- 1Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2Department of General Surgery, Peking Union Medical College Hospital (CAMS), Beijing, China
Interferon-gamma (IFN-γ) has been identified as a crucial factor in determining the responsiveness to immunotherapy. Produced primarily by natural killer (NK) and T cells, IFN-γ promotes activation, maturation, proliferation, cytokine expression, and effector function in immune cells, while simultaneously inducing antigen presentation, growth arrest, and apoptosis in tumor cells. However, tumor cells can hijack the IFN-γ signaling pathway to mount IFN-γ resistance: rather than increasing antigenicity and succumbing to death, tumor cells acquire stemness characteristics and express immunosuppressive molecules to defend against antitumor immunity. In this review, we summarize the potential mechanisms of IFN-γ resistance occurring at two critical stages: disrupted signal transduction along the IFNG/IFNGR/JAK/STAT pathway, or preferential expression of specific interferon-stimulated genes (ISGs). Elucidating the molecular mechanisms through which tumor cells develop IFN-γ resistance help identify promising therapeutic targets to improve immunotherapy, with broad application value in conjugation with targeted, antibody or cellular therapies.
Introduction
To date, over twenty distinct interferon (IFN) genes and proteins have been identified, typically categorized into three classes: Type I IFN (IFN-α and IFN-β), Type II IFN (IFN-γ), and Type III IFN (IFN-λ). IFN-γ is a notably different member characterized by unique receptor activity and distinct intracellular signaling pathway: type I IFNs depends on the interferon-stimulated gene factor-3 (ISGF3) complex containing STAT1/STAT2/IRF9, which binds to interferon-sensitive response elements (ISREs), whereas the phosphorylated signal transducer and activator of transcription 1 (STAT1) homodimer downstream of IFN-γ binds to interferon-gamma activation sites (GASs). However, these members share common interferon-stimulated genes (ISGs), but also have distinct gene profiles (1). For example, IFN-γ preferentially induces the expression of IRF1, while HIF-1 responses primarily to IFN-β (2). IFN-γ signaling and ISG expression is marked by extensive interaction with regulatory molecules, epigenetic modifications and chromatin remodeling, constituting multiple positive and negative feedbacks (3, 4). IFN-γ also produce variable effects at different duration and concentration of exposure, with acute exposure to high concentrations of IFN-γ leading to growth arrest and apoptosis, while chronic exposure to low concentrations promotes cell survival (5). The IFN-γ signaling pathway has extensive crosstalks with the PI3K, MAPK/p38, and other cellular pathways (6). Collectively, these mechanisms maintain immunological homeostasis, ensuring an effective immune response while keeping the immune activity in check to avoid damage from over activation. However, these regulatory mechanisms can also be exploited by tumors to mount IFN-γ resistance.
IFN-γ in tumor
Source
IFN-γ in tumors is primarily produced by activated immune cells, particularly NK, cytotoxic CD8+ and Th1 cells (7, 8). Evidence also suggests that macrophages, dendritic cells (DC), and innate B cells can produce IFN-γ (9–13). While tumor cells have been reported to rely on autocrine secretion of IFN-β (5, 14), further investigation is required to determine whether tumor cells secrete IFN-γ (15). IFN-γ production is stimulated by specific cytokines, including interleukin-12 (IL-12), interleukin-18 (IL-18), and macrophage colony-stimulating factor (M-CSF) (9, 16, 17), as well as receptor signaling by activating natural killer cell receptors (NKRs) and CD16 in NK cells and the T cell receptor (TCR) in T cells (17). These stimulations activate the Src/MAPK/ERK/p38 pathway (18), ultimately inducing IFNG expression through transcription factors STAT4, T-bet, AP-1, Eomes, Fos, and Jun (18, 19). Notably, GAS is present in the promoter IRF1 (20, 21), STAT1, IFNGR, and IFN-γ (22), imposing positive feedback and self-sustained inflammation (3, 19). The most recent report identified the interaction between GATA3 and CNS-28 silencer as a restrainer of IFN-y mRNA transcription by diminishing enhancer-promoter interactions within the gene locus (23).
Effect
IFN-γ is known to regulate the expression of hundreds of ISGs, whose differential expression patterns confer various effects including growth suppression, induction of apoptosis, but also activation and secretion of pro-inflammatory cytokines, and other functions such as expression of immunosuppressive genes. However, the mechanisms underlying these opposing effects under the same set of signals, receptors, and main signaling molecules require further investigation.
Immune cells
In CD8 T cells, IFN-γ is crucial in T cell expansion (24), induces the differentiation of precursor T cells into effector T cells (25), further enhances their cytotoxicity and motility (26), and participates in the formation of immunological memory (27). In CD4 T cells, IFN-γ is pro-Th1 and antitumor, repressing Th2 and Th17 polarization (7, 28). In NK cells, IFN-γ induces CXCR3 expression to allow tumor infiltration (29) and TRAIL expression to promote cytotoxicity (30). In myeloid cells, IFN-γ promotes polarization toward the inflammatory cDC1 (31) and M1 (7, 32, 33). In B cells, IFN-γ not only cooperates with IL-12 to mediate class switching (34), but also aids germinal center formation through BCL6 (35).
Interestingly, IFN-γ also exerts suppressive effects on certain immune cells, producing both wanted and unwanted effects. IFN-γ stimulation generates ‘fragile’ regulatory T cells (Tregs) with impaired suppressive activity yet maintained Treg phenotype in terms of FOXP3+ (7, 36). However, chronic exposure and constitutive signaling of IFN-γ impair T cell functions and lead to exhaustion (37), not only through the well-studied induction of IDO1 and PD-L1 expression, but also through direct inhibition of T cell stemness, proliferation, clonal diversity, and maintenance (38–40), along with pro-apoptotic effect in the contraction phase through FAS and BIM (41, 42). Additionally, IFN-γ inhibits memory formation by limiting IL-7Ra in models of influenza virus infection (42).
Tumor cells
In general, IFN-γ exerts antitumor actions in two ways: directly through growth inhibition and indirectly through increased killing by immune cells (43). IFN-γ has growth-inhibitory effect through cell death induction by apoptosis (44–46) and necroptosis (47), and growth arrest by senescence (48–51). The immunostimulatory effects of increased MHC-I, antigen presentation, and antigenicity has been extensively characterized in both infection and tumor models (52).
On the other hand, IFN-γ itself and ISG signatures are pro-tumor in various cancer types including breast cancer (46, 53) and glioblastoma (54). Mechanistically, IFN-γ directly acts on tumor cells to maintain survival (BCL2 and surviving expression) and stemness (55–57), decreases antigen presentation (58), enhances the expression of immunosuppressive molecules markedly ICBs and IDO (58–60), and leads to infiltration of tumor-associated neutrophil (46). Additionally, chronic exposure to IFN-γ inevitably induce immunosuppressive cells, such as myeloid-derived suppressor cells (MDSCs), Tregs and exhausted T cells, ultimately leading to immune evasion and tumor growth.
Resistance
Ideally, IFN-γ stimulation would force the tumor cells to increase their antigenicity and secrete pro-inflammatory cytokines, along with growth arrest and pro-apoptosis. However, tumor cells can suppress these immunogenic ISGs and favor the expression immunosuppressive and pro-tumor ISGs instead. IFN-γ resistance is a major obstacle in cancer immunotherapy, offering protection against T cell cytotoxicity and ICBs, and allowing tumor cells to exploit IFN-γ signaling for survival advantage rather than succumb to antitumor immunity. Elucidating the mechanisms of interferon resistance and developing new strategies to tackle it are important areas of research, with great potential to identify effective therapeutic target as adjuvant to current cancer immunotherapy. Briefly, there are two principle mechanisms though which tumor cells mount IFN-γ resistance: 1) disruption in the primary signaling pathway, IFNGR/JAK/STAT, responsible for effector functions of IFN-γ stimulation; and 2) epigenetic or regulatory background leading to favorable expression of some ISGs over others. Therefore, we next summarize recent findings regarding the molecular mechanisms behind the aberrant IFN-γ response in tumor cells. It is important note the limitation in generalizability across different experimental models, as these mechanisms are often context-specific.
IFN-γ signaling
The canonical pathway of IFNGR/JAK1/STAT1/ISGs is the main effector downstream of IFN-γ signaling. Signal transduction along this pathway is affected by genomic mutation, transcriptional regulation of expression, post-translational modifications and protein-protein interaction, and sub-cellular localization (Figure 1), along with further complexity due to homologous members of the same family with overlapping but distinct functions (7, 61–63).
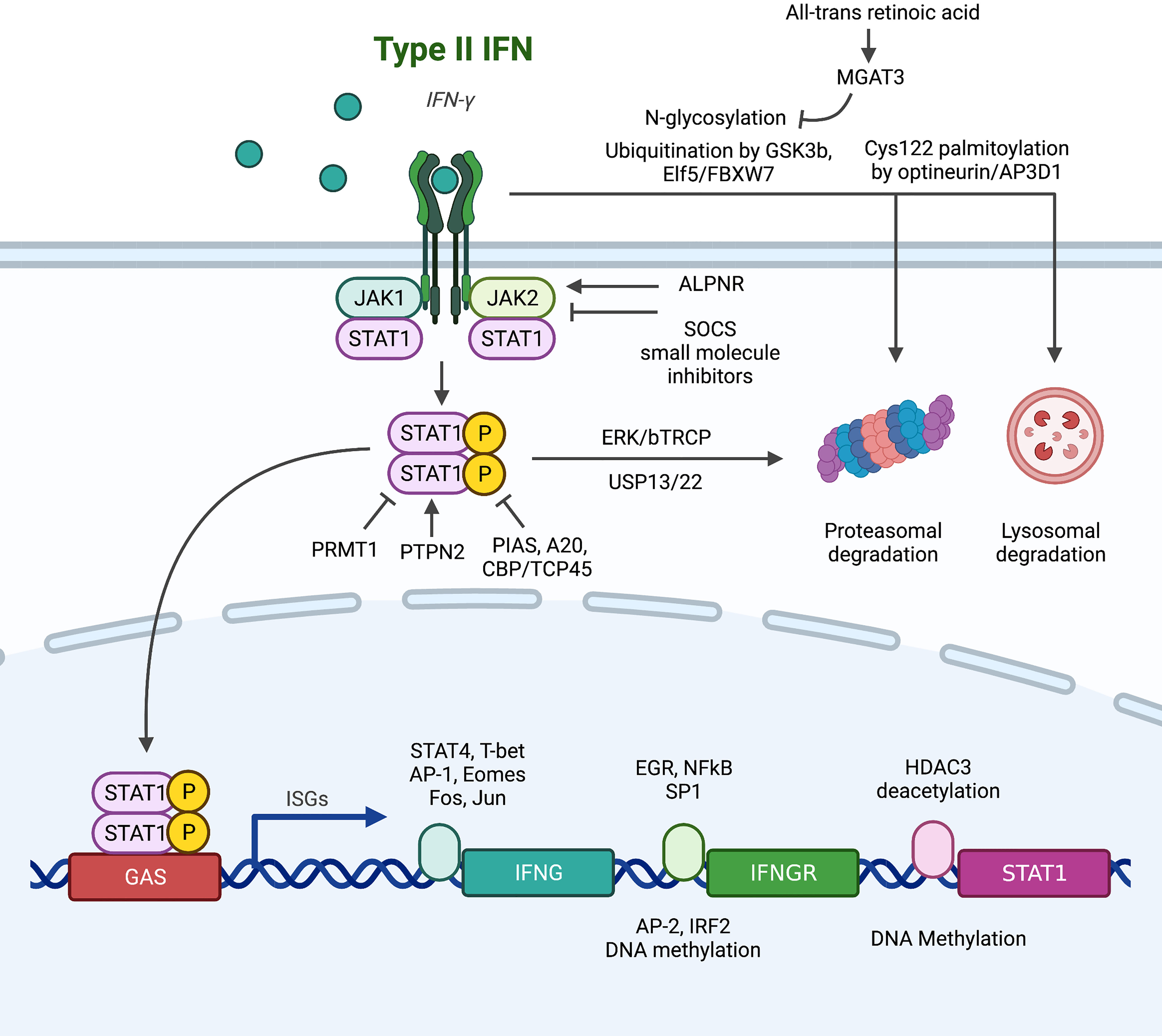
Figure 1 Summary of alterations in the conventional downstream signaling pathway of IFNGR/JAK/STAT that lead to IFN-γ resistance. (1) At the IFNGR level: transcriptionally, EGR, NFκB and SP1 promote the transcription of IFNGR mRNA, while AP2 and IRF2 are suppressive transcription factors. Post-transcriptionally, N-glycosylation stabilizes plasma membrane IFNGR, while degradation is promoted by ubiquitination and palmitoylation. (2) At the JAK level, SOCS and small molecule inhibitors abrogate signaling, and ALPNR is crucial for normal functioning. (3) At the STAT1 level, post-translational modification by PIAS, A20, and CBP/TCP45 suppress signaling activity through deposphorylation of p-STAT1, while PRMT1, ERK/bTRCP, and USP13/22 regulate STAT1 activity or stability independent of its phosphorylation status.
