- 1School of Marine Science, Ningbo University, Ningbo, China
- 2Key Laboratory of Aquacultural Biotechnology Ministry of Education, Ningbo University, Ningbo, China
- 3Aquatic Technology Promotion Station, Sanmen Rural Bureau, Taizhou, China
- 4Guangxi Engineering Technology Research Center of Breeding of New Mariculture Varieties, Guangxi Institute of Oceanology Co., Ltd., Beihai, China
- 5Aquatic Technology Promotion Station, Yanjin Rural Bureau, Xinxiang, China
Scylla paramamosain is an important marine aquaculture crustacean on the southeastern coast of China. Given the difficulties of overcrowded coastal aquaculture space and insufficient production, inland saline-alkaline water culture has the potential to alleviate this bottleneck. This study separated 600 crabs into four groups: normal salinity (12-18 ppt, NS), coastal low salinity (3-5 ppt, CS), inland low salinity saline-alkaline (1.5 ppt, IS), and acute low salinity (23 ppt down to 3 ppt, AS), followed by a transcriptomic analysis of the gills. CS-vs-NS, IS-vs-NS, and AS-vs-NS obtained 1154, 1012, and 707 DEGs, respectively. GO analysis showed that the DEGs of the three comparison groups were mainly involved in cellular process, metabolic process, biological regulation, organelle, membrane, extracellular region, binding, and catalytic activity. The findings demonstrate that a significant number of genes are engaged in controlling metabolic processes in the osmoregulation process, and that cell membrane catalysis and different enzymes play a vital part in the environmental adaption process. KEGG enrichment analysis revealed that IS possesses a considerable number of signaling pathways that play important roles in osmoregulation when compared to NS. The cAMP signaling pathway increased the expression of CaM and Na+/K+-ATPase. These findings show that cations like Ca2+, Na+, and K+ are critical for S. paramamosain to adapt to inland low salinity saline-alkaline water, and that the cAMP signaling pathway regulates their levels. This research provides a theoretical foundation for improving the saline-alkaline aquaculture technology of S. paramamosain.
Introduction
Soil salinization is a global problem that affects both developing and developed countries. It affects around 20% of the world’s agricultural land and nearly half of the world’s irrigated lands (Kumari et al., 2021). Salinization not only affects the productivity of the land and threatens the sustainability of agriculture, but it can also lead to wetland degradation, resulting in imbalances in ecosystems that affect biodiversity (Herbert et al., 2015). This is particularly important where irrigation waters are compromised by salinity as this directly affects food production. Salinity thus intersects with major global concerns, including food security, desertification, and biodiversity protection. (Bal et al., 2021; Harper et al., 2021). Developing strategies to make use of saline land will be crucial for addressing the problem of meeting the challenge of providing food security for the projected global population of 9.3 billion people by 2050 (Liu and Wang, 2021). The advent of aquaculture technology has opened up new options for the conversion of underutilized saline land natural resources into productive resources. The growing demand for aquatic products, however, has pushed aquaculturists to favor inland aquaculture because of the restricted yield of caught fisheries, as well as the expensive cost of coastal land, tight regulations, and environmental concerns. Furthermore, inland ground saline water is unsuitable for traditional agriculture in many regions of the world, piquing aquaculturists’ interest in inland saline aquaculture (Li et al., 2017).
There are approximately 99.13 million hectares of saline-alkaline land areas in China, which are primarily situated in the north, northeast, and northwest portions of the country, yet, these limited land resources could be transformed into great potential for fisheries development (Zhang and Wang, 2021). In China’s Henan, Shandong, Ningxia, etc., there have been fishery development and utilization of saline-alkaline water resources, which has created a new way for the development and utilization of saline-alkaline water resources (Chen et al., 2020). Inland saline-alkaline waters are rich in resources, but conventional breeding species struggle to survive due to the high salinity, alkalinity, pH values, and complex ionic composition of such waters, severely limiting the development and utilization of saline-alkaline waters. Land-based aquaculture employing inland saline-alkaline groundwater is referred to as inland saline-alkaline waters culture. The ionic composition of inland saline-alkaline waters differs from that of coastal saltwater, and identifying species that are tolerant of these changes is one of the most difficult aspects of the inland saline-alkaline water culture. Litopenaeus Vannamei (Roy et al., 2010) and Nile tilapia (Zhao et al., 2015; Zhao et al., 2020) are currently the most popular marine species for inland saline-alkaline water farming.
Mud crab, Scylla paramamosain is distributed along the coast of the South China Sea (Chung and Lin, 2006). Because of its abundance, fast growth rate and high market value, the species is an important aquaculture crab in China. mud crab inhabits waters of salinity around 5-33 ppt (Niu et al., 2020). Even though the mud crab adapted to survive in a wide range of salinity, changes in salinity beyond its range of osmoregulation capacity of it can lead to individual death, resulting in severe production losses (Xu et al., 2017). Changes in salinity had a significant effect on the growth and moltin of mud crab (Qi et al., 2013; Ji et al., 2022). Zhang et al. (2020) previously reported transcriptome analysis of S. paramamosain under salinity stress. Expression patterns under low and high salinity conditions has been compared with normal salinity to absorb the salinity stress and recognize osmoregulation-related genes of the mud crab, indicated that the mud crab might regulate some mechanisms such as metabolism, immunity responses, and osmoregulation to adapt to the alteration of the low salinity environment. Salinity also affects the nutritional quality of mud crab, it reared at high salinity had better nutritional value than that at low salinity (Wu et al., 2019). The adaptability of mud crab to environmental salinity is mainly reflected in the ability to regulate osmotic pressure and ion concentration. The gills are the most essential organ in the osmoregulation process (Romano and Zeng, 2012). The osmoregulation of the posterior gills allows the mud crab to maintain normal physiological function when the water salinity changes (Wang et al., 2018a; Wang et al., 2018b).
In 2017, our research team began investigating the aquaculture mode of S. paramamosain in the saline-alkaline land of the Yellow River basin, and in 2018 they successfully established the inland low salinity saline-alkaline water (1.5-2ppt) aquaculture technology system, with yield per acre reaching the coastal medium level, marking the first time that S. paramamosain has been successfully cultured in inland saline-alkaline. However, the molecular mechanism of the adaptation of S. paramamosain to inland low-salinity saline-alkaline water has yet to be discovered. Therefore, in the present study, S. paramamosain was cultured in normal salinity (12-18ppt), coastal low salinity (3-5ppt), and inland low salinity saline-alkaline water (1.5-2ppt), and acute low salinity (23ppt dropped to 3ppt), respectively. Then, the molecular mechanism of gill responses to inland low salinity saline-alkaline water stress by transcriptomic analysis, to further improve inland saline-alkaline water aquaculture technology.
Materials and methods
Animals and experimental conditions
A total of 600 crabs weighing 30 ± 5g were randomly selected from Sanmen Bay (Zhejiang, China), and each group of 50 crabs (n=3) was transferred to the following 4 experimental ponds with the same domestication conditions: normal salinity group (NS), coastal low salinity group (CS), inland low salinity saline-alkaline group (IS) and acute low salinity group (AS). NS, located in Sanmen Bay on the eastern coast of Zhejiang, China, is one of the main producing areas of mud crabs, with a salinity of 12-13ppt (as control). CS, located in Hangzhou Bay, northeastern Zhejiang, China, is affected by river inflow and precipitation, the seawater salinity is low and unstable, with a salinity of 3-5ppt. IS, located in Yanjin on the north bank of the Yellow River in Henan, China, the breeding water is saline-alkaline water, and the mud crab is successfully adapted to inland low salinity saline-alkaline water culture after domestication, with a salinity of 1.5-2ppt. AS, located at Ningbo University, Zhejiang, China, normal salinity (12-13ppt) acclimated crabs were directly transferred to diluted 3ppt seawater. Crabs were held in a 30 m3 cement pond with disinfected seawater and fed once daily in the afternoon (5:00 pm) with razor clam Sinonovacula constricta at approximately 10% body weight. The experiment was conducted at ambient temperature and natural photoperiod. Water quality parameters were monitored every morning. The parameters were as followed: water temperature 25-30°C, dissolved oxygen 5.24-7.52 mg L-1, ammonia nitrogen concentration 0.16-1.58 mg L-1, nitrite concentration 0.009-0.102 mg L-1, pH 7.8-8.6, alkalinity 115-160 mg/L CaCO3.
NS, CS and IS crabs were long-term cultured and sampled on day 90, respectively. According to a previous study (Wang et al., 2018a), AS was sampled at 120 h. On each sampling day, crabs were collected from the respective pond with traps and transferred to the laboratory for sampling. the crabs were soaked in iced water until anesthetized and then dissected promptly. The posterior gills of each individual were separated immediately with sterile forceps, frozen in liquid nitrogen, and stored at −80°C for further analysis of transcriptomic. For the collection of the posterior gills, three replicates were set up at the treatment.
