- UMR CNRS 7276, INSERM 1262 and Université de Limoges: Contrôle de la Réponse Immune B et des Lymphoproliférations, 2 Rue du Pr. Descottes, Limoges, France
B cells undergo genetic rearrangements at immunoglobulin gene (Ig) loci during B cell maturation. First V(D)J recombination occurs during early B cell stages followed by class switch recombination (CSR) and somatic hypermutation (SHM) which occur during mature B cell stages. Given that RAG1/2 induces DNA double strand breaks (DSBs) during V(D)J recombination and AID (Activation-Induced Deaminase) leads to DNA modifications (mutations during SHM or DNA DSBs during CSR), it is mandatory that IgH rearrangements be tightly regulated to avoid any mutations or translocations within oncogenes. Ig loci contain various cis-regulatory elements that are involved in germline transcription, chromatin modifications or RAG/AID recruitment. Ig cis-regulatory elements are increasingly recognized as being involved in nuclear positioning, heterochromatin addressing and chromosome loop regulation. In this review, we examined multiple data showing the critical interest of studying Ig gene regulation at the whole nucleus scale. In this context, we highlighted the essential function of Ig gene regulatory elements that now have to be considered as nuclear organizers in B lymphocytes.
Introduction
To produce highly specific antibodies, B cells undergo genetic modifications of their immunoglobulin (Ig) genes. Among these events, V(D)J recombination takes place in the bone marrow during the early steps of B cell development and occurs in an antigen-independent manner. Mature B cells migrate towards secondary lymphoid organs and continue their differentiation once stimulated by antigens. This process integrates secondary beneficial DNA remodeling events including class switch recombination (CSR) and somatic hypermutation (SHM) but can also induce B cell death through locus suicide recombination (LSR), a detrimental rearrangement that abrogates surface B cell receptor expression (1). These events, all mediated by the activation-induced deaminase (AID) enzyme, characterize the late antigen-dependent phase of developing cells (Figure 1).
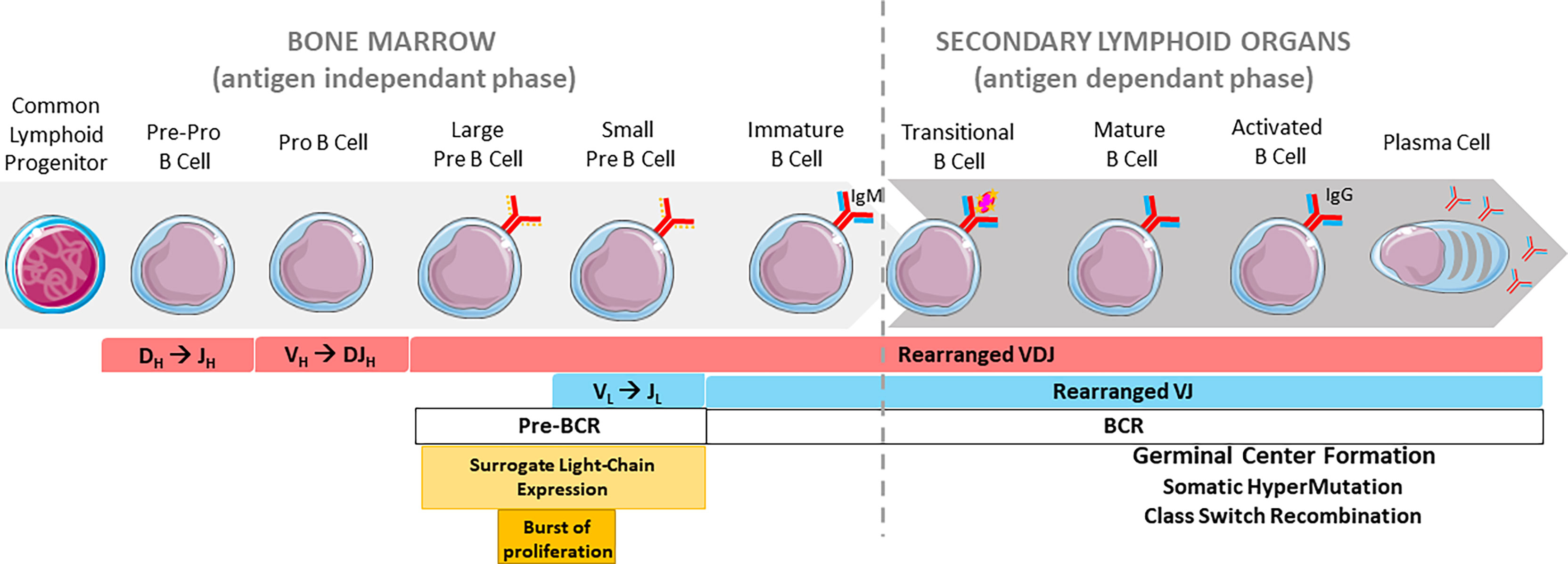
Figure 1 Overview of B cell development. Chronological order of B cell stages in the bone marrow (left) and secondary lymphoid organs (right). The pre-BCR is represented by a continuous red line and a discontinuous orange line (surrogate light chain). The BCR is represented by continuous red and blue lines.
Nuclear organization and chromosome distribution in B-lineage cells have to be considered as important parameters for the control of all these events. Chromosomes are not randomly scattered in the nucleus; their positions change in a dynamic fashion especially during the B cell lifespan. Indeed, distinct organizational levels co-exist in the genome on nucleosomal (genes and loci), supranucleosomal (chromatin domains and compartments) and nuclear (chromosome territories) scales (2–4). In addition to histone mark patterns that reflect the nucleosomal context, the emergence of new molecular biology approaches led to the discovery of Topologically Associated Domains (TADs) and the dynamic loop extrusion model (5). Such methods, based on chromosome capture conformation (6) (3C, 4C and Hi-C), make it possible to evaluate additional levels of gene regulation at the supranucleosomal scale. This particular level of nuclear topology includes TAD structures, A and B chromatin compartments, DNA loops and interchromatin space. Gene transcription takes place in the A-euchromatin compartment whereas B-heterochromatin prevents it. In a simplified scheme, A and B compartments are respectively positioned at the center of nuclei and at the nuclear periphery (4). More recently, the Volk’s group showed that heterochromatin and euchromatin, respectively defined as B and A compartments, are localized at the nucleus periphery, leaving the nucleus center devoid of nucleosomes (7). As a structural unit of genome organization, a TAD is a large chromosomal region in which the contact frequencies between genes or regulatory regions are higher than elsewhere in the genome. TADs themselves are subdivided into multiple sub-TAD structures (chromatin loops) that undergo dynamic cell-type specific connections. The mouse genome contains around 2000 TADs, each with an approximate mean size of 1 megabase (Mb) (8). Indeed, some dynamic processes drive chromatin regions into a free space termed the interchromatin compartment in order to permit gene segment interactions. Such contacts between gene portions occur either in active (A) or inactive (B) chromatin compartments and it is widely recognized that interactions take place within the same TAD. Among chromatin compartments, long-range homologous contacts (A–A or B–B) are largely favored over heterologous contacts (A–B) (9). Moreover, additional TAD interactions exist since chromosome portions are not only able to establish close contacts in cis, but also in trans with other chromosomes (10). Some of these trans interactions have been documented in the case of olfactory receptor (11) and Th2 cytokines genes (12). At TAD extremities, TAD borders are enriched in CTCF (CCCTC-binding Factor) insulator protein, mediator complexes (MED1, MED12) as well as active histone marks (H3K4me3 and H3K36me3) (8). TAD borders display specific “insulating” features, preventing loci located on each side of this border to establish contacts (8).
By considering genome nuclear topology, these emerging models are particularly relevant for the tightly-regulated Ig gene loci. Most Ig gene regulation studies have so far been performed at the nucleosomal scale (epigenetic modifications and regulatory transcription of loci and gene segments). The increasing interest in understanding gene regulation at the whole nucleus scale prompted B cell scientists to revisit previous models at both supranucleosomal (DNA loops and TADs) and nuclear (chromosome territories and nuclear position) levels (13–22). To provide a clearer picture of how B cell development is tightly regulated by the nuclear location of Ig genes, including chromosome looping and loci positioning in the mouse, we will begin with an overview of Ig genes and their enhancers and then focus on the role of their main enhancers on 3D-nuclear organization. The relationship between Ig and respective enhancers will be discussed in this review.
Overview of Immunoglobulin Genes and Their Regulatory Elements
In mice, Ig genes are encoded by three loci located on three distinct chromosomes. The immunoglobulin heavy chain (IgH) locus lies on chromosome 12. Immunoglobulin light (IgL) chain loci contain either kappa immunoglobulin light chain (Igκ) or lambda immunoglobulin light chain (Igλ) genes and are respectively located on chromosomes 6 and 16. In mice, at least 95% of B cells express Igκ light chains (13, 14).
