- 1Engineering School of Sustainable Infrastructure and Environment, Department of Environmental Engineering Sciences, University of Florida, Gainesville, FL, United States
- 2Department of Soil, Water, and Ecosystem Sciences, University of Florida, Gainesville, FL, United States
- 3Smithsonian Marine Station, Fort Pierce, FL, United States
- 4Red Sea Research Center, Biological and Environmental Science and Engineering Division, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia
- 5The Elizabeth Moore International Center for Coral Reef Research and Restoration, Mote Marine Laboratory, Summerland Key, FL, United States
Ocean deoxygenation is intensifying globally due to human activities – and is emerging as a grave threat to coral reef ecosystems where it can cause coral bleaching and mass mortality. However, deoxygenation is one of many threats to coral reefs, making it essential to understand how prior environmental stress may influence responses to deoxygenation. To address this question, we examined responses of the coral holobiont (i.e., the coral host, Symbiodiniaceae, and the microbiome) to deoxygenation in corals with different environmental stress backgrounds. We outplanted Acropora cervicornis fragments of known genotypes from an in situ nursery to two sites in the Florida Keys spanning an inshore-offshore gradient. After four months, fragments from the outplanted corals were transferred to the laboratory, where we tested differences in survivorship, tissue loss, photosynthetic efficiency, Symbiodiniaceae cell density, and coral microbiome composition after persistent exposure to one of four oxygen treatments ranging from extreme deoxygenation (0.5 mg L-1) to normoxia (6 mg L-1). We found that, for the short duration of exposure tested in this study (four days), the entire coral holobiont was resistant to dissolved oxygen (DO) concentrations as low as 2.0 mg L-1, but that the responses of members of the holobiont decoupled at 0.5 mg L-1. In this most extreme treatment, the coral host showed decreased photosynthetic efficiency, tissue loss, and mortality, and lower Symbiodiniaceae densities in a bleaching response, but most microbial taxa remained stable. Although deoxygenation did not cause major community shifts in microbiome composition, the population abundance of some microbial taxa did respond. Site history influenced some responses of the coral host and endosymbiont, but not the coral microbiome, with corals from the more stressful inshore site showing greater susceptibility to subsequent deoxygenation. Our study reveals that coral holobiont members respond differently to deoxygenation, with greater sensitivity in the coral host and Symbiodiniaceae and greater resistance in the coral microbiome, and that prior stress exposure can decrease host tolerance to deoxygenation.
1 Introduction
Coral reefs are susceptible to climate change-related stressors such as ocean warming (Hughes et al., 2017; Hughes et al., 2018), acidification (Hoegh-Guldberg et al., 2007; Fabricius et al., 2011; Allemand and Osborn, 2019; Cornwall et al., 2021), and deoxygenation (Altieri et al., 2017; Nelson and Altieri, 2019; Johnson et al., 2021a; Hughes et al., 2022). Ocean warming, the best recognized of these stressors, decreases habitat suitability for corals in their current latitudinal range (Couce et al., 2013) and causes coral bleaching, the physiological decoupling of the coral-algal symbiosis (Glynn, 1993). Mass bleaching events can cause loss of coral cover and biodiversity globally (Loya et al., 2001; Baker et al., 2008; Hughes et al., 2022). It is important to consider warming and other stressors, such as deoxygenation, together to understand the potential for climate change to elicit responses that scale from the holobiont to ecosystem level.
Deoxygenation in the ocean is linked to climate change (through a variety of metabolic, biogeochemical, and physical processes) and eutrophication (through a proliferation of algae and bacteria that depletes oxygen) and acts on multiple spatial scales (Rabalais et al., 2010; Altieri and Gedan, 2015; Breitburg et al., 2018). In recent years, deoxygenation has gained attention as a threat to coral reefs (Altieri et al., 2017; Altieri and Diaz, 2019; Nelson and Altieri, 2019; Hughes et al., 2020) that is underestimated and expected to increase in severity as ocean warming intensifies (Pezner et al., 2023). Deoxygenation can lead to conditions considered hypoxic, or life-limiting, for many marine organisms (around 3.0 mg L-1), but this threshold is highly variable across taxa and climates (Laffoley and Baxter, 2019). Because of oxygen’s importance for many of the essential processes on coral reefs (Nelson and Altieri, 2019), it is critical that we understand how escalating deoxygenation will impact these ecosystems.
Recent research has revealed variable responses of corals to decreased dissolved oxygen (DO) and implicates a variety of mechanisms underpinning these responses. For example, some studies have found different tolerances and physiological responses to deoxygenation across coral species (Johnson et al., 2021b; Pontes et al., 2023). The health and photophysiology of the coral Acropora yongei was found to be sensitive to oxygen concentrations of 2-4 mg L-1 after only three days of exposure (Haas et al., 2014). Acute hypoxic events (down to 0.18 ± 0.12 SD mg L−1) have resulted in coral community shifts driven by bleaching and mortality (Altieri et al., 2017; Johnson et al., 2021a). Some research suggests Symbiodiniaceae clades and microbial communities may underpin coral resilience to deoxygenation (Camp et al., 2020). The discovery of a hypoxia-inducible factor (HIF)-mediated hypoxia response system in corals revealed a mechanism that is activated during times of hypoxic stress in some corals, while corals that fail to activate this system bleach (Alderdice et al., 2021). These studies point to a suite of mechanisms, acting in concert, resulting in variable tolerance of corals to deoxygenation.
The coral microbiome may be an important intermediary between the coral and its environment (Smith et al., 2006; Grottoli et al., 2018; van Oppen and Blackall, 2019) and can be influenced by oxygen dynamics (Howard et al., 2023). Diurnal oxygen fluctuation can shape microbial communities at the coral’s surface (Kühl et al., 1995) as corals experience nightly localized deoxygenation and daytime oxygen availability when their algal symbionts photosynthesize (Shashar et al., 1993; Haas et al., 2010). This fluctuation may create a diverse community with facultative aerobic and anaerobic microbes that can tolerate both extremes (Kellogg, 2004). When deoxygenation persists for days to weeks in hypoxia events, coral reef-associated bacterial communities have been shown to shift but return to assemblages resembling normoxic conditions shortly after (Johnson et al., 2021b). Some coral populations have adapted to extreme environments where deoxygenation is common (Camp et al., 2018) and possess very different bacterial and endosymbiont communities relative to the same species in less harsh environments (Camp et al., 2020). This adaptability of the microbiome may benefit corals, as both anaerobic and aerobic bacteria play a crucial role in the coral’s nitrogen cycle (Babbin et al., 2021). Because of the coral microbiome’s adaptability and the frequent and dramatic oxygen fluxes that occur naturally on coral reefs (Altieri et al., 2021), coral microbiomes may have more natural resistance to hypoxia than to other types of stress.
Warming commonly co-occurs with or precedes hypoxia in the ocean (Keeling et al., 2010), as on coral reefs where ocean warming is expected to increase the severity, duration, and spatial extent of hypoxia events through increased stratification and decreased oxygen solubility (Pezner et al., 2023). This co-occurrence of temperature and oxygen stress could trigger synergistic interactions. For example, corals that have experienced deoxygenation bleach at lower temperatures than those naïve to low-DO stress (Alderdice et al., 2022). In a variety of marine benthic organisms, increasing temperatures decreased organismal hypoxia tolerance (Vaquer-Sunyer and Duarte, 2010). On the other hand, stress hardening can occur, such as when corals are repeatedly exposed to warming (Hackerott et al., 2021), yet the extent to which thermal history influences subsequent hypoxia tolerance has not yet been explored.
In the present study, we used both field and laboratory-based techniques to examine the response of the coral holobiont to deoxygenation following exposure at two sites along an inshore-offshore gradient, where sites differed primarily in temperature. The cnidarian host, Symbiodiniaceae, and a small subset of prokaryotic taxa showed hypoxia-sensitivity, with varying influences of site history. These divergent responses suggest that within the coral holobiont, responses to environmental stress can be complex and occur on different timescales.
2 Materials and methods
Our combined field and laboratory experimental approach tested coral tolerance to a range of dissolved oxygen (DO) concentrations in experimental aquaria following in situ exposure to inshore or offshore conditions. We evaluated responses of the coral holobiont, including the cnidarian host, photosynthetic symbionts (Symbiodiniaceae), and the coral’s surface microbiome. For the purposes of this paper, the term “microbiome” is used to refer to Bacteria and Archaea associated with the cnidarian host.
2.1 Field exposure to different environmental regimes
To expose corals in situ to different environmental regimes, fragments of Acropora cervicornis of five known genotypes were outplanted to two restoration sites off Summerland Key, Florida Keys National Marine Sanctuary in June 2019 (Figure 1A). We chose A. cervicornis for its widespread use in restoration, historical dominance in the region, threatened status (National Marine Fisheries Service and National Oceanic and Atmospheric Association, 2006), and the availability of known genotypes from Mote Marine Laboratory’s in-water nursery. Coral fragments were collected from the in-water nursery on the day of outplanting and transported to the study sites. Genotypes were kept in separate containers during transit. The study sites were selected based on observed differences in environmental regimes. One study site, hereafter “inshore,” is approximately 3.6 km from shore and experienced warmer average and peak summer temperatures than the second study site, hereafter “offshore,” which was approximately 6.5 km from shore (Figures 1A, B). At each site, 25 plots, each consisting of five ~15 cm coral fragments outplanted within 15 cm of one another, were attached with cable ties to masonry nails that had been hammered into dead reef substrate. All coral fragments within a given plot belonged to the same genotype, and genotypes of plots were randomly assigned across sites, for a total of five plots of each of the five genotypes at each site.
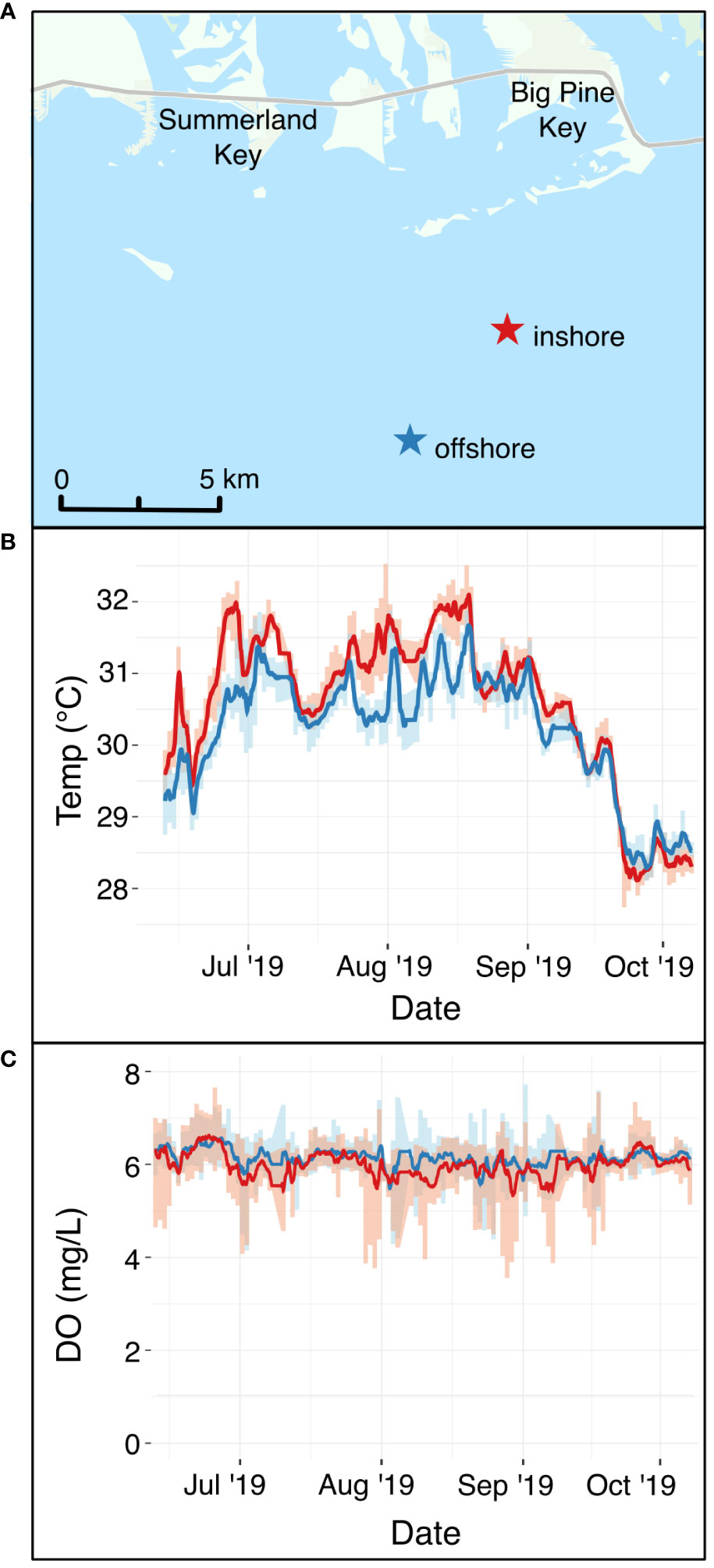
Figure 1 (A) Field sites where A. cervicornis was outplanted at an inshore site (red star) and an offshore site (blue star) in the Florida Keys. (B) In situ continuous temperature at the two sites (red line = inshore site, blue line = offshore site) during the period that corals were outplanted. (C) In situ continuous dissolved oxygen in mg L-1 at the two sites (red line = inshore site, blue line = offshore site) during the same time period. The map in Panel (A) uses an ArcGIS basemap with the following attribution: “FDEP, Esri, HERE, Garmin, SafeGraph, METI/ NASA, USGS, EPA, NPS, USDA.