RNA extraction and RNA-seq
RNA was extracted from crabs in NS, CS, IS and AS groups, respectively. Total RNA was extracted using the mirVana miRNA Isolation Kit (Ambion) following the manufacturer’s protocol. RNA integrity was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The samples with RNA Integrity Number (RIN) ≥ 7 were subjected to the subsequent analysis. The libraries were constructed using TruSeq Stranded mRNA LTSample Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Then these libraries were sequenced on the Illumina sequencing platform (HiSeqTM 2500) and 125bp/150bp paired-end reads were generated.
Analysis of sequencing data
Raw data (raw reads) were processed using Trimmomatic (Bolger et al., 2014). The reads containing ploy-N and the low-quality reads were removed to obtain the clean reads. Then the clean reads were mapped to the reference genome using hisat2 (Kim et al., 2015).
Analysis of differentially expressed transcript (DEGs)
FPKM (Kim et al., 2015) and read counts value of each transcript (protein_coding) was calculated using bowtie2 (Langmead and Salzberg, 2012) and eXpress (Roberts and Pachter, 2013). DEGs were identified using the DESeq (Anders and Huber, 2013) functions estimateSizeFactors and nbinomTest. p value < 0.05 and foldchange >2 or foldchange < 0.5 was set as the threshold for significantly differential expression.
Cluster analysis, GO, and KEGG enrichment
Hierarchical cluster analysis of DEGs was performed to explore transcripts expression patterns. GO enrichment and KEGG (Kanehisa et al., 2008) pathway enrichment analysis of DEGs were respectively performed using R based on the hypergeometric distribution.
Quantitative real-time PCR for RNA-seq results validation
Nine genes were screened out for RNA-seq validation using quantitative real-time PCR (qRT-PCR) with TB Green® Premix Ex Taq™ II (Takara Biomedical Technology Co., Ltd, Beijing, China) according to the manufacturer’s instructions.
Results
Sequencing analysis
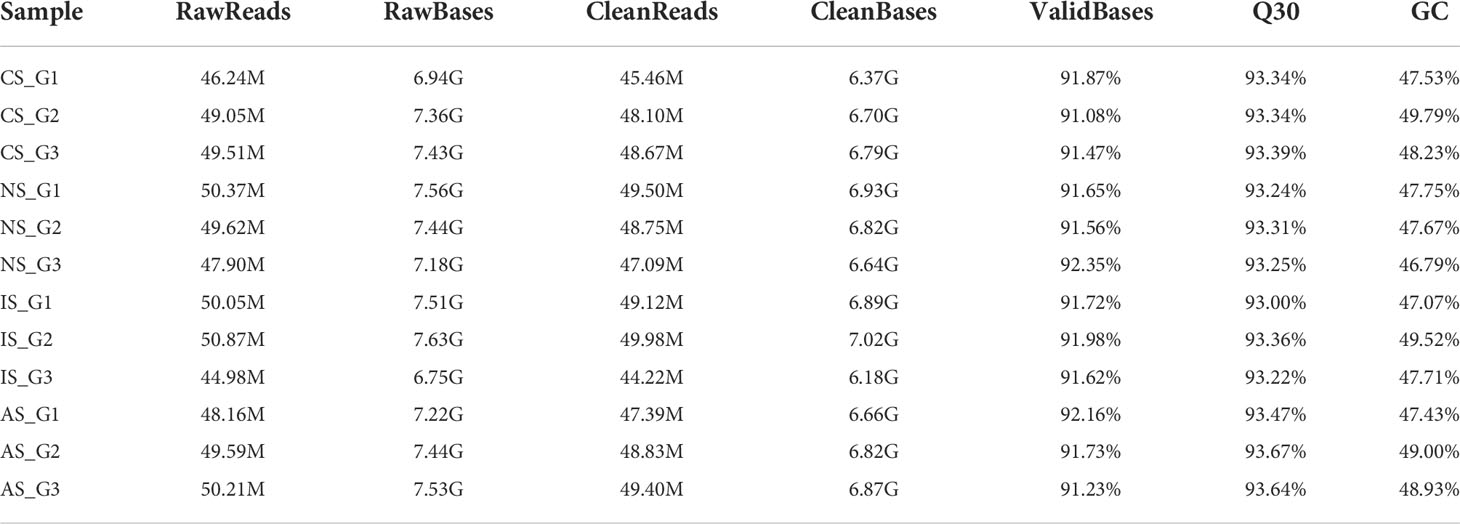
A total of 12 samples were sequenced in this study, and a total of 80.69 G of clean data was obtained through the Illumina sequencing platform. The effective data volume of each sample is distributed at 6.18-7.02 G, the Q30 base is distributed at 93.0-93.67%, and the average GC content is 48.12% (Table 1). The clean reads were aligned to the reference genome, and the genome alignment of each sample was obtained. Based on the comparison results, protein-coding gene expression analysis was performed. DEGs were screened according to the expression levels of protein-coding genes in different samples, and there were three differential groups (CS-vs-NS, IS-vs-NS, AS-vs-NS).
DEGs expression analysis
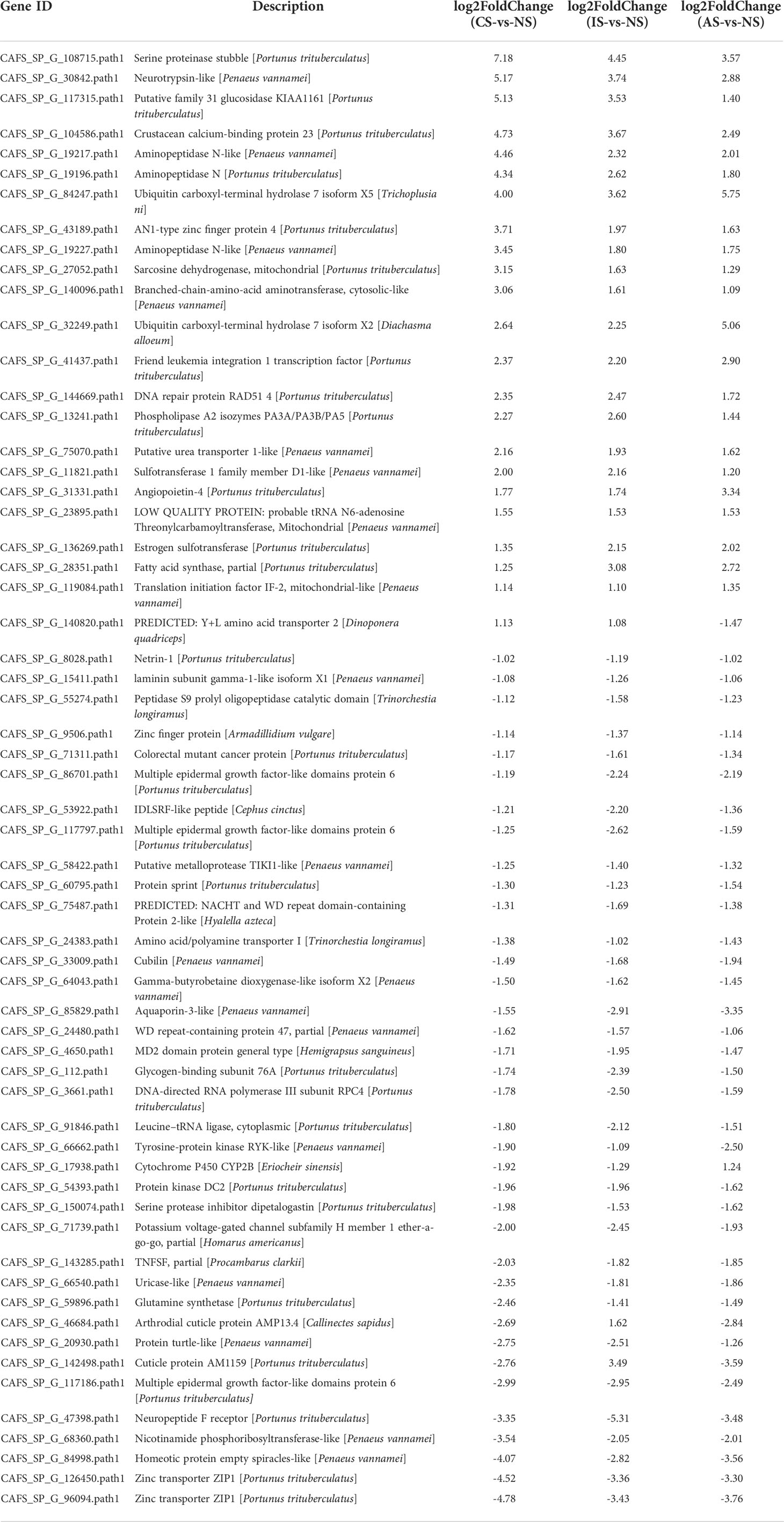
In terms of the degree of aggregation of the samples, the distribution distance between IS and the NS, AS, and CS groups is relatively large, but the NS, AS, and CS samples are more clustered (Figure 1A). When combined with the sample correlation analysis, the gene expression of NS, CS, and AS is similar, but there is a big difference with IS (Figure 1B). To elucidate gene expression patterns at different salinities, the numbers of DEGs were compared among the four treatment groups. Compared with NS, CS, IS, and AS obtained 1154 DEGs (689 up-regulated genes and 465 down-regulated genes), 1012 DEGs (580 up-regulated genes and 432 down-regulated genes), and 707 DEGs (340 up-regulated genes and 367down-regulated genes) (Figure 1C), respectively. Interestingly, 117 common DEGs were found in the three comparison groups (Figure 1D). Of the 117 DEGs, 11 were found without any description, 12 were not annotated in GO and KEGG, 28 were described as “hypothetical”, and 6 were described as uncharacterized protein, so these 57 DEGs were excluded. Finally, 60 DEGs with GO annotations were screened and plotted as a heatmap.
