- 1BK21 Center for Precision Medicine & Smart Engineering, Inha University, Incheon, Republic of Korea
- 2Division of Ecoscience, Ewha Womans University, Seoul, Republic of Korea
- 3Institute of Evolution & Marine Biodiversity, Ocean University of China, Qingdao, China
- 4Key Laboratory of Marine Ecosystem Dynamics, Second Institute of Oceanography, Ministry of Natural Resources, Hangzhou, China
The hairy snails of the genus Alviniconcha are representative deep-sea hydrothermal vent animals distributed across the Western Pacific and Indian Ocean. Out of six known species in the genus Alviniconcha, only one nominal species of A. marisindica was found in the Indian Ocean from the Carlsberg Ridge (CR), Central Indian Ridge (CIR) to the northern part of Southwest Indian Ridge (SWIR) and Southeast Indian Ridge (SEIR). Recently, the Alviniconcha snails were found at three new vent fields, named Onnare, Onbada, and Onnuri, in the northern CIR, which promotes a more comprehensive phylogeographic study of this species. Here, we examined the phylogeography and connectivity of the Alviniconcha snails among seven vent fields representing the CR and CIR based on DNA sequence data of a mitochondrial COI gene and two protein-coding nuclear genes. Phylogenetic inferences revealed that the Alviniconcha snails of the newly found in the northern CIR and two vent fields of Wocan and Tianxiu in the CR were divergent with the previously identified A. marisindica in the southern CIR and mitochondrial COI data supported the divergence with at least greater than 3% sequence divergence. Population structure analyses based on the three genetic markers detected a phylogeographic boundary between Onnuri and Solitaire that divides the whole snail populations into northern and southern groups with a low migration rate. The high degree of genetic disconnection around the ‘Onnuri’ boundary suggests that the Alviniconcha snails in the Indian Ocean may undergo allopatric speciation. The border may similarly act as a dispersal barrier to many other vent species co-distributed in the CIR. This study would expand understanding the speciation and connectivity of vent species in the Indian Ocean.
Introduction
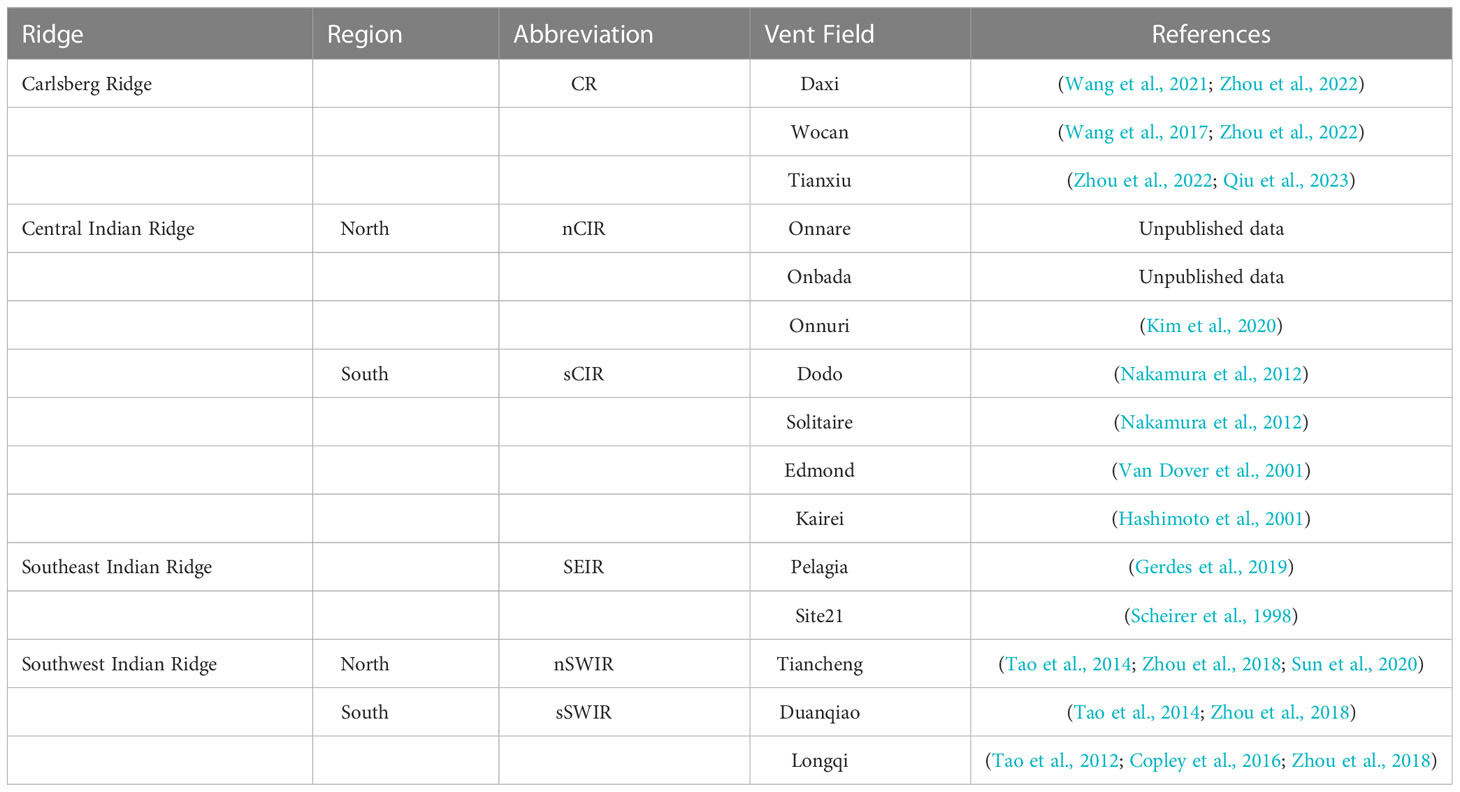
Historically, deep-sea exploration in the Indian Ocean is rare and delayed compared to other Oceans (Hashimoto et al., 2001; Van Dover et al., 2001). But recently, deep-sea exploration in the Indian Ocean region getting enormous interest (Thaler and Amon, 2019). Result of this, more biogeographical provinces were discovered in the Indian Ocean regions. Earlier, according to the eleven biogeographic provinces model of global hydrothermal vents (Rogers et al., 2012), there was only one province with two close vent fields in the Central Indian Ridge (CIR) regions. However, recently two more biogeographical provinces were identified with discoveries of new active vent fields and biological communities (Table 1): the northernmost Carlsberg Ridge (CR) and the southern Southwest Indian Ridge (sSWIR) (Zhou et al., 2018; Zhou et al., 2022). Besides, new active vent fields have been discovered along the CIR since the first discovery of the Kairei field around the Rodrigues Triple junction (Hashimoto et al., 2001): Edmond, Solitaire, Dodo, and Onnuri (Van Dover et al., 2001; Nakamura et al., 2012; Kim et al., 2020). Recently, in 2021, two more vent fields, Onnare and Onbada (Kim et al., unpublished data), were discovered towards the north of Onnuri. These two vent fields have active chimneys quite different from Onnuri with diffusive vent fluids rich in methane (Kim et al., 2020).
Because of the lack of enough sampling in the CIR region, the vent fauna is not described entirely (Watanabe and Beedessee, 2015). However, due to the recent 20 years of exploration and research in the CIR region, the biogeography and phylogeography of hydrothermal organisms in the region have been roughly outlined despite low resolution by limited information of genetic variations on the mitochondrial fragment (Perez et al., 2021). Most recently, Zhou et al. (2022) studied biogeography and genetic connectivity of organisms for expanded regions ranging from the CIR to the CR (Daxi, Wocan, and Tianxiu vent fields) and the SWIR (Taincheng, Duanqiao, and Longqi vent fields). Biogeographically the vent fields along the CIR were clustered into the same province extending to the Tiancheng vent field in the northern SWIR (nSWIR) region (Sun et al., 2020; Zhou et al., 2022). The community composition of the CR (Wocan and Tianxiu vent fields) was quite similar to the CIR communities, including the common species such as the scaly-foot snails (Chrysomallon squamiferum), hairy snails (Alviniconcha marisindica), and mussels (Bathymodiolus septemdierum.) observed in the CIR and SWIR regions, which create a biogeography dendrogram pattern along the Indian Ocean from north to south ((CR, (CIR & nSWIR)), sSWIR) (Zhou et al., 2022). These three common species were not observed at the Daxi vent field of the CR (Wang et al., 2021). Besides, A. marisindica was not found at sSWIR but observed along with bathymodioline mussels at the Pelagia vent field in SEIR (Gerdes et al., 2019). Such variation in the community composition along the hydrothermal vent fields of the Indian Ocean indicates the possibility of ridge offset along the Indian Ocean ridges that probably act as barriers for the dispersal of vent species, similar to the community contrast observed in the eastern Pacific region (Perez et al., 2021). In the eastern Pacific Ocean, the barrier around the equator and Easter microplate resulted in the population divergence and speciation of several polychaetes and bathymodioline mussels (Coykendall et al., 2011; Johnson et al., 2013; Zhang et al., 2015; Jang et al., 2016). Interestingly, genetically admixed populations of provannid snails (A. marisindica), bythogreid crabs (Austinograea rodriguezensis), alvinocaridid shrimp (Rimicaris kairei), bathymodioline mussels (B. septemdierum.) were observed among vent fields from the Solitaire to the Kairei in the southern CIR (sCIR) region (Nakamura et al., 2012; Beedessee et al., 2013; Chen et al., 2015; Sun et al., 2020; Zhou et al., 2022). A recent phylogenetic study in the CIR region showed that the Bathymodiolus mussel and its symbiotic bacterial populations of Onnuri had diverged from the ancestral lineages of the southern vent populations of Solitare and Kairei (Jang et al., 2022), which certainly indicates a presence of geographical structure between Onnuri and the others in the sCIR region.
In this study, we investigated the genetic connectivity of snails within the genus Alviniconcha from the CIR to CR, including the most recently discovered vent fields, Onnare and Onbada, in the northern CIR (nCIR). The nCIR are geographically located in the middle the sCIR and the CR. Especially Alviniconcha species is one of the representative species broadly distributed in the Indian Ocean region. Therefore, the present study will be helpful to prove or narrow down the location of the possible geographic boundary between nCIR and sCIR, as suggested by Jang et al. (2022), and deepen our understanding of the overall population subdivision pattern of vent species in the Indian Ocean.
