- 1Traditional Chinese Medicine Integrated Department of Nephrology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 2Institute of Nephrology, Zhengzhou University, Zhengzhou, China
- 3Henan Province Research Center for Kidney Disease, Zhengzhou, China
- 4Key Laboratory of Precision Diagnosis and Treatment for Chronic Kidney Disease in Henan Province, Zhengzhou, China
Diabetic kidney disease (DKD) is one of the major microvascular complications of diabetes mellitus and is also one of the serious risk factors in cardiovascular events, end-stage renal disease, and mortality. DKD is associated with the diversified, compositional, and functional alterations of gut microbiota. The interaction between gut microbiota and host is mainly achieved through metabolites, which are small molecules produced by microbial metabolism from exogenous dietary substrates and endogenous host compounds. The gut microbiota plays a critical role in the pathogenesis of DKD by producing multitudinous metabolites. Nevertheless, detailed mechanisms of gut microbiota and its metabolites involved in the occurrence and development of DKD have not been completely elucidated. This review summarizes the specific classes of gut microbiota-derived metabolites, aims to explore the molecular mechanisms of gut microbiota in DKD pathophysiology and progression, recognizes biomarkers for the screening, diagnosis, and prognosis of DKD, as well as provides novel therapeutic strategies for DKD.
Introduction
Diabetic kidney disease (DKD) is a pivotal complication of diabetes mellitus and significantly increases the risk of cardiovascular disease and end-stage renal disease (ESRD), that ultimately results in dialysis or high-mortality and economic burdens (1). The increased number of DKD and ESRD is partially attributed to lifestyle and dietary habits associated with diabetes and hypertension (2). Management and treatment strategy of patients with DKD includes controlling blood glucose, blood lipid, and blood pressure as well as blockade of the renin-angiotensin system (RAS); however, the risk of DKD still remains to be high (3) indicating the presence of unrecognized factors and mechanisms involved. The occurrence and progression of DKD is correlated to the interaction between gene and environment (4). Despite that hyperglycemia-induced metabolic alterations, hemodynamics changes, RAS activation, podocyte injury or loss, epithelial dysfunction, inflammation, and immunoreaction contributed to disease progression, specific molecular mechanisms and pathogenesis need to be explored (5).
The gut microbiota is powerful for maintaining host internal environmental homeostasis. For one thing, microbiome prevents infection caused by pathogens, promotes the digestion and absorption of nutrients, and synthesizes essential vitamins and amino acids (6). For another thing, it exerts an anti-inflammatory function (6), regulates fat metabolism (7), and participates in immune system development (8). And thirdly, gut microbiota-derived metabolites such as short-chain fatty acids (SCFAs), bile acids (BAs), lipopolysaccharide (LPS), and trimethylamine N-oxide (TMAO) are essential mediators of microbial-host crosstalk by interacting with host environment (9). The diversified, compositional, and functional alterations of gut microbiome are termed dysbiosis (10), which leads to a reduction in SCFAs and an increase in uremic toxins, activation of RAS, inflammation, and aggravated immune response. Nonetheless, specific mechanisms by which gut microbiota affects DKD have not been fully elucidated. This review summarized the role of gut microbiota and its metabolites in DKD, discussed underlying mechanisms of gut microbiota involved in DKD progression, and explored its potentials in DKD management and treatment.
Gut microbiota and its metabolites
Gut microbiota
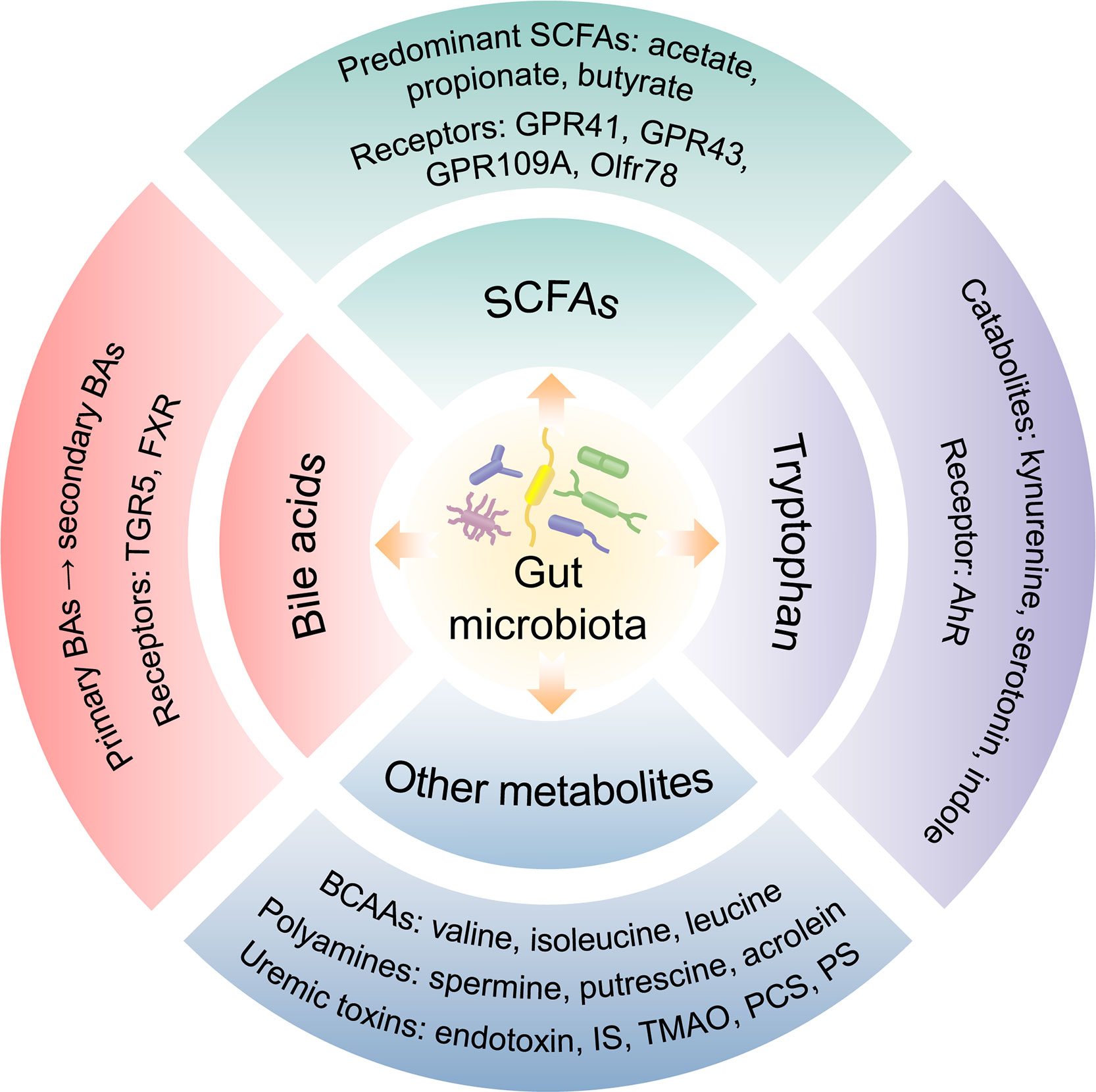
The human gastrointestinal tract possesses a plentiful microbial community which collects approximately 100 trillion microorganisms, including bacteria, fungi, viruses, phages, and archaea (11). Commonly, the gut microbiota is comprised of 6 phyla incorporating with Bacteroidetes, Frimicutes, Verrucomicrobia, Proteobacteria, Actinobacteria, and Fusobacteria, in which Bacteroidetes and Frimicutes are the majority components (12). The stability of intestinal microbiota is closely related to host health and disease. What is more, gut microbiota is symbiotic with the host and participates in a variety of physiological activities, such as fermenting food, resisting pathogens and regulating immune function (13). The gut microbiota contributes to host physiology by producing a multitude of metabolites (14) (Figure 1). Numerous metabolites derived from gut microbiota fermentation are vital factors in host-microbiota cross-talk and have been shown to be correlated with kidney function.
16S rDNA, metagenomics, and mass spectrometry can be utilized to explore the diversity, composition, and function of gut microbiota as well as microbiota-related serum metabolites in patients with DKD. Interaction studies between plasma metabolomics and gut microbiome in experimental DKD mouse/rat model provided evidence for the gut-metabolism-kidney axis, and verified the involvement of gut microbiota and circulating metabolites in DKD progression (15, 16). DKD patients displayed dysbiosis with composition, richness and diversity in gut microbiota (17–19). Roseburia intestinalis was significantly decreased while Bacteroides stercoris was increased in DKD patients (20). Furthermore, studies in early DKD caused by type 1 diabetes indicated that differences in gut microbiota and serum metabolite profiles were dependent on albuminuria levels (21). Several studies also revealed diversity and species differences in gut microbiota between DKD patients and non-DKD patients (22–24).
SCFAs
SCFAs are produced by the fermentation of polysaccharides with the assistance of gut microbiota and are the main source of nutrition for colon epithelial cells. Acetate, propionate, and butyrate generated from the bacterial fermentation of dietary fiber are the predominant SCFAs (25). SCFAs have been shown to inhibit the activity of histone deacetylase (HDAC) and involve in G protein-coupled receptors (GPRs) mediated signaling pathway (26, 27). SCFAs can bind to GPRs such as GPR41, GPR43, GPR109A, and olfactory receptors (Olfr) 78, and then were absorbed into system circulation after reaching distant tissues. Furthermore, SCFAs were demonstrated to participate in the sustainment of intestinal barrier integrity (28), enhance glucose and lipid metabolism, restraint energy expenditure (29), and modulate immunoreaction and inflammatory responses (30). The reduction of SCFAs-producing bacteria as well as low serum and fecal SCFAs level may be correlated with kidney injury (31–33). Butyrate was reported to improve the intestinal barrier function by promoting the production of colonic mucin and tight junction proteins (ZO-1) (34). It could also mitigate oxidative stress, inflammation, and fibrosis in kidney disease through GPRs or HDAC (35–37). Serum valerate and caproate levels were negatively correlated with the progression of DKD to ESRD (38). It has been shown that acetate mediated the dysregulation of cholesterol homeostasis by activation of GPR43, thereby contributing to the tubulointerstitial injury of DKD (39).