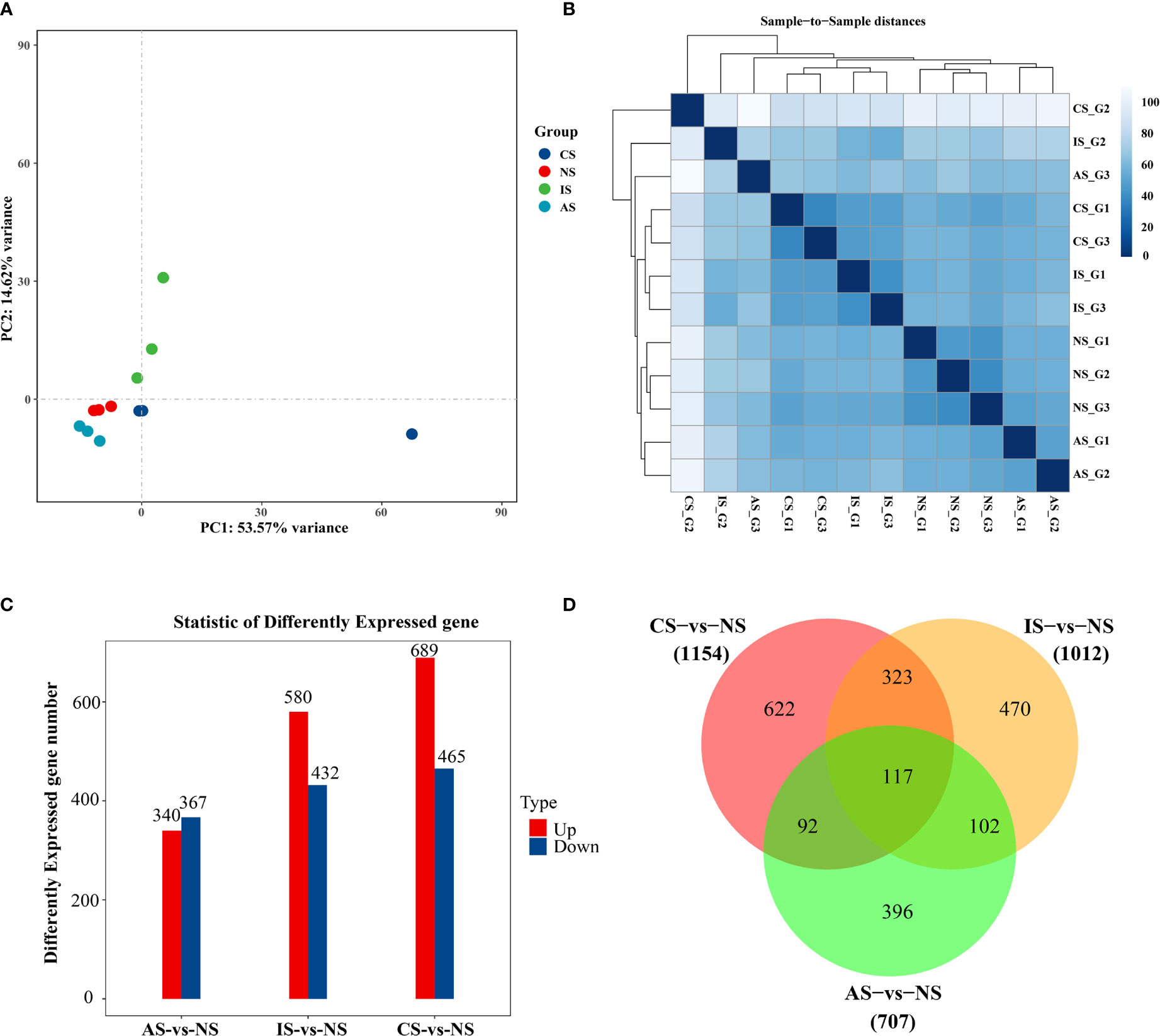
Figure 1 Differential gene expression analysis. (A) PCA diagram. (B) Cluster analysis results. (C) Bar Chart. (D) VennGraph. In (A), dots represent samples, colors represent groups. In (B), the horizontal axis represents the sample name, the vertical axis represents the corresponding sample name, and the color represents the size of the correlation coefficient. Foldchange> = 2.00 and adjusted p-value <= 0.05.
The heatmap shows the expression of common DEGs between NS and CS, IS, and AS. Among the 60 common DEGs, 22 were up-regulated and 34 were down-regulated. In addition, cuticle protein AM1159 (CAFS_SP_G_142498.path1) and arthrodial cuticle protein AMP13.4 (CAFS_SP_G_46684.path1) down-regulated in CS-vs-NS and AS-vs-NS, but up-regulated in IS-vs-NS. Y+L amino acid transporter 2 (CAFS_SP_G_140820.path1) up-regulated in CS-vs-NS and IS-vs-NS, but down-regulated in AS-vs-NS. Cytochrome P450 CYP2B (CAFS_SP_G_17938.path1) down-regulated in CS-vs-NS and IS-vs-NS and up-regulated in AS-vs-NS (Figure 2A, Table 2). In IS-vs-NS, serine proteinase stubble (CAFS_SP_G_108715.path1, 4.45-fold) was the most up-regulated, followed by neurotrypsin-like (CAFS_SP_G_30842.path1, 3.74-fold) and crustacean calcium-binding protein 23 (CAFS_SP_G_104586.path1, 3.67-fold) (Figure 2B, Table 2). The expression of these common DEGs indicated that they played an important role in the adaptation of S. paramamosain to low salinity.