Immunoglobulin Heavy (IgH) Chain Loci
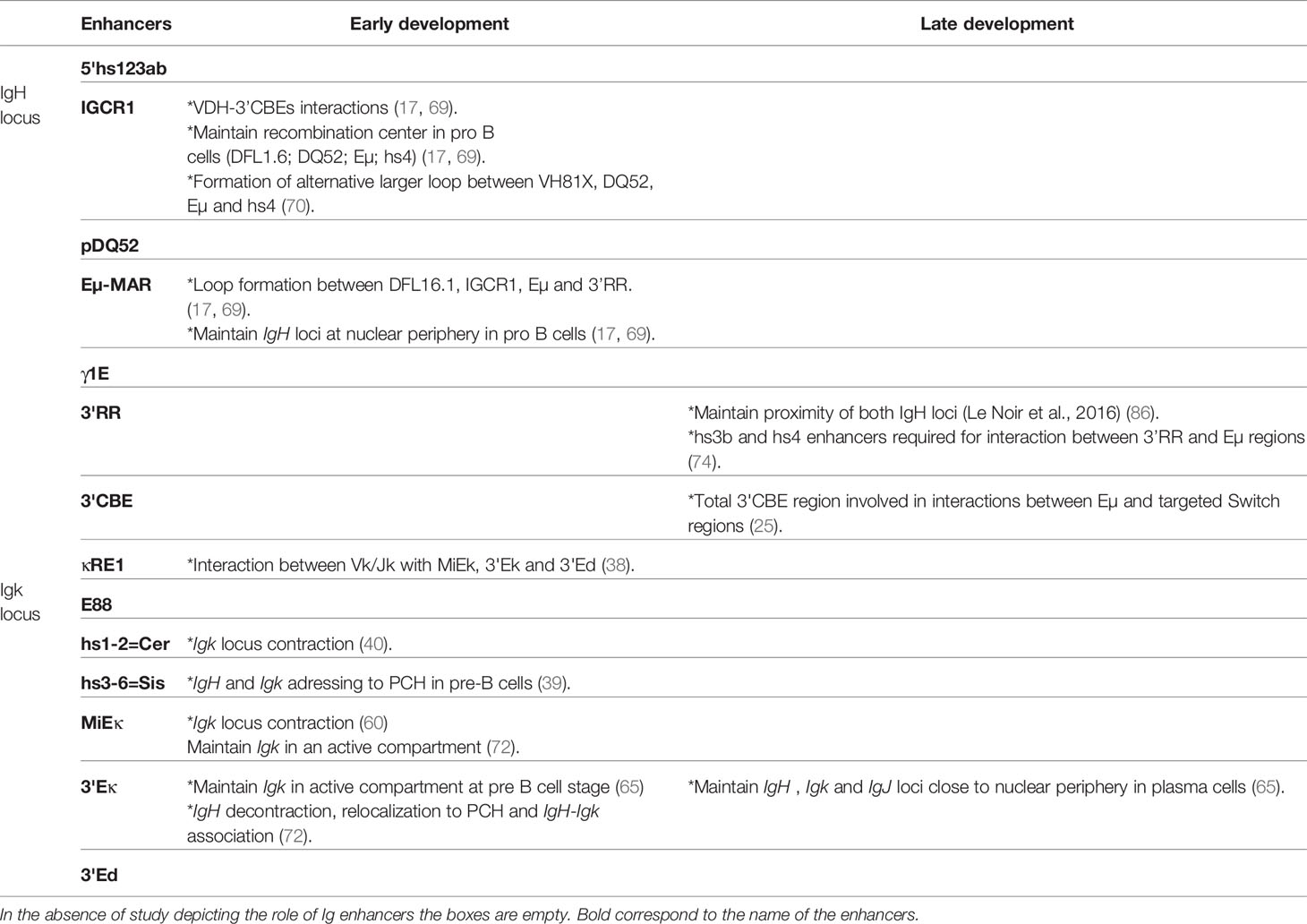
The IgH locus spans approximately 3Mb, in its germline configuration, and contains various cis-regulatory regions (Figure 2A). From 5’ to 3’, the 5’hs123ab elements, are situated upstream from the first VH segment (15). The intergenic VHDH region, located between the most distal DH segment, DFL16, and the most proximal VH segment, VH7183a.2.3, contains six DNase I sensitive sites (hs1 to 6) among which hs4 and hs5 are CTCF binding sites (17, 18). This set is also called Intergenic Control Region 1 (IGCR1) (16, 17). The promoter/enhancer pDQ52, is located just upstream from DQ52 (18). The Eµ-MARs region, spanning about 1kb, is composed of a 220-base pair (bp) core enhancer element (cEµ) flanked by two matrix attachment regions (MARs) located between the last JH exon and the Sµ region. Between the Cγ1 and Cγ2b constant genes, two transcriptional enhancers hRE1 and hRE2 (19) form the γ1E regulatory element (20). At the 3’extremity, the 3’Regulatory Region (3’RR), spanning more than 30 kb, is composed of four enhancers, called hs3a, hs1.2, hs3b and hs4 (21). The central hs1.2 enhancer is flanked by inverted repeated intervening sequences (IRIS) that form a 25kb-long quasi-palindrome (22). Downstream from this palindrome, hs4 is the most distal element harboring an enhancer activity within the 3’RR (23). Although highly divergent in different species, IRIS sequences always stand as inverted copies on both sides of hs1.2, conserving a singular symmetry within the 3’RR (24). Downstream from the 3’RR, four hs elements (hs5, hs6, hs7 and hs8) lie in a region containing ten CTCF Binding Elements (3’CBEs) (25, 26). This region acts as an insulator and delimits the 3’ IgH TAD border (25, 26).
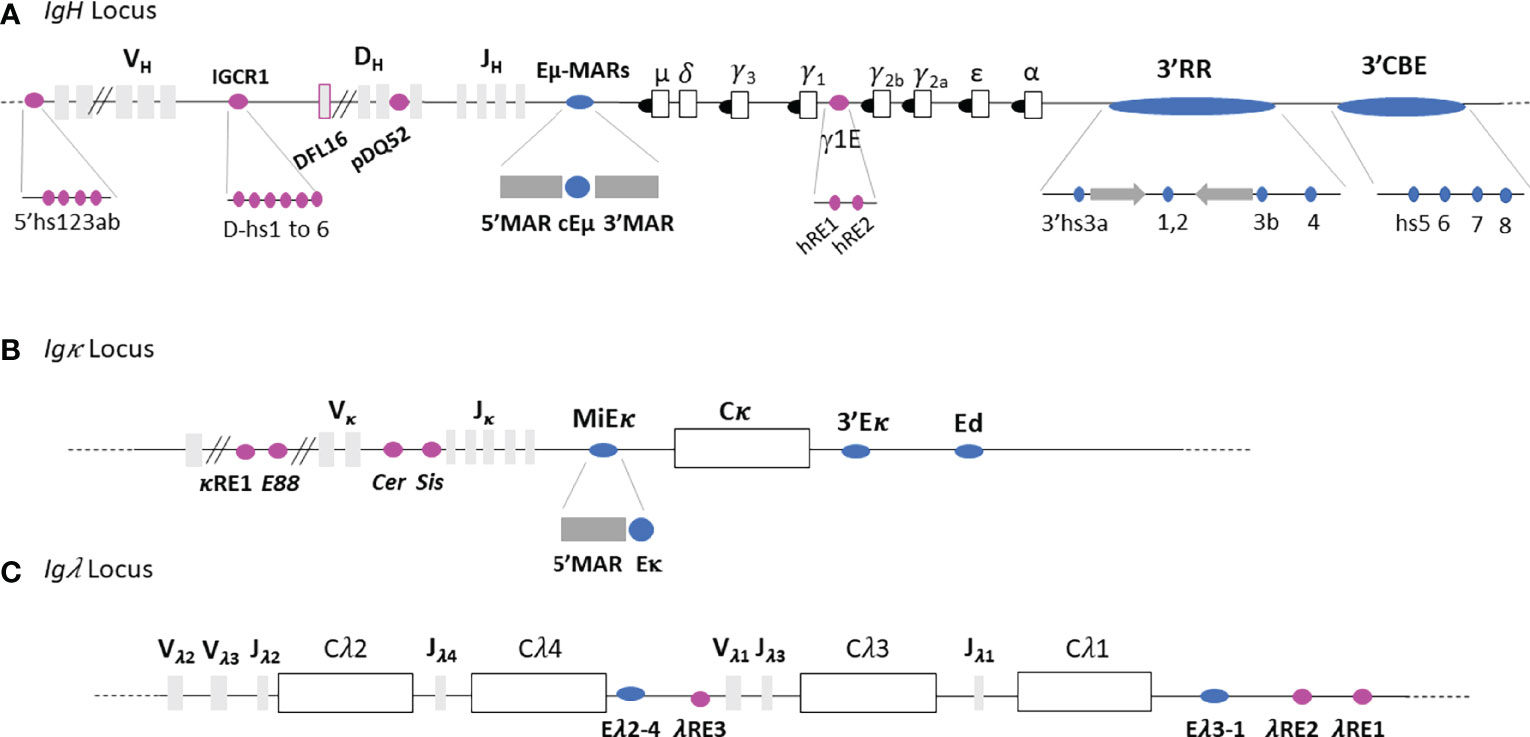
Figure 2 Immunoglobulin loci. (A) IgH locus. (B) Igκ locus. (C) Igλ locus. Light grey and white rectangles represent V, D and J gene segments and constant genes respectively. Black circles represent switch regions and blue and pink ovals represent respectively the main Ig loci regulatory elements and other enhancers. Not to scale.
Our group and others developed numerous mouse models carrying deletions within the IgH regulatory elements that helped elucidate Eµ and 3’RR super-enhancer functions during early and late B cell development (20, 27–37).
Immunoglobulin Light Chain Loci: Igκ and Igλ
The 3.2 Mb Igκ locus contains regulatory elements within the Vκ segments (Figure 2B) which are partially homologous to those within the IgH: κRE1 (19) and E88 (38). Six DNase I hypersensitive sites (hs), hs1 to 6, are located in the intervening Vκ-Jκ region (39, 40). Hs1-2 forms Cer (Contracting element for recombination) and hs3-6 comprises the Sis (Silencer in the intervening sequence) elements. A MAR-intronic Eκ enhancer (MiEκ) (41–43), located downstream from the last Jκ segment, is composed of a 5’flanking matrix attachment region (MARκ) and an intronic enhancer. The 3’Eκ enhancer region is situated downstream from the unique Cκ gene (44). A third regulatory region, called Ed (due to its downstream location within the Igκ locus), is located distal to 3’Eκ (45).
