- 1Fishery College, Guangdong Ocean University, Zhanjiang, China
- 2Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang), Zhanjiang, China
- 3Guangdong Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Economic Animals, Zhanjiang, China
Large-scale fish farming faces many environmental stresses, which affect their immune systems, growth performance, and physiological homeostasis, resulting in increase in their susceptibility to infections. Some of the most common bacterial infections of cobia fish (Rachycentron canadum) include streptococcosis, vibriosis, furunculosis and mycobacteriosis, and pastelleurosis. Probiotics could be helpful in reducing or limiting the incidence of severe disease infections or outbreaks. Therefore, the present study aimed to isolate the indigenous bacterial species from healthy cobia fish and then selected 3 strains, including Bacillus sp. RCS1 (MW560712), Pantoea agglomerans RCS2 (MW560713), and Bacillus cereus RCS3 (MW560714) from the gut of juvenile’s cobia having advantageous assets or positive characteristics. Their analysis indicated the presence of similar biochemical profiles and all could effectively utilize carbon sources. The biosafety assessment did not show any pathological symptoms after 10 days of injecting the fish with isolated bacteria. The results showed that all the isolated bacteria in the present study had low auto-aggregation capacity within the first 3 h of incubation. The isolated bacteria showed strong tolerance when exposed to a range of pH. Although asymmetrically, a slow rise in the growth of isolated bacteria was observed within the pH range of 1–8 for RC1, 1–7 for RC2, and 1–6 for RC3. The antagonistic effects of isolated bacterial strains on the development of pathogens, including Vibrio alginolyticus, Vibrio harveyi, Streptococcus iniae, and Streptococcus agalactiae, were investigated using Luria-Bertani (LB) agar plates. All the isolated bacteria exhibited inhibitory effects against the pathogens, including V. alginolyticus, V. harveyi, S. iniae, and S. agalactiae. These isolated bacteria were characterized with a wide range of antagonistic activities, non-hemolytic activities, high survivability after heat-treatments and safety confidence, and antibiotic susceptibility. Generally, the characteristics displayed by these strains indicated that they could be used as potential probiotics in the aquaculture industry.
Introduction
Aquaculture is a reliable production source for sustenance and nourishment of the global population’s increasing demand for animal and fish proteins. Ensuring a suitable environment, healthy fish seeds and juveniles, and quality feed are necessary for successful aquaculture. Cobia (Rachycentron canadum) is appreciated for its characteristic features, which make it an important culture fish. It has a fast growth rate and inadequate availability in the wild, along with high market values due to its excellent flesh quality. These characteristics have inspired its production in aquaculture all over Asia, especially in China and other parts of the globe as well (Holt et al., 2007; Benetti et al., 2008, 2010; Zhang et al., 2013; Chen et al., 2016; Huang et al., 2020; Xie et al., 2021). Its production reached to approximately 40,000 tons worldwide in 2015 and 59538 tons in 2018 [Food and Agriculture Organization (FAO), 2015, 2020; Wang et al., 2021]. The cobia aquaculture faces challenges due the frequent incidences of disease infections in fish farming, high risk of disease infection and inadequate knowledge of its gut microbiota. Large-scale fish farming face many environmental stresses, which affect their immune systems, growth performance, and physiological homeostasis, resulting in their susceptibility to infections. Antibiotics have been used previously, and have effectively decreased the incidence of disease infection but the excessive use of many antibiotics has resulted in bacterial resistance (Montes et al., 2006; Dlamini et al., 2019). Some of the most common bacterial infections that affect cobia include streptococcosis, vibriosis, furunculosis, and mycobacteriosis. The opportunistic species from Photobacterium genus causes a disease pastelleurosis that has been identified as a significant emerging problem for the aquaculture of cobia throughout its production cycle (Lopez et al., 2002; Liu et al., 2003; Rajan et al., 2003; Liao et al., 2004; Chen and Hsu, 2005).
The indigenous gut microbiota of fish plays a significant role in the growth, development, and health of host (Vine et al., 2004) by safeguarding and protecting against pathogens in gut and assisting in host’s digestive function by producing exogenous digestive enzymes and vitamins (Ray et al., 2012; Bhatnagar and Dhillon, 2019).
The utilization of valuable microbes and probiotics as a substitute approach to antimicrobial compounds for the control and prevention of diseases in aquaculture is increasing, which has become a hot-spot topic for research. Therefore, the probiotics can be utilized in aquatic farming, especially in fish farming, for the establishment of disease-free aquaculture (Bhatnagar and Dhillon, 2019). In order to fight diseases and promote growth, weight gain, and size, a range of valuable diet supplements and probiotics, which are advantageous to aquatic organisms, especially for fish, are being used in fish farming (Irianto and Austin, 2002; Amenyogbe et al., 2020), and encourage the host’s immune response. Probiotics can improve host’s health. Therefore, the Food and Agriculture Organization (FAO) has defined probiotics as foods or supplements, containing live microorganisms, which are intended to maintain or improve host’s health [Food and Agriculture Organization (FAO), 2001]. Probiotics are useful in reducing or limiting the incidence of severe disease infections or outbreaks and enhance digestive function and non-specific immunity in aquaculture species (Carnevali et al., 2006; Knackstedt and Gatherwright, 2019; Ringo, 2019).
Many indigenous microbial species from fish gut, such as Agarivorans spp. (Liu et al., 2017), Pseudomonas spp. (Nayak and Mukherjee, 2011), Lactobacillus spp. (Balcázar et al., 2007), and Bacillus spp. (Bandyopadhyay and Patra, 2004; Bhatnagar et al., 2012; Bhatnagar and Raparia, 2014; Bhatnagar and Lamba, 2015, 2017; Yang et al., 2015; Kuebutornye et al., 2019; Li et al., 2019; Wang et al., 2019) and Aeromonas spp. (Hao et al., 2014) have been isolated and utilized as feed additives in order to study their effects on immunity, growth performance and development, and nutritional superiority. Among the probiotics utilized in feed additives and disease control, the species in Bacillus genus, including B. subtilis, B. licheniformis, and B. cereus, are the dominant species (Aleti et al., 2015; Wang et al., 2015; Chen et al., 2019; Park et al., 2020). Many of these indigenous species have been proven to be beneficial for fish species by improving immunity, performance of growth and development, and nutritional superiority.
Pantoea, a Gram-negative bacterial genus that belongs to Enterobacteriaceae family, currently includes 21 identified species (Walterson and Stavrinides, 2015; Nawrath et al., 2020). The strains of this genus, including P. agglomerans BSL 2, are commercially utilized as biological control agents (Smits et al., 2010; Nawrath et al., 2020). Many studies have discussed and reviewed the harmful and beneficial effects of Pantoea species (Dutkiewicz et al., 2015, 2016b,c). Nevertheless, Pantoea is a diverse and versatile bacterium genus that displays some mutual traits associated with extraordinary biochemical actions and adaptation to a wild range of hosts and ecological conditions (Völksch et al., 2009; Nadarasah and Stavrinides, 2014; Walterson and Stavrinides, 2015; Dutkiewicz et al., 2016a). This makes an exclusive opportunity to utilize P. agglomerans as an effective candidate for the control of bio-remediation, and as drug, and bio-control agent. Interestingly, some of their potential traits include their capability of controlling several functions of animal-pathogen interactions, subject to their density of population, which is acknowledged as quorum sensing (Chalupowicz et al., 2008; Dutkiewicz et al., 2015, 2016b,c). These potential traits can be competently utilized for the treatment of human infections (Kohchi et al., 2006) and advancement of plant/animal growth (Jiang et al., 2015).
The most efficient approach to have probiotics is to isolate them from host’s gut (O’Sullivan, 2001; Bhatnagar and Dhillon, 2019). Generally, the selection of probiotics depends on their growth and colonization in gastric mucus, adhesion, or in vitro antagonism (Vine et al., 2004). Therefore, the present study isolated indigenous bacteria (Bacillus sp. RCS1, Pantoea agglomerans RCS2, and Bacillus cereus strain RCS3) having advantageous assets or positive characteristics from the gut of juvenile’s cobia. The antagonistic effects of these isolated indigenous bacteria on the growth of pathogenic Vibrio alginolyticus, Vibrio harveyi, Streptococcus iniae, and Streptococcus agalactiae were characterized and preliminary scrutinized.
Materials and Methods
Collection of Samples
Eight (8) cobia, carnivorous marine fish (mix gender), from the wild with an average body weight of 202 ± 04 g (Mean ± SD) and average standard length of 28 ± 9 cm, having no symptoms of infection were obtained from Donghai Island. The live fish were transported in polythene bags containing oxygenated water with the DO level in the water (>6 mg/L), water temperature (26.3 ± 2.5°C) to Fish Seed Engineering and Healthy Farming laboratory, Fisheries College of Guangdong Ocean University, China, for immediate use.
Isolation of Gut Microbiota
The fish were anesthetized using ethyl 3-aminobenzoate methanesulfonate, tricaine methanesulfonate (Sigma-Aldrich, 150 mg L-1MS-222), in order to minimize pain, which were then killed through a blow to their head. The fish were cleaned externally with cotton dipped into 75% ethanol to eliminate any external microbes on their bodies. Fish intestines were removed and cautiously stripped to eradicate all digestive content after the fish were dissected with sterile scissors. The intestine sections were washed with sterile phosphate buffer saline (PBS) three times. The intestinal sections were weighed and equal amounts of PBS by volume were added to them. The intestinal samples were homogenized using 15 mL borosilicate glass tissue homogenizer (Shanghai Lenggu Instrument Company, Shanghai, China) under sterile ice conditions. After homogenization, to a volume of 0.5 mL homogenized mixture, 4.5 mL of PBS solution was added to dilute the mixture. A volume of 0.1 mL of the diluted mixture was spread on Luria-Bertani (LB) agar plates. The LB plates were incubated for 24 h at 30°C. After 24 h of incubation, individual colonies were selected randomly and cultured into Luria-Bertani media to grow on a large scale under similar incubation conditions. Streaking of the isolated mixture was done repeatedly in order to obtain pure bacterial colonies. Based on their morphology, the potential probiotic strains were selected and identified using 16S rRNA gene sequencing. For this, polymerase chain reaction (PCR) (Weisburg et al., 1991) was carried out using universal bacterial primers 1492R (5′-GGTTACCTTGTTACGACTT-3′) and 27F (5′-AGAGTTTGATCCTGGCTCAG-3′). The PCR reaction mixture included 2.5 μL of each isolate, 2.5 μL of each primer at 0.2 μ M concentration (27F and 1492R), 25 μL of rTaq buffers, and 17.5 μL of double-distilled water. Streptococcus agalactiae, acquired from Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Animals, College of Fisheries, Guangdong Ocean University, Huguang Yan East, Zhanjiang 524088, Guangdong, China, was used as positive control. Double distilled water was used as negative control (Kuebutornye et al., 2018). The PCR conditions were as follows: initial denaturation at 95°C for 5 min; followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 45 s, and extension at 72°C for 1 min 30 s; and final extension at 72°C for 10 min (Kuebutornye et al., 2019). The PCR products were analyzed using agarose (1% w/v) gel electrophoresis and later sent to Sangon Biotech Co., Ltd. (Guangzhou, China) for sequencing. The basic local alignment search tool (BLAST) from National Center for Biotechnology Information (NCBI) was used for comparing the sequences with available 16S rRNA gene sequences in database and the isolated strains were identified. Mega 7 software was employed to construct a phylogenetic tree with other sequences obtained from NCBI for establishing evolutionary relationships among different isolates. The confidence of the resultant phylogenetic tree branch topology was set to bootstrap value of 10,000. The 16S rRNA gene sequences of the isolated microorganisms were submitted to NCBI for accession numbers.
Biochemical Characterization
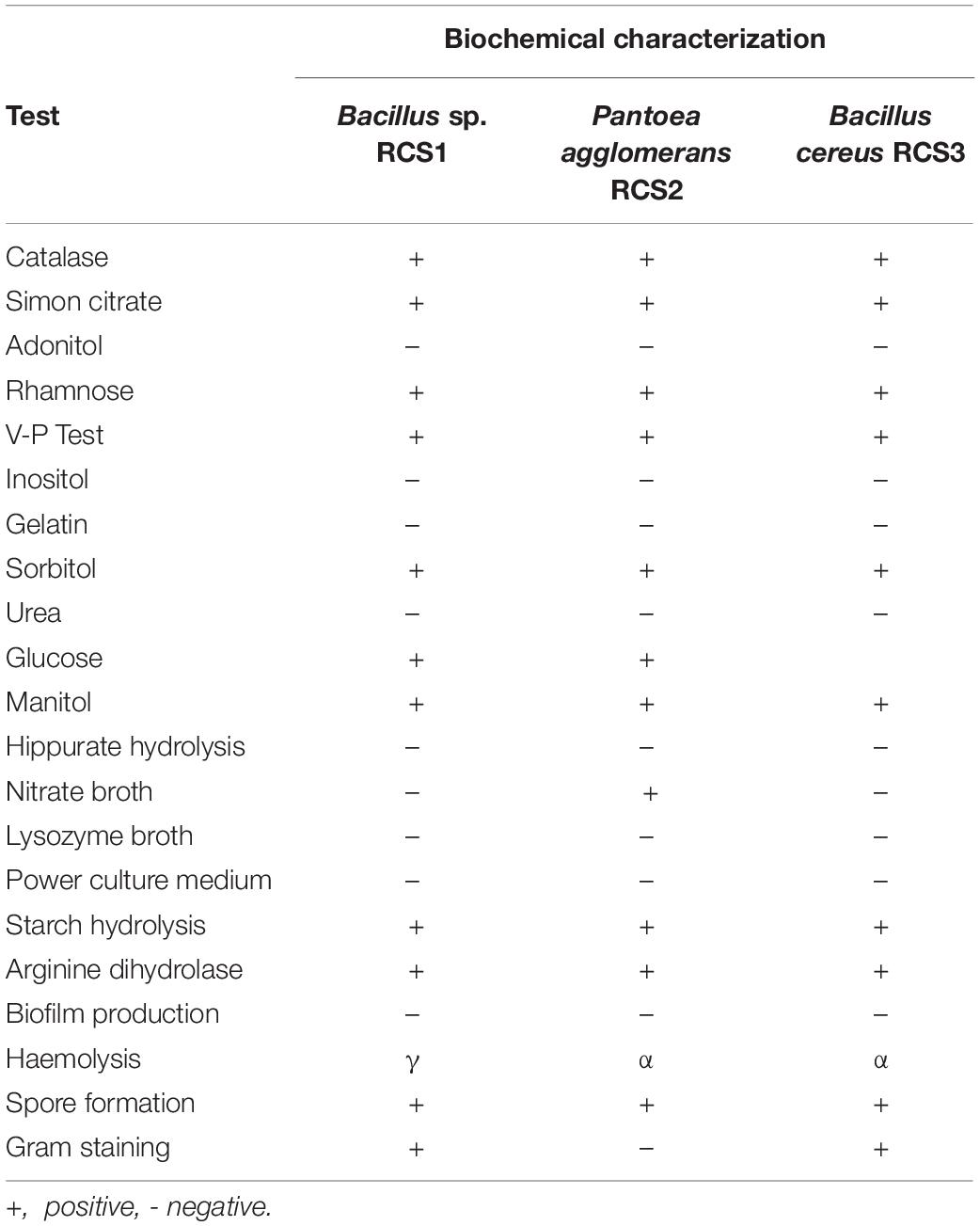
For biochemical characterization (Table 1) and confirmation, the commercial kits from Huankai Microbial, Guangzhou, China, and Bacillus cereus (HBIG07-1) identification bar from Qingdao Hope Bio-Technology Co., Ltd., Qingdao, China were used, following the manufacturer’s protocol.
