- State Key Laboratory of Veterinary Biotechnology, National Poultry Laboratory Animal Resource Center, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin, China
The conserved protective epitopes of hemagglutinin (HA) are essential to the design of a universal influenza vaccine and new targeted therapeutic agents. Over the last 15 years, numerous broadly neutralizing antibodies (bnAbs) targeting the HA of influenza A viruses have been isolated from B lymphocytes of human donors and mouse models, and their binding epitopes identified. This work has brought new perspectives for identifying conserved protective epitopes of HA. In this review, we succinctly analyzed and summarized the antigenic epitopes and functions of more than 70 kinds of bnAb. The highly conserved protective epitopes are concentrated on five regions of HA: the hydrophobic groove, the receptor-binding site, the occluded epitope region of the HA monomers interface, the fusion peptide region, and the vestigial esterase subdomain. Our analysis clarifies the distribution of the conserved protective epitope regions on HA and provides distinct targets for the design of novel vaccines and therapeutics to combat influenza A virus infection.
Introduction
Influenza A viruses are negative-sense RNA viruses, belongs to the family Orthomyxoviridae. Their genome consists of eight single-stranded negative-sense RNA fragments, that encode 10 essential proteins. Currently, 18 different hemagglutinin (HA) (H1–H18) and 11 different neuraminidase (NA) (N1–N11) subtypes have been identified or detected (1). Influenza A viruses evolve rapidly and can cause pandemics and epidemics of acute respiratory disease in domestic poultry, lower mammals, and humans, continuously challenging the poultry industry and human health (2). According to the World Health Organization (WHO), annual influenza epidemics result in an estimated about 3 to 5 million cases of severe illness and 290,000 to 650,000 respiratory deaths worldwide (3). Influenza A viruses have also caused pandemics, including the 1918 H1N1, 1957 H2N2, 1968 H3N2 and 2009 H1N1 pandemics, which caused millions of human deaths (4–7). Occasionally, zoonotic influenza A subtypes, such as H5Nx and H7N9, also infect humans through cross-species transmission, with a mortality rate of up to 52% (8).
Vaccination remains the best strategy for preventing influenza infections. Currently, vaccines are available against seasonal influenza viruses, the vaccines contain either three (trivalent) or four (tetravalent) influenza virus components, and are formulated every year based on worldwide influenza surveillance (9). However, the effectiveness of seasonal influenza vaccines is often quite low, only 10%–60% for the influenza seasons from 2004 to 2020 (10). Two types of influenza antiviral drugs that target the viral membrane protein (M2) ion channel and inhibitors of NA also have been approved for prophylaxis and therapy. However, the use of these antivirals is still limited (11). Therefore, a universal influenza vaccine that can elicit more broadly cross-reactive and long-term protection, and novel therapeutic agents would be highly desirable. Since HA is the most important and abundant surface glycoprotein of influenza viruses and the target of almost all neutralizing antibodies (12), the HA protein is a major target for the development of universal influenza vaccine and therapeutic agents.
Over the last 15 years, numerous broadly neutralizing antibodies (bnAbs) that cross-react and neutralize a wide range of subtype HAs of influenza viruses have been isolated from B lymphocytes of human donors and mouse models, and the epitopes recognized by these antibodies have mapped through the use of escape mutants and Cryo-electron microscopy. These works have figured out the conserved protective epitope region of HA, and provide hope for development of universal influenza vaccines and new targeted therapeutic agents. Multiple efforts have therefore been made to develop broad-spectrum, universal vaccines, such as sequential vaccination with chimeric HA (13, 14), and HA stem-based immunogens (15–17). At the same time, several bnAbs themselves have been used as passive immunotherapy (18). In addition, guided by structural knowledge of the interactions and mechanism of bnAb, series of therapeutic agents such as small proteins, peptides and molecules have been designed to mimic the function of bnAb (19–23). Here, we analyzed and summarized the antigenic epitopes and functions of more than 70 kinds of bnAbs reported since 1980s. Our analysis clarifies the distribution of the conserved protective epitope regions on HA and provides new insights for the design of novel vaccines and therapeutics against influenza A virus infections.
Overview of the HA protein
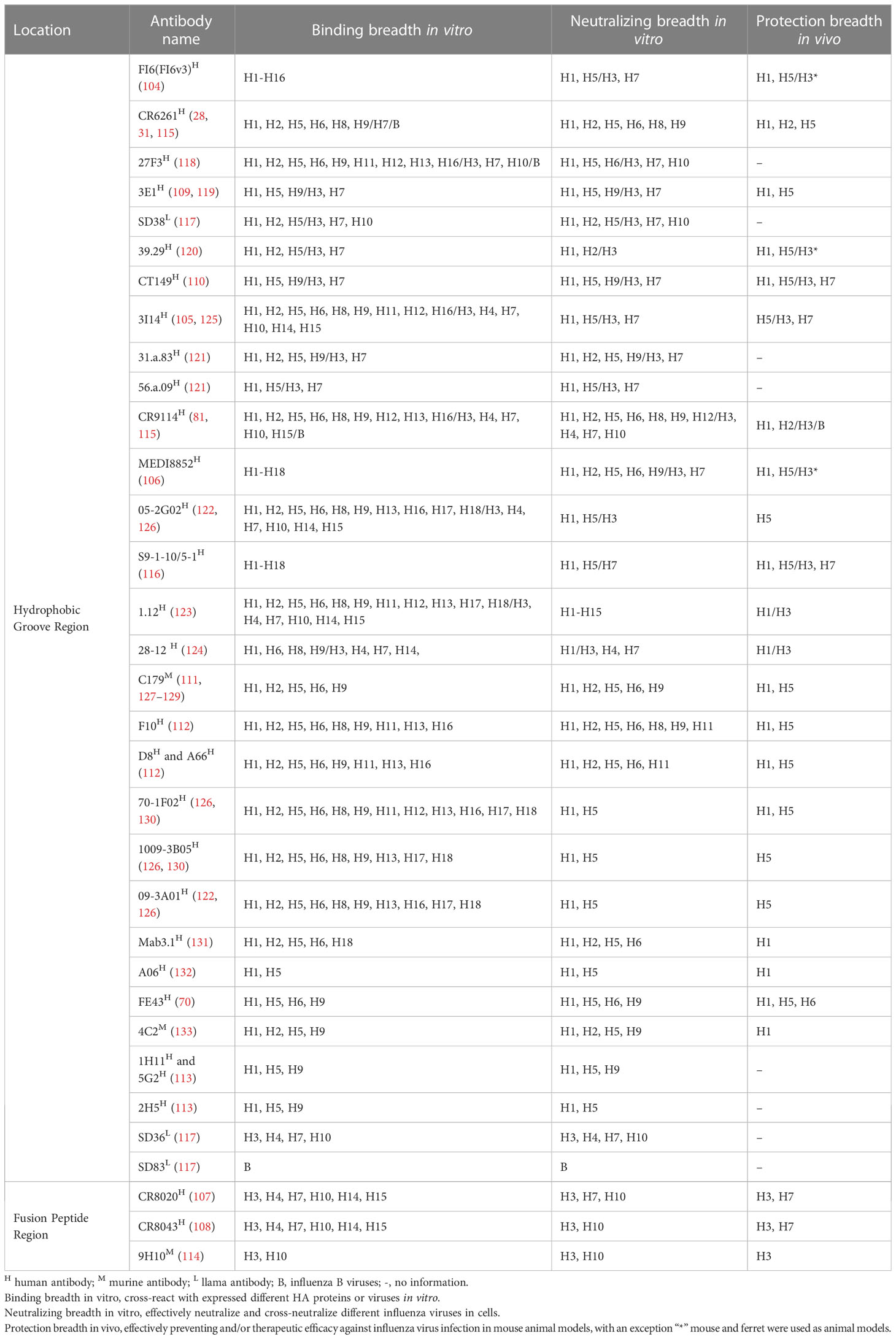
The structure of HA was identified in 1981 (24). Although the amino acid sequence homology of different subtype HAs can be as low as about 40%, HA always adopts the same protein folding and its architecture is highly conserved (25). However, the surface properties and glycosylation patterns of HA vary extensively between influenza subtypes. Influenza A viruses are divided into two phylogenic groups based on their HA, group 1 (H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, H17 and H18 subtypes) and group 2 (H3, H4, H7, H10, H14, and H15 subtypes) (26, 27). Group 1 HAs has similar stem structures, whereas group 2 HAs display intra-group similarity in the stem (28). Mature HA is a trimer composed of three identical monomeric subunits (24) (Figure 1A). Every monomeric HA is synthesized as an immature single polypeptide chain (HA0) in the endoplasmic reticulum, and is cleaved at its cleavage site by host cell proteases to yield two subunits, HA1 and HA2, which are linked via a single disulfide bond (32). Each HA monomer subunit is divided into a head domain and a stem domain. The membrane-distal globular head domain composed of HA1, and contains the receptor-binding (RB) subdomain and the vestigial esterase (VE) subdomain (Figures 1A, B, C, H). The membrane-proximal stem domain is primarily composed of HA2 with some HA1 residues, and contains the F’ subdomains, the F subdomain, and the fusion peptide subdomain (33) (Figures 1A, D–H). The head domain mediates attachment of the virus to host cell surface receptors, and the stem domain mediates liberation of the viral genome into the cytoplasm through membrane fusion.
