- 1Department of Biological Sciences, California State University (CSU) Sacramento, Sacramento, CA, United States
- 2DNA Learning Center, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, United States
- 3Biology Department, James Madison University, Harrisonburg, VA, United States
In this era of climate change there is an urgent need to better understand the mechanisms that allow organisms to thrive vs. fail in thermally stressful environments. In particular, there is growing evidence that the “holobiont” (host animal + microbiome community of bacteria, fungi, and archaea that live in an organism) affects how organisms respond to environmental stressors such as temperature and thus should be studied further. Rocky intertidal species such as Tegula snails are ideal organisms for these types of studies because closely related species exhibit variability in heat tolerance. Here, we assess potential microbiome bacterial contributions to thermal tolerance in Tegula eiseni, Tegula funebralis, and Tegula gallina that co-occur in southern California but occupy different intertidal heights that vary in thermal stress exposure. 16S sequencing of the V4 region of individuals of each species exposed to control conditions (ambient temperature = 15°C) or a single short duration 5.5-hour heat stress (maximum temperature = 34°C) revealed distinct bacterial communities across species. Moreover, unique bacterial genera of the microbiome were significantly enriched (more abundant) in each Tegula species. Lutimonas, Polaribacter, and the exopolysaccharide (EPS)-producing bacteria Pelagicoccus were most abundant in T. gallina, the species that occupies the highest intertidal heights and thus experiences heat stress most frequently. These results suggest that microbiome-derived metabolites such as EPS could be contributing to the higher thermal tolerance of T. gallina. Overall, this study demonstrates that the bacterial microbiome should be considered when examining mechanisms of thermal tolerance in marine invertebrates.
Introduction
Climate change is on track to cause a sixth mass extinction event. Warming temperatures have thus far caused the extirpation of 400 species (IPCC, 2022), and one third of marine animals could become extinct in the next 300 years (Penn and Deutsch, 2022). Thus, there is an urgent need to better understand the mechanisms that differentiate organisms that thrive vs. fail in thermally stressful environments. In particular, there is growing recognition among organismal biologists that the “holobiont” (host animal + microbiome community of bacteria, fungi, and archaea that live in an organism) functions as an integrated unit (Lynch and Hsiao, 2019) and affects how organisms respond to environmental stressors such as temperature (Alberdi et al., 2016; Hector et al., 2022). Specifically, the microbiome is hypothesized to be associated with thermal tolerance in a diversity of organisms including lizards (Moeller et al., 2020), flies (Moghadam et al., 2017; Price et al., 2025), aphids (Dunbar et al., 2007), corals (de Breuyn et al., 2025; Ziegler et al., 2017), frogs (Fontaine et al., 2022), and several types of algae (Quigley et al., 2020; Xie et al., 2013).
Specific bacteria of the microbiome can affect thermal tolerance of the host by stimulating increased expression of stress response pathways genes, such as heat shock proteins (Brumin et al., 2011; Nakagawa et al., 2016; Porras et al., 2020) or by producing protective metabolites and proteins (Burke et al., 2010; Dunbar et al., 2007; Fontaine and Kohl, 2023). For example, some bacteria produce exopolysaccharides (EPS), complex sugar polymers excreted into the external environment that can protect against extreme stress conditions such as high temperatures, low nutrients, drought, salinity stress, and antimicrobial agents (Liu et al., 2013; Nichols et al., 2005; Wagh et al., 2022; Wang et al., 2019). There is evidence in tomato plants that EPS produced by plant-associated plant-growth promoting rhizobacteria reduce the negative effects of heat stress in the host plant (Morcillo and Manzanera, 2021). Whether EPS produced by the microbiome could also be playing a role in the thermal tolerance of marine invertebrate hosts remains unknown.
Rocky intertidal organisms are commonly studied when examining mechanisms of thermal tolerance because temperatures in the intertidal vary across small spatial scales and microhabitats, enabling comparisons of the same or related species that regularly experience different degrees of thermal stress. Previous work investigating microbiome differences in the intertidal has demonstrated that the host microbiome varies across these microhabitats and across temperature exposures. For example, 80% of the bacterial assemblage differs across bivalve clams Ruditapes philippinarum outplanted to three different intertidal levels that vary in emersion time (Offret et al., 2020). The cirri microbiome of the intertidal barnacle Semibalanus balanoides also varied across low vs. high intertidal microhabitats, and Fucus algae congeners occupying different heights of the intertidal zone vary in their microbiome composition and structure (Quigley et al., 2020). Similar differences have also been observed when comparing the microbiome of intertidal mollusks across temperature treatments: the bacterial community composition of the intertidal Sydney rock oyster Saccostrea glomerata changes following exposure to elevated heat wave temperatures (Scanes et al., 2023). Microbial communities also changed in response to elevated temperature in the mussels Mytilus galloprovincialis, Perna canaliculus, and Mytilus coruscus (Ericson et al., 2024; Li et al., 2019; Zhu et al., 2024).