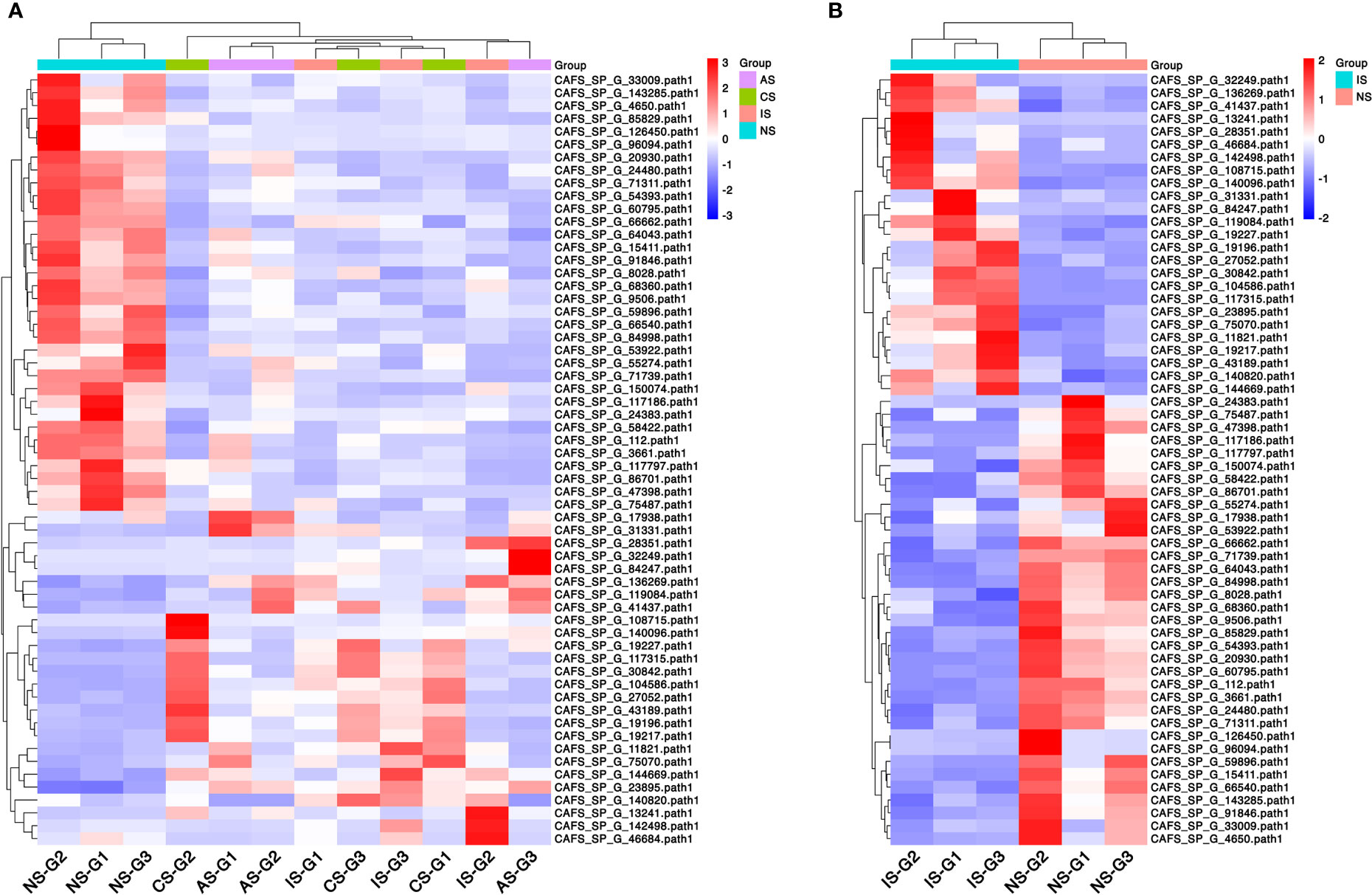
Figure 2 Clustered heatmap of common differentially expressed genes (DEGs). (A) Common DEGs expression between the three comparison groups. (B) DEGs expression of IS-vs-NS. Rows represent DEGs and each column represents one sample. The colors represent the expression level, and red and blue indicate higher and lower expression levels, respectively.
GO functional annotation
To better understand how different low salinity affects the osmoregulation process of S. paramamosain, all DEGs (CS-vs-NS, IS-vs-NS, AS-vs-NS) were subjected to GO analysis to determine their main functions. GO annotations were divided into three types: biological process, cellular component and molecular function.
The main significant processes of DEGs in CS-vs-NS group, IS-vs-NS group and AS-vs-NS group involved involved cellular process, metabolic process, biological regulation, organelle, membrane, extracellular region, binding, and catalytic activity (Figure S1). This indicates that there are many genes involved in regulating metabolic activities in the osmoregulation process, and the catalysis of the cell membrane and various enzymes plays an important role in the process of environmental adaptation.
KEGG enrichment analysis of DEGs
We performed comparative analysis through the KEGG pathway to identify the enrichment pathways for DEGs. Differential genes are classified into six categories according to their functions, including metabolism, genetic information processing, environmental information processing, cellular processes, organismal systems, and human diseases.
In the CS-vs-NS group, the pathways significantly enriched in the KEGG Enrichment top 20 are mainly related to Energy metabolism, Amino acid metabolism, Carbohydrate metabolism, Carbohydrate metabolism and Lipid metabolism, such as Oxidative phosphorylation, Valine, leucine and isoleucine degradation, propanoate metabolism, citrate cycle (TCA cycle), fatty acid degradation, pyruvate metabolism, glyoxylate and dicarboxylate metabolism, glycine, serine and threonine metabolism, glycolysis/gluconeogenesis, glycerolipid metabolism and fatty acid biosynthesis (Figure 3A). In the IS-vs-NS group, the top 20 pathways showed that eight pathways were directly related to the expression regulation of the Na+/K+-ATPase: cardiac muscle contraction, thyroid hormone synthesis, pancreatic secretion, salivary secretion, protein digestion and absorption, proximal tubule bicarbonate reclamation, carbohydrate digestion and absorption, and bile secretion (Figure 3B). In the AS-vs-NS group, the pathways significantly enriched mainly include aminoacyl-tRNA biosynthesis, Nitrogen metabolism, renin-angiotensin system, antifolate resistance, ECM-receptor interaction, ABC transporters, AGE-RAGE signaling pathway in diabetes, alanine, aspartate and glutamate complications metabolism, cytosolic DNA-sensing pathway, and PI3K-Akt signaling pathway, etc. (Figure 3C).
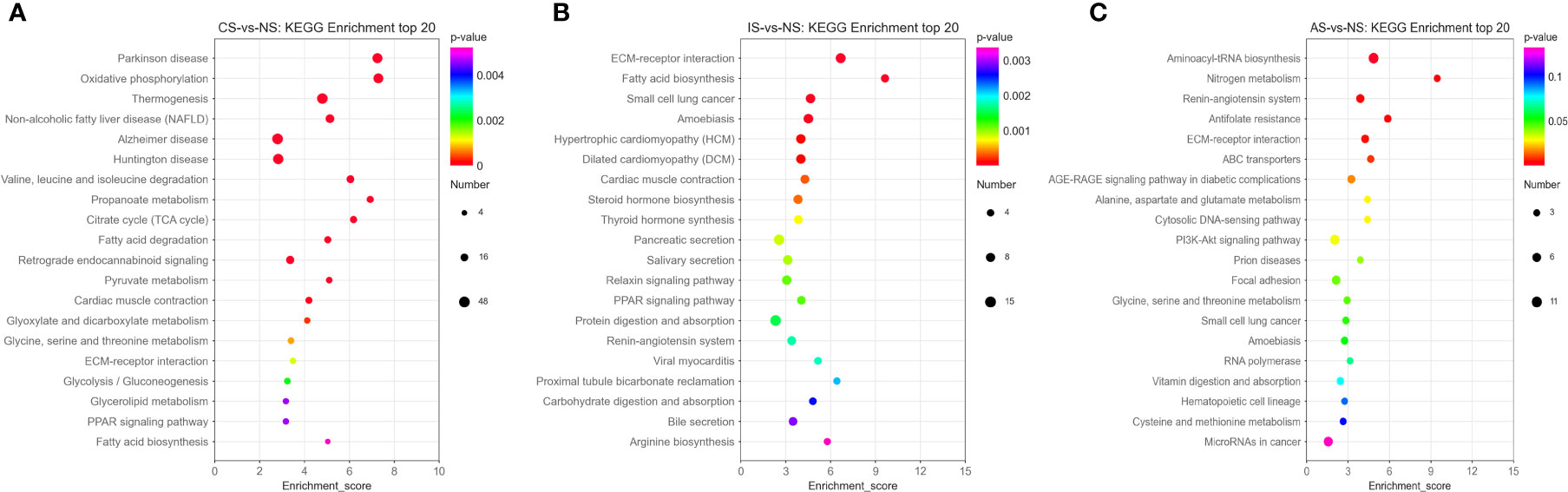
Figure 3 Pathway functional enrichment of differentially expressed genes (DEGs). (A) CS-vs-NS group. (B) IS-vs-NS group. (C) AS-vs-NS group. X axis represents enrichment score. Y axis represents pathway name. The color indicates the q value, the lower p-value indicates the more significant enrichment. Point size indicates DEG number (The larger dots refer to a larger amount).
