- 1Department of Medical Oncology, Affiliated Hospital of Jiangsu University, Zhenjiang, China
- 2Department of Gastroenterology, Affiliated Hospital of Jiangsu University, Zhenjiang, Jiangsu, China
- 3Institute of Digestive Diseases, Jiangsu University, Zhenjiang, China
N6-methyladenosine (m6A) methylation modification is a ubiquitous RNA modification involved in the regulation of various cellular processes, including regulation of RNA stability, metabolism, splicing and translation. Gastrointestinal (GI) cancers are some of the world’s most common and fatal cancers. Emerging evidence has shown that m6A modification is dynamically regulated by a complex network of enzymes and that the catalytic subunit m6A-METTL complex (MAC)-METTL3/14, a core component of m6A methyltransferases, participates in the development and progression of GI cancers. Furthermore, it has been shown that METTL3/14 modulates immune cell infiltration in an m6A-dependent manner in TIME (Tumor immune microenvironment), thereby altering the response of cancer cells to ICIs (Immune checkpoint inhibitors). Immunotherapy has emerged as a promising approach for treating GI cancers. Moreover, targeting the expression of METTL3/14 and its downstream genes may improve patient response to immunotherapy. Therefore, understanding the role of MAC in the pathogenesis of GI cancers and its impact on immune cell infiltration may provide new insights into the development of effective therapeutic strategies for GI cancers.
1 Introduction
Gastrointestinal (GI) cancers currently rank as the most common and deadliest cancers, accounting for 26% of global cancer incidence and 35% of all cancer-related deaths. The spectrum of GI cancers encompasses esophageal, gastric, liver, pancreatic, and colorectal cancers, manifesting as a complex multi-step process (1). While current GI cancer treatment mainly includes surgery, radiation therapy, chemotherapy, and targeted therapy, the effectiveness has plateaued. In recent years, immunotherapy has emerged as a prominent topic in GI cancer treatment. Immunotherapy refers to the treatment of tumors through the regulation, activation, or suppression of the patient’s immune system, thereby enhancing their anti-cancer immunity (2). Immune Checkpoint Inhibitors (ICIs) targeting the Cytotoxic T Lymphocyte-associated antigen 4 (CTLA-4) or Programmed cell death protein 1 (PD-1) pathways have demonstrated early success in treating various cancers. However, the subset of patients benefiting from ICIs in GI cancers remains relatively limited (3). Currently, researchers are actively exploring immune mechanisms beyond immune checkpoints to improve the response rate and duration of ICI therapy in GI cancer patients. Considerable attention is also directed towards further investigating m6A methylation modification to enhance the efficacy of immunotherapy and screen personalized treatment strategies.
N6-methyladenosine (m6A) refers to a methylation modification placed on the sixth nitrogen atom of RNA adenine. It is one of the most common RNA modifications in eukaryotic cells (4) and is widely distributed in eukaryotic and viral mRNAs (5). Sequencing analysis of m6A has revealed that it is primarily deposited in the protein-coding sequence (CDS), long internal exons, upstream of the stop codon, and in the untranslated region at the 3’ end of mRNA (6–8). m6A methylation modifications exhibit dynamic, reversible, and diversified characteristics, regulated by three m6A regulators: methyltransferases, demethylases, and binding proteins (9). As a key regulator of transcriptional expression, m6A is closely linked to various aspects of cancer, including development, proliferation, growth, invasion, metastasis, immune system evasion, and other oncogenic or inhibitory functions, through its regulation of RNA processing, splicing, nucleation, translation, and other aspects (10). This paper explores the role of Methyltransferase-Like 3/14 (METTL3/14), components of the catalytic subunit m6A-METTL complex (MAC), in regulating tumor immune infiltration. It specifically focuses on their potential and current research progress in clinical applications as biomarkers and immunotherapeutic targets for GI cancers.
2 m6A methyltransferase complex
The methylation of m6A is catalyzed by the m6A methyltransferase complex (MTC), also commonly referred to as writers, and it is composed of m6A methyltransferases located in the nuclear speckles (11, 12). MTC catalyzes the transfer of methyl groups from the cofactor S-adenosyl-L-methionine (SAM) to the N6 end of adenine by binding to SAM (11). The crystal structure of MTC has been determined, revealing its compact globular protein with a highly organized and asymmetric configuration. The active site is located in the hydrophobic pocket at the center of the protein (13). MTC primarily comprises the catalytic subunit, m6A-METTL complex (MAC), which is catalytically active, and the regulatory subunit, m6A-METTL-associated complex (MACOM), which plays a role in regulating enzymatic activity. This complex is cross-linked through the interaction between Wilms tumor 1-associated protein (WTAP) and METTL3 (Figure 1). Subdividing further, METTL3 and METTL14 are the major components of MAC, while HAKAI (also known as CBLL1), WTAP, VIRMA (vir like m6A methyltransferase associated), and ZC3H13 (zinc finger CCCH type containing 13) combine to form MACOM (14).
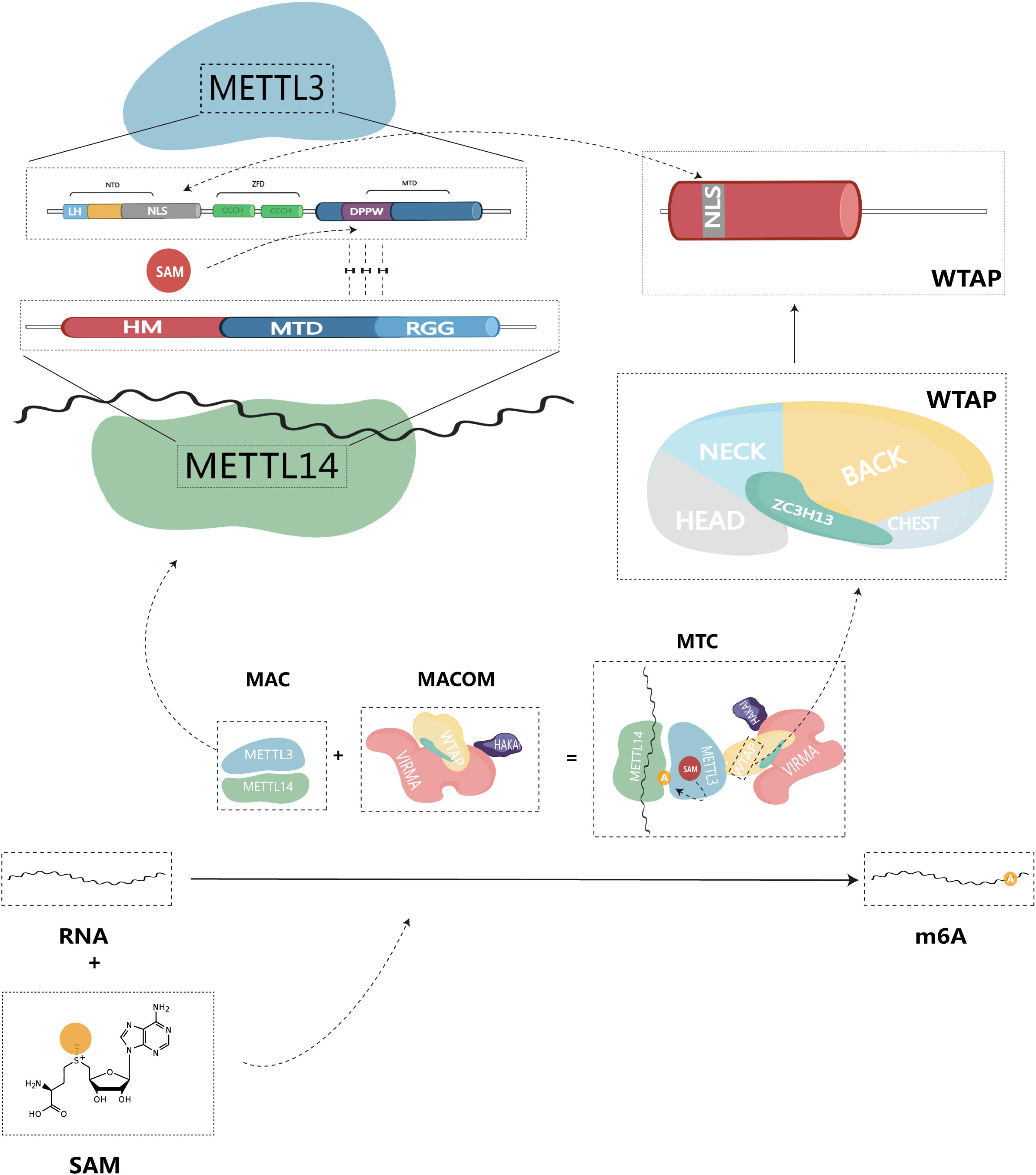
Figure 1 Structure of m6A methyltransferase complex (MTC), and composition of structural domain of MAC-METTL3/14.
2.1 Catalytic subunit m6A-METTL complex
As the first identified m6A methyltransferase, METTL3 is a 70 kDa protein with a full-length amino acid count of 580, and it plays a crucial role in catalytic methylation (13, 15). The METTL3 protein consists of three primary structural domains: the N-terminal structural domain (NTD), the zinc finger structural domain (ZFD), and the methyltransferase structural domain (MTD), serving as the primary catalytic structure.
NTD: This structural domain is located at the beginning of the protein sequence and contains a leader helix (LH) and a nuclear localization signal (NLS) region (16). The NTD plays a key role in regulating catalytic activity and substrate specificity. Specifically, the NTD has been shown to influence enzymatic activity by methylating different RNA structures based on their stability and folding patterns, thus altering the dependency of secondary structure on yield levels (17). Additionally, the NTD collaborates with other elements in MAC, such as the RGG structural domain of METTL14, to improve the efficiency of the methyltransferase reaction (17, 18). This synergistic interaction of the NTD with other structural domains in METTL3 highlights the complex interaction of various elements within the enzyme, ultimately governing its catalytic activity and substrate specificity.
ZFD: Located near the C-terminal end of the protein, the ZFD contains two CCCH-type zinc fingers, ZnF1 and ZnF2, connected by an anti-parallel β-sheet (19). This structural domain is responsible for recognizing and binding RNA molecules, particularly the GGACU motif found in the region where methylation occurs. The ZFD stabilizes the protein structure and allows it to interact with RNA molecules, a requirement for maintaining MTC’s enzymatic activity (20).
