- 1Department of Thyroid Head and Neck Surgery, Affiliated Cancer Hospital of Zhengzhou University, Henan Cancer Hospital, Zhengzhou, Henan, China
- 2Public Laboratory, Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center for Cancer, Key Laboratory of Cancer Prevention and Therapy, Tianjin’s Clinical Research Center for Cancer, Tianjin, China
Metabolic reprogramming is a process by which cells adapt to the nutrient microenvironment by regulating energy metabolism. Compared with normal cells, tumor cells tend to undergo metabolic reprogramming, which is one of the hallmarks of concurrent genomic instability, and immune evasion in tumor cells. The microbial community, known as “second genome” of human beings, can cause systemic disease by predisposing cells to tumors, and modulating immune responses to cancer. Metabolic reprogramming and microorganisms can crosstalk with each other in multiple ways to influence various physiological and pathological responses in cancer progression. The products of increased synthesis by tumor cells can reach the intestinal tract via the circulation and act on the microorganisms, promoting mucosal inflammation, causing systemic disorders, and may also regulate the immune response to cancer. In addition, the metabolites of the microorganisms can in turn be transported to the tumor microenvironment (TME) through the systemic circulation and participate in the process of tumor metabolic reprogramming. Different molecular mechanisms related to metabolic reprogramming and microbiota imbalance control the outcome of tumor or anti-tumor responses, depending on the type of cancer, stage of the disease and the TME. In this review, we focus on the fundamental role of metabolic reprogramming in the interaction between microorganisms and cancers and explore the molecular mechanisms by which metabolic reprogramming modulates this complex biological process. This comment aims to provide valuable resources for clinicians and researchers and promote further research in the field.
Background
Metabolism encompasses complex biochemical networks that convert nutrients into metabolites (1), enabling cells to generate energy, synthesize macromolecules, and maintain cellular functions (2). The most prominent biological characteristic of tumor cells is their uncontrolled proliferation. To meet the biosynthetic demands of rapid growth, cancer cells exhibit distinct metabolic patterns compared with normal cells. Under the influence of factors such as the harsh tumor microenvironment (TME), the metabolic characteristics of cancer cells undergo adaptive changes, which is called metabolic reprogramming (3), and it is one of a hallmark of malignancy (4). This phenomenon was first described by Otto Warburg, who observed that cancer cells preferentially utilize aerobic glycolysis even under oxygen-rich conditions (the Warburg effect) (5). Subsequent research has revealed that metabolic reprogramming extends far beyond the Warburg effect, involving diverse pathways including fatty acid synthesis, glutamate metabolism, and other complex biochemical processes (2, 6). Importantly, these metabolic alterations are dynamic, evolving throughout cancer progression (4). Furthermore, emerging evidence indicates that cancer cells are able to reshape the TME and suppress anti-tumor immune response by depleting essential nutrients (7, 8).
Despite years of research and significant advances in cancer prevention, diagnosis, and treatment, cancer remains a major health burden worldwide (9). As a complex disease, cancer development involves dynamic interactions between tumor cells and the TME (10), with emerging evidence highlighting the critical role of microorganisms (11). It has been found that multiply types of cancers are associated with microbiota, including in breast cancer, lung cancer, gastric cancer, ovarian cancer, and et. al (12–15). The correlation between cancer and microbiota has become a focal point in oncology research, with remarkable progress has been made revealing the functional roles and therapeutic potential of microbiota in cancer progression (16). Clinically, human papillomavirus vaccination has demonstrated remarkable success in reducing gynecological cancers, such as cervical cancer (17). Meanwhile, Helicobacter pylori screening and eradication programs have shown efficacy in gastric cancer (GC) prevention (18). In addition, intratumor microbiota have been implicated in influencing both tumor initiation and metastatic processes (19). These findings have spurred the development of innovative treatment strategies targeting cancer-associated microorganisms, including approaches that modulate microbial communities to enhance therapeutic responses, which is an emerging paradigm with significant clinical potential.
The therapeutic potential of microorganisms against solid tumors was documented firstly over a century ago (20), yet significant progress in this field remained limited until recently. Advances in detection technologies, microbial cultivation methods, and our growing understanding of the TME have now provided compelling evidence of microbial influences on host metabolism and cancer biology, revitalizing research in this area (16). In addition to their individual roles in physiological and pathological processes, the interaction between microbiota and the metabolic reprogramming has emerged as a critical factor in tumorigenesis and therapeutic response (21, 22). For instance, Akkermansia muciniphila promotes lung cancer progression by modulating glycolytic, glutaminolytic, and nucleotide metabolism to shape the TME (23). Conversely, certain commensals like Bifidobacterium pseudolongum, Lactobacillus johnsonii, and Olsenella species enhance anti-tumor immunity through inosine-mediated T cell activation (24). Further studies on the crosstalk between metabolic reprogramming and the microorganisms are needed to better examine the correlations between them, and the potential mechanisms by which they influence cancer progression. Therefore, a systematic treatment strategy is urgently needed to effectively identify the current cancer phase and the crosstalk between metabolic reprogramming and the microorganisms, then provide appropriate and effective interventions to discusses their potential for clinical translation to provide new insights into cancer treatment.
In this review, we systematically examine the critical roles of metabolic reprogramming and microorganisms in cancer progression, highlighting their distinct contributions to tumor development. We further analyze how specific bacteria regulate metabolic pathways to influence tumor behavior, as well as how metabolic reprogramming may drive microbiota-mediated carcinogenesis. Our discussion underscores the central role of metabolic reprogramming in mediating the dynamic interplay between microbiota and human tumors. Moreover, we summarized the crosstalk between them and proposed a therapeutic concept based on the theory. That is a set of clinical therapeutic interventions tailored for different cancer stages by targeting the metabolic reprogramming/microbiota axis. In addition, multiple drugs were summarized, and clinical trials or animal experiments were evaluated to assess the therapeutic potential of targeting the metabolic reprogramming/microbiota axis as anticancer strategies.
Overview of metabolic reprogramming and microbiota
Metabolic reprogramming and cancer
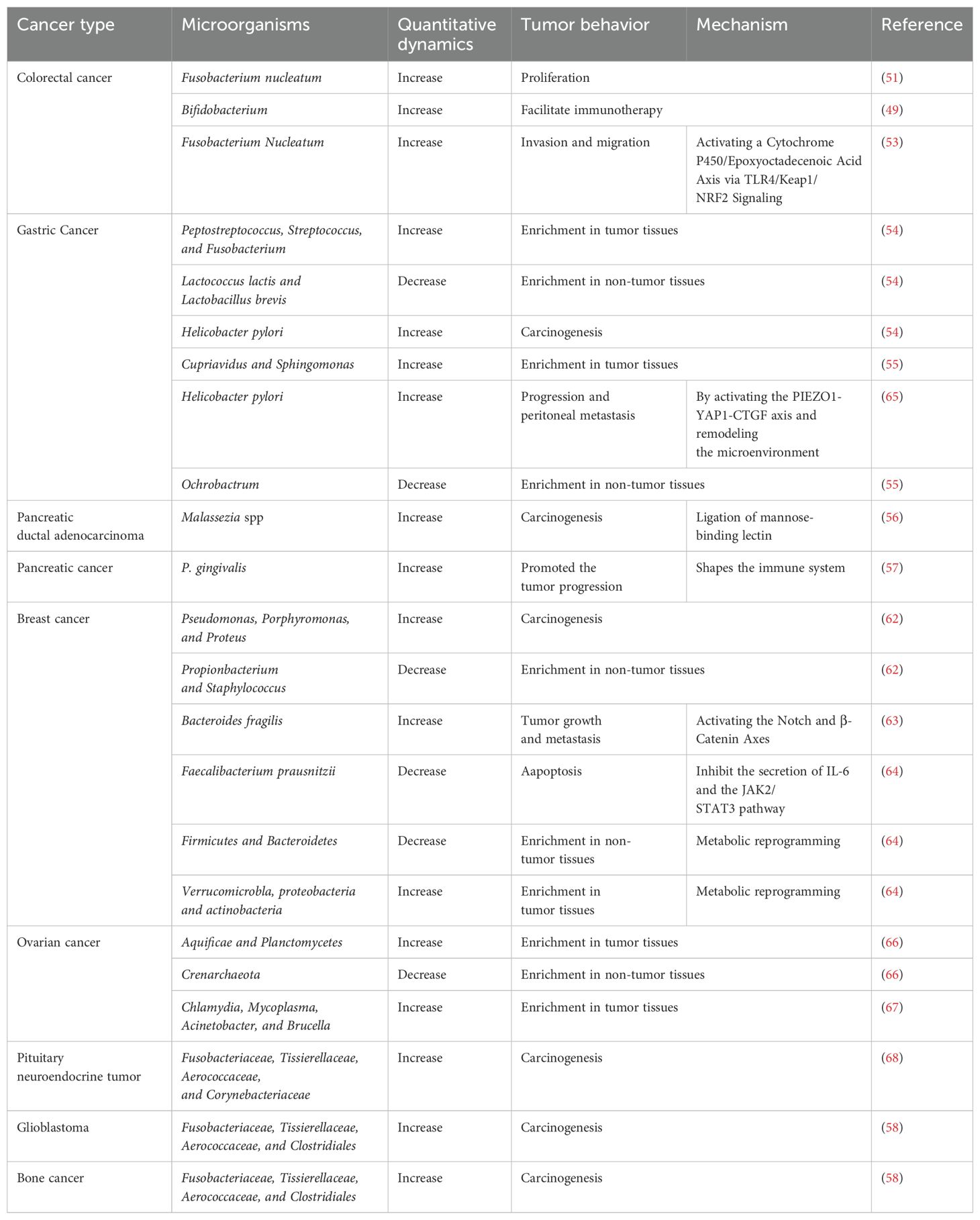
Metabolic reprogramming plays a critical role in the maintenance of increased nutrient demands while producing oncometabolites and coping with the demanding the TME (Figure 1). Metabolic reprogramming has expanded to cover almost entirely metabolic progressions, including in the metabolism of glucose, fatty acid, and amino acid. This process involves a variety of mechanisms, including regulation of gene expression, the activity of metabolic enzymes, metabolite accumulation and activating signaling pathways (25). In general, there are three main mechanisms in which changes occur: first, the transcriptional level including the activation of transcription factors and the regulation of metabolic pathways by epigenetic modifications (26, 27); second, regulation of metabolic enzyme activities by post-translational modifications, such as phosphorylation and ubiquitination (28, 29); third, changes of metabolites in the TME may also lead to inhibition or activation specific signaling pathways resulting in metabolic reprogramming (30). A large number of studies have found that metabolic reprogramming is closely but complexly related to tumor development, metastasis and drug resistance (4, 25, 31). circRNA circSLIT2 is highly expressed in pancreatic ductal adenocarcinoma and promoted the aerobic glycolysis by targeting miR-510-5p/c-Myc/LDHA axis, ultimately promoting proliferation of cancer cells (32). In colorectal cancer, SIRT1, a hub of metabolic glucolipid conversion, is upregulated and increases the level of deacetylated β-catenin in response to oxidative stress (33). Sodium butyrate can suppress glycolysis by downregulation the expression of HK2 through the c-myc signaling pathway, which results in the suppression of proliferation of the hepatocellular carcinoma (34). Inoue et al. found that pyruvate dehydrogenase (PDH) component X expression is necessary for PDH activity and inhibition of its activity is involved with glycolysis via conversion of pyruvate to lactate, essential for the development of esophageal squamous cell carcinoma (35). Lipids are important components of biological membranes and the type and saturation of fatty acids in the membrane affect its stability and function (8). The key enzyme SCD1 reduces the fatty acid ratio and contributes to the protection of ovarian cancer cells from ferroptosis (36). Lipids are also involved in signal transduction. Prostaglandins (PGs) reduce oxidative stress and prevent lipid peroxidation in tumor cells (37). cPLA2 inhibition shows remarkable synergy restriction to inhibit growth of mutant PIK3CA-bearing breast tumors through influencing the secretion of arachidonic acid (AA) (30). Furthermore, in luminal breast cancer, the mutant of PIK3CA gene initiated the AA metabolic reprogramming through 5-LOX (38). Nelson et al. reported that the USP25/HIF-1α axis is an essential mechanism of metabolic reprogramming and survival in pancreatic ductal adenocarcinoma (39). In addition, the enhanced glycolysis and pentose phosphate pathway were associated with increased HIF-1α expression in colorectal cancer (26). Metabolic phenotypes vary among different types of tumor cells and different stages of tumorigenesis and even in different parts of tumor tissue. Faubert et al. reported that metabolic phenotypes develop as cancer progresses from precancerous lesions to localized, clinically apparent malignancies to metastatic cancer (4). Firstly, metabolic reprogramming can provide malignancy cells with energy and metabolites needed for growth and maintain the stability of the microenvironment (40). Secondly, tumor cells can undergo metabolic reprogramming due to DNA damage, inactivation of genes, and activation of the signaling pathway, thus promoting cancer (41). The above molecules and their mechanisms of regulating metabolic reprogramming are summarized in Table 1.