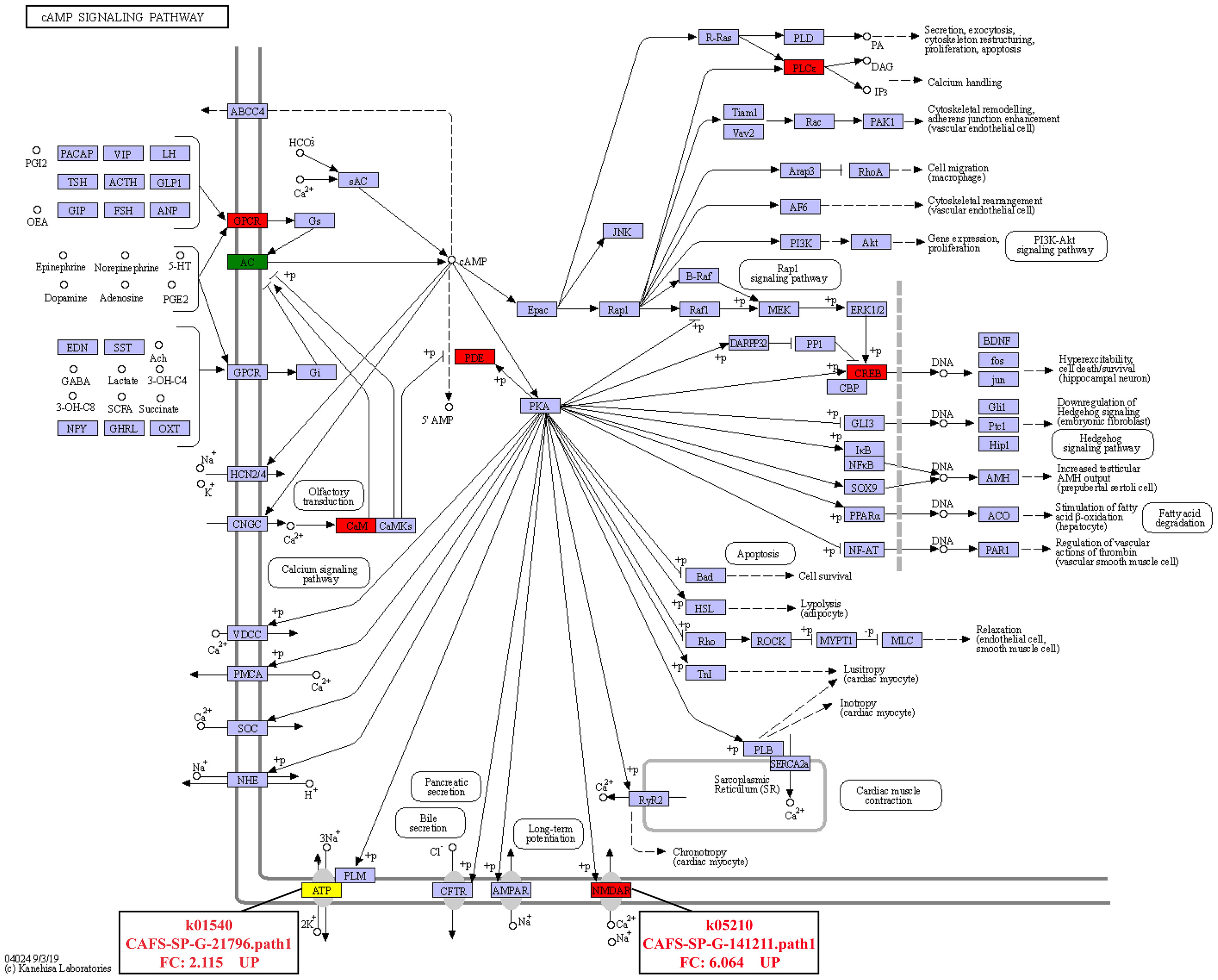
In addition, it was found that the DEGs involved in signal transduction were the most among the three comparison groups (Figure 4). In the IS-vs-NS group, the analysis of the DEGs involved in Signal transduction showed that four significantly signaling pathways were directly related to the expression regulation of the calmodulin (CaM): cGMP-PKG signaling pathway, calcium signaling pathway, cAMP signaling pathway, apelin signaling pathway (Figure S2). Interestingly, the cAMP signaling pathway is also involved in the regulation of Na+/K+-ATPase and NMDAR, and its expression levels were significantly up-regulated by 2.115-fold and 6.064-fold, respectively (Figure 5).
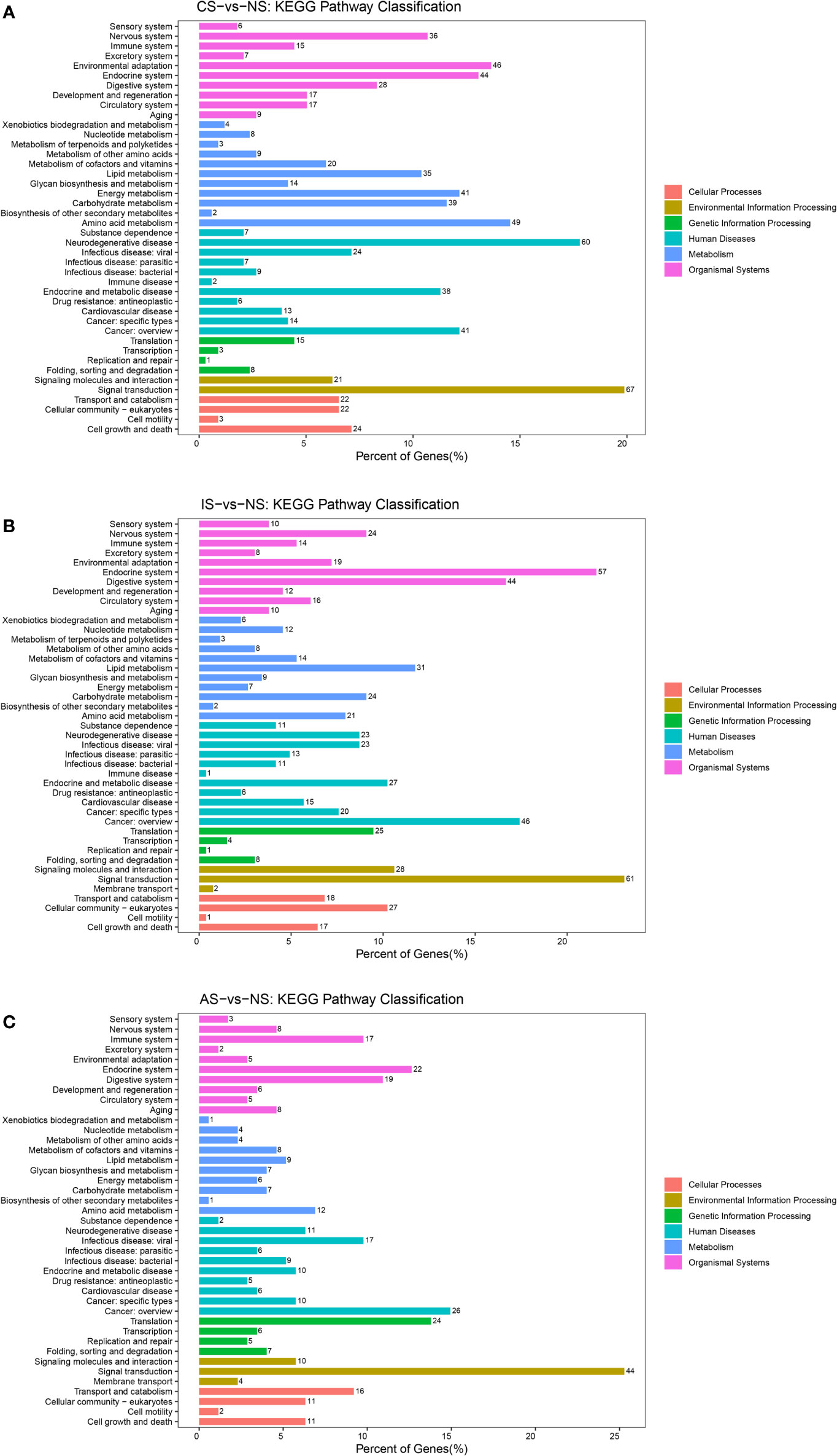
Figure 4 KEGG pathways classification. (A) CS-vs-NS group. (B) IS-vs-NS group. (C) AS-vs-NS group. There were six branches for KEGG pathways: Cellular Processes, Environmental Information Processing, Genetic Information Processing, Human Disease (For animals only), Metabolism and Organismal Systems.
Validity of DEGs in transcriptomic data
To validate the RNA sequencing, nine genes were selected and quantified using qPCR, namely Uricase-like (UL), TNFSF, partial (TNFSF), Homeotic protein empty spiracles-like (HPES), Amino acid/polyamine transporter I (AATI), Cytochrome P450 CYP2B(CP450), Aminopeptidase N-like (ANL), Estrogen sulfotransferase (ES), WD repeat-containing protein 47 (WDRCP), Laminin subunit gamma-1-like isoform X1 (LSGIX). The relative expression tendencies of the nine genes were in accordance with the RNA-seq (Figure 6), validating the results of the mRNA sequencing analysis.