Methods & materials
Sampling & DNA extraction
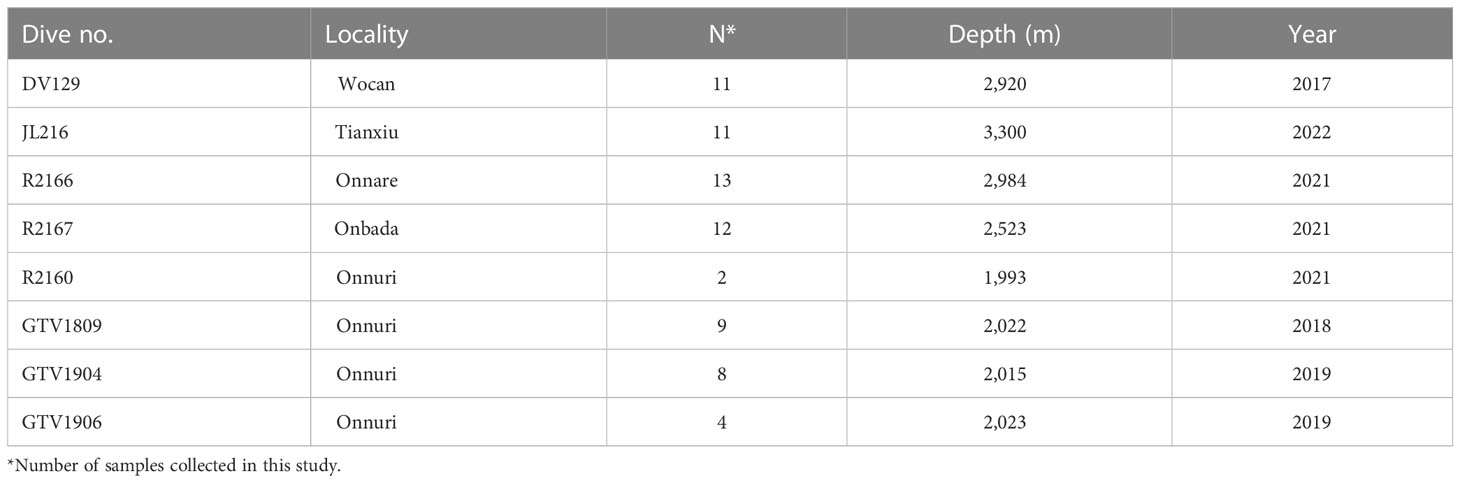
Hairy snails (Alviniconcha sp.) were obtained from five vent fields, Tianxiu and Wocan on the CR and Onnare, Onbada, and Onnuri on the CIR (Table 2). The specimens were collected using remote operated vehicle (ROV) Ropos, Xiangyanghong 9 and ShenhaiYihao, human occupied vehicle (HOV) Jiaolong and video-guided hydraulic grab (Oktopu, Germany) from 2017 to 2022. The Onnare and Onbada are newly discovered vent fields near 9°S with depths of about 2,984 and 2,523 m, respectively and are situated at the northern side of the Onnuri vent field (Figure 1, Table 2). More active vents with black smoker chimneys were observed around the new vent fields, which are distinct from the diffusing vents in the Onnuri fields. At those sites, the hairy snail assemblages were surrounded by bathymodioline mussel assemblages. Upon arrival on deck, specimens of the Onnare and Onbada fields were preserved in 99% ethanol and −20°C, and specimens of the Onnuri, Wocan and Tianxiu fields were immediately preserved in −80°C after collection on board. Genomic DNA was extracted from the foot tissue using QIAamp Fast DNA Tissue Kit (Qiagen, Inc., Hilden, Germany) or DNeasy Blood & Tissue Kits (Qiagen, Inc., Hilden, Germany). Two specimens of the Onnuri field were used to observe the community composition of symbiotic bacteria in gill tissue. Gill tissues of the specimens were immediately dissected after arrival on the deck and fixed in RNAlater at 4°C overnight and preserved at −20°C. Subsequently, genomic DNA was extracted using FastDNA Spin Kit for soil (MP Biomedicals, USA).
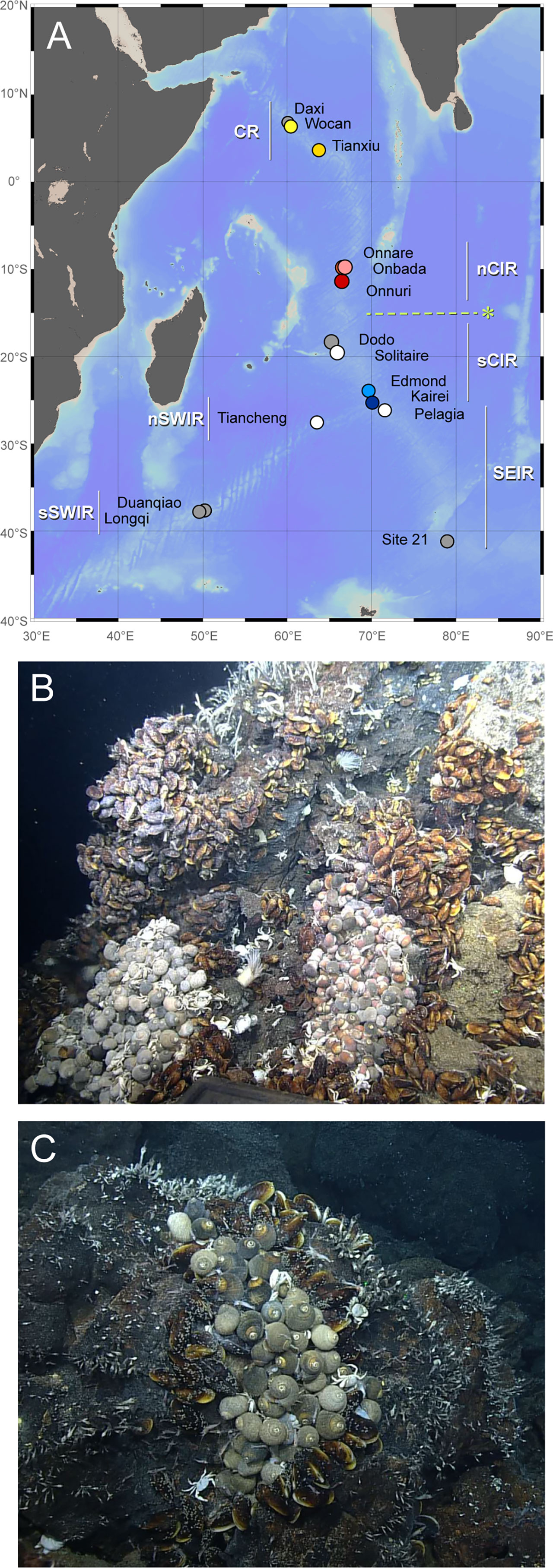
Figure 1 Sampling locations and hydrothermal vent fields in Indian Ocean reported to date. (A) The sampling sites of Alviniconcha snails along Carlsberg Ridge and Central Indian Ridge. The map was created using Ocean Data View v. 5.6.3 with the General Bathymetric Chart of the Oceans (GEBCO) 2014 grid. Colored circles represent sampling locations. White and gray circles represent the vent fields where Alviniconcha snails were observed but not analyzed in this study and not observed to date, respectively (Nakamura et al., 2012; Zhou et al., 2018; Gerdes et al., 2019; Sun et al., 2020). The abbreviations are described in Table 1. The dashed line with “*” represents the phylogeographic boundary observed from Alviniconcha snails in this study. Vent community with Alviniconcha snails and Bathymodiolus mussels at the Onnare vent field (B) and Onbada vent field (C).
We targeted three mitochondrial genes (mtCOI, mt12S rRNA, and mt16S rRNA) and five nuclear genes (EF1α, ATPSα, H3, 18S rRNA, and 28S rRNA) of Alviniconcha snails. Partial fragment of each gene was amplified using previously developed or newly designed primer sets and primer-specific PCR programs (Supplementary Table 1). PCR was performed by IP-Taq polymerase (COSMO genetech, South Korea) with 10×buffer, dNTP, the extracted genomic DNA, and gene-specific primer set according to a protocol of the manufacturer. All amplified products were sequenced bidirectionally using 3730xL DNA Analyzer (Thermo Fisher Scientific). Nuclear gene fragments with two or more heterozygous sites were phased using PHASE v. 2.1 (Stephens et al., 2001; Stephens and Scheet, 2005). The two nuclear genes (EF1α and ATPSα) of several specimens from the Wocan and Tianxiu fields were sequenced on Illumina Novaseq platform: six samples of the Wocan and two samples of the Tianxiu. DNA library was prepared with the insert length of 350 bp and sequenced with paired-end mode and read length of 150bp. Approximately 10Gb of raw Illumina reads were generated. Low-quality reads and adapter-contaminated reads were trimmed by Trimmomatic version 0.39 with the settings of ‘LEADING:20 TRAILING:20 SLIDINGWINDOW:4:20 MINLEN:50’, and the clean reads were assembled by SPAdes version 3.15.3 (Prjibelski et al., 2020). Target gene sequences were manually picked up from the assembled contigs through the blast result. All sequences generated from this study were submitted to NCBI (GenBank accession ID: OQ160352–OQ160421 for mtCOI gene; OQ149997–OQ150020 for mt12S rRNA, mt16S rRNA, 18S rRNA and 28S rRNA; OQ161779–OQ161988 for EF1α, ATPSα, and H3).
Phylogenetic analyses and molecular analyses
Phylogenetic analyses were conducted using concatenated sequences of the three mitochondrial and five nuclear genes. Two specimens were randomly chosen from three vent fields, Onnare, Onbada and Onnuri, on the nCIR. Sequences of Alviniconcha species and outgroup, Ifremeria nautilei, were downloaded from GenBank (Supplementary Table 2). Alignment of sequences was conducted using the MUSCLE algorithm (Edgar, 2004) in the Geneious Prime 2022.2.1. In the case of ribosomal RNA sequences, their ambiguously aligned positions of them were trimmed using Gblocks v. 0.91b on the Phylogeny.fr website, http://phylogeny.lirmm.fr/ (Castresana, 2000; Dereeper et al., 2008). The poorly aligned sites were eliminated by the setting for a less stringent selection allowing smaller final blocks, gap positions, and less strict flanking positions. Finally, we concatenated multi-locus sequencing data into two different datasets: 1) concatenated sequences of 981 bp including mtCOI (459 bp), ATPSα (297 bp), and EF1α (225 bp) and 2) concatenated sequences of 2,880 bp including mtCOI (459 bp), mt12S rRNA (303 bp), mt16S rRNA (470 bp), ATPSα (297 bp), EF1α (225 bp), H3 (273 bp), 18S rRNA (538 bp), and 28S rRNA (315 bp).