Bacterial Growth in Luria-Bertani Broth
From the LB agar plates, a distinct colony of the isolated probiotic bacteria was carefully chosen and incubated at 37°C overnight in 5.0 mL of LB broth. A 500-mL Erlenmeyer flask containing 100 mL sterile LB broth and 1 mL of the culture was incubated at 37°C with constant shaking at 150 rpm. Their growth was monitored by measuring the absorbance at 600 nm for 24 h at 2 h intervals (Xie et al., 2014).
Biosafety Assay
In order to analyze the pathogenic effects of isolated microorganisms in cobia, 0.1 mL (108 CFU/mL) of each sample was injected intra-peritoneally into two groups, each containing 6 cobia fish with an average body weight of 150 g. The same volume of sterile PBS (pH 7.2) was injected into sixth fish as a control group. The conditions for fish culture were monitored as described previously (Abarike et al., 2018; Wang et al., 2021). Briefly, the fiberglass was a flowing water aquaculture system with continuous aeration for 24 h, and plastic tanks of 500 L capacity of filling with water. A dissolved oxygen meter (Hengxin, Taiwan, AZ8403) was used to monitor changes in dissolved oxygen. The experimental fish was fed with compound feed (Guangdong Yuequn Marine Biology Research and Development Co., Ltd., Jieyang, China) twice a day. The feed composed of about 430 g.kg–1 crude protein and 80 g.kg–1 crude lipid using the fish meal and soybean as protein sources, and fish oil and soybean oil as lipid sources (Geng et al., 2011). The feed was calculated as 10% of their body weights. Their feces were cleaned regularly by siphoning off with water. The fish were observed daily for clinical signs and mortality rate for 2 weeks (14 days).
Antibiotic Susceptibility
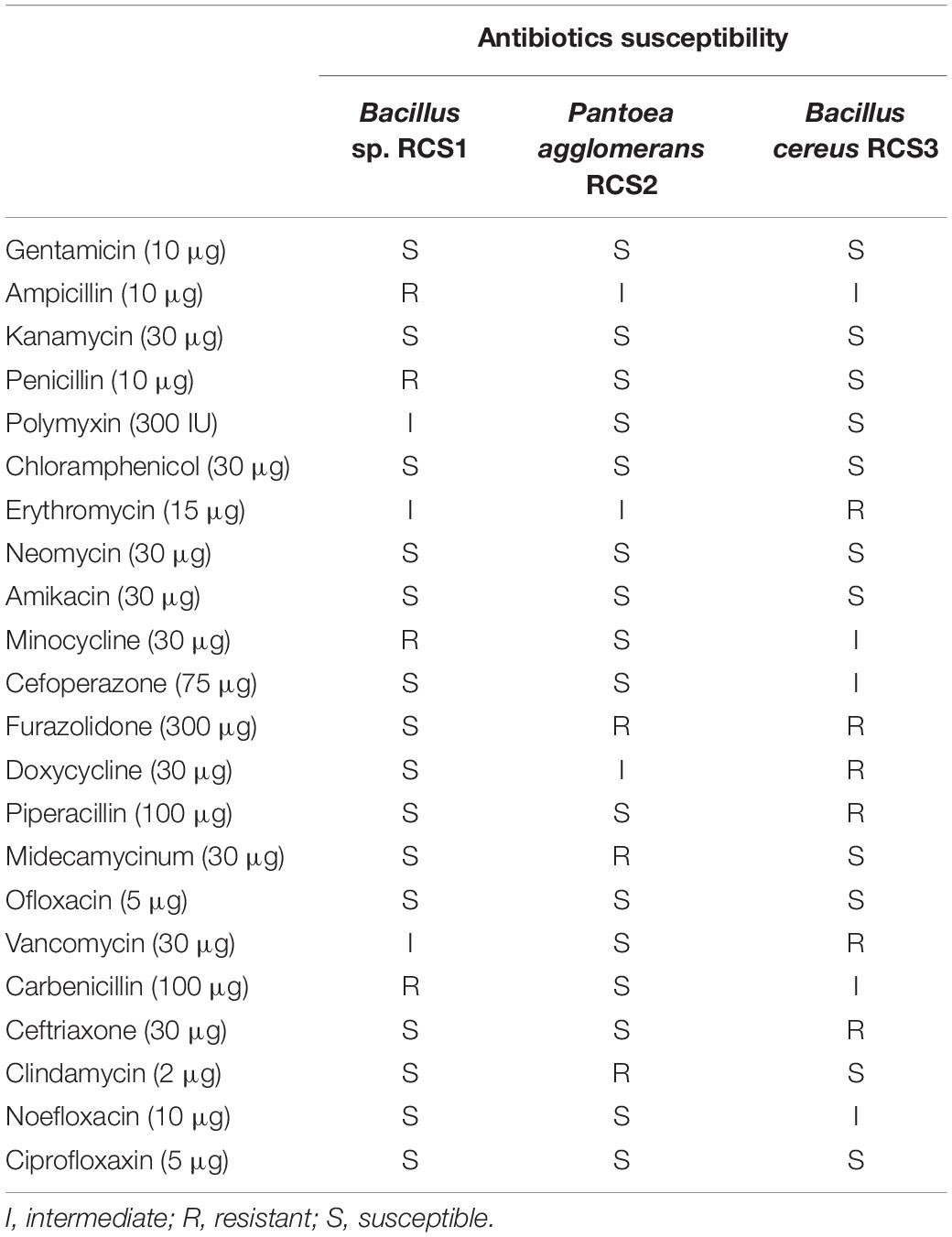
The isolated bacteria were assessed for their antibiotic susceptibility against some antibiotics (Table 3), using the commercially available antibiotics discs obtained from Hangzhou Microbial Reagent Co., Ltd., Hangzhou, China. Using the disk diffusion susceptibility method (Murray et al., 2007; Clinical and Laboratory Standards Institute, 2009), the isolated bacteria (100 μL) were spread on LB agar plates with the commercial antibiotics discs carefully placed on them and incubated for 24 h at 37°C. Their antibiotic susceptibility was calculated by measuring (mm) their inhibition zone as previously described (Patel et al., 2009; Kuebutornye et al., 2019).
Bile Salts Resistance
The resistance of isolated bacteria to bile salts was analyzed using the modified methods, as described by Argyri et al. (2013); Kuebutornye et al. (2019). In brief, the bacteria were cultured in LB broth overnight, harvested the following morning (9,000 g, 5 min, 4°C), washed with PBS buffer (pH 7.2) twice and then kept in PBS solution (pH 7.4), containing 0.5% (w/v) bile salts (BBI Life Sciences, Shanghai, China). Their resistance to bile salts was assessed by counting the colonies at 1 h interval during the incubation at 37°C for 4 h (counting viable colonies at 1, 2, 3, and 4 h of incubation).
High-Temperature Resistance
The resistance of isolated microorganisms to different temperatures was evaluated using the previously described procedures with slight modifications (Guo et al., 2016; Kuebutornye et al., 2019). Since the fish feed processing at times demands high temperatures. After culturing the isolated bacteria in LB broth overnight at 37°C and 150 rpm, the isolates were harvested by centrifuging at 9,000g for 5 min, followed by washing with PBS (pH 7.4) twice. The isolates were then exposed to 80, 90, and 100°C temperature for 2, 5, and 10 min, respectively. After exposure, sterile LB broth of equal volume was added to the temperature-exposed isolated bacteria in order to evaluate their growth capacity after heat treatments. The bacterial growth was observed by measuring the absorbance at 600 nm after 12 h of incubation at 37°C with constant shaking at the speed of 150 rpm.
Compatibility Test
According to literature, the mono-probiotics species are used in food products. Hence, the compatibility test was carried out for the utilization of multispecies probiotics (Saarela et al., 2000; Rajyalakshmi et al., 2016; Kuebutornye et al., 2019). The compatibility test was performed as described previously by Rajyalakshmi et al. (2016); Kuebutornye et al. (2019). Briefly, the isolated bacteria were streaked vertically and perpendicularly on LB agar plates at a distance of 5 and 10 mm from each other. After 24 h of incubation at 37°C, their compatibility was determined by observing and measuring the inhibition zone of isolated bacteria.
Antimicrobial Activity
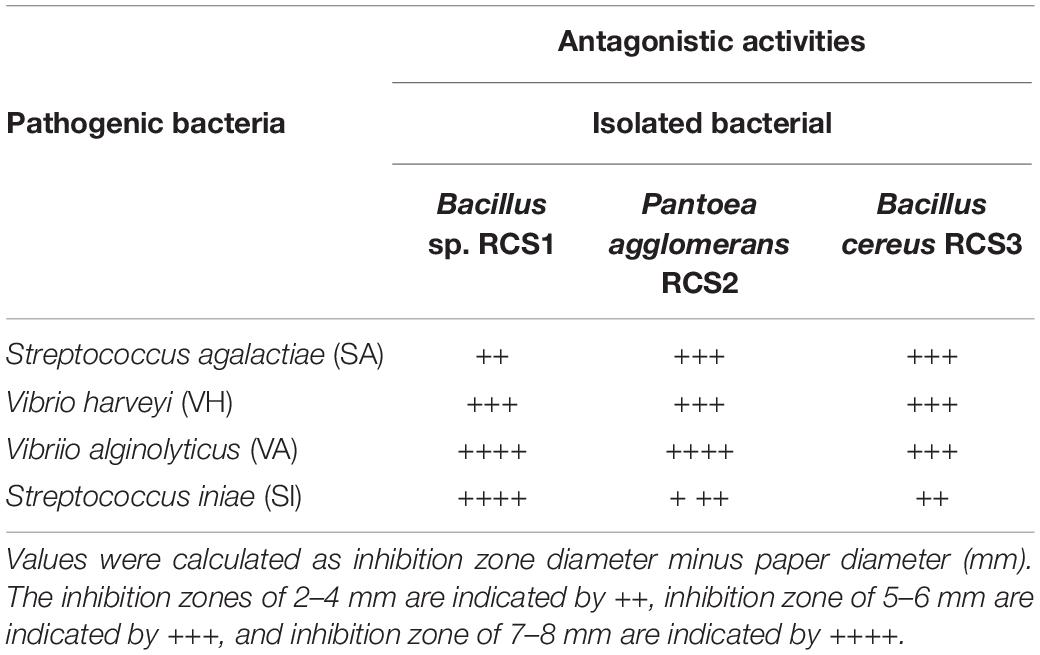
The pathogenic bacterial strains, including V. alginolyticus, V. harveyi, S. iniae, and S. agalactiae, were obtained from Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Animals, College of Fisheries, Guangdong Ocean University, Huguang Yan East, Zhanjiang 524088, Guangdong, China, for laboratory use. These pathogenic bacterial strains were tested against the isolated bacteria using the agar well diffusion method and cross-streak methods (Lertcanawanichakul and Sawangnop, 2008).
Auto-Aggregation
The auto-aggregation of the isolated bacteria was evaluated using the previously described method by Lee et al. (2017); Kuebutornye et al. (2019) with slight modifications. After centrifugation at 9,400 g for 5 min, the cells of isolated bacteria were harvested. The harvested cells were washed twice with PBS, kept back into the supernatant, and then mixed using vortex for 30 s. The absorbance was measured at 600 nm using a spectrophotometer (Shanghai Inesa Analytical Instrument Company, shanghai, China), at 0, 1, 2, 3, and 24 h of harvesting. The auto-aggregation was measured using Eq. (1).
where A0 = Absorbance at 0 h at 600 nm and At = Absorbance at 1, 2, 3, and 24 h at 600 nm.