MTD: The MTD construct of METTL3 spans residues 358-580 and contains the conserved DPPW motif, which is essential for enzyme activity (1). MTD primarily adopts a classical α -β -α sandwich fold structure, comprising a mixed eight-stranded β-sheet, four α-helices, and three 310 helices on either side (13). This region binds to the methyltransferase structural domain of METTL14, forming a heterodimeric complex responsible for the specific recognition of the m6A modification site in RNA (19).
In summary, the three structural domains of METTL3 work in concert to ensure the proper recognition, binding, and methylation of RNA molecules, which are essential for the regulation of gene expression and various other cellular processes.
Similar to METTL3, METTL14 also belongs to the family of proteins homologous to the MT-A70 subunit (21). METTL14 is a monomeric protein with a molecular weight of approximately 52 kDa (16). It consists of three structural domains: N-terminal extension (denoted HM), methyltransferase structural domain (MTD), and Arginine-Glycine repeats (RGG) at the C-terminus (22). The RGG repeats of METTL14 can contribute to maintaining the catalytic activity of MTC during methylation. Since the RGG repeats are present in RNA-binding proteins, they can bind to the target RNA, thus playing an important role in the methylation reaction (23). MAC can be directly recruited to specific methylation sites through the preferential and specific binding of RNA G-quadruplex (rG4) structures by the RGG repeats of METTL14 (18). In vitro, MAC demonstrates high activity with the RRACH site, preferentially binding to it and methylating it (24). In contrast to METTL3, METTL14 lacks residues that form hydrogen bonds with SAM. Therefore, the MTD domain of METTL14 does not have an active site accommodating donor and acceptor substrates. Instead, it enhances methyltransferase activity by recognizing RNA substrates and methyl localization (13, 24, 25).
METTL3 possesses binding sites for SAM and S-adenosylhomocysteine (SAH), mainly located at the carboxyl terminus of the β1, β7, and β8 chains (19). SAM donor binding occurs through multiple hydrogen bonding interactions. Consequently, METTL3 acts as the primary catalytically active subunit in MAC, while METTL14 mainly contributes to m6A modification as a metastable activator of METTL3 activity (26). Given that both METTL3 and METTL14 possess a methyltransferase structural domain (MTD), METTL14 forms a stable heterodimer with the MTD of METTL3 in a 1:1 ratio. METTL14 serves as an RNA binding platform, facilitating MAC’s binding to RNA substrates (13, 19, 24). Therefore, we believe that MAC is a core component mediating m6A methylation.
2.2 Regulatory subunit m6A-METTL-associated complex
As a subunit responsible for regulating enzyme activity, MACOM possesses a stronger RNA binding ability than MAC and can directly bind to RNA substrates (14). In the absence of MACOM, the catalytic activity of MAC is significantly reduced (27). Among its components, WTAP lacks any recognizable structural domain by itself (23). Upon binding to MAC, WTAP can recruit other m6A methyltransferases to target RNAs, thus regulating intracellular m6A deposition (28). Additionally, WTAP plays a crucial role in localizing MAC to nuclear speckles through the potential nuclear localization signal (NLS) found in both WTAP and METTL3 N terminus (29). VIRMA, also known as KIAA1429, is the largest component of the MACOM complex and forms the core framework of MACOM by cross-linking with WTAP (14). Similar to WTAP, VIRMA is also localized in nuclear speckles (30). VIRMA predominantly acts on the 3’UTR and near-termination codon attachment, selectively and preferentially mediating m6A methylation modifications in this region (31). During the methylation process, VIRMA mediates regioselective methylation by recruiting core methyltransferases, METTL3/14. ZC3H13, a key component of MACOM, can induce conformational changes in MACOM through interactions with VIRMA (14). ZC3H13 contributes to the nuclear localization of MACOM by localizing to nuclear patches via a low-complexity (LC) region (occupying 80% of the ZC3H13 protein sequence) (32). In the study by Philip et al, ZC3H13 maintains the connection between the RNA-binding protein RBM15 and WTAP. By bridging these factors, ZC3H13 plays a role in regulating the specificity and selectivity of m6A modification, ensuring it occurs at the appropriate location in the mRNA (33). HAKAI is not a core of the MACOM structure. It has a flexible but relatively conservative presence in the MACOM complex, serving to maintain the stability and integrity of the MACOM components (14). The ubiquitination domain of HAKAI plays a decisive role in this process (34). By regulating RNA-binding components that interact with the m6A methyltransferase complex, HAKAI influences the site-specific placement of m6A modifications on mRNA and regulates mRNA stability, translation, and other processes (35).
3 MAC and GI cancer progression
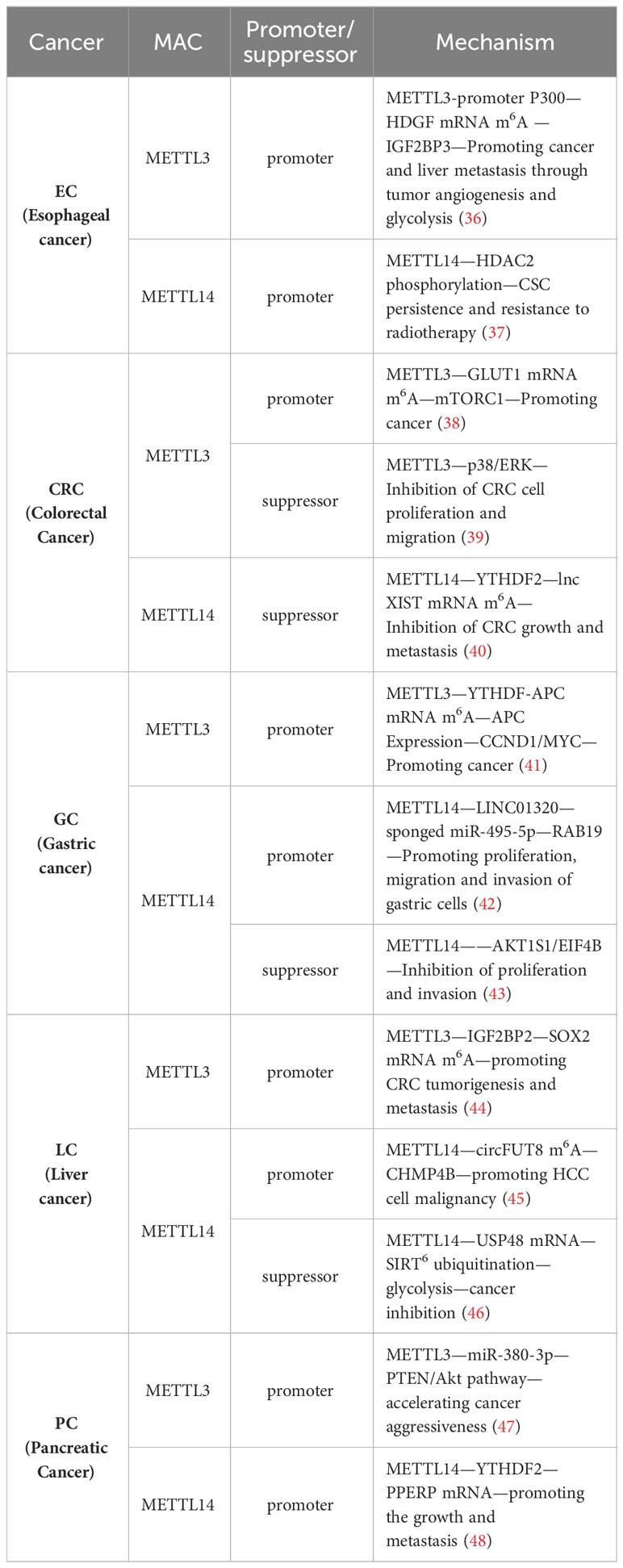
Numerous studies have previously demonstrated the close association between MAC and cancer, particularly in GI cancers, where METTL3/14, a component of MAC, exerts various effects on tumor progression, including proliferation, growth, invasion, and metastasis. METTL3/14 can act as either a tumor promoter or a tumor suppressor in GI cancers. Different studies have indicated that METTL3/14 may play opposing roles within the same type of tumor. This variability may be attributed to differences in the targeted genes or pathways (Table 1). Given this intricate and paradoxical relationship, a comprehensive understanding of the role of METTL3/14 in tumor immunity is necessary, with a specific emphasis on deciphering the potential mechanisms of METTL3/14 within the TIME.