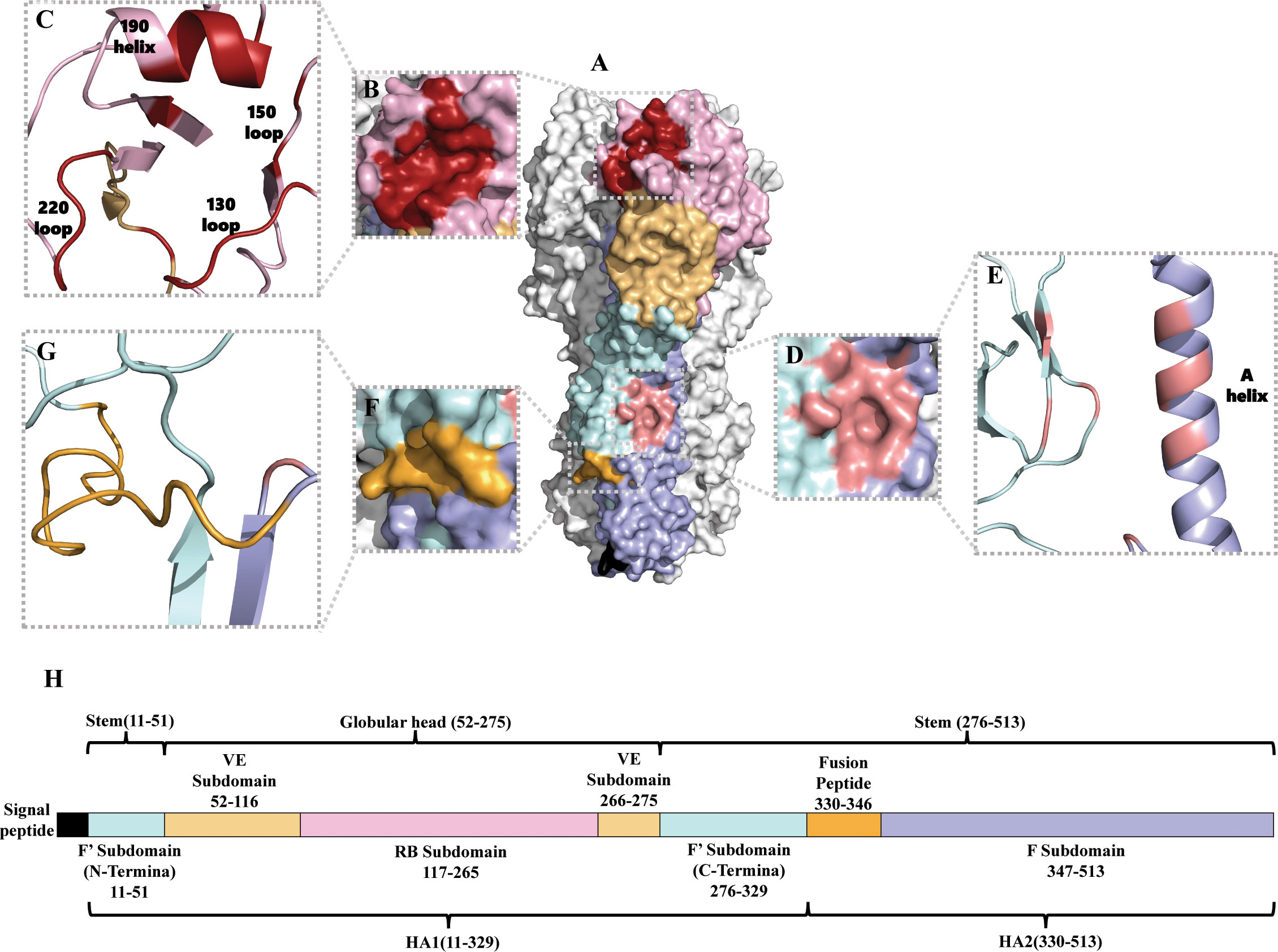
Figure 1 Structure of the HA of influenza A virus. (A) Protein molecular model of H3 HA (A/Aichi/2/68). The RB subdomain is shown in pink, the RBS in dark red, the VE subdomain in yellow, the F’ subdomain in bright blue, the F subdomain in light purple, the fusion peptide in orange-yellow, and the hydrophobic groove region in orange-red (29). (B, C) are the RBS surface and cartoon representation, respectively (30). (D, E) are the hydrophobic groove region surface and cartoon representation, respectively (31). (F, G) are the fusion peptide surface and cartoon representation, respectively (29). (H) Amino acid sequence of HA (H3 numbering) (29).
The receptor-binding site (RBS) is a shallow hydrophobic pocket at the tip of the RB subdomain, and comprises four secondary structural elements: the 130-loop, the 150-loop, the 190-helix, and the 220-loop (Figures 1A–C). Several key residues in the base of the pocket that interact with sialic acid (SA) are conserved, including W153, H183, L194, and Y195 (34). HA binds to the host cell via RBS recognizing receptors–glycolipids or glycoproteins containing terminal SA moieties with van der Waals interactions and initiates membrane fusion (35–37). Most regions of the RB subdomain besides the RBS are highly variable and prone to antigenic drift. The VE subdomain is located at the junction between the RB subdomain and the stem domain, but their functions are not well-known. The sequence of the VE subdomain is highly conserved within subtypes, and variable between different subtypes (29, 38).
The stem domain anchors HA in the viral envelope. After HA binding to the host cell receptor, endocytosis transports the influenza viral particle to the endosome, where the low pH triggers conformational changes in HA2 that mediate fusion of the viral and endosomal membranes and release of the viral genome into the cytoplasm, establishing the onset of the replication cycle (39). During this process, the HA structure is dynamic and undergoes spontaneous and reversible transitions between multiple conformations (40). The stem domain of HA is also functionally conserved, with much less sequence variation across strains and subtypes.
Conserved protective epitopes on the head domain of HA
Despite the higher variation in the HA head domain, a series of bnAbs capable of binding and neutralizing multiple subtypes or subtype-specific influenza A virus were isolated. Their protective epitopes on the HA head domain are mainly concentrated in three regions: the RBS pocket and its surrounding area, the VE subdomain, and the occluded epitope region, which is hidden in the HA monomer interface of HA1. Anti-head bnAbs generally inhibit virus attachment to the host cell receptor, thereby blocking viral entry (41–50). However, some bnAbs targeting the VE subdomain and the occluded epitope can inhibit membrane fusion (51–53) or mediate Fc-Fcγ receptor (Fc-FcγR) interactions, antibody dependent cell-mediated cytotoxicity (ADCC) or complement dependent cytotoxicity (CDC) (54–58).
Epitopes of the RBS pocket and surrounding area
Residues at the rim of the RBS pocket are highly conserved, and the reactive breadth of bnAbs targeting the RBS is limited by the region of epitopes beyond the RBS pocket. Most of these antibodies show hemagglutination-inhibition (HAI) activity (41, 42, 50, 59–67), and inhibit viral entry by preventing HA binding to host receptors (41–50). They may also prevent HA conformational changes during membrane fusion by cross-linking neighboring subunits of the HA trimer, as has been reported for bnAbs HC63 (63, 68). To date, there have been no reports of anti-RBS bnAbs that can mediate Fc-FcγR responses.
Of the bnAbs that bind to the RBS pocket, the following seven possess cross-group or cross-subtype binding or neutralizing activities: S139/1 (41), C05 (42), F045-092 (60), K03.12 (69), 2G1 (59), FE17 (70) and 12H5 (71) (Table 1). S139/1, C05, and F045-092 can neutralize both group 1 and group 2 viruses in vitro. S139/1 provides heterosubtypic protection against H1N1 and H3N2 influenza virus passive immunization of mice (41, 72). C05 completely protects mice from a lethal challenge with H1N1 or H3N2 virus (42). F045-092 possesses broadly binding activity against H3 viruses that across five decades (1963–2011), also neutralized H1N1, H2N2, and H5N1 viruses (43, 60). The antibodies K03.12 and 2G1 also show broad binding activity against both group 1 and group 2 viruses (69). 2G1 was isolated from 1957 H2N2 pandemic healthy donors, inhibits the 2006 swine H2N3 influenza virus, and protects mice from a lethal challenge with H2N2 viruses (59, 73). The epitope footprints of these bnAbs are mainly concentrated within the RBS pocket, having little to no contact with the surrounding variable positions (Figures 2A, B). These bnAbs commonly insert a single heavy-chain complementarity-determining region (CDR) loop with hydrophobic residues into the RBS pocket.
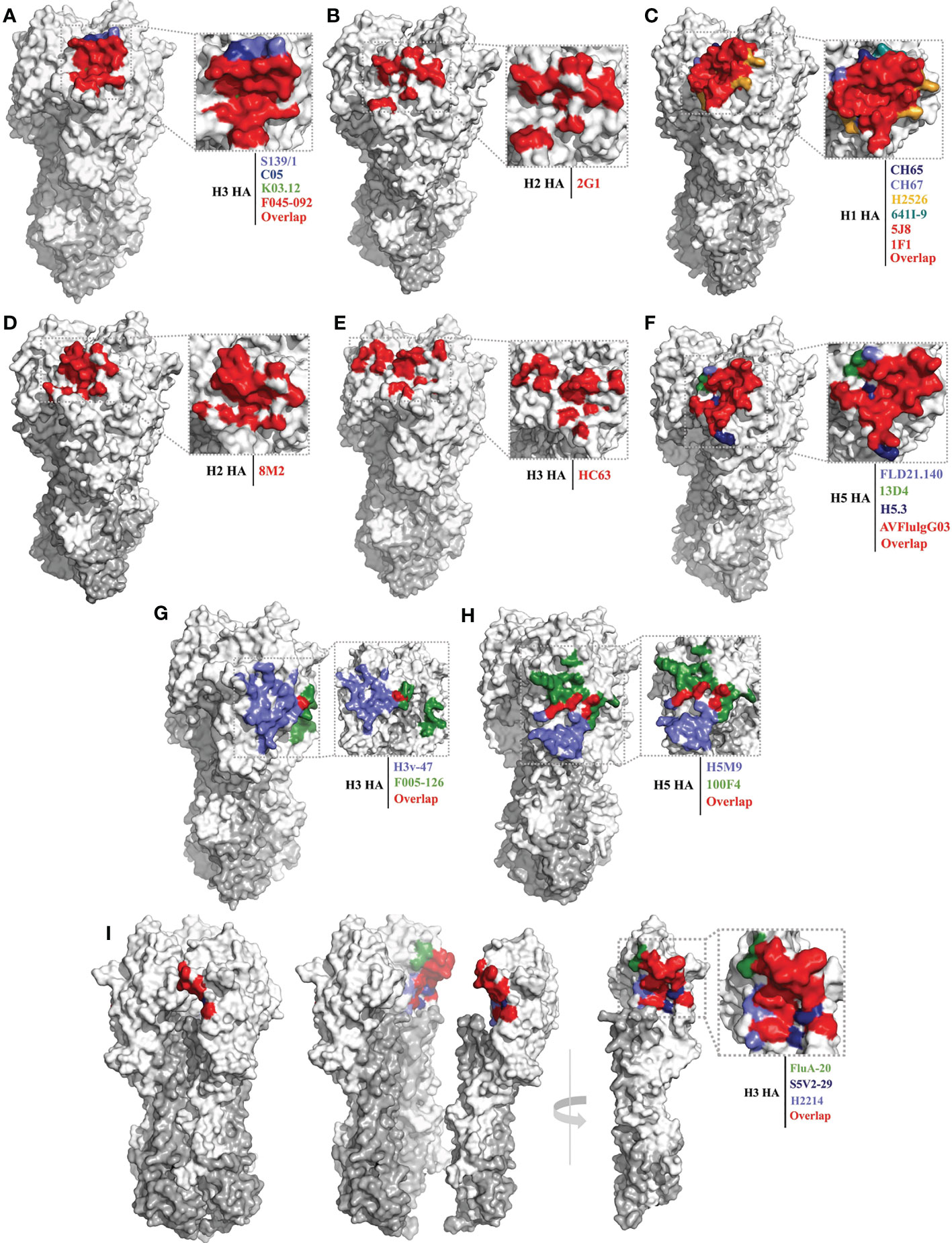
Figure 2 Conserved epitopes of the HA head domain. HA1 is shown in white, HA2 in gray. (A, B) are epitope footprints of cross-subtype bnAbs binding to the RBS. (A) H3 HA (PDB ID: 4FNK) as a model, the epitope of F045-092 (PDB ID: 4O58) is all overlapped shown in red, the non-overlapping residues are shown in light purple (S139/1, PDB ID:4GMS), dark blue (C05, PDB ID:4FQR), and green (K03.12, PDB ID: 5W08), respectively. (B) H2 HA (PDB ID: 4HLZ) as a model, the epitope of 2G1 (PDB ID: 4HG4). (C–F) are epitope footprints of subtype specific bnAbs binding to the RBS. (C) H1 HA (PDB ID: 4M4Y) as a model, the epitopes of 5J8 (PDB ID:4M5Z) and 1F1 (PDB ID: 4GXU) are overlapped in red. The non-overlapping residues are shown in dark blue (CH65, PDB ID: 5UGY), light purple (CH67, PDB ID: 4HKX), yellow (H2526, PDB ID: 4YJZ), blue-green (641I-9, PDB ID: 4YK4), respectively. (D) H2 HA (PDB ID: 4HLZ) as a model, the epitope of 8M2 (PDB ID: 4HFU). (E) H3 HA (PDB ID: 4FNK) as a model, the epitope of HC63 (PDB ID: 1KEN). (F) H5 HA (PDB ID: 4MHH) as a model, the epitope of AVFlulgG03 (PDB ID: 5DUP) is overlapped in red. The non-overlapping residues are shown in light purple (FLD21.140, PDB ID: 6A67), green (13D4, PDB ID: 6A0Z), and dark blue (H5.3, PDB ID: 4XNM), respectively. (G, H) are epitope footprints of bnAbs binding to the VE subdomain. (G) H3 HA (PDB ID: 4FNK) as a model, non-overlapping residues are shown in light purple (H3v-47, PDB ID: 5W42) and green (F005-126, PDB ID: 3WHE). (H) H5 HA (PDB ID: 4MHH) as a model, non-overlapping residues are shown in light purple (H5M9, PDB ID: 4MHH) and green (100F4, PDB ID:5DUR). (I) Epitope footprint of bnAbs binding to the occluded epitope region of the HA monomers interface. H3 HA (PDB ID: 2VIU) as a model, non-overlapping residues are shown in green (FluA20, PDB ID: 6OCB), dark blue (S5V2-29, PDB ID:6E4X), and light purple (H2214, PDB ID: 6E56), respectively.