Phylogenetic analyses were carried out based on Maximum Likelihood (ML) and Bayesian Inference (BI) approaches. The selection of the best-fitting substitution model of the data was estimated using Smart Model Selection, SMS, based on the AIC (Akaike Information Criterion) and ML approach were implemented in PhyML v. 3.0 (Guindon et al., 2010) on the website, ATGC, http://www.atgc-montpellier.fr/phyml/ (Guindon and Gascuel, 2003). The ‘GTR+ Γ + I’ model was used for the concatenated sequences. The ML tree was constructed with 1,000 bootstrap replicates. The BI tree was inferred with 30,000,000 iteration and 25% burnin using the MrBayes (Huelsenbeck and Ronquist, 2001) on the Geneious Prime. The fragments of mtCOI gene from one specimen per each vent field in the CR and CIR were used to calculate the genetic distance, which was carried out based on the Kimura 2 parameter model using MEGA v. 11 (Tamura et al., 2021).
The mtCOI gene plus nuclear ATPSα and EF1α genes were used for the population genetic analyses. Sequences of the A. marisindica in the Edmond and Kairei fields were obtained from previous population genetic studies, Johnson et al. (2015) and Breusing et al. (2020) (GenBank accession ID: KF467897–KF467921 for mtCOI; MT147664–MT147723 for EF1α; MT148274–MT148329 for ATPSα). Sequences for each gene were aligned using the MUSCLE algorithm on the Genious Prime. Overlapped fragments were used for subsequent analyses after elimination of indel sites: 582 bp for mtCOI, 219 bp for EF1α, and 296 bp for ATPSα. The individual named as “JL301-8” from the Edmond was ruled out for subsequent molecular analyses due to very low sequence similarity on ATPSα (< 92%) and the limit for identification of species by no data for mtCOI gene (MT148328–9 for ATPSα gene; MT147720–21 for EF1α gene). Estimation of genetic diversity and genetic differentiation (FST), and analysis of molecular variance (AMOVA) were conducted using Arlequin v. 3.5 (Excoffier and Lischer, 2010). For mantel test, the geographic distance between vent fields were estimated based on longitude and latitude from the webserver of National Hurricane Center and Central Pacific Hurricane Center (https://www.nhc.noaa.gov/gccalc.shtml). The GPS locations of the Edmond and Kairei fields were based on the Van Dover et al. (2001). The correlation between the pairwise FST values for each genetic marker and the geographic distances were examined through Mantel tests using Genodive v. 3.0 (Meirmans, 2020). In addition, stratified mantel tests were performed to examine the effect of the hierarchical grouping of populations using the same program. Median-joining network (Bandelt et al., 1999) was estimated and drawn using PopART v. 1.7 (Leigh and Bryant, 2015). Assignment of individuals into population was examined based on genotypes of mtCOI and nuclear ATPSα and EF1α sequences using STRUCTURE v. 2.3.4 (Pritchard et al., 2000). The posterior probability of the number of discrete cluster (K) ranging from 1 to 7 was estimated through 10 independent runs including 1,000,000 iterations and 10% burnin. The most probable of K was determined by the delta K method of Evanno et al. (2005) on website (https://taylor0.biology.ucla.edu/structureHarvester/), Structure Harvester (Earl and Vonholdt, 2012). Finally, the mean Q-matrix for individuals were produced by CLUMPP v. 1.1.2 (Jakobsson and Rosenberg, 2007). The genotypes of two nuclear genes were used to analyze recent migration based on the bayesian approach using BayesAss v. 3.04 (Wilson and Rannala, 2003). The individuals with only mtCOI data were ruled out from this analysis. It assumed the linkage disequilibrium of loci within parental populations, which were tested using GenePop v. 4.7.5 on the webserver and based on the Bonferroni corrected significant level (https://genepop.curtin.edu.au/) (Rousset, 2008). The posterior probability was estimated from 250,000,000 iterations after burnin of 1,000,000 and a sampling frequency of 5,000. Mixing parameters for allele frequencies and inbreeding coefficient were set as 0.3 and 0.8, respectively. The convergence diagnostics of MCMC estimates were tested based on several diagnostics using R-script, including methods of Geweke (Geweke, 1992), Raftery and Lewis (Raftery and Lewis, 1992), and Heidelberg and Welch (Heidelberger and Welch, 1983).
Relative abundance and phylogenetic analysis of symbiotic bacteria
We conducted amplicon sequencing of 16S rRNA targeting the V3–V4 region using dual indexing approach (Fadrosh et al., 2014) based on universal primer for bacteria, 341F (5’-CCTACGGGNGGCWGCAG-3’) and 805R (5’-GACTACHVGGGTATCTAATCC-3’) (Herlemann et al., 2011). Sequences were generated from Illumina MiSeq platform using 2×250 paired-end protocol. Raw sequencing data were analyzed through the microbiome taxonomic profiling pipeline in EzBioCloud (https://www.ezbiocloud.net, Chunlab, Inc., Seoul, Korea). Raw sequencing data of paired-end reads was filtered using Trimmomatic (Bolger et al., 2014), merged using PANDAseq (Bartram et al., 2011; Masella et al., 2012). Subsequently, the primers were trimmed. The filtered reads were aligned using HMMER and denoised using DUDESeq (Lee et al., 2017). The reads were assigned according to taxonomic level (Yarza et al., 2014) using USEARCH (Edgar, 2010) based on the EzBioCloud 16S rRNA database (Yoon et al., 2017; Park and Won, 2018), and chimeric sequences among reads not assigned were removed using UCHIME algorithm (Edgar et al., 2011). The most dominant contig among reads was chosen and used to examine the phylogenetic relationship with other gill-associated bacteria of Alviniconcha species. The sequence used for the phylogenetic analysis were submitted to NCBI (GenBank accession ID: OQ198964). The sequences of sister taxa and out group were obtained from Genbank. After MUSCLE alignment, ML and BI inferences were carried out with same processes used for the host species. The ‘GTR’ with FreeRate for variation across sites (‘R’ model) was figured out as the best-fitting model and used for the ML approach. For BI approach, no variation across sites (‘equal’) was assumed instead of the FreeRate model, which was unavailable on the MrBayes.
Results
Phylogenetic relationship
Phylogenetic analysis with concatenated sequences of multi loci suggests that Alviniconcha snails from the nCIR, including Onnare, Onbada, and Onnuri fields, clustered into the well-supported monophyletic clade and formed a sister clade with A. marisindica (Figure 2). Among species in the Western Pacific (WP), A. boucheti was the most closely related to Alviniconcha species of the Indian Ocean based on high bootstrap value and probability from both ML and BI inferences. The mtCOI gene sequences of Alviniconcha snails from CR and nCIR diverged 0.2–0.5% intra- and inter-region but at least greater than 3% from A. marisindica in Edmond and Kairei fields (Table 3). According to the phylogenetic relationship and genetic distance estimation, we, hereafter, refer to the Alviniconcha snails from nCIR and CR as A. aff. marisindica. Species in the WP exhibited at least approximately 4% of inter-species genetic distance (Table 3).
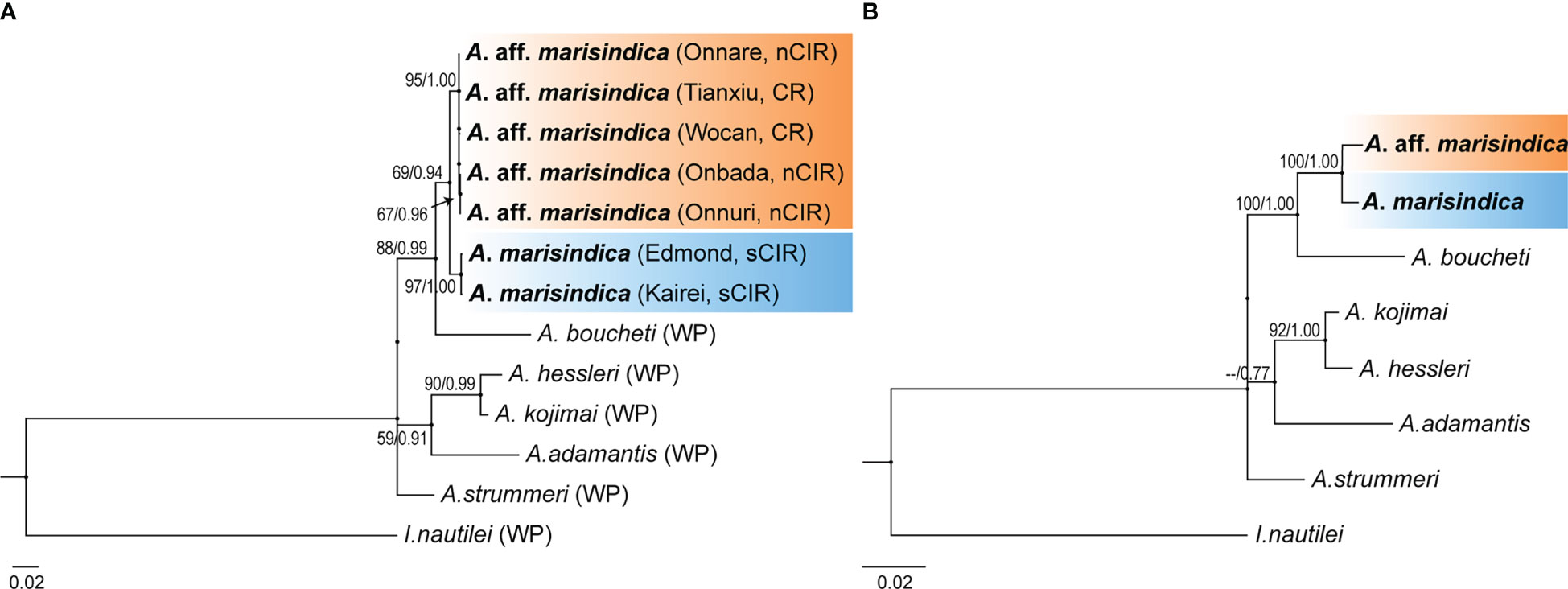
Figure 2 Phylogenetic relationship of Alviniconcha snail species. ML tree based on concatenated data set of three genetic markers (mtCOI, ATPSα, and EF1α) in (A) and eight genetic markers (mtCOI, mt12S rRNA, mt16S rRNA, ATPSα, EF1α, H3, 18S rRNA, and 28S rRNA) in (B). The bootstrap value (>50%) and posterior probabilities (>0.70) estimated from ML and BI analyses, respectively, are shown on the nodes. (A) Vent habitats are enclosed in parentheses next to each taxon name.
