- Department of Biology, State University of New York at Fredonia, Fredonia, NY, United States
The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for the current coronavirus disease 2019 (COVID-19) pandemic. Majority of COVID-19 patients have mild disease but about 20% of COVID-19 patients progress to severe disease. These patients end up in the intensive care unit (ICU) with clinical manifestations of acute respiratory distress syndrome (ARDS) and sepsis. The formation of neutrophil extracellular traps (NETs) has also been associated with severe COVID-19. Understanding of the immunopathology of COVID-19 is critical for the development of effective therapeutics. In this article, we discuss evidence indicating that severe COVID-19 has clinical presentations consistent with the definitions of viral sepsis. We highlight the role of neutrophils and NETs formation in the pathogenesis of severe COVID-19. Finally, we highlight the potential of therapies inhibiting NETs formation for the treatment of COVID-19.
Introduction
Coronavirus disease 2019 (COVID-19) was first reported in the city of Wuhan, Hubei province in mainland China in late 2019. The disease spread rapidly around the globe and was declared a pandemic by the World Health Organization on March 11, 2020 (1). In 2021, at the peak of the surge, COVID-19 was the number one cause of death in the United States (US) surpassing heart disease and cancer with an average of more than 3000 deaths per day (2). In fact, COVID-19 has led to the biggest drop in life expectancy in the US in more than seven decades (3). The successful rollout of vaccines has significantly halted mortality from the disease in the US. However, the emergence of more virulent strains of the virus remains a public health concern. As of January 2022, COVID-19 resulted in more than 800,000 deaths in the US and more than five million deaths globally with experts suggesting the number is significantly higher (4).
The causative agent is severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), whose origin is unknown. The closest human coronavirus related to SARS-CoV-2 is SARS-CoV which caused the SARS outbreak from 2002-2004 with 79% genetic similarity (5). However, SARS-CoV-2 bears the greatest genetic similarity to bat coronavirus RaTG13, with 96% similarity (6), fueling a suspicion that the virus originated from bats.
Most patients with COVID-19 have mild disease. Roughly 20% of patients exhibit exaggerated immune responses, including a hyper-inflammatory state and cytokine storm that leads to acute respiratory distress syndrome (ARDS) and eventually resulting in multi-organ damage and death. Several clinical observations indicate that severe COVID-19 meets the criteria to be classified as viral sepsis (7).
Although the cause of aberrant host immune response in severe COVID-19 is not completely understood, accumulating evidence indicates that immune dysfunction contributes to disease severity. The adaptive immune system plays a crucial role in host defense following SARS-CoV-2 infection. Antigen presenting cells (APCs) present viral antigens to CD4+ T cells which induce robust neutralizing antibody responses by B cells (8). In addition, CD8+ Cytotoxic T lymphocytes (CTLs) produce perforins and granzymes which mediate killing of virally infected cells and are important for antiviral immunity (9). Studies have shown that severe COVID-19 is associated with significant decrease in numbers of CD4+ T cells, CD8+ T cells and B cells (10, 11). Severe SARS-CoV-2 infection is also associated with an overwhelming inflammatory phenotype (12, 13). Inflammatory CD4+ Th17 cells have been shown to mediate lung damage in COVID-19 patients (14). Likewise, innate immune cells like macrophages and neutrophils have been shown to be skewed towards an inflammatory phenotype in SARS-CoV-2 infection (15, 16). In particular, the production of neutrophil extracellular traps has been shown to propagate severe COVID-19 (17–19). The role of T and B cells in COVID-19 has been extensively reviewed (8, 20, 21) and we will focus on the role of neutrophils in the pathology of severe COVID-19.
In this article, we highlight important observations which indicate that severe COVID-19 has clinical presentations consistent with the definitions of viral sepsis. We discuss the significant contribution of neutrophils in driving disease pathology following infection with SARS-CoV-2 via formation of neutrophil extracellular traps (NETs). Furthermore, we highlight the potential of therapies inhibiting NETs formation for the treatment of severe COVID-19.
NETs and Inflammation
Polymorphonuclear neutrophils (PMNs) are the most abundant white blood cells in circulation and are rapidly deployed to the site of bacterial, fungal or viral infection as a critical part of host defense (22, 23). The role of neutrophils in host defense is widely appreciated and defective neutrophil function is associated with recurrent infections or occurrence of rare diseases (24). For several decades, neutrophils have been known to kill pathogens through phagocytosis and oxidative burst accompanied by granular release of potent antimicrobials (25). Recently, neutrophils were shown to kill microbes through the release of neutrophil extracellular traps (NETs). NETs are web-like extrusions, composed of a DNA framework and decorated with granular proteins like neutrophil elastase (NE) and myeloperoxidase (MPO) (26).
The molecular mechanisms involved in NET formation is incompletely understood and the processes that lead to the release of DNA by neutrophils is still a subject of debate. It has been reported that neutrophils form NETs through a tightly regulated cell death pathway called NETosis that involves collapse of the nuclear envelope and rupture of the cytoplasmic membrane (27). Studies have also shown that neutrophils release NETs in the absence of cell death (28, 29). These discrepancies may be due to the use of different stimulants for NET induction. Nevertheless, the critical role of certain enzymes and molecules in NET formation including NE, NADPH oxidase complex, peptidylarginine deiminase 4 (PAD4) and the protein kinase C (PKC) pathway have been highlighted and reviewed elsewhere (30–32). NETs have been shown to kill bacteria, fungi, viruses, and parasites (26, 33–35) and there is significant interest in the role of NETs in SARS-CoV-2 infection.
Although NET formation is a mechanism of host defense, excessive NET formation or defective clearance of NETs triggers sustained inflammatory response that can lead to organ damage and drive disease pathology. For example, histones released during NET formation have been shown to be cytotoxic and damage endothelial cells (36). NET formation leads to the production of autoantibodies that damage important organs (37) and inhibition of NETs formation has been shown to be protective in several models of inflammatory diseases (38, 39). Accumulating evidence indicates that NETs contributes to the pathophysiology of severe COVID-19 (18, 19). The role of NETs in the pathophysiology of COVID-19 constitutes a major focus of this review and will be discussed in later sections.
Viral Sepsis
Despite decades of research and treatment, sepsis still constitutes a major challenge in modern medicine and is a leading cause of death in the intensive care unit (ICU). Sepsis is a heterogeneous and dynamic syndrome, due to a complex interplay between the host immune response and the invading microbe. The Third International Consensus Definitions Task Force defined sepsis as life-threatening organ dysfunction caused by a dysregulated host response to infection (40). This definition implies the general notion that bacteria, fungi and viruses can equally cause sepsis. However, there has been concerns that physicians are reluctant to designate viral infections as a case of sepsis (7). Although, bacteria accounts for more than 70% of documented sepsis (41, 42), the role of viruses in sepsis should not be ignored and this knowledge is important to tailor adequate treatment to culture negative patients.
The global burden of viral sepsis is huge with an estimated occurrence of 200 million cases of viral community-acquired pneumonia (CAP) each year (43). Pneumonia is the most common clinical syndrome in patients with sepsis (41, 42). Interestingly, studies have shown that viruses are the most common causes of CAP (44, 45). Therefore, the strict association of sepsis with bacterial infection can be costly given that early antiviral therapy is associated with better outcome in viral sepsis (46). It is also concerning that antibiotics have been administered in culture negative cases of pneumonia (47) indicating the bias of physicians to ignore viruses as a veritable cause of sepsis. It must be stated that the presence of a virus is not sufficient for the diagnosis of viral sepsis. This is due to the possibility of bacterial co-infection or bacterial sepsis resulting from virus-induced immunosuppression. However, among patients with a diagnosis of pure viral CAP, 61% and 7% presented with sepsis and septic shock respectively upon admission to the clinic (47).
Several viruses have been reported to cause sepsis including influenza viruses, rhinoviruses, respiratory syncytial viruses, adenoviruses, herpes simplex viruses, human enteroviruses, dengue viruses and coronaviruses (7, 47). Importantly, the betacoronaviruses – Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV, and SARS-CoV-2 that threaten global health have also been known to cause sepsis. For example, patients with severe COVID-19 have clinical symptoms of viral sepsis. In one study, 59% of patients with COVID-19 were diagnosed with sepsis (48). Importantly, 76% of COVID-19 patients diagnosed with sepsis were negative for bacterial or fungal infections (48). Another study diagnosed sepsis in 100% of patients who died of COVID-19 (49). More studies are required for the diagnosis of sepsis in critically ill patients with COVID-19. However, taking into consideration several clinical observations and the above definition of sepsis, the authors agree that severe COVID-19 is a typical case of dysregulated host response to infection and therefore qualifies as sepsis caused by SARS-CoV-2 infection.
Pathophysiology of Sepsis
The normal immune response to microbial invasion leads to the activation of host defense mechanisms to counter the microbe and prevent colonization of the host by the microbe. This involves cellular activation, vasodilation, leukocyte recruitment and increased endothelial permeability (50, 51). This complex and well-choreographed mechanism of immune activation describes the inflammatory response. Overwhelming infection caused by a virulent microbe or dysregulated immune response to an infection can lead to an overtly exaggerated immune activation or hyper-inflammatory state causing tissue injury and collateral damage to the host.