Compared with the above-mentioned bnAbs, more bnAbs with subtype-specific reactivity have been reported and identified, including: the H1 subtype-specific antibodies 1F1 (62), 5J8 (61), CH65 (74), CH67 (45), H2526 (46), 641 I-9 (46) and 3D11 (75); the H2 subtype-specific antibodies 8M2 and 8F8 (59); the H3 subtype-specific antibodies HC63 (63) and A2.91.3 (64); and the H5 subtype-specific antibodies AVFlulgG03 (65), FLD21.140 (78), 13D4 (66), HAb21 (50), and H5.3 (79) (Table 1). The epitope footprints recognized by these bnAbs are also concentrated in the RBS pocket, but have more contact with the surrounding variable residues beyond the outer edges of the RBS pocket compared with the cross-subtype bnAbs (Figures 2C–F). This is because the surface area of the antigenic epitopes recognized by these antibodies (1200–1500 Å) is generally larger than that of the RBS pocket (800 Å) (93). Most of these subtype-specific bnAbs insert a single CDR loop into the RBS pocket, with more CDRs contacting the variable residues outside the RBS pocket; hence, these bnAbs have limited reactivity breadth (42).
The 1F1 antibody was isolated from a 1918 influenza pandemic survivor, aged 91–101 years (2–12 years in 1918), inhibits and neutralizes human H1 viruses (1918, 1930, 1943 and 1977 isolate strains), and protects mice from lethal challenge with 1918 H1 virus (47, 62). Antibody 5J8 has broadly neutralizing activity against 20th century H1N1 viruses and the 2009 pandemic H1N1 (44, 61). Antibodies CH65 and CH67 have naturalized seasonal H1 strains across three decades (1977–2007/2009) (45, 74). The H1 subtype-specific antibodies 1F1, 5J8, CH65, CH67, H2526, and 641I-9 approach the HA head from different directions and have somewhat different peripheral contact footprints outside the RBS pocket (Figure 2C).
Like 2G1, antibodies 8M2 and 8F8 were isolated from 1957 H2N2 pandemic healthy donors; however, 8M2 and 8F8 only react with human H2N2 viruses and a swine H2N3 strain (59, 73) (Figure 2D). Antibody HC63 was the earliest reported bnAb targeting the HA head in 1987; it reacts with most of the H3N2 viruses isolated between 1968 and 1982 (63). The epitope of HC63 is similar to that of 1F1 (47), but HC63 simultaneously binds two HA monomers (Figure 2E), effectively cross-linking them and blocking the pH-induced conformational changes in HA that drive membrane fusion (68).
Antibodies AVFluIgG03, FLD21.140, 13D4, HAb21, and H5.3 have widely cross-neutralizing activity with different clade of H5N1 viruses, and 13D4 protects mice against lethal challenge with H5N1 viruses of clades 1, 2.1, 2.2, and 2.3, even at the stage of infection when H5N1 virus has disseminated beyond the pulmonary system (66) (Table 1 and Figure 2F). Antibody CR8033, which was isolated from volunteers vaccinated with the seasonal influenza vaccine, protects mice against lethal challenge with either of the Yamagata and Victoria lineages of influenza B viruses; its epitope also targets the RBS pocket (81).
Till now, a series of viral attachment inhibitors have been developed targeting RBS, such as PAA-YDS (94, 95), 6SL-PAMAM (96), S2(1–5) (97), A22 (98), D-26 (99, 100). Some of these inhibitors have been shown to protect mice from influenza virus infection, such as PAA-YDS and A22 (94, 95, 98).
Epitopes of the VE subdomain
BnAbs binding the VE subdomain only possess subtype-specific neutralizing activity. They include: the H1 subtype-specific antibody PR8-23 (82); the H3 subtype-specific antibodies H3v-47 (57), F005-126 (51), and A2.4.1 (64); the H5 subtype-specific antibodies H5M9 (83), 9F4 (52), HA-7 (53), 100F4 (101), and 4F5 (89); and the H7 subtype-specific antibodies 1H5 and 1H10 (58) (Table 1). Their epitope footprints are mainly located on the VE subdomain, but some extend into the RB subdomain (Figures 2G, H). These bnAbs can play a role in inhibiting virus binding to the host receptor (89) or membrane fusion (51–53), and may possess dual functions, as is the case with antibody H5M9 (83, 84). Some of them can block viral egress from infected cells (57), and mediate Fc-FcγR responses (ADCC) (57, 58).
Antibody H3v-47 exhibits potent neutralizing activity against multiple human and swine H3N2 viruses that circulated from 1989 to 2014. The H3v-47 epitope spans the VE and RB subdomains (57). Antibody F005-126 can broadly neutralize H3N2 viruses, binds to the VE subdomain (site R) and RB subdomain (site L), spans a cleft formed by two HA monomers in the HA trimer, and cross-links them (51) (Figure 2G).
Antibody H5M9 can neutralize different clades of H5N1 viruses (Clades 0, 1, 2.3.4, and 7), and protects mice from lethal H5N1 viral challenge both prophylactically and therapeutically in vivo (83, 84) (Figure 2H). Antibody 9F4 can neutralize H5N1 viruses of clade 1, 2.1, 2.2, 2.2.2, 2.3.2.1a, and 2.3.4 in vitro, and neutralizes H5N1 viruses of clade 2.2.2 in vivo. Antibodies HA-7 and 100F4 can potently inhibit or neutralize multiple clades of H5N1 viruses, and completely protect mice from lethal challenges of H5N1 (53). Antibody 100F4 also protects mice from lethal challenges of H5N6 and H5N8 viruses (87, 88). HA-7 specifically targets the VE subdomain and does not inhibit virus binding to the host cell receptor but does affect membrane fusion (53).
Antibodies 1H5 and 1H10 can bind to a wide range of H7 strains, but lack HAI and neutralizing activity in vitro. Both antibodies can engage Fc-FcγR responses, and provide protection in vivo upon passive transfer in the mouse model (58). Antibody CR8071 also target the VE subdomain of influenza B viruses, can protect mice against lethal challenge from the Yamagata and Victoria lineages, but is less potent in vivo than CR8033, and lacks HAI activity (81).
Epitopes of the occluded epitope region of the HA monomer interface
During the adsorption and endocytosis of influenza virus, HA can undergo spontaneous and reversible transitions between multiple conformations. Acidification and receptor binding can shift the dynamic equilibrium of HA conformation (40). The interface, occluded at the contact surface between the HA monomers of the head domain, can then be exposed to antibodies. Antibodies bind to these temporarily exposed epitopes, disrupt the HA trimeric structure and affect influenza virus replication (54). Early to 1993, Yewdell and colleagues demonstrated that the epitope of murine antibody Y8-10C2 is located at the interface of adjacent subunits of the HA head of H1 (102), but the reactive range was not identified. More recently, several bnAbs targeting this region were identified, including antibodies FluA-20 (54), S5V2-29 (55), H2214 (55), 8H10 (56), FL-1066 (56), H7-200 (90) and H7.5 (91) (Table 1). Most of these antibodies possess cross-group HA binding activity, and generally lack neutralizing activity in vitro, but confer robust protection in vivo against multi-subtype lethal virus challenge by mediating Fc-FcγR responses (ADCC or CDC) (54–56). The epitope footprints recognized by FluA-20, S5V2-29, H2214, 8H10, and FL-1066 are located in a similar region in the contact surface between the HA monomers of the head domain (Figure 2I).
Antibody FluA-20 shows extraordinary reactive breadth and affinity for recombinant HA trimers from subtypes H1 through H15, except for H13, and protects mice from lethal challenge with group 1 and group 2 viruses when as prophylaxis or therapy. FluA-20 rapidly disrupts HA trimers, inhibits the cell-to-cell spread of virus, and mediates ADCC activity in vivo (54). Antibodies S5V2-29 and H2214 can also bind multiple HAs of group 1 and group 2 viruses, and provide protection against lethal challenges with H1N1 and H3N2 viruses in mice (55). Antibodies 8H10 and FL-1066 possess broad reactivity with historical H3 HAs across over 30 years, and can bind a representative H4 (56). The epitope of antibody H7.5 also includes residues in the inter-HA head contact region, which allows H7.5 to simultaneously bind two separate surfaces of two adjacent HA protomers, thereby blocking HA binding to SA (91, 92).
Conserved protective epitopes on the HA stem domain
In recent decades, tremendous effort has been invested in isolating and structurally characterizing bnAbs that target the HA stem domain. The conserved protective epitopes that these bnAbs recognize are mainly located in two regions: the hydrophobic groove and the fusion peptide. These bnAbs are generally encoded by a relatively restricted set of variable-heavy (VH) gene segments (103). Due to the high conservation of the stem domain, anti-stem bnAbs usually exhibit more widely reactive and neutralizing breadth to influenza A virus strains than anti-head bnAbs. All of the anti-stem bnAbs lack HAI activity. The mechanism of protection involves inhibiting the host cell protease cleavage of HA0 (104–108), or preventing membrane fusion via antibody binding to cleaved HA, inhibiting its low pH-induced conformational change (28, 81, 104–114). Anti-stem bnAbs mediating Fc-FcγR responses also play a critical role in vivo in the protection (104–106, 110, 115). In addition, antibodies S9-1-10/5-1 and 9H10 inhibit virus particle release from infected cells (114, 116).