In this study we focus on the Tegula genus of intertidal marine snails on the west coast of the United States, looking specifically at T. eiseni, T. funebralis, and T. gallina. These species are well positioned to address questions regarding biological responses to climate change because they occupy overlapping but distinct geographic ranges and ecological niches (Hellberg, 1998). All three species co-occur in southern California. T. eiseni occupies the shallow subtidal zone (Schmitt, 1982) and is thus submerged underwater more and exposed to extreme high air temperatures less than the other two species. Conversely, T. gallina occupies the high intertidal zone, coinciding with more frequent and prolonged exposure to thermally stressful high air temperatures. T. funebralis, whose tidal height ranges from +0.4 to +2.0 m above mean lower low water (MLLW) in southern California (Gleason and Burton, 2016b), occupies the mid to high intertidal zone. T. funebralis and T. gallina co-occur at roughly the same tidal heights in La Jolla and Bird Rock in San Diego County, California, but the range of T. gallina extends higher (authors’ unpubl. data). The phylogeny (Hellberg, 1998), heat shock response (Tomanek, 2002, 2005; Tomanek and Sanford, 2003; Tomanek and Somero, 1999, 2000), and transcriptome-wide response to heat stress (Gleason and Burton, 2015) of these species are well characterized, and the field microbiomes of T. eiseni and T. funebralis in southern California have also been examined (Neu et al., 2019). However, to date no work has been done comparing the microbiome of these species exposed to varying temperatures. Thus, our knowledge about how host species and heat stress affect the Tegula microbiome remains limited.
The objective of this study was to examine whether the microbiome contributes to thermal tolerance in Tegula intertidal snail species that occupy different tidal heights and thus experience distinct levels of thermal stress in the field. We exposed individuals of each Tegula species (T. eiseni, T. funebralis, and T. gallina) to control vs. heat stress conditions simulating a single low tide event and performed 16S sequencing to characterize the bacterial community present in each condition. Specifically, we used the microbiome datasets to address the following questions: (1) does the bacterial community differ across Tegula species?; (2) does the bacterial community differ across control vs. heat stress treatments?; and (3) which specific bacteria are enriched (i.e., significantly more abundant) in thermally tolerant and thermally sensitive Tegula species?
Methods
Microbiome sample preparation
Medium sized Tegula eiseni, Tegula funebralis, and Tegula gallina adults 15-20mm in shell diameter (n = 50 per species) were collected in the summer of 2022 from the southern California site Bird Rock in San Diego County (32°48’N, 117°15’W; Figure 1). Ideally Tegula would be collected from multiple southern California locations, but as of 2022, sites that have been used for previous research (Gleason and Burton, 2013, 2016b) no longer support robust enough populations of T. funebralis for collection from the field (Sato, 2001). Within 12 hours of collection from Bird Rock, snails were transported to California State University, Sacramento (Sac State) in plastic Nalgene bottles ~one quarter filled with seawater from the collection site. Bottles were stored in a cooler on ice to prevent extreme temperature fluctuations during transport. At Sac State snails of all three species were moved to a flow-through recirculating saltwater aquarium system containing artificial seawater created using Instant Ocean Sea Salt mixed with non-sterilized DI water to 32 ppt. All tanks were filled approximately 80% with water, which allowed Tegula individuals of all species to position themselves either fully submerged underwater or towards the top portion of the tank out of water, in air. The aquarium system was set to 15°C (with an allowable offset of 2°C) and no temperature deviations above 17°C occurred. Snails of all three species were regularly fed dried green algae sheets ad libitum. Before the start of any experiments, snails (n = 50 per species) were kept at these temperature and food conditions for a common garden acclimation period of three weeks. Snails not used in the experiments for this current study were used for other projects in the Gleason Lab.

Figure 1. Graphical representation of the experimental set up from field collection of T. eiseni, T. funebralis, and T. gallina snails to tissue preservation following exposure to heat stress or control conditions. Picture of aquaria tanks by Andrea Price. Created in BioRender. Gleason, L. (2025) https://BioRender.com/bijkuqa.
Following the common garden period T. eiseni, T. funebralis, and T. gallina individuals were exposed to either 1) control 15°C conditions (n = 3 per species) or 2) a 34°C heat stress over 5.5 hours to simulate a single low tide event, following the same 3°C increase per 30 min ramping protocol as described in Gleason and Burton, 2013 (n = 3 per species). Heat stress and control exposures were conducted in air to simulate low tide conditions Tegula individuals experience in the field. Each individual snail was placed in an empty, sterile, capped 50 mL falcon tube that was then placed in a temperature-controlled water bath to reach the desired experimental temperatures (either 15°C for controls or 34°C for heat stress). A foam tube holder was used to float each falcon tube at the top of the water line and to ensure that each tube was held underwater and thus exposed to the desired temperatures. At the conclusion of the control or heat stress exposures, snails were frozen in liquid nitrogen and kept at -80°C. Samples were collected immediately after the 5.5-hour temperature exposure because a heat stress event of this duration is sufficient to cause mortality for all Tegula species, albeit at different maximum temperatures (authors’ unpubl. data). Therefore, this study examines the potential microbiome responses correlated with these survival differences. For sample processing, the shell of each frozen Tegula individual was removed with an ethanol-cleaned woodworking vise, the remaining body was rinsed with ethanol to remove surface bacteria, the gonads and muscular foot were removed, and DNA was extracted from 250 mg of the remaining whole-body tissue using a Qiagen DNeasy PowerSoil Pro kit. This tissue processing ensured that 1) microbiome differences based on gonadal sex differences did not influence the data, and that 2) the microbiome living in/on organs such as the gill, stomach, heart, etc. were examined (as opposed to the exterior surface bacteria on the muscular foot). A previous study found no differences in the microbiome of individual organs in T. funebralis and T. gallina (Neu et al., 2019), and thus whole-body tissue samples were used here. Using whole body samples, as opposed to a single organ such as gill tissue that is in direct contact with the surrounding seawater, also minimizes the likelihood that the microbiome recovered from the Tegula hosts is heavily influenced by the ambient seawater microbiome (Li et al., 2022). No negative controls were used during DNA extraction because swabbing the air or an empty microfuge tube is not a true negative control for a whole-body tissue sample (Hornung et al., 2019); however, we were consistent when taking all samples, which is important for minimizing technical variation and is especially important when a true negative control is difficult to obtain (Vandeputte et al., 2017). All extracted DNA from 18 total samples was quantified with a UV-Vis Nanodrop spectrophotometer (range 51.2 – 329.9 ng/uL).