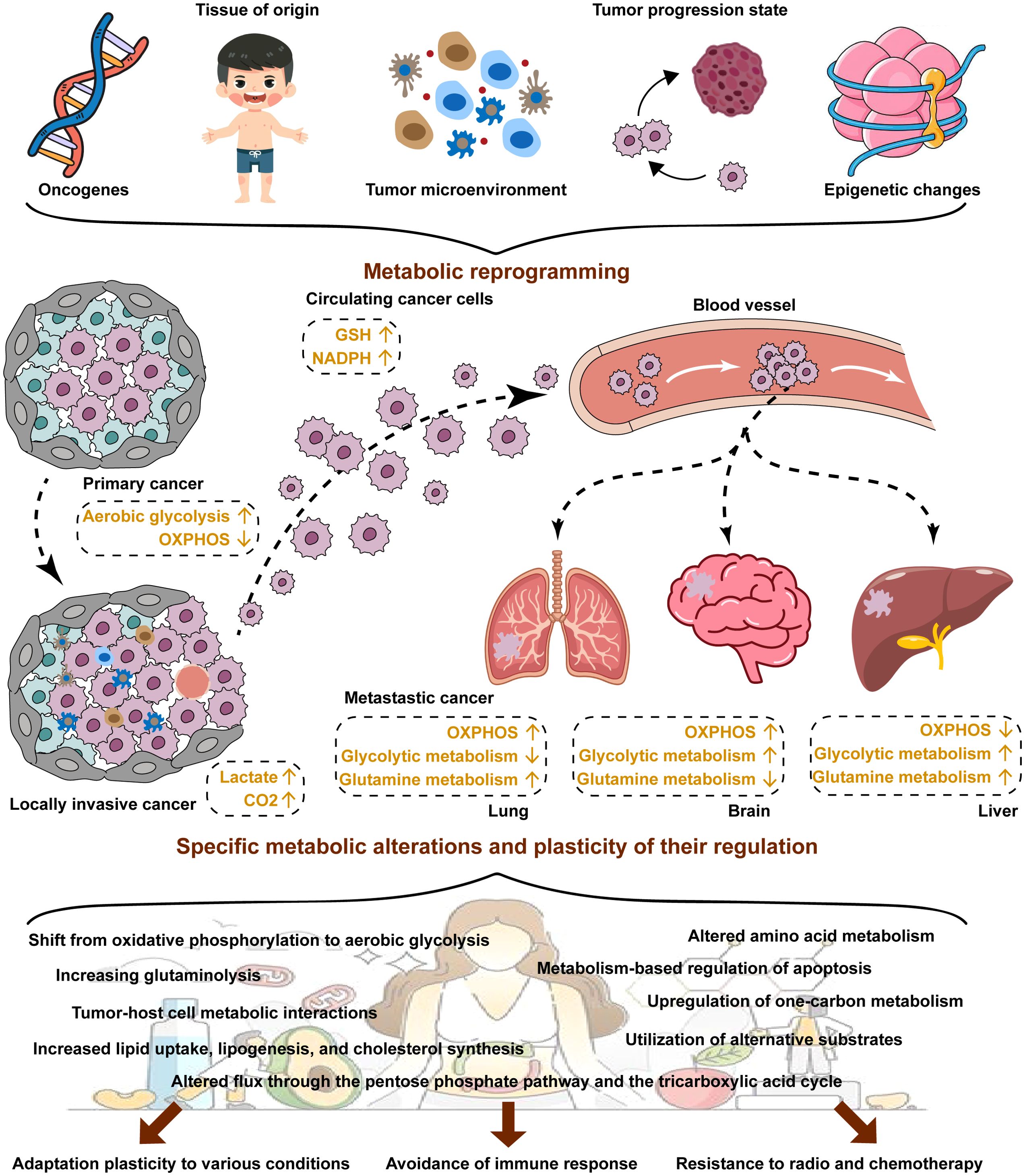
Figure 1. Metabolic reprogramming in cancer. Cancer cells usually exhibit aberrant metabolism resulting from metabolic reprogramming. Metabolic reprogramming is dependent on many factors, including oncogenes, tissue of origins, the TME, tumor progression stage and epigenetic changes. Metabolic reprogramming plays a key role in the reprogramming of adaptation plasticity to various conditions, avoidance of immune response, and resistance to radio and chemotherapy for cancer cells.
Microbiota and cancer
So far, many important discoveries about microbiota have been reported (Figure 2). The microbial community may have an indirect or direct carcinogenic function to regulate cancer initiation, progression and response to therapies by regulation of oncogenic pathways, or modulation of the immune system (42). Bacteria, viruses and fungi are the main microorganisms that regulate promoting mucosal inflammation and human immunity (43–45), where most studies focused on the extra-tumoral microbiota and intra-tumoral microbiota.
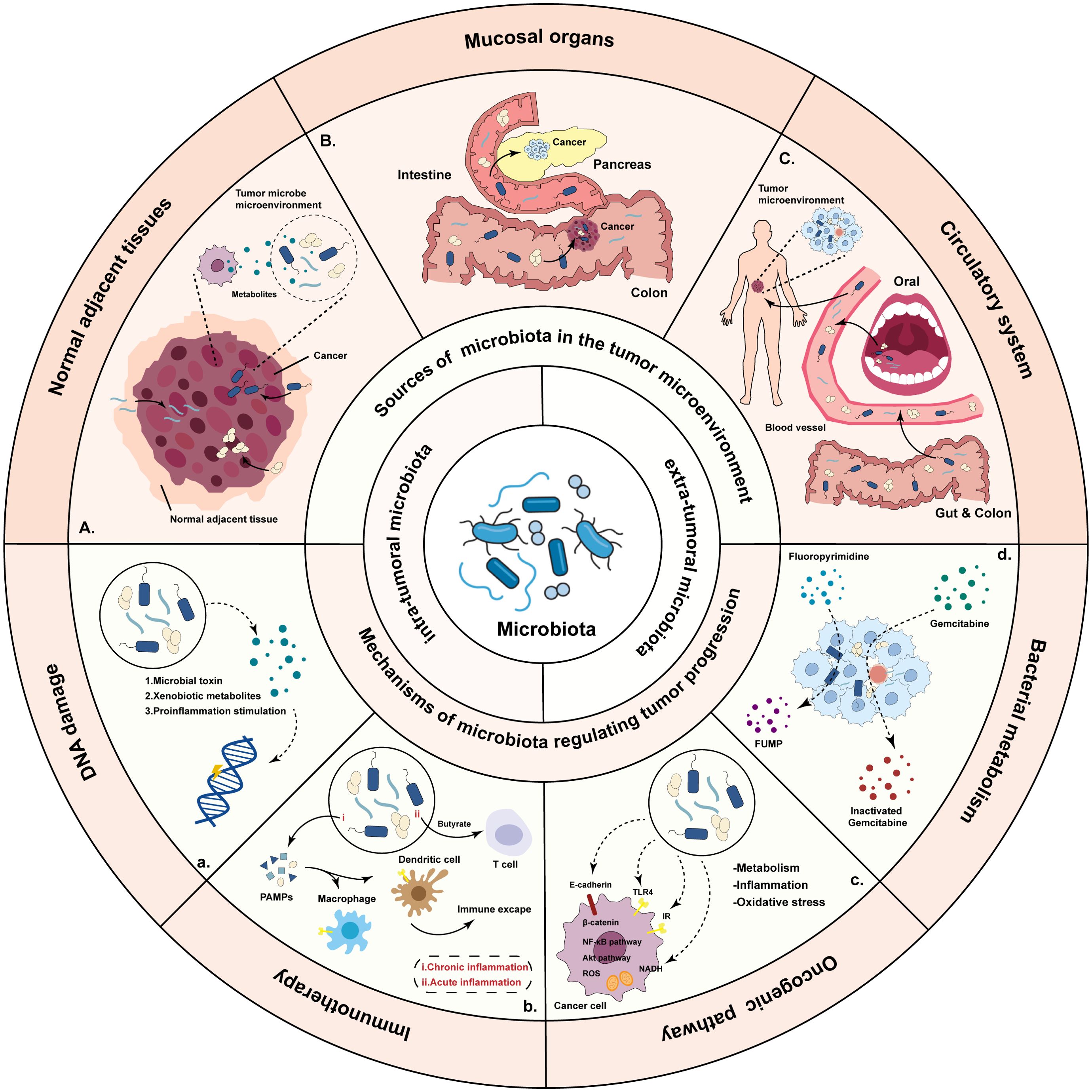
Figure 2. Overview of the sources of the microbiota and mechanisms by which bacteria regulate tumor progression. There are three potential sources of microbiota: microbiota originating from normal adjacent tissues, microbiota from mucosal organs through mucosal barriers, and microbiota which are the result of hematogenous spread. Microbiota influence tumorigenesis and treatment through DNA damage, activating the signaling pathway, influencing the anti-tumor immunity and metabolize drugs.
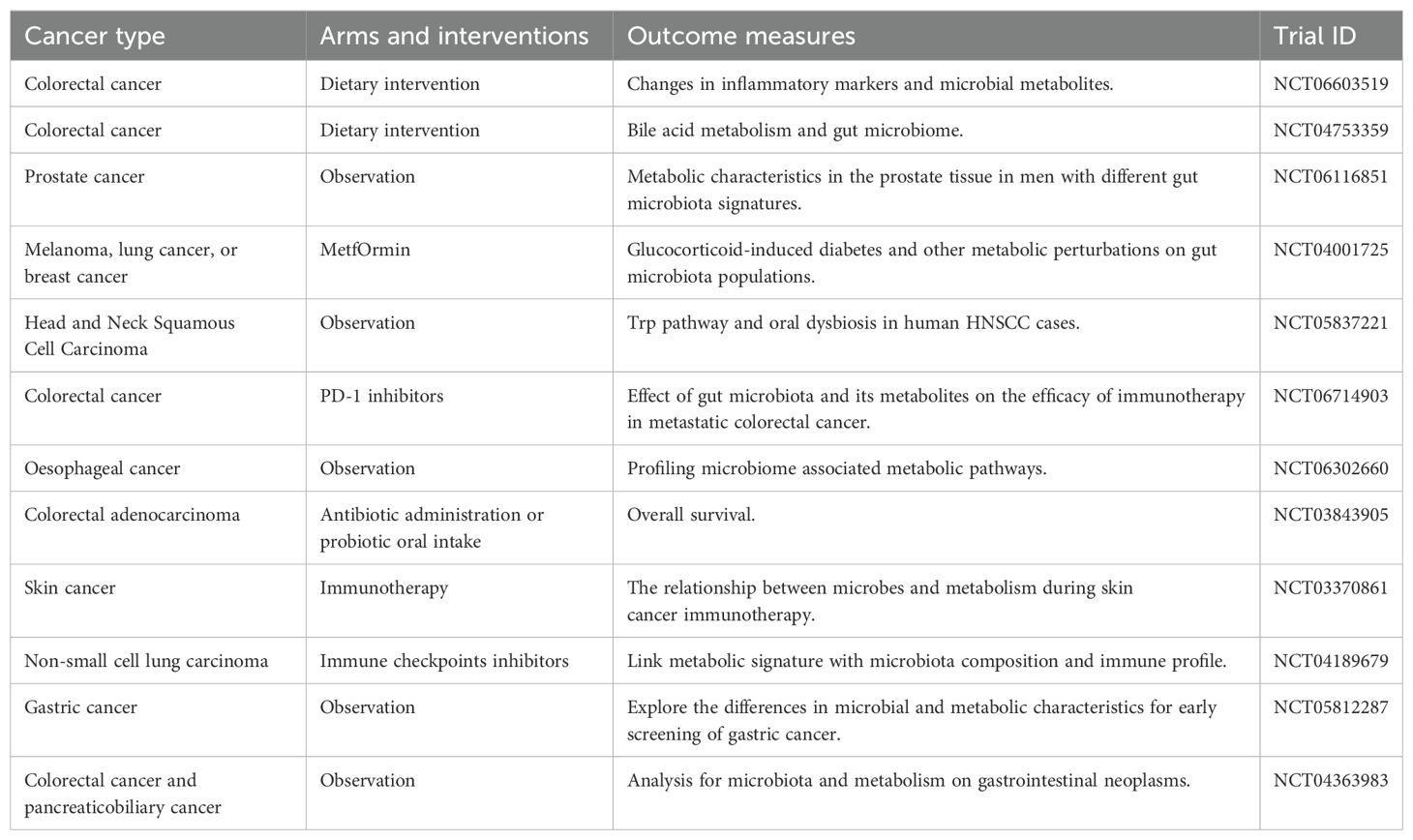
Abundant microbes (approximately 4×1013 microbial cells) exist and colonize on and inside human (46). The most abundant microorganisms are harbored in the mucosal organs of human bodies, including in the intestinal tract, oral cavity, and skin (11, 16). The microbiota and the mucosal organs form a symbiotic, holistic system together, and maintain the homeostasis as a biological barrier. Interestingly, the mice which are bred and housed in an environment devoid of microorganisms were immune deficient and exhibited a flimsy gut barrier (47). Indeed, studies have shown that pathogenic bacteria in mucosal organs are associated with multiple cancers, such as head and neck cancer, lung cancer, colorectal cancer, esophageal cancer, and pancreatic cancer (48–51). The progression of skin cancer has been reported to be related to bacteria through the TLR-5 signaling pathway (52). Fusobacterium nucleatum might utilize the TLR4/Keap1/NRF2 signaling to promote colorectal cancer development and metastasis (53). The detection rate of Streptococcus and Clostridium in gastric cancer are higher than those in normal tissue, whereas Lactobacillus brevis is more enriched in normal controls (54). Shi et al. reported that microbial richness is significantly decreased in gastric cancer tissues compared with adjacent normal tissues, and some microbes, such as Cupriavidus and Sphingomonas, are enriched in cancer tissues, while Ochrobactrum are enriched in normal tissues (55). In addition, the mycobiome also plays an important role in tumorigenesis of pancreatic ductal adenocarcinoma. The cancer tissues displayed an increase in fungi of about 3,000-fold compared to normal pancreatic tissue, and the Malassezia species were found to be associated with oncogenesis (56). Researchers also found Porphyromonas gingivalis, which is highly epidemically connected with pancreatic cancer, promoted pancreatic cancer progression via elevating the secretion of neutrophilic chemokines and neutrophil elastase (57).
