- 1Department of Orthopedics, Zibo Central Hospital, Zibo, China
- 2Department of Cardiology, Zibo Central Hospital, Zibo, China
- 3Department of Infectious Diseases, Zibo Central Hospital, Zibo, China
1 Introduction: the need for a new paradigm in drug efficacy evaluation
The evaluation of drug efficacy remains a central pillar in the development and clinical application of immunotherapies. Traditionally, therapeutic success has been assessed using static and endpoint-based biomarkers, such as tumor size reduction, survival extension, or changes in a limited set of immune markers (e.g., PD-L1 expression or circulating cytokine levels) (1–3). While these metrics have guided many clinical decisions, they often fail to capture the full complexity and heterogeneity of dynamic immune responses, particularly in the context of immuno-oncology, where therapeutic effects can be delayed, indirect, or spatially restricted (4).
Recent advances in immunotherapy, including immune checkpoint inhibitors, CAR-T cells, and tumor vaccines, demand a new framework for evaluating drug efficacy—one that accounts for the spatiotemporal dynamics of immune responses (5, 6). Immune cells may transiently infiltrate tumors, reorganize spatially, or engage in local interactions that are critical for therapeutic outcomes but remain undetectable using conventional assays. Conventional assays typically employed for immune response assessment include IHC, enzyme-linked immunosorbent assay (ELISA), flow cytometry, and quantitative polymerase chain reaction (qPCR). Although informative, these assays provide endpoint measurements and generally lack the resolution to detect transient, spatially restricted, or dynamic interactions of immune cells within the tumor microenvironment (TME) (4). The immune landscape is not static, and responses can evolve rapidly over time and vary widely between tumor regions (7).
In this evolving therapeutic landscape, emerging imaging technologies—ranging from multiplexed spatial imaging at the tissue level to real-time in vivo imaging platforms—offer a transformative opportunity (6). Emerging imaging technologies that are significantly enhancing our understanding of the tumor-immune microenvironment include Multiplexed Ion Beam Imaging (MIBI), Imaging Mass Cytometry (IMC), Cyclic Immunofluorescence (CycIF), CO-Detection by Indexing (CODEX), Positron Emission Tomography (PET), Single-Photon Emission Computed Tomography (SPECT), Magnetic Resonance Imaging (MRI), and Intravital Microscopy. These advanced methods provide detailed spatial, temporal, and molecular resolution, enabling visualization of immune cell dynamics and interactions within the tumor microenvironment at levels previously unattainable by conventional assays (4, 8, 9). These tools allow researchers and clinicians to visualize immune activity where it happens and as it unfolds. By directly observing how drugs engage their targets, modulate the immune microenvironment, and impact immune cell behavior, imaging can provide a richer and more accurate representation of therapeutic efficacy (9).
This opinion article argues that drug efficacy evaluation must shift beyond static biomarkers toward integrated, image-guided approaches that combine spatial, temporal, and functional insights. Such a paradigm shift could greatly enhance precision medicine and improve therapeutic outcomes in immunotherapy.
2 The rise of multiplex and spatial imaging for tissue-level analysis
Tissue-level drug efficacy evaluation has historically relied on basic histological techniques and immunohistochemistry (IHC), offering limited information on the complex spatial relationships that define immune response (10). While traditional biomarkers like PD-L1 or CD8+ T cell counts remain clinically relevant, they offer a static and often incomplete snapshot (11). As our understanding of tumor–immune dynamics deepens, the ability to analyze immune responses in spatial context has become indispensable.
Multiplexed spatial imaging technologies have emerged as powerful tools to overcome these limitations (8). Techniques such as Imaging Mass Cytometry (IMC), Multiplexed Ion Beam Imaging (MIBI), Cyclic Immunofluorescence (CycIF), and CO-Detection by Indexing (CODEX) allow simultaneous visualization of 30 to over 60 proteins within intact tissue sections, preserving spatial architecture (12–14). The multiplex spatial imaging pipeline is illustrated in Figure 1, which includes both the experimental workflow (top panel) and a representative imaging output (bottom panel). The workflow begins with tissue preparation steps including paraffin removal and antigen retrieval, followed by iterative rounds of antibody staining and image acquisition (4). Each cycle involves the application of a primary antibody and a fluorescently labeled secondary antibody, after which the tissue is imaged and the signal is chemically stripped. This process is repeated multiple times (Cycle 1 to Cycle N), each targeting a distinct set of protein markers.