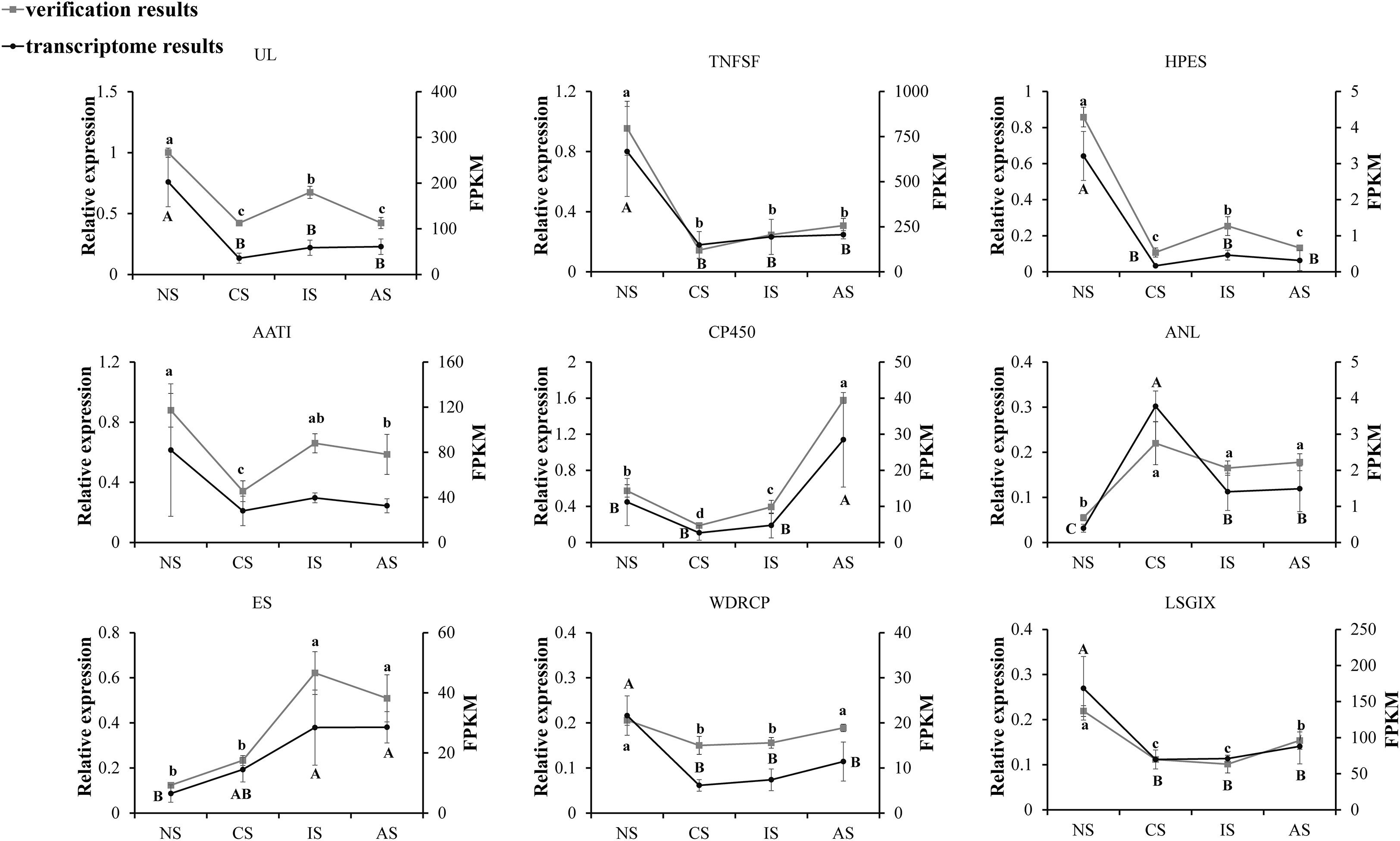
Figure 6 Candidate gene expression levels revealed by qRT-PCR and RNA-seq. NS represents normal salinity of 12-13ppt, CS represents coastal low salinity of 3-5ppt, IS represents inland low salinity saline-alkaline water of 1.5-2ppt, AS represents acute low salinity, 3ppt from normal salinity (12-13ppt) directly, which dropped. The different letters above the line chart indicate significant differences.
Discussion
S. paramamosain is an important marine aquaculture crustacean (Tang et al., 2020). In 2017, our research team began investigating the aquaculture mode of S. paramamosain in the saline-alkaline water of the Yellow River basin, and in 2018 we successfully established the inland low salinity saline-alkaline water (1.5-2ppt) breeding technology system, with yield per acre reaching the coastal medium level, marking the first time that S. paramamosain has been successfully cultured in inland saline-alkaline. However, the molecular mechanism by which S. paramamosain responds to saline-alkaline water stress with reduced salinity has yet to be discovered. As a result, we investigated the transcriptomic analysis of S. paramamosain gills cultivated in normal salinity, coastal low salinity, inland low salinity saline-alkaline water, and acute low salinity, respectively. To analyze the molecular mechanism of S. paramamosain responds to inland low salinity saline-alkaline water stress at the transcriptional level, and to provide theoretical reference for improving inland saline-alkaline water aquaculture technology.
Under low salinity seawater stress, S. paramamosain can acquire adaptive capacity through osmoregulation and reach an adaptive state after 120 h (Wang et al., 2018a). The osmoregulation of S. paramamosain is to compensate and transport ions through the posterior gills to regulate hemolymph ion concentration (Yao et al., 2020), which determines hemolymph osmotic pressure (Lignot et al., 2000), so the ionic composition and concentration of the water environment will affect its osmoregulation capacity. However, the ionic composition of saline-alkaline water differs greatly from that of seawater, and the posterior gill response mechanism may differ. The results of PCA analysis revealed that there were clear distribution differences between the IS group and the NS, AS, and CS groups, but the NS, AS, and CS groups with the same seawater background but different salinity were more clustered. When combined with the sample correlation analysis, the gene expression of NS, CS, and AS is similar, but there is a significant difference when compared to IS. The high degree of sample dispersion may be due to the differences in the osmoregulation capacity of S. paramamosain individuals under the degree of salinity stress. This indicates that the molecular mechanism of S. paramamosain adapting to saline-alkaline water may be different from the osmoregulation mechanism of adapting to low salinity seawater stress, but the osmoregulation mechanism of seawater with different salinity has a certain similarity.
Osmoregulation in crustaceans is an energy-consuming process (Yao et al., 2020). Free amino acids are used as energy substrates for physiological stress compensation (Zhou et al., 2011). L-serine levels in the hemolymph and muscle of Litopenaeus vannamei increased dramatically with decreasing salinity in earlier research, and osmoregulation by consuming L-serine as an energy source was demonstrated (Shinji et al., 2012). Serine is one of the major amino acids in Serine proteinase’s catalytic active site (Shi et al., 2008). We discovered that in a low salinity environment, the expression of serine proteinase stubble was dramatically elevated. CS group was the most up-regulated (7.18-fold) compared to NS group, followed by IS group (4.45-fold) and AS group (3.45-fold) (3.57-fold). This finding may indicate serine proteinase stubble of the gills is the energy source for osmoregulation of S. paramamosain.
In crustaceans, gill tissue has been found to modulate ion transport, acclimatization to various salinities, and significant active areas for osmoregulation (Nikapitiya et al., 2015). Gill filament is the most fundamental functional unit of a gill, and its basic structure is made up of cuticle, subcuticular space, and gill epithelial cells, with cuticle being the most important for osmoregulation (Barra et al., 1983). The major component of the cuticle, cuticle protein, interacts with chitin to maintain the cuticle’s structural integrity (Andersen, 1999). Cuticle protein is extremely permeable to the ions Na+ and Cl-. The major ions that make up hemolymph’s osmotic pressure are Na+ and Cl-, which together determine the osmotic pressure of hemolymph (Lignot et al., 2000).
Cuticle protein AM1159 and cuticle protein AMP13.4 were down-regulated by 2.69-fold and 2.76-fold in CS group, down-regulated by 2.84-fold and 3.59-fold in AS group, but up-regulated by 3.49-fold and 1.62-fold in IS group, respectively, compared to NS group. The findings imply that cuticle protein is significant in the adaptation of S. paramamosain to inland low salinity saline-alkaline water, but its molecular mechanism needs to be investigated further.
Calcium signaling is important for cell growth, death, and metabolism (Wang et al., 2015; Dubois et al., 2016). Calmodulin (CaM) modulates cellular processes such as stress response, apoptosis, and calcium homeostasis by rapidly increasing the concentration of Ca2+ on the cell membrane when cells are stimulated (Tadross et al., 2008; Berchtold and Villalobo, 2014; Wang et al., 2015; Huang et al., 2020). Additionally, Ca2+ signal transduction may be a key signaling route for gills to cope with osmotic stress (Marshall et al., 2000; Fiol et al., 2006). CaM is involved in the regulation of Na+ and Cl- transport in the posterior gills of Eriocheir sinensis, resulting in osmoregulation, according to Péqueux and Gilles (1992). Na+ and Cl— transport is the most important osmoregulation method in crustaceans, which is driven by the Na+/K+-ATPase (Huang et al., 2020). The Na+/K+-ATPase is a membrane protein that connects the exchange of two external K+ for three internal Na+ linked to the hydrolysis of a single ATP molecule (Post et al., 1972). Jia and Liu (2018) found that the activity of the Na+/K+-ATPase can be controlled by cAMP. Injecting cAMP into Leptograpsus variegatus causes the gill filament Na+/K+-ATPase to activate quickly (Lucu et al., 2000). IS up-regulated CaM expression in comparison to NS via numerous signaling pathways, including the cGMP-PKG signaling system, calcium signaling pathway, cAMP signaling pathway, and apelin signaling pathway. In the cAMP signaling pathway, the expressions of Na+/K+-ATPase and NMDAR were dramatically up-regulated. It is no surprise that the osmoregulation related genes including Na+/K+-ATPase and NMDAR were upregulated under the low salinity condition. Under salinity stress, the genes related to ion transport will play a very important role in S. paramamosain. Zhang et al. (2020) analyzed transcriptomic changes in ambient salinity challenges of different salinities, indicated that under low salinity stress, S. paramamosain might cause induce changes in genes related to ion transport, such as the significant up regulation of Na+/K+-ATPase. NMDAR ion channels are highly permeable to Ca2+, Na+ and K+ ions (Hansen et al., 2018). Based on the above findings, it is hypothesized that the process of adapting to inland low salinity saline-alkaline water in S. paramamosain regulates the expression changes of Na+/K+-ATPase and NMDAR via the cAMP signaling pathway, improving the transport efficiency of Ca2+, Na+, and K+, and finally achieving osmotic pressure balance inside and outside the cell membrane, but the molecular mechanism needs to be investigated further. At the same time, this study also shows that cations such as Ca2+, Na+, and K+ are extremely important for S. paramamosain to adapt to inland low salinity saline water.
Conclusion
In conclusion, we analyzed the differences in gene expression between normal salinity, coastal low salinity, inland saline-alkaline low salinity, and acute low salinity in the gills of S. paramamosain. CS-vs-NS, IS-vs-NS, and AS-vs-NS obtained 1154 (689 up-regulated genes and 465 down-regulated genes), 1012 (580 up-regulated genes and 432 down-regulated genes), and 707 (340 up-regulated genes and 367 down-regulated genes) DEGs, respectively. KEGG enrichment analysis showed that IS has numerous signaling pathways involved in the regulation of CaM and Na+/K+-ATPase when compared to NS. The results suggest that the adaptation of S. paramamosain to inland low salinity saline-alkaline water regulates the expression changes of Na+/K+-ATPase and NMDAR via the cAMP signaling pathway, improving Ca2+, Na+, and K+ transport efficiency and achieving osmotic pressure balance inside and outside the cell membrane, but the molecular mechanism needs to be investigated further. At the same time, our research reveals that cations like Ca2+, Na+, and K+ are critical for S. paramamosain to adapt to inland low salinity saline-alkaline water. In this study, the molecular mechanism of S. paramamosain adapting to inland low salinity saline-alkaline water was investigated for the first time, and numerous potential genes and KEGG pathways involved in regulation were revealed, to provide a scientific basis for guiding the aquaculture technology of crustaceans aquaculture.
Data availability statement
The data presented in the study are deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) repository, accession number PRJNA851163.
Author contributions
HW conceived and designed the study. GL performed the cultivation of experimental animals. GL, KQ, YC, MN, CW, CM, LC, FW, QS, and RZ performed and analyzed all other experiments. GL and HW wrote the manuscript with support from all authors. All authors read and approved the final manuscript.
Funding
This study was sponsored by the National Key R&D Program of China (2020YFD0900203), the Sanmen County Agricultural and Rural Bureau, Zhejiang Province (2020-GK088-1), Key Scientific and Technological Grant of Zhejiang for Breeding New Agricultural Varieties (2021C02069-6), Zhejiang Agricultural Science and Technology Cooperation Project (2021SNLF029).
Conflict of interest
Author QS is employed by Guangxi Institute of Oceanology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.974501/full#supplementary-material
Supplementary Figure 1 | GO annotation of differentially expressed genes (DEGs). (A) CS-vs-NS group. (B) IS-vs-NS group. (C) AS-vs-NS group. DEGs were assigned to second-tier GO categories associated with three parent terms: biological process, cellular component, and molecular function.
Supplementary Figure 2 | The signaling pathway involved in regulating CaM in IS-vs-NS. (A) Calcium signaling pathway, ko04020. (B) cGMP-PKG signaling pathway, ko04022. (C) Apelin signaling pathway, ko04371.
References
Andersen S. O. (1999). Exoskeletal proteins from the crab, cancer pagurus. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 123 (2), 203–211. doi: 10.1016/S1095-6433(99)00051-3
Anders S., Huber W. (2013). Differential expression of RNA-seq data at the gene level – the DESeq package Vol. 1843 (EMBL), 398–435.
Bal A., Panda F., Pati S. G., Das K., Agrawal P. K., Paital B. (2021). Modulation of physiological oxidative stress and antioxidant status by abiotic factors especially salinity in aquatic organisms. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 241, 108971. doi: 10.1016/j.cbpc.2020.108971
Barra J. A., Pequeux A., Humbert W. (1983). A morphological study on gills of a crab acclimated to fresh water. Tissue Cell. 15 (4), 583–596. doi: 10.1016/0040-8166(83)90009-5
Berchtold M. W., Villalobo A. (2014). The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochim. Biophys. Acta 1843 (2), 398–435. doi: 10.1016/j.bbamcr.2013.10.021
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170
Chen X. Z., Lai Q., Mo Z. L., Gao H. Y., Han Z. X. (2020). Saline alkali water green breeding technology mode. China Fish. 9, 61–63.
Chung K. F., Lin H. C. (2006). Osmoregulation and Na,K-ATPase expression in osmoregulatory organs of Scylla paramamosain. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 144 (1), 48–57. doi: 10.1016/j.cbpa.2006.02.003
Dubois C., Prevarskaya N., Vanden Abeele F. (2016). The calcium-signaling toolkit: Updates needed. Biochim. Biophys. Acta 1863 (6 Pt B), 1337–1343. doi: 10.1016/j.bbamcr.2015.11.033
Fiol D. F., Chan S. Y., Kültz D. (2006). Identification and pathway analysis of immediate hyperosmotic stress responsive molecular mechanisms in tilapia (Oreochromis mossambicus) gill. Comp. Biochem. Physiol. Part D Genomics Proteomics 1 (3), 344–356. doi: 10.1016/j.cbd.2006.08.002
Hansen K. B., Yi F., Perszyk R. E., Furukawa H., Wollmuth L. P., Gibb A. J., et al (2018). Structure, function, and allosteric modulation of NMDA receptors. J. Gen. Physiol. 150 (8), 1081–1105. doi: 10.1085/jgp.201812032
Harper R. J., Dell B., Ruprecht J. K., Sochacki S. J., Smettem K. R. J. (2021). Salinity and the reclamation of salinized lands. Soils Landscape Restor. 7, 193–208. doi: 10.1016/B978-0-12-813193-0.00007-2
Herbert E. R., Boon P., Burgin A. J., Neubauer S. C., Franklin R. B., Ardón M., et al (2015). A global perspective on wetland salinization: ecological consequences of a growing threat to freshwater wetlands. Ecosphere 6, 10, 1–43. Wiley-Blackwell. doi: 10.1890/ES14-00534.1
Huang S. J., Jiang S., Xu Q. H. (2020). Acute salinity stress in gill Na+/K+-ATPase in the swimming crab, Portunus trituberculatus (Brachyura, portunidae). Crustaceana 93 (6), 587–609. doi: 10.1163/15685403-bja10012
Jia Y., Liu X. (2018). Expression of Na+/K+-ATPase was affected by salinity change in pacific abalone haliotis discus hannai. Front. Physiol. 9, 1244. doi: 10.3389/fphys.2018.01244
Ji Z. L., Li R. H., Wang C. L., Zhang Z. Y., Zhang W. J., Mu C. K., et al (2022). Effect of long-term low-salinity culture on the survival, growth, and nutrient composition of mud crab Scylla paramamosain. J. Ocean Univ. China Vol. 21, 179–185. doi: 10.1007/s11802-022-4812-x
Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, 480–484. doi: 10.1093/nar/gkm882D480-4
Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12 (4), 357–360. doi: 10.1038/nmeth.3317
Kumari S., Harikrishna V., Surasani V., Balange A. K., Rani B. (2021). Growth, biochemical indices and carcass quality of red tilapia reared in zero water discharge based biofloc system in various salinities using inland saline ground water. Aquaculture 540 (9), 736730. doi: 10.1016/j.aquaculture.2021.736730
Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9 (4), 357–359. doi: 10.1038/nmeth.1923
Lignot J. H., Spanings-Pierrot C., Charmantier G. (2000). Osmoregulatory capacity as a tool in monitoring the physiological condition and the effect of stress in crustaceans - ScienceDirect. Aquaculture 191 (1–3), 209–245. doi: 10.1016/S0044-8486(00)00429-4
Liu L., Wang B. (2021). Protection of halophytes and their uses for cultivation of saline-alkali soil in China. Biology (Basel) 10 (5), 353. doi: 10.3390/biology10050353
Li E., Wang X. D., Chen K., Xu C., Qin J. G., Chen L. Q. (2017). Physiological change and nutritional requirement of pacific white shrimp Litopenaeus vannamei at low salinity. Rev. Aquac. 9, 57–75. doi: 10.1111/raq.12104
Lucu C., Devescovi M., Skaramuca B., Kozul V. V. (2000). Gill Na, K-ATPase in the spiny lobster Palinurus elephas and other marine osmoconformers: Adaptiveness of enzymes from osmoconformity to hyperregulation. J. Exp. Mar. Biol. Ecol. 246 (2), 163–178. doi: 10.1016/S0022-0981(99)00179-3
Marshall W. S., Bryson S. E., Luby T. (2000). Control of epithelial cl(-) secretion by basolateral osmolality in the euryhaline teleost fundulus heteroclitus. J. Exp. Biol. 203 (Pt 12), 1897–1905. doi: 10.1242/jeb.203.12.1897
Nikapitiya C., Kim W. S., Park K., Kim J., Lee M. O., Kwak I. S. (2015). Chitinase gene responses and tissue sensitivity in an intertidal mud crab (Macrophthalmus japonicus) following low or high salinity stress. Cell Stress Chaperones 20 (3), 517–526. doi: 10.1007/s12192-015-0576-1
Niu J., Hu X. L., Ip J. C. H., Ma K. Y., Chu K. H. (2020). Multi-omic approach provides insights into osmoregulation and osmoconformation of the crab Scylla paramamosain. Sci. Rep. 10 (1), 21771. doi: 10.1038/s41598-020-78351-w
Péqueux A., Gilles R. (1992). Calmodulin as a modulator of NaCl transport in the posterior salt-transporting gills of the Chinese crab eriocheir sinensis. Mar. Biol. 113 (1), 65–69. doi: 10.1007/BF00367639
Post R. L., Hegyvary C., Kume S. (1972). Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J. Biol. Chem. 247 (20), 6530–6540. doi: 10.1016/S0021-9258(19)44725-X
Qi L., Gu X. L., Jiang K. J., Qiao Z. G. (2013). Effect of salinity on the survival,growth and Na+/K+-ATPase activity of early juvenile mud crabs, Scylla paramamosain. Mar. Sci. 37 (2), 56–60.
Roberts A., Pachter L. (2013). Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods 10 (1), 71–73. doi: 10.1038/nmeth.2251
Romano N., Zeng C. S. (2012). Osmoregulation in decapod crustaceans: implications to aquaculture productivity, methods for potential improvement and interactions with elevated ammonia exposure. Aquaculture 334–337, 12–23. doi: 10.1016/j.aquaculture.2011.12.035
Roy L. A., Davis D. A., Saoud I. P., Boyd C. A., Pine H. J., Boyd C. E. (2010). Shrimp culture in inland low salinity waters. Rev. Aquac. 2 (4), 191–208. doi: 10.1111/j.1753-5131.2010.01036.x
Shinji J., Okutsu T., Jayasankar V., Jasmani S., Wilder M. N. (2012). Metabolism of amino acids during hyposmotic adaptation in the whiteleg shrimp, litopenaeus vannamei. Amino Acids 43 (5), 1945–1954. doi: 10.1007/s00726-012-1266-2
Shi X. Z., Zhao X. F., Wang J. X. (2008). Molecular cloning and expression analysis of chymotrypsin-like serine protease from the Chinese shrimp, fenneropenaeus chinensis. Fish Shellfish Immunol. 25 (5), 589–597. doi: 10.1016/j.fsi.2008.07.011
Tadross M. R., Dick I. E., Yue D. T. (2008). Mechanism of local and global Ca2+ sensing by calmodulin in complex with a Ca2+ channel. Cell 133 (7), 1228–1240. doi: 10.1016/j.cell.2008.05.025
Tang L., Wang H., Wang C. L., Mu C. K., Wei H. L., Yao H. Z., et al (2020). Temperature potentially induced distinctive flavor of mud crab Scylla paramamosain mediated by gut microbiota. Sci. Rep. 10 (1), 3720. doi: 10.1038/s41598-020-60685-0
Wang H., Tang L., Wei H. L., Lu J. K., Mu C. K., Wang C. L. (2018a). Transcriptomic analysis of adaptive mechanisms in response to sudden salinity drop in the mud crab, Scylla paramamosain. BMC Genomics 19 (1), 421. doi: 10.1186/s12864-018-4803-x
Wang H., Wei H. L., Tang L., Lu J. K., Mu C. K., Wang C. L. (2018b). Identification and characterization of miRNAs in the gills of the mud crab (Scylla paramamosain) in response to a sudden drop in salinity. BMC Genomics 19 (1), 609. doi: 10.1186/s12864-018-4981-6
Wang J., Zhang P., Liu N., Wang Q., Luo J., Wang L. (2015). Cadmium induces apoptosis in freshwater crab Sinopotamon henanense through activating calcium signal transduction pathway. PloS One 10 (12), e0144392. doi: 10.1371/journal.pone.0144392
Wu Q. Y., Shi C., Fang S. B., Xie Z. F., Guan M. Y., Li S. K., et al (2019). Different biochemical composition and nutritional value attribute to salinity and rearing period in male and female mud crab Scylla paramamosain. Aquaculture 513, 734417. doi: 10.1016/j.aquaculture.2019.734417
Xu B. P., Tu D. D., Yan M. C., Shu M. A., Shao Q. J. (2017). Molecular characterization of a cDNA encoding Na+/K+/2Cl- cotransporter in the gill of mud crab (Scylla paramamosain) during the molt cycle: Implication of its function in osmoregulation. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 203, 115–125. doi: 10.1016/j.cbpa.2016.08.019
Yao H. Z., Li X., Tang L., Wang H., Wang C. L., Mu C. K., et al (2020). Metabolic mechanism of the mud crab (Scylla paramamosain) adapting to salinity sudden drop based on GC-MS technology. Aquac. Rep. 18, 100533. doi: 10.1016/j.aqrep.2020.100533
Zhang B. Q., Wang N. (2021). Study on the harm of saline alkali land and its improvement technology in China. IOP Conf. Ser.: Earth Environ. Sci. 692, 042053. doi: 10.1088/1755-1315/692/4/042053
Zhang Y., Wu Q. Y., Fang S. B., Li S. K., Zheng H. P., Zhang Y. L., et al (2020). mRNA profile provides novel insights into stress adaptation in mud crab megalopa, Scylla paramamosain after salinity stress. BMC Genomics 21 (1), 559. doi: 10.1186/s12864-020-06965-5
Zhao Y., Wang J., Thammaratsuntorn J., Wu J. W., Wei J. H., Wang Y., et al (2015). Comparative transcriptome analysis of Nile tilapia (Oreochromis niloticus) in response to alkalinity stress. Genet. Mol. Res. 14 (4), 17916–17926. doi: 10.4238/2015.December.22.16
Zhao Y., Zhang C. S., Zhou H., Song L. Y., Wang J., Zhao J. L. (2020). Transcriptome changes for Nile tilapia (Oreochromis niloticus) in response to alkalinity stress. Comp. Biochem. Physiol. Part D Genomics Proteomics 33, 100651. doi: 10.1016/j.cbd.2019.100651
Keywords: scylla paramamosain, inland saline-alkaline water, transcriptomic analysis, low salinity, adaptive mechanisms
Citation: Liang G, Qin K, Chen Y, Niu M, Wang H, Wang C, Mu C, Chen L, Wang F, Su Q and Zhu R (2022) Transcriptomic analysis of adaptive mechanisms in response to inland saline-alkaline water in the mud crab, Scylla paramamosain. Front. Mar. Sci. 9:974501. doi: 10.3389/fmars.2022.974501
Received: 21 June 2022; Accepted: 11 July 2022;
Published: 02 August 2022.
Edited by:
Hongyu Ma, Shantou University, ChinaReviewed by:
Ardavan Farhadi, Hainan University, ChinaBiswaranjan Paital, Orissa University of Agriculture and Technology, India
Copyright © 2022 Liang, Qin, Chen, Niu, Wang, Wang, Mu, Chen, Wang, Su and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huan Wang, d2FuZ2h1YW4xQG5idS5lZHUuY24=; Chunlin Wang, d2FuZ2NodW5saW5AbmJ1LmVkdS5jbg==
 Guoling Liang1
Guoling Liang1 Huan Wang
Huan Wang Chunlin Wang
Chunlin Wang Changkao Mu
Changkao Mu