The tumor tissues once considered sterile before, however, with the development of technology, researchers actually discovered a variety of microorganisms are also found in tumors that do not arise from mucosal sites (58, 59). Therefore, the concept of intra-tumoral microbiota present in tumor tissues is proposed (60), and intra-tumoral microorganisms have been found in at least 33 major cancer types, such as breast cancer (61, 62). Certain bacteria in the breast tissues are associated with cell dysplasia and carcinogenic effects. The levels of Pseudomonas, Porphyromonas, and Proteus in breast cancer tissues are significantly higher than those in normal tissue, while Propionbacterium and Staphylococcus are less enriched than that in the non-tumor tissue conversely (62). Enterotoxigenic Bacteroides fragilis have been shown to be cancer-causing bacteria in breast cancer, it activates the notch and beta-catenin axes and induces growth and metastasis (63). Furthermore, Faecalibacterium prausnitzii is less abundant in breast cancer patients, which could suppress the growth of breast cancer cells through inhibition the JAK2/STAT3 signaling pathway (64). Recent studies have revealed that H. pylori‐NF‐κB activates the PIEZO1‐YAP1‐CTGF axis to remodel the GC microenvironment by promoting CAF infiltration (65). The detection rate of Aquificae and Planctomycetes in ovarian carcinoma is higher than that in adjacent tissue, and the level of Crenarchaeota is decreased (66). Researchers also found that the development of ovarian carcinoma is related to Chlamydia, Mycoplasma, Acinetobacter, and Brucella (67). A recent study analyzed the composition of intra-tumoral microbiota in pituitary neuroendocrine tumors. In this study, researchers found that Fusobacteriaceae, Tissierellaceae, Aerococcaceae, and Corynebacteriaceae may correlate with the pathogenesis and development of the tumors (68). Nejman et al. reported that microbiota (such as Fusobacteriaceae, Tissierellaceae, Aerococcaceae, and Clostridiales) was detected in tumors that have no direct connection with the external environment, such as glioblastoma multiforme and bone cancer (58). The abovementioned bacteria and other microbiota in a few cancers are all summarized to understand their role cancer progression (Table 2).
Crosstalk between metabolic reprogramming and microbiota in cancer development
Specific microbiota intricately regulates tumor progression through dynamic interactions with the metabolic pathways in the host through three key mechanisms: regulation the metabolism of cancer cells, nutrient competition with immune cells and reshaping the TME, and immune-metabolic reprogramming (69–71) (Figure 3). Commensal bacteria like Clostridium butyricum and Bacteroides ferment dietary fiber to produce short-chain fatty acids (SCFAs), where butyrate triggered superoxidative stress and intracellular lipid accumulation, which enhanced ferroptosis susceptibility in cancer cells (72, 73). Concurrently, species such as Clostridium transform primary bile acids into tumor-promoting secondary bile acids that activate the expression of urokinase-type plasminogen activator receptor (uPAR) to drive ERK and AP-1 signaling in colon cancer cells (74). The TME is further shaped through microbial nutrient metabolism, where tryptophan metabolism by Lactobacillus activates the aryl hydrocarbon receptor to enhanced T cell production of IL-17 (75), while Bifidobacterium breve-derived indole-3-lactic acid (ILA) ameliorates tumorigenesis by directing the differentiation of macrophages (76). Microbiota also orchestrate immune-metabolic crosstalk, with streptococcal pyrogenic exotoxin A (SPEA) of Streptococcus promoting CD25+/Foxp3+ Treg expansion via PD-L1 and kynurenine (77), whereas Lactobacillus gallinarum-derived metabolites inhibit the function of Tregs through modulating IDO1/Kyn/AhR axis (78). Epigenetic modifications, including Lactobacillus plantarum and its metabolite enhancing H3K27ac binding at the enhancer regions of IL-12a, further priming CD8+ T cell immunity against tumor growth (79). These findings highlight the microbiome as a master regulator of tumor metabolism, offering novel avenues for intercepting cancer progression through microbial-metabolic reprogramming. The crosstalk between metabolic reprogramming and microbiota varies substantially across different types and stages of malignancy. Here, we summarized the microbiota in a few cancers to understand the role in cancer development (Figure 4).
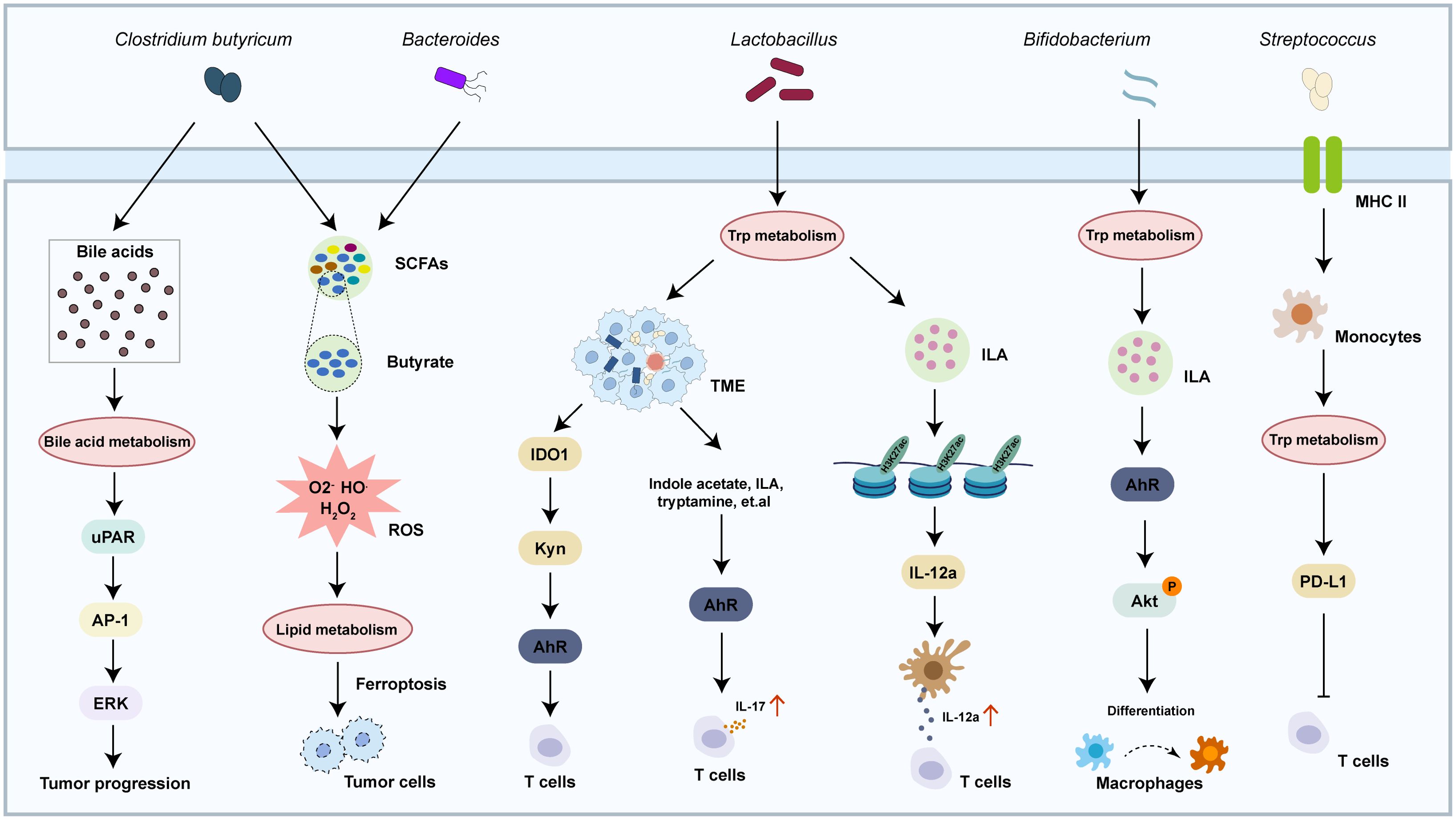
Figure 3. Crosstalk between metabolism pathway and microbiota in cancer. The crosstalk between metabolism pathway and microbiota regulates multiple physiological and pathological responses, including cancer progression and reshaping the TME.
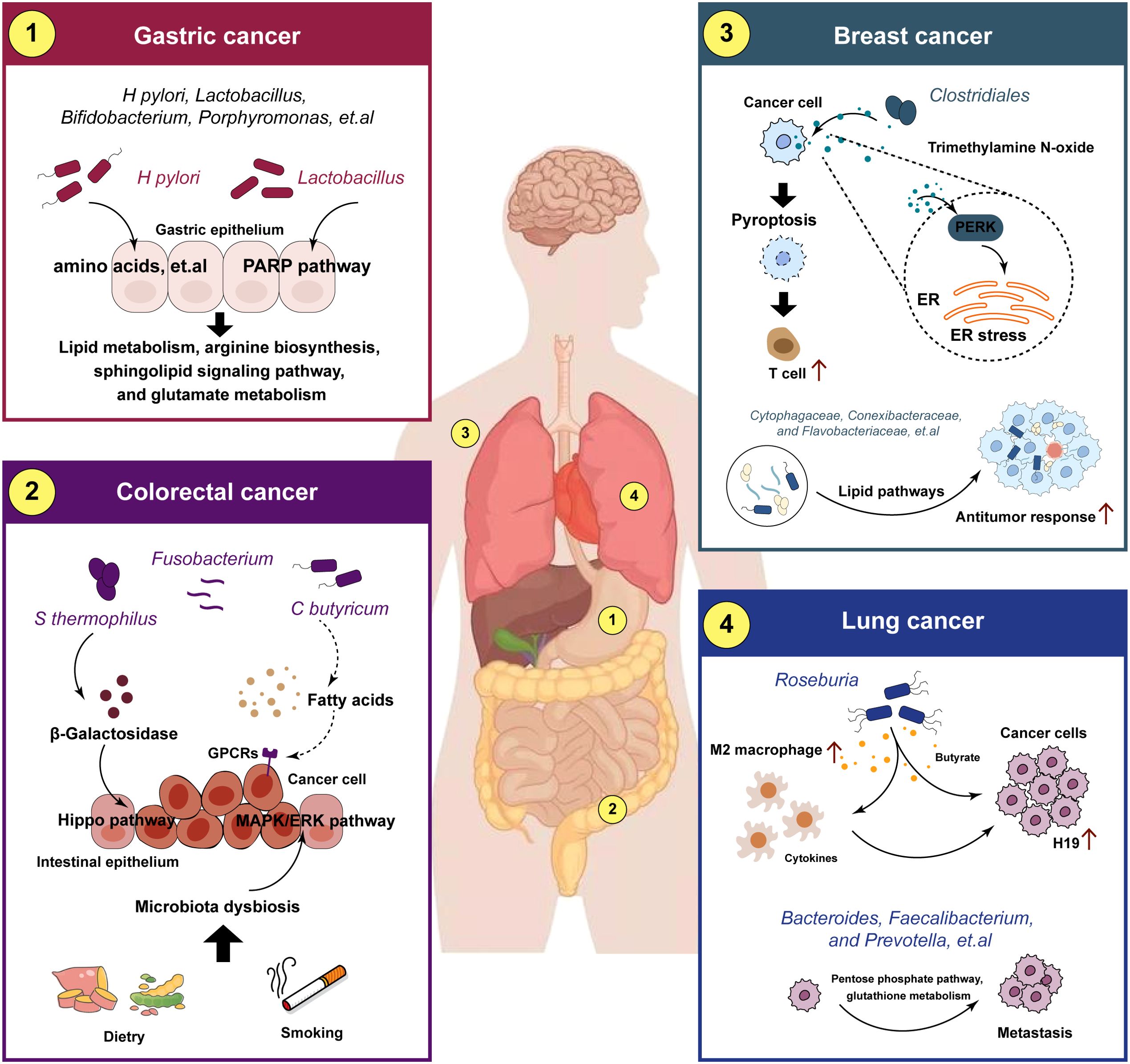
Figure 4. Crosstalk between metabolic reprogramming and microbiota in different tumors. The crosstalk between metabolic reprogramming and microbiota regulates cancer progression. metabolic reprogramming can play a role in microbiota-mediated tumorigenesis, metastasis and drug resistance in different cancers.
Gastric cancer
The gastric microbiota and their metabolites are the major risk factors for the development and progression of gastric cancer (GC). Helicobacter pylori is the most predominant microorganism detected in GC, and it has been associated with precancerous lesions that can eventually promote the development of GC (80). The specific mechanisms by which H. pylori mediate the carcinogenesis and progression of GC are still unclear, and is associated with multiple factors, including in inflammatory responses, host genetic diversity, and environmental influences (11). Recent studies have revealed that the reason for the differences in metabolome profiles between GC tumor and non-tumor tissues may due to the collective activities of H. pylori, Lactobacillus, and other microorganisms (81). The metabolome analysis demonstrated that H. pylori is negatively and positively correlated with the majority of differential metabolites in the classes of amino acids, carbohydrates, nucleosides, nucleotides, and glycerophospholipids, respectively, suggesting that it might play a key role in degradation and synthesis of the majority of differential metabolites (82). Moreover, alterations in gut microbiota and metabolism are potentially linked to chronic inflammation and GC (83). In precancerous lesions of GC rat model, Lactobacillus and Bifidobacterium increased significantly while Turicibacter and Romboutsia reduced significantly, mechanically, the microbiota and the metabolites are related to the lipid metabolism and PPAR signaling pathway (84). Additionally, Wang et al. reported that Lactobacillus could enhance the production of N-nitroso compounds, thus, these enriched bacteria could participate in the carcinogenesis (85). Yang et al. observed that Methylobacterium-Methylorubrum was significantly increased in distal gastric cancer tissues, positively correlated with cancer-promoting metabolites, including in arginine biosynthesis, sphingolipid signaling pathway, and glutamate metabolism (86). Meanwhile, Porphyromonas, Catonella, Proteus, Oribacterium, and Moraxella were significantly correlated with hormone metabolism in proximal gastric cancer (86).
