- 1Key Laboratory of Artificial Organs and Computational Medicine in Zhejiang Province, Shulan International Medical College, Zhejiang Shuren University, Hangzhou, China
- 2Ocean College, Beibu Gulf University, Qinzhou, China
Fibrinogen-related proteins (FREPs) and Down syndrome cell adhesion molecules (Dscams) are important immune-related molecules in invertebrates. Although they are found in different taxonomic groups and possess unique functions, both exhibit high diversity and adaptability. FREPs are characterized by their fibrinogen-related domains and have been primarily studied in mollusks, such as Biomphalaria glabrata. Through mechanisms of diversity generation, such as gene conversion and point mutations, BgFREP plays a critical role in the host’s defense against parasites. Dscams are immunoglobulin-like transmembrane proteins, mainly studied in arthropods, such as Drosophila melanogaster and Anopheles gambiae. Through alternative splicing, Dscams generate multiple isoforms that participate in pathogen recognition and the precise wiring of neural circuits. In D. melanogaster, DmDscam plays a role not only in neuronal self-recognition but also in pathogen recognition. In A. gambiae, AgDscam defends against parasite infections, by binding to pathogens and mediating phagocytosis. This paper highlights the key roles of FREPs and Dscams in the immunity of two major invertebrate groups—mollusks and arthropods—and summarizes the main advancements in current research. These studies not only deepen the understanding of invertebrate immune mechanisms but also lay a solid foundation for future exploration of their potential applications in the biomedical field.
1 Introduction
In the biological world, immunological memory refers to the immune system’s capability to retain and remember information about previously encountered pathogens, allowing it to mount a specific response upon subsequent exposure. Immunological memory is thought to be exclusive to jawed vertebrates, achieved through somatic recombination and clonal expansion of lymphocytes (1). Although they lack acquired immunity, invertebrates possess some highly diverse immune molecules that can be used to recognize different antigens, maintain homeostasis, and promote the organism’s adaptability. For example, fibrinogen-related protein (BgFREP) in Biomphalaria glabrata (2, 3), Variable chitin-binding proteins (VCBPs) in protochordates (4) and tunicates (5), Down syndrome cell adhesion molecules (Dscam) in arthropods (6–9)—these proteins each have their own corresponding functions, reflecting the unique immune mechanisms that have evolved in invertebrates. In recent years, there has been increasing attention on the highly diverse immune molecules in invertebrates and their roles in immunity as well as other functions.
Fibrinogen-related proteins, or FREPs, are a family of proteins that contain fibrinogen-related domains (FreD). They are widely distributed across various animal species and exhibit diverse forms and functions. Among them, a unique class of FREPs (BgFREP) with distinct structures exists in the immune system of B. glabrata (10). Freshwater snails of the genus Biomphalaria, especially B. glabrata, play a crucial role as intermediate hosts of Schistosoma mansoni, significantly influencing the transmission and geographical distribution of schistosomes (11). S. mansoni causes human intestinal schistosomiasis, with symptoms including hepatosplenomegaly, portal hypertension, anemia, and eosinophilia (12). After S. mansoni miracidium penetrates the intermediate host B. glabrata, they transform into mother sporocysts and subsequently develop into daughter sporocysts. This process is a crucial stage in larval infection and the establishment of the parasitic relationship (Figure 1A). During this period, they need to successfully defend against or evade the snail’s immune system (12). B. glabrata possesses a pathogen recognition system, composed of humoral and cellular receptors, specifically designed to recognize invading schistosomes (13). In humoral immunity, BgFREP acts as a soluble immune factor with lectin properties, classified as a calcium-dependent lectin (10), and plays a key role in the immune response to pathogens. The N-terminus of BgFREP typically contains one or two immunoglobulin superfamily (IgSF) domains (11), while the C-terminus contains a FreD domain. These two regions are connected by an α-helix linker (14, 15), as shown in Figure 1B. Molecules with a similar unique structure to BgFREP have also been found in certain other gastropod species (16, 17).
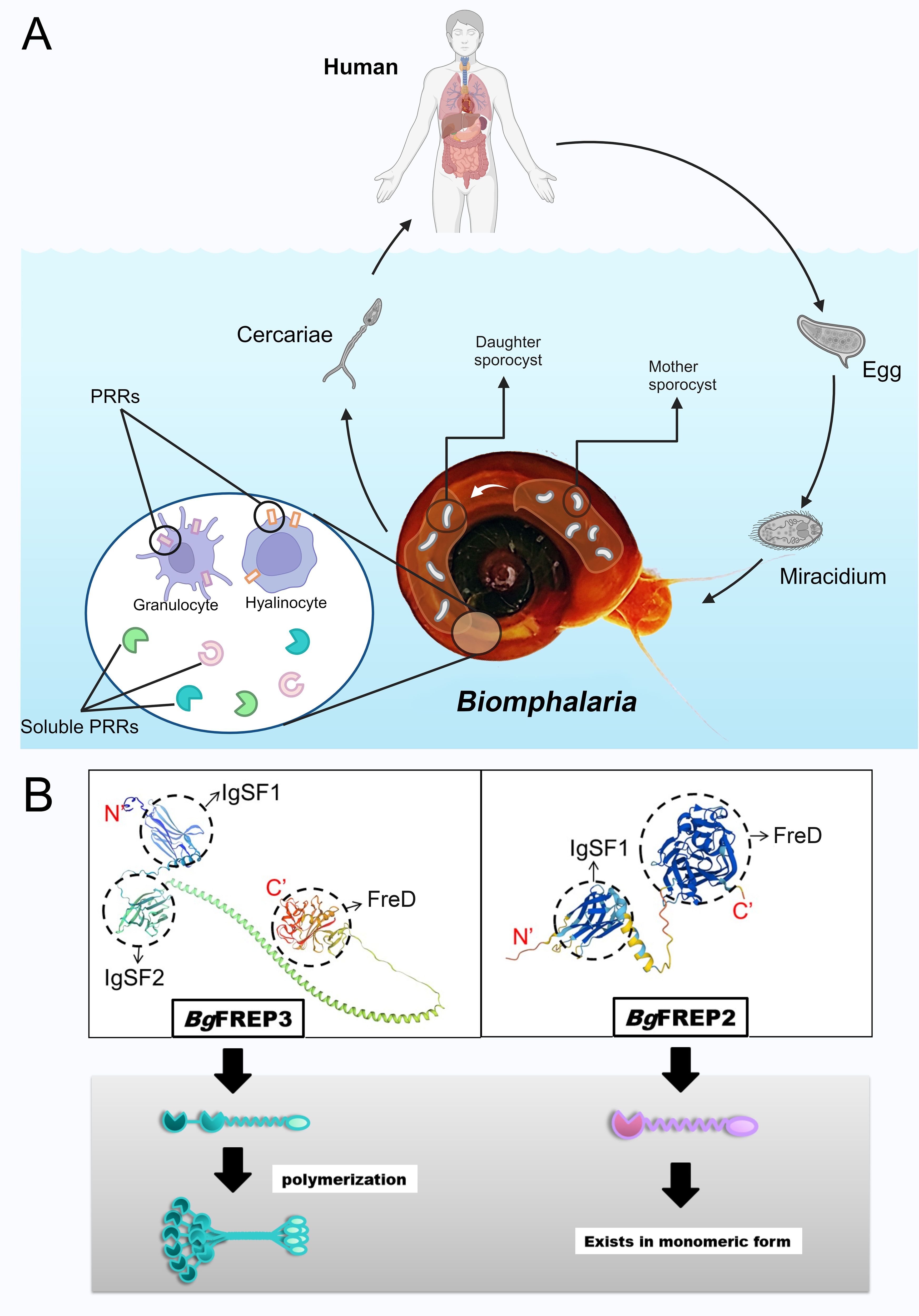
Figure 1. The life cycle of S. mansoni and the structural characteristics of the key immune molecules BgFREPs. (A) Key stages involved in the life cycle of S. mansoni and the immune response of B. glabrata. Adult schistosomes reproduce within the definitive host (typically a human), and the eggs are excreted through the host’s feces. In a suitable freshwater environment, the eggs hatch into miracidia, which typically invade the snail through the head-foot region. There, they shed their ciliated epidermal plates and develop into mother sporocysts, which then give rise to daughter sporocysts. The daughter sporocysts further replicate, ultimately producing cercariae, which are released from the snail. Once released into the water, the cercariae swim to locate a mammalian host (such as a human). The figure also illustrates the pathogen recognition system of B. glabrata, which consists of humoral pattern recognition receptors and cellular pattern recognition receptors. (B) Structural differences between the BgFREP3 and BgFREP2 proteins. On the left, the 3D modeled molecular structure of BgFREP3 (Uniprot ID: Q8WQX8, URL: Fibrinogen-related protein 3-2 – B. glabrata (Bloodfluke planorb)UniProtKBUniProt) is shown along with the schematic diagram below (green), which consists of two IgSF domains (IgSF1 and IgSF2) and a FreD domain. These domains are connected by a coiled-coil helical structure, which is hypothesized to facilitate the formation of a multimeric structure. On the right, the 3D modeled molecular structure of BgFREP2 (Uniprot ID: Q95UV9, URL: BgMFREP2 - BgMFREP2 – B. glabrata (Bloodfluke planorb)UniProtKBUniProt) is shown along with the schematic diagram below (light purple). BgFREP2 has only one IgSF domain, which is connected to the FreD domain by a shorter α-helix structure. According to the literature, it primarily exists in monomeric form. Created with BioRender.com.
Dscam is widely found across various animal species, particularly in insects, where it was first extensively studied in Drosophila melanogaster (DmDscam) (18). It is a unique neuronal adhesion protein and a member of the immunoglobulin (Ig) superfamily (19, 20). AgDscam has been identified in Anopheles gambiae, where it functions as a hypervariable pattern recognition receptor (PRR) in the immune system (21). Dscam is a complex transmembrane protein with extracellular, transmembrane, and intracellular domains, playing significant roles primarily in nervous system development (20) and the immune system. Dscam has relatively conserved domains in both D. melanogaster and vertebrates, including 10 Ig domains, 6 fibronectin type III (FNIII) domains, and a transmembrane domain (19). In A. gambiae, each AgDscam consists of 10 Ig domains, 6 fibronectin repeat domains, and a transmembrane domain (22). The DmDscam receptor is a typical example of affinity-based binding specificity and is involved in important biological processes (23).