Cell Hydrophobicity
The cell hydrophobicity is the most generally used method to determine the microbial cell surface adhesion. Since the assay is based on adhesion, its efficacy to determine the cell surface hydrophobicity is questionable. The cell hydrophobicity of isolated bacteria was analyzed using the previously described methods by Lee et al. (2017) with slight modifications. In brief, the isolates were cultured for 24 h and then centrifuged at 9,000 g for 5 min to harvest the bacterial cells. The harvested cells were washed with 2 mL of PBS (PBS, pH 7.4) twice. The absorbance was measured at 600 nm wavelength using a spectrophotometer in order to evaluate the percentage hydrophobicity and recorded as A0. Then, the harvested cells were mixed with ethyl acetate (a basic solvent), xylene (a non-polar solvent) and chloroform (an acidic solvent) using vortex for 5 min. The solution was kept for 30 min to allow it to separate into two phases. The absorbance was measured and recorded as A1. Hydrophobicity (%) could be calculated as given in Eq. (2).
where A0 = absorbance before mixing with solvent at 600 nm and A1 = absorbance after mixing with solvent at 600 nm.
Hemolytic Activity
The isolated bacteria were exposed to a hemolytic assay by streaking onto agar plates supplemented with 7% sheep blood. The agar plates were incubated at 37°C for 48 h followed by the observation and measurement of hemolytic zones. Subsequently, the isolated microbes were classified as β, α, or γ-hemolysis. The isolated bacteria with clear zone were denoted as β-hemolysis, those with green zone were denoted as α-hemolysis, and those without any zone were denoted as without hemolysis (Engel et al., 1972; Lee et al., 2017; Kuebutornye et al., 2019).
Optimal Growth and pH Determination
The determination of optimal growth and pH was carried out following the method described by Kavitha et al. (2018) with slight modifications. Briefly, the isolates were cultured for 24 h at 37°C in LB broth with adjustable pH (1–10). HCl and NaOH were used to adjust pH. The growth was measured by measuring the OD (optical density) using spectrophotometer (Shanghai Inesa Analytical Instrument Company, shanghai, China) at 600 nm. The LB broth without any bacteria was used as control.
Biofilm Formation and Detection (Congo Red Agar Method)
In order to measure the production of biofilm, the methods described by Kavitha et al. (2018) were followed with slight modifications. Briefly, the isolates were cultured at 37°C for 24 h. The cultured bacteria were streaked and incubated at 37°C for 48 h on Mueller Hinton agar medium, containing 0.8 g/l of Congo red dye. The red colonies were identified as non-biofilm-producing strains, while the black colonies with consistent dry crystalline structures were identified as biofilm-producing strains.
Statistical Analysis
In this study, the data were analyzed with one-way analysis of variance (ANOVA) using SPSS (IBM SPSS STATISTICS, 16.0 package, IBM Corporation, New York, United States) for Windows version 7.0 (SPSS, Chicago, United States). The data were previously tested for normal distribution and equal variances between treatments and homogeneity before ANOVA analysis and expressed in percentages. Data expressed as percentage were angularly transformed before being statistically assessed. The data were expressed as mean ± standard error (SE). The differences in mean values were identified using Tukey’s HSD tests (P < 0.05). Different letters were used to indicate the statistical significant differences.
Results
Identification of Gut Bacteria
Following biochemical and morphological characterization, three prospective probiotic bacteria were isolated and identified, namely, Bacillus sp. RCS1, Pantoea agglomerans RCS2, and Bacillus cereus RCS3, which were selected as microorganisms to be investigated in this study (Table 1). The gene sequencing analysis of 16S rRNA gene indicated that the isolated bacteria were Bacillus sp., Bacillus cereus strain, and Pantoea agglomerans strain. The Bacillus sp. RCS1, and Bacillus cereus strain RCS3 exhibited neighboring sequence homology (99. 52%) with Bacillus cereus strains while Pantoea agglomerans RCS2 exhibited neighboring sequence homology (98. 39%) with the Pantoea agglomerans strains. The analysis of the constructed phylogenetic tree (Figure 1) indicated that the isolates Bacillus sp. RCS1, and Bacillus cereus RCS3 had the highest similarity to Bacillus cereus strain NBRC 15305 (NR_112630.1) while the Pantoea agglomerans RCS2 had the highest similarity to Pantoea agglomerans strain CZ-BHG003, Pantoea agglomerans strain TPD7001 and Pantoea agglomerans strain NSH. The 16S rRNA gene sequences of the isolated strains were submitted to the NCBI for GenBank accession numbers as follows; Bacillus sp. RCS1 (MW560712), Bacillus cereus RCS3 (MW560714), and Pantoea agglomerans RCS2 (MW560713), respectively.
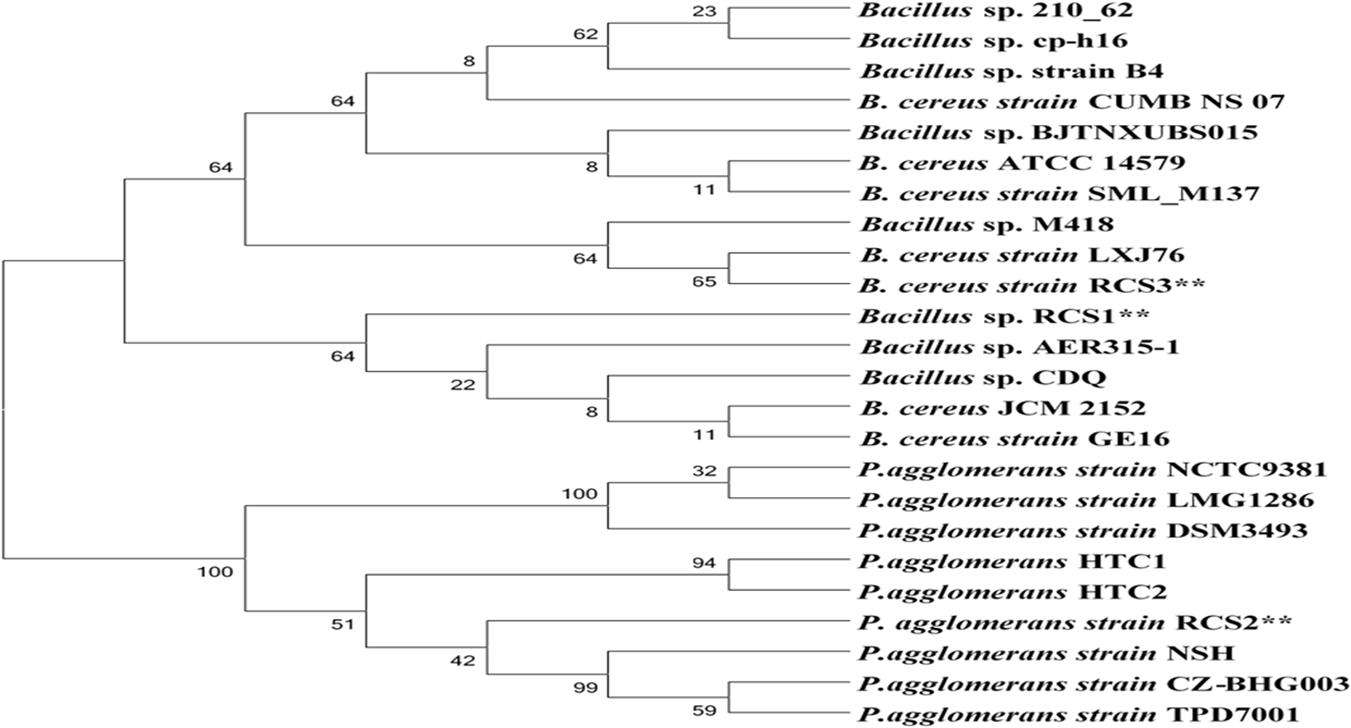
Figure 1. A neighbor joining method was used to construct phylogenetic tree using the sequences of isolated bacterial strains and others from NCBI database in order to identify genetic relatedness.
The biochemical characteristics of the isolated bacteria are listed in Table 1. Their analysis showed that they had comparable biochemical characteristics and hence can all virtually utilize carbon sources. The three isolated microbes were tested negative for gelatin liquefaction, inositol, urea, lysozyme broth, and hippuric acid, while tested positive for mannitol; hence they were halophiles. Nonetheless, they had different morphological features.
Generally, microbes reproduce via binary fission, which include the establishment two equal-sized progeny cells and hence amplifying their number with each division. A process of cell division of this kind, is known as exponential phase. The unit of bacterial growth is “generation time.” In order to identify the exponential growth phase of isolates in the present study, the growth of potential probiotics were determined. The exponential growth phase for all the three isolated bacteria started at roughly 2 h subsequently incubation at 37°C with uninterrupted shaking (150 rpm) (Figure 2).
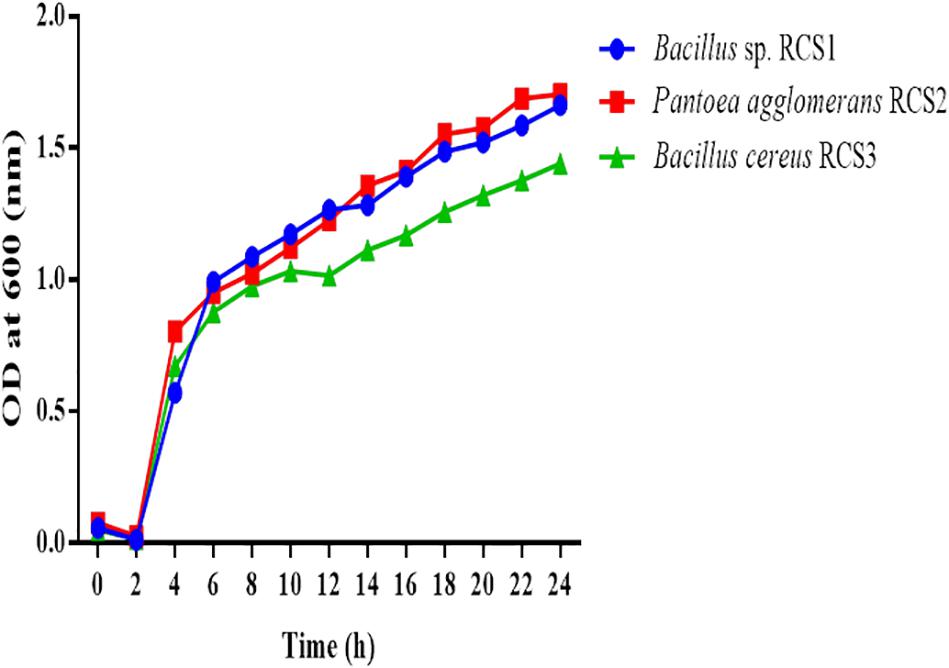
Figure 2. Growth curves of isolated microorganisms. The growth curves were observed to determine the growth of these bacteria. The curves of the microbial growth were measured at 600 nm.
The biosafety assessment showed that after 10 days of injecting the fish with isolated bacteria, they showed no pathological symptoms (such as mucus, lesions, edema, loss of scale and hemorrhage) as witnessed in both the control and experimental groups. There were no mortalities recorded during the biosafety assay, which actually confirmed that these isolated bacteria were not pathogenic.
The outcomes of the antibiotic susceptibility tests for isolated bacteria are listed in Table 2. Out of the 22 antibiotics tested, the isolates Bacillus sp. RCS1, Pantoea agglomerans RCS2, and Bacillus cereus RCS3 were susceptible to 15, 16, and 11 of the antibiotics, respectively. They showed intermediate susceptibility to 3, 3, and 5 antibiotics, respectively and resistance to 4, 3, and 6 antibiotics, respectively.
In order to analyze the bile resistance, the isolated microbes were subjected to 0.5% bile salt stress and resistance assay. The resistance was observed by counting the unit colonies formed subsequently after 4 h of exposure and calculated as a percentage. After 3 h of exposure, the result indicated that 76.16% of the Pantoea agglomerans RCS3 survived, while 54.06 and 65.38% of Bacillus sp. RCS1 and Bacillus cereus RCS2 survived, respectively. Nonetheless, the percentage of survivability of all the isolated bacteria dropped subsequently after 4 h of exposure to bile salt but still remained above 50% (Figure 3).
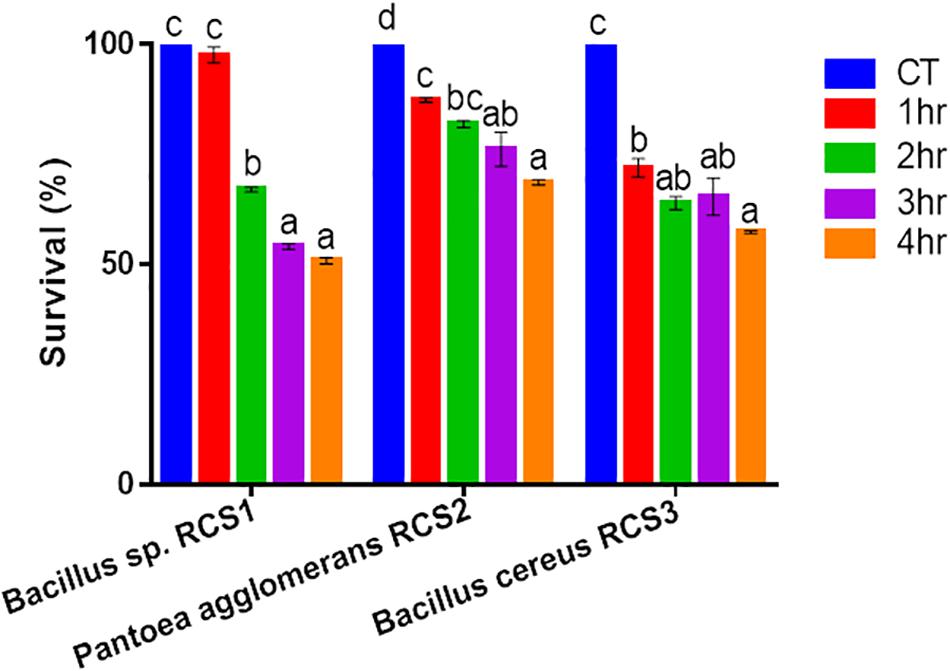
Figure 3. Bile endurance of bacteria after 4 h at 37°C. Values are presented as mean ± SE. Different letters show significant differences (P < 0.05).
