- Department of Rheumatology and Clinical Immunology, National Clinical Research Center for Dermatologic and Immunologic Diseases (NCRC-DID), Ministry of Science and Technology, State Key Laboratory of Complex Severe and Rare Diseases, Peking Union Medical College Hospital, Key Laboratory of Rheumatology and Clinical Immunology, Ministry of Education, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
Sjögren’s syndrome (SS) is a common chronic systemic autoimmune disease and its main characteristic is lymphoid infiltration of the exocrine glands, particularly the salivary and lacrimal glands, leading to sicca symptoms of the mouth and eyes. Growing evidence has shown that SS is also characterized by microbial perturbations like other autoimmune diseases. Significant alterations in diversity, composition, and function of the microbiota were observed in SS. The dysbiosis of the microbiome correlates with worse symptoms and higher disease severity, suggesting that dysbiosis may be of great importance in the pathogenesis of SS. In this review, we provide a general view of recent studies describing the microbiota alterations of SS, the possible pathways that may cause microbiota dysbiosis to trigger SS, and the existence of the gut-ocular/gut-oral axis in SS.
Introduction
Sjögren’s Syndrome (SS) is a chronic autoimmune disease with a female-to-male predominance of 9:1, of which the peak incidence is about 50 years of age (1). SS primarily causes inflammation of the exocrine glands (predominantly the salivary and lacrimal glands), resulting in severe oral and ocular dryness. The clinical manifestation of SS is heterogeneous, which can differ from sicca symptoms to systemic disease and lymphoma (2). The significant immunologic abnormalities of SS, including the presence of serum anti-SSA antibodies and focal lymphocytic sialadenitis on biopsy of labial salivary glands, is the basis of formal criteria for the diagnosis (3). To date, little is known about the pathogenesis of SS. A wide range of environmental factors interacting with the genetic predisposition of an individual may affect the development of SS.
The Human Microbiome Project has revealed that the microbiome plays an outstanding role in the pathogenesis of autoimmune diseases (4). Numerous studies of human autoimmune disease consistently report a state of microbial dysbiosis. For instance, dysbiosis of gut microbiota has been demonstrated as a characteristic of some rheumatic diseases including, rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), and systemic sclerosis (5–9). Recently, many studies have described the alterations due to SS of microbial communities and the composition of the intestinal, oral, and ocular areas, though some results were heterogeneous. Furthermore, dysbiosis is associated with the worsening of symptoms and disease activity in SS patients. However, the cause or effect role that microbiome plays in the pathogenesis of SS remains unclear.
This review aims to describe the microbiome alterations of SS, the clinical association of microbiome dysbiosis, and substantial mechanisms of the microbiota involved in the pathology of SS.
Diversity changes in microbiota
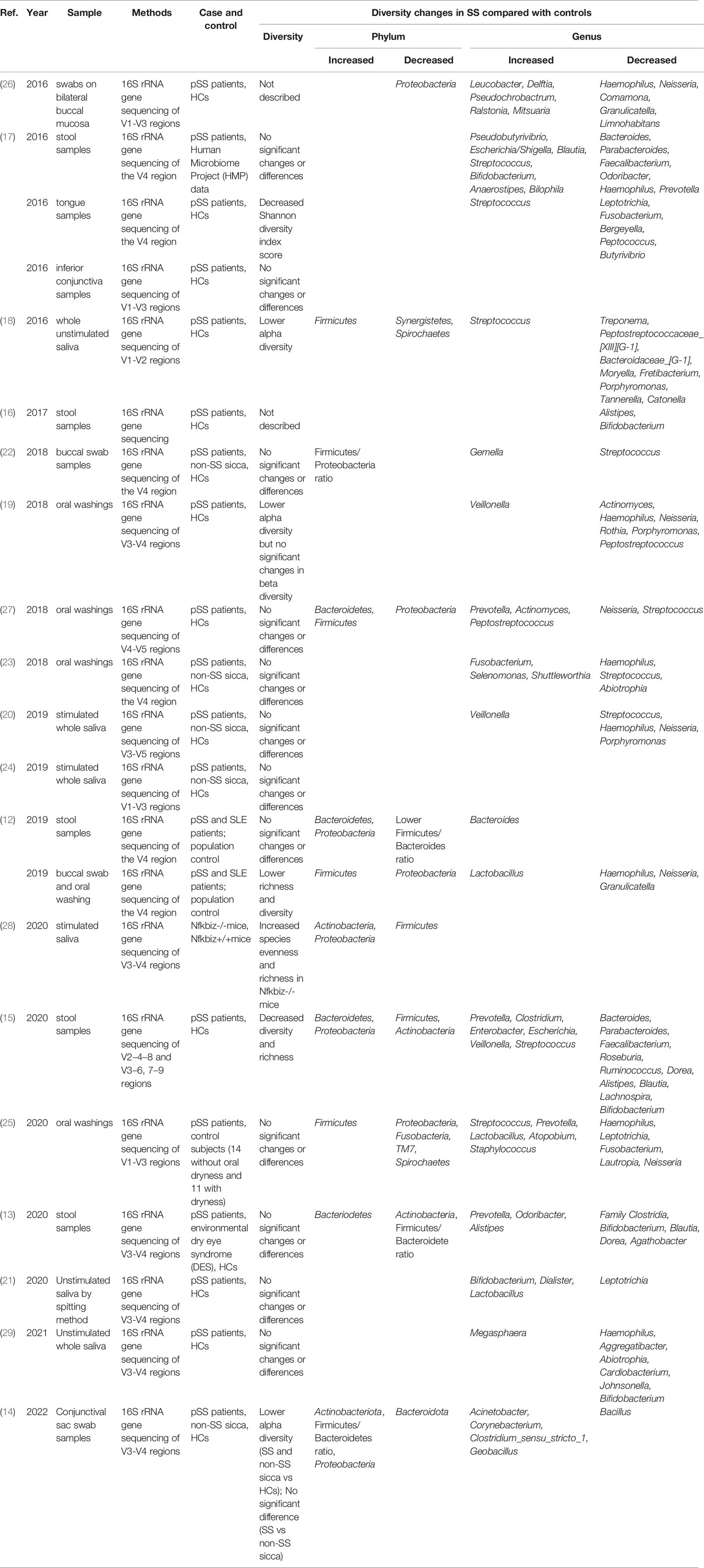
Microbiota diversity is essential to maintain the stability and efficiency of the ecosystem. The ratio of Firmicutes to Bacteroidetes (F/B ratio) has a close connection with the composition of the intestinal microbiota, which is frequently used to evaluate the imbalance of intestinal microbiota (7, 10). The F/B ratio has been found to be lower than the normal level in several diseases, including inflammatory bowel disease, RA, SLE, and obesity (11). Interestingly, recent studies demonstrated that a lower F/B ratio in SS patients was a main characteristic of the gut microbiota composition compared with HCs, which means Firmicutes were suppressed and Bacteroidetes became conditional pathogens and also indicate the existence of gut microbial dysbiosis in SS (12–14). In addition, significantly decreased diversity of SS was found when compared with healthy controls (15–21), implying that microbiota dysbiosis may participate in the pathogenesis. However, the mechanism of microbiome alteration in SS was still known.