To deepen our understanding of the diversity of FREPs in gastropods and Dscams in insects, and to further comprehend the immune mechanisms in invertebrates, this paper summarizes recent research progress on FREPs and Dscams, particularly on BgFREP in B. glabrata, DmDscam in D. melanogaster, and AgDscam in A. gambiae. A comparative analysis of FREPs and Dscams is presented. The article mainly describes the species distribution, molecular structure and diversity, immune-related functions, and other important functions of gastropod FREPs (BgFREP) and insect Dscams (DmDscam and AgDscam), along with the latest research developments. Additionally, by comparing FREPs and Dscams—two structurally and evolutionarily distinct immune molecules—this paper highlights their respective roles in invertebrate immunity and their unique mechanisms of pathogen recognition. Such a comparison not only deepens our understanding of innate immune diversity, but also offers new perspectives for biomedical research, particularly in designing novel immune-based therapies and exploring evolutionary principles of immune recognition.
2 Fibrinogen-related proteins
2.1 FREPs in species
FREP is a broad concept referring to all proteins that contain a FreD domain. In gastropod mollusks, a class of FREPs with unique structural features has been identified in species from certain subgroups of Heterobranchia and Caenogastropoda (24). These proteins have an IgSF domain at the N-terminus and a FreD domain at the C-terminus, playing an important role in the immune defense mechanisms of these mollusks.
These types of FREPs have been identified in various gastropod mollusks, including B. glabrata, Helisoma trivolvis, Lymnaea stagnalis, Bulinus truncatus, Biomphalaria pfeifferi, Helix aspersa, Limax flavus, Littorina littorea, and Aplysia californica (16). In vertebrates, no FREPs with similar structural features have been found to date. However, proteins containing FreD domains also exist in vertebrates and are involved in immune defense. Examples include those in Danio rerio (25), Oncorhynchus mykiss (26), Xenopus laevis (27), mice (28), chickens (29), and bovines (30).
2.2 Exploration of FREPs
Since the 1960s, agglutination molecules have been discovered in the plasma of mollusks (16). In oysters, these lectin-like molecules exhibit opsonization effects. When rabbit red blood cells are incubated in oyster plasma including lectins from Crassostrea virginica, the opsonization significantly enhances the phagocytosis of rabbit red blood cells by hemocytes (31). Later, in the 1970s and 1980s, similar lectins were found in the gastropods B. glabrata (2) and Helix pomatia (32). These lectin-like factors have been shown to participate in non-self-recognition through interaction with carbohydrate targets on the membranes of hemocytes and pathogen-related surfaces in other species (33). In a series of experiments conducted in the mid-1980s, scientists discovered agglutination reactions between snail plasma and schistosome sporocysts. Subsequent studies revealed changes in agglutination factors in snail plasma after exposure to trematode parasites, including upregulation of specific peptides and increased agglutination factor titers (34–38). Scientists isolated and purified agglutination factors from snail plasma and found that these factors are related to schistosome secretion/excretion products (SEP). Further research showed that these agglutination factors are encoded by multiple genes and their expression levels significantly increase after schistosome infection. In 1997, scientists finally discovered and characterized BgFREPs through the analysis of precipitates in snail plasma (10).
As FREP continues to evolve, it reflects the invertebrates’ ability to adapt to environmental pressures. Through gene expansion, domain recombination, and functional diversification, invertebrates can effectively enhance their immune defense capabilities. Research has long shown that after B. glabrata is infected with the trematode Echinostoma paraensei, its hemolymph contains a lectin composed of a 65 kDa subunit (10). In uninfected snails, three different cDNAs were obtained that were similar to the peptide sequences of the 65 kDa lectin. It was unexpectedly discovered that these encode fibrinogen-related proteins (BgFREP), which also include regions similar to those found in Ig superfamily members (10). This indicates the role of BgFREP in recognizing parasite-derived molecules and provides a paradigm for studying the diversity of non-self-recognition functions in invertebrates (10).
2.3 Functional diversity of BgFREPs and their role in immune responses
2.3.1 Variability and polymorphism of BgFREPs
BgFREP plays a vital role in immune recognition and defense during the invasion of trematodes in flatworms, exhibiting high levels of diversity and polymorphism. The diversity and polymorphism of BgFREP are primarily achieved through gene conversion and point mutations, which are essential for covering a wide range of parasite diversity and providing varying levels of infection resistance to snails. This diversity and polymorphism help to understand the evolutionary and adaptive mechanisms between snails and parasites. Recent studies have shown that there are currently 40 known types of the BgFREP family (39), and the mechanisms responsible for generating the diversity of BgFREP at the genomic DNA and mRNA levels have been a focal point of research (3, 11). The BgFREP gene family can serve as a model to study invertebrate immune responses, mechanisms of immune-related gene diversification, and host-parasite interaction molecules (3). Different gene families also exhibit selective immune responses to various pathogens (40). These gene families diversify through alternative splicing, exon loss, and random somatic mutations (41).
Many members of B. glabrata, such as BgFREP2, BgFREP3, BgFREP4, and BgFREP12, exhibit somatic diversity (40). Selective splicing is recognized as the most likely mechanism for generating FREP diversity. A large body of transcriptomic research supports this view, revealing different transcript variants of the same BgFREP in mollusks (42). Experiments have shown that transcripts from 20–40 hemocyte pools of B. glabrata include different BgFREP sequence modifications (43), confirming that the hemocyte pool is updated during mollusk development and the lineages of BgFREP expressed in these cells also change (40). Pathogen stimulation may enhance the hematopoietic function of B. glabrata, thereby promoting the mutation process and increasing BgFREP diversity (44).
Future studies could explore how specific BgFREP variants influence immune response and infection resistance. Investigating how pathogen exposure and environmental factors regulate BgFREP diversity may further illuminate the adaptive mechanisms involved in host-parasite interactions.
2.3.2 Pathogen recognition role
The IgSF domain at the N-terminus and the FreD at the C-terminus of BgFREPs are both considered to be involved in immune recognition (14). Some BgFREPs can form multimers (Figure 1B) (14, 45), which affects their biological functions during immune challenges. For example, non-denaturing PAGE analysis shows that BgFREPs in the 65–70 kDa range can form covalent hexamers, with these hexamers observed as complexes of approximately 400 kDa on SDS-PAGE gels (46). These 400 kDa complexes can further associate non-covalently to form complexes observed at around 1600 kDa (46). However, the specific BgFREPs that compose each complex are largely unclear, and research into this facet of BgFREP biology has shown that not every BgFREP can polymerize (16). This capacity for polymerization may depend on their domain architecture, disulfide bond-forming regions, or external stimuli such as pathogen exposure, although the underlying molecular mechanisms remain to be clarified.
The S. mansoni polymorphic mucins (SmPoMucs) act as pathogen-associated molecular patterns (PAMPs) to initiate immune recognition and are targets for certain BgFREPs (13). After infection with S. mansoni, snails produce large amounts of BgFREPs with lectin-like characteristics. These BgFREPs use their upstream IgSF domains and C-terminal FreD to participate in the response against Schistosoma parasite parasites (47). A study showed that BgFREP2 expression significantly increases in resistant snails, while no such increase was observed in susceptible M-line snails (48). This indicates that two different BgFREP defensive responses may occur in resistant and susceptible snails, leading to different outcomes for the parasites (11).
Despite the immune recognition functions of BgFREPs, certain pathogens have evolve mechanisms to evade these responses. Genomic analyses suggest that parasites may avoid recognition by rapidly evolving their surface proteins (49). Moreover, pathogens can secrete substantial quantities of excretory-secretory products (ESPs), including enzymes and polymorphic mucins, which may further aid in escaping host immune detection (50). Elucidating these evasion strategies will not only deepen our understanding of immune evolution in invertebrates but also inform the development of novel approaches to control infections in susceptible host species.
2.3.3 Synergy
The plasma of B. glabrata contains a variety of soluble immune recognition proteins, including numerous PRRs with high polymorphism and diversity. Prominent among these are BgFREPs (49), galectin-related proteins (BgGREPs), C-type lectin-related proteins (BgCREPs) (47), and thioester-containing proteins (BgTEPs) (13, 51, 52). The plasma also contains other types of lectins, endotoxin-binding proteins, and antimicrobial/permeability-increasing proteins, which are capable of recognizing and binding to PAMPs (53) (Figure 2). BgFREP, BgGREP, and BgCREPs belong to the variable immunoglobulin and lectin domain (VIgL) family. This family consists of glycoproteins containing IgSF and lectin domains (2, 47). FREPs, CREPs, and GREPs share strong similarity at the amino acid levels, suggesting that these sequences may derive from a shared ancestral gene and/or that these molecules may engage in similar or related biological pathways. The VIgL family is the most significant group of immune lectins targeting schistosome larvae in the plasma (53).