4 MAC and tumor immune cell infiltration
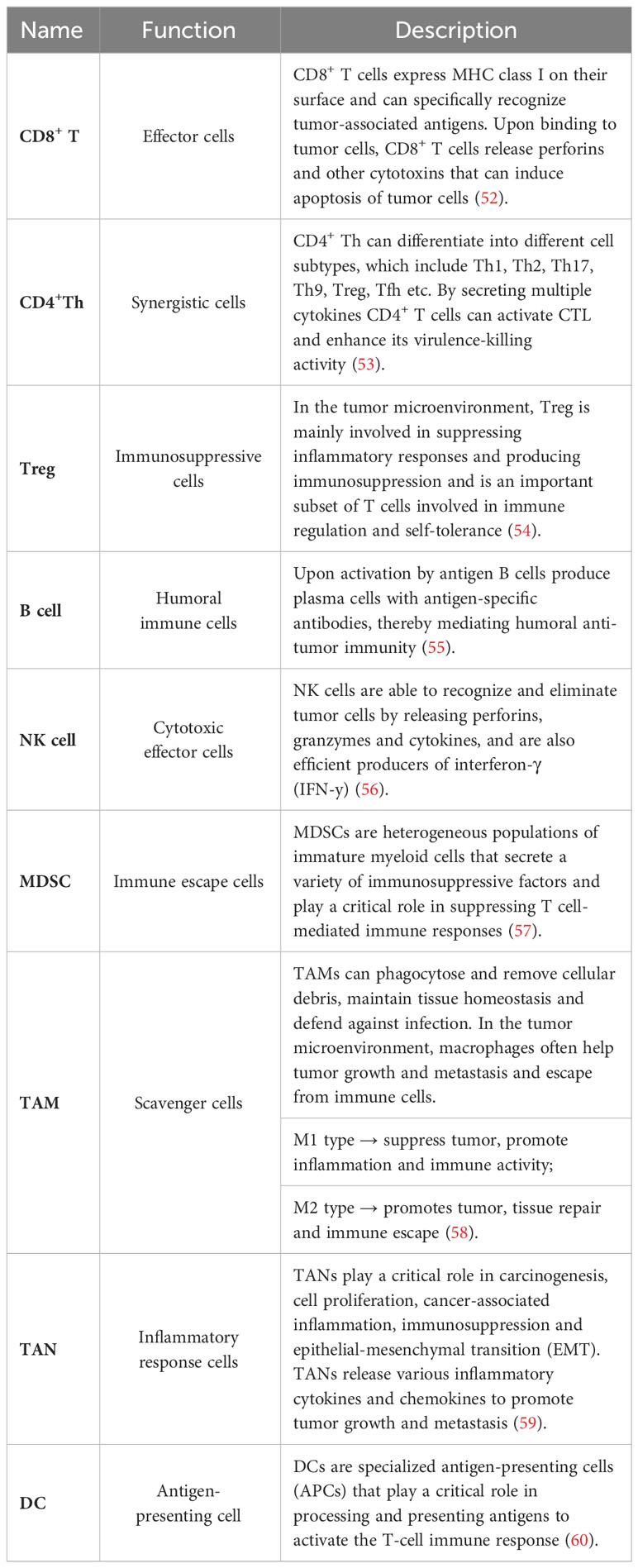
In recent years, as our understanding of the immune system has expanded, an increasing number of researchers have recognized the important role played by the TIME in various tumor biological processes, including tumor development and metastasis (49). TIME refers to all the immune components within it, which, through their interactions, exert influence on the dynamics and complexity of the tumor microenvironment (50). TIME comprises a diverse array of components, including immune infiltrating cells, cytokines, and regulatory proteins (51). Among these constituents, immune infiltrating cells are the primary components of TIME and include CD8+ T cells, CD4+ helper T cells, regulatory T cells (Treg), B cells, natural killer cells, myeloid-derived suppressor cells (MDSCs), tumor-associated macrophages (TAMs), tumor-associated neutrophils (TANs), and dendritic cells (Table 2). One of the main mechanisms of immunotherapy involves immune checkpoint blockade, which targets inhibitory signals on T cells to enhance their activity against cancer cells (61). Tumor cells can upregulate immune checkpoint molecules, such as PD-L1, which inhibit T cell activity and evade immune surveillance. By blocking these inhibitory signals, ICIs enhance the infiltration and activation of immune cells within the tumor microenvironment (62). The level of immune cell infiltration within the tumor microenvironment also plays a key in determining the response to immunotherapy. Tumors with high levels of T-cell infiltration, often referred to as “hot” tumors, are more likely to respond to immunotherapy compared to tumors with low levels of infiltration known as “cold” tumors (63). Therefore, strategies aimed at increasing immune cell infiltration in tumors, including combination therapies that target the tumor microenvironment, are under investigation to improve the response to immunotherapy. In this context, we provide a detailed description of the regulation of immune infiltrating cells within tumors by MAC.
4.1 T cells
METTL3 plays a crucial role in maintaining the proper balance of T cells in vivo. Mechanistically, METTL3-dependent m6A methylation of specific mRNA transcripts within T cells can modulate the stability and translation of these transcripts, leading to changes in gene expression that are critical for T cell survival, proliferation, and differentiation. In particular, METTL3/14 promotes the degradation of SOCS mRNA and reduces the expression of SOCS family proteins through mediated methylation of SOCS family mRNAs such as SOCS1, SOCS3, and CISH. This leads to sustained activation of the IL-7/STAT5 signaling pathway, which promotes T-cell differentiation and proliferation (64). A recent study identified the effect of METTL3 on the expression of PD-L1. They found that METTL3 mediated the m6A modification of PD-L1 mRNA, leading to increased expression of PD-L1. When PD-L1 binds to PD-1 on T cells, this elevated expression of PD-L1 suppresses the immune response against tumors by curbing T cell activation and function (65). This process can be regulated by JNK signaling (66). Wang and colleagues share a similar perspective, suggesting that METTL3/14-mediated m6A modifications affect the stability and translation efficiency of Stat1 and Irf1 mRNAs. This, in turn, results in the suppression of CD8+ T cell infiltration within the TIME, resulting in decreased secretion of cytokines such as IFN-γ (67). Therefore, based on the aforementioned study, Wang et al. discovered that the phytochemical Delicaflavone inhibits the expression of METTL3/14, subsequently reducing the secretion of cytokines and chemokines. This inhibition also led to the downregulation of STAT1-IRF1, thereby activating antitumor immunity. Additionally, it triggers oxidative stress, amplifies the recruitment of CD8+ T cells into tumor tissues, increases the production of Th1 cytokines, and ultimately inhibits tumor growth (68).
Treg, as a subset of T cells, plays an essential role within the TIME by suppressing the immune response of other T cells and secreting immunosuppressive factors that promote tumor escape (54). Conversely, the deletion of METTL14 expression in T cells disrupts the differentiation of naive T cells into Treg cells, resulting in increased secretion of Th1 and Th17 cytokines (69). Tong et al. elevated SOCS mRNA and protein levels through the inhibition of METTL3-mediated m6A modification in regulatory T cells. This resulted in the inhibition of the JAK-STAT signaling pathway, causing Treg to lose its ability to suppress effector T cells and naive T cells (70). Similar findings were observed in METTL14, where the knockdown of METTL14 inhibited Treg cell growth while promoting the infiltration of CD4+ and CD8+ cells (71). Additionally, METTL3-mediated modification of circQSOX1 leads to the sponging of miR-326 and miR-330-5p, thus promoting PGAM1 expression. This process leads to an upregulation of glycolysis levels and increased lactate production in colorectal cancer (CRC) cells. The accumulation of lactate within the TIME facilitates Treg infiltration while suppressing the infiltration of CD8+ T cells, ultimately promoting immune evasion (72).
T follicle helper (Tfh) cells, a subset of CD4+ T cells, are responsible for directing and activating tumor-specific B cells and T cells, thereby promoting the formation and enhancement of anti-tumor immunity. However, tumors can exploit these cells to suppress the immune response, promoting tumor growth and metastasis (73). METTL14 promotes Th-17 cell differentiation by mediating m6A methylation of exosomal miR-149-3p (74). Similarly, METTL3 promotes TFH differentiation by stabilizing transcripts of key TFH signature genes, such as Tcf7, and mediating their m6A methylation (75). A study by Zhu et al. elucidated the mechanism by which the E3 ubiquitin ligase VHL regulates Tfh cell differentiation. Specifically, the authors identified GAPDH as a key player in Tfh cell differentiation and demonstrated that VHL deficiency leads to the upregulation of GAPDH. This, in turn, led to reduced expression of the Tfh cell marker ICOS and attenuated Tfh cell differentiation, attributed to METTL3/14-mediated modification of ICOS mRNA m6A. This mechanism plays a crucial role in regulating metabolic reprogramming during the early stages of Tfh cell differentiation, with the HIF-1α axis serving as the primary regulator (76). Th1 and Th2 cells are the two distinct classes of T cells involved in immune responses. Th1 cells are responsible for rejecting foreign molecules, whereas Th2 cells contribute to tolerance toward foreign molecules. Knockdown of METTL3 effectively counteracts LPS-induced Th1 differentiation while significantly increasing the proportion of Th2 (77).
4.2 B cells
Currently, there exists limited research exploring the mechanisms through which METTL3 regulates B cells within the tumor microenvironment. Some studies have suggested that METTL3 has only marginal effects on B cell development in normal tissue environments (78). However, Zheng’s study demonstrated that METTL3/14 is necessary for normal B-cell development because it induces cell proliferation of Pro-B and facilitates the differentiation of Pro-B into large-Pre-B/small-Pre-B by promoting IL-17 secretion and IL-17 receptor expression. Additionally, the activation of key transcription factors such as IKZF3, which promotes the conversion of large Pre-B to small Pre-B, is intricately linked to METTL14 (79). Moreover, METTL14-mediated m6A modification upregulates genes crucial for the germinal centre response, thus positively contributing to the positive selection and proliferation of germinal centre B cells (80). While these investigations weren’t specifically carried out in a tumor setting, they offer valuable insights into how METTL3/14 regulates B cells. These findings hint at how METTL3/14 could potentially influence B cells within the tumor microenvironment, conducting additional experiments is imperative to gain a comprehensive understanding of the precise mechanisms by which METTL3/14 governs B cell behavior within this specific context.
4.3 DCs
Dendritic cells play an important immunoregulatory role within the tumor microenvironment. They capture and process antigens from tumor cells and secrete various cytokines, thereby activating T cell-mediated anti-tumor immune responses (60). METTL3 knockout impairs DC activation, characterized by reduced surface expression of costimulatory molecules, decreased cytokine production, and impaired T-cell activation. Further mechanistic studies suggest that METTL3 mediates m6A methylation modification of key signaling molecules (Tirap, CD40, and CD80) involved in DC activation and function. This enhances the stability and translational efficiency of these signaling molecule mRNAs, ultimately promoting DC activation and maturation, and improving DC function both in vitro and in vivo (81). Wu et al. also observed significantly higher levels of METTL3 mRNA and protein in mature DCs compared to immature DCs. Knocking down the METTL3 gene in DCs resulted in reduced expression of related cytokines, such as IFN-γ and IL-12, and reduced T cell proliferation, ultimately inducing immune tolerance (81).
4.4 NK cells
The expression of METTL3 plays a pivotal role in the anti-tumor immunity of NK cells, which are core immune cells responsible for cytotoxic effects. METTL3 promotes elevated expression of SHP-2, a key ERK activator, by mediating the m6A modification of PTPN11. This, in turn, promotes IL-15 signaling, and in turn, leads to increased proliferation and enhanced effector functions of NK cells (82).