Bile acids
BAs are synthesized from cholesterol in the hepatocytes and participates in the absorption of lipid as well as metabolic or inflammatory signaling pathways (40). The primary BAs including chenodeoxycholic acid (CDCA) and cholic acid (CA), are indispensable for lipid and vitamin digestion and absorption by conjugating to glycine or taurine (41). Primary BAs could transform and decompose into secondary BAs via gut microbiota. The gut microbiota modulates BA metabolism process through deconjugation, dehydrogenation, and dihydroxylation of primary BAs (42). Additionally, the synthesis of BAs is influenced by cholesterol 7α-hydroxylase (CYP7A1) and sterol 27-hydroxylase (CYP27A1) regulating via gut microbiota (14). BAs are ligands for G protein-coupled bile acid receptor (TGR5) and nuclear hormone receptor farnesoid X receptor (FXR). Moreover, the profiles of BAs and gut microbiota influence each other. BAs could alter the composition of intestinal microbiota. Conversely, microbiota modulates the size and composition of the BA pool as well as BA signaling (43). BAs combine with TGR5 to improve insulin sensitivity via glucagon-like peptide-1 (GLP-1) and regulate energy expenditure in muscle or brown adipose tissue (44). The activation of FXR decreases lipogenesis and hepatic gluconeogenesis, and inhibits bacterial overgrowth and translocation by producing antimicrobial peptides (45). FXR and TGR5 play a renal protective role in diabetes and obesity-related kidney disease by regulating renal signaling pathways (46). Gentiopicroside inhibits the NF-κB signaling pathway via TGR5 activation, thereby alleviating inflammation and fibrosis in DKD (47).
Tryptophan
An essential aromatic amino-acid, tryptophan, generally originates from daily diet such as fish, milk, oats, cheese. Besides the synthesis of proteins, dietary tryptophan could act as a precursor of critical metabolites including kynurenine, serotonin, indole, and its derivatives (48). Kynurenine, a tryptophan-derived metabolite produced by tryptophan 2,3-dioxygenase and indoleamine (2, 3)-dioxygenase, is correlated with kidney function (49, 50). Tryptophan is decomposed by bacterial tryptophanase into indole, which is a compound responsible for intercellular signal transduction, participating in the gene expression of intestinal epithelium connections and anti-inflammatory factors in intestinal epithelial cells, as well as maintaining host-microbiota homeostasis on the mucosa surface (51). As downstream critical metabolites, 3-(2-Hydroxyethyl) indole, 3-methylindole, and indoleacrylic acid were downregulated in the DKD model and were reinstated after treatment with Tangshen Formula (15). Some compounds produced by tryptophan metabolism are ligands for the aryl hydrocarbon receptor (AhR) and could induce AhR conformational changes. Moreover, these compounds are involved in the gene expression of pro-inflammatory factors, the metabolism of cytochrome P450 (CYP) superfamily CYP1A1, CYP1A2, CYP1B1 and cyclooxygenase-2 (COX-2), or the degradation of selective proteins (52). The deficient activation of AhR pathway could reduce the production of GLP-1 and interleukin (IL)-22, increase intestinal permeability and LPS translocation, which contribute to inflammation and insulin resistance (53). Based on the combined analysis of gut microbiota, serum metabolites and clinical indicators in DKD patients, phenylalanine and tryptophan metabolic pathways were demonstrated to be associated with the progression of DKD (54).
Other metabolites
Branched-chain amino acids (BCAAs) are essential amino-acids synthesized by gut microbiota, including valine, isoleucine, and leucine. BCAAs modulate protein synthesis, glucose/lipid metabolism, insulin resistance, and immunity, as well as maintain homeostasis (55). Polyamines, such as spermine, putrescine, polyamine oxidase and acrolein, are participated in the development of kidney disease by altering the metabolism of intestinal microbiota (56). The dysbiosis of gut microbiota promotes the production of bacteria-derived uremic toxins, such as indoxyl sulfate (IS), endotoxin, TMAO, and p-cresyl sulfate (PCS), which increase intestinal permeability and transfer into the systemic circulation through the damaged intestinal barrier. Accumulation of uremic toxins in kidneys could lead to kidney dysfunction (57). TMAO, a gut microbiota-derived metabolite, was associated with mortality and renal outcome in type 1 diabetes (58). Higher serum TMAO levels increased the risk of abdominal aortic venture in hemodialysis patients (59). Phenyl sulfate (PS) contributed to podocyte damage and albuminuria and was shown to be related to the progression of DKD (60). Imidazole propionate, a metabolite produced by the breakdown of histidine via gut microbiota, was increased in type 2 diabetes, affecting host inflammation and metabolism (61). Both PS and TMAO could be involved in the development of DKD through a secretory associated senescence phenotype and chronic low-grade inflammation (62). IS and PCS contributed to the nephrology and cardiovascular toxicities via the activation of inflammation and oxidative stress (63). Additionally, several uremic toxins such as urea, TMAO, PCS, and 3-carboxylic acid 4-methyl-5-propyl-2-furan propionic (CMPF) were associated with glucose homeostasis abnormalities and diabetes incidence (64). The dysbiosis of Gram-negative bacteria and increased LPS level were detected in type 2 diabetes related DKD (65).
Gut microbiota-related factors in DKD progression
Insulin resistance
DKD originates from metabolic dysregulation including hyperglycemia, hyperlipidemia, and insulin resistance (4). Hyperglycemia increases the generation of advanced glycation end products. The variance in insulin levels and insulin resistance might be a significant factor in DKD. Severe albuminuria and glomerulosclerosis were occurred in animals with complete deletion of podocyte insulin receptor (66). The dysbiosis of gut microbiota is linked to insulin resistance (67, 68) (Figure 2). A few species of microbiota, especially Prevotella copri and Bacteroides vulgatus are associated with insulin resistance and then impact host metabolism (69). Gut commensal Bacteroides acidifaciens could improve insulin sensitivity and may have therapeutic potential for diabetes and obesity (70). Microbiota depletion such as antibiotic-treated or germ-free mice could enhance insulin sensitivity and glucose tolerance (71). Podocyte insulin resistance caused podocyte injury and led to albuminuria in early DKD. Dysregulated GPR43 by gut microbiota dysbiosis resulted in podocyte insulin resistance through the inhibition of adenosine monophosphate-activated protein kinase (AMPK)-α activity (72). Butyrate enhanced AMPK phosphorylation and increased GLP-1 secretion, thereby alleviating insulin resistance and renal failure (34). Imidazole propionate, a microbial histidine-derived metabolite, may contribute to insulin resistance through activation of mechanistic target of rapamycin complex1 (mTORC1) (73).
RAS
RAS is critical in the pathogenesis and progression of DKD. Moreover, local RAS might play a greater role than the circulating RAS (74). The secretion of renin in the juxtaglomerular apparatus plays an important role in the activation of intrarenal RAS by hyperglycemia. Olfr78 expressed in the renal juxtaglomerular afferent arteriole responded to signals from intestinal microbiota by mediating renin secretion, after that SCFAs could modulate blood pressure through Olfr78 and GPR41 (75). Succinate accumulated in the distal nephron-collecting duct, and activation of GPR91 responded to hyperglycemia through the stored (pro)renin and provoked tissue injury in DKD (76). The activation of intrarenal RAS by gut microbiota dysbiosis-derived excessive acetate was involved in the kidney injury of early DKD (77). Gut microbiota could promote angiotensin II (Ang II)-induced vascular dysfunction and hypertension by facilitating CCL2/IL-17-driven vascular immune cell infiltration and inflammation (78). Conversely, butyrate exerted an improvement for Ang II-induced renal injury and an antihypertension action by attenuating expression of (pro)renin receptor and renin as well as suppressing the (pro)renin receptor-mediated intrarenal RAS (79). During the fermentation of probiotics, angiotensin converting enzyme (ACE) inhibitory peptide and renin inhibitory peptide could be released, which are beneficial for lowering blood pressure (80, 81). In addition, ACE2 was associated with tryptophan metabolism and was sensitive to intestinal inflammation (82). A few uremic toxins such as IS and PCS are important stimulator of local RAS. Moreover, the inhibition of RAS ameliorated IS and PCS induced renal fibrosis (83).