IFNGR
IFN-γR is composed of two subunits: IFNGR1 (α-subunit) has high affinity to IFN-γ and a major role in ligand binding, while IFNGR2 (β-subunit) is predominantly responsible for downstream signaling via JAK recruitment (61, 64). Upregulation of IFNGR on colorectal cancer stem cell confers sensitivity to chemotherapy through apoptosis (56), but increased IFNGR expression on CD8+ T cells leads to T cell apoptosis and ICB resistance (38). Th1 cells can downregulate IFNGR to enhance survival and maintain antitumor effects (28), but tumor cells also developed multiple mechanisms to abrogate IFNGR function. At the genomic level, IFNGR has high mutation frequency of 12%, comparable with JAK1, JAK2, and IRF1, in melanoma patients resistant to anti-CTLA4 therapy (65). At the transcription level, IFN-γR is under dynamic regulation by transcription factors. EGR and NFkB (66) and SP1 in breast cancer (67) have been reported to induce the mRNA expression of IFNGR, while AP2 in breast cancer (67) and IRF2-mediated negative feedback in esophageal cancer (68) have suppressive roles. Chronic IFN-γ stimulation in colorectal cancer cell line mounts IFNGR resistance through DNA methylation, a process fully reversible by 5-Aza-deoxycytidine (69). At the post-translational level, IFNGR dynamically localizes between plasma, endosomal and nuclear membranes, leading to three potential fates: 1) recycling back to plasma membrane for further signaling, 2) degradation to attenuates function, 3) nuclear translocation. The degradation of IFNGR is under extensive regulation. IFNGR is subjected to degradation by the proteasome through GSK3b (70), ELF5/FBXW7 (46) and N-glycosylation (reversible by all-trans retinoic acid induces MGAT expression) (71) mediated ubiquitination in monocytic cell lines, breast cancer, and colorectal cancer respectively. Optineurin/AP3D1 mediated Cys122 palmitoylation targets IFNGR for lysosomal degradation in colorectal cancer (72). Furthermore, the nuclear import of IFNGR has been recently reported be functionally significant in breast cancer (51), mechanistically by bringing STAT1 into the nucleus (73–75) and direct GAS binding (76, 77).
JAKs
As IFNGR lacks intrinsic kinase activity and only functions as a scaffold upon dimerization induced by IFN-γ, the four members of tyrosine kinase adaptors, JAK1, JAK2, JAK3, and TYK2, are needed to carry out subsequent signaling by recruitment and phosphorylation of STAT1. Loss-of-function mutations in JAK1 and JAK2 are extensively reported to be the cause of ICB resistance in melanoma patients (65, 78–82) and gynecologic cancers (83). JAKs are also the primary brake for negative regulation of IFN-γ activity by protein-protein interactions. The well-studied suppressor of cytokine signaling (SOCS) proteins bind to activated JAK catalytic sites (84) and are constantly active in melanoma cell lines to suppress interferon response (85). Disruption of APLNR-JAK1 interaction (81) both capable of abrogating IFN-γ activity. However, JAKs are often reported to have pro-tumor roles, highly expressed and correlated with poor survival in pancreatic cancer (86). The activating mutation (S703I) of JAK1 elevates p-STAT3 and STAT5 in liver cancer, driving tumor progression (87). Hence, lots of JAK inhibitors have been developed and put under clinical testing (88, 89). A phase II study (NCT01423604) of ruxolitinib and capecitabine/gemcitabine improved survival of metastatic pancreatic cancer patients with CRP>13 mg/L (90), and a phase Ib/II study (NCT01858883) of itacitinib and nab-paclitaxel and gemcitabine achieved objective response rate of 24% in patients with solid tumors (91). However, subsequent Phase III trials JANUS 1 (NCT02117479) and JANUS 2 (NCT02119663) (92), along with phase I trials of other agents including the selective JAK1 inhibitor INCB047986 (NCT01929941) (89) and momelotinib + gemcitabine and nab-paclitaxel (NCT02101021) (93) or capecitabine and oxaliplatin (NCT02244489), have all been terminated due to lack of efficacy. Despite much failure, there are still ongoing studies awaiting results, such as the phase Ib study of ruxolitinib and trametinib (MEK inhibitor) in patients with RAS mutations (NCT04303403). Another agent, AG490, was effective in preclinical model of mouse pancreatic cancer but awaits clinical validation (94). The challenge in clinical application of JAK inhibitors suggested the need for more specific interventions such as STAT3 inhibitors (95, 96), or better selection of patients that may benefit from JAK inhibitors.
STATs
The seven members of the STAT family, STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6, are often simultaneously activated, leading to diverse, cell-specific effects depending on abundance and availability of each member. Among them, STAT3 and STAT5 are often considered pro-tumor (97), while STAT1 is generally considered the key downstream effector of IFN-γ, capable binding GAS and inducing ISG transcriptions. STAT1 has been reported to be protective in gastric cancer (98), liver cancer (99), and melanoma (100), but hazardous in glioblastoma (101) and sarcoma (102), and controversial in breast (103–106) and pancreatic cancer. STAT1 is under extensive post-translational modifications: 1) STAT1 is targeted for proteasomal degradation by ERK/bTRCP (107, 108) and stabilized by USP13 (109) and USP22 (110) mediated deubiquitination; 2) aberrant phosphorylation of STAT1 due to negative regulators PIAS (111), A20 (112) and CBP/TCP45 (113); and 3) functional impairment due to arginine methylation by PRMT1 (114) and loss of the helper PTPN2 (115). Interestingly, unphosphorylated STAT1 (u-STAT1) is also functional (116). Marked by persistence for several days post-IFN-γ stimulation, u-STAT1 are reservoir for phosphorylation-activation, allowing IFN-γ stimulation to prime for IFN-α response (117, 118). u-STAT1 is also responsible for the constitutive baseline expression caspases, conferring sensitivity to apoptotic signals (119). These functions of u-STAT1 further suggest the importance of expression control by epigenetic modifications of the STAT1 promoter: suppressed by methylation (120) and enhanced by HDAC3 mediated deacetylation (121).
ISGs
IFN-γ stimulation changes the expression of hundred of genes, collectively producing a wide spectrum of effects. The large number and dynamic nature of ISGs render a consensus definition challenging. It is important to acknowledge the context-specific expression pattern of ISGs, who spearhead changes in cellular pathways and activities, directly corresponding to the effector functions of IFN-γ (Figure 2). Several key members of ISGs with important functional consequences, such as IRF1, CXCL9/10, MHC-I/II, and PDL1, have been extensively studied. The general pattern and mechanism behind the simultaneous expression of multiple ISGs must also be kept in mind, as simple protein-protein interactions may be insufficient to characterize such extensive regulation. Epigenetic remodeling such as histone and promoter modifications, chromatin conformation and accessibility, transcription-factor binding, latent enhancer and promoter regulations have crucial but rather unelucidated roles (122).
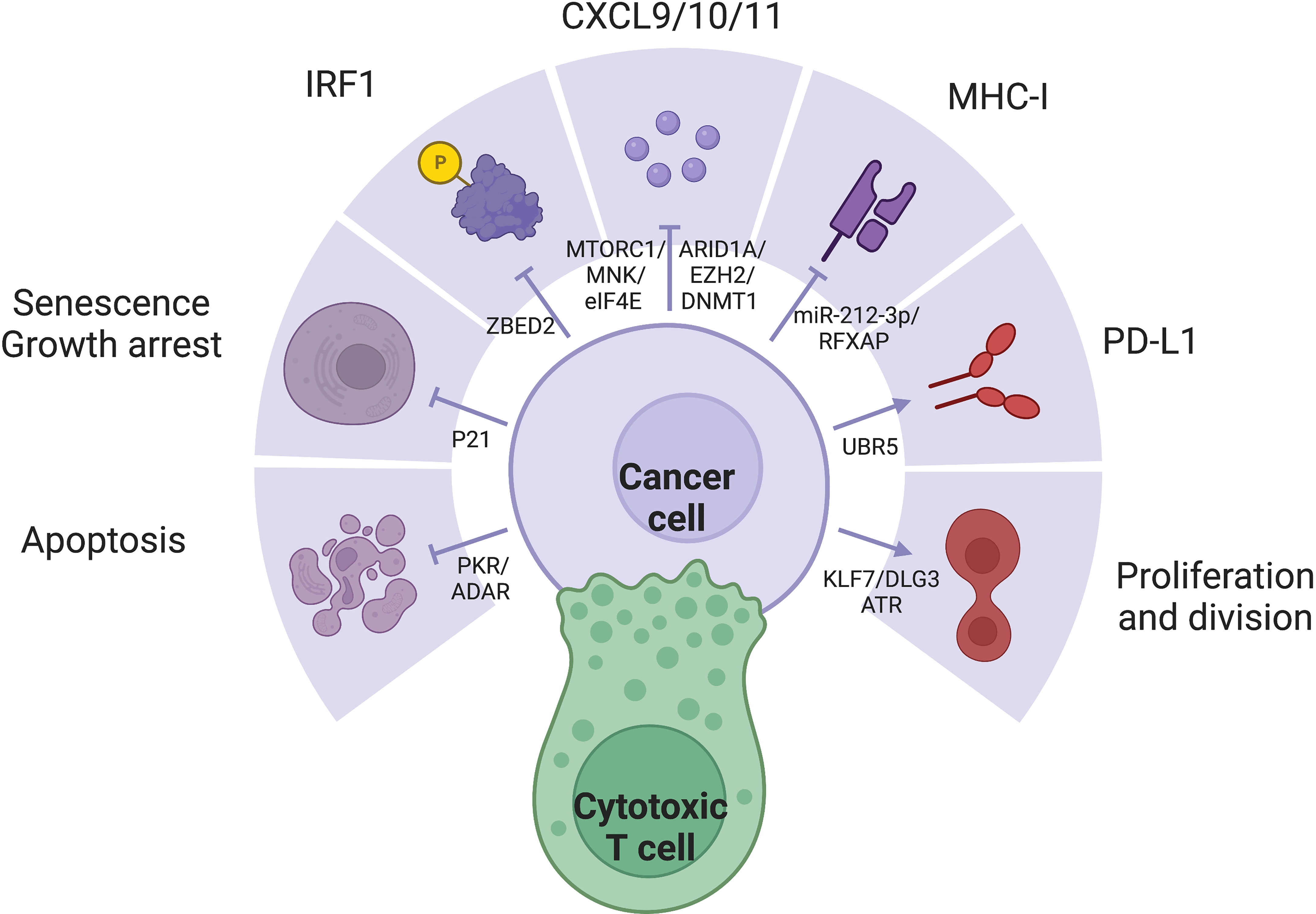
Figure 2 IFN-γ exerts a variety of effects through diverse expression pattern of ISGs, which is often dysregulated in cancer cells: (1) IRF1 is a major downstream effector of IFN-γ with diverse functions. (2) IFN-y represses tumor through apoptosis, senescence, growth arrest, secretion of inflammatory cytokines, and MHC-I expression. (3) IFN-γ signaling may also promote tumor through proliferation and PD-L1 expression.