4.5 Macrophages
When tumor cells evade clearance and surveillance by macrophages, these macrophages polarize and differentiate into two subtypes: M1 and M2. M1 macrophages are capable of killing tumor cells and recognizing foreign antigens, while M2 macrophages produce immunosuppressive factors in the tumor microenvironment, acting as suppressors of the immune response (58). Defective expression of METTL14 on macrophages results in increased EBI3 expression, leading to CD8+ T cell dysfunction (83). Liu et al. suggested that METTL3 modifies STAT1 mRNA through m6A methylation, influencing the expression of genes associated with the M1 macrophage phenotype and decreasing the expression of genes associated with the M2 macrophage phenotype. This makes METTL3 a key regulator of macrophage polarization and function (84). Tong et al. also suggested that METTL3 positively regulates M1 macrophages primarily by targeting IRAKM mRNA, promoting its degradation and thereby regulating TNF-α levels through TLR4 signaling in macrophages (85). Furthermore, inhibiting METTL3 activity with LXA4 promotes macrophage polarization toward the M2 phenotype (86). Shu et al. proposed an innovative signaling axis consisting of METTL3/MALAT1/PTBP1/USP8/TAK1, where METTL3-mediated m6A modification of MALAT1 leads to macrophage pyroptosis by interacting with PTBP1, subsequently triggering downstream signals, including USP8 and TAK1 (87). Recent studies by Yin et al. have shown that METTL3 depletion in macrophages activates the NF-κB pathway and STAT3 signaling, resulting in increased infiltration of M1 and M2 TAMs and Treg cells into the tumor, ultimately remodeling the tumor microenvironment (88). In response, M2-TAMs significantly enhance METTL3 expression in lung adenocarcinoma cells, leading to increased m6A methylation levels and overall levels of immune resistance, thus promoting tumor progression (89).
4.6 MDSCs
MDSCs are a type of immune cell that exhibits strong immunosuppressive effects. They employ various pathways to inhibit the body’s immune response within the TIME (57). Studies have revealed a positive correlation between the density of CD33+ MDSCs in tumor tissues and METTL3 expression (90). Research indicates that METTL3 plays a crucial role in advancing MDSC migration in colorectal cancer by exerting control over the m6A-BHLHE41-CXCL1/CXCR2 axis. This involves METTL3 reducing the expression of BHLHE41, a transcription factor associated with tumor immunity suppression, by methylating m6A sites on its mRNA. Consequently, this promotes the migration of MDSCs to tumor sites (91). In clinical practice, arterial infusion chemotherapy with cisplatin has been shown to reduce the methylation status of METTL3-regulated genes and decrease MDSCs in tumors (92). This suggests the potential for combining immunotherapy and chemotherapy in cancer treatment.
Derived from the bone marrow, tumor-infiltrating myeloid cells (TIMs) can indirectly influence MDSC differentiation and function by regulating the activity of immune cells within the TIME. METTL3 expression was found to be upregulated in various tumor tissues, particularly in TIMs compared to normal adjacent tissues. The loss of the METTL3 gene in myeloid cells led to a reduction in the infiltration of exhausted CD8+ T cells, a decrease in the number of Treg cells, and an increase in the number of tumor-infiltrating CD8+ T cells (93).
4.7 TANs
Tumor-associated neutrophils (TANs) can adopt various roles within the tumor microenvironment, including promoting tumor growth and metastasis and suppressing immune cell activity (59). Elevated METTL3 expression progressively promotes the expression of c-Rel, a transcription factor involved in immune response regulation, leading to increased IL-8 production and enhanced infiltration of TANs. This, in turn, results in a more robust immune response against cancer cells (94). When TANs are activated by cancer cells in the tumor microenvironment or by inflammatory factors such as G-CSF/IL-8, they undergo a process known as NETosis, where they release DNA and other granular proteins into the extracellular space, forming Neutrophil extracellular traps (NETs) (95). By releasing NETs, neutrophils participate in the immune defense against tumors, yet this action potentially contributes to immunotherapy resistance and immune suppression. Nevertheless, the removal of NETs or the application of specific treatment strategies like DNase I or adeno-associated virus (AAV) vector could enhance immunotherapy efficacy, thereby preventing tumor recurrence (96). Qu et al. suggested that METTL3-mediated modification of SIRT1 mRNA m6A leads to autophagosome formation and impaired autophagic flux, regulating the formation and release of NETs (97). Another study found that METTL3 promotes NET-induced cellular iron death by mediating the m6A modification of GPX4 (98).
5 MAC and GI cancer immunotherapy
Although ICIs have significantly improved the prognosis of patients with advanced GI cancers, a significant proportion of patients remain unresponsive to ICIs. Emerging evidence indicates that both intrinsic factors of tumor cells (e.g., immune checkpoints expression levels, tumor mutational load) and external factors (e.g., immune cells infiltration levels within the TIME), are closely associated with ICIs efficacy (99). Therefore, a deeper exploration of the impact of MAC on the TIME (Figure 2), including immune cell infiltration levels and immune checkpoint expression such as PD-1, can provide valuable insights for developing novel immunotherapies.
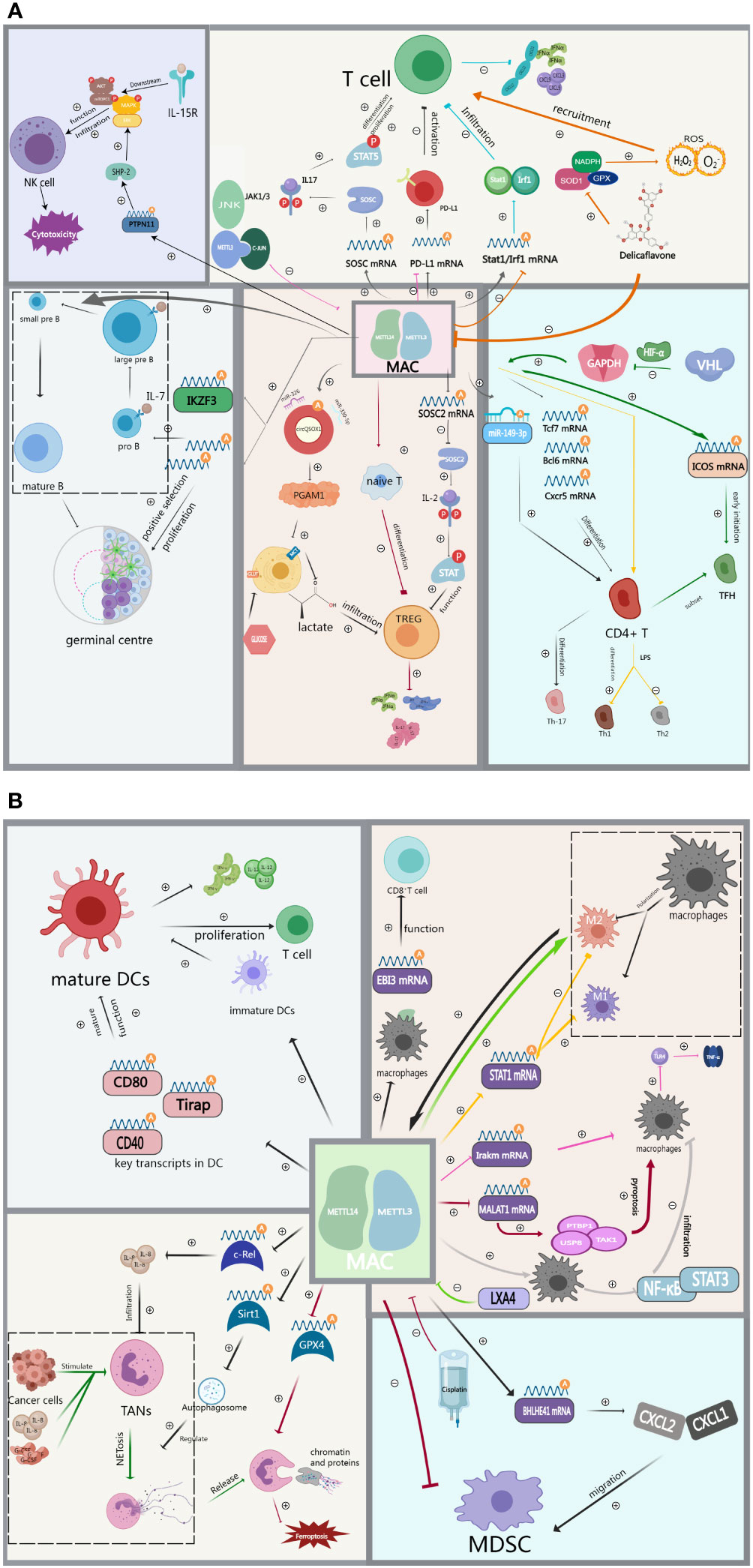
Figure 2 (A) MAC regulates immune cell infiltration including T cells, Th cells, TREG, B cells and NK cells in the tumor immune microenvironment. (B) MAC regulates immune cell infiltration including DCs, TAMs, TANs and MDSCs in the tumor immune microenvironment.
5.1 Esophageal cancer
Ranked as the eighth most common cancer type worldwide, the overall prognosis of EC patients under current conventional treatments remains unfavorable. Immunotherapy has shown promise in enhancing the survival of advanced esophageal cancer patients. However, the clinical efficacy of current ICI monotherapy is limited due to the complex immune evasion mechanism involved in EC (100). Guo et al. studied 20 m6A methylation regulators and discovered a significant negative correlation between the expression levels of METTL3/METL14 and PD-L1 in patients with esophageal squamous cell carcinoma (ESCC) (101). Additionally, Zhou et al. found that METTL3 expression levels negatively correlated with the infiltration levels of T lymphocytes, B lymphocytes, and neutrophils in ESCC, suggesting that METTL3 plays a key role in the immune regulation of esophageal cancer (102). Therefore, MAC could potentially enhance the sensitivity of ESCC patients to immunotherapy by modulating the TIME and PD-L1 expression. Moreover, shRNA knockdown experiments demonstrated that IFIT2 is a downstream target gene regulated by METTL3 and is closely associated with prognostic immune infiltration. Both factors can serve as immunotherapy targets (103); and prognostic or diagnostic indicators for esophageal cancer, offering novel insights for immunotherapy selection in ESSC patients.