Epitopes of the hydrophobic groove region
Among the bnAbs that bind to the hydrophobic groove region, some exhibit extremely broad binding properties to all subtype HAs from H1 to H18 (cross-group). Many antibodies exhibit group 1 virus-specific reactivity, whereas only one antibody (SD36) exhibits group 2 virus-specific reactivity (117).
In 2008, antibody CR6261 was the first bnAb reported to exhibit group 1 and group 2 reactivity (31). Since then, a series of this type of bnAb was isolated and identified, such as antibodies FI6(FI6v3) (104), 27F3 (118), 3E1 (119), SD38 (117), 39.29 (120), CT149 (110), 3I14 (105), 31.a.83 (121), 56.a.09 (121), CR9114 (81), MEDI8852 (106), 05-2G02 (122), S9-1-10/5-1 (116), 1.12 (116, 123) and 28-12 (124) (Table 2). Most of these antibodies exhibit broad neutralizing activity and protection against influenza A virus in vivo.
The epitope footprints of antibodies CR6261, 27F3, 3E1, SD38, and Mab 3.1 are shown in the H1 HA model of Figure 3A. All of these antibodies bind to the hydrophobic groove region with CDRs of heavy and light chains. Antibody 3E1 also bind to the fusion peptide region (109, 119). CR6261 neutralizes viruses by blocking conformational rearrangements of HA associated with membrane fusion (28, 31, 115). The epitope footprints of antibodies FI6(FI6v3), 39.29, CT149, and 3I14 are shown in the H3 HA model of Figure 3C. FI6(FI6v3) inhibits the conformational changes of HA, prevents membrane fusion, and inhibits HA0 processing (104). CT149 binds residues of two adjacent protomers of HA, and CT149 and 3I14 neutralize viruses by inhibiting low pH-induced membrane fusion (105, 110). The epitope footprints of antibodies CR9114 and MEDI8852 are shown in the H5 HA model of Figure 3D. CR9114 exhibits extremely broad reactivity, including against 14 influenza A subtypes and both influenza B virus lineages, and it can neutralize 11 of these subtypes of virus. CR9114 also protected mice from lethal challenge with H1N1, H2N2, H2N3, H3N2, and influenza B viruses in prophylaxis studies (81, 115). MEDI8852 binds to the hydrophobic groove and a large portion of the fusion peptide through a coordinated movement of CDRs (106).
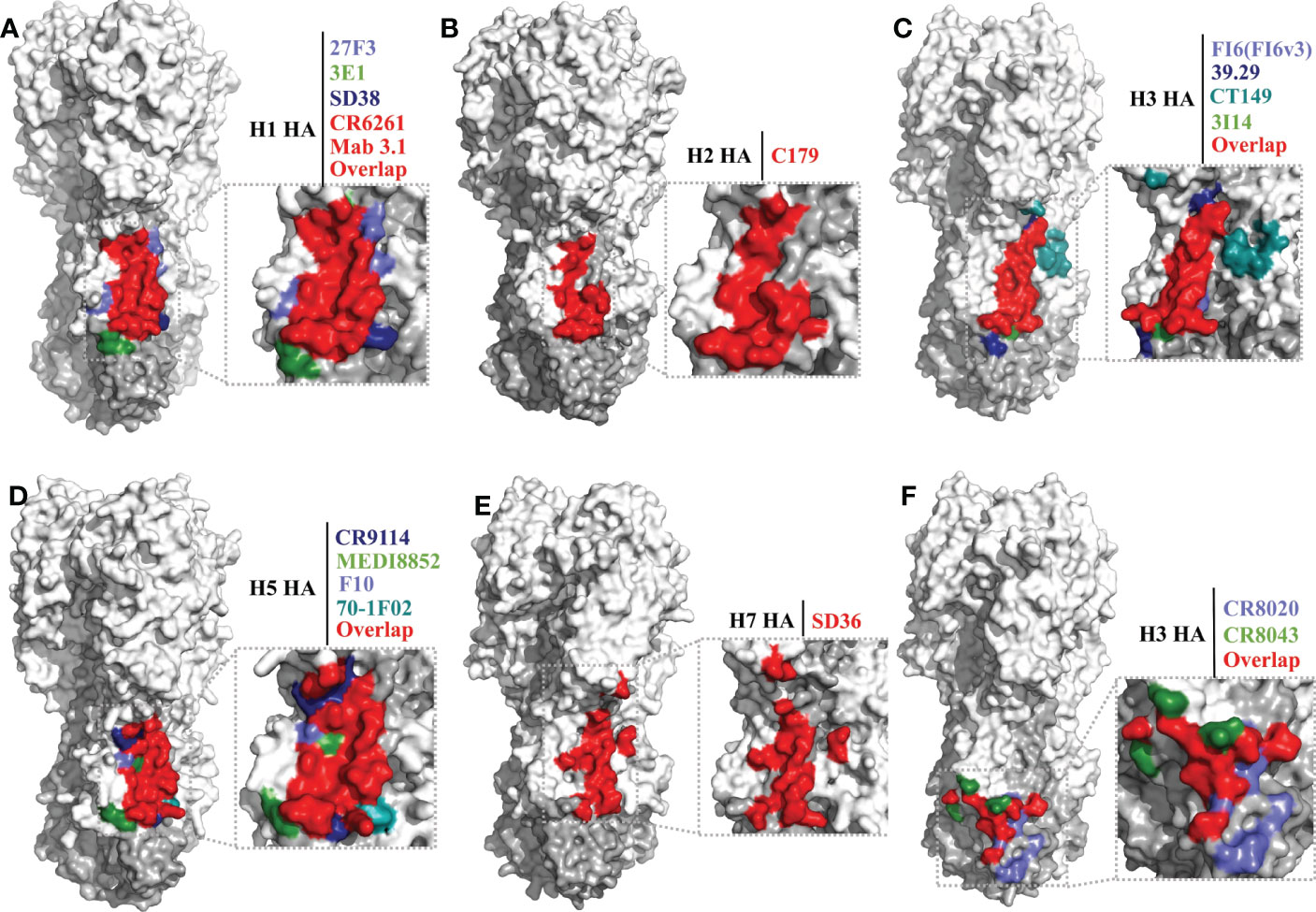
Figure 3 Conserved epitopes of the HA stem domain. HA1 is shown in white, HA2 in gray. (A–E) are epitope footprints of bnAbs binding the hydrophobic groove region. (A) H1 HA (PDB ID: 4M4Y) as a model, the epitopes of CR6261 (PDB ID:3GBN) and Mab 3.1 (PDB ID: 4PY8) overlap in red. The non-overlapping residues are shown in light purple (27F3, PDB ID: 5WKO), green (3E1 PDB ID: 5GJT) and dark blue (SD38, PDB ID: 6FYT), respectively. (B) H2 HA (PDB ID: 4HLZ) as a model, the epitope of C179 (PDB ID: 4HLZ). (C) H3 HA (PDB ID: 4FNK) as a model, the non-overlapping residues are shown in light purple (FI6 (FI6v3), PDB ID: 3ZTJ), dark blue (39.29, PDB ID: 4KVN), blue-green (CT149, PDB ID:4UBD), and green (3I14, PDB ID: 6WF0), respectively. (D) H5 HA (PDB ID: 4MHH) as a model, the non-overlapping residues are shown in dark blue (CR9114, PDB ID: 4FQI), green (MEDI8852, PDB ID: 5JW4), light purple (F10, PDB ID: 3FKU), and blue-green (70-1F02, PDB ID: 6B3M), respectively. (E) H7 HA (A/tree sparrow/Shanghai/01/2013) as a model, the epitope of SD36 (PDB ID: 6FYU). (F) Epitope footprint of bnAbs binding to the fusion peptide region. H3 HA (PDB ID: 4FNK) as a model, the non-overlapping residues are shown in light purple (CR8020, PDB ID: 3SDY) and green (CR8043, PDB ID: 4NM8).
Antibody C179 was the first reported bnAb to neutralize influenza A viruses, and was isolated from a mouse in 1993 (111). In the last decade, many bnAbs with group 1 virus-specific reactivity targeting the hydrophobic groove region have been isolated, including antibodies F10 (112), D8 (112), A66 (112), 70-1F02 (130), 1009-3B05 (130), 09-3A01 (122), Mab3.1 (131), A06 (132), FE43 (70), 4C2 (133), 1H11 (113), 5G2 (113), and 2H5 (113) (Table 2). C179 cross-neutralizes H1, H2, and H5 subtype viruses, and protects mice from lethal challenge with H5N1 and 2009 H1N1 pandemic viruses (111, 127, 128). The epitope of C179 bound to the 1957 H2N2 HA is similar to that of bnAbs CR6261, F10, CR9114, and FI6(FI6v3) (Figures 3A–D) (127). F10 bound to H5 HA with heavy-chain CDRs inserts into the hydrophobic groove pocket and locks the fusion peptide (112) (Figure 3D).
As mentioned previously, of the bnAbs that bind to the hydrophobic groove region, only one exhibits group 2 HA reactivity, the llama single-domain antibody (sdAb) SD36 (Table 2). SD36 recognizes conserved epitopes that partially overlap with those of bnAbs CR9114, CR6261, and FI6(FI6v3) (117) (Figures 3A, C–E). Why are bnAbs with group 2 virus-specific reactivity that target the hydrophobic groove region so rare? It may be that a conserved Asn38 glycan in the HA1 of group 2 viruses may interfere with the accessibility of the conserved antigenic site on helix A (28, 112, 134). However, some antibodies, such as 27F3, can navigate around this N38 glycan to achieve cross-group neutralization (118, 135).
Llama sdAb SD83 can neutralize both influenza B virus lineages (Table 2), and its epitope is also in the hydrophobic groove region. This epitope is highly conserved, with the residues being >99% identical in influenza B viruses (117).
Till now, a series of small protein or peptide viral fusion inhibitors targeting conserved epitopes of HA stem have been developed, such as HB36 and HB80 (19), HB36.3 and HB80.4 (23), HB36.6 (21), JNJ4796 (22), P7 (20). Some of these inhibitors have been shown to protect mice from influenza virus infection, such as HB36.6 and JNJ4796 (21, 22), and four antibody drugs (CR6261, 39.29, CR8020 and MEDI8852) have entered phase II clinical trial (136–138). In addition, several universal vaccines based on the HA stem, such as headless HA and HA mini-stems, have also shown promising prospects, and chimeric HA has entered phase I clinical trial (136, 137).
Epitopes of the fusion peptide region
The bnAbs that bind to the fusion peptide region of the HA stem are only reactive with group 2 HA; these bnAbs include CR8020 (107), CR8043 (108), and 9H10 (114) (Table 2). The epitopes recognized by these antibodies are located lower down on the stem domain, close to the virus membrane, but accessible on virions (Figure 3F). Electron microscopy reconstructions shows that the three antibodies bind to a similar epitope footprint, but their sensitivities to mutations are distinct, due to their slightly different approach angles to the HA (114). These antibodies inhibit viral replication by blocking membrane fusion and mediating Fc-FcγR responses. In addition, CR8020 inhibits the host cell protease cleavage of HA0 (107) and 9H10 disrupts viral particle egress in the late stage of infection (114).