16S sequencing
A 350 bp region of the bacterial V4 region of the 16S rRNA gene was amplified using indexed forward (5’-GTGCCAGCMGCCGCGGTAA-3’) and reverse (5’-GGACTACHVGGGTWTCTAAT-3’) primers (Kozich et al., 2013) using the following protocol: 94°C for 3 min, 35 cycles of 94°C for 45 sec, 50°C for 60 sec, 72°C for 90 sec, and finally 72°C for 10 min. PCR reactions were performed in 25 uL volumes and contained 12.5 uL 2x Phusion High-Fidelity DNA Polymerase Master Mix (NEB), 5 uL ddH2O, 1.25 uL of each primer (10 uM), and 5 uL of template DNA. Amplifications were verified on a 1% agarose gel with GelRed and pooled. A double-sided bead cleanup using Cytiva Sera-Mag SpeedBeads Carboxyl Magnetic Beads (Fisher Scientific) was carried out to remove primer-dimers and a low amount of off-target larger PCR products. Quality and concentration of the pooled library was checked using a Bioanalyzer (Agilent, Santa Clara, CA, USA) and NEB’s Library Quant Kit for Illumina following the manufacturer’s standard protocol. The library representing 18 total samples was then sequenced on an Illumina MiniSeq, yielding 300bp PE reads, using a mid-output reagent cartridge at the James Madison University Center for Genome and Metagenome Studies (CGEMS). Before loading, the library was combined with Illumina’s PhiX control (30:70 16S:PhiX) to ensure a high-quality run despite the low diversity of the 16S library.
Sequence read processing and bacterial identification
All 16S rRNA amplicon sequences were processed using the QIIME2 bioinformatics pipeline as implemented in the Purple Line workflow of DNA Subway that is designed for analyzing microbiome metabarcoding data (Bolyen et al., 2019). Before bioinformatics analysis all primers and sequencing adapters were removed. DADA2 (Callahan et al., 2016) was used to trim low quality reads using the following parameters: truncLenF = 250; truncLenR = 231. After trimming to retain only high-quality reads, all samples were rarefied to a maximum depth of 33,529 sequences. We used a sampling depth of 3933 and the classifier Greengenes2 (515F/806R) to identify bacterial taxa in each sample. The Greengenes2 classifier was chosen due to its compatibility with NCBI taxonomy, optimization for 16S rRNA amplicon sequencing, and design to work seamlessly with QIIME 2 workflows and plugins. The plot_bar function of the phyloseq package was used to visualize the relative abundances of the 25 most common bacterial genera with abundance counts of 400 and above.
Assessment of microbiome differences
Alpha diversity was calculated in the metabarcoding Purple Line workflow of DNA Subway using Pielou’s Evenness (Pielou, 1966) and Faith’s Phylogenetic Diversity (Faith, 1992), following the approach of previous microbiome papers such as O’Connell et al., 2018. Pielou’s Evenness assesses, independent of sample size, the relative abundance distribution of species, with a higher value indicating more evenness (i.e., a relatively equal number of bacteria in each taxon). Faith’s Phylogenetic Diversity assesses how many phylogenetically diverse bacterial taxa are present, with a higher value indicating more richness (i.e., more evolutionarily distinct bacterial taxa). Pielou’s Evenness and Faith’s Phylogenetic Diversity are appropriate alpha diversity metrics for our dataset because, unlike alternative metrics such as Chao1 and Observed ASVs, these chosen metrics do not depend on singleton values that are removed as part of the denoising algorithm during DADA2 processing (Cassol et al., 2025). Moreover, given that our sample size per treatment group is relatively small, Faith’s Phylogenetic Diversity is an appropriate metric for this study because it requires a lower sample size to identify statistical significance compared to alternative alpha diversity metrics such as Shannon and Chao1 indices (Kers and Saccenti, 2022).
We used Bray-Curtis distances to assess beta diversity of samples. NMDS plots were created using the metaMDS function in the R package vegan (Oksanen et al., 2001). To determine whether species or treatment significantly affected the microbiome, we conducted a Permutational multivariate analysis of variance (PERMANOVA) using the adonis2 function in the R package vegan with 9,999 permutations. We used the betadisper function of the vegan package to assess PERMANOVA results for heterogeneity of variance. We used the linear discriminant analysis Effect Size (LEfSe) tool (Segata et al., 2011) as implemented in the R package yingtools2 version 0.0.1.174 (Taur, 2024) to identify specific microbial taxa that are enriched, or found significantly more often, in each Tegula species compared to the others. The R package ggplot2 was used to generate and customize all figures. Lastly, for each treatment (control vs. heat stress), bacterial genera that are found 1) only in a single species, 2) in two of the three species, and 3) in all three species were identified and visualized in Venn diagrams using the online tool available at https://bioinformatics.psb.ugent.be/webtools/Venn/.