To document water quality differences between the two sites, YSI EXO2 multiparameter sondes were deployed, logging temperature, dissolved oxygen, pH, salinity, and total dissolved solids every 15 minutes for the duration that corals were outplanted (Figure 1; Supplementary Figure S1). Light availability was not measured directly, but inshore reefs in the Florida Keys often have lower photosynthetic active radiation than offshore sites (Manzello et al., 2015; Manzello et al., 2019). Divers visited each site monthly to retrieve sondes for calibration and to visually monitor coral performance. On these visits, coral fragments were evaluated for survivorship (live or dead) and the presence of bleaching (Table 1).
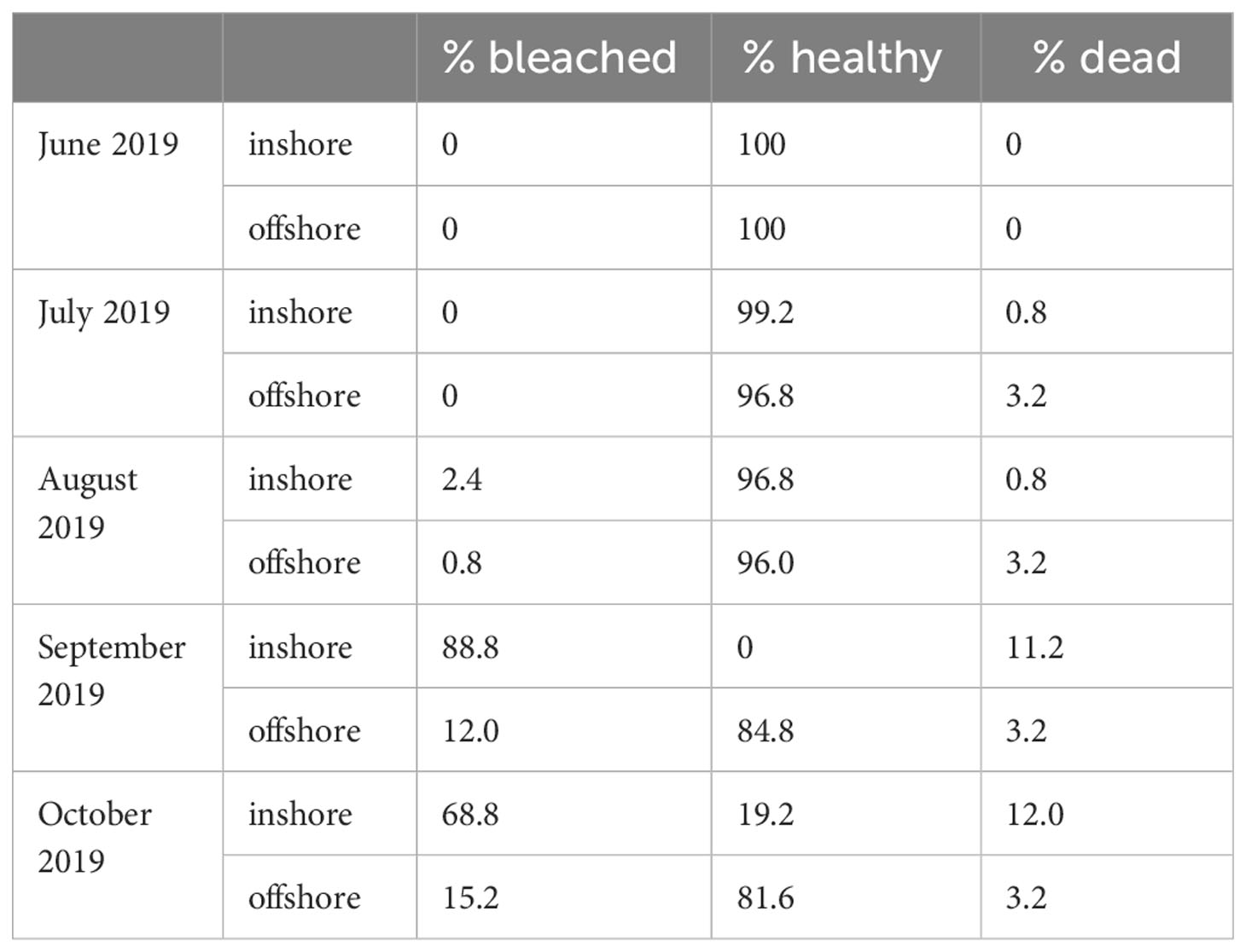
Table 1 Condition data for corals outplanted at the inshore and offshores sites during the field exposure period of 2019.
Research activities were conducted under permit number FKNMS-2018-129 issued by the National Oceanic and Atmospheric Administration’s Office of National Marine Sanctuaries. Corals were outplanted under permit FKNMS-2015-163-A2.
2.2 Laboratory experimental conditions
In October 2019, 10 cm fragments including 1-2 apical tips were collected from living coral outplants and transported to the Smithsonian Marine Station in Fort Pierce, FL to conduct the laboratory portion of the study. Twelve fragments from each of the five genotypes were collected from each site, resulting in one fragment of each of the five genotypes from each of the two sites in each aquarium, and a total of ten fragments in each of the 12 aquaria (n=120 fragments total in the laboratory experiment).
Corals were maintained under ambient conditions (6.0 mg L-1 DO and 27°C) in the experimental aquaria for one week before starting the laboratory experiment to allow a recovery period from potential stress associated with fragmentation and transport.
Experiments were conducted in a mesocosm system consisting of twelve, independently controlled, 50 L aquaria. Each aquarium was filled with 42 L of 0.35 μm filtered seawater from a proximal ocean source and adjusted daily with reverse-osmosis (RO) water to maintain a salinity of 35 ppt. Each aquarium was equipped with one LED aquarium light (Aquaillumination Hydra 52), programmed to a 12:12 hour diel cycle, reaching 300 µmol photon m-2 sec-1 of photosynthetic active radiation (PAR) at the peak of the light period. PAR was measured using a light meter equipped with an underwater spherical quantum sensor (LI-COR, LI-1400 and LI-193SA) placed underwater at the center of each aquarium where coral fragments were positioned.
Dissolved oxygen treatments of 0.5, 2.0, 4.0, and 6.0 mg L-1 (control) were assigned randomly to the twelve aquaria, with three aquarium replicates per DO treatment. The oxygen manipulation system is described in detail in Johnson et al. (2021b). Briefly, a Neptune Apex aquarium controller system, equipped with OxyGuard oxygen probes, used nitrogen gas and air to maintain DO levels within 0.1 mg L-1 of treatment settings (Supplementary Figure S2). Temperature was maintained at 27°C (+/- 1°C) throughout the experiment by Aqualogic Inc. temperature control units, which monitored seawater temperatures with internal temperature probes and adjusted temperatures by activating titanium heaters or chilled coils. Temperature, pH, and salinity were spot-checked daily with a handheld YSI ProDSS multiparameter water quality meter that was calibrated according to manufacturer protocols.
Percent tissue loss was assessed visually throughout the experiment. Photographs of each coral fragment were taken daily to document condition. The experimental period ended after four days, at which point corals in the most severe DO treatment (0.5 mg L-1) had suffered 50% mortality. Mortality was defined by at least 95% tissue loss, as determined by daily visual inspections. Any corals that reached mortality prior to the end of the experiment were removed from their aquaria immediately.
2.3 Pulse-amplitude modulated fluorometry
Pulse-amplitude modulated (PAM) fluorometry measures the capacity of the coral holobiont to photosynthesize and can detect photochemical stress in corals (Ralph et al., 2016). PAM measurements were taken on the day prior to the onset of experimental treatments and each day for the duration of the experiment using the red light Walz Diving PAM. Because we measured maximum quantum efficiency of PSII (Fv/Fm), measurements were taken at 0600 h, prior to the end of the 12 hour dark period, when symbiont photosystems are inactivated by the absence of light. Each coral fragment was measured at a standardized position on its central branch using the Walz dark acclimation probe attachment to maintain a perpendicular angle and 0.5 cm distance from the coral surface.
2.4 Sample preservation and processing
Surface microbial samples were collected at the end of the experiment, prior to preserving coral fragments for tissue analysis. Microbial samples were only collected from corals that were still living at the end of the experiment. Because of this, the sample size from the most severe DO treatment (0.5 mg L-1) was 12 replicates, 20 for corals in the 2mg L-1 treatment, and 21 in both the 4 mg L-1 and 6 mg L-1 treatments. To collect a surface sample, several polyps were agitated with a 50 mL sterile plastic syringe tip to initiate mucus production. Mucus and aspirated tissue were drawn into the syringe, with care taken to collect minimal seawater. The sample was then transferred to a microcentrifuge tube and centrifuged at 15,000 x g for ten minutes before decanting seawater. Concentrated mucus/tissue samples of ~200-500 μL were preserved in 1 mL of RNAlater and stored in a -80°C freezer for downstream analyses.
Following microbial sampling, coral fragments were frozen at -20°C for tissue preservation. Tissue was stripped from the coral skeleton using an air compressor and airbrush (AS-196) filled with 0.2 μm filtered seawater. The resulting tissue slurry was homogenized (Tissue-Tearor, Biospec) and stored at -20°C. Remaining bare coral skeleton was oven-dried at 60°C and surface area was approximated using the wax-dipping method (Veal et al., 2010).
To further quantify photophysiological responses to deoxygenation, Symbiodiniaceae densities were quantified microscopically. Tissue slurry aliquots were diluted with filtered seawater, and six 1 µL replicates of each sample were loaded onto a hemocytometer for manual counting under a compound microscope. Symbiodiniaceae were identified visually based on their size, shape, and color. The Symbiodiniaceae cell density of each sample was then corrected by dilution factor and skeletal surface area. Cell densities were normalized to live tissue area at the time of sampling; therefore, this response represents density of algal endosymbionts within the surface area of living tissue.
2.5 16S rRNA sequencing
The coral microbiome was characterized using procedures based on the Earth Microbiome Project pipeline for 16S rRNA genes (Gilbert et al., 2018). First, DNA was extracted from mucus/tissue samples using the Qiagen DNeasy PowerSoil kit. The V4 region of the 16S rRNA gene was amplified with barcoded primers 515F (Parada et al., 2016) and 806R (Apprill et al., 2015) using Polymerase Chain Reaction (PCR). Samples were prepared in triplicate 25 µl reactions for each sample using Phusion High-fidelity Master Mix (New England Biolabs, Ipswich, MA), 0.25 μM of each primer, 3% dimethyl sulfoxide, and 2 μl of DNA template. Negative controls were included for each unique forward primer, the purity of which was verified using gel electrophoresis. Samples were cleaned and concentrated using the Qiagen MinElute PCR Purification Kit. The amount of DNA in resulting PCR products was quantified using a Denovix DS-11 FX+ spectrophotometer, and 240 ng of each amplicon library was pooled before submitting to the Interdisciplinary Center for Biotechnology Research (RRID : SCR_019152) at the University of Florida. Sequencing was conducted using the Illumina MiSeq system with a 2x150bp v.2 cycle format.