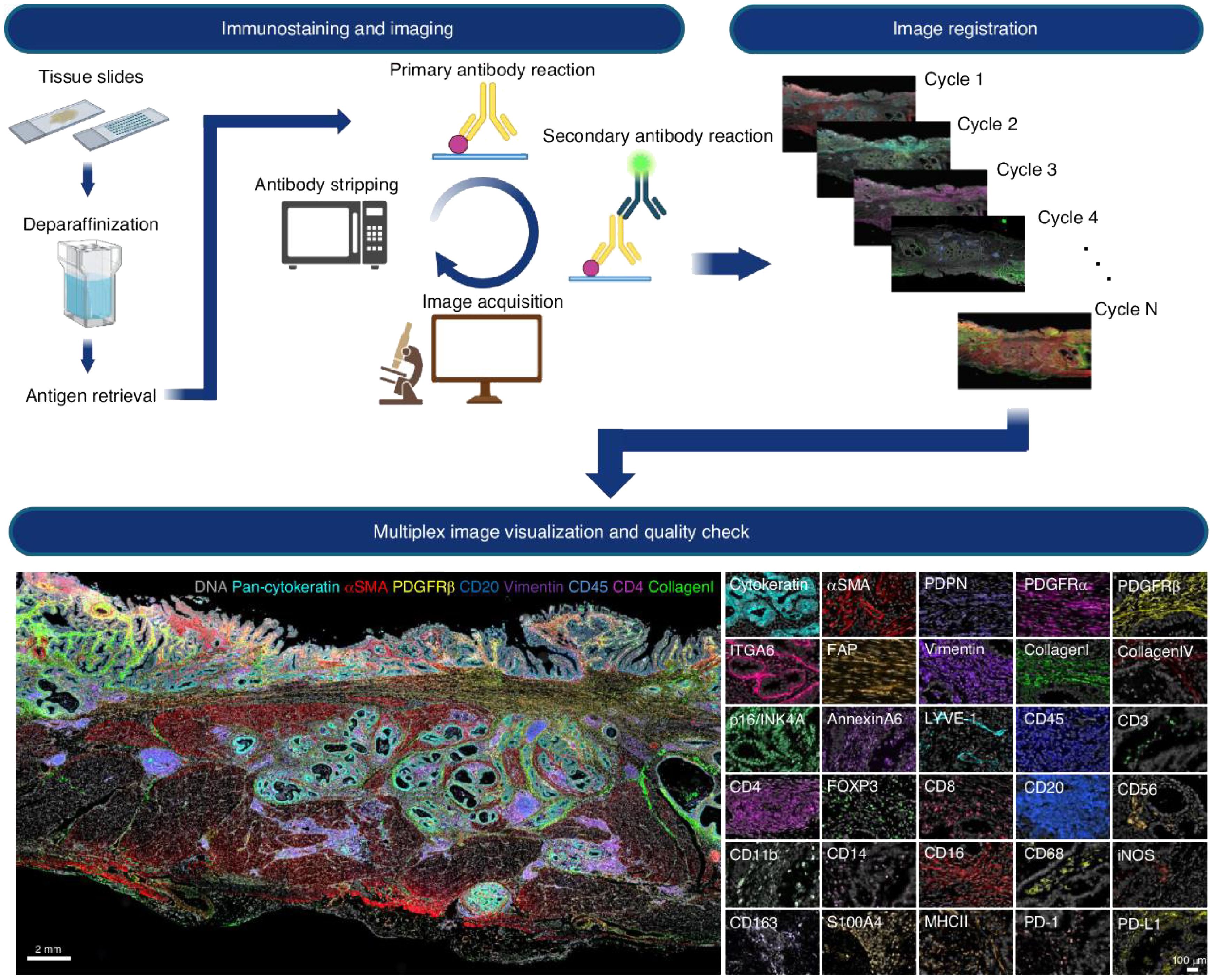
Figure 1. Cyclic multiplex immunofluorescence imaging workflow and representative results. Top panel: Iterative imaging process begins with formalin-fixed paraffin-embedded (FFPE) tissue slides undergoing deparaffinization and antigen retrieval. Each cycle consists of primary and secondary antibody labeling, image acquisition, and fluorophore stripping. The resulting images from multiple cycles are computationally aligned (registered) to generate a spatially resolved dataset containing dozens of protein markers. Bottom panel: Composite visualization of a multiplex image from tumor tissue, displaying color-coded spatial expression of markers such as DNA (blue), pan-cytokeratin (cyan), αSMA (red), PDGFRβ (green), CD20, CD45, CD4, CD8, vimentin, and collagen. The right side presents selected single-marker images (e.g., ITGA6, CD3, CD8, MHC-II, PD-L1), enabling detailed analysis of immune cell subsets and structural compartments within the tumor microenvironment. Reproduced with permission from ref (4).
Once imaging is complete, all cycle images are computationally registered to produce a high-dimensional, spatially resolved composite image. The bottom panel shows an example of such a multiplex image, highlighting distinct cell types and structures within the TME. Markers such as DNA, pan-cytokeratin, αSMA, PDGFRβ, CD20, CD45, CD4, CD8, vimentin, and collagen reveal a rich tissue architecture that includes epithelial structures, stromal fibroblasts, immune infiltrates, and extracellular matrix components. Additional single-channel panels demonstrate high-resolution staining of over 20 individual markers, facilitating the detailed classification of immune and stromal subtypes and their spatial distribution.