Innate immune cells like neutrophils and macrophages express molecular receptors called pattern recognition receptors (PRRs) that recognize pathogen-associated molecular patterns (PAMPs) on microbes (52). Several PRRs have been described and among them, TLRs are the most studied.
SARS-CoV-2 is an enveloped virus, with a single-stranded, positive-sense RNA genome (53). During replication, RNA viruses produce double-stranded RNA (dsRNA) as an intermediate (53). Both ssRNA and dsRNA can activate TLRs leading to the production of proinflammatory cytokines via MyD88 and NFk-B activation (54, 55).
Innate immune cells also play a role in the maintenance of antiviral state by the activation of Stimulator of Interferon Genes (STING) pathway (56–58). Upon activation, STING recruits TANK binding kinase 1 (TBK1) and the STING-TBK1 complex subsequently phosphorylates Interferon Regulatory Factor 3 (IRF3) (58). STING can also stimulate IKK leading to NF-κB activation (58). The transcription factors, IRF3 and NF-κB induce the production of type I IFNs and other pro-inflammatory cytokines important for antiviral immunity (58). For example, activation of STING pathway has been shown to block human coronavirus infection (59) and defective type I IFN production is associated with severe COVID-19 (60, 61).
The production of cytokines via NF-κB activation is an important step for the recruitment of neutrophils and other immune cells. However, a major hallmark of sepsis and severe COVID-19 is the excessive production of pro-inflammatory cytokines termed cytokine storm (CS) (7, 49). Cytokines like tumor necrosis factor (TNF), Interleukin (IL)-1, IL-6, IL-8, IL-12 and IL-17 propagate the inflammatory response through leukocyte recruitment, release of secondary inflammatory mediators, endothelial dysfunction and NETs formation (7, 18). For example, TNF and IL-1 induce vasodilation, facilitate the release of secondary mediators such as nitric oxide (NO), platelet activation factor (PAF), prostaglandins, leukotrienes and the activation of the complement system (62). Indeed, CS has been implicated in the pathogenesis of sepsis, viral diseases, autoimmune diseases, cancer and COVID-19 (18, 62–65).
CS also promotes leukocyte recruitment and endothelial permeability in the pulmonary capillaries resulting in lung injury and acute respiratory distress syndrome (ARDS) (64). Microbes associated with pulmonary infection will induce neutrophil migration to the lungs. The lumen of the pulmonary capillaries are more narrow and this leads to extended transit time along the pulmonary endothelium. Neutrophil accumulation and sequestration in the lungs leads to prolonged release of proteolytic enzymes that results in acute lung injury (ALI) and ARDS (66). Sepsis is the most common cause of ARDS and sepsis-related ARDS is associated with overall higher disease severity, longer ICU stays and mortality (67, 68).
Additionally, cytokine activity also activates the coagulation pathway, which can lead to disseminated intravascular coagulation (DIC) and/or coagulopathy which is a hallmark of sepsis (62). Aberrant activation of the coagulation pathway leads to capillary microthrombi, tissue hypoperfusion and end-organ ischemia (69).
Overall, there is consensus that sepsis is driven by the host immune response to infection rather than the pathogen itself (63). However, several clinical trials of therapies targeting important steps in the host immune response during sepsis have not been successful (62). We anticipate that advances in technology will increase our knowledge of sepsis pathogenesis leading to more novel therapeutic interventions.
NETs, Sepsis and Severe COVID-19
Neutrophils are the first immune cells to arrive at the site of bacterial infection and play an important role in host defense. These cells are equipped with antimicrobial granular content that is rapidly deployed to eliminate the invading microbe. However, there is unequivocal experimental evidence that neutrophils contribute to sepsis pathology by release of cytolytic granular content, vaso-occlusion, and NET formation (66, 70).
The discovery of the process of NET formation by neutrophils highlighted a novel mechanism of innate immune defense against microbes. NETs have been shown to trap and kill a wide range of microbes including bacteria, fungi and viruses (26, 33–35). NETs formation can be beneficial during sepsis because NETs spatially restrict the dissemination of microbes during infection (26). To prevent physical containment by NETs, some bacteria have evolved to degrade NETs and NET degradation promotes bacterial virulence (71). Patients with chronic granulomatous disease (CGD) caused by mutations in genes encoding NADPH oxidase subunits do not make NETs and are susceptible to recurrent life-threatening infections (72). Gene therapy in a CGD patient restored NET forming ability of neutrophils resulting in clearance of refractory fungal infection (72). Additionally, NET proteins like histones, NE, MPO and proteinase 3 (PR3) have potent antimicrobial properties and help in bacterial killing (73).
However, accumulating evidence suggests that NETs formation is a double-edged sword (74) that contributes to the pathogenesis of several diseases including sepsis (70), rheumatoid arthritis (75), vasculitis (76), diabetes (77), lupus (78), cancer (79) and COVID-19 (18, 80). For example, studies have shown that levels of circulating cell-free DNA that are released during NET formation is a strong predictor of sepsis mortality (81). Also, histones which are the most abundant proteins in NETs (82) are cytotoxic towards epithelial and endothelial cells (36, 83). Histone administration to mice resulted in neutrophil accumulation in the lungs, microvascular thrombosis and death (83). Additionally, in non-human primates challenged with lethal concentration of E. coli, histone levels correlate with onset of renal failure. Furthermore, using three different models of sepsis: injection of LPS, injection of TNF, and CLP, the authors showed that antibodies against H4 improved animal survival (83). Consistent with this, we recently showed that inhibition of NE produced during NET formation reduced lung neutrophil accumulation, systemic levels of proinflammatory cytokines and improved survival in a mouse model of endotoxic shock (38).
A major complication attributed to NETs formation is thrombosis resulting in multi-organ failure (84–87). Due to their ability to form scaffolds, NETs can occlude blood vessels and cause thrombosis. NET scaffolds also promote adhesion of platelets leading to thrombus formation (85, 86). Importantly, serine proteases released by NETs like neutrophil elastase enhance tissue factor and factor XII dependent coagulation thereby leading to intravascular thrombus formation (88). Histones produced by NETs can promote platelet aggregation and thrombin generation via toll-like receptor (TLR) 2 and 4 (89). Interestingly, activated platelets have been shown to induce de novo NETs formation thereby propagating the vicious circle of platelet-neutrophil interaction in coagulopathy (90–92). Indeed, dysregulated NETs formation is associated with coagulopathy in sepsis and severe COVID-19 (80, 92–95).
We have drawn comparisons between sepsis and severe COVID-19 and conclude that the clinical presentations of sepsis and severe COVID-19 intersect at so many levels. Sepsis and severe COVID-19 commonly affect the pulmonary, cardiovascular, and renal systems. Many patients with severe COVID-19 exhibited clinical manifestations of shock-like cold extremities, weak peripheral pulses, dysfunction in microcirculation and organ damage notably in the lungs, kidney and liver (96). Like sepsis, ARDS and respiratory failure is the most common cause of death in COVID-19 patients (49, 97). Additionally, like sepsis, mortality in severe COVID-19 is driven by risk factors like age and presence of predisposing conditions (49). Severe COVID-19 is also characterized by excessive inflammatory cytokine production (19, 98, 99). Moreover, C-reactive protein, a biomarker for sepsis severity has also been shown to predict poor prognosis in COVID-19 (100). Furthermore, similar to sepsis, patients with severe COVID-19 show evidence of coagulopathy and dysregulated thrombus formation (80, 101). Indeed, one study showed that 100% of patients who died from COVID-19 were diagnosed with sepsis (49). In line with the evidence given above, we argue that severe COVID-19 is a typical case of viral sepsis.
Since NETs have been shown to contribute to sepsis pathology, it is conceivable that NETs may contribute to the pathogenesis of severe COVID-19 (Figure 1). Indeed, several studies have implicated NETs in the pathogenesis of severe COVID-19. For example, it was shown that SARS-CoV-2 replicates in neutrophils and triggers NETosis which contributes to COVID-19 pathology by killing lung epithelial cells (102). Sera from patients with COVID-19 have elevated levels of markers of NET formation including cell-free DNA, MPO-DNA, citrullinated histone H3, and neutrophil elastase (Table 1) and these markers are associated with disease severity (18, 111–113). Neutrophilia and NETosis is a major cause of ARDS and lung injury in severe COVID-19 (80, 114, 115). NETs formation is associated with systemic inflammation and cytokine storm which contributes to mortality in severe COVID-19 (116, 117). Additionally, dysregulated thrombus formation which contributes to mortality in severe COVID-19 has been associated with NET formation (80, 118, 119). Furthermore, COVID-19 has been shown to induce the production of autoantibodies associated with NET production (101). These observations have led to an overwhelming scientific support for targeting NETs formation as a veritable approach for the treatment of severe COVID-19 (19, 120, 121).