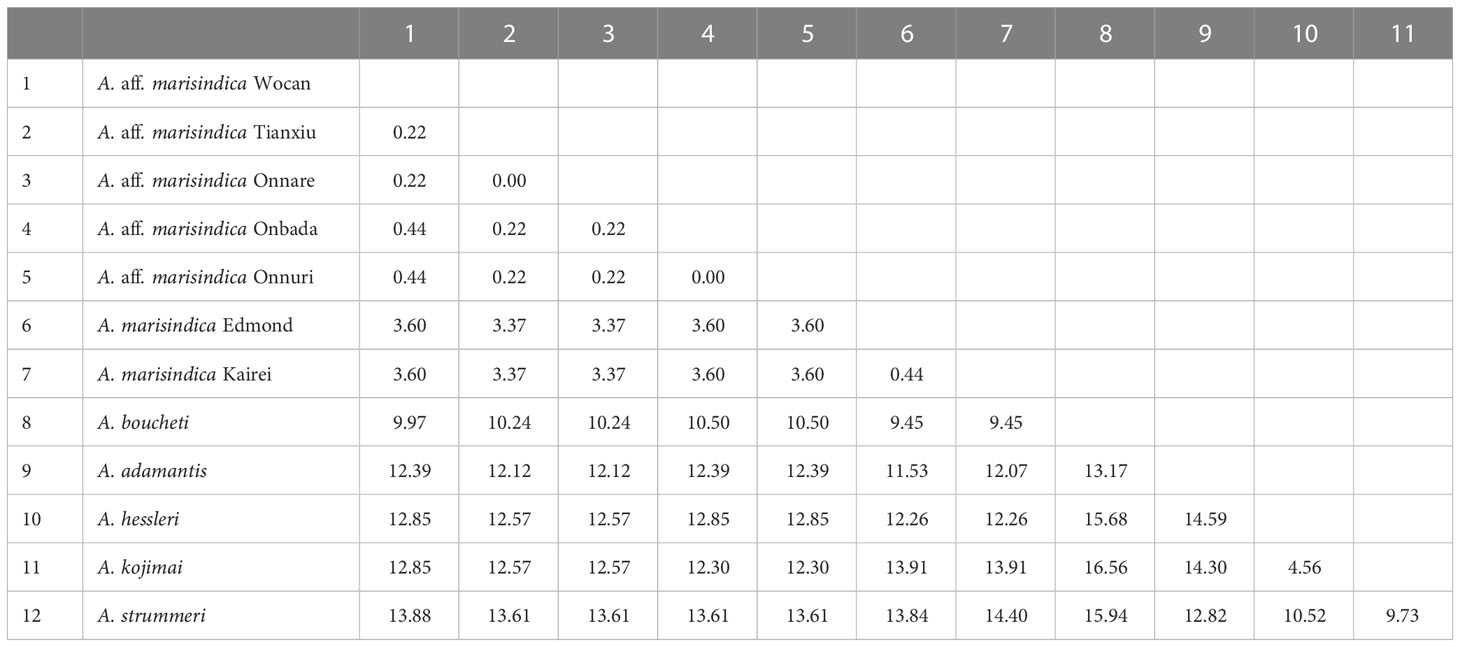
Table 3 Pairwise genetic distance (%) of mtCOI sequences (459 bp) of A. aff. marisindica with other Alviniconcha species based on Kimura 2 parameter model.
Population structure
The median-joining networks for the mtCOI and nuclear ATPSα genes represented genetic segregation between geographically northern and southern clusters of A. aff. marisindica and A. marisindica, respectively (Figure 3): The CR (Wocan and Tianxiu) and nCIR (Onnare, Onbada, and Onnuri) populations were separated from the southern CIR (sCIR; Edmond and Kairei) populations. While distinct differentiation of northern and southern haplotypes and alleles for mtCOI and ATPSα genes can be observed, the allele of EF1α gene was distributed broadly from the CR to sCIR with a few rare genetic variations in CIR populations. The northern and southern haplotypes for mtCOI genes were differentiated by at least 17 bp, 2.92% of the total length of 582 bp (Figure 3). Also, some southern alleles for ATPSα genes were distributed in nCIR with low frequency.
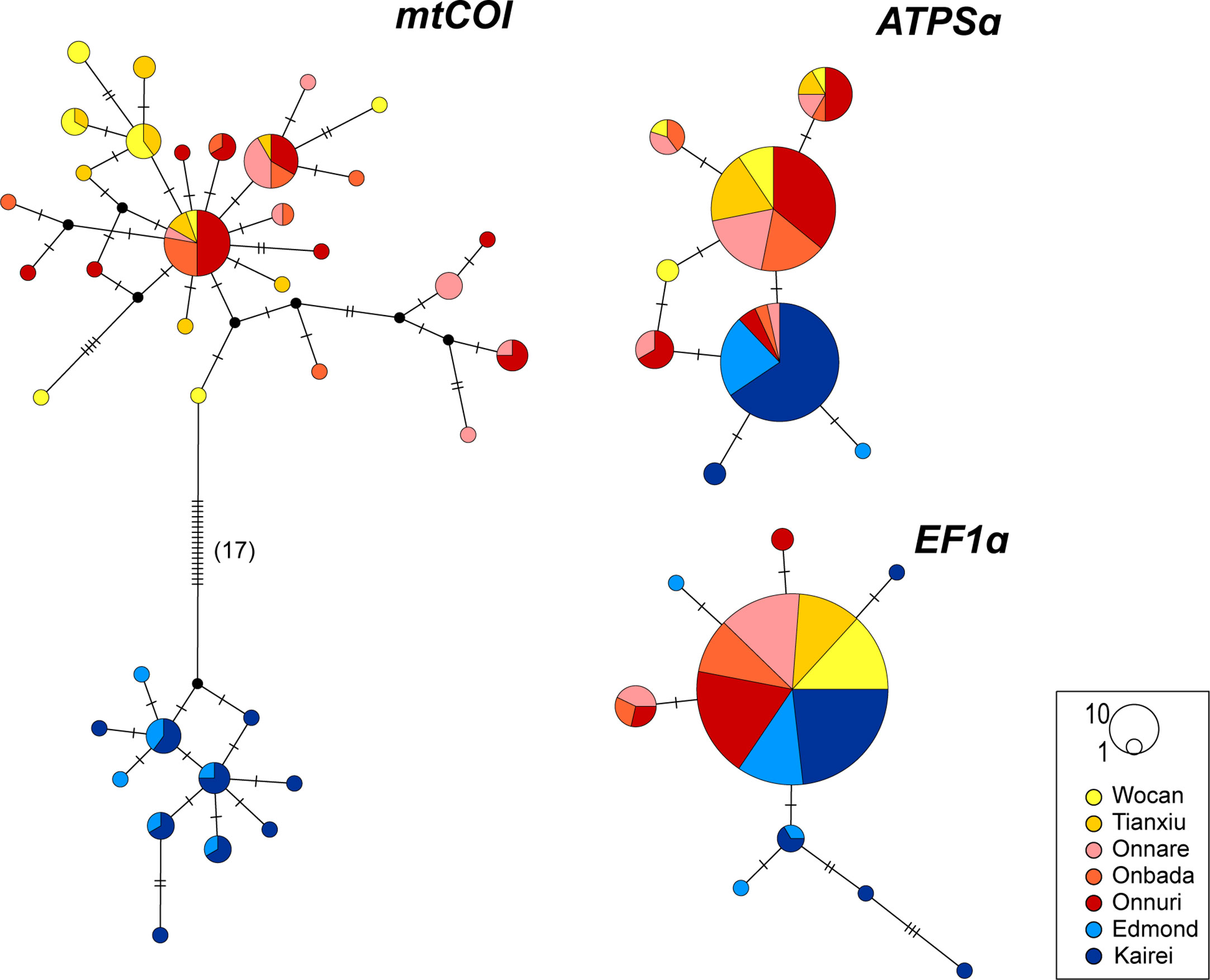
Figure 3 Median-joining network of mtCOI, ATPSα and EF1α genes. Each circle represents a distinct haplotype or allele, and its size is proportional to the frequency. Colors in the circle are consistent with the colors of vent fields on the map in the Figure 1. Black circle is the hypothetical haplotype. Hatch mark represents a single nucleotide mutation.
STRUCTURE also assigned Alviniconcha snails into two geographically distinct groups, corresponding to the clustering pattern from the network, based on the second-order rate of change of the log probability, delta K (K = 2; Figure 4A). The northern individuals were relatively more heterogeneous than the southern group. BayesAss revealed that contemporary migrants barely contributed to the admixed ancestries from relatively low migration rates between the two groups, about 0.01–0.03: The posterior mean and median values of migration rate were 0.0243 (s.d. = 0.0201) and 0.0189 from southern to northern groups, and 0.0158 (s.d. = 0.0144) and 0.0116 from north to southern groups. The proportion of immigrant ancestries in each individual also appeared to be limited, given admixed portion inferred from the STRUCTURE result (Figure 4B). Most immigrants were identified as post-1st generation immigrants regardless of source group. The STRUCTURE of K = 3 did not add more subclusters (Supplementary Figure 1). Additionally, the recent migration rates among three regions (CR, nCIR and sCIR populations) were estimated (Supplementary Table 3). The bidirectional migration rate was assessed between the CR and CIR populations, but the extent of southward migration was much larger than northward migration: posterior mean = 0.1031 (s.d. = 0.1005) and median = 0.0594 for northward migration; posterior mean = 0.2350 (s.d. = 0.1110) and median = 0.2912 for southward migration.