Building upon workflows like the one shown in Figure 1, these technologies offer unprecedented resolution into the tumor immune microenvironment (TIME), revealing how immune cells are distributed relative to tumor cells, vasculature, and each other. For example, the presence of tertiary lymphoid structures (TLS), spatial clustering of CD8+ T cells near tumor nests, or exclusion of effector T cells from tumor cores are spatial features that have all been correlated with response or resistance to immune checkpoint inhibitors (15). Such findings underscore the need to incorporate spatial biomarkers into drug evaluation pipelines. Moreover, spatial imaging enables retrospective evaluation of clinical trial specimens, helping explain heterogeneous responses. In trials where traditional biomarkers fail to predict outcomes, spatial immune phenotypes—such as myeloid-rich immunosuppressive niches or immune deserts—can offer mechanistic insights and support patient stratification strategies (16). Emerging applications also include the study of therapeutic interventions themselves, such as evaluating immune infiltration post-vaccination or CAR-T cell localization after infusion. Multiplex imaging allows researchers to quantify how drugs reshape the TIME, revealing shifts in cell phenotypes, activation states, or the emergence of suppressive cell types (17).
Despite their promise, integration of these technologies into clinical workflows remains limited due to high cost, labor intensity, and analytical complexity. Nonetheless, advances in automation, cloud-based analysis platforms, and machine learning-driven interpretation are making spatial imaging increasingly accessible. When paired with clinical endpoints, spatial imaging offers not only correlative insights but the potential for spatial biomarker-driven patient selection and real-time therapy monitoring.
In sum, multiplex and spatial imaging have redefined how immune activity within tissues can be visualized, quantified, and interpreted. They offer a much-needed bridge between molecular data and tissue-level functional context, laying the foundation for more nuanced and effective immunotherapy evaluation.
3 In vivo and real-time imaging: capturing immune dynamics beyond the slide
The evaluation of drug efficacy in immunotherapy has traditionally relied on static biomarkers and endpoint assessments, which often fail to capture the dynamic nature of immune responses (18). Recent advancements in in vivo and real-time imaging technologies have revolutionized our ability to monitor immune dynamics, providing deeper insights into therapeutic mechanisms and facilitating the development of more effective treatment strategies (19, 20).
3.1 Advancements in in vivo and real-time imaging technologies
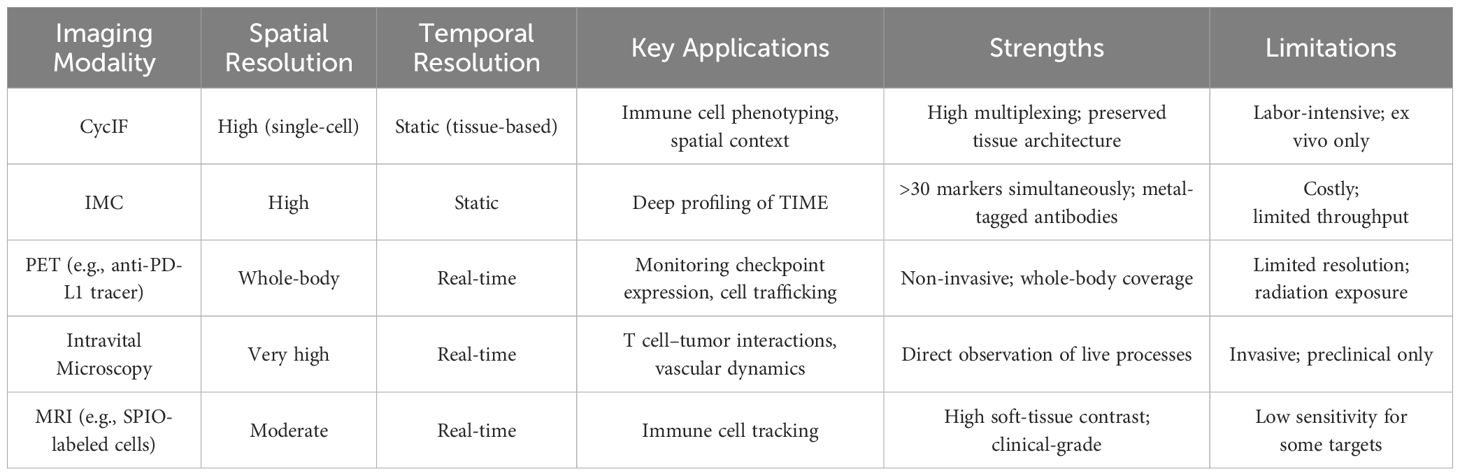
In vivo imaging techniques have evolved to allow non-invasive visualization of immune cells within their native environments, enabling the study of cellular behaviors and interactions over time (21, 22). Table 1 provides an overview of key imaging modalities employed in immunotherapy research, highlighting their spatial and temporal resolution, primary applications, as well as their respective advantages and limitations. Key technologies include intravital microscopy, positron emission tomography (PET), single-photon emission computed tomography (SPECT), and magnetic resonance imaging (MRI). These modalities offer unique advantages in tracking immune cell migration, activation, and function in response to immunotherapies.