The isolated bacteria showed promising results after exposing them to different temperatures of 100, 90, and 80°C for different duration of 10, 5, and 2 min, respectively. Though the multiplying capacity of the strains declined differentially with accumulative temperature, they could still proliferate at 80, 90, and 100°C, demonstrating that these strains could tolerate temperature up to 100°C. In comparison with controls, an increase in the growth (OD) of all the three microbes was observed at all the three temperature exposures (Figure 4). The compatibility tests showed that there was no convincing sign of dominance of one isolated bacterium over another, suggesting their compatible nature.
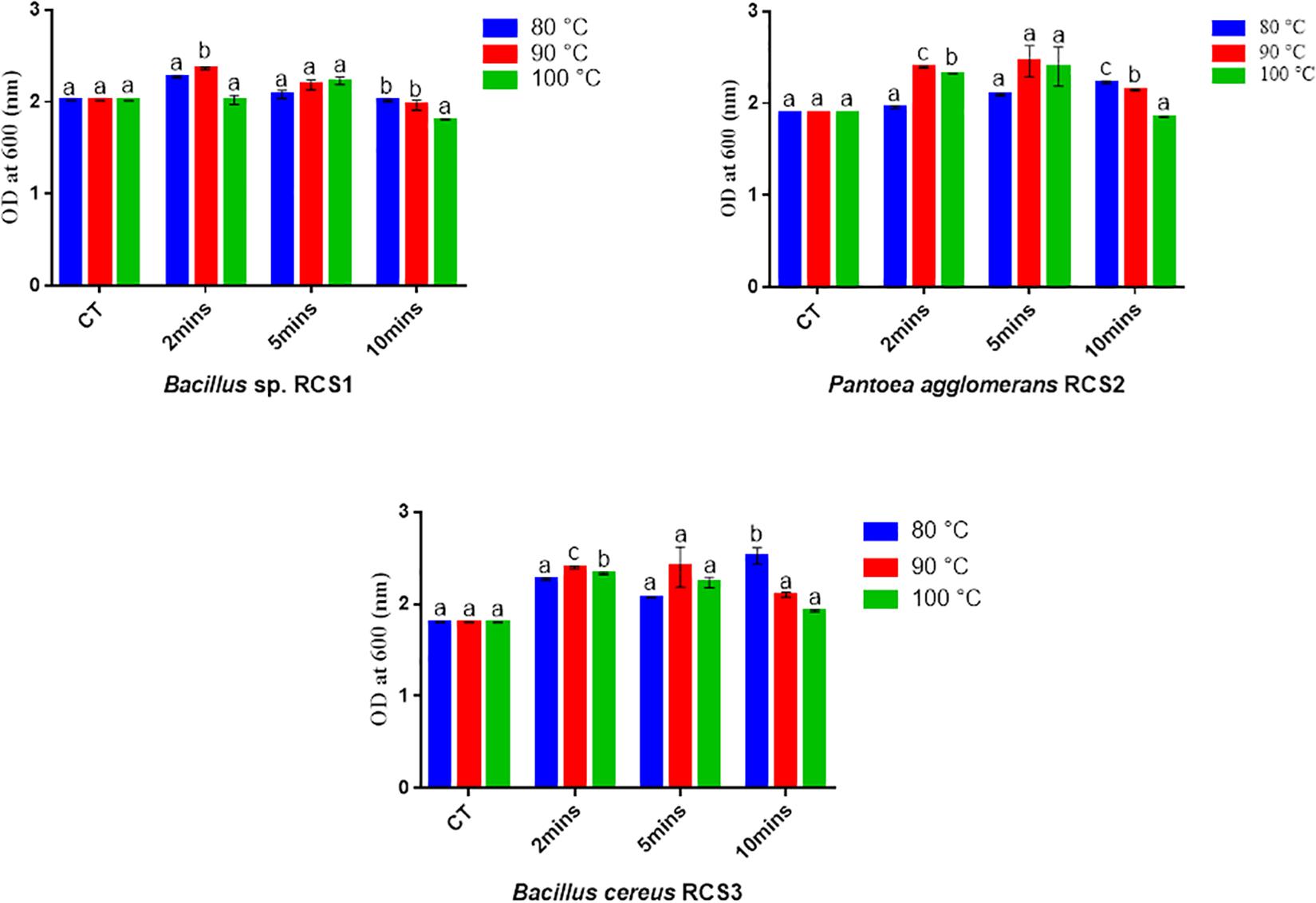
Figure 4. Bacterial endurance to high temperatures. Values are presented as mean ± SE. Different letters show significant differences (P < 0.05). CT indicates control.
Both isolated bacteria were assessed for their antimicrobial activities against four fish pathogens namely; Vibrio alginolyticus, Vibrio haeveyi, Streptococcus iniae, and Streptococcus agalactiae in this study. The isolates were observed to inhibit all the pathogenic bacteria tested in this study (Table 3) and in the agar well diffusion method (Supplementary Figure 1).
The results showed that all the isolated bacteria in the present study had low auto-aggregation capacity within the first 3 h of incubation. Nonetheless, their auto-aggregation capacity increased after 24 h–79 ± 0.44, 80.1 ± 0.25, and 80.6 ± 0.13%, respectively (Figure 5).
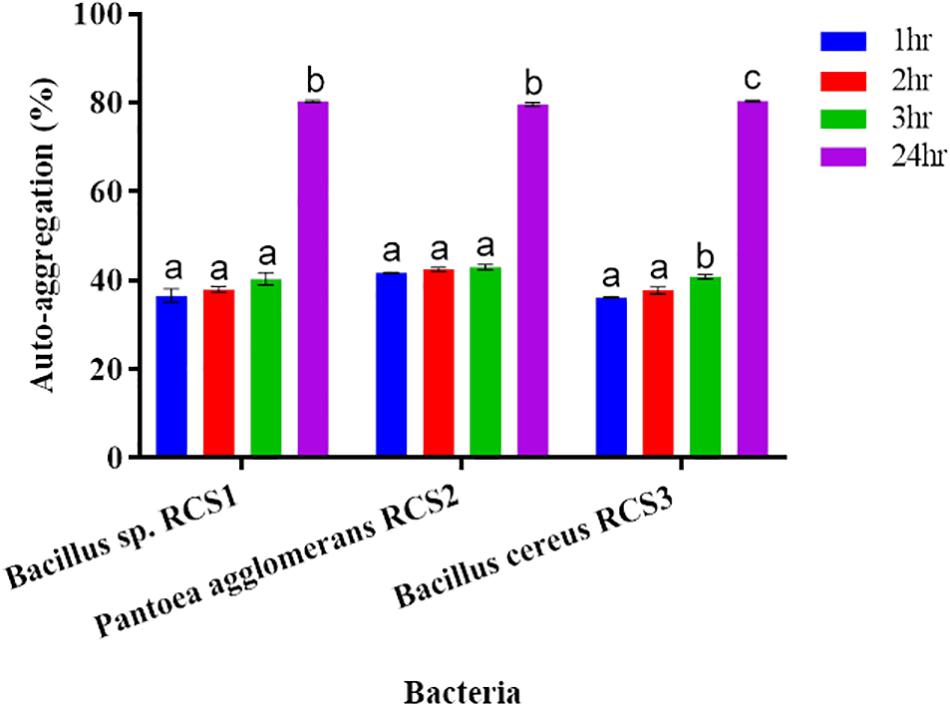
Figure 5. Auto-aggregation of bacteria after 24 h. Values are presented as mean ± SE. Different letters show significant differences (P < 0.05).
The adhesion of bacterial isolates to the ethyl acetate (a basic solvent), chloroform (an acidic solvent), and xylene (a non-polar solvent) was assessed, and the percentages of their cell surface hydrophobicity competence were consequently calculated. All the three isolated bacterial strains showed excellent adherence to ethyl acetate, chloroform, and xylene. The adhesion of the isolated bacteria to xylene, chloroform and ethyl acetate were investigated in the present study to determine their adhesion competence to cell surfaces. The results indicated that all the three isolated bacteria showed good cell surface hydrophobicity to the three solvents tested as mentioned above, thereby qualifying the adhesion effectiveness (Figure 6).
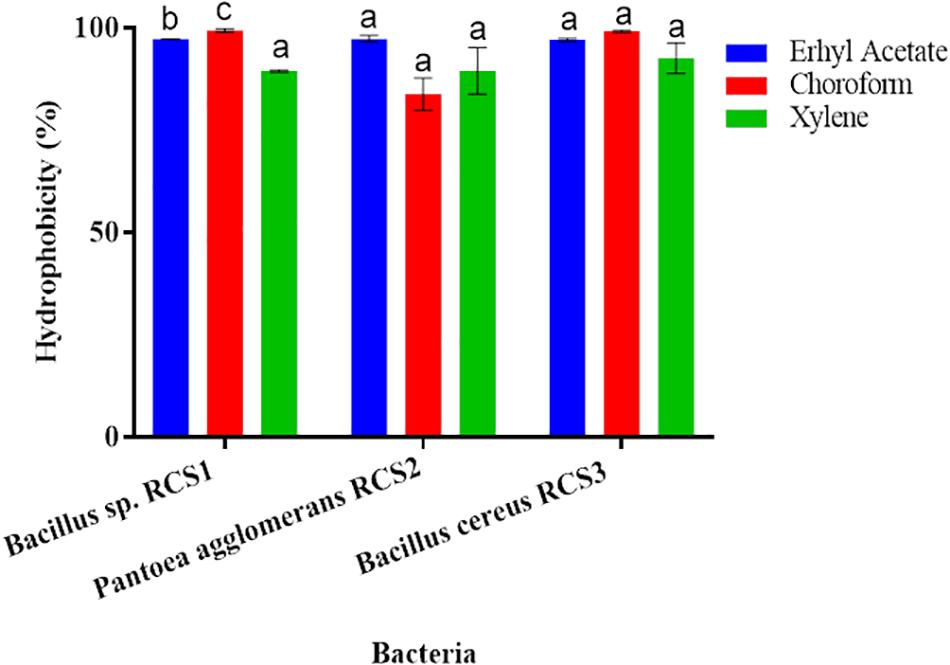
Figure 6. Cell surface hydrophobicity of bacterial strains to various solvents (such as Ethyl acetate, Chloroform and Xylene). Values are presented as mean ± SE. Different letters (a, b, and c) indicate significant differences (P < 0.05).
The results of hemolytic activities showed that Pantoea agglomerans RCS2 and Bacillus cereus RCS3 displayed α-hemolysis, while Bacillus sp. RCS1 demonstrated γ-hemolysis (Table 1).
The isolated bacteria showed strong tolerance when exposed to a range of pH. Although asymmetrically, a slow rise in the growth of isolated microbes was observed within the pH range of 1–8 for RC1, 1–7 for RC2, and 1–6 for RC3. The bacterial growth was observed to be declined at pH of 10.0. This indicated that the isolated microbes could endure both the alkaline as well as highly acidic conditions. At some points in different pH conditions, the three isolated bacteria exhibited significant differences (P < 0.05) (Figure 7).
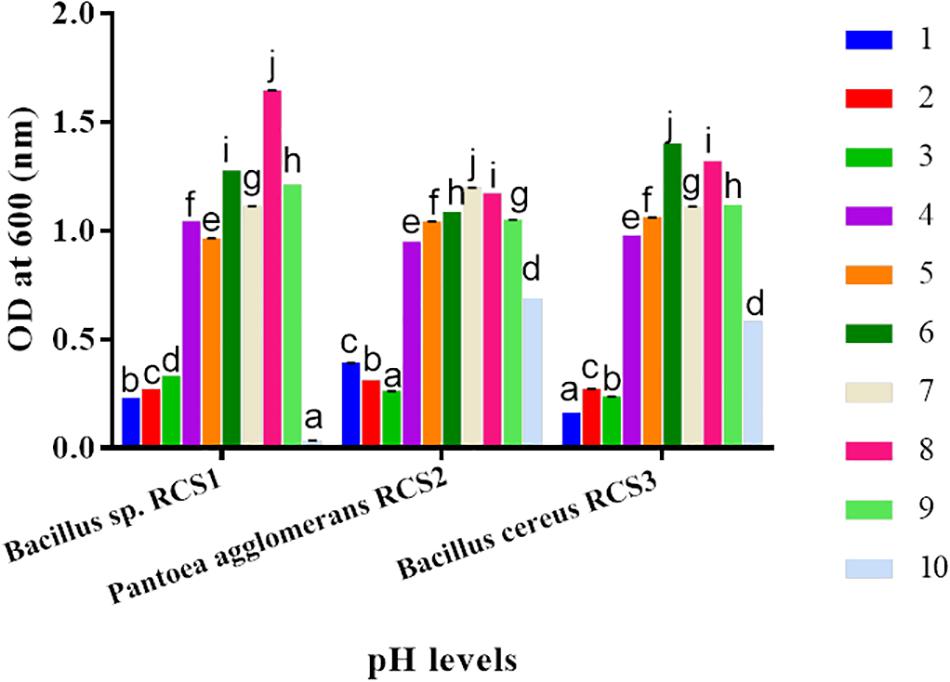
Figure 7. Bacteria growths at different pH (1.0–10.0) levels. Values are shown as mean ± SE. Different letters (a, b, c, d, e, f, g, h, i, and j) indicate significant differences (P < 0.05).