Definition
Being able to clearly define ISGs would aid in selecting the appropriate therapeutic target. There are currently two approaches: in silicon or in vitro. The in silicon definition of ISGs rely on GAS, a consensus, palindromic DNA pattern (TTCN2-4 GAA) found in the promoters of ISGs. Its recognition and binding by p-STAT1 dimer produce profound effect in modulating the expression of its downstream genes. Hence, the SABioscience website, the Transcription Factor binding site search tools and the REFINEMENT program can identify GAS distributed throughout the human and mouse genome, theoretically denoting the downstream genes as ISGs (123). In vitro definitions of ISGs are primarily based on high-throughput characterization of gene expression changes after IFN-γ stimulation. Microarray analysis identified more than 300 ISGs assigned into categories (124). Human fibrosarcoma cell line treated with IFN-α, IFN-β and IFN-γ identified shared and distinct ISGs between these classes (2). ISGs have also been characterized in the Huh7 and Huh7.5 hepatocellular cell lines (125). However, these results are heavily dependent on the model and background, limiting their generalizability. The large number of ISGs, dynamic and context-dependent expression profile affected by epigenetic modification and chromatin remodeling, the degeneration and non-redundancy between the three classes of IFNs (126), and different STAT and IRF isoforms all pose challenges to a consensus definition of ISGs (127).
Key members
Interferon regulatory factors (IRFs) are major ISGs (128). The nine members of the IRF family share similar structures. The homologous N-terminal DNA-binding domain (DBD) recognizes the helix-loop-helix motif of IRF-element (IRFE) motifs upstream of their effector genes; while the C-terminal, responsible for protein interaction confer the diversity of regulation. IRF1/5/6/8/9 have been reported to be protective and pro-apoptotic, while IRF2/3/4/7 are often considered hazardous (20). Among the IRF members, IRF1 is the major driver of the expression of many ISGs (129, 130). With a GAS but not ISRE in its promoter, IRF1 is induced by IFN-γ rather than IFN-α and IFN-β, as a major distinction between the effects of type I and II IFNs (131). IRF8, whose expression is often jointed with IRF1, enhances the pro-apoptotic effect of IRF1 (132). IFN-γ exerts antiproliferative effects through cell cycle control and cell death induction. IFN-γ mainly depends on STAT1 activity to induce pro-death molecules including p53 in ovarian cancer (133, 134), caspase 1/3/8 in pancreatic cancer (135–137) and FAS/FASL (138), and the necroptotic RIP1 (47). IFN-γ also arrest growth through p16/p21-induced senescence (48, 51, 139) and inhibition of PI3K/AKT pathway (140). IFN-γ increases antigenicity through antigen processing and presentation, upregulating both MHC-I/II themselves (141, 142), as well as antigen processing machinery including TAP1/2, invariant chain, and the expression and activity of the proteasome (143), and the MHC class II transactivator (CIITA) (144). IFN-γ pathway directly transcribes pro-inflammatory cytokines especially CXCL9/10/11 (145). However, IFN-γ also induces the expression of the ICBs including but not limited to PDL1 and CTLA4 (146, 147), along with other immunosuppressive mechanisms such as IDO (148). Favorable expression of these key ISGs with drastically different functions profoundly influence the outcome of IFN-γ stimulation.
Regulation
The importance of the SWI/SNF complex mediated chromatin remodeling in IFN-y response was further supported by its crucial rolel in the coordination of T cell activation and exhaustion (149). RNF138 mediated K48-linked polyubiquitination at position Lys643 of SMARCC1 drives proteasomal degradation, inhibiting chromatin remodeling at SWI/SNF-regulated gene loci, suppress transcription of late inflammatory genes of macrophages in innate immune activation (150). On a global scale, differences in genomic accessibility due to chromatin remodeling allow the preferential expression of some ISGs over others. In melanoma cell lines, the PBAF form of the SWI/SNF chromatin remodeling complex suppresses IFN-γ signaling and cytokine gene transcription to resist T cell infiltration and cytotoxicity (151). Employment of CRISPR screening, Chip-seq and ATAC-seq may further identify other regulatory mechanisms of chromatin remodeling in different models (126), shedding light on comprehensive. More specifically, many molecules have been reported to selectively affect key ISGs, rendering them potential therapeutic targets for precise modulation of IFN-γ response. ZBED2 antagonizes IRF1 and drives T cell dysfunction (4, 152). Important antitumor cytokines CXCL9 and CXCL10 are negatively regulated by epigenetic silencing by EZH2/DNMT1 in ovarian cancer and antagonized by ARID1A (153, 154), but activated by mTORC1/MNK/eIF4E (32). Inability to upregulate MHC I expression has been the earliest reported sign to IFN resistance reported in 33% melanoma cell lines and 24% lung adenocarcinoma cell lines (155). MHC-II is suppressed by c-myc activity (156), miR-212-3p through inhibition of RFXAP (157) and DNMT1/3B mediated DNA methylation that epigenetically inactivates CIITA (158–160), as a crucial limiting factor to tumor antigenicity (161). UBR5 promotes IFN-γ and STAT1 dependent PDL1 expression (162, 163), leading to combination of IFN-γ and nivolumab achieved the best response (164). KLF7/DLG3 maintains Golgi integrity and expression of pro-tumor ISGs (165). U-STAT1 expression without Y701 phosphorylation induce MUC4 expression in PDAC cells and exerts pro-EMT functions (166). ATR mediated nucleotide metabolism help maintain survival despite IFN stimulation (167). The dsRNA-activated kinase PKR confers lethality by IFN stimulation, a process antagonized by the dsRNA editing enzyme ADAR, which maintains survival despite the expression of death-inducing ISGs (168). IFN-γ activates the RhoGDI2/Rac1/NF-κB pathway in tumor cells to reduce the production of the pro-tumor cytokine CXCL8, promoting tumor apoptosis (169) and reducing CXCR2+ M2 macrophages (170). Targeting of tumor cell–intrinsic resistance mechanisms to T cell-mediated cytotoxicity is important and complementary to checkpoint inhibitors.
Therapeutics
Prognostic
IFN-γ activity is necessary and prognostic in immunotherapy of multiple cancer types (171). At the genomic level, mutation of key molecules along the IFN-γ signaling pathway are common in ICB refractory patients with melanoma (65), lung cancer (172) and gastric cancer (173). At the transcriptomic level, high expression of IFN-γ itself and IFN-γ responsive genes, reflective of a T cell-inflamed microenvironment, correspond to better survival and ICB response across different cancer types (174), especially lung cancer (175) and melanoma (176, 177). Aberrant attenuation of the IFN-γ pathway confers ICB resistance through several mechanisms, including both primary resistance by suppression of PDL1 expression (79) and acquired resistance by MHC-I downregulation (78).
Cellular therapy
IFN-γ is especially important in cellular immunity and adoptive cell transfer. Patients with pancreatic cancer have decreased plasma IFN-γ compared to normal people (averaged 16.4 fg/L versus 27.4 fg/L), potentially leading to low CD4:CD8 ratio and systemic immunosuppression (178). Higher plasma IFN-γ after TILs administration is associated with prolonged T cell persistence and better survival (179). The cytotoxicity of T and NK cells heavily depends on IFN-γ secretion (180–182). In the context of CAR-T cells, IFN-γ secretion upon activation (183) not only exerts self-sustaining effects but also establishes cytokine crosstalk with endogenous T and NK cells (26, 184). Interestingly, tumor response to IFN-γ, such as APLNR interaction with JAK1 in melanoma cells (81), is crucial for CAR-T cytotoxicity only in solid tumors but not hematological tumors (185). IFN-γ abrogation offers protection against CRS (186) without impairing the therapeutic efficacy of anti-CD19 CAR-T in leukemias or lymphomas (187). CAR-T with an excessively high IFN-γ expression has been reported to upregulate PD-L1 expression in cancer cells, leading to their own dysfunction (188).
As therapeutic agent
The clinical application of IFN-γ covered a wide variety of diseases such as cancer, infectious diseases, and autoimmune disorders (189). The antitumor efficacy of direct IFN-γ administration has long been debated. Initial attempts in renal cell carcinoma was disappointing (190), but later studies in prostate cancer (191) and bladder cancer had promising results (73.4% versus 57.2% with no recurrence during follow-up) (192). IFN-γ in ovarian cancer had controversial results: 2/6 patients experienced a 90% reduction in tumor cells in ascites (193), but limited responses have also been reported (194, 195).
The short half-life and cost of protein production led to much effort into the optimization of IFN-γ products, aiming to minimize adverse effect, amplify antitumor activity, and optimize cost and convenience of delivery. Apart from clinically available forms including the recombinant protein (IFN-γ1b, Actimmune), adenovirus vectors that express IFN-γ cDNA (TG-1041, TG-1042), and neutralizing antibodies against IFN-γ (HuZaf and AMG811), researchers have also reported: 1) engineered chimeric proteins such as chTNT3/muIFN-y (196), PLGF2/IFN-α (197), PDGFbR/IFN-γ (198) aiming to achieve specific and targeted delivery; 2) liposome- and nonmaterial-aided delivery (199) to reduce DNA damage and oxidative stress in lymphocytes; 3) as adjuvant treatment in conjugation with oncolytic viruses (200) and adoptive cell therapy; 4) loaded into cellular therapy, such as IFN-γ-secreting stromal cells with TME-homing capacities (201), and fourth generation CAR-T cells (TRUNKS) engineered to secrete IFN-γ constitutively or upon activation.