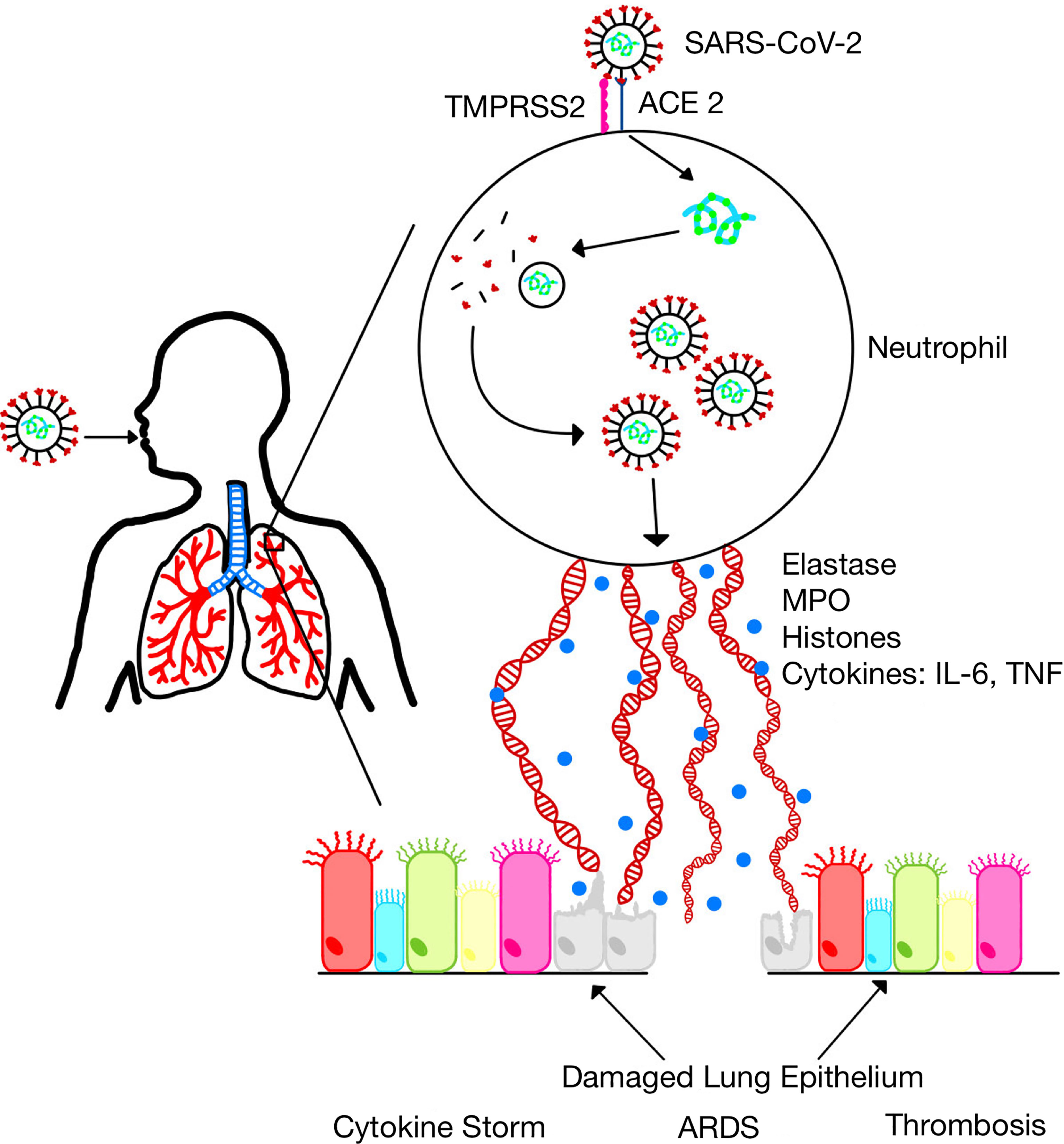
Figure 1 SARS-CoV-2 infection induces neutrophil extracellular traps. SARS-CoV-2 replicates in neutrophils and induces the formation of NETs which leads to the release of inflammatory cytokines and several proteins that damage lung epithelium resulting in acute lung injury and acute respiratory distress syndrome (ARDS).
Targeting NETs in COVID-19
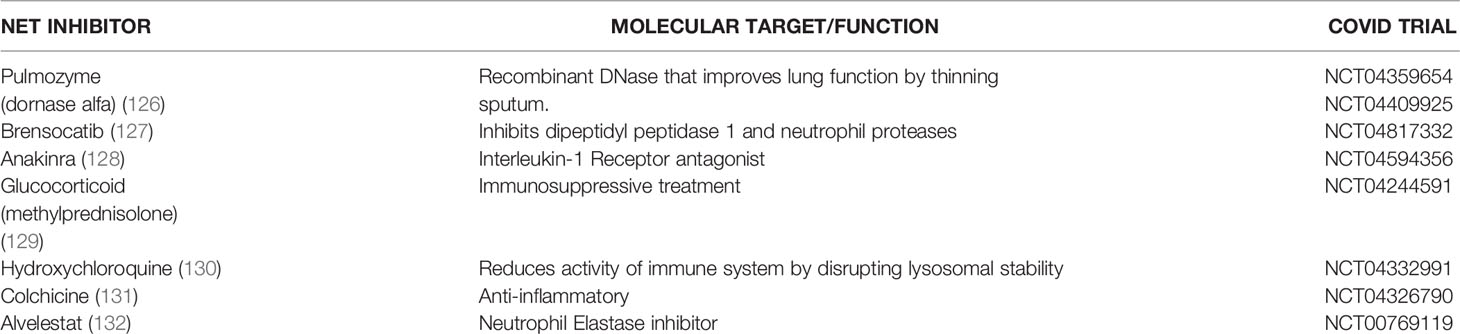
Recently, there has been concerted efforts to develop therapies targeting NETs in several diseases. Therapies targeting NETs have shown excellent success in mitigating lung inflammation and ARDS in preclinical models (19, 38). Since ARDS is the major cause of death in COVID-19, we advocate for the investigation of NET therapies in the treatment of COVID-19 patients. Different approaches to targeting NETs have shown remarkable success in preclinical models. Such approaches include dissolving NET backbone, for example using DNAse (122), blocking molecules relevant in NET formation for example ROS, PAD4 and gasdermin D (39, 123) or blocking the activity of NET proteins like neutrophil elastase (38). Some of these NET therapeutics are currently available in the clinic and should be considered for the treatment of patients critically ill with COVID-19. For example, DNAse treatment is used in the clinic for patients with cystic fibrosis and the NE inhibitor sivelestat is clinically approved for the treatment of ARDS in Japan and South Korea (124, 125). Indeed, clinical trials of several NET inhibitors are underway for the treatment of COVID-19 (Table 2) and some of them have already been adopted as the standard of care for COVID-19 patients. For example, glucocorticoid therapy which is one of the earliest anti-inflammatory treatments available for sepsis patients has been shown to be beneficial for COVID-19 patients and dexamethasone is routinely given to COVID-19 patients (133). Importantly, dexamethasone has been shown to reduce NETs formation (134). Heparin, another NET inhibitor has also been shown to be beneficial for the treatment of COVID-19 patients (135, 136).
Anti-inflammatory therapies and anti-cytokine therapies can also be beneficial in reducing neutrophilia, NETs formation and NET-induced thrombosis. For example, elevated levels of IL-6 has been associated with severe COVID-19 thereby highlighting IL-6 as a therapeutic target. IL-6 signaling has been shown to promote NET formation and lung inflammation (137). We recently showed that inhibition of NETs formation led to decrease in systemic levels of IL-6 and improved survival in a mouse model of endotoxic shock (38). Indeed, Tocilizumab, a recombinant humanized monoclonal anti-IL-6 antibody targeting the human IL-6 receptor was recently approved for the treatment of COVID-19 (138). Previous studies showed that Tocilizumab is also associated with decrease in NET formation (139).
As our understanding of the molecular mechanisms of NET formation increases, more therapies targeting NETs will become available and may hold promise for the effective treatment of severe COVID-19.
Concluding Remarks
The management of sepsis has been a challenge in modern medicine and the launch of surviving sepsis campaign was aimed to curtail the unacceptably high mortality of sepsis patients in the ICU (40). The mortality induced by the novel SARS-CoV-2 responsible for the current global pandemic has been attributed to sepsis (49). In this regard, biomarkers used for sepsis can be used for the early identification of COVID-19 patients that are at risk of progressing to severe disease. There is consensus that mortality in sepsis and COVID-19 is due to host immune response. Hence, modulating dysfunctional immune response in COVID-19 is critical for improving survival.
The formation of neutrophil extracellular traps has emerged as a contributing factor to the pathogenesis of COVID-19 (102). Importantly, SARS-CoV-2 has been shown to infect neutrophils and promote NETosis (102). Understanding of the role of NETs in the pathogenesis of severe COVID-19 holds potential for improving survival of patients. NET biomarkers can be easily detected in the blood and has been shown to indicate disease severity in COVID-19 (18). Hence, biomarkers of NET formation can be used to stratify COVID-19 patients at risk of progressing to severe disease. Since therapies targeting NETs have shown success in experimental models of ARDS, we propose that therapies targeting NETs have great potential for the treatment of COVID-19.