After 24 h of incubation at 37°C, the morphology of bacterial colonies changed to different colors, which indicated the absence or presence of biofilm production. The screening and analysis of the biofilm-producing capability of the isolated bacteria were investigated using Congo red agar method. The results showed that none of the isolated microorganisms formed black colonies, suggesting that they all were non-biofilm producing strains.
Discussion
In an endeavor to curtail the incidence of fish infections in aquaculture, the isolation of probiotics from host’s tract and then utilization as probiotics is a promising alternative to antibiotics, which have extensively utilized but have adversarial effects as well (Magnadottir, 2010; Resende et al., 2012). The isolation of probiotics from host’s tract is also use for promoting the condition and health of the host as well. In the present study, the indigenous Bacillus sp. RCS1, Pantoea agglomerans RCS2, and Bacillus cereus strain RCS3 were isolated from the intestines of cobia fish through culture and in vitro approaches, and their probiotic potentials were evaluated. These isolated indigenous potential probiotics bacteria were assessed using different biochemical and morphological tests and identified using 16S rRNA gene sequencing. For the identification of different microbial species, their specific reactions or biochemical responses are very important. The biochemical tests depend on some enzymes, such as gelatinase, urease and catalase, which are produced by these microbes. Different kinds of bacteria produce different spectra of enzymes. For instance, certain enzymes are essential for the metabolism of individual bacteria, while others expedite the bacterium’s capacity to compete with other microorganisms or launch an infection. Traditionally, the living organisms have been classified, according to similarities and differences in their phenotypic characteristics. However, the objective of taxonomic classification by these methods can be difficult because of variations in their phenotypic characteristics. Nowadays, the 16S rRNA gene sequencing has been widely used for the bacterial identification (Woese et al., 1990).
Several strains from Bacillus genus have been previously reported to effectively protect cultured aquaculture species from pathogens (Mishra, 2011; Silva et al., 2012; Laranja et al., 2017; Mukherjee et al., 2019; Hu et al., 2021). Besides, other studies have demonstrated that the dietary species of Bacillus species, including B. cereus, B. subtilis, and B. subtilis T13 (Wu et al., 2014), have improved the sea cucumber’s growth and quality of water against pathogens (Galagarza et al., 2018). Bacillus sp. JL47 (Silva et al., 2012), B. subtilis AQAHBS001 (Zhang et al., 2010), and B. cereus BC-1 (Yang et al., 2015) have been reported to fight against pathogens and regulate microbiota to improve the growth performance of cultured species. Nevertheless, the inhibitory effects of B. cereus are still not wholly studied in many species, including cobia.
In the present study, the growth of V. alginolyticus, V. haeveyi, S. iniae, and S. agalactiae was inhibited by Bacillus sp. RCS1, Pantoea agglomerans RCS2, and Bacillus cereus strain RCS3. The potential probiotics bacteria display antagonism against pathogenic microorganisms by producing antibiotics, which affect the colon microbiota. The production of “antibacterial compounds” is a key property of probiotics, which distinguish them from the other bacteria. The ability to produce “antibacterial compounds” is an actual or fundamental property for the competitive inhibition of bacterial pathogens and is a vital characteristic of potential probiotic bacterial strains.
The antagonistic activities of Bacillus sp. RCS1 and Bacillus cereus strain RCS3 have inhibition zones within the region of 2.0 cm. The antagonistic elements could impede the reproduction and growth of pathogens of cultured species in aquaculture and efficiently inhibited and regulated the incidence of infections in culture fish species (Marianeto et al., 2012; Sumi et al., 2015). The isolated bacteria Bacillus sp. RCS1 and Bacillus cereus strain RCS3 in the present study exploited a wide range of carbon sources, including starch, adonitol, sorbitol, rhamnose, glucose, mannitol, citrate, and inositol, as well as amino acid arginine, which indicated that these isolates might be useful in the assimilation of carbohydrates and hydrolysis of amino acids (arginine) (Ramesh et al., 2015; Lee et al., 2017; Kavitha et al., 2018). Bacillus species are characterized with heat tolerance (Nicholson et al., 2000; Guo et al., 2016) and resistance to low pH and a high proportion of bile concentration (Spinosa et al., 2000; Barbosa et al., 2005). They are also capable of evolving and enduring in the intestines of fish (Hoa et al., 2000; Barbosa et al., 2005; Hong et al., 2009). The probiotics need to tolerate the intestinal (high bile concentration) and gastric (low pH) environments to colonize there and endure intestine and yield valuable traits for host (Guglielmotti et al., 2007; Cartman et al., 2008). A probiotic bacterial strain should be capable of surviving gastric acidic environment, which makes the acid tolerance as an essential selection standard for a bacterial strain to be probiotic. In order to reach colon, the probiotic microbes are more resilient to stomach acidity than other microbes. They are generally exposed to intestinal acids with pH ranges from 2.5 to 3.5. Similarly, the heat-treatment is an indispensable procedure in the course of feed preparation to kill pathogens and increase palatability (Guo et al., 2016). The isolated potential probiotics in the present study demonstrated that they were capable of enduring low pH, higher temperatures (100, 90, and 80°C) and 0.5% bile concentration. Therefore, it can be suggested that the high temperatures triggered the microbe’s strains, which resulted in increase in their growth. The greater capability of the isolated strains to withstand heat treatments strongly indicated that the isolates can be utilized as feed supplements.
Pantoea agglomerans have revealed exceptional metabolic competences (Smith et al., 2013). They can be utilized against human, plant, and animal pathogens (Dutkiewicz et al., 2016a). In the present study, Pantoea agglomerans RCS2 was isolated from the intestines of cobia fish through culture and in vitro approaches, their probiotic potentials were evaluated. This isolated indigenous probiotic microbe was assessed using different biochemical and morphological tests and identified using 16S rRNA gene sequencing. It also exploited a wide range of carbon sources, including starch, adonitol, sorbitol, rhamnose, glucose, mannitol, citrate, and inositol, as well as amino acid arginine, which indicated that the isolate might be useful in the assimilation of carbohydrates and hydrolysis of amino acids (Ramesh et al., 2015; Lee et al., 2017; Kavitha et al., 2018). The growth of V. alginolyticus, V. haeveyi, S. iniae, and S. agalactiae was inhibited by Pantoea agglomerans RCS2, having the inhibition zones within the region of 2.0 cm.
Researchers in Japan have demonstrated an exceptionally wide spectrum of curative properties of P. agglomerans LPS (IP-PA1), mostly owing to its macrophage-triggering ability, which plays a significant role in the maintenance of homeostasis in all multi-cellular animals and averts several categories of stresses, such as chronic psychological stress (Kohchi et al., 2006; Dutkiewicz et al., 2016a). Skalli et al. (2013) also reported that the LPS derived from the cell walls of Gram-negative bacteria Pantoea agglomerans stimulated the growth and immune status of rainbow trout (Oncorhynchus mykiss) juveniles. An additional valuable asset of IP-PA1 is its efficacy in the treatment of wide range of diseases (Inagawa et al., 1992a, b, 2011; Nishizawa et al., 1992; Hebishima et al., 2010a, b). Hebishima et al. (2010b) showed that IP-PA1 is an effective edible immuno-modulator that can be utilized to treat and prevent a wide range of infections, either triggered or aggravated by stress-induced immunosuppression in humans and several other animals. According to Nakata et al. (2011), a major reason of the development of infection is the destruction of macrophage’s function that plays a crucial role in the maintenance of homeostasis and innate immunity. Although the above-mentioned functions of P. agglomerans LPS (IP-PA1) were observed in human, we presumed that the same functions could be performed in fish. The Pantoea agglomerans RCS2 showed promising characteristics, which were comparable to the one mentioned above.
The physiological concentration of bile salts ranges between 0.3 and 0.5% in intestinal tract (Begley et al., 2005). The ability of probiotics bacteria to withstand the bile salts is related to the action of bile salt hydrolases, which alleviate the inhibitory effect of bile by hydrolyzing the conjugated bile salts (Oh et al., 2000; Mourad and Nour-Eddine, 2006). After 3 h of exposure, the results indicated that these potential probiotics bacteria could endure bile salts up to 0.5%.
Hydrophobicity could be beneficial for the strains that compete with other microorganisms in digestive system (Todorova et al., 2007; Yerlikaya, 2018). The adhesion to and colonization of mucosal surfaces and epithelial cells (Conventional enterocytes “colonocytes in colon,” of prominence are goblet cells) are the essential features of potential probiotics, as it ensure their ability to resist the vacillation of gastric contents, as well as besides inhibits the adhesion of pathogenic microbes and inflammatory reactions (Kos et al., 2003; Guo et al., 2010; Sim et al., 2015). The adhesion capability of probiotics can be indirectly evaluated by finding their hydrophobicity and auto-aggregation (Collado et al., 2008; Meidong et al., 2017; Kuebutornye et al., 2019). Per Wasko et al. (2014) reported bacterial hydrophobicity as vital for adhesion, while others found no connection between the microorganisms’ adhesive properties and hydrophobicity (Iturralde et al., 1993). Microorganisms “favor a substrate for adhesion resembling their surface charge” (An and Friedman, 1998). In the present study, all the isolates (Bacillus sp. RCS1, Pantoea agglomerans RCS2, and Bacillus cereus RCS3), exhibited considerable high hydrophobicity: 97.2, 97.3, and 97.1%, respectively, in ethyl acetate; 99.1, 83.8, and 99.1%, respectively, in chloroform; and 89.4, 89.5, and 92.5%, respectively, in xylene. This demonstrated their adhesion potential to hydrocarbons. There is no standard requirement for hydrophobicity value in bacteria but high hydrophobicity is favored for probiotic properties (Yerlikaya, 2018).
The results of hydrophobicity in the present study were similar to those reported by Kuebutornye et al. (2019) with respect to Bacillus species, but were comparatively higher as reported by Lee et al. (2017); Manhar et al. (2015). This indicated higher affinity for electron acceptance (ethyl acetate) and electron donation (chloroform) of the current isolates, thereby suggesting their higher epithelial cells adhesion potential and qualifying their adhesion effectiveness. The in vitro assessment of auto-aggregation could be utilized for the initial selection and screening of the most refined probiotic strains. There is also a solid connection between the adhesion and auto-aggregation of probiotics to the gastrointestinal tract, as a precondition for potential probiotics (Kuebutornye et al., 2019). All the isolates (Bacillus sp. RCS1, and Pantoea agglomerans RCS2, Bacillus cereus RCS3) showed high auto-aggregation (80.2, 79, and 80.4%, respectively) after 24 h of incubation. The in vitro assessment of auto-aggregation could be utilized for the initial selection and screening of the finest probiotics strain. The results of the in vitro assessment of auto-aggregation showed the competence of isolated potential probiotics to self-aggregate efficiently.
The important preconditions for the selection of probiotic strains are their lack of antibiotic resistance and hemolytic activity (Argyri et al., 2013). The hemolysis is well-known virulent factor, which triggers infections by entering into trivial lesions in skin and mucous membranes of host (Ramesh et al., 2015; Nandi et al., 2017). The safe hemolysis includes α-hemolysis, γ-hemolysis, and no hemolysis, whilst the β-hemolysis is thought to be dangerous (Prescott, 2005; Shin et al., 2012; Pelczar, 2017)1. Pantoea agglomerans RCS2 and Bacillus cereus RCS3 exhibited α-hemolysis while Bacillus sp. RCS1 exhibited γ- hemolysis in the present study. Comparable observations were reported by Lee et al. (2017) and Kavitha et al. (2018) for Bacillus strains. In this study, 22 antibiotics were tested, among which, Bacillus sp. RCS1 was susceptible to 15, intermediate susceptible to 3 and resistant to 4 (Table 3), Pantoea agglomerans RCS2 was susceptible to 16, intermediate susceptible to 3 and resistant to 3, while Bacillus cereus RCS3 was susceptible to 11, intermediate susceptible to 5 and resistant to 6. These results indicated that these isolates are susceptible to a sufficient number of antibiotics tested (Table 2), which were similar to the results reported for Bacillus species by Kuebutornye et al. (2019).
Biofilm producing microbes cause nosocomial and recurring infections. The formation of biofilm starts with the adhesion of microorganisms to abiotic surfaces, such as a host cell. After attachment, the aggregation of microbes is initiated by cell-cell adhesion. Congo red agar method is a qualitative assay for the detection of biofilm producing microbes, indicated by the differences in the colors of colonies on Congo red agar medium (Kırmusaoğlu, 2019). A study has shown that the infections could be accompanied by the formation of microbial biofilm (Schönborn and Krömker, 2016). Donlan (2002) described biofilm as the grouping of microbes permanently enclosed in a matrix and attached to a surface. A number of studies relate biofilms with the failure of antibiotic therapy and persistent infections. The biofilm can escape hosts’ immune system (An and Friedman, 1998) and decrease the influence of valuable antibiotics (Anderl et al., 2000; Zahller and Stewart, 2002). Biofilms possess a great importance for public wellbeing. For instance, the biofilm-producing microbes show reduced susceptibility to antagonistic agents (Donlan, 2001), despite the benefits related to biofilm formation (O’Toole et al., 2000; Morikawa, 2006). Meanwhile, the antibiotics-resistant species are thought to be dangerous for being used as probiotics. Frola et al. (2012) reported that the formation of biofilm can be beneficial due to the colonization of the internal surfaces of udder, thereby building a fence against pathogenic microbes, an imperative factor of the potential probiotics strains. All the three isolated microorganisms in this study tested negative for biofilm formation, as reported by Kavitha et al. (2018).
