- 1Department of Gastroenterology, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou Medical University, Guangzhou, China
- 2Department of Neurology, Peking University Third Hospital, Beijing, China
- 3Deartment of Thyroid Surgery, First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 4Institute of Therapeutic Innovations and Outcomes, College of Pharmacy, The Ohio State University, Columbus, OH, United States
Background: To assess the causal role of lipid traits and lipid-lowering agents in inflammatory bowel disease (IBD).
Methods: Univariable mendelian randomization (MR) and multivariable MR (MVMR) analyses were conducted to evaluate the causal association between low-density lipoprotein cholesterol (LDL-C), triglycerides (TG), high-density lipoprotein cholesterol (HDL-C) and IBD. Drug-targeted MR analyzed the effects of lipid-lowering drugs on IBD, and network MR was used to analyze potential mediation effects.
Results: The levels of HDL-C had an inverse relationship with the risk of Crohn’s disease (CD, OR: 0.85, 95% CI: 0.73-0.98, P = 0.024). In MVMR, the inverse relationships were found in all three outcomes. Drug-targeted MR analyses showed that with one-SD LDL-C decrease predicted by variants at or near proprotein convertase subtilisin/kexin type 9 (PCSK9), the OR values of people diagnosed with IBD, ulcerative colitis (UC) and CD were 1.75 (95%CI: 1.13-2.69, P = 0.011), 2.1 (95%CI: 1.28-3.42, P = 0.003) and 2.24 (95%CI: 1.11-4.5, P = 0.024), respectively. With one-SD LDL-C decrease predicted by variants at or near cholesteryl ester transfer protein (CETP), the OR value of people diagnosed with CD was 0.12 (95%CI: 0.03-0.51, P = 0.004). Network-MR showed that HDL-C mediated the causal pathway from variants at or near CETP to CD.
Conclusion: Our study suggested a causal association between HDL-C and IBD, UC and CD. Genetically proxied inhibition of PCSK9 increased the risk of IBD, UC and CD, while inhibition of CETP decreased the risk of CD. Further studies are needed to clarify the long-term effect of lipid-lowering drugs on the gastrointestinal disorders.
1 Introduction
Inflammatory bowel disease (IBD) is chronic intestinal inflammation disease with an incidence of over 0.3%. It includes ulcerative colitis (UC) and Crohn’s disease (CD), causing a significant disease burden (1). Patients with IBD often have high levels of low-density lipoprotein cholesterol (LDL-C), triglycerides (TG), and low level of high-density lipoprotein cholesterol (HDL-C) (2, (3). Dyslipidemia may also contribute to the development of IBD. Koutroumpakis et al. found that persistent dyslipidemia was associated with disease activity in patients with IBD (3). Lower HDL-C was associated with an increased risk of poor outcomes, such as surgery and tumors in IBD patients (4). Additionally, evidence suggests that lower HDL-C in childhood may be associated with the subsequent diagnosis of IBD (5).
In recent years, some researchers have recommended early evaluation and intervention of lipid levels to prevent new-onset IBD and improve the prognosis in patients with IBD. Statins, a widely used medication to lower LDL-C, have been found to have a protective effect against new-onset IBD, CD, and UC (6). Similarly, a population-based case-control study in Sweden showed that statins were associated with a lower risk of CD (7). In addition, the use of statins in UC patients was found to be associated with a reduction in steroid hormone dosage (8). Furthermore, transcriptomic analysis found that atorvastatin had the highest negative correlation with the UC gene signature, suggesting that statins may be potential therapies for IBD (9). However, the therapeutic effect of statins on IBD is controversial (10). Khalil et al. found that statins did not prevent new-onset IBD (11). Dhamija et al. reported that statin therapy was not associated with beneficial effects in patients with UC (10).
The contradictory findings in these studies highlight the need for further investigation into the potential role of statins in IBD. However, observational studies are limited by inherent constraints, including residual confounding and reverse causality (12). Additionally, lipid-lowering drugs have extensive immunomodulatory effects (13). which raises the question of whether their effect on IBD is mediated through lipid lowering or immunomodulatory mechanisms. Therefore, it is essential to elucidate the underlying mechanisms of the potential therapeutic effect of statins in IBD.
Mendelian randomization (MR) is a powerful tool for assessing the causal effects of exposure factors on outcomes. It utilizes genetic variation randomly allocated at conception as instrumental variables (IVs), allowing for the evaluation of causality in a way that is less susceptible to confounding factors (12). Univariate MR requires that the instrumental variable satisfies three assumptions: first, it must be associated with the exposure; second, it must be independent of the outcome given the exposure; and third, it must be independent of all known confounders (14). Multivariate MR (MVMR) is an extension of MR that leverages genetic variation associated with multiple potentially relevant exposures to estimate the direct effect of each exposure on a single outcome (14). Drug-target MR analysis uses the genetic variants within or near the gene that encodes the targeted protein as IVs to predict efficacy (15). In this study, we applied the latest genome-wide association study (GWAS) from UK Biobank on lipids as IVs to study the relationship between lipids and IBD. Additionally, we employed drug-target MR analysis to investigate the effect of lipid-lowering drugs on IBD.
2 Materials and methods
2.1 GWAS data source
The data used in this study on IBD patients were obtained from the FinnGen study conducted in 2021 (16). The study included patients with UC, CD, and indeterminate colitis, with UC and CD diagnosed by ICD codes. Information of participants, genotype platforms are available at the FinnGen website [https://www.finngen.fi/en/]. The dataset used comprised of 5,673 IBD patients and 213,119 control patients, including 4,320 patients with UC and 210,300 controls, as well as 2,056 patients with CD and 210,300 controls To minimize bias resulting from racial differences, the SNPs identified as instrumental variables (IVs) associated with lipid traits were obtained from the UK Biobank (17), and only SNPs from studies based on European ancestry were selected.