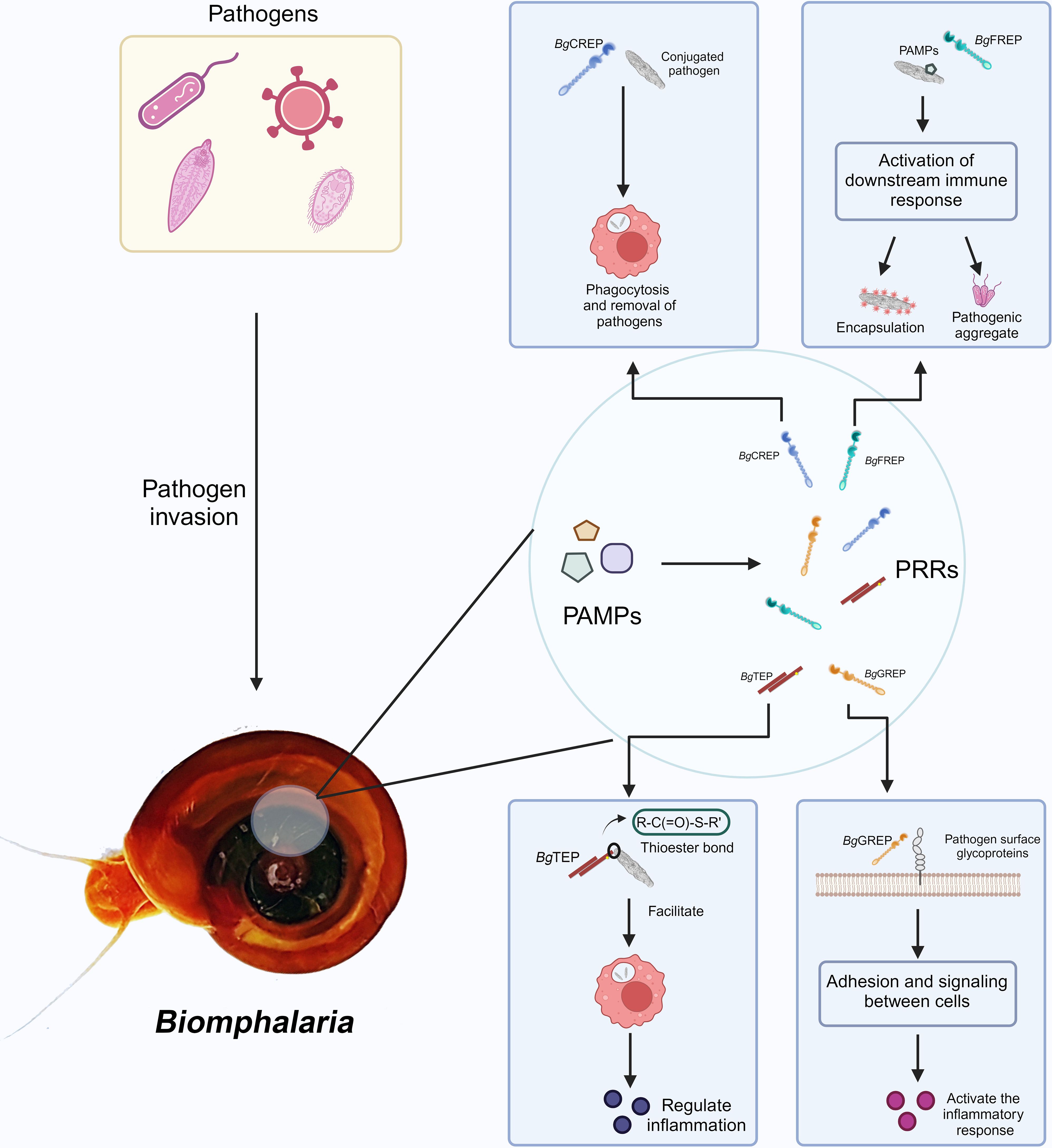
Figure 2. VIgL Family and TEP Family involved in immune recognition and response mechanisms in B. glabrata. After pathogen invasion into the B. glabrata, PAMPs are recognized by the snail’s PRRs. The figure shows several key PRRs and their functions: BgCREP binds to pathogens, promoting phagocytosis and aiding in pathogen clearance (54); BgFREP recognizes PAMPs and activates downstream immune responses, including encapsulation and formation of pathogenic aggregates; BgTEP regulates the inflammatory response by promoting the binding of molecules with thioester bonds to targets; BgGREP is involved in cell adhesion and signal transduction, activating the inflammatory response. Created with BioRender.com.
BgTEPs are a key component of the immune system (55), and are secretory glycoproteins distinguished by the presence of a thioester domain featuring a unique intra-chain β-cysteinyl-γ-glutamyl thioester bond (56). TEPs are divided into three main families: the invertebrate-specific TEP family, the complement C3-related family, and the α-2-macroglobulin (A2M) family (57). In freshwater snails, BgTEP is found within a precisely characterized immune complex that includes FREP and SmPoMuc. BgTEP shares several similarities with other invertebrate TEPs, including a domain specialized for A2M receptor binding and a typical thioester motif. Additionally, these TEPs are processed prior to binding to the mucin of schistosome larvae (51). Studies have shown that BgTEP is specifically exhibited in the immune-specialized hemocytes of invertebrates and is released into the hemolymph. BgTEP can bind to the surfaces of different microbes and parasites in either its full-length or processed form. During schistosome infection, immune localization studies have revealed that BgTEP is expressed by only one hemocyte subtype (embryonic cells) (55). This hemocyte subtype is located in the cell protective capsule enveloping the parasite, indicating that it may play an important role in encapsulating and clearing the parasite (55). Whether the specific expression of BgTEP and the splice diversity of BgFREP are involved in immune memory remains unclear, but these mechanisms may contribute to more efficient responses upon repeated pathogen exposure.
BgFREP collaborates with other immune proteins, such as BgTEPs and Biomphalysin, to enhance the immune response. For example, BgFREP2 and BgTEP1 can bind to S. mansoni sporocysts, forming immune complexes that increase the production of reactive oxygen species (ROS) by hemocytes, thereby effectively combating the infection. In B. glabrata resistant to Schistosoma infection, BgTEP1 interacts with BgFREP2 to mediate the recognition of SmPoMucs and enhance resistance to S. mansoni infection (13, 14, 55). Immune complexes composed of BgFREP2, BgFREP3, and BgTEP1 can bind to SmPoMucs and promote ROS production by B. glabrata hemocytes (53, 58).
2.3.4 BgFREP and immune memory
BgFREP plays a crucial role in the similar immune memory of B. glabrata by recognizing and binding to PAMPs, thus stimulating immune responses. The immune function of BgFREP is demonstrated through its differential expression during infection and specific interactions with SmPoMucs (47). Experiments have shown that after an initial infection, B. glabrata’s immune system shifts from cellular immune responses, such as phagocytosis and encapsulation, to humoral immune responses, including the secretion of antimicrobial peptides and other immune molecules. After initial infection, B. glabrata shows complete protection against subsequent infections with homologous pathogens, which lasts throughout the snail’s entire lifespan (59). Additionally, research has found that this innate immune memory is genotype-dependent, meaning that the protective effect decreases as the neutral genetic distance between the initial and subsequent pathogen infections increases (60). After the first exposure to a pathogen, the polymorphism and expression patterns of BgFREPs can be adjusted and optimized to enhance the response to specific pathogens. Studies have shown that FREPs not only recognize and bind to pathogens during the initial infection but also provide a faster and more effective response during reinfection, including the recognition and binding of pathogens and the activation of immune cells (47).
Future studies could explore whether specific polymorphisms in the BgFREP gene contribute to the differential strength of immune memory responses and whether the expression patterns of BgFREP evolve to optimize pathogen recognition over time. Furthermore, investigating how the immune memory of B. glabrata can be modulated by environmental factors or prior exposure to different pathogen strains may provide deeper insights into the mechanisms that govern immune memory in invertebrates. Research into these areas could also reveal potential targets for enhancing immune responses in B. glabrata to improve disease resistance.
3 Down syndrome cell adhesion molecule in arthropods
3.1 Dscams in species
Dscams in Invertebrates are highly diverse cell adhesion molecules that play crucial roles in neural development and immune function. Dscams have been discovered in arthropods (such as D. melanogaster (61), honey bees (62), A. mosquitoes (63), Marsupenaeus japonicus (64), Scylla paramamosain (65), Penaeus monodon (66)), annelids (Hirudo medicinalis (67)), mollusks (B. glabrata (58), Crassostrea virginica (68)), as well as Caenorhabditis elegans (69), sponges (70). Dscam has also been discovered in vertebrates, including amphibians such as Xenopus laevis (71), birds (72), fishes (Danio rerio (73)), mammals (humans (74), mice (75)) as well as Canis lupus familiaris (69).
3.2 Exploration of Dscam
The precursor of Dscam proteins in invertebrates may have existed before the evolution of Bilateria and diploblastic organisms, as seen in cnidarians like Nemastostella vectensis and sponges like Amphimedon queenslandica (76). Although these organisms lack the typical Dscam structural organization, some of their genes contain highly conserved Ig-like domains with amino acid sequences similar to those of Dscam (76). N. vectensis and human DSCAM share similar signaling pathways (7), suggesting that some features of Dscam in complex groups like vertebrates may have evolved early in metazoan evolution. Phylogenetic reconstruction of Dscam molecules across major animal groups indicates that Dscam was initially identified for its role in the nervous system, with its immune properties evolving later (7, 77).
As the Dscam gene evolved across various animal species, it demonstrated the ability to generate diverse isoforms through gene recombination and alternative splicing mechanisms. This diversity plays a crucial role in neural development, immune function, and environmental adaptability. In arthropods, the Dscam gene can generate a variety of different protein isoforms through alternative splicing (77). These isoforms exhibit specificity for various pathogens, enhancing phagocytosis and operating in a manner akin to antibodies (78). Additionally, the Dscam gene regulates connections and recognition between neurons, ensuring the proper formation of neural networks, a mechanism observed in many arthropods (79). In arthropods, unlike in mammals, the Dscam gene exhibits a high level of complexity, suggesting differences in the mechanisms of Dscam function across organisms. While the diversity of the Dscam gene is a unique feature of arthropods, genetic studies on vertebrate DSCAM genes have revealed that the functions of certain molecules in the neural connectivity process are conserved (18).
3.3 Diversity and functions of AgDscam and DmDscam
3.3.1 The diversity of Dscam
DmDscam has a notable feature: it generates highly diverse protein isoforms through alternative splicing (Figure 3). D. melanogaster possesses four genes encoding DmDscam: DmDscam1, DmDscam2, DmDscam3, and DmDscam4. Among these, DmDscam2 and DmDscam3 undergo a relatively modest scale of alternative splicing, while DmDscam1 can produce over 38,000 different isoforms through extensive alternative splicing (Figure 3A); When considering the independent alternative splicing of internal domain exons, this number increases to 152,064 (Figure 3C). Each of these isoforms has a unique extracellular domain (19), formed by distinct combinations of three variable immunoglobulin-like domains, which are linked to one of two alternative transmembrane domains. The variable exon segments are encoded by 12, 48, and 33 variants of exons 4, 6, and 9, respectively, while the transmembrane domain is encoded by two variant exons 17 (81) (Figures 3A, B). DmDscam gene endows each neuron with a unique identity, allowing it to distinguish its own projections from those of other neurons. This self-recognition mechanism is crucial for the proper formation of neural connections in the D. melanogaster brain (81).