5.2 Gastric cancer
Previous clinical studies have demonstrated the clear therapeutic potential of immunotherapy for patients with advanced gastric cancer. However, the highly heterogeneous nature of GC makes the selection of sensitive predictive biological targets a primary focus in current immunotherapy research (104). Zhang et al. classified gastric cancer samples into three categories based on the expression levels of 21 m6A regulators through unsupervised clustering analysis. These categories corresponded to three distinct tumor immune patterns: the immune-rejection phenotype, immune-inflammatory phenotype, and immune desert phenotype. These patterns can predict the intensity of the anti-tumor immune response in patients (105). Subsequently, METTL3/14 was found to be highly expressed in m6ACluster B, representing an immunoinflammatory phenotype associated with adaptive immune activation. Notably, patients with microsatellite instability (MSI), characteristic of m6ACluster B, tend to exhibit better immunotherapy efficacy (106).
METTL3, a major component of MAC, has been shown to influence the response to immunotherapy in GC patients through METTL3-mediated m6A methylation. This is evident when analyzing its downstream methylation core molecules (107). Similarly, it was observed that immune cells such as CD8+ T cells and T follicular helper cells were significantly more abundant in patient population-Cluster 2, characterized by overall high METTL3/14 expression. Additionally, METTL3 was associated with elevated PD-L1 expression in gastric cancer cells (108). In summary, this study suggests that quantitative analysis of METTL3/14 and its downstream molecules could help in predicting the response of gastric cancer patients to immunotherapy. The potential of using METTL3/14 as a direct target for anti-tumor immunotherapy in gastric cancer patients to promote treatment efficacy warrants further exploration.
5.3 Liver cancer
LC continues to have a persistently high mortality rate worldwide due to its unique vascular and neurological distribution. Immunotherapy offers hope for patients with advanced liver cancer who have limited treatment options such as surgery, liver transplantation, and local ablative therapy (109). However, overcoming LC patient resistance to immunotherapy remains a pressing challenge. Gu et al. found a negative correlation between the expression of writers represented by METTL3/14 and the infiltration of pDC, DC, and cytotoxic cells in Hepatocellular carcinoma (HCC). They concluded that METTL3 expression is positively correlated with sensitivity to multiple immune drugs, suggesting potential biomarkers for drug screening (110). Conversely, Jiang et al. discovered a significant correlation between the expression of METTL3/14 and immune checkpoint markers such as PD-L1 and CTLA4. This implies that METTL3/14 could serve as a novel biomarker for predicting the response to anti-PD-1/L1 immunotherapy in HCC (111, 112).
Zhen et al. reported that ZNF320, a member of the Kruppel-like zinc finger gene family, can regulate immune cell infiltration in HCC by affecting the expression levels of related chemokines through positive regulation of METTL3 Correlations with immune checkpoint-related genes revealed that patients with high ZNF320 expression also had elevated PDL-1 expression, suggesting potentially improved anti-PDL-L1 immunotherapy efficacy (113). In contrast, Peng et al. conducted lipopolysaccharide induction experiments in HCC cells and found that high METTL14 expression mediates m6A methylation of MIR155HG. MIR155HG can regulate PD-L1 expression through RNA binding protein ELAVL1 or via the miR-223/STAT1 axis, potentially affecting immunotherapy (114). Cholangiocarcinoma (CCA), the second most common form of liver cancer, is characterized by its high recurrence rate, low survival rate, and poor prognosis (115). In a recent study by Zhen et al., METTL14 was found to induce the degradation of SIAH2 mRNA and protein in a YTHDF2-dependent manner in cholangiocarcinoma. Subsequent in vitro and in vivo experiments revealed that SIAH2 inhibits T-cell expansion and cytotoxicity by enhancing the K63-linked ubiquitination of PD-L1 and its interaction with PD-L1, consequently reducing the efficacy of immunotherapy in patients (116). In summary, m6A and other related epigenetic modifications can reactivate immunotherapy-related genes in combination with immune therapy. The expression of METTL3/14 can synergize with ICI therapy by regulating METTL3/14 expression, potentially enhancing the anti-tumor response. This holds significant promise for improving the clinical efficacy of immunotherapy in HCC patients.
5.4 Pancreatic cancer
Pancreatic cancer (PC), characterized by a poor prognosis, continues to pose a significant clinical challenge. Classical therapies, including chemotherapy, radiotherapy, and targeted therapy have limited long-term benefits for patients. Furthermore, early-stage pancreatic cancer often exhibits suppressed tumor immunity, hindering the effectiveness of immunotherapy in improving survival outcomes for PC patients (117). Liu et al. found that in pancreatic ductal adenocarcinoma (PDAC), high METTL3/14 expression was significantly and positively associated with the enrichment of activated NK cells and mast cells (118). Specifically, within the CD4+ T cell subpopulation, METTLE3 was found to be negatively correlated with Th2 and Th17 cells, which regulate inflammatory responses, while it was positively correlated with Th1 and Tfh cells, known for their crucial role in tumor immunotherapy. This suggests that METTL3/14 could potentially regulate the immune microenvironment in pancreatic ductal adenocarcinoma. Wang et al. constructed an m6A score model using METTL3 as a prognostic protective gene and found that patients with low scores exhibited higher Immune cell Proportion Scores (IPS) for both anti-PD-1/CTLA-4 therapy alone and immune combination therapy (119). This study underscores the potential of METTL3/14 as a predictor of immunotherapy response in pancreatic cancer patients.
Additionally, Song et al. suggested that LncRNA MALAT1 could serve as a downstream target of METTL3. Inhibiting the expression of METTL3 would lead to a further reduction in MALAT1 expression in pancreatic cancer cells, consequently inhibiting the expression of PD-L1 (120). Conversely, METTL3 increases the expression level of circMYO1C and induces its cyclization. circMYO1C, through its binding to the 3’UTR of PD-L1 mRNA via IGF2BP2 protein at its methylation site, stabilizes PD-L1 expression. This ultimately promotes tumor growth by increasing the expression of immunosuppressive molecules such as PD-L1, promoting immune evasion by pancreatic cancer cells (121). Therefore, the direct targeting of m6A-modifying regulators to modulate immune checkpoint expression emerges as a potential strategy to improve the efficacy of immunotherapy and extend the prognosis of pancreatic cancer patients.
5.5 Colorectal cancer
CRC presents a complex profile and wide range of mutational characteristics. For this reason, immunotherapy has not shown good results in other types of CRC despite yielding satisfactory outcomes in some patients (122). Therefore, there is a need to find effective combinations with enhanced response to immunotherapy in patients with other types of CRC. In Xiong’s study, METTL3 improved immunosuppressive function of myeloid cells by mediating m6A modification of JAK1 mRNA and promoting its protein translation (93). Chen et al. reported that METTL3 can regulate the BHLHE41-CXCL1/CXCR2 axis in an m6A-dependent manner to promote MDSC migration. Based on this theory, inclusion of METTL3 inhibitors in anti-PD1 therapy showed improved sensitivity of CRC patients to anti-PD1 therapy (93). In addition, as another immune checkpoint inhibitor, anti-CTLA-4 therapy is currently used to treat CRC. The METTL3-mediated m6A modification of circQSOX1 results in heightened glycolysis levels and elevated lactate production in CRC cells. This, in turn, facilitates immune evasion by CRC cells. Consequently, the inhibition of these processes, in conjunction with anti-CTLA-4 therapy, holds the potential to diminish resistance to immunotherapy in CRC patients (72).
As another component of MAC, deletion of METTL14 expression in macrophages leads to insufficient activation of CD8+ T cell function, which in turn inhibits T-cell anti-tumor responses (83). This concept can be used to guide the development of potential immunotherapies against macrophages. In rectal cancer, Cai et al. found that downregulation of METTL14 resulted in reduced immune cell infiltration indicating that it may be used as an immune infiltration-related prognostic biomarker in rectal cancer (123). On the other hand, it was found that knockdown of METTL3/METTL14 in CRC cells increased CD8+ T cell infiltration and promoted IFN-γ, CXCL9 and CXCL10 cytokine secretion, leading to enhanced patient’s response to anti-PD-1 therapy (67). In summary, optimizing the response of CRC patients to ICIs by directly targeting METTL3/14 or its downstream mechanisms may be a new avenue for developing future immunotherapy of colorectal cancer.
6 Discussion
Given the role of m6A modifications in the regulation of gene expression, the catalytic subunit m6A-METTL complex (MAC)-METTL3/14 has emerged as a key player in various biological processes, including cancer development and progression. Recent studies have shown that METTL3/14 can regulate immune cell infiltration in the TIME, highlighting its importance in cancer immunology. In particular, the expression levels of METTL3/14 have been observed to regulate immune function in individuals with GI cancers and impact the effectiveness of tumor immunotherapy. These findings support the clinical potential of METTL3/14 as an immune marker for GI tumors. However, investigations into m6A methylation in immunotherapy are still in early stages and many questions remain to be answered. Further studies on METTL3/14 and its downstream mechanisms are needed to provide new possibilities for developing immune precision therapy for GI cancers.