Results
Microbiome comparison across species and treatments
A total of 348,662 reads were sequenced across all 18 samples (9 total control samples plus 9 total heat stress samples), and 284,649 reads were retained after all filtering steps. From these filtered reads 1406 total amplicon sequence variants (ASVs) were identified (Figure 2). The dominant taxa identified in Tegula samples were the phylum Proteobacteria. Regarding alpha diversity, average (± SEM) Pielou’s Evenness values for T. eiseni, T. funebralis, and T. gallina were 0.63 ± 0.042, 0.07 ± 0.041, and 0.69 ± 0.027, respectively (Figure 3A). Average (± SEM) Faith’s Phylogenetic Diversity values for T. eiseni, T. funebralis, and T. gallina were 11.27 ± 0.59, 15.19 ± 1.09, and 14.19 ± 1.42, respectively (Figure 3B). There were no significant differences in alpha diversity across the three Tegula species using either Pielou’s Eveness or Faith’s Phylogenetic Diversity indices (Kruskal-Wallis [pairwise], p>0.05; Figure 3). Therefore, there are no significant differences in the relative abundance distribution of bacteria (Pielou’s Evenness) or in the amount of evolutionarily distinct bacterial taxa (Faith’s Phylogenetic Diversity) across the three Tegula species. PERMANOVA results indicate that the microbiome communities were significantly different across the three Tegula species (df = 2, F = 2.758, p = 0.0016, and betadisper p = 0.392). Control vs. heat stress treatments were not significantly different from each other (df = 1, F = 1.958, p = 0.0511, and betadisper p = 0.0568; Figure 4), although the interaction between species and treatment was significant (df = 2, F = 1.789, p = 0.0370).
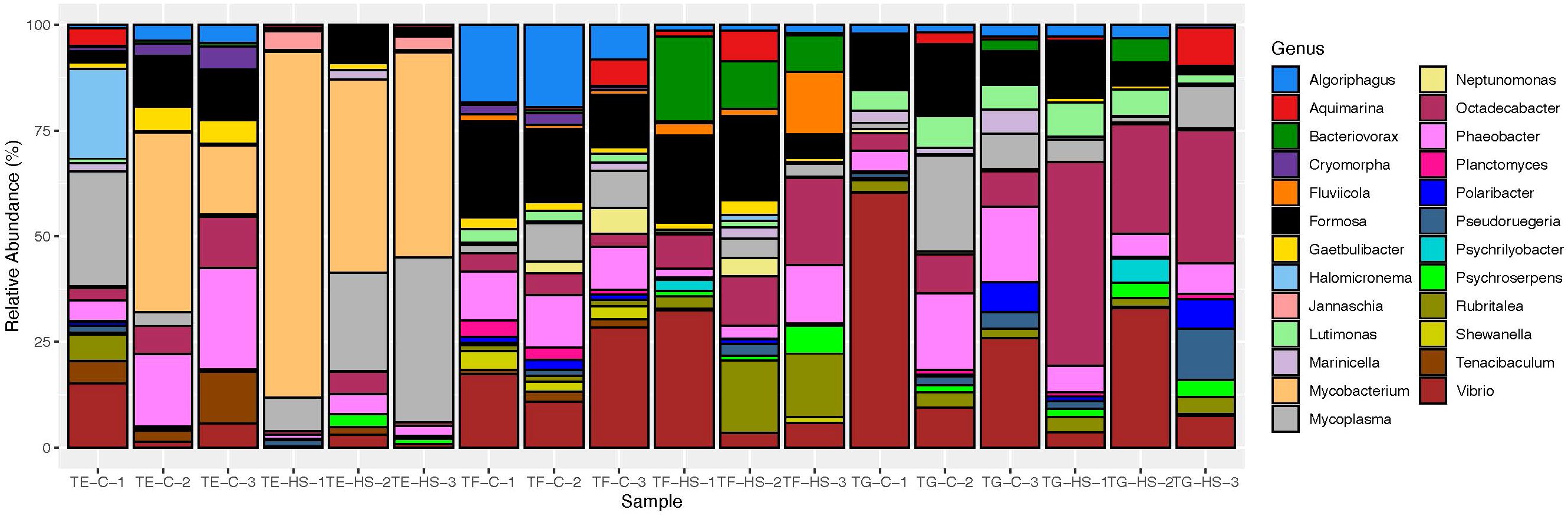
Figure 2. Taxa abundance plot of Tegula microbiomes under control (C) and heat stress (HS) conditions. Each vertical bar represents a different individual (TE, T. eiseni; TF, T. funebralis; TG, T. gallina), with bars grouped according to 1) species and 2) condition (n = 3). Each bar represents the relative abundance of bacterial taxa in that individual’s microbiome. Only taxa identifiable down to the genus level and with more than 400 counts are included here.