Figure 3. Variable splicing and protein structural diversity of the DmDscam1 gene. (A) DmDscam1 Gene intro/exon Structure: The DmDscam1 gene comprises multiple exons, with 20 fixed exons indicated by black vertical lines. Specific exons 4, 6, 9, and 17 (marked with orange, light purple, light green, and pink vertical lines, respectively) are involved in encoding through mutually exclusive splicing. Each of these exons can produce multiple isoforms: exon 4 has 12 variants, exon 6 has 48 variants, exon 9 has 33 variants, and exon 17 has 2 options, resulting in a total of 38,016 (12×48×33×2) potential protein isoforms. Red stars in the diagram highlight the regions of these diverse splicing exons. The pink vertical line represents the transmembrane region. (B) DmDscam1 mRNA organization: All fixed exons are represented by white rectangles. Exons with mutually exclusive splicing capabilities are color-coded to match their DNA counterparts, illustrating their diverse selection in the mRNA. Additionally, internal domain exons 19 and 23 (highlighted by orange triangles in the diagram) can be selectively included or excluded, further increasing the diversity of mRNA isoforms. Considering the four possible combinations of internal domain exons (including exon 19, including exon 23, including both, or including neither) along with the 38,016 mutually exclusive splicing combinations, a total of 152,064 (38,016×4) distinct mRNA isoforms can be generated. Red stars in the diagram indicate the regions of exons with selective diversity. The pink area also represents the transmembrane region. (C) DmDscam1 Protein Structure: The described diversity is reflected in the protein structure, particularly evident in the second (orange), third (light purple), and seventh (light green) immunoglobulin-like domains (depicted as small circles). Detailed Ig protein information can be found on the InterPro website, with domains labeled as Ig1, Ig2, Ig3, Ig4, Ig5, Ig6, and Ig7 (with corresponding numbers: cd20955, cd20953, cd20957, cd20956, cd20958, cd20959, cd20954). Six longer ovals represent the relatively conserved fibronectin type III domains, with detailed FNIII protein information available on the InterPro website (ID: PF18447). The pink vertical bar indicates the transmembrane domain and the black vertical line to its right points to the intracellular region. Red stars in the diagram mark the regions of the amino acid sequences translated from selectively diversified exons. (D) DmDscam1 Protein structure: The figure shows the crystal structure of DmDscam1 (80). (RCSB PDB ID: 2V5M), more detailed information can be found on the RCSB PDB website (https://www.rcsb.org/structure/2v5m). Created with BioRender.com.
Similar to DmDscam, the genomic organization of AgDscam is also highly complex. It contains 101 exons, including 16 constant exons present in all splice forms and three exon cassettes encoding Ig domains (exon cassettes 4, 6, and 10), which are composed of 14, 30, and 38 alternatively spliced Ig exons, respectively (21). Through alternative splicing, each exon cassette can contribute one Ig exon to a single mature messenger RNA (mRNA), thereby generating potential splice forms with distinct adhesive domains and interaction specificities (21).
Although the structural diversity of Dscam isoforms is well characterized, their role in immune specificity remains unclear. Future research could explore whether specific splice variants are activated by pathogen-specific signals and contribute to immune recognition. This line of investigation could further enhance our understanding of the broader biological roles of Dscam, especially in neurodevelopment and immune function.
3.3.2 Neurodevelopment
Dscam plays a crucial role in the development of the nervous systems of both D. melanogaster and vertebrates, particularly through various signaling pathways (Figure 4). The alternative splicing of Dscam not only generates diverse protein isoforms but also stands out due to the complexity of its splicing mechanisms (18). The molecular diversity resulting from this splicing may contribute to the specificity of neuronal connections (20). Additionally, Dock is an adaptor protein containing three Src homology 3 (SH3) domains and one Src homology 2 (SH2) domain and is closely related to the non-catalytic tyrosine-phosphorylated protein (Nck) in mammals (82–84). It plays a crucial role in the guidance and targeting of R-cell axons in D. melanogaster (19). DmDscam interacts with Dock by directly binding to its SH2 and SH3 domains, exhibiting an affinity similar to that of human DSCAM with Dock (20). Studies have also shown that P21-activated kinase (Pak), a p21-activated serine/threonine kinase, acts on Dock’s downstream signaling pathways in adult photoreceptor neurons (85). Genetic research further indicates that DmDscam, Dock, and Pak work together in the guidance of Bolwig’s nerve, with DmDscam playing a crucial role in the formation of axonal pathways in the embryonic central nervous system (20).
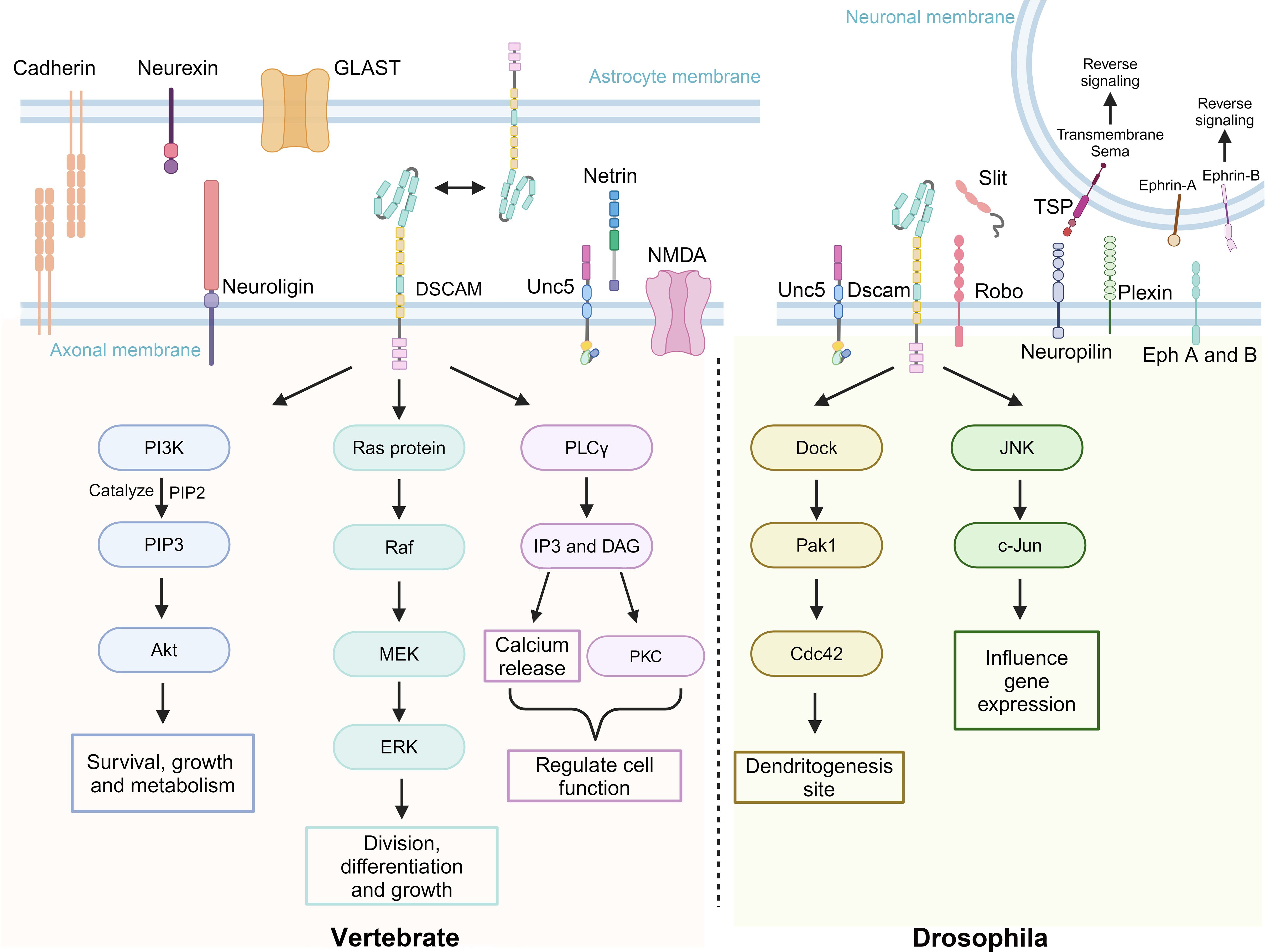
Figure 4. Dscam and its associated transmembrane molecules in signaling pathways of vertebrates and D. melanogaster. Dscam exhibits functional conservation and biological diversity across different species, playing a key role, particularly in neural development. The upper part of the image depicts a group of transmembrane molecules associated with Dscam function, whether they directly interact or not. The left side of the lower part illustrates how DSCAM and its related molecules in vertebrates regulate cell growth, differentiation, survival, and function through signaling pathways such as Phosphoinositide 3-kinase - Protein kinase B (PI3K-Akt), Ras - Rapidly accelerated fibrosarcoma - Mitogen-activated protein kinase - Extracellular signal-regulated kinase (Ras-Raf-MEK-ERK), Phospholipase C gamma (PLCγ), and Protein kinase C (PKC). The right side of the lower part shows how DmDscam in D. melanogaster primarily affects gene expression and dendritic development through the Dedicator of cytokinesis - P21-activated kinase 1 - Cell division control protein 42 (Dock-Pak1-Cdc42) and c-Jun N-terminal kinase (JNK) signaling pathways. Created with BioRender.com.
In D. melanogaster, the homologous interactions of the DmDscam gene require that the interacting isoforms be identical to each other. As previously mentioned, the DmDscam gene is subjected to extensive alternative splicing, showcasing its diversity (20, 86, 87). This diversity is primarily reflected in the variable regions of the Ig2, Ig3, Ig7 domains and transmembrane domains. Within these regions, homologous interactions are formed through the matching of Ig2, Ig3, and Ig7 domains of identical isoforms (23, 87). Studies have shown that DmDscam interactions between identical isoforms largely depend on the consistency of the Ig2, Ig3, and Ig7 domains. If there is any variation in these three domains, the interaction cannot occur (88).
DmDscam plays a unique role in D. melanogaster by participating in the mechanisms of “tiling” and “self-avoidance.” The self-avoidance mechanism ensures that the dendrites of a neuron do not intersect with those of other neurons, allowing for precise processing of sensory or synaptic inputs. On the other hand, tiling refers to the phenomenon where certain neurons avoid overlapping with other similar neurons, thereby respecting each other’s territory (89). DmDscam proteins mediate this tiling process through homologous interactions, allowing these proteins to recognize and prevent adhesion between neurons of the same type or function. This mechanism ensures that dendrites from neurons of the same type can cover sensory and postsynaptic regions with minimal overlap. Additionally, homologous interactions also mediate self-avoidance, where sister neurites from the same neuron repel each other, ensuring uniform coverage of the receptive field and effective processing of sensory information (19). DmDscam 1 plays a crucial role in the self-avoidance mechanism (90). During development, the axons of mushroom body (MB) neurons bifurcate to form two sister branches, known as the medial and dorsal lobes. Although MB neurons lacking DmDscam 1 can still bifurcate normally, the process of separating the sister branches is disrupted (19). DmDscam 2, on the other hand, is involved in the visual system of D. melanogaster (91). In this system, neurons include a pair of invariant intermediate neurons, L1/L2, and two pairs of selective neurons, L3. L1 and L2, along with the terminal ends of R cells (R1-R6), form postsynaptic tetrad synapses surrounded by glial cells. This system ensures that axon terminals of the same type of photoreceptor cells remain separated across different ommatidia and do not overlap or make excessive contact within the target area (19).