Both the specific symptoms (dry mouth and dry eye) and autoimmunity (the specific disease background of rheumatoid disease) are potential factors that may influence the constitution of the microbiome. Between SS and symptom controls, most of the studies observed no significant difference in the gut, oral, and ocular microbiota (13, 22–25), which indicates that the changes in the microbiome might be only associated with dry eye or dry mouth (Table 1).
Since the correlation between the common clinical symptom and autoimmunity is almost definite, it seems that this specific disease background of SS should also be closely associated with microbiome alteration. However, no difference was found in the richness of gut microbiota but significantly lower diversity of oral microbiota was observed in SS than in SLE (12), which was counted in dry mouth by the researchers (Table 1). Along with these reports that questioned the role autoimmunity plays in the microbiota dysbiosis of SS, one possible reason to explain the microbiota changes is that the alteration of the microbiome is the leading factor of clinical symptoms as well as the breakdown of autoimmune system. After all, other studies also reported that microbial dysbiosis can occur independent of salivary deficiency and SS can be developed independent of microbiome changes (18, 30).
Many factors were found to be related to microbiome changes. For example, the oral microbiota composition could be affected by oral conditions such as periodontitis, caries, and age (31, 32). However, the triggers that result in altered microbiota in SS have not yet been clarified.
Because the standard of normal microbiome has not been established, we could not make a definite conclusion on the point that excluding pSS patients via characteristics of microbiome is correct. Considering that the pathogenesis of SS is complex, it is possible that not all the patients exert microbiota alteration, which is also observed in some studies (19, 26, 27).
Correlation between altered microbiota and SS
Microbial diversity has been proven to change in SS as described above. The nature of these changes is one of the most important questions to be answered.
Growing evidence of microbiota dysbiosis in SS confirmed the trend that lower diversity could lead to higher disease activity. An early study reported that oral antibiotic treatment and desiccating stress leads to great alterations in the gut microbiota (17). In addition, the diversity of fecal microbiota was negatively associated with combined ocular and systemic disease index. Similarly, SS patients with severe dysbiosis had a higher ESSDAI total score, ClinESSDAI total score, and levels of fecal calprotectin, while having lower levels of complement component 4 (16). Furthermore, a strong positive relation was found between tear break-up time and both Actinobacteria and Bifidobacteria (13) (Table 1).
To further understand the mechanisms related to these observed correlations, researchers also studied microscopic evidence. In SS, the levels of proinflammation factors IL-6, IL-12, IL-17, and TNF-α were positively associated with the richness of Enterobacter, while negatively associated with the abundance of Lachnospira, Roseburia, Bifidobacterium, Ruminococcus, Blautia, and Roseburia. Compared with SS, a significant inverse correlation was only observed between the abundance of Parabacteroides distasonis and the levels of IL-6 and TNF-α in controls (15) (Table 1). It is intuitive that the germs involved in inflammation were more prevalent in SS and the bacterial communities that participated in the immune regulation were totally different between SS and the controls.
The research noted above made an impression that the microbial alteration of SS has a close association with severe clinical symptoms via some inflammation pathways. Regarding the link between microbiota dysbiosis and inflammation pathways, little is known as research regarding the cause in SS is limited. Based on present studies, we summarize three potential mechanisms: molecular mimicry, metabolite changes, and epithelial tolerance breakdown.