Colorectal cancer
Metabolic reprogramming are important components in the crosstalk between the gut microbiota and the human body and play key roles in the development of colorectal cancer (CRC) (87). Mechanistically, microorganisms produce metabolites after the uptake of certain nutrients, which include lipids, proteins, secondary bile products, biogenic amines, oligosaccharides, glycolipids, organic acids, and amino acids, and then the metabolites act as signaling elements and substrates in metabolic reactions (88, 89). Metabolic reprogramming serves vary functions in the progression of CRC, and further studies have shown that metabolic reprogramming plays a dual role (90). Coker et al. demonstrated that gut metabolites and their association with amino acids metabolic pathways in the colorectum are altered in the early the stages leading to CRC (91). Bile acid was recently been demonstrated to be bio-transformed by the microbiota and play a role in the microenvironment in Inflammatory bowel disease and CRC (92). Clostridium Butyricum is one of the original gut microbiota species found in human. C. butyricum can produce SCFAs, which can directly activate G-coupled-receptors, inhibit histone deacetylases, and serve as a protector in CRC (93). Li ert.al verified that Streptococcus Thermophilus could secret β-Galactosidase to activate oxidative phosphorylation and downregulate the Hippo signaling pathway, which lead to the suppressive effects of CRC cells (94). Fusobacterium nucleatum enrichment and short-chain fatty acid depletion characterizes late-onset CRC, while early-onset CRC tended to be associated with increased tryptophan, bile acid and choline metabolism and enriched Flavonifractor plauti (95). Furthermore, high-fat diet drives CRC through modulating microbiota and metabolites (96). Yang et al. reported that the increased bacteria of Alistipessp.Marseille-P5997 and Alistipessp.5CPEGH6, and depleted probiotic Parabacteroides distasonis, along with elevated lysophosphatidic acid in CRC mice model (97). In addition, the smoke-induced gut microbiota dysbiosis, including the enrichment of Eggerthella lenta and depletion of Parabacteroides distasonis and Lactobacillus spp, increased the bile acid metabolism and impaired gut barrier function, through activating the MAPK/ERK signaling pathway in CRC cells (98).
Breast cancer
Breast cancer (BC) is the most widespread malignant cancer among women worldwide and presents significant challenges to female health (99). The microorganisms exert a profound impact on the host and has emerged as a pivotal frontier in the BC pathogenesis (100). In recent years, a growing number of scholars have reported that BC tissues have a great diversity and abundance of microorganisms (58). Hieken et al. discovered that the differences of Lactobacillus between the breast tissue in benign and malignancy, which are associated with metabolic pathway involving cysteine and methionine metabolism, glycosyltransferases, fatty acid biosynthesis (101). Xuan et al. demonstrated that genus Cytophagaceae, Conexibacteraceae, and Flavobacteriaceae possibly modulate the BC immune microenvironment and elicit an antitumor response through lipid pathways (102). Another study revealed that metabolite of Clostridiales, trimethylamine N-oxide, induced pyroptosis by activating the endoplasmic reticulum stress kinase PERK in triple-negative BC (103). Heath et al. reported that gut microbiota-derived metabolites alter estrogen receptor activity and endocrine therapy responsiveness in ER+ BC, and targeting metabolic pathways through diet or drugs could be useful to improve endocrine therapy efficacy in the clinic (104). Interestingly, women with malignant breast carcinoma are found to exhibit enriched level of Citrobacter in their gut, which is associated with elevated glycan and lipopolysaccharide biosynthesis, in comparison to the group with benign tumors (105). One of the pivotal independent risk factor for breast tumorigenesis is hormonal deregulation, and multiple studies have shown a direct role of gut microbiota in hormonal deregulation through influence the expression of β-glucuronidase (100). Furthermore, indicated the relationship between microorganisms and metabolism, and found their potential roles in the prognosis value, indicating that patients with both high Campylobacter abundance and inositol phosphate metabolic activity had the worst survival probability (106).
Lung cancer
The lung is another organ with an abundant microbiome, which leads to the exposure of lung cancer to numerous microorganisms (107). It is certain that the composition of lung microbiota is altered in lung cancers, which promoted tumor growth and metastatic progression (16). Smoking is the most important environmental risk factor associated with lung cancer (108). A recent study analyzed the correlation of microbes and smoking‐related metabolic pathways (109). In this study, researchers found that Acidovorax is enriched in smokers. Additionally, Ma et al. found that Roseburia-derived butyrate promotes lung cancer metastasis by increasing expression of H19 in tumor cells through inhibiting HDAC2 and increasing H3K27 acetylation at the H19 promoter and inducing M2 macrophage polarization (110). Notably, deep microbiome sequencing and targeted bacterial culture confirmed that the tumor-resident Lactobacillus in the TME in lung cancers can alter tumor metabolism and lactate signaling pathways (111). A retrospective study showed that the characteristics of the microbiome and metabolite presented significantly differences between lung cancer patients and benign pulmonary nodules patients (112). The abundance of Subdoligranulum and Romboutsia increased in lung cancer patients, combined with the enrichment in beta-Alanine metabolism, styrene degradation and pyrimidine metabolism pathway. Liu et al. found that the microbiome of lung cancer patients, such as Enterobacteriaceae and Streptococcus, had increased expression of gene modules involved in metabolism and amino acid metabolism (113). Microorganisms have been shown to participate in the metabolism of bile acids and proteins and to help form aromatic amines and sulfides, subsequently promoting lung cancer progression (114). Moreover, microbial metabolites were shown to be significantly altered in lung cancer (115). Recently, Wang and colleagues reported evidence that Bacteroides, Faecalibacterium, and Prevotella may participate in regulating metabolism-related pathways, such as the pentose phosphate pathway and glutathione metabolism, in lung adenocarcinoma (116).
Pancreatic cancer
The incidence of and number of deaths caused by pancreatic cancer have been gradually rising, and it is a leading cause of cancer death worldwide (117). The role of microbiota and metabolic reprogramming in pancreatic cancers have been proven in several studies. Tintelnot et al. reported that the tryptophan metabolite, 3-IAA, derived from microbiota as a key amplifier of the response to chemotherapy in pancreatic ductal adenocarcinoma (PDAC) (118). Furthermore, a recent study showed that Lactobacillus participate in the process that tryptophan-derived metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress immunotherapy (119). Alam et al. discover that intratumor mycobiome, such as Malassezia and Alternaria, activates the secretion of IL-33, which can induce metabolic reprogramming and accelerate the development of PDAC (45). High-fat diet has been confirmed as one of a risk factor for the development of pancreatic cancer (120). The disturbance of lipid metabolism could induce changes the intestinal environment and further leading to the dysbiosis of internal microflora in mice model of pancreatic cancer (121). Ruze and colleagues reported that the main mechanisms involved in the pancreatic carcinogenesis include microbiome dysfunction further compromise immunometabolic regulation while also aggravating mutagenic and carcinogenic metabolic disorders by affecting multiple pathways (122).
Other cancers
In addition to the cancers reported above, multiple studies have also shown the crosstalk between microbiota and metabolic reprogramming in other tumors. A clinical study showed that the existence of microorganisms in the tumor tissue of head and neck squamous cell carcinoma (HNSCC) patients (123). Sun et al. reported that F. nucleatum affects the tumor immune microenvironment via modulating via GLUT1-driven glycolysis and extracellular lactate deposition (124). This is consistent with a study by Colbert et al. showed that lactic acid bacteria in the TME can alter tumor metabolism and lactate signaling pathways, affecting chemoradiation response in patients with cervical cancer (111). Yost et al. showed that the microorganisms of OSCC patients are associated with the increased activities of iron ion transport-related enzymes, tryptophanase, glutamate dehydrogenase, starch synthase, and superoxide dismutase (125). Additionally, recent studies have demonstrated that microbiota, which drastically alter the metabolome of cervical cancer, is involved in HPV persistence, progression of cervical neoplasia, and genital inflammation (126). Ilhan and colleagues showed that cervicovaginal metabolic profiles, charactered by amino acid and nucleotide metabolisms, were driven by genital inflammation, HPV infection, and vaginal microbiota, including Sneathia, Streptococcus, Prevotella, and Gardnerella (127). 16S rRNA gene sequencing and untargeted metabolomics revealed that Prevotella is involved in the synthesis of fatty acyl, carboxylic acids and derivatives, benzenes and substituted derivatives, organic oxygenates, and indoles and derivatives as metabolites. Fusicatenibacter and Lachnospira are involved in the degradation of indoles and derivatives. Alistipes, Agathobacter, and Parabacteroides are involved in the synthesis of indoles in esophageal squamous cell carcinoma (ESCC) (128). Further analysis revealed that Prevotella, Alistipes, Agathobacter, and Parabacteroides might regulate the synthesis of indoles and promote ESCC. Cheung et al. observed that an enrichment of carcinogenic bacteria, such as Butyricimonas, Veillonella, and Streptococcus, and a depletion of Butyricicoccus of ESCC patients (129). Recently, Lau et al. reported that L. acidophilus exhibits anti-tumorigenic effect in mice by secreting valeric acid, and probiotic supplementation is a potential prophylactic of hepatocellular carcinoma (130). The relationships between metabolic reprogramming and microorganisms of cancers have not been thoroughly studied.
Targeting metabolism/microbiota as an emerging therapy in cancer
Since metabolism plays an important role in the development of human malignancy, it may promote tumor formation, or inhibit tumor cell growth. Alternatively, the abovementioned microorganisms may cause cancer through certain mechanisms. In recent years, breakthrough results have been obtained in clinical trials assessing the relationship between metabolic reprogramming and microorganisms in cancer. In the following section, we summarized the drugs, as well as the corresponding clinical trials, that target metabolic reprogramming or microbiota and exert anticancer effects and their mechanisms. More importantly, we emphasized the critical role of metabolism reprogramming in mediating microbial communities and cancer. A variety of bacteria regulate tumor development, metastasis and treatment-resistant through metabolic reprogramming. Therefore, the simultaneous use of drugs targeting metabolism and microorganisms may play a synergistic anticancer role. However, few such studies have been performed, and more clinical trials are urgently needed to better understand the therapeutic potential of drugs targeting the metabolic reprogramming/microbiota axis, which could help to develop new drugs to prevent or treat human cancer.
Targeting metabolism for cancer treatment
Metabolic reprogramming is not only a biological hallmark of cancer but also reveals treatment vulnerabilities. The vulnerability caused by metabolism rewiring may present therapeutic opportunities as malignancy cells become more and more dependent on specific metabolic pathways. Farber and Diamond reported that the anti-folate drug, aminopterin, could induce remission in pediatric acute lymphocytic leukemia as early as 1948, and has been used as an anti-tumor therapy for many years (131). Xiao Y. indicated that the metabolic targets for cancer treatment could be categorized into three groups: (i) targeting the metabolic vulnerability of tumor cells, (ii) targeting metabolism of the TME, and (iii) regulating body metabolism (132). Considering the active nucleotide synthesis and high proliferation rate of cancer cells, disrupting nucleotide metabolism is considered a promising anti-tumor therapeutic strategy (133). Except the aminopterin, many purine and pyrimidine analogs have been widely used in the clinic, including in 5-fluorouracil, 6-mercaptopurine, gemcitabine, capecitabine, and fludarabine (132, 134, 135). Jin et al. reported that the treatment of leflunomide, a drug that inhibits dihydroorotate dehydrogenase, prevented lung metastasis in the mouse lung cancer metastasis model (136). Studies have shown that inhibitors of dihydroorotate dehydrogenase can exhibit antitumor effects in a variety of preclinical models, such as glioma (137, 138) and lymphoma (139). Furthermore, studies have shown that some promising metabolic targets in energy metabolism for cancer treatment, including in glycolysis, fatty acid metabolism, glutamine metabolism, the tricarboxylic acid (TCA) cycle, and oxidative phosphorylation (OXPHOS) (140, 141). Tran et al. reported that α-Ketoglutarate promote stemness and leads to CRC formation through Wnt signaling pathway (142). Inhibitors targeting glucose transporters (GLUT), hexokinase, lactate dehydrogenase, and lactate-proton symporters have shown promise in impairing tumor growth (132). Additionally, enzymes involved in fatty acid oxidation (FAO) and fatty acid synthesis and desaturation are being actively studied (143). Inhibitors of glutamine transporters and glutaminases have also been investigated (144). Han et al. demonstrated that activation of polyamine catabolism promotes glutamine metabolism and creates a targetable vulnerability in lung cancer cells (145). A phase I clinical trial and pharmacodynamic study of complex I inhibitor of oxidative phosphorylation in advanced solid tumors and acute myeloid leukemia patients showed that strategies targeting the TCA cycle and OXPHOS, such as inhibitors targeting electron transport chain (ETC) complexes, have displayed antitumor effects (146). Metformin, a drug known to inhibit ETC complex I, has been extensively studied in clinical trials for cancer treatment (147). Moreover, except for tumor cells, immune cells also have metabolic reprogramming during tumor progression to facilitate the escape of tumor cells from immune surveillance (148, 149). Myeloid cells play a pivotal role in tumor biology, and they play key roles on tumor growth and antitumor immune responses (150). Geiger et al. reported that L-arginine concentrations directly impact the metabolic fitness and survival capacity of T cells, which are crucial for anti-tumor responses (151). In addition, the metabolic reprogramming also could happen in the stromal cells, such as cancer-associated fibroblasts (CAFs). CAFs require proline synthesis by PYCR1 for the deposition of pro-tumorigenic extracellular matrix (152). Beyond metabolic reprogramming at the tumor site, the metabolism of the whole body also presents as a potential therapeutic target. Physical exercise and dietary interventions are the two most important and easily controlled factor that can reduce cancer progression (153, 154). For instance, cPLA2 inhibition shows remarkable synergy with dietary fat restriction to restore tumoral immune cell infiltration and inhibit growth of mutant PIK3CA-bearing breast tumors (30).