Perspectives
IFN-γ is the central cytokine in anti-tumor immunity, not only indispensable for its supportive role in immune cell maturation and activation, but more importantly as an effector function. Increased antigenicity and apoptosis due to IFN-γ stimulation are the foundation of T cell cytotoxicity. However, tumor cells can take advantage the extensive regulatory mechanisms along the IFN-γ signaling pathway and the diversity of ISGs, favoring the expression of pro-tumor ISGs over others. Elucidating the mechanisms by which tumor cells hijack the IFN-γ pathway to suppress antitumor response including MHC-I and CXCL9/10/11expression, while preferentially enhancing the expression of immunosuppressive molecules such as PD-L1 and IDO1, would shed light on the conflicting identify of IFN-γ in tumor progression, along with providing therapeutic targets to improve current immunotherapies. In summary, tumor cells can mount IFN-γ resistance at two stages: disruption of the main signal transduction pathway responsible for all downstream effects of IFN-γ stimulation, and the alternative expression/suppression of specific key ISG or a group of ISGs with synergistic functions. Tumors cells can abrogate the IFNGR/JAK/STAT pathway through the following mechanisms: 1) loss of function mutation of key genes, most commonly IFNGR1/IFNGR2/JAK1/JAK2; 2) suppressed mRNA transcription of IFNG, IFNGR, and STAT1 leading to lower expression; 3) loss of crucial protein cooperators such as ALPNR/JAK1, PTPN2/STAT1; 3) aberrant hyperactivity of protein inhibitors such as SOCS, PIAS, and PRMT1; 4) post-translational modification, especially ubiquitination, subjecting the protein to degradation by the proteasome or lysosome. Further downstream, tumor cells can preferentially express some ISGs over others through 1) global chromatin remodeling and 2) specific modulations of key ISGs.
IFN-γ based therapies face several challenges that limit effectiveness: the development of resistance by tumor cells, and general adverse effects due to non-specific, systemic delivery. These mechanisms of IFN-γ resistance described above offer promising therapeutic targets to fine-tune IFN-γ response and improve immunotherapy. However, as these results were mostly based on experimental models, further confirmation and validations in real-world clinical data would provide valuable insights on the prevalence and significance of these potential targets.
Author contributions
JH and MW contributed equally to this work. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci U S A. (2012) 109(11):4239–44. doi: 10.1073/pnas.1114981109
2. Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. (1998) 95(26):15623–8. doi: 10.1073/pnas.95.26.15623
3. Michalska A, Blaszczyk K, Wesoly J, Bluyssen HAR. A positive feedback amplifier circuit that regulates interferon (IFN)-stimulated gene expression and controls type I and type II IFN responses. Front Immunol (2018) 9:1135. doi: 10.3389/fimmu.2018.01135
4. Somerville TDD, Xu Y, Wu XS, Maia-Silva D, Hur SK, de Almeida LMN, et al. ZBED2 is an antagonist of interferon regulatory factor 1 and modifies cell identity in pancreatic cancer. Proc Natl Acad Sci U S A. (2020) 117(21):11471–82. doi: 10.1073/pnas.1921484117
5. Cheon H, Wang Y, Wightman SM, Jackson MW, Stark GR. How cancer cells make and respond to interferon-I. Trends Cancer. (2023) 9(1):83–92. doi: 10.1016/j.trecan.2022.09.003
6. Platanias LC. Mechanisms of type-i- and type-II-interferon-mediated signalling. Nat Rev Immunol (2005) 5(5):375–86. doi: 10.1038/nri1604
7. Gocher AM, Workman CJ, Vignali DAA. Interferon-gamma: teammate or opponent in the tumour microenvironment? Nat Rev Immunol (2022) 22(3):158–72. doi: 10.1038/s41577-021-00566-3
8. Murray PD, McGavern DB, Pease LR, Rodriguez M. Cellular sources and targets of IFN-gamma-mediated protection against viral demyelination and neurological deficits. Eur J Immunol (2002) 32(3):606–15. doi: 10.1002/1521-4141(200203)32:3<606::AID-IMMU606>3.0.CO;2-D
9. Darwich L, Coma G, Pena R, Bellido R, Blanco EJ, Este JA, et al. Secretion of interferon-gamma by human macrophages demonstrated at the single-cell level after costimulation with interleukin (IL)-12 plus IL-18. Immunology. (2009) 126(3):386–93. doi: 10.1111/j.1365-2567.2008.02905.x
10. Mezouar S, Mege JL. Changing the paradigm of IFN-gamma at the interface between innate and adaptive immunity: macrophage-derived IFN-gamma. J Leukoc Biol (2020) 108(1):419–26. doi: 10.1002/JLB.4MIR0420-619RR
11. Bogdan C, Schleicher U. Production of interferon-gamma by myeloid cells–fact or fancy? Trends Immunol (2006) 27(6):282–90. doi: 10.1016/j.it.2006.04.004
12. Taieb J, Chaput N, Menard C, Apetoh L, Ullrich E, Bonmort M, et al. A novel dendritic cell subset involved in tumor immunosurveillance. Nat Med (2006) 12(2):214–9. doi: 10.1038/nm1356
13. Bao Y, Liu X, Han C, Xu S, Xie B, Zhang Q, et al. Identification of IFN-gamma-producing innate b cells. Cell Res (2014) 24(2):161–76. doi: 10.1038/cr.2013.155
14. Yu R, Zhu B, Chen D. Type I interferon-mediated tumor immunity and its role in immunotherapy. Cell Mol Life Sci (2022) 79(3):191. doi: 10.1007/s00018-022-04219-z
15. von Locquenghien M, Rozalen C, Celia-Terrassa T. Interferons in cancer immunoediting: sculpting metastasis and immunotherapy response. J Clin Invest. (2021) 131(1):e143296. doi: 10.1172/JCI143296
16. Harris DP, Goodrich S, Gerth AJ, Peng SL, Lund FE. Regulation of IFN-gamma production by b effector 1 cells: essential roles for T-bet and the IFN-gamma receptor. J Immunol (2005) 174(11):6781–90. doi: 10.4049/jimmunol.174.11.6781
17. Mah AY, Cooper MA. Metabolic regulation of natural killer cell IFN-gamma production. Crit Rev Immunol (2016) 36(2):131–47. doi: 10.1615/CritRevImmunol.2016017387
18. Schoenborn JR, Wilson CB. Regulation of interferon-gamma during innate and adaptive immune responses. Adv Immunol (2007) 96:41–101. doi: 10.1016/S0065-2776(07)96002-2
19. Jorgovanovic D, Song M, Wang L, Zhang Y. Roles of IFN-gamma in tumor progression and regression: a review. biomark Res (2020) 8:49. doi: 10.1186/s40364-020-00228-x
20. Feng H, Zhang YB, Gui JF, Lemon SM, Yamane D. Interferon regulatory factor 1 (IRF1) and anti-pathogen innate immune responses. PLoS Pathog (2021) 17(1):e1009220. doi: 10.1371/journal.ppat.1009220
21. Horvath CM, Wen Z, Darnell JE Jr. A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev (1995) 9(8):984–94. doi: 10.1101/gad.9.8.984
22. Decker T, Kovarik P, Meinke A. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J Interferon Cytokine Res (1997) 17(3):121–34. doi: 10.1089/jir.1997.17.121
23. Cui K, Chen Z, Cao Y, Liu S, Ren G, Hu G, et al. Restraint of IFN-γ expression through a distal silencer CNS-28 for tissue homeostasis. Immunity. (2023) 4:S1074–7613(23)00126–7. doi: 10.1016/j.immuni.2023.03.006
24. Whitmire JK, Tan JT, Whitton JL. Interferon-gamma acts directly on CD8+ T cells to increase their abundance during virus infection. J Exp Med (2005) 201(7):1053–9. doi: 10.1084/jem.20041463
25. Curtsinger JM, Agarwal P, Lins DC, Mescher MF. Autocrine IFN-gamma promotes naive CD8 T cell differentiation and synergizes with IFN-alpha to stimulate strong function. J Immunol (2012) 189(2):659–68. doi: 10.4049/jimmunol.1102727
26. Bhat P, Leggatt G, Waterhouse N, Frazer IH. Interferon-gamma derived from cytotoxic lymphocytes directly enhances their motility and cytotoxicity. Cell Death Dis (2017) 8(6):e2836. doi: 10.1038/cddis.2017.67
27. Whitmire JK, Eam B, Benning N, Whitton JL. Direct interferon-gamma signaling dramatically enhances CD4+ and CD8+ T cell memory. J Immunol (2007) 179(2):1190–7. doi: 10.4049/jimmunol.179.2.1190
28. Pernis A, Gupta S, Gollob KJ, Garfein E, Coffman RL, Schindler C, et al. Lack of interferon gamma receptor beta chain and the prevention of interferon gamma signaling in TH1 cells. Science. (1995) 269(5221):245–7. doi: 10.1126/science.7618088
29. Wendel M, Galani IE, Suri-Payer E, Cerwenka A. Natural killer cell accumulation in tumors is dependent on IFN-gamma and CXCR3 ligands. Cancer Res (2008) 68(20):8437–45. doi: 10.1158/0008-5472.CAN-08-1440
30. Park SY, Seol JW, Lee YJ, Cho JH, Kang HS, Kim IS, et al. IFN-gamma enhances TRAIL-induced apoptosis through IRF-1. Eur J Biochem (2004) 271(21):4222–8. doi: 10.1111/j.1432-1033.2004.04362.x
31. Pan J, Zhang M, Wang J, Wang Q, Xia D, Sun W, et al. Interferon-gamma is an autocrine mediator for dendritic cell maturation. Immunol Lett (2004) 94(1-2):141–51. doi: 10.1016/j.imlet.2004.05.003
32. Su X, Yu Y, Zhong Y, Giannopoulou EG, Hu X, Liu H, et al. Interferon-gamma regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat Immunol (2015) 16(8):838–49. doi: 10.1038/ni.3205
33. Ivleva E, Andreeva N, Grivennikov S. IFN-γ signaling in myeloid cells regulates pancreatic cancer growth and progression. Cancer Res (2022) 82(12_Supplement):2117–. doi: 10.1158/1538-7445.AM2022-2117
34. Metzger DW, McNutt RM, Collins JT, Buchanan JM, Van Cleave VH, Dunnick WA. Interleukin-12 acts as an adjuvant for humoral immunity through interferon-gamma-dependent and -independent mechanisms. Eur J Immunol (1997) 27(8):1958–65. doi: 10.1002/eji.1830270820
35. Jackson SW, Jacobs HM, Arkatkar T, Dam EM, Scharping NE, Kolhatkar NS, et al. B cell IFN-gamma receptor signaling promotes autoimmune germinal centers via cell-intrinsic induction of BCL-6. J Exp Med (2016) 213(5):733–50. doi: 10.1084/jem.20151724
36. Overacre-Delgoffe AE, Chikina M, Dadey RE, Yano H, Brunazzi EA, Shayan G, et al. Interferon-gamma drives t(reg) fragility to promote anti-tumor immunity. Cell. (2017) 169(6):1130–41.e11. doi: 10.1016/j.cell.2017.05.005
37. Tau GZ, Cowan SN, Weisburg J, Braunstein NS, Rothman PB. Regulation of IFN-gamma signaling is essential for the cytotoxic activity of CD8(+) T cells. J Immunol (2001) 167(10):5574–82. doi: 10.4049/jimmunol.167.10.5574
38. Pai CS, Huang JT, Lu X, Simons DM, Park C, Chang A, et al. Clonal deletion of tumor-specific T cells by interferon-gamma confers therapeutic resistance to combination immune checkpoint blockade. Immunity. (2019) 50(2):477–92.e8. doi: 10.1016/j.immuni.2019.01.006
39. Mazet JM, Mahale JN, Tong O, Watson RA, Lechuga-Vieco AV, Pirgova G, et al. IFNgamma signaling in cytotoxic T cells restricts anti-tumor responses by inhibiting the maintenance and diversity of intra-tumoral stem-like T cells. Nat Commun (2023) 14(1):321. doi: 10.1038/s41467-023-35948-9
40. Berner V, Liu H, Zhou Q, Alderson KL, Sun K, Weiss JM, et al. IFN-gamma mediates CD4+ T-cell loss and impairs secondary antitumor responses after successful initial immunotherapy. Nat Med (2007) 13(3):354–60. doi: 10.1038/nm1554
41. Ghosh PK, Antretter H. Left coronary artery system source diseases. Surg issues. J Cardiovasc Surg (Torino). (1990) 31(3):298–304.