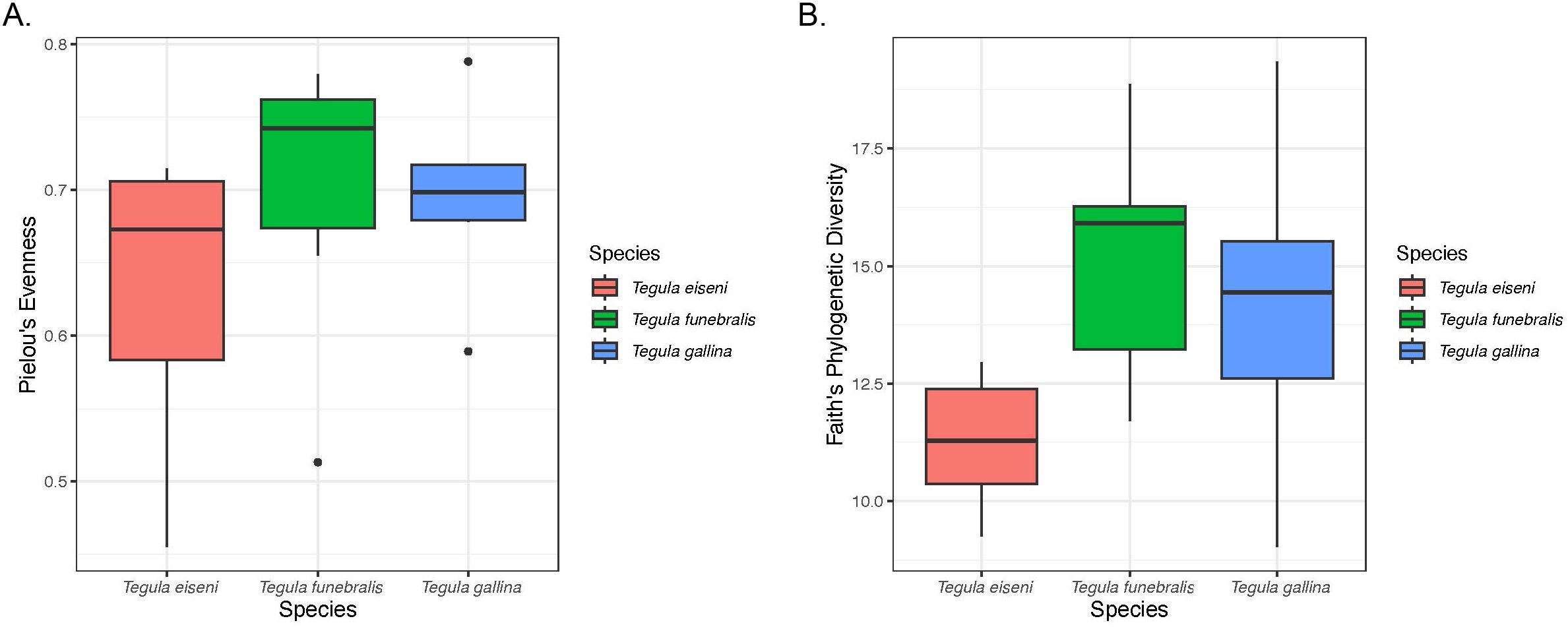
Figure 3. Alpha diversity across the three Tegula species calculated using Pielou’s Evenness (A) and Faith’s Phylogenetic Diversity (B) metrics.
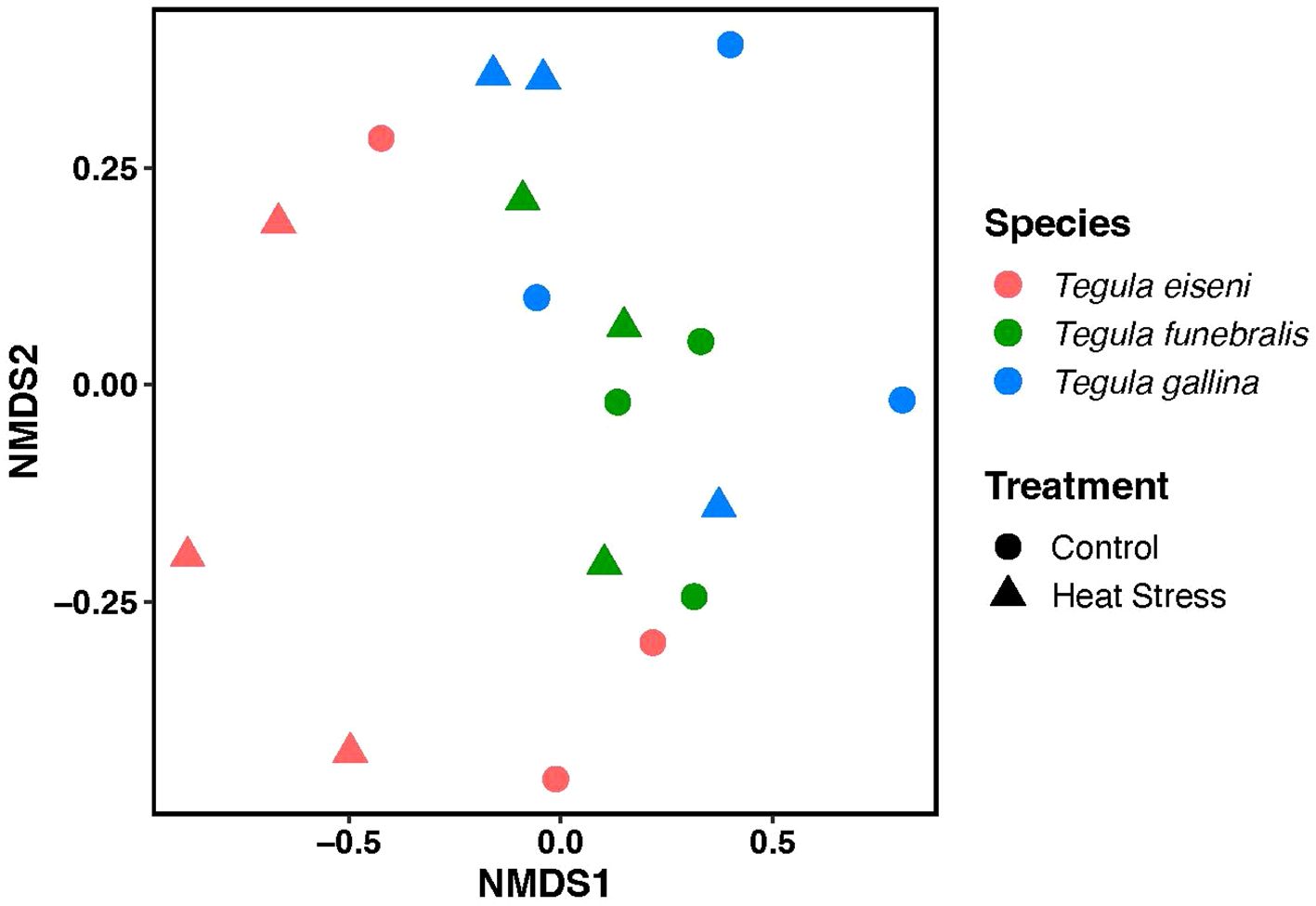
Figure 4. Non-metric multidimensional scaling (NMDS) plot of Tegula microbiomes under control (circles) and heat stress (triangles) conditions. n = 3 for each treatment group (e.g., T. eiseni control). Each data point represents an individual snail, with T. eiseni shown in pink, T. funebralis in green, and T. gallina in blue.