Targeting microorganisms for cancer treatment
Traditionally, some specific microorganisms may act as direct factors responsible for the development of cancer (61). However, the survival benefits of tumor bacteria on tumor cells are most prominent during metastasis rather than primary tumor growth in recent studies (155). Antibiotics have been used clinically to eradicate microorganisms to prevent cancer as much as possible and to assist in the treatment of cancer. For instance, the eradication of H. pylori may inhibit the occurrence of gastric cancer (156). Additionally, a retrospective investigation found that antibiotic therapy improves the survival of pancreatic adenocarcinoma patients (157). Recent studies demonstrated that reduction of Staphylococcus in the mammary tumor microbiota induces antitumor immunity and decreases breast cancer aggressiveness in mice models (158). Another mouse study indicated that a direct influence of long-term ampicillin exposure can alter the lung microbiota of rats (159). In addition, the treatment of mice bearing a colon cancer xenograft with the antibiotic metronidazole reduced Fusobacterium load, cancer cell proliferation, and overall tumor growth (160). Furthermore, some studies indicated that the use of antibiotics can disturb the balance of the gut microbiome, which further might ultimately lead to an unpredictable array of consequences (161). A study indicated that wide-spectrum antibiotics treatment prior to CD19-targeted chimeric antigen receptor T-cell immunotherapy (CAR-T) is associated with adverse outcomes for B cell lymphoma patients (162). In addition to antibiotics treatment, using microorganisms to active anti-tumor immune response and using bacteria as carriers that can release drugs are the main frontier in the treatment of malignancy (16). One example is the use of the BCG vaccine, which is one of the most successful immunotherapies in oncology to date (163). Treatment with BCG is effective for many cancers in clinical therapies, especially bladder cancer (164, 165). Routy et al. demonstrated that fecal microbiota transplant and sufficient dietary fiber intake could improve immunotherapy response rates, and was associated with improved progression-free survival of melanoma patients (166). Besides, some microbiota such as Listeria and Salmonella typhimurium are used as delivery platforms for drug targeting (167–169). Furthermore, Wu and the colleagues constructed a bacteria-inspired microbots for the treatment of metastatic triple negative breast cancer (170). Recently, a group of researchers developed an engineered probiotic Escherichia coli that improved the anti‐tumor effect (171). We summarized representative clinical trials that focused on metabolism and microbiota (Table 3). Together, therapies based on microorganisms have attracted a lot of attention.
Antitumor effects of microorganisms combined with metabolic targets
At present, there are few antitumor studies evaluating the effect of combined therapy with antibiotics and metabolic targets, and most current studies are based on gut microbes. It remains unknown whether changing the gut microbes will affect the characteristics of the host metabolism and the progression of cancer; these aspects need to be explored. Gut microbiota‐derived metabolites are key hubs connecting the gut microbiome and cancer progression (172). The study by Canale et al. can provide a way forward (171). Increase intra-tumoral L-arginine levels is a key determinant of an efficient anti-tumor T cell response (173). Canale et al. reported that they used the synthetic biology approach to develop an engineered probiotic Escherichia coli Nissle 1917 strain that colonizes tumors and continuously converts ammonia to L-arginine. There is a growing recognition that using live engineered bacteria for metabolic modulation is a new strategy for cancer immunotherapy (174). Moreover, He et al. reported that mice infused back with butyric acid or supplemented with intestinal flora were also able to promote CD8+ T-cell infiltration and function in tumors and rescue the efficacy of the chemotherapeutic agent oxaliplatin (175). Moreover, a recent study showed that Pien Tze Huang suppresses colorectal tumorigenesis through restoring gut microbiota and metabolites (176). Traditional medicine regulated the crosstalk between microbiota and metabolites, which provided novel ideas for clinical treatment of cancer. Some scholars attempted to use dietary improvement (177, 178), utilizing phages to eliminate certain microorganisms which secret metabolites (179), fecal microbiota transplantation (180), and so on for cancer treatment by modulating the crosstalk between microbiota and metabolic reprogramming (Figure 5). Understanding the complex relationship between microbes and metabolic reprogramming could provide valuable insights into potential cancer treatment options.
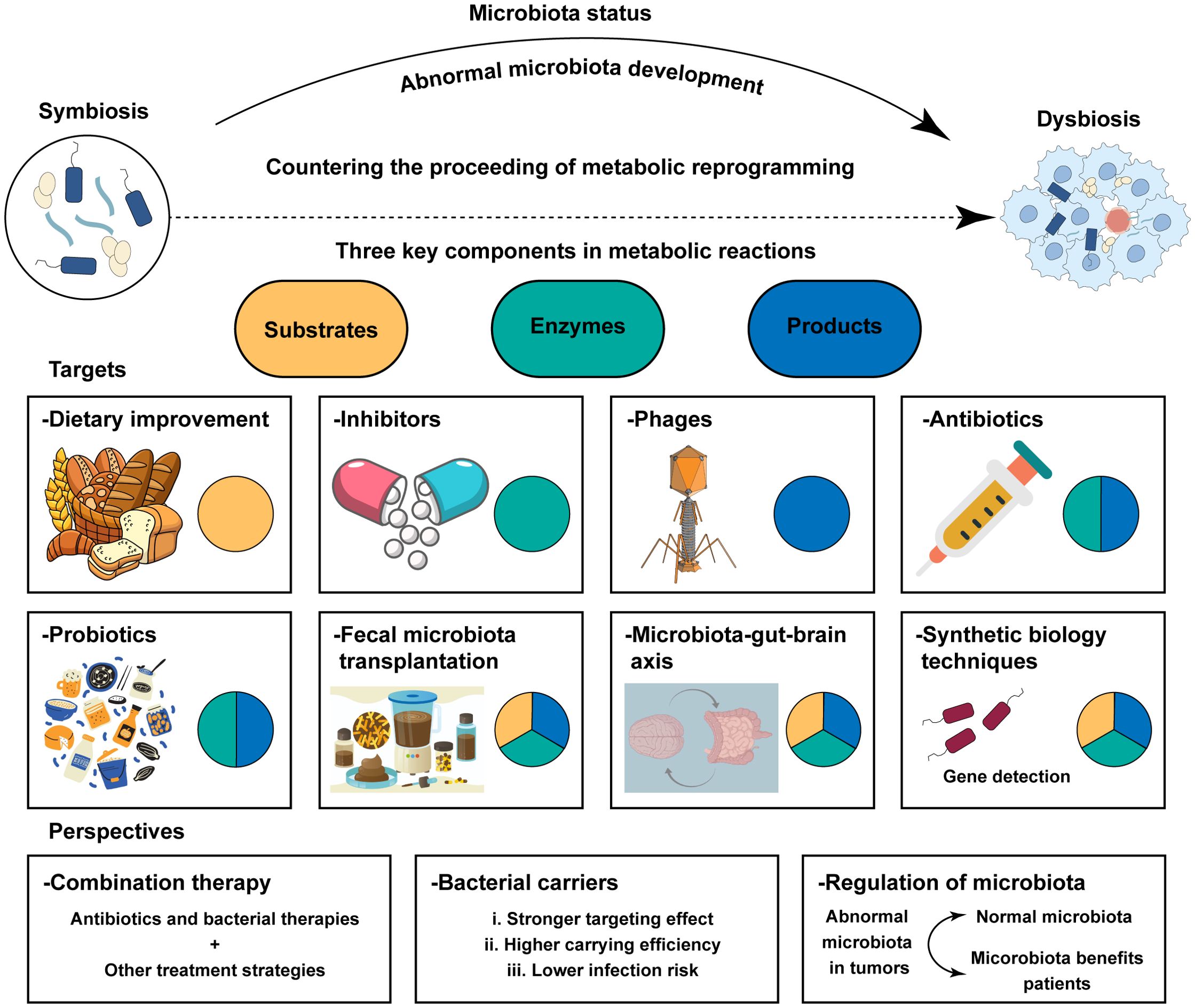
Figure 5. Treatment strategies and prospect of clinical application based on the crosstalk between metabolic reprogramming and microbiota. Metabolic reactions imply three components: substrates, enzymes, and products. The regulation and alteration of each component will affect the metabolic process and have an impact on cancer treatment. Potential strategies enhance the efficiency of anti‐tumor therapies including in researching better bacterial carries, normalizing microbiota in tumor patients, and bacterial therapies combined with other anti‐tumor therapies.
Concluding remarks
Metabolic reprogramming plays a critical role in the maintenance of increased nutrient demands for cancer cells. Metabolic reprogramming is a hallmark of cancer and often observed in human malignancy. Therefore, it is speculated that certain metabolism can play a regulatory role, as it can promote tumor growth or inhibit tumor cells by providing metabolic energy or activating certain intracellular signaling pathways. The complex community of microorganisms found in the human body has the potential to regulate tumor progression and the treatment response of multiple types of cancer. More importantly, bacteria have been globally proven to be involved in the progression of cancer via metabolic regulation, suggesting a complex interaction between metabolic reprogramming and microbiota. Despite significant progress, studies on microbiota and metabolic reprogramming face several critical limitations. Firstly, while high-throughput sequencing (16S rRNA or metagenomics) reveals associations between microbial composition and metabolic phenotypes, it cannot prove direct mechanistic links. Mice models often fail to fully recapitulate human host-microbe interactions due to interspecies differences and individual variability. Secondly, the factors like age, diet, geography, and genetics lead to substantial baseline variations, making it difficult to define a “healthy” microbiome. Additionally, microbial communities are highly dynamic and sensitive to short-term perturbations (such as antibiotic use or dietary changes), requiring complex longitudinal study designs. Thirdly, a single bacterial species may influence the host through multiple metabolites, and host-microbe co-metabolism is challenging to replicate in vitro. Additionally, the current technology cannot distinguish live/dead bacteria and lacks sensitivity for low-abundance taxa. Finally, clinical translation faces barriers, with current interventions showing inconsistent efficacy and safety risks that need to be carefully evaluated. Thus, prolonged efforts will be required to solve the problems to create the best therapeutic protocols that can bring a new hope for patients with cancer.
This review explains how microbiota regulate the development of malignancy by the metabolism and summarizes the influence of various bacteria-related metabolic reprogramming mechanisms on the biological behavior of cancer. Considering that both metabolic reprogramming and microbiota play a key role in tumor progression and that crosstalk between them has been discovered recently, targeting metabolism and/or microbiota with drugs may be beneficial for controlling tumor development. We believe that the unique metabolic pathways related to the certain microbiota; identifying the crosstalk between them and exploring their mechanisms will advance the field of cancer research.
Author contributions
XL: Data curation, Funding acquisition, Visualization, Writing – original draft. YJ: Data curation, Visualization, Writing – original draft. YL: Funding acquisition, Writing – original draft. HH: Writing – review & editing. SZ: Writing – review & editing. JQ: Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by The National Natural Science Foundation of China, No.82403448, No.82302965 and Medical Science and Technology Research Project of Henan Province, No.SBGJ202403018.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Rodriguez C, Puente-Moncada N, Reiter RJ, Sanchez-Sanchez AM, Herrera F, Rodriguez-Blanco J, et al. Regulation of cancer cell glucose metabolism is determinant for cancer cell fate after melatonin administration. J Cell Physiol. (2021) 236:27–40. doi: 10.1002/jcp.29886
2. Xia L, Oyang L, Lin J, Tan S, Han Y, Wu N, et al. The cancer metabolic reprogramming and immune response. Mol Cancer. (2021) 20:28. doi: 10.1186/s12943-021-01316-8
3. Li Z, Sun C, and Qin Z. Metabolic reprogramming of cancer-associated fibroblasts and its effect on cancer cell reprogramming. Theranostics. (2021) 11:8322–36. doi: 10.7150/thno.62378
4. Faubert B, Solmonson A, and DeBerardinis RJ. Metabolic reprogramming and cancer progression. Science. (2020) 368. doi: 10.1126/science.aaw5473
5. Callao V and Montoya E. Toxohormone-like factor from microorganisms with impaired respiration. Science. (1961) 134:2041–2. doi: 10.1126/science.134.3495.2041
6. Altman BJ, Stine ZE, and Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. (2016) 16:619–34. doi: 10.1038/nrc.2016.71
7. Guerra L, Bonetti L, and Brenner D. Metabolic modulation of immunity: A new concept in cancer immunotherapy. Cell Rep. (2020) 32:107848. doi: 10.1016/j.celrep.2020.107848
8. Yang K, Wang X, Song C, He Z, Wang R, Xu Y, et al. The role of lipid metabolic reprogramming in tumor microenvironment. Theranostics. (2023) 13:1774–808. doi: 10.7150/thno.82920
9. Crosby D, Bhatia S, Brindle KM, Coussens LM, Dive C, Emberton M, et al. Early detection of cancer. Science. (2022) 375:eaay9040. doi: 10.1126/science.aay9040
11. Wang Y, Du J, Wu X, Abdelrehem A, Ren Y, Liu C, et al. Crosstalk between autophagy and microbiota in cancer progression. Mol Cancer. (2021) 20:163. doi: 10.1186/s12943-021-01461-0
12. Urbaniak C, Gloor GB, Brackstone M, Scott L, Tangney M, and Reid G. The microbiota of breast tissue and its association with breast cancer. Appl Environ Microbiol. (2016) 82:5039–48. doi: 10.1128/AEM.01235-16
13. Martins D, Mendes F, and Schmitt F. Microbiome: A supportive or a leading actor in lung cancer? Pathobiology. (2021) 88:198–207. doi: 10.1159/000511556
14. Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, MaChado JC, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. (2018) 67:226–36. doi: 10.1136/gutjnl-2017-314205
15. Sipos A, Ujlaki G, Miko E, Maka E, Szabo J, Uray K, et al. The role of the microbiome in ovarian cancer: mechanistic insights into oncobiosis and to bacterial metabolite signaling. Mol Med. (2021) 27:33. doi: 10.1186/s10020-021-00295-2
16. Xie Y, Xie F, Zhou X, Zhang L, Yang B, Huang J, et al. Microbiota in tumors: from understanding to application. Adv Sci (Weinh). (2022) 9:e2200470. doi: 10.1002/advs.202200470
17. Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. (2003) 16:1–17. doi: 10.1128/CMR.16.1.1-17.2003