42. Prabhu N, Ho AW, Wong KH, Hutchinson PE, Chua YL, Kandasamy M, et al. Gamma interferon regulates contraction of the influenza virus-specific CD8 T cell response and limits the size of the memory population. J Virol (2013) 87(23):12510–22. doi: 10.1128/JVI.01776-13
43. Zaidi MR. The interferon-gamma paradox in cancer. J Interferon Cytokine Res (2019) 39(1):30–8. doi: 10.1089/jir.2018.0087
44. Fang C, Weng T, Hu S, Yuan Z, Xiong H, Huang B, et al. IFN-gamma-induced ER stress impairs autophagy and triggers apoptosis in lung cancer cells. Oncoimmunology. (2021) 10(1):1962591. doi: 10.1080/2162402X.2021.1962591
45. Yu M, Peng Z, Qin M, Liu Y, Wang J, Zhang C, et al. Interferon-gamma induces tumor resistance to anti-PD-1 immunotherapy by promoting YAP phase separation. Mol Cell (2021) 81(6):1216–30.e9. doi: 10.1016/j.molcel.2021.01.010
46. Singh S, Kumar S, Srivastava RK, Nandi A, Thacker G, Murali H, et al. Loss of ELF5-FBXW7 stabilizes IFNGR1 to promote the growth and metastasis of triple-negative breast cancer through interferon-gamma signalling. Nat Cell Biol (2020) 22(5):591–602. doi: 10.1038/s41556-020-0495-y
47. Thapa RJ, Basagoudanavar SH, Nogusa S, Irrinki K, Mallilankaraman K, Slifker MJ, et al. NF-kappaB protects cells from gamma interferon-induced RIP1-dependent necroptosis. Mol Cell Biol (2011) 31(14):2934–46. doi: 10.1128/MCB.05445-11
48. Chin YE, Kitagawa M, Su WC, You ZH, Iwamoto Y, Fu XY. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. (1996) 272(5262):719–22. doi: 10.1126/science.272.5262.719
49. Brenner E, Schorg BF, Ahmetlic F, Wieder T, Hilke FJ, Simon N, et al. Cancer immune control needs senescence induction by interferon-dependent cell cycle regulator pathways in tumours. Nat Commun (2020) 11(1):1335. doi: 10.1038/s41467-020-14987-6
50. Chen W, Wang W, Chen L, Chen J, Lu X, Li Z, et al. Long-term G(1) cell cycle arrest in cervical cancer cells induced by co-immobilized TNF-alpha plus IFN-gamma polymeric drugs. J Mater Chem B (2018) 6(2):327–36. doi: 10.1039/C7TB02608K
51. Garcia-Tunon I, Ricote M, Ruiz AA, Fraile B, Paniagua R, Royuela M. Influence of IFN-gamma and its receptors in human breast cancer. BMC Cancer. (2007) 7:158. doi: 10.1186/1471-2407-7-158
52. Martini M, Testi MG, Pasetto M, Picchio MC, Innamorati G, Mazzocco M, et al. IFN-gamma-mediated upmodulation of MHC class I expression activates tumor-specific immune response in a mouse model of prostate cancer. Vaccine. (2010) 28(20):3548–57. doi: 10.1016/j.vaccine.2010.03.007
53. Heimes AS, Hartner F, Almstedt K, Krajnak S, Lebrecht A, Battista MJ, et al. Prognostic significance of interferon-gamma and its signaling pathway in early breast cancer depends on the molecular subtypes. Int J Mol Sci (2020) 21(19):7178. doi: 10.3390/ijms21197178
54. Khan S, Mahalingam R, Sen S, Martinez-Ledesma E, Khan A, Gandy K, et al. Intrinsic interferon signaling regulates the cell death and mesenchymal phenotype of glioblastoma stem cells. Cancers (Basel) (2021) 13(21):5284. doi: 10.3390/cancers13215284
55. Qadir AS, Ceppi P, Brockway S, Law C, Mu L, Khodarev NN, et al. CD95/Fas increases stemness in cancer cells by inducing a STAT1-dependent type I interferon response. Cell Rep (2017) 18(10):2373–86. doi: 10.1016/j.celrep.2017.02.037
56. Ni C, Wu P, Zhu X, Ye J, Zhang Z, Chen Z, et al. IFN-gamma selectively exerts pro-apoptotic effects on tumor-initiating label-retaining colon cancer cells. Cancer Lett (2013) 336(1):174–84. doi: 10.1016/j.canlet.2013.04.029
57. Song M, Ping Y, Zhang K, Yang L, Li F, Zhang C, et al. Low-dose IFNγ induces tumor cell stemness in tumor microenvironment of non–small cell lung CancerLow-dose IFNγ induces tumor stemness in NSCLC. Cancer Res (2019) 79(14):3737–48. doi: 10.1158/0008-5472.CAN-19-0596
58. Le Poole IC, Riker AI, Quevedo ME, Stennett LS, Wang E, Marincola FM, et al. Interferon-gamma reduces melanosomal antigen expression and recognition of melanoma cells by cytotoxic T cells. Am J Pathol (2002) 160(2):521–8. doi: 10.1016/S0002-9440(10)64871-7
59. Beatty GL, Paterson Y. IFN-gamma can promote tumor evasion of the immune system in vivo by down-regulating cellular levels of an endogenous tumor antigen. J Immunol (2000) 165(10):5502–8. doi: 10.4049/jimmunol.165.10.5502
60. Mo X, Zhang H, Preston S, Martin K, Zhou B, Vadalia N, et al. Interferon-gamma signaling in melanocytes and melanoma cells regulates expression of CTLA-4. Cancer Res (2018) 78(2):436–50. doi: 10.1158/0008-5472.CAN-17-1615
61. Green DS, Young HA, Valencia JC. Current prospects of type II interferon gamma signaling and autoimmunity. J Biol Chem (2017) 292(34):13925–33. doi: 10.1074/jbc.R116.774745
62. Du W, Frankel TL, Green M, Zou W. IFNgamma signaling integrity in colorectal cancer immunity and immunotherapy. Cell Mol Immunol (2022) 19(1):23–32. doi: 10.1038/s41423-021-00735-3
63. Owen KL, Brockwell NK, Parker BS. JAK-STAT signaling: a double-edged sword of immune regulation and cancer progression. Cancers (Basel). (2019) 11(12):2002. doi: 10.3390/cancers11122002
64. Farrar MA, Schreiber RD. The molecular cell biology of interferon-gamma and its receptor. Annu Rev Immunol (1993) 11:571–611. doi: 10.1146/annurev.iy.11.040193.003035
65. Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, et al. Loss of IFN-gamma pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. (2016) 167(2):397–404.e9. doi: 10.1016/j.cell.2016.08.069
66. Ebensperger C, Rhee S, Muthukumaran G, Lembo D, Donnelly R, Pestka S, et al. Genomic organization and promoter analysis of the gene ifngr2 encoding the second chain of the mouse interferon-gamma receptor. Scand J Immunol (1996) 44(6):599–606. doi: 10.1046/j.1365-3083.1996.d01-353.x
67. Chen C, Guo L, Shi M, Hu M, Hu M, Yu M, et al. Modulation of IFN-gamma receptor 1 expression by AP-2alpha influences IFN-gamma sensitivity of cancer cells. Am J Pathol (2012) 180(2):661–71. doi: 10.1016/j.ajpath.2011.10.040
68. Wang Y, Liu D, Chen P, Koeffler HP, Tong X, Xie D. Negative feedback regulation of IFN-gamma pathway by IFN regulatory factor 2 in esophageal cancers. Cancer Res (2008) 68(4):1136–43. doi: 10.1158/0008-5472.CAN-07-5021
69. Britzen-Laurent N, Straube J, Waldner M, Becker C, Croner R, Pilarsky C, et al. Loss of IFN-γ pathway gene expression in tumor cells as mechanism of immune escape of colorectal carcinoma. Cancer Res (2018) 78(13_Supplement):4047–. doi: 10.1158/1538-7445.AM2018-4047
70. Londino JD, Gulick DL, Lear TB, Suber TL, Weathington NM, Masa LS, et al. Post-translational modification of the interferon-gamma receptor alters its stability and signaling. Biochem J (2017) 474(20):3543–57. doi: 10.1042/BCJ20170548
71. Krug J, Rodrian G, Petter K, Yang H, Khoziainova S, Guo W, et al. N-glycosylation regulates intrinsic IFN-gamma resistance in colorectal cancer: implications for immunotherapy. Gastroenterology. (2023) 164(3):392–406.e5. doi: 10.1053/j.gastro.2022.11.018
72. Du W, Hua F, Li X, Zhang J, Li S, Wang W, et al. Loss of optineurin drives cancer immune evasion via palmitoylation-dependent IFNGR1 lysosomal sorting and degradation. Cancer Discovery (2021) 11(7):1826–43. doi: 10.1158/2159-8290.CD-20-1571
73. Larkin J 3rd, Johnson HM, Subramaniam PS. Differential nuclear localization of the IFNGR-1 and IFNGR-2 subunits of the IFN-gamma receptor complex following activation by IFN-gamma. J Interferon Cytokine Res (2000) 20(6):565–76. doi: 10.1089/10799900050044769
74. Ahmed CM, Burkhart MA, Mujtaba MG, Subramaniam PS, Johnson HM. The role of IFNgamma nuclear localization sequence in intracellular function. J Cell Sci (2003) 116(Pt 15):3089–98. doi: 10.1242/jcs.00528
75. Bader T, Weitzerbin J. Nuclear accumulation of interferon gamma. Proc Natl Acad Sci U S A. (1994) 91(25):11831–5. doi: 10.1073/pnas.91.25.11831
76. Subramaniam PS, Green MM, Larkin J 3rd, Torres BA, Johnson HM. Nuclear translocation of IFN-gamma is an intrinsic requirement for its biologic activity and can be driven by a heterologous nuclear localization sequence. J Interferon Cytokine Res (2001) 21(11):951–9. doi: 10.1089/107999001753289569
77. Ahmed CM, Johnson HM. IFN-gamma and its receptor subunit IFNGR1 are recruited to the IFN-gamma-activated sequence element at the promoter site of IFN-gamma-activated genes: evidence of transactivational activity in IFNGR1. J Immunol (2006) 177(1):315–21. doi: 10.4049/jimmunol.177.1.315
78. Sucker A, Zhao F, Pieper N, Heeke C, Maltaner R, Stadtler N, et al. Acquired IFNgamma resistance impairs anti-tumor immunity and gives rise to T-cell-resistant melanoma lesions. Nat Commun (2017) 8:15440. doi: 10.1038/ncomms15440
79. Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discovery (2017) 7(2):188–201. doi: 10.1158/2159-8290.CD-16-1223
80. Hargadon KM, Gyorffy B, McGee TJ. Genomic and transcriptional changes in IFNgamma pathway genes are putative biomarkers of response to ipilimumab immunotherapy in melanoma patients. Expert Rev Clin Immunol (2020) 16(12):1099–103. doi: 10.1080/1744666X.2021.1847644
81. Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, et al. Identification of essential genes for cancer immunotherapy. Nature. (2017) 548(7669):537–42. doi: 10.1038/nature23477
82. Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med (2016) 375(9):819–29. doi: 10.1056/NEJMoa1604958
83. Ren Y, Zhang Y, Liu RZ, Fenstermacher DA, Wright KL, Teer JK, et al. JAK1 truncating mutations in gynecologic cancer define new role of cancer-associated protein tyrosine kinase aberrations. Sci Rep (2013) 3:3042. doi: 10.1038/srep03042
84. Liau NPD, Laktyushin A, Lucet IS, Murphy JM, Yao S, Whitlock E, et al. The molecular basis of JAK/STAT inhibition by SOCS1. Nat Commun (2018) 9(1):1558. doi: 10.1038/s41467-018-04013-1
85. Lesinski GB, Zimmerer JM, Kreiner M, Trefry J, Bill MA, Young GS, et al. Modulation of SOCS protein expression influences the interferon responsiveness of human melanoma cells. BMC Cancer. (2010) 10:142. doi: 10.1186/1471-2407-10-142
86. Song Y, Tang MY, Chen W, Wang Z, Wang SL. High JAK2 protein expression predicts a poor prognosis in patients with resectable pancreatic ductal adenocarcinoma. Dis Markers. (2020) 2020:7656031. doi: 10.1155/2020/7656031
87. Yang S, Luo C, Gu Q, Xu Q, Wang G, Sun H, et al. Activating JAK1 mutation may predict the sensitivity of JAK-STAT inhibition in hepatocellular carcinoma. Oncotarget. (2016) 7(5):5461–9. doi: 10.18632/oncotarget.6684
88. Lin CM, Cooles FA, Isaacs JD. Basic mechanisms of JAK inhibition. Mediterr J Rheumatol (2020) 31(Suppl 1):100–4. doi: 10.31138/mjr.31.1.100
89. Qureshy Z, Johnson DE, Grandis JR. Targeting the JAK/STAT pathway in solid tumors. J Cancer Metastasis Treat (2020) 6.