Some of the well-known fish diseases, reported in aquaculture, are caused by Vibrio (Bluford et al., 2017), Aeromonas (Amal et al., 2018), and Streptococcus (Shoemaker et al., 2001) species. Particularly, in cobia fish, streptococcosis, vibriosis, furunculosis, and mycobacteriosis, and pasteurellosis, are considered as emerging fish diseases in aquaculture (Lopez et al., 2002; Liu et al., 2003; Rajan et al., 2003; Liao et al., 2004; Chen and Hsu, 2005). Several studies have reported that a number of Bacillus species exhibit antagonistic effects against a number of Gram-negative and Gram-positive pathogenic bacteria. All the isolated microbes in this study, including Bacillus sp. RCS1, Pantoea agglomerans RCS2, and Bacillus cereus RCS3 were all effective against V. alginolyticus, V. harveyi, S. iniae, and S. agalactiae. These results indicated that these isolated bacterial strains are potential probiotics and can be utilized to combat fish diseases in aquaculture.
Conclusion
In conclusion, the bacterial strains, including Bacillus sp. RCS1, Pantoea agglomerans RCS2, and Bacillus cereus RCS3 were isolated from the intestine of a healthy juvenile cobia fish (Rachycentron canadum). All the isolated microorganisms exhibited inhibitory effects against the pathogens, including V. alginolyticus, V. harveyi, S. iniae, and S. agalactiae. These microbes are characterized with a wide range of antagonistic activities, non-hemolytic activities, high survivability after heat-treatments and safety confidence as well antibiotic susceptibility. Generally, the characteristics displayed by these microorganisms indicated that they could be potentially used as probiotics in aquaculture industry.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, (MW560712, MW560713, and MW560714).
Ethics Statement
This animal study was reviewed and approved by the Guangdong Ocean University Research Council (approval number: GDOU-LAE-2020-013).
Author Contributions
EA participated in data curation, data analysis, writing, reviewing, and editing of the original article. W-ZW participated in data collection. GC and J-SH planned and designed the experiments, supervised the experiments, and acquired funding. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the grants from Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang) (ZJW-2019-06) and China Agriculture Research System (CARS-47).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.672213/full#supplementary-material
Supplementary Figure 1 | Graphical representation of the morphological and antagonistic activities of the three isolated bacterial strains. Lane 1: morphology of RCS1, RCS2, RCS3. Lane 2: antagonistic activities of RCS1, RCS2, and RCS3 against Vibrio haeveyi. Lane 3: antagonistic activities of RCS1, RCS2, and RCS3 against Vibrio alginolyticus. Lane 4: antagonistic activities of RCS1, RCS2, and RCS3 against Streptococcus iniae. Lane 5: antagonistic activities of RCS1, RCS2, and RCS3 against Streptococcus agalactiae. Note: A represents Bacillus spp.RCS1; C represents Pantoea agglomerans strain RCS2 and F represents Bacillus Cereus strain RCS3; VH indicates Vibrio haeveyi; VA indicates Vibrio alginolyticus; SI indicates Streptococcus iniae, and SA indicates Streptococcus agalactiae.
Footnotes
References
Abarike, E. D., Jian, J., Tang, J., Cai, J., Yu, H., Lihua, C., et al. (2018). Influence of traditional Chinese medicine and Bacillus species (TCMBS) on growth, immune response and disease resistance in Nile tilapia, Oreochromis niloticus. Aquac. Res. 49, 2366–2375. doi: 10.1111/are.13691
Aleti, G., Sessitsch, A., and Brader, G. (2015). Genome mining: prediction of lipopeptides and polyketides from Bacillus and related firmicutes. Comput. Struct. Biotechnol. J. 15, 192–203. doi: 10.1016/j.csbj.2015.03.003
Resende, A. J. L., Silva, V., Oliveira Fontes, C., Alves Souza-Filho, J., Rocha de Oliveira, T. L., et al. (2012). Multidrug-resistance and toxic metal tolerance of medically important bacteria isolated from an aquaculture system. Microb. Environ. 27, 449–455. doi: 10.1264/jsme2.me12049
Amal, M. N. A., Koh, C. B., Nurliyana, M., Suhaiba, M., Nor-Amalina, Z., Santha, S., et al. (2018). A case of natural co-infection of Tilapia Lake virus and Aeromonas veronii in a Malaysian red hybrid tilapia (Oreochromis niloticus×O. mossambicus) farm experiencing high mortality. Aquaculture 485, 12–16. doi: 10.1016/j.aquaculture.2017.11.019
Amenyogbe, E., Chen, G., Wang, Z., Huang, J.-S., Huang, B., and Li, H.-J. (2020). The exploitation of probiotics, prebiotics and synbiotics in aquaculture: present study, limitations and future directions: a review. Aquacult Int. 28, 1017–1041. doi: 10.1007/s10499-020-00509-0
An, Y. H., and Friedman, R. J. (1998). Concise review of mechanisms of bacterial adhesion to biomaterial surfaces. J. Biomed. Mater. Res. 43, 338–348. doi: 10.1002/(sici)1097-4636(199823)43:3<338::aid-jbm16>3.0.co;2-b
Anderl, J. N., Franklin, M. J., and Stewart, P. S. (2000). Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Ch. 44, 1818–1824. doi: 10.1128/aac.44.7.1818-1824.2000
Argyri, A. A., Zoumpopoulou, G., Karatzas, K. A. G., Tsakalidou, E., Nychas, G. J. E., Panagou, E. Z., et al. (2013). Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 33, 282–291. doi: 10.1016/j.fm.2012.10.005
Balcázar, J. L., Vendrell, D., de Blas, I., Ruiz-Zarzuela, I., Gironés, O., and José Luis Múzquiz, J. L. (2007). In vitro competitive adhesion and production of antagonistic compounds by lactic acid bacteria against fish pathogens. Vet. Microbiol. 122, 373–380. doi: 10.1016/j.vetmic.2007.01.023
Bandyopadhyay, P., and Patra, B. C. (2004). Probiotics for Sustainable Aquaculture and its Utilization Trial: Course Manual of the Summer School on Development of Sustainable Aquaculture Technology for Fresh and Saline Waters, Department of Zoology. India: CCS Haryana Agricultural University, 61–66.
Barbosa, T. M., Serra, C. R., La Ragione, R. M., Woodward, M. J., and Henriques, A. O. (2005). Screening for Bacillus isolates in the broiler gastrointestinal tract. Appl. Environ. Microbiol. 71, 968–978. doi: 10.1128/AEM.71.2.968-978.2005
Begley, M., Gahan, C. G., and Hill, C. (2005). The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651. doi: 10.1016/j.femsre.2004.09.003
Benetti, D. D., O’Hanlon, B., Rivera, J. A., Welch, A. W., Maxey, C., and Orhun, M. R. (2010). Growth rates of cobia (Rachycentron canadum) cultured in open ocean submerged cages in the Caribbean. Aquaculture 302, 195–201. doi: 10.1016/j.aquaculture.2010.02.021
Benetti, D. D., Orhun, M. R., Sardenberg, B., O’Hanlon, B., Welch, A. W., Hoenig, R., et al. (2008). Advances in hatchery and grow-out technology of cobia Rachycentron canadum (Linnaeus). Aquacult. Res. 39, 701–711. doi: 10.1111/j.1365-2109.2008.01922.x
Bhatnagar, A., and Dhillon, O. (2019). Characterization, screening and application of bacteria with probiotic adequacy isolated from the gut of Labeo calbasu (Hamilton, 1822). Fisher. Aquat. Life 27, 178–189. doi: 10.2478/aopf-2019-0020
Bhatnagar, A., and Lamba, R. (2015). Antimicrobial ability and growth promoting effects of feed supplemented probiotic bacterium isolated from gut microflora of Cirrhinus mrigala. J. Integrat. Agricult. 14, 583–592. doi: 10.1016/s2095-3119(14)60836-4
Bhatnagar, A., and Lamba, R. (2017). Molecular characterization and dosage application of autochthonous potential probiotic bacteria in Cirrhinus mrigala. J. Fisher. Sci. Com 11, 46–56.
Bhatnagar, A., and Raparia, S. (2014). Optimum dietary inclusion level of Bacillus coagulans for growth and digestibility improvement for Catla catla (Hamilton). Int. J. Curr. Res. Rev. 6, 1–10.
Bhatnagar, A., Raparia, S., and Kumari, S. (2012). Influence of isolated Bacillus coagulans on growth performance and digestive enzyme activities of Catla catla. J. Nat. Sci. Sustain. Technol. 6, 225–235.
Bluford, J., Gauthier, D., Colasanto, M., Rhodes, M., Vogelbein, W., and Haines, A. (2017). Identification of virulence genes in Vibrio spp. isolates from the 2009 Bermuda reef fish mortality event. J. Fish Dis. 40, 597–600. doi: 10.1111/jfd.12532
Carnevali, O., de Vivo, L., Sulpizio, R., Gioacchini, G., Olivotto, I., Silvi, S., et al. (2006). Growth improvement by probiotic in European sea brass juveniles (Dicentrachus labrax, L.), with particular attention to IGF-1, myostatin and cortiosol gene expression. Aquaculture 258, 430–438. doi: 10.1016/j.aquaculture.2006.04.025
Cartman, S. T., La Ragione, R. M., and Woodward, M. J. (2008). Bacillus subtilis spores germinate in the chicken gastrointestinal tract. Appl. Environ. Microbiol. 74, 5254–5258. doi: 10.1128/AEM.00580-08
Chalupowicz, L., Manulis-Sasson, S., Itkin, M., Sacher, A., Sessa, G., and Barash, I. (2008). Quorum-sensing system affects gall development incited by Pantoea agglomerans pv. gypsophilae. Mol. Plant Microbe. Interact. 21, 1094–1105. doi: 10.1094/mpmi-21-8-1094
Chen, B., Peng, M., Tong, W., Zhang, Q., and Song, Z. (2019). The quorum quenching bacterium Bacillus licheniformis T-1 protects zebrafish against Aeromonas hydrophila infection. Probiot. Antimicrob. Proteins 12, 160–171. doi: 10.1007/s12602-018-9495-7
Chen, Q., Liu, H., Tan, B. P., Dong, X. H., Chi, S. Y., Yang, Q. H., et al. (2016). Effects of dietary cholesterol level on growth performance, blood biochemical parameters and lipid metabolism of juvenile cobia (Rachycentron canadu). J. Guangd. Ocean Univ. 36, 35–43.
Chen, S. C., and Hsu, C. (2005). Studies on the pathogenicity and pathology of photobacterium damselae subsp. piscicida on Rachycentron canadum. J. Fish. Soc. Taiwan 32:4.
Clinical and Laboratory Standards Institute (2009). Clinical and Laboratory Standards Institute, Performance Standards for Antimicrobial Disk Susceptibility Tests. Approved Standard M2-A10. Wayne, PA: Clinical and Laboratory Standards Institute.
Collado, M. C., Meriluoto, J., and Salminen, S. (2008). Adhesion and aggregation properties of probiotic and pathogen strains. Eur. Food Res. Technol. 226, 1065–1073. doi: 10.1007/s00217-007-0632-x
Dlamini, M. T., Lessells, R. J., Iketleng, T., and Oliveira, T. (2019). Whole genome sequencing for drug-resistant tuberculosis management in South Africa: what gaps would this address and what are the challenges to implementation. J. Clin. Tubercul. Other Mycobacter. Dis. 16:100115. doi: 10.1016/j.jctube.2019.100115
Donlan, R. M. (2001). Biofilm formation: a clinically relevant microbiological process. Clin. Infect. Dis. 33, 1387–1392. doi: 10.1086/322972
Donlan, R. M. (2002). Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8, 881–890. doi: 10.3201/eid0809.020063
Dutkiewicz, J., Mackiewicz, B., Lemieszek, M. K., Golec, M., and Milanowski, J. (2016a). Pantoea agglomerans: a mysterious bacterium of evil and good. Part IV. Beneficial effects. Ann. Agric. Environ. Med. 23, 206–222. doi: 10.5604/12321966.1203879
Dutkiewicz, J., Mackiewicz, B., Lemieszek, M. K., Golec, M., and Milanowski, J. (2015). Pantoea agglomerans: a mysterious bacterium of evil and good. Part I. Deleterious effects: dust-borne endotoxins and allergens – focus on cotton dust. Ann. Agric. Environ. Med. 22, 576–588. doi: 10.5604/12321966.1185757
Dutkiewicz, J., Mackiewicz, B., Lemieszek, M. K., Golec, M., and Milanowski, J. (2016b). Pantoea agglomerans: a mysterious bacterium of evil and good. Part III. Deleterious effects: Infections of humans, animals and plants. Ann. Agric. Environ. Med. 23, 197–205. doi: 10.5604/12321966.1203878
Dutkiewicz, J., Mackiewicz, B., Lemieszek, M. K., Golec, M., Skórska, C., Góra- Florek, A., et al. (2016c). Pantoea agglomerans: a mysterious bacterium of evil and good. Part II. Deleterious effects: Dust-borne endotoxins and allergens – focus on grain dust, other agricultural dusts and wood dust. Ann. Agric. Environ. Med. 23, 110–133.
Engel, R. R., Matsen, J. M., Chapman, S. S., and Schwartz, S. (1972). Carbon monoxide production from heme compounds by bacteria. J. Bacteriol. 112, 1310–1315. doi: 10.1128/jb.112.3.1310-1315.1972
Food and Agriculture Organization (FAO) (2001). Food and Agricultural Organization 815 of the United Nations and World Health Organization. Health and Nutritional Properties of Probiotics in Food Including Powder Milk With Live Lactic Acid Bacteria. Rome: RAO.
Food and Agriculture Organization (FAO) (2015). Global Aquaculture Production 1950-2013. Available online at: http://www.fao.org/fishery/statistics/global-aquaculture-production/query/en (accessed November 15, 2015).