2.2 Instrumental variable selection
In the univariable MR analyses, we applied rigorous quality control procedures to identify independent, eligible, and genome-wide significant SNPs (linkage disequilibrium, LD clumping r2 threshold = 0.001, window size = 10 Mb and p < 5×10-8) associated with each trait, including HDL-C, LDL-C and TG. For MVMR analyses, SNPs were clumped with respect to the lowest P-value corresponding to each exposure in a multivariable model using a 1-Mb window and pairwise LD R2 < 0.001. F-statistic was used to access the strength of instruments in univariable MR, and conditional F-statistic to access the strength in MVMR. The value of a statistic less than 10 were excluded to reduce weak instrument bias. We obtained the lipid-lowering drug targets related IVs from the Global Lipid Genetics Consortium (GLGC) for Drug-target MR analyses.
The IVs for lipid-lowering drug targets were from a GWAS of LDL-C conducted by the Global Lipid Genetics Consortium (GLGC) (linkage disequilibrium, LD r2 ≤ 0.2, physical distance = 250 kb, window size =100 kb and p < 5×10-8) (18). The targets included 3-Hydroxy-3-Methylglutaryl-CoA Reductase (HMGCR), NPC1 Like Intracellular Cholesterol Transporter 1(NPC1L1), Proprotein convertase subtilisin/kexin type 9 (PCSK9), Apolipoprotein B (APOB), Cholesteryl ester transfer protein (CETP), with each gene including 5,4,11,15,7 SNP that were used as IVs. Each lipid-lowering drug target corresponds to statins, ezetimibe, evolocumab & alirocumab, anacetrapib and mipomersen, separately. For each SNP, the effect allele is significantly associated with lower concentrations of LDL-C. We ensured that there were no strong correlations between IVs of each trait (R2 < 0.4 or R2 < 0.3).
2.3 MR analyses
Figure 1 illustrates the different methods we employed to conduct univariable and multivariable Mendelian randomization (MR) analyses, including simple median, weighted median, MR-Egger, MR robust adjusted profile score (MR.RAPS) (19) and inverse-variance weighted (IVW) method. Fixed-effect model IVW, which offers high efficiency and statistical power, was the primary method used (20). To assess heterogeneity and pleiotropy, we used Cochran’s Q statistic and the MR-Egger test (intercept). In cases where significant heterogeneity was present, we utilized the multiplicative random effects IVW method. Additionally, we employed the MR-radial method, robust regression, and outlying variants penalized to investigate the impact of outliers on the outcome (21, (22). In instances where horizontal pleiotropy was detected, we employed the MR-Egger test as the primary analysis