90. Hurwitz HI, Uppal N, Wagner SA, Bendell JC, Beck JT, Wade SM 3rd, et al. Randomized, double-blind, phase II study of ruxolitinib or placebo in combination with capecitabine in patients with metastatic pancreatic cancer for whom therapy with gemcitabine has failed. J Clin Oncol (2015) 33(34):4039–47. doi: 10.1200/JCO.2015.61.4578
91. Beatty GL, Shahda S, Beck T, Uppal N, Cohen SJ, Donehower R, et al. A phase Ib/II study of the JAK1 inhibitor, itacitinib, plus nab-paclitaxel and gemcitabine in advanced solid tumors. Oncologist. (2019) 24(1):14–e0. doi: 10.1634/theoncologist.2017-0665
92. Hurwitz H, Van Cutsem E, Bendell J, Hidalgo M, Li CP, Salvo MG, et al. Ruxolitinib + capecitabine in advanced/metastatic pancreatic cancer after disease progression/intolerance to first-line therapy: JANUS 1 and 2 randomized phase III studies. Invest New Drugs (2018) 36(4):683–95. doi: 10.1007/s10637-018-0580-2
93. Ng K, Hendifar A, Starodub A, Chaves J, Yang Y, Koh B, et al. Phase 1 dose-escalation study of momelotinib, a janus kinase 1/2 inhibitor, combined with gemcitabine and nab-paclitaxel in patients with previously untreated metastatic pancreatic ductal adenocarcinoma. Invest New Drugs (2019) 37(1):159–65. doi: 10.1007/s10637-018-0650-5
94. Palagani V, Bozko P, El Khatib M, Belahmer H, Giese N, Sipos B, et al. Combined inhibition of notch and JAK/STAT is superior to monotherapies and impairs pancreatic cancer progression. Carcinogenesis. (2014) 35(4):859–66. doi: 10.1093/carcin/bgt394
95. Li X, Jiang W, Dong S, Li W, Zhu W, Zhou W. STAT3 inhibitors: a novel insight for anticancer therapy of pancreatic cancer. Biomolecules. (2022) 12(10):1450. doi: 10.3390/biom12101450
96. Shan H, Yao S, Ye Y, Yu Q. 3-Deoxy-2beta,16-dihydroxynagilactone e, a natural compound from podocarpus nagi, preferentially inhibits JAK2/STAT3 signaling by allosterically interacting with the regulatory domain of JAK2 and induces apoptosis of cancer cells. Acta Pharmacol Sin (2019) 40(12):1578–86. doi: 10.1038/s41401-019-0254-4
97. Verhoeven Y, Tilborghs S, Jacobs J, De Waele J, Quatannens D, Deben C, et al. The potential and controversy of targeting STAT family members in cancer. Semin Cancer Biol (2020) 60:41–56. doi: 10.1016/j.semcancer.2019.10.002
98. Deng H, Zhen H, Fu Z, Huang X, Zhou H, Liu L. The antagonistic effect between STAT1 and survivin and its clinical significance in gastric cancer. Oncol Lett (2012) 3(1):193–9. doi: 10.3892/ol.2011.423
99. Hosui A, Klover P, Tatsumi T, Uemura A, Nagano H, Doki Y, et al. Suppression of signal transducers and activators of transcription 1 in hepatocellular carcinoma is associated with tumor progression. Int J Cancer. (2012) 131(12):2774–84. doi: 10.1002/ijc.27580
100. Meyer S, Wild PJ, Vogt T, Bataille F, Ehret C, Gantner S, et al. Methylthioadenosine phosphorylase represents a predictive marker for response to adjuvant interferon therapy in patients with malignant melanoma. Exp Dermatol (2010) 19(8):e251–7. doi: 10.1111/j.1600-0625.2010.01072.x
101. Duarte CW, Willey CD, Zhi D, Cui X, Harris JJ, Vaughan LK, et al. Expression signature of IFN/STAT1 signaling genes predicts poor survival outcome in glioblastoma multiforme in a subtype-specific manner. PLoS One (2012) 7(1):e29653. doi: 10.1371/journal.pone.0029653
102. Zimmerman MA, Rahman NT, Yang D, Lahat G, Lazar AJ, Pollock RE, et al. Unphosphorylated STAT1 promotes sarcoma development through repressing expression of fas and bad and conferring apoptotic resistance. Cancer Res (2012) 72(18):4724–32. doi: 10.1158/0008-5472.CAN-12-1347
103. Widschwendter A, Tonko-Geymayer S, Welte T, Daxenbichler G, Marth C, Doppler W. Prognostic significance of signal transducer and activator of transcription 1 activation in breast cancer. Clin Cancer Res (2002) 8(10):3065–74.
104. Khodarev N, Ahmad R, Rajabi H, Pitroda S, Kufe T, McClary C, et al. Cooperativity of the MUC1 oncoprotein and STAT1 pathway in poor prognosis human breast cancer. Oncogene. (2010) 29(6):920–9. doi: 10.1038/onc.2009.391
105. Magkou C, Giannopoulou I, Theohari I, Fytou A, Rafailidis P, Nomikos A, et al. Prognostic significance of phosphorylated STAT-1 expression in premenopausal and postmenopausal patients with invasive breast cancer. Histopathology. (2012) 60(7):1125–32. doi: 10.1111/j.1365-2559.2011.04143.x
106. Tymoszuk P, Charoentong P, Hackl H, Spilka R, Muller-Holzner E, Trajanoski Z, et al. High STAT1 mRNA levels but not its tyrosine phosphorylation are associated with macrophage infiltration and bad prognosis in breast cancer. BMC Cancer. (2014) 14:257. doi: 10.1186/1471-2407-14-257
107. Soond SM, Townsend PA, Barry SP, Knight RA, Latchman DS, Stephanou A. ERK and the f-box protein betaTRCP target STAT1 for degradation. J Biol Chem (2008) 283(23):16077–83. doi: 10.1074/jbc.M800384200
108. Zhang Y, Chen Y, Liu Z, Lai R. ERK is a negative feedback regulator for IFN-gamma/STAT1 signaling by promoting STAT1 ubiquitination. BMC Cancer. (2018) 18(1):613. doi: 10.1186/s12885-018-4539-7
109. Yeh HM, Yu CY, Yang HC, Ko SH, Liao CL, Lin YL. Ubiquitin-specific protease 13 regulates IFN signaling by stabilizing STAT1. J Immunol (2013) 191(6):3328–36. doi: 10.4049/jimmunol.1300225
110. Li M, Xu Y, Liang J, Lin H, Qi X, Li F, et al. USP22 deficiency in melanoma mediates resistance to T cells through IFNgamma-JAK1-STAT1 signal axis. Mol Ther (2021) 29(6):2108–20. doi: 10.1016/j.ymthe.2021.02.018
111. Niu GJ, Xu JD, Yuan WJ, Sun JJ, Yang MC, He ZH, et al. Protein inhibitor of activated STAT (PIAS) negatively regulates the JAK/STAT pathway by inhibiting STAT phosphorylation and translocation. Front Immunol (2018) 9:2392. doi: 10.3389/fimmu.2018.02392
112. Yin L, Fang Z, Shen NJ, Qiu YH, Li AJ, Zhang YJ. Downregulation of A20 increases the cytotoxicity of IFN-gamma in hepatocellular carcinoma cells. Drug Des Devel Ther (2017) 11:2841–50. doi: 10.2147/DDDT.S135993
113. Kramer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Guhrs KH, et al. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev (2009) 23(2):223–35. doi: 10.1101/gad.479209
114. Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, et al. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. (2001) 104(5):731–41. doi: 10.1016/S0092-8674(01)00269-0
115. Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. (2017) 547(7664):413–8. doi: 10.1038/nature23270
116. Cheon H, Stark GR. Unphosphorylated STAT1 prolongs the expression of interferon-induced immune regulatory genes. Proc Natl Acad Sci U S A. (2009) 106(23):9373–8. doi: 10.1073/pnas.0903487106
117. Tassiulas I, Hu X, Ho H, Kashyap Y, Paik P, Hu Y, et al. Amplification of IFN-alpha-induced STAT1 activation and inflammatory function by syk and ITAM-containing adaptors. Nat Immunol (2004) 5(11):1181–9. doi: 10.1038/ni1126
118. Lehtonen A, Matikainen S, Julkunen I. Interferons up-regulate STAT1, STAT2, and IRF family transcription factor gene expression in human peripheral blood mononuclear cells and macrophages. J Immunol (1997) 159(2):794–803. doi: 10.4049/jimmunol.159.2.794
119. Kumar A, Commane M, Flickinger TW, Horvath CM, Stark GR. Defective TNF-alpha-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science. (1997) 278(5343):1630–2. doi: 10.1126/science.278.5343.1630
120. Xi S, Dyer KF, Kimak M, Zhang Q, Gooding WE, Chaillet JR, et al. Decreased STAT1 expression by promoter methylation in squamous cell carcinogenesis. J Natl Cancer Inst (2006) 98(3):181–9. doi: 10.1093/jnci/djj020
121. Chen X, Barozzi I, Termanini A, Prosperini E, Recchiuti A, Dalli J, et al. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc Natl Acad Sci U S A. (2012) 109(42):E2865–74. doi: 10.1073/pnas.1121131109
122. Barrat FJ, Crow MK, Ivashkiv LB. Interferon target-gene expression and epigenomic signatures in health and disease. Nat Immunol (2019) 20(12):1574–83. doi: 10.1038/s41590-019-0466-2
123. Tsukahara T, Kim S, Taylor MW. REFINEMENT: a search framework for the identification of interferon-responsive elements in DNA sequences–a case study with ISRE and GAS. Comput Biol Chem (2006) 30(2):134–47. doi: 10.1016/j.compbiolchem.2006.01.002
124. de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol (2001) 69(6):912–20. doi: 10.1189/jlb.69.6.912