Author contributions
CP: Writing – original draft, Writing – review & editing. FX: Writing – original draft. XP: Writing – original draft. ZH: Writing – review & editing. YY: Writing – review & editing. XQ: Data curation, Writing – review & editing. YJ: Investigation, Writing – review & editing. MH: Investigation, Writing – review & editing. DW: Writing – review & editing. XL: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The study was funded by the China Postdoctoral Science Foundation (2021M693272 to DW), the Science and Technology Planning Social Development Project of Zhenjiang City (SH2021068 to DW), Innovation Funds from the Chinese Society Of Clinical Oncology Youth Committee (Y-Young2021-0107 to DW), and 5123 Scholar Program of the Affiliated Hospital of Jiangsu University (51232017301 to DW).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Arnold M, Abnet CC, Neale RE, Vignat J, Giovannucci EL, McGlynn KA, et al. Global burden of 5 major types of gastrointestinal cancer. Gastroenterology (2020) 159(1):335–349.e15. doi: 10.1053/j.gastro.2020.02.068
2. Abdul-Latif M, Townsend K, Dearman C, Shiu KK, Khan K. Immunotherapy in gastrointestinal cancer: The current scenario and future perspectives. Cancer Treat Rev (2020) 88:102030. doi: 10.1016/j.ctrv.2020.102030
3. Smyth EC, Moehler M. Late-line treatment in metastatic gastric cancer: today and tomorrow. Ther Adv Med Oncol (2019) 11:1758835919867522. doi: 10.1177/1758835919867522
4. Zhao Y, Peng H. The role of N(6)-methyladenosine (m(6)A) methylation modifications in hematological Malignancies. Cancers (Basel) (2022) 14(2):332. doi: 10.3390/cancers14020332
5. Zhang W, Qian Y, Jia G. The detection and functions of RNA modification m(6)A based on m(6)A writers and erasers. J Biol Chem (2021) 297(2):100973. doi: 10.1016/j.jbc.2021.100973
6. Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer (2019) 18(1):103. doi: 10.1186/s12943-019-1033-z
7. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell (2012) 149(7):1635–46. doi: 10.1016/j.cell.2012.05.003
8. Bodi Z, Bottley A, Archer N, May ST, Fray RG. Yeast m6A Methylated mRNAs Are Enriched on Translating Ribosomes during Meiosis, and under Rapamycin Treatment. PloS One (2015) 10(7):e0132090. doi: 10.1371/journal.pone.0132090
9. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature (2012) 485(7397):201–6. doi: 10.1038/nature11112
10. Lan Q, Liu PY, Haase J, Bell JL, Hüttelmaier S, Liu T. The critical role of RNA m(6)A methylation in cancer. Cancer Res (2019) 79(7):1285–92. doi: 10.1158/0008-5472.CAN-18-2965
11. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m⁶A RNA methylation. Nat Rev Genet (2014) 15(5):293–306. doi: 10.1038/nrg3724
12. Huang Q, Mo J, Liao Z, Chen X, Zhang B. The RNA m(6)A writer WTAP in diseases: structure, roles, and mechanisms. Cell Death Dis (2022) 13(10):852. doi: 10.1038/s41419-022-05268-9
13. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature (2016) 534(7608):575–8. doi: 10.1038/nature18298
14. Su S, Li S, Deng T, Gao M, Yin Y, Wu B, et al. Cryo-EM structures of human m(6)A writer complexes. Cell Res (2022) 32(11):982–94. doi: 10.1038/s41422-022-00725-8
15. Tuck MT. Partial purification of a 6-methyladenine mRNA methyltransferase which modifies internal adenine residues. Biochem J (1992) 288(Pt 1):): 233–40. doi: 10.1042/bj2880233
16. Oerum S, Meynier V, Catala M, Tisné C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res (2021) 49(13):7239–55. doi: 10.1093/nar/gkab378
17. Meiser N, Mench N, Hengesbach M. RNA secondary structure dependence in METTL3-METTL14 mRNA methylation is modulated by the N-terminal domain of METTL3. Biol Chem (2020) 402(1):89–98. doi: 10.1515/hsz-2020-0265
18. Yoshida A, Oyoshi T, Suda A, Futaki S, Imanishi M. Recognition of G-quadruplex RNA by a crucial RNA methyltransferase component, METTL14. Nucleic Acids Res (2022) 50(1):449–57. doi: 10.1093/nar/gkab1211
19. Śledź P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife (2016) 5:e18434. doi: 10.7554/eLife.18434
20. Huang J, Dong X, Gong Z, Qin L-Y, Yang S, Zhu Y-L, et al. Solution structure of the RNA recognition domain of METTL3-METTL14 N(6)-methyladenosine methyltransferase. Protein Cell (2019) 10(4):272–84. doi: 10.1007/s13238-018-0518-7
21. Bujnicki JM, Feder M, Radlinska M, Blumenthal RM. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m(6)A methyltransferase. J Mol Evol (2002) 55(4):431–44. doi: 10.1007/s00239-002-2339-8
22. Zhou H, Yin K, Zhang Y, Tian J, Wang S. The RNA m6A writer METTL14 in cancers: Roles, structures, and applications. Biochim Biophys Acta Rev Cancer (2021) 1876(2):188609. doi: 10.1016/j.bbcan.2021.188609
23. Schöller E, Weichmann F, Treiber T, Ringle S, Treiber N, Flatley A, et al. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA (2018) 24(4):499–512. doi: 10.1261/rna.064063.117
24. Zhou KI, Pan T. Structures of the m(6)A Methyltransferase Complex: Two Subunits with Distinct but Coordinated Roles. Mol Cell (2016) 63(2):183–5. doi: 10.1016/j.molcel.2016.07.005
25. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol (2014) 10(2):93–5. doi: 10.1038/nchembio.1432
26. He PC, He C. m(6) A RNA methylation: from mechanisms to therapeutic potential. EMBO J (2021) 40(3):e105977. doi: 10.15252/embj.2020105977
27. Xie W, Patel DJ. A 'warhorse-shaped' topology adopted by the m(6)A RNA writer complex. Cell Res (2023) 33(1):9–10. doi: 10.1038/s41422-022-00748-1
28. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang W-J, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res (2014) 24(2):177–89. doi: 10.1038/cr.2014.3
29. Fan Y, Li X, Sun H, Gao Z, Zhu Z, Yuan K. Role of WTAP in cancer: from mechanisms to the therapeutic potential. Biomolecules (2022) 12(9):1224. doi: 10.3390/biom12091224
30. Little NA, Hastie ND, Davies RC. Identification of WTAP, a novel Wilms' tumour 1-associating protein. Hum Mol Genet (2000) 9(15):2231–9. doi: 10.1093/oxfordjournals.hmg.a018914
31. Yue Y, Liu J, Cui X, et al. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discovery (2018) 4:10. doi: 10.1038/s41421-018-0019-0
32. Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol Cell (2018) 69(6):1028–1038.e6. doi: 10.1016/j.molcel.2018.02.015
33. Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev (2018) 32(5-6):415–29. doi: 10.1101/gad.309146.117
34. Bawankar P, Lence T, Paolantoni C, Haussmann IU, Kazlauskiene M, Jacob D, et al. Hakai is required for stabilization of core components of the m(6)A mRNA methylation machinery. Nat Commun (2021) 12(1):3778. doi: 10.1038/s41467-021-23892-5
35. Wang Y, Zhang L, Ren H, Ma L, Guo J, Mao D, et al. Role of Hakai in m(6)A modification pathway in Drosophila. Nat Commun (2021) 12(1):2159. doi: 10.1038/s41467-021-22424-5
36. Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut (2020) 69(7):1193–205. doi: 10.1136/gutjnl-2019-319639
37. Liu Z, Wu K, Gu S, Wang W, Xie S, Lu T, et al. A methyltransferase-like 14/miR-99a-5p/tribble 2 positive feedback circuit promotes cancer stem cell persistence and radioresistance via histone deacetylase 2-mediated epigenetic modulation in esophageal squamous cell carcinoma. Clin Transl Med (2021) 11(9):e545. doi: 10.1002/ctm2.545
38. Chen H, Gao S, Liu W, Wong C-C, Wu J, Wu J, et al. RNA N(6)-methyladenosine methyltransferase METTL3 facilitates colorectal cancer by activating the m(6)A-GLUT1-mTORC1 axis and is a therapeutic target. Gastroenterology (2021) 160(4):1284–1300.e16. doi: 10.1053/j.gastro.2020.11.013
39. Deng R, Cheng Y, Ye S, Zhang J, Huang R, Li P, et al. m(6)A methyltransferase METTL3 suppresses colorectal cancer proliferation and migration through p38/ERK pathways. Onco Targets Ther (2019) 12:4391–402. doi: 10.2147/OTT.S201052
40. Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer (2020) 19(1):46. doi: 10.1186/s12943-020-1146-4
41. Wang W, Shao F, Yang X, Wang J, Zhu R, Yang Y, et al. METTL3 promotes tumour development by decreasing APC expression mediated by APC mRNA N(6)-methyladenosine-dependent YTHDF binding. Nat Commun (2021) 12(1):3803. doi: 10.1038/s41467-021-23501-5
42. Hu N, Ji H. N6-methyladenosine (m6A)-mediated up-regulation of long noncoding RNA LINC01320 promotes the proliferation, migration, and invasion of gastric cancer via miR495-5p/RAB19 axis. Bioengineered (2021) 12(1):4081–91. doi: 10.1080/21655979.2021.1953210
43. Fan HN, Chen ZY, Chen XY, Chen M, Yi Y-C, Zhu J-S, et al. METTL14-mediated m(6)A modification of circORC5 suppresses gastric cancer progression by regulating miR-30c-2-3p/AKT1S1 axis. Mol Cancer (2022) 21(1):51. doi: 10.1186/s12943-022-01521-z
44. Li T, Hu PS, Zuo Z, Lin J-F, Li X, Wu Q-N, et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer (2019) 18(1):112. doi: 10.1186/s12943-019-1038-7
45. Wang L, Yi X, Xiao X, Zheng Q, Ma L, Li B. Exosomal miR-628-5p from M1 polarized macrophages hinders m6A modification of circFUT8 to suppress hepatocellular carcinoma progression. Cell Mol Biol Lett (2022) 27(1):106. doi: 10.1186/s11658-022-00406-9