From correlation to potential causality
The microbiome alteration has a negative effect on the pathogenesis of SS, but the causality between them remained unknown. Based on the diversity and correlations described above, we presume that an unknown variant leads to microbiota changes which directly affects the function of organs, for example, salivation. Also, an altered microbiome further activates the immune system which results in the damage of a specific site via inflammation. Three pathways might help explain these presumptions (Figure 1).
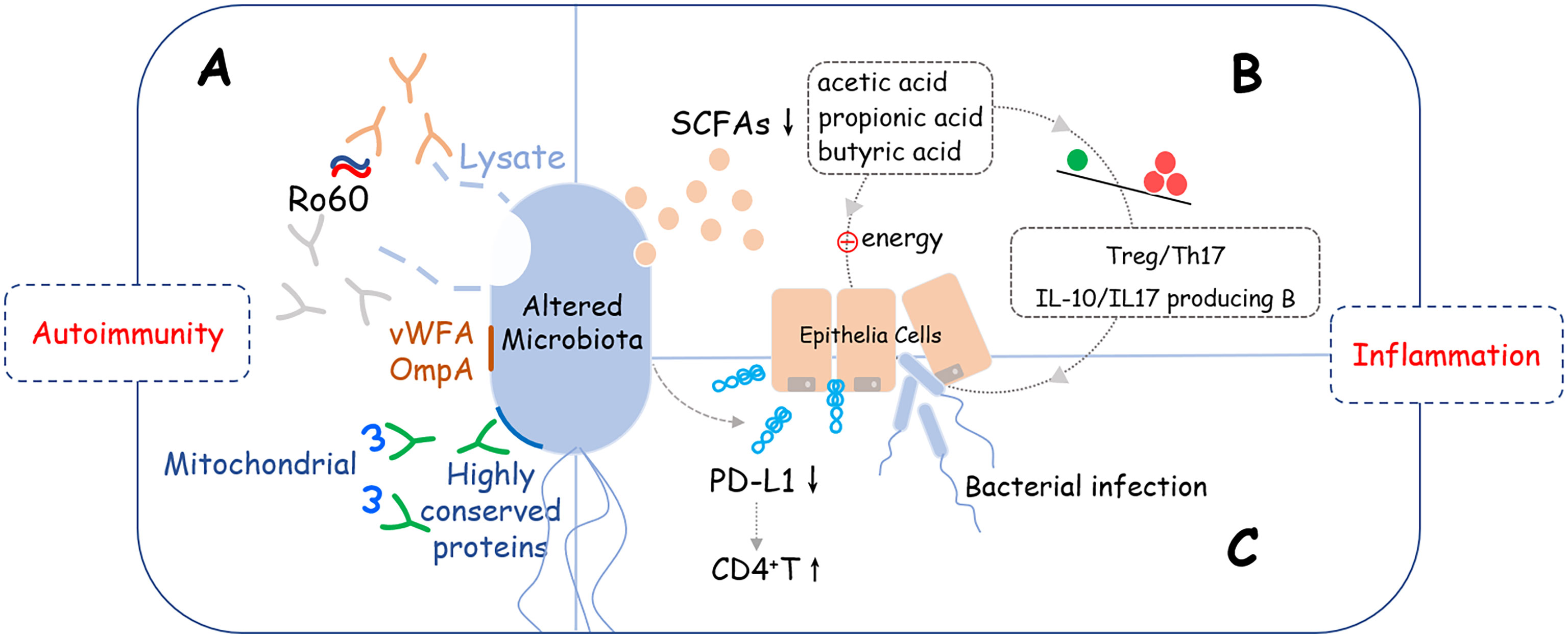
Figure 1 Potential crosslinks between microbiome alteration and pathogenesis of SS. Altered microbiome may induce development of SS via three potential pathways: (A) Molecular mimicry: Lysates, proteins, or peptides that originate from bacteria, including von Willebrand factor type A domain protein (vWFA), OmpA, and some other highly conserved proteins could react with anti-Ro60 antibodies or SSA/Ro60-reactive T cells, which are involved in the activation of excessive autoimmune response; (B) Metabolite changes: Short-chain fatty acids (SCFAs) produced by bacteria were decreased that led to reducing the immune checkpoint molecules and imbalance of T cells (Treg/Th17) and B cells (IL-10/IL-17 producing B cells), which provides a pro-inflammation microenvironment is SS. (C) Epithelial tolerance breakdown: Reducing the supply of energy for epithelial cells, disrupting the immunomodulatory function such as down-regulating the expression level of PD-L1 on epithelial cells, and interfering with the mucus barrier are possible mechanisms that epithelial tolerance was broken down.
Molecular Mimicry
Molecular mimicry has been widely acknowledged as a mechanism of autoimmune diseases caused by infectious agents with epitope intermolecular spreading (33, 34). B cell cross-reactivity between Coxsackie virus 2B protein and Ro60 has been reported to have significant association with the initiation and perpetuation of anti-Ro60 autoantibody response, which is essential in the pathology of SS (35). Recently, Greiling et al. found that the serum of anti-Ro60-positive patients can react with the lysates of Bacteroides thetaiotaomicron (36). Also, Yanagisawa et al. found a new antigen named OmpA from E. coli, which is highly antigenic to elicit antibodies against SSA/Ro and SSB/La and cause inflammation of the Harderian and the salivary glands (37), suggesting that microbe-originated molecules elicit inflammation in exocrine glands with specificity.