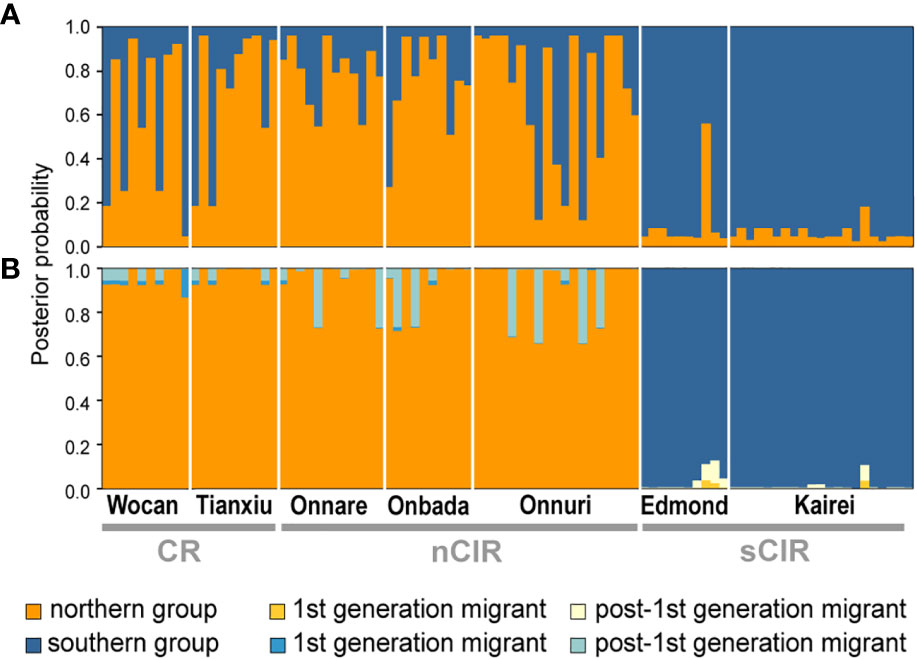
Figure 4 Genetic structure and recent migration of Alviniconcha populations in the Indian Ocean. (A) STRUCTURE analysis (K = 2). Colors of bar represent assigned clusters K of each individual into the northern group (orange) and southern group (blue). (B) BayesAss estimation. The colors of the bar represent the posterior probabilities of native and migrant ancestries estimated from the genotypes of each individual. The migrant ancestries were colored with a similar color tone to the native group.
Pairwise FST for mtCOI and ATPSα genes supported the geographical population structure between northern and southern groups (Table 4). The pairwise FST values between the two groups were significant and ranged from 0.88 to 0.92 for mtCOI and from 0.51 to 0.88 for ATPSα. For the mtCOI gene, pairwise FST inter-populations within the northern group, between CR and nCIR, were also statistically significant despite less genetic differentiation than that inter groups (FST = 0.128–0.24, p-value=0.0005–0.008). The Onnare population differed significantly from the Onbada population (FST = 0.109, p-value = 0.046). For the EF1α gene, a significant FST value was estimated between the Onnuri and sCIR populations (FST = 0.040, p-value = 0.046 between the Onnuri and Kairei; FST = 0.030, p-value = 0.039 between the Onnuri and Edmond). The pairwise FST values were significantly correlated with geographic distance between vent habitats (Mantel’s r = 0.668, p-value = 0.044 for mtCOI; Mantel’s r = 0.699, p-value = 0.015 for ATPSα). However, the stratified Mantel test conducting permutation within clusters based on the STRUCTURE result exhibited a significant correlation only for the ATPSα, not for the mtCOI gene (Stratified Mantel’s r = 0.699, p-value = 0.043 for ATPSα).
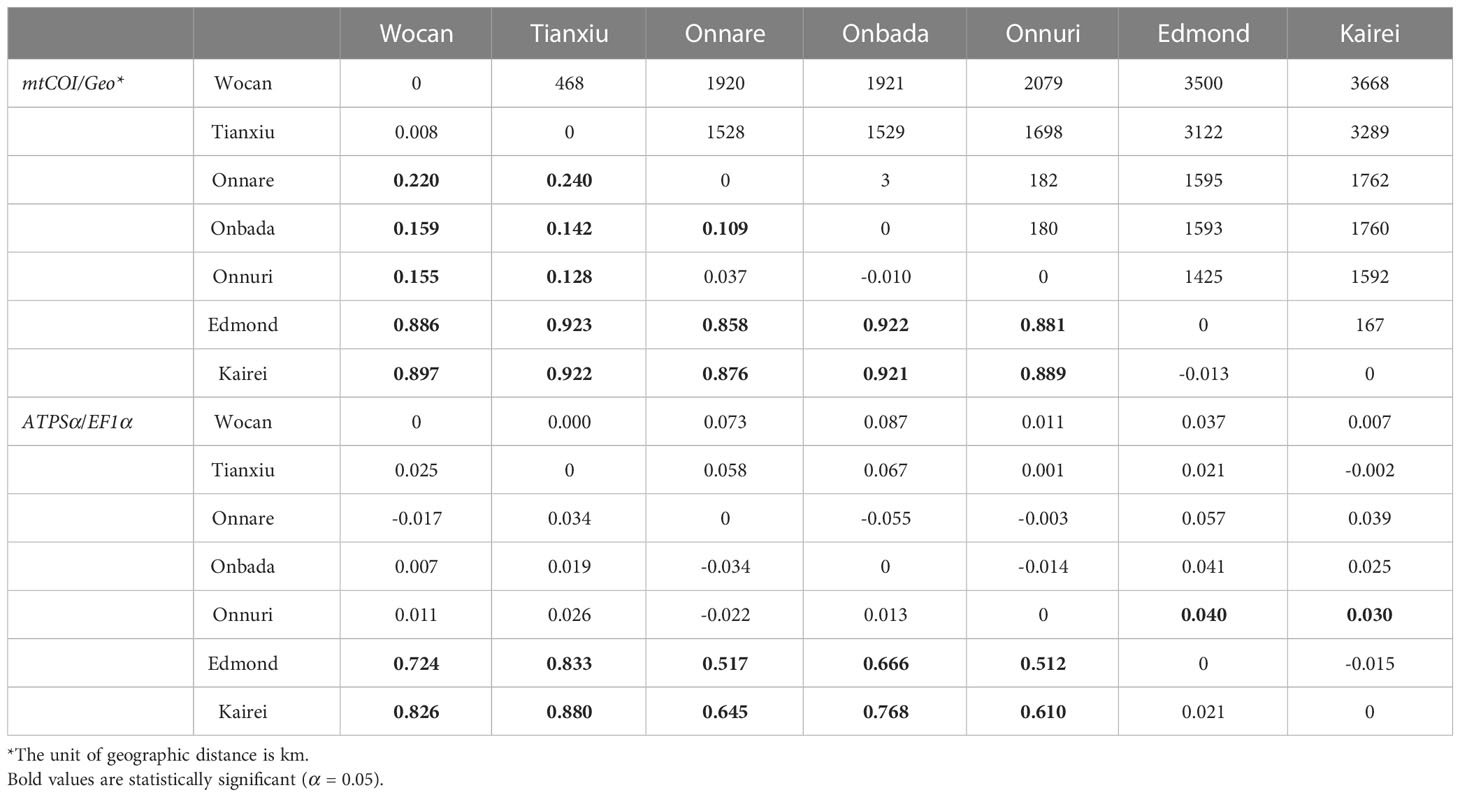
Table 4 Geographic distance (Geo) and pairwise genetic differentiation (FST) between vent localities. First listed matrix is below and second matrix is above the diagonal.
Gill-associated symbiotic bacteria
An average of 25,220 reads were obtained in each host individual collected from Onnuri vent field. Gill-associated symbiotic bacteria of A. aff. marisindica was predominantly identified as Sulfurovum-related Campylobacteria based on 16S rRNA gene fragments (>99%; Figure 5A). The most abundant 16S rRNA contig of two A. aff. marisindica specimens were identical (sequence similarity = 100%). Phylogenetic analysis revealed that gill-associated bacteria of A. aff. marisindica formed a monophyletic cluster with the Sulfurovum lineage along with the gill symbiotic bacteria of A. marisindica and A. boucheti, but this cluster was weakly supported with less than 50% of bootstrap value and approximately 0.74 posterior probability (Figure 5B). We found two distinct lineages of bacteria associated with Alviniconcha snails, some are clustered with Sulfurovum and others are with Sulfurimonas lineages (Figure 5B).