method, while using MR-PRESSO to further correct for pleiotropy (23). For multivariable MR (MVMR) analyses, we utilized the multivariable IVW, multivariable MR-Egger, multivariable median-based, and multivariable MR-Lasso methods. We used the multivariable MR-Egger method for pleiotropy testing and Cochran’s Q statistic for heterogeneity testing. All MR analyses adhered to the STROBE-MR Statement (24).
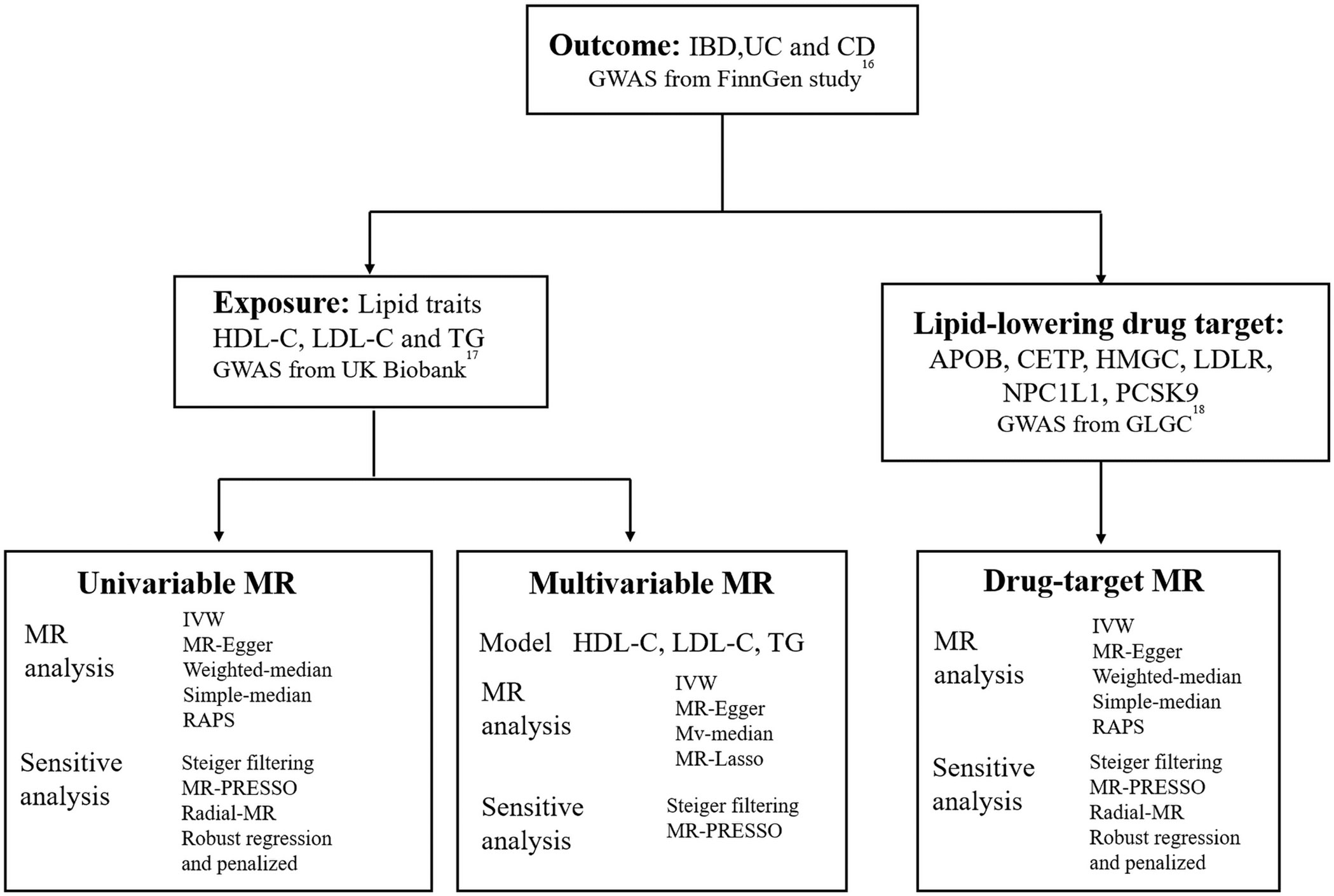
Figure 1 Overview of the study design. GWAS, genome-wide association study; IBD, inflammatory bowel disease; UC, ulcerative colitis; CD, Crohn’s disease; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TG, triglycerides; IVW, inverse-variance weighted; RAPS, MR robust adjusted profile score; HMGCR, 3-Hydroxy-3-Methylglutaryl-CoA Reductase; NPC1L1, NPC1 Like Intracellular Cholesterol Transporter 1; PCSK9, Proprotein convertase subtilisin/kexin type 9; APOB, Apolipoprotein B; CETP, Cholesteryl ester transfer protein; GLGC, Global Lipid Genetics Consortium; MR, Mendelian randomization.
To investigate the causality of lipid-lowering drug targets in IBD,UC and CD, as well as the potential mediators involved, we performed network MR by conducting at least three two-sample MR tests among lipid-lowering drug targets, IBD/UC/CD, and potential mediators (25).
We used publicly available genome-wide association study (GWAS) data. Relevant informed consent and ethical approval had already been obtained, so additional ethical approval was not required for this study. We conducted all statistical analyses using the “TwoSample MR” (version 0.5.6) and “MendelianRandomization” (version 0.5.1) packages in R software (version 4.1.1), with a statistical significance threshold of P value < 0.05.
3 Results
We identified 525, 218, and 435 SNPs as instrumental variables (IVs) for HDL-C, LDL-C, and TG, respectively (Supplementary Table 1). The median F statistic of these IVs was 47.49 (with a quartile range of 35.56-84.49), indicating that our data were not susceptible to weak instrument bias.
3.1 Causal effect of lipids on IBD, UC and CD
Significant heterogeneity was observed between the independent variables (IVs) and the outcomes of IBD, UC, and CD, as indicated by Cochran’s Q test (P < 0.05) (Supplementary Table 2, Supplemental Figures 1–3).
Therefore, the multiplicative random effects model, known as the inverse variance weighted (IVW) method, was used as the main model. Univariate MR analyses revealed a significant association between HDL-C and CD, with an OR of 0.85 (95% CI: 0.73-0.98, P = 0.024) for a one-standard deviation increase in HDL-C, as shown in Figure 2.