Interestingly, Szymula et al. also identified a peptide originating from C. ochracea, named von Willebrand factor type A domain protein (vWFA), which has been proven to be the most potent activator of SSA/Ro60-reactive T cells, consequently, inducing robust IL-2 production. In addition, recombinant vWFA protein and whole E. coli expressing vWFA are both capable of initiating the activation of Ro60-reactive T cells (38).
Besides, the highly-conserved microbial proteins of Staphylococcus aureus and E. coli can induce patients with SS to develop corresponding antibodies, and as a consequence of molecular mimicry, these antibodies cross-react against mitochondrial self-antigens, which may account for their fatigue symptoms (39).
These observations affirmed the concept of molecular mimicry between microbiota and the pathology of SS. Target genera that participate in this pathway still need to be clarified for various genera of the Enterobacteriaceae possessed proteins that have high amino-acid sequence similarities with OmpA and vWFA.
Metabolite Changes
Short-chain fatty acids (SCFAs) are one of the important metabolites of intestinal microorganisms and are mainly composed of acetic acid, propionic acid, and butyric acid. As signaling molecules, SCFAs play a key role in regulating host metabolism, immune system and cell proliferation (40, 41). In systemic autoimmune diseases, several SCFA-producing bacteria, including Lachnoclostridium, Lachnospira, and Sutterella were reduced. Of note, these bacteria played a highly pro-regulatory, tolerogenic role in immune functions (42). In SS, butyrate is the most commonly mentioned bacterial metabolite. Several studies have found butyrate-producing bacteria (including Faecalibacterium prausnitzii, Bacteroides fragilis, Lachnoclostridium, Roseburia, Lachnospira, and Ruminococcus) were significantly decreased in SS (13, 15–17, 28). As a vital microbiota-derived metabolite, butyrate provides energy for colonic epithelial cells and promotes intestinal barrier functions. Moreover, recent research observed that butyrate-producing bacteria, Bacteroides spp. and the Clostridia clusters XIVa and IV were important for maintaining the Treg/Th17 balance. To be noted, the differentiation of Treg cells is not only promoted by butyrate but also by the immunomodulatory molecule polysaccharide A derived from the Bacteroides fragilis (43–45). Interrupting the Treg/Th17 balance will lose the protection of the mucosa barrier from colonization by pathogenic microorganisms (15). Actually, butyrate could both affect the balances in T cells and regulate the frequency of IL-10 and/or IL-17 producing B cells by regulating circadian-clock-related genes to perform the anti-inflammation function (46).
In other respects, butyrate could elevate the flow rate of salivation and the relief of salivary gland inflammation (46). All these clues suggest that the reduced SCFAs or butyrate-producing bacteria might affect the permeability of the mucosa barrier, the frequency or function of immune cells, and even secretion of the salivary gland, so as to be involved in the pathogenesis of SS.
Epithelial Tolerance Breakdown
Breakdown of epithelial tolerance exists in SS but the mechanism has not yet been clarified. Some studies observed that salivary gland epithelial cells (SGECs) were expressed with immune checkpoint molecules that may have a potential role in the immune response (47) and bacteria involvement may be an important reason for this phenomenon. H. parainfluenzae was considered to act as an immunomodulatory commensal bacterium in SS, as its reduction may result in decrease of the PD-L1 on SGECs, whose inhibitory control on the proliferation of CD4+T may be loosened according to the observation found in the pretreatment of A253 cells (from human submandibular gland squamous cell carcinoma) with H. parainfluenzae (29).
Interestingly, Jehan et al. reported the involved salivary gland of SS was infected by bacteria including Prevotella melaninogenica (25). Prevotella is an SS-associated oral bacteria that could upregulate MHC molecules and CD80 in human submandibular gland tumor cells, which may initiate IFN pathway-related inflammation (13). Interestingly, the significantly higher abundance of Prevotella was found in a family that contains key enzymes for mucin degradation that could also affect the colonic mucus barrier (48). Other than reducing the supply of energy for epithelial cells, disrupting the immunomodulatory function of epithelial cells and interfering with the mucus barrier that protects the epithelial cells are the two other possible ways for the microbiota in SS to achieve the breaking down goal.
Besides molecular mimicry, metabolite changes, and epithelial tolerance breakdown, some correlation between bacterial alteration and pathological changes was observed but the causality needs further exploration. For example, in TSP-1-/- mice, Staphylococcus aureus and coagulase-negative staphylococci sp. species were significantly increased and associated with increased neutrophil infiltration into the conjunctiva (49). Importantly, the three links we presumed were not absolutely independent but might cross with each other, which makes the exploration of the causality between microbiota and SS more complicated.
Gut-ocular and gut-oral axis
Exploring the clinical relationship/pathogenesis and the microbiome changes among the microbiota from gut, ocular, and oral, also called gut-ocular-oral axis, helps illustrate the role that bacteria plays in the development of SS. Mahira et al. observed that germ-free CD25 knockout mice have worsened involvement of gut, ocular, and oral. Also, the levels of pro-inflammation factors were increased and the percentage of CD4+IFN-γ+ cells was greater in this germ-free model. Noteworthy, fecal matter from normal mice being transplanted into CD25 knockout mice could help reverse the pathological changes in the gut, ocular, and oral areas (50). Therefore, the gut-ocular-oral axis might exist in SS.