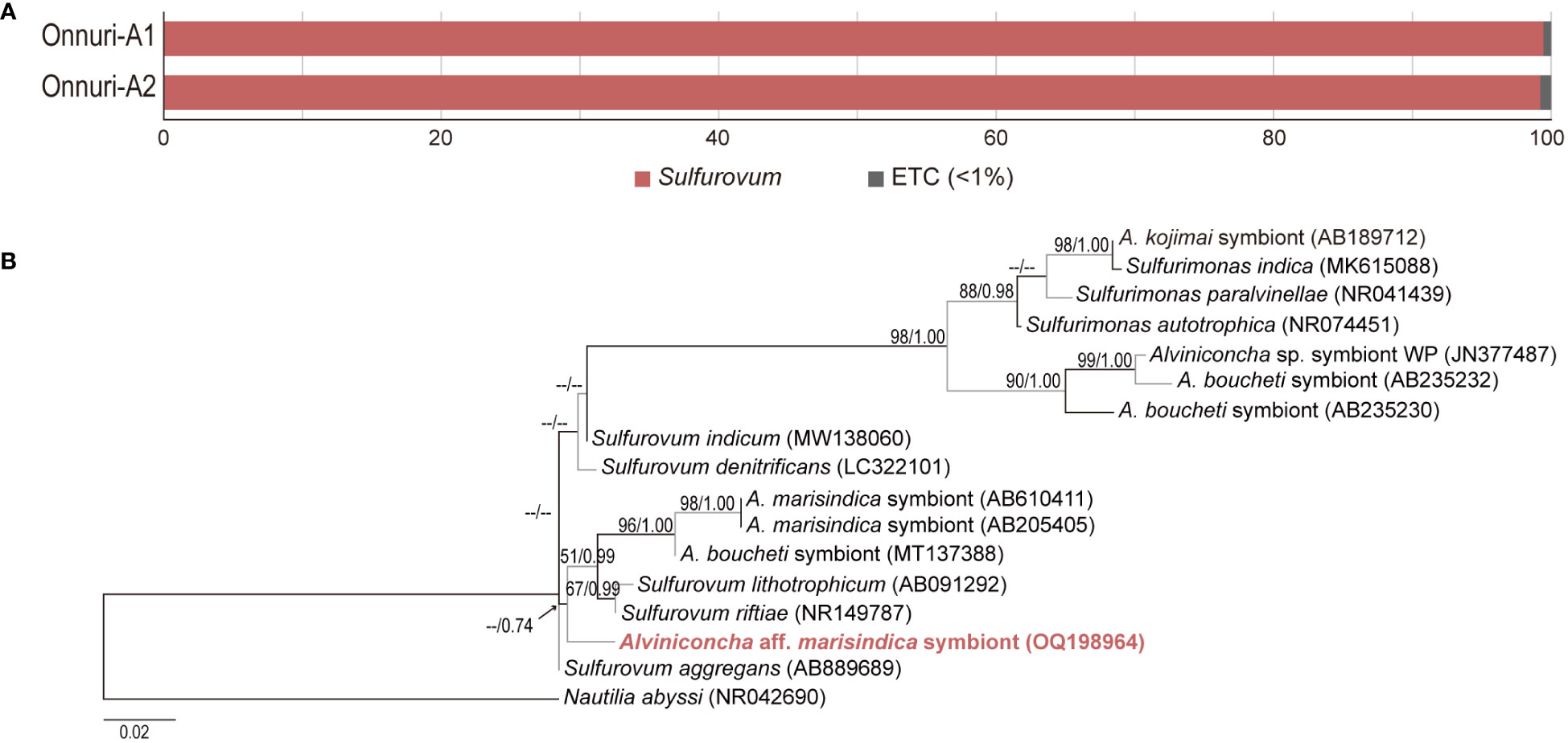
Figure 5 Relative abundance of gill-associated symbiotic bacteria and phylogenetic tree of dominant symbiont based on 16S rRNA sequences. (A) At the genus level, bacterial community compositions in gill tissue of Alviniconcha snails from the Onnuri vent field. (B) Phylogenetic relationship among gill-association bacteria of Alviniconcha snails.
Discussion
Biogeographical discontinuity
Earlier it was observed that the Mid-Ocean Ridge system of the Indian Ocean consists of three distinguished biogeographical provinces, CR, CIR-nSWIR and sSWIR (Zhou et al., 2022). The addition of three vent sites, Onnare, Onbada and Onnuri, in the northern vent sites helped find the location of potential boundaries between CIR and CR. In this study, we observed the biogeographical discontinuity in nCIR and sCIR regions. Particularly multi loci sequences including mitochondrial and nuclear genetic markers showed that two phylogenetically distinct Alviniconcha snail lineages exist across the boundary in allopatry. The high level of genetic distance for mtCOI sequences suggests the divergence beyond population-level to speciation level. Genotypic assignment based on three genetic loci supported a genetic subdivision of A. aff. marisindica populations in CR and nCIR region from the A. marisindica populations in sCIR region. The high genetic connectivity of A. marisindica in the sCIR region is coincide with the findings observed among three vents of sCIR (Solitaire, Edmond and Kairei) (Beedessee et al., 2013). Taken together the observation by Beedessee et al., 2013, the limited migration among northern and southern CIR vent populations observed in this study suggested that the putative dispersal barrier is expected to be located between the Onnuri and Solitaire vent fields. Statistically significant genetic differentiation between two geographical groups also supported the divergence by geographic isolation, that is, allopatric divergence between the two Alviniconcha snail lineages. The stratified mantel test for the mtCOI and ATPSα genes supported the differentiation of population clustering into north and south regions due to the isolation by geographical distance on the divergence process of Alviniconcha lineages.
The genetic structure and haplotype networks of the three genetic markers show that Alviniconcha individuals from the CR and nCIR ridges are genetically very close. Therefore, the intermediate region between the sampled vents of the two ridges does not seem to have any dispersal barrier to the snails that could potentially subdivide them. However, given the limited genetic variation from just several loci of the present study, we cannot entirely rule out the possibility of a geographic boundary that could be seen with only sufficient genetic data, such as genome-wide single nucleotide polymorphisms (SNPs) between the two ridges. The high genetic connectivity between the CR and nCIR is somewhat unexpected since the Tianxiu vent field in the CR, and the Onnare in the nCIR are separated by more than 1,500 km. Though the genetic differentiation (FST) of mtCOI between these two vents was statistically significant, the genetic distances of DNA sequences were minimal or nothing (Figure 3). Furthermore, the other nuclear gene makers (ATPSα and EF1α) showed no genetic differentiation between them (Table 4). This result may be because there are many undiscovered hydrothermal vents in the 1,500 km gap. These hidden habitats likely act as stepping stones linking the spread and distribution of Alviniconcha snails along the CR and nCIR. In other words, the unexpectedly high similarity compared to the distance apart may be due to the undetected intermediate habitats between the two hydrothermal regions. Therefore, future deep-sea exploration in this intermediate region between the Tianxiu and Onnare vent fields will likely lead to the discovery of new potential hydrothermal vents.
Gene flow and effect of environmental and ecological factors on the divergence
The genotype assignment based on mitochondrial and nuclear genes suggested that many admixed individuals in the northern region originated from the southern region. This finding extended the observation of the gene flow from the south to the north ridge of the Indian Ocean (Perez et al., 2021). The Bayesian approach based on genotypes of nuclear genes indicates the current gene flow scarcely contributed. Estimation at the individual level suggested a relatively higher degree of gene flow from south to north in the past, as an allele type of post-1st generation immigrant was observed in a relatively large number of individuals in the northern region compared to the southern region. This directionality matches well with the northward flow of the deep current at 2500–3000 m depth predicted by the modeling approach (Reid, 2003).
The dispersal of bathymodiolin mussel populations was limited due to lateral offsets by transform faults (TF), such as Marie Celeste FZ, of about 100 km long and about 5 km deep between the Onnuri and Solitaire vent fields (Jang et al., 2022). The lateral offsets by the TF disrupt larval dispersal, especially in the slow-spreading ridge system, which induces the cross-axis current to interrupt passively dispersing larvae (Johnson et al., 2006; Young et al., 2008). Likewise, the TFs between Tiancheng and Longqi along the SWIR were interpreted to result in low connectivity of the scaly-foot snail, C. squamiferum (Sun et al., 2020). The dispersal distance of Alviniconcha snails was more limited than the bathymodiolin mussels, even though Alviniconcha species were expected to possess a planktotrophic larval stage like the vent mussels during larvae (Waren and Bouchet, 1993). Although the behavior and physiology of Alviniconcha larvae are poorly known, several in-situ observations revealed a predominant distribution of gastropod larvae near the bottom, probably driven by negative buoyancy or downward swimming (Mullineaux et al., 2005; Metaxas, 2011; Mullineaux et al., 2013). Because the vertical distribution of larvae affects the dispersal process related to the current depending on the depth (Mcgillicuddy et al., 2010; Gary et al., 2020), the dispersal of larvae near the bottom is more likely to be impedded by the lateral offset of the ridge axis. Consequently, the geographical subdivision of the Alviconcha snails in the Indian Ocean might be associated with the spatial distribution pattern of larvae near the bottom. Future investigations for larva duration and dispersal depth are needed to fully understand the geographical connectivity and diverging process identified by genetic studies.
In addition, the unique geomorphological features of the Onnuri vent field are also suggested to contribute to accelerating the geographic subdivision between nCIR and sCIR. The Onnuri vent field attracted attention with several different characteristics from the general hydrothermal vents. First, the Onnuri hydrothermal area was formed at the top of the topography rising from the main surface, not near the expansion axis of the Mid-Oceanic Ridge, where volcanic activity is concentrated. The Onnuri field locates on the summit of ocean core complexes of dome-shaped and is relatively distant from the ridge axis, approximately 12 km apart (Pak et al., 2017; Kim et al., 2020; Lim et al., 2022). This topography is composed of the lower ocean crust and upper mantle, which have been raised to the sea floor by long-lasting faulting activities. As a result, the Onnuri field is ultramafic-hosted hydrothermal system with non-magmatic heat source (Lim et al., 2022). When olivine, a mantle rock, is altered by hydrothermal circulation, it turns into serpentine, and hydrogen and methane are generated in this process. Therefore, the hydrothermal plume of Onnuri is characterized by high hydrogen and methane concentration. Therefore, it is presumed that these geological and chemical characteristics affect the vent environment and organisms and ecosystem of Onnuri, possibly slightly differentiating its populations from those of other vents located near areas of ridge axes. Comparative studies involving vent organisms in this area and its neighboring vent fields, such as Onnare and Onbada, are urgently needed.
On the other hand, bidirectional migration was observed between the CR and nCIR populations, and in particular, the migration rate southward was relatively higher. The limited information on the oceanic bottom current did not allow for linking the biological and abiotic factors to understand the dispersal pattern of vent snails. However, the migration pattern of Alviniconcha snails would serve as a reference for inferring the geographical connectivity of other vent invertebrates in the Indian Ocean region.
Alviniconcha snail gill-associated bacteria
Gill-associated bacteria from A. aff. marisindica were identified as taxa of the genus Sulfurovum within Campylobacteria. In WP, Alviniconcha snails’ divergence was explained by geographical separation and ecological isolation due to niche segregation through the acquisition of different phylotypes of chemosynthetic bacterial symbionts in Gammaproteobacteria and Campylobacteria at different vent fields through horizontal transmission of symbionts (Breusing et al., 2020; Breusing et al., 2022). However, as dominant symbiont species, we could only detect the Sulfurovum genus within the phylum Camphylobacteria from the Onnuri vent Alviniconcha snails. Due to the small sample size in this study, we might not detect multiple phylogenetic lineages of symbionts as equivalent as seen in the West Pacific. A more extensive data set with more samples covering the various vents are needed to investigate symbionts’ role in the divergence of Alviniconcha snails in the Indian Ocean.