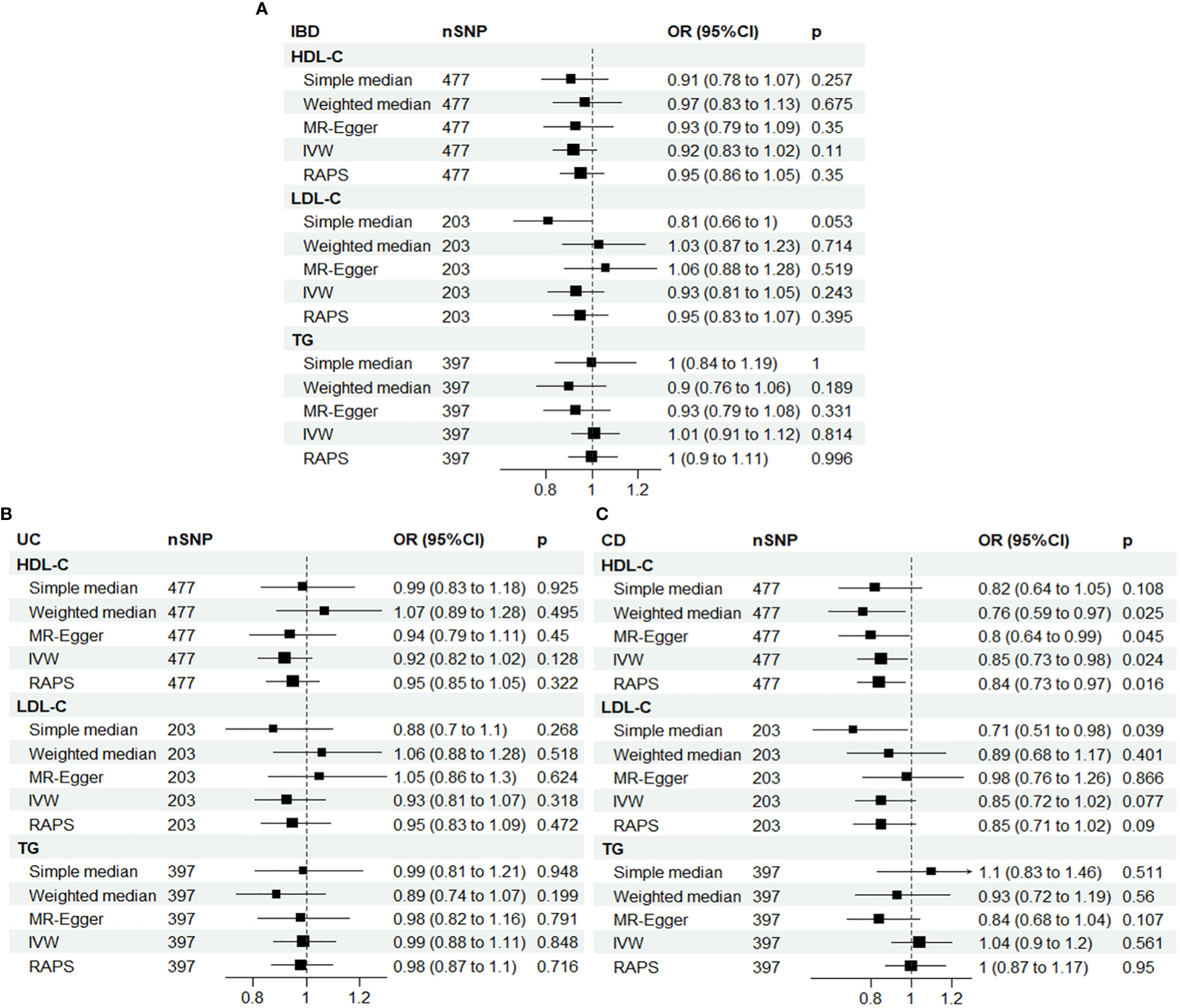
Figure 2 Univariable Mendelian randomization results using different methods. (A) IBD; (B) UC; (C) CD. IBD, inflammatory bowel disease; UC, ulcerative colitis; CD, Crohn’s disease; SNP N, number of single nucleotide polymorphisms; OR, odds ratio; CI, confidence interval; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TG, triglycerides; IVW, inverse-variance weighted; RAPS, MR robust adjusted profile score.
To ensure the robustness of our results, we applied MR-radial method and robust regression and outlying variants (MR-RAPS) penalized analyses, which also showed a significant inverse association between HDL-C and CD, with ORs of 0.84 (95% CI: 0.73-0.97, P = 0.019) and 0.83 (95% CI: 0.72-0.96, P = 0.012), respectively, for a one-standard deviation increase in HDL-C. These findings are presented in Supplementary Tables 3, 4 and Supplemental Figures 4–7.
Although some horizontal pleiotropy was detected in the MR analyses of HDL-C and IBD, and TG and CD, as shown by the MR-Egger test, the results of this test and the IVW analysis were consistent, indicating minimal impact of pleiotropy on the results. Furthermore, further analysis using the MR-PRESSO package yielded similar results, confirming the robustness of our findings (Supplementary Table 5).
3.2 Direct causal effect of lipids on IBD, UC and CD using multivariable MR
The present study used a multivariable Mendelian randomization (MVMR) approach, which incorporated three variables (HDL-C, LDL-C, and TG) to evaluate potential weak instrumental variable bias. The conditional F-statistic values corresponding to these variables were 68.7, 35.46, and 62.2, respectively, indicating no significant bias. Our MVMR analyses further revealed that higher HDL-C levels were inversely associated with the risk of IBD (OR: 0.87, 95% CI: 0.78-0.98, P = 0.026), UC (OR: 0.87, 95% CI: 0.77-0.99, P = 0.04), and CD (OR: 0.82, 95% CI: 0.69-0.97, P = 0.02), as shown in Figure 3A and Supplementary Table 6. Notably, pleiotropy test did not detect significant pleiotropy, as summarized in Supplementary Table 7.
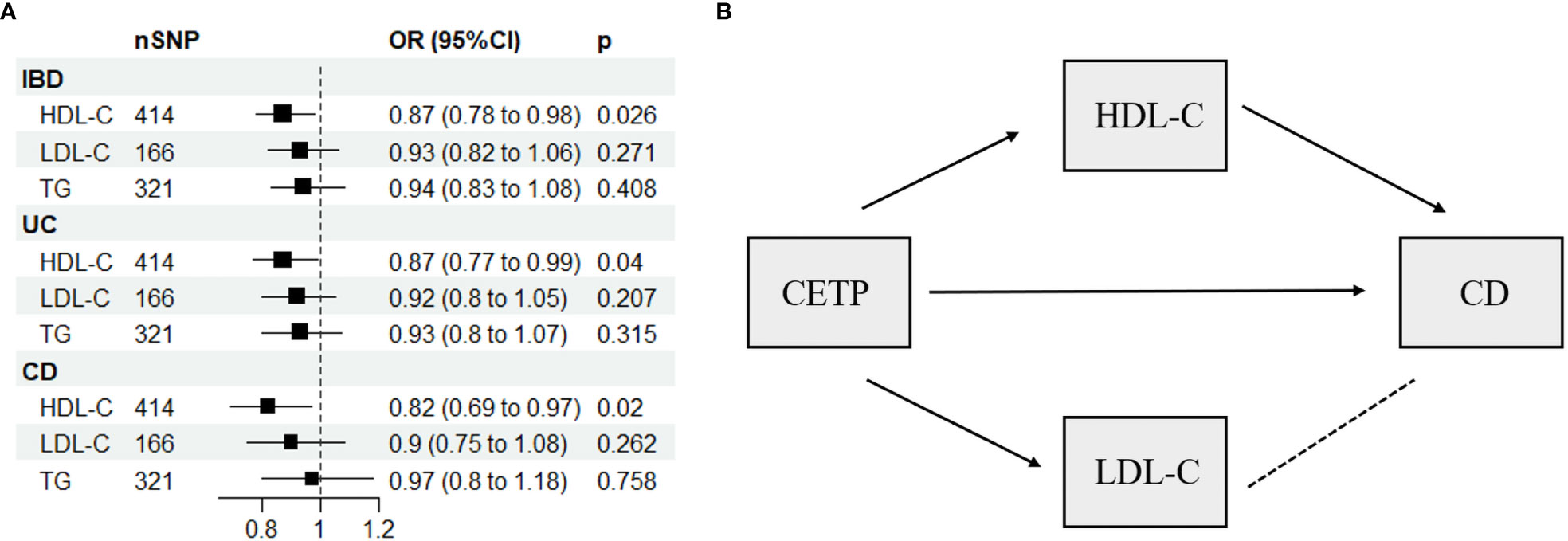
Figure 3 (A) Multivariable Mendelian randomization results using the inverse-variance weighted method. (B) Network-MR diagram of CETP and CD. IBD, inflammatory bowel disease; UC, ulcerative colitis; CD, Crohn’s disease; SNP N, number of single nucleotide polymorphisms; OR, odds ratio; CI, confidence interval; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TG, triglycerides; CETP, Cholesteryl ester transfer protein, →, causal association; – – – –, no causal association.
3.3 Gene-specific analyses for drug proxy variants on IBD, UC and CD
Supplementary Table 8 displays the IVs related to lipid-lowering drug targets. In Figure 4, drug-targeted MR analyses revealed that a one-SD decrease in LDL-C predicted by variants at or near PCSK9 was associated with higher ORs of being diagnosed with IBD, UC, and CD with values of 1.75 (95%CI: 1.13-2.69, P = 0.011), 2.1 (95%CI: 1.28-3.42, P = 0.003), and 2.24 (95%CI: 1.11-4.5, P = 0.024), respectively. Conversely, a one-SD decrease in LDL-C predicted by variants at or near CETP was associated with a lower OR of CD with a value of 0.12 (95%CI: 0.03-0.51, P = 0.004), while no such association was observed for IBD or UC patients. Figure 3B and Supplementary Table 9 demonstrated that HDL-C mediated the causal pathway from variants at or near CETP to CD in network-MR. Additionally, the heterogeneity analysis and pleiotropy test revealed no significant heterogeneity or pleiotropy (Supplementary Table 10). Furthermore, the leave-one-out analysis demonstrated that the drug-targeted MR results were robust, even after excluding a single single-nucleotide polymorphism (Supplemental Figure 8).
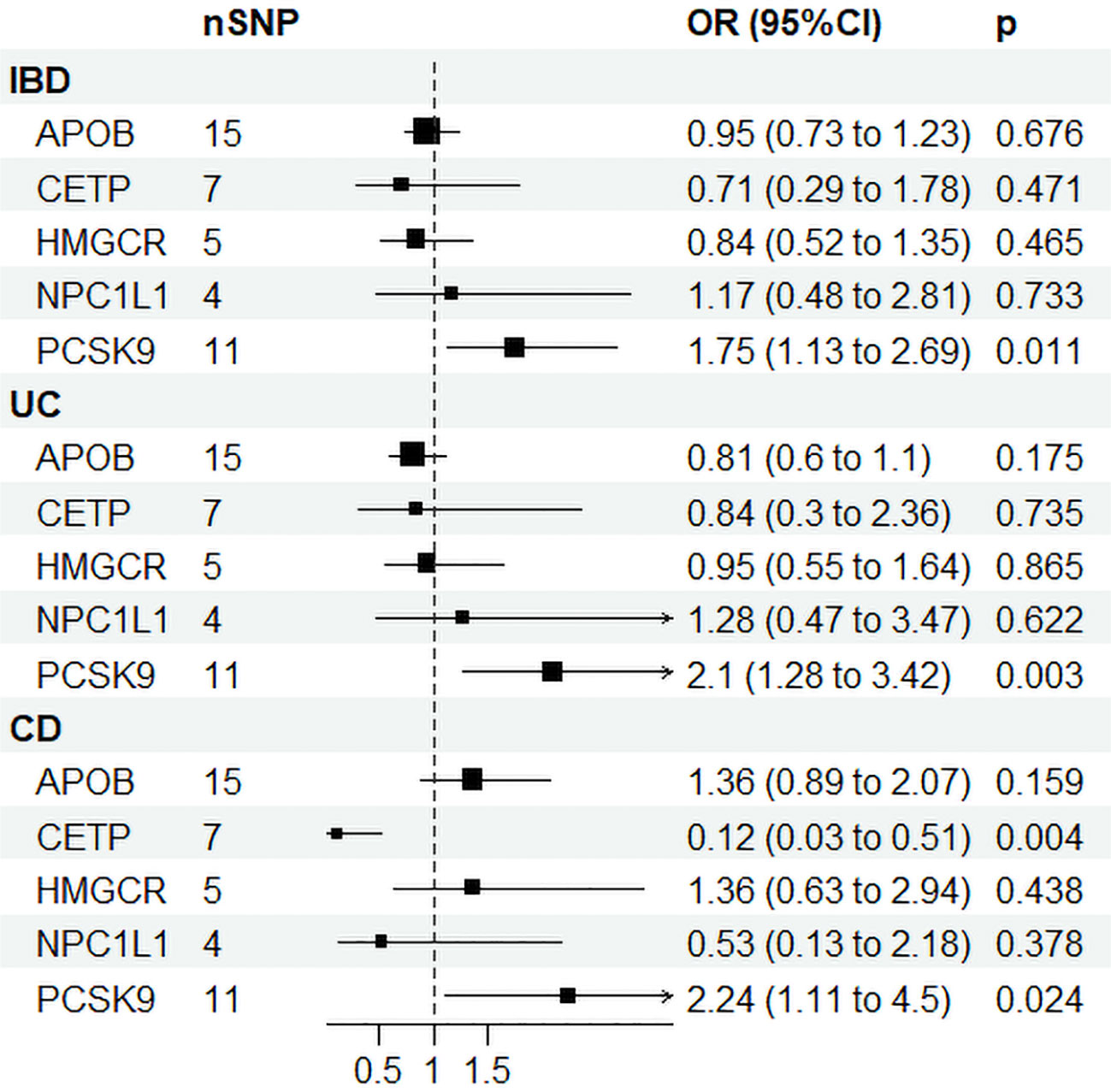
Figure 4 Drug target Mendelian randomization results. IBD, inflammatory bowel disease; UC, ulcerative colitis; CD, Crohn’s disease; SNP N, number of single nucleotide polymorphisms; OR, odds ratio; CI, confidence interval; HMGCR, 3-Hydroxy-3-Methylglutaryl-CoA Reductase; NPC1L1, NPC1 Like Intracellular Cholesterol Transporter 1; PCSK9, Proprotein convertase subtilisin/kexin type 9; APOB, Apolipoprotein B; CETP, Cholesteryl ester transfer protein.
4 Discussion
To the best of our knowledge, this study is the first to utilize MR analysis to investigate the relationship between lipids, lipid-lowering drugs, and IBD. Our findings suggest a causal association between elevated levels of HDL-C and a reduced risk of all three IBD diseases, including UC and CD. Additionally, the drug-targeted MR analysis indicates that inhibiting PCSK9 leads to an increased risk of all three diseases, while suppressing CETP reduces the risk of CD.
Our findings are consistent with previous studies that observed significant reductions in HDL-C levels in IBD patients (3), UC patients (26) and CD patients (27), indicating that HDL-C may play a role in the development of IBD. However, large prospective studies that evaluate the causal relationship between HDL-C and IBD are lacking. HDL-C is widely recognized as a complex and pleiotropic anti-inflammatory particle that plays a critical role in protecting the cardiovascular system by promoting cholesterol efflux in vascular endothelial cells, stimulating prostacyclin synthesis, and inhibiting platelet-activating factor synthesis (28). Whether HDL-C plays a protective role in the pathogenesis of IBD by inhibiting inflammation requires further investigation.
CETP primarily binds to HDL in the circulatory system, transporting cholesterol esters from HDL to LDL and VLDL (29). Our study found that inhibition of CETP could reduce the risk of CD. However, since variants in or near CETP are related to both HDL-C and LDL-C levels, it is challenging to differentiate whether the protective effect is due to changes in HDL-C or LDL-C. To address this issue, we conducted a network-MR analysis that indicated HDL-C, rather than LDL-C, mediated the causal relationship between CETP and CD. We did not find evidence of a mediating effect of CETP inhibitors on HDL-C in IBD and UC. This may be related to the complex metabolic pathways of HDL-C, which involve multiple components, including ATP-binding cassette transporter G1, ATP-binding cassette transporter A1, and lecithin cholesterol acyltransferase, in addition to CETP (30). Our study at least confirms that CETP does not regulate HDL-C in IBD and UC, suggesting the involvement of other molecules in its regulation that require further investigation.
In a previous MR study, no causal relationship was found between CETP and PCSK9 inhibitors and IBD, UC, and CD (31). The discrepancy in results may be due to the different IVs used in our study, with our study using SNPs associated with LDL-C levels. Our study found a causal relationship between PCSK9 inhibitors and IBD, UC, and CD. Interestingly, our study did not find a causal relationship between LDL-C and the three diseases, indicating that PCSK9 affects IBD, UC, and CD through means other than by reducing LDL-C. Although PCSK9 was initially linked to elevated LDL-C, recent studies have found its involvement in atherosclerosis, viral infections, immune activation, and tumor progression (32, 33). While PCSK9 is less expressed in the gastrointestinal tract, serum PCSK9 levels were significantly higher in UC patients and associated with disease activity (34). The PCSK9 inhibitor could inhibit the activity of TLR4/NF-κB to ameliorate colitis induced by 2,4,6-trinitrobenzene sulfonic acid (35). These findings appear to contradict our study, but further research is needed to fully understand the role of PCSK9 in the gastrointestinal tract. Studies using PCSK9 knockout mice have provided insight into interpreting our results. For instance, PCSK9-/-mice had more severe fibrosing steatohepatitis and were more prone to hepatocellular carcinoma (36). More severe oxidative stress was found in PCSK99-/-mice (37). In these studies, the variants of PCSK9 variants involve a lifelong process that starts with embryonic development, which is different from the short-term effect of clinical drug use. Therefore, future research is necessary to explore the role of PCSK9 in the gastrointestinal tract fully. Finally, our study did not establish a direct causal relationship between the HMGCR inhibitor and the three outcomes, suggesting that there is no direct causality between statins and IBD, UC, and CD.
Our study offers several notable strengths. Firstly, we utilized the largest lipid-related GWAS database to date, providing a robust theoretical basis for future treatment of IBD, UC, and CD by demonstrating the causal effect of HDL-C. Secondly, we employed drug target MR and network MR analysis to demonstrate the mediation effect of HDL-C in CETP on CD, and the causal effect of the PCSK9 inhibitor on IBD, UC, and CD. These findings highlight the need for further research into the potential gastrointestinal side effects of long-term use of PCSK9 inhibitors. Finally, we conducted detailed heterogeneity and pleiotropy tests, ensuring the reliability of our results. However, our study also has some limitations. The FinnGen study included only individuals of European ancestry, limiting the generalizability of our conclusions to other populations. Additionally, our drug target MR analysis reflects the impact of lifelong application of lipid-lowering drugs on outcomes, which may not accurately reflect the relationship between short-term use of these drugs and related outcomes.
5 Conclusions
In conclusion, this study provided strong evidence for the causal impact of HDL-C on IBD, UC, and CD. Furthermore, we have identified HDL-C as a mediator in the causal pathway linking CETP inhibitors to CD. Genetically proxied inhibition of PCSK9 increased the risk of IBD, UC and CD, while inhibition of CETP decreased the risk of CD. However, further research is needed to investigate the potential role of PCSK9 inhibitors in gastrointestinal disorders.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by The ethics committee of The First Affiliated Hospital of Guangzhou Medical University. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
HT, ZY and YD planned and designed the study, analyzed and interpreted the data, and wrote the manuscript. LL and XC analyzed the data and made the tables presented in the manuscript. LP and YD, as corresponding authors, made important advice in the study design, supervised and coordinated the study conduct process, revised the manuscript and tables, as well as reviewed and verified all the data, methods, and results. All authors contributed to the article and approved the submitted version.
Funding
This study was supported in part by a grant from the Guangdong Natural Science Foundation(2018A030313970) and (2021A1515110491) managed by LP and HT, National Natural Science Foundation of the P.R. China (81900521) and Programs for Science and Technology Development of Henan province (SBJ202003033) managed by YD. The funders of the study had no role in the study design, data collection, analysis, interpretation, writing of the report, or decision to submit for publication.
Acknowledgments
We want to acknowledge the participants and investigators of the original studies that provided the GWAS data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1160312/full#supplementary-material
References
1. Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet (2017) 390:2769–78. doi: 10.1016/s0140-6736(17)32448-0
2. Sappati Biyyani RS, Putka BS, Mullen KD. Dyslipidemia and lipoprotein profiles in patients with inflammatory bowel disease. J Clin Lipidol (2010) 4:478–82. doi: 10.1016/j.jacl.2010.08.021
3. Koutroumpakis E, Ramos-Rivers C, Regueiro M, Hashash JG, Barrie A, Swoger J, et al. Association between long-term lipid profiles and disease severity in a Large cohort of patients with inflammatory bowel disease. Dig Dis Sci (2016) 61:865–71. doi: 10.1007/s10620-015-3932-1
4. Liu Z, Tang H, Liang H, Bai X, Zhang H, Yang H, et al. Dyslipidaemia is associated with severe disease activity and poor prognosis in ulcerative colitis: a retrospective cohort study in China. Nutrients (2022) 14. doi: 10.3390/nu14153040
5. Voutilainen M, Hutri-Kähönen N, Tossavainen P, Sipponen T, Pitkänen N, Laitinen T, et al. Low childhood high density lipoprotein cholesterol levels and subsequent risk for chronic inflammatory bowel disease. Dig Liver Dis (2018) 50:348–52. doi: 10.1016/j.dld.2018.01.121
6. Ungaro R, Chang HL, Côté-Daigneault J, Mehandru S, Atreja A, Colombel JF, et al. Statins associated with decreased risk of new onset inflammatory bowel disease. Am J Gastroenterol (2016) 111:1416–23. doi: 10.1038/ajg.2016.233
7. Lochhead P, Khalili H, Sachs MC, Chan AT, Olén O, Ludvigsson JF. Association between statin use and inflammatory bowel diseases: results from a Swedish, nationwide, population-based case-control study. J Crohns Colitis (2021) 15:757–65. doi: 10.1093/ecco-jcc/jjaa235
8. Crockett SD, Hansen RA, Stürmer T, Schectman R, Darter J, Sandler RS, et al. Statins are associated with reduced use of steroids in inflammatory bowel disease: a retrospective cohort study. Inflammation Bowel Dis (2012) 18:1048–56. doi: 10.1002/ibd.21822
9. Bai L, Scott MKD, Steinberg E, Kalesinskas L, Habtezion A, Shah NH, et al. Computational drug repositioning of atorvastatin for ulcerative colitis. J Am Med Inform Assoc (2021) 28:2325–35. doi: 10.1093/jamia/ocab165
10. Dhamija P, Hota D, Kochhar R, Sachdev A, Chakrabarti A. Randomized clinical trial: atorvastatin versus placebo in patients with acute exacerbation of mild to moderate ulcerative colitis. Indian J Gastroenterol (2014) 33:151–6. doi: 10.1007/s12664-013-0420-4
11. Khalil D, Boktor M, Mortensen EM, Frei CR, Mansi I. Comparison of frequency of inflammatory bowel disease and noninfectious gastroenteritis among statin users versus nonusers. Am J Cardiol (2015) 115:1396–401. doi: 10.1016/j.amjcard.2015.02.035
12. Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. Jama (2017) 318:1925–6. doi: 10.1001/jama.2017.17219
13. Côté-Daigneault J, Mehandru S, Ungaro R, Atreja A, Colombel JF. Potential immunomodulatory effects of statins in inflammatory bowel disease. Inflammation Bowel Dis (2016) 22:724–32. doi: 10.1097/mib.0000000000000640
14. Sanderson E. Multivariable mendelian randomization and mediation. Cold Spring Harb Perspect Med (2021) 11. doi: 10.1101/cshperspect.a038984
15. Schmidt AF, Finan C, Gordillo-Marañón M, Asselbergs FW, Freitag DF, Patel RS, et al. Genetic drug target validation using mendelian randomisation. Nat Commun (2020) 11:3255. doi: 10.1038/s41467-020-16969-0
16. Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner K, et al. FinnGen: unique genetic insights from combining isolated population and national health register data. medRxiv (2022).
17. Richardson TG, Sanderson E, Palmer TM, Ala-Korpela M, Ference BA, Davey Smith G, et al. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: a multivariable mendelian randomisation analysis. PloS Med (2020) 17:e1003062. doi: 10.1371/journal.pmed.1003062
18. Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet (2013) 45:1274–83. doi: 10.1038/ng.2797
19. Zhao Q, Chen Y, Wang J, Small DS. Powerful three-sample genome-wide design and robust statistical inference in summary-data mendelian randomization. Int J Epidemiol (2019) 48:1478–92. doi: 10.1093/ije/dyz142
20. Burgess S, Davey Smith G, Davies NM, Dudbridge F, Gill D, Glymour MM, et al. Guidelines for performing mendelian randomization investigations. Wellcome Open Res (2019) 4:186. doi: 10.12688/wellcomeopenres.15555.2
21. Burgess S, Bowden J, Fall T, Dudbridge F, Gill D, Glymour MM, et al. Sensitivity analyses for robust causal inference from mendelian randomization analyses with multiple genetic variants. Epidemiology (2017) 28:30–42. doi: 10.1097/ede.0000000000000559
22. Bowden J, Spiller W, Del Greco MF, Sheehan N, Thompson J, Minelli C, et al. Improving the visualization, interpretation and analysis of two-sample summary data mendelian randomization via the radial plot and radial regression. Int J Epidemiol (2018) 47:1264–78. doi: 10.1093/ije/dyy101
23. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat Genet (2018) 50:693–8. doi: 10.1038/s41588-018-0099-7
24. Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the STROBE-MR statement. Jama (2021) 326:1614–21. doi: 10.1001/jama.2021.18236
25. Morgan RG. Network mendelian randomization study design to assess factors mediating the causal link between telomere length and heart disease. Circ Res (2017) 121:200–2. doi: 10.1161/circresaha.117.311387
26. Vaghari Tabari M, Moein S, Qujeq D, Kashifard M, Shokri Shirvani J, Hajian Tilaki K, et al. Evaluation of the potential antioxidant role of high-density lipoprotein-cholesterol (HDL-c) in patients with ulcerative colitis. Iranian J Colorectal Res (2017) 5. doi: 10.5812/acr.13699
27. Soh H, Chun J, Han K, Park S, Kang EA, Im JP, et al. P797 crohn’s disease and ulcerative colitis was associated with different lipid profile disorders: a nationwide population-based study. J Crohn’s Colitis (2019) 13:S521–1. doi: 10.1093/ecco-jcc/jjy222.921
28. Barter PJ, Nicholls S, Rye KA, Anantharamaiah GM, Navab M, Fogelman AM. Antiinflammatory properties of HDL. Circ Res (2004) 95:764–72. doi: 10.1161/01.res.0000146094.59640.13
29. Charles MA, Kane JP. New molecular insights into CETP structure and function: a review. J Lipid Res (2012) 53:1451–8. doi: 10.1194/jlr.R027011
30. Poznyak AV, Sukhorukov VN, Eremin II, Nadelyaeva II, Gutyrchik NA, Orekhov AN. HDL-based therapy: vascular protection at all stages. Biomedicines (2023) 11. doi: 10.3390/biomedicines11030711
31. Schmidt AF, Hunt NB, Gordillo-Marañón M, Charoen P, Drenos F, Kivimaki M, et al. Cholesteryl ester transfer protein (CETP) as a drug target for cardiovascular disease. Nat Commun (2021) 12:5640. doi: 10.1038/s41467-021-25703-3
32. Urban D, Pöss J, Böhm M, Marin R, D'Incà R, Gubbiotti A, et al. Targeting the proprotein convertase subtilisin/kexin type 9 for the treatment of dyslipidemia and atherosclerosis. J Am Coll Cardiol (2013) 62:1401–8. doi: 10.1016/j.jacc.2013.07.056
33. Seidah NG, Garçon D. Expanding biology of PCSK9: roles in atherosclerosis and beyond. Curr Atheroscler Rep (2022) 24:821–30. doi: 10.1007/s11883-022-01057-z
34. Marinelli C, Zingone F, Lupo MG, Marin R, D'Incà R, Gubbiotti A, et al. Serum levels of PCSK9 are increased in patients with active ulcerative colitis representing a potential biomarker of disease activity: a cross-sectional study. J Clin Gastroenterol (2022) 56:787–93. doi: 10.1097/mcg.0000000000001607
35. Lei L, Li X, Yuan YJ, Chen ZL, He JH, Wu JH, et al. Inhibition of proprotein convertase subtilisin/kexin type 9 attenuates 2,4,6-trinitrobenzenesulfonic acid-induced colitis via repressing toll-like receptor 4/nuclear factor-kappa b. Kaohsiung J Med Sci (2020) 36:705–11. doi: 10.1002/kjm2.12225
36. Ioannou GN, Lee SP, Linsley PS, Gersuk V, Yeh MM, Chen YY, et al. Pcsk9 deletion promotes murine nonalcoholic steatohepatitis and hepatic carcinogenesis: role of cholesterol. Hepatol Commun (2022) 6:780–94. doi: 10.1002/hep4.1858
Keywords: lipids, lipid-lowering, inflammatory bowel disease, Mendelian randomization, PCSK9, CTEP 2
Citation: Tao H, Yu Z, Dong Y, Liu L, Peng L and Chen X (2023) Lipids, lipid-lowering agents, and inflammatory bowel disease: a Mendelian randomization study. Front. Immunol. 14:1160312. doi: 10.3389/fimmu.2023.1160312
Received: 07 February 2023; Accepted: 25 May 2023;
Published: 07 June 2023.
Edited by:
Murugaiyan Gopal, Harvard Medical School, United StatesReviewed by:
Sunil Martin, Rajiv Gandhi Centre for Biotechnology, IndiaYing Liang, Florida International University, United States
Shrishti Saxena, Brigham and Women’s Hospital and Harvard Medical School, United States
Copyright © 2023 Tao, Yu, Dong, Liu, Peng and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liang Peng, d3NmaXJlZmx5QDEyNi5jb20=; Yongqiang Dong, ZWR3aW5AYmptdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Heqing Tao1†
Heqing Tao1† Yongqiang Dong
Yongqiang Dong Ligang Liu
Ligang Liu Liang Peng
Liang Peng