3.3.3 Immune function
The A. gambiae has a complex life cycle, diverse breeding environments, and blood-feeding habits, which expose it to a variety of pathogens, including bacteria, fungi, and viruses (92). AgDscam can respond to infections by generating splice variant combinations specific to the pathogen challenge (21). Evidence suggests that the specificity can occur at the level of species or even broader pathogen classes, such as bacteria, fungi, and parasites, though strain-level discrimination has not yet been clearly demonstrated (21). The expression levels of these splice variants containing Ig domains (either overexpressed or underexpressed) depend on the type of pathogen encountered by the host cells. This specificity is demonstrated by the observation that two species of Plasmodium, which cause malaria, as well as different bacterial pathogens such as Escherichia coli and Staphylococcus aureus (21), induce significantly different splice variant combinations in adult A. gambiae (93). When AgDscam is transiently silenced, the A. gambiae’s ability to resist bacterial and malaria parasite infections is significantly impaired. AgDscam can bind to bacteria through specific splice variants and mediate phagocytosis, thereby contributing to pathogen defense (21). Previous studies have shown that the immune response in mosquitoes is significantly regulated by the Immune deficiency (IMD)and Toll-like receptor (Toll) pathways. Activation of the immune response mediated by the IMD pathway is more effective in inhibiting Plasmodium falciparum infection, while activation of the Toll pathway exhibits a more specific defense against Plasmodium berghei (92–94). Additionally, the IMD and Toll pathways also regulate the selective splicing of AgDscam. To confirm this, a study analyzed the transcriptional changes of AgDscam under basal conditions or upon immune activation by lipopolysaccharide (LPS) and peptidoglycan (PGN). The results showed that activation of either the IMD or Toll pathway significantly altered the mRNA levels of AgDscam’s variable Ig domains, with significant differences compared to the control group (22). AgDscam can generate different splice variant pools in response to at least eight different immune elicitors, thereby enabling pathogen-specific defense (21, 22).
DmDscam is involved in pathogen recognition and is associated with the phagocytosis and clearance functions of immune cells in D. melanogaster. A potential model for pathogen immune responses is illustrated in Figure 5. DmDscam is a single-pass transmembrane receptor that not only functions in axonal pathways but is also expressed in phagocytic hemocytes (95). The DmDscam gene encodes proteins containing immunoglobulin (Ig) domains through alternative splicing, potentially generating thousands of protein isoforms (Dscam Hypervariable, Dscam-hv). This splicing process involves mutually exclusive selection of exons encoding half of the Ig2 domain, half of the Ig3 domain, and the entire Ig7 domain (96).
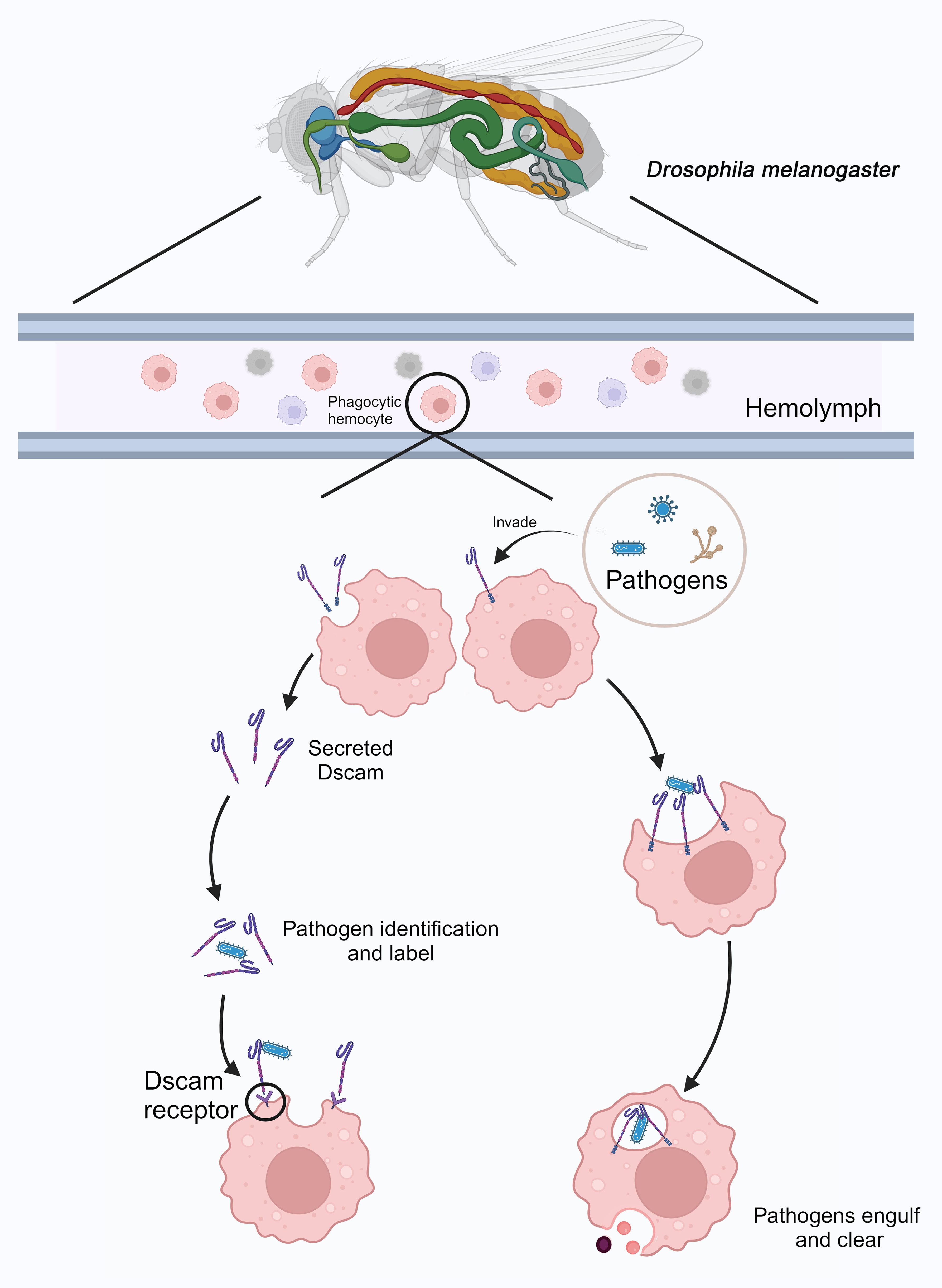
Figure 5. Immune response mediated by DmDscam in D. melanogaster. In D. melanogaster, phagocytic hemocytes patrol the hemolymph. When pathogens invade, some of these phagocytic cells secrete DmDscam molecules to recognize and tag the pathogens. Pathogens labeled with DmDscam are then identified, bound, and cleared by other phagocytic cells through the DmDscam receptors. Additionally, DmDscam on the surface of certain phagocytic hemocytes can directly recognize and bind to pathogens, leading to their phagocytosis and elimination. Created with BioRender.com.
Recombinant DmDscam proteins can bind to E. coli, and hemocytes extracted from DmDscam gene-knockdown D. melanogaster larvae or S2 cells (a D. melanogaster cell line) pre-incubated with DmDscam antibodies show reduced bacterial phagocytic activity, indicating that DmDscam may function as a receptor or regulatory factor in phagocytes (96). Although increasing evidence suggests that DmDscam-hv might act as a pathogen-specific recognition molecule, with different DmDscam-hv variants recognizing different pathogens and participating in immune memory similar to adaptive immunity in vertebrates, its role in invertebrate immunity remains controversial (96). For example, some studies have shown that short-term expression of DmDscam genes in D. melanogaster larvae hemocytes and fat bodies does not significantly change after exposure to E. coli, Pseudomonas fluorescens, and Bacillus thuringiensis (97). Similarly, other research has found that the overall splice variant diversity of Dscam1 in D. melanogaster does not significantly change after early exposure to bacteria such as E. coli (98), while the effects of viral or fungal infections on this diversity remain to be fully elucidated. However, in A. gambiae, the diversity of Ig2 and Ig3 variants of AgDscam increases after pathogen infection (98, 99). This dynamic shift in splice variant usage may also reflect an aspect of immune response plasticity (100). Therefore, further research is needed to validate the immune functions of Dscam. Future studies could test the hypothesis that specific Dscam isoforms are selectively induced by distinct pathogens and mediate differential immune responses through regulated alternative splicing.
Despite its role in pathogen recognition and immune defense, some pathogens have evolved strategies to evade Dscam-mediated immune responses. These strategies may include antigenic variation, whereby pathogens alter their surface proteins to avoid detection (21); or suppression of the host’s complement-like immune system through their own surface molecules (101). A deeper understanding of these evasion mechanisms is crucial for elucidating the complexities of host-pathogen interactions and enhancing immune defense strategies in invertebrates.
3.4 Dscam and disease
In D. melanogaster, the absence of DmDscam leads to abnormal neural development (20), increased susceptibility to infections, and reduced phagocytic activity (9). Conversely, overexpression of DmDscam can affect the intestinal nervous system of D. melanogaster. Studies have found that overexpression of DmDscam1 in the intestinal nervous system of D. melanogaster leads to excessive growth of the foregut and hindgut neurons, reducing the efficiency with which the larvae clear food from the gut (102). Additionally, a systematic study has tested the effects of DmDscam1 on dendrite growth and spacing in eight different types of central nervous system neurons in D. melanogaster. It was observed that knockdown of DmDscam1 resulted in severe dendritic clumping and reduced length in output glutamatergic and aminergic neurons (103).
Abnormal expression of DSCAM is associated with human diseases such as Down syndrome (DS), autism, and certain immune disorders. Research based on the location of DSCAM on chromosome 21, its specific expression in the central nervous system and neural crest, and its involvement in neural migration, differentiation, and synaptic function suggests that DSCAM is involved in neural differentiation and is related to defects in the central and peripheral nervous systems observed in DS (104). Additionally, some studies have concluded that DSCAM may act as an inhibitory factor that prevents premature maturation of the spinal cord and excessive glutamatergic transmission, with its deficiency potentially leading to autism-like behaviors (105).
4 Summary and outlook
In invertebrates, FREPs and Dscams are two important immune molecules. Both contain immunoglobulin domains and are closely related to development and the innate immune response. They each exhibit unique immune functions and mechanisms. BgFREP is a soluble pattern recognition receptor in B. glabrata, which enhances the immune response to parasites (such as schistosomes) by binding PAMPs and forming multimers. Its diversity primarily arises from gene conversion and point mutations. In contrast, Dscam has been mainly studied in D. melanogaster and A. gambiae. It is a transmembrane protein containing immunoglobulin-like domains, which generates specific isoforms to respond to pathogen infections through an extremely complex process of selective splicing. In D. melanogaster, DmDscam is more involved in pathogen recognition and phagocytosis regulation, while in A. gambiae, AgDscam’s function is more focused on binding to pathogens and blocking parasite development through the defense mechanisms of midgut epithelial cells (106). This functional difference reflects the adaptive evolutionary strategies of the two model organisms in responding to different immune challenges.