Conclusion
In this study, we confirmed a phylogeographic boundary in the CIR region using the endemic Alviniconcha vent snails that distributes broadly in the Indian Ocean. We have extended sample collection from the initially found southern Central Indian Ridge up to the Carlsberg Ridge north of the Indian Ocean. While previous genetic studies have shown high genetic connectivity of this species among the vents in the sCIR, this study suggested the existence of a phylogeographic boundary between the Onnuri and Solitaire vent fields. The Alviniconcha snails of the newly found Onnare, Onnuri, and Onbada vent fields in the nCIR and two other vent fields of Wocan and Tianxiu in the CR formed a monophyletic but divergent clade with the previously identified A. marisindica in the sCIR, suggesting that the degree of separation between the two major clades goes beyond population-level to speciation. The location of the phylogeographic boundary is generally consistent with other studies on invertebrate vent species. The phylogeographic border was likely formed by the geographical habitat gaps that impede the connectivity of Alviniconcha snails. In future works, comparative studies with multiple vent species around the boundary are required to further re-confirm the findings in this study. In addition, examinations of morphological characteristics of Alviniconcha snails in the CR and nCIR are needed to clarify taxonomic identification.
Data availability statement
The datasets presented in this study can be found in online repositories. The sequence data were deposited in GenBank under accession number(s): OQ160352–OQ160421, OQ149997–OQ150020, OQ161779–OQ161988, OQ198964. 16S rRNA amplicon reads were deposited in Sequence Read Archive (SRA) under BioProject number: PRJNA903468.
Ethics statement
Ethical review and approval was not required for the animal study because Alviniconcha snails are not subject to prior review as invertebrates and are neither an endangered species in Korea nor elsewhere.
Author contributions
S-JJ and Y-JW designed the study. S-JJ, S-YC, CL, YZ, HW and JS collected samples and performed the laboratory experiments and sequencing analyses. S-JJ analyzed phylogeny and population genetics. S-JJ, AP and Y-JW drafted the manuscript and edited with contributions from all authors. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by a project titled ‘Understanding the deep-sea biosphere on seafloor hydrothermal vents in the Indian Ridge (No. 20170411)’ funded by the Ministry of Oceans and Fisheries, Korea, the Basic Science Research Program through National Research Foundation of Korea (NRF) funded by the Ministry of Education (No. 2021R1I1A1A01060419), National Natural Science Foundation of China (No. 42176110), and the Foundation of China Ocean Mineral Resources R & D Association (No. DY135-E2-1-02), and the National Key Research and Development Program of China (No. 2017YFC0306603).
Acknowledgments
We appreciate reviewers for helpful comments which improved the manuscript. We are grateful to all scientist and operation teams during 2018 to 2021 cruise of Korea Institute of Ocean Science & Technology to the CIR for assistance and thank the chief scientist and crew member of R/V Xiangyanghong 9 and ShenhaiYihao and the pilots of HOV Jiaolong during the cruise of DY38-I and DY72 for the sample collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1139190/full#supplementary-material. All supplementary files are available at Figshare: https://doi.org/10.6084/m9.figshare.21829017
References
Bandelt H.-J., Forster P., Röhl A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Bartram A. K., Lynch M. D., Stearns J. C., Moreno-Hagelsieb G., Neufeld J. D. (2011). Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end illumina reads. Appl. Environ. Microbiol. 77, 3846–3852. doi: 10.1128/AEM.02772-10
Beedessee G., Watanabe H., Ogura T., Nemoto S., Yahagi T., Nakagawa S., et al. (2013). High connectivity of animal populations in deep-sea hydrothermal vent fields in the Central Indian Ridge relevant to its geological setting. PloS One 8, e81570. doi: 10.1371/journal.pone.0081570
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Breusing C., Genetti M., Russell S. L., Corbett-Detig R. B., Beinart R. A. (2022). Horizontal transmission enables flexible associations with locally adapted symbiont strains in deep-sea hydrothermal vent symbioses. Proc. Natl. Acad. Sci. 119, e2115608119. doi: 10.1073/pnas.2115608119
Breusing C., Johnson S. B., Tunnicliffe V., Clague D. A., Vrijenhoek R. C., Beinart R. A. (2020). Allopatric and sympatric drivers of speciation in Alviniconcha hydrothermal vent snails. Mol. Biol. Evol. 37, 3469–3484. doi: 10.1093/molbev/msaa177
Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Chen C., Copley J. T., Linse K., Rogers A. D. (2015). Low connectivity between ‘scaly-foot gastropod’(Mollusca: Peltospiridae) populations at hydrothermal vents on the southwest Indian Ridge and the Central Indian Ridge. Organisms. Diversity Evol. 15, 663–670. doi: 10.1007/s13127-015-0224-8
Copley J. T., Marsh L., Glover A. G., Hühnerbach V., Nye V. E., Reid W. D., et al. (2016). Ecology and biogeography of megafauna and macrofauna at the first known deep-sea hydrothermal vents on the ultraslow-spreading Southwest Indian Ridge. Sci. Rep. 6, 1–13. doi: 10.1038/srep39158
Coykendall D. K., Johnson S. B., Karl S. A., Lutz R. A., Vrijenhoek R. C. (2011). Genetic diversity and demographic instability in Riftia pachyptila tubeworms from eastern pacific hydrothermal vents. BMC Evol. Biol. 11, 1–12. doi: 10.1186/1471-2148-11-96
Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., et al. (2008). Phylogeny. fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. doi: 10.1093/nar/gkn180
Earl D. A., Vonholdt B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Edgar R. C. (2004). MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 5, 1–19. doi: 10.1186/1471-2105-5-113
Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Evanno G., Regnaut S., Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Excoffier L., Lischer H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Fadrosh D. W., Ma B., Gajer P., Sengamalay N., Ott S., Brotman R. M., et al. (2014). An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the illumina MiSeq platform. Microbiome 2, 1–7. doi: 10.1186/2049-2618-2-6
Gary S. F., Fox A. D., Biastoch A., Roberts J. M., Cunningham S. A. (2020). Larval behaviour, dispersal and population connectivity in the deep sea. Sci. Rep. 10, 1–12. doi: 10.1038/s41598-020-67503-7
Gerdes K., Martínez Arbizu P., Schwarz-Schampera U., Schwentner M., Kihara T. C. (2019). Detailed mapping of hydrothermal vent fauna: A 3D reconstruction approach based on video imagery. Front. Mar. Sci. 6, 96. doi: 10.3389/fmars.2019.00096
Geweke J. (1992). “Evaluating the Accuracy of Sampling-Based Approaches to the Calculation of Posterior Moments” in Bernardo J. M., Berger J. O., Dawid A. P., Smith A. F. M. (Eds.). Bayesian Statistics 4. Oxford: Clarendon Press, 169–193.
Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Hashimoto J., Ohta S., Gamo T., Chiba H., Yamaguchi T., Tsuchida S., et al. (2001). First hydrothermal vent communities from the Indian Ocean discovered. Zool. Sci. 18, 717–721. doi: 10.2108/zsj.18.717
Heidelberger P., Welch P. D. (1983). Simulation run length control in the presence of an initial transient. Operations. Res. 31, 1109–1144. doi: 10.1287/opre.31.6.1109
Herlemann D. P., Labrenz M., Jürgens K., Bertilsson S., Waniek J. J., Andersson A. F. (2011). Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME. J. 5, 1571–1579. doi: 10.1038/ismej.2011.41
Huelsenbeck J. P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Jakobsson M., Rosenberg N. A. (2007). CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806. doi: 10.1093/bioinformatics/btm233
Jang S.-J., Chung Y., Jun S., Won Y.-J. (2022). Connectivity and divergence of symbiotic bacteria of deep-sea hydrothermal vent mussels in relation to the structure and dynamics of mid-ocean ridges. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.845965
Jang S.-J., Park E., Lee W.-K., Johnson S. B., Vrijenhoek R. C., Won Y.-J. (2016). Population subdivision of hydrothermal vent polychaete Alvinella pompejana across equatorial and Easter microplate boundaries. BMC Evol. Biol. 16, 1–15. doi: 10.1186/s12862-016-0807-9
Johnson S. B., Warén A., Tunnicliffe V., Dover C. V., Wheat C. G., Schultz T. F., et al. (2015). Molecular taxonomy and naming of five cryptic species of Alviniconcha snails (Gastropoda: Abyssochrysoidea) from hydrothermal vents. Syst. Biodivers. 13, 278–295. doi: 10.1080/14772000.2014.970673
Johnson S. B., Won Y.-J., Harvey J. B., Vrijenhoek R. C. (2013). A hybrid zone between Bathymodiolus mussel lineages from eastern pacific hydrothermal vents. BMC Evol. Biol. 13, 1–18. doi: 10.1186/1471-2148-13-21
Johnson S. B., Young C. R., Jones W. J., Warén A., Vrijenhoek R. C. (2006). Migration, isolation, and speciation of hydrothermal vent limpets (Gastropoda; lepetodrilidae) across the blanco transform fault. Biol. Bull. 210, 140–157. doi: 10.2307/4134603
Kim J., Son S. K., Kim D., Pak S. J., Yu O. H., Walker S. L., et al. (2020). Discovery of active hydrothermal vent fields along the Central Indian Ridge, 8–12°S. Geochem. Geophys. Geosyst. 21, e2020GC009058. doi: 10.1029/2020GC009058