In comparison to IgH and Igκ loci, the Igλ locus is smaller (200kb) and uniquely organized. It comprises four families which contain a pair of Jλ and Cλ segments with only three Vλ segments (46) (Figure 2C). The Igλ locus contains two main enhancers called Eλ2-4, located between Cλ4 and Vλ1, and Eλ3-1 located downstream from Cλ1 (47). Three supplementary elements featuring enhancer activity have been described: λRE3, λRE2 and λRE1 (19). λRE3 lies between Eλ2-4 and Vλ1 while λRE1 and λRE2 are located close to the Eλ3-1 enhancer. Both Igλ enhancers, Eλ2-4 and Eλ3-1 are involved in transcription and VJ rearrangement regulation (47). λRE elements, especially λRE1 and λRE3, have been shown to potentiate the enhancer activity of Eλ3-1 and Eλ2-4, in pro-B and plasma cells respectively (19).
Immunoglobulin Joining Chain Loci: IgJ
Located on chromosome 5 in mice, the IgJ loci encodes the joining peptide (J) chain which promotes active IgA (48) or IgM (49) secretion by ensuring efficient assembly of these Ig subtypes in plasma cells.
Nuclear Organization Driven by Ig Enhancers at Early B Cell Stages
Early B cell development takes place in bone marrow where a lymphoid precursor progresses through different stages to the final immature stage. This progression is concomitant with V(D)J recombination. Once engaged in the B cell lineage after expression of B cell specific transcription factors (50), the common lymphoid precursor (CLP) differentiates into a pre-pro B and then pro-B cell that undergoes DH-JH rearrangement at the IgH locus (51). Once the DJH segment rearranges (52), the pro-B cell joins a VH segment to the previously rearranged DJH segment and progresses to the large pre-B cell stage. At this stage, the IgH locus is completely rearranged and the cell expresses a pre-B Cell Receptor (pre-BCR) at its surface. The pre-BCR, indispensable for B cell development (53), is composed of a functional IgH chain linked to an invariant surrogate light chain, altogether associated with the Igα-Igβ transmembrane heterodimer signaling component. Pre-BCR signaling stops IgH rearrangement and triggers a burst of proliferation leading to the small pre-B cell stage and the occurrence of VL-JL rearrangements at IgL loci. Successful IgL chain rearrangement and production leads to membrane IgM expression and consequently a functional BCR (50), on the immature B cell. These cells then migrate towards secondary lymphoid organs and continue their maturation (Figure 1).
IgH Sub-Nuclear Positioning and Chromatin Loops
Ig loci positioning within the nucleus is dynamic throughout early B cell development and it has been clearly shown that nuclear organization provides a critical level of regulation during V(D)J recombination, particularly for loci contraction/decontraction, nuclear positioning (center vs periphery) and heterochromatin addressing (Figure 3).
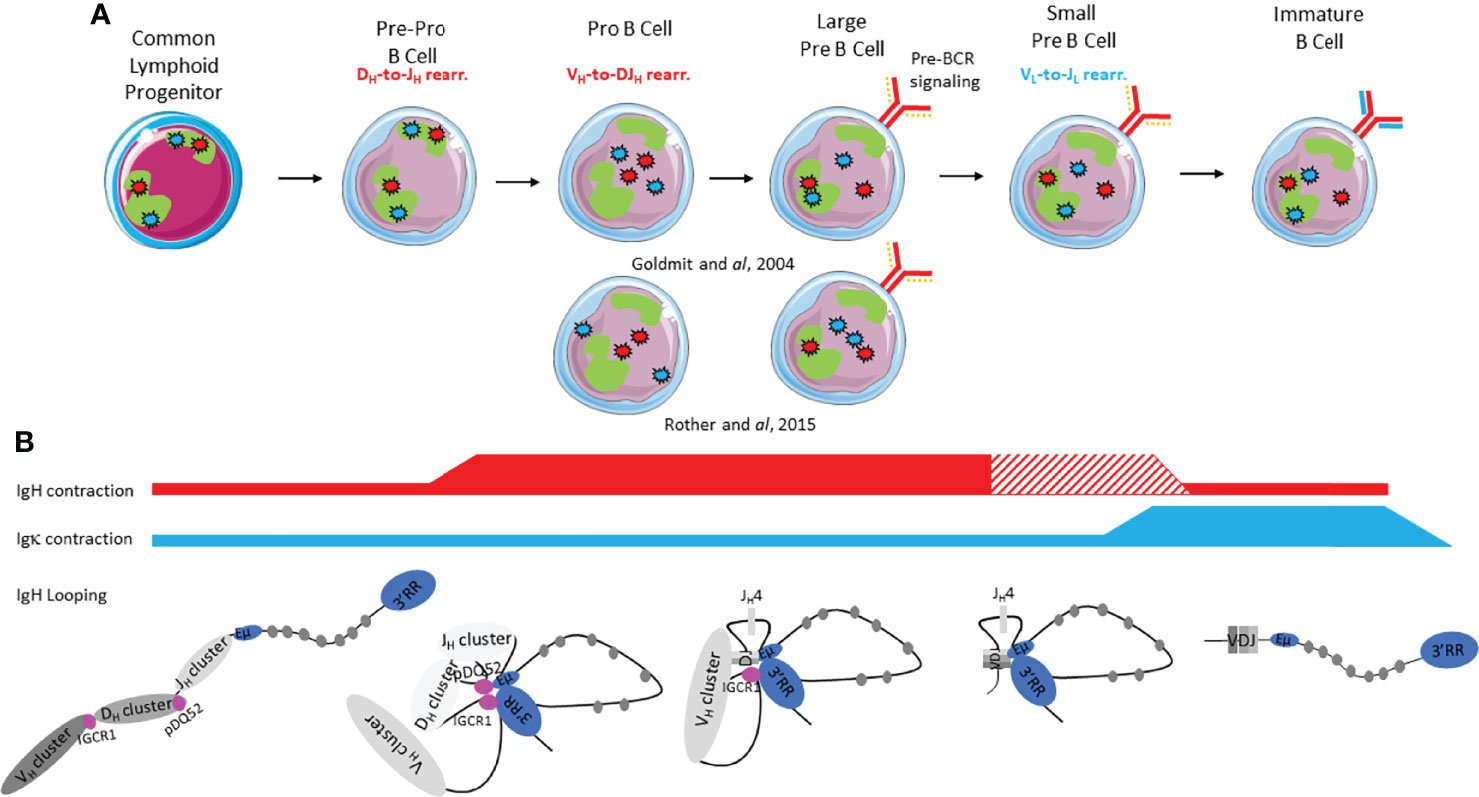
Figure 3 Immunoglobulin loci contraction, conformation, heterochromatin addressing and nuclear positioning during early B cell development. (A) IgH and IgL alleles are respectively represented by red stars and blue stars; the green form represents the pericentromeric heterochromatin compartment (PCH). The pre-BCR is represented by a continuous red line and a discontinuous orange line (surrogate light chain). The BCR is represented by continuous red and blue lines. (B) Ig loci contraction is shown by red and blue lines for IgH and Igk respectively: thickness of the line indicates degree of contraction. The hatched lines correspond to controversial studies.
IgH Sub-Nuclear Positioning
Dynamic repositioning of the IgH locus throughout early stages is involved in IgH accessibility in order to correctly orchestrate V(D)J recombination (Figure 3A). In CLP and pre-pro-B cells, IgH loci are preferentially located at the nuclear periphery within a repressive compartment (54). While only few changes occur at immature stages (55), the pro-B cell stage undergoes, in contrast, global genome reorganization since around 20% of genes switch to A-B or B-A compartments (56). Indeed, both IgH alleles are relocated to the active A compartment to allow DH-JH recombination on both alleles. At the large pre-B cell stage, once VDJH recombination completed, pre-BCR signaling induces repositioning of one IgH allele into a repressive centromeric domain. Although pericentromeric recruitment has been associated with allelic exclusion (57), this nuclear location does not preclude transcriptional expression of the non-productive allele (58). Monoallelic IgH addressing to pericentromeric heterochromatin (PCH) remains until the immature B cell stage (57).
IgH Locus Contraction and Chromatin Loops
Beyond nuclear positioning, IgH allele contraction/decontraction, as well as TAD and loop reorganization represent an additional level of regulation, considered as critical for early B cell development (59). At the pre-pro B cell stage, both IgH alleles are decontracted (60) and at the pro-B cell stage both IgH loci contract to bring DH et JH clusters closer to promote DH to JH rearrangement (Figure 3B). 3C (Chromosome Capture Conformation) and 4C (Circular Chromosome Capture Conformation) experiments revealed that IgH locus compaction was mediated by large loops in which Eµ interacts with 3’RR, IGCR1 and VH regions (5’ VH7183 and 5’ VH558) (60). During the transition from pro-B to large pre-B cell stages, the entire VH region is brought into juxtaposition with DH elements due to extensive IgH locus contraction (60). This contraction is completed by looping within the VH region to facilitate VDJ recombination (57, 61). This builds a rosette-like structure and allows equal usage between proximal, middle and distal VH genes, essential for the generation of a diversified immunoglobulin repertoire (62). After productive IgH recombination and in response to pre-BCR signaling, locus contraction is reversed in small pre-B cells (60) or in large pre-B cells (57) depending on the studies. This decontracted state remains until the immature stage (57) Discrepancies between Rother’s (60) and Roldan’s (57) reports could be due to different probes used in 3D-FISH. Rother used three different probes encompassing distal VH, proximal VH and CH genes whereas Roldan employed only two probes including one spanning Cγ1 and the other VHJ558 (distal). Ultimately, IgH looping plays a critical role in V(D)J recombination by forming a “recombination center” containing pDQ52, all four JH segments and cEµ (17). These loops permit RAG scanning between two recognition signal sequences (RSS) and therefore focal RAG targeting activity during DH to JH joining. This loop extrusion, is thought to ensure better sequence recognition by the RAG complex, avoiding off-targeting (63).