3.2 Intravital microscopy
Intravital microscopy provides high-resolution, real-time visualization of cellular processes in live animals (23). This technique has been instrumental in elucidating the dynamics of T cell infiltration into tumors, mechanisms of cancer cell killing, and the role of myeloid cells in tumor progression. For instance, Intravital microscopy uniquely enables direct visualization of dynamic immune cell behaviors such as T-cell migration patterns, stable versus transient interactions with tumor cells, and their real-time cytotoxic effects in vivo. For instance, studies using intravital imaging have illustrated how stable, long-lasting interactions between cytotoxic T lymphocytes (CTLs) and tumor cells correlate with enhanced tumor cell apoptosis, providing mechanistic insights that static assays fail to capture (24, 25). Conversely, transient interactions may indicate ineffective immune responses and tumor evasion strategies (26).
3.3 Positron emission tomography and single-photon emission computed tomography
PET and SPECT generate imaging contrast through the use of radiolabeled probes. In PET imaging, positron-emitting isotopes such as 18F, 68Ga, or 89Zr emit positrons upon decay, which interact with electrons to produce gamma photons detectable by the PET scanner. SPECT imaging utilizes gamma-emitting isotopes like 99mTc or 111In, directly detecting gamma photons via gamma cameras. These radiotracers can be conjugated to antibodies, peptides, or metabolic substrates, enabling the specific visualization of immune cells, tumor markers, or molecular processes non-invasively with high sensitivity (27, 28). PET and SPECT imaging utilize radiolabeled tracers to detect specific molecular targets, offering whole-body insights into immune cell distribution and activity (29). These modalities have been employed to monitor the expression of immune checkpoints, such as PD-1/PD-L1, and to assess the biodistribution of therapeutic antibodies. For example, PET imaging with radiolabeled anti-PD-L1 antibodies has enabled the non-invasive assessment of PD-L1 expression in tumors, providing valuable information for patient stratification and treatment planning (30).
3.4 Magnetic resonance imaging and cancer vaccine imaging: tracking immune activation in vivo
MRI offers high-resolution anatomical imaging with excellent soft-tissue contrast, making it suitable for tracking labeled immune cells in vivo (31, 32). Superparamagnetic iron oxide (SPIO) nanoparticles have been used to label various immune cell populations, allowing their migration and accumulation in tumors to be visualized. SPIO nanoparticles label immune cells through ex vivo incubation followed by reinfusion or via antibody-mediated targeting of surface markers in vivo. Upon administration, SPIO-labeled cells disturb local magnetic fields detectable by MRI, producing contrast enhancement in images. However, the inherent limitation is the passive accumulation of SPIO nanoparticles in tumors via the enhanced permeability and retention (EPR) effect, which may obscure the precise identification of specific immune populations. Recent advances involve coupling SPIO nanoparticles with specific antibodies or ligands to improve targeting specificity, reducing nonspecific tumor uptake, and enhancing cellular resolution in MRI (33, 34). This approach has been applied to monitor the recruitment of cytotoxic T lymphocytes (CTLs) and regulatory T cells (Tregs) following immunotherapy, providing insights into the mechanisms underlying therapeutic responses (35).
Moreover, the efficacy of cancer vaccines relies on the activation and recruitment of antigen-specific T cells to tumor sites (36). In vivo imaging has been employed to monitor these processes, providing insights into vaccine-induced immune responses. MRI tracking of SPIO-labeled dendritic cells, used as vaccine adjuvants, has demonstrated successful migration to lymph nodes and subsequent T cell activation, correlating with tumor regression in preclinical models (37). Together, these applications illustrate how MRI serves as a powerful platform to visualize and quantify immune activation triggered by cancer vaccines, complementing conventional biomarker-based evaluation.
3.5 Tracking CAR T-cell therapy
Chimeric antigen receptor (CAR) T-cell therapy has shown promise in treating certain hematologic malignancies (38, 39). In vivo imaging has been pivotal in tracking the migration, expansion, and persistence of CAR T cells post-infusion. For instance, PET imaging using 89Zr-labeled CAR T cells has allowed researchers to monitor the trafficking of these cells to tumor sites, correlating their accumulation with therapeutic outcomes (40). While CAR T cell expansion and persistence can indeed be quantitatively monitored through blood sampling, imaging modalities such as PET, SPECT, or MRI provide complementary insights into spatial biodistribution, trafficking, and infiltration of CAR T cells into solid tumor masses or sanctuary sites not readily accessible via peripheral blood analysis. Imaging approaches thus are invaluable for assessing CAR T cell targeting efficacy, understanding resistance mechanisms, and optimizing therapy regimens, especially in scenarios involving solid tumors or metastatic niches beyond hematologic contexts (41, 42).