In summary, the highly conserved protective epitopes of HA in influenza A viruses are concentrated in five regions: the RBS pocket, the VE subdomain, the occluded epitope region between the HA heads, the hydrophobic groove region, and the fusion peptide region. The breadth of bnAbs targeting these five conserved protective epitope regions is summarized in Figure 4. The hydrophobic groove region is the most conserved protective epitope region of HA. Most bnAbs targeting these regions exhibit extremely broad reactivity to both group 1 and/or group 2 HAs of influenza A virus, and even to influenza B virus. The epitopes of the RBS pocket and the occluded epitope region are also conserved; some bnAbs targeting these two regions are broadly reactive, but bnAbs targeting the RBS pocket having higher potency than bnAbs targeting the occluded epitope region. To date, bnAbs targeting the fusion peptide region only neutralize group 2 HAs, and bnAbs targeting the VE subdomain are subtype-specific.
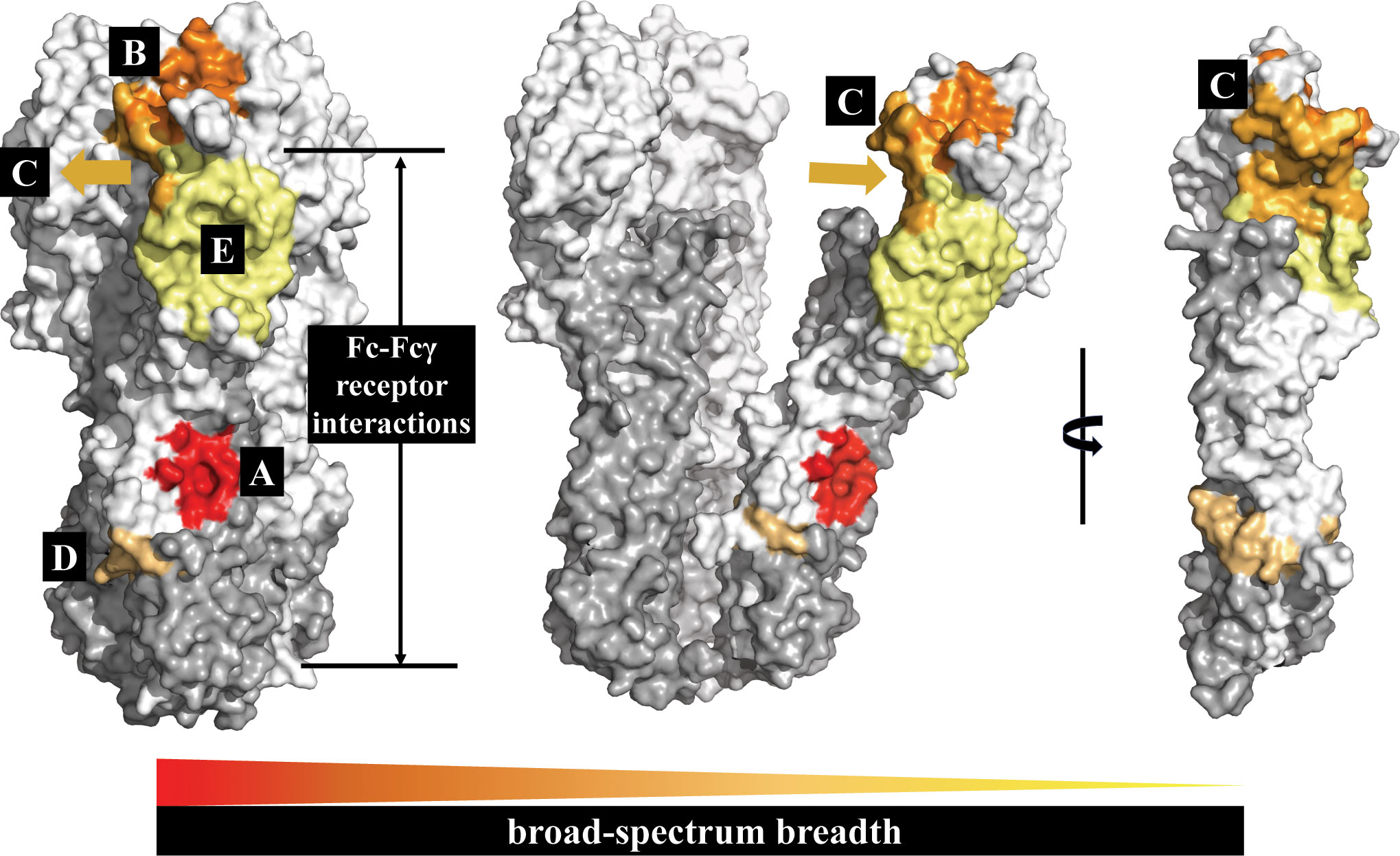
Figure 4 The HA conserved epitope regions of influenza A virus. HA1 is shown in white, HA2 in gray. The five conserved protective epitope regions summarized in this review: (A) The hydrophobic groove region is shown in red; (B) the RBS in orange; (C) the occluded epitope region of the HA monomers interface in light orange; (D) the fusion peptide region in orange-yellow; and (E) the VE subdomain in yellow. The broad-spectrum breadth of the five conserved protective epitope regions are ranked A>B>C>D>E.
Conclusions
The constant antigenic drift and antigenic shift of influenza viruses and the outbreak of the SARS-CoV-2 pandemic since 2020 have further emphasized the urgent need for a universal influenza vaccine and therapeutic agents. The isolation of new bnAbs and identification of highly conserved protective epitopes of HA have identified more distinct targets for the development of novel vaccines and therapeutic based on HA. Because the epitopes of the HA head are more accessible, humoral responses to the head region of HA are more robust than those to the HA stem. In one study, about 14% of HA-specific memory B cells from healthy human donors, 76% B cell receptor were specific for epitopes present on the HA head (69). Generally, antibodies targeting the head of HA shown more potency by directly or indirectly blocking viral adsorption (61, 74). Such antibodies can neutralize infectious viruses at a low concentration, in contrast with antibodies to the stem (70, 112). So, the characteristics of the conserved protective epitopes on the HA head are more suited to the requirements of targets of novel vaccine development than those on the stem. The antibodies that target the stem of HA offer broader but less potent reactivity. The epitopes of HA stem are intrinsically less permissive for mutations, due to the need to maintain interchain packing and to undergo conformational changes during the fusion process (139). So, the characteristics of the epitopes on the HA stem are still compatible with therapeutic development.
In recent years, significant advances have been made in universal influenza vaccine research, and multiple strategies are currently being explored based on HA, including chimeric HA, mosaic HA, computationally optimized broadly reactive antigens (COBARs), Mini-HA, and mosaic nanoparticle vaccination approaches (140, 141). To date, series of universal influenza vaccine (136, 142, 143) and therapeutic agents (137, 144) that targeting HA have been tested in clinical trials. We are confident that as the highly conserved protective epitopes of HA are more clearly elucidated and the antiviral mechanism of bnAbs becomes clearer, universal influenza virus vaccines that provide higher vaccine efficacy and more novel therapeutic candidates that mimic the function of bnAbs will be developed.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by the National Natural Science Foundation of China (32072878) and the National Key Research and Development Program of China (2022YFC2604204).
Acknowledgments
We thank Prof. Hualan Chen for revising and editing the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, et al. New world bats harbor diverse influenza a viruses. PloS Pathog (2013) 9(10):e1003657. doi: 10.1371/journal.ppat.1003657
2. Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza a viruses. Microbiol Rev (1992) 56(1):152–79. doi: 10.1128/mr.56.1.152-179.1992
3. World Health Organization. Influenza (seasonal), in: Fact sheet (2023) (Accessed February 5, 2023).
4. Viboud C, Simonsen L, Fuentes R, Flores J, Miller MA, Chowell G. Global mortality impact of the 1957-1959 influenza pandemic. J Infect Dis (2016) 213(5):738–45. doi: 10.1093/infdis/jiv534
5. Johnson NP, Mueller J. Updating the accounts: global mortality of the 1918-1920 “Spanish” influenza pandemic. Bull Hist Med (2002) 76(1):105–15. doi: 10.1353/bhm.2002.0022
6. Center for Disease Control and Prevention (CDC). 1968 pandemic (H3N2 virus) (2019) (Accessed May 13, 2022).
7. Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza a epidemic. Nature (2009) 459(7250):1122–5. doi: 10.1038/nature08182
8. Shi J, Zeng X, Cui P, Yan C, Chen H. Alarming situation of emerging H5 and H7 avian influenza and effective control strategies. Emerg Microbes Infect (2023) 12(1):2155072. doi: 10.1080/22221751.2022.2155072
9. Coughlan L, Palese P. Overcoming barriers in the path to a universal influenza virus vaccine. Cell Host Microbe (2018) 24(1):18–24. doi: 10.1016/j.chom.2018.06.016
10. Centers for Disease Control Prevention (CDC). Seasonal flu vaccine effectiveness studies (2022) (Accessed May 13, 2022).