The comparison between BgFREP and Dscam in terms of amplification and selection reveals the diversity and flexibility of the invertebrate immune system. BgFREP generates a broad antigen recognition capacity through gene recombination and somatic diversification, adapting to the complex and dynamic parasitic environment. In contrast, Dscam in D. melanogaster and A. gambiae generates tens of thousands of different isoforms through selective splicing, reflecting the high precision of immune function. This mechanistic difference not only highlights the flexibility of the invertebrate immune system but also suggests that PRRs may evolve highly specialized functions in different species. Furthermore, regarding the specificity of PRRs, studies in some invertebrates indicate that these molecules can generate specific receptor repertoires to respond to different pathogens (107). This phenomenon is particularly evident in the Dscam of D. melanogaster and A. gambiae, further challenging the traditional paradigm that invertebrate immune responses are ‘simple’ and ‘lacking specificity’. These findings suggest that invertebrates may form complex immune memory mechanisms through genetic diversity and selective splicing, offering new directions and insights for research in the field of immunology.
Future research could further explore the regulatory networks and molecular mechanisms of FREP and Dscam in invertebrates. For example, do FREP and Dscam exhibit similar regulatory mechanisms across different species? Do other mollusks also possess polymorphism-generating mechanisms similar to BgFREP? Additionally, through modern molecular biology techniques such as single-cell sequencing and structural biology, it will be possible to gain deeper insights into the distribution and dynamic functions of these molecules at the cellular and tissue levels. At the same time, expanding the study of BgFREP and Dscam to other invertebrates may reveal new types of PRRs or expand existing functional patterns. These studies will lead to new breakthroughs in the research of invertebrate immune mechanisms and contribute to innovations in biomedical and biotechnological fields.
Author contributions
HL: Conceptualization, Writing – original draft, Writing – review & editing. QZ: Conceptualization, Writing – original draft, Writing – review & editing. JX: Methodology, Writing – review & editing. XL: Methodology, Writing – review & editing. XC: Supervision, Writing – review & editing. YiZ: Supervision, Writing – review & editing. HL: Formal Analysis, Writing – review & editing. YuZ: Supervision, Writing – review & editing. ML: Supervision, Writing – review & editing. LZ: Methodology, Writing – review & editing. DH: Supervision, Writing – review & editing. XZ: Validation, Writing – review & editing. KC: Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the National Natural Science Foundation of China (Grant No: 32000293); Guangxi Natural Science Foundation (Grant Nos: 2020JJA130077 and 2018JJB140423); the University Level Scientific Research Project of Zhejiang Shuren University (Grant No: 2022R064); and Zhejiang Shuren University Basic Scientific Research Special Funds (Grant No: 2024XZ014); the National Innovation and Entrepreneurship Training Program for College Students in 2023 (Grant No: 202311842052X).
Acknowledgments
We thank all the participants in this study. We sincerely thank BioRender for providing the tools and support that helped us create the figures in this review. Their platform enabled us to effectively visualize complex biological concepts.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Lanz-Mendoza H, Contreras-Garduño J. Innate immune memory in invertebrates: Concept and potential mechanisms. Dev Comp Immunol. (2022) 127:104285. doi: 10.1016/j.dci.2021.104285
2. Zhang S-M, Loker ES. The FREP gene family in the snail Biomphalaria glabrata: additional members, and evidence consistent with alternative splicing and FREP retrosequences. Dev Comp Immunol. (2003) 27:175–87. doi: 10.1016/S0145-305X(02)00091-5
3. Zhang S-M, Loker ES. Representation of an immune responsive gene family encoding fibrinogen-related proteins in the freshwater mollusc Biomphalaria glabrata, an intermediate host for Schistosoma mansoni. Gene. (2004) 341:255–66. doi: 10.1016/j.gene.2004.07.003
4. Cannon JP, Haire RN, Litman GW. Identification of diversified genes that contain immunoglobulin-like variable regions in a protochordate. Nat Immunol. (2002) 3:1200–7. doi: 10.1038/ni849
5. Dishaw LJ, Giacomelli S, Melillo D, Zucchetti I, Haire RN, Natale L, et al. A role for variable region-containing chitin-binding proteins (VCBPs) in host gut–bacteria interactions. Proc Natl Acad Sci. (2011) 108:16747–52. doi: 10.1073/pnas.1109687108
6. Brites D, McTaggart S, Morris K, Anderson J, Thomas K, Colson I, et al. The Dscam homologue of the crustacean Daphnia is diversified by alternative splicing like in insects. Mol Biol evolution. (2008) 25:1429–39. doi: 10.1093/molbev/msn087
7. Brites D, Brena C, Ebert D, Du Pasquier L. More than one way to produce protein diversity: duplication and limited alternative splicing of an adhesion molecule gene in basal arthropods. Evolution. (2013) 67:2999–3011. doi: 10.1111/evo.12179
8. Neves G, Chess A eds. Dscam-mediated self-versus non-self-recognition by individual neurons. In: Cold Spring Harbor symposia on quantitative biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
9. Watson FL, Püttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, et al. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. (2005) 309:1874–8. doi: 10.1126/science.1116887
10. Adema CM, Hertel LA, Miller RD, Loker ES. A family of fibrinogen-related proteins that precipitates parasite-derived molecules is produced by an invertebrate after infection. Proc Natl Acad Sci. (1997) 94:8691–6. doi: 10.1073/pnas.94.16.8691
11. Hanington PC, Zhang SM. The primary role of fibrinogen-related proteins in invertebrates is defense, not coagulation. J Innate Immun. (2011) 3:17–27. doi: 10.1159/000321882
12. Abou-El-Naga IF, Mogahed NMFH. Immuno-molecular profile for Biomphalaria glabrata/Schistosoma mansoni interaction. Dev Comp Immunol. (2023) 105083. doi: 10.1016/j.dci.2023.105083
13. Moné Y, Gourbal B, Duval D, Du Pasquier L, Kieffer-Jaquinod S, Mitta G. A large repertoire of parasite epitopes matched by a large repertoire of host immune receptors in an invertebrate host/parasite model. PloS neglected Trop diseases. (2010) 4:e813. doi: 10.1371/journal.pntd.0000813
14. Li H, Hambrook JR, Pila EA, Gharamah AA, Fang J, Wu X, et al. Coordination of humoral immune factors dictates compatibility between Schistosoma mansoni and Biomphalaria glabrata. Elife. (2020) 9:e51708. doi: 10.7554/eLife.51708
15. Pila EA, Li H, Hambrook JR, Wu X, Hanington PC. Schistosomiasis from a snail’s perspective: advances in snail immunity. Trends parasitology. (2017) 33:845–57. doi: 10.1016/j.pt.2017.07.006
16. Gordy MA, Pila EA, Hanington PC. The role of fibrinogen-related proteins in the gastropod immune response. Fish shellfish Immunol. (2015) 46:39–49. doi: 10.1016/j.fsi.2015.03.005
17. Gorbushin A, Panchin Y, Iakovleva N. In search of the origin of FREPs: characterization of Aplysia californica fibrinogen-related proteins. Dev Comp Immunol. (2010) 34:465–73. doi: 10.1016/j.dci.2009.12.007
18. Schmucker D, Chen B. Dscam and DSCAM: complex genes in simple animals, complex animals yet simple genes. Genes Dev. (2009) 23:147–56. doi: 10.1101/gad.1752909
19. Hizawa K, Sasaki T, Arimura N. A comparative overview of DSCAM and its multifunctional roles in Drosophila and vertebrates. Neurosci Res. (2023) 202:1–7. doi: 10.1016/j.neures.2023.12.005
20. Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. (2000) 101:671–84. doi: 10.1016/S0092-8674(00)80878-8
21. Dong Y, Taylor HE, Dimopoulos G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles Gambiae innate immune system. PloS Biol. (2006) 4:e229. doi: 10.1371/journal.pbio.0040229
22. Dong Y, Cirimotich CM, Pike A, Chandra R, Dimopoulos G. Anopheles NF-κB-regulated splicing factors direct pathogen-specific repertoires of the hypervariable pattern recognition receptor AgDscam. Cell host Microbe. (2012) 12:521–30. doi: 10.1016/j.chom.2012.09.004
23. Wojtowicz WM, Wu W, Andre I, Qian B, Baker D, Zipursky SL. A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell. (2007) 130:1134–45. doi: 10.1016/j.cell.2007.08.026
24. Gorbushin AM. Derivatives of the lectin complement pathway in Lophotrochozoa. Dev Comp Immunol. (2019) 94:35–58. doi: 10.1016/j.dci.2019.01.010
25. Zhou Y, Chen L, Hao S, Cao X, Ni S. Zebrafish ANGPT4, member of fibrinogen-related proteins, is an LTA-, LPS-and PGN-binding protein with a bacteriolytic activity. Fish Shellfish Immunol. (2024) 147:109451. doi: 10.1016/j.fsi.2024.109451
26. Russell S, Young K, Smith M, Hayes M, Lumsden J. Identification, cloning and tissue localization of a rainbow trout (Oncorhynchus mykiss) intelectin-like protein that binds bacteria and chitin. Fish shellfish Immunol. (2008) 25:91–105. doi: 10.1016/j.fsi.2008.02.018
27. Sun J-J, Lan J-F, Shi X-Z, Yang M-C, Yang H-T, Zhao X-F, et al. A fibrinogen-related protein (FREP) is involved in the antibacterial immunity of Marsupenaeus japonicus. Fish shellfish Immunol. (2014) 39:296–304. doi: 10.1016/j.fsi.2014.05.005
28. Zhang H, Wang L, Song L, Song X, Wang B, Mu C, et al. A fibrinogen-related protein from bay scallop Argopecten irradians involved in innate immunity as pattern recognition receptor. Fish shellfish Immunol. (2009) 26:56–64. doi: 10.1016/j.fsi.2008.07.019
29. Pindyck J, Mosesson MW, Bannerjee D, Galanakis D. The structural characteristics of chicken fibrinogen. Biochim Biophys Acta (BBA)-Protein Struct. (1977) 492:377–86. doi: 10.1016/0005-2795(77)90089-7
30. Krakow W, Endres G, Siegbl B, Schekaga H. An electron microscopic investigation of the polymerization of bovine fibrin monomer. J Mol Biol. (1972) 71:95–103. doi: 10.1016/0022-2836(72)90403-2
31. Tripp M. Hemagglutinin in the blood of the oyster Crassostrea virginica. J Invertebrate Pathology. (1966) 8:478–84. doi: 10.1016/0022-2011(66)90074-7
32. Moné Y, Gourbal B, Duval D, Du Pasquier L, Kieffer-Jaquinod S. A large repertoire of parasite epitopes matched by a large repertoire of. PLOS Negl Trop Dis. (2010) 4(9):e813. doi: 10.1371/journal.pntd.0000813
33. Renwrantz L. Lectins in molluscs and arthropods: Their occurrence, origin and roles in immunity. Symp Zool Soc Lond. (1986) 56:81–93.