3.6 Assessing immune checkpoint inhibitor therapy
Immune checkpoint inhibitors targeting PD-1/PD-L1 and CTLA-4 have revolutionized cancer treatment. In vivo imaging has facilitated the evaluation of these therapies by enabling the visualization of dynamic changes in immune cell infiltration and activation within the tumor microenvironment (43). For example, PET imaging with radiolabeled PD-1 antibodies has been used to assess PD-1 expression levels in tumors, aiding in the prediction of patient responses to checkpoint blockade therapies (44).
3.7 From technical barriers to imaging innovation
Despite the advancements, several challenges hinder the widespread clinical adoption of in vivo immune imaging. Technical limitations, such as the need for highly specific and sensitive imaging agents, and the potential for tracer-induced alterations in cell behavior, must be addressed (45). Additionally, standardization of imaging protocols and data interpretation is essential to ensure reproducibility and comparability across studies (20).
Future research should focus on developing novel imaging probes with enhanced specificity for immune cell subsets and activation states. Combining multiple imaging modalities, such as PET/MRI, could leverage the strengths of each technique, providing comprehensive insights into immune dynamics. Furthermore, integrating in vivo imaging data with other biomarkers and clinical parameters may enhance predictive models for treatment responses, ultimately guiding personalized immunotherapy strategies (46).
In summary, in vivo and real-time imaging technologies have significantly advanced our ability to monitor immune dynamics beyond traditional histological methods. By providing spatiotemporal insights into immune responses, these techniques offer valuable tools for evaluating and optimizing immunotherapies, paving the way for more effective and personalized cancer treatments.
4 Challenges and integration into clinical practice
Despite the significant promise of advanced imaging technologies in drug efficacy evaluation, several critical challenges continue to limit their widespread adoption in clinical settings. These challenges span technical, operational, analytical, and regulatory domains (47). One of the foremost technical challenges lies in the development and standardization of imaging agents and protocols. Many imaging platforms, especially in vivo real-time modalities such as PET, SPECT, and intravital microscopy, rely on customized tracers, labeled antibodies, or nanoparticles that require rigorous validation. These reagents often lack regulatory approval for routine human use and may suffer from variability in synthesis, stability, or immunogenicity (45). Additionally, achieving sufficient resolution, sensitivity, and specificity in a clinical setting—while maintaining patient safety—remains an ongoing challenge, particularly in deep-tissue imaging.
From an operational perspective, the infrastructure required for advanced imaging is substantial. High-end platforms such as imaging mass cytometry, multiplexed ion beam imaging, or hybrid PET/MRI systems are costly to install and maintain. Furthermore, the execution of multi-modal imaging studies demands highly skilled personnel, cross-disciplinary coordination (e.g., pathology, radiology, immunology), and extended processing times, all of which strain hospital resources and reduce scalability. Data analysis and interpretation pose further hurdles. Imaging datasets are large, multidimensional, and complex, requiring bioinformatics expertise, machine learning pipelines, and standardized analytic workflows. Currently, there is a lack of consensus on how to translate spatial or dynamic imaging findings into clinical decisions. While some spatial biomarkers have shown predictive power in trials, few have undergone prospective validation or regulatory qualification as companion diagnostics (48).
To successfully integrate these technologies into clinical immunotherapy practice, several steps are needed. These include the development of standardized imaging protocols, harmonization of analysis tools across platforms, and validation of predictive imaging biomarkers in large, multicenter cohorts. Moreover, regulatory frameworks must evolve to accommodate dynamic and spatial biomarkers, with pathways for the approval of imaging-based diagnostics and clinical decision tools. With strategic investment and collaboration, imaging can shift from an academic asset to a routine pillar of personalized cancer care.
5 Outlook and future perspectives
As immunotherapy continues to reshape the oncology landscape, there is a growing consensus that traditional, static methods of drug efficacy assessment are no longer sufficient. The future of immunotherapy evaluation lies in integrating imaging technologies that can provide comprehensive spatial, temporal, and functional information—enabling a more dynamic and nuanced understanding of immune responses at both tissue and whole-body levels. The convergence of tissue-level multiplex imaging and in vivo real-time imaging marks a major step forward. Multiplex platforms like CODEX and IMC offer unprecedented granularity in characterizing the tumor immune microenvironment, while non-invasive modalities such as PET, MRI, and intravital microscopy allow longitudinal monitoring of immune activity and therapeutic impact. Together, these technologies offer the potential to build a unified, high-resolution view of drug–immune system interactions that can guide real-time clinical decisions.