2.6 Statistical analyses
Bayesian multilevel models were used for all univariate statistical tests in this study. Bayesian methods were chosen for their ability to incorporate prior information and uncertainty that aligned with the hierarchical nature of our data. Variable selection was conducted using leave-one-out (loo) cross validation, Watanabe-Akaike information criterion (WAIC) scores, and the model_weights function in R package brms (Bürkner, 2017). Predictors that improved model fit, loo scores, and WAIC were included in models, even if their credible intervals overlapped with zero. DO treatment was included as a categorical variable because preliminary data exploration revealed seemingly nonlinear physiological responses to DO. Genotype, experimental aquarium, and individual sample (when appropriate) were included as random effects to control for variation associated with these factors. Because Symbiodiniaceae enumeration and microbiome characterization required destructive sampling at the end of the experiment, there is only one timepoint in the analysis of these variables, but all the other responses were measured daily throughout the experiment. Model convergence and fit were assessed using Rhat values, effective sample sizes, and posterior predictive checks. Effect sizes and credible intervals were used to compare the magnitude and direction of each level of each predictor variable on a given response.
For 16S rRNA amplicon analyses, paired reads were first demultiplexed by the sequencing center. Illumina adapters and 16S primer sequences were removed using the program cutadapt (Martin, 2011). The DADA2 pipeline was used to visually assess read quality, remove low-quality reads, denoise, merge paired reads, and remove chimeras (Callahan et al., 2016). Taxonomy was assigned using the SILVA 132 database (Quast et al., 2013). Using the R package phyloseq (McMurdie and Holmes, 2013), samples were filtered to exclude chloroplasts and mitochondria, singletons, doubletons, and any taxa comprising less than 0.001% of sequencing reads. Centered log-ratio transformation was used to account for the compositional nature of microbiome data and to account for uneven numbers of reads across samples (Gloor et al., 2017). The Aitchison distance matrix was created using the package coda.base in R for multivariate analyses (Aitchison et al., 2000). Alpha diversity was assessed using the Shannon diversity index because it incorporates both richness and evenness. Differences in alpha diversity were tested across samples using linear models. Beta diversity of microbial taxa across samples was assessed with a PERMANOVA analysis using the R package vegan and by analyzing dispersion of beta diversity. Dispersion of beta diversity was tested by calculating distances to the centroid in multivariate space and using linear models to test for significant differences in dispersion across treatments. The core microbiome was assessed using the package microbiome in R and was defined as species prevalent in 90% of all samples at a detection level of 0.001% within each sample (Lahti and Shetty, 2019). After assessing differences in overall community composition with analyses of alpha diversity, beta diversity, and the core microbiome, differential abundance of families was tested using an analysis of composition of microbiomes (ANCOM) analysis (Mandal et al., 2015). All statistical analyses for this study were performed in R version 4.2.2. Raw sequences are available in NCBI Sequence Read Archives SRR26222001 - SRR26222075 under BioProject # PRJNA1019720. All other data and code are available in the publicly available GitHub repository: https://github.com/saraswaminathan/acropora_deoxygenation_microbiome.git.
3 Results
3.1 Environmental conditions at field sites
Outplanted fragments of Acropora cervicornis performed differently at the two restoration sites. In October 2019, when fragments were collected prior to the laboratory experiment, 12.0% of outplants at the inshore site had died, compared to 3.2% offshore. At this time, 68.8% of all corals originally outplanted at the inshore site were bleached, relative to 15.2% at the offshore site (Table 1). To assess environmental differences between the two restoration sites (Figure 1A), we compared in situ temperature (Figure 1B) and dissolved oxygen (Figure 1C) at each site during the field deployment. Temperatures were significantly warmer at the inshore site during the field deployment period (Figure 1B), with a credible interval of [0.35, 0.36]. The best model of temperature, which included site and time, had an R2 of 0.91. Temperatures of 31°C and above are generally considered stressful for corals and are commonly associated with stress responses such as bleaching (Fitt et al., 2001). Over the duration of the summer, the corals at the inshore site experienced 50 days (44.2% of the field deployment period) in which the mean daily temperature exceeded 31°C. In contrast, the mean temperature at the offshore site exceeded 31°C on 14 days (12.4% of field deployment period). From September to October 2019, temperatures began a seasonal decline at both sites, and began to overlap. Although the inshore site was consistently warmer, diel temperature fluctuation was greater on average at the inshore site, with a mean diel temperature range of 0.61°C ± 0.26°C compared to 0.55°C ± 0.29°C offshore.
Dissolved oxygen (DO) was slightly, but significantly, lower at the at the inshore site (µ = 5.99 ± 0.04) mg L-1) relative to the offshore site (µ = 6.14 ± 0.03 mg L-1) throughout the field deployment, with a 95% credible interval of [-0.15, -0.14] (Figure 1C). The best DO model, which included site and time, had an R2 of 0.23. Although DO did not reach levels known to be stressful to corals at either site, DO at the inshore site dropped below 4 mg L-1 on rare occasions (18 total observations throughout the summer deployment, with the longest consecutive period of <4 mg L-1 DO lasting only one hour), whereas the offshore site never dropped below 4 mg L-1 during the 2019 field deployment.
Salinity, total dissolved solids (TDS), and pH were also monitored at both sites (Supplementary Figure S1). On average, pH was lower inshore (μ = 8.15) compared to offshore (μ = 8.21) (Supplementary Figure S1). TDS was also elevated inshore (μ = 35848.28 mg L-1) compared to offshore (μ = 35700.94 mg L-1) (Supplementary Figure S1B). Salinity appeared correlated with TDS, with marginally higher salinity inshore (μ = 36.38 ppt) compared to offshore (μ = 36.22 ppt) (Supplementary Figure S1C). These environmental parameters demonstrate that in this study, “site” represents a regime of environmental variables that differ between inshore and offshore reefs in the Florida Keys.
3.2 Laboratory responses to deoxygenation and site history
3.2.1 Photophysiology
Coral photosynthetic efficiency was negatively impacted by all deoxygenation treatments in the laboratory experiment (Figures 2A, B and Table 2). Dark-adapted, maximum quantum yield (Fv/Fm) was best explained by DO treatment and “Day” of the experiment, with “individual” included as a random effect. The 4.0 mg L-1, 2.0 mg L-1, and 0.5 mg L-1 DO treatments all had statistically significant, negative influences on Fv/Fm. The 0.5 mg L-1 DO treatment had the largest negative effect size, with a 95% credible interval of [-0.19, -0.13], followed by the 2.0 mg L-1 and 4.0 mg L-1 DO treatments, which had similar 95% credible intervals of [-0.07, -0.01] and [-0.06, -0.01], respectively (Table 2 and Figure 2B). Exposure time, represented by “Day”, had a statistically significant, but nonlinear, influence on Fv/Fm. Fv/Fm increased significantly on the first day of the experiment, was lowest on Day 2, and was also significantly lower than the pre-experiment values on Days 3 and 4 of the experiment (Table 2 and Figure 2B). Site, and the interaction of DO treatment and site, increased the WAIC of the model, indicating they were not significant predictors of Fv/Fm, and were therefore not included in the final model.
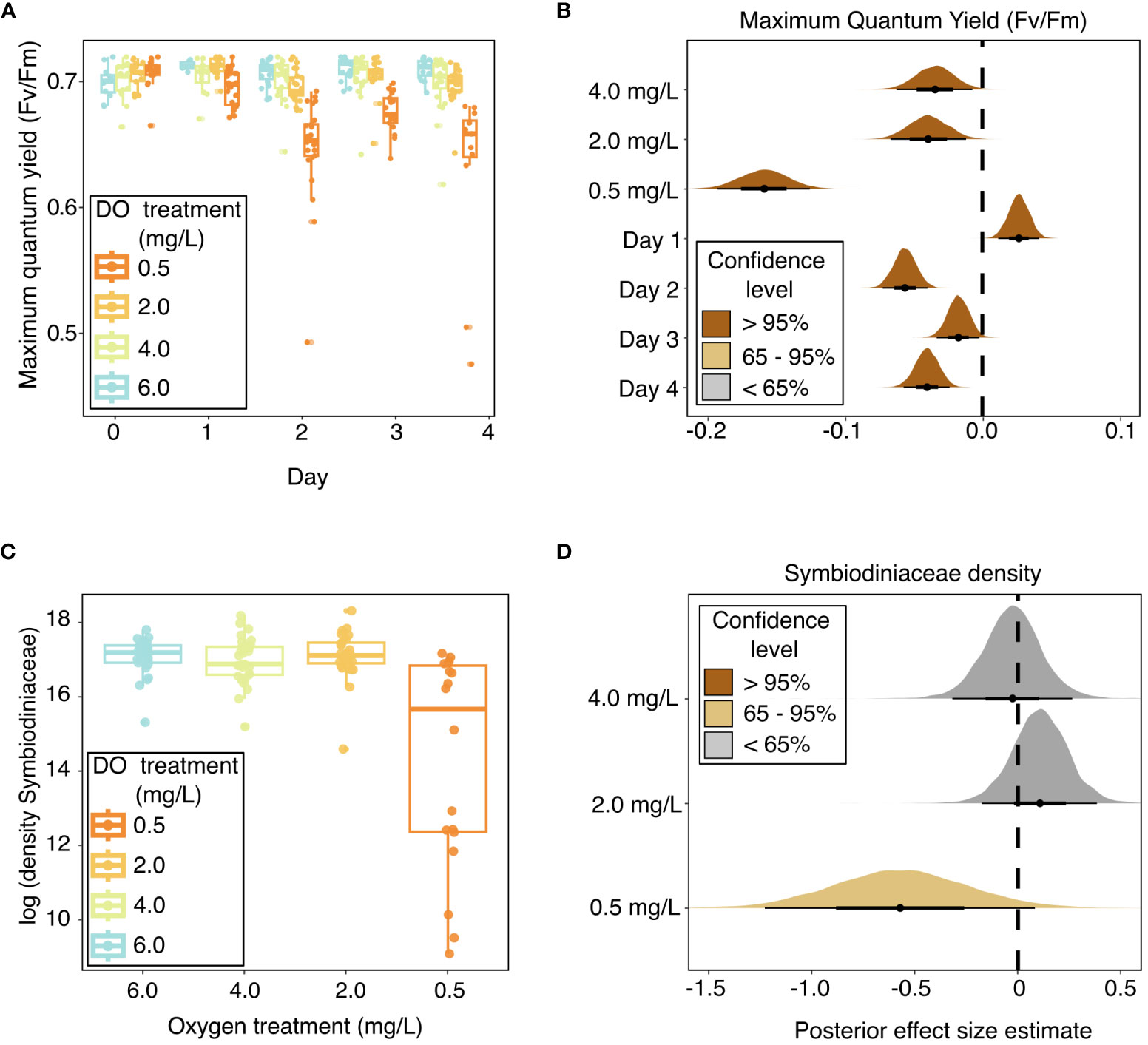
Figure 2 Coral photophysiology throughout experimental exposure to dissolved oxygen treatments in the laboratory experiment, as indicated by (A) box and whisker plots of maximum quantum yield over the experiment duration, with raw values shown in points; (B) stat-eye plots showing posterior effect size estimates of all population-level predictors included in the best model of maximum quantum yield; (C) box and whisker plots of log- transformed Symbiodiniaceae cell densities across oxygen treatments; and (D) stat-eye plots showing posterior effect size estimates of all population-level predictors included in the best model of Symbiodiniaceae cell densities. The lines on the box plots represent the median and first and third quartiles, with the whiskers extending to the smallest and largest values within 1.5 times the inter-quartile range (IQR). The points within the stat-eye plots show the median effect size. The thin and thick lines within each stat-eye plot represent the 95% and 65% credible intervals, respectively. The colors of the stat-eye plots in panels (B, D) show which part of the distribution overlaps with 0: Dark brown shows distributions where neither the 95% nor the 65% credible interval overlaps with 0; light brown represents distributions where the 65% credible interval does not overlap with 0, but the 95% credible interval does; and grey represents distributions where both the 65% and 95% credible intervals overlap with 0. Site is not shown in these plots because it was not found to be a significant predictor of maximum quantum yield or Symbiodiniaceae cell densities.