While IgH locus contraction and looping are important to permit equal usage of all VH genes, nuclear positioning of IgH loci seems more determinant than locus contraction in the orchestration of ordered V(D)J rearrangements (60).
Igκ Nuclear Positioning and Chromosome Loops
In contrast to the IgH locus, the exact kinetics of Igk loci contraction and relocation within the nucleus remain unclear but it is admitted that dynamic changes also occur at Igκ loci. In CLP and pre-pro B cells, both Igκ alleles are located in PCH. At the pro-B cell stage, Igk loci relocate to a more central and active area within the nucleus (54, 64, 65). From large pre-B cells and until the immature stage, one Igκ allele becomes more closely associated with active chromatin and the other Igκ allele stays in the PCH (64) (Figure 3A). This chronology is still debated. Rother et al. described relocation of both Igκ loci only at pre-B cell stages (60). In small pre-B cells, contraction occurs at the Igκ locus, in preparation for Vκ-Jκ recombination and this contracted status persists until the immature B cell stage (39, 57) (Figure 3B). While some controversy still remains concerning contraction of Igκ locus in all early developmental stages, it is clear that redistribution of intra-loci interactions occurs at the small pre-B cell stage (60, 66). This redistribution, mediated by pre-BCR signaling, results in Igκ looping through ordered coordination between MiEκ, 3’Eκ and Sis regulatory elements spread throughout the locus (67).
Role of Ig Enhancers in B Cell Nuclear Organization
IgH V-DH Intergenic Region and IGCR1
Beyond the observation that IGCR1 interacts with the 3’CBEs insulator region in pro-B cells (68, 69); recent studies suggest an insulation function for IGCR1 itself. In an IGCR1 deficient mouse model, interaction between the whole V-DH intergenic region and 3’CBEs was abolished while Eµ/3’RR interaction was not (69). In IGCR1 deficient pro-B cells, Eµ/IGCR1 interaction was decreased as expected but Eµ/VH81 and Eµ/3’RR interactions still occurred (70). Moreover, the recombination center, in which DFL1.6/DQ52/Eµ/hs4 interact in a wt context, was disturbed in IGCR1 deficient pro-B cells. The absence of IGCR1 allows an alternative bigger loop to occur between VH81X/DQ52/Eµ/hs4. By including proximal VH segments in the former loop, this new chromosome conformation alters V(D)J repertoire by decreasing distal VH usage (70). According to the actual model, IGCR1 seems to work as an insulator to delimit Eμ action/function. These findings are contradicted by Busslinger’s group who carried out 4C-Seq experiments in IGCR1 deficient pro-B cells where interaction patterns remain unchanged compared to Rag-/- pro-B cells (71). This discrepancy between studies could be explained by the restriction enzymes used in 4C experiments. When Busslinger’s group used sequentially HindIII (6pb cutter) and Sau3AI enzymes, Sen’s group used two 4pb cutter enzymes (MseI and NlaIII) that generate smaller fragments and probably offer higher resolution (Table 1).
IgH DQ52 Enhancer and Eµ Region
Many studies have contributed to the identification and unraveling of the role of Ig gene enhancers on 3D loci conformation during early B cell development proving that such regulatory elements are clearly involved in subnuclear positioning and chromosome looping (intra-TAD modification). To decipher the role of pDQ52 and the Eµ enhancer, Guo and colleagues generated two mouse models (17): P-E- (both pDQ52/DQ52 and Eµ enhancers are deleted) and P-E+ (only promotor pDQ52 and DQ52 segments are deleted). Using 3D-FISH, authors demonstrated that the Eμ enhancer is required for IgH locus contraction in pro-B cells. Indeed, in P-E-, but not in the P-E+ model, the large loops between V-DH intergenic regions (DFL16.1, IGCR1), Eµ and the 3’RR are drastically reduced underlining the important role of Eµ in loop formation. Moreover, ChIP-Seq experiments coupled with 3C experiments demonstrated that such large domain interactions are shaped by CTCF and cohesin proteins (68). In this way, CTCF-mediated IgH looping facilitates the generation of a diversified repertoire by juxtaposing distal VH to DJH regions (71). It has also been shown that, in pro-B cells, cEµ deletion leads to IgH relocation in close proximity to the nuclear periphery (17). Taken together, these studies show that the Eµ region has a pivotal role during early V(D)J recombination by regulating both long range interactions and IgH sub-nuclear positioning. While Eµ seems to facilitate IgH loops in developing B cells, the presence of this enhancer is not strictly necessary since another study showed, by 4C-Seq, that several long-range interactions remain detectable in the absence of Eµ (71) (Table 1).
Igκ E88
The E88 element located in the intervening V-Jκ region also participates in the generation of a diversified Ig repertoire (38). Endowed with enhancer activity starting at pre-B cell stages, E88 regulates long-range Igκ chromatin interactions and participates in sub-TAD determination of the Igκ locus. E88 deletion in pro-B cells disrupts interactions between Vκ/Jκ genes and three other known enhancers, especially MiEκ, but also to some extent 3’Eκ and 3’Ed (38). It is then reasonable to consider E88 as a major hub of Igk locus interactions critical for regulation of Igk repertoire diversity.
Igk Cer and Sis Elements
While of critical interest to generate a diversified antibody repertoire of the Igκ chain (39, 40); Cer and Sis regulatory elements located within the Igκ locus are also involved in Ig loci PCH addressing. It is now established that IgH and Igκ loci are monoallelically repositioned into PCH in pre-B cells (72). In Sis-/- pre-B cells, both IgH and Igk alleles do not relocate to PCH. While Igκ locus contraction and looping still occur normally in the absence of this element, Sis-deficient B cells harbor a biased Igκ repertoire with increased proximal Vκ gene usage accompanied by decreased distal Vκ utilization. Taken together, these results show that the Sis element is required for Igκ (cis-acting) and IgH (trans-acting) monoallelic positioning within the nucleus and somehow promotes a diversified antibody repertoire (39). Cer deletion leads to a strong increase in proximal Vκ usage with decreased distal Vκ usage. Cer-/- pre-B cells show normal epigenetic marks throughout the Igκ locus but the compaction level is clearly decreased suggesting that Cer regulates Igκ locus contraction (40). As a whole, Sis seems to be more involved in nuclear Ig loci positioning and Cer in Igκ locus conformation. The idea that nuclear positioning, rather than loci contraction, promotes Ig gene recombination (60) is thereby questioned by the reciprocal functions of Sis and Cer elements in the Igκ locus.
Igk MiEκ Element
Igκ locus contraction, mediated by MiEκ, seems to occur at the pro-B cell stage and remains until the pre-B cell stage. This observation indicates that locus contraction is lineage, but not stage specific (60). In pro-B cells, MiEκ deletion both provokes positioning of both Igκ alleles to PCH and reduces physical distance between IgH and Igκ loci evidence of Ig loci crosstalk at this stage. In pre-B cells, MiEκ deletion enforces both Igk and IgH allele positioning to PCH (76). The current model proposes that MiEk is more implicated in Ig loci sub-nuclear organization than in locus looping.
3’ Eκ Region
Involvement of 3’Eκ in early rearrangement of the Igκ locus is clearly established. This enhancer region is necessary for Igκ germline transcription activation, which is a prerequisite for Vκ-Jκ recombination (77). In line with the previously observed transcription defect, deletion of the3’Eκ enhancer induces premature repositioning of Igκ into PCH at the pre-B cell stage. Moreover, according to Park and colleagues, in pro-B cells, Igκ seems to be distant from the IgH locus although both IgH and Igκ appear to be located at the nuclear periphery (65). More strikingly, in 3’Eκ deficient mice, Skok and colleagues also documented multiple defects in IgH decontraction, relocalization to PCH and IgH-Igκ association (76). Altogether these results suggest that IgH decontraction is dependent on PCH repositioning and Igκ-IgH colocalization. This particular model highlights an unexpected trans – acting effect upon deletion of an Ig gene enhancer. This observation supposes strong crosstalk between Ig loci in developing B cells and supports the hypothesis that regulatory regions are involved in interactions between loci. In parallel, crosstalk between Igκ and IgH is a little more frequent in pre-B cells than in pro-B cells and becomes almost inexistent at immature stages (65).
Nuclear Organization Driven by Ig Enhancers in Late Developing B Cells
In secondary lymphoid organs, mature B cells can encounter soluble or membrane antigens able to engage their BCR and induce proper B cell activation for secondary remodeling events. Within the germinal center (GC), B cells undergo to two major genetic rearrangements initiated by AID (78): SHM and CSR at IgH loci whereas only SHM occurs at IgL loci. By inducing frequent point mutations into the variable regions of Ig loci, SHM is a driving force for antibody affinity maturation. CSR results in µ heavy chain replacement by another subtype (IgG, IgA or IgE) after initiating DNA double strand breaks and recombination. In addition, locus suicide recombination (LSR) is another rearrangement which leads to deletion of all IgH constant genes and therefore induces B cell death by abrogating surface B cell receptor expression (1). In parallel to such secondary remodeling events, activated B cells differentiate into antibody secreting plasma cel or memory B cells (73).