11. Sarker A, Gu Z, Mao L, Ge Y, Hou D, Fang J, et al. Influenza-existing drugs and treatment prospects. Eur J Med Chem (2022) 232:114189. doi: 10.1016/j.ejmech.2022.114189
12. Skehel J. An overview of influenza haemagglutinin and neuraminidase. Biologicals (2009) 37(3):177–8. doi: 10.1016/j.biologicals.2009.02.012
13. Krammer F, Pica N, Hai R, Margine I, Palese P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol (2013) 87(12):6542–50. doi: 10.1128/JVI.00641-13
14. Krammer F, Hai R, Yondola M, Tan GS, Leyva-Grado VH, Ryder AB, et al. Assessment of influenza virus hemagglutinin stalk-based immunity in ferrets. J Virol (2014) 88(6):3432–42. doi: 10.1128/JVI.03004-13
15. Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RM, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science (2015) 349(6254):1301–6. doi: 10.1126/science.aac7263
16. Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med (2015) 21(9):1065–70. doi: 10.1038/nm.3927
17. Bommakanti G, Citron MP, Hepler RW, Callahan C, Heidecker GJ, Najar TA, et al. Design of an HA2-based escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc Natl Acad Sci U.S.A. (2010) 107(31):13701–6. doi: 10.1073/pnas.1007465107
18. Sparrow E, Friede M, Sheikh M, Torvaldsen S, Newall AT. Passive immunization for influenza through antibody therapies, a review of the pipeline, challenges and potential applications. Vaccine (2016) 34(45):5442–8. doi: 10.1016/j.vaccine.2016.08.057
19. Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch EM, et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science (2011) 332(6031):816–21. doi: 10.1126/science.1202617
20. Kadam RU, Juraszek J, Brandenburg B, Buyck C, Schepens WBG, Kesteleyn B, et al. Potent peptidic fusion inhibitors of influenza virus. Science (2017) 358(6362):496–502. doi: 10.1126/science.aan0516
21. Koday MT, Nelson J, Chevalier A, Koday M, Kalinoski H, Stewart L, et al. A computationally designed hemagglutinin stem-binding protein provides In vivo protection from influenza independent of a host immune response. PloS Pathog (2016) 12(2):e1005409. doi: 10.1371/journal.ppat.1005409
22. van Dongen MJP, Kadam RU, Juraszek J, Lawson E, Brandenburg B, Schmitz F, et al. A small-molecule fusion inhibitor of influenza virus is orally active in mice. Science (2019) 363(6431):eaar6221. doi: 10.1126/science.aar6221
23. Whitehead TA, Chevalier A, Song Y, Dreyfus C, Fleishman SJ, De Mattos C, et al. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat Biotechnol (2012) 30(6):543–8. doi: 10.1038/nbt.2214
24. Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 a resolution. Nature (1981) 289(5796):366–73. doi: 10.1038/289366a0
25. Nobusawa E, Aoyama T, Kato H, Suzuki Y, Tateno Y, Nakajima K. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza a viruses. Virology (1991) 182(2):475–85. doi: 10.1016/0042-6822(91)90588-3
26. Air GM. Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza a virus. Proc Natl Acad Sci U.S.A. (1981) 78(12):7639–43. doi: 10.1073/pnas.78.12.7639
27. Hashem AM. Prospects of HA-based universal influenza vaccine. BioMed Res Int (2015) 2015:414637. doi: 10.1155/2015/414637
28. Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, et al. Antibody recognition of a highly conserved influenza virus epitope. Science (2009) 324(5924):246–51. doi: 10.1126/science.1171491
29. Zheng Z, Paul SS, Mo X, Yuan YA, Tan YJ. The vestigial esterase domain of haemagglutinin of H5N1 avian influenza a virus: Antigenicity and contribution to viral pathogenesis. Vaccines (Basel) (2018) 6(3):53. doi: 10.3390/vaccines6030053
30. Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature (1988) 333(6172):426–31. doi: 10.1038/333426a0
31. Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory b cells. PloS One (2008) 3(12):e3942. doi: 10.1371/journal.pone.0003942
32. Skehel JJ, Waterfield MD. Studies on the primary structure of the influenza virus hemagglutinin. Proc Natl Acad Sci U.S.A. (1975) 72(1):93–7. doi: 10.1073/pnas.72.1.93
33. Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem (2000) 69:531–69. doi: 10.1146/annurev.biochem.69.1.531
34. Wu NC, Wilson IA. Influenza hemagglutinin structures and antibody recognition. Cold Spring Harb Perspect Med (2020) 10(8):a038778. doi: 10.1101/cshperspect.a038778
35. Gambaryan AS, Tuzikov AB, Piskarev VE, Yamnikova SS, Lvov DK, Robertson JS, et al. Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: non-egg-adapted human H1 and H3 influenza a and influenza b viruses share a common high binding affinity for 6’-sialyl(N-acetyllactosamine). Virology (1997) 232(2):345–50. doi: 10.1006/viro.1997.8572
36. Sauter NK, Bednarski MD, Wurzburg BA, Hanson JE, Whitesides GM, Skehel JJ, et al. Hemagglutinins from two influenza virus variants bind to sialic acid derivatives with millimolar dissociation constants: a 500-MHz proton nuclear magnetic resonance study. Biochemistry (1989) 28(21):8388–96. doi: 10.1021/bi00447a018
37. Takemoto DK, Skehel JJ, Wiley DC. A surface plasmon resonance assay for the binding of influenza virus hemagglutinin to its sialic acid receptor. Virology (1996) 217(2):452–8. doi: 10.1006/viro.1996.0139
38. Ha Y, Stevens DJ, Skehel JJ, Wiley DC. H5 avian and H9 swine influenza virus haemagglutinin structures: possible origin of influenza subtypes. EMBO J (2002) 21(5):865–75. doi: 10.1093/emboj/21.5.865
39. Fontana J, Steven AC. Influenza virus-mediated membrane fusion: Structural insights from electron microscopy. Arch Biochem Biophys (2015) 581:86–97. doi: 10.1016/j.abb.2015.04.011
40. Das DK, Govindan R, Nikic-Spiegel I, Krammer F, Lemke EA, Munro JB. Direct visualization of the conformational dynamics of single influenza hemagglutinin trimers. Cell (2018) 174(4):926–37 e12. doi: 10.1016/j.cell.2018.05.050
41. Yoshida R, Igarashi M, Ozaki H, Kishida N, Tomabechi D, Kida H, et al. Cross-protective potential of a novel monoclonal antibody directed against antigenic site b of the hemagglutinin of influenza a viruses. PloS Pathog (2009) 5(3):e1000350. doi: 10.1371/journal.ppat.1000350
42. Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, et al. Cross-neutralization of influenza a viruses mediated by a single antibody loop. Nature (2012) 489(7417):526–32. doi: 10.1038/nature11414
43. Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, et al. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun (2014) 5:3614. doi: 10.1038/ncomms4614
44. Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, Laursen NS, et al. Antibody recognition of the pandemic H1N1 influenza virus hemagglutinin receptor binding site. J Virol (2013) 87(22):12471–80. doi: 10.1128/JVI.01388-13
45. Schmidt AG, Xu H, Khan AR, O’Donnell T, Khurana S, King LR, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci U.S.A. (2013) 110(1):264–9. doi: 10.1073/pnas.1218256109
46. Schmidt AG, Therkelsen MD, Stewart S, Kepler TB, Liao HX, Moody MA, et al. Viral receptor-binding site antibodies with diverse germline origins. Cell (2015) 161(5):1026–34. doi: 10.1016/j.cell.2015.04.028
47. Tsibane T, Ekiert DC, Krause JC, Martinez O, Crowe JE Jr., Wilson IA, et al. Influenza human monoclonal antibody 1F1 interacts with three major antigenic sites and residues mediating human receptor specificity in H1N1 viruses. PloS Pathog (2012) 8(12):e1003067. doi: 10.1371/journal.ppat.1003067
48. Portnoff AD, Patel N, Massare MJ, Zhou H, Tian JH, Zhou B, et al. Influenza hemagglutinin nanoparticle vaccine elicits broadly neutralizing antibodies against structurally distinct domains of H3N2 HA. Vaccines (Basel) (2020) 8(1):99. doi: 10.3390/vaccines8010099
49. Lin Q, Li T, Chen Y, Lau SY, Wei M, Zhang Y, et al. Structural basis for the broad, antibody-mediated neutralization of H5N1 influenza virus. J Virol (2018) 92(17):e00547–18. doi: 10.1128/JVI.00547-18
50. Wu R, Li X, Leung HC, Cao Z, Qiu Z, Zhou Y, et al. A novel neutralizing antibody against diverse clades of H5N1 influenza virus and its mutants capable of airborne transmission. Antiviral Res (2014) 106:13–23. doi: 10.1016/j.antiviral.2014.03.005
51. Iba Y, Fujii Y, Ohshima N, Sumida T, Kubota-Koketsu R, Ikeda M, et al. Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses. J Virol (2014) 88(13):7130–44. doi: 10.1128/JVI.00420-14
52. Oh HL, Akerstrom S, Shen S, Bereczky S, Karlberg H, Klingstrom J, et al. An antibody against a novel and conserved epitope in the hemagglutinin 1 subunit neutralizes numerous H5N1 influenza viruses. J Virol (2010) 84(16):8275–86. doi: 10.1128/JVI.02593-09
53. Du L, Jin L, Zhao G, Sun S, Li J, Yu H, et al. Identification and structural characterization of a broadly neutralizing antibody targeting a novel conserved epitope on the influenza virus H5N1 hemagglutinin. J Virol (2013) 87(4):2215–25. doi: 10.1128/JVI.02344-12
54. Bangaru S, Lang S, Schotsaert M, Vanderven HA, Zhu X, Kose N, et al. A site of vulnerability on the influenza virus hemagglutinin head domain trimer interface. Cell (2019) 177(5):1136–52 e18. doi: 10.1016/j.cell.2019.04.011
55. Watanabe A, McCarthy KR, Kuraoka M, Schmidt AG, Adachi Y, Onodera T, et al. Antibodies to a conserved influenza head interface epitope protect by an IgG subtype-dependent mechanism. Cell (2019) 177(5):1124–35 e16. doi: 10.1016/j.cell.2019.03.048
56. Bajic G, Maron MJ, Adachi Y, Onodera T, McCarthy KR, McGee CE, et al. Influenza antigen engineering focuses immune responses to a subdominant but broadly protective viral epitope. Cell Host Microbe (2019) 25(6):827–35 e6. doi: 10.1016/j.chom.2019.04.003
57. Bangaru S, Zhang H, Gilchuk IM, Voss TG, Irving RP, Gilchuk P, et al. A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA. Nat Commun (2018) 9(1):2669. doi: 10.1038/s41467-018-04704-9
58. Tan GS, Leon PE, Albrecht RA, Margine I, Hirsh A, Bahl J, et al. Broadly-reactive neutralizing and non-neutralizing antibodies directed against the H7 influenza virus hemagglutinin reveal divergent mechanisms of protection. PloS Pathog (2016) 12(4):e1005578. doi: 10.1371/journal.ppat.1005578
59. Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Albrecht R, Blum DL, et al. Human monoclonal antibodies to pandemic 1957 H2N2 and pandemic 1968 H3N2 influenza viruses. J Virol (2012) 86(11):6334–40. doi: 10.1128/JVI.07158-11