18. Collatuzzo G, Pelucchi C, Negri E, Lopez-Carrillo L, Tsugane S, Hidaka A, et al. Exploring the interactions between Helicobacter pylori (Hp) infection and other risk factors of gastric cancer: A pooled analysis in the Stomach cancer Pooling (StoP) Project. Int J Cancer. (2021) 149:1228–38. doi: 10.1002/ijc.33678
20. Coley WB. The Treatment of Inoperable Sarcoma by Bacterial Toxins (the Mixed Toxins of the Streptococcus erysipelas and the Bacillus prodigiosus). Proc R Soc Med. (1910) 3:1–48. doi: 10.1177/003591571000301601
21. Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, and Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. (2013) 8:e70803. doi: 10.1371/journal.pone.0070803
22. Quante M, Bhagat G, Abrams JA, Marache F, Good P, Lee MD, et al. Bile acid and inflammation activate gastric cardia stem cells in a mouse model of Barrett-like metaplasia. Cancer Cell. (2012) 21:36–51. doi: 10.1016/j.ccr.2011.12.004
23. Zhu Z, Cai J, Hou W, Xu K, Wu X, Song Y, et al. Microbiome and spatially resolved metabolomics analysis reveal the anticancer role of gut Akkermansia muciniphila by crosstalk with intratumoral microbiota and reprogramming tumoral metabolism in mice. Gut Microbes. (2023) 15:2166700. doi: 10.1080/19490976.2023.2166700
24. Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. (2020) 369:1481–9. doi: 10.1126/science.abc3421
25. Yang J, Shay C, Saba NF, and Teng Y. Cancer metabolism and carcinogenesis. Exp Hematol Oncol. (2024) 13:10. doi: 10.1186/s40164-024-00482-x
26. Dong S, Liang S, Cheng Z, Zhang X, Luo L, Li L, et al. ROS/PI3K/Akt and Wnt/beta-catenin signalings activate HIF-1alpha-induced metabolic reprogramming to impart 5-fluorouracil resistance in colorectal cancer. J Exp Clin Cancer Res. (2022) 41:15. doi: 10.1186/s13046-021-02229-6
27. An Y and Duan H. The role of m6A RNA methylation in cancer metabolism. Mol Cancer. (2022) 21:14. doi: 10.1186/s12943-022-01500-4
28. Ma M, Zhang C, Cao R, Tang D, Sang X, Zou S, et al. UBE2O promotes lipid metabolic reprogramming and liver cancer progression by mediating HADHA ubiquitination. Oncogene. (2022) 41:5199–213. doi: 10.1038/s41388-022-02509-1
29. Sun T, Liu Z, and Yang Q. The role of ubiquitination and deubiquitination in cancer metabolism. Mol Cancer. (2020) 19:146. doi: 10.1186/s12943-020-01262-x
30. Koundouros N, Karali E, Tripp A, Valle A, Inglese P, Perry NJS, et al. Metabolic fingerprinting links oncogenic PIK3CA with enhanced arachidonic acid-derived eicosanoids. Cell. (2020) 181:1596–1611.e27. doi: 10.1016/j.cell.2020.05.053
31. Carlos-Reyes A, Romero-Garcia S, and Prado-Garcia H. Metabolic responses of lung adenocarcinoma cells to survive under stressful conditions associated with tumor microenvironment. Metabolites. (2024) 14. doi: 10.3390/metabo14020103
32. Guan H, Luo W, Liu Y, and Li M. Novel circular RNA circSLIT2 facilitates the aerobic glycolysis of pancreatic ductal adenocarcinoma via miR-510-5p/c-Myc/LDHA axis. Cell Death Dis. (2021) 12:645. doi: 10.1038/s41419-021-03918-y
33. Wei Z, Xia J, Li J, Cai J, Shan J, Zhang C, et al. SIRT1 promotes glucolipid metabolic conversion to facilitate tumor development in colorectal carcinoma. Int J Biol Sci. (2023) 19:1925–40. doi: 10.7150/ijbs.76704
34. Yu Q, Dai W, Ji J, Wu L, Feng J, Li J, et al. Sodium butyrate inhibits aerobic glycolysis of hepatocellular carcinoma cells via the c-myc/hexokinase 2 pathway. J Cell Mol Med. (2022) 26:3031–45. doi: 10.1111/jcmm.17322
35. Inoue J, Kishikawa M, Tsuda H, Nakajima Y, Asakage T, and Inazawa J. Identification of PDHX as a metabolic target for esophageal squamous cell carcinoma. Cancer Sci. (2021) 112:2792–802. doi: 10.1111/cas.14938
36. Tesfay L, Paul BT, Konstorum A, Deng Z, Cox AO, Lee J, et al. Stearoyl-coA desaturase 1 protects ovarian cancer cells from ferroptotic cell death. Cancer Res. (2019) 79:5355–66. doi: 10.1158/0008-5472.CAN-19-0369
37. Wang Q, Morris RJ, Bode AM, and Zhang T. Prostaglandin pathways: opportunities for cancer prevention and therapy. Cancer Res. (2022) 82:949–65. doi: 10.1158/0008-5472.CAN-21-2297
38. Li X, Chen G, Wang F, Guo X, Zhang R, Liu P, et al. Oncogenic PIK3CA recruits myeloid-derived suppressor cells to shape the immunosuppressive tumour microenvironment in luminal breast cancer through the 5-lipoxygenase-dependent arachidonic acid pathway. Clin Transl Med. (2023) 13:e1483. doi: 10.1002/ctm2.1483
39. Nelson JK, Thin MZ, Evan T, Howell S, Wu M, Almeida B, et al. USP25 promotes pathological HIF-1-driven metabolic reprogramming and is a potential therapeutic target in pancreatic cancer. Nat Commun. (2022) 13:2070. doi: 10.1038/s41467-022-29684-9
40. Sun L, Zhang H, and Gao P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell. (2022) 13:877–919. doi: 10.1007/s13238-021-00846-7
41. Chakraborty S, Balan M, Sabarwal A, Choueiri TK, and Pal S. Metabolic reprogramming in renal cancer: Events of a metabolic disease. Biochim Biophys Acta Rev Cancer. (2021) 1876:188559. doi: 10.1016/j.bbcan.2021.188559
42. Wong-Rolle A, Wei HK, Zhao C, and Jin C. Unexpected guests in the tumor microenvironment: microbiome in cancer. Protein Cell. (2021) 12:426–35. doi: 10.1007/s13238-020-00813-8
43. Shiao SL, Kershaw KM, Limon JJ, You S, Yoon J, Ko EY, et al. Commensal bacteria and fungi differentially regulate tumor responses to radiation therapy. Cancer Cell. (2021) 39:1202–1213.e6. doi: 10.1016/j.ccell.2021.07.002
44. Kalaora S, Nagler A, Nejman D, Alon M, Barbolin C, Barnea E, et al. Identification of bacteria-derived HLA-bound peptides in melanoma. Nature. (2021) 592:138–43. doi: 10.1038/s41586-021-03368-8
45. Alam A, Levanduski E, Denz P, Villavicencio HS, Bhatta M, Alhorebi L, et al. Fungal mycobiome drives IL-33 secretion and type 2 immunity in pancreatic cancer. Cancer Cell. (2022) 40:153–167.e11. doi: 10.1016/j.ccell.2022.01.003
46. Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, and Knight R. The microbiome and human cancer. Science. (2021) 371. doi: 10.1126/science.abc4552
47. Spiljar M, Merkler D, and Trajkovski M. The immune system bridges the gut microbiota with systemic energy homeostasis: focus on TLRs, mucosal barrier, and SCFAs. Front Immunol. (2017) 8:1353. doi: 10.3389/fimmu.2017.01353
48. Sharma VR, Singh M, Kumar V, Yadav M, Sehrawat N, Sharma DK, et al. Microbiome dysbiosis in cancer: Exploring therapeutic strategies to counter the disease. Semin Cancer Biol. (2021) 70:61–70. doi: 10.1016/j.semcancer.2020.07.006
49. Shi Y, Zheng W, Yang K, Harris KG, Ni K, Xue L, et al. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J Exp Med. (2020) 217. doi: 10.1084/jem.20192282
50. Sun J, Tang Q, Yu S, Xie M, Xie Y, Chen G, et al. Role of the oral microbiota in cancer evolution and progression. Cancer Med. (2020) 9:6306–21. doi: 10.1002/cam4.3206
51. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, and Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. (2013) 14:195–206. doi: 10.1016/j.chom.2013.07.012
52. Hoste E, Arwert EN, Lal R, South AP, Salas-Alanis JC, Murrell DF, et al. Innate sensing of microbial products promotes wound-induced skin cancer. Nat Commun. (2015) 6:5932. doi: 10.1038/ncomms6932
53. Kong C, Yan X, Zhu Y, Zhu H, Luo Y, Liu P, et al. Fusobacterium nucleatum promotes the development of colorectal cancer by activating a cytochrome P450/epoxyoctadecenoic acid axis via TLR4/keap1/NRF2 signaling. Cancer Res. (2021) 81:4485–98. doi: 10.1158/0008-5472.CAN-21-0453
54. Chen XH, Wang A, Chu AN, Gong YH, and Yuan Y. Mucosa-associated microbiota in gastric cancer tissues compared with non-cancer tissues. Front Microbiol. (2019) 10:1261. doi: 10.3389/fmicb.2019.01261
55. Shi L, Fan Q, Zhou B, Wu J, Jin M, Yu D, et al. The composition and functional profile of the microbial communities in human gastric cancer tissues and adjacent normal tissues. Acta Biochim Biophys Sin (Shanghai). (2022) 54:47–54. doi: 10.3724/abbs.2021010
56. Aykut B, Pushalkar S, Chen R, Li Q, Abengozar R, Kim JI, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. (2019) 574:264–7. doi: 10.1038/s41586-019-1608-2
57. Tan Q, Ma X, Yang B, Liu Y, Xie Y, Wang X, et al. Periodontitis pathogen Porphyromonas gingivalis promotes pancreatic tumorigenesis via neutrophil elastase from tumor-associated neutrophils. Gut Microbes. (2022) 14:2073785. doi: 10.1080/19490976.2022.2073785
58. Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. (2020) 368:973–80. doi: 10.1126/science.aay9189
59. Poore GD, Kopylova E, Zhu Q, Carpenter C, Fraraccio S, Wandro S, et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. (2020) 579:567–74. doi: 10.1038/s41586-020-2095-1
60. Gagliani N, Hu B, Huber S, Elinav E, and Flavell RA. The fire within: microbes inflame tumors. Cell. (2014) 157:776–83. doi: 10.1016/j.cell.2014.03.006
61. Cao Y, Xia H, Tan X, Shi C, Ma Y, Meng D, et al. Intratumoural microbiota: a new frontier in cancer development and therapy. Signal Transduct Target Ther. (2024) 9:15. doi: 10.1038/s41392-023-01693-0
62. Tzeng A, Sangwan N, Jia M, Liu CC, Keslar KS, Downs-Kelly E, et al. Human breast microbiome correlates with prognostic features and immunological signatures in breast cancer. Genome Med. (2021) 13:60. doi: 10.1186/s13073-021-00874-2
63. Parida S, Wu S, Siddharth S, Wang G, Muniraj N, Nagalingam A, et al. A procarcinogenic colon microbe promotes breast tumorigenesis and metastatic progression and concomitantly activates notch and beta-catenin axes. Cancer Discov. (2021) 11:1138–57. doi: 10.1158/2159-8290.CD-20-0537
64. Ma J, Sun L, Liu Y, Ren H, Shen Y, Bi F, et al. Alter between gut bacteria and blood metabolites and the anti-tumor effects of Faecalibacterium prausnitzii in breast cancer. BMC Microbiol. (2020) 20:82. doi: 10.1186/s12866-020-01739-1
65. Chen B, Liu X, Yu P, Xie F, Kwan JSH, Chan WN, et al. pylori-induced NF-kappaB-PIEZO1-YAP1-CTGF axis drives gastric cancer progression and cancer-associated fibroblast-mediated tumour microenvironment remodelling. Clin Transl Med. (2023) 13:e1481. doi: 10.1002/ctm2.1481
66. Wang Q, Zhao L, Han L, Fu G, Tuo X, Ma S, et al. The differential distribution of bacteria between cancerous and noncancerous ovarian tissues in situ. J Ovarian Res. (2020). doi: 10.1186/s13048-019-0603-4
67. Laniewski P, Ilhan ZE, and Herbst-Kralovetz MM. The microbiome and gynaecological cancer development, prevention and therapy. Nat Rev Urol. (2020) 17:232–50. doi: 10.1038/s41585-020-0286-z
68. Ye L, Wu BS, Xu ZA, Ji XF, Guan L, Li PP, et al. Evidence for an intra-tumoral microbiome in pituitary neuroendocrine tumors with different clinical phenotypes. J Neurooncol. (2023) 163:133–42. doi: 10.1007/s11060-023-04318-2
69. Lin Y, Xie M, Lau HC, Zeng R, Zhang R, Wang L, et al. Effects of gut microbiota on immune checkpoint inhibitors in multi-cancer and as microbial biomarkers for predicting therapeutic response. Med. (2025) 6:100530. doi: 10.1016/j.medj.2024.10.007
70. Guo X and Shao Y. Role of the oral-gut microbiota axis in pancreatic cancer: a new perspective on tumor pathophysiology, diagnosis, and treatment. Mol Med. (2025) 31:103. doi: 10.1186/s10020-025-01166-w
71. Chang Y, Long M, Shan H, Liu L, Zhong S, and Luo JL. Combining gut microbiota modulation and immunotherapy: A promising approach for treating microsatellite stable colorectal cancer. Crit Rev Oncol Hematol. (2025) 208:104629. doi: 10.1016/j.critrevonc.2025.104629
72. Yang X, Zhang Z, Shen X, Xu J, Weng Y, Wang W, et al. Clostridium butyricum and its metabolite butyrate promote ferroptosis susceptibility in pancreatic ductal adenocarcinoma. Cell Oncol (Dordr). (2023) 46:1645–58. doi: 10.1007/s13402-023-00831-8
73. Shin JH, Tillotson G, MacKenzie TN, Warren CA, Wexler HM, and Goldstein EJC. Bacteroides and related species: The keystone taxa of the human gut microbiota. Anaerobe. (2024) 85:102819. doi: 10.1016/j.anaerobe.2024.102819