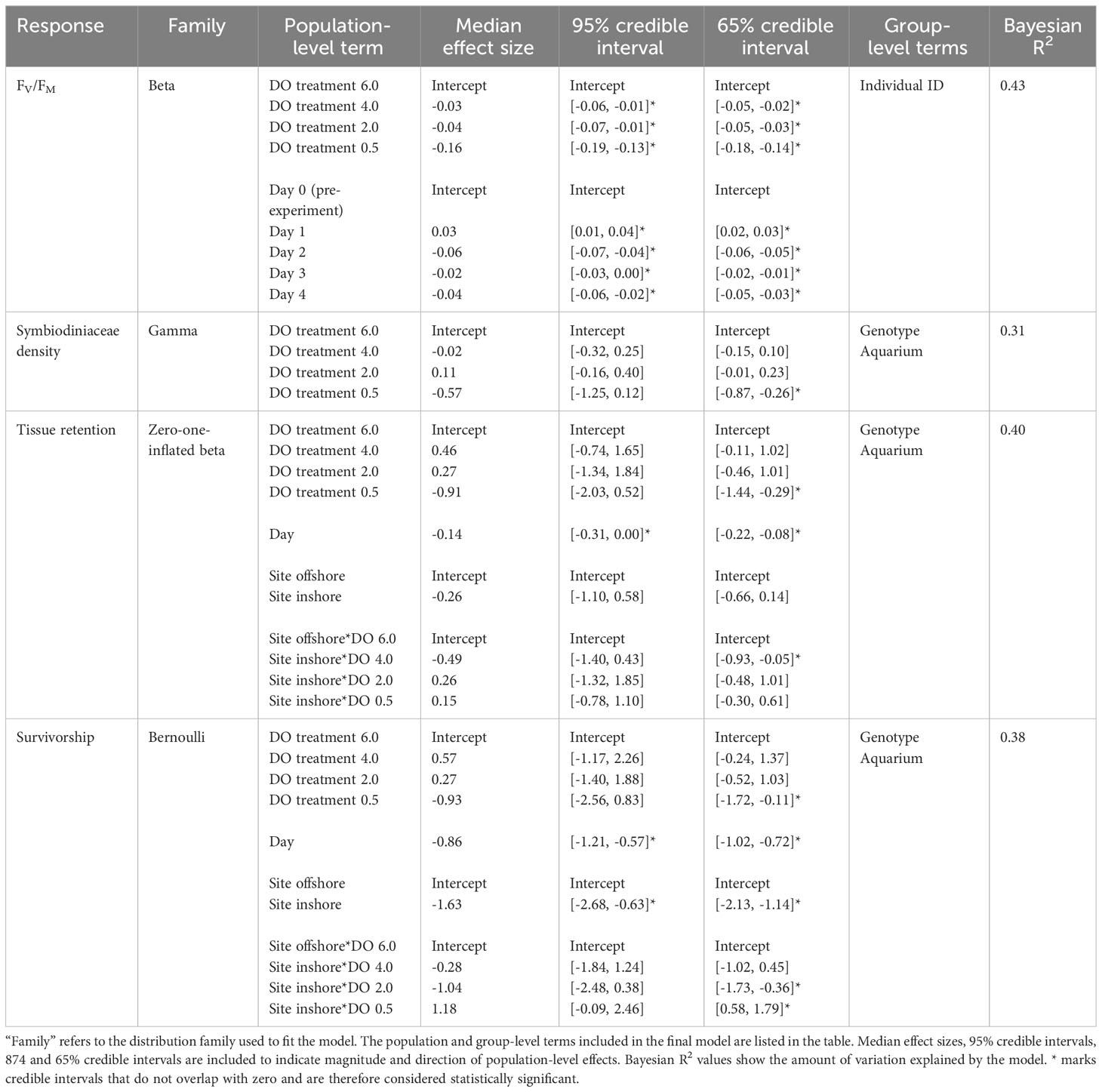
Table 2 Results of the Bayesian multilevel models of maximum quantum yield (FV/Fm), Symbiodiniaceae density, tissue retention, and survivorship.
Symbiodiniaceae cell density was negatively impacted by severe (0.5 mg L-1) deoxygenation in the laboratory experiment but did not respond to either the 2.0 mg L-1 or the 4.0 mg L-1 DO treatment (Figures 2C, D and Table 2). The best model of Symbiodiniaceae cell density included DO treatment, with genotype and aquarium included as random effects. The only DO treatment with a statistically significant influence on Symbiodiniaceae cell density was the 0.5 mg L-1 DO treatment, with a 65% credible interval of [-0.87, -0.26] (Table 2 and Figure 2D). Site, and the interaction of DO treatment and site, increased the WAIC of the model, indicating they were not significant predictors of Symbiodiniaceae cell density, and were therefore not included in the final model.
3.2.2 Coral host survivorship and tissue retention
Coral host tissue retention (percentage of live tissue remaining) was negatively influenced by severe deoxygenation (0.5 mg L-1) and exposure time (represented by “Day” of the experiment) (Figures 3A, B and Table 2). The best model of tissue retention included DO treatment, “Day” of the experiment, site, and the interaction of DO and site, with genotype and aquarium included as random effects. Tissue retention was significantly lowered in the most severe (0.5 mg L-1) DO treatment, with a 65% credible interval of [-1.44, -0.29], but was not significantly different from the control group in the 2.0 mg L-1 or 4.0 mg L-1 DO treatments (Table 2 and Figure 3B). While site alone did not significantly influence tissue retention (Figure 3B), there was a significant interaction between DO treatment and site that was detectable in the 4.0 mg L-1 DO treatment, suggesting that a history of exposure to inshore conditions worsened the effects of deoxygenation on tissue retention in this treatment (65% credible interval of [-0.93, -0.05]) (Figure 3B).
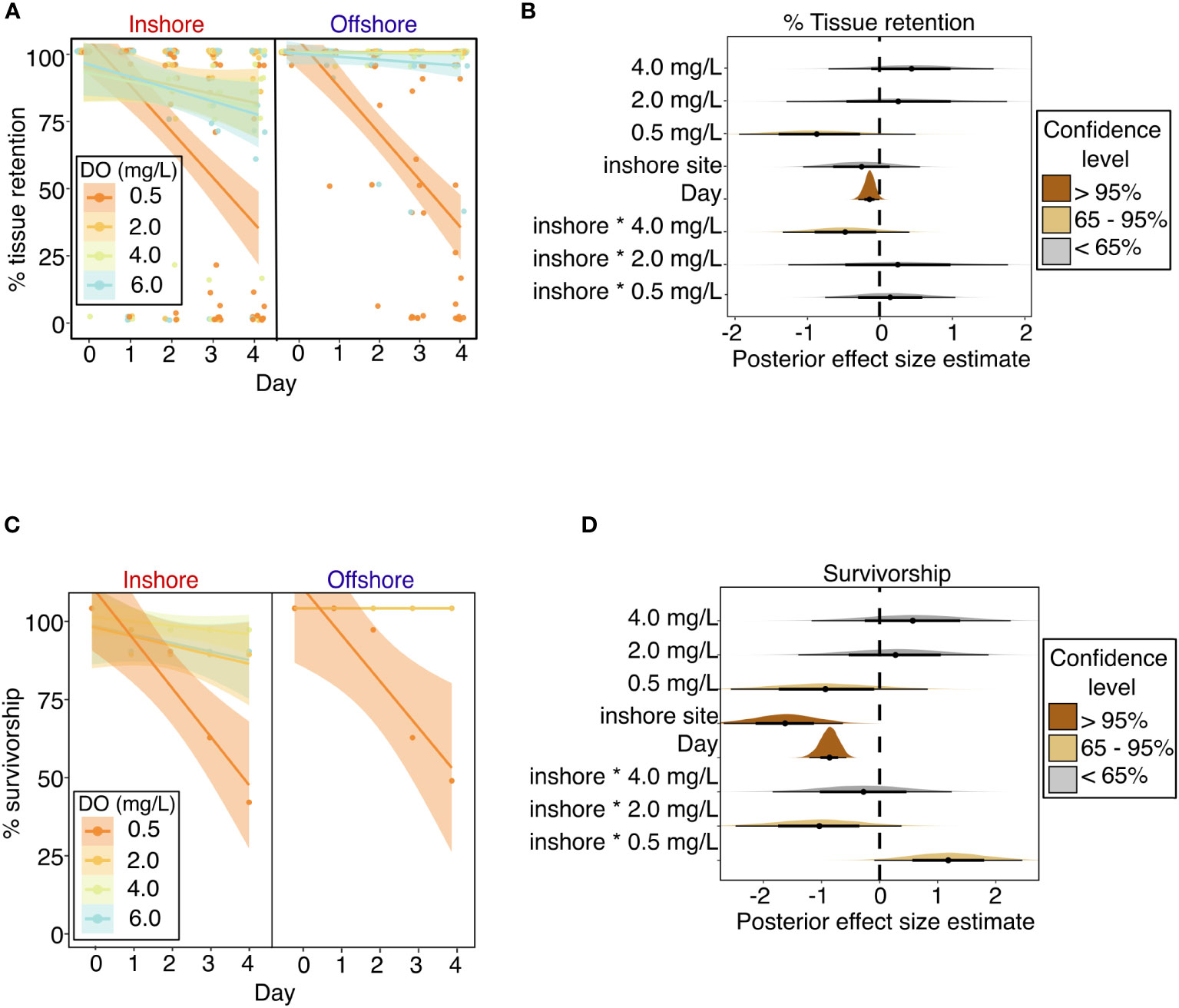
Figure 3 Coral host performance throughout experimental exposure to dissolved oxygen treatments in the laboratory experiment, as indicated by (A) Percentage of remaining tissue on individual coral colonies over the experiment duration, with points showing percentage values for individual coral colonies and lines showing best fit linear regressions for each treatment with shading showing 95% confidence intervals; (B) stat-eye plots showing posterior effect size estimates of all population-level predictors included in the best model of tissue retention; (C) Percentage of surviving coral colonies over the experiment duration, with points showing % survivorship on each day, for each treatment, among corals from each site, and lines showing best fit linear regressions for each treatment with shading showing 95% confidence intervals; and (D) stat-eye plots showing posterior effect size estimates of all population-level predictors included in the best model of coral survivorship. The thin and thick lines within each stat-eye plot represent the 95% and 65% credible intervals, respectively.
Coral survivorship was negatively influenced by severe deoxygenation (0.5 mg L-1), exposure time, and prior exposure to the inshore site (Figures 3C, D and Table 2). The best model for coral survivorship included DO treatment, “Day” of the experiment, site, and the interaction of DO and site, with genotype and aquarium included as random effects. Survivorship was significantly lowered in the most severe (0.5 mg L-1) DO treatment, with a 65% credible interval of [-1.72, -0.11], but was not significantly different from the control group in the 2.0 mg L-1 or 4.0 mg L-1 DO treatments (Table 2 and Figure 3D). Corals from the inshore site had significantly lower survivorship than those from the offshore site, with a 95% credible interval of [-2.68, -0.63] (Figure 3D). There was a significant interaction between site and DO treatment, as exposure to the inshore site seemed to exacerbate negative effects of deoxygenation in the 2.0 mg L-1 DO treatment, but seemed to have a positive effect in the 0.5 mg L-1 DO treatment (Table 2 and Figure 3D).
3.2.3 Surface microbiome composition
High-throughput sequencing of the 74 coral surface microbiomes characterized in this study resulted in a total of 6.36 million sequencing reads. Quality-filtering and merging resulted in a total of 3.95 million sequencing reads, with an average of 53,382 (2,730 – 203,826) sequencing reads per sample. A total of 5,461 Amplicon Sequence Variants (ASVs) were observed across all samples.
Analyses of microbiome diversity and composition revealed overall resilience to deoxygenation and site history (Figure 4). Alpha diversity, described by the Shannon index, was statistically similar across DO treatments (Figure 4A), with a 95% credible interval of [-0.05, 0.05] (Table 3). The best model of Shannon index included aquarium as a random effect and DO treatment but did not include site. The best model of the dispersion of beta diversity included DO and site, with aquarium and genotype as random effects. However, there were no statistically significant differences in dispersion of beta diversity across DO treatments (95% credible interval [-1.94, 1.11]) or site (95% credible interval [-1.87, 2.01] (Table 3). Principal Component Analysis (PCA) plots showed minimal clustering across DO treatments, suggesting DO treatment did not structure community composition (Figure 4B). PERMANOVA analyses revealed no statistically significant differences between corals in the same DO treatments or sites (Table 4).