46. Du L, Li Y, Kang M, Feng M, Ren Y, Dai H, et al. USP48 is upregulated by mettl14 to attenuate hepatocellular carcinoma via regulating SIRT6 stabilization. Cancer Res (2021) 81(14):3822–34. doi: 10.1158/0008-5472.CAN-20-4163
47. Jiang Z, Song X, Wei Y, Li Y, Kong D, Sun J. N(6)-methyladenosine-mediated miR-380-3p maturation and upregulation promotes cancer aggressiveness in pancreatic cancer. Bioengineered (2022) 13(6):14460–71. doi: 10.1080/21655979.2022.2088497
48. Wang M, Liu J, Zhao Y, He R, Xu X, Guo X, et al. Upregulation of METTL14 mediates the elevation of PERP mRNA N(6) adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol Cancer (2020) 19(1):130. doi: 10.1186/s12943-020-01249-8
49. Lv B, Wang Y, Ma D, Cheng W, Liu J, Yong T, et al. Immunotherapy: reshape the tumor immune microenvironment. Front Immunol (2022) 13:844142. doi: 10.3389/fimmu.2022.844142
50. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med (2018) 24(5):541–50. doi: 10.1038/s41591-018-0014-x
51. Liu Q, Zhang H, Jiang X, Qian C, Liu Z, Luo D. Factors involved in cancer metastasis: a better understanding to "seed and soil" hypothesis. Mol Cancer (2017) 16(1):176. doi: 10.1158/1538-7445.EPSO16-B17
52. Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol (2013) 14(10):1014–22. doi: 10.1038/ni.2703
53. Basu A, Ramamoorthi G, Albert G, Gallen C, Beyer A, Snyder C, et al. Differentiation and regulation of T(H) cells: A balancing act for cancer immunotherapy. Front Immunol (2021) 12:669474. doi: 10.3389/fimmu.2021.669474
54. Li C, Jiang P, Wei S, Xu X, Wang J. Regulatory T cells in tumor microenvironment: new mechanisms, potential therapeutic strategies and future prospects. Mol Cancer (2020) 19(1):116. doi: 10.1158/1557-3125.HIPPO19-B11
55. Downs-Canner SM, Meier J, Vincent BG, Serody JS. B cell function in the tumor microenvironment. Annu Rev Immunol (2022) 40:169–93. doi: 10.1146/annurev-immunol-101220-015603
56. Terrén I, Orrantia A, Vitallé J, Zenarruzabeitia O, Borrego F. NK cell metabolism and tumor microenvironment. Front Immunol (2019) 10:2278. doi: 10.3389/fimmu.2019.02278
57. Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol (2016) 37(3):208–20. doi: 10.1016/j.it.2016.01.004
58. Boutilier AJ, Elsawa SF. Macrophage polarization states in the tumor microenvironment. Int J Mol Sci (2021) 22(13):6995. doi: 10.3390/ijms22136995
59. Giese MA, Hind LE, Huttenlocher A. Neutrophil plasticity in the tumor microenvironment. Blood (2019) 133(20):2159–67. doi: 10.1182/blood-2018-11-844548
60. Wculek SK, Cueto FJ, Mujal AM, Melero I, Krummel MF, Sancho D. Dendritic cells in cancer immunology and immunotherapy. Nat Rev Immunol (2020) 20(1):7–24. doi: 10.1038/s41577-019-0210-z
61. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer (2012) 12(4):252–64. doi: 10.1038/nrc3239
62. Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell (2015) 27(4):450–61. doi: 10.1016/j.ccell.2015.03.001
63. Zhang J, Huang D, Saw PE, Song E. Turning cold tumors hot: from molecular mechanisms to clinical applications. Trends Immunol (2022) 43(7):523–45. doi: 10.1016/j.it.2022.04.010
64. Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature (2017) 548(7667):338–42. doi: 10.1038/nature23450
65. Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W, et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N(6)-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol Cancer (2022) 21(1):60. doi: 10.1186/s12943-021-01447-y
66. Ni Z, Sun P, Zheng J, Wu M, Yang C, Cheng M, et al. JNK Signaling Promotes Bladder Cancer Immune Escape by Regulating METTL3-Mediated m6A Modification of PD-L1 mRNA. Cancer Res (2022) 82(9):1789–802. doi: 10.1158/0008-5472.CAN-21-1323
67. Wang L, Hui H, Agrawal K, Kang Y, Li N, Tang R, et al. m(6) A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J (2020) 39(20):e104514. doi: 10.15252/embj.2020104514
68. Wang X, Xu D, Chen B, Huang D, Li Z, Sui Y, et al. Delicaflavone represses lung cancer growth by activating antitumor immune response through N6-methyladenosine transferases and oxidative stress. Oxid Med Cell Longev (2022) 2022:8619275. doi: 10.1155/2022/8619275
69. Lu TX, Zheng Z, Zhang L, Sun H-L, Bissonnette M, Huang H, et al. A new model of spontaneous colitis in mice induced by deletion of an RNA m(6)A methyltransferase component METTL14 in T cells. Cell Mol Gastroenterol Hepatol (2020) 10(4):747–61. doi: 10.1016/j.jcmgh.2020.07.001
70. Tong J, Cao G, Zhang T, Sefik E, Vesely MCA, Broughton JP, et al. m(6)A mRNA methylation sustains Treg suppressive functions. Cell Res (2018) 28(2):253–6. doi: 10.1038/cr.2018.7
71. Liu Y, Yuan Y, Zhou Z, Cui Y, Teng Y, Huang H, et al. Mettl14-mediated m6A modification enhances the function of Foxp3(+) regulatory T cells and promotes allograft acceptance. Front Immunol (2022) 13:1022015. doi: 10.3389/fimmu.2022.1022015
72. Liu Z, Zheng N, Li J, Li C, Zheng D, Jiang X, et al. N6-methyladenosine-modified circular RNA QSOX1 promotes colorectal cancer resistance to anti-CTLA-4 therapy through induction of intratumoral regulatory T cells. Drug Resist Update (2022) 65:100886. doi: 10.1016/j.drup.2022.100886
73. Hou Y, Cao Y, Dong L, Huang Y, Zhang Z, Bi Y, et al. Regulation of follicular T helper cell differentiation in antitumor immunity. Int J Cancer (2023) 153(2):265–77. doi: 10.1002/ijc.34368
74. Cao Y, Wang Z, Yan Y, Ji L, He J, Xuan B, et al. Enterotoxigenic bacteroidesfragilis promotes intestinal inflammation and Malignancy by inhibiting exosome-packaged miR-149-3p. Gastroenterology (2021) 161(5):1552–1566.e12. doi: 10.1053/j.gastro.2021.08.003
75. Yao Y, Yang Y, Guo W, Xu L, You M, Zhang Y-C, et al. METTL3-dependent m(6)A modification programs T follicular helper cell differentiation. Nat Commun (2021) 12(1):1333. doi: 10.1038/s41467-021-21594-6
76. Zhu Y, Zhao Y, Zou L, Zhang D, Aki D, Liu YC. The E3 ligase VHL promotes follicular helper T cell differentiation via glycolytic-epigenetic control. J Exp Med (2019) 216(7):1664–81. doi: 10.1084/jem.20190337
77. Wu H, Xu Z, Wang Z, Ren Z, Li L, Ruan Y. Dendritic cells with METTL3 gene knockdown exhibit immature properties and prolong allograft survival. Genes Immun (2020) 21(3):193–202. doi: 10.1038/s41435-020-0099-3
78. Kang X, Chen S, Pan L, Liang X, Lu D, Chen H, et al. Deletion of mettl3 at the pro-B stage marginally affects B cell development and profibrogenic activity of B cells in liver fibrosis. J Immunol Res (2022) 2022:8118577. doi: 10.1155/2022/8118577
79. Zheng Z, Zhang L, Cui XL, Yu X, Hsu PJ, Lyu R, et al. Control of early B cell development by the RNA N(6)-methyladenosine methylation. Cell Rep (2020) 31(13):107819. doi: 10.1016/j.celrep.2020.107819
80. Huang H, Zhang G, Ruan GX, Li Y, Chen W, Zou J, et al. Mettl14-mediated m6A modification is essential for germinal center B cell response. J Immunol (2022) 208(8):1924–36. doi: 10.4049/jimmunol.2101071
81. Wang H, Hu X, Huang M, Liu J, Gu Y, Ma L, et al. Mettl3-mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat Commun (2019) 10(1):1898. doi: 10.1038/s41467-019-09903-6
82. Song H, Song J, Cheng M, Zheng M, Wang T, Tian S, et al. METTL3-mediated m(6)A RNA methylation promotes the anti-tumour immunity of natural killer cells. Nat Commun (2021) 12(1):5522. doi: 10.1038/s41467-021-25803-0
83. Dong L, Chen C, Zhang Y, Guo P, Wang Z, Li J, et al. The loss of RNA N(6)-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8(+) T cell dysfunction and tumor growth. Cancer Cell (2021) 39(7):945–957.e10. doi: 10.1016/j.ccell.2021.04.016
84. Liu Y, Liu Z, Tang H, Shen Y, Gong Z, Xie N, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 facilitates M1 macrophage polarization through the methylation of STAT1 mRNA. Am J Physiol Cell Physiol (2019) 317(4):C762–75. doi: 10.1152/ajpcell.00212.2019
85. Tong J, Wang X, Liu Y, Ren X, Wang A, Chen Z, et al. Pooled CRISPR screening identifies m(6)A as a positive regulator of macrophage activation. Sci Adv (2021) 7(18):eabd4742. doi: 10.1126/sciadv.abd4742
86. Jia G, Wang X, Wu W, Zhang Y, Chen S, Zhao J, et al. LXA4 enhances prostate cancer progression by facilitating M2 macrophage polarization via inhibition of METTL3. Int Immunopharmacol (2022) 107:108586. doi: 10.1016/j.intimp.2022.108586
87. Shu B, Zhou YX, Li H, Zhang RZ, He C, Yang X. The METTL3/MALAT1/PTBP1/USP8/TAK1 axis promotes pyroptosis and M1 polarization of macrophages and contributes to liver fibrosis. Cell Death Discov (2021) 7(1):368. doi: 10.1038/s41420-021-00756-x
88. Yin H, Zhang X, Yang P, Zhang X, Peng Y, Li D, et al. RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat Commun (2021) 12(1):1394. doi: 10.1038/s41467-021-21514-8