Lee B., Moon T., Yoon S., Weissman T. (2017). DUDE-seq: Fast, flexible, and robust denoising for targeted amplicon sequencing. PloS One 12, e0181463. doi: 10.1371/journal.pone.0181463
Leigh J. W., Bryant D. (2015). POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
Lim D., Kim J., Kim W., Kim J., Kim D., Zhang L., et al. (2022). Characterization of geochemistry in hydrothermal sediments from the newly discovered Onnuri vent field in the middle region of the Central Indian Ridge. Front. Mar. Sci. 9, 810949. doi: 10.3389/fmars.2022.810949
Masella A. P., Bartram A. K., Truszkowski J. M., Brown D. G., Neufeld J. D. (2012). PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinf. 13, 1–7. doi: 10.1186/1471-2105-13-31
Mcgillicuddy D. Jr., Lavelle J., Thurnherr A., Kosnyrev V., Mullineaux L. (2010). Larval dispersion along an axially symmetric mid-ocean ridge. Deep. Sea. Res. Part I.: Oceanogr. Res. Papers. 57, 880–892. doi: 10.1016/j.dsr.2010.04.003
Meirmans P. G. (2020). Genodive version 3.0: Easy-to-use software for the analysis of genetic data of diploids and polyploids. Mol. Ecol. Resour. 20, 1126–1131. doi: 10.1111/1755-0998.13145
Metaxas A. (2011). Spatial patterns of larval abundance at hydrothermal vents on seamounts: Evidence for recruitment limitation. Mar. Ecol. Prog. Ser. 437, 103–117. doi: 10.3354/meps09283
Mullineaux L. S., Mcgillicuddy D. Jr., Mills S. W., Kosnyrev V., Thurnherr A. M., Ledwell J. R., et al. (2013). Active positioning of vent larvae at a mid-ocean ridge. Deep. Sea. Res. Part II.: Topical. Stud. Oceanogr. 92, 46–57. doi: 10.1016/j.dsr2.2013.03.032
Mullineaux L. S., Mills S. W., Sweetman A. K., Beaudreau A. H., Metaxas A., Hunt H. L. (2005). Vertical, lateral and temporal structure in larval distributions at hydrothermal vents. Mar. Ecol. Prog. Ser. 293, 1–16. doi: 10.3354/meps293001
Nakamura K., Watanabe H., Miyazaki J., Takai K., Kawagucci S., Noguchi T., et al. (2012). Discovery of new hydrothermal activity and chemosynthetic fauna on the Central Indian Ridge at 18–20 s. PloS One 7, e32965. doi: 10.1371/journal.pone.0032965
Pak S.-J., Moon J.-W., Kim J., Chandler M. T., Kim H.-S., Son J., et al. (2017). Widespread tectonic extension at the Central Indian Ridge between 8°S and 18°S. Gondwana. Res. 45, 163–179. doi: 10.1016/j.gr.2016.12.015
Park S.-C., Won S. (2018). Evaluation of 16S rRNA databases for taxonomic assignments using a mock community. Genomics Inf. 16, 4. doi: 10.5808/GI.2018.16.4.e24
Perez M., Sun J., Xu Q., Qian P.-Y. (2021). Structure and connectivity of hydrothermal vent communities along the mid-ocean ridges in the west Indian Ocean: A review. Front. Mar. Sci. 8, 744874. doi: 10.3389/fmars.2021.744874
Pritchard J. K., Stephens M., Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Prjibelski A., Antipov D., Meleshko D., Lapidus A., Korobeynikov A. (2020). Using SPAdes de novo assembler. Curr. Protoc. Bioinf. 70, e102. doi: 10.1002/cpbi.102
Qiu Z., Fan W., Han X., Chen X., Yin X. (2023). Distribution, speciation and mobility of metals in sediments of the Tianxiu hydrothermal field, Carlsberg Ridge, Northwest Indian Ocean. J. Mar. Syst. 237, 103826. doi: 10.1016/j.jmarsys.2022.103826
Raftery A. E., Lewis S. M. (1992). [Practical Markov Chain Monte carlo]: comment: One long run with diagnostics: Implementation strategies for Markov Chain Monte Carlo. Stat. Sci. 7, 493–497. doi: 10.1214/ss/1177011143
Reid J. L. (2003). On the total geostrophic circulation of the Indian Ocean: Flow patterns, tracers, and transports. Prog. Oceanogr. 56, 137–186. doi: 10.1016/S0079-6611(02)00141-6
Rogers A. D., Tyler P. A., Connelly D. P., Copley J. T., James R., Larter R. D., et al. (2012). The discovery of new deep-sea hydrothermal vent communities in the southern ocean and implications for biogeography. PloS Biol. 10, e1001234. doi: 10.1371/journal.pbio.1001234
Rousset F. (2008). GENEPOP’007: a complete re-implementation of the genepop software for windows and Linux. Mol. Ecol. Resour. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Scheirer D. S., Baker E. T., Johnson K. T. (1998). Detection of hydrothermal plumes along the southeast Indian Ridge near the Amsterdam-st. Paul Plateau. Geophys. Res. Lett. 25, 97–100. doi: 10.1029/97GL03443
Stephens M., Scheet P. (2005). Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 76, 449–462. doi: 10.1086/428594
Stephens M., Smith N. J., Donnelly P. (2001). A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989. doi: 10.1086/319501
Sun J., Zhou Y., Chen C., Kwan Y. H., Sun Y., Wang X., et al. (2020). Nearest vent, dearest friend: Biodiversity of Tiancheng vent field reveals cross-ridge similarities in the Indian Ocean. R. Soc. Open Sci. 7, 200110. doi: 10.1098/rsos.200110
Tamura K., Stecher G., Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tao C., Li H., Jin X., Zhou J., Wu T., He Y., et al. (2014). Seafloor hydrothermal activity and polymetallic sulfide exploration on the Southwest Indian Ridge. Chin. Sci. Bull. 59, 2266–2276. doi: 10.1007/s11434-014-0182-0
Tao C., Lin J., Guo S., Chen Y. J., Wu G., Han X., et al. (2012). First active hydrothermal vents on an ultraslow-spreading center: Southwest Indian Ridge. Geology 40, 47–50. doi: 10.1130/G32389.1
Thaler A. D., Amon D. (2019). 262 voyages beneath the Sea: a global assessment of macro-and megafaunal biodiversity and research effort at deep-sea hydrothermal vents. PeerJ 7, e7397. doi: 10.7717/peerj.7397
Van Dover C. L., Humphris S. E., Fornari D., Cavanaugh C. M., Collier R., Goffredi S. K., et al. (2001). Biogeography and ecological setting of Indian Ocean hydrothermal vents. Science 294, 818–823. doi: 10.1126/science.1064574
Wang Y., Han X., Petersen S., Frische M., Qiu Z., Li H., et al. (2017). Mineralogy and trace element geochemistry of sulfide minerals from the Wocan hydrothermal field on the slow-spreading Carlsberg Ridge, Indian Ocean. Ore. Geol. Rev. 84, 1–19. doi: 10.1016/j.oregeorev.2016.12.020
Wang Y., Han X., Zhou Y., Qiu Z., Yu X., Petersen S., et al. (2021). The Daxi vent field: an active mafic-hosted hydrothermal system at a non-transform offset on the slow-spreading Carlsberg Ridge, 6° 48′ n. Ore. Geol. Rev. 129, 103888. doi: 10.1016/j.oregeorev.2020.103888
Waren A., Bouchet P. (1993). New records, species, genera, and a new family of gastropods from hydrothermal vents and hydrocarbon seeps. Zool. Scripta. 22, 1–90. doi: 10.1111/j.1463-6409.1993.tb00342.x
Watanabe H., Beedessee G. (2015). “Vent fauna on the Central Indian Ridge,” in Ishibashi J.-I., Okino K., Sunamura M. (Eds.). Subseafloor biosphere linked to hydrothermal systems: TAIGA Concept (Tokyo: Springer), 205–212.
Wilson G. A., Rannala B. (2003). Bayesian Inference of recent migration rates using multilocus genotypes. Genetics 163, 1177–1191. doi: 10.1093/genetics/163.3.1177
Yarza P., Yilmaz P., Pruesse E., Glöckner F. O., Ludwig W., Schleifer K.-H., et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645. doi: 10.1038/nrmicro3330
Yoon S.-H., Ha S.-M., Kwon S., Lim J., Kim Y., Seo H., et al. (2017). Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613. doi: 10.1099/ijsem.0.001755
Young C., Fujio S., Vrijenhoek R. (2008). Directional dispersal between mid-ocean ridges: deep-ocean circulation and gene flow in Ridgeia Piscesae. Mol. Ecol. 17, 1718–1731. doi: 10.1111/j.1365-294X.2008.03609.x
Zhang H., Johnson S. B., Flores V. R., Vrijenhoek R. C. (2015). Intergradation between discrete lineages of Tevnia jerichonana, a deep-sea hydrothermal vent tubeworm. Deep. Sea. Res. Part II.: Topical. Stud. Oceanogr. 121, 53–61. doi: 10.1016/j.dsr2.2015.04.028
Zhou Y., Chen C., Zhang D., Wang Y., Watanabe H. K., Sun J., et al. (2022). Delineating biogeographic regions in Indian Ocean deep-sea vents and implications for conservation. Diversity Distributions. 28, 2858–2870. doi: 10.1111/ddi.13535
Zhou Y., Zhang D., Zhang R., Liu Z., Tao C., Lu B., et al. (2018). Characterization of vent fauna at three hydrothermal vent fields on the Southwest Indian Ridge: Implications for biogeography and interannual dynamics on ultraslow-spreading ridges. Deep. Sea. Res. Part I.: Oceanogr. Res. Papers. 137, 1–12. doi: 10.1016/j.dsr.2018.05.001
Keywords: hydrothermal vent, Alviniconcha snail, allopatric divergence, metapopulation, Central Indian Ridge, Carlsberg Ridge, gill-associated bacteria
Citation: Jang S-J, Cho S-Y, Li C, Zhou Y, Wang H, Sun J, Patra AK and Won Y-J (2023) Geographical subdivision of Alviniconcha snail populations in the Indian Ocean hydrothermal vent regions. Front. Mar. Sci. 10:1139190. doi: 10.3389/fmars.2023.1139190
Received: 06 January 2023; Accepted: 15 March 2023;
Published: 03 April 2023.
Edited by:
Paris Vasileios Stefanoudis, University of Oxford, United KingdomReviewed by:
Elin Angharad Thomas, Queen’s University Belfast, United KingdomGenki Kobayashi, Ishinomaki Senshu University, Japan
Copyright © 2023 Jang, Cho, Li, Zhou, Wang, Sun, Patra and Won. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong-Jin Won, d29uQGV3aGEuYWMua3I=
 Sook-Jin Jang
Sook-Jin Jang Soo-Yeon Cho
Soo-Yeon Cho Chuyu Li
Chuyu Li Yadong Zhou
Yadong Zhou Hui Wang
Hui Wang Jin Sun
Jin Sun Ajit Kumar Patra
Ajit Kumar Patra Yong-Jin Won
Yong-Jin Won