Food and Agriculture Organization (FAO) (2020). “Fishery and aquaculture statistics. Global production by production source 1950-2018 (FishstatJ),” in FAO Fisheries and Aquaculture Department [online], Rome. Available online at: www.fao.org/fishery/statistics/software/fishstatj/en
Frola, I. D., Pellegrino, M. S., Espeche, M. C., Giraudo, J. A., Nader-Macias, M. E., and Bogni, C. I. (2012). Effects of intramammary inoculation of Lactobacillus perolens CRL1724 in lactating cows’ udders. J. Dairy Res. 79, 84–92. doi: 10.1017/S0022029911000835
Galagarza, O. A., Smith, S. A., Drahos, D. J., Eifert, J. D., Williams, R. C., and Kuhn, D. D. (2018). Modulation of innate immunity in nile tilapia (Oreochromis niloticus) by dietary supplementation of Bacillus subtilis endospores. Fish Shellf. Immunol. 83, 171–179. doi: 10.1016/j.fsi.2018.08.062
Geng, X., Dong, X.-H., Tan, B.-P., Yang, Q.-H., Chi, S.-Y., Liu, H.-Y., et al. (2011). Effects of dietary probiotic on the growth performance, non-specific immunity and disease resistance of cobia, Rachycentron canadum. Aquacult. Nutri. 18, 46–55. doi: 10.1111/j.1365-2095.2011.00875.x
Guglielmotti, D. M., Marcó, M. B., Golowczyc, M., Reinheimer, J. A., and Quiberoni, A. L. (2007). Probiotic potential of Lactobacillus delbrueckii strains and their phage resistant mutants. Int. Dairy J. 17, 916–925. doi: 10.1016/j.idairyj.2006.11.004
Guo, X. H., Kim, J. M., Nam, H. M., Park, S. Y., and Kim, J. M. (2010). Screening lactic acid bacteria from swine origins for multistrain probiotics based on in vitro functional properties. Anaerobe 16, 321–326. doi: 10.1016/j.anaerobe.2010.03.006
Guo, X., Chen, D. D., Peng, K. S., Cui, Z. W., Zhang, X. J., Li, S., et al. (2016). Identification and characterization of Bacillus subtilis from grass carp (Ctenopharynodon idellus) for use as probiotic additives in aquatic feed. Fish Shellf. Immunol. 52, 74–84. doi: 10.1016/j.fsi.2016.03.017
Hao, K., Liu, J., Ling, F., Liu, X., Lu, L., Xia, L., et al. (2014). Effects of dietary administration of Shewanella haliotis D4, Bacillus cereus D7 and Aeromonas bivalvium D15, single or combined, on the growth, innate immunity and disease resistance of shrimp, Litopenaeus vannamei. Aquaculture 428, 141–149. doi: 10.1016/j.aquaculture.2014.03.016
Hebishima, T., Matsumoto, Y., Watanabe, G., Soma, G., Kohchi, C., Taya, K., et al. (2010a). Protective effects of the immunopotentiator from Pantoea agglomerans 1 on chemotherapeutic agent-induced macrophage growth inhibition. Anticancer Res. 30, 2033–2040.
Hebishima, T., Matsumoto, Y., Watanabe, G., Soma, G., Kohchi, C., Taya, K., et al. (2010b). Recovery from immunosuppression-related disorders in humans and animals by IP-PA1, an edible lipopolysaccharide. Anticancer Res. 30, 3113–3118.
Hoa, N. T., Baccigalupi, L., Huxham, A., Smertenko, A., van, P. H., Ammendola, S., et al. (2000). Characterization of Bacillus species used for oral bacteriotherapy and bacterioprophylaxis of gastrointestinal disorders. Appl. Environ. Microbiol. 66, 5241–5247. doi: 10.1128/AEM.66.12.5241-5247.2000
Holt, G. J., Faulk, C. K., and Schwarz, M. H. (2007). A review of the larviculture of cobia Rachycentron canadum, a warm water marine fish. Aquaculture 268, 181–187. doi: 10.1016/j.aquaculture.2007.04.039
Hong, H. A., To, E., Fakhry, S., Baccigalupi, L., Ricca, E., and Cutting, S. M. (2009). Defining the natural habitat of Bacillus spore-formers. Res. Microbiol. 160, 375–379. doi: 10.1016/j.resmic.2009.06.006
Hu, Z., Zhang, W., Liang, W., Zhang, Z., Guo, M., and Li, C. (2021). Bacillus cereus LS2 from Apostichopus japonicus antagonizes Vibrio splendidus growth. Aquaculture 531:735983. doi: 10.1016/j.aquaculture.2020.735983
Huang, J.-S., Amenyogbe, E., Chen, G., and Wang, W.-Z. (2020). Biochemical composition and activities of digestive and antioxidant enzymes during the egg and yolk-sac larval development of the cobia (Rachycentron canadum). Aquacult. Res. 20, 1–14. doi: 10.1111/are.15017
Inagawa, H., Kohchi, C., and Soma, G. (2011). Oral administration of lipopolysaccharides for the prevention of various diseases: benefit and usefulness. Anticancer Res. 31, 2431–2436.
Inagawa, H., Nishizawa, T., Tsukioka, D., Suda, T., Chiba, Y., Okutomi, T., et al. (1992a). Homeostasis as regulated by activated macrophage. II. LPS of plant origin other than wheat flour and their concomitant bacteria. Chem. Pharm. Bull. (Tokyo) 40, 994–997. doi: 10.1248/cpb.40.994
Inagawa, H., Saitoh, F., Iguchi, M., Nishizawa, T., Okutomi, T., Morikawa, A., et al. (1992b). Homeostasis as regulated by activated macrophage. III. Protective effect of LPSw (lipopolysaccharide (LPS) of wheat flour) on gastric ulcer in mice as compared with those of other LPS from various sources. Chem. Pharm. Bull. (Tokyo) 40, 998–1000. doi: 10.1248/cpb.40.998
Iturralde, M., Aguilar, B., Baselga, R., and Amorena, B. (1993). Adherence of ruminant mastitis Staphylococcus aureus strains to epithelial cells from ovine mammry gland primary cultures and from a rat intestinal cell line. Vet. Microbiol. 38, 115–127. doi: 10.1016/0378-1135(93)90079-m
Jiang, J., Wu, S., Wang, J., and Feng, Y. (2015). AHL-type quorum sensing and its regulation on symplasmata formation in Pantoea agglomerans YS19. J. Basic Microbiol. 55, 607–616. doi: 10.1002/jobm.201400472
Kavitha, M., Raja, M., and Perumal, P. (2018). Evaluation of probiotic potential of Bacillus spp. isolated from the digestive tract of freshwater fish Labeo calbasu (Hamilton, 1822). Aquac. Rep. 11, 59–69. doi: 10.1016/j.aqrep.2018.07.001
Kırmusaoğlu, S. (2019). The Methods for Detection of Biofilm and Screening Antibiofilm Activity of Agents. Antimicrobials, Antibiotic Resistance, Antibiofilm Strategies and Activity Methods. London: IntechOpen. doi: 10.5772/intechopen.84411
Knackstedt, R., and Gatherwright, J. (2019). The role of thermal injury on intestinal bacterial translocation and the mitigating role of probiotics: a review of animal and human studies. Burns 46, 1005–1012. doi: 10.1016/j.burns.2019.07.007
Kohchi, C., Inagawa, H., Nishizawa, T., Yamaguchi, T., Nagai, S., and Soma, G. (2006). Applications of lipopolysaccharide derived from Pantoea agglomerans (IP-PA1) for health care based on macrophage network theory. J. Biosci. Bioeng. 102, 485–496. doi: 10.1263/jbb.102.485
Kos, B., Šušković, J., Vuković, S., Šimpraga, M., Frece, J., and Matošić, S. (2003). Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 94, 981–987. doi: 10.1046/j.1365-2672.2003.01915.x
Kuebutornye, F. K. A., Liao, J., Pang, H., Lu, Y., Ayiku, S., and Sakyi, M. E. (2018). Molecular cloning and bioinformatics analysis of T3SS inner membrane ring HrpQ from Vibrio harveyi. Genom. Appl. Biol. 9, 40–47. doi: 10.5376/gab.2018.09.0007
Kuebutornye, F. K. A., Lu, Y., Abarike, E. D., Wang, Z., Li, Y., and Sakyi, M. E. (2019). In vitro Assessment of the probiotic characteristics of three Bacillus species from the gut of nile tilapia, Oreochromis niloticus. Probiot. Antimicrob. Prot. 12, 412–424. doi: 10.1007/s12602-019-09562-5
Laranja, J. L. Q., Amar, E. C., Ludevese-Pascual, G. L., Niu, Y. F., Geaga, M. J., De Schryver, P., et al. (2017). A probiotic Bacillus strain containing amorphous poly-betahydroxybutyrate (PHB) stimulates the innate immune response of Penaeus monodon postlarvae. Fish Shellf. Immunol. 68, 202–210. doi: 10.1016/j.fsi.2017.07.023
Lee, S., Lee, J., Jin, Y. I., Jeong, J. C., Chang, Y. H., Lee, Y., et al. (2017). Probiotic characteristics of Bacillus strains isolated from Korean traditional soy sauce. LWT Food Sci. Technol. 79, 518–524. doi: 10.1016/j.lwt.2016.08.040
Lertcanawanichakul, M., and Sawangnop, S. (2008). A comparison of two methods used for measuring the antagonistic activity of Bacillus species. Walailak J. Sci. Technol. 5, 161–171. doi: 10.2004/wjst.v5i2.86
Li, A., Wang, Y., Pei, L., Mehmood, K., Li, K., Qamar, H., et al. (2019). Influence of dietary supplementation with Bacillus velezensis on intestinal microbial diversity of mice. Microb. Pathog. 136:103671. doi: 10.1016/j.micpath.2019.103671
Liao, I. C., Huang, T. S., Tsai, W. S., Hseueh, C. M., Chang, S. L., and Leano, E. M. (2004). Cobia culture in Taiwan: current status and problems. Aquaculture 237, 155–165. doi: 10.1016/j.aquaculture.2004.03.007
Liu, N., Zhang, S., Zhang, W., and Li, C. (2017). Vibrio sp. 33 a potential bacterial antagonist of Vibrio splendidus pathogenic to sea cucumber (Apostichopus japonicus). Aquaculture 470, 68–73. doi: 10.1016/j.aquaculture.2016.12.028
Liu, P. C., Liu, J. Y., and Lee, K. K. (2003). Virulence of Photobacterium damselae subsp piscicida in cultured cobia Rachycentron canadum. J. Basic Microbiol. 43, 499–507.
Lopez, C., Rajan, P. R., Lin, J. H., Kuo, T., and Yang, H. (2002). Disease outbreak in seafarmed cobia (Rachycentron canadum) associated with Vibrio spp., Photobacterium damselae ssp. piscicida, mongenean and myxosporean parasites. Bull. Eur. Assoc. Fish Pathol. 22, 206–211.
Magnadottir, B. (2010). Immunological control of fish diseases. Mar. Biotechnol. 12, 361–379. doi: 10.1007/s10126-010-9279-X
Manhar, A. K., Saikia, D., Bashir, Y., Mech, R. K., Nath, D., Konwar, B. K., et al. (2015). In vitro evaluation of celluloytic Bacillus amyloliquefaciens AMS1 isolated from traditional fermented soybean (Churpi) as an animal probiotic. Res. Vet. Sci. 99, 149–156. doi: 10.1016/j.rvsc.2015.01.008
Marianeto, S., Candido, E. D., Rodrigues, D. R., Sousa, D. A., Silva, E. M., Moraes, L. M., et al. (2012). Deciphering the magainin resistance process of Escherichia coli strains in light of the cytosolic proteome. Antimicrob. Agents Chemother. 56, 1714–1724. doi: 10.1128/aac.05558-11
Meidong, R., Doolgindachbaporn, S., Jamjan, W., Sakai, K., Tashiro, Y., Okugawa, Y., et al. (2017). A novel probiotic Bacillus siamensis B44v isolated from Thai pickled vegetables (Phakdong) for potential use as a feed supplement in aquaculture. J. Gen. Appl. Microbiol. 63, 246–253. doi: 10.2323/jgam.2016.12.002
Mishra, G. P. (2011). Treatment of drug-resistant tuberculosis. Lancet Infect. Dis. 4, 129–135. doi: 10.2147/idr.s10332
Montes, M., Farto, R., Maria, J. P., Armada, S. P., and Nieto, T. P. (2006). Genotypic diversity of Vibrio isolates associated with turbot (Scophthalmus maximus) culture. Res. Microbiol. 157, 495.
Morikawa, M. (2006). Beneficial biofilm formation by industrial bacteria Bacillus subtilis and related species. J. Biosci. Bioeng. 101, 1–8. doi: 10.1263/jbb.101.1
Mourad, K., and Nour-Eddine, K. (2006). Microbiological study of naturally fermented Algerian green olives: Isolation and identification of lactic acid bacteria and yeasts along with the effects of brine solutions obtained at the end of olive fermentation on Lactobacillus plantarum. Grasas Aceites 57, 292–300.
Mukherjee, A., Chandra, G., and Ghosh, K. (2019). Single or conjoint application of autochthonous Bacillus strains as potential probiotics: effects on growth, feed utilization, immunity and disease resistance in Rohu Labeo rohita (Hamilton). Aquaculture 512:734302. doi: 10.1016/j.aquaculture.2019.734302
Murray, P. R., Baron, E. J., Jorgensen, J. H., Landry, M. L., and Pfaller, M. A. (2007). Antibacterial Susceptibility Tests: Dilution and Disk Diffusion Methods, Manual of Clinical Microbiolog, 9th Edn. Washington, DC: American Society for Microbiology, 1152–1172.