74. Baek MK, Park JS, Park JH, Kim MH, Kim HD, Bae WK, et al. Lithocholic acid upregulates uPAR and cell invasiveness via MAPK and AP-1 signaling in colon cancer cells. Cancer Lett. (2010) 290:123–8. doi: 10.1016/j.canlet.2009.08.030
75. Montgomery TL, Eckstrom K, Lile KH, Caldwell S, Heney ER, Lahue KG, et al. Lactobacillus reuteri tryptophan metabolism promotes host susceptibility to CNS autoimmunity. Microbiome. (2022) 10:198. doi: 10.1186/s40168-022-01408-7
76. Li Y, Li Q, Yuan R, Wang Y, Guo C, and Wang L. Bifidobacterium breve-derived indole-3-lactic acid ameliorates colitis-associated tumorigenesis by directing the differentiation of immature colonic macrophages. Theranostics. (2024) 14:2719–35. doi: 10.7150/thno.92350
77. Giesbrecht K, Former S, Sahr A, Heeg K, and Hildebrand D. Streptococcal pyrogenic exotoxin A-stimulated monocytes mediate regulatory T-cell accumulation through PD-L1 and kynurenine. Int J Mol Sci. (2019) 20. doi: 10.3390/ijms20163933
78. Fong W, Li Q, Ji F, Liang W, Lau HCH, Kang X, et al. Lactobacillus gallinarum-derived metabolites boost anti-PD1 efficacy in colorectal cancer by inhibiting regulatory T cells through modulating IDO1/Kyn/AHR axis. Gut. (2023) 72:2272–85. doi: 10.1136/gutjnl-2023-329543
79. Zhang Q, Zhao Q, Li T, Lu L, Wang F, Zhang H, et al. Lactobacillus plantarum-derived indole-3-lactic acid ameliorates colorectal tumorigenesis via epigenetic regulation of CD8(+) T cell immunity. Cell Metab. (2023) 35:943–960.e9. doi: 10.1016/j.cmet.2023.04.015
80. Zhao Y, Zhang J, Cheng ASL, Yu J, To KF, and Kang W. Gastric cancer: genome damaged by bugs. Oncogene. (2020) 39:3427–42. doi: 10.1038/s41388-020-1241-4
81. Liu Z, Zhang D, and Chen S. Unveiling the gastric microbiota: implications for gastric carcinogenesis, immune responses, and clinical prospects. J Exp Clin Cancer Res. (2024) 43:118. doi: 10.1186/s13046-024-03034-7
82. Dai D, Yang Y, Yu J, Dang T, Qin W, Teng L, et al. Interactions between gastric microbiota and metabolites in gastric cancer. Cell Death Dis. (2021) 12:1104. doi: 10.1038/s41419-021-04396-y
83. Visconti A, Le Roy CI, Rosa F, Rossi N, Martin TC, Mohney RP, et al. Interplay between the human gut microbiome and host metabolism. Nat Commun. (2019) 10:4505. doi: 10.1038/s41467-019-12476-z
84. Chu F, Li Y, Meng X, Li Y, Li T, Zhai M, et al. Gut microbial dysbiosis and changes in fecal metabolic phenotype in precancerous lesions of gastric cancer induced with N-methyl-N’-nitro-N-nitrosoguanidine, sodium salicylate, ranitidine, and irregular diet. Front Physiol. (2021) 12:733979. doi: 10.3389/fphys.2021.733979
85. Wang L, Zhou J, Xin Y, Geng C, Tian Z, Yu X, et al. Bacterial overgrowth and diversification of microbiota in gastric cancer. Eur J Gastroenterol Hepatol. (2016) 28:261–6. doi: 10.1097/MEG.0000000000000542
86. Yang Y, Dai D, Jin W, Huang Y, Zhang Y, Chen Y, et al. Microbiota and metabolites alterations in proximal and distal gastric cancer patients. J Transl Med. (2022) 20:439. doi: 10.1186/s12967-022-03650-x
87. Wong SH and Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. (2019) 16:690–704. doi: 10.1038/s41575-019-0209-8
88. Krautkramer KA, Fan J, and Backhed F. Gut microbial metabolites as multi-kingdom intermediates. Nat Rev Microbiol. (2021) 19:77–94. doi: 10.1038/s41579-020-0438-4
89. Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. (2017) 551:648–52. doi: 10.1038/nature24661
90. Qu R, Zhang Y, Ma Y, Zhou X, Sun L, Jiang C, et al. Role of the gut microbiota and its metabolites in tumorigenesis or development of colorectal cancer. Adv Sci (Weinh). (2023) 10:e2205563. doi: 10.1002/advs.202205563
91. Coker OO, Liu C, Wu WKK, Wong SH, Jia W, Sung JJY, et al. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome. (2022) 10:35. doi: 10.1186/s40168-021-01208-5
92. Cai J, Sun L, and Gonzalez FJ. Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe. (2022) 30:289–300. doi: 10.1016/j.chom.2022.02.004
93. Koh A, De Vadder F, Kovatcheva-Datchary P, and Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. (2016) 165:1332–45. doi: 10.1016/j.cell.2016.05.041
94. Li Q, Hu W, Liu WX, Zhao LY, Huang D, Liu XD, et al. Streptococcus thermophilus Inhibits Colorectal Tumorigenesis Through Secreting beta-Galactosidase. Gastroenterology. (2021) 160. doi: 10.1053/j.gastro.2020.09.003
95. Kong C, Liang L, Liu G, Du L, Yang Y, Liu J, et al. Integrated metagenomic and metabolomic analysis reveals distinct gut-microbiome-derived phenotypes in early-onset colorectal cancer. Gut. (2023) 72:1129–42. doi: 10.1136/gutjnl-2022-327156
96. Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. (2019) 25:968–76. doi: 10.1038/s41591-019-0458-7
97. Yang J, Wei H, Zhou Y, Szeto CH, Li C, Lin Y, et al. High-fat diet promotes colorectal tumorigenesis through modulating gut microbiota and metabolites. Gastroenterology. (2022) 162:135–149.e2. doi: 10.1053/j.gastro.2021.08.041
98. Bai X, Wei H, Liu W, Coker OO, Gou H, Liu C, et al. Cigarette smoke promotes colorectal cancer through modulation of gut microbiota and related metabolites. Gut. (2022) 71:2439–50. doi: 10.1136/gutjnl-2021-325021
99. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2024) 74:229–63. doi: 10.3322/caac.21834
100. Nandi D, Parida S, and Sharma D. The gut microbiota in breast cancer development and treatment: The good, the bad, and the useful! Gut Microbes. (2023) 15:2221452. doi: 10.1080/19490976.2023.2221452
101. Hieken TJ, Chen J, Hoskin TL, Walther-Antonio M, Johnson S, Ramaker S, et al. The microbiome of aseptically collected human breast tissue in benign and Malignant disease. Sci Rep. (2016) 6:30751. doi: 10.1038/srep30751
102. Wang Y, Qu D, Zhang Y, Jin Y, Feng Y, Zhang H, et al. Intra-tumoral microbial community profiling and associated metabolites alterations of TNBC. Front Oncol. (2023) 13:1143163. doi: 10.3389/fonc.2023.1143163
103. Wang H, Rong X, Zhao G, Zhou Y, Xiao Y, Ma D, et al. The microbial metabolite trimethylamine N-oxide promotes antitumor immunity in triple-negative breast cancer. Cell Metab. (2022) 34:581–594.e8. doi: 10.1016/j.cmet.2022.02.010
104. Heath H, Mogol AN, Santaliz Casiano A, Zuo Q, and Madak-Erdogan Z. Targeting systemic and gut microbial metabolism in ER(+) breast cancer. Trends Endocrinol Metab. (2024) 35:321–30. doi: 10.1016/j.tem.2023.12.008
105. Yang P, Wang Z, Peng Q, Lian W, and Chen D. Comparison of the gut microbiota in patients with benign and Malignant breast tumors: A pilot study. Evol Bioinform Online. (2021) 17:11769343211057573. doi: 10.1177/11769343211057573
106. Chen F, Yang J, Guo Y, Su D, Sheng Y, and Wu Y. Integrating bulk and single-cell RNA sequencing data reveals the relationship between intratumor microbiome signature and host metabolic heterogeneity in breast cancer. Front Immunol. (2023) 14:1140995. doi: 10.3389/fimmu.2023.1140995
107. Sommariva M, Le Noci V, Bianchi F, Camelliti S, Balsari A, Tagliabue E, et al. The lung microbiota: role in maintaining pulmonary immune homeostasis and its implications in cancer development and therapy. Cell Mol Life Sci. (2020) 77:2739–49. doi: 10.1007/s00018-020-03452-8
108. Li Y, Xiao X, Han Y, Gorlova O, Qian D, Leighl N, et al. Genome-wide interaction study of smoking behavior and non-small cell lung cancer risk in Caucasian population. Carcinogenesis. (2018) 39:336–46. doi: 10.1093/carcin/bgx113
109. Greathouse KL, White JR, Vargas AJ, Bliskovsky VV, Beck JA, von Muhlinen N, et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. (2018) 19:123. doi: 10.1186/s13059-018-1501-6
110. Ma Y, Chen H, Li H, Zheng M, Zuo X, Wang W, et al. Intratumor microbiome-derived butyrate promotes lung cancer metastasis. Cell Rep Med. (2024) 5:101488. doi: 10.1016/j.xcrm.2024.101488
111. Colbert LE, El Alam MB, Wang R, Karpinets T, Lo D, Lynn EJ, et al. Tumor-resident Lactobacillus iners confer chemoradiation resistance through lactate-induced metabolic rewiring. Cancer Cell. (2023) 41:1945–1962.e11. doi: 10.1016/j.ccell.2023.09.012
112. Luan J, Zhang F, Suo L, Zhang W, Li Y, Yu X, et al. Analyzing lung cancer risks in patients with impaired pulmonary function through characterization of gut microbiome and metabolites. BMC Pulm Med. (2024) 24:1. doi: 10.1186/s12890-023-02825-6
113. Liu F, Li J, Guan Y, Lou Y, Chen H, Xu M, et al. Dysbiosis of the gut microbiome is associated with tumor biomarkers in lung cancer. Int J Biol Sci. (2019) 15:2381–92. doi: 10.7150/ijbs.35980
114. Miao S and Qiu H. The microbiome in the pathogenesis of lung cancer: The role of microbiome in lung cancer pathogenesis. APMIS. (2024) 132:68–80. doi: 10.1111/apm.13359
115. Zhang L, Zheng J, Ahmed R, Huang G, Reid J, Mandal R, et al. A high-performing plasma metabolite panel for early-stage lung cancer detection. Cancers (Basel). (2020) 12. doi: 10.3390/cancers12030622
116. Wang S, Chen H, Yang H, Zhou K, Bai F, Wu X, et al. Gut microbiome was highly related to the regulation of metabolism in lung adenocarcinoma patients. Front Oncol. (2022) 12:790467. doi: 10.3389/fonc.2022.790467
117. Klein AP. Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat Rev Gastroenterol Hepatol. (2021) 18:493–502. doi: 10.1038/s41575-021-00457-x
118. Tintelnot J, Xu Y, Lesker TR, Schonlein M, Konczalla L, Giannou AD, et al. Microbiota-derived 3-IAA influences chemotherapy efficacy in pancreatic cancer. Nature. (2023) 615:168–74. doi: 10.1038/s41586-023-05728-y
119. Hezaveh K, Shinde RS, Klotgen A, Halaby MJ, Lamorte S, Ciudad MT, et al. Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity. Immunity. (2022) 55:324–340.e8. doi: 10.1016/j.immuni.2022.01.006
120. Liu H, Li X, Zhu Y, Huang Y, Zhang Q, Lin S, et al. Effect of plant-derived n-3 polyunsaturated fatty acids on blood lipids and gut microbiota: A double-blind randomized controlled trial. Front Nutr. (2022) 9:830960. doi: 10.3389/fnut.2022.830960
121. Liu B, Wang F, Chen L, Xin Y, Liu L, Wu D, et al. Effects of high-fat diet on carcinogen-induced pancreatic cancer and intestinal microbiota in C57BL/6 wild-type mice. Pancreas. (2021) 50:564–70. doi: 10.1097/MPA.0000000000001797
122. Ruze R, Song J, Yin X, Chen Y, Xu R, Wang C, et al. Mechanisms of obesity- and diabetes mellitus-related pancreatic carcinogenesis: a comprehensive and systematic review. Signal Transduct Target Ther. (2023) 8:139. doi: 10.1038/s41392-023-01376-w
123. Wang H, Funchain P, Bebek G, Altemus J, Zhang H, Niazi F, et al. Microbiomic differences in tumor and paired-normal tissue in head and neck squamous cell carcinomas. Genome Med. (2017) 9:14. doi: 10.1186/s13073-017-0405-5
124. Sun J, Tang Q, Yu S, Xie M, Zheng W, Chen G, et al. nucleatum facilitates oral squamous cell carcinoma progression via GLUT1-driven lactate production. EBioMedicine. (2023) 88:104444. doi: 10.1016/j.ebiom.2023.104444
125. Yost S, Stashenko P, Choi Y, Kukuruzinska M, Genco CA, Salama A, et al. Increased virulence of the oral microbiome in oral squamous cell carcinoma revealed by metatranscriptome analyses. Int J Sci. (2018) 10:32. doi: 10.1038/s41368-018-0037-7
126. Shen J, Sun H, Chu J, Gong X, and Liu X. Cervicovaginal microbiota: a promising direction for prevention and treatment in cervical cancer. Infect Agent Cancer. (2024) 19:13. doi: 10.1186/s13027-024-00573-8
127. Ilhan ZE, Laniewski P, Thomas N, Roe DJ, Chase DM, and Herbst-Kralovetz MM. Deciphering the complex interplay between microbiota, HPV, inflammation and cancer through cervicovaginal metabolic profiling. EBioMedicine. (2019) 44:675–90. doi: 10.1016/j.ebiom.2019.04.028
128. Gao M, Wu J, Zhou S, Chen Y, Wang M, He W, et al. Combining fecal microbiome and metabolomics reveals diagnostic biomarkers for esophageal squamous cell carcinoma. Microbiol Spectr. (2024) 12:e0401223. doi: 10.1128/spectrum.04012-23