Regarding the ocular-gut axis, several studies have shown that intestinal dysbiosis also has a connection with the severity of ocular mucosal diseases in SS. Moon et al. reported that the relative abundance of Bacteroidetes, Actinobacteria, and Bifidobacterium in the SS gut was significantly associated with dry eye symptoms (13). Animal models, pretreated with desiccating stress and antibiotics, were found to have extreme changes in gut microbiota, which was partly counting on the Proteobacteria found to be associated with the more severe ocular phenotype (17). These findings suggest that intestinal dysbiosis has a significant clinical association with the severity of dry eye. It is speculated that dysbiosis of the gut microbiota might aggravate germinal center loss in dry eyes by reducing the frequency of resident NK/NKT cells and changing their cytokine profile, leading to suppressed Th2 tone (17).
Worth mention, buccal epithelial cells inhabited by intracellular bacteria can be an important reservoir. The intracellular bacteria may be transmitted to the gut through the shedding process of buccal epithelial cells (51, 52). In SS, the relative abundance of Actinomyces and Lactobacillus in oral samples was associated with their abundance in stool samples (12). Moreover, we have described above that several studies have suggested the cross-reactivity of commensal oral and gut bacteria with SSA/Ro60 is of great importance in the pathogenesis of SS (36, 38). Therefore, it is plausible that there is a link between oral and gut bacteria but the related mechanism is still unknown.
It has been found that Firmicutes, Actinobacteria, Proteobacteria, and Bacteroidetes phyla are dominant in conjunctival samples from patients with SS and controls (17). However, as yet, no significant difference has been found in the diversity and composition of the ocular surface microbiome between SS and controls. Also, research on the ocular-oral axis is limited and no related result was observed.
Hints for treatment
In recent years, targeted manipulation of the microbiome to restore immunity and improve disease outcomes has drawn close attention from researchers. Regulating the abnormal diversity of the microbiome was the main direction for SS therapy. Several studies have applied cohousing therapy, fecal transplant, and probiotic therapy among SS animal models. For example, Lee et al. found that cohousing young Nfkbiz+/+ with Nfkbiz−/− mice synchronized their oral microbiome toward that of young Nfkbiz+/+ mice, preventing dysbiosis of Nfkbiz−/− mice (28). Further, Zaheer et al. showed that fecal bacteria transplantation decreased the generation of pathogenic CD4+IFN-γ+ cells and reversed the spontaneous dry eye phenotype of germ-free CD25KO mice (50). Similarly, another study of an SS animal model showed that ocular surface disease was alleviated by probiotic therapy with 5 ingredients of Lactobacillus reuteri, Streptococcus thermophilus, Bifidobacterium bifidum, Lactobacillus casei, and Lactobacillus acidophilus (53). Future clinical trials performed in SS patients will be needed to confirm these exciting results.
Though the effect of therapeutic strategies involving the microbiome was not valid, the shift of microbiota during traditional treatment helps enrich the cognition of these therapies. Lu et al. demonstrated that treating SS patients with total glucoside of paeony (TGP) improved the diversity and the composition of the intestinal microbiome, including increasing the abundance of Firmicutes, thereby inhibiting the pathogenic role of Bacteroides (11). Moreover, TGP combined with hydroxychloroquine promoted the thriving of several commensal bacteria (Lactobacillus, Incertae, and Desulfovibrio) and limited the expansion of dominant pathogenic bacteria (Bacteroides and Alloprevotella) (11). To a certain extent, supervision of the microbial diversity and the composition of microbiota may not only help explain the pharmacological mechanism of drugs but also be a potential marker to evaluate the effectiveness of treatment.
Summary
Microbiota of SS has been a popular issue in recent years. Generally, studies have shown that the SS microbiome is mainly characterized by reduced gut and oral diversity. In addition, a negative correlation was observed between the diversity and clinical symptoms. Based on current research, microbiota alteration may play their pathological role via at least three pathways: molecular mimicry, metabolite changes, and epithelial tolerance breakdown, which ultimately result in a dysregulated immune system and abnormal clinical symptoms such as dry eye and dry mouth. Causality studies are direly needed in this field, not only for the explanation of disease pathogenesis but also for the application of microbiota changes in clinical decision-making.