34. Stein P, Basch P. Purification and binding properties of hemagglutinin from Biomphalaria glabrata. J Invertebrate Pathology. (1979) 33:10–8. doi: 10.1016/0022-2011(79)90125-3
35. Loker ES, Yui MA, Bayne CJ. Schistosoma mansoni: agglutination of sporocysts, and formation of gels on miracidia transforming in plasma of Biomphalaria glabrata. Exp parasitology. (1984) 58:56–62. doi: 10.1016/0014-4894(84)90020-1
36. Bayne C, Loker E, Yui MA. Interactions between the plasma proteins of Biomphalaria glabrata (Gastropoda) and the sporocyst tegument of Schistosoma mansoni (Trematoda). Parasitology. (1986) 92:653–64. doi: 10.1017/S0031182000065513
37. Couch L, Hertel LA, Loker ES. Humoral response of the snail Biomphalaria glabrata to trematode infection: observations on a circulating hemagglutinin. J Exp Zoology. (1990) 255:340–9. doi: 10.1002/jez.1402550310
38. Loker ES, Hertel LA. Alterations in Biomphalaria glabrata plasma induced by infection with the digenetic trematode Echinostoma paraensei. J parasitology. (1987) 73(3):503–13. doi: 10.2307/3282128
39. Zhong D, Bu L, Habib MR, Lu L, Yan G, Zhang S-M. A haplotype-like, chromosome-level assembled and annotated genome of Biomphalaria glabrata, an important intermediate host of schistosomiasis and the best studied model of schistosomiasis vector snails. PloS Neglected Trop Diseases. (2024) 18:e0011983. doi: 10.1371/journal.pntd.0011983
40. Prokhorova E, Ataev G. Fibrinogen-related proteins of gastropoda molluscs. Biol Bull Rev. (2023) 13:S184–S98. doi: 10.1134/S2079086423080091
41. Adema CM. Fibrinogen-related proteins (FREPs) in mollusks. Pathogen-Host Interactions: Antigenic Variation v Somatic Adaptations. (2015), 111–29.
42. Galinier R, Roger E, Moné Y, Duval D, Portet A, Pinaud S, et al. A multistrain approach to studying the mechanisms underlying compatibility in the interaction between Biomphalaria glabrata and Schistosoma mansoni. PloS neglected Trop diseases. (2017) 11:e0005398. doi: 10.1371/journal.pntd.0005398
43. Hanington PC, Lun C-M, Adema CM, Loker ES. Time series analysis of the transcriptional responses of Biomphalaria glabrata throughout the course of intramolluscan development of Schistosoma mansoni and Echinostoma paraensei. Int J parasitology. (2010) 40:819–31. doi: 10.1016/j.ijpara.2009.12.005
44. Hanington PC, Forys MA, Loker ES. A somatically diversified defense factor, FREP3, is a determinant of snail resistance to schistosome infection. PloS neglected Trop diseases. (2012) 6:e1591. doi: 10.1371/journal.pntd.0001591
45. Li H, Chen Y, Zhu Y, Feng Y, Qian Y, Ye X, et al. Exploring the immune interactions between Oncomelania hupensis and Schistosoma japonicum, with a cross-comparison of immunological research progress in other intermediate host snails. Parasites Vectors. (2023) 16:453. doi: 10.1186/s13071-023-06011-9
46. Adema CM, Hertel LA, Loker ES. Infection with Echinostoma paraensei (Digenea) induces parasite-reactive polypeptides in the hemolymph of the gastropod host Biomphalaria glabrata. In: Parasites and Pathogens: Effects on Host Hormones and Behavior. New York, NY: Springer (1997). p. 76–98.
47. Dheilly NM, Duval D, Mouahid G, Emans R, Allienne J-F, Galinier R, et al. A family of variable immunoglobulin and lectin domain containing molecules in the snail Biomphalaria glabrata. Dev Comp Immunol. (2015) 48:234–43. doi: 10.1016/j.dci.2014.10.009
48. Hertel LA, Adema CM, Loker ES. Differential expression of FREP genes in two strains of Biomphalaria glabrata following exposure to the digenetic trematodes Schistosoma mansoni and Echinostoma paraensei. Dev Comp Immunol. (2005) 29:295–303. doi: 10.1016/j.dci.2004.08.003
49. Adema CM, Hillier LW, Jones CS, Loker ES, Knight M, Minx P, et al. Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat communications. (2017) 8:15451. doi: 10.1038/ncomms15451
50. Bayne CJ. Successful parasitism of vector snail Biomphalaria glabrata by the human blood fluke (trematode) Schistosoma mansoni: a 2009 assessment. Mol Biochem parasitology. (2009) 165:8–18. doi: 10.1016/j.molbiopara.2009.01.005
51. Duval D, Pichon R, Lassalle D, Laffitte M, Gourbal B, Galinier R. A new assessment of thioester-containing proteins diversity of the freshwater snail Biomphalaria glabrata. Genes. (2020) 11:69. doi: 10.3390/genes11010069
52. Marquez J, Dinguirard N, Gonzalez A, Kane A, Joffe N, Yoshino T, et al. Molecular characterization of thioester-containing proteins in Biomphalaria glabrata and their differential gene expression upon Schistosoma mansoni exposure. Front Immunol. (2022) 13:903158. doi: 10.3389/fimmu.2022.903158
53. Wu X-J, Dinguirard N, Sabat G, H-d L, Gonzalez L, Gehring M, et al. Proteomic analysis of Biomphalaria glabrata plasma proteins with binding affinity to those expressed by early developing larval Schistosoma mansoni. PloS pathogens. (2017) 13:e1006081. doi: 10.1371/journal.ppat.1006081
54. Tetreau G, Pinaud S, Portet A, Galinier R, Gourbal B, Duval D. Specific pathogen recognition by multiple innate immune sensors in an invertebrate. Front Immunol. (2017) 8:1249. doi: 10.3389/fimmu.2017.01249
55. Portet A, Galinier R, Pinaud S, Portela J, Nowacki F, Gourbal B, et al. BgTEP: an antiprotease involved in innate immune sensing in Biomphalaria glabrata. Front Immunol. (2018) 9:1206. doi: 10.3389/fimmu.2018.01206
56. Williams M, Baxter R. The structure and function of thioester-containing proteins in arthropods. Biophys Rev. (2014) 6:261–72. doi: 10.1007/s12551-014-0142-6
57. Sekiguchi R, Fujito NT, Nonaka M. Evolution of the thioester-containing proteins (TEPs) of the arthropoda, revealed by molecular cloning of TEP genes from a spider, Hasarius adansoni. Dev Comp Immunol. (2012) 36:483–9. doi: 10.1016/j.dci.2011.05.003
58. Li H, Gharamah AA, Hambrook JR, Wu X, Hanington PC. Single-cell RNA-seq profiling of individual Biomphalaria glabrata immune cells with a focus on immunologically relevant transcripts. Immunogenetics. (2022) 74:77–98. doi: 10.1007/s00251-021-01236-3
59. Pinaud S, Portela J, Duval D, Nowacki FC, Olive M-A, Allienne J-F, et al. A shift from cellular to humoral responses contributes to innate immune memory in the vector snail Biomphalaria glabrata. PloS pathogens. (2016) 12:e1005361. doi: 10.1371/journal.ppat.1005361
60. Portela J, Duval D, Rognon A, Galinier R, Boissier J, Coustau C, et al. Evidence for specific genotype-dependent immune priming in the lophotrochozoan Biomphalaria glabrata snail. J innate immunity. (2013) 5:261–76. doi: 10.1159/000345909
61. Celotto AM, Graveley BR. Alternative splicing of the Drosophila Dscam pre-mRNA is both temporally and spatially regulated. Genetics. (2001) 159:599–608. doi: 10.1093/genetics/159.2.599
62. Ustaoglu P, McQuarrie DW, Rochet A, Dix TC, Haussmann IU, Arnold R, et al. Memory consolidation in honey bees is enhanced by down-regulation of Down syndrome cell adhesion molecule and changes its alternative splicing. Front Mol Neurosci. (2024) 16:1322808. doi: 10.3389/fnmol.2023.1322808
63. Smith PH, Mwangi JM, Afrane YA, Yan G, Obbard DJ, Ranford-Cartwright LC, et al. Alternative splicing of the Anopheles Gambiae Dscam gene in diverse Plasmodium falciparum infections. Malaria J. (2011) 10:1–7. doi: 10.1186/1475-2875-10-156
64. Zheng J, Wang P, Mao Y, Su Y, Wang J. Full-length transcriptome analysis provides new insights into the innate immune system of Marsupenaeus japonicus. Fish Shellfish Immunol. (2020) 106:283–95. doi: 10.1016/j.fsi.2020.07.018
65. Li W, Tang X, Chen Y, Sun W, Liu Y, Gong Y, et al. Characterize a typically Dscam with alternative splicing in mud crab Scylla paramamosain. Fish Shellfish Immunol. (2017) 71:305–18. doi: 10.1016/j.fsi.2017.10.023
66. Chou P-H, Chang H-S, Chen I-T, Lee C-W, Hung H-Y, Wang KH-C. Penaeus monodon Dscam (PmDscam) has a highly diverse cytoplasmic tail and is the first membrane-bound shrimp Dscam to be reported. Fish shellfish Immunol. (2011) 30:1109–23. doi: 10.1016/j.fsi.2011.02.009
67. Kym ES. Characterization of two Dscam orthologues in Hirudo medicinalis. La Jolla, CA: UC San Diego (2011).
68. Arzeta-Pino L, Acosta A, Sarmiento ME, Rojas-Contreras M, Rodríguez-Jaramillo C, Vázquez-Juárez R. Herpes virus OsHV-1 and the protist Perkinsus marinus modify the expression of the Down syndrome cell adhesion molecule gene in gill and mantle of Crassostrea spp. Aquaculture Res. (2018) 49:3638–46. doi: 10.1111/are.2018.49.issue-11
69. Armitage SA, Freiburg RY, Kurtz J, Bravo IG. The evolution of Dscam genes across the arthropods. BMC evolutionary Biol. (2012) 12:1–15. doi: 10.1186/1471-2148-12-53
70. Ma Y, Bu D, Long J, Chai W, Dong J. LncRNA DSCAM-AS1 acts as a sponge of miR-137 to enhance Tamoxifen resistance in breast cancer. J Cell Physiol. (2019) 234:2880–94. doi: 10.1002/jcp.v234.3
71. Santos RA, Fuertes AJ, Short G, Donohue KC, Shao H, Quintanilla J, et al. DSCAM differentially modulates pre-and postsynaptic structural and functional central connectivity during visual system wiring. Neural Dev. (2018) 13:1–19. doi: 10.1186/s13064-018-0118-5
72. Friocourt F, Lafont A-G, Kress C, Pain B, Manceau M, Dufour S, et al. Recurrent DCC gene losses during bird evolution. Sci Reports. (2017) 7:37569. doi: 10.1038/srep37569
73. Julien DP, Chan AW, Barrios J, Mathiaparanam J, Douglass A, Wolman MA, et al. Zebrafish expression reporters and mutants reveal that the IgSF cell adhesion molecule Dscamb is required for feeding and survival. J neurogenetics. (2018) 32:336–52. doi: 10.1080/01677063.2018.1493479
74. Huang J, Wang Y, Raghavan S, Feng S, Kiesewetter K, Wang J. Human down syndrome cell adhesion molecules (DSCAMs) are functionally conserved with Drosophila Dscam [TM1] isoforms in controlling neurodevelopment. Insect Biochem Mol Biol. (2011) 41:778–87. doi: 10.1016/j.ibmb.2011.05.008
75. Schramm RD, Li S, Harris BS, Rounds RP, Burgess RW, Ytreberg FM, et al. A novel mouse Dscam mutation inhibits localization and shedding of DSCAM. PloS One. (2012) 7:e52652. doi: 10.1371/journal.pone.0052652
76. Armitage SA, Brites D. The immune-related roles and the evolutionary history of Dscam in arthropods. In: The Evolution of the Immune System. London, UK: Elsevier (2016). p. 241–74.
77. Crayton ME, Powell BC, Vision TJ, Giddings MC. Tracking the evolution of alternatively spliced exons within the Dscam family. BMC evolutionary Biol. (2006) 6:1–15. doi: 10.1186/1471-2148-6-16
78. Li W. Dscam in arthropod immune priming: What is known and what remains unknown. Dev Comp Immunol. (2021) 125:104231. doi: 10.1016/j.dci.2021.104231
79. Jin Y, Li H. Revisiting Dscam diversity: lessons from clustered protocadherins. Cell Mol Life Sci. (2019) 76:667–80. doi: 10.1007/s00018-018-2951-4
80. Meijers R, Puettmann-Holgado R, Skiniotis G, Liu J-H, Walz T, Wang J-H, et al. Structural basis of Dscam isoform specificity. Nature. (2007) 449:487–91. doi: 10.1038/nature06147
81. Hattori D, Demir E, Kim HW, Viragh E, Zipursky SL, Dickson BJ. Dscam diversity is essential for neuronal wiring and self-recognition. Nature. (2007) 449:223–7. doi: 10.1038/nature06099
82. Clemens JC, Ursuliak Z, Clemens KK, Price JV, Dixon JE. A Drosophila protein-tyrosine phosphatase associates with an adapter protein required for axonal guidance. J Biol Chem. (1996) 271:17002–5. doi: 10.1074/jbc.271.29.17002
83. Garrity PA, Rao Y, Salecker I, McGlade J, Pawson T, Zipursky SL. Drosophila photoreceptor axon guidance and targeting requires the dreadlocks SH2/SH3 adapter protein. Cell. (1996) 85:639–50. doi: 10.1016/S0092-8674(00)81231-3
84. Rao Y, Zipursky SL. Domain requirements for the Dock adapter protein in growth-cone signaling. Proc Natl Acad Sci. (1998) 95:2077–82. doi: 10.1073/pnas.95.5.2077
85. Hing H, Xiao J, Harden N, Lim L, Zipursky SL. Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila. Cell. (1999) 97:853–63. doi: 10.1016/S0092-8674(00)80798-9
86. Neves G, Zucker J, Daly M, Chess A. Stochastic yet biased expression of multiple Dscam splice variants by individual cells. Nat Genet. (2004) 36:240–6. doi: 10.1038/ng1299
87. Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. (2004) 118:619–33. doi: 10.1016/j.cell.2004.08.021
88. Sawaya MR, Wojtowicz WM, Andre I, Qian B, Wu W, Baker D, et al. A double S shape provides the structural basis for the extraordinary binding specificity of Dscam isoforms. Cell. (2008) 134:1007–18. doi: 10.1016/j.cell.2008.07.042
89. Soba P, Zhu S, Emoto K, Younger S, Yang S-J, Yu H-H, et al. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron. (2007) 54:403–16. doi: 10.1016/j.neuron.2007.03.029
90. Zipursky SL, Wojtowicz WM, Hattori D. Got diversity? Wiring the fly brain with Dscam. Trends Biochem Sci. (2006) 31:581–8. doi: 10.1016/j.tibs.2006.08.003
91. Millard SS, Flanagan JJ, Pappu KS, Wu W, Zipursky SL. Dscam2 mediates axonal tiling in the Drosophila visual system. Nature. (2007) 447:720–4. doi: 10.1038/nature05855
92. Cirimotich CM, Dong Y, Garver LS, Sim S, Dimopoulos G. Mosquito immune defenses against Plasmodium infection. Dev Comp Immunol. (2010) 34:387–95. doi: 10.1016/j.dci.2009.12.005
93. Kurtz J, Armitage SA. Alternative adaptive immunity in invertebrates. Trends Immunol. (2006) 27:493–6. doi: 10.1016/j.it.2006.09.001
94. Garver LS, Dong Y, Dimopoulos G. Caspar controls resistance to Plasmodium falciparum in diverse anopheline species. PloS pathogens. (2009) 5:e1000335. doi: 10.1371/journal.ppat.1000335
95. Cherry S, Silverman N. Host-pathogen interactions in drosophila: new tricks from an old friend. Nat Immunol. (2006) 7:911–7. doi: 10.1038/ni1388
96. Lu Y, Su F, Li Q, Zhang J, Li Y, Tang T, et al. Pattern recognition receptors in Drosophila immune responses. Dev Comp Immunol. (2020) 102:103468. doi: 10.1016/j.dci.2019.103468
97. Peuß R, Wensing KU, Woestmann L, Eggert H, Milutinović B, Sroka MG, et al. Down syndrome cell adhesion molecule 1: testing for a role in insect immunity, behaviour and reproduction. R Soc Open science. (2016) 3:160138. doi: 10.1098/rsos.160138
98. Armitage SA, Sun W, You X, Kurtz J, Schmucker D, Chen W. Quantitative profiling of Drosophila melanogaster Dscam1 isoforms reveals no changes in splicing after bacterial exposure. PloS One. (2014) 9:e108660. doi: 10.1371/journal.pone.0108660
99. Smith PH. Dscam gene expression in invertebrate immunity: alternative splicing in response to diverse pathogens. Malar J. (2011) 10(1):156.
100. Lanz-Mendoza H, Gálvez D, Contreras-Garduño J. The plasticity of immune memory in invertebrates. J Exp Biol. (2024) 227:jeb246158. doi: 10.1242/jeb.246158
101. Molina-Cruz A, Garver LS, Alabaster A, Bangiolo L, Haile A, Winikor J, et al. The human malaria parasite Pfs47 gene mediates evasion of the mosquito immune system. Science. (2013) 340:984–7. doi: 10.1126/science.1235264
102. Hernández K, Godoy L, Newquist G, Kellermeyer R, Alavi M, Mathew D, et al. Dscam1 overexpression impairs the function of the gut nervous system in Drosophila. Dev Dynamics. (2023) 252:156–71. doi: 10.1002/dvdy.v252.1
103. Wilhelm N, Kumari S, Krick N, Rickert C, Duch C. Dscam1 has diverse neuron type-specific functions in the developing Drosophila CNS. ENeuro. (2022) 9. doi: 10.1523/ENEURO.0255-22.2022
104. Yamakawa K, Huo Y-K, Haendel MA, Hubert R, Chen X-N, Lyons GE, et al. DSCAM: a novel member of the immunoglobulin superfamily maps in a Down syndrome region and is involved in the development of the nervous system. Hum Mol Genet. (1998) 7:227–37. doi: 10.1093/hmg/7.2.227
105. Chen P, Liu Z, Zhang Q, Lin D, Song L, Liu J, et al. DSCAM deficiency leads to premature spine maturation and autism-like behaviors. J Neurosci. (2022) 42:532–51. doi: 10.1523/JNEUROSCI.1003-21.2021
106. Clayton AM, Dong Y, Dimopoulos G. The Anopheles innate immune system in the defense against malaria infection. J innate immunity. (2014) 6:169–81. doi: 10.1159/000353602
Keywords: invertebrates, immune molecules, BgFREP, DmDscam, AgDscam
Citation: Li H, Zhao Q, Xu J, Li X, Chen X, Zhang Y, Li H, Zhu Y, Liu M, Zhao L, Hua D, Zhang X and Chen K (2025) From Biomphalaria glabrata to Drosophila melanogaster and Anopheles gambiae: the diversity and role of FREPs and Dscams in immune response. Front. Immunol. 16:1579905. doi: 10.3389/fimmu.2025.1579905
Received: 19 February 2025; Accepted: 10 April 2025;
Published: 30 April 2025.
Edited by:
Leon Grayfer, George Washington University, United StatesReviewed by:
Ioannis Eleftherianos, George Washington University, United StatesJorge Contreras-Garduño, National Autonomous University of Mexico, Mexico
Copyright © 2025 Li, Zhao, Xu, Li, Chen, Zhang, Li, Zhu, Liu, Zhao, Hua, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyu Li, aG9uZ3l1ODg5MjZAempzcnUuZWR1LmNu; Keda Chen, Y2hlbmtkQHpqc3J1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Hongyu Li
Hongyu Li Qingzhi Zhao1†
Qingzhi Zhao1† Yijie Zhang
Yijie Zhang Keda Chen
Keda Chen