89. Wu L, Cheng D, Yang X, Zhao W, Fang C, Chen R, et al. M2-TAMs promote immunoresistance in lung adenocarcinoma by enhancing METTL3-mediated m6A methylation. Ann Transl Med (2022) 10(24):1380. doi: 10.21037/atm-22-6104
90. Ni HH, Zhang L, Huang H, Dai SQ, Li J. Connecting METTL3 and intratumoural CD33(+) MDSCs in predicting clinical outcome in cervical cancer. J Transl Med (2020) 18(1):393. doi: 10.1186/s12967-020-02553-z
91. Chen H, Pan Y, Zhou Q, Liang C, Wong C-C, Zhou Y, et al. METTL3 inhibits antitumor immunity by targeting m(6)A-BHLHE41-CXCL1/CXCR2 axis to promote colorectal cancer. Gastroenterology (2022) 163(4):891–907. doi: 10.1053/j.gastro.2022.06.024
92. Mu X, Wu K, Zhu Y, Zhu Y, Wang Y, Xiao L, et al. Intra-arterial infusion chemotherapy utilizing cisplatin inhibits bladder cancer by decreasing the fibrocytic myeloid-derived suppressor cells in an m6A-dependent manner. Mol Immunol (2021) 137:28–40. doi: 10.1016/j.molimm.2021.06.012
93. Xiong J, He J, Zhu J, Pan J, Liao W, Ye H, et al. Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol Cell (2022) 82(9):1660–1677.e10. doi: 10.1016/j.molcel.2022.02.033
94. He J, Zhou M, Yin J, Wan J, Chu J, Jia J, et al. METTL3 restrains papillary thyroid cancer progression via m(6)A/c-Rel/IL-8-mediated neutrophil infiltration. Mol Ther (2021) 29(5):1821–37. doi: 10.1016/j.ymthe.2021.01.019
95. Masucci MT, Minopoli M, Del Vecchio S, Carriero MV. The emerging role of neutrophil extracellular traps (NETs) in tumor progression and metastasis. Front Immunol (2020) 11:1749. doi: 10.3389/fimmu.2020.01749
96. Fang Q, Stehr AM, Naschberger E, Knopf J, Herrmann M, Stürzl M. No NETs no TIME: Crosstalk between neutrophil extracellular traps and the tumor immune microenvironment. Front Immunol (2022) 13:1075260. doi: 10.3389/fimmu.2022.1075260
97. Qu M, Chen Z, Qiu Z, Nan K, Wang Y, Shi Y, et al. Neutrophil extracellular traps-triggered impaired autophagic flux via METTL3 underlies sepsis-associated acute lung injury. Cell Death Discov (2022) 8(1):375. doi: 10.1038/s41420-022-01166-3
98. Zhang H, Liu J, Zhou Y, Qu M, Wang Y, Guo K, et al. Neutrophil extracellular traps mediate m(6)A modification and regulates sepsis-associated acute lung injury by activating ferroptosis in alveolar epithelial cells. Int J Biol Sci (2022) 18(8):3337–57. doi: 10.7150/ijbs.69141
99. Zhang M, Song J, Yuan W, Zhang W, Sun Z. Roles of RNA methylation on tumor immunity and clinical implications. Front Immunol (2021) 12:641507. doi: 10.3389/fimmu.2021.641507
100. Huang TX, Fu L. The immune landscape of esophageal cancer. Cancer Commun (Lond) (2019) 39(1):79. doi: 10.1186/s40880-019-0427-z
101. Guo W, Tan F, Huai Q, Wang Z, Shao F, Zhang G, et al. Comprehensive analysis of PD-L1 expression, immune infiltrates, and m6A RNA methylation regulators in esophageal squamous cell carcinoma. Front Immunol (2021) 12:669750. doi: 10.3389/fimmu.2021.669750
102. Zhou Y, Guo S, Li Y, Chen F, Wu Y, Xiao Y, et al. METTL3 is associated with the Malignancy of esophageal squamous cell carcinoma and serves as a potential immunotherapy biomarker. Front Oncol (2022) 12:824190. doi: 10.3389/fonc.2022.824190
103. Ge F, Li Z, Hu J, Pu Y, Zhao F, Kong L. METTL3/m(6)A/IFIT2 regulates proliferation, invasion and immunity in esophageal squamous cell carcinoma. Front Pharmacol (2022) 13:1002565. doi: 10.3389/fphar.2022.1002565
104. Zhao Q, Cao L, Guan L, Bie L, Wang S, Xie B, et al. Immunotherapy for gastric cancer: dilemmas and prospect. Brief Funct Genomics (2019) 18(2):107–12. doi: 10.1093/bfgp/ely019
105. Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer (2020) 19(1):53. doi: 10.1186/s12943-020-01170-0
106. Yang J, Wu Z, Wu X, Chen S, Xia X, Zeng J. Constructing and validating of m6a-related genes prognostic signature for stomach adenocarcinoma and immune infiltration: Potential biomarkers for predicting the overall survival. Front Oncol (2022) 12:1050288. doi: 10.3389/fonc.2022.1050288
107. Chen S, Su X, Wang J, Zheng N, Tang Y, Peng G, et al. Identification and validation of METTL3-related molecules for predicting prognosis and efficacy of immunotherapy in gastric cancer based on m6A methylome and transcriptome sequencing analysis. Front Oncol (2022) 12:935239. doi: 10.3389/fonc.2022.935239
108. Xu Z, Chen Q, Shu L, Zhang C, Liu W, Wang P. Expression profiles of m6A RNA methylation regulators, PD-L1 and immune infiltrates in gastric cancer. Front Oncol (2022) 12:970367. doi: 10.3389/fonc.2022.970367
109. Feng GS, Hanley KL, Liang Y, Lin X. Improving the efficacy of liver cancer immunotherapy: the power of combined preclinical and clinical studies. Hepatology (2021) 73 Suppl 1(Suppl 1):104–14. doi: 10.1002/hep.31479
110. Gu Z, Du Y, Zhao X, Wang C. Diagnostic, therapeutic, and prognostic value of the m(6)A writer complex in hepatocellular carcinoma. Front Cell Dev Biol (2022) 10:822011. doi: 10.3389/fcell.2022.822011
111. Huang X, Qiu Z, Li L, Chen B, Huang P. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in hepatocellular carcinoma. Aging (Albany NY) (2021) 13(16):20698–715. doi: 10.18632/aging.203456
112. Jiang H, Ning G, Wang Y, Lv W. Identification of an m6A-related signature as biomarker for hepatocellular carcinoma prognosis and correlates with sorafenib and anti-PD-1 immunotherapy treatment response. Dis Markers (2021) 2021:5576683. doi: 10.1155/2021/5576683
113. Zhen J, Ke Y, Pan J, Zhou M, Zeng H, Song G, et al. ZNF320 is a hypomethylated prognostic biomarker involved in immune infiltration of hepatocellular carcinoma and associated with cell cycle. Aging (Albany NY) (2022) 14(20):8411–36. doi: 10.18632/aging.204350
114. Peng L, Pan B, Zhang X, Wang Z, Qiu J, Wang X, et al. Lipopolysaccharide facilitates immune escape of hepatocellular carcinoma cells via m6A modification of lncRNA MIR155HG to upregulate PD-L1 expression. Cell Biol Toxicol (2022) 38(6):1159–73. doi: 10.1007/s10565-022-09718-0
115. Rizvi S, Khan SA, Hallemeier CL, Kelley RK, Gores GJ. Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat Rev Clin Oncol (2018) 15(2):95–111. doi: 10.1038/nrclinonc.2017.157
116. Zheng H, Zheng WJ, Wang ZG, Tao Y-P, Huang Z-P, Yang L, et al. Decreased expression of programmed death ligand-L1 by seven in absentia homolog 2 in cholangiocarcinoma enhances T-cell-mediated antitumor activity. Front Immunol (2022) 13:845193. doi: 10.3389/fimmu.2022.845193
117. Bear AS, Vonderheide RH, O'Hara MH. Challenges and opportunities for pancreatic cancer immunotherapy. Cancer Cell (2020) 38(6):788–802. doi: 10.1016/j.ccell.2020.08.004
118. Liu Y, Li G, Yang Y, Lu Z, Wang T, Wang X, et al. Analysis of N6-methyladenosine modification patterns and tumor immune microenvironment in pancreatic adenocarcinoma. Front Genet (2021) 12:752025. doi: 10.3389/fgene.2021.752025
119. Wang L, Zhang S, Li H, Xu Y, Wu Q, Shen J, et al. Quantification of m6A RNA methylation modulators pattern was a potential biomarker for prognosis and associated with tumor immune microenvironment of pancreatic adenocarcinoma. BMC Cancer (2021) 21(1):876. doi: 10.1186/s12885-021-08550-9
120. Song Z, Wang X, Chen F, Chen Q, Liu W, Yang X, et al. LncRNA MALAT1 regulates METTL3-mediated PD-L1 expression and immune infiltrates in pancreatic cancer. Front Oncol (2022) 12:1004212. doi: 10.3389/fonc.2022.1004212
121. Guan H, Tian K, Luo W, Li M. m(6)A-modified circRNA MYO1C participates in the tumor immune surveillance of pancreatic ductal adenocarcinoma through m(6)A/PD-L1 manner. Cell Death Dis (2023) 14(2):120. doi: 10.1038/s41419-023-05570-0
122. Lichtenstern CR, Ngu RK, Shalapour S, Karin M. Immunotherapy, inflammation and colorectal cancer. Cells (2020) 9(3):618. doi: 10.3390/cells9030618
Keywords: N 6 -methyladenosine (m 6 A) methylation modification, catalytic subunit m 6 A-METTL complex, METTL3/14, immune cell infiltration, immunotherapy, gastrointestinal cancers
Citation: Peng C, Xiong F, Pu X, Hu Z, Yang Y, Qiao X, Jiang Y, Han M, Wang D and Li X (2023) m6A methylation modification and immune cell infiltration: implications for targeting the catalytic subunit m6A-METTL complex in gastrointestinal cancer immunotherapy. Front. Immunol. 14:1326031. doi: 10.3389/fimmu.2023.1326031
Received: 22 October 2023; Accepted: 04 December 2023;
Published: 15 December 2023.
Edited by:
Zong Sheng Guo, University at Buffalo, United StatesReviewed by:
Yongfei Song, Hangzhou Medical College, ChinaHester Doyle, Yale University, United States
Christian Augsberger, AstraZeneca, Germany
Fanen Yuan, University of Pittsburgh, United States
Copyright © 2023 Peng, Xiong, Pu, Hu, Yang, Qiao, Jiang, Han, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoqin Li, MTAwMDAxMDMwNEB1anMuZWR1LmNu; Deqiang Wang, MTAwMDAxMTQ1MkB1anMuZWR1LmNu
†These authors have contributed equally to this work
‡ORCID: Chen Peng, orcid.org/0009-0005-0494-9269
 Chen Peng
Chen Peng Fen Xiong
Fen Xiong Xi Pu2†
Xi Pu2† Zhangmin Hu
Zhangmin Hu Yufei Yang
Yufei Yang Xiaoqin Li
Xiaoqin Li