Looking ahead, the integration of these platforms with computational tools—particularly artificial intelligence (AI) and machine learning—will be key. These approaches can help process vast, multidimensional datasets to identify predictive patterns, generate response signatures, and even forecast resistance. Additionally, combining imaging data with other omics layers (e.g., genomics, transcriptomics, proteomics) will further enhance our ability to stratify patients and tailor therapies.
To fully realize this potential, future efforts must focus on standardization, scalability, and clinical validation. Imaging protocols should be harmonized across institutions, and regulatory frameworks must evolve to recognize imaging-based spatial and functional biomarkers as legitimate endpoints in clinical trials. Equally important is the development of user-friendly analytical platforms that can democratize access to high-content imaging, even in resource-limited settings. In conclusion, imaging technologies are poised to transition from passive diagnostic tools to active drivers of precision immunotherapy. Their ability to visualize immune dynamics in space and time offers a powerful avenue to improve therapeutic evaluation, optimize patient selection, and ultimately enhance clinical outcomes in cancer immunotherapy.
Author contributions
XZ: Writing – original draft, Writing – review & editing. CC: Writing – original draft. XQ: Writing – original draft. PW: Writing – original draft. DZ: Writing – original draft, Writing – review & editing. SD: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by Natural Science Foundation of Shandong Province (ZR2021QH032) and The Medical and Health Science and Technology Project of Shandong Province (202304070941).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Maleki Vareki S, Garrigs C, and Duran I. Biomarkers of response to PD-1/PD-L1 inhibition. Crit Rev Oncology/Hematology. (2017) 116:116–24. doi: 10.1016/j.critrevonc.2017.06.001
2. Li H, van der Merwe PA, and Sivakumar S. Biomarkers of response to PD-1 pathway blockade. Br J Cancer. (2022) 126:1663–75. doi: 10.1038/s41416-022-01743-4
3. Zhong Y, Zhang W, Liu D, Zeng Z, Liao S, Cai W, et al. Screening biomarkers for Siogrengaos Syndrome by computer analysis and evaluating the expression correlations with the levels of immune cells. Front Immunol. (2023) 14:1023248. doi: 10.3389/fimmu.2023.1023248
4. Semba T and Ishimoto T. Spatial analysis by current multiplexed imaging technologies for the molecular characterisation of cancer tissues. Br J Cancer. (2024) 131:1737–47. doi: 10.1038/s41416-024-02882-6
5. Wang B, Yu W, Jiang H, Meng X, Tang D, and Liu D. Clinical applications of STING agonists in cancer immunotherapy: current progress and future prospects. Front Immunol. (2024) 15:1485546. doi: 10.3389/fimmu.2024.1485546
6. Lv Y, Luo X, Xie Z, Qiu J, Yang J, Deng Y, et al. Prospects and challenges of CAR-T cell therapy combined with ICIs. Front Oncol. (2024) 14:1368732. doi: 10.3389/fonc.2024.1368732
7. Sun K, Tempia S, Kleynhans J, von Gottberg A, McMorrow ML, Wolter N, et al. P.-C.g. the, Rapidly shifting immunologic landscape and severity of SARS-CoV-2 in the Omicron era in South Africa. Nat Commun. (2023) 14:246. doi: 10.1038/s41467-022-35652-0
8. de Souza N, Zhao S, and Bodenmiller B. Multiplex protein imaging in tumour biology. Nat Rev Cancer. (2024) 24:171–91. doi: 10.1038/s41568-023-00657-4
9. Rabiee N. Super-resolution microscopy for protein imaging: Unraveling cellular architecture and function. TrAC Trends Analytical Chem. (2025) 184:118140. doi: 10.1016/j.trac.2025.118140
10. Giesen C, Wang HAO, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. (2014) 11:417–22. doi: 10.1038/nmeth.2869
11. He Y, Ren T, Ji C, Zhao L, and Wang X. The baseline hemoglobin level is a positive biomarker for immunotherapy response and can improve the predictability of tumor mutation burden for immunotherapy response in cancer, Frontiers in Pharmacology 15. (2024). doi: 10.3389/fphar.2024.1456833