Author Contributions
YF supervised the work. CD and QX collected the data and wrote the manuscript. CD was also responsible for the outline and modification of this review. All authors have read and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (grant numbers 81971545, 82172343) and the CAMS Innovation Fund for Medical Sciences (CIFMS, 2020-I2M-C & T-A-002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Qin B, Wang J, Yang Z, Yang M, Ma N, Huang F, et al. Epidemiology of Primary Sjögren’s Syndrome: A Systematic Review and Meta-Analysis. Ann Rheum Dis (2015) 74(11):1983–9. doi: 10.1136/annrheumdis-2014-205375
2. Brito-Zerón P, Baldini C, Bootsma H, Bowman SJ, Jonsson R, Mariette X, et al. Sjögren Syndrome. Nat Rev Dis Primers (2016) 2:16047. doi: 10.1038/nrdp.2016.47
3. Shiboski CH, Shiboski SC, Seror R, Criswell LA, Labetoulle M, Lietman TM, et al. 2016 American College of Rheumatology/European League Against Rheumatism Classification Criteria for Primary Sjögren’s Syndrome: A Consensus and Data-Driven Methodology Involving Three International Patient Cohorts. Arthritis Rheumatol (2017) 69(1):35–45. doi: 10.1002/art.39859
4. Rosenbaum JT, Asquith MJ. The Microbiome: A Revolution in Treatment for Rheumatic Diseases? Curr Rheumatol Rep (2016) 18(10):62. doi: 10.1007/s11926-016-0614-8
5. van der Meulen TA, Harmsen H, Bootsma H, Spijkervet F, Kroese F, Vissink A. The Microbiome-Systemic Diseases Connection. Oral Dis (2016) 22(8):719–34. doi: 10.1111/odi.12472
6. Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of Intestinal Prevotella Copri Correlates With Enhanced Susceptibility to Arthritis. Elife (2013) 2:e01202. doi: 10.7554/eLife.01202
7. Hevia A, Milani C, López P, Cuervo A, Arboleya S, Duranti S, et al. Intestinal Dysbiosis Associated With Systemic Lupus Erythematosus. mBio (2014) 5(5):e01548–14. doi: 10.1128/mBio.01548-14
8. Shahi SK, Freedman SN, Mangalam AK. Gut Microbiome in Multiple Sclerosis: The Players Involved and the Roles They Play. Gut Microbes (2017) 8(6):607–15. doi: 10.1080/19490976.2017.1349041
9. Tsigalou C, Stavropoulou E, Bezirtzoglou E. Current Insights in Microbiome Shifts in Sjogren’s Syndrome and Possible Therapeutic Interventions. Front Immunol (2018) 9:1106. doi: 10.3389/fimmu.2018.01106
10. Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The Oral and Gut Microbiomes are Perturbed in Rheumatoid Arthritis and Partly Normalized After Treatment. Nat Med (2015) 21(8):895–905. doi: 10.1038/nm.3914
11. Lu WW, Fu TX, Wang Q, Chen YL, Li TY, Wu GL. The Effect of Total Glucoside of Paeony on Gut Microbiota in NOD Mice With Sjögren’s Syndrome Based on High-Throughput Sequencing of 16srrna Gene. Chin Med (2020) 15:61. doi: 10.1186/s13020-020-00342-w
12. van der Meulen TA, Harmsen HJM, Vila AV, Kurilshikov A, Liefers SC, Zhernakova A, et al. Shared Gut, But Distinct Oral Microbiota Composition in Primary Sjögren’s Syndrome and Systemic Lupus Erythematosus. J Autoimmun (2019) 97:77–87. doi: 10.1016/j.jaut.2018.10.009
13. Moon J, Choi SH, Yoon CH, Kim MK. Gut Dysbiosis is Prevailing in Sjögren’s Syndrome and is Related to Dry Eye Severity. PloS One (2020) 15(2):e0229029. doi: 10.1371/journal.pone.0229029
14. Song H, Xiao K, Chen Z, Long Q. Analysis of Conjunctival Sac Microbiome in Dry Eye Patients With and Without Sjögren’s Syndrome. Front Med (Lausanne) (2022) 9:841112. doi: 10.3389/fmed.2022.841112
15. Cano-Ortiz A, Laborda-Illanes A, Plaza-Andrades I, Membrillo Del Pozo A, Villarrubia Cuadrado A, Rodríguez Calvo de Mora M, et al. Connection Between the Gut Microbiome, Systemic Inflammation, Gut Permeability and FOXP3 Expression in Patients With Primary Sjögren’s Syndrome. Int J Mol Sci (2020) 21(22):8733. doi: 10.3390/ijms21228733
16. Mandl T, Marsal J, Olsson P, Ohlsson B, Andréasson K. Severe Intestinal Dysbiosis is Prevalent in Primary Sjögren’s Syndrome and is Associated With Systemic Disease Activity. Arthritis Res Ther (2017) 19(1):237. doi: 10.1186/s13075-017-1446-2
17. de Paiva CS, Jones DB, Stern ME, Bian F, Moore QL, Corbiere S, et al. Altered Mucosal Microbiome Diversity and Disease Severity in Sjögren Syndrome. Sci Rep (2016) 6:23561. doi: 10.1038/srep23561
18. Siddiqui H, Chen T, Aliko A, Mydel PM, Jonsson R, Olsen I. Microbiological and Bioinformatics Analysis of Primary Sjogren’s Syndrome Patients With Normal Salivation. J Oral Microbiol (2016) 8:31119. doi: 10.3402/jom.v8.31119