Inflammation
Inflammation accompanies the pathogenesis and progression of DKD whereas anti-inflammatory therapies might be beneficial for alleviating renal damage in DKD. Several inflammatory pathways participate in the complicated molecular networks and processes in DKD, including chemokines (CCL2, CX3CL1 and CCL5), inflammatory cytokines (IL-1, IL-6, IL-18), adhesion molecules, E-selectin, α-actinin 4, transcription factor nuclear factor-kappa B (NF-κB), and tumor necrosis factor (84). The initial stage of the inflammatory response to injury or metabolic dysfunction involves the release of proinflammatory mediators and the recruitment of leukocytes. Therefore, targeting inflammatory-resolution pathways might contribute to impede the progression of DKD (85). SCFAs could be involved in the modulation of pro-inflammatory and anti-inflammatory responses by inhibiting HDAC directly and binding GPRs indirectly (86). SCFAs produced by dietary fiber fermentation decreased the expression of inflammatory cytokines, chemokines, and fibrosis-promoting proteins in experimental DKD, thereby reducing albuminuria, glomerular hypertrophy, podocyte injury, and interstitial fibrosis. Moreover, this process required the involvement of GPR43 or GPR109A (87). Host/gut microbiota-derived tryptophan metabolites regulated AhR and then affected oxidative stress and inflammation in DKD (88). TMAO and PS accelerated kidney inflammation and fibrosis, resulting in development of DKD (60, 89). LPS, combined with toll-like receptors (TLRs) TLR2 and TLR4, participated in the inflammatory process of DKD through NF-κB activation and pro-inflammatory cytokines release, leading to the renal injury (90). Obesity enhanced intestinal permeability and chronic low-grade inflammation by inducing gut microbiota dysbiosis, ultimately causing the exasperation of DKD (91)
Immunity
The activation of innate immunity through immune cells and resident renal cells contributed to the initiation and maintenance of inflammation (92). TLRs induced sterile tubulointerstitial inflammatory responses via NF-κB signaling pathway. The nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain-containing 3 (NLRP3) inflammasome were associated with the connection of metabolic stress and pro-inflammatory cascades by inducing IL-1β and IL-18. The kallikrein-kinin system contributed to inflammatory progression by generating bradykinin and activating bradykinin receptors. Furthermore, coagulation enzymes promoted the activation of protease-activated receptors on kidney cells, leading to renal inflammation and fibrosis in DKD. Gut microbiota plays a significant role in maintaining host homeostasis as well as in modulating immune system (93). There have several studies characterizing the complex interaction between DKD, microbes and its metabolites, and immune responses. The microbiota colonized the intestinal tract after birth and regulated the antigenic responsiveness of lymphatic tissue (94). With the involvement of gut microbiota, the intestinal immune system started to build up and to be matured gradually. The dysbiosis of gut microbiota attracted immune cell activation and proinflammatory factors secretion, which led to immune dysregulation and inflammation (95). Mitochondrial antiviral signaling protein (MAVS), a component of innate immunity, was involved in maintaining intestinal integrity and barrier function. Damaged MAVS was conducive to the disrupted intestinal homeostasis, contributing to DKD progression (96). Microbiome-host interactions cooperatively maintained microbial community stability through metabolite-mediated innate immune modulation. What’s more, metabolites could influence the host’s immune homeostasis (97). Gut microbiota-derived metabolites passed through the intestinal barrier, accumulated in the circulation, recognized by immune system, and performed functions through gut-microbiome-immune axis (98). Bacteroids-derived SCFAs contributed to the activation of immune system by promoting neutrophil chemotaxis and inducing differentiation and proliferation of natural killer cells and Tregs (99).
Management and treatment options for gut microbiota in DKD
Clinical drugs
Various kinds of drug may alleviate DKD by affecting intestinal microbiota. Metformin was shown to contribute to several SCFAs-producing microbiota and increase the production of butyrate and propionate, thus participating in glucose homeostasis (100). Sodium-glucose cotransporter 2 inhibitor, as emerging antidiabetic drugs including empagliflozin, canagliflozin and dapagliflozin, restored the diversity of gut microbiota in experimental DKD mouse model. Moreover, reduced LPS production and increased SCFAs production by regulating the microbiota were observed in patients after inhibition of SGLT2 (101–103). Pirfenidone treatment increased gut microbial diversity in diabetic mouse model and reversed gut microbial dysbiosis and diabetic ketoacidosis biomarkers (104). Magnesium lithospermate B was found to ameliorate kidney injury by modulating gut microbiome dysbiosis and BAs metabolism (105). Abundant polysaccharides are beneficial for DKD. Polysaccharide from Armillariella tabescens mycelia, Cordyceps cicadae polysaccharide, and Bupleurum polysaccharide were demonstrated to modulate gut microbiota dysbiosis and inflammatory response (106–108). Traditional Chinese medicine such as Zicuiyin (109), Moutan Cortex polysaccharide (110), QiDiTangShen granules (111), Shenyan Kangfu tablet (112), and Tangshen Formula (113), have been used clinically to treat DKD. They had a significant curative role in regulating gut microbiota, eliminating intestinal toxins, inhibiting renal inflammation and immunity, alleviating renal injury, and protecting kidney function.
Dietary intervention
Diet is fundamental to support human growth, health, and reproduction. Furthermore, diet was also shown to modulate and maintain the symbiotic gut microbiota communities colonized the intestinal tract (114). Under multiple host-containing endogenous and exogenous factors, diet becomes a pivotal determinant of the structure and function in gut microbiota (115) (Figure 3). The latest review regarding the effect of dietary nutrient intake on gut microbiota indicated that diet-microbiota crosstalk and personalized nutrition strategies are associated with chronic kidney disease progression (116). Moreover, the variation in dietary protein sources affected the gut microbiota, microbiota-derived metabolites, immune cell activation, and production of inflammatory cytokines (117). Studies from human population with different diets showed that Bacteroides was enriched in a protein-rich diets while Prevotella was enriched in a carbohydrate-based diets (118). Whole-plant fibers from fresh vegetables contained a lot of necessary micronutrients compared with highly processed fibers or fibers from seed coats (119). Plant-based low-protein diets seemingly contributed to postpone kidney replacement therapy by disturbing RAS, reducing proteinuria, and decreasing insulin resistance (120). Fermented and germinated foxtail millet whole grain diet raised the bacterial diversity especially probiotics, thereby ameliorating kidney injury in experimental DKD mouse model through inhibition of inflammation and immunity signaling pathways (121). A high linolenic acid diet aggravated gut microbiota dysbiosis and inflammatory responses in diabetes mouse model. Conversely, a low n-6/n-3 ratio diet improved glucose homeostasis, inhibited systematic inflammation, and ameliorated DKD (122). Punicalagin from pomegranates, a prospective bioactive polyphenol, was shown to alleviate diabetic kidney injury through gut-kidney axis (123).
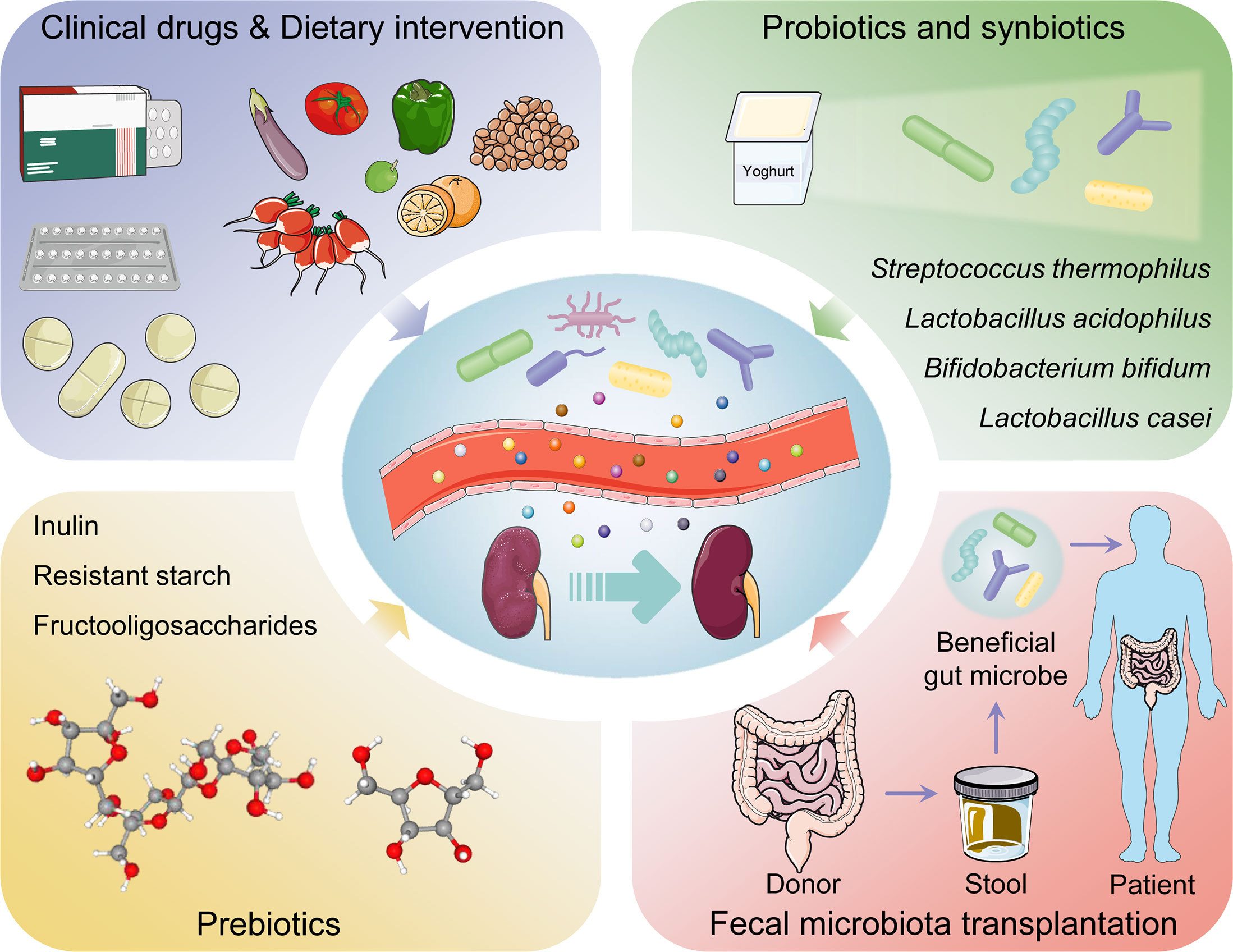
Figure 3 The management and therapeutic strategies of diabetic kidney disease based on gut microbiota.
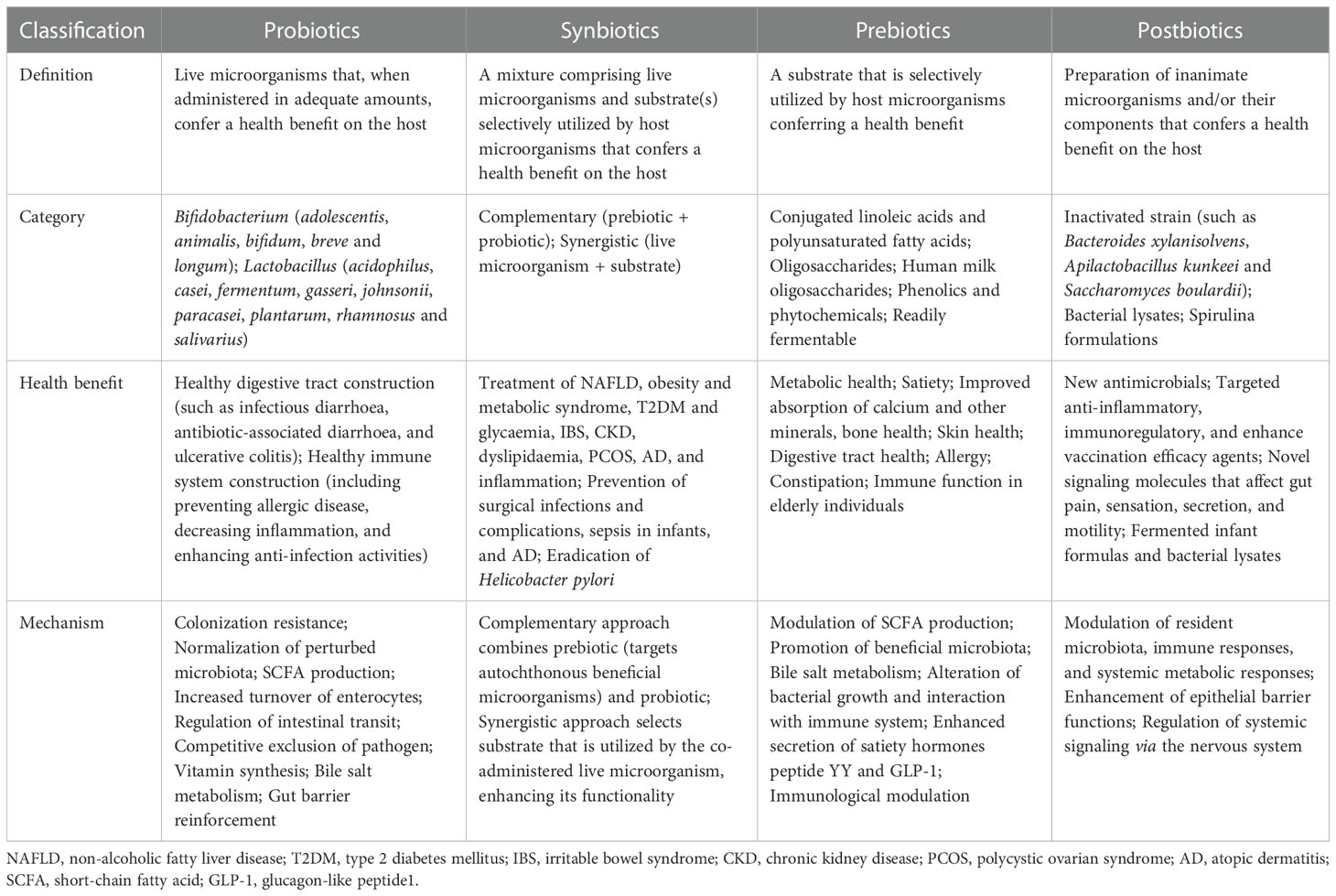
Probiotics and synbiotics
Probiotics contain live microorganisms that can change composition of microbiota and are supposed to provide health benefits to host (124). Synbiotics, a mixture comprising live microorganisms and substrates selectively utilized by host microorganisms confer a health benefit on the host (125) (Table 1). Probiotic and synbiotic supplementation, such as Lactobacillus acidophilus, Lactobacillus casei and Bifidobacterium bifidum, had beneficial effects on blood glucose and intestinal imbalance, production of uremic toxins, and inflammation or oxidative stress in diabetic hemodialysis patients (126–128). Furthermore, probiotics could ameliorate insulin resistance, stabilize fasting blood glucose levels, and improve antioxidant status (90, 129). Addition of probiotics such as Lactobacillus acidophilus, Streptococcus thermophilus and Bifidobacterium longum reduced the blood urea nitrogen level and uric acid concentration in patients with stage 3 and stage 4 chronic kidney disease (130, 131). Systematic review and meta-analysis demonstrated that probiotics might ameliorate high sensitivity-C reactive protein and oxidative stress biomarkers, as well as regulate lipid profile and anthropometric indices in DKD patients (132, 133).
Prebiotics and postbiotics
Prebiotics such as noncarbohydrate food components, are substrates that are selectively used by host microorganisms for health benefits (134). The supplementation of prebiotics in daily dietary could exterminate pathogens, facilitate the growth of beneficial microorganisms, and regulate host intestinal microbiota (135). Moreover, prebiotic supplements might increase SCFAs levels (notably butyrate), restore intestinal barrier function, and relieve inflammatory response (136). Fructooligosaccharides could alleviate pathological changes in diabetes related kidney disease (137). Inulin-type fructans, a type of dietary fiber, was demonstrated to improve kidney diseases via modulating gut microbiota and SCFAs profile (138). Additionally, inulin-type fructans also decreased insulin resistance, serum insulin and fasting blood glucose levels, and increased fasting serum GLP-1 level in diabetes rats (139, 140). Resistant starch is a prebiotic compound that accelerates proliferation of health-promoting gut microbiota such as Bifidobacteria and Lactobacilli, increases the production of SCFAs, decreases the concentrations of uremic toxins and alleviates renal dysfunction (141). Postbiotics, defined as “preparation of inanimate microorganisms and/or their components that confers a health benefit on the host” in 2019 (142), have appeared increasingly in the literature and products; however, their effects on DKD are insufficient in research. Postbiotics exert immunomodulatory and intestinal barrier protective roles by increasing anti-inflammatory cytokine secretion and ZO-1 expression (143). Postbiotic-GABA-salt, spirulina formulations, sonicated Lactobacillus paracasei and O. formigenes lysates contribute to improve renal outcomes (144, 145).
Fecal microbiota transplantation
Fecal microbiota transplantation (FMT) is a treatment in which the microbial community from a healthy donor’s stool was minimally transplanted into the patient’s intestinal tract (146). FMT is implemented with the purpose of restoring normal function of the gut microbiota and has generally been adapted into treatment for Clostridium difficile infection (147). Faecal microbiota is separated cautiously from selected donor’s stool, quantified in accordance with viable bacteria, and cryopreservation (148). Transplantable materials can be delivered in the form of encapsulated oral medication (149). As a true organ, gut microbiota is indispensable to human pathophysiology, suggesting that FMT might be an advantageous treatment for problems with metabolism, autoimmunity, and system development (150). Body weight gain, insulin resistance, albuminuria, and tumor necrosis factor-α levels in experimental DKD mouse model could be prevented by FMT (151). After six weeks post-FMT using stool derived from lean donors, the peripheral insulin sensitivity was significantly improved in male patients with metabolic syndrome, although the result was not sustained in following few weeks (152). Another double blind randomized controlled trial demonstrated that TMAO or proxies of vascular inflammation was undifferentiated in patients with metabolic syndrome received FMT from either lean donors or autologous (153). Hence, abundant experiments are needed to explore these potential therapeutic indications.
Conclusion and perspective
The pathogenesis and pathophysiology of DKD incorporate not only hyperglycemia-induced metabolic alterations, hemodynamics changes, RAS activation, podocyte injury or loss, epithelial dysfunction, inflammation, and immune dysregulation, but also the influences of environmental factors and interactions between host and gut microbiota as well as its metabolites. Gut microbiota is associated with kidney disease, confirming the presence of gut-kidney axis through the involvement of genetic, immunity and dietary approaches. The gut microbiota participates in host homeostasis by producing a myriad of metabolites, which act ss key signaling molecules and substrates for metabolic reactions. The combination of metagenomics and metabolomics could help to investigate the relationship between dysbiosis of gut microbiota and metabolic disorders. Nonetheless, there are still complexities to overcome in identifying the potential causality of some metabolites from fully microbiota-derived or diet and host itself. High-quality microbiome analysis workflow is important to obtain reliable and repeatable results (154).
Dietary intervention, probiotics, synbiotics, and prebiotics are widely acceptable to patients in relative safety and traditional concept. However, various intestinal bacteria and metabolites have heterogeneous effects on host, some of which are beneficial to human health and others contribute to pathophysiology of diseases. Hence, it is necessary to investigate the signals and effects mediated by different bacteria and metabolites as well as reasonable application of bacteria community in the treatment strategies. Gut Microbiota-derived metabolites could act as biomarkers of DKD. Identification of biomarkers for screening, diagnosis, and prognosis of DKD as well as exploration of molecular mechanisms or pathways involved in DKD can facilitate individualized prevention and treatment. However, further studies involving human trials are needed to investigate the beneficial role of prebiotics, probiotics, synbiotics or FMT in DKD management by regulating gut microbiota. The therapeutic strategy targeting intestinal microbiota has prodigious potential in the future and will open an emerging perspective and orientation for DKD treatment.
Author contributions
PW and Z-SL conceived the idea. Z-HM prepared the figures and tables, and drafted the manuscript. PW, Z-XG, D-WL and Z-SL revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China [No. 31971065, No. 81900651, No.81970633], Natural Science Foundation of Henan Province [No. 222300420089] and Talents Project of Health Science and Technology Innovation for Young and Middle-aged Investigators in Henan Province [YXKC2020038].
Acknowledgments
The authors acknowledge smart.servier.com for some art elements in Figures 2 and 3.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Thomas MC, Brownlee M, Susztak K, Sharma K, Jandeleit-Dahm KAM, Zoungas S, et al. Diabetic kidney disease. Nat Rev Dis Primers (2015) 1:15018. doi: 10.1038/nrdp.2015.18
2. Jha V, Garcia-Garcia G, Iseki K, Li Z, Naicker S, Plattner B, et al. Chronic kidney disease: global dimension and perspectives. Lancet (2013) 382:260–72. doi: 10.1016/S0140-6736(13)60687-X
3. Lytvyn Y, Bjornstad P, van Raalte DH, Heerspink HL, Cherney DZI. The new biology of diabetic kidney disease-mechanisms and therapeutic implications. Endocr Rev (2020) 41:202–231. doi: 10.1210/endrev/bnz010
4. Reidy K, Kang HM, Hostetter T, Susztak K. Molecular mechanisms of diabetic kidney disease. J Clin Invest (2014) 124:2333–40. doi: 10.1172/JCI72271
5. DeFronzo RA, Reeves WB, Awad AS. Pathophysiology of diabetic kidney disease: Impact of SGLT2 inhibitors. Nat Rev Nephrol (2021) 17:319–34. doi: 10.1038/s41581-021-00393-8
6. Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature (2009) 461:1282–6. doi: 10.1038/nature08530
7. Frick J-S, Autenrieth IB. The gut microflora and its variety of roles in health and disease. Curr Top Microbiol Immunol (2013) 358:273–89. doi: 10.1007/82_2012_217
8. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science (2016) 352:539–44. doi: 10.1126/science.aad9378
9. Blander JM, Longman RS, Iliev ID, Sonnenberg GF, Artis D. Regulation of inflammation by microbiota interactions with the host. Nat Immunol (2017) 18:851–60. doi: 10.1038/ni.3780
10. Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol (2017) 17:219–32. doi: 10.1038/nri.2017.7
11. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature (2012) 489:220–30. doi: 10.1038/nature11550
12. Shanahan F, Ghosh TS, O’Toole PW. The healthy microbiome-what is the definition of a healthy gut microbiome? Gastroenterology (2021) 160:483–94. doi: 10.1053/j.gastro.2020.09.057
13. Hou K, Wu Z-X, Chen X-Y, Wang J-Q, Zhang D, Xiao C, et al. Microbiota in health and diseases. Signal Transduct Target Ther (2022) 7:135. doi: 10.1038/s41392-022-00974-4
14. Krautkramer KA, Fan J, Bäckhed F. Gut microbial metabolites as multi-kingdom intermediates. Nat Rev Microbiol (2021) 19:77–94. doi: 10.1038/s41579-020-0438-4
15. Zhang B, Wan Y, Zhou X, Zhang H, Zhao H, Ma L, et al. Characteristics of serum metabolites and gut microbiota in diabetic kidney disease. Front Pharmacol (2022) 13:872988. doi: 10.3389/fphar.2022.872988
16. Wu C, Fei J, Xu Q, Tao Y, Zhou Z, Wang Y, et al. Interaction between plasma metabolomics and intestinal microbiome in db/db mouse, an animal model for study of type 2 diabetes and diabetic kidney disease. Metabolites (2022) 12:775. doi: 10.3390/metabo12090775
17. Du X, Liu J, Xue Y, Kong X, Lv C, Li Z, et al. Alteration of gut microbial profile in patients with diabetic nephropathy. Endocrine (2021) 73:71–84. doi: 10.1007/s12020-021-02721-1
18. Tao S, Li L, Li L, Liu Y, Ren Q, Shi M, et al. Understanding the gut-kidney axis among biopsy-proven diabetic nephropathy, type 2 diabetes mellitus and healthy controls: An analysis of the gut microbiota composition. Acta Diabetol (2019) 56:581–92. doi: 10.1007/s00592-019-01316-7
19. Wang Y, Zhao J, Qin Y, Yu Z, Zhang Y, Ning X, et al. The specific alteration of gut microbiota in diabetic kidney diseases-a systematic review and meta-analysis. Front Immunol (2022) 13:908219. doi: 10.3389/fimmu.2022.908219
20. Zhang L, Wang Z, Zhang X, Zhao L, Chu J, Li H, et al. Alterations of the gut microbiota in patients with diabetic nephropathy. Microbiol Spectr (2022) 10:e0032422. doi: 10.1128/spectrum.00324-22
21. Winther SA, Henriksen P, Vogt JK, Hansen TH, Ahonen L, Suvitaival T, et al. Gut microbiota profile and selected plasma metabolites in type 1 diabetes without and with stratification by albuminuria. Diabetologia (2020) 63:2713–24. doi: 10.1007/s00125-020-05260-y
22. Chen R, Zhu D, Yang R, Wu Z, Xu N, Chen F, et al. Gut microbiota diversity in middle-aged and elderly patients with end-stage diabetic kidney disease. Ann Transl Med (2022) 10:750. doi: 10.21037/atm-22-2926
23. He X, Sun J, Liu C, Yu X, Li H, Zhang W, et al. Compositional alterations of gut microbiota in patients with diabetic kidney disease and type 2 diabetes mellitus. Diabetes Metab Syndr Obes (2022) 15:755–65. doi: 10.2147/DMSO.S347805
24. Han S, Chen M, Cheng P, Zhang Z, Lu Y, Xu Y, et al. A systematic review and meta-analysis of gut microbiota in diabetic kidney disease: Comparisons with diabetes mellitus, non-diabetic kidney disease, and healthy individuals. Front Endocrinol (Lausanne) (2022) 13:1018093. doi: 10.3389/fendo.2022.1018093
25. Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol (2015) 11:577–91. doi: 10.1038/nrendo.2015.128
26. Nogal A, Valdes AM, Menni C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes (2021) 13:1–24. doi: 10.1080/19490976.2021.1897212
27. Baxter NT, Schmidt AW, Venkataraman A, Kim KS, Waldron C, Schmidt TM. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio (2019) 10:e02566–18. doi: 10.1128/mBio.02566-18
28. Kim CH. Microbiota or short-chain fatty acids: which regulates diabetes? Cell Mol Immunol (2018) 15:88–91. doi: 10.1038/cmi.2017.57
29. Hu J, Lin S, Zheng B, Cheung PCK. Short-chain fatty acids in control of energy metabolism. Crit Rev Food Sci Nutr (2018) 58:1243–9. doi: 10.1080/10408398.2016.1245650
30. Kim CH. Control of lymphocyte functions by gut microbiota-derived short-chain fatty acids. Cell Mol Immunol (2021) 18:1161–71. doi: 10.1038/s41423-020-00625-0
31. Zhong C, Dai Z, Chai L, Wu L, Li J, Guo W, et al. The change of gut microbiota-derived short-chain fatty acids in diabetic kidney disease. J Clin Lab Anal (2021) 35:e24062. doi: 10.1002/jcla.24062
32. Cai K, Ma Y, Cai F, Huang X, Xiao L, Zhong C, et al. Changes of gut microbiota in diabetic nephropathy and its effect on the progression of kidney injury. Endocrine (2022) 76:294–303. doi: 10.1007/s12020-022-03002-1
33. Li Y, Su X, Gao Y, Lv C, Gao Z, Liu Y, et al. The potential role of the gut microbiota in modulating renal function in experimental diabetic nephropathy murine models established in same environment. Biochim Biophys Acta Mol Basis Dis (2020) 1866:165764. doi: 10.1016/j.bbadis.2020.165764
34. Gonzalez A, Krieg R, Massey HD, Carl D, Ghosh S, Gehr TWB, et al. Sodium butyrate ameliorates insulin resistance and renal failure in CKD rats by modulating intestinal permeability and mucin expression. Nephrol Dial Transplant (2019) 34:783–94. doi: 10.1093/ndt/gfy238
35. Cheng X, Zhou T, He Y, Xie Y, Xu Y, Huang W. The role and mechanism of butyrate in the prevention and treatment of diabetic kidney disease. Front Microbiol (2022) 13:961536. doi: 10.3389/fmicb.2022.961536
36. Lu Y, Fan C, Li P, Lu Y, Chang X, Qi K. Short chain fatty acids prevent high-fat-diet-induced obesity in mice by regulating G protein-coupled receptors and gut microbiota. Sci Rep (2016) 6:37589. doi: 10.1038/srep37589
37. Lin MY, de Zoete MR, van Putten JPM, Strijbis K. Redirection of epithelial immune responses by short-chain fatty acids through inhibition of histone deacetylases. Front Immunol (2015) 6:554. doi: 10.3389/fimmu.2015.00554
38. Zhong C, Bai X, Chen Q, Ma Y, Li J, Zhang J, et al. Gut microbial products valerate and caproate predict renal outcome among the patients with biopsy-confirmed diabetic nephropathy. Acta Diabetol (2022) 59:1469–77. doi: 10.1007/s00592-022-01948-2
39. Hu ZB, Lu J, Chen PP, Lu CC, Zhang JX, Li XQ, et al. Dysbiosis of intestinal microbiota mediates tubulointerstitial injury in diabetic nephropathy via the disruption of cholesterol homeostasis. Theranostics (2020) 10:2803–16. doi: 10.7150/thno.40571
40. Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discovery (2008) 7:678–93. doi: 10.1038/nrd2619
41. Just S, Mondot S, Ecker J, Wegner K, Rath E, Gau L, et al. The gut microbiota drives the impact of bile acids and fat source in diet on mouse metabolism. Microbiome (2018) 6:134. doi: 10.1186/s40168-018-0510-8
42. Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall H-U, Bamberg K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab (2013) 17:225–35. doi: 10.1016/j.cmet.2013.01.003
43. Jia W, Xie G, Jia W. Bile acid–microbiota cross-talk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol (2018) 15:111–28. doi: 10.1038/nrgastro.2017.119
44. Schaap FG, Trauner M, Jansen PLM. Bile acid receptors as targets for drug development. Nat Rev Gastroenterol Hepatol (2014) 11:55–67. doi: 10.1038/nrgastro.2013.151
45. Huang W, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, et al. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science (2006) 312:233–6. doi: 10.1126/science.1121435
46. Wang XX, Wang D, Luo Y, Myakala K, Dobrinskikh E, Rosenberg AZ, et al. FXR/TGR5 dual agonist prevents progression of nephropathy in diabetes and obesity. J Am Soc Nephrol (2018) 29:118–37. doi: 10.1681/ASN.2017020222
47. Xiao H, Sun X, Liu R, Chen Z, Lin Z, Yang Y, et al. Gentiopicroside activates the bile acid receptor Gpbar1 (TGR5) to repress NF-kappaB pathway and ameliorate diabetic nephropathy. Pharmacol Res (2020) 151:104559. doi: 10.1016/j.phrs.2019.104559
48. Comai S, Bertazzo A, Brughera M, Crotti S. Tryptophan in health and disease. Adv Clin Chem (2020) 95:165–218. doi: 10.1016/bs.acc.2019.08.005
49. Liu J-R, Miao H, Deng D-Q, Vaziri ND, Li P, Zhao Y-Y. Gut microbiota-derived tryptophan metabolism mediates renal fibrosis by aryl hydrocarbon receptor signaling activation. Cell Mol Life Sci (2021) 78:909–22. doi: 10.1007/s00018-020-03645-1
50. Pawlak K, Myśliwiec M, Pawlak D. Kynurenine pathway - a new link between endothelial dysfunction and carotid atherosclerosis in chronic kidney disease patients. Adv Med Sci (2010) 55:196–203. doi: 10.2478/v10039-010-0015-6
51. Lee J-H, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev (2010) 34:426–44. doi: 10.1111/j.1574-6976.2009.00204.x
52. Safe S, Han H, Goldsby J, Mohankumar K, Chapkin RS. Aryl hydrocarbon receptor (AhR) ligands as selective AhR modulators: Genomic studies. Curr Opin Toxicol (2018) 11–12:10–20. doi: 10.1016/j.cotox.2018.11.005
53. Taleb S. Tryptophan dietary impacts gut barrier and metabolic diseases. Front Immunol (2019) 10:2113. doi: 10.3389/fimmu.2019.02113
54. Zhang Q, Zhang Y, Zeng L, Chen G, Zhang L, Liu M, et al. The role of gut microbiota and microbiota-related serum metabolites in the progression of diabetic kidney disease. Front Pharmacol (2021) 12:757508. doi: 10.3389/fphar.2021.757508
55. Tajiri K, Shimizu Y. Branched-chain amino acids in liver diseases. Transl Gastroenterol Hepatol (2018) 3:47. doi: 10.21037/tgh.2018.07.06
56. Feng Y-L, Cao G, Chen D-Q, Vaziri ND, Chen L, Zhang J, et al. Microbiome-metabolomics reveals gut microbiota associated with glycine-conjugated metabolites and polyamine metabolism in chronic kidney disease. Cell Mol Life Sci (2019) 76:4961–78. doi: 10.1007/s00018-019-03155-9
57. Nallu A, Sharma S, Ramezani A, Muralidharan J, Raj D. Gut microbiome in chronic kidney disease: challenges and opportunities. Transl Res (2017) 179:24–37. doi: 10.1016/j.trsl.2016.04.007
58. Winther SA, Øllgaard JC, Tofte N, Tarnow L, Wang Z, Ahluwalia TS, et al. Utility of plasma concentration of trimethylamine n-oxide in predicting cardiovascular and renal complications in individuals with type 1 diabetes. Diabetes Care (2019) 42:1512–20. doi: 10.2337/dc19-0048
59. He L, Yang W, Yang P, Zhang X, Zhang A. Higher serum trimethylamine-n-oxide levels are associated with increased abdominal aortic calcification in hemodialysis patients. Ren Fail (2022) 44:2019–27. doi: 10.1080/0886022X.2022.2145971
60. Kikuchi K, Saigusa D, Kanemitsu Y, Matsumoto Y, Thanai P, Suzuki N, et al. Gut microbiome-derived phenyl sulfate contributes to albuminuria in diabetic kidney disease. Nat Commun (2019) 10:1835. doi: 10.1038/s41467-019-09735-4
61. Molinaro A, Lassen PB, Henricsson M, Wu H, Adriouch S, Belda E, et al. Imidazole propionate is increased in diabetes and associated with dietary patterns and altered microbial ecology. Nat Commun (2020) 11:5881. doi: 10.1038/s41467-020-19589-w
62. Fernandes R, Viana SD, Nunes S, Reis F. Diabetic gut microbiota dysbiosis as an inflammaging and immunosenescence condition that fosters progression of retinopathy and nephropathy. Biochim Biophys Acta Mol Basis Dis (2019) 1865:1876–97. doi: 10.1016/j.bbadis.2018.09.032
63. Rossi M, Campbell KL, Johnson DW, Stanton T, Vesey DA, Coombes JS, et al. Protein-bound uremic toxins, inflammation and oxidative stress: a cross-sectional study in stage 3-4 chronic kidney disease. Arch Med Res (2014) 45:309–17. doi: 10.1016/j.arcmed.2014.04.002
64. Koppe L, Fouque D, Soulage CO. Metabolic abnormalities in diabetes and kidney disease: Role of uremic toxins. Curr Diabetes Rep (2018) 18:97. doi: 10.1007/s11892-018-1064-7
65. Salguero MV, Al-Obaide MAI, Singh R, Siepmann T, Vasylyeva TL. Dysbiosis of gram-negative gut microbiota and the associated serum lipopolysaccharide exacerbates inflammation in type 2 diabetic patients with chronic kidney disease. Exp Ther Med (2019) 18:3461–9. doi: 10.3892/etm.2019.7943
66. Welsh GI, Hale LJ, Eremina V, Jeansson M, Maezawa Y, Lennon R, et al. Insulin signaling to the glomerular podocyte is critical for normal kidney function. Cell Metab (2010) 12:329–40. doi: 10.1016/j.cmet.2010.08.015
67. Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol (2016) 14:20–32. doi: 10.1038/nrmicro3552
68. Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature (2018) 562:589–94. doi: 10.1038/s41586-018-0620-2
69. Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BAH, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature (2016) 535:376–81. doi: 10.1038/nature18646
70. Yang J-Y, Lee Y-S, Kim Y, Lee S-H, Ryu S, Fukuda S, et al. Gut commensal bacteroides acidifaciens prevents obesity and improves insulin sensitivity in mice. Mucosal Immunol (2017) 10:104–16. doi: 10.1038/mi.2016.42
71. Suárez-Zamorano N, Fabbiano S, Chevalier C, Stojanović O, Colin DJ, Stevanović A, et al. Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat Med (2015) 21:1497–501. doi: 10.1038/nm.3994
72. Lu J, Chen PP, Zhang JX, Li XQ, Wang GH, Yuan BY, et al. GPR43 deficiency protects against podocyte insulin resistance in diabetic nephropathy through the restoration of AMPKα activity. Theranostics (2021) 11:4728–42. doi: 10.7150/thno.56598
73. Koh A, Molinaro A, Ståhlman M, Khan MT, Schmidt C, Mannerås-Holm L, et al. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell (2018) 175:947–961.e17. doi: 10.1016/j.cell.2018.09.055
74. Wysocki J, Ye M, Khattab AM, Fogo A, Martin A, David NV, et al. Angiotensin-converting enzyme 2 amplification limited to the circulation does not protect mice from development of diabetic nephropathy. Kidney Int (2017) 91:1336–46. doi: 10.1016/j.kint.2016.09.032
75. Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci U.S.A. (2013) 110:4410–5. doi: 10.1073/pnas.1215927110
76. Peti-Peterdi J. High glucose and renin release: the role of succinate and GPR91. Kidney Int (2010) 78:1214–7. doi: 10.1038/ki.2010.333
77. Lu C-C, Hu Z-B, Wang R, Hong Z-H, Lu J, Chen P-P, et al. Gut microbiota dysbiosis-induced activation of the intrarenal renin-angiotensin system is involved in kidney injuries in rat diabetic nephropathy. Acta Pharmacol Sin (2020) 41:1111–8. doi: 10.1038/s41401-019-0326-5
78. Karbach SH, Schönfelder T, Brandão I, Wilms E, Hörmann N, Jäckel S, et al. Gut microbiota promote angiotensin II-induced arterial hypertension and vascular dysfunction. J Am Heart Assoc (2016) 5:e003698. doi: 10.1161/JAHA.116.003698
79. Wang L, Zhu Q, Lu A, Liu X, Zhang L, Xu C, et al. Sodium butyrate suppresses angiotensin II-induced hypertension by inhibition of renal (pro)renin receptor and intrarenal renin-angiotensin system. J Hypertens (2017) 35:1899–908. doi: 10.1097/HJH.0000000000001378
80. Dave LA, Hayes M, Montoya CA, Rutherfurd SM, Moughan PJ. Human gut endogenous proteins as a potential source of angiotensin-i-converting enzyme (ACE-i)-, renin inhibitory and antioxidant peptides. Peptides (2016) 76:30–44. doi: 10.1016/j.peptides.2015.11.003
81. Gonzalez-Gonzalez C, Gibson T, Jauregi P. Novel probiotic-fermented milk with angiotensin I-converting enzyme inhibitory peptides produced by bifidobacterium bifidum MF 20/5. Int J Food Microbiol (2013) 167:131–7. doi: 10.1016/j.ijfoodmicro.2013.09.002
82. Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature (2012) 487:477–81. doi: 10.1038/nature11228
83. Sun C-Y, Chang S-C, Wu M-S. Uremic toxins induce kidney fibrosis by activating intrarenal renin-angiotensin-aldosterone system associated epithelial-to-mesenchymal transition. PloS One (2012) 7:e34026. doi: 10.1371/journal.pone.0034026
84. Navarro-González JF, Mora-Fernández C, Muros de Fuentes M, García-Pérez J. Inflammatory molecules and pathways in the pathogenesis of diabetic nephropathy. Nat Rev Nephrol (2011) 7:327–40. doi: 10.1038/nrneph.2011.51
85. Brennan E, Kantharidis P, Cooper ME, Godson C. Pro-resolving lipid mediators: regulators of inflammation, metabolism and kidney function. Nat Rev Nephrol (2021) 17:725–39. doi: 10.1038/s41581-021-00454-y
86. Dupraz L, Magniez A, Rolhion N, Richard ML, Da Costa G, Touch S, et al. Gut microbiota-derived short-chain fatty acids regulate IL-17 production by mouse and human intestinal γδ T cells. Cell Rep (2021) 36:109332. doi: 10.1016/j.celrep.2021.109332
87. Li YJ, Chen X, Kwan TK, Loh YW, Singer J, Liu Y, et al. Dietary fiber protects against diabetic nephropathy through short-chain fatty acid-mediated activation of G protein-coupled receptors GPR43 and GPR109A. J Am Soc Nephrol (2020) 31:1267–81. doi: 10.1681/ASN.2019101029
88. Tan Y-Q, Wang Y-N, Feng H-Y, Guo Z-Y, Li X, Nie X-L, et al. Host/microbiota interactions-derived tryptophan metabolites modulate oxidative stress and inflammation via aryl hydrocarbon receptor signaling. Free Radic Biol Med (2022) 184:30–41. doi: 10.1016/j.freeradbiomed.2022.03.025
89. Fang Q, Zheng B, Liu N, Liu J, Liu W, Huang X, et al. Trimethylamine n-oxide exacerbates renal inflammation and fibrosis in rats with diabetic kidney disease. Front Physiol (2021) 12:682482. doi: 10.3389/fphys.2021.682482
90. He C, Shan Y, Song W. Targeting gut microbiota as a possible therapy for diabetes. Nutr Res (2015) 35:361–7. doi: 10.1016/j.nutres.2015.03.002
91. Li J, Lv J-L, Cao X-Y, Zhang H-P, Tan Y-J, Chu T, et al. Gut microbiota dysbiosis as an inflammaging condition that regulates obesity-related retinopathy and nephropathy. Front Microbiol (2022) 13:1040846. doi: 10.3389/fmicb.2022.1040846
92. Tang SCW, Yiu WH. Innate immunity in diabetic kidney disease. Nat Rev Nephrol (2020) 16:206–22. doi: 10.1038/s41581-019-0234-4
93. Mertowska P, Mertowski S, Wojnicka J, Korona-Głowniak I, Grywalska E, Błażewicz A, et al. A link between chronic kidney disease and gut microbiota in immunological and nutritional aspects. Nutrients (2021) 13:3637. doi: 10.3390/nu13103637
94. Cerf-Bensussan N, Eberl G. The dialog between microbiota and the immune system: shaping the partners through development and evolution. Semin Immunol (2012) 24:1–2. doi: 10.1016/j.smim.2011.11.007
95. Chi M, Ma K, Wang J, Ding Z, Li Y, Zhu S, et al. The immunomodulatory effect of the gut microbiota in kidney disease. J Immunol Res (2021) 2021:5516035. doi: 10.1155/2021/5516035
96. Linh HT, Iwata Y, Senda Y, Sakai-Takemori Y, Nakade Y, Oshima M, et al. Intestinal bacterial translocation contributes to diabetic kidney disease. J Am Soc Nephrol (2022) 33:1105–19. doi: 10.1681/ASN.2021060843
97. Levy M, Thaiss CA, Zeevi D, Dohnalová L, Zilberman-Schapira G, Mahdi JA, et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell (2015) 163:1428–43. doi: 10.1016/j.cell.2015.10.048
98. Ma N, Ma X. Dietary amino acids and the gut-Microbiome-Immune axis: Physiological metabolism and therapeutic prospects. Compr Rev Food Sci Food Saf (2019) 18:221–42. doi: 10.1111/1541-4337.12401
99. Kim CH, Park J, Kim M. Gut microbiota-derived short-chain fatty acids, T cells, and inflammation. Immune Netw (2014) 14:277–88. doi: 10.4110/in.2014.14.6.277
100. Vallianou NG, Stratigou T, Tsagarakis S. Metformin and gut microbiota: their interactions and their impact on diabetes. Hormones (Athens) (2019) 18:141–4. doi: 10.1007/s42000-019-00093-w
101. Deng L, Yang Y, Xu G. Empagliflozin ameliorates type 2 diabetes mellitus-related diabetic nephropathy via altering the gut microbiota. Biochim Biophys Acta Mol Cell Biol Lipids (2022) 1867:159234. doi: 10.1016/j.bbalip.2022.159234
102. Mishima E, Fukuda S, Kanemitsu Y, Saigusa D, Mukawa C, Asaji K, et al. Canagliflozin reduces plasma uremic toxins and alters the intestinal microbiota composition in a chronic kidney disease mouse model. Am J Physiol Renal Physiol (2018) 315:F824–33. doi: 10.1152/ajprenal.00314.2017
103. Lee DM, Battson ML, Jarrell DK, Hou S, Ecton KE, Weir TL, et al. SGLT2 inhibition via dapagliflozin improves generalized vascular dysfunction and alters the gut microbiota in type 2 diabetic mice. Cardiovasc Diabetol (2018) 17:62. doi: 10.1186/s12933-018-0708-x
104. Singh H, Miyamoto S, Darshi M, Torralba MG, Kwon K, Sharma K, et al. Gut microbial changes in diabetic db/db mice and recovery of microbial diversity upon pirfenidone treatment. Microorganisms (2020) 8:E1347. doi: 10.3390/microorganisms8091347
105. Zhao J, Zhang Q-L, Shen J-H, Wang K, Liu J. Magnesium lithospermate b improves the gut microbiome and bile acid metabolic profiles in a mouse model of diabetic nephropathy. Acta Pharmacol Sin (2019) 40:507–13. doi: 10.1038/s41401-018-0029-3
106. Yang R, Li Y, Mehmood S, Yan C, Huang Y, Cai J, et al. Polysaccharides from armillariella tabescens mycelia ameliorate renal damage in type 2 diabetic mice. Int J Biol Macromol (2020) 162:1682–91. doi: 10.1016/j.ijbiomac.2020.08.006
107. Yang J, Dong H, Wang Y, Jiang Y, Zhang W, Lu Y, et al. Cordyceps cicadae polysaccharides ameliorated renal interstitial fibrosis in diabetic nephropathy rats by repressing inflammation and modulating gut microbiota dysbiosis. Int J Biol Macromol (2020) 163:442–56. doi: 10.1016/j.ijbiomac.2020.06.153
108. Feng Y, Weng H, Ling L, Zeng T, Zhang Y, Chen D, et al. Modulating the gut microbiota and inflammation is involved in the effect of bupleurum polysaccharides against diabetic nephropathy in mice. Int J Biol Macromol (2019) 132:1001–11. doi: 10.1016/j.ijbiomac.2019.03.242
109. Liu J, Gao L-D, Fu B, Yang H-T, Zhang L, Che S-Q, et al. Efficacy and safety of zicuiyin decoction on diabetic kidney disease: A multicenter, randomized controlled trial. Phytomedicine (2022) 100:154079. doi: 10.1016/j.phymed.2022.154079
110. Zhang M, Yang L, Zhu M, Yang B, Yang Y, Jia X, et al. Moutan cortex polysaccharide ameliorates diabetic kidney disease via modulating gut microbiota dynamically in rats. Int J Biol Macromol (2022) 206:849–60. doi: 10.1016/j.ijbiomac.2022.03.077
111. Wei H, Wang L, An Z, Xie H, Liu W, Du Q, et al. QiDiTangShen granules modulated the gut microbiome composition and improved bile acid profiles in a mouse model of diabetic nephropathy. BioMed Pharmacother (2021) 133:111061. doi: 10.1016/j.biopha.2020.111061
112. Chen Q, Ren D, Wu J, Yu H, Chen X, Wang J, et al. Shenyan kangfu tablet alleviates diabetic kidney disease through attenuating inflammation and modulating the gut microbiota. J Nat Med (2021) 75:84–98. doi: 10.1007/s11418-020-01452-3
113. Zhao T, Zhang H, Yin X, Zhao H, Ma L, Yan M, et al. Tangshen formula modulates gut microbiota and reduces gut-derived toxins in diabetic nephropathy rats. BioMed Pharmacother (2020) 129:110325. doi: 10.1016/j.biopha.2020.110325
114. Makki K, Deehan EC, Walter J, Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe (2018) 23:705–15. doi: 10.1016/j.chom.2018.05.012
115. Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol (2019) 16:35–56. doi: 10.1038/s41575-018-0061-2
116. Koppe L, Soulage CO. The impact of dietary nutrient intake on gut microbiota in the progression and complications of chronic kidney disease. Kidney Int (2022) 102:728–39. doi: 10.1016/j.kint.2022.06.025
117. Mattson DL, Dasinger JH, Abais-Battad JM. Gut-Immune-Kidney axis: Influence of dietary protein in salt-sensitive hypertension. Hypertension (2022) 79:2397–408. doi: 10.1161/HYPERTENSIONAHA.122.18556
118. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature (2012) 486:222–7. doi: 10.1038/nature11053
119. Padayachee A, Netzel G, Netzel M, Day L, Zabaras D, Mikkelsen D, et al. Binding of polyphenols to plant cell wall analogues - part 2: Phenolic acids. Food Chem (2012) 135:2287–92. doi: 10.1016/j.foodchem.2012.07.004
120. Mocanu C-A, Simionescu TP, Mocanu AE, Garneata L. Plant-based versus animal-based low protein diets in the management of chronic kidney disease. Nutrients (2021) 13:3721. doi: 10.3390/nu13113721
121. Liu X, Qiu B, Liu W, Zhang Y, Wang X, Li X, et al. The preventive effects of fermented and germinated foxtail millet whole grain on kidney damage in a diabetic mouse model. Front Nutr (2022) 9:940404. doi: 10.3389/fnut.2022.940404
122. Lee H-C, Yu S-C, Lo Y-C, Lin I-H, Tung T-H, Huang S-Y. A high linoleic acid diet exacerbates metabolic responses and gut microbiota dysbiosis in obese rats with diabetes mellitus. Food Funct (2019) 10:786–98. doi: 10.1039/c8fo02423e
123. Hua Q, Han Y, Zhao H, Zhang H, Yan B, Pei S, et al. Punicalagin alleviates renal injury via the gut-kidney axis in high-fat diet-induced diabetic mice. Food Funct (2022) 13:867–79. doi: 10.1039/d1fo03343c
124. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol (2014) 11:506–14. doi: 10.1038/nrgastro.2014.66
125. Swanson KS, Gibson GR, Hutkins R, Reimer RA, Reid G, Verbeke K, et al. The international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat Rev Gastroenterol Hepatol (2020) 17:687–701. doi: 10.1038/s41575-020-0344-2
126. Soleimani A, Zarrati Mojarrad M, Bahmani F, Taghizadeh M, Ramezani M, Tajabadi-Ebrahimi M, et al. Probiotic supplementation in diabetic hemodialysis patients has beneficial metabolic effects. Kidney Int (2017) 91:435–42. doi: 10.1016/j.kint.2016.09.040
127. Liu S, Liu H, Chen L, Liang S-S, Shi K, Meng W, et al. Effect of probiotics on the intestinal microbiota of hemodialysis patients: a randomized trial. Eur J Nutr (2020) 59:3755–66. doi: 10.1007/s00394-020-02207-2
128. Zheng HJ, Guo J, Jia Q, Huang YS, Huang W-J, Zhang W, et al. The effect of probiotic and synbiotic supplementation on biomarkers of inflammation and oxidative stress in diabetic patients: A systematic review and meta-analysis of randomized controlled trials. Pharmacol Res (2019) 142:303–13. doi: 10.1016/j.phrs.2019.02.016
129. Ejtahed HS, Mohtadi-Nia J, Homayouni-Rad A, Niafar M, Asghari-Jafarabadi M, Mofid V. Probiotic yogurt improves antioxidant status in type 2 diabetic patients. Nutrition (2012) 28:539–43. doi: 10.1016/j.nut.2011.08.013
130. Ranganathan N, Friedman EA, Tam P, Rao V, Ranganathan P, Dheer R. Probiotic dietary supplementation in patients with stage 3 and 4 chronic kidney disease: a 6-month pilot scale trial in Canada. Curr Med Res Opin (2009) 25:1919–30. doi: 10.1185/03007990903069249
131. Miranda Alatriste PV, Urbina Arronte R, Gómez Espinosa CO, de los Ángeles Espinosa Cuevas M. Effect of probiotics on human blood urea levels in patients with chronic renal failure. Nutr Hosp (2014) 29:582–90. doi: 10.3305/nh.2014.29.3.7179
132. Bohlouli J, Namjoo I, Borzoo-Isfahani M, Hojjati Kermani MA, Balouch Zehi Z, Moravejolahkami AR. Effect of probiotics on oxidative stress and inflammatory status in diabetic nephropathy: A systematic review and meta-analysis of clinical trials. Heliyon (2021) 7:e05925. doi: 10.1016/j.heliyon.2021.e05925
133. Moravejolahkami AR, Hojjati Kermani MA, Balouch Zehi Z, Mirenayat SMS, Mansourian M. The effect of probiotics on lipid profile & anthropometric indices in diabetic nephropathy; a systematic review and meta-analysis of clinical trials. J Diabetes Metab Disord (2021) 20:893–904. doi: 10.1007/s40200-021-00765-8
134. Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, et al. Expert consensus document: The international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol (2017) 14:491–502. doi: 10.1038/nrgastro.2017.75
135. Zoumpopoulou G, Pot B, Tsakalidou E, Papadimitriou K. Dairy probiotics: Beyond the role of promoting gut and immune health. Int Dairy J (2017) 67:46–60. doi: 10.1016/j.idairyj.2016.09.010
136. Snelson M, de Pasquale C, Ekinci EI, Coughlan MT. Gut microbiome, prebiotics, intestinal permeability and diabetes complications. Best Pract Res Clin Endocrinol Metab (2021) 35:101507. doi: 10.1016/j.beem.2021.101507
137. Pengrattanachot N, Thongnak L, Lungkaphin A. The impact of prebiotic fructooligosaccharides on gut dysbiosis and inflammation in obesity and diabetes related kidney disease. Food Funct (2022) 13:5925–45. doi: 10.1039/d1fo04428a
138. Luo L, Luo J, Cai Y, Fu M, Li W, Shi L, et al. Inulin-type fructans change the gut microbiota and prevent the development of diabetic nephropathy. Pharmacol Res (2022) 183:106367. doi: 10.1016/j.phrs.2022.106367
139. Zaky A, Glastras SJ, Wong MYW, Pollock CA, Saad S. The role of the gut microbiome in diabetes and obesity-related kidney disease. Int J Mol Sci (2021) 22:9641. doi: 10.3390/ijms22179641
140. Zhang Q, Yu H, Xiao X, Hu L, Xin F, Yu X. Inulin-type fructan improves diabetic phenotype and gut microbiota profiles in rats. PeerJ (2018) 6:e4446. doi: 10.7717/peerj.4446
141. Snelson M, Kellow NJ, Coughlan MT. Modulation of the gut microbiota by resistant starch as a treatment of chronic kidney diseases: Evidence of efficacy and mechanistic insights. Adv Nutr (2019) 10:303–20. doi: 10.1093/advances/nmy068
142. Salminen S, Collado MC, Endo A, Hill C, Lebeer S, Quigley EMM, et al. The international scientific association of probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat Rev Gastroenterol Hepatol (2021) 18:649–67. doi: 10.1038/s41575-021-00440-6
143. Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: Pathophysiological basis for therapy. J Hepatol (2020) 72:558–77. doi: 10.1016/j.jhep.2019.10.003
144. Gargouri M, Hamed H, Akrouti A, Dauvergne X, Magné C, El Feki A. Effects of spirulina platensis on lipid peroxidation, antioxidant defenses, and tissue damage in kidney of alloxan-induced diabetic rats. Appl Physiol Nutr Metab (2018) 43:345–54. doi: 10.1139/apnm-2017-0461
145. Favero C, Giordano L, Mihaila SM, Masereeuw R, Ortiz A, Sanchez-Niño MD. Postbiotics and kidney disease. Toxins (Basel) (2022) 14:623. doi: 10.3390/toxins14090623
146. Smith MB, Kelly C, Alm EJ. Policy: How to regulate faecal transplants. Nature (2014) 506:290–1. doi: 10.1038/506290a
147. Khoruts A, Sadowsky MJ. Understanding the mechanisms of faecal microbiota transplantation. Nat Rev Gastroenterol Hepatol (2016) 13:508–16. doi: 10.1038/nrgastro.2016.98
148. Hamilton MJ, Weingarden AR, Sadowsky MJ, Khoruts A. Standardized frozen preparation for transplantation of fecal microbiota for recurrent clostridium difficile infection. Am J Gastroenterol (2012) 107:761–7. doi: 10.1038/ajg.2011.482
149. Youngster I, Russell GH, Pindar C, Ziv-Baran T, Sauk J, Hohmann EL. Oral, capsulized, frozen fecal microbiota transplantation for relapsing clostridium difficile infection. JAMA (2014) 312:1772–8. doi: 10.1001/jama.2014.13875
150. Smits LP, Bouter KEC, de Vos WM, Borody TJ, Nieuwdorp M. Therapeutic potential of fecal microbiota transplantation. Gastroenterology (2013) 145:946–53. doi: 10.1053/j.gastro.2013.08.058
151. Bastos RMC, Simplício-Filho A, Sávio-Silva C, Oliveira LFV, Cruz GNF, Sousa EH, et al. Fecal microbiota transplant in a pre-clinical model of type 2 diabetes mellitus, obesity and diabetic kidney disease. Int J Mol Sci (2022) 23:3842. doi: 10.3390/ijms23073842
152. Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JFWM, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology (2012) 143:913–916.e7. doi: 10.1053/j.gastro.2012.06.031
153. Smits LP, Kootte RS, Levin E, Prodan A, Fuentes S, Zoetendal EG, et al. Effect of vegan fecal microbiota transplantation on carnitine- and choline-derived trimethylamine-N-Oxide production and vascular inflammation in patients with metabolic syndrome. J Am Heart Assoc (2018) 7:e008342. doi: 10.1161/JAHA.117.008342
Keywords: gut microbiota, metabolite, diabetic kidney disease, immunity, therapy
Citation: Mao Z-H, Gao Z-X, Liu D-W, Liu Z-S and Wu P (2023) Gut microbiota and its metabolites – molecular mechanisms and management strategies in diabetic kidney disease. Front. Immunol. 14:1124704. doi: 10.3389/fimmu.2023.1124704
Received: 15 December 2022; Accepted: 06 January 2023;
Published: 19 January 2023.
Edited by:
Zhe-Sheng Chen, St. John’s University, United StatesReviewed by:
Jie Luo, Nanchang University, ChinaNaheed Mojgani, Razi Vaccine and Serum Research Institute, Iran
Copyright © 2023 Mao, Gao, Liu, Liu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Wu, d3VwZW5nY2dAenp1LmVkdS5jbg==; Zhang-Suo Liu, emhhbmdzdW9saXVAenp1LmVkdS5jbg==
 Zi-Hui Mao
Zi-Hui Mao Zhong-Xiuzi Gao1,2,3,4
Zhong-Xiuzi Gao1,2,3,4 Dong-Wei Liu
Dong-Wei Liu Zhang-Suo Liu
Zhang-Suo Liu Peng Wu
Peng Wu