Enriched and unique microbial taxa
Significantly enriched bacterial genera with higher relative abundance were identified in each of the three Tegula species (Table 1). For T. eiseni, the genera Tunicatimonas, Halomicronema, and Mycobacterium were enriched (linear discriminant analysis [LDA] log scores = 2.89, 3.24, and 4.90, respectively; Figure 5). T. funebralis had four enriched bacterial genera: Polynucleobacter, Shewanella, Fluviicola, and Rubritalea (LDA log scores = 2.67, 3.59, 3.73, and 3.89, respectively). Lastly, the three bacterial genera Pelagicoccus, Polaribacter, and Lutimonas (LDA log scores = 2.12, 3.26, and 3.52, respectively) were significantly more abundant in T. gallina. These enriched bacteria are unlikely to be a sampling artifact of the ambient environment because they occur at relatively low frequencies (Figure 2); in previous Tegula microbiome studies, most bacterial OTUs were not shared between T. funebralis and algal and water samples, and the OTUs that were shared were the most abundant ones (Neu et al., 2019).
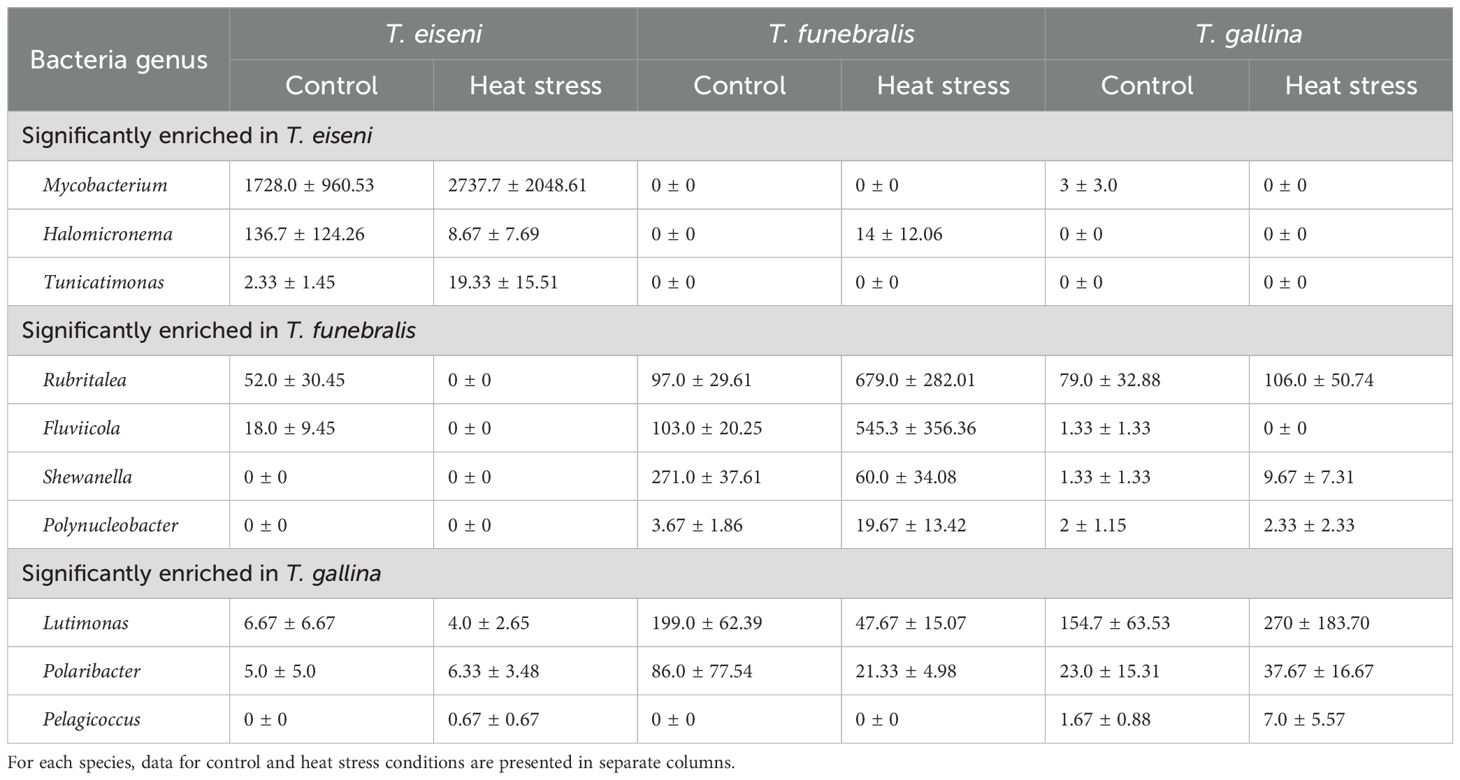
Table 1. Average abundance in raw number of sequences (± SEM) of bacteria genera identified in linear discriminant analysis (LDA) to be significantly enriched in (i) T. eiseni, (ii) T. funebralis, and (iii) T. gallina..