IgH Nuclear Positioning and Loops in Mature Naive and Activated B Cells
Mature B cell differentiation and its accompanying late genetic remodeling events are also characterized by changes in nuclear organization and chromatin loops. Pioneering studies used 3D-FISH to show that IgH alleles were not located in similar positions within nuclei of resting naive and activated B cells. At the resting mature B cell stage, both IgH alleles are localized in euchromatin whereas their respective nuclear position upon in vitro activation is still debated. According to Skok’s lab, the IgH allele colocalized with PCH and replicated later, suggesting that this allele is the unproductive allele (72) (Figure 4). These elements imply that the non-productive IgH allele is “tagged and maintained as excluded” by nuclear location. Although when using the same 3D-FISH approach in stimulated cells, the De Latt group showed that both IgH loci are sitting in an active compartment (79) (Figure 4A). This observation is in agreement with another study in which IgH alleles relocalized to the nuclear periphery in proliferating splenic B cells (80). By using mouse models carrying IgM of “a” and “b” allotypes (respectively from C57Bl6 and SV129 backgrounds), Holwerda and colleagues also showed, by 4C-Seq, that tardive replication in splenic B cells was lymphoid specific but independent of nuclear location and topology of Ig loci (79). The fact that allelic exclusion might not be driven by nuclear location is in agreement with previous studies showing IgH biallelic expression in mature B cells (58, 81). Evidence for loop formation was first provided by the Kenter group that described high frequencies of interaction between Eµ and 3’RR in mature resting B cells. Loop conformation within IgH changes upon in vitro stimulation by lipopolysaccharide (LPS) with or without interleukin (IL-4), which respectively induce CSR mostly towards IgG3 and IgG1; by acquiring additional contact between previously interacting enhancers and acceptor switch region (Sγ3 or Sγ1) involved in CSR (75) (Figure 4B). The advantage of this architectural scaffolding, promoting synapsis between S regions, has been mechanistically demonstrated to facilitate CSR by the loop extrusion mechanism (74, 82). Briefly, cohesin is loaded at the 3’RR end and the extrusion mechanism bring together 3’RR and Eµ regions. An additional internal loop is further formed to juxtapose the transcriptionally active S regions. This particular conformation first allows AID recruitment at both Sµ and S acceptor regions to induce DSB and second maintains S regions together for ligation by the NHEJ pathway [for review (83)].
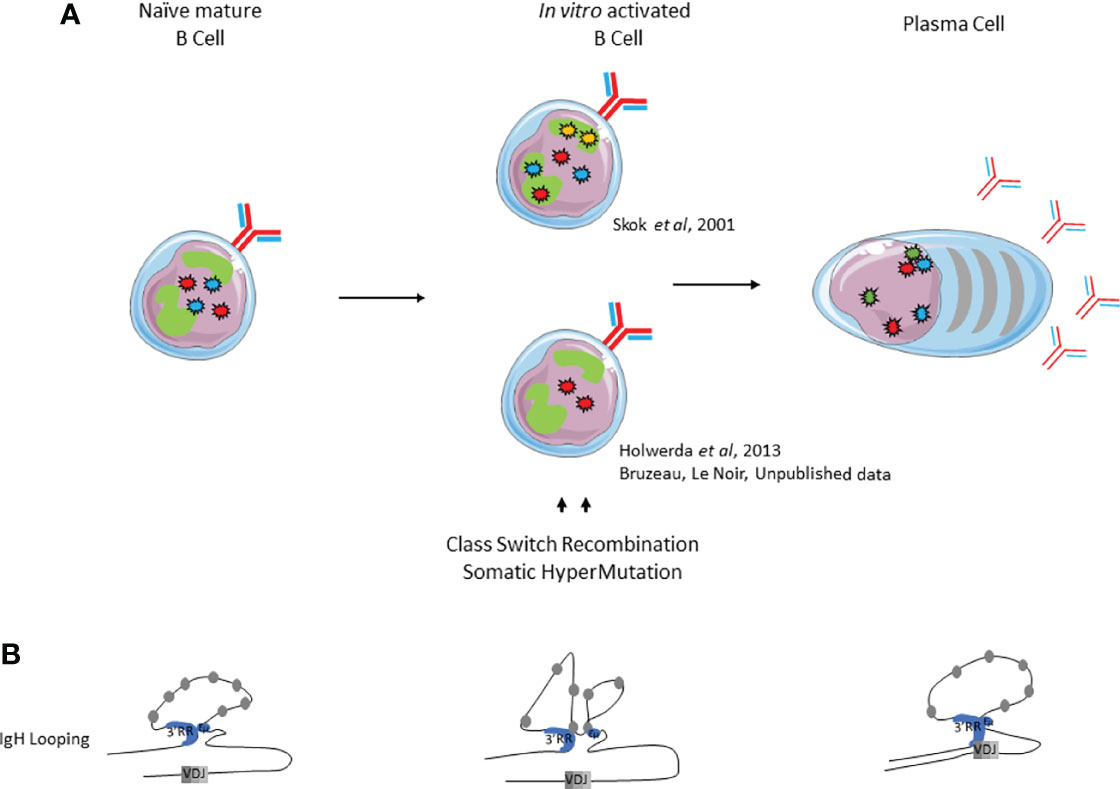
Figure 4 Immunoglobulin loci positioning and conformation within the nucleus during late B cell development. (A). Nuclear positioning of IgH, Igk and IgJ chains. (B). IgH looping. IgH alleles are represented by red stars, Igκ alleles by blue stars, Igλ alleles by yellow stars and J chain alleles by green stars.
Igκ Nuclear Positioning in Mature Naive and Activated B Cells
Similar observations can be made for IgL loci (Figure 4). Igκ monoallelic positioning in PCH is often observed in activated splenic B cells and this PCH-localized allele replicates later compared to the other one (64, 84). The current model proposes that this monoallelic positioning drives both light chain allelic and isotypic exclusion. Effectively, both Igλ alleles are located in repressive areas in the nuclear periphery of in vitro stimulated murine B cells, which mostly express Igκ.When Igλ expression is forced, one Igλ allele is recruited to a central permissive compartment within the nucleus and both Igk alleles are repositioned to the nuclear periphery, thus inhibiting Igκ expression (72). Moreover, colocalization of Igκ with IgH loci is observed more frequently upon LPS stimulation of B cells (65).
Ig Nuclear Positioning and Chromosome Looping in Plasma Cells
The intense antibody secreting function of plasma cells requires high levels of immunoglobulin gene transcription. For this purpose, nuclear organization can now be considered as one important level of regulation as described by Garrard and colleagues (65). Although located on three different chromosomes, Ig genes (IgH, Igκ and Igλ) in plasma cells often undergo physical clustering by forming pairs or triplets. Such clusters, often composed of functional alleles, preferentially localize within the same transcription factory near the nuclear periphery (Figure 4A). In this study, ChIP-3C-seq experiments performed with anti-RNA PolII antibodies indicate that physical interactions within transcription factories are mediated by Ig gene enhancers: cEµ and 3’RR for IgH, Eiκ and 3’Eκ for Igκ loci. Nuclear location also facilitates transport of Ig transcripts from the nucleus towards the endoplasmic reticulum (65). Moreover, 3C experiments performed by Birshtein’s group underscored the importance of IgH chromatin conformation in a plasma cell line. In this study, authors showed that physical interactions between VH genes and the 3’RR were involved in efficient IgH transcription, heavy chain expression and ensuing antibody synthesis (85). All of these elements demonstrate that nuclear organization contributes favorably to massive antibody synthesis (65).
Role of Ig Enhancers on B Cell Nuclear Organization and Loops (Table 1)
While globally less documented, the implication of Ig gene enhancers in B cell nuclear organization (Ig positioning and loop formation) has been proposed in the context of the first-described IgH locus loop bringing the 3’RR and Eµ regions into close contact before CSR. Upon LPS-stimulation, this configuration acquires additional contact between the acceptor S region involved in CSR and previous interacting IgH enhancers (75). In plasma cells, Ig loci “coordination” seems to be mediated by the 3’Eκ regulatory element since its deletion leads to a decrease in cohabitation of all Ig loci with decreases in IgH-Igk, IgH-IgJ and Igk-IgJ communication. Likewise, nuclear localization of these loci seems to be modified with the relocation of IgH, Igk and IgJ alleles further from the nuclear periphery in 3’Eκ-/- plasma cells compared to wt plasma cells. Together, these results suggest that interactions between Ig genes (including interaction between IgH and IgJ) are mediated by Igk, especially by 3’Ek enhancer. Moreover, relocation and mis-cohabitation of Ig loci correlate with decreased transcription of each Ig gene, as observed in 3’Eκ-/- plasma cells in comparison with wt cells (65).
IgH positioning within the nucleus might also be supported by the 3’RR. Complete 3’RR deletion leads to an increase in distance between both IgH loci in activated B cells (86). The pioneering study by Kenter’s group showed that IgH looping requires an intact 3’RR. This statement is supported by the model devoid of its two last enhancers, hs3b and hs4, that led to a decrease in interactions between 3’RR and Eµ regions in resting and LPS ± IL4 stimulated splenic B cells (75). In contrast, partial deletion of 3’CBEs (hs5 to 7) does not impair loop formation at resting and activated B cell stages (26). However, Alt’s group reported that total 3’CBEs deletion of led to significant decreases in interactions between Eµ and targeted switch regions in activated B cells (25). Deletion of the cEµ enhancer seems to have no impact on interactions between 3’RR and Eµ regions since the contact frequencies in cEµ KO B cells are comparable to wt B cells (75). Changes in IgH locus conformation mediated by interactions between promoters and enhancers, could impair CSR by limiting the activity of promoters located upstream from each constant gene, and therefore, their ability to initiate prerequisite germline transcription (87).
Discussion
Nuclear positioning and chromosome looping have clearly been shown to display a functional role in early developing B cells and in B cells in general. Beyond some discrepancies, studies overall showed that positioning and conformation of Ig loci play a major role in ordered H and L chains rearrangements. Furthermore, at such early stages, nuclear repositioning of Ig loci to PCH is moreover integral to allelic exclusion mechanism while chromosome looping optimizes antibody repertoire constitution. Regulatory regions of Ig gene loci largely participate in positioning and looping. At both IgH and Igk, Eµ and 3’RR as well as Cer elements are essential for the proper loop formation in their respective loci. In contrast, Igk Sis element is supposed to ensure crosstalk between IgH and Igk loci since influencing cis- as well as trans-positioning. Similarly, 3’Eκ also promotes, at early stages, temporal association between IgH and Igκ.
In contrast, in mature B cells, the role of Ig loci position within the nucleus remains more elusive since studies unveil various differences. Nevertheless, IgH chromosome looping has been distinctly shown to play a role in CSR, especially by bringing donor Sµ and acceptor switch regions in close proximity to optimize switch recombination events. During SHM, primary transcription of the V exon, potentially in both sense and antisense direction, is an important prerequisite for AID-induced mutations. In KO mouse models in which the 3’RRpalindromic structure is disrupted, the observed SHM defects correlate to a decrease in VH primary transcription (29, 35, 36). As transcription is an essential step to initiate loop extrusion prior CSR, establishing a link between SHM and loop extrusion would be of critical interest. In addition, the function of ncRNA in chromosome topology during CSR (88) and SHM (89) has been elucidated. In plasma cells, only few studies reported that Ig loci are colocalized to transcription factories, surely to improve antibody production. Similarly, the role of Ig loci conformation throughout terminal B cell development is also of major interest.
IgH locus positioning within the nucleus during CSR seems to be important for legitimate maintenance. While IgH nuclear location in close proximity to an oncogene is a contributing factor for translocation (90); its distal 3’RR enhancers hs3b and hs4 have be shown to be only involved in oncogene activation but not in translocation (91). Given that AID activity initiates DNA modifications, it is mandatory that IgH rearrangements be tightly regulated to avoid any mutations or translocations within oncogenes (so called “AID off targets”). Several AID off target genes (ie: Pax5, Il4ra and Inf8) lie in close proximity to IgH during CSR and altogether are located in a chromosomal territory containing a high AID concentration (90). As a consequence, the non-random positioning and conformation of Ig loci could be widely considered as critical to maintain B cell genome integrity. In this way, their respective regulatory elements could play the role of B cell genome guardians thereby avoiding illegitimate events that contribute to lymphomagenesis.
Author Contributions
CB wrote the manuscript and prepared the figures. SN wrote the manuscript. EP provided critical feedback. All authors contributed to the article and approved the submitted version.
Funding
CB was supported by a Ph.D. fellowship of the french Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation. ANR-21-CE15-0001-01.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Laffleur B, Denis-Lagache N, Cook-Moreau J, Tinguely A, Delpy L, et al. AID-Driven Deletion Causes Immunoglobulin Heavy Chain Locus Suicide Recombination in B Cells. Science (2012) 336:931–4. doi: 10.1126/science.1218692
2. Dekker J, Rippe K, Dekker M, Kleckner N. Capturing Chromosome Conformation. Science (2002) 295:1306–11. doi: 10.1126/science.1067799
3. Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and Interaction Between Hypersensitive Sites in the Active Beta-Globin Locus. Mol Cell (2002) 10:1453–65. doi: 10.1016/S1097-2765(02)00781-5
4. Ea V, Baudement M-O, Lesne A, Forné T. Contribution of Topological Domains and Loop Formation to 3D Chromatin Organization. Genes (2015) 6:734–50. doi: 10.3390/genes6030734
5. Wutz G, Várnai C, Nagasaka K, Cisneros DA, Stocsits RR, Tang W, et al. Topologically Associating Domains and Chromatin Loops Depend on Cohesin and are Regulated by CTCF, WAPL, and PDS5 Proteins. EMBO J (2017) 36:3573–99. doi: 10.15252/embj.201798004
6. Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. Spatial Partitioning of the Regulatory Landscape of the X-Inactivation Center. Nature (2012) 485:381–5. doi: 10.1038/nature11049
7. Amiad-Pavlov D, Lorber D, Bajpai G, Reuveny A, Roncato F, Alon R, et al. Live Imaging of Chromatin Distribution Reveals Novel Principles of Nuclear Architecture and Chromatin Compartmentalization. Sci Adv (2021) 7:eabf6251. doi: 10.1126/sciadv.abf6251
8. Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, et al, et al. Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions. Nature (2012) 485:376–80. doi: 10.1038/nature11082
9. Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science (2009) 326:289–93. doi: 10.1126/science.1181369
10. Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, et al. Nuclear Organization of Active and Inactive Chromatin Domains Uncovered by Chromosome Conformation Capture–on-Chip (4C). Nat Genet (2006) 38:1348–54. doi: 10.1038/ng1896
11. Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel, et al. Interchromosomal Interactions and Olfactory Receptor Choice. Cell (2006) 126:403–13. doi: 10.1016/j.cell.2006.06.035
12. Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal Associations Between Alternatively Expressed Loci. Nature (2005) 435:637–45. doi: 10.1038/nature03574
13. Arakawa H, Shimizu T, Takeda S. Re-Evaluation of the Probabilities for Productive Rearrangements on the K and X Loci. Int Immunol (1996) 9(8):91–9. doi: 10.1093/intimm/8.1.91
14. Engel H, Rolink A, Weiss S. B Cells are Programmed to Activate B and Ø for Rearrangement at Consecutive Developmental Stages. Eur J Immunol (1999) 10(29):2167–76. doi: 10.1002/(SICI)1521-4141(199907)29:07<2167::AID-IMMU2167>3.0.CO;2-H
15. Pawlitzky I, Angeles CV, Siegel AM, Stanton ML, Riblet R, Brodeur PH. Identification of a Candidate Regulatory Element Within the 5’ Flanking Region of the Mouse Igh Locus Defined by Pro-B Cell-Specific Hypersensitivity Associated With Binding of PU.1, Pax5, and E2A. J Immunol Baltim Md 1950 (2006) 176:6839–51. doi: 10.4049/jimmunol.176.11.6839
16. Featherstone K, Wood AL, Bowen AJ, Corcoran AE. The Mouse Immunoglobulin Heavy Chain V-D Intergenic Sequence Contains Insulators That May Regulate Ordered V(D)J Recombination. J Biol Chem (2010) 285:9327–38. doi: 10.1074/jbc.M109.098251
17. Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, et al. Two Forms of Loops Generate the Chromatin Conformation of the Immunoglobulin Heavy-Chain Gene Locus. Cell (2011) 147:332–43. doi: 10.1016/j.cell.2011.08.049
18. Alessandrini A, Desiderio SV. Coordination of Immunoglobulin DJH Transcription and D-To-JH Rearrangement by Promoter-Enhancer Approximation. Mol Cell Biol (1991) 11:2096–107. doi: 10.1128/mcb.11.4.2096-2107.1991
19. Predeus AV, Gopalakrishnan S, Huang Y, Tang J, Feeney AJ, Oltz EM, et al. Targeted Chromatin Profiling Reveals Novel Enhancers in Ig H and Ig L Chain Loci. J Immunol Baltim Md 1950 (2014) 192:1064–70. doi: 10.4049/jimmunol.1302800
20. Amoretti-Villa R, Rogier M, Robert I, Heyer V, Reina-San-Martin B. A Novel Regulatory Region Controls IgH Locus Transcription and Switch Recombination to a Subset of Isotypes. Cell Mol Immunol (2019) 16:887–9. doi: 10.1038/s41423-019-0267-4
21. Lieberson R, Giannini SL, Birshtein BK, Eckhardt LA. An Enhancer at the 3’ End of the Mouse Immunoglobulin Heavy Chain Locus. Nucleic Acids Res (1991) 19:933–7. doi: 10.1093/nar/19.4.933
22. Chauveau C, Cogné M. Palindromic Structure of the IgH 3’locus Control Region. Nat Genet (1996) 14:15–6. doi: 10.1038/ng0996-15
23. Madisen L, Groudine M. Identification of a Locus Control Region in the Immunoglobulin Heavy-Chain Locus That Deregulates C-Myc Expression in Plasmacytoma and Burkitt’s Lymphoma Cells. Genes Dev (1994) 8:2212–26. doi: 10.1101/gad.8.18.2212
24. Sepulveda M, Garrett F, Pricewhelan A, Birshtein B. Comparative Analysis of Human and Mouse 3? Regulatory Regions Identifies Distinctive Structural Features. Mol Immunol (2005) 42:605–15. doi: 10.1016/j.molimm.2004.09.006
25. Zhang X, Yoon HS, Chapdelaine-Williams AM, Kyritsis N, Alt FW. Physiological Role of the 3′IgH CBEs Super-Anchor in Antibody Class Switching. Proc Natl Acad Sci (2021) 118:e2024392118. doi: 10.1073/pnas.2024392118
26. Volpi SA, Verma-Gaur J, Hassan R, Ju Z, Roa S, Chatterjee S, et al. Germline Deletion of Igh 3′ Regulatory Region Elements Hs 5, 6, 7 (Hs5–7) Affects B Cell-Specific Regulation, Rearrangement, and Insulation of the Igh Locus. J Immunol (2012) 188:2556–66. doi: 10.4049/jimmunol.1102763