While we have focused on the role of extracellular traps produced by neutrophils in this review, macrophages also produce macrophage extracellular traps (METs) which propagate inflammation (140, 141). Interestingly, macrophages have been shown to contribute to inflammation in COVID-19 (12). Moreover, neutrophil extracellular traps from COVID-19 patients induce a proinflammatory response in monocyte-derived macrophages thereby linking NET formation to inflammatory macrophage activity (17). It is worthy of note that similar to neutrophils, macrophages also release elastase, histones and MPO during MET formation (142, 143). Hence, it is conceivable that mechanisms inhibiting the formation of NETs as highlighted here will also inhibit the formation of METs. Studies investigating the role of macrophage extracellular traps in severe COVID-19 will help unravel its role in the condition.
As with the case in sepsis, it is likely that one drug may not be sufficient to improve survival in COVID-19. Rather, a combinatorial approach may be necessary to reverse mortality in COVID-19. For example, the recently approved Tocilizumab showed benefit for COVID-19 patients who received it in conjunction with corticosteroids (144). We advocate for clinical trials investigating such combinations of NET therapeutics for the treatment of COVID-19. As another example, although sivelestat did not improve mortality in patients with ARDS (145), combination of sivelestat with antiviral therapy or another NET inhibitor may be beneficial for COVID-19 patients.
The intelligent design of clinical trials of therapies targeting NETs is essential and several factors including timing of intervention is critical for success. For example, there are concerns that DNase may enhance the dispersal of free histones and promote inflammation thereby leading to worse outcome in sepsis. In line with this, Meng et al, showed that early administration of DNase led to hyper-susceptibility to polymicrobial sepsis in mice (146). In a follow-up study, Mai et al showed that delayed administration of DNase is necessary for improved outcome in sepsis (147).
More research is needed to understand neutrophil behavior during SARS-CoV-2 infection. For example, an interesting question is whether different viral strains that have varying degrees of immunogenicity differ in their degree of NET induction, and this remains an important subject of investigation in our laboratory. Increase in our knowledge and understanding of the pathogenesis of COVID-19 will widen the availability of molecular targets that will yield the desired therapeutic benefit. With concerted research efforts, the menace of severe COVID-19 in the ICU will be curtailed.
Author Contributions
EV did literature search and wrote portions of the manuscript. JN also did literature search and wrote portions of the manuscript. EO corrected and edited the manuscript for publication. CS did literature review and table for manuscript revision. All authors contributed to the article and approved the submitted version.
Funding
Funding for this work was provided by the State University of New York at Fredonia.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Cucinotta D, Vanelli M. WHO Declares COVID-19 a Pandemic. Acta BioMed (2020) 91:157–60. doi: 10.23750/ABM.V91I1.9397
2. Woolf SH, Chapman DA, Lee JH. COVID-19 as the Leading Cause of Death in the United States. JAMA (2021) 325:123–4. doi: 10.1001/JAMA.2020.24865
3. Dyer O. US Life Expectancy Plunged in 2020, Especially in Hispanic and African Americans. BMJ (2021) 374:n1873. doi: 10.1136/BMJ.N1873
4. CDC COVID Data Tracker. Available at: https://covid.cdc.gov/covid-data-tracker/#datatracker-home (Accessed August 9, 2021).
5. Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, et al. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-Ncov and Naming it SARS-CoV-2. Nat Microbiol (2020) 5:536–44. doi: 10.1038/S41564-020-0695-Z
6. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A Pneumonia Outbreak Associated With a New Coronavirus of Probable Bat Origin. Nature (2020) 579:270–3. doi: 10.1038/S41586-020-2012-7
7. Lin GL, McGinley JP, Drysdale SB, Pollard AJ. Epidemiology and Immune Pathogenesis of Viral Sepsis. Front Immunol (2018) 9:2147. doi: 10.3389/FIMMU.2018.02147
8. Cox RJ, Brokstad KA. Not Just Antibodies: B Cells and T Cells Mediate Immunity to COVID-19. Nat Rev Immunol 2020 2010 (2020) 20:581–2. doi: 10.1038/s41577-020-00436-4
9. Mallajosyula V, Ganjavi C, Chakraborty S, McSween AM, Pavlovitch-Bedzyk AJ, Wilhelmy J, et al. Cd8+T Cells Specific for Conserved Coronavirus Epitopes Correlate With Milder Disease in COVID-19 Patients. Sci Immunol (2021) 6:5669. doi: 10.1126/SCIIMMUNOL.ABG5669/SUPPL_FILE/SCIIMMUNOL.ABG5669_MDAR_CHECKLIST.ZIP
10. Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and Immunological Features of Severe and Moderate Coronavirus Disease 2019. J Clin Invest (2020) 130:2620–9. doi: 10.1172/JCI137244
11. Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front Immunol (2020) 11:827. doi: 10.3389/FIMMU.2020.00827
12. Merad M, Martin JC. Author Correction: Pathological Inflammation in Patients With COVID-19: A Key Role for Monocytes and Macrophages (Nature Reviews Immunology, (2020), 20, 6, (355-362), 10.1038/S41577-020-0331-4). Nat Rev Immunol (2020) 20:448. doi: 10.1038/S41577-020-0353-Y
13. Bhaskar S, Sinha A, Banach M, Mittoo S, Weissert R, Kass JS, et al. Cytokine Storm in COVID-19—Immunopathological Mechanisms, Clinical Considerations, and Therapeutic Approaches: The REPROGRAM Consortium Position Paper. Front Immunol (2020) 11:1648/BIBTEX. doi: 10.3389/FIMMU.2020.01648/BIBTEX
14. Zhao Y, Kilian C, Turner JE, Bosurgi L, Roedl K, Bartsch P, et al. Clonal Expansion and Activation of Tissue-Resident Memory-Like Th17 Cells Expressing GM-CSF in the Lungs of Severe COVID-19 Patients. Sci Immunol (2021) 6:eabf6692. doi: 10.1126/SCIIMMUNOL.ABF6692
15. Wen W, Su W, Tang H, Le W, Zhang X, Zheng Y, et al. Immune Cell Profiling of COVID-19 Patients in the Recovery Stage by Single-Cell Sequencing. Cell Discovery (2020) 6:31. doi: 10.1038/S41421-020-0168-9
16. Shaath H, Vishnubalaji R, Elkord E, Alajez NM. Single-Cell Transcriptome Analysis Highlights a Role for Neutrophils and Inflammatory Macrophages in the Pathogenesis of Severe COVID-19. Cells (2020) 9:2374. doi: 10.3390/CELLS9112374
17. Torres-Ruiz J, Absalón-Aguilar A, Nuñez-Aguirre M, Pérez-Fragoso A, Carrillo-Vázquez DA, Maravillas-Montero JL, et al. Neutrophil Extracellular Traps Contribute to COVID-19 Hyperinflammation and Humoral Autoimmunity. Cells (2021) 10:2545. doi: 10.3390/CELLS10102545/S1
18. Zuo Y, Yalavarthi S, Shi H, Gockman K, Zuo M, Madison JA, et al. Neutrophil Extracellular Traps in COVID-19. JCI Insight (2020) 5:e138999. doi: 10.1172/JCI.INSIGHT.138999
19. Barnes BJ, Adrover JM, Baxter-Stoltzfus A, Borczuk A, Cools-Lartigue J, Crawford JM, et al. Targeting Potential Drivers of COVID-19: Neutrophil Extracellular Traps. J Exp Med (2020) 217:e20200652. doi: 10.1084/JEM.20200652/151683
20. Sette A, Crotty S. Adaptive Immunity to SARS-CoV-2 and COVID-19. Cell (2021) 184:861. doi: 10.1016/J.CELL.2021.01.007