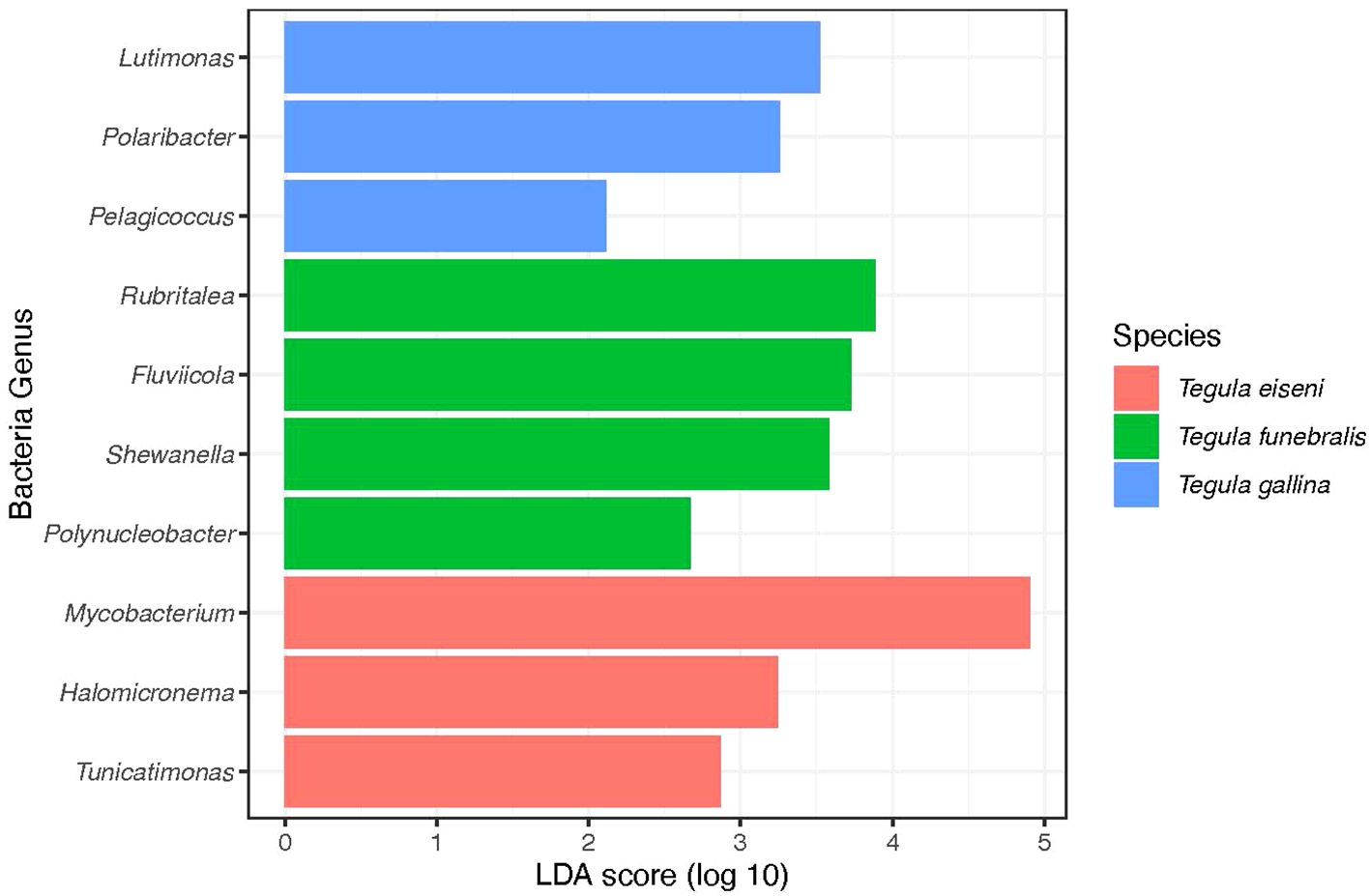
Figure 5. Linear discriminant analysis (LDA) log scores for the significantly enriched (i.e., more abundant) bacterial genera in each Tegula species (T. eiseni in pink, T. funebralis in green, and T. gallina in blue).
As shown in Figure 6A, under control conditions T. eiseni had the least unique bacterial taxa (36 genera, 26.5% of the total genera identified; Supplementary Table 1), including three members of the Flammeovirgaceae family. In contrast, T. gallina had the most unique bacterial taxa (50 genera, 38.2% of the total genera identified). Unique taxa in T. gallina include two members of each of the following families: Alcaligenaceae, Cytophagaceae, Peptostreptococcaceae, and Rhodobacteraceae. Under heat stress conditions this pattern was different: T. gallina had the least unique bacterial taxa (20 genera, 15.4% of the total genera identified; Supplementary Table 2), including two members each of the Flavobacteriaceae and Verrumicrobiaceae families. Heat-stressed T. funebralis had the most unique bacterial taxa (61 genera, 32.3% of the total genera identified), including three members each of the families Moraxellaceae and Pirellulaceae (Figure 6B). There were three bacterial genera found only in T. eiseni under both control and heat stress conditions: Owenweeksia, Tunicatimonas, and Jannaschia. Five bacterial genera were only found in T. funebralis under both control and heat stress conditions: Clostridium, Aquicella, Croninitomix, Plantomycete, and Roseovarius. There were no taxa present only in T. gallina under both control and heat stress conditions.