60. Ohshima N, Iba Y, Kubota-Koketsu R, Asano Y, Okuno Y, Kurosawa Y. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J Virol (2011) 85(21):11048–57. doi: 10.1128/JVI.05397-11
61. Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, Crowe JE Jr. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol (2011) 85(20):10905–8. doi: 10.1128/JVI.00700-11
62. Yu X, Tsibane T, McGraw PA, House FS, Keefer CJ, Hicar MD, et al. Neutralizing antibodies derived from the b cells of 1918 influenza pandemic survivors. Nature (2008) 455(7212):532–6. doi: 10.1038/nature07231
63. Daniels PS, Jeffries S, Yates P, Schild GC, Rogers GN, Paulson JC, et al. The receptor-binding and membrane-fusion properties of influenza virus variants selected using anti-haemagglutinin monoclonal antibodies. EMBO J (1987) 6(5):1459–65. doi: 10.1002/j.1460-2075.1987.tb02387.x
64. Smith G, Liu Y, Flyer D, Massare MJ, Zhou B, Patel N, et al. Novel hemagglutinin nanoparticle influenza vaccine with matrix-m adjuvant induces hemagglutination inhibition, neutralizing, and protective responses in ferrets against homologous and drifted A(H3N2) subtypes. Vaccine (2017) 35(40):5366–72. doi: 10.1016/j.vaccine.2017.08.021
65. Sun L, Lu X, Li C, Wang M, Liu Q, Li Z, et al. Generation, characterization and epitope mapping of two neutralizing and protective human recombinant antibodies against influenza a H5N1 viruses. PloS One (2009) 4(5):e5476. doi: 10.1371/journal.pone.0005476
66. Chen Y, Qin K, Wu WL, Li G, Zhang J, Du H, et al. Broad cross-protection against H5N1 avian influenza virus infection by means of monoclonal antibodies that map to conserved viral epitopes. J Infect Dis (2009) 199(1):49–58. doi: 10.1086/594374
67. Li J, Yang Y, Wang M, Ren X, Yang Z, Liu L, et al. Rapid isolation of a potent human antibody against H7N9 influenza virus from an infected patient. Antiviral Res (2019) 170:104564. doi: 10.1016/j.antiviral.2019.104564
68. Barbey-Martin C, Gigant B, Bizebard T, Calder LJ, Wharton SA, Skehel JJ, et al. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology (2002) 294(1):70–4. doi: 10.1006/viro.2001.1320
69. McCarthy KR, Watanabe A, Kuraoka M, Do KT, McGee CE, Sempowski GD, et al. Memory b cells that cross-react with group 1 and group 2 influenza a viruses are abundant in adult human repertoires. Immunity (2018) 48(1):174–84 e9. doi: 10.1016/j.immuni.2017.12.009
70. Corti D, Suguitan AL Jr., Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest (2010) 120(5):1663–73. doi: 10.1172/JCI41902
71. Li T, Chen J, Zheng Q, Xue W, Zhang L, Rong R, et al. Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses. Nat Commun (2022) 13(1):5182. doi: 10.1038/s41467-022-32926-5
72. Lee PS, Yoshida R, Ekiert DC, Sakai N, Suzuki Y, Takada A, et al. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci U.S.A. (2012) 109(42):17040–5. doi: 10.1073/pnas.1212371109
73. Xu R, Krause JC, McBride R, Paulson JC, Crowe JE Jr., Wilson IA. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol (2013) 20(3):363–70. doi: 10.1038/nsmb.2500
74. Whittle JR, Zhang R, Khurana S, King LR. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of manischewitz J, Golding h, et al. influenza virus hemagglutinin. Proc Natl Acad Sci U.S.A. (2011) 108(34):14216–21. doi: 10.1073/pnas.1111497108
75. Yang F, Yan S, Zhu L, Wang FXC, Liu F, Cheng L, et al. Evaluation of panel of neutralising murine monoclonal antibodies and a humanised bispecific antibody against influenza A(H1N1)pdm09 virus infection in a mouse model. Antiviral Res (2022) 208:105462. doi: 10.1016/j.antiviral.2022.105462
76. Zuo T, Sun J, Wang G, Jiang L, Zuo Y, Li D, et al. Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection. Nat Commun (2015) 6:8855. doi: 10.1038/ncomms9855
77. Zuo Y, Wang P, Sun J, Guo S, Wang G, Zuo T, et al. Complementary recognition of the receptor-binding site of highly pathogenic H5N1 influenza viruses by two human neutralizing antibodies. J Biol Chem (2018) 293(42):16503–17. doi: 10.1074/jbc.RA118.004604
78. Simmons CP, Bernasconi NL, Suguitan AL, Mills K, Ward JM, Chau NV, et al. Prophylactic and therapeutic efficacy of human monoclonal antibodies against H5N1 influenza. PloS Med (2007) 4(5):e178. doi: 10.1371/journal.pmed.0040178
79. Thornburg NJ, Nannemann DP, Blum DL, Belser JA, Tumpey TM, Deshpande S, et al. Human antibodies that neutralize respiratory droplet transmissible H5N1 influenza viruses. J Clin Invest (2013) 123(10):4405–9. doi: 10.1172/JCI69377
80. Winarski KL, Thornburg NJ, Yu Y, Sapparapu G, Crowe JE Jr., Spiller BW. Vaccine-elicited antibody that neutralizes H5N1 influenza and variants binds the receptor site and polymorphic sites. Proc Natl Acad Sci U.S.A. (2015) 112(30):9346–51. doi: 10.1073/pnas.1502762112
81. Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, et al. Highly conserved protective epitopes on influenza b viruses. Science (2012) 337(6100):1343–8. doi: 10.1126/science.1222908
82. Yan L, Sun L, Guo C, Li L, Sun J, Huang X, et al. Neutralizing antibody PR8-23 targets the footprint of the sialoglycan receptor binding site of H1N1 hemagglutinin. J Med Virol (2021) 93(6):3508–15. doi: 10.1002/jmv.26779
83. Li J, Wang Y, Liang Y, Ni B, Wan Y, Liao Z, et al. Fine antigenic variation within H5N1 influenza virus hemagglutinin’s antigenic sites defined by yeast cell surface display. Eur J Immunol (2009) 39(12):3498–510. doi: 10.1002/eji.200939532
84. Zhu X, Guo YH, Jiang T, Wang YD, Chan KH, Li XF, et al. A unique and conserved neutralization epitope in H5N1 influenza viruses identified by an antibody against the A/Goose/Guangdong/1/96 hemagglutinin. J Virol (2013) 87(23):12619–35. doi: 10.1128/JVI.01577-13
85. Mak TM, Hanson BJ, Tan YJ. Chimerization and characterization of a monoclonal antibody with potent neutralizing activity across multiple influenza a H5N1 clades. Antiviral Res (2014) 107:76–83. doi: 10.1016/j.antiviral.2014.04.011
86. Paul SS, Mok CK, Mak TM, Ng OW, Aboagye JO, Wohlbold TJ, et al. A cross-clade H5N1 influenza a virus neutralizing monoclonal antibody binds to a novel epitope within the vestigial esterase domain of hemagglutinin. Antiviral Res (2017) 144:299–310. doi: 10.1016/j.antiviral.2017.06.012
87. Wang S, Ren H, Jiang W, Chen H, Hu H, Chen Z, et al. Divergent requirement of fc-fcgamma receptor interactions for In vivo protection against influenza viruses by two pan-H5 hemagglutinin antibodies. J Virol (2017) 91(11):e02065–16. doi: 10.1128/JVI.02065-16
88. Ren H, Wang G, Wang S, Chen H, Chen Z, Hu H, et al. Cross-protection of newly emerging HPAI H5 viruses by neutralizing human monoclonal antibodies: A viable alternative to oseltamivir. MAbs (2016) 8(6):1156–66. doi: 10.1080/19420862.2016.1183083
89. Zhang X, Qi X, Zhang Q, Zeng X, Shi Z, Jin Q, et al. Human 4F5 single-chain fv antibody recognizing a conserved HA1 epitope has broad neutralizing potency against H5N1 influenza a viruses of different clades. Antiviral Res (2013) 99(2):91–9. doi: 10.1016/j.antiviral.2013.05.001
90. Dong J, Gilchuk I, Li S, Irving R, Goff MT, Turner HL, et al. Anti-influenza H7 human antibody targets antigenic site in hemagglutinin head domain interface. J Clin Invest (2020) 130(9):4734–9. doi: 10.1172/JCI136032
91. Thornburg NJ, Zhang H, Bangaru S, Sapparapu G, Kose N, Lampley RM, et al. H7N9 influenza virus neutralizing antibodies that possess few somatic mutations. J Clin Invest (2016) 126(4):1482–94. doi: 10.1172/JCI85317
92. Turner HL, Pallesen J, Lang S, Bangaru S, Urata S, Li S, et al. Potent anti-influenza H7 human monoclonal antibody induces separation of hemagglutinin receptor-binding head domains. PloS Biol (2019) 17(2):e3000139. doi: 10.1371/journal.pbio.3000139
93. Knossow M, Skehel JJ. Variation and infectivity neutralization in influenza. Immunology (2006) 119(1):1–7. doi: 10.1111/j.1365-2567.2006.02421.x
94. Gambaryan AS, Tuzikov AB, Chinarev AA, Juneja LR, Bovin NV, Matrosovich MN. Polymeric inhibitor of influenza virus attachment protects mice from experimental influenza infection. Antiviral Res (2002) 55(1):201–5. doi: 10.1016/s0166-3542(02)00020-7
95. Lauster D, Klenk S, Ludwig K, Nojoumi S, Behren S, Adam L, et al. Phage capsid nanoparticles with defined ligand arrangement block influenza virus entry. Nat Nanotechnol (2020) 15(5):373–9. doi: 10.1038/s41565-020-0660-2
96. Kwon SJ, Na DH, Kwak JH, Douaisi M, Zhang F, Park EJ, et al. Nanostructured glycan architecture is important in the inhibition of influenza a virus infection. Nat Nanotechnol (2017) 12(1):48–54. doi: 10.1038/nnano.2016.181
97. Matsubara T, Onishi A, Saito T, Shimada A, Inoue H, Taki T, et al. Sialic acid-mimic peptides as hemagglutinin inhibitors for anti-influenza therapy. J Med Chem (2010) 53(11):4441–9. doi: 10.1021/jm1002183
98. Jeon SH, Kayhan B, Ben-Yedidia T, Arnon R. A DNA aptamer prevents influenza infection by blocking the receptor binding region of the viral hemagglutinin. J Biol Chem (2004) 279(46):48410–9. doi: 10.1074/jbc.M409059200
99. Gopinath SC, Kumar PK. Aptamers that bind to the hemagglutinin of the recent pandemic influenza virus H1N1 and efficiently inhibit agglutination. Acta Biomat (2013) 9(11):8932–41. doi: 10.1016/j.actbio.2013.06.016
100. Li W, Feng X, Yan X, Liu K, Deng L. A DNA aptamer against influenza a virus: An effective inhibitor to the hemagglutinin-glycan interactions. Nucleic Acid Ther (2016) 26(3):166–72. doi: 10.1089/nat.2015.0564
101. Hu H, Voss J, Zhang G, Buchy P, Zuo T, Wang L, et al. A human antibody recognizing a conserved epitope of H5 hemagglutinin broadly neutralizes highly pathogenic avian influenza H5N1 viruses. J Virol (2012) 86(6):2978–89. doi: 10.1128/JVI.06665-11
102. Yewdell JW, Taylor A, Yellen A, Caton A, Gerhard W, Bachi T. Mutations in or near the fusion peptide of the influenza virus hemagglutinin affect an antigenic site in the globular region. J Virol (1993) 67(2):933–42. doi: 10.1128/jvi.67.2.933-942.1993
103. Chen F, Tzarum N, Wilson IA, Law M. VH1-69 antiviral broadly neutralizing antibodies: genetics, structures, and relevance to rational vaccine design. Curr Opin Virol (2019) 34:149–59. doi: 10.1016/j.coviro.2019.02.004
104. Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza a hemagglutinins. Science (2011) 333(6044):850–6. doi: 10.1126/science.1205669
105. Fu Y, Zhang Z, Sheehan J, Avnir Y, Ridenour C, Sachnik T, et al. A broadly neutralizing anti-influenza antibody reveals ongoing capacity of haemagglutinin-specific memory b cells to evolve. Nat Commun (2016) 7:12780. doi: 10.1038/ncomms12780
106. Kallewaard NL, Corti D, Collins PJ, Neu U, McAuliffe JM, Benjamin E, et al. Structure and function analysis of an antibody recognizing all influenza a subtypes. Cell (2016) 166(3):596–608. doi: 10.1016/j.cell.2016.05.073
107. Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, et al. A highly conserved neutralizing epitope on group 2 influenza a viruses. Science (2011) 333(6044):843–50. doi: 10.1126/science.1204839
108. Friesen RH, Lee PS, Stoop EJ, Hoffman RM, Ekiert DC, Bhabha G, et al. A common solution to group 2 influenza virus neutralization. Proc Natl Acad Sci U.S.A. (2014) 111(1):445–50. doi: 10.1073/pnas.1319058110
109. Wang W, Sun X, Li Y, Su J, Ling Z, Zhang T, et al. Human antibody 3E1 targets the HA stem region of H1N1 and H5N6 influenza a viruses. Nat Commun (2016) 7:13577. doi: 10.1038/ncomms13577
110. Wu Y, Cho M, Shore D, Song M, Choi J, Jiang T, et al. A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza a virus. Nat Commun (2015) 6:7708. doi: 10.1038/ncomms8708
111. Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza a virus H1 and H2 strains. J Virol (1993) 67(5):2552–8. doi: 10.1128/jvi.67.5.2552-2558.1993
112. Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza a viruses. Nat Struct Mol Biol (2009) 16(3):265–73. doi: 10.1038/nsmb.1566
113. Pan Y, Sasaki T, Kubota-Koketsu R, Inoue Y, Yasugi M, Yamashita A, et al. Human monoclonal antibodies derived from a patient infected with 2009 pandemic influenza a virus broadly cross-neutralize group 1 influenza viruses. Biochem Biophys Res Commun (2014) 450(1):42–8. doi: 10.1016/j.bbrc.2014.05.060
114. Tan GS, Lee PS, Hoffman RM, Mazel-Sanchez B, Krammer F, Leon PE, et al. Characterization of a broadly neutralizing monoclonal antibody that targets the fusion domain of group 2 influenza a virus hemagglutinin. J Virol (2014) 88(23):13580–92. doi: 10.1128/JVI.02289-14
115. Sutton TC, Lamirande EW, Bock KW, Moore IN, Koudstaal W, Rehman M, et al. In vitro neutralization is not predictive of prophylactic efficacy of broadly neutralizing monoclonal antibodies CR6261 and CR9114 against lethal H2 influenza virus challenge in mice. J Virol (2017) 91(24):e01603–17. doi: 10.1128/JVI.01603-17
116. Yamayoshi S, Uraki R, Ito M, Kiso M, Nakatsu S, Yasuhara A, et al. A broadly reactive human anti-hemagglutinin stem monoclonal antibody that inhibits influenza a virus particle release. EBioMedicine (2017) 17:182–91. doi: 10.1016/j.ebiom.2017.03.007
117. Laursen NS, Friesen RHE, Zhu X, Jongeneelen M, Blokland S, Vermond J, et al. Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science (2018) 362(6414):598–602. doi: 10.1126/science.aaq0620
118. Lang S, Xie J, Zhu X, Wu NC, Lerner RA, Wilson IA. Antibody 27F3 broadly targets influenza a group 1 and 2 hemagglutinins through a further variation in VH1-69 antibody orientation on the HA stem. Cell Rep (2017) 20(12):2935–43. doi: 10.1016/j.celrep.2017.08.084
119. Hu W, Chen A, Miao Y, Xia S, Ling Z, Xu K, et al. Fully human broadly neutralizing monoclonal antibodies against influenza a viruses generated from the memory b cells of a 2009 pandemic H1N1 influenza vaccine recipient. Virology (2013) 435(2):320–8. doi: 10.1016/j.virol.2012.09.034
120. Nakamura G, Chai N, Park S, Chiang N, Lin Z, Chiu H, et al. An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza a antibodies. Cell Host Microbe (2013) 14(1):93–103. doi: 10.1016/j.chom.2013.06.004
121. Joyce MG, Wheatley AK, Thomas PV, Chuang GY, Soto C, Bailer RT, et al. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza a viruses. Cell (2016) 166(3):609–23. doi: 10.1016/j.cell.2016.06.043
122. Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory b cells. Proc Natl Acad Sci U.S.A. (2012) 109(23):9047–52. doi: 10.1073/pnas.1118979109
123. Wyrzucki A, Bianchi M, Kohler I, Steck M, Hangartner L. Heterosubtypic antibodies to influenza a virus have limited activity against cell-bound virus but are not impaired by strain-specific serum antibodies. J Virol (2015) 89(6):3136–44. doi: 10.1128/JVI.03069-14
124. Sun X, Liu C, Lu X, Ling Z, Yi C, Zhang Z, et al. Unique binding pattern for a lineage of human antibodies with broad reactivity against influenza a virus. Nat Commun (2022) 13(1):2378. doi: 10.1038/s41467-022-29950-w
125. Harshbarger WD, Deming D, Lockbaum GJ, Attatippaholkun N, Kamkaew M, Hou S, et al. Unique structural solution from a VH3-30 antibody targeting the hemagglutinin stem of influenza a viruses. Nat Commun (2021) 12(1):559. doi: 10.1038/s41467-020-20879-6
126. Nachbagauer R, Shore D, Yang H, Johnson SK, Gabbard JD, Tompkins SM, et al. Broadly reactive human monoclonal antibodies elicited following pandemic H1N1 influenza virus exposure protect mice against highly pathogenic H5N1 challenge. J Virol (2018) 92(16):e00949–18. doi: 10.1128/JVI.00949-18
127. Dreyfus C, Ekiert DC, Wilson IA. Structure of a classical broadly neutralizing stem antibody in complex with a pandemic H2 influenza virus hemagglutinin. J Virol (2013) 87(12):7149–54. doi: 10.1128/JVI.02975-12
128. Sakabe S, Iwatsuki-Horimoto K, Horimoto T, Nidom CA, Le M, Takano R, et al. A cross-reactive neutralizing monoclonal antibody protects mice from H5N1 and pandemic (H1N1) 2009 virus infection. Antiviral Res (2010) 88(3):249–55. doi: 10.1016/j.antiviral.2010.09.007
129. Smirnov Iu A, Lipatov AS, Okuno I, Gitel’man AK. [A common antigenic epitope in influenza a virus (H1, H2, H5, H6) hemagglutinin]. Vopr Virusol (1999) 44(3):111–5.
130. Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, et al. Broadly cross-reactive antibodies dominate the human b cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med (2011) 208(1):181–93. doi: 10.1084/jem.20101352
131. Wyrzucki A, Dreyfus C, Kohler I, Steck M, Wilson IA, Hangartner L. Alternative recognition of the conserved stem epitope in influenza a virus hemagglutinin by a VH3-30-encoded heterosubtypic antibody. J Virol (2014) 88(12):7083–92. doi: 10.1128/JVI.00178-14
132. Kashyap AK, Steel J, Rubrum A, Estelles A, Briante R, Ilyushina NA, et al. Protection from the 2009 H1N1 pandemic influenza by an antibody from combinatorial survivor-based libraries. PloS Pathog (2010) 6(7):e1000990. doi: 10.1371/journal.ppat.1000990
133. Wang W, Vassell R, Song HS, Chen Q, Keller PW, Verma S, et al. Generation of a protective murine monoclonal antibody against the stem of influenza hemagglutinins from group 1 viruses and identification of resistance mutations against it. PloS One (2019) 14(9):e0222436. doi: 10.1371/journal.pone.0222436
134. Wu NC, Yamayoshi S, Ito M, Uraki R, Kawaoka Y, Wilson IA. Recurring and adaptable binding motifs in broadly neutralizing antibodies to influenza virus are encoded on the D3-9 segment of the ig gene. Cell Host Microbe (2018) 24(4):569–78 e4. doi: 10.1016/j.chom.2018.09.010
135. Wu NC, Wilson IA. Structural biology of influenza hemagglutinin: An amaranthine adventure. Viruses (2020) 12(9):1053. doi: 10.3390/v12091053
136. Ostrowsky J, Arpey M, Moore K, Osterholm M, Friede M, Gordon J, et al. Tracking progress in universal influenza vaccine development. Curr Opin Virol (2020) 40:28–36. doi: 10.1016/j.coviro.2020.02.003
137. Zhang Q, Liang T, Nandakumar KS, Liu S. Emerging and state of the art hemagglutinin-targeted influenza virus inhibitors. Expert Opin Pharmacother (2021) 22(6):715–28. doi: 10.1080/14656566.2020.1856814
138. McBride JM, Lim JJ, Burgess T, Deng R, Derby MA, Maia M, et al. Phase 2 randomized trial of the safety and efficacy of MHAA4549A, a broadly neutralizing monoclonal antibody, in a human influenza a virus challenge model. Antimicrob Agents Chemother (2017) 61(11):e01154–17. doi: 10.1128/AAC.01154-17
139. Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu Rev Immunol (2013) 31:705–42. doi: 10.1146/annurev-immunol-032712-095916
140. Nachbagauer R, Palese P. Is a universal influenza virus vaccine possible? Annu Rev Med (2020) 71:315–27. doi: 10.1146/annurev-med-120617-041310
141. Krammer F, Palese P. Universal influenza virus vaccines that target the conserved hemagglutinin stalk and conserved sites in the head domain. J Infect Dis (2019) 219(Suppl_1):S62–S7. doi: 10.1093/infdis/jiy711
142. Bernstein DI, Guptill J, Naficy A, Nachbagauer R, Berlanda-Scorza F, Feser J, et al. Immunogenicity of chimeric haemagglutinin-based, universal influenza virus vaccine candidates: interim results of a randomised, placebo-controlled, phase 1 clinical trial. Lancet Infect Dis (2020) 20(1):80–91. doi: 10.1016/S1473-3099(19)30393-7
143. Liu WC, Nachbagauer R, Stadlbauer D, Strohmeier S, Solorzano A, Berlanda-Scorza F, et al. Chimeric hemagglutinin-based live-attenuated vaccines confer durable protective immunity against influenza a viruses in a preclinical ferret model. Vaccines (Basel) (2021) 9(1):40. doi: 10.3390/vaccines9010040
Keywords: influenza A viruses, hemagglutinin, broadly neutralizing antibodies, conserved protective epitopes, universal influenza vaccines, therapeutic agents
Citation: Jiao C, Wang B, Chen P, Jiang Y and Liu J (2023) Analysis of the conserved protective epitopes of hemagglutinin on influenza A viruses. Front. Immunol. 14:1086297. doi: 10.3389/fimmu.2023.1086297
Received: 01 November 2022; Accepted: 07 February 2023;
Published: 17 February 2023.
Edited by:
Mahbuba Rahman, McMaster University, CanadaReviewed by:
Al Hakim, Jagannath University, BangladeshSaba Ismail, National University of Medical Sciences (NUMS), Pakistan
Copyright © 2023 Jiao, Wang, Chen, Jiang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinxiong Liu, bGl1amlueGlvbmdAY2Fhcy5jbg==
 Chenchen Jiao
Chenchen Jiao Bo Wang
Bo Wang Jinxiong Liu
Jinxiong Liu