12. Scuiller Y, Hemon P, Rochais M, Pers J-O, Jamin C, and Foulquier N. YOUPI: Your powerful and intelligent tool for segmenting cells from imaging mass cytometry data. Front Immunol. (2023) 14:1072118. doi: 10.3389/fimmu.2023.1072118
13. Baharlou H, Canete NP, Cunningham AL, Harman AN, and Patrick E. Mass cytometry imaging for the study of human diseases Applications and data analysis strategies. Front Immunol. (2019) 10:02657. doi: 10.3389/fimmu.2019.02657
14. Lin J-R, Fallahi-Sichani M, Chen J-Y, and Sorger PK. Cyclic immunofluorescence (CycIF), A highly multiplexed method for single-cell imaging. Curr Protoc Chem Biol. (2016) 8:251–64. doi: 10.1002/9780470559277.2016.8.issue-4
15. Gobbini E, Hubert M, Doffin A-C, Eberhardt A, L.o. Hermet D, Duplouye LP, et al. The spatial organization of cDC1 with CD8+ T cells is critical for the response to immune checkpoint inhibitors in patients with melanoma. Cancer Immunol Res. (2025) 13:517–26. doi: 10.1158/2326-6066.CIR-24-0421
16. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. (2018) 24:541–50. doi: 10.1038/s41591-018-0014-x
17. Reinhard K, Rengstl B, Oehm P, Michel K, Billmeier A, Hayduk N, et al. An RNA vaccine drives expansion and efficacy of claudin-CAR-T cells against solid tumors. Science. (2020) 367:446–53. doi: 10.1126/science.aay5967
18. van de Donk PP, Kist de Ruijter L, Lub-de Hooge MN, Brouwers AH, van der Wekken AJ, Oosting SF, et al. Molecular imaging biomarkers for immune checkpoint inhibitor therapy. Theranostics. (2020) 10:1708–18. doi: 10.7150/thno.38339
19. Abdulreda MH, Faleo G, Molano RD, Lopez-Cabezas M, Molina J, Tan Y, et al. High-resolution, noninvasive longitudinal live imaging of immune responses. Proc Natl Acad Sci. (2011) 108:12863–8. doi: 10.1073/pnas.1105002108
20. Hor JL and Germain RN. Intravital and high-content multiplex imaging of the immune system. Trends Cell Biol. (2022) 32:406–20. doi: 10.1016/j.tcb.2021.11.007
21. Liu J, Cheng P, Xu C, and Pu K. Molecular probes for in vivo optical imaging of immune cells, Nature Biomedical Engineering. (2025). doi: 10.1038/s41551-024-01275-7
22. Heaton AR, Rehani PR, Hoefges A, Lopez AF, Erbe AK, Sondel PM, et al. ingle cell metabolic imaging of tumor and immune cells in vivo in melanoma bearing mice. Front Oncol. (2023) 13:1110503. doi: 10.3389/fonc.2023.1110503
23. Pittet MJ and Weissleder R. Intravital imaging. Cell. (2011) 147:983–91. doi: 10.1016/j.cell.2011.11.004
24. B.a. Breart F, Celli LS, and Bousso P. Two-photon imaging of intratumoral CD8+ T cell cytotoxic activity during adoptive T cell therapy in mice. J Clin Invest. (2008) 118:1390–7. doi: 10.1172/JCI34388
25. Nolz JC and Hill AB. Strength in numbers: visualizing CTL-mediated killing in Vivo. Immunity. (2016) 44:207–8. doi: 10.1016/j.immuni.2016.01.026
26. Boissonnas A, Fetler L, Zeelenberg IS, Hugues S, and Amigorena S. In vivo imaging of cytotoxic T cell infiltration and elimination of a solid tumor. J Exp Med. (2007) 204:345–56. doi: 10.1084/jem.20061890
27. Weissleder R and Pittet MJ. Imaging in the era of molecular oncology. Nature. (2008) 452:580–9. doi: 10.1038/nature06917
28. Wierstra P, Sandker G, Aarntzen E, Gotthardt M, Adema G, Bussink J, et al. Tracers for non-invasive radionuclide imaging of immune checkpoint expression in cancer. EJNMMI Radiopharmacy Chem. (2019) 4:29. doi: 10.1186/s41181-019-0078-z
29. Jiemy WF, Heeringa P, Kamps JAAM, van der Laken CJ, Slart RHJA, and Brouwer E. Positron emission tomography (PET) and single photon emission computed tomography (SPECT) imaging of macrophages in large vessel vasculitis: Current status and future prospects. Autoimmun Rev. (2018) 17:715–26. doi: 10.1016/j.autrev.2018.02.006
30. Bansal A, Barham W, Liu X, Harrington S, Lucien-Matteoni F, Dong H, et al. PET imaging of PD-L1 using anti-PD-L1-B11 antibody in breast cancer tumor model. J Nuclear Med 62(supplement. (2021) 1):1486.
31. Zhao Y, Ding Y, Lau V, Man C, Su S, Xiao L, et al. Whole-body magnetic resonance imaging at 0.05 Tesla. Science. (2024) 384:eadm7168.