27. Vincent-Fabert C, Fiancette R, Pinaud E, Denizot Y. Genomic Deletion of the Whole IgH 3Ј Regulatory Region (Hs3a, Hs1,2, Hs3b, and Hs4) Dramatically Affects Class Switch Recombination and Ig Secretion to All Isotypes. Blood (2010) 116:5. doi: 10.1182/blood-2010-01-264689
28. Perlot T, Alt FW, Bassing CH, Suh H, Pinaud E. Elucidation of IgH Intronic Enhancer Functions via Germ-Line Deletion. Proc Natl Acad Sci (2005) 102:14362–7. doi: 10.1073/pnas.0507090102
29. Garot A, Marquet M, Saintamand A, Bender S, Le Noir S, Rouaud P, et al. Sequential Activation and Distinct Functions for Distal and Proximal Modules Within the IgH 3′ Regulatory Region. Proc Natl Acad Sci (2016) 113:1618–23. doi: 10.1073/pnas.1514090113
30. Vincent-Fabert C, Truffinet V, Fiancette R, Cogné N, Cogné M, Denizot Y, et al. Ig Synthesis and Class Switching Do Not Require the Presence of the Hs4 Enhancer in the 3′ IgH Regulatory Region. J Immunol (2009) 182:6926–32. doi: 10.4049/jimmunol.0900214
31. Bébin A-G, Carrion C, Marquet M, Cogné N, Lecardeur S, Cogné M, et al. In Vivo Redundant Function of the 3′ IgH Regulatory Element HS3b in the Mouse. J Immunol (2010) 184:3710–7. doi: 10.4049/jimmunol.0901978
32. Pinaud E, Khamlichi AA, Le Morvan C, Drouet M, Nalesso V, Le Bert M, et al. Localization of the 3′ IgH Locus Elements That Effect Long-Distance Regulation of Class Switch Recombination. Immunity (2001) 15:187–99. doi: 10.1016/S1074-7613(01)00181-9
33. Manis JP, van derStoep N, Tian M, Ferrini R, Davidson L, Bottaro A, et al. Class Switching in B Cells Lacking 3′ Immunoglobulin Heavy Chain Enhancers. J Exp Med (1998) 188:1421–31. doi: 10.1084/jem.188.8.1421
34. Cogné M, Lansford R, Bottaro A, Zhang J, Gorman J, Young, et al. A Class Switch Control Region at the 3′ End of the Immunoglobulin Heavy Chain Locus. Cell (1994) 77:737–47. doi: 10.1016/0092-8674(94)90057-4
35. Le Noir S, Boyer F, Lecardeur S, Brousse M, Oruc Z, Cook-Moreau J, et al. Functional Anatomy of the Immunoglobulin Heavy Chain 3' Super-Enhancer Needs Not Only Core Enhancer Elements But Also Their Unique DNA Context. Nucleic Acids Res (2017) 45:5829–37. doi: 10.1093/nar/gkx203
36. Saintamand A, Vincent-Fabert C, Garot A, Rouaud P, Oruc Z, Magnone V, et al. Deciphering the Importance of the Palindromic Architecture of the Immunoglobulin Heavy-Chain 3’ Regulatory Region. Nat Commun (2016) 7:10730. doi: 10.1038/ncomms10730
37. Bruzeau C, Moreau J, Le Noir S, Pinaud E. Panorama of Stepwise Involvement of the IgH 3’ Regulatory Region in Murine B Cells. Adv Immunol (2021) 149:95–114. doi: 10.1016/bs.ai.2021.03.004
38. Barajas-Mora EM, Kleiman E, Xu J, Carrico NC, Lu H, Oltz EM, et al. A B-Cell-Specific Enhancer Orchestrates Nuclear Architecture to Generate a Diverse Antigen Receptor Repertoire. Mol Cell (2019) 73:48–60.e5. doi: 10.1016/j.molcel.2018.10.013
39. Xiang Y, Zhou X, Hewitt SL, Skok JA, Garrard WT. A Multifunctional Element in the Mouse Igκ Locus That Specifies Repertoire and Ig Loci Subnuclear Location. J Immunol (2011) 186:5356–66. doi: 10.4049/jimmunol.1003794
40. Xiang Y, Park S-K, Garrard WT. Vκ Gene Repertoire and Locus Contraction Are Specified by Critical DNase I Hypersensitive Sites Within the Vκ-Jκ Intervening Region. J Immunol (2013) 190:1819–26. doi: 10.4049/jimmunol.1203127
41. Cockerill PN, Garrard WT. Chromosomal Loop Anchorage of the Kappa Immunoglobulin Gene Occurs Next to the Enhancer in a Region Containing Topoisomerase II Sites. Cell (1986) 44:273–82. doi: 10.1016/0092-8674(86)90761-0
42. Emorine L, Kuehl M, Weir L, Leder P, Max EE. A Conserved Sequence in the Immunoglobulin Jκ–Cκ Intron: Possible Enhancer Element. Nature (1983) 304:447–9. doi: 10.1038/304447a0
43. Bergman Y, Rice D, Grosschedl R, Baltimore D. Two Regulatory Elements for Immunoglobulin Kappa Light Chain Gene Expression. Proc Natl Acad Sci U S A (1984) 81:7041–5. doi: 10.1073/pnas.81.22.7041
44. Meyer KB, Neuberger MS. The Immunoglobulin Kappa Locus Contains a Second, Stronger B-Cell-Specific Enhancer Which is Located Downstream of the Constant Region. EMBO J (1989) 8:1959–64. doi: 10.1002/j.1460-2075.1989.tb03601.x
45. Liu Z-M, George-Raizen JB, Li S, Meyers KC, Chang MY, Garrard WT, et al. Chromatin Structural Analyses of the Mouse Igκ Gene Locus Reveal New Hypersensitive Sites Specifying a Transcriptional Silencer and Enhancer. J Biol Chem (2002) 277:32640–9. doi: 10.1074/jbc.M204065200
46. Carson S, Wu GE. A Linkage Map of the Mouse Immunoglobulin Lambda Light Chain Locus. Immunogenetics (1989) 29:173–9. doi: 10.1007/BF00373642
47. Hagman J, Rudin CM, Haasch D, Chaplin D, Storb U. A Novel Enhancer in the Immunoglobulin Lambda Locus is Duplicated and Functionally Independent of NF Kappa B. Genes Dev (1990) 4:978–92. doi: 10.1101/gad.4.6.978
48. Hendrickson BA, Conner DA, Ladd DJ, Kendall D, Casanova JE, Corthesy B, et al. Altered Hepatic Transport of Immunoglobulin A in Mice Lacking the J Chain. J Exp Med (1995) 182:1905–11. doi: 10.1084/jem.182.6.1905
49. Erlandsson L, Andersson K, Sigvardsson M, Lycke N, Leanderson T. Mice With an Inactivated Joining Chain Locus Have Perturbed IgM Secretion. Eur J Immunol (1998) 28:2355–65. doi: 10.1002/(SICI)1521-4141(199808)28:08<2355::AID-IMMU2355>3.0.CO;2-L
50. Hardy RR, Kincade PW, Dorshkind K. The Protean Nature of Cells in the B Lymphocyte Lineage. Immunity (2007) 26:703–14. doi: 10.1016/j.immuni.2007.05.013
51. Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-Lymphoid Lineage Depends on the Transcription Factor Pax5. Nature (1999) 401:7. doi: 10.1038/44076
52. Chakraborty T, Chowdhury D, Keyes A, Jani A, Subrahmanyam R, Ivanova, et al. Repeat Organization and Epigenetic Regulation of the DH-Cμ Domain of the Immunoglobulin Heavy-Chain Gene Locus. Mol Cell (2007) 27:842–50. doi: 10.1016/j.molcel.2007.07.010
53. Mårtensson I-L, Rolink A, Melchers F, Mundt C, Licence S, Shimizu T, et al. The Pre-B Cell Receptor and its Role in Proliferation and Ig Heavy Chain Allelic Exclusion. Semin Immunol (2002) 14:335–42. doi: 10.1016/S1044-5323(02)00066-0
54. Kosak ST. Subnuclear Compartmentalization of Immunoglobulin Loci During Lymphocyte Development. Science (2002) 296:158–62. doi: 10.1126/science.1068768
55. Johanson TM, Lun ATL, Coughlan HD, Tan T, Smyth GK, Nutt SL, et al. Transcription-Factor-Mediated Supervision of Global Genome Architecture Maintains B Cell Identity. Nat Immunol (2018) 19:1257–64. doi: 10.1038/s41590-018-0234-8
56. Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, et al. Global Changes in the Nuclear Positioning of Genes and Intra- and Interdomain Genomic Interactions That Orchestrate B Cell Fate. Nat Immunol (2012) 13:1196–204. doi: 10.1038/ni.2432
57. Roldán E, Fuxa M, Chong W, Martinez D, Novatchkova M, Busslinger M, et al. Locus ‘Decontraction’ and Centromeric Recruitment Contribute to Allelic Exclusion of the Immunoglobulin Heavy-Chain Gene. Nat Immunol (2005) 6:31–41. doi: 10.1038/ni1150
58. Daly J, Licence S, Nanou A, Morgan G, Mårtensson I-L. Transcription of Productive and Nonproductive VDJ-Recombined Alleles After IgH Allelic Exclusion. EMBO J (2007) 26:4273–82. doi: 10.1038/sj.emboj.7601846
59. Kumari G, Sen R. Chromatin Interactions in the Control of Immunoglobulin Heavy Chain Gene Assembly. Adv Immunol (2015) 128:41–92. doi: 10.1016/bs.ai.2015.08.001
60. Rother MB, Palstra R-J, Jhunjhunwala S, van Kester KAM, van IJcken WFJ, Hendriks RW, et al. Nuclear Positioning Rather Than Contraction Controls Ordered Rearrangements of Immunoglobulin Loci. Nucleic Acids Res (2016) 44:175–86. doi: 10.1093/nar/gkv928
61. Sayegh C. Visualization of Looping Involving the Immunoglobulin Heavy-Chain Locus in Developing B Cells. Genes Dev (2005) 19:322–7. doi: 10.1101/gad.1254305
62. Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJM, et al. The 3d Structure of the Immunoglobulin Heavy-Chain Locus: Implications for Long-Range Genomic Interactions. Cell (2008) 133:265–79. doi: 10.1016/j.cell.2008.03.024
63. Zhang Y, Zhang X, Ba Z, Liang Z, Dring EW, Hu H, et al. The Fundamental Role of Chromatin Loop Extrusion in Physiological V(D)J Recombination. Nature (2019) 573:600–4. doi: 10.1038/s41586-019-1547-y
64. Goldmit M, Ji Y, Skok J, Roldan E, Jung S, Cedar H, et al. Epigenetic Ontogeny of the Igk Locus During B Cell Development. Nat Immunol (2005) 6:198–203. doi: 10.1038/ni1154
65. Park S-K, Xiang Y, Feng X, Garrard WT. Pronounced Cohabitation of Active Immunoglobulin Genes From Three Different Chromosomes in Transcription Factories During Maximal Antibody Synthesis. Genes Dev (2014) 28:1159–64. doi: 10.1101/gad.237479.114
66. Fitzsimmons SP, Bernstein RM, Max EE, Skok JA, Shapiro MA. Dynamic Changes in Accessibility, Nuclear Positioning, Recombination, and Transcription at the Ig Kappa Locus. J Immunol Baltim Md 1950 (2007) 179:5264–73. doi: 10.4049/jimmunol.179.8.5264
67. Stadhouders R, de Bruijn MJW, Rother MB, Yuvaraj S, Ribeiro de Almeida C, Kolovos P, et al. Pre-B Cell Receptor Signaling Induces Immunoglobulin κ Locus Accessibility by Functional Redistribution of Enhancer-Mediated Chromatin Interactions. PloS Biol (2014) 12:e1001791.