21. Chen Z, John Wherry E. T Cell Responses in Patients With COVID-19. Nat Rev Immunol (2020) 20:529–36. doi: 10.1038/s41577-020-0402-6
22. Bardoel BW, Kenny EF, Sollberger G, Zychlinsky A. The Balancing Act of Neutrophils. Cell Host Microbe (2014) 15:526–36. doi: 10.1016/j.chom.2014.04.011
23. Ley K, Hoffman HM, Kubes P, Cassatella MA, Zychlinsky A, Hedrick CC, et al. Neutrophils: New Insights and Open Questions. Sci Immunol (2018) 3:eaat4579. doi: 10.1126/SCIIMMUNOL.AAT4579
24. Klein C. Genetic Defects in Severe Congenital Neutropenia: Emerging Insights Into Life and Death of Human Neutrophil Granulocytes. Annu Rev Immunol (2011) 29:399–413. doi: 10.1146/ANNUREV-IMMUNOL-030409-101259
25. Nathan C. Neutrophils and Immunity: Challenges and Opportunities. Nat Rev Immunol (2006) 6:173–82. doi: 10.1038/nri1785
26. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil Extracellular Traps Kill Bacteria. Science (2004) 303:1532–5. doi: 10.1126/science.1092385
27. Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, et al. Novel Cell Death Program Leads to Neutrophil Extracellular Traps. J Cell Biol (2007) 176:231–41. doi: 10.1083/JCB.200606027
28. Yousefi S, Mihalache C, Kozlowski E, Schmid I, Simon HU. Viable Neutrophils Release Mitochondrial DNA to Form Neutrophil Extracellular Traps. Cell Death Differ (2009) 16:1438–44. doi: 10.1038/cdd.2009.96
29. Amini P, Stojkov D, Felser A, Jackson CB, Courage C, Schaller A, et al. Neutrophil Extracellular Trap Formation Requires OPA1-Dependent Glycolytic ATP Production. Nat Commun (2018) 9:2958. doi: 10.1038/S41467-018-05387-Y
30. Brinkmann V, Zychlinsky A. Neutrophil Extracellular Traps: Is Immunity the Second Function of Chromatin? J Cell Biol (2012) 198:773–83. doi: 10.1083/JCB.201203170
31. Papayannopoulos V, Metzler KD, Hakkim A, Zychlinsky A. Neutrophil Elastase and Myeloperoxidase Regulate the Formation of Neutrophil Extracellular Traps. J Cell Biol (2010) 191:677–91. doi: 10.1083/jcb.201006052
32. Hakkim A, Fuchs TA, Martinez NE, Hess S, Prinz H, Zychlinsky A, et al. Activation of the Raf-MEK-ERK Pathway is Required for Neutrophil Extracellular Trap Formation. Nat Chem Biol (2011) 7:75–7. doi: 10.1038/nchembio.496
33. Urban CF, Reichard U, Brinkmann V, Zychlinsky A. Neutrophil Extracellular Traps Capture and Kill Candida Albicans Yeast and Hyphal Forms. Cell Microbiol (2006) 8:668–76. doi: 10.1111/J.1462-5822.2005.00659.X
34. Guimarães-Costa AB, Nascimento MTC, Froment GS, Soares RPP, Morgado FN, Conceição-Silva F, et al. Leishmania Amazonensis Promastigotes Induce and are Killed by Neutrophil Extracellular Traps. Proc Natl Acad Sci USA (2009) 106:6748–53. doi: 10.1073/PNAS.0900226106
35. Saitoh T, Komano J, Saitoh Y, Misawa T, Takahama M, Kozaki T, et al. Neutrophil Extracellular Traps Mediate a Host Defense Response to Human Immunodeficiency Virus-1. Cell Host Microbe (2012) 12:109–16. doi: 10.1016/J.CHOM.2012.05.015
36. Saffarzadeh M, Juenemann C, Queisser MA, Lochnit G, Barreto G, Galuska SP, et al. Neutrophil Extracellular Traps Directly Induce Epithelial and Endothelial Cell Death: A Predominant Role of Histones. PLoS One (2012) 7:e32366. doi: 10.1371/journal.pone.0032366
37. Khandpur R, Carmona-Rivera C, Vivekanandan-Giri A, Gizinski A, Yalavarthi S, Knight JS, et al. NETs are a Source of Citrullinated Autoantigens and Stimulate Inflammatory Responses in Rheumatoid Arthritis. Sci Transl Med (2013) 5:178ra40. doi: 10.1126/scitranslmed.3005580
38. Okeke EB, Louttit C, Fry C, Najafabadi AH, Han K, Nemzek J, et al. Inhibition of Neutrophil Elastase Prevents Neutrophil Extracellular Trap Formation and Rescues Mice From Endotoxic Shock. Biomaterials (2020) 238:119836. doi: 10.1016/j.biomaterials.2020.119836
39. Knight JS, Zhao W, Luo W, Subramanian V, O’Dell AA, Yalavarthi S, et al. Peptidylarginine Deiminase Inhibition is Immunomodulatory and Vasculoprotective in Murine Lupus. J Clin Invest (2013) 123:2981–93. doi: 10.1172/JCI67390
40. Singer M, Deutschman CS, Seymour C, Shankar-Hari M, Annane D, Bauer M, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA (2016) 315:801–10. doi: 10.1001/JAMA.2016.0287
41. Zahar JR, Timsit JF, Garrouste-Orgeas M, Français A, Vesim A, Descorps-Declere A, et al. Outcomes in Severe Sepsis and Patients With Septic Shock: Pathogen Species and Infection Sites are Not Associated With Mortality. Crit Care Med (2011) 39:1886–95. doi: 10.1097/CCM.0B013E31821B827C
42. Phua J, Ngerng WJ, See KC, Tay CK, Kiong T, Lim HF, et al. Characteristics and Outcomes of Culture-Negative Versus Culture-Positive Severe Sepsis. Crit Care (2013) 17:R202. doi: 10.1186/CC12896
43. Ruuskanen O, Lahti E, Jennings LC, Murdoch DR. Viral Pneumonia. Lancet (London England) (2011) 377:1264–75. doi: 10.1016/S0140-6736(10)61459-6
44. Jain S, Self WH, Wunderink RG, Fakhran S, Balk R, Bramley AM, et al. Community-Acquired Pneumonia Requiring Hospitalization Among U.S. Adults. N Engl J Med (2015) 373:415–27. doi: 10.1056/NEJMOA1500245
45. Cilla G, Oñate E, Perez-Yarza EG, Montes M, Vicente D, Perez-Trallero E. Viruses in Community-Acquired Pneumonia in Children Aged Less Than 3 Years Old: High Rate of Viral Coinfection. J Med Virol (2008) 80:1843–9. doi: 10.1002/JMV.21271
46. Gu X, Zhou F, Wang Y, Fan G, Cao B. Respiratory Viral Sepsis: Epidemiology, Pathophysiology, Diagnosis and Treatment. Eur Respir Rev (2020) 29:1–12. doi: 10.1183/16000617.0038-2020
47. Cillóniz C, Dominedò C, Magdaleno D, Ferrer M, Gabarrús A, Torres A. Pure Viral Sepsis Secondary to Community-Acquired Pneumonia in Adults: Risk and Prognostic Factors. J Infect Dis (2019) 220:1166–71. doi: 10.1093/INFDIS/JIZ257
48. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical Course and Risk Factors for Mortality of Adult Inpatients With COVID-19 in Wuhan, China: A Retrospective Cohort Study. Lancet (London England) (2020) 395:1054–62. doi: 10.1016/S0140-6736(20)30566-3
49. Chen T, Wu D, Chen H, Yan W, Yang D, Chen G, et al. Clinical Characteristics of 113 Deceased Patients With Coronavirus Disease 2019: Retrospective Study. BMJ (2020) 368:m1091. doi: 10.1136/BMJ.M1091
50. Sommers MS. The Cellular Basis of Septic Shock. Crit Care Nurs Clin North Am (2003) 15:13–25. doi: 10.1016/S0899-5885(02)00046-1
51. Brodsky IE, Medzhitov R. Targeting of Immune Signalling Networks by Bacterial Pathogens. Nat Cell Biol (2009) 11:521–6. doi: 10.1038/NCB0509-521
52. Aderem A, Ulevitch RJ. Toll-Like Receptors in the Induction of the Innate Immune Response. Nature (2000) 406:782–7. doi: 10.1038/35021228
53. V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus Biology and Replication: Implications for SARS-CoV-2. Nat Rev Microbiol 2020 193 (2020) 19:155–70. doi: 10.1038/s41579-020-00468-6
54. Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, et al. Recognition of Single-Stranded RNA Viruses by Toll-Like Receptor 7. Proc Natl Acad Sci (2004) 101:5598–603. doi: 10.1073/PNAS.0400937101
55. Lee SMY, Yip TF, Yan S, Jin DY, Wei HL, Guo RT, et al. Recognition of Double-Stranded RNA and Regulation of Interferon Pathway by Toll-Like Receptor 10. Front Immunol (2018) 9:516/BIBTEX. doi: 10.3389/FIMMU.2018.00516/BIBTEX
56. Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, et al. Pan-Viral Specificity of IFN-Induced Genes Reveals New Roles for cGAS in Innate Immunity. Nature (2014) 505:691–5. doi: 10.1038/nature12862