32. Zhang C, Hsu P, Wang D, Zhang W, Zhang C, Guo S, et al. Superparamagnetic iron oxide (SPIO) nanoparticles labeled endothelial progenitor cells (EPCs) administration inhibited heterotopic ossification in rats. Nanomedicine: Nanotechnology Biol Med. (2019) 21:102078. doi: 10.1016/j.nano.2019.102078
33. Cho EC, Glaus C, Chen J, Welch MJ, and Xia Y. Inorganic nanoparticle-based contrast agents for molecular imaging. Trends Mol Med. (2010) 16:561–73. doi: 10.1016/j.molmed.2010.09.004
34. Daldrup-Link HE. Ten things you might not know about iron oxide nanoparticles. Radiology. (2017) 284:616–29. doi: 10.1148/radiol.2017162759
35. Beer AJ, Holzapfel K, Neudorfer J, Piontek G, Settles M, Krinig H, et al. Visualization of antigen-specific human cytotoxic T lymphocytes labeled with superparamagnetic iron-oxide particles. Eur Radiol. (2008) 18:1087–95. doi: 10.1007/s00330-008-0874-4
36. Zhou H, Ma Y, Liu F, Li B, Qiao D, Ren P, et al. Current advances in cancer vaccines targeting NY-ESO-1 for solid cancer treatment. Front Immunol. (2023) 14. doi: 10.3389/fimmu.2023.1255799
37. Xu Y, Wu C, Zhu W, Xia C, Wang D, Zhang H, et al. Superparamagnetic MRI probes for in vivo tracking of dendritic cell migration with a clinical 3T scanner. Biomaterials. (2015) 58:63–71. doi: 10.1016/j.biomaterials.2015.04.016
38. Butler SE, Hartman CJ, Huang YH, and Ackerman ME. Toward high-throughput engineering techniques for improving CAR intracellular signaling domains. Front Bioengineering Biotechnol. (2023) 11:1101122. doi: 10.3389/fbioe.2023.1101122
39. Requejo Cier CJ, Valentini N, and Lamarche C. Unlocking the potential of Tregs: innovations in CAR technology. Front Mol Biosci. (2023) 10:1267762. doi: 10.3389/fmolb.2023.1267762
40. Lee SH, Soh H, Chung J, Cho E, Lee S, Ju J-M, et al. Feasibility of real-time in vivo Zr-DFO-labeled CAR T-cell trafficking using PET imaging. PloS One. (2020) 15:e0223814.
41. Van Hoeck J, Vanhove C, De Smedt SC, and Raemdonck K. Non-invasive cell-tracking methods for adoptive T cell therapies. Drug Discov Today. (2022) 27:793–807. doi: 10.1016/j.drudis.2021.10.012
42. Sakemura R, Cox MJ, Bansal A, Roman CM, Hefazi M, Vernon CJ, et al. Dynamic imaging of chimeric antigen receptor T cells with Tetrafluoroborate positron emission tomography/computed tomography. JoVE. (2022) 180):e62334.
43. Heskamp S, Hobo W, Molkenboer-Kuenen JDM, Olive D, Oyen WJG, Dolstra H, et al. Noninvasive imaging of tumor PD-L1 expression using radiolabeled anti PD-L1 antibodies. Cancer Res. (2015) 75:2928–36. doi: 10.1158/0008-5472.CAN-14-3477
44. England CG, Ehlerding EB, Hernandez R, Rekoske BT, Graves SA, Sun H, et al. Preclinical pharmacokinetics and biodistribution studies of -labeled pembrolizumab. J Nuclear Med. (2017) 58:162. doi: 10.2967/jnumed.116.177857
45. Kircher MF, Hricak H, and Larson SM. Molecular imaging for personalized cancer care. Mol Oncol. (2012) 6:182–95. doi: 10.1016/j.molonc.2012.02.005
46. Turley SJ, Cremasco V, and Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol. (2015) 15:669–82. doi: 10.1038/nri3902
47. Rowe SP and Pomper MG. Molecular imaging in oncology: Current impact and future directions. CA: A Cancer J Clinicians. (2022) 72:333–52. doi: 10.3322/caac.21713
Keywords: tumor immune microenvironment, spatiotemporal imaging, immunotherapy evaluation, in vivo imaging, multiplexed tissue imaging
Citation: Zhang X, Cheng C, Qu X, Wang P, Zhang D and Dai S (2025) Spatiotemporal imaging of immune dynamics: rethinking drug efficacy evaluation in cancer immunotherapy. Front. Immunol. 16:1609606. doi: 10.3389/fimmu.2025.1609606
Received: 10 April 2025; Accepted: 25 April 2025;
Published: 16 May 2025.
Edited by:
Kelsey P. Kubelick, University of Virginia, United StatesReviewed by:
Myeongsoo Kim, Georgia Institute of Technology, United StatesCopyright © 2025 Zhang, Cheng, Qu, Wang, Zhang and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dawei Zhang, emR3MTcwMzIwQDE2My5jb20=; Shuhong Dai, c2h1aG9uZzA0MDRAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Xiaowei Zhang
Xiaowei Zhang Cheng Cheng
Cheng Cheng Xiaoyang Qu3
Xiaoyang Qu3