Nadarasah, G., and Stavrinides, J. (2014). Quantitative evaluation of the host-colonizing capabilities of the enteric bacterium Pantoea using plant and insect hosts. Microbiology. 160, 602–615. doi: 10.1099/mic.0.073452-0
Nakata, K., Inagawa, H., and Soma, G. (2011). Lipopolysaccharide IP-PA1 from Pantoea agglomerans prevents suppression of macrophage function in stress-induced diseases. Anticancer Res. 31, 2437–2440.
Nandi, A., Dan, S. K., Banerjee, G., Ghosh, P., Ghosh, K., Ringø, E., et al. (2017). Probiotic potential of autochthonous bacteria isolated from the gastrointestinal tract of four freshwater teleosts. Probiot. Antim. Prot. 9, 12–21. doi: 10.1007/s12602-016-9228-8
Nawrath, M. M., Ottenheim, C., Wu, J. C., and Zimmermann, W. (2020). Pantoea sp. P37 as a novel nonpathogenic host for the heterologous production of rhamnolipids. Microbiol. Open 9:e1019. doi: 10.1002/mbo3.1019
Nayak, S. K., and Mukherjee, S. C. (2011). Screening of gastrointestinal bacteria of Indian major carps. Aquacult. Res. 42, 1034–1041. doi: 10.1111/j.1365-2109.2010.02686.x
Nicholson, W. L., Munakata, N., Horneck, G., Melosh, H. J., and Setlow, P. (2000). Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64, 548–572. doi: 10.1128/MMBR.64.3.548-572.2000
Nishizawa, T., Inagawa, H., Oshima, H., Okutomi, T., Tsukioka, D., Iguchi, M., et al. (1992). Homeostasis as regulated by activated macrophage. I. Lipopolysaccharide (LPS) from wheat flour: isolation, purification and some biological activities. Chem. Pharm. Bull. (Tokyo) 40, 479–483. doi: 10.1248/cpb.40.479
O’Sullivan, D. J. (2001). Screening of intestinal microflora for effective probiotic bacteria. J. Agric. Food Chem. 49, 1751–1760. doi: 10.1021/jf0012244
O’Toole, G., Kaplan, H. B., and Kolter, R. (2000). Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79. doi: 10.1146/annurev.micro.54.1.49
Oh, S., Kim, S. H., and Worobo, R. W. (2000). Characterization and purification of a bacteriocin produced by a potential probiotic culture, Lactobacillus acidophilus 30SC. J. Dairy Sci. 83, 2747–2752. doi: 10.3168/jds.s0022-0302(00)75169-1
Park, Y., Kim, H., Won, S., Hamidoghli, A., Hasan, T., Kong, I., et al. (2020). Effects of two dietary probiotics (Bacillus subtilis or licheniformis) with two prebiotics (mannan or fructo oligosaccharide) in Japanese eel, Anguilla japonica. Aquac. Nutr. 26, 316–327. doi: 10.1111/anu.12993
Patel, A. K., Ahire, J. J., Pawar, S. P., Chaudhari, B. L., and Chincholkar, S. B. (2009). Comparative accounts of probiotic characteristics of Bacillus spp. isolated from food wastes. Food Res. Int. 42, 505–510. doi: 10.1016/j.foodres.2009.01.013
Rajan, P. R., Lin, J. H., Ho, M. S., and Yang, H. L. (2003). Simple and rapid detection of Photobacterium damselae ssp. piscicida by a PCR technique and plating method. J. Appl. Microbiol. 95, 1375–1380. doi: 10.1046/j.1365-2672.2003.02119.x
Rajyalakshmi, K., Roopa, B., Saikat, D. M., Priyanka, D., Vadlamudi, S., and Subramaniam, G. (2016). Characterization of potential probiotic bacteria isolated from sorghum and pearl millet of the semi-arid tropics. Afr. J. Biotechnol. 15, 613–621. doi: 10.5897/AJB2016.15212
Ramesh, D., Vinothkanna, A., Rai, A. K., and Vignesh, V. S. (2015). Isolation of potential probiotic Bacillus spp. and assessment of their subcellular components to induce immune responses in Labeo rohita against Aeromonas hydrophila. Fish Shellf. Immunol. 45, 268–276. doi: 10.1016/j.fsi.2015.04.018
Ray, A. K., Ghosh, K., and Ringo, E. (2012). Enzyme-producing bacteria isolated from fish gut, A review. Aquac. Nutr. 18, 465–492. doi: 10.1111/j.1365-2095.2012.00943.x
Ringo, E. (2019). Probiotics in shellfish aquaculture. Aquacult. Fish. 5, 1–27. doi: 10.1016/j.aaf.2019.12.001
Saarela, M., Mogensen, G., Fondén, R., Mättö, J., and Mattila-Sandholm, T. (2000). Probiotic bacteria: safety, functional and technological properties. J. Biotechnol. 84, 197–205. doi: 10.1016/S0168-1656(00)00375-8
Schönborn, S., and Krömker, V. (2016). Detection of the biofilm component polysaccharide intercellular adhesin in Staphylococcus aureus infected cow udders. Vet. Microbiol. 196, 126–128. doi: 10.1016/j.vetmic.2016.10.023
Shin, H. J., Choi, H., and Kim, D. W. (2012). Probiotic potential of Pediococcus pentosaceus BCNU 9070. J. Life Sci. 22, 1194–1200. doi: 10.5352/JLS.2012.22.9.1194
Shoemaker, C. A., Klesius, P. H., and Evans, J. J. (2001). Prevalence of Streptococcus iniae in tilapia, hybrid striped bass, and channel catfish on commercial fish farms in the United States. Am. J. Vet. Res. 62, 174–177. doi: 10.2460/ajvr.2001.62.174
Silva, E. F., Soares, M. A., Calazans, N. F., Vogeley, J. L., do Valle, B. C., Soares, R., et al. (2012). Effect of probiotic (Bacillus spp.) addition during larvae and postlarvae culture of the white shrimp Litopenaeus vannamei. Aquac. Res. 44, 13–21. doi: 10.1111/j.1365-2109.2011.03001.x
Sim, I., Koh, J. H., Kim, D. J., Gu, S. H., Park, A., and Lim, Y. H. (2015). In vitro assessment of the gastrointestinal tolerance and immunomodulatory function of Bacillus methylotrophicus isolated from a traditional Korean fermented soybean food. J. Appl. Microbiol. 118, 718–726. doi: 10.1111/jam.12719
Skalli, A., Castillo, M., Andree, K. B., Tort, L., Furones, D., and Gilbert, E. (2013). The LPS derived from the cell walls of the Gram-negative bacteria Pantoea agglomerans stimulates growth and immune status of rainbow trout (Oncorhynchus mykiss) juveniles. Aquaculture 416-417, 272–279. doi: 10.1016/j.aquaculture.2013.09.037
Smith, D. D., Kirzinger, M. W., and Stavrinides, J. (2013). Draft genome sequence of the antibiotic-producing cystic fibrosis isolate Pantoea agglomerans Tx10. Genome Announc. 1, e904–e913.
Smits, T. H. M., Rezzonico, F., Kamber, T., Goesmann, A., Ishimaru, C. A., Stockwell, V. O., et al. (2010). Genome sequence of the biocontrol agent Pantoea vagans strain C9–1. J. Bacteriol. 192, 6486–6487. doi: 10.1128/JB.01122-10
Spinosa, M. R., Braccini, T., Ricca, E., de Felice, M., Morelli, L., Pozzi, G., et al. (2000). On the fate of ingested Bacillus spores. Res. Microbiol. 151, 361–368. doi: 10.1016/S0923-2508(00)00159-5
Sumi, C. D., Yang, B. W., Yeo, I. C., and Hahm, Y. T. (2015). Antimicrobial peptides of the genus Bacillus: a new era for antibiotics. Can. J. Microbiol. 61, 93–103. doi: 10.1139/cjm-2014-0613
Todorova, S. D., Nyati, H., Meincken, M., and Dicks, L. M. T. (2007). Partial characterization of bacteriocin AMA-K, produced by Lactobacillus plantarum AMA-K isolated from naturally fermented milk from Zimbabwe. Food Control 18, 656–664. doi: 10.1016/j.foodcont.2006.03.003
Vine, N. G., Leukes, W. D., Kaiser, H., Daya, S., Baxter, J., and Hecht, T. (2004). Competition for attachment of aquaculture candidate probiotic and pathogenic bacteria on fish intestinal mucus. J. Fish. Dis. 27, 319–326. doi: 10.1111/j.1365-2761.2004.00542.x
Völksch, B., Thon, S., Jacobsen, I. D., and Gube, M. (2009). Polyphasic study of plant- and clinic-associated Pantoea agglomerans strains reveals indistinguishable virulence potential. Infect. Genet. Evol. 9, 1381–1391. doi: 10.1016/j.meegid.2009.09.016
Walterson, A. M., and Stavrinides, J. (2015). Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 39, 968–984. doi: 10.1093/femsre/fuv027
Wang, C., Liu, Z., Huang, Y., Zhang, Y., Wang, X., and Hu, Z. (2019). Cadmium-resistant rhizobacterium Bacillus cereus M4 promotes the growth and reduces cadmium accumulation in rice (Oryza sativa L). Environ. Toxicol. Pharmacol. 72:103265. doi: 10.1016/j.etap.2019.103265
Wang, T., Liang, Y., Wu, M., Chen, Z., Lin, J., and Yang, L. (2015). Natural products from Bacillus subtilis with antimicrobial properties. Chin. J. Chem. Eng. 23, 744–754. doi: 10.1016/j.cjche.2014.05.020
Wang, W.-Z., Huang, J.-S., Zhang, J.-D., Wang, Z.-L., Li, H.-J., Amenyogbe, E., et al. (2021). Effects of hypoxia stress on the intestinal microflora of juvenile of cobia (Rachycentron canadum). Aquaculture 536:736419. doi: 10.1016/j.aquaculture.2021.736419
Wasko, A., Polak-Berecka, M., Paduch, R., and Jóźwiak, K. (2014). The effect of moonlighting proteins on the adhesion and aggregation ability of Lactobacillus helveticus. Anaerobe 30, 161–168. doi: 10.1016/j.anaerobe.2014.10.002
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Woese, C. R., Kandler, O., and Wheelis, M. L. (1990). Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U.S.A. 87, 4576–4579. doi: 10.1073/pnas.87.12.4576
Wu, H., Sun, L., Li, C., Li, Z., Zhang, Z., Wen, X., et al. (2014). Enhancement of the immune response and protection against Vibrio parahaemolyticus by indigenous probiotic Bacillus strains in mud crab (Scylla paramamosain). Fish Shellf. Immunol. 41, 156–162. doi: 10.1016/j.fsi.2014.08.027
Xie, F., Quan, S., Liu, D., Ma, H., Li, F., and Zhou, F. (2014). Purification and characterization of a novel α-amylase from a newly isolated Bacillus methylotrophicus strain P11-2. Process Biochem. 49, 47–53. doi: 10.1016/j.procbio.2013.09.025
Xie, R. t., Amenyogbe, E., Wang, W. z., Guo, Z.-X., Chen, G., and Huang, J.-S. (2021). Cloning and expression analysis of hypoxia-related gene HO in cobia. Aquacult. Int. 1, 75–89. doi: 10.1007/s10499-020-00611-3
Yang, G., Tian, X., Dong, S., Peng, M., and Wang, D. (2015). Effects of dietary Bacillus cereus G19, B. cereus BC-01, and Paracoccus marcusii DB11 supplementation on the growth, immune response, and expression of immune-related genes in coelomocytes and intestine of the sea cucumber (Apostichopus japonicus Selenka). Fish Shellf. Immunol. 45, 800–807. doi: 10.1016/j.fsi.2015.05.032
Yerlikaya, O. (2018). Probiotic potential and biochemical and technological properties of Lactococcus lactis ssp. lactis strains isolated from raw milk and kefir grains. J. Dairy Sci. 102, 124–134. doi: 10.3168/jds.2018-14983
Zahller, J., and Stewart, P. S. (2002). Transmission electron microscopic study of antibiotic action on Klebsiella pneumoniae biofilm. Antimicrob Agents Ch. 46, 2679–2683. doi: 10.1128/aac.46.8.2679-2683.2002
Zhang, Q., Ma, H., Mai, K., Zhang, W., Liufu, Z., and Xu, W. (2010). Interaction of dietary Bacillus subtilis and fructooligosaccharide on the growth performance, non-specific immunity of sea cucumber, Apostichopus japonicus. Fish Shellf. Immunol. 29:211.
Keywords: Rachycentron canadum, Bacillus spp. RCS1, Pantoea agglomerans strain RCS2, Bacillus cereus RCS3, antagonistic effects
Citation: Amenyogbe E, Huang J-s, Chen G and Wang W-z (2021) Probiotic Potential of Indigenous (Bacillus sp. RCS1, Pantoea agglomerans RCS2, and Bacillus cereus strain RCS3) Isolated From Cobia Fish (Rachycentron canadum) and Their Antagonistic Effects on the Growth of Pathogenic Vibrio alginolyticus, Vibrio harveyi, Streptococcus iniae, and Streptococcus agalactiae. Front. Mar. Sci. 8:672213. doi: 10.3389/fmars.2021.672213
Received: 25 February 2021; Accepted: 23 April 2021;
Published: 17 May 2021.
Edited by:
Enric Gisbert, Institute of Agrifood Research and Technology (IRTA), SpainReviewed by:
Paola Navarrete, University of Chile, ChileDariel Tovar-Ramírez, Centro de Investigación Biológica del Noroeste (CIBNOR), Mexico
Copyright © 2021 Amenyogbe, Huang, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-sheng Huang, aHVhbmdqc0BnZG91LmVkdS5jbg==; Gang Chen, Z2RvdWNnQDEyNi5jb20=
 Eric Amenyogbe
Eric Amenyogbe Jian-sheng Huang1,2,3*
Jian-sheng Huang1,2,3*