68. Degner SC, Verma-Gaur J, Wong TP, Bossen C, Iverson GM, Torkamani A, et al. CCCTC-Binding Factor (CTCF) and Cohesin Influence the Genomic Architecture of the Igh Locus and Antisense Transcription in Pro-B Cells. Proc Natl Acad Sci (2011) 108:9566–71. doi: 10.1073/pnas.1019391108
69. Guo C, Chunguang Yoon HS, Franklin A, Jain S, Ebert A, Cheng H-L, et al. CTCF-Binding Elements Mediate Control of V(D)J Recombination. Nature (2011) 477:424–30. doi: 10.1038/nature10495
70. Qiu X, Kumari G, Gerasimova T, Du H, Labaran L, Singh A, et al. Sequential Enhancer Sequestration Dysregulates Recombination Center Formation at the IgH Locus. Mol Cell (2018) 70:21–33.e6. doi: 10.1016/j.molcel.2018.02.020
71. Medvedovic J, Ebert A, Tagoh H, Tamir IM, Schwickert TA, Novatchkova M, et al. Flexible Long-Range Loops in the VH Gene Region of the Igh Locus Facilitate the Generation of a Diverse Antibody Repertoire. Immunity (2013) 39:229–44. doi: 10.1016/j.immuni.2013.08.011
72. Skok JA, Brown KE, Azuara V, Caparros M-L, Baxter J, Takacs K, et al. Nonequivalent Nuclear Location of Immunoglobulin Alleles in B Lymphocytes. Nat Immunol (2001) 2:848–54. doi: 10.1038/ni0901-848
73. Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, et al. Blimp-1 Orchestrates Plasma Cell Differentiation by Extinguishing the Mature B Cell Gene Expression Program. Immunity (2002) 17:51–62. doi: 10.1016/S1074-7613(02)00335-7
74. Zhang X, Zhang Y, Ba Z, Kyritsis N, Casellas R, Alt FW, et al. Fundamental Roles of Chromatin Loop Extrusion in Antibody Class Switching. Nature (2019) 575:385–9. doi: 10.1038/s41586-019-1723-0
75. Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T, et al. S-S Synapsis During Class Switch Recombination Is Promoted by Distantly Located Transcriptional Elements and Activation-Induced Deaminase. Immunity (2007) 27:711–22. doi: 10.1016/j.immuni.2007.09.007
76. Hewitt SL, Farmer D, Marszalek K, Cadera E, Liang H-E, Xu Y, et al. Association Between the Igk and Igh Immunoglobulin Loci Mediated by the 3′ Igk Enhancer Induces ‘Decontraction’ of the Igh Locus in Pre–B Cells. Nat Immunol (2008) 9:396–404. doi: 10.1038/ni1567
77. Inlay M, Alt FW, Baltimore D, Xu Y. Essential Roles of the κ Light Chain Intronic Enhancer and 3′ Enhancer in κ Rearrangement and Demethylation. Nat Immunol (2002) 3:463–8. doi: 10.1038/ni790
78. Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T, et al. Class Switch Recombination and Hypermutation Require Activation-Induced Cytidine Deaminase (AID), a Potential RNA Editing Enzyme. Cell (2000) 102:553–63. doi: 10.1016/S0092-8674(00)00078-7
79. Holwerda SJB, van deWerken HJG, Ribeiro deAlmeida C, Bergen IM, de Bruijn MJW, Verstegen MJAM, et al. Allelic Exclusion of the Immunoglobulin Heavy Chain Locus is Independent of its Nuclear Localization in Mature B Cells. Nucleic Acids Res (2013) 41:6905–16. doi: 10.1093/nar/gkt491
80. Yang Q, Riblet R, Schildkraut CL. Sites That Direct Nuclear Compartmentalization Are Near the 5Ј End of the Mouse Immunoglobulin Heavy-Chain Locus. Mol Cell Biol (2005) 25:10. doi: 10.1128/MCB.25.14.6021-6030.2005
81. Tinguely A, Chemin G, Péron S, Sirac C, Reynaud S, Cogné M, et al. Cross Talk Between Immunoglobulin Heavy-Chain Transcription and RNA Surveillance During B Cell Development. Mol Cell Biol (2012) 32:107–17. doi: 10.1128/MCB.06138-11
82. Shen HM, Wuerffel R, Cantillo JF, Priyadarshi S, Lei X, Liang J, et al. Loop Extrusion Promotes an Alternate Pathway for Isotype Switching. Cell Rep (2021) 37:110059. doi: 10.1016/j.celrep.2021.110059
83. Zhang Y, Zhang X, Dai H-Q, Hu H, Alt FW. The Role of Chromatin Loop Extrusion in Antibody Diversification. Nat Rev Immunol (2022). doi: 10.1038/s41577-022-00679-3
84. Mostoslavsky R, Singh N, Tenzen T, Goldmit M, Gabay C, Elizur S, et al. Asynchronous Replication and Allelic Exclusion in the Immune System. Nature (2001) 414:5. doi: 10.1038/35102606
85. Ju Z, Volpi SA, Hassan R, Martinez N, Giannini SL, Gold T, et al. Evidence for Physical Interaction Between the Immunoglobulin Heavy Chain Variable Region and the 3′ Regulatory Region. J Biol Chem (2007) 282:35169–78. doi: 10.1074/jbc.M705719200
86. Le Noir S, Laffleur B, Carrion C, Garot A, Lecardeur S, Pinaud E, et al. The IgH Locus 3’ Cis-Regulatory Super-Enhancer Co-Opts AID for Allelic Transvection. Oncotarget (2017) 8:12929–40. doi: 10.18632/oncotarget.14585
87. Feldman S, Achour I, Wuerffel R, Kumar S, Gerasimova T, Sen R, et al. Constraints Contributed by Chromatin Looping Limit Recombination Targeting During Ig Class Switch Recombination. J Immunol Baltim Md 1950 (2015) 194:2380–9. doi: 10.4049/jimmunol.1401170
88. Rothschild G, Zhang W, Lim J, Giri PK, Laffleur B, Chen Y, et al. Noncoding RNA Transcription Alters Chromosomal Topology to Promote Isotype-Specific Class Switch Recombination. Sci Immunol (2020) 5:eaay5864. doi: 10.1126/sciimmunol.aay5864
89. Laffleur B, Lim J, Zhang W, Chen Y, Pefanis E, Bizarro J, et al. Noncoding RNA Processing by DIS3 Regulates Chromosomal Architecture and Somatic Hypermutation in B Cells. Nat Genet (2021) 53:230–42. doi: 10.1038/s41588-020-00772-0
90. Rocha PP, Micsinai M, Kim JR, Hewitt SL, Souza PP, Trimarchi T, et al. Close Proximity to Igh Is a Contributing Factor to AID-Mediated Translocations. Mol Cell (2012) 47:873–85. doi: 10.1016/j.molcel.2012.06.036
Keywords: immunoglobuline genes, enhancers, nuclear organization, B lymphocytes, chromatin loops
Citation: Bruzeau C, Cook-Moreau J, Pinaud E and Le Noir S (2022) Contribution of Immunoglobulin Enhancers to B Cell Nuclear Organization. Front. Immunol. 13:877930. doi: 10.3389/fimmu.2022.877930
Received: 17 February 2022; Accepted: 26 May 2022;
Published: 24 June 2022.
Edited by:
Paolo Casali, The University of Texas Health Science Center at San Antonio, United StatesReviewed by:
Jayanta Chaudhuri, Memorial Sloan Kettering Cancer Center, United StatesUttiya Basu, Columbia University, United States
Copyright © 2022 Bruzeau, Cook-Moreau, Pinaud and Le Noir. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sandrine Le Noir, c2FuZHJpbmUubGUtbm9pckB1bmlsaW0uZnI=
 Charlotte Bruzeau
Charlotte Bruzeau Jeanne Cook-Moreau
Jeanne Cook-Moreau Eric Pinaud
Eric Pinaud Sandrine Le Noir
Sandrine Le Noir