129. Cheung MK, Yue GGL, Lauw S, Li CSY, Yung MY, Ng SC, et al. Alterations in gut microbiota of esophageal squamous cell carcinoma patients. J Gastroenterol Hepatol. (2022) 37:1919–27. doi: 10.1111/jgh.15941
130. Lau HC, Zhang X, Ji F, Lin Y, Liang W, Li Q, et al. Lactobacillus acidophilus suppresses non-alcoholic fatty liver disease-associated hepatocellular carcinoma through producing valeric acid. EBioMedicine. (2024) 100:104952. doi: 10.1016/j.ebiom.2023.104952
131. Diamond LK. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N Engl J Med. (1948) 238:787–93. doi: 10.1056/NEJM194806032382301
132. Xiao Y, Yu TJ, Xu Y, Ding R, Wang YP, Jiang YZ, et al. Emerging therapies in cancer metabolism. Cell Metab. (2023) 35:1283–303. doi: 10.1016/j.cmet.2023.07.006
133. Mullen NJ and Singh PK. Nucleotide metabolism: a pan-cancer metabolic dependency. Nat Rev Cancer. (2023) 23:275–94. doi: 10.1038/s41568-023-00557-7
134. Yu Z, Zhou X, and Wang X. Metabolic reprogramming in hematologic Malignancies: advances and clinical perspectives. Cancer Res. (2022) 82:2955–63. doi: 10.1158/0008-5472.CAN-22-0917
135. Huff SE, Winter JM, and Dealwis CG. Inhibitors of the cancer target ribonucleotide reductase, past and present. Biomolecules. (2022) 12. doi: 10.3390/biom12060815
136. Jin R, Liu B, Liu X, Fan Y, Peng W, Huang C, et al. Leflunomide suppresses the growth of LKB1-inactivated tumors in the immune-competent host and attenuates distant cancer metastasis. Mol Cancer Ther. (2021) 20:274–83. doi: 10.1158/1535-7163.MCT-20-0567
137. Shi DD, Savani MR, Levitt MM, Wang AC, Endress JE, Bird CE, et al. De novo pyrimidine synthesis is a targetable vulnerability in IDH mutant glioma. Cancer Cell. (2022) 40:939–956.e16. doi: 10.1016/j.ccell.2022.07.011
138. Pal S, Kaplan JP, Nguyen H, Stopka SA, Savani MR, Regan MS, et al. A druggable addiction to de novo pyrimidine biosynthesis in diffuse midline glioma. Cancer Cell. (2022) 40:957–972.e10. doi: 10.1016/j.ccell.2022.07.012
139. McDonald G, Chubukov V, Coco J, Truskowski K, Narayanaswamy R, Choe S, et al. Selective vulnerability to pyrimidine starvation in hematologic Malignancies revealed by AG-636, a novel clinical-stage inhibitor of dihydroorotate dehydrogenase. Mol Cancer Ther. (2020) 19:2502–15. doi: 10.1158/1535-7163.MCT-20-0550
140. DeBerardinis RJ and Chandel NS. Fundamentals of cancer metabolism. Sci Adv. (2016) 2:e1600200. doi: 10.1126/sciadv.1600200
141. Chen Y, Wang B, Zhao Y, Shao X, Wang M, Ma F, et al. Metabolomic machine learning predictor for diagnosis and prognosis of gastric cancer. Nat Commun. (2024) 15:1657. doi: 10.1038/s41467-024-46043-y
142. Tran TQ, Hanse EA, Habowski AN, Li H, Ishak Gabra MB, Yang Y, et al. alpha-Ketoglutarate attenuates Wnt signaling and drives differentiation in colorectal cancer. Nat Cancer. (2020) 1:345–58. doi: 10.1038/s43018-020-0035-5
143. Li B, Li Y, Zhou H, Xu Y, Cao Y, Cheng C, et al. Multiomics identifies metabolic subtypes based on fatty acid degradation allocating personalized treatment in hepatocellular carcinoma. Hepatology. (2024) 79:289–306. doi: 10.1097/HEP.0000000000000553
144. Yang WH, Qiu Y, Stamatatos O, Janowitz T, and Lukey MJ. Enhancing the efficacy of glutamine metabolism inhibitors in cancer therapy. Trends Cancer. (2021) 7:790–804. doi: 10.1016/j.trecan.2021.04.003
145. Han X, Wang D, Yang L, Wang N, Shen J, Wang J, et al. Activation of polyamine catabolism promotes glutamine metabolism and creates a targetable vulnerability in lung cancer. Proc Natl Acad Sci U S A. (2024) 121:e2319429121. doi: 10.1073/pnas.2319429121
146. Yap TA, Daver N, Mahendra M, Zhang J, Kamiya-Matsuoka C, Meric-Bernstam F, et al. Complex I inhibitor of oxidative phosphorylation in advanced solid tumors and acute myeloid leukemia: phase I trials. Nat Med. (2023) 29:115–26. doi: 10.1038/s41591-022-02103-8
147. Bridges HR, Blaza JN, Yin Z, Chung I, Pollak MN, and Hirst J. Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides. Science. (2023) 379:351–7. doi: 10.1126/science.ade3332
148. Stine ZE, Schug ZT, Salvino JM, and Dang CV. Targeting cancer metabolism in the era of precision oncology. Nat Rev Drug Discov. (2022) 21:141–62. doi: 10.1038/s41573-021-00339-6
149. Gao W, Wang X, Zhou Y, Wang X, and Yu Y. Autophagy, ferroptosis, pyroptosis, and necroptosis in tumor immunotherapy. Signal Transduct Target Ther. (2022) 7:196. doi: 10.1038/s41392-022-01046-3
150. Dussold C, Zilinger K, Turunen J, Heimberger AB, and Miska J. Modulation of macrophage metabolism as an emerging immunotherapy strategy for cancer. J Clin Invest. (2024) 134. doi: 10.1172/JCI175445
151. Geiger R, Rieckmann JC, Wolf T, Basso C, Feng Y, Fuhrer T, et al. L-arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell. (2016) 167:829–842.e13. doi: 10.1016/j.cell.2016.09.031
152. Kay EJ, Paterson K, Riera-Domingo C, Sumpton D, Dabritz JHM, Tardito S, et al. Cancer-associated fibroblasts require proline synthesis by PYCR1 for the deposition of pro-tumorigenic extracellular matrix. Nat Metab. (2022) 4:693–710. doi: 10.1038/s42255-022-00582-0
153. Ligibel JA, Bohlke K, May AM, Clinton SK, Demark-Wahnefried W, Gilchrist SC, et al. Exercise, diet, and weight management during cancer treatment: ASCO guideline. J Clin Oncol. (2022) 40:2491–507. doi: 10.1200/JCO.22.00687
154. Wang Q and Zhou W. Roles and molecular mechanisms of physical exercise in cancer prevention and treatment. J Sport Health Sci. (2021) 10:201–10. doi: 10.1016/j.jshs.2020.07.008
155. Fu A, Yao B, Dong T, Chen Y, Yao J, Liu Y, et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell. (2022) 185:1356–1372.e26. doi: 10.1016/j.cell.2022.02.027
156. Moss SF, Shah SC, Tan MC, and El-Serag HB. Evolving concepts in helicobacter pylori management. Gastroenterology. (2024) 166:267–83. doi: 10.1053/j.gastro.2023.09.047
157. Mohindroo C, Hasanov M, Rogers JE, Dong W, Prakash LR, Baydogan S, et al. Antibiotic use influences outcomes in advanced pancreatic adenocarcinoma patients. Cancer Med. (2021) 10:5041–50. doi: 10.1002/cam4.3870
158. Bernardo G, Le Noci V, Ottaviano E, De Cecco L, Camisaschi C, Guglielmetti S, et al. Reduction of Staphylococcus epidermidis in the mammary tumor microbiota induces antitumor immunity and decreases breast cancer aggressiveness. Cancer Lett. (2023) 555:216041. doi: 10.1016/j.canlet.2022.216041
159. Chen P, Hu T, Jiang H, Li B, Li G, Ran P, et al. Chronic exposure to ampicillin alters lung microbial composition in laboratory rat. Exp Lung Res. (2023) 49:116–30. doi: 10.1080/01902148.2023.2219790
160. Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. (2017) 358:1443–8. doi: 10.1126/science.aal5240
161. Lurienne L, Cervesi J, Duhalde L, de Gunzburg J, Andremont A, Zalcman G, et al. NSCLC Immunotherapy Efficacy and antibiotic use: A systematic review and meta-analysis. J Thorac Oncol. (2020) 15:1147–59. doi: 10.1016/j.jtho.2020.03.002
162. Stein-Thoeringer CK, Saini NY, Zamir E, Blumenberg V, Schubert ML, Mor U, et al. A non-antibiotic-disrupted gut microbiome is associated with clinical responses to CD19-CAR-T cell cancer immunotherapy. Nat Med. (2023) 29:906–16. doi: 10.1038/s41591-023-02234-6
163. Ortiz-Bonilla CJ, Uccello TP, Gerber SA, Lord EM, Messing EM, and Lee YF. Bladder cancer extracellular vesicles elicit a CD8 T cell-mediated antitumor immunity. Int J Mol Sci. (2022) 23. doi: 10.3390/ijms23062904
164. Morales A, Eidinger D, and Bruce AW. Intracavitary Bacillus Calmette-Guerin in the treatment of superficial bladder tumors. J Urol. (1976) 116:180–3. doi: 10.1016/s0022-5347(17)58737-6
165. Pettenati C and Ingersoll MA. Mechanisms of BCG immunotherapy and its outlook for bladder cancer. Nat Rev Urol. (2018) 15:615–25. doi: 10.1038/s41585-018-0055-4
166. Routy B, Jackson T, Mahlmann L, Baumgartner CK, Blaser M, Byrd A, et al. Melanoma and microbiota: Current understanding and future directions. Cancer Cell. (2024) 42:16–34. doi: 10.1016/j.ccell.2023.12.003
167. Ikryannikova LN, Gorokhovets NV, Belykh DA, Kurbatov LK, and Zamyatnin AA Jr. Bacterial therapy of cancer: A way to the dustbin of history or to the medicine of the future? Int J Mol Sci. (2023) 24. doi: 10.3390/ijms24119726
168. Zheng JH, Nguyen VH, Jiang SN, Park SH, Tan W, Hong SH, et al. Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci Transl Med. (2017) 9. doi: 10.1126/scitranslmed.aak9537
169. Guo Y, Chen Y, Liu X, Min JJ, Tan W, and Zheng JH. Targeted cancer immunotherapy with genetically engineered oncolytic Salmonella typhimurium. Cancer Lett. (2020) 469:102–10. doi: 10.1016/j.canlet.2019.10.033
170. Wu H, Zhong D, Zhang Z, Wu Y, Li Y, Mao H, et al. A bacteria-inspired morphology genetic biomedical material: self-propelled artificial microbots for metastatic triple negative breast cancer treatment. ACS Nano. (2021) 15:4845–60. doi: 10.1021/acsnano.0c09594
171. Canale FP, Basso C, Antonini G, Perotti M, Li N, Sokolovska A, et al. Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature. (2021) 598:662–6. doi: 10.1038/s41586-021-04003-2
172. Yang Q, Wang B, Zheng Q, Li H, Meng X, Zhou F, et al. A review of gut microbiota-derived metabolites in tumor progression and cancer therapy. Adv Sci (Weinh). (2023) 10:e2207366. doi: 10.1002/advs.202207366
173. Marti i Lindez AA, Dunand-Sauthier I, Conti M, Gobet F, Nunez N, Hannich JT, et al. Mitochondrial arginase-2 is a cell−autonomous regulator of CD8+ T cell function and antitumor efficacy. JCI Insight. (2019) 4. doi: 10.1172/jci.insight.132975
174. Li T and Wang W. New strategy for cancer immunotherapy: using live engineered bacteria for metabolic modulation. Signal Transduct Target Ther. (2021) 6:415. doi: 10.1038/s41392-021-00829-4
175. He Y, Fu L, Li Y, Wang W, Gong M, Zhang J, et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8(+) T cell immunity. Cell Metab. (2021) 33:988–1000.e7. doi: 10.1016/j.cmet.2021.03.002
176. Gou H, Su H, Liu D, Wong CC, Shang H, Fang Y, et al. Traditional medicine pien tze huang suppresses colorectal tumorigenesis through restoring gut microbiota and metabolites. Gastroenterology. (2023) 165:1404–19. doi: 10.1053/j.gastro.2023.08.052
177. Kazmierczak-Siedlecka K, Skonieczna-Zydecka K, Hupp T, Duchnowska R, Marek-Trzonkowska N, and Polom K. Next-generation probiotics - do they open new therapeutic strategies for cancer patients? Gut Microbes. (2022) 14:2035659. doi: 10.1080/19490976.2022.2035659
178. Rizvi ZA, Dalal R, Sadhu S, Kumar Y, Kumar S, Gupta SK, et al. High-salt diet mediates interplay between NK cells and gut microbiota to induce potent tumor immunity. Sci Adv. (2021) 7:eabg5016. doi: 10.1126/sciadv.abg5016
179. Zheng DW, Dong X, Pan P, Chen KW, Fan JX, Cheng SX, et al. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat BioMed Eng. (2019) 3:717–28. doi: 10.1038/s41551-019-0423-2
Keywords: anti-tumor therapy, metabolic reprogramming, microbiota, tumor microenvironment, cancer progression
Citation: Li X, Jia Y, Li Y, Hei H, Zhang S and Qin J (2025) Crosstalk between metabolic reprogramming and microbiota: implications for cancer progression and novel therapeutic opportunities. Front. Immunol. 16:1582166. doi: 10.3389/fimmu.2025.1582166
Received: 24 February 2025; Accepted: 30 April 2025;
Published: 20 May 2025.
Edited by:
Kai Li, Sichuan University, ChinaReviewed by:
Udayan Bhattacharya, NewYork-Presbyterian, United StatesYuanyuan Cui, Henan University, China
Copyright © 2025 Li, Jia, Li, Hei, Zhang and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianwu Qin, emx5eXFpbmppYW53dTAzOTdAenp1LmVkdS5jbg==; Songtao Zhang, emhzdDE0M0AxMjYuY29t; Hu Hei, dGlnZXJibGFja0AxMjYuY29t
†These authors have contributed equally to this work
 Xingchen Li
Xingchen Li Yidi Jia2†
Yidi Jia2† Jianwu Qin
Jianwu Qin