19. Zhou Z, Ling G, Ding N, Xun Z, Zhu C, Hua H, et al. Molecular Analysis of Oral Microflora in Patients With Primary Sjögren’s Syndrome by Using High-Throughput Sequencing. PeerJ (2018) 6:e5649. doi: 10.7717/peerj.5649
20. Rusthen S, Kristoffersen AK, Young A, Galtung HK, Petrovski B, Palm Ø, et al. Dysbiotic Salivary Microbiota in Dry Mouth and Primary Sjögren’s Syndrome Patients. PloS One (2019) 14(6):e0218319. doi: 10.1371/journal.pone.0218319
21. Sharma D, Sandhya P, Vellarikkal SK, Surin AK, Jayarajan R, Verma A, et al. Saliva Microbiome in Primary Sjögren’s Syndrome Reveals Distinct Set of Disease-Associated Microbes. Oral Dis (2020) 26(2):295–301. doi: 10.1111/odi.13191
22. van der Meulen TA, Harmsen HJM, Bootsma H, Liefers SC, Vich Vila A, Zhernakova A, et al. Dysbiosis of the Buccal Mucosa Microbiome in Primary Sjögren’s Syndrome Patients. Rheumatol (Oxford) (2018) 57(12):2225–34. doi: 10.1093/rheumatology/key215
23. van der Meulen TA, Harmsen HJM, Bootsma H, Liefers SC, Vich Vila A, Zhernakova A, et al. Reduced Salivary Secretion Contributes More to Changes in the Oral Microbiome of Patients With Primary Sjögren’s Syndrome Than Underlying Disease. Ann Rheum Dis (2018) 77(10):1542–4. doi: 10.1136/annrheumdis-2018-213026
24. Sembler-Møller ML, Belstrøm D, Locht H, Enevold C, Pedersen AML. Next-Generation Sequencing of Whole Saliva From Patients With Primary Sjögren’s Syndrome and non-Sjögren’s Sicca Reveals Comparable Salivary Microbiota. J Oral Microbiol (2019) 11(1):1660566. doi: 10.1080/20002297.2019.1660566
25. Alam J, Lee A, Lee J, Kwon DI, Park HK, Park JH, et al. Dysbiotic Oral Microbiota and Infected Salivary Glands in Sjögren’s Syndrome. PloS One (2020) 15(3):e0230667. doi: 10.1371/journal.pone.0230667
26. Li M, Zou Y, Jiang Q, Jiang L, Yu Q, Ding X, et al. A Preliminary Study of the Oral Microbiota in Chinese Patients With Sjögren’s Syndrome. Arch Oral Biol (2016) 70:143–8. doi: 10.1016/j.archoralbio.2016.06.016
27. Zhou S, Cai Y, Wang M, Yang WD, Duan N. Oral Microbial Flora of Patients With Sicca Syndrome. Mol Med Rep (2018) 18(6):4895–903. doi: 10.3892/mmr.2018.9520
28. Lee J, Alam J, Choi E, Ko YK, Lee A, Choi Y. Association of a Dysbiotic Oral Microbiota With the Development of Focal Lymphocytic Sialadenitis in Iκb-ζ-Deficient Mice. NPJ Biofilms Microbiomes (2020) 6(1):49. doi: 10.1038/s41522-020-00158-4
29. Tseng YC, Yang HY, Lin WT, Chang CB, Chien HC, Wang HP, et al. Salivary Dysbiosis in Sjögren’s Syndrome and a Commensal-Mediated Immunomodulatory Effect of Salivary Gland Epithelial Cells. NPJ Biofilms Microbiomes (2021) 7(1):21. doi: 10.1038/s41522-021-00192-w
30. Sato M, Arakaki R, Tawara H, Tsunematsu T, Ishimaru N. Formation of Autoimmune Lesions Is Independent of Antibiotic Treatment in NOD Mice. Int J Mol Sci (2021) 22(6):3239. doi: 10.3390/ijms22063239
31. Singh M, Teles F, Uzel NG, Papas A. Characterizing Microbiota From Sjögren’s Syndrome Patients. JDR Clin Trans Res (2021) 6(3):324–32. doi: 10.1177/2380084420940623
32. Leung KC, Leung WK, McMillan AS. Supra-Gingival Microbiota in Sjögren’s Syndrome. Clin Oral Investig (2007) 11(4):415–23. doi: 10.1007/s00784-007-0132-1
33. Yamazaki M, Kitamura R, Kusano S, Eda H, Sato S, Okawa-Takatsuji M, et al. Elevated Immunoglobulin G Antibodies to the Proline-Rich Amino-Terminal Region of Epstein-Barr Virus Nuclear Antigen-2 in Sera From Patients With Systemic Connective Tissue Diseases and From a Subgroup of Sjögren’s Syndrome Patients With Pulmonary Involvements. Clin Exp Immunol (2005) 139(3):558–68. doi: 10.1111/j.1365-2249.2004.02704.x
34. Vanderlugt CL, Miller SD. Epitope Spreading in Immune-Mediated Diseases: Implications for Immunotherapy. Nat Rev Immunol (2002) 2(2):85–95. doi: 10.1038/nri724
35. Stathopoulou EA, Routsias JG, Stea EA, Moutsopoulos HM, Tzioufas AG. Cross-Reaction Between Antibodies to the Major Epitope of Ro60 kD Autoantigen and a Homologous Peptide of Coxsackie Virus 2B Protein. Clin Exp Immunol (2005) 141(1):148–54. doi: 10.1111/j.1365-2249.2005.02812.x
36. Greiling TM, Dehner C, Chen X, Hughes K, Iñiguez AJ, Boccitto M, et al. Commensal Orthologs of the Human Autoantigen Ro60 as Triggers of Autoimmunity in Lupus. Sci Transl Med (2018) 10(434):eaan2306. doi: 10.1126/scitranslmed.aan2306