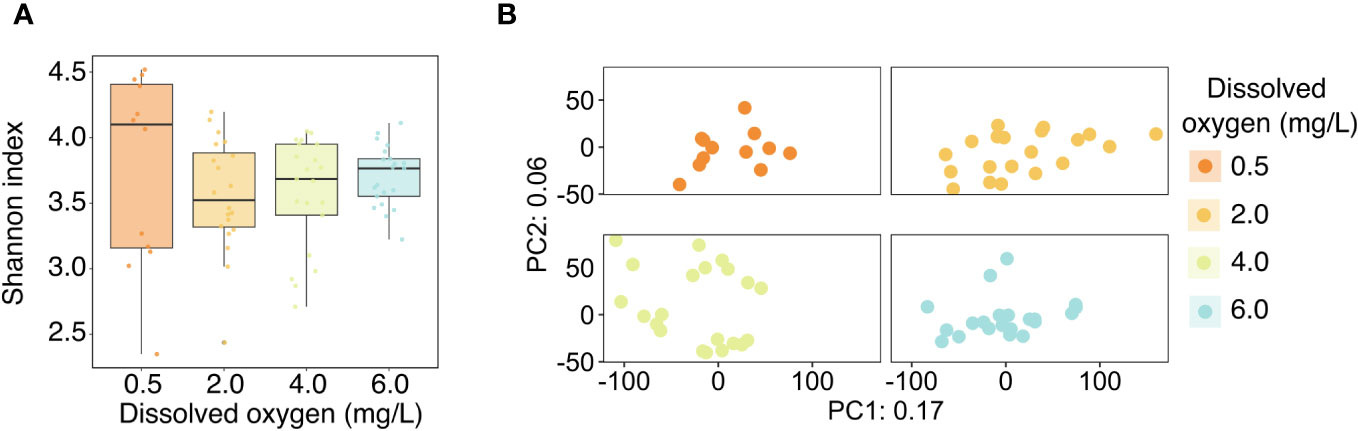
Figure 4 (A) Alpha diversity of the Acropora cervicornis microbiome across the four experimental dissolved oxygen treatments, as represented by the Shannon index. Points represent the Shannon index of individual coral samples, and boxplots show medians and inter-quartile ranges. (B) PCA ordination of the Aitchison distance between microbial communities. The ordination is faceted by dissolved oxygen treatment for improved resolution of points.
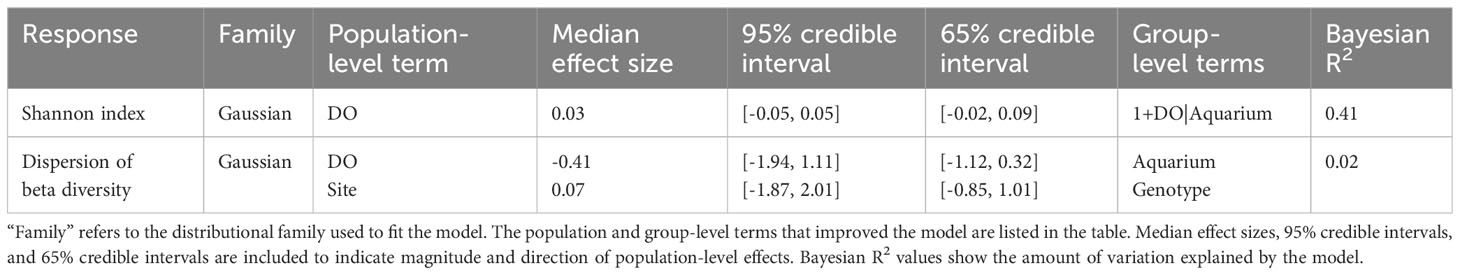
Table 3 Results of the Bayesian linear models of Shannon index and dispersion of beta diversity of coral microbiomes characterized in this study.
A subset of microbial taxa comprised a “core microbiome” across all the corals in the experiment (Figure 5). This suggests that regardless of experimental treatments, some bacteria are ubiquitous in the A. cervicornis surface microbiome. There were ten ASVs found in at least 90% of samples (at a detection level of 0.001%) from the following eight taxonomic groups: Oceanospirillum, Aestuariibacter, Rhodobacteraceae, Alteromonas, Saprospiraceae, Oleibacter, MD3-55 (also known as Candidatus Aquarickettsia rohweri (Klinges et al., 2019)), and Alteromonadaceae. The heatmap in Figure 5 shows that although the ten core ASVs are present in all samples, they occur in different relative abundances across DO treatments. For example, ASV 4 from the genus Alteromonas is present in all samples but comprised a greater fraction of the coral microbiome in the 0.5 mg L-1 DO treatment (μ = 0.094 ± 0.024) compared to the 2.0 mg L-1 (μ = 0.021 ± 0.004), 4.0 mg L-1 (μ = 0.028 ± 0.004), and 6.0 mg L-1 (μ = 0.016 ± 0.003) DO treatments. Alternatively, Candidatus Aquarickettsia rohweri appears in lowest concentrations in the 0.5 mg L-1 DO treatment.
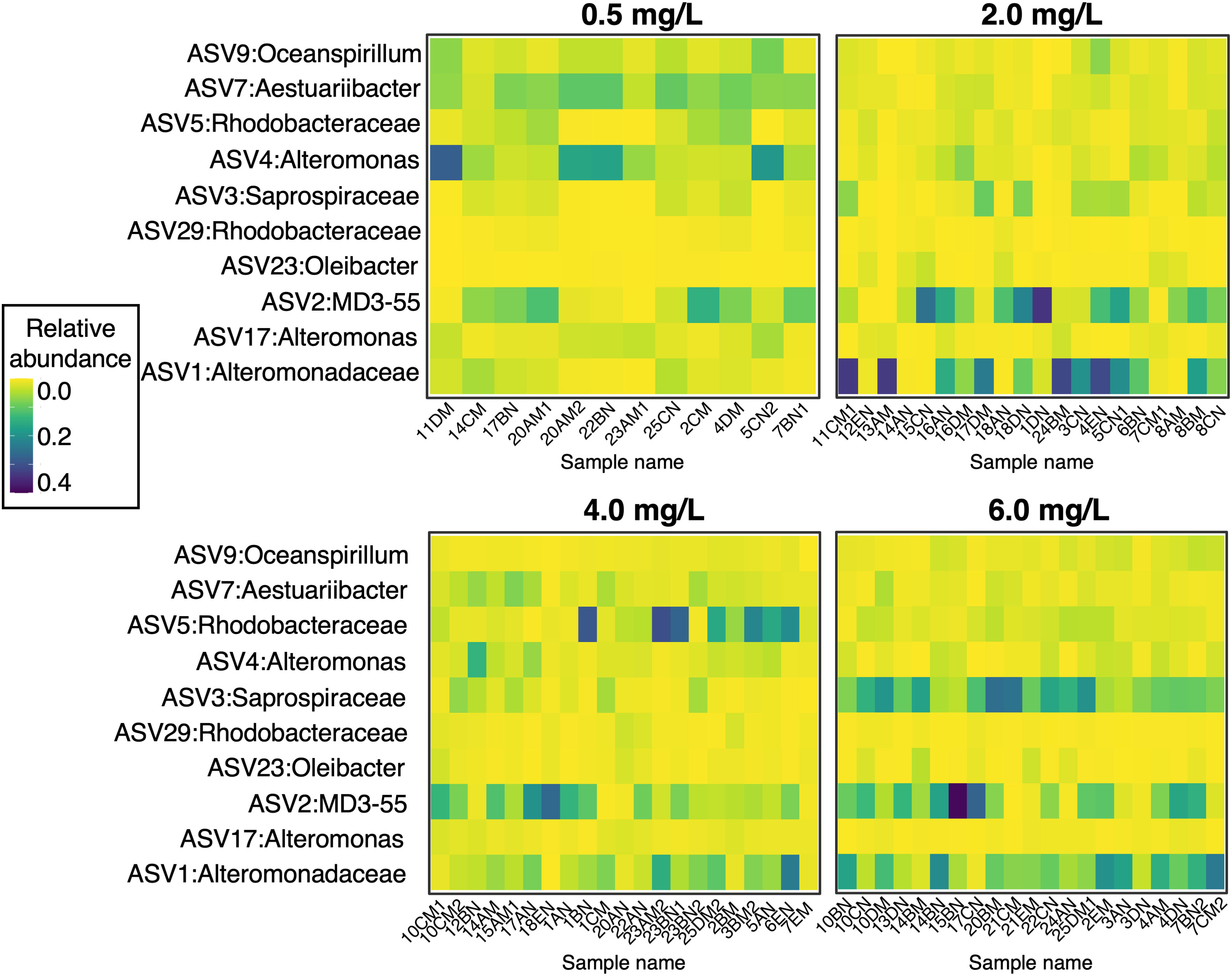
Figure 5 Relative abundance (ASV abundance normalized to total number of reads in a sample) of the ten core microbiome ASVs across corals in the four oxygen treatments. At a detection level of 0.001% relative abundance, these taxa were present in 90% of samples.
Although community structure was not correlated with experimental treatments, some microbial families were differentially abundant across oxygen treatments. Using the ANCOM function (Mandal et al., 2015), we identified 22 differentially abundant families (Supplementary Figure S3), ten of which had a rank order abundance correlated with dissolved oxygen treatments (Figure 6), whereas the other 12 families had their highest or lowest abundance at some intermediate level of dissolved oxygen (Supplementary Figure S3). Of the taxa with relative abundances that correlated with DO treatments, families PB19, an unclassified family of Caenarcaniphilales, Beggiatoaceae, Shewanellaceae, Colwelliaceae, Arcobacteraceae, Nitrincolaceae, and Holosporaceae had greater relative abundances in samples exposed to deoxygenation (Figure 6). Conversely, families Flavobacteriaceae and Rhodospirillaceae had greater relative abundances with higher DO levels.
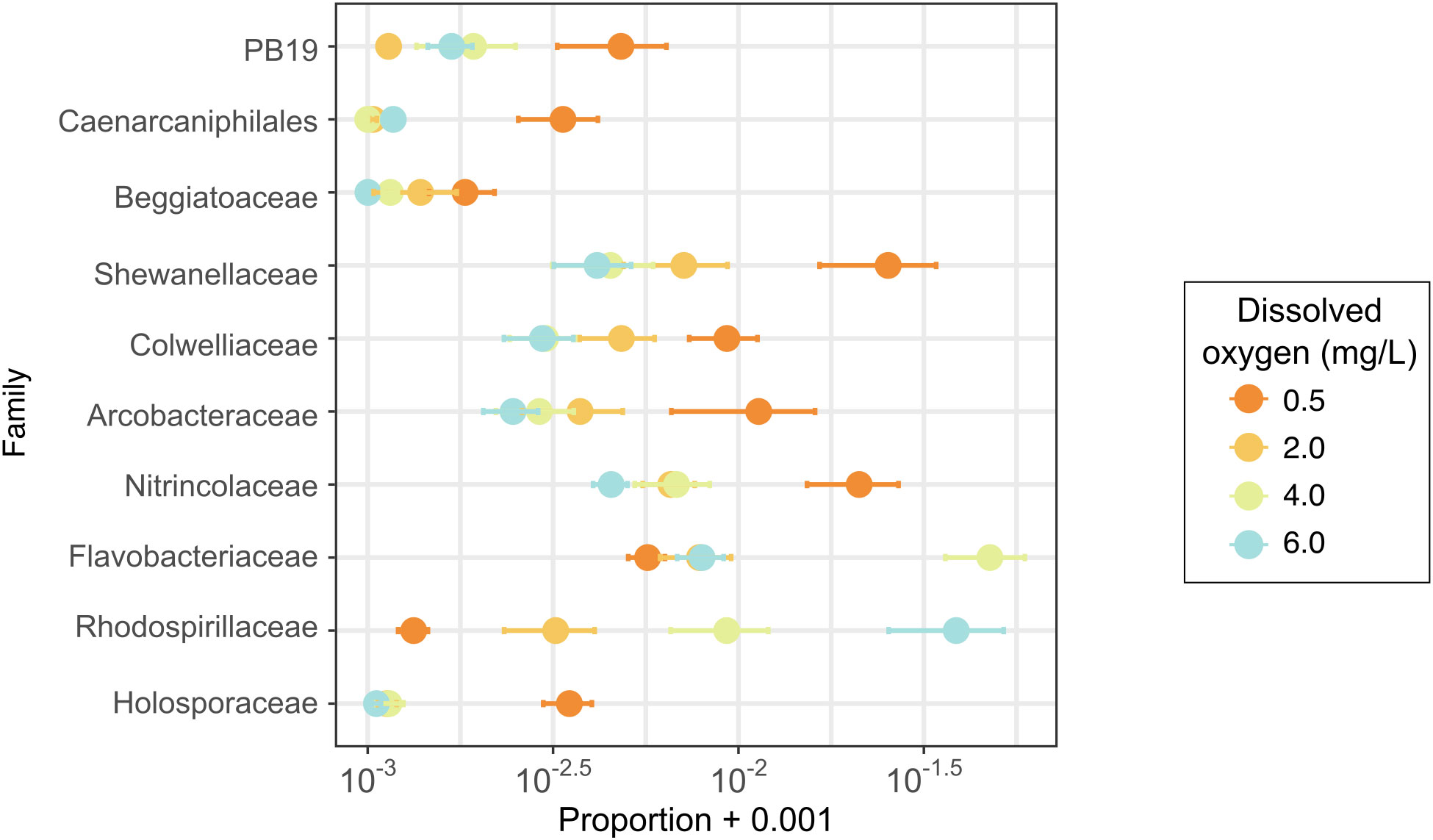
Figure 6 Proportions of bacterial families identified as differentially abundant with an ANCOM (Analysis of Composition of Microbiomes) analysis. This figure shows only families with differential abundance occurring along the oxygen treatment gradient.
4 Discussion
Our study revealed divergent responses among the coral host and its symbionts to deoxygenation and environmental history. The coral host and Symbiodiniaceae had a strong response to the lowest level of deoxygenation tested, 0.5 mg L-1. Although 2.0 and 4.0 mg L-1 dissolved oxygen (DO) may eventually harm this coral species, the duration of exposure tested in this study (four days) was not enough to cause significant damage to the coral host in these DO treatments. We found no evidence that site history influenced photophysiology, but there was higher mortality in corals from the inshore site, and there was an interactive effect of site and DO on both coral survivorship and tissue retention. Overall diversity and community composition metrics of the coral surface microbiome indicated resistance to both deoxygenation and prior site history, however, some potentially important bacterial taxa responded through differential abundances. General resilience of the surface microbiome during hypoxic stress may show it can persist independently of the coral host’s health with the potential to continue functions that may contribute to host stress resistance. However, further investigation is required to elucidate the role of the microbiome in coral tolerance to deoxygenation.
We found clear photophysiological responses of Acropora cervivornis to severe deoxygenation. This finding is consistent with another study of deoxygenation with Acropora cervicornis (Johnson et al., 2021b), which found greater physiological sensitivity to DO in this species compared to another coral species, Orbicella faveolata. Although our study ran for only four days, the dark-adapted, maximum quantum yield values in our study were similar to Haas et al. (2014), which found recovery in Fv/Fm in all but their most severe (2-4 mg L-1) DO treatments after ten days. Due to the dependence of coral growth and reproduction on photobiology health (Goreau and Macfarlane, 1990; Muscatine et al., 1998; Roth, 2014), damage to, or reduction in, the algal symbionts due to low DO may lead to decreased growth and reproductive capacity. However, our study only supports the idea that short-term exposure to deoxygenation incurs photophysiological damage in cases of extreme deoxygenation, as would occur in a hypoxic event. Although DO-linked photobiological reduction represents a threat to the ecological functioning of tropical corals, it appears to be a sublethal response during intermediate exposure to deoxygenation. Short-term (hours) low levels of oxygen may not be sufficient to trigger loss of Symbiodiniaceae (Deleja et al., 2022), whereas sustained exposure in our study led to decreased symbiont densities. Notably, declines in Symbiodiniaceae densities were observed only in the most extreme deoxygenation treatment nearing anoxia (0.5 mg L-1), indicating resistance of the symbiosis to moderate levels of hypoxia (2 and 4 mg L-1) over the duration of our experiment.
In the coral host, extreme deoxygenation led to tissue loss and, in some cases, mortality after four days of exposure. Acute tissue loss is a serious consequence and can be difficult to overcome due to competitive pressures including algal overgrowth of coral skeletal tissue or can lead to secondary mortality of coral tissue due to disease or predators (Bender et al., 2012). Differential mortality across species or genotypes, in some contexts, can represent the first step in selection toward genotypes or species that are more tolerant of environmental stressors in a region (Bell, 2013). Because of their sensitivity to severe hypoxia, and because deoxygenation events on coral reefs can last longer than four days (Johnson et al., 2021a), A. cervicornis populations are likely to decline in areas that experience acute hypoxia events.
Variation in surface microbiome composition was not significantly associated with deoxygenation or site history. This finding is provisional due to two main features of this study. First, because corals were sampled at the end of the experiment, the microbiomes of corals with the most severe responses to hypoxia (those with >95% tissue loss) were not sampled. Therefore, the results represent those with sublethal physiological responses and may be a conservative estimate of microbial responses to hypoxia. Furthermore, our microbiome sampling captured primarily coral mucus, with a small amount of aspirated tissue. The mucus layer is known to be more reflective of the abiotic environment and less subject to host differences than the tissue and skeletal microbes (Ritchie, 2006; Pollock et al., 2018). Because of this, we expected surface samples to be especially sensitive to deoxygenation. However, alpha and beta diversity did not vary as a function of the experimental oxygen treatments. This contrasts with many other coral microbiome studies that found increased alpha and beta diversity in response to environmental stress (Zaneveld et al., 2017; Gardner et al., 2019; McDevitt-Irwin et al., 2019). Rather than contradicting the diversity-stress relationship commonly seen in coral microbiomes, our finding suggests that for the duration tested in our experiment, low-DO exposure, while stressful for the cnidarian host, did not represent a driver of change in surface microbiome community composition.
Because coral microbial communities were resistant to changes in dissolved oxygen, they may have the potential to provide the holobiont with some resistance to stress. Moreover, the coral microbiome may be well-adapted to the ‘front lines’ of deoxygenation stress due to the hypoxic conditions that are commonly encountered in the diffusive boundary layer of the organism due to their nocturnal respiration (Shashar et al., 1993). This microbiome resistance may be an important mechanism supporting the apparent resilience of tropical marine systems to deoxygenation (Altieri et al., 2021).
We found ten microbial taxa present across all coral samples, which represent a “core microbiome”, or taxa that are prevalent regardless of environmental context (Ainsworth et al., 2015). Characterizing the core microbiome can help identify symbiotic taxa across species or environmental conditions (Chu and Vollmer, 2016). One of the core taxa in this study, MD3-55 (Candidatus Aquarickettsia rohweri) has been identified as an obligate parasite of A. cervicornis in Florida and the broader Caribbean (Gignoux-Wolfsohn and Vollmer, 2015; Godoy-Vitorino et al., 2017; Klinges et al., 2020; Miller et al., 2020; Aguirre et al., 2022). We detected Ca. A. rohweri in highest relative abundances in the control DO treatment (6 mg L-1), suggesting that, as with increasingly stressful temperatures (Klinges et al., 2020), deoxygenation may suppress Ca. A. rohweri. Because the role of Ca. A. rohweri is not fully understood, the implications of its decreased prevalence for its host remain unknown. Another core taxa, Alteromonas ASV 4, was found in highest abundance in the hypoxic DO treatment (0.5 mg L-1). Members of the genus Alteromonas have been identified in the core microbiome of other coral species (Hernandez-Agreda et al., 2016). Although Alteromonas species have been associated with disease states (Sweet et al., 2013), bleaching, and tissue loss in Acropora spp. (Brown et al., 2013), this may be due to their beneficial functions in times of stress. Alteromonas species have been shown to metabolize dimethylsulfide (Raina et al., 2009), which is produced by Symbiodiniaceae and associated with coral stress (Deschaseaux et al., 2014). Because an Alteromonas ASV was enriched in the hypoxic DO treatment, this may indicate that, although universally present in the core microbiome, this taxon may become enriched as a response to oxidative stress in the coral host.
Our differential abundance analysis revealed microbial families that co-varied with deoxygenation. Some of the differentially abundant families have been associated with deoxygenation in other studies. Nitrincolaceae, which became enriched with deoxygenation in our study, has been found to proliferate in the water column above hypoxic coral reefs (Johnson et al., 2021a) as well as the coral mucus itself in other coral species (Howard et al., 2023). Nitrincolaceae, which contains genes for nitrite reduction (Mori et al., 2019), has been associated with increases in phytoplankton-derived exudates (Valdés-Castro et al., 2022), which is a common precursor to deoxygenation. We also observed enrichment of Holosporaceae, which are known to be endosymbiotic parasites of Paramecium, a prokaryotic ciliate, in the lowest DO treatment (Santos and Massard, 2014; Castelli et al., 2022). The presence of Holosporaceae may indicate ciliate proliferation in this treatment. Ciliates can proliferate during hypoxic events in marine environments (Stauffer et al., 2013), have been associated with coral diseases (Sweet and Bythell, 2012), and are known to control bacterial communities (Vargas and Hattori, 1990). Further investigation into these and other differentially abundant taxa is warranted given their potential to modulate coral resistance to deoxygenation.
In the present study, we tested whether prior environmental stress conveyed tolerance or susceptibility to deoxygenation. Corals have demonstrated “environmental memory,” wherein prior exposure to a stressor improves tolerance to that stressor later (Boyd et al., 2016). Trans-priming, or stress memory across multiple stressors, has only been examined in a handful of coral studies, such as a reciprocal transplant study where prior exposure to high irradiance improved tolerance to subsequent thermal stress (Brown et al., 2015). Prior exposure to deoxygenation was recently shown to lower thermal thresholds in corals (Alderdice et al., 2022). Likewise, it is possible that corals from different environmental regimes could have different DO thresholds. Rather than priming the corals for hypoxia, we found that prior environmental stress weakened the coral and its symbionts’ tolerance to deoxygenation, with lower tissue retention and survivorship in corals from the more stressful inshore site when exposed to hypoxia in laboratory aquaria.
Our study showed varied interactions between site and DO treatment, suggesting prior environmental stress can modulate the effects of deoxygenation differently based on the intensity of subsequent deoxygenation. Prior exposure to the inshore site appeared to worsen the effects of the 4.0 mg L-1 treatment on tissue retention (Figure 3B) and the 2.0 mg L-1 treatment on survivorship (Figure 3D). However, in the most severe, 0.5 mg L-1 deoxygenation treatment, prior exposure to inshore conditions appeared to mitigate the effects of deoxygenation on survivorship (Figure 3D). This suggests that at low DO levels, prior environmental stress may worsen the effects of deoxygenation; but that there may be a point of deoxygenation at which prior exposure to environmental stress can slightly mitigate effects of deoxygenation. Further study of prolonged exposure to different levels of deoxygenation, crossed with prior environmental stress, is necessary to determine thresholds at which prior environmental stress exposure may become an asset that provides benefits through ‘stress hardening’.
While deoxygenation did not appear to be a stressor at our field sites, the field-exposure phase of this study allowed us to test how prior exposure to different environmental regimes can influence tolerance to subsequent deoxygenation. Both pH and temperature at the inshore site reached levels known to be stressful for corals (Fitt et al., 2001; Fabricius et al., 2011). Although corals’ high-temperature and low-DO tolerance mechanisms are related (Alderdice et al., 2021), it is possible that inshore conditions were too extreme to elicit a stress-hardening response. Instead, the inshore corals’ defenses appeared weakened by environmental stress, making deoxygenation more difficult to tolerate. Our study revealed a synergism between environmental history and deoxygenation on A. cervicornis, adding to the list of multiple stressor synergisms, such as those between UV and temperature (Bhagooli and Hidaka, 2004) and macroalgae and temperature (Donovan et al., 2021), documented on coral reefs. Additional multiple-stressor laboratory experiments including temperature and pH are necessary to elucidate the relationships between the environmental stressors associated with inshore environments and deoxygenation (Hughes et al., 2020).
Some coral species are more tolerant to hypoxia than A. cervicornis, with species ranging in their ability to tolerate extreme deoxygenation treatments from days to weeks (Johnson et al., 2021b; Alva García et al., 2022). Given that A. cervicornis is among the more sensitive species to hypoxia, we examined the members of its holobiont, following exposure to inshore and offshore environments to better understand the factors that limit hypoxia tolerance. This study highlights the importance of considering the coral holobiont as a complex entity in response to deoxygenation whose members may respond to multiple stressors in different ways and on different timescales. As restoration is concerned, we show that exposure to harsh inshore environments can weaken tolerance to deoxygenation in sensitive species. Because deoxygenation can go unnoticed, oxygen monitoring at potential restoration sites is critical to determine suitability for coral species that are sensitive to deoxygenation, including A. cervicornis. We suggest that additional studies comparing sensitive species, such as A. cervicornis, with more tolerant species will provide additional insights as to the role of holobiont members in explaining the variation in tolerance to deoxygenation that exists among species.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1019720; https://github.com/saraswaminathan/acropora_deoxygenation_microbiome
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
SS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing. JM: Formal analysis, Methodology, Resources, Supervision, Visualization, Writing – review & editing. MJ: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – review & editing. VP: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing. EB: Funding acquisition, Investigation, Methodology, Resources, Supervision, Writing – review & editing. AA: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by an award from the National Oceanic and Atmospheric Administration’s National Centers for Coastal Ocean Science Competitive Research Program to AA, MJ, and VP (NA18NOS4780170) through the University of Florida. SS was supported by a National Science Foundation Graduate Research Fellowship (Grant No. DGE-1842473). MJ was funded by postdoctoral fellowship awards from the Smithsonian’s MarineGEO and Tennenbaum Marine Observatories Network and the Smithsonian Marine Station at Fort Pierce.
Acknowledgments
The authors thank Emily Nixon, Tessa Vekich, Alyssa Guariniello, Woody Lee, and Blake Ushijima for laboratory support. Joe Kuehl, Todd Van Natta, and Patrick Norby were instrumental in the field component of this study. We thank Will Ferrell, Delaney Harnish, Claire Hiaasen, Hunter Kaminski, Mikee Orense, and Adrian Sakr for their help with Symbiodiniaceae enumeration. We also thank Mote Marine Laboratory for facilities and housing during the field component of this study. This is contribution 265 from the Coastal Hypoxia Research Program, 136 from the Smithsonian’s MarineGEO and Tennenbaum Marine Observatories Network, and 1210 from the Smithsonian Marine Station at Fort Pierce.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1301474/full#supplementary-material
References
Aguirre E. G., Million W. C., Bartels E., Krediet C. J., Kenkel C. D. (2022). Host-specific epibiomes of distinct Acropora cervicornis genotypes persist after field transplantation. Coral Reefs 41, 265–276. doi: 10.1007/s00338-022-02218-x
Ainsworth T. D., Krause L., Bridge T., Torda G., Raina J.-B., Zakrzewski M., et al. (2015). The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 9, 2261–2274. doi: 10.1038/ismej.2015.39
Aitchison J., Barceló-Vidal C., Martín-Fernández J. A., Pawlowsky-Glahn V. (2000). Logratio analysis and compositional distance. Math. Geology 32, 271–275. doi: 10.1023/A:1007529726302
Alderdice R., Perna G., Cárdenas A., Hume B. C. C., Wolf M., Kühl M., et al. (2022). Deoxygenation lowers the thermal threshold of coral bleaching. Sci. Rep. 12, 18273. doi: 10.1038/s41598-022-22604-3
Alderdice R., Suggett D. J., Cárdenas A., Hughes D. J., Kühl M., Pernice M., et al. (2021). Divergent expression of hypoxia response systems under deoxygenation in reef-forming corals aligns with bleaching susceptibility. Global Change Biol. 27, 312–326. doi: 10.1111/gcb.15436
Allemand D., Osborn D. (2019). Ocean acidification impacts on coral reefs: From sciences to solutions. Regional Stud. Mar. Sci. 28, 100558. doi: 10.1016/j.rsma.2019.100558
Altieri A. H., Diaz R. J. (2019). “Dead zones: Oxygen depletion in coastal ecosystems,” in World seas: an environmental evaluation (Cambridge, MA: Academic Press), 453–473.
Altieri A. H., Gedan K. B. (2015). Climate change and dead zones. Global Change Biol. 21, 1395–1406. doi: 10.1111/gcb.12754
Altieri A. H., Harrison S. B., Seemann J., Collin R., Diaz R. J., Knowlton N. (2017). Tropical dead zones and mass mortalities on coral reefs. Proc. Natl. Acad. Sci. 114, 3660–3665. doi: 10.1073/pnas.1621517114
Altieri A. H., Johnson M. D., Swaminathan S. D., Nelson H. R., Gedan K. B. (2021). Resilience of tropical ecosystems to ocean deoxygenation. Trends Ecol. Evol. 36, 227–238. doi: 10.1016/j.tree.2020.11.003
Alva García J. V., Klein S. G., Alamoudi T., Arossa S., Parry A. J., Steckbauer A., et al. (2022). Thresholds of hypoxia of two Red Sea coral species (Porites sp. and Galaxea fascicularis). Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.945293
Apprill A., McNally S., Parsons R., Weber L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microbial Ecol. 75, 129–137. doi: 10.3354/ame01753
Babbin A. R., Tamasi T., Dumit D., Weber L., Rodríguez M. V. I., Schwartz S. L., et al. (2021). Discovery and quantification of anaerobic nitrogen metabolisms among oxygenated tropical Cuban stony corals. ISME J. 15, 1222–1235. doi: 10.1038/s41396-020-00845-2
Baker A. C., Glynn P. W., Riegl B. (2008). Climate change and coral reef bleaching: An ecological assessment of long-term impacts, recovery trends and future outlook. Estuarine Coast. Shelf Sci. 80, 435–471. doi: 10.1016/j.ecss.2008.09.003
Bell G. (2013). Evolutionary rescue and the limits of adaptation. Philos. Trans. R. Soc. B: Biol. Sci. 368, 20120080. doi: 10.1098/rstb.2012.0080
Bender D., Diaz-Pulido G., Dove S. (2012). Effects of macroalgae on corals recovering from disturbance. J. Exp. Mar. Biol. Ecol. 429, 15–19. doi: 10.1016/j.jembe.2012.06.014
Bhagooli R., Hidaka M. (2004). Photoinhibition, bleaching susceptibility and mortality in two scleractinian corals, Platygyra ryukyuensis and Stylophora pistillata, in response to thermal and light stresses. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 137, 547–555. doi: 10.1016/j.cbpb.2003.11.008
Boyd P. W., Cornwall C. E., Davison A., Doney S. C., Fourquez M., Hurd C. L., et al. (2016). Biological responses to environmental heterogeneity under future ocean conditions. Global Change Biol. 22, 2633–2650. doi: 10.1111/gcb.13287
Breitburg D., Levin L. A., Oschlies A., Grégoire M., Chavez F. P., Conley D. J., et al. (2018). Declining oxygen in the global ocean and coastal waters. Science 359, eaam7240. doi: 10.1126/science.aam7240
Brown B. E., Dunne R. P., Edwards A. J., Sweet M. J., Phongsuwan N. (2015). Decadal environmental ‘memory’ in a reef coral? Mar. Biol. 162, 479–483. doi: 10.1007/s00227-014-2596-2
Brown T., Bourne D., Rodriguez-Lanetty M. (2013). Transcriptional Activation of c3 and hsp70 as Part of the Immune Response of Acropora millepora to Bacterial Challenges. PloS One 8, e67246. doi: 10.1371/journal.pone.0067246
Bürkner P.-C. (2017). brms : An R package for Bayesian multilevel models Using Stan. J. Stat. Software 80, 1–28. doi: 10.18637/jss.v080.i01
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Camp E. F., Schoepf V., Mumby P. J., Hardtke L. A., Rodolfo-Metalpa R., Smith D. J., et al. (2018). The Future of coral reefs subject to rapid climate change: lessons from natural extreme environments. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00004
Camp E. F., Suggett D. J., Pogoreutz C., Nitschke M. R., Houlbreque F., Hume B. C. C., et al. (2020). Corals exhibit distinct patterns of microbial reorganisation to thrive in an extreme inshore environment. Coral Reefs 39, 701–716. doi: 10.1007/s00338-019-01889-3
Castelli M., Lanzoni O., Giovannini M., Lebedeva N., Gammuto L., Sassera D., et al. (2022). ‘Candidatus Gromoviella agglomerans’, a novel intracellular Holosporaceae parasite of the ciliate Paramecium showing marked genome reduction. Environ. Microbiol. Rep. 14, 34–49. doi: 10.1111/1758-2229.13021
Chu N. D., Vollmer S. V. (2016). Caribbean corals house shared and host-specific microbial symbionts over time and space. Environ. Microbiol. Rep. 8, 493–500. doi: 10.1111/1758-2229.12412
Cornwall C. E., Comeau S., Kornder N. A., Perry C. T., van Hooidonk R., DeCarlo T. M., et al. (2021). Global declines in coral reef calcium carbonate production under ocean acidification and warming. Proc. Natl. Acad. Sci. 118, e2015265118. doi: 10.1073/pnas.2015265118
Couce E., Ridgwell A., Hendy E. (2013). Future habitat suitability for coral reef ecosystems under global warming and ocean acidification. Global Change Biol 19, 3592–3606. doi: 10.1111/gcb.12335
Deleja M., Paula J. R., Repolho T., Franzitta M., Baptista M., Lopes V., et al. (2022). Effects of hypoxia on coral photobiology and oxidative stress. Biology 11, 1068. doi: 10.3390/biology11071068
Deschaseaux E. S. M., Jones G. B., Deseo M. A., Shepherd K. M., Kiene R. P., Swan H. B., et al. (2014). Effects of environmental factors on dimethylated sulfur compounds and their potential role in the antioxidant system of the coral holobiont. Limnology Oceanography 59, 758–768. doi: 10.4319/lo.2014.59.3.0758
Donovan M. K., Burkepile D. E., Kratochwill C., Shlesinger T., Sully S., Oliver T. A., et al. (2021). Local conditions magnify coral loss after marine heatwaves. Science 372, 977–980. doi: 10.1126/science.abd9464
Fabricius K. E., Langdon C., Uthicke S., Humphrey C., Noonan S., De’ath G., et al. (2011). Losers and winners in coral reefs acclimatized to elevated carbon dioxide concentrations. Nat. Climate Change 1, 165–169. doi: 10.1038/nclimate1122
Fitt W. K., Brown B. E., Warner M. E., Dunne R. P. (2001). Coral bleaching: interpretation of thermal tolerance limits and thermal thresholds in tropical corals. Coral Reefs 20, 51–65. doi: 10.1007/s003380100146
Gardner S. G., Camp E. F., Smith D. J., Kahlke T., Osman E. O., Gendron G., et al. (2019). Coral microbiome diversity reflects mass coral bleaching susceptibility during the 2016 El Niño heat wave. Ecol. Evol. 9, 938–956. doi: 10.1002/ece3.4662
Gignoux-Wolfsohn S. A., Vollmer S. V. (2015). Identification of candidate coral pathogens on White Band Disease-infected staghorn coral. PloS One 10, e0134416. doi: 10.1371/journal.pone.0134416
Gilbert J. A., Jansson J. K., Knight R. (2018). Earth microbiome project and global systems biology. mSystems 3, e00217–e00217. doi: 10.1128/mSystems.00217-17
Gloor G. B., Macklaim J. M., Pawlowsky-Glahn V., Egozcue J. J. (2017). Microbiome datasets are compositional: and this is not optional. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02224
Glynn P. W. (1993). Coral reef bleaching: ecological perspectives. Coral Reefs 12, 1–17. doi: 10.1007/BF00303779
Godoy-Vitorino F., Ruiz-Diaz C. P., Rivera-Seda A., Ramírez-Lugo J. S., Toledo-Hernández C. (2017). The microbial biosphere of the coral Acropora cervicornis in Northeastern Puerto Rico. PeerJ 5, e3717. doi: 10.7717/peerj.3717
Goreau T. J., Macfarlane A. H. (1990). Reduced growth rate of Montastrea annularis following the 1987–1988 coral-bleaching event. Coral Reefs 8, 211–215. doi: 10.1007/BF00265013
Grottoli A. G., Martins P. D., Wilkins M. J., Johnston M. D., Warner M. E., Cai W.-J., et al. (2018). Coral physiology and microbiome dynamics under combined warming and ocean acidification. PloS One 13, e0191156. doi: 10.1371/journal.pone.0191156
Haas A., Jantzen C., Naumann M., Iglesias-Prieto R., Wild C. (2010). Organic matter release by the dominant primary producers in a Caribbean reef lagoon: implication for in situ O2 availability. Mar. Ecol. Prog. Ser. 409, 27–39. doi: 10.3354/meps08631
Haas A. F., Smith J. E., Thompson M., Deheyn D. D. (2014). Effects of reduced dissolved oxygen concentrations on physiology and fluorescence of hermatypic corals and benthic algae. PeerJ 2, e235. doi: 10.7717/peerj.235
Hackerott S., Martell H. A., Eirin-Lopez J. M. (2021). Coral environmental memory: causes, mechanisms, and consequences for future reefs. Trends in Ecology & Evolution 36, 1011–1023.
Hernandez-Agreda A., Leggat W., Bongaerts P., Ainsworth T. D. (2016). The Microbial Signature Provides Insight into the Mechanistic Basis of Coral Success across Reef Habitats. mBio 7. doi: 10.1128/mbio.00560-16
Hoegh-Guldberg O., Mumby P. J., Hooten A. J., Steneck R. S., Greenfield P., Gomez E., et al. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742. doi: 10.1126/science.1152509
Howard R. D., Schul M. D., Rodriguez Bravo L. M., Altieri A. H., Meyer J. L. (2023). Shifts in the coral microbiome in response to in situ experimental deoxygenation. Appl. Environ. Microbiol. 0, e00577–e00523. doi: 10.1128/aem.00577-23
Hughes D. J., Alderdice R., Cooney C., Kühl M., Pernice M., Voolstra C. R., et al. (2020). Coral reef survival under accelerating ocean deoxygenation. Nat. Climate Change 10, 296–307. doi: 10.1038/s41558-020-0737-9
Hughes D. J., Alexander J., Cobbs G., Kühl M., Cooney C., Pernice M., et al. (2022). Widespread oxyregulation in tropical corals under hypoxia. Mar. pollut. Bull. 179, 113722. doi: 10.1016/j.marpolbul.2022.113722
Hughes T. P., Kerry J. T., Álvarez-Noriega M., Álvarez-Romero J. G., Anderson K. D., Baird A. H., et al. (2017). Global warming and recurrent mass bleaching of corals. Nature 543, 373–377H. doi: 10.1038/nature21707
Hughes T. P., Kerry J. T., Baird A. H., Connolly S. R., Dietzel A., Eakin C. M., et al. (2018). Global warming transforms coral reef assemblages. Nature 556, 492–496. doi: 10.1038/s41586-018-0041-2
Johnson M. D., Scott J. J., Leray M., Lucey N., Bravo L. M. R., Wied W. L., et al. (2021a). Rapid ecosystem-scale consequences of acute deoxygenation on a Caribbean coral reef. Nat. Commun. 12, 4522. doi: 10.1038/s41467-021-24777-3
Johnson M. D., Swaminathan S. D., Nixon E. N., Paul V. J., Altieri A. H. (2021b). Differential susceptibility of reef-building corals to deoxygenation reveals remarkable hypoxia tolerance. Sci. Rep. 11, 23168. doi: 10.1038/s41598-021-01078-9
Keeling R. F., Körtzinger A., Gruber N. (2010). Ocean deoxygenation in a warming world. Annu. Rev. Mar. Sci. 2, 199–229. doi: 10.1146/annurev.marine.010908.163855
Kellogg C. A. (2004). Tropical Archaea: diversity associated with the surface microlayer of corals. Mar. Ecol. Prog. Ser. 273, 81–88. doi: 10.3354/meps273081
Klinges G., Maher R. L., Thurber R. L. V., Muller E. M. (2020). Parasitic ‘Candidatus Aquarickettsia rohweri’ is a marker of disease susceptibility in Acropora cervicornis but is lost during thermal stress. Environ. Microbiol. 22, 5341–5355. doi: 10.1111/1462-2920.15245
Klinges J. G., Rosales S. M., McMinds R., Shaver E. C., Shantz A. A., Peters E. C., et al. (2019). Phylogenetic, genomic, and biogeographic characterization of a novel and ubiquitous marine invertebrate-associated Rickettsiales parasite, Candidatus Aquarickettsia rohweri, gen. nov. sp. nov. ISME J. 13, 2938–2953. doi: 10.1038/s41396-019-0482-0
Kühl M., Cohen Y., Jørgensen B. B., Revsbech N. P. (1995). Microenvironment and photosynthesis of zooxanthellae in scleractinian corals studied with microsensors for O2, pH and light. Mar. Ecology-Progress Ser. 117, 159–172. doi: 10.3354/meps117159
Laffoley D., Baxter J. M. (2019). “Ocean deoxygenation: everyone’s problem,” in Causes, impacts, consequences and solutions (Gland, Switzerland: IUCN, International Union for Conservation of Nature).
Loya Y., Sakai K., Yamazato K., Nakano Y., Sambali H., van Woesik R. (2001). Coral bleaching: the winners and the losers. Ecol. Lett. 4, 122–131. doi: 10.1046/j.1461-0248.2001.00203.x
Mandal S., Van Treuren W., White R. A., Eggesbø M., Knight R., Peddada S. D. (2015). Analysis of composition of microbiomes: a novel method for studying microbial composition. Microbial Ecol. Health Dis. 26, 27663. doi: 10.3402/mehd.v26.27663
Manzello D., Enochs I., Kolodziej G., Carlton R. (2015). Recent decade of growth and calcification of Orbicella faveolata in the Florida Keys: an inshore-offshore comparison. Mar. Ecol. Prog. Ser. 521, 81–89. doi: 10.3354/meps11085
Manzello D. P., Matz M. V., Enochs I. C., Valentino L., Carlton R. D., Kolodziej G., et al. (2019). Role of host genetics and heat-tolerant algal symbionts in sustaining populations of the endangered coral Orbicella faveolata in the Florida Keys with ocean warming. Global Change Biol. 25, 1016–1031. doi: 10.1111/gcb.14545
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17. doi: 10.14806/ej.17.1.200
McDevitt-Irwin J. M., Garren M., McMinds R., Vega Thurber R., Baum J. K. (2019). Variable interaction outcomes of local disturbance and El Niño-induced heat stress on coral microbiome alpha and beta diversity. Coral Reefs 38, 331–345. doi: 10.1007/s00338-019-01779-8
McMurdie P. J., Holmes S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Miller N., Maneval P., Manfrino C., Frazer T. K., Meyer J. L. (2020). Spatial distribution of microbial communities among colonies and genotypes in nursery-reared Acropora cervicornis. PeerJ 8, e9635. doi: 10.7717/peerj.9635
Mori J. F., Chen L.-X., Jessen G. L., Rudderham S. B., McBeth J. M., Lindsay M. B. J., et al. (2019). Putative mixotrophic nitrifying-denitrifying Gammaproteobacteria implicated in nitrogen cycling within the ammonia/oxygen transition zone of an oil sands pit lake. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02435
Muscatine L., Ferrier-Pagès C., Blackburn A., Gates R. D., Baghdasarian G., Allemand D. (1998). Cell-specific density of symbiotic dinoflagellates in tropical anthozoans. Coral Reefs 17, 329–337. doi: 10.1007/s003380050133
National Marine Fisheries Service and National Oceanic and Atmospheric Association (2006). Endangered and Threatened Species: Final listing determinations for elkhorn coral and staghorn coral. Federal Register 71.
Nelson H. R., Altieri A. H. (2019). Oxygen: the universal currency on coral reefs. Coral Reefs 38, 177–198. doi: 10.1007/s00338-019-01765-0
Parada A. E., Needham D.M., Fuhrman J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Pezner A. K., Courtney T. A., Barkley H. C., Chou W.-C., Chu H.-C., Clements S. M., et al. (2023). Increasing hypoxia on global coral reefs under ocean warming. Nat. Climate Change 13, 403–409. doi: 10.1038/s41558-023-01619-2
Pollock F. J., McMinds R., Smith S., Bourne D. G., Willis B. L., Medina M., et al. (2018). Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat. Commun. 9, 4921. doi: 10.1038/s41467-018-07275-x
Pontes E., Langdon C., Al-Horani F., Johnson M., Alderdice R., Arossa S., et al. (2023). Caribbean scleractinian corals exhibit highly variable tolerances to acute hypoxia. Front. Mar. Sci. 10. doi: 10.3389/fmars.2023.1120262
Rabalais N. N., Díaz R. J., Levin L. A., Turner R. E., Gilbert D., Zhang J. (2010). Dynamics and distribution of natural and human-caused hypoxia. Biogeosciences 7, 585–619. doi: 10.5194/bg-7-585-2010
Raina J.-B., Tapiolas D., Willis B. L., Bourne D. G. (2009). Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol. 75, 3492–3501. doi: 10.1128/AEM.02567-08
Ralph P. J., Hill R., Doblin M. A., Davy S. K. (2016). Theory and application of pulse amplitude modulated chlorophyll fluorometry in coral health assessment. Diseases of Coral, 506–523.
Ritchie K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14. doi: 10.3354/meps322001
Roth M. S. (2014). The engine of the reef: photobiology of the coral-algal symbiosis. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00422
Santos H. A., Massard C. L. (2014). “The family holosporaceae,” in The prokaryotes: alphaproteobacteria and betaproteobacteria. Eds. Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F. (Berlin, Heidelberg: Springer), 237–246.
Shashar N., Cohen Y., Loya Y. (1993). Extreme diel fluctuations of oxygen in diffusive boundary layers surrounding stony corals. Biol. Bull. 185, 455–461. doi: 10.2307/1542485
Smith J. E., Shaw M., Edwards R. A., Obura D., Pantos O., Sala E., et al. (2006). Indirect effects of algae on coral: algae-mediated, microbe-induced coral mortality. Ecol. Lett. 9, 835–845. doi: 10.1111/j.1461-0248.2006.00937.x
Stauffer B. A., Schnetzer A., Gellene A. G., Oberg C., Sukhatme G. S., Caron D. A. (2013). Effects of an acute hypoxic event on microplankton community structure in a coastal harbor of southern California. Estuaries Coasts 36, 135–148. doi: 10.1007/s12237-012-9551-6
Sweet M., Bythell J. (2012). Ciliate and bacterial communities associated with White Syndrome and Brown Band Disease in reef-building corals. Environ. Microbiol. 14, 2184–2199. doi: 10.1111/j.1462-2920.2012.02746.x
Sweet M. J., Bythell J. C., Nugues M. M. (2013). Algae as reservoirs for coral pathogens. PloS One 8, e69717. doi: 10.1371/journal.pone.0069717
Valdés-Castro V., González H. E., Giesecke R., Fernández C., Molina V. (2022). Assessment of microbial community composition changes in the presence of phytoplankton-derived exudates in two contrasting areas from Chilean Patagonia. Diversity 14, 195. doi: 10.3390/d14030195
van Oppen M. J. H., Blackall L. L. (2019). Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 17, 557–567. doi: 10.1038/s41579-019-0223-4
Vaquer-Sunyer R., Duarte C. M. (2010). Temperature effects on oxygen thresholds for hypoxia in marine benthic organisms. Global Change Biol 17, 1788–1797. doi: 10.3389/fmars.2018.00004
Vargas R., Hattori T. (1990). The distribution of protozoa among soil aggregates. FEMS Microbiol. Lett. 74, 73–77. doi: 10.1111/j.1574-6968.1990.tb04053.x
Veal C. J., Carmi M., Fine M., Hoegh-Guldberg O. (2010). Increasing the accuracy of surface area estimation using single wax dipping of coral fragments. Coral Reefs 29, 893–897.
Keywords: coral reef, hypoxia, temperature, microbiome, multi-stressor, resilience, symbiosis, environmental stress
Citation: Swaminathan SD, Meyer JL, Johnson MD, Paul VJ, Bartels E and Altieri AH (2024) Divergent responses of the coral holobiont to deoxygenation and prior environmental stress. Front. Mar. Sci. 10:1301474. doi: 10.3389/fmars.2023.1301474
Received: 25 September 2023; Accepted: 27 December 2023;
Published: 01 February 2024.
Edited by:
Verena Schoepf, University of Amsterdam, NetherlandsReviewed by:
Trent Haydon, New York University Abu Dhabi, United Arab EmiratesAndreas Florian Haas, Royal Netherlands Institute for Sea Research (NIOZ), Netherlands
Copyright © 2024 Swaminathan, Meyer, Johnson, Paul, Bartels and Altieri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sara D. Swaminathan, c2FyYWRldmlzd2FtaW5hdGhhbkBnbWFpbC5jb20=
 Sara D. Swaminathan
Sara D. Swaminathan Julie L. Meyer
Julie L. Meyer Maggie D. Johnson
Maggie D. Johnson Valerie J. Paul
Valerie J. Paul Erich Bartels
Erich Bartels Andrew H. Altieri
Andrew H. Altieri