125. Cheroni C, Manganaro L, Donnici L, Bevilacqua V, Bonnal RJP, Rossi RL, et al. Novel interferon-sensitive genes unveiled by correlation-driven gene selection and systems biology. Sci Rep (2021) 11(1):18043. doi: 10.1038/s41598-021-97258-8
126. Mostafavi S, Yoshida H, Moodley D, LeBoite H, Rothamel K, Raj T, et al. Parsing the interferon transcriptional network and its disease associations. Cell. (2016) 164(3):564–78. doi: 10.1016/j.cell.2015.12.032
127. Brierley MM, Marchington KL, Jurisica I, Fish EN. Identification of GAS-dependent interferon-sensitive target genes whose transcription is STAT2-dependent but ISGF3-independent. FEBS J (2006) 273(7):1569–81. doi: 10.1111/j.1742-4658.2006.05176.x
128. Jefferies CA. Regulating IRFs in IFN driven disease. Front Immunol (2019) 10:325. doi: 10.3389/fimmu.2019.00325
129. Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. (2011) 472(7344):481–5. doi: 10.1038/nature09907
130. Frontini M, Vijayakumar M, Garvin A, Clarke N. A ChIP-chip approach reveals a novel role for transcription factor IRF1 in the DNA damage response. Nucleic Acids Res (2009) 37(4):1073–85. doi: 10.1093/nar/gkn1051
131. Forero A, Ozarkar S, Li H, Lee CH, Hemann EA, Nadjsombati MS, et al. Differential activation of the transcription factor IRF1 underlies the distinct immune responses elicited by type I and type III interferons. Immunity. (2019) 51(3):451–64.e6. doi: 10.1016/j.immuni.2019.07.007
132. Horiuchi M, Itoh A, Pleasure D, Ozato K, Itoh T. Cooperative contributions of interferon regulatory factor 1 (IRF1) and IRF8 to interferon-gamma-mediated cytotoxic effects on oligodendroglial progenitor cells. J Neuroinflammation. (2011) 8:8. doi: 10.1186/1742-2094-8-8
133. Kano A, Haruyama T, Akaike T, Watanabe Y. IRF-1 is an essential mediator in IFN-gamma-induced cell cycle arrest and apoptosis of primary cultured hepatocytes. Biochem Biophys Res Commun (1999) 257(3):672–7. doi: 10.1006/bbrc.1999.0276
134. Chawla-Sarkar M, Lindner DJ, Liu YF, Williams BR, Sen GC, Silverman RH, et al. Apoptosis and interferons: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis. (2003) 8(3):237–49. doi: 10.1023/A:1023668705040
135. Detjen KM, Farwig K, Welzel M, Wiedenmann B, Rosewicz S. Interferon gamma inhibits growth of human pancreatic carcinoma cells via caspase-1 dependent induction of apoptosis. Gut. (2001) 49(2):251–62. doi: 10.1136/gut.49.2.251
136. Fulda S, Debatin KM. IFNgamma sensitizes for apoptosis by upregulating caspase-8 expression through the Stat1 pathway. Oncogene. (2002) 21(15):2295–308. doi: 10.1038/sj.onc.1205255
137. Chin YE, Kitagawa M, Kuida K, Flavell RA, Fu XY. Activation of the STAT signaling pathway can cause expression of caspase 1 and apoptosis. Mol Cell Biol (1997) 17(9):5328–37. doi: 10.1128/MCB.17.9.5328
138. Xu X, Fu XY, Plate J, Chong AS. IFN-gamma induces cell growth inhibition by fas-mediated apoptosis: requirement of STAT1 protein for up-regulation of fas and FasL expression. Cancer Res (1998) 58(13):2832–7.
139. Bromberg JF, Horvath CM, Wen Z, Schreiber RD, Darnell JE Jr. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc Natl Acad Sci U S A. (1996) 93(15):7673–8. doi: 10.1073/pnas.93.15.7673
140. Zhang R, Banik NL, Ray SK. Combination of all-trans retinoic acid and interferon-gamma suppressed PI3K/Akt survival pathway in glioblastoma T98G cells whereas NF-kappaB survival signaling in glioblastoma U87MG cells for induction of apoptosis. Neurochem Res (2007) 32(12):2194–202. doi: 10.1007/s11064-007-9417-7
141. Shirayoshi Y, Burke PA, Appella E, Ozato K. Interferon-induced transcription of a major histocompatibility class I gene accompanies binding of inducible nuclear factors to the interferon consensus sequence. Proc Natl Acad Sci U S A. (1988) 85(16):5884–8. doi: 10.1073/pnas.85.16.5884
142. Amaldi I, Reith W, Berte C, Mach B. Induction of HLA class II genes by IFN-gamma is transcriptional and requires a trans-acting protein. J Immunol (1989) 142(3):999–1004. doi: 10.4049/jimmunol.142.3.999
143. Cramer LA, Nelson SL, Klemsz MJ. Synergistic induction of the tap-1 gene by IFN-gamma and lipopolysaccharide in macrophages is regulated by STAT1. J Immunol (2000) 165(6):3190–7. doi: 10.4049/jimmunol.165.6.3190
144. Steimle V, Siegrist CA, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. (1994) 265(5168):106–9. doi: 10.1126/science.8016643
145. Melero I, Rouzaut A, Motz GT, Coukos G. T-Cell and NK-cell infiltration into solid tumors: a key limiting factor for efficacious cancer immunotherapy. Cancer Discovery (2014) 4(5):522–6. doi: 10.1158/2159-8290.CD-13-0985
146. Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. (2016) 167(6):1540–54.e12. doi: 10.1016/j.cell.2016.11.022
147. Abiko K, Matsumura N, Hamanishi J, Horikawa N, Murakami R, Yamaguchi K, et al. IFN-gamma from lymphocytes induces PD-L1 expression and promotes progression of ovarian cancer. Br J Cancer. (2015) 112(9):1501–9. doi: 10.1038/bjc.2015.101
148. Brody JR, Costantino CL, Berger AC, Sato T, Lisanti MP, Yeo CJ, et al. Expression of indoleamine 2,3-dioxygenase in metastatic malignant melanoma recruits regulatory T cells to avoid immune detection and affects survival. Cell Cycle (2009) 8(12):1930–4. doi: 10.4161/cc.8.12.8745
149. Battistello E, Hixon KA, Comstock DE, Collings CK, Chen X, Rodriguez Hernaez J, et al. Stepwise activities of mSWI/SNF family chromatin remodeling complexes direct T cell activation and exhaustion. Mol Cell. (2023) 83(8):1216–36.e12. doi: 10.1016/j.molcel.2023.02.026
150. Liu W, Wang Z, Liu S, Zhang X, Cao X, Jiang M. RNF138 inhibits late inflammatory gene transcription through degradation of SMARCC1 of the SWI/SNF complex. Cell Rep. (2023) 42(2):112097. doi: 10.1016/j.celrep.2023.112097
151. Pan D, Kobayashi A, Jiang P, Ferrari de Andrade L, Tay RE, Luoma AM, et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science. (2018) 359(6377):770–5. doi: 10.1126/science.aao1710
152. Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell. (2019) 176(4):775–89.e18. doi: 10.1016/j.cell.2018.11.043
153. Peng D, Kryczek I, Nagarsheth N, Zhao L, Wei S, Wang W, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. (2015) 527(7577):249–53. doi: 10.1038/nature15520
154. Li J, Wang W, Zhang Y, Cieslik M, Guo J, Tan M, et al. Epigenetic driver mutations in ARID1A shape cancer immune phenotype and immunotherapy. J Clin Invest. (2020) 130(5):2712–26. doi: 10.1172/JCI134402
155. Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, et al. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci U S A. (1998) 95(13):7556–61. doi: 10.1073/pnas.95.13.7556
156. Matsushita K, Takenouchi T, Shimada H, Tomonaga T, Hayashi H, Shioya A, et al. Strong HLA-DR antigen expression on cancer cells relates to better prognosis of colorectal cancer patients: possible involvement of c-myc suppression by interferon-gamma in situ. Cancer Sci (2006) 97(1):57–63. doi: 10.1111/j.1349-7006.2006.00137.x
157. Ding G, Zhou L, Shen T, Cao L. IFN-gamma induces the upregulation of RFXAP via inhibition of miR-212-3p in pancreatic cancer cells: a novel mechanism for IFN-gamma response. Oncol Lett (2018) 15(3):3760–5. doi: 10.3892/ol.2018.7777
158. Satoh A, Toyota M, Ikeda H, Morimoto Y, Akino K, Mita H, et al. Epigenetic inactivation of class II transactivator (CIITA) is associated with the absence of interferon-gamma-induced HLA-DR expression in colorectal and gastric cancer cells. Oncogene. (2004) 23(55):8876–86. doi: 10.1038/sj.onc.1208144
159. van der Stoep N, Biesta P, Quinten E, van den Elsen PJ. Lack of IFN-gamma-mediated induction of the class II transactivator (CIITA) through promoter methylation is predominantly found in developmental tumor cell lines. Int J Cancer. (2002) 97(4):501–7. doi: 10.1002/ijc.1623
160. Croce M, De Ambrosis A, Corrias MV, Pistoia V, Occhino M, Meazza R, et al. Different levels of control prevent interferon-gamma-inducible HLA-class II expression in human neuroblastoma cells. Oncogene. (2003) 22(49):7848–57. doi: 10.1038/sj.onc.1207054
161. Galindo Escobedo LV. Interferon gamma induces MHC class I/II expression on primary pancreatic cancer cells and enhances their recognition by autologous cytotoxic and helper T cells. (2006).