37. Yanagisawa N, Ueshiba H, Abe Y, Kato H, Higuchi T, Yagi J. Outer Membrane Protein of Gut Commensal Microorganism Induces Autoantibody Production and Extra-Intestinal Gland Inflammation in Mice. Int J Mol Sci (2018) 19(10):3241. doi: 10.3390/ijms19103241
38. Szymula A, Rosenthal J, Szczerba BM, Bagavant H, Fu SM, Deshmukh US. T Cell Epitope Mimicry Between Sjögren’s Syndrome Antigen A (SSA)/Ro60 and Oral, Gut, Skin and Vaginal Bacteria. Clin Immunol (2014) 152(1-2):1–9. doi: 10.1016/j.clim.2014.02.004
39. Zhang P, Minardi LM, Kuenstner JT, Zekan SM, Kruzelock R. Anti-Microbial Antibodies, Host Immunity, and Autoimmune Disease. Front Med (Lausanne) (2018) 5:153. doi: 10.3389/fmed.2018.00153
40. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites Produced by Commensal Bacteria Promote Peripheral Regulatory T-Cell Generation. Nature (2013) 504(7480):451–5. doi: 10.1038/nature12726
41. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal Microbe-Derived Butyrate Induces the Differentiation of Colonic Regulatory T Cells. Nature (2013) 504(7480):446–50. doi: 10.1038/nature12721
42. Bellocchi C, Fernández-Ochoa Á, Montanelli G, Vigone B, Santaniello A, Quirantes-Piné R, et al. Identification of a Shared Microbiomic and Metabolomic Profile in Systemic Autoimmune Diseases. J Clin Med (2019) 8(9):1291. doi: 10.3390/jcm8091291
43. Masui R, Sasaki M, Funaki Y, Ogasawara N, Mizuno M, Iida A, et al. G Protein-Coupled Receptor 43 Moderates Gut Inflammation Through Cytokine Regulation From Mononuclear Cells. Inflammation Bowel Dis (2013) 19(13):2848–56. doi: 10.1097/01.MIB.0000435444.14860.ea
44. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, Receptor for Niacin and the Commensal Metabolite Butyrate, Suppresses Colonic Inflammation and Carcinogenesis. Immunity (2014) 40(1):128–39. doi: 10.1016/j.immuni.2013.12.007
45. Fernando MR, Saxena A, Reyes JL, McKay DM. Butyrate Enhances Antibacterial Effects While Suppressing Other Features of Alternative Activation in IL-4-Induced Macrophages. Am J Physiol Gastrointest Liver Physiol (2016) 310(10):G822–31. doi: 10.1152/ajpgi.00440.2015
46. Kim DS, Woo JS, Min HK, Choi JW, Moon JH, Park MJ, et al. Short-Chain Fatty Acid Butyrate Induces IL-10-Producing B Cells by Regulating Circadian-Clock-Related Genes to Ameliorate Sjögren’s Syndrome. J Autoimmun (2021) 119:102611. doi: 10.1016/j.jaut.2021.102611
47. Verstappen GM, Pringle S, Bootsma H, Kroese FGM. Epithelial-Immune Cell Interplay in Primary Sjögren Syndrome Salivary Gland Pathogenesis. Nat Rev Rheumatol (2021) 17(6):333–48. doi: 10.1038/s41584-021-00605-2
48. Wright DP, Rosendale DI, Robertson AM. Prevotella Enzymes Involved in Mucin Oligosaccharide Degradation and Evidence for a Small Operon of Genes Expressed During Growth on Mucin. FEMS Microbiol Lett (2000) 190(1):73–9. doi: 10.1111/j.1574-6968.2000.tb09265.x
49. Terzulli M, Contreras-Ruiz L, Kugadas A, Masli S, Gadjeva M. TSP-1 Deficiency Alters Ocular Microbiota: Implications for Sjögren’s Syndrome Pathogenesis. J Ocul Pharmacol Ther (2015) 31(7):413–8. doi: 10.1089/jop.2015.0017
50. Zaheer M, Wang C, Bian F, Yu Z, Hernandez H, de Souza RG, et al. Protective Role of Commensal Bacteria in Sjögren Syndrome. J Autoimmun (2018) 93:45–56. doi: 10.1016/j.jaut.2018.06.004
51. Rudney JD, Chen R, Zhang G. Streptococci Dominate the Diverse Flora Within Buccal Cells. J Dent Res (2005) 84(12):1165–71. doi: 10.1177/154405910508401214
52. Rudney JD, Chen R, Sedgewick GJ. Intracellular Actinobacillus Actinomycetemcomitans and Porphyromonas Gingivalis in Buccal Epithelial Cells Collected From Human Subjects. Infect Immun (2001) 69(4):2700–7. doi: 10.1128/IAI.69.4.2700-2707.2001
Keywords: microbiome, autoimmunity, inflammation, Sjogren’s syndrome, pathogenesis
Citation: Deng C, Xiao Q and Fei Y (2022) A Glimpse Into the Microbiome of Sjögren’s Syndrome. Front. Immunol. 13:918619. doi: 10.3389/fimmu.2022.918619
Received: 12 April 2022; Accepted: 20 June 2022;
Published: 14 July 2022.
Edited by:
Aleksandar David Kostic, Harvard Medical School, United StatesCopyright © 2022 Deng, Xiao and Fei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunyun Fei, ZmVpeXVueXVuQHB1bWNoLmNu
†These authors have contributed equally to this work
 Chuiwen Deng
Chuiwen Deng Qiufeng Xiao
Qiufeng Xiao Yunyun Fei
Yunyun Fei