57. Glück S, Guey B, Gulen MF, Wolter K, Kang TW, Schmacke NA, et al. Innate Immune Sensing of Cytosolic Chromatin Fragments Through cGAS Promotes Senescence. Nat Cell Biol (2017) 19:1061–70. doi: 10.1038/ncb3586
58. Li T, Chen ZJ. The cGAS–cGAMP–STING Pathway Connects DNA Damage to Inflammation, Senescence, and Cancer. J Exp Med (2018) 215:1287–99. doi: 10.1084/JEM.20180139
59. Liu W, Reyes HM, Yang JF, Li Y, Stewart KM, Basil MC, et al. Activation of STING Signaling Pathway Effectively Blocks Human Coronavirus Infection. J Virol (2021) 95: e00490–21. doi: 10.1128/JVI.00490-21
60. Zhang Q, Liu Z, Moncada-Velez M, Chen J, Ogishi M, Bigio B, et al. Inborn Errors of Type I IFN Immunity in Patients With Life-Threatening COVID-19. Science (2020) 370:eabd4570. doi: 10.1126/SCIENCE.ABD4570
61. Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired Type I Interferon Activity and Inflammatory Responses in Severe COVID-19 Patients. Science (2020) 369:718–24. doi: 10.1126/SCIENCE.ABC6027
62. Okeke EB, Uzonna JE. In Search of a Cure for Sepsis: Taming the Monster in Critical Care Medicine. J Innate Immun (2016) 8:156–70. doi: 10.1159/000442469
63. Chousterman BG, Swirski FK, Weber GF. Cytokine Storm and Sepsis Disease Pathogenesis. Semin Immunopathol (2017) 39:517–28. doi: 10.1007/S00281-017-0639-8
64. Fajgenbaum DC, June CH. Cytokine Storm. N Engl J Med (2020) 383:2255–73. doi: 10.1056/NEJMRA2026131/SUPPL_FILE/NEJMRA2026131_DISCLOSURES.PDF
65. Díelia RV, Harrison K, Oyston PC, Lukaszewski RA, Clark GC. Targeting the “Cytokine Storm” for Therapeutic Benefit. Clin Vaccine Immunol (2013) 20:319–27. doi: 10.1128/CVI.00636-12/ASSET/C6C0D2AB-72D5-4549-8FED-F8B5A53D7B36/ASSETS/GRAPHIC/ZCD9990946740002.JPEG
66. Grommes J, Soehnlein O. Contribution of Neutrophils to Acute Lung Injury. Mol Med (2010) 17:293–307. doi: 10.2119/MOLMED.2010.00138
67. Sheu CC, Gong MN, Zhai R, Chen F, Bajwa EK, Clardy PF, et al. Clinical Characteristics and Outcomes of Sepsis-Related vs non-Sepsis-Related ARDS. Chest (2010) 138:559–67. doi: 10.1378/CHEST.09-2933
68. Kim WY, Hong SB. Sepsis and Acute Respiratory Distress Syndrome: Recent Update. Tuberc Respir Dis (Seoul) (2016) 79:53–7. doi: 10.4046/TRD.2016.79.2.53
69. Gando S, Kameue T, Matsuda N, Sawamura A, Hayakawa M, Kato H. Systemic Inflammation and Disseminated Intravascular Coagulation in Early Stage of ALI and ARDS: Role of Neutrophil and Endothelial Activation. Inflammation 2004 284 (2004) 28:237–44. doi: 10.1023/B:IFLA.0000049049.81688.FE
70. Czaikoski PG, Mota JMSC, Nascimento DC, Sônego F, Castanheira FV e S, Melo PH, et al. Neutrophil Extracellular Traps Induce Organ Damage During Experimental and Clinical Sepsis. PLoS One (2016) 11:e0148142. doi: 10.1371/journal.pone.0148142
71. Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, Kotb M, et al. DNase Expression Allows the Pathogen Group A Streptococcus to Escape Killing in Neutrophil Extracellular Traps. Curr Biol (2006) 16:396–400. doi: 10.1016/J.CUB.2005.12.039
72. Bianchi M, Niemiec MJ, Siler U, Urban CF, Reichenbach J. Restoration of Anti-Aspergillus Defense by Neutrophil Extracellular Traps in Human Chronic Granulomatous Disease After Gene Therapy is Calprotectin-Dependent. J Allergy Clin Immunol (2011) 127:1243–52.e7. doi: 10.1016/J.JACI.2011.01.021
73. Korkmaz B, Moreau T, Gauthier F. Neutrophil Elastase, Proteinase 3 and Cathepsin G: Physicochemical Properties, Activity and Physiopathological Functions. Biochimie (2008) 90:227–42. doi: 10.1016/j.biochi.2007.10.009
74. Kaplan MJ, Radic M. Neutrophil Extracellular Traps: Double-Edged Swords of Innate Immunity. J Immunol (2012) 189:2689–95. doi: 10.4049/jimmunol.1201719
75. Sur Chowdhury C, Giaglis S, Walker UA, Buser A, Hahn S, Hasler P. Enhanced Neutrophil Extracellular Trap Generation in Rheumatoid Arthritis: Analysis of Underlying Signal Transduction Pathways and Potential Diagnostic Utility. Arthritis Res Ther (2014) 16:R122. doi: 10.1186/ar4579
76. Kambas K, Chrysanthopoulou A, Vassilopoulos D, Apostolidou E, Skendros P, Girod A, et al. Tissue Factor Expression in Neutrophil Extracellular Traps and Neutrophil Derived Microparticles in Antineutrophil Cytoplasmic Antibody Associated Vasculitis may Promote Thromboinflammation and the Thrombophilic State Associated With the Disease. Ann Rheum Dis (2014) 73:1854–63. doi: 10.1136/annrheumdis-2013-203430
77. Wong SL, Demers M, Martinod K, Gallant M, Wang Y, Goldfine AB, et al. Diabetes Primes Neutrophils to Undergo NETosis, Which Impairs Wound Healing. Nat Med (2015) 21:815–9. doi: 10.1038/nm.3887
78. Hakkim A, Fürnrohr BG, Amann K, Laube B, Abed UA, Brinkmann V, et al. Impairment of Neutrophil Extracellular Trap Degradation is Associated With Lupus Nephritis. Proc Natl Acad Sci U S A (2010) 107:9813–8. doi: 10.1073/pnas.0909927107
79. Albrengues J, Shields MA, Ng D, Park CG, Ambrico A, Poindexter ME, et al. Neutrophil Extracellular Traps Produced During Inflammation Awaken Dormant Cancer Cells in Mice. Science (2018) 361:eaao4227. doi: 10.1126/science.aao4227
80. Middleton EA, He XY, Denorme F, Campbell RA, Ng D, Salvatore SP, et al. Neutrophil Extracellular Traps Contribute to Immunothrombosis in COVID-19 Acute Respiratory Distress Syndrome. Blood (2020) 136:1169–79. doi: 10.1182/blood.2020007008
81. Dwivedi DJ, Toltl LJ, Swystun LL, Pogue J, Liaw KL, Weitz JI, et al. Prognostic Utility and Characterization of Cell-Free DNA in Patients With Severe Sepsis. Crit Care (2012) 16:R151. doi: 10.1186/CC11466
82. Urban CF, Ermert D, Schmid M, Abu-Abed U, Goosmann C, Nacken W, et al. Neutrophil Extracellular Traps Contain Calprotectin, a Cytosolic Protein Complex Involved in Host Defense Against Candida Albicans. PLoS Pathog (2009) 5:e1000639. doi: 10.1371/JOURNAL.PPAT.1000639
83. Xu J, Zhang X, Pelayo R, Monestier M, Ammollo CT, Semeraro F, et al. Extracellular Histones are Major Mediators of Death in Sepsis. Nat Med (2009) 15:1318–21. doi: 10.1038/NM.2053
84. Brill A, Fuchs TA, Savchenko AS, Thomas GM, Martinod K, De Meyer SF, et al. Neutrophil Extracellular Traps Promote Deep Vein Thrombosis in Mice. J Thromb Haemost (2012) 10:136–44. doi: 10.1111/j.1538-7836.2011.04544.x
85. Fuchs TA, Brill A, Duerschmied D, Schatzberg D, Monestier M, Myers DD, et al. Extracellular DNA Traps Promote Thrombosis. Proc Natl Acad Sci U S A (2010) 107:15880–5. doi: 10.1073/PNAS.1005743107
86. Laridan E, Martinod K, De Meyer SF. Neutrophil Extracellular Traps in Arterial and Venous Thrombosis. Semin Thromb Hemost (2019) 45:86–93. doi: 10.1055/S-0038-1677040/ID/JR02600-32
87. Gould TJ, Vu TT, Swystun LL, Dwivedi DJ, Mai SHC, Weitz JI, et al. Neutrophil Extracellular Traps Promote Thrombin Generation Through Platelet-Dependent and Platelet-Independent Mechanisms. Arterioscler Thromb Vasc Biol (2014) 34:1977–84. doi: 10.1161/ATVBAHA.114.304114
88. Massberg S, Grahl L, Von Bruehl ML, Manukyan D, Pfeiler S, Goosmann C, et al. Reciprocal Coupling of Coagulation and Innate Immunity via Neutrophil Serine Proteases. Nat Med (2010) 16:887–96. doi: 10.1038/NM.2184