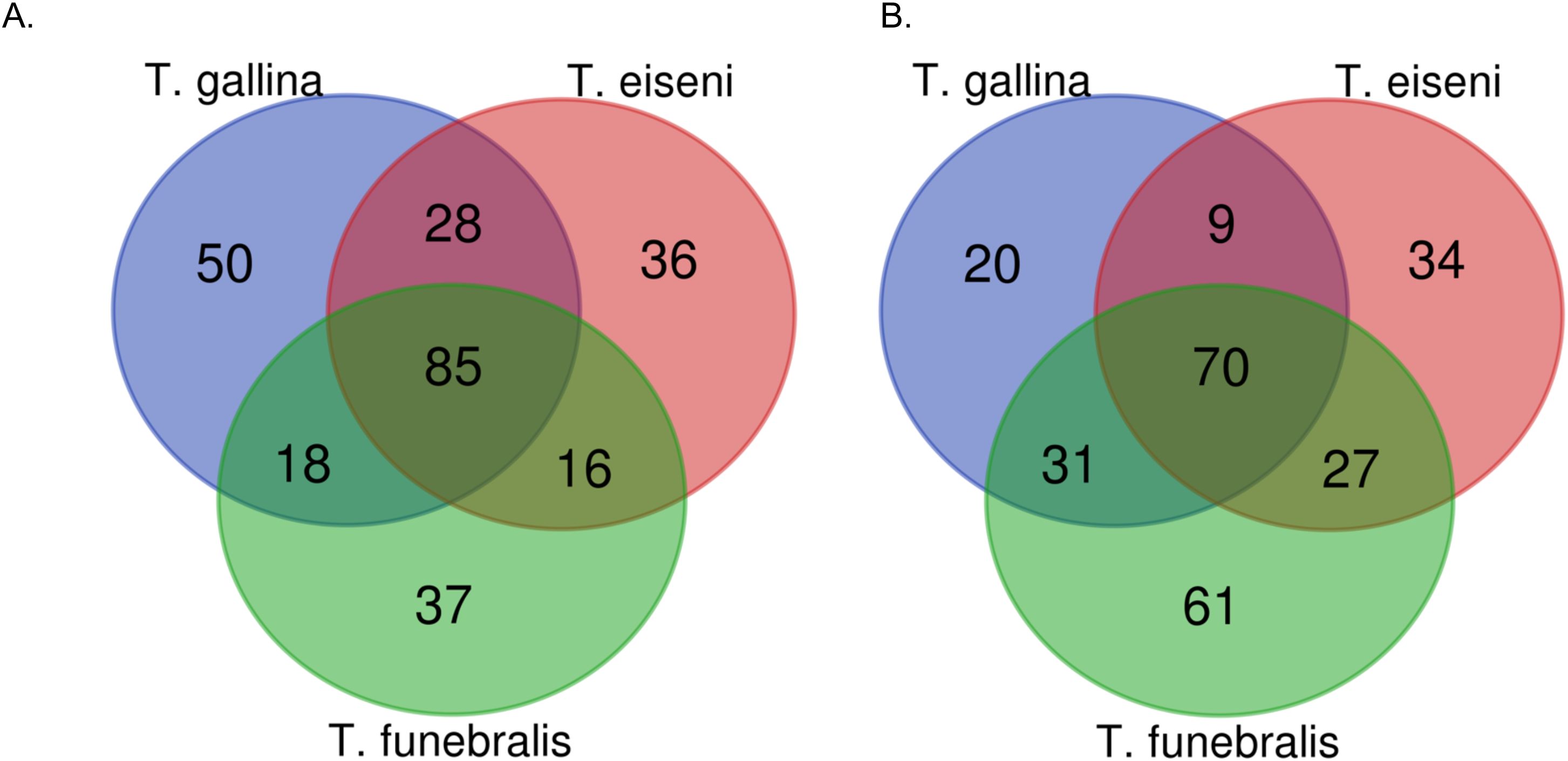
Figure 6. Number of unique vs. shared bacteria genera identified in each Tegula species under control (A) and heat stress (B) conditions.
Discussion
The microbiome of the intertidal marine snails T. eiseni, T. funebralis, and T. gallina significantly differs across species. These differences are driven by both unique (i.e., bacteria that are only present in one of the three Tegula species) and enriched (i.e., bacteria that are significantly more abundant in one of the three Tegula species) bacterial genera.
Microbiome communities significantly differ across species
Proteobacteria is the most common bacterial phyla across all three Tegula species – this is consistent with previous work in marine animals such as snails, limpets, red abalone, corals, copepods, fish, and barnacles (Bayer et al., 2013; Brown et al., 2020; Dorosz et al., 2016; Dudek et al., 2014; Givens et al., 2015; Neu et al., 2019; Ousley, 2023). However, at the genus level the microbial community composition significantly varies across Tegula species. Notably, these differences persisted after a common garden acclimation period in which all individuals were housed in the same flowthrough aquarium system, ate the same dried algae diet, and were exposed to the same submersion times in the same recirculating artificial seawater. Overall, these differences across species are consistent with previous work in Tegula (Chlorostoma) eiseni and Tegula (Chlorostoma) funebralis examined directly from the field from La Jolla, California (Neu et al., 2019). Our results also match findings in other marine species such as sponges and algae. De Castro-Fