- 1Department of Pharmacology, College of Pharmacy, Chongqing Medical University, Chongqing, China
- 2Chongqing Key Laboratory of Drug Metabolism, Chongqing, China
- 3Key Laboratory for Biochemistry and Molecular Pharmacology of Chongqing, Chongqing, China
- 4Medical Research Center, Southwest Hospital, Third Military Medical University, Chongqing, China
Sepsis is a major life-threatening syndrome of organ dysfunction caused by a dysregulated host response due to infection. Dysregulated immunometabolism is fundamental to the onset of sepsis. Particularly, short-chain fatty acids (SCFAs) are gut microbes derived metabolites serving to drive the communication between gut microbes and the immune system, thereby exerting a profound influence on the pathophysiology of sepsis. Protein post-translational modifications (PTMs) have emerged as key players in shaping protein function, offering novel insights into the intricate connections between metabolism and phenotype regulation that characterize sepsis. Accumulating evidence from recent studies suggests that SCFAs can mediate various PTM-dependent mechanisms, modulating protein activity and influencing cellular signaling events in sepsis. This comprehensive review discusses the roles of SCFAs metabolism in sepsis associated inflammatory and immunosuppressive disorders while highlights recent advancements in SCFAs-mediated lysine acylation modifications, such as substrate supplement and enzyme regulation, which may provide new pharmacological targets for the treatment of sepsis.
1 Introduction
Sepsis, according to the latest criteria (Sepsis 3.0) is defined a life-threatening organ dysfunction syndrome caused by a dysregulated host response due to infection or infectious agents. It serves as one of the most common complications in patients with clinical trauma/burns and infections while ranks a leading cause of death in critically ill units (1). An estimated number of 31.5 million cases and 5.3 million deaths are recorded worldwide in a single year, making sepsis as a prominent health problem for the global medical community (2). Sepsis is traditionally recognized as a two- stage syndrome which manifests with hyperinflammation and immune suppression. More recently, it is suggested that sepsis may manifest concurrent hyperinflammation and immune suppression (3). In recent years, substantial progress has been made in revealing the mechanisms that drives the development of sepsis, especially in the areas of metabolisms and epigenetics (4). Remarkably, profound alterations in intracellular metabolites and protein epigenetic markers have been identified as key regulatory mechanisms for the initiation of inflammation and immunosuppression as well as their phenotypic transformation. These findings hold significant implications for understanding the immunopathology of sepsis and the search for potential drug intervention targets.
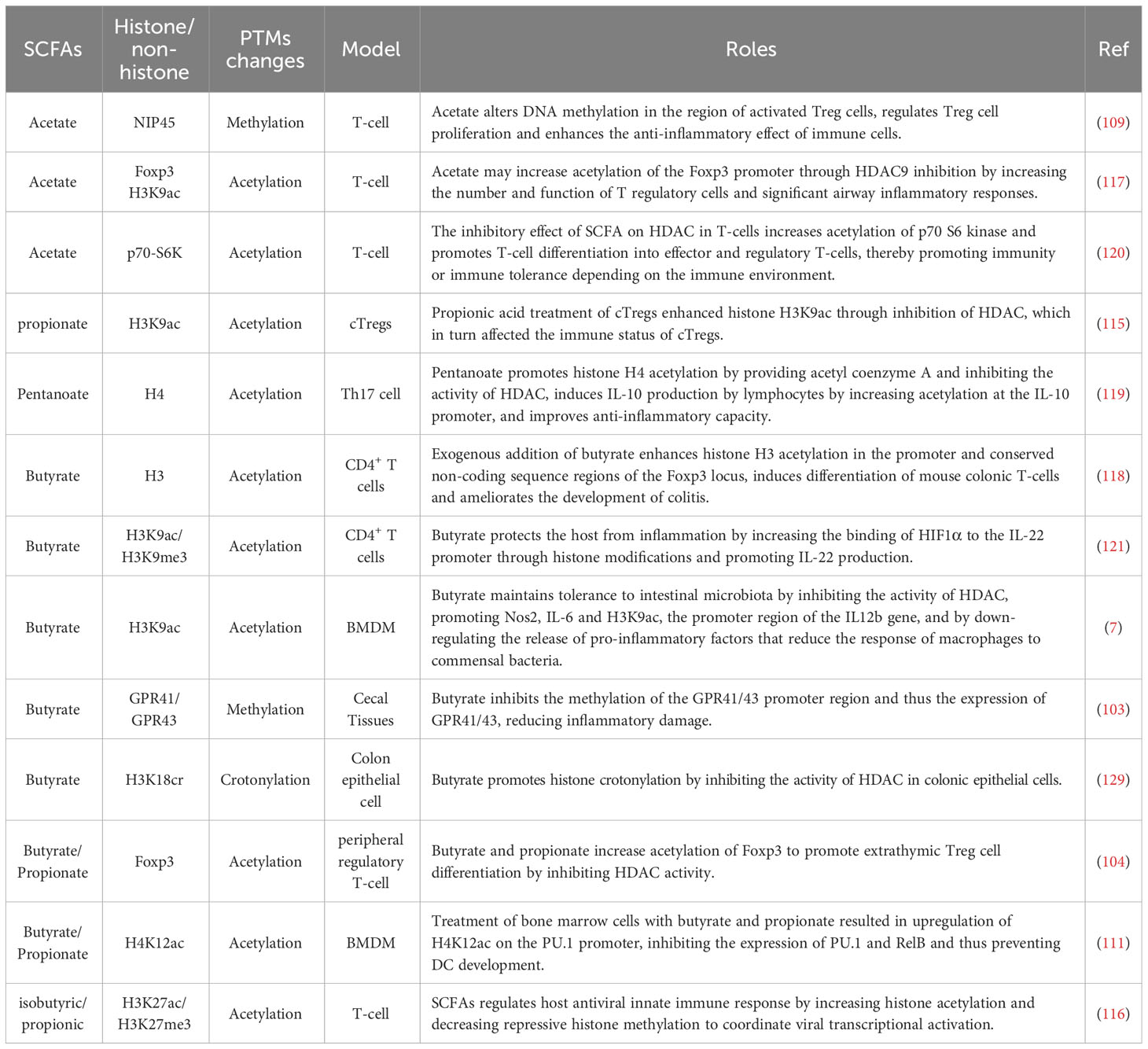
The primary events of sepsis include infection, inflammation induced by infection, immune dysregulation, and organ dysfunction. Sepsis also involves various events that are secondary to those primary changes. During sepsis, notably changes in host metabolism have been detected, such as hyperglycemia, aberrant lipid metabolism, and amino acid metabolism disorders, leading to significant alterations in the level and activity of cellular metabolites that result in immune dysfunction (5, 6). The modified levels of metabolites can affect the pathophysiological process of sepsis by either affecting the activity of enzymes or otherwise altering protein structures and interactions (7, 8). The protein post-translational modifications (PTMs), as a result of an altered metabolism, represent one of the main causes of functional diversity of mammalian protein molecules, and are thus involved in the regulation of protein function and activity by inducing covalent binding of proteins to different functional metabolites (such as acetate, butyrate and lactate) (9). The regulatory machinery of protein PTMs mainly include substrates, modifying enzymes and de-modifying enzymes, among which the substrates are directly derived from the transformation of cell-related metabolites (10, 11). For instance, lactate can serve as a substrate for lactylation, while acetyl-CoA can be utilized as a substrate for acetylation modification (12, 13). Additionally, certain metabolites act as endogenous inhibitors of chromatin-modifying enzymes, thereby influencing the levels of PTMs by modulating the activity of modifying and de-modifying enzymes (14, 15). For instance, butyrate is a metabolite that regulates PTMs by inhibiting histone deacetylase (HDAC) activity(16). Importantly, PTMs can occur in histones, affecting the regulation of gene expression, as well as in non-histone proteins, influencing their function or interactions with other proteins (17, 18). Histone modification includes methylation, acetylation, phosphorylation and other forms, which can regulate gene transcriptional activity and chromatin structure by changing the chemical structure of histone (19–21). The role of histone modification in the pathogenesis and development of sepsis is multifaceted, encompassing the regulation of inflammatory factor expression, modulation of immune cell function and differentiation, and regulation of cell apoptosis and survival (22–24). PTMs can also influence the activity of themselves or interactive proteins by acting on non-histone proteins, thereby regulating the level of autophagy and nuclear translocation that may intervene in the progression of sepsis (25, 26). Furthermore, metabolite-dependent PTMs may affect the short- and long-term immunosuppressive state by providing rapid and prolonged responses that counteract the overreaction of circulating acute pro-inflammatory cytokines in sepsis patients (27).
In recent years, the significance of intestinal flora and their metabolites in inflammatory and metabolism-related diseases has gained increasing recognition(28). Among these, short-chain fatty acids (SCFAs), obtained by the flora through fermentation of food fibers, have received considerable attention, particularly in their regulatory roles in metabolism, maintenance of the intestinal mucosal barrier, and immune homeostasis (29, 30). SCFAs and its corresponding acyl coenzyme A are located at the crossroads of metabolic pathways and play an important role in various cellular processes. It has been observed that SCFAs can play a signal transduction role through covalent or non-covalent binding to proteins and can also bind to modifying enzymes to mediate post-translational modification of proteins (31). Moreover, SCFAs have been found to exert major regulatory effects in sepsis, including affecting gene expression, enhancing phagocytosis of macrophages, altering cell proliferation and function, and inhibiting the activity of HDAC (32–36). Several recent studies have demonstrated that there were significant differences in SCFAs levels in the feces of severe septic patients and in murine models of sepsis induced by cecal ligation and puncture (CLP) (37, 38). Therefore, the disorder of SCFAs is secondary to changes in the primary event of sepsis, and changes in SCFAs levels may further affect the formation of PTMs, which in turn may mediate the primary event of sepsis.
In this review, we have provided a summary of the metabolic mechanisms involved in the inflammatory and immunosuppressive phases of sepsis, with a particular focus on the roles of SCFAs metabolism. We have also discussed recent advances in SCFAs-mediated lysine acylation modifications.
2 The intricate pathophysiology of sepsis and the underlying metabolic mechanisms
The pathogenesis of sepsis is a complex phenomenon, involving a multitude of factors spanning from the molecular to the organ level. This intricate interplay encompasses issues such as infection, inflammation, immunity, coagulation, and tissue damage (3). Cytokine storm is an early manifestation of sepsis, which primarily arises from macrophages engulfing pathogens and subsequently releasing a myriad of inflammatory cytokines, thereby triggering the body’s inflammatory response (39). In the later stages of sepsis, immunosuppression assumes a dominant role, characterized by the downregulation of pro-inflammatory factors, secondary infections, and increased apoptosis and autophagy of immune cells (40–42). Apoptosis of immune cells is a major factor in the development of immunosuppression in sepsis(43). Notably, sepsis is also suggested to manifest concurrent hyperinflammation and immune suppression that further increase the intricateness of this syndrome (3).
The alterations in the levels of relevant metabolites during sepsis can interfere with inflammatory and immune processes by regulating the levels of PTMs. Clinical investigations have shown that metabolic reprogramming during sepsis leads to profound changes in glucose, lipid, and fatty acid metabolism (44). Among these, glucose metabolic reprogramming occurs during sepsis when glycolytic pathways are enhanced in response to cellular biosynthesis and bioenergy requirements, thereby promoting cell growth, differentiation, and effector functions (45). Second, lipid production is an important adaptive response to normal tissue function. Clinical studies have found that cholesterol and lipoprotein levels vary markedly in patients with inflammation, with patients with severe sepsis having lower levels of cholesterol, including high-density lipoprotein (HDL), low-density lipoprotein (LDL) and apolipoprotein A-I (Apo A-I), as well as high levels of triglycerides (TGs) and free fatty acids (FFA) (46). Consequently, HDL and LDL are considered to be important modulators of the host immune response during sepsis. At the same time, sepsis induces the release of lipid mediators, many of which activate nuclear receptors. For instance, peroxisome proliferator-activated receptor (PPAR) α, a nuclear receptor activated by fatty acids (FAs), controls lipid metabolism and inflammation. Paumelle et al. found that PPARα deficiency led to a pro-inflammatory response and reduced survival in the CLP model, which impaired the adaptive metabolic shift from glucose to fatty acid (FAs) utilization (47).
Beyond the major metabolic pathways, some specific metabolic pathways and their products are also significantly altered in sepsis, such as SCFAs. During sepsis, increased inflammatory factors lead to intestinal epithelial cell apoptosis and increased intestinal wall permeability (48). In addition, sepsis induces kidney injury, with a sharp increase in urea, sodium, and water leading to intestinal wall edema and disruption of tight junction proteins in colonic epithelial cells. Inflammatory cytokines as well as urea retention damage the intestinal barrier, leading to bacterial translocation through the portal vein or mesenteric lymphatic system, causing a systemic inflammatory response and eventually leading to multiple organ failure and death (49, 50). Due to the effects of antibiotics, parenteral nutrition and systemic inflammation, the number of anaerobic bacteria in the intestinal flora is significantly down-regulated, accompanied by the significant down-regulation of intestinal metabolites SCFAs (48, 51). Studies have shown that SCFAs play an important role in immune regulation and inflammation suppression (52). For example, SCFAs can regulate the function of immune cells by activating G protein-coupled receptors, inhibit the release of inflammatory mediators, and mitigating the inflammatory response (53). SCFAs can also promote intestinal barrier function and regulate intestinal flora balance, potentially conferring effects against the occurrence and progression of sepsis (54). Furthermore, both murine models and in vitro experiments have demonstrated notable alterations in SCFAs levels within immune cells, strongly correlating with sepsis progression. For example, one study revealed fecal SCFAs levels were significantly downregulated in a rat sepsis model induced by CLP, while the levels of TNF-α, IL-1β, IL-6, and other inflammatory factors in the hippocampus were markedly upregulated (55). Additionally, exogenous supplementation of SCFAs in lipopolysaccharide (LPS)-stimulated primary rat neutrophils not only significantly suppressed the levels of TNF-α and nitric oxide synthase by inhibiting the activation of nuclear factor κB (NF-κB), but also inhibited the activity of HDAC, potentially influencing inflammation progression through the modulation of specific PTMs (56).
3 Gut microbiota and microbiota derived SCFAs
Gut microbiota refers to the community of various microorganisms present in the human gut, including bacteria, archaea, fungi, and viruses, plays a crucial role in human health (57). Recent studies have highlighted its involvement in various biological processes, such as the regulation of tumor cell immunogenicity and innate immune functions, particularly in tumor cells and in macrophages (58, 59). In addition, the interplay between the intestinal microbiota, local immunity, and gut integrity has emerged as a key determinant in disease development (60). During sepsis, factors like intestinal hypoperfusion, intestinal cell apoptosis, systemic cytokine storms, and intestinal dysbiosis contribute to the disruption of intestinal cell permeability, promoting the migration of flora and the transfer of inflammatory mediators, leading to multiple organ dysfunction syndrome and systemic inflammatory response (61, 62). Gai et al. found that there was an imbalance of flora 12 hours after CLP in the mouse model, with significant reductions in Firmicutes and Bacteroidetes (63). Giridharan et al. found that acetate, propionate and butyrate were significantly down-regulated in the CLP model group compared with the control group (55). Thus, disruption of gut microbiota during sepsis leads to secondary down-regulation of SCFAs levels. Meanwhile, SCFAs, metabolites of the gut microbiota, could reshape intestinal dysbiosis, reduce the release of inflammatory cytokines, and improve survival rates in CLP-induced model (54). Therefore, the stability of gut microbiota is crucial for the production of SCFAs and host immune regulation. Meanwhile, SCFAs as products in turn maintain the intestinal ecological balance and participate in the regulation of immune response.
Acetate, propionate, and butyrate are the primary SCFAs found in the gut, accounting for more than 95% of SCFAs, of the total SCFAs composition, with an estimated ratio of approximately 3:1:1 (64). The relative abundance of these SCFAs varies depending on factors such as the host’s diet, microbiome composition, and tissue location, as they are produced by specific microbial communities in different regions of the gut (65). Acetate, the most abundant SCFAs, is extensively produced in the gut and can be metabolized and transported by the intestinal epithelium and liver, serving as an energy source (66). Gut bacteria can convert pyruvate to acetate by either acetyl-CoA or the Wood-Ljungdahl pathway (67, 68). The production of propionate mainly occurs in the large intestine and is produced by bacteria such as Bacteroides and Clostridium anaerobes (69). In addition, the succinate pathway is an important pathway for the production of propionic acid by human gut microbiota, and methylmalonyl-CoA is the key enzyme in the succinate pathway. Propionic acid can also be generated by the acrylate pathway and the propylene glycol pathway (70, 71). Propionate can be absorbed by the intestinal epithelium and transported to the liver, where it is further metabolized into glucose or fatty acids (72). Butyrate is formed by condensation and subsequent reduction of two acetyl-CoA molecules to butyrate, which can be converted to butyrate by phospho-transbutyrylase and butyrate kinase (73). Butyrate can be absorbed by intestinal epithelial cells and metabolized in the liver as ketone bodies or carbon dioxide (74). Studies have shown that the concentration of SCFAs in the intestine ranges from 20 to 140 mM, with extremely high concentrations in the proximal colon (70 to 140 mM), and relatively low concentrations in the distal colon (20 to 70 mM) and distal ileum (20 to 40 mM), depending on the presence of infection/inflammation in the host (32, 75). In addition, studies have found that the production of gut-derived SCFAs can be regulated by changing the proportion of SCFAs -producing flora in the gut (76). At present, the main ways to regulate the structure of gut microbiota include changing the host dietary structure (Mediterranean diet), direct intake of probiotics or prebiotics to regulate gut microbiota (77–79). In conclusion, gut microbiota plays a crucial role in the generation of SCFAs.
As the most abundant metabolite of the intestinal microbiota in the intestinal lumen, SCFAs play a multifaceted role in the regulation of the host immune system (80). Firstly, SCFAs serve as a vital energy source for colon and ileal cells, influencing the expression of genes involved in intestinal epithelial barrier function and defense mechanisms (32). Second, modulate the function of key immune cells such as macrophages, neutrophils, dendritic cells, and even adipocytes, thereby impacting inflammation progression through the regulation of TNF-α, IL-10, and IL-4 levels in adipose tissue (81). Thirdly, SCFAs exert their immunomodulatory effects by inhibiting HDAC activity and activating specific receptors, namely, free fatty acid receptors type 2 and 3 (FFA2 and FFA3 receptors) and G protein-coupled receptor 109A (GPR109A) (82). These mechanisms collectively contribute to the regulation of immune responses and the formation of various post-translational modifications involved in cellular processes.
4 The relationship between sepsis and SCFAs metabolism
4.1 Changes in SCFAs levels in sepsis and their relationship to disease progression or regression
The pivotal pathway responsible for the production of SCFAs has been found to be intricately linked with the presence of microbiota (80). Notably, a recent study has demonstrated that individuals with diminished gut microbiota diversity face an elevated susceptibility to sepsis (83, 84). Furthermore, in an observational investigation involving critically ill patients with non-abdominal infections, perturbations in the gut microbiome have been shown to predispose individuals to sepsis by enabling the proliferation of pathogens, promoting an aberrant immune response, and impeding the production of beneficial SCFAs (85). Additionally, the concentration of propionic acid in serum has been found to increase in tandem with the severity of sepsis, suggesting that serum propionate holds significant potential as a predictor and prognostic biomarker for septic patients (86). Consequently, alterations in SCFAs levels during sepsis may result from an imbalance in intestinal flora, thereby inducing immune dysregulation and damage to the intestinal epithelial barrier. This, in turn, exacerbates the detrimental cycle of immune dysregulation prompted by the inflammatory response to sepsis.
Furthermore, Liao et al. have observed a significant downregulation of acetic acid and propionic acid in the CLP-induced model relative to the sham group. Remarkably, exogenous supplementation of SCFAs has been shown to substantially increase acetic acid and propionic acid levels while significantly downregulating the levels of IL-1β, IL-6, and TNF-α (38). Similarly, Li et al. have discovered that in mice subjected to the CLP model, SCFAs levels are significantly reduced, accompanied by considerable alterations in the gut microbiota. Notably, exogenous SCFAs administration has been found to enhance intestinal barrier integrity and significantly downregulate IL-1β, TNF-α, and IL-6 levels in mice (87). Although SCFAs may have potential value in the prevention and treatment of sepsis, its feasibility for the treatment of clinical patients should be verified by human trials.
4.2 The regulatory effect of SCFAs on sepsis
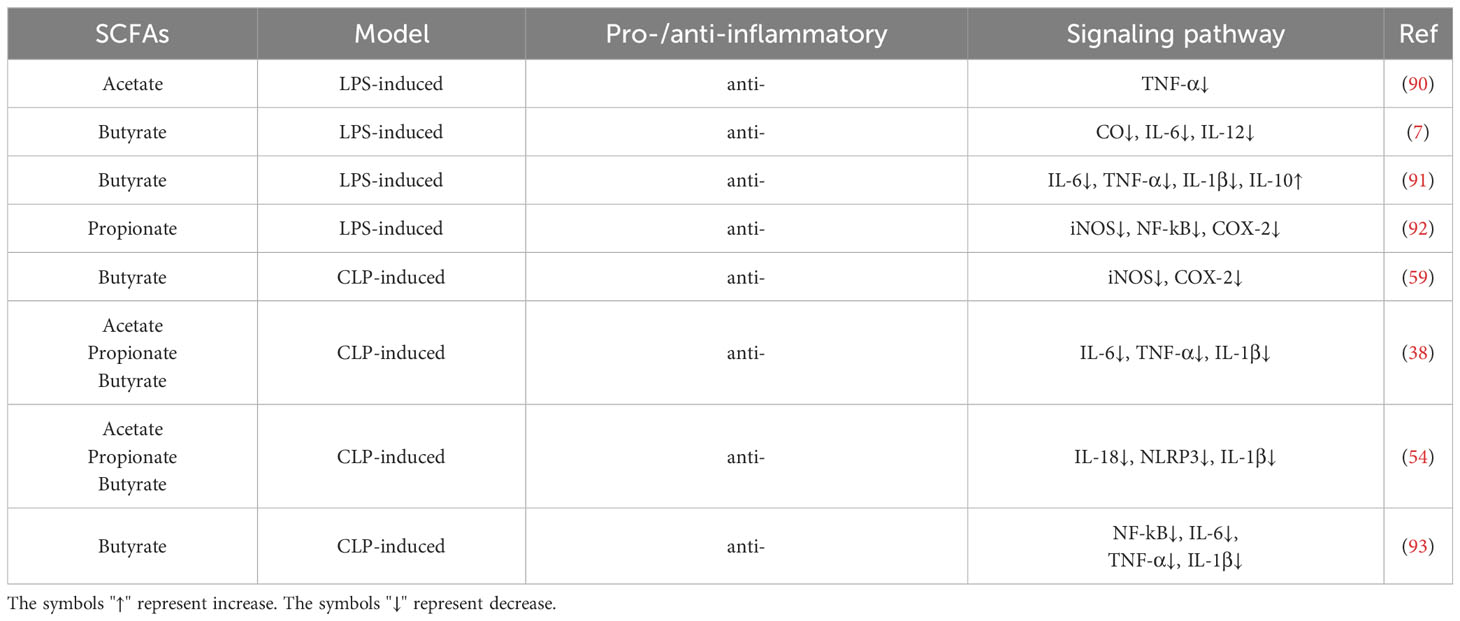
SCFAs exert multiple modulatory effects on the pathophysiology of sepsis. First, SCFAs are a key source of energy for colon and ileal cells and affect intestinal epithelial barrier and defense functions by regulating related gene expression (68). Wang et al. found that butyrate provides energy to colonic intestinal epithelial cells, regulates intestinal gene expression, inhibits the intestinal inflammation induced by LPS (88). Zhan et al. have demonstrated that SCFAs hinder pathogen invasion by supplying energy and regulating the barrier function and immune status of the host intestine (89). Secondly, SCFAs intervene in the inflammatory storm phase during sepsis by regulating the production of immune cytokines (Table 1). At present, a number of studies have found that SCFAs can significantly down-regulate the levels of a variety of pro-inflammatory mediators (38, 90, 92, 93). For example, Vinolo et al. found that butyrate and propionate can reduce the expression of TNF-α and nitric oxide synthase (NOS) in primary mouse neutrophils induced by LPS (56). Similarly, Wang et al. demonstrated that butyrate significantly decreased the levels of TNF-α, IL-1β and IL-6 in LPS-injected mice (91). In another study, butyrate treatment was shown to inhibit the levels of nitric oxide, IL-6, and IL-12 inflammatory mediators in LPS-stimulated bone marrow-derived macrophage (BMDM) cells by inhibiting HDAC (7). Thirdly, SCFAs could regulate sepsis by regulating PTMs. For example, in CD8+ T cells within a low-glucose tumor environment, acetate facilitates histone acetylation to enhance the transcription of the IFN-γ gene and cytokine production, thereby modulating the progression of inflammation (94). In addition, acetate can supplement acetyl-CoA, which promotes the acetylation of GAPDH to enhance its activity, thereby promoting glycolysis, so as to promote rapid memory CD8(+) T cell response and enhance the immune response (95). Taken together, these findings suggest that SCFAs can suppress the inflammatory response, and that the absence of SCFAs and the reaction of SCFAs with PTMs are secondary events of the inflammatory response in sepsis.
4.3 The mechanism of SCFAs regulating post-translational modification of proteins in sepsis
PTMs represent a crucial pathway for regulating protein function, and significant changes in phosphorylation, ubiquitination, methylation, and acylation have been observed during and after sepsis (96–98). Lysine acylation includes long chain acylation and short chain acylation. Protein acylation is involved in a variety of cellular processes, such as protein stability, protein subcellular localization, enzyme activity, transcriptional activity, protein-protein interaction and protein-DNA interaction (99). Among them, protein lysine acylation exerts important and unique regulatory roles. Studies have indicated that lysine acylation can regulate the development of sepsis by regulating the activity of modifying enzymes, targeting innate sensors and downstream signaling molecules (100, 101).
Interestingly, the most substrates of these novel lysine acylation modification are SCFAs, such as acetate, malonate, crotonic acid and 2-hydroxyisobutyric acid, among which lysine acetylation (Kac), lysine malonylation (Kmal), lysine crotonylation (Kcr) and lysine 2-hydroxyisobutyrylation (Khib) play crucial roles in inflammation and immunity. Consequently, SCFAs may participate in regulating the immune response during sepsis by influencing the corresponding acylation modification levels (102–105). To simplify the intricate regulation of SCFAs-mediated PTMs on the immune system, this review will combine current research to elucidate the mechanism by which SCFAs affect PTMs through two primary pathways: i: SCFAs as inhibitors of HDAC that regulate the level of lysine acylation modification and participate in the regulation of sepsis; ii. SCFAs as substrates for lysine post-translational modification that regulate the level of modification and participate in the regulation of sepsis.
4.3.1 SCFAs as inhibitors of HDAC
4.3.1.1 Methylation
Protein lysine methylation is a dynamic process that plays a vital role in various biological processes, such as DNA damage repair, cell growth, metabolism, and signal transduction (106). The dynamic lysine methylation system comprises three key components: protein lysine methyltransferases (PKMTs), which add methylation marks; methyl-binding domains (MBDs), which participate in methylation events with biological outcomes; and lysine-specific demethylases (KDMs), responsible for the removal of methylation marks (107).
In the context of early sepsis, protein lysine methylation assumes a critical role in the epigenetic regulation of innate immunity. Xia et al. discovered that Ash1l, an H3K4 methyltransferase, suppressed the expression of TNF-α and IL-6 by promoting H3K4 methylation at the Tnfaip3 promoter in mice injected with LPS (108). Interestingly, SCFAs also regulate inflammation progression and immune cell functional expression by modulating lysine methylation modifications. Kaye et al. observed that in hypertensive mice, acetate activated DNA methylation in Treg cell regions, enhancing the anti-inflammatory effects of immune cells by regulating Treg cell proliferation (109). Additionally, Chang et al. found that in LPS-stimulated BMDM cells, exogenous addition of butyrate could inhibit the methylation modification of the GPR41/43 promoter region, thereby suppressing the expression of GPR41/43 and reducing host inflammatory damage (7). Collectively, the methylation of histone and non-histone lysine significantly influences the function of immune cells during sepsis. SCFAs regulate the levels of inflammatory and anti-inflammatory factors by inhibiting HDAC activity, thereby affecting methylation levels and interfering with the transcriptional activities of transcription factors. Hence, SCFAs hold promise as potential therapeutic agents for early sepsis treatment.
4.3.1.2 Acetylation
Lysine acetylation modifications are a universally recognized group of PTMs that that have the potential to impact protein function through various mechanisms. These mechanisms include the regulation of protein stability, enzymatic activity, subcellular localization, interplay with other post-translational modifications, and the control of protein-protein and protein-DNA interactions(110). Among these, the diverse HAT/KAT and HDAC/KDAC enzymes are widely involved in biological cellular processes, and the lysine acetylation or deacetylation they induce in cells may contribute to the development of several diseases.
SCFAs play a role in the regulation of lysine acetylation modifications by inhibiting HDAC activity. This interference with HDAC activity has been shown to impact the progression of inflammation in various diseases (111–118). Notably, Luu et al. discovered that in experimental mouse models of colitis, valerate promotes the acetylation of histone H4 at the IL-10 promoter by providing acetyl-CoA and inhibiting HDAC activity. This, in turn, induces the production of IL-10 in lymphocytes, playing an anti-inflammatory immunomodulatory role (119). Moreover, Park et al. found that acetate upregulates the acetylation modification of p70 S6 kinase by inhibiting HDAC activity. This promotes the differentiation of T cells into effector T cells and regulatory T cells, consequently reducing anti-CD3-induced inflammation in an IL-10-dependent manner (120). Meanwhile, Yang et al. discovered that in anti-CD3-activated CD4+ cells, butyrate upregulates histone acetylation on the IL-22 promoter, facilitating HIF1α binding to this region. This ultimately promotes the production of IL-22, safeguarding the host from inflammation (121). Therefore, SCFAs can regulate inflammatory and immune processes by regulating protein acetylation modification, which may potentially exert a similar effect on inhibiting hyperinflammation in sepsis. In conclusion, facilitate the acetylation of both histone and non-histone proteins by inhibiting HDAC activity. This regulation of lysine acetylation contributes to the innate immune response of the host and participates in the immune pathogenesis of sepsis. Consequently, targeting key lysine acetylation sites to inhibit inflammation progression represents an important approach to the treatment and intervention of sepsis.
4.3.1.3 Crotonylation
Lysine crotonylation (Kcr) modification is a highly conserved protein modification that has evolved from histone lysine acetylation (Kac). Although crotonylation and acetylation have distinct biological characteristics, they share the same regulatory enzymes. Crotonylation modification plays a role in regulating diverse biological processes and the development of various diseases, including gene expression, spermatogenesis, cell cycle regulation, and the pathogenesis of conditions ranging from depression to cancer (122–125). In addition to intracellular crotonyl-CoA, the regulation of Kcr also involves the addition and removal of modified writers and erasers, respectively, contributing to homeostasis (126). Histone acetyltransferases (HATs), such as p300/CBP, possess significant histone crotonyl transferase (HCT) activity, highlighting their role in crotonylation modification (127).
It has been also demonstrated that crotonylation is involved in sepsis. Sabari et al. found that crotonyl-CoA supplementation in macrophages led to a substantial increase in histone H3K18Cr. Additionally, knockdown of ACSS2, an enzyme involved in crotonyl-CoA synthesis, reduced the expression of histone H3K18Cr and inflammatory genes in LPS-stimulated RAW264.7 cells (128). This suggests a close association between crotonylation and sepsis, with the possibility of modulating inflammatory progression through the regulation of crotonylation. Furthermore, SCFAs are involved in regulating Kcr modification levels by inhibiting HDAC activity. Fellows et al. demonstrated that exogenous butyrate supplementation inhibited HDAC activity in colonic epithelial cells, resulting in the upregulation of histone H3K18cr modification and influencing the cell cycle. This finding further underscores the connection between SCFAs and chromatin signaling (129). Collectively, the levels of lysine Kcr modification undergo significant changes following LPS stimulation, and SCFAs participate in the regulation of Kcr by inhibiting HDAC activity. This suggests that SCFAs may impact sepsis through the modulation of Kcr modification. Therefore, it is crucial to further investigate the mechanisms underlying histone and non-histone Kcr modification targets in sepsis for the development of effective treatments.
4.3.2 SCFAs as substrates for lysine acylation modification
4.3.2.1 Lysine 2-hydroxyisobutyrylation
Lysine 2-hydroxyisobutyrylation (Khib) is a novel histone acylation modification first identified in 2014 with 2-hydroxybutyrate and its cozymoylated form 2-hydroxyisobutyl CoA as the substrates (130–132). The 2-hydroxybutyric acid is a typical SCFAs detected in micromolar concentrations in a variety of human biological fluids. Of note, its high occurrence in the urine of obese patients was found associated with the abundance of specific taxa of the gut microbiota (133, 134). Similar to other types of PTMs, khib also maintains a dynamic balance by adding and removing writers and erasers respectively. Huang et al. demonstrated that Esa1p and its human homolog TIP60 from yeast cells can regulate khib proteinogenesis and act as writers for khib (130). Additionally, the lysine acetyltransferase p300, mentioned earlier, exhibits enzymatic activity not only for lysine acetylation but also for lysine butyrylation, propionylation, and crotonylation (135–137a).
Although khib occurs in diverse cellular proteins, it has only emerged to realize that it may exert mutifaced roles physiological and pathological conditions. For instance, Yamazaki et al. found that the serum level of 2-HIBA was significantly up-regulated in a mouse model of periodontitis through metabolomics (138). Similarly, Tsoukalas et al. found statistically significant differences in the metabolite levels of 2-hydroxyisobutyric acid in urine of rheumatoid arthritis patients through metabolomics (139). Therefore, as a SCFAs, the level of 2-hydroxyisobutyric acid is significantly up-regulated in a variety of diseases involving inflammation, suggesting that the change of its level has an important relationship with the development of inflammation. Notably, the occurrence of khib is also strongly associated with inflammatory regulation. Ge et al. found that in the skin tissue of patients with psoriasis, the proteins encoded by S100A9, FUBP1 and SERPINB2 genes have significant khib with proteins in the PI3K-Akt signaling pathway (137b). Among them, S100A9 is a potential target for the treatment of sepsis, and further investigation is needed to determine if its khib is involved in sepsis progression (140). In addition, Dong et al. analyzed khib occurrence in key enzymes of the glycolytic pathway in blood monocytes from end-stage renal disease patients, which may affect immune cell numbers and induce immune senescence by influencing glycolytic function (141). Furthermore, Xie et al. found high enrichment of khib in blood monocytes from lupus erythematosus patients during the antigen processing and presentation process and leukocyte migration pathways. This suggests that khib is involved in inflammation regulation by affecting the khib level of key proteins in these pathways (142). Although there is no direct reported relationship between khib and sepsis, khib is essentially involved in the regulation of various inflammation-related diseases, implicating the necessity to further elucidate the specific mechanism of immune function regulated by khib-modified histone or non-histone proteins and discover new targets for sepsis treatment.
4.3.2.2 Lysine malonylation
Lysine malonylation (Kmal) is a highly conserved acylation modification that was initially discovered in 2011 (143). Its modified substrates include malonate and malonyl-CoA. The occurrence of Kmal relies on the addition of malonyl groups to lysine by malonyl-CoA, resulting in its charge changed from +1 to -1. Of note, malonate is also a typical SCFAs which participates in the regulation of mammalian glycolytic physiological processes (144, 145). Similar to other acylation modifications, the extent of Kmal modification is governed by the de-modifying enzyme. For example, Nishida et al. unveiled that SIRT5 functions as a global regulator of lysine malonylation and orchestrates the energy cycle through the glycolytic pathway (146).
Kmal is also implicated in regulating intracellular inflammatory and immune events. In a study conducted by Lee et al., it was discovered that malonate profoundly hindered the activation of the p38 MAPK/NF-κB pathway in LPS-stimulated microglia, thereby exerting an anti-inflammatory effect (147). Additionally, Park et al. observed that malonate significantly downregulated the expression of ROS-induced NF-κB and inflammation-related cytokines (IL-6, COX-2, and TNF-α) in HaCaT cells(148). Hence, it is plausible that malonate, acting as a SCFAs, modulates the innate immune response through the inhibition of the inflammatory cascade. Interestingly, Galván-Peña et al. demonstrated that the level of Kmal was significantly upregulated in LPS-stimulated BMDM cells. Notably, malonylation mass spectrometry revealed the presence of malonylation modifications at the lysine 213 site on GAPDH. Under normal circumstances, GAPDH binds to and represses the translation of various mRNAs associated with inflammation, including the one encoding TNFα. However, upon malonylation modification of GAPDH, it dissociates from the TNFα mRNA, thereby promoting its translation (149). Furthermore, Qu et al. (150) found that atractylodin inhibits the malonylation of GAPDH and subsequently suppresses the level of TNF-α in LPS-induced RAW264.7 cells. Consequently, Kmal modification may play a pivotal role in the regulation of sepsis. The identification of novel targets of Kmal-modified histones or non-histones holds great significance for the treatment of sepsis.
Figure 1 An overview on the effects of SCFAs on signaling pathways and protein post-translational modifications during sepsis. The binding of LPS or TNF-α to their respective receptors triggers the activation of Toll-like receptor 4 (TLR4), MAPK and nuclear factor kappa B (NF-κB) signaling pathways, leading to the release of cytokines, which plays a crucial role in sepsis development. However, SCFAs, predominantly acetate, propionate, and butyrate, exert anti-inflammatory effects by activating G protein-coupled receptor (GPR) 41, GPR43, and GPR109A receptors or inhibiting the activation of NF-κB and mitogen-activated protein kinase (MAPK) signaling pathways. Furthermore, SCFAs can serve as substrates for protein post-translational modifications and inhibit the activity of HDAC, thereby regulating the levels of various lysine acylation modifications. This inhibition of HDAC activity leads to the transcriptional downregulation of inflammatory factors and actively participates in immune regulation within the body (By Figdraw).
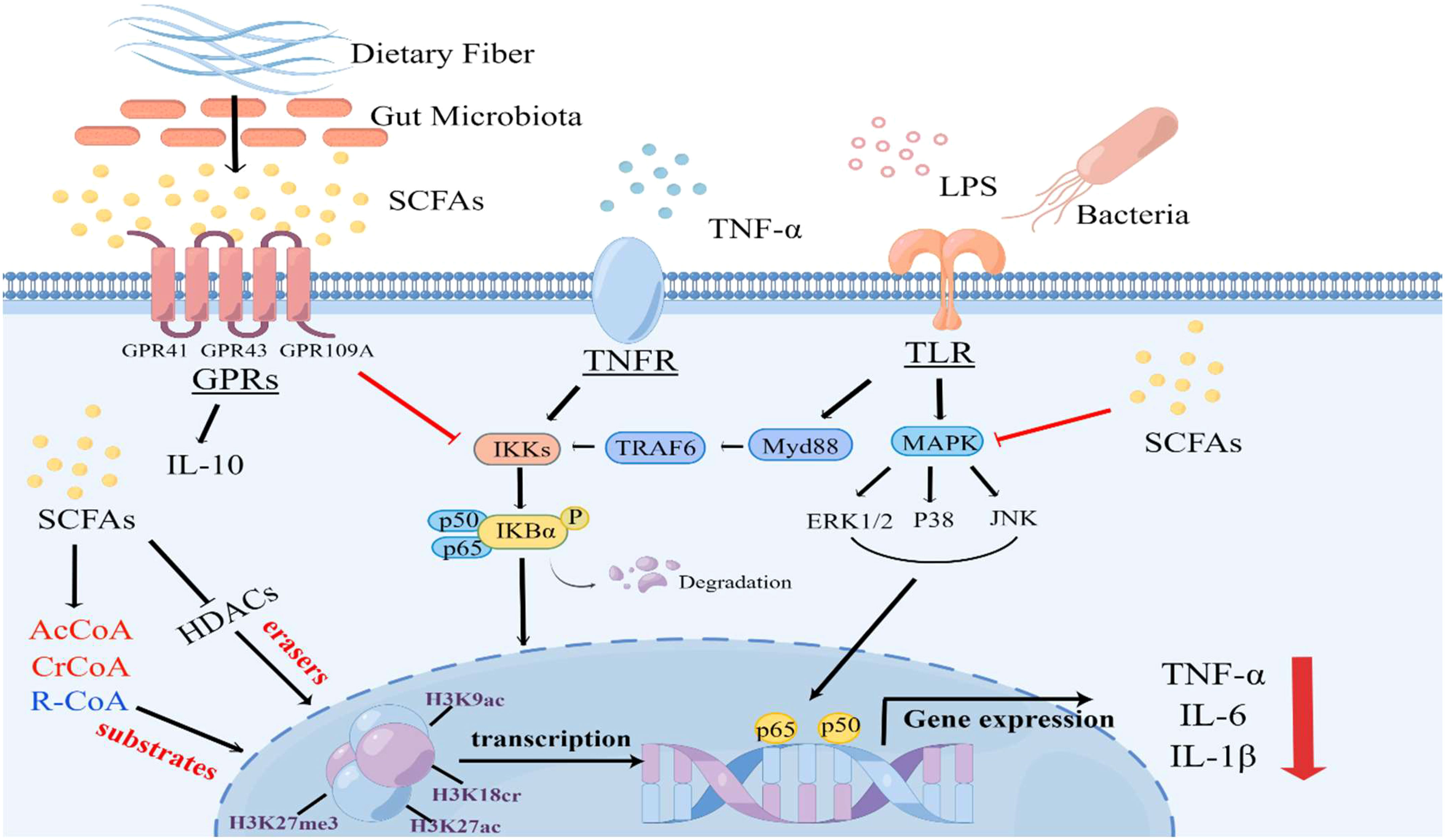
Figure 1 An overview on the effects of SCFAs on signaling pathways and protein post-translational modifications during LPS or TNFα-induced sepsis.
5 Summary and perspectives
As a byproduct of intestinal bacteria and a metabolite, SCFAs actively participate in the regulation of the host’s immune function, playing a crucial role in sepsis and inflammatory diseases. Furthermore, PTMs are closely associated with key stages of sepsis, exerting their influence on the regulation of cytokine storms and immune suppression by inhibiting or reversing the transcription of pro-inflammatory genes. Interestingly, both the epigenetic changes caused by PTMs and the immunosuppressive state of sepsis are long-lasting conditions. In our review, we have focused on the role of SCFAs, which may have a significant impact on the regulation of lysine acylation modifications in sepsis. There is mounting evidence suggesting that SCFAs can modulate innate and adaptive immune responses by regulating lysine acylation modifications (Table 2).
In this review, we have summarized the research progress regarding SCFAs in the pathological processes of inflammation and sepsis by their impact on PTMs. While host metabolism and immune regulation have garnered significant attention, further experiments are required to elucidate the specific mechanisms through which SCFAs regulate different lysine acylation modifications and the interplay between these modifications in sepsis-induced inflammation and immune suppression. Building upon the current research progress, we propose a scientific hypothesis that exogenous supplementation of SCFAs can regulate the levels of lysine acylation modifications, thereby restoring the immune response function of host immune cells and reversing host immunosuppression. With the advancement of metabolomics and proteomics technologies, it becomes increasingly feasible to explore the molecular targets of different lysine acylation modifications regulated by SCFAs during the immunosuppression stage of sepsis. Consequently, targeting lysine acylation regulated by SCFAs emerges as a potential therapeutic strategy for treating immunosuppression in sepsis, although further clinical trials are necessary to validate its feasibility in the future.
Author contributions
LZ and XS collected documents, wrote the manuscript and drew the mechanism figures. HQ collected documents and made tables. SL and TY polished the language and revised the manuscript. XLL and XL conceived ideas, designed the structure of the manuscript and revised the manuscript. All authors read and approved the submitted version. All authors contributed to the article.
Funding
This project is supported by grants from the National Natural Science Foundation of China (81873955 and 82172133) and Chongqing National Natural Science Program (csct2020jcyj-msxmX0223 and csct2022nscq-msx0214).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Cecconi M, Evans L, Levy M, Rhodes A. Sepsis and septic shock. Lancet (2018) 392(10141):75–87. doi: 10.1016/s0140-6736(18)30696-2
2. van Vught LA, Klein Klouwenberg PM, Spitoni C, Scicluna BP, Wiewel MA, Horn J, et al. Incidence, risk factors, and attributable mortality of secondary infections in the intensive care unit after admission for sepsis. Jama (2016) 315(14):1469–79. doi: 10.1001/jama.2016.2691
3. van der Poll T, Shankar-Hari M, Wiersinga WJ. The immunology of sepsis. Immunity (2021) 54(11):2450–64. doi: 10.1016/j.immuni.2021.10.012
4. Domínguez-Andrés J, Novakovic B, Li Y, Scicluna BP, Gresnigt MS, Arts RJW, et al. The itaconate pathway is a central regulatory node linking innate immune tolerance and trained immunity. Cell Metab (2019) 29(1):211–220.e215. doi: 10.1016/j.cmet.2018.09.003
5. Spitzer JJ, Bagby GJ, Mészáros K, Lang CH. Alterations in lipid and carbohydrate metabolism in sepsis. JPEN J Parenter Enteral Nutr (1988) 12(6 Suppl):53s–8s. doi: 10.1177/014860718801200604
6. Marik PE, Raghavan M. Stress-hyperglycemia, insulin and immunomodulation in sepsis. Intensive Care Med (2004) 30(5):748–56. doi: 10.1007/s00134-004-2167-y
7. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA (2014) 111(6):2247–52. doi: 10.1073/pnas.1322269111
8. Yang K, Fan M, Wang X, Xu J, Wang Y, Tu F, et al. Lactate promotes macrophage HMGB1 lactylation, acetylation, and exosomal release in polymicrobial sepsis. Cell Death Differ (2022) 29(1):133–46. doi: 10.1038/s41418-021-00841-9
9. Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol (2006) 7(7):473–83. doi: 10.1038/nrm1960
10. Zheng YG, Wu J, Chen Z, Goodman M. Chemical regulation of epigenetic modifications: opportunities for new cancer therapy. Med Res Rev (2008) 28(5):645–87. doi: 10.1002/med.20120
11. Zhao Y, Garcia BA. Comprehensive catalog of currently documented histone modifications. Cold Spring Harb Perspect Biol (2015) 7(9):a025064. doi: 10.1101/cshperspect.a025064
12. Shen Y, Wei W, Zhou DX. Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci (2015) 20(10):614–21. doi: 10.1016/j.tplants.2015.07.005
13. Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, et al. Metabolic regulation of gene expression by histone lactylation. Nature (2019) 574(7779):575–80. doi: 10.1038/s41586-019-1678-1
14. Yang M, Soga T, Pollard PJ. Oncometabolites: linking altered metabolism with cancer. J Clin Invest (2013) 123(9):3652–8. doi: 10.1172/jci67228
15. Diehl KL, Muir TW. Chromatin as a key consumer in the metabolite economy. Nat Chem Biol (2020) 16(6):620–9. doi: 10.1038/s41589-020-0517-x
16. He J, Zhang P, Shen L, Niu L, Tan Y, Chen L, et al. Short-chain fatty acids and their association with signalling pathways in inflammation, glucose and lipid metabolism. Int J Mol Sci (2020) 21(17):6356. doi: 10.3390/ijms21176356
17. Millán-Zambrano G, Burton A, Bannister AJ, Schneider R. Histone post-translational modifications - cause and consequence of genome function. Nat Rev Genet (2022) 23(9):563–80. doi: 10.1038/s41576-022-00468-7
18. Lee JM, Hammarén HM, Savitski MM, Baek SH. Control of protein stability by post-translational modifications. Nat Commun (2023) 14(1):201. doi: 10.1038/s41467-023-35795-8
19. Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature (2005) 438(7071):1176–80. doi: 10.1038/nature04254
20. Isbel L, Iskar M, Durdu S, Weiss J, Grand RS, Hietter-Pfeiffer E, et al. Readout of histone methylation by Trim24 locally restricts chromatin opening by p53. Nat Struct Mol Biol (2023) 30(7):948–57. doi: 10.1038/s41594-023-01021-8
21. Wu K, Fan D, Zhao H, Liu Z, Hou Z, Tao W, et al. Dynamics of histone acetylation during human early embryogenesis. Cell Discovery (2023) 9(1):29. doi: 10.1038/s41421-022-00514-y
22. Weiterer S, Uhle F, Lichtenstern C, Siegler BH, Bhuju S, Jarek M, et al. Sepsis induces specific changes in histone modification patterns in human monocytes. PloS One (2015) 10(3):e0121748. doi: 10.1371/journal.pone.0121748
23. Ostuni R, Natoli G, Cassatella MA, Tamassia N. Epigenetic regulation of neutrophil development and function. Semin Immunol (2016) 28(2):83–93. doi: 10.1016/j.smim.2016.04.002
24. Patel BV, Lee T, O'Dea K. CLUSTERINg circulating histones in sepsis. Am J Respir Crit Care Med (2023) 208(2):125–7. doi: 10.1164/rccm.202305-0935ED
25. Wei S, Gao Y, Dai X, Fu W, Cai S, Fang H, et al. SIRT1-mediated HMGB1 deacetylation suppresses sepsis-associated acute kidney injury. Am J Physiol Renal Physiol (2019) 316(1):F20–31. doi: 10.1152/ajprenal.00119.2018
26. Sun M, Li J, Mao L, Wu J, Deng Z, He M, et al. p53 deacetylation alleviates sepsis-induced acute kidney injury by promoting autophagy. Front Immunol (2021) 12:685523. doi: 10.3389/fimmu.2021.685523
27. McCall CE, Yoza B, Liu T, El Gazzar M. Gene-specific epigenetic regulation in serious infections with systemic inflammation. J Innate Immun (2010) 2(5):395–405. doi: 10.1159/000314077
28. Saad MJ, Santos A, Prada PO. Linking gut microbiota and inflammation to obesity and insulin resistance. Physiol (Bethesda) (2016) 31(4):283–93. doi: 10.1152/physiol.00041.2015
29. Yue X, Wen S, Long-Kun D, Man Y, Chang S, Min Z, et al. Three important short-chain fatty acids (SCFAs) attenuate the inflammatory response induced by 5-FU and maintain the integrity of intestinal mucosal tight junction. BMC Immunol (2022) 23(1):19. doi: 10.1186/s12865-022-00495-3
30. Wang J, Zhu N, Su X, Gao Y, Yang R. Gut-microbiota-derived metabolites maintain gut and systemic immune homeostasis. Cells (2023) 12(5):2296. doi: 10.3390/cells12050793
31. Huang H, Tang S, Ji M, Tang Z, Shimada M, Liu X, et al. p300-mediated lysine 2-hydroxyisobutyrylation regulates glycolysis. Mol Cell (2018) 70(4):663–678.e666. doi: 10.1016/j.molcel.2018.04.011
32. Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol (2014) 121:91–119. doi: 10.1016/b978-0-12-800100-4.00003-9
33. Lin MY, de Zoete MR, van Putten JP, Strijbis K. Redirection of epithelial immune responses by short-chain fatty acids through inhibition of histone deacetylases. Front Immunol (2015) 6:554. doi: 10.3389/fimmu.2015.00554
34. Sanford JA, Zhang LJ, Williams MR, Gangoiti JA, Huang CM, Gallo RL. Inhibition of HDAC8 and HDAC9 by microbial short-chain fatty acids breaks immune tolerance of the epidermis to TLR ligands. Sci Immunol (2016) 1(4):eaah4609. doi: 10.1126/sciimmunol.aah4609
35. Wu T, Li H, Su C, Xu F, Yang G, Sun K, et al. Microbiota-derived short-chain fatty acids promote LAMTOR2-mediated immune responses in macrophages. mSystems (2020) 5(6):eaah4609. doi: 10.1128/mSystems.00587-20
36. Liu J, Jin Y, Ye Y, Tang Y, Dai S, Li M, et al. The neuroprotective effect of short chain fatty acids against sepsis-associated encephalopathy in mice. Front Immunol (2021) 12:626894. doi: 10.3389/fimmu.2021.626894
37. Valdés-Duque BE, Giraldo-Giraldo NA, Jaillier-Ramírez AM, Giraldo-Villa A, Acevedo-Castaño I, Yepes-Molina MA, et al. Stool short-chain fatty acids in critically ill patients with sepsis. J Am Coll Nutr (2020) 39(8):706–12. doi: 10.1080/07315724.2020.1727379
38. Liao H, Li H, Bao H, Jiang L, Du J, Guo Y, et al. Short chain fatty acids protect the cognitive function of sepsis associated encephalopathy mice via GPR43. Front Neurol (2022) 13:909436. doi: 10.3389/fneur.2022.909436
39. Boyd JH, Russell JA, Fjell CD. The meta-genome of sepsis: host genetics, pathogens and the acute immune response. J Innate Immun (2014) 6(3):272–83. doi: 10.1159/000358835
40. Borouchaki A, de Roquetaillade C, Barthelemy R, Mebazaa A, Chousterman BG. Immunotherapy to treat sepsis induced-immunosuppression: Immune eligibility or outcome criteria, a systematic review. J Crit Care (2022) 72:154137. doi: 10.1016/j.jcrc.2022.154137
41. Liu D, Huang SY, Sun JH, Zhang HC, Cai QL, Gao C, et al. Sepsis-induced immunosuppression: mechanisms, diagnosis and current treatment options. Mil Med Res (2022) 9(1):56. doi: 10.1186/s40779-022-00422-y
42. Torres LK, Pickkers P, van der Poll T. Sepsis-induced immunosuppression. Annu Rev Physiol (2022) 84:157–81. doi: 10.1146/annurev-physiol-061121-040214
43. Yao RQ, Ren C, Zheng LY, Xia ZF, Yao YM. Advances in immune monitoring approaches for sepsis-induced immunosuppression. Front Immunol (2022) 13:891024. doi: 10.3389/fimmu.2022.891024
44. Preau S, Vodovar D, Jung B, Lancel S, Zafrani L, Flatres A, et al. Energetic dysfunction in sepsis: a narrative review. Ann Intensive Care (2021) 11(1):104. doi: 10.1186/s13613-021-00893-7
45. Iida J, Ishii S, Nakajima Y, Sessler DI, Teramae H, Kageyama K, et al. Hyperglycaemia augments lipopolysaccharide-induced reduction in rat and human macrophage phagocytosis via the endoplasmic stress-C/EBP homologous protein pathway. Br J Anaesth (2019) 123(1):51–9. doi: 10.1016/j.bja.2019.03.040
46. Cappi SB, Noritomi DT, Velasco IT, Curi R, Loureiro TC, Soriano FG. Dyslipidemia: a prospective controlled randomized trial of intensive glycemic control in sepsis. Intensive Care Med (2012) 38(4):634–41. doi: 10.1007/s00134-011-2458-z
47. Paumelle R, Haas JT, Hennuyer N, Baugé E, Deleye Y, Mesotten D, et al. Hepatic PPARα is critical in the metabolic adaptation to sepsis. J Hepatol (2019) 70(5):963–73. doi: 10.1016/j.jhep.2018.12.037
48. Zhang J, Ankawi G, Sun J, Digvijay K, Yin Y, Rosner MH, et al. Gut-kidney crosstalk in septic acute kidney injury. Crit Care (2018) 22(1):117. doi: 10.1186/s13054-018-2040-y
49. Dominguez JA, Coopersmith CM. Can we protect the gut in critical illness? The role of growth factors and other novel approaches. Crit Care Clin (2010) 26(3):549–65. doi: 10.1016/j.ccc.2010.04.005
50. Vaziri ND, Yuan J, Rahimi A, Ni Z, Said H, Subramanian VS. Disintegration of colonic epithelial tight junction in uremia: a likely cause of CKD-associated inflammation. Nephrol Dial Transplant (2012) 27(7):2686–93. doi: 10.1093/ndt/gfr624
51. Hayakawa M, Asahara T, Henzan N, Murakami H, Yamamoto H, Mukai N, et al. Dramatic changes of the gut flora immediately after severe and sudden insults. Dig Dis Sci (2011) 56(8):2361–5. doi: 10.1007/s10620-011-1649-3
52. Kim CH. Complex regulatory effects of gut microbial short-chain fatty acids on immune tolerance and autoimmunity. Cell Mol Immunol (2023) 20(4):341–50. doi: 10.1038/s41423-023-00987-1
53. Wang A, Li Z, Sun Z, Zhang D, Ma X. Gut-derived short-chain fatty acids bridge cardiac and systemic metabolism and immunity in heart failure. J Nutr Biochem (2023) 120:109370. doi: 10.1016/j.jnutbio.2023.109370
54. Lou X, Xue J, Shao R, Yang Y, Ning D, Mo C, et al. Fecal microbiota transplantation and short-chain fatty acids reduce sepsis mortality by remodeling antibiotic-induced gut microbiota disturbances. Front Immunol (2022) 13:1063543. doi: 10.3389/fimmu.2022.1063543
55. Giridharan VV, Generoso JS, Lence L, Candiotto G, Streck E, Petronilho F, et al. A crosstalk between gut and brain in sepsis-induced cognitive decline. J Neuroinflamm (2022) 19(1):114. doi: 10.1186/s12974-022-02472-4
56. Vinolo MA, Rodrigues HG, Hatanaka E, Sato FT, Sampaio SC, Curi R. Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J Nutr Biochem (2011) 22(9):849–55. doi: 10.1016/j.jnutbio.2010.07.009
57. Sorboni SG, Moghaddam HS, Jafarzadeh-Esfehani R, Soleimanpour S. A comprehensive review on the role of the gut microbiome in human neurological disorders. Clin Microbiol Rev (2022) 35(1):e0033820. doi: 10.1128/cmr.00338-20
58. Deng H, Li Z, Tan Y, Guo Z, Liu Y, Wang Y, et al. A novel strain of Bacteroides fragilis enhances phagocytosis and polarises M1 macrophages. Sci Rep (2016) 6:29401. doi: 10.1038/srep29401
59. Zhang H, Xu J, Wu Q, Fang H, Shao X, Ouyang X, et al. Gut microbiota mediates the susceptibility of mice to sepsis-associated encephalopathy by butyric acid. J Inflammation Res (2022) 15:2103–19. doi: 10.2147/jir.S350566
60. Yang W, Cong Y. Gut microbiota-derived metabolites in the regulation of host immune responses and immune-related inflammatory diseases. Cell Mol Immunol (2021) 18(4):866–77. doi: 10.1038/s41423-021-00661-4
61. Mittal R, Coopersmith CM. Redefining the gut as the motor of critical illness. Trends Mol Med (2014) 20(4):214–23. doi: 10.1016/j.molmed.2013.08.004
62. Chancharoenthana W, Sutnu N, Visitchanakun P, Sawaswong V, Chitcharoen S, Payungporn S, et al. Critical roles of sepsis-reshaped fecal virota in attenuating sepsis severity. Front Immunol (2022) 13:940935. doi: 10.3389/fimmu.2022.940935
63. Gai X, Wang H, Li Y, Zhao H, He C, Wang Z, et al. Fecal microbiota transplantation protects the intestinal mucosal barrier by reconstructing the gut microbiota in a murine model of sepsis. Front Cell Infect Microbiol (2021) 11:736204. doi: 10.3389/fcimb.2021.736204
64. Fernandes J, Su W, Rahat-Rozenbloom S, Wolever TM, Comelli EM. Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutr Diabetes (2014) 4(6):e121. doi: 10.1038/nutd.2014.23
65. Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther (2008) 27(2):104–19. doi: 10.1111/j.1365-2036.2007.03562.x
66. MaChado MG, Patente TA, Rouillé Y, Heumel S, Melo EM, Deruyter L, et al. Acetate Improves the Killing of Streptococcus pneumoniae by Alveolar Macrophages via NLRP3 Inflammasome and Glycolysis-HIF-1α Axis. Front Immunol (2022) 13:773261. doi: 10.3389/fimmu.2022.773261
67. Ragsdale SW, Pierce E. Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim Biophys Acta (2008) 1784(12):1873–98. doi: 10.1016/j.bbapap.2008.08.012
68. Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell (2016) 165(6):1332–45. doi: 10.1016/j.cell.2016.05.041
69. Nogal A, Valdes AM, Menni C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes (2021) 13(1):1–24. doi: 10.1080/19490976.2021.1897212
70. Hetzel M, Brock M, Selmer T, Pierik AJ, Golding BT, Buckel W. Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and electron-transferring flavoprotein. Eur J Biochem (2003) 270(5):902–10. doi: 10.1046/j.1432-1033.2003.03450.x
71. Scott KP, Martin JC, Campbell G, Mayer CD, Flint HJ. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium "Roseburia inulinivorans". J Bacteriol (2006) 188(12):4340–9. doi: 10.1128/jb.00137-06
72. Xie L, Alam MJ, Marques FZ, Mackay CR. A major mechanism for immunomodulation: Dietary fibres and acid metabolites. Semin Immunol (2023) 66:101737. doi: 10.1016/j.smim.2023.101737
73. Louis P, Duncan SH, McCrae SI, Millar J, Jackson MS, Flint HJ. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J Bacteriol (2004) 186(7):2099–106. doi: 10.1128/jb.186.7.2099-2106.2004
74. Wang X, Sun Z, Yang T, Lin F, Ye S, Yan J, et al. Sodium butyrate facilitates CRHR2 expression to alleviate HPA axis hyperactivity in autism-like rats induced by prenatal lipopolysaccharides through histone deacetylase inhibition. mSystems (2023) 8(4):e0041523. doi: 10.1128/msystems.00415-23
75. Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev (2001) 81(3):1031–64. doi: 10.1152/physrev.2001.81.3.1031
76. Lecerf JM, Dépeint F, Clerc E, Dugenet Y, Niamba CN, Rhazi L, et al. Xylo-oligosaccharide (XOS) in combination with inulin modulates both the intestinal environment and immune status in healthy subjects, while XOS alone only shows prebiotic properties. Br J Nutr (2012) 108(10):1847–58. doi: 10.1017/s0007114511007252
77. Le Leu RK, Winter JM, Christophersen CT, Young GP, Humphreys KJ, Hu Y, et al. Butyrylated starch intake can prevent red meat-induced O6-methyl-2-deoxyguanosine adducts in human rectal tissue: a randomised clinical trial. Br J Nutr (2015) 114(2):220–30. doi: 10.1017/s0007114515001750
78. Ghosh TS, Shanahan F, O'Toole PW. The gut microbiome as a modulator of healthy ageing. Nat Rev Gastroenterol Hepatol (2022) 19(9):565–84. doi: 10.1038/s41575-022-00605-x
79. Haskey N, Estaki M, Ye J, Shim RK, Singh S, Dieleman LA, et al. A Mediterranean Diet Pattern improves intestinal inflammation concomitant with reshaping of the bacteriome in ulcerative colitis: A randomized controlled trial. J Crohns Colitis (2023) jjad073. doi: 10.1093/ecco-jcc/jjad073
80. Yao Y, Cai X, Fei W, Ye Y, Zhao M, Zheng C. The role of short-chain fatty acids in immunity, inflammation and metabolism. Crit Rev Food Sci Nutr (2022) 62(1):1–12. doi: 10.1080/10408398.2020.1854675
81. Al-Lahham S, Roelofsen H, Rezaee F, Weening D, Hoek A, Vonk R, et al. Propionic acid affects immune status and metabolism in adipose tissue from overweight subjects. Eur J Clin Invest (2012) 42(4):357–64. doi: 10.1111/j.1365-2362.2011.02590.x
82. Fellows R, Varga-Weisz P. Chromatin dynamics and histone modifications in intestinal microbiota-host crosstalk. Mol Metab (2020) 38:100925. doi: 10.1016/j.molmet.2019.12.005
83. MacFie J, O'Boyle C, Mitchell CJ, Buckley PM, Johnstone D, Sudworth P. Gut origin of sepsis: a prospective study investigating associations between bacterial translocation, gastric microflora, and septic morbidity. Gut (1999) 45(2):223–8. doi: 10.1136/gut.45.2.223
84. Graspeuntner S, WasChina S, Künzel S, Twisselmann N, Rausch TK, Cloppenborg-Schmidt K, et al. Gut dysbiosis with bacilli dominance and accumulation of fermentation products precedes late-onset sepsis in preterm infants. Clin Infect Dis (2019) 69(2):268–77. doi: 10.1093/cid/ciy882
85. Xu W, Zhong M, Pan T, Qu H, Chen E. Gut microbiota and enteral nutrition tolerance in non-abdominal infection septic ICU patients: an observational study. Nutrients (2022) 14(24):5342. doi: 10.3390/nu14245342
86. Weng J, Wu H, Xu Z, Xi H, Chen C, Chen D, et al. The role of propionic acid at diagnosis predicts mortality in patients with septic shock. J Crit Care (2018) 43:95–101. doi: 10.1016/j.jcrc.2017.08.009
87. Li Z, Zhang F, Sun M, Liu J, Zhao L, Liu S, et al. The modulatory effects of gut microbes and metabolites on blood-brain barrier integrity and brain function in sepsis-associated encephalopathy. PeerJ (2023) 11:e15122. doi: 10.7717/peerj.15122
88. Wang H, Chen H, Lin Y, Wang G, Luo Y, Li X, et al. Butyrate glycerides protect against intestinal inflammation and barrier dysfunction in mice. Nutrients (2022) 14(19):3991. doi: 10.3390/nu14193991
89. Zhan Z, Tang H, Zhang Y, Huang X, Xu M. Potential of gut-derived short-chain fatty acids to control enteric pathogens. Front Microbiol (2022) 13:976406. doi: 10.3389/fmicb.2022.976406
90. Masui R, Sasaki M, Funaki Y, Ogasawara N, Mizuno M, Iida A, et al. G protein-coupled receptor 43 moderates gut inflammation through cytokine regulation from mononuclear cells. Inflammation Bowel Dis (2013) 19(13):2848–56. doi: 10.1097/01.MIB.0000435444.14860.ea
91. Wang F, Liu J, Weng T, Shen K, Chen Z, Yu Y, et al. The Inflammation Induced by Lipopolysaccharide can be Mitigated by Short-chain Fatty Acid, Butyrate, through Upregulation of IL-10 in Septic Shock. Scand J Immunol (2017) 85(4):258–63. doi: 10.1111/sji.12515
92. Filippone A, Lanza M, Campolo M, Casili G, Paterniti I, Cuzzocrea S, et al. The anti-inflammatory and antioxidant effects of sodium propionate. Int J Mol Sci (2020) 21(8):3026. doi: 10.3390/ijms21083026
93. Fu J, Li G, Wu X, Zang B. Sodium butyrate ameliorates intestinal injury and improves survival in a rat model of cecal ligation and puncture-induced sepsis. Inflammation (2019) 42(4):1276–86. doi: 10.1007/s10753-019-00987-2
94. Qiu J, Villa M, Sanin DE, Buck MD, O'Sullivan D, Ching R, et al. Acetate promotes T cell effector function during glucose restriction. Cell Rep (2019) 27(7):2063–74.e2065. doi: 10.1016/j.celrep.2019.04.022
95. Balmer ML, Ma EH, Bantug GR, Grählert J, Pfister S, Glatter T, et al. Memory CD8(+) T cells require increased concentrations of acetate induced by stress for optimal function. Immunity (2016) 44(6):1312–24. doi: 10.1016/j.immuni.2016.03.016
96. Sun Y, Varambally S, Maher CA, Cao Q, Chockley P, Toubai T, et al. Targeting of microRNA-142-3p in dendritic cells regulates endotoxin-induced mortality. Blood (2011) 117(23):6172–83. doi: 10.1182/blood-2010-12-325647
97. Cao L, Zhu T, Lang X, Jia S, Yang Y, Zhu C, et al. Inhibiting DNA methylation improves survival in severe sepsis by regulating NF-κB pathway. Front Immunol (2020) 11:1360. doi: 10.3389/fimmu.2020.01360
98. Eyenga P, Rey B, Eyenga L, Sheu SS. Regulation of oxidative phosphorylation of liver mitochondria in sepsis. Cells (2022) 11(10):1598. doi: 10.3390/cells11101598
99. Shang S, Liu J, Hua F. Protein acylation: mechanisms, biological functions and therapeutic targets. Signal Transduct Target Ther (2022) 7(1):396. doi: 10.1038/s41392-022-01245-y
100. Mowen KA, David M. Unconventional post-translational modifications in immunological signaling. Nat Immunol (2014) 15(6):512–20. doi: 10.1038/ni.2873
101. Zhang F, Qi L, Feng Q, Zhang B, Li X, Liu C, et al. HIPK2 phosphorylates HDAC3 for NF-κB acetylation to ameliorate colitis-associated colorectal carcinoma and sepsis. Proc Natl Acad Sci USA (2021) 118(28):e2021798118. doi: 10.1073/pnas.2021798118
102. Schliehe C, Flynn EK, Vilagos B, Richson U, Swaminanthan S, Bosnjak B, et al. The methyltransferase Setdb2 mediates virus-induced susceptibility to bacterial superinfection. Nat Immunol (2015) 16(1):67–74. doi: 10.1038/ni.3046
103. Chang G, Ma N, Zhang H, Wang Y, Huang J, Liu J, et al. Sodium butyrate modulates mucosal inflammation injury mediated by GPR41/43 in the cecum of goats fed a high concentration diet. Front Physiol (2019) 10:1130. doi: 10.3389/fphys.2019.01130
104. Campbell C, McKenney PT, Konstantinovsky D, Isaeva OI, Schizas M, Verter J, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature (2020) 581(7809):475–9. doi: 10.1038/s41586-020-2193-0
105. Yamada N, Karasawa T, Kimura H, Watanabe S, Komada T, Kamata R, et al. Ferroptosis driven by radical oxidation of n-6 polyunsaturated fatty acids mediates acetaminophen-induced acute liver failure. Cell Death Dis (2020) 11(2):144. doi: 10.1038/s41419-020-2334-2
106. Wu Z, Connolly J, Biggar KK. Beyond histones - the expanding roles of protein lysine methylation. FEBS J (2017) 284(17):2732–44. doi: 10.1111/febs.14056
107. Kaniskan HU, Martini ML, Jin J. Inhibitors of protein methyltransferases and demethylases. Chem Rev (2018) 118(3):989–1068. doi: 10.1021/acs.chemrev.6b00801
108. Xia M, Liu J, Wu X, Liu S, Li G, Han C, et al. Histone methyltransferase Ash1l suppresses interleukin-6 production and inflammatory autoimmune diseases by inducing the ubiquitin-editing enzyme A20. Immunity (2013) 39(3):470–81. doi: 10.1016/j.immuni.2013.08.016
109. Kaye DM, Shihata WA, Jama HA, Tsyganov K, Ziemann M, Kiriazis H, et al. Deficiency of prebiotic fiber and insufficient signaling through gut metabolite-sensing receptors leads to cardiovascular disease. Circulation (2020) 141(17):1393–403. doi: 10.1161/circulationaha.119.043081
110. Wang M, Lin H. Understanding the function of mammalian sirtuins and protein lysine acylation. Annu Rev Biochem (2021) 90:245–85. doi: 10.1146/annurev-biochem-082520-125411
111. Singh N, Thangaraju M, Prasad PD, Martin PM, Lambert NA, Boettger T, et al. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J Biol Chem (2010) 285(36):27601–8. doi: 10.1074/jbc.M110.102947
112. Feng T, Cao AT, Weaver CT, Elson CO, Cong Y. Interleukin-12 converts Foxp3+ regulatory T cells to interferon-γ-producing Foxp3+ T cells that inhibit colitis. Gastroenterology (2011) 140(7):2031–43. doi: 10.1053/j.gastro.2011.03.009
113. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature (2013) 504(7480):451–5. doi: 10.1038/nature12726
114. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature (2013) 504(7480):446–50. doi: 10.1038/nature12721
115. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science (2013) 341(6145):569–73. doi: 10.1126/science.1241165
116. Das B, Dobrowolski C, Shahir AM, Feng Z, Yu X, Sha J, et al. Short chain fatty acids potently induce latent HIV-1 in T-cells by activating P-TEFb and multiple histone modifications. Virology (2015) 474:65–81. doi: 10.1016/j.virol.2014.10.033
117. Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun (2015) 6:7320. doi: 10.1038/ncomms8320
118. Takahashi D, Hoshina N, Kabumoto Y, Maeda Y, Suzuki A, Tanabe H, et al. Microbiota-derived butyrate limits the autoimmune response by promoting the differentiation of follicular regulatory T cells. EBioMedicine (2020) 58:102913. doi: 10.1016/j.ebiom.2020.102913
119. Luu M, Pautz S, Kohl V, Singh R, Romero R, Lucas S, et al. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nat Commun (2019) 10(1):760. doi: 10.1038/s41467-019-08711-2
120. Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol (2015) 8(1):80–93. doi: 10.1038/mi.2014.44
121. Yang W, Yu T, Huang X, Bilotta AJ, Xu L, Lu Y, et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Commun (2020) 11(1):4457. doi: 10.1038/s41467-020-18262-6
122. Liu S, Yu H, Liu Y, Liu X, Zhang Y, Bu C, et al. Chromodomain protein CDYL acts as a crotonyl-coA hydratase to regulate histone crotonylation and spermatogenesis. Mol Cell (2017) 67(5):853–866.e855. doi: 10.1016/j.molcel.2017.07.011
123. Chan JC, Maze I. Histone crotonylation makes its mark in depression research. Biol Psychiatry (2019) 85(8):616–8. doi: 10.1016/j.biopsych.2019.01.025
124. Tang X, Chen XF, Sun X, Xu P, Zhao X, Tong Y, et al. Short-chain enoyl-coA hydratase mediates histone crotonylation and contributes to cardiac homeostasis. Circulation (2021) 143(10):1066–9. doi: 10.1161/circulationaha.120.049438
125. Wang S, Mu G, Qiu B, Wang M, Yu Z, Wang W, et al. The function and related diseases of protein crotonylation. Int J Biol Sci (2021) 17(13):3441–55. doi: 10.7150/ijbs.58872
126. Jiang G, Li C, Lu M, Lu K, Li H. Protein lysine crotonylation: past, present, perspective. Cell Death Dis (2021) 12(7):703. doi: 10.1038/s41419-021-03987-z
127. Liu X, Wei W, Liu Y, Yang X, Wu J, Zhang Y, et al. MOF as an evolutionarily conserved histone crotonyltransferase and transcriptional activation by histone acetyltransferase-deficient and crotonyltransferase-competent CBP/p300. Cell Discov (2017) 3:17016. doi: 10.1038/celldisc.2017.16
128. Sabari BR, Tang Z, Huang H, Yong-Gonzalez V, Molina H, Kong HE, et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol Cell (2015) 58(2):203–15. doi: 10.1016/j.molcel.2015.02.029
129. Fellows R, Denizot J, Stellato C, Cuomo A, Jain P, Stoyanova E, et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat Commun (2018) 9(1):105. doi: 10.1038/s41467-017-02651-5
130. Huang H, Luo Z, Qi S, Huang J, Xu P, Wang X, et al. Landscape of the regulatory elements for lysine 2-hydroxyisobutyrylation pathway. Cell Res (2018) 28(1):111–25. doi: 10.1038/cr.2017.149
131. Huang S, Tang D, Dai Y. Metabolic functions of lysine 2-hydroxyisobutyrylation. Cureus (2020) 12(8):e9651. doi: 10.7759/cureus.9651
132. Wang N, Jiang Y, Peng P, Liu G, Qi S, Liu K, et al. Quantitative proteomics reveals the role of lysine 2-hydroxyisobutyrylation pathway mediated by tip60. Oxid Med Cell Longev (2022) 2022:4571319. doi: 10.1155/2022/4571319
133. Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA (2008) 105(6):2117–22. doi: 10.1073/pnas.0712038105
134. Calvani R, Miccheli A, Capuani G, Tomassini Miccheli A, Puccetti C, Delfini M, et al. Gut microbiome-derived metabolites characterize a peculiar obese urinary metabotype. Int J Obes (Lond) (2010) 34(6):1095–8. doi: 10.1038/ijo.2010.44
135. Zhao S, Zhang X, Li H. Beyond histone acetylation-writing and erasing histone acylations. Curr Opin Struct Biol (2018) 53:169–77. doi: 10.1016/j.sbi.2018.10.001
136. Dong H, Zhai G, Chen C, Bai X, Tian S, Hu D, et al. Protein lysine de-2-hydroxyisobutyrylation by CobB in prokaryotes. Sci Adv (2019) 5(7):eaaw6703. doi: 10.1126/sciadv.aaw6703
137. Ge H, Li B, Chen W, Xu Q, Chen S, Zhang H, et al. Differential occurrence of lysine 2-hydroxyisobutyrylation in psoriasis skin lesions. J Proteomics (2019) 205:103420. doi: 10.1016/j.jprot.2019.103420
138. Yamazaki K, Miyauchi E, Kato T, Sato K, Suda W, Tsuzuno T, et al. Dysbiotic human oral microbiota alters systemic metabolism via modulation of gut microbiota in germ-free mice. J Oral Microbiol (2022) 14(1):2110194. doi: 10.1080/20002297.2022.2110194
139. Tsoukalas D, Fragoulakis V, Papakonstantinou E, Antonaki M, Vozikis A, Tsatsakis A, et al. Prediction of autoimmune diseases by targeted metabolomic assay of urinary organic acids. Metabolites (2020) 10(12):502. doi: 10.3390/metabo10120502
140. Zhao B, Lu R, Chen J, Xie M, Zhao X, Kong L. S100A9 blockade prevents lipopolysaccharide-induced lung injury via suppressing the NLRP3 pathway. Respir Res (2021) 22(1):45. doi: 10.1186/s12931-021-01641-y
141. Dong J, Li Y, Zheng F, Chen W, Huang S, Zhou X, et al. Co-occurrence of protein crotonylation and 2-hydroxyisobutyrylation in the proteome of end-stage renal disease. ACS Omega (2021) 6(24):15782–93. doi: 10.1021/acsomega.1c01161
142. Xie T, Dong J, Zhou X, Tang D, Li D, Chen J, et al. Proteomics analysis of lysine crotonylation and 2-hydroxyisobutyrylation reveals significant features of systemic lupus erythematosus. Clin Rheumatol (2022) 41(12):3851–8. doi: 10.1007/s10067-022-06254-4
143. Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics (2011) 10(12):M111.012658. doi: 10.1074/mcp.M111.012658
144. Li J, Zhao S, Zhou X, Zhang T, Zhao L, Miao P, et al. Inhibition of lipolysis by mercaptoacetate and etomoxir specifically sensitize drug-resistant lung adenocarcinoma cell to paclitaxel. PloS One (2013) 8(9):e74623. doi: 10.1371/journal.pone.0074623
145. Chen XF, Chen X, Tang X. Short-chain fatty acid, acylation and cardiovascular diseases. Clin Sci (Lond) (2020) 134(6):657–76. doi: 10.1042/cs20200128
146. Nishida Y, Rardin MJ, Carrico C, He W, Sahu AK, Gut P, et al. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol Cell (2015) 59(2):321–32. doi: 10.1016/j.molcel.2015.05.022
147. Lee H, Jang JH, Kim SJ. Malonic acid suppresses lipopolysaccharide-induced BV2 microglia cell activation by inhibiting the p38 MAPK/NF-κB pathway. Anim Cells Syst (Seoul) (2021) 25(2):110–8. doi: 10.1080/19768354.2021.1901781
148. Park C, Park J, Kim WJ, Kim W, Cheong H, Kim SJ. Malonic acid isolated from pinus densiflora inhibits UVB-induced oxidative stress and inflammation in haCaT keratinocytes. Polymers (Basel) (2021) 13(5):816. doi: 10.3390/polym13050816
149. Galván-Peña S, Carroll RG, Newman C, Hinchy EC, Palsson-McDermott E, Robinson EK, et al. Malonylation of GAPDH is an inflammatory signal in macrophages. Nat Commun (2019) 10(1):338. doi: 10.1038/s41467-018-08187-6
Keywords: sepsis, short-chain fatty acids (SCFAs), protein post-translational modifications (PTMs), immunity, inflammation
Citation: Zhang L, Shi X, Qiu H, Liu S, Yang T, Li X and Liu X (2023) Protein modification by short-chain fatty acid metabolites in sepsis: a comprehensive review. Front. Immunol. 14:1171834. doi: 10.3389/fimmu.2023.1171834
Received: 22 February 2023; Accepted: 15 September 2023;
Published: 06 October 2023.
Edited by:
Alessandra Stasi, University of Bari Aldo Moro, ItalyReviewed by:
Gianvito Caggiano, University of Bari Aldo Moro, ItalyUndurti Narasimha Das, UND Life Sciences LLC, United States
Copyright © 2023 Zhang, Shi, Qiu, Liu, Yang, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Liu, bGl1eDA3MDRAdG1tdS5lZHUuY24=; Xiaoli Li, bGl4aWFvbGlAY3FtdS5lZHUuY24=
†These authors have contributed equally to this work
 Liang Zhang
Liang Zhang Xinhui Shi
Xinhui Shi Hongmei Qiu1,2,3
Hongmei Qiu1,2,3 Sijia Liu
Sijia Liu Ting Yang
Ting Yang Xiaoli Li
Xiaoli Li Xin Liu
Xin Liu