162. Imai D, Yoshizumi T, Okano S, Itoh S, Ikegami T, Harada N, et al. IFN-gamma promotes epithelial-mesenchymal transition and the expression of PD-L1 in pancreatic cancer. J Surg Res (2019) 240:115–23. doi: 10.1016/j.jss.2019.02.038
163. Wu B, Song M, Dong Q, Xiang G, Li J, Ma X, et al. UBR5 promotes tumor immune evasion through enhancing IFN-gamma-induced PDL1 transcription in triple negative breast cancer. Theranostics. (2022) 12(11):5086–102. doi: 10.7150/thno.74989
164. Ding G, Shen T, Yan C, Zhang M, Wu Z, Cao L. IFN-gamma down-regulates the PD-1 expression and assist nivolumab in PD-1-blockade effect on CD8+ T-lymphocytes in pancreatic cancer. BMC Cancer. (2019) 19(1):1053. doi: 10.1186/s12885-019-6145-8
165. Gupta R, Malvi P, Parajuli KR, Janostiak R, Bugide S, Cai G, et al. KLF7 promotes pancreatic cancer growth and metastasis by up-regulating ISG expression and maintaining golgi complex integrity. Proc Natl Acad Sci U S A. (2020) 117(22):12341–51. doi: 10.1073/pnas.2005156117
166. Andrianifahanana M, Singh AP, Nemos C, Ponnusamy MP, Moniaux N, Mehta PP, et al. IFN-gamma-induced expression of MUC4 in pancreatic cancer cells is mediated by STAT-1 upregulation: a novel mechanism for IFN-gamma response. Oncogene. (2007) 26(51):7251–61. doi: 10.1038/sj.onc.1210532
167. Abt ER, Le TM, Dann AM, Capri JR, Poddar S, Lok V, et al. Reprogramming of nucleotide metabolism by interferon confers dependence on the replication stress response pathway in pancreatic cancer cells. Cell Rep (2022) 38(2):110236. doi: 10.1016/j.celrep.2021.110236
168. Liu H, Golji J, Brodeur LK, Chung FS, Chen JT, deBeaumont RS, et al. Tumor-derived IFN triggers chronic pathway agonism and sensitivity to ADAR loss. Nat Med (2019) 25(1):95–102. doi: 10.1038/s41591-018-0302-5
169. Zhang M, Ding G, Zhou L, Shen T, Xu X, Zhao T, et al. Interferon gamma inhibits CXCL8-induced proliferation and migration of pancreatic cancer BxPC-3 cell line via a RhoGDI2/Rac1/NF-kappaB signaling pathway. J Interferon Cytokine Res (2018) 38(9):413–22. doi: 10.1089/jir.2018.0070
170. Zhang M, Huang L, Ding G, Huang H, Cao G, Sun X, et al. Interferon gamma inhibits CXCL8-CXCR2 axis mediated tumor-associated macrophages tumor trafficking and enhances anti-PD1 efficacy in pancreatic cancer. J Immunother Cancer (2020) 8(1):e000308. doi: 10.1136/jitc-2019-000308
171. Paschen A, Melero I, Ribas A. Central role of the antigen-presentation and interferon-γ pathways in resistance to immune checkpoint blockade. Annu Rev Cancer Biol (2022) 6:85–102. doi: 10.1146/annurev-cancerbio-070220-111016
172. Karachaliou N, Crespo G, Aldeguer E, Drozdowskyj A, Gimenez Capitan A, Teixido C, et al. Interferon-gamma (INFG), an important marker of response to immune checkpoint blockade (ICB) in non-small cell lung cancer (NSCLC) and melanoma patients. Am Soc Clin Oncol (2017) 10:1758834017749748. doi: 10.1177/1758834017749748
173. Li S, Li K, Tian F, Li H, Xia Q, Li T, et al. A high interferon gamma signature of CD8+ T cells predicts response to neoadjuvant immunotherapy plus chemotherapy in gastric cancer. Front Immunol. (2022) 13:1056144. doi: 10.3389/fimmu.2022.1056144
174. Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. (2017) 127(8):2930–40. doi: 10.1172/JCI91190
175. Karachaliou N, Gonzalez-Cao M, Crespo G, Drozdowskyj A, Aldeguer E, Gimenez-Capitan A, et al. Interferon gamma, an important marker of response to immune checkpoint blockade in non-small cell lung cancer and melanoma patients. Ther Adv Med Oncol (2018) 10:1758834017749748. doi: 10.1177/1758834017749748
176. Zhao Q, Bi Y, Sun H, Xiao M. Serum IL-5 and IFN-gamma are novel predictive biomarkers for anti-PD-1 treatment in NSCLC and GC patients. Dis Markers. (2021) 2021:5526885. doi: 10.1155/2021/5526885
177. Grasso CS, Tsoi J, Onyshchenko M, Abril-Rodriguez G, Ross-Macdonald P, Wind-Rotolo M, et al. Conserved interferon-gamma signaling drives clinical response to immune checkpoint blockade therapy in melanoma. Cancer Cell (2020) 38(4):500–15.e3. doi: 10.1016/j.ccell.2020.11.015
178. Chen J, Du M, Li WB, Lei C, Yang B, Tang HK, et al. [The relationship between CD4(-) CD8(-) T cells in the peripheral blood of patients with pancreatic carcinoma and IL-4, IFN-gamma levels]. Zhonghua Wai Ke Za Zhi. (2009) 47(13):995–8. doi: 10.3760/cma.j.issn.0529-5815.2009.13.011
179. Ishikawa T, Kokura S, Sakamoto N, Okayama T, Endo M, Tsuchiya R, et al. Whole blood interferon-gamma levels predict the therapeutic effects of adoptive T-cell therapy in patients with advanced pancreatic cancer. Int J Cancer. (2013) 133(5):1119–25. doi: 10.1002/ijc.28117
180. Prioli R, Arvidson L, Jensen S, Smith M, White K, Heil-Chapdelaine R, et al. 1 capturing the spatial landscape of tumor and immune cell lineages in the microenvironment of human cancer tissues. BMJ Specialist Journals (2022). doi: 10.1136/jitc-2022-SITC2022.0001
181. Lerner E, D'Anniballe V, Tomaszewski W, Perera J, Cui X, Waibl-Polania J, et al. CD8 T cell mediated killing of MHC class 1 negative tumors requires antigen presenting myeloid cells and interferon gamma. Cancer Res (2022) 82(12_Supplement):1378. doi: 10.1158/1538-7445.AM2022-1378
182. Takeda K, Smyth MJ, Cretney E, Hayakawa Y, Yamaguchi N, Yagita H, et al. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in NK cell-mediated and IFN-gamma-dependent suppression of subcutaneous tumor growth. Cell Immunol (2001) 214(2):194–200. doi: 10.1006/cimm.2001.1896
183. Fang Y, Zhang Y, Guo C, Chen C, Gao H, Zhou X, et al. Safety and efficacy of an immune cell-specific chimeric promoter in regulating anti-PD-1 antibody expression in CAR T cells. Mol Ther Methods Clin Dev (2020) 19:14–23. doi: 10.1016/j.omtm.2020.08.008
184. Boulch M, Cazaux M, Loe-Mie Y, Thibaut R, Corre B, Lemaitre F, et al. A cross-talk between CAR T cell subsets and the tumor microenvironment is essential for sustained cytotoxic activity. Sci Immunol (2021) 6(57):eabd4344. doi: 10.1126/sciimmunol.abd4344
185. Larson RC, Kann MC, Bailey SR, Haradhvala NJ, Llopis PM, Bouffard AA, et al. CAR T cell killing requires the IFNgammaR pathway in solid but not liquid tumours. Nature. (2022) 604(7906):563–70. doi: 10.1038/s41586-022-04585-5
186. Zhang H, Lv X, Kong Q, Tan Y. IL-6/IFN-gamma double knockdown CAR-T cells reduce the release of multiple cytokines from PBMCs in vitro. Hum Vaccin Immunother (2022) 18(1):1–14. doi: 10.1080/21645515.2021.2016005
187. Bailey SR, Vatsa S, Larson R, Bouffard AA, Scarfò I, Gallagher KM, et al. Blocking IFNγ in CAR-T reduces checkpoint inhibitors and cell-mediated toxicity without compromising therapeutic efficacy in CD19+ malignancies. Blood. (2021) 138:1723. doi: 10.1182/blood-2021-146042
188. An Z, Hu Y, Bai Y, Zhang C, Xu C, Kang X, et al. Antitumor activity of the third generation EphA2 CAR-T cells against glioblastoma is associated with interferon gamma induced PD-L1. Oncoimmunology. (2021) 10(1):1960728. doi: 10.1080/2162402X.2021.1960728
189. Miller CH, Maher SG, Young HA. Clinical use of interferon-gamma. Ann N Y Acad Sci (2009) 1182:69–79. doi: 10.1111/j.1749-6632.2009.05069.x
190. Gleave ME, Elhilali M, Fradet Y, Davis I, Venner P, Saad F, et al. Interferon gamma-1b compared with placebo in metastatic renal-cell carcinoma. Canadian urologic oncology group. N Engl J Med (1998) 338(18):1265–71. doi: 10.1056/NEJM199804303381804
191. Korentzelos D, Wells A, Clark AM. Interferon-gamma increases sensitivity to chemotherapy and provides immunotherapy targets in models of metastatic castration-resistant prostate cancer. Sci Rep (2022) 12(1):6657. doi: 10.1038/s41598-022-10724-9
192. Giannopoulos A, Constantinides C, Fokaeas E, Stravodimos C, Giannopoulou M, Kyroudi A, et al. The immunomodulating effect of interferon-gamma intravesical instillations in preventing bladder cancer recurrence. Clin Cancer Res (2003) 9(15):5550–8.
193. Wall L, Burke F, Barton C, Smyth J, Balkwill F. IFN-gamma induces apoptosis in ovarian cancer cells in vivo and in vitro. Clin Cancer Res (2003) 9(7):2487–96.
194. Windbichler GH, Hausmaninger H, Stummvoll W, Graf AH, Kainz C, Lahodny J, et al. Interferon-gamma in the first-line therapy of ovarian cancer: a randomized phase III trial. Br J Cancer. (2000) 82(6):1138–44. doi: 10.1054/bjoc.1999.1053
195. Alberts DS, Marth C, Alvarez RD, Johnson G, Bidzinski M, Kardatzke DR, et al. Randomized phase 3 trial of interferon gamma-1b plus standard carboplatin/paclitaxel versus carboplatin/paclitaxel alone for first-line treatment of advanced ovarian and primary peritoneal carcinomas: results from a prospectively designed analysis of progression-free survival. Gynecol Oncol (2008) 109(2):174–81. doi: 10.1016/j.ygyno.2008.01.005
196. Mizokami MM, Hu P, Khawli LA, Li J, Epstein AL. Chimeric TNT-3 antibody/murine interferon-gamma fusion protein for the immunotherapy of solid malignancies. Hybrid Hybridomics. (2003) 22(4):197–207. doi: 10.1089/153685903322328929
197. Yin H, Chen N, Guo R, Wang H, Li W, Wang G, et al. Antitumor potential of a synthetic interferon-alpha/PLGF-2 positive charge peptide hybrid molecule in pancreatic cancer cells. Sci Rep (2015) 5:16975. doi: 10.1038/srep16975
198. Bansal R, Prakash J, Post E, Beljaars L, Schuppan D, Poelstra K. Novel engineered targeted interferon-gamma blocks hepatic fibrogenesis in mice. Hepatology. (2011) 54(2):586–96. doi: 10.1002/hep.24395
199. Alhawmdeh M, Isreb M, Aziz A, Jacob BK, Anderson D, Najafzadeh M. Interferon-gamma liposome: a new system to improve drug delivery in the treatment of lung cancer. ERJ Open Res (2021) 7(3):00555-2020. doi: 10.1183/23120541.00555-2020
200. Grekova SP, Aprahamian M, Daeffler L, Leuchs B, Angelova A, Giese T, et al. Interferon gamma improves the vaccination potential of oncolytic parvovirus h-1PV for the treatment of peritoneal carcinomatosis in pancreatic cancer. Cancer Biol Ther (2011) 12(10):888–95. doi: 10.4161/cbt.12.10.17678
Keywords: interferon, immunotherapy, immunoresistance, ISG (interferon stimulated genes), IFNGR, JAK - STAT signaling pathway, IFN-γ
Citation: Han J, Wu M and Liu Z (2023) Dysregulation in IFN-γ signaling and response: the barricade to tumor immunotherapy. Front. Immunol. 14:1190333. doi: 10.3389/fimmu.2023.1190333
Received: 20 March 2023; Accepted: 14 April 2023;
Published: 18 May 2023.
Edited by:
Bryan E. Strauss, Institute of Cancer of São Paulo, University of São Paulo, BrazilReviewed by:
Francesca Maria Consonni, University of Milan, ItalyCopyright © 2023 Han, Wu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ziwen Liu, bGl1eml3ZW5AcHVtY2guY24=
†These authors have contributed equally to this work
 Jiashu Han1,2†
Jiashu Han1,2† Mengwei Wu
Mengwei Wu Ziwen Liu
Ziwen Liu