89. Semeraro F, Ammollo CT, Morrissey JH, Dale GL, Friese P, Esmon NL, et al. Extracellular Histones Promote Thrombin Generation Through Platelet-Dependent Mechanisms: Involvement of Platelet TLR2 and TLR4. Blood (2011) 118:1952–61. doi: 10.1182/BLOOD-2011-03-343061
90. Maugeri N, Campana L, Gavina M, Covino C, De Metrio M, Panciroli C, et al. Activated Platelets Present High Mobility Group Box 1 to Neutrophils, Inducing Autophagy and Promoting the Extrusion of Neutrophil Extracellular Traps. J Thromb Haemost (2014) 12:2074–88. doi: 10.1111/JTH.12710
91. Dyer MR, Chen Q, Haldeman S, Yazdani H, Hoffman R, Loughran P, et al. Deep Vein Thrombosis in Mice is Regulated by Platelet HMGB1 Through Release of Neutrophil-Extracellular Traps and DNA. Sci Rep (2018) 8:2068. doi: 10.1038/S41598-018-20479-X
92. Clark SR, Ma AC, Tavener SA, McDonald B, Goodarzi Z, Kelly MM, et al. Platelet TLR4 Activates Neutrophil Extracellular Traps to Ensnare Bacteria in Septic Blood. Nat Med (2007) 13:463–9. doi: 10.1038/NM1565
93. McDonald B, Davis RP, Kim S-J, Tse M, Esmon CT, Kolaczkowska E, et al. Platelets and Neutrophil Extracellular Traps Collaborate to Promote Intravascular Coagulation During Sepsis in Mice. Blood (2017) 129:1357–67. doi: 10.1182/blood-2016-09-741298
94. Blasco A, Coronado MJ, Hernández-Terciado F, Martín P, Royuela A, Ramil E, et al. Assessment of Neutrophil Extracellular Traps in Coronary Thrombus of a Case Series of Patients With COVID-19 and Myocardial Infarction. JAMA Cardiol (2021) 6:469–74. doi: 10.1001/JAMACARDIO.2020.7308
95. Bautista-Becerril B, Campi-Caballero R, Sevilla-Fuentes S, Hernández-Regino LM, Hanono A, Flores-Bustamante A, et al. Immunothrombosis in COVID-19: Implications of Neutrophil Extracellular Traps. Biomol (2021) 11:694. doi: 10.3390/BIOM11050694
96. Li H, Liu L, Zhang D, Xu J, Dai H, Tang N, et al. SARS-CoV-2 and Viral Sepsis: Observations and Hypotheses. Lancet (London England) (2020) 395:1517–20. doi: 10.1016/S0140-6736(20)30920-X
97. Contou D, Cally R, Sarfati F, Desaint P, Fraissé M, Plantefève G. Causes and Timing of Death in Critically Ill COVID-19 Patients. Crit Care (2021) 25:79. doi: 10.1186/S13054-021-03492-X
98. Liu J, Li S, Liu J, Liang B, Wang X, Wang H, et al. Longitudinal Characteristics of Lymphocyte Responses and Cytokine Profiles in the Peripheral Blood of SARS-CoV-2 Infected Patients. EBioMedicine (2020) 55:102763. doi: 10.1016/J.EBIOM.2020.102763
99. Hu B, Huang S, Yin L. The Cytokine Storm and COVID-19. J Med Virol (2021) 93:250–6. doi: 10.1002/JMV.26232
100. Sahu BR, Kampa RK, Padhi A, Panda AK. C-Reactive Protein: A Promising Biomarker for Poor Prognosis in COVID-19 Infection. Clin Chim Acta (2020) 509:91–4. doi: 10.1016/J.CCA.2020.06.013
101. Zuo Y, Estes SK, Ali RA, Gandhi AA, Yalavarthi S, Shi H, et al. Prothrombotic Autoantibodies in Serum From Patients Hospitalized With COVID-19. Sci Transl Med (2020) 12:eabd3876. doi: 10.1126/SCITRANSLMED.ABD3876
102. Veras FP, Pontelli MC, Silva CM, Toller-Kawahisa JE, de Lima M, Nascimento DC, et al. SARS-CoV-2-Triggered Neutrophil Extracellular Traps Mediate COVID-19 Pathology. J Exp Med (2020) 217:e20201129. doi: 10.1084/JEM.20201129
103. Huckriede J, de Vries F, Hultström M, Wichapong K, Reutelingsperger C, Lipcsey M, et al. Histone H3 Cleavage in Severe COVID-19 ICU Patients. Front Cell Infect Microbiol (2021) 11:694186. doi: 10.3389/FCIMB.2021.694186
104. Akgun E, Tuzuner MB, Sahin B, Kilercik M, Kulah C, Cakiroglu HN, et al. Proteins Associated With Neutrophil Degranulation are Upregulated in Nasopharyngeal Swabs From SARS-CoV-2 Patients. PloS One (2020) 15:e0240012. doi: 10.1371/JOURNAL.PONE.0240012
105. Uppal NN, Kello N, Shah HH, Khanin Y, De Oleo IR, Epstein E, et al. De Novo ANCA-Associated Vasculitis With Glomerulonephritis in COVID-19. Kidney Int Rep (2020) 5:2079. doi: 10.1016/J.EKIR.2020.08.012
106. Felzer JR, Fogwe DT, Samrah S, Michet CJ, Specks U, Baqir M, et al. Association of COVID-19 Antigenicity With the Development of Antineutrophilic Cytoplasmic Antibody Vasculitis. Respirol Case Rep (2022) 10:e0894. doi: 10.1002/RCR2.894
107. Beloglazov V, Yatskov I, Nikolaeva A, Lavrenchuk E, DuBuske L. Cathepsin G in Patients With SARS-Cov-2 Infection of Various Degrees of Severity. J Allergy Clin Immunol (2022) 149:AB59. doi: 10.1016/J.JACI.2021.12.223
108. Mellhammar L, Thelaus L, Elen S, Fisher J, Linder A. Heparin Binding Protein in Severe COVID-19-A Prospective Observational Cohort Study. PloS One (2021) 16:e0249570. doi: 10.1371/JOURNAL.PONE.0249570
109. Bojkova D, Costa R, Reus P, Bechtel M, Jaboreck MC, Olmer R, et al. Targeting the Pentose Phosphate Pathway for SARS-CoV-2 Therapy. Metabolites (2021) 11:699. doi: 10.3390/METABO11100699
110. Shrivastava S, Chelluboina S, Jedge P, Doke P, Palkar S, Mishra AC, et al. Elevated Levels of Neutrophil Activated Proteins, Alpha-Defensins (DEFA1), Calprotectin (S100A8/A9) and Myeloperoxidase (MPO) Are Associated With Disease Severity in COVID-19 Patients. Front Cell Infect Microbiol (2021) 11:751232. doi: 10.3389/FCIMB.2021.751232
111. Leppkes M, Knopf J, Naschberger E, Lindemann A, Singh J, Herrmann I, et al. Vascular Occlusion by Neutrophil Extracellular Traps in COVID-19. EBioMedicine (2020) 58:102925. doi: 10.1016/J.EBIOM.2020.102925
112. Wang J, Li Q, Yin Y, Zhang Y, Cao Y, Lin X, et al. Excessive Neutrophils and Neutrophil Extracellular Traps in COVID-19. Front Immunol (2020) 11:2063. doi: 10.3389/FIMMU.2020.02063
113. Karampoor S, Hesamizadeh K, Maleki F, Farahmand M, Zahednasab H, Mirzaei R, et al. A Possible Pathogenic Correlation Between Neutrophil Elastase (NE) Enzyme and Inflammation in the Pathogenesis of Coronavirus Disease 2019 (COVID-19). Int Immunopharmacol (2021) 100:108137. doi: 10.1016/J.INTIMP.2021.108137
114. Radermecker C, Detrembleur N, Guiot J, Cavalier E, Henket M, d’Emal C, et al. Neutrophil Extracellular Traps Infiltrate the Lung Airway, Interstitial, and Vascular Compartments in Severe COVID-19. J Exp Med (2020) 217:e20201012. doi: 10.1084/JEM.20201012
115. Narasaraju T, Tang BM, Herrmann M, Muller S, Chow VTK, Radic M. Neutrophilia and NETopathy as Key Pathologic Drivers of Progressive Lung Impairment in Patients With COVID-19. Front Pharmacol (2020) 11:870. doi: 10.3389/FPHAR.2020.00870
116. Ng H, Havervall S, Rosell A, Aguilera K, Parv K, Von Meijenfeldt FA, et al. Circulating Markers of Neutrophil Extracellular Traps Are of Prognostic Value in Patients With COVID-19. Arterioscler Thromb Vasc Biol (2021) 41:988–94. doi: 10.1161/ATVBAHA.120.315267
117. Arcanjo A, Logullo J, Menezes CCB, de Souza Carvalho Giangiarulo TC, dos Reis MC, de Castro GMM, et al. The Emerging Role of Neutrophil Extracellular Traps in Severe Acute Respiratory Syndrome Coronavirus 2 (COVID-19). Sci Rep (2020) 10:19630. doi: 10.1038/S41598-020-76781-0
118. Zuo Y, Zuo M, Yalavarthi S, Gockman K, Madison JA, Shi H, et al. Neutrophil Extracellular Traps and Thrombosis in COVID-19. J Thromb Thrombolysis (2021) 51:446–53. doi: 10.1007/S11239-020-02324-Z
119. Skendros P, Mitsios A, Chrysanthopoulou A, Mastellos DC, Metallidis S, Rafailidis P, et al. Complement and Tissue Factor-Enriched Neutrophil Extracellular Traps are Key Drivers in COVID-19 Immunothrombosis. J Clin Invest (2020) 130:6151–7. doi: 10.1172/JCI141374
