection and mortality related to infectious diseases. However, the main reason would be related to insufficient and/or transient activation of the CMI response induced by conventional vaccines (1, 10–13, 25, 29, 38, 97, 105). Thus, translating recent knowledge about fish-P. salmonis interactions, specifically about the immune response and its pathogenesis, into better vaccines or better vaccination strategies is not an easy task and remains one of the main challenges in fish vaccinology.
The kinetics of the antibody response are transient after freshwater vaccination, and antibody levels begin to decline approximately 1800 days postvaccination (22). However, the mechanisms that could explain this fact are the natural challenge and exposure of fish to P. salmonis and, consequently, the consumption of antibodies, as has been observed in salmon vaccinated with IPN (140). These results supported the idea of complementing the classical parenteral vaccination strategy with the application of a vaccine booster administered orally before the fall of the antibody titer (22). However, the field results obtained from several generations of farmed salmon were not as expected, and this strategy did not contribute significantly to disease control (1, 25, 29, 38). The intraperitoneal route is usually chosen for the administration of vaccines in salmon, but there is little knowledge about its immune response. Recently, a significant increase in leukocytes, total IgM antibody-secreting cells (ASCs) and P. salmonis-specific ASCs in the peritoneal cavity at 3 and 6 weeks after infection with P. salmonis has been described (157). Hence, the authors suggest a putative role for adipose tissue in the peritoneal cavity immune response.
The microbiome plays an important role in the maturation of the vertebrate adaptive immune system and stimulation of the immune response and can directly enhance the host pathogen defense via colonization resistance and the production of inhibitory compounds (158). Complementarily, the mucosal immune response based on IgT+ B cells and secreted IgT plays a key role, as IgT is highly induced by pathogens on mucosal surfaces and coats much of the fish microbiota (34, 159, 160). However, there is an important gap regarding how to complement systemic immunity against P. salmonis with advances in the knowledge of activating local immunity at the site(s) of P. salmonis entry, such as the gills and skin, as well as the gut, which could help limit the success of infection.
Complementarily, selective breeding for improved resistance to infectious diseases is a potentially a more sustainable strategy for the long-term control of disease outbreaks in aquaculture (161), especially when vaccination strategies have not been as effective as expected. Previous studies have demonstrated significant genetic variation for resistance to P. salmonis in Atlantic salmon (h2 = 0.11 to 0.41), rainbow trout (h2 = 0.45 to 0.62) and Coho salmon (h2 = 0.16) using disease challenge data (162–167). There is an important genetic component of SRS resistance in Atlantic salmon supported by a polygenic architecture. When comparing the response between fish with high and low resistance to P. salmonis, changes in the expression of genes related to the cytoskeleton, apoptosis and cell survival, bacterial invasion/intracellular trafficking and the inflammasome are observed, which correlate with genetic resistance (168). Thus, the possible mechanisms leading to genetic resistance are likely heterogeneous and vary among different families and individuals.
One SNP related to B cell development was identified as a potential functional candidate associated with resistance to P. salmonis defined as days to death (162). B cells are critically important in the humoral immune response, but they are also dedicated phagocytes, so the latter innate function could be important in the immune response against P. salmonis in Coho salmon. Early growth is positively correlated genetically with resistance to P. salmonis, measured by the day to death and by the harvest weight in Coho salmon, so selective breeding for early growth could indirectly improve the harvest weight and resistance to P. salmonis in the population (169).
According to Figueroa et al. (155), the between-familiar variation is a strong intrinsic factor that determines the variable protection of vaccines for SRS. In some full-sibling families, the added protection by vaccination increased the survival time compared to their unvaccinated siblings; in other families, there was no added protection by vaccination. However, further studies are needed to assess whether variations in the host immune response to vaccination for SRS control could explain the differences in resistance observed among vaccinated fish and what biological mechanisms could explain it, because vaccination and selective breeding have always been considered supplementary strategies in disease control in aquaculture. In addition, although cohabitation challenge models provide little information on whether fish with higher genetic resistance and/or vaccinated fish are less likely to transmit infection when infected, they are more suitable models than intraperitoneal challenge models for evaluating disease resistance and/or vaccine efficacy.
Mortality is the main phenotype for disease resistance in fish, but there is emerging evidence that directly targeting survival by breeding or vaccination without considering the epidemiological effects does not reduce disease transmission or the disease prevalence at the population level (170–172). Three key epidemiological host traits affect the infectious disease prevalence and population mortality rates are susceptibility, infectivity, and endurance (170). Chase-Topping et al. (173) reported that both vaccination and genetic selection reduce but do not prevent ISA transmission. The authors suggest that it would be very beneficial to assess the combined effects of genetic selection and vaccination on ISA transmission and to evaluate the mucus viral load as a potential in vivo indicator for individual’s infectivity. These results should be applied to SRS transmission experiments to understand the effects of vaccination strategies and genetic resistance on SRS control under field conditions. Similarly, since host genetics and vaccines provide only partial protection and raise questions about their efficacy in the field, the control plan for SRS must consider the epidemiological edge, as little is known about the impact of genetic management and/or vaccines on the transmission and prevalence of SRS. Based on this scenario, it could be hypothesized that relative control of SRS is based on regulatory and production changes designed and implemented by industry due to an evolving public–private roadmap, which includes vaccination strategies but also a low relative contribution.
Concluding Remarks and Future Avenues
Infectious diseases caused by intracellular bacteria in different farmed fish species worldwide represent a great challenge, especially for their control by vaccination and genetic resistance to disease. One of these diseases is SRS, which has been described in several salmon-producing countries, but which especially affects Chilean salmon farming. In practice, given the industry’s collective imperative need to advance in the control of SRS in order to increase its long-term sustainability and competitiveness, it seems a frivolity to discuss whether P. salmonis is a pathogenic or environmental bacterium (174, 175), since, in either case, reality shows us that: (1) P. salmonis is a very complex bacterium that we need to more about, especially its interaction with host cells; (2) SRS is the main infectious disease of Chilean salmon farming, generating economic losses conservatively estimated at approximately 700 million dollars per year; and (3) an average of 93.28% of total antibiotics used in the Chilean salmon industry in the last 5 years has been only for the control of SRS. That said, it is undoubtedly the main mission of all members of the salmon industry to continue working together to fill the knowledge gaps that will allow us to optimize the control of SRS; with a specific focus on the interaction of the components of the epidemiological triad: the pathogen, the fish and the aquatic environment (including production management).
Significant knowledge has been generated in recent years on different virulence factors of P. salmonis that could explain the difficulties in its control under field conditions: (1) Sec-dependent pathway and Type 4 Dot/Icm secretion system; (2) expression of ClpB and BipA proteins; (3) siderophore production; (4) genes for vibroferrin biosynthesis; (5) pathogenicity islands; (6) membrane vesicles; (7) large cryptic plasmids; (8) Sec dependent pathway; (9) type IV pili and biofilms; and (10) expression of virulence-associated genes during stringent response, among others. However, more specific knowledge is still required regarding the mechanisms of action of the virulence factors of P. salmonis that give it a high capacity to evade the host response. New methods in genetics and molecular biology have contributed to the in vitro study of the basic biology of P. salmonis. Although not all genes of pathogenic microorganisms play a role in their pathogenicity or virulence, nor can they be confirmed only in a cell culture invasion model. Methods such as subcellular fractionation and single-molecule super-resolution microscopy should be applied to tag molecules inside bacteria and host cells to follow their fate during early and late infection, and even to find P. salmonis genes or proteins that are activated only when they are inside cells.
Although effective control of infectious diseases in aquaculture requires the integration of several management measures and interventions, there is a consensus that the availability of an effective vaccine(s) should be one of the most important pillars to support control. However, although there is evidence that efforts have been made in the development of vaccines against SRS, unfortunately, the objective has not yet been achieved, and the SRS control plan basically lacks an effective vaccine. Existing evidence shows that the production of CMI-based vaccines would be the most effective approach to reduce the prevalence and severity of SRS outbreaks. Given that this is one of the most virulent bacterial pathogens for salmon, continued research on how P. salmonis evades fish adaptive responses is necessary to lead to new insights that will improve our understanding of host-pathogen interactions.
Basically, the strategy for developing effective vaccines against P. salmonis must change, starting with understanding the biological pathways that the bacterium uses to infect and replicate in host cells and then defining strategies to block them. The conventional approach historically used for vaccine development against P. salmonis has not been the solution to the problem, which is why salmon producers have no other alternatives in the field when faced with SRS outbreaks other than to use the last card they have available, chemotherapy. Consequently, the time has come for new generation vaccines and reverse vaccinology in aquaculture: DNA, mRNA, viral vectors, multiepitope and multiantigenic chimeric vaccines, among others. However, it is necessary to generate deeper and more specific knowledge about the interaction between P. salmonis and fish cells and, of course, to generate the appropriate regulatory instances for the formal registration of these new vaccines. The governmental agencies in charge of regulating the vaccine registration system and the control of its application in field conditions must advance as fast as the new knowledge and products from research and innovation are generated. It is not prudent to develop new generation vaccines without a regulatory structure in place to authorize and regulate their commercial use in advance.
From a bacterial-host cell interaction point of view, more is known about P. salmonis-induced changes in cell invasion and intracellular survival, but we are only beginning to understand how the immune system detects and responds to these changes. Considering these new data, several important gaps emerge: (1) Does the role of the cytoskeleton in cell-autonomous immunity differ between phagocytes or epithelial cells? Further study is required to determine how cytoskeletal components in cell-autonomous immunity—actin, microtubules, intermediate filaments and septins—work together during P. salmonis infection. (2) Immunological speaking, more specific knowledge should be promoted to understand the mechanisms of processing and presentation of P. salmonis antigens in APCs, macrophages and DCs, to T-cells. Further knowledge about fish macrophages and DCs biology is needed to better understand how P. salmonis modulates their biology to its own advantage. Does P. salmonis prevent the presentation of its antigens by MHC-I? Do APCs activate T cells in the quantity and quality necessary to detect and eliminate P. salmonis-infected cells? How do APCs activate salmon T cells, and how do we quantitatively define the efficacy of the T cell activation process to be considered a protective response? What is the role of B cells as phagocytes and professional antigen-presenting cells in P. salmonis infection? How can the in vivo mechanism of action of the main virulence factors of P. salmonis be blocked? What is the mucosal immune response against P. salmonis and the role of the microbiota?. (3) What is the genetic variability of fish in terms of their immune response against P. salmonis? How do genetic resistance management and vaccines complement each other in the control of SRS? Why do these two strategies not reduce the transmission and prevalence of SRS? Which epidemiological variables are we not considering in the control of SRS, or which variables are we not giving enough relevance to? The million-dollar question will remain whether effective control of SRS will be achieved even if vaccines that trigger effective CMI responses are developed. The challenges of inducing CMI responses against P. salmonis are considerable, as are the challenges presented by other intracellular pathogens in aquaculture, such as Francisella, Renibacterium and Edwarsiella. The importance of CMI, which consists of the production of natural killer cells, CD8 cytotoxic T cells and CD4+ Th1 cells, is critical for protection against P. salmonis through proper activation of mononuclear phagocytes. Strategies to increase resistance to the bacterium could focus on altering its modulation of cellular homeostasis (cytoskeleton, apoptosis, or cell cycle progression) or enhancing immune processes that prevent or slow infection (inflammasome, antigen recognition and presentation, T cell activation and proliferation, increased cytotoxic capacity of CD8+ T cells, oxidative stress), among other pathways. These strategies may include knockout or CRISPR/Cas modulation in cell line models or, ultimately, in vivo to interrogate the impact of disruption of identified genes on genetic resistance.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
The author declare that this study received funding from Pathovet Labs. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Conflict of Interest
Author MR-S was employed by the company Pathovet Labs.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Rozas M, Enriquez R. Piscirickettsiosis and Piscirickettsia Salmonis in Fish: A Review. J Fish Dis (2014) 37:163–88. doi: 10.1111/jfd.12211
2. Sernapesca. Informe Sanitario De La Salmonicultura En Centros Marinos Año 2020. Valparaíso, Chile (2021).
3. Bravo F, Sidhu JPS, Bernal P, Bustamante RH, Condie S, Gorton B, et al. Hydrodynamic Connectivity, Water Temperature, and Salinity are Major Drivers of Piscirickettsiosis Prevalence and Transmission Among Salmonid Farms in Chile. Aquacult Environ Interact (2020) 12:263–79. doi: 10.3354/aei00368
4. Wada M, Lam CT, Rosanowski S, Patanasatienkul T, Price D, St-Hilaire S. Development of Simulation Models for Transmission of Salmonid Rickettsial Septicaemia Between Salt Water Fish Farms in Chile. Transbound Emerg Dis (2021) 68:1586–600. doi: 10.1111/tbed.13830
5. Sernapesca, Informe Sobre Uso De Antimicrobianos En La Salmonicultura Nacional Año 2020. (2021).
6. Almendras FE, Jones SR, Fuentealba C, Wright GM. In Vitro Infection of a Cell Line From Ictalurus Nebulosus With Piscirickettsia Salmonis. Can J Vet Res (1997) 61:66–8.
7. Smith PA, Pizarro P, Ojeda P, Contreras J, Oyanedel S, Larenas J. Routes of Entry of Piscirickettsia Salmonis in Rainbow Trout Oncorhynchus Mykiss. Dis Aquat Organ (1999) 37:165–72. doi: 10.3354/dao037165
8. Rozas-Serri M, Ildefonso R, Pena A, Enriquez R, Barrientos S, Maldonado L. Comparative Pathogenesis of Piscirickettsiosis in Atlantic Salmon (Salmo Salar L.) Post-Smolt Experimentally Challenged With LF-89-Like and EM-90-Like Piscirickettsia Salmonis Isolates. J Fish Dis (2017) 40:1451–72. doi: 10.1111/jfd.12671
9. Levipan HA, Avendano-Herrera R. Assessing the Impacts of Skin Mucus From Salmo Salar and Oncorhynchus Mykiss on the Growth and In Vitro Infectivity of the Fish Pathogen Piscirickettsia Salmonis. J Fish Dis (2021) 44:181–90. doi: 10.1111/jfd.13275
10. Rise ML, Jones SR, Brown GD, von Schalburg KR, Davidson WS, Koop BF. Microarray Analyses Identify Molecular Biomarkers of Atlantic Salmon Macrophage and Hematopoietic Kidney Response to Piscirickettsia Salmonis Infection. Physiol Genomics (2004) 20:21–35. doi: 10.1152/physiolgenomics.00036.2004
11. Rozas-Serri M, Pena A, Arriagada G, Enriquez R, Maldonado L. Comparison of Gene Expression in Post-Smolt Atlantic Salmon Challenged by LF-89-Like and EM-90-Like Piscirickettsia Salmonis Isolates Reveals Differences in the Immune Response Associated With Pathogenicity. J Fish Dis (2018) 41:539–52. doi: 10.1111/jfd.12756
12. Rozas-Serri M, Pena A, Maldonado L. Transcriptomic Profiles of Post-Smolt Atlantic Salmon Challenged With Piscirickettsia Salmonis Reveal a Strategy to Evade the Adaptive Immune Response and Modify Cell-Autonomous Immunity. Dev Comp Immunol (2018) 81:348–62. doi: 10.1016/j.dci.2017.12.023
13. Tacchi L, Bron JE, Taggart JB, Secombes CJ, Bickerdike R, Adler MA, et al. Multiple Tissue Transcriptomic Responses to Piscirickettsia Salmonis in Atlantic Salmon (Salmo Salar). Physiol Genomics (2011) 43:1241–54. doi: 10.1152/physiolgenomics.00086.2011
14. Meza K, Inami M, Dalum AS, Bjelland AM, Sorum H, Lovoll M. Development of Piscirickettsiosis in Atlantic Salmon (Salmo Salar L.) Smolts After Intraperitoneal and Cohabitant Challenge Using an EM90-Like Isolate: A Comparative Study. J Fish Dis (2019) 42:1001–11. doi: 10.1111/jfd.13004
15. Long A, Goodall A, Jones SRM. Development of a Piscirickettsia Salmonis Immersion Challenge Model to Investigate the Comparative Susceptibility of Three Salmon Species. J Fish Dis (2021) 44:1–9. doi: 10.1111/jfd.13261
16. Smith PA, Contreras JR, Larenas JJ, Aguillon JC, Garces LH, Perez B, et al. Immunization With Bacterial Antigens: Piscirickettsiosis. Dev Biol Stand (1997) 90:161–6.
17. Kuzyk MA, Burian J, Thornton JC, Kay WW. OspA, a Lipoprotein Antigen of the Obligate Intracellular Bacterial Pathogen Piscirickettsia Salmonis. J Mol Microbiol Biotechnol (2001) 3:83–93.
18. Miquel A, Muller I, Ferrer P, Valenzuela PD, Burzio LO. Immunoresponse of Coho Salmon Immunized With a Gene Expression Library From Piscirickettsia Salmonis. Biol Res (2003) 36:313–23.
19. Birkbeck TH, Rennie S, Hunter D, Laidler LA, Wadsworth S. Infectivity of a Scottish Isolate of Piscirickettsia Salmonis for Atlantic Salmon Salmo Salar and Immune Response of Salmon to This Agent. Dis Aquat Organ (2004) 60:97–103.
20. Wilhelm V, Soza C, Martinez R, Rosemblatt M, Burzio LO, Valenzuela PD. Production and Immune Response of Recombinant Hsp60 and Hsp70 From the Salmon Pathogen Piscirickettsia Salmonis. Biol Res (2005) 38:69–82.
21. Wilhelm V, Miquel A, Burzio LO, Rosemblatt M, Engel E, Valenzuela S, et al. A Vaccine Against the Salmonid Pathogen Piscirickettsia Salmonis Based on Recombinant Proteins. Vaccine (2006) 24:5083–91. doi: 10.1016/j.vaccine.2006.03.027
22. Tobar I, Arancibia S, Torres C, Vera V, Soto P, Carrasco C, et al. Successive Oral Immunizations Against Piscirickettsia Salmonis and Infectious Salmon Anemia Virus are Required to Maintain a Long-Term Protection in Farmed Salmonids. Front Immunol (2015) 6:244. doi: 10.3389/fimmu.2015.00244
23. Caruffo M, Vidal S, Santis L, Siel D, Perez O, Huenchullan PR, et al. Effectiveness of a Proteoliposome-Based Vaccine Against Salmonid Rickettsial Septicaemia in Oncorhynchus Mykiss. Vet Res (2021) 52:111.
24. Pontigo JP, Espinoza C, Hernandez M, Nourdin G, Oliver C, Avendano-Herrera R, et al. Protein-Based Vaccine Protect Against Piscirickettsia Salmonis in Atlantic Salmon (Salmo Salar). Front Immunol (2021) 12:602689.
25. Maisey K, Montero R, Christodoulides M. Vaccines for Piscirickettsiosis (Salmonid Rickettsial Septicaemia, SRS): The Chile Perspective. Expert Rev Vaccines (2017) 16:215–28. doi: 10.1080/14760584.2017.1244483
26. Happold J, Sadler R, Meyer A, Hillman A, Cowled B, Mackenzie C, et al. Effectiveness of Vaccination for the Control of Salmonid Rickettsial Septicaemia in Commercial Salmon and Trout Farms in Chile. Aquaculture (2020) 520:734968. doi: 10.1016/j.aquaculture.2020.734968
27. Jakob E, Stryhn H, Yu J, Medina MH, Rees EE, Sanchez J, et al. Epidemiology of Piscirickettsiosis on Selected Atlantic Salmon (Salmo Salar) and Rainbow Trout (Oncorhynchus Mykiss) Salt Water Aquaculture Farms in Chile. Aquaculture (2014) 433:288–94. doi: 10.1016/j.aquaculture.2014.06.018
28. Tobar JA, Jerez S, Caruffo M, Bravo C, Contreras F, Bucarey SA, et al. Oral Vaccination of Atlantic Salmon (Salmo Salar) Against Salmonid Rickettsial Septicaemia. Vaccine (2011) 29:2336–40. doi: 10.1016/j.vaccine.2010.12.107
29. Rozas-Serri M, Pena A, Maldonado L. Gene Expression Associated With Immune Response in Atlantic Salmon Head-Kidney Vaccinated With Inactivated Whole-Cell Bacterin of Piscirickettsia Salmonis and Pathogenic Isolates. Fish Shellfish Immunol (2019) 93:789–95. doi: 10.1016/j.fsi.2019.08.031
30. Valenzuela-Miranda D, Gallardo-Escarate C. Dual RNA-Seq Uncovers Metabolic Amino Acids Dependency of the Intracellular Bacterium Piscirickettsia Salmonis Infecting Atlantic Salmon. Front Microbiol (2018) 9:2877. doi: 10.3389/fmicb.2018.02877
31. Rozas-Serri M, Lobos C, Correa R, Ildefonso R, Vasquez J, Munoz A, et al. Atlantic Salmon Pre-Smolt Survivors of Renibacterium Salmoninarum Infection Show Inhibited Cell-Mediated Adaptive Immune Response and a Higher Risk of Death During the Late Stage of Infection at Lower Water Temperatures. Front Immunol (2020) 11:1378. doi: 10.3389/fimmu.2020.01378
32. Yamasaki M, Araki K, Nakanishi T, Nakayasu C, Yoshiura Y, Iida T, et al. Adaptive Immune Response to Edwardsiella Tarda Infection in Ginbuna Crucian Carp, Carassius Auratus Langsdorfii. Vet Immunol Immunopathol (2013) 153:83–90. doi: 10.1016/j.vetimm.2013.02.004
33. Munang’andu HM, Evensen O. A Review of Intra- and Extracellular Antigen Delivery Systems for Virus Vaccines of Finfish. J Immunol Res (2015) 2015:960859. doi: 10.1155/2015/960859
34. Xu Z, Parra D, Gomez D, Salinas I, Zhang YA, von Gersdorff Jorgensen L, et al. Teleost Skin, an Ancient Mucosal Surface That Elicits Gut-Like Immune Responses. Proc Natl Acad Sci U.S.A. (2013) 110:13097–102. doi: 10.1073/pnas.1304319110
35. Yamasaki M, Araki K, Nakanishi T, Nakayasu C, Yamamoto A. Role of CD4(+) and CD8alpha(+) T Cells in Protective Immunity Against Edwardsiella Tarda Infection of Ginbuna Crucian Carp, Carassius Auratus Langsdorfii. Fish Shellfish Immunol (2014) 36:299–304. doi: 10.1016/j.fsi.2013.11.016
36. Abram QH, Dixon B, Katzenback BA. Impacts of Low Temperature on the Teleost Immune System. Biol (Basel) (2017) 6:39. doi: 10.3390/biology6040039
37. Sanhueza N, Fuentes R, Aguilar A, Carnicero B, Vega K, Munoz D, et al. Behavioural Fever Promotes an Inflammatory Reflex Circuit in Ectotherms. Int J Mol Sci (2021) 22:8860. doi: 10.3390/ijms22168860
38. Vargas D, Vallejos-Vidal E, Reyes-Cerpa S, Oyarzun-Arrau A, Acuna-Castillo C, Imarai M, et al. The Analysis of Live-Attenuated Piscirickettsia Salmonis Vaccine Reveals the Short-Term Upregulation of Innate and Adaptive Immune Genes in Atlantic Salmon (Salmo Salar): An In Situ Open-Sea Cages Study. Microorganisms (2021) 9:703. doi: 10.3390/microorganisms9040703
39. McCarthy UM, Bron JE, Brown L, Pourahmad F, Bricknell IR, Thompson KD, et al. Survival and Replication of Piscirickettsia Salmonis in Rainbow Trout Head Kidney Macrophages. Fish Shellfish Immunol (2008) 25:477–84. doi: 10.1016/j.fsi.2008.07.005
40. Rojas V, Galanti N, Bols NC, Marshall SH. Productive Infection of Piscirickettsia Salmonis in Macrophages and Monocyte-Like Cells From Rainbow Trout, a Possible Survival Strategy. J Cell Biochem (2009) 108:631–7. doi: 10.1002/jcb.22295
41. Yanez AJ, Silva H, Valenzuela K, Pontigo JP, Godoy M, Troncoso J, et al. Two Novel Blood-Free Solid Media for the Culture of the Salmonid Pathogen Piscirickettsia Salmonis. J Fish Dis (2013) 36:587–91. doi: 10.1111/jfd.12034
42. Cortes MP, Mendoza SN, Travisany D, Gaete A, Siegel A, Cambiazo V, et al. Analysis of Piscirickettsia Salmonis Metabolism Using Genome-Scale Reconstruction, Modeling, and Testing. Front Microbiol (2017) 8:2462. doi: 10.3389/fmicb.2017.02462
43. Pulgar R, Travisany D, Zuniga A, Maass A, Cambiazo V. Complete Genome Sequence of Piscirickettsia Salmonis LF-89 (ATCC VR-1361) a Major Pathogen of Farmed Salmonid Fish. J Biotechnol (2015) 212:30–1. doi: 10.1016/j.jbiotec.2015.07.017
44. Nourdin-Galindo G, Sanchez P, Molina CF, Espinoza-Rojas DA, Oliver C, Ruiz P, et al. Comparative Pan-Genome Analysis of Piscirickettsia Salmonis Reveals Genomic Divergences Within Genogroups. Front Cell Infect Microbiol (2017) 7:459. doi: 10.3389/fcimb.2017.00459
45. Ortiz-Severin J, Travisany D, Maass A, Chavez FP, Cambiazo V. Piscirickettsia Salmonis Cryptic Plasmids: Source of Mobile DNA and Virulence Factors. Pathogens (2019) 8:269. doi: 10.3390/pathogens8040269
46. Mauel MJ, Giovannoni SJ, Fryer JL. Phylogenetic Analysis of Piscirickettsia Salmonis by 16S, Internal Transcribed Spacer (ITS) and 23S Ribosomal DNA Sequencing. Dis Aquat Organ (1999) 35:115–23. doi: 10.3354/dao035115
47. Bohle H, Henriquez P, Grothusen H, Navas E, Sandoval A, Bustamante F, et al. Comparative Genome Analysis of Two Isolates of the Fish Pathogen Piscirickettsia Salmonis From Different Hosts Reveals Major Differences in Virulence-Associated Secretion Systems. Genome Announc (2014) 2:e01219-14. doi: 10.1128/genomeA.01219-14
48. Bravo C, Martinez V. Whole-Genome Comparative Analysis of the Pathogen Piscirickettsia Salmonis. Vet Microbiol (2016) 196:36–43. doi: 10.1016/j.vetmic.2016.10.015
49. Mandakovic D, Glasner B, Maldonado J, Aravena P, Gonzalez M, Cambiazo V, et al. Genomic-Based Restriction Enzyme Selection for Specific Detection of Piscirickettsia Salmonis by 16S rDNA PCR-RFLP. Front Microbiol (2016) 7:643. doi: 10.3389/fmicb.2016.00643
50. Otterlei A, Brevik OJ, Jensen D, Duesund H, Sommerset I, Frost P, et al. Phenotypic and Genetic Characterization of Piscirickettsia Salmonis From Chilean and Canadian Salmonids. BMC Vet Res (2016) 12:55. doi: 10.1186/s12917-016-0681-0
51. Aravena P, Pulgar R, Ortiz-Severin J, Maza F, Gaete A, Martinez S, et al. PCR-RFLP Detection and Genogroup Identification of Piscirickettsia Salmonis in Field Samples. Pathogens (2020) 9:358. doi: 10.3390/pathogens9050358
52. Isla A, Martinez-Hernandez JE, Levipan HA, Haussmann D, Figueroa J, Rauch MC, et al. Development of a Multiplex PCR Assay for Genotyping the Fish Pathogen Piscirickettsia Salmonis Through Comparative Genomics. Front Microbiol (2021) 12:673216. doi: 10.3389/fmicb.2021.673216
53. Saavedra J, Hernandez N, Osses A, Castillo A, Cancino A, Grothusen H, et al. Prevalence, Geographic Distribution and Phenotypic Differences of Piscirickettsia Salmonis EM-90-Like Isolates. J Fish Dis (2017) 40:1055–63. doi: 10.1111/jfd.12581
54. Isla A, Saldarriaga-Cordoba M, Fuentes DE, Albornoz R, Haussmann D, Mancilla-Schulz J, et al. Multilocus Sequence Typing Detects New Piscirickettsia Salmonis Hybrid Genogroup in Chilean Fish Farms: Evidence for Genetic Diversity and Population Structure. J Fish Dis (2019) 42:721–37. doi: 10.1111/jfd.12976
55. Perez-Stuardo D, Morales-Reyes J, Tapia S, Ahumada DE, Espinoza A, Soto-Herrera V, et al. Non-Lysosomal Activation in Macrophages of Atlantic Salmon (Salmo Salar) After Infection With Piscirickettsia Salmonis. Front Immunol (2019) 10:434. doi: 10.3389/fimmu.2019.00434
56. Perez-Stuardo D, Espinoza A, Tapia S, Morales-Reyes J, Barrientos C, Vallejos-Vidal E, et al. Non-Specific Antibodies Induce Lysosomal Activation in Atlantic Salmon Macrophages Infected by Piscirickettsia Salmonis. Front Immunol (2020) 11:544718. doi: 10.3389/fimmu.2020.544718
57. Caruffo M, Mandakovic D, Mejias M, Chavez-Baez I, Salgado P, Ortiz D, et al. Pharmacological Iron-Chelation as an Assisted Nutritional Immunity Strategy Against Piscirickettsia Salmonis Infection. Vet Res (2020) 51:134. doi: 10.1186/s13567-020-00845-2
58. Diaz R, Troncoso J, Jakob E, Skugor S. Limiting Access to Iron Decreases Infection of Atlantic Salmon SHK-1 Cells With Bacterium Piscirickettsia Salmonis. BMC Vet Res (2021) 17:155. doi: 10.1186/s12917-021-02853-6
59. Vadovic P, Fodorova M, Toman R. Structural Features of Lipid A of Piscirickettsia Salmonis, the Etiological Agent of the Salmonid Rickettsial Septicemia. Acta Virol (2007) 51:249–59.
60. Fodorova M, Vadovic P, Toman R. Structural Features of Lipid A of Rickettsia Typhi. Acta Virol (2011) 55:31–44.
61. Vinogradov E, Frimmelova M, Toman R. Chemical Structure of the Carbohydrate Backbone of the Lipopolysaccharide From Piscirickettsia Salmonis. Carbohydr Res (2013) 378:108–13.
62. Figueroa J, Villagran D, Cartes C, Solis C, Nourdin-Galindo G, Haussmann D. Analysis of Genes Encoding for Proteolytic Enzymes and Cytotoxic Proteins as Virulence Factors of Piscirickettsia Salmonis in SHK-1 Cells. J Fish Dis (2021) 44:495–504. doi: 10.1111/jfd.13333
63. Isla A, Haussmann D, Vera T, Kausel G, Figueroa J. Identification of the clpB and bipA Genes and an Evaluation of Their Expression as Related to Intracellular Survival for the Bacterial Pathogen Piscirickettsia Salmonis. Vet Microbiol (2014) 173:390–4. doi: 10.1016/j.vetmic.2014.08.014
64. Oliver C, Hernandez MA, Tandberg JI, Valenzuela KN, Lagos LX, Haro RE, et al. The Proteome of Biologically Active Membrane Vesicles From Piscirickettsia Salmonis LF-89 Type Strain Identifies Plasmid-Encoded Putative Toxins. Front Cell Infect Microbiol (2017) 7:420. doi: 10.3389/fcimb.2017.00420
65. Oliver C, Sanchez P, Valenzuela K, Hernandez M, Pontigo JP, Rauch MC, et al. Subcellular Location of Piscirickettsia Salmonis Heat Shock Protein 60 (Hsp60) Chaperone by Using Immunogold Labeling and Proteomic Analysis. Microorganisms (2020) 8:117. doi: 10.3390/microorganisms8010117
66. Marshall SH, Henriquez V, Gomez FA, Cardenas C. ISPsa2, the First Mobile Genetic Element to be Described and Characterized in the Bacterial Facultative Intracellular Pathogen Piscirickettsia Salmonis. FEMS Microbiol Lett (2011), 18–24.
67. Gomez FA, Cardenas C, Henriquez V, Marshall SH. Characterization of a Functional Toxin-Antitoxin Module in the Genome of the Fish Pathogen Piscirickettsia Salmonis. FEMS Microbiol Lett (2011) 317:83–92.
68. Marshall SH, Gomez FA, Ramirez R, Nilo L, Henriquez V. Biofilm Generation by Piscirickettsia Salmonis Under Growth Stress Conditions: A Putative In Vivo Survival/Persistence Strategy in Marine Environments. Res Microbiol (2012) 163:557–66.
69. Levipan HA, Irgang R, Yanez A, Avendano-Herrera R. Improved Understanding of Biofilm Development by Piscirickettsia Salmonis Reveals Potential Risks for the Persistence and Dissemination of Piscirickettsiosis. Sci Rep (2020) 10:12224. doi: 10.1038/s41598-020-68990-4
70. Abu Kwaik Y, Gao L-Y, Stone Barbara J, Venkataraman C, Harb Omar S. Invasion of Protozoa by Legionella Pneumophila and Its Role in Bacterial Ecology and Pathogenesis. Appl Environ Microbiol (1998) 64:3127–33. doi: 10.1128/AEM.64.9.3127-3133.1998
71. Rojas ME, Galleguillos M, Diaz S, Machuca A, Carbonero A, Smith PA. Evidence of Exotoxin Secretion of Piscirickettsia Salmonis, the Causative Agent of Piscirickettsiosis. J Fish Dis (2013) 36:703–9.
72. Smith PA, Diaz FE, Rojas ME, Diaz S, Galleguillos M, Carbonero A. Effect of Piscirickettsia Salmonis Inoculation on the ASK Continuous Cell Line. J Fish Dis (2015) 38:321–4.
73. Lagos F, Cartes C, Vera T, Haussmann D, Figueroa J. Identification of Genomic Islands in Chilean Piscirickettsia Salmonis Strains and Analysis of Gene Expression Involved in Virulence. J Fish Dis (2017) 40:1321–31. doi: 10.1111/jfd.12604
74. Gomez FA, Tobar JA, Henriquez V, Sola M, Altamirano C, Marshall SH. Evidence of the Presence of a Functional Dot/Icm Type IV-B Secretion System in the Fish Bacterial Pathogen Piscirickettsia Salmonis. PloS One (2013) 8:e54934. doi: 10.1371/journal.pone.0054934
75. Labra A, Arredondo-Zelada O, Flores-Herrera P, Marshall SH, Gomez FA. In Silico Identification and Characterization of Putative Dot/Icm Secreted Virulence Effectors in the Fish Pathogen Piscirickettsia Salmonis. Microb Pathog (2016) 92:11–8. doi: 10.1016/j.micpath.2015.12.002
76. Cortes M, Sanchez P, Ruiz P, Haro R, Saez J, Sanchez F, et al. In Vitro Expression of Sec-Dependent Pathway and Type 4B Secretion System in Piscirickettsia Salmonis. Microb Pathog (2017) 110:586–93. doi: 10.1016/j.micpath.2017.08.003
77. Bohle H, Henríquez P, Grothusen H, Navas E, Sandoval A, Bustamante F, et al. Comparative Genome Analysis of Two Isolates of the Fish Pathogen Piscirickettsia Salmonis From Different Hosts Reveals Major Differences in Virulence-Associated Secretion Systems. Genome Announc (2014) 2.
78. Ortiz-Severin J, Travisany D, Maass A, Cambiazo V, Chavez FP. Global Proteomic Profiling of Piscirickettsia Salmonis and Salmon Macrophage-Like Cells During Intracellular Infection. Microorganisms (2020) 8:1845. doi: 10.20944/preprints202009.0106.v1
79. Lagos L, Tandberg J, Kashulin-Bekkelund A, Colquhoun DJ, Sorum H, Winther-Larsen HC. Isolation and Characterization of Serum Extracellular Vesicles (EVs) From Atlantic Salmon Infected With Piscirickettsia Salmonis. Proteomes (2017) 5:34. doi: 10.3390/proteomes5040034
80. Tandberg J, Oliver C, Lagos L, Gaarder M, Yanez AJ, Ropstad E, et al. Membrane Vesicles From Piscirickettsia Salmonis Induce Protective Immunity and Reduce Development of Salmonid Rickettsial Septicemia in an Adult Zebrafish Model. Fish Shellfish Immunol (2017) 67:189–98. doi: 10.1016/j.fsi.2017.06.015
81. Calquin P, Ruiz P, Oliver C, Sanchez P, Haro R, Oliva H, et al. Physiological Evidence That Piscirickettsia Salmonis Produces Siderophores and Uses Iron From Different Sources. J Fish Dis (2018) 41:553–8. doi: 10.1111/jfd.12745
82. Sanchez P, Oliver C, Hernandez M, Cortes M, Cecilia Rauch M, Valenzuela K, et al. In Vitro Genomic and Proteomic Evidence of a Type IV Pili-Like Structure in the Fish Pathogen Piscirickettsia Salmonis. FEMS Microbiol Lett (2018) 365:1–9. doi: 10.1093/femsle/fny169
83. Zuniga A, Aravena P, Pulgar R, Travisany D, Ortiz-Severin J, Chavez FP, et al. Transcriptomic Changes of Piscirickettsia Salmonis During Intracellular Growth in a Salmon Macrophage-Like Cell Line. Front Cell Infect Microbiol (2019) 9:426. doi: 10.3389/fcimb.2019.00426
84. Zuniga A, Solis C, Cartes C, Nourdin G, Yanez A, Romero A, et al. Transcriptional Analysis of Metabolic and Virulence Genes Associated With Biofilm Formation in Piscirickettsia Salmonis Strains. FEMS Microbiol Lett (2020) 367:1–9. doi: 10.1093/femsle/fnaa180
85. Thi EP, Lambertz U, Reiner NE. Sleeping With the Enemy: How Intracellular Pathogens Cope With a Macrophage Lifestyle. PloS Pathog (2012) 8:e1002551. doi: 10.1371/journal.ppat.1002551
86. Sgro GG, Oka GU, Souza DP, Cenens W, Bayer-Santos E, Matsuyama BY, et al. Bacteria-Killing Type IV Secretion Systems. Front Microbiol (2019) 10:1078. doi: 10.3389/fmicb.2019.01078
87. Segal G, Feldman M, Zusman T. The Icm/Dot Type-IV Secretion Systems of Legionella Pneumophila and Coxiella Burnetii. FEMS Microbiol Rev (2005) 29:65–81. doi: 10.1016/j.femsre.2004.07.001
88. Oliver C, Valenzuela K, Hernandez M, Sandoval R, Haro RE, Avendano-Herrera R, et al. Characterization and Pathogenic Role of Outer Membrane Vesicles Produced by the Fish Pathogen Piscirickettsia Salmonis Under In Vitro Conditions. Vet Microbiol (2016) 184:94–101. doi: 10.1016/j.vetmic.2015.09.012
89. Leiva F, Bravo S, Garcia KK, Moya J, Guzman O, Bascunan N, et al. Temporal Genome-Wide DNA Methylation Signature of Post-Smolt Pacific Salmon Challenged With Piscirickettsia Salmonis. Epigenetics (2021) 16:1335–46. doi: 10.1080/15592294.2020.1864166
90. Santibanez N, Vega M, Perez T, Yanez A, Gonzalez-Stegmaier R, Figueroa J, et al. Biofilm Produced In Vitro by Piscirickettsia Salmonis Generates Differential Cytotoxicity Levels and Expression Patterns of Immune Genes in the Atlantic Salmon Cell Line SHK-1. Microorganisms (2020) 8:1609. doi: 10.3390/microorganisms8101609
91. Declerck P, Behets J, van Hoef V, Ollevier F. Detection of Legionella Spp. And Some of Their Amoeba Hosts in Floating Biofilms From Anthropogenic and Natural Aquatic Environments. Water Res (2007) 41:3159–67. doi: 10.1016/j.watres.2007.04.011
92. Labra Á, Bravo S, Marshall SH. Defining the Role of Caligus Rogercresseyi in Transmission and Spreading of Piscirickettsia Salmonis. Aquaculture (2020) 528:735489. doi: 10.1016/j.aquaculture.2020.735489
93. Alvarez CA, Gomez FA, Mercado L, Ramirez R, Marshall SH. Piscirickettsia Salmonis Imbalances the Innate Immune Response to Succeed in a Productive Infection in a Salmonid Cell Line Model. PloS One (2016) 11:e0163943. doi: 10.1371/journal.pone.0163943
94. Morales-Lange B, Nombela I, Ortega-Villaizan MDM, Imarai M, Schmitt P, Mercado L. Induction of Foxp3 During the Crosstalk Between Antigen Presenting Like-Cells MHCII(+)CD83(+) and Splenocytes CD4(+)IgM(-) in Rainbow Trout. Biol (Basel) (2021) 10:324. doi: 10.3390/biology10040324
95. Ramirez R, Gomez FA, Marshall SH. The Infection Process of Piscirickettsia Salmonis in Fish Macrophages is Dependent Upon Interaction With Host-Cell Clathrin and Actin. FEMS Microbiol Lett (2015) 362:1–8. doi: 10.1093/femsle/fnu012
96. Valenzuela-Miranda D, Gallardo-Escarate C. Novel Insights Into the Response of Atlantic Salmon (Salmo Salar) to Piscirickettsia Salmonis: Interplay of Coding Genes and lncRNAs During Bacterial Infection. Fish Shellfish Immunol (2016) 59:427–38. doi: 10.1016/j.fsi.2016.11.001
97. Pulgar R, Hodar C, Travisany D, Zuniga A, Dominguez C, Maass A, et al. Transcriptional Response of Atlantic Salmon Families to Piscirickettsia Salmonis Infection Highlights the Relevance of the Iron-Deprivation Defence System. BMC Genomics (2015) 16:495. doi: 10.1186/s12864-015-1716-9
98. Morales-Lange B, Ramirez-Cepeda F, Schmitt P, Guzman F, Lagos L, Overland M, et al. Interferon Gamma Induces the Increase of Cell-Surface Markers (CD80/86, CD83 and MHC-II) in Splenocytes From Atlantic Salmon. Front Immunol (2021) 12:666356. doi: 10.3389/fimmu.2021.666356
99. Xue X, Caballero-Solares A, Hall JR, Umasuthan N, Kumar S, Jakob E, et al. Transcriptome Profiling of Atlantic Salmon (Salmo Salar) Parr With Higher and Lower Pathogen Loads Following Piscirickettsia Salmonis Infection. Front Immunol (2021) 12. doi: 10.3389/fimmu.2021.789465
100. Valenzuela-Miranda D, Valenzuela-Munoz V, Farlora R, Gallardo-Escarate C. MicroRNA-Based Transcriptomic Responses of Atlantic Salmon During Infection by the Intracellular Bacterium Piscirickettsia Salmonis. Dev Comp Immunol (2017) 77:287–96. doi: 10.1016/j.dci.2017.08.016
101. Carrizo V, Valenzuela CA, Aros C, Dettleff P, Valenzuela-Munoz V, Gallardo-Escarate C, et al. Transcriptomic Analysis Reveals a Piscirickettsia Salmonis-Induced Early Inflammatory Response in Rainbow Trout Skeletal Muscle. Comp Biochem Physiol Part D Genomics Proteomics (2021) 39:100859. doi: 10.1016/j.cbd.2021.100859
102. Carrizo V, Valenzuela CA, Zuloaga R, Aros C, Altamirano C, Valdes JA, et al. Effect of Cortisol on the Immune-Like Response of Rainbow Trout (Oncorhynchus Mykiss) Myotubes Challenged With Piscirickettsia Salmonis. Vet Immunol Immunopathol (2021) 237:110240. doi: 10.1016/j.vetimm.2021.110240
103. Zuloaga R, Dettleff P, Bastias-Molina M, Meneses C, Altamirano C, Valdes JA, et al. RNA-Seq-Based Analysis of Cortisol-Induced Differential Gene Expression Associated With Piscirickettsia Salmonis Infection in Rainbow Trout (Oncorhynchus Mykiss) Myotubes. Anim (Basel) (2021) 11:2399. doi: 10.3390/ani11082399
104. Dettleff P, Bravo C, Patel A, Martinez V. Patterns of Piscirickettsia Salmonis Load in Susceptible and Resistant Families of Salmo Salar. Fish Shellfish Immunol (2015) 45:67–71. doi: 10.1016/j.fsi.2015.03.039
105. Machuca A, Martinez V. Transcriptome Analysis of the Intracellular Facultative Pathogen Piscirickettsia Salmonis: Expression of Putative Groups of Genes Associated With Virulence and Iron Metabolism. PloS One (2016) 11:e0168855. doi: 10.1371/journal.pone.0168855
106. Pontigo JP, Yanez A, Sanchez P, Vargas-Chacoff L. Characterization and Expression Analysis of Nod-Like Receptor 3 (NLRC3) Against Infection With Piscirickettsia Salmonis in Atlantic Salmon. Dev Comp Immunol (2021) 114:103865. doi: 10.1016/j.dci.2020.103865
107. Fuentealba P, Aros C, Latorre Y, Martinez I, Marshall S, Ferrer P, et al. Genome-Scale Metabolic Reconstruction for the Insidious Bacterium in Aquaculture Piscirickettsia Salmonis. Bioresour Technol (2017) 223:105–14. doi: 10.1016/j.biortech.2016.10.024
108. Italiani P, Boraschi D. From Monocytes to M1/M2 Macrophages: Phenotypical vs. Functional Differentiation. Front Immunol (2014) 5:514. doi: 10.3389/fimmu.2014.00514
109. Yang S, Li Q, Mu Y, Ao J, Chen X. Functional Activities of Interferon Gamma in Large Yellow Croaker Larimichthys Crocea. Fish Shellfish Immunol (2017) 70:545–52. doi: 10.1016/j.fsi.2017.09.051
110. Grayfer L, Kerimoglu B, Yaparla A, Hodgkinson JW, Xie J, Belosevic M. Mechanisms of Fish Macrophage Antimicrobial Immunity. Front Immunol (2018) 9:1105. doi: 10.3389/fimmu.2018.01105
111. Hodgkinson JW, Fibke C, Belosevic M. Recombinant IL-4/13A and IL-4/13B Induce Arginase Activity and Down-Regulate Nitric Oxide Response of Primary Goldfish (Carassius Auratus L.) Macrophages. Dev Comp Immunol (2017) 67:377–84. doi: 10.1016/j.dci.2016.08.014
112. Wiegertjes GF, Wentzel AS, Spaink HP, Elks PM, Fink IR. Polarization of Immune Responses in Fish: The ‘Macrophages First’ Point of View. Mol Immunol (2016) 69:146–56. doi: 10.1016/j.molimm.2015.09.026
113. Smith NC, Christian SL, Woldemariam NT, Clow KA, Rise ML, Andreassen R. Characterization of miRNAs in Cultured Atlantic Salmon Head Kidney Monocyte-Like and Macrophage-Like Cells. Int J Mol Sci (2020) 21:3989. doi: 10.3390/ijms21113989
114. Smith NC, Wajnberg G, Chacko S, Woldemariam NT, Lacroix J, Crapoulet N, et al. Characterization of miRNAs in Extracellular Vesicles Released From Atlantic Salmon Monocyte-Like and Macrophage-Like Cells. Front Immunol (2020) 11:587931. doi: 10.3389/fimmu.2020.587931
115. Smith NC, Umasuthan N, Kumar S, Woldemariam NT, Andreassen R, Christian SL, et al. Transcriptome Profiling of Atlantic Salmon Adherent Head Kidney Leukocytes Reveals That Macrophages Are Selectively Enriched During Culture. Front Immunol (2021) 12:709910. doi: 10.3389/fimmu.2021.709910
116. Thakur A, Mikkelsen H, Jungersen G. Intracellular Pathogens: Host Immunity and Microbial Persistence Strategies. J Immunol Res (2019) 2019:1356540. doi: 10.1155/2019/1356540
117. Latomanski EA, Newton HJ. Taming the Triskelion: Bacterial Manipulation of Clathrin. Microbiol Mol Biol Rev (2019) 83:1–25. doi: 10.1128/MMBR.00058-18
118. Farris M, Grant A, Richardson TB, O’Connor CD. BipA: A Tyrosine-Phosphorylated GTPase That Mediates Interactions Between Enteropathogenic Escherichia Coli (EPEC) and Epithelial Cells. Mol Microbiol (1998) 28:265–79. doi: 10.1046/j.1365-2958.1998.00793.x
119. Das B, Bhadra RK. (P)Ppgpp Metabolism and Antimicrobial Resistance in Bacterial Pathogens. Front Microbiol (2020) 11:563944–4. doi: 10.3389/fmicb.2020.563944
120. Kumar Y, Valdivia RH. Leading a Sheltered Life: Intracellular Pathogens and Maintenance of Vacuolar Compartments. Cell Host Microbe (2009) 5:593–601. doi: 10.1016/j.chom.2009.05.014
121. Oliva G, Sahr T, Buchrieser C. The Life Cycle of L. Pneumophila: Cellular Differentiation Is Linked to Virulence and Metabolism. Front Cell Infect Microbiol (2018) 8:3. doi: 10.3389/fcimb.2018.00003
122. Newton HJ, Ang DKY, van Driel IR, Hartland EL. Molecular Pathogenesis of Infections Caused by Legionella Pneumophila. Clin Microbiol Rev (2010) 23:274–98. doi: 10.1128/CMR.00052-09
123. Carril GP, Gomez FA, Marshall SH. Expression of Flagellin and Key Regulatory Flagellar Genes in the non-Motile Bacterium Piscirickettsia Salmonis. Dis Aquat Organ (2017) 123:29–43. doi: 10.3354/dao03079
124. Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting Edge: Bacterial Flagellin Activates Basolaterally Expressed TLR5 to Induce Epithelial Proinflammatory Gene Expression. J Immunol (2001) 167:1882–5. doi: 10.4049/jimmunol.167.4.1882
125. Broekman DC, Guethmundsson GH, Maier VH. Differential Regulation of Cathelicidin in Salmon and Cod. Fish Shellfish Immunol (2013) 35:532–8. doi: 10.1016/j.fsi.2013.05.005
126. Munoz-Flores C, Astuya-Villalon A, Romero A, Acosta J, Toledo JR. Salmonid MyD88 is a Key Adapter Protein That Activates Innate Effector Mechanisms Through the TLR5M/TLR5S Signaling Pathway and Protects Against Piscirickettsia Salmonis Infection. Fish Shellfish Immunol (2022) 121:387–94. doi: 10.1016/j.fsi.2021.12.030
127. Liu X, Zhang H, Jiao C, Liu Q, Zhang Y, Xiao J. Flagellin Enhances the Immunoprotection of Formalin-Inactivated Edwardsiella Tarda Vaccine in Turbot. Vaccine (2017) 35:369–74. doi: 10.1016/j.vaccine.2016.11.031
128. Gonzalez-Stegmaier R, Pena A, Villarroel-Espindola F, Aguila P, Oliver C, MacLeod-Carey D, et al. Full Recombinant Flagellin B From Vibrio Anguillarum (rFLA) and its Recombinant D1 Domain (Rnd1) Promote a Pro-Inflammatory State and Improve Vaccination Against P. Salmonis in Atlantic Salmon (S. Salar). Dev Comp Immunol (2021) 117:103988. doi: 10.1016/j.dci.2020.103988
129. Wang J, Wang T, Benedicenti O, Collins C, Wang K, Secombes CJ, et al. Characterisation of ZBTB46 and DC-SCRIPT/ZNF366 in Rainbow Trout, Transcription Factors Potentially Involved in Dendritic Cell Maturation and Activation in Fish. Dev Comp Immunol (2018) 80:2–14. doi: 10.1016/j.dci.2016.11.007
130. Wang Y, Sun HY, Kumar S, Puerta MDM, Jo H, Rezvan A. ZBTB46 is a Shear-Sensitive Transcription Factor Inhibiting Endothelial Cell Proliferation via Gene Expression Regulation of Cell Cycle Proteins. Lab Invest (2019) 99:305–18. doi: 10.1038/s41374-018-0060-5
131. Mostowy S, Shenoy AR. The Cytoskeleton in Cell-Autonomous Immunity: Structural Determinants of Host Defence. Nat Rev Immunol (2015) 15:559–73. doi: 10.1038/nri3877
132. Leiva F, Bravo S, Garcia KK, Moya J, Guzman O, Vidal R. Temporal Gene Expression Signature of Plasma Extracellular Vesicles-MicroRNAs From Post-Smolt Coho Salmon Challenged With Piscirickettsia Salmonis. Marine Biotechnol (2021) 23:602–14.
133. Nakanishi T, Shibasaki Y, Matsuura Y. T Cells in Fish. Biol (Basel) (2015) 4:640–63. doi: 10.3390/biology4040640
134. Fischer U, Koppang EO, Nakanishi T, Teleost T. And NK Cell Immunity. Fish Shellfish Immunol (2013) 35:197–206. doi: 10.1016/j.fsi.2013.04.018
135. Toda H, Araki K, Moritomo T, Nakanishi T. Perforin-Dependent Cytotoxic Mechanism in Killing by CD8 Positive T Cells in Ginbuna Crucian Carp, Carassius Auratus Langsdorfii. Dev Comp Immunol (2011) 35:88–93. doi: 10.1016/j.dci.2010.08.010
136. Toda H, Shibasaki Y, Koike T, Ohtani M, Takizawa F, Ototake M, et al. Alloantigen-Specific Killing is Mediated by CD8-Positive T Cells in Fish. Dev Comp Immunol (2009) 33:646–52. doi: 10.1016/j.dci.2008.11.008
137. Partula S, de Guerra A, Fellah JS, Charlemagne J. Structure and Diversity of the TCR Alpha-Chain in a Teleost Fish. J Immunol (1996) 157:207–12.
138. Bernard D, Riteau B, Hansen JD, Phillips RB, Michel F, Boudinot P, et al. Costimulatory Receptors in a Teleost Fish: Typical CD28, Elusive CTLA4. J Immunol (2006) 176:4191–200. doi: 10.4049/jimmunol.176.7.4191
139. Yoshiura Y, Kiryu I, Fujiwara A, Suetake H, Suzuki Y, Nakanishi T, et al. Identification and Characterization of Fugu Orthologues of Mammalian Interleukin-12 Subunits. Immunogenetics (2003) 55:296–306. doi: 10.1007/s00251-003-0582-9
140. Munang’andu HM, Fredriksen BN, Mutoloki S, Dalmo RA, Evensen O. The Kinetics of CD4+ and CD8+ T-Cell Gene Expression Correlate With Protection in Atlantic Salmon (Salmo Salar L) Vaccinated Against Infectious Pancreatic Necrosis. Vaccine (2013) 31:1956–63. doi: 10.1016/j.vaccine.2013.02.008
141. Jørgensen SM, Lyng-Syvertsen B, Lukacs M, Grimholt U, Gjøen T. Expression of MHC Class I Pathway Genes in Response to Infectious Salmon Anaemia Virus in Atlantic Salmon (Salmo Salar L.) Cells. Fish Shellfish Immunol (2006) 21:548–60. doi: 10.1016/j.fsi.2006.03.004
142. Hewitt EW. The MHC Class I Antigen Presentation Pathway: Strategies for Viral Immune Evasion. Immunology (2003) 110:163–9. doi: 10.1046/j.1365-2567.2003.01738.x
143. Tortorella D, Gewurz B, Schust D, Furman M, Ploegh H. Down-Regulation of MHC Class I Antigen Presentation by HCMV; Lessons for Tumor Immunology. Immunol Invest (2000) 29:97–100. doi: 10.3109/08820130009062289
144. Grimholt U. MHC and Evolution in Teleosts. Biol (Basel) (2016) 5:6. doi: 10.3390/biology5010006
145. Grimholt U, Tsukamoto K, Hashimoto K, Dijkstra JM. Discovery of a Novel MHC Class I Lineage in Teleost Fish Which Shows Unprecedented Levels of Ectodomain Deterioration While Possessing an Impressive Cytoplasmic Tail Motif. Cells (2019) 8:1056. doi: 10.3390/cells8091056
146. Adams EJ, Luoma AM. The Adaptable Major Histocompatibility Complex (MHC) Fold: Structure and Function of Nonclassical and MHC Class I-Like Molecules. Annu Rev Immunol (2013) 31:529–61. doi: 10.1146/annurev-immunol-032712-095912
147. Svenning S, Gondek-Wyrozemska AT, van der Wal YA, Robertsen B, Jensen I, Jorgensen JB, et al. Microbial Danger Signals Control Transcriptional Induction of Distinct MHC Class I L Lineage Genes in Atlantic Salmon. Front Immunol (2019) 10:2425. doi: 10.3389/fimmu.2019.02425
148. Gudding R, Goodrich T. The History of Fish Vaccination. In: Fish Vaccination. John Wiley & Sons, Lda (2014). p. 1–11. doi: 10.1002/9781118806913
149. Iliev DB, Lagos L, Thim HL, Jorgensen SM, Krasnov A, Jorgensen JB. CpGs Induce Differentiation of Atlantic Salmon Mononuclear Phagocytes Into Cells With Dendritic Morphology and a Proinflammatory Transcriptional Profile But an Exhausted Allostimulatory Activity. Front Immunol (2019) 10:378. doi: 10.3389/fimmu.2019.00378
150. Bhattacharya M, Malick RC, Mondal N, Patra P, Pal BB, Patra BC, et al. Computational Characterization of Epitopic Region Within the Outer Membrane Protein Candidate in Flavobacterium Columnare for Vaccine Development. J Biomolec Struct Dynam (2020) 38:450–9. doi: 10.1080/07391102.2019.1580222
151. Mahendran R, Jeyabaskar S, Sitharaman G, Michael RD, Paul AV. Computer-Aided Vaccine Designing Approach Against Fish Pathogens Edwardsiella Tarda and Flavobacterium Columnare Using Bioinformatics Softwares. Drug Des Devel Ther (2016) 10:1703–14. doi: 10.2147/DDDT.S95691
152. Pumchan A, Krobthong S, Roytrakul S, Sawatdichaikul O, Kondo H, Hirono I, et al. Novel Chimeric Multiepitope Vaccine for Streptococcosis Disease in Nile Tilapia (Oreochromis Niloticus Linn.). Sci Rep (2020) 10:603. doi: 10.1038/s41598-019-57283-0
153. Machimbirike VI, Pornputtapong N, Senapin S, Wangkahart E, Srisapoome P, Khunrae P, et al. A Multi-Epitope Chimeric Protein Elicited a Strong Antibody Response and Partial Protection Against Edwardsiella Ictaluri in Nile Tilapia. J Fish Dis (2022) 45:1–18. doi: 10.1111/jfd.13525
154. Figueroa C, Bustos P, Torrealba D, Dixon B, Soto C, Conejeros P, et al. Coinfection Takes its Toll: Sea Lice Override the Protective Effects of Vaccination Against a Bacterial Pathogen in Atlantic Salmon. Sci Rep (2017) 7:17817. doi: 10.1038/s41598-017-18180-6
155. Figueroa C, Veloso P, Espin L, Dixon B, Torrealba D, Elalfy IS, et al. Host Genetic Variation Explains Reduced Protection of Commercial Vaccines Against Piscirickettsia Salmonis in Atlantic Salmon. Sci Rep (2020) 10:18252. doi: 10.1038/s41598-020-70847-9
156. Everson JL, Jones DR, Taylor AK, Rutan BJ, Leeds TD, Langwig KE, et al. Genetic Line, and Vaccination Affect Rainbow Trout (Oncorhynchus Mykiss) Disease Susceptibility and Infection Dynamics. Front Immunol (2021) 12:721048. doi: 10.3389/fimmu.2021.721048
157. van der Wal YA, Jenberie S, Nordli H, Greiner-Tollersrud L, Kool J, Jensen I, et al. The Importance of the Atlantic Salmon Peritoneal Cavity B Cell Response: Local IgM Secreting Cells are Predominant Upon Piscirickettsia Salmonis Infection. Dev Comp Immunol (2021) 123:104125. doi: 10.1016/j.dci.2021.104125
158. Kelly C, Salinas I. Under Pressure: Interactions Between Commensal Microbiota and the Teleost Immune System. Front Immunol (2017) 8:559. doi: 10.3389/fimmu.2017.00559
159. Xu Z, Takizawa F, Casadei E, Shibasaki Y, Ding Y, Sauters TJC, et al. Specialization of Mucosal Immunoglobulins in Pathogen Control and Microbiota Homeostasis Occurred Early in Vertebrate Evolution. Sci Immunol (2020) 5:1–17. doi: 10.1126/sciimmunol.aay3254
160. Xu Z, Takizawa F, Parra D, Gomez D, von Gersdorff Jorgensen L, LaPatra SE, et al. Mucosal Immunoglobulins at Respiratory Surfaces Mark an Ancient Association That Predates the Emergence of Tetrapods. Nat Commun (2016) 7:10728. doi: 10.1038/ncomms10728
161. Bishop SC, Woolliams JA. Genomics and Disease Resistance Studies in Livestock. Livest Sci (2014) 166:190–8. doi: 10.1016/j.livsci.2014.04.034
162. Barría A, Christensen KA, Yoshida GM, Correa K, Jedlicki A, Lhorente JP, et al. Genomic Predictions and Genome-Wide Association Study of Resistance Against Piscirickettsia Salmonis in Coho Salmon (Oncorhynchus Kisutch) Using ddRAD Sequencing. G3 (Bethesda) (2018) 8:1183–94. doi: 10.1534/g3.118.200053
163. Correa K, Lhorente JP, López ME, Bassini L, Naswa S, Deeb N, et al. Genome-Wide Association Analysis Reveals Loci Associated With Resistance Against Piscirickettsia Salmonis in Two Atlantic Salmon (Salmo Salar L.) Chromosomes. BMC Genomics (2015) 16:854–4. doi: 10.1186/s12864-015-2038-7
164. Yáñez JM, Bangera R, Lhorente JP, Barría A, Oyarzún M, Neira R, et al. Negative Genetic Correlation Between Resistance Against Piscirickettsia Salmonis and Harvest Weight in Coho Salmon (Oncorhynchus Kisutch). Aquaculture (2016) 459:8–13. doi: 10.1016/j.aquaculture.2016.03.020
165. Yáñez JM, Bangera R, Lhorente JP, Oyarzún M, Neira R. Quantitative Genetic Variation of Resistance Against Piscirickettsia Salmonis in Atlantic Salmon (Salmo Salar). Aquaculture (2013) 414-415:155–9. doi: 10.1016/j.aquaculture.2013.08.009
166. Yáñez JM, Lhorente JP, Bassini LN, Oyarzún M, Neira R, Newman S. Genetic Co-Variation Between Resistance Against Both Caligus Rogercresseyi and Piscirickettsia Salmonis, and Body Weight in Atlantic Salmon (Salmo Salar). Aquaculture (2014) 433:295–8. doi: 10.1016/j.aquaculture.2014.06.026
167. Yoshida GM, Bangera R, Carvalheiro R, Correa K, Figueroa R, Lhorente JP, et al. Genomic Prediction Accuracy for Resistance Against Piscirickettsia Salmonis in Farmed Rainbow Trout. G3 Genes|Genomes|Genetics (2018) 8:719–26. doi: 10.1534/g3.117.300499
168. Moraleda CP, Robledo D, Gutiérrez AP, del-Pozo J, Yáñez JM, Houston RD. Investigating Mechanisms Underlying Genetic Resistance to Salmon Rickettsial Syndrome in Atlantic Salmon Using RNA Sequencing. BMC Genomics (2021) 22:156. doi: 10.1186/s12864-021-07443-2
169. Barría A, Christensen KA, Yoshida G, Jedlicki A, Leong JS, Rondeau EB, et al. Whole Genome Linkage Disequilibrium and Effective Population Size in a Coho Salmon (Oncorhynchus Kisutch) Breeding Population Using a High-Density SNP Array. Front Genet (2019) 10:498–8. doi: 10.3389/fgene.2019.00498
170. Anacleto O, Cabaleiro S, Villanueva B, Saura M, Houston RD, Woolliams JA, et al. Genetic Differences in Host Infectivity Affect Disease Spread and Survival in Epidemics. Sci Rep (2019) 9:4924. doi: 10.1038/s41598-019-40567-w
171. Anche MT, de Jong MC, Bijma P. On the Definition and Utilization of Heritable Variation Among Hosts in Reproduction Ratio R0 for Infectious Diseases. Heredity (Edinb) (2014) 113:364–74. doi: 10.1038/hdy.2014.38
172. Bitsouni V, Lycett S, Opriessnig T, Doeschl-Wilson A. Predicting Vaccine Effectiveness in Livestock Populations: A Theoretical Framework Applied to PRRS Virus Infections in Pigs. PloS One (2019) 14:e0220738. doi: 10.1371/journal.pone.0220738
173. Chase-Topping ME, Pooley C, Moghadam HK, Hillestad B, Lillehammer M, Sveen L, et al. Impact of Vaccination and Selective Breeding on the Transmission of Infectious Salmon Anemia Virus. Aquaculture (2021) 535:736365. doi: 10.1016/j.aquaculture.2021.736365
174. Cabello FC, Godfrey HP. Salmon Aquaculture, Piscirickettsia Salmonis Virulence, and One Health: Dealing With Harmful Synergies Between Heavy Antimicrobial Use and Piscine and Human Health. Aquaculture (2019) 507:451–6. doi: 10.1016/j.aquaculture.2019.04.048
Keywords: piscirickettsiosis, Piscirickettsia salmonis, immunology, vaccines, control, SRS, salmonids
Citation: Rozas-Serri M (2022) Why Does Piscirickettsia salmonis Break the Immunological Paradigm in Farmed Salmon? Biological Context to Understand the Relative Control of Piscirickettsiosis. Front. Immunol. 13:856896. doi: 10.3389/fimmu.2022.856896
Received: 17 January 2022; Accepted: 22 February 2022;
Published: 21 March 2022.
Edited by:
Carmen G. Feijoo, Andres Bello University, ChileReviewed by:
Kevin R. Maisey, University of Santiago of Chile, ChileByron Morales-Lange, Norwegian University of Life Sciences, Norway
Copyright © 2022 Rozas-Serri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marco Rozas-Serri, bWFyY28ucm96YXNAcGF0aG92ZXQuY2w=
 Marco Rozas-Serri
Marco Rozas-Serri