120. Zuo Y, Kanthi Y, Knight JS, Kim AHJ. The Interplay Between Neutrophils, Complement, and Microthrombi in COVID-19. Best Pract Res Clin Rheumatol (2021) 35:101661. doi: 10.1016/J.BERH.2021.101661
121. Ackermann M, Anders HJ, Bilyy R, Bowlin GL, Daniel C, De Lorenzo R, et al. Patients With COVID-19: In the Dark-NETs of Neutrophils. Cell Death Differ (2021) 28:3125–39. doi: 10.1038/s41418-021-00805-z
122. Park J, Wysocki RW, Amoozgar Z, Maiorino L, Fein MR, Jorns J, et al. Cancer Cells Induce Metastasis-Supporting Neutrophil Extracellular DNA Traps. Sci Transl Med (2016) 8:361ra138–361ra138. doi: 10.1126/scitranslmed.aag1711
123. Sollberger G, Choidas A, Burn GL, Habenberger P, Di Lucrezia R, Kordes S, et al. Gasdermin D Plays a Vital Role in the Generation of Neutrophil Extracellular Traps. Sci Immunol (2018) 3:eaar6689. doi: 10.1126/sciimmunol.aar6689
124. Papayannopoulos V, Staab D, Zychlinsky A. Neutrophil Elastase Enhances Sputum Solubilization in Cystic Fibrosis Patients Receiving DNase Therapy. PloS One (2011) 6:e28526. doi: 10.1371/journal.pone.0028526
125. Aikawa N, Ishizaka A, Hirasawa H, Shimazaki S, Yamamoto Y, Sugimoto H, et al. Reevaluation of the Efficacy and Safety of the Neutrophil Elastase Inhibitor, Sivelestat, for the Treatment of Acute Lung Injury Associated With Systemic Inflammatory Response Syndrome; a Phase IV Study. Pulm Pharmacol Ther (2011) 24:549–54. doi: 10.1016/j.pupt.2011.03.001
126. Fisher J, Mohanty T, Karlsson CAQ, Khademi SMH, Malmström E, Frigyesi A, et al. Proteome Profiling of Recombinant DNase Therapy in Reducing NETs and Aiding Recovery in COVID-19 Patients. Mol Cell Proteomics (2021) 20:100113. doi: 10.1016/J.MCPRO.2021.100113
127. Korkmaz B, Lesner A, Marchand-Adam S, Moss C, Jenne DE. Lung Protection by Cathepsin C Inhibition: A New Hope for COVID-19 and ARDS? J Med Chem (2020) 63:13258–65. doi: 10.1021/ACS.JMEDCHEM.0C00776
128. Huet T, Beaussier H, Voisin O, Jouveshomme S, Dauriat G, Lazareth I, et al. Anakinra for Severe Forms of COVID-19: A Cohort Study. Lancet Rheumatol (2020) 2:e393–400. doi: 10.1016/S2665-9913(20)30164-8
129. Edalatifard M, Akhtari M, Salehi M, Naderi Z, Jamshidi A, Mostafaei S, et al. Intravenous Methylprednisolone Pulse as a Treatment for Hospitalised Severe COVID-19 Patients: Results From a Randomised Controlled Clinical Trial. Eur Respir J (2020) 56:2002808. doi: 10.1183/13993003.02808-2020
130. Kashour Z, Riaz M, Garbati MA, AlDosary O, Tlayjeh H, Gerberi D, et al. Efficacy of Chloroquine or Hydroxychloroquine in COVID-19 Patients: A Systematic Review and Meta-Analysis. J Antimicrob Chemother (2021) 76:30–42. doi: 10.1093/JAC/DKAA403
131. Mikolajewska A, Fischer AL, Piechotta V, Mueller A, Metzendorf MI, Becker M, et al. Colchicine for the Treatment of COVID-19. Cochrane Database Syst Rev (2021) 10:CD015045. doi: 10.1002/14651858.CD015045
132. Mereo BioPharma Sees Positive Results in Trial for Covid-19 Treatment - MarketWatch . Available at: https://www.marketwatch.com/story/mereo-biopharma-sees-positive-results-in-trial-for-covid-19-treatment-271640175318 (Accessed January 17, 2022).
133. Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, et al. Dexamethasone in Hospitalized Patients With Covid-19. N Engl J Med (2021) 384:693–704. doi: 10.1056/NEJMOA2021436
134. Vargas A, Boivin R, Cano P, Murcia Y, Bazin I, Lavoie JP. Neutrophil Extracellular Traps are Downregulated by Glucocorticosteroids in Lungs in an Equine Model of Asthma. Respir Res (2017) 18:207. doi: 10.1186/S12931-017-0689-4
135. Sun Y, Chen C, Zhang X, Wang S, Zhu R, Zhou A, et al. Heparin Improves Alveolarization and Vascular Development in Hyperoxia-Induced Bronchopulmonary Dysplasia by Inhibiting Neutrophil Extracellular Traps. Biochem Biophys Res Commun (2020) 522:33–9. doi: 10.1016/J.BBRC.2019.11.041
136. Nadkarni GN, Lala A, Bagiella E, Chang HL, Moreno PR, Pujadas E, et al. Anticoagulation, Bleeding, Mortality, and Pathology in Hospitalized Patients With COVID-19. J Am Coll Cardiol (2020) 76:1815–26. doi: 10.1016/J.JACC.2020.08.041
137. Keir HR, Chalmers JD. IL-6 Trans-Signalling: How Haemophilus Surfs the NET to Amplify Inflammation in COPD. Eur Respir J (2021) 58:2102143. doi: 10.1183/13993003.02143-2021
138. FDA Approves Actemra for Hospitalised COVID-19 Patients . Available at: https://www.europeanpharmaceuticalreview.com/news/157399/fda-approves-actemra-for-emergency-use-in-hospitalised-covid-19-patients/ (Accessed January 16, 2022).
139. Ruiz-Limón P, Ortega R, Arias de la Rosa I, Abalos-Aguilera M del C, Perez- Sanchez C, Jimenez- Gomez Y, et al. Tocilizumab Improves the Proatherothrombotic Profile of Rheumatoid Arthritis Patients Modulating Endothelial Dysfunction, NETosis, and Inflammation. Transl Res (2017) 183:87–103. doi: 10.1016/J.TRSL.2016.12.003
140. Rasmussen KH, Hawkins CL. Role of Macrophage Extracellular Traps in Innate Immunity and Inflammatory Disease. Biochem Soc Trans (2022) 50:21–32. doi: 10.1042/BST20210962
141. Doster RS, Rogers LM, Gaddy JA, Aronoff DM. Macrophage Extracellular Traps: A Scoping Review. J Innate Immun (2018) 10:3–13. doi: 10.1159/000480373
142. Halder LD, Abdelfatah MA, Jo EAH, Jacobsen ID, Westermann M, Beyersdorf N, et al. Factor H Binds to Extracellular DNA Traps Released From Human Blood Monocytes in Response to Candida Albicans. Front Immunol (2017) 7:671. doi: 10.3389/FIMMU.2016.00671
143. Je S, Quan H, Yoon Y, Na Y, Kim BJ, Seok SH. Mycobacterium Massiliense Induces Macrophage Extracellular Traps With Facilitating Bacterial Growth. PLoS One (2016) 11:e0155685. doi: 10.1371/JOURNAL.PONE.0155685
144. Abani O, Abbas A, Abbas F, Abbas M, Abbasi S, Abbass H, et al. Tocilizumab in Patients Admitted to Hospital With COVID-19 (RECOVERY): A Randomised, Controlled, Open-Label, Platform Trial. Lancet (London England) (2021) 397:1637–45. doi: 10.1016/S0140-6736(21)00676-0
145. Tagami T, Tosa R, Omura M, Fukushima H, Kaneko T, Endo T, et al. Effect of a Selective Neutrophil Elastase Inhibitor on Mortality and Ventilator-Free Days in Patients With Increased Extravascular Lung Water: A Post Hoc Analysis of the PiCCO Pulmonary Edema Study. J Intensive Care (2014) 2:67. doi: 10.1186/S40560-014-0067-Y
146. Meng W, Paunel-Görgülü A, Flohé S, Hoffmann A, Witte I, MacKenzie C, et al. Depletion of Neutrophil Extracellular Traps In Vivo Results in Hypersusceptibility to Polymicrobial Sepsis in Mice. Crit Care (2012) 16:R137. doi: 10.1186/CC11442
Keywords: cytokines, inflammation, lymphocyte, septic shock, homeostasis, acute respiratory distress syndrome, pneumonia, cytokine storm
Citation: Ventura-Santana E, Ninan JR, Snyder CM and Okeke EB (2022) Neutrophil Extracellular Traps, Sepsis and COVID-19 – A Tripod Stand. Front. Immunol. 13:902206. doi: 10.3389/fimmu.2022.902206
Received: 22 March 2022; Accepted: 11 May 2022;
Published: 10 June 2022.
Edited by:
Vijay Kumar, Duke University, United StatesReviewed by:
Marko Radic, University of Tennessee College of Medicine, United StatesAhmed Yaqinuddin, Alfaisal University, Saudi Arabia
Copyright © 2022 Ventura-Santana, Ninan, Snyder and Okeke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emeka B. Okeke, b2tla2VAZnJlZG9uaWEuZWR1
 Esmeiry Ventura-Santana
Esmeiry Ventura-Santana Emeka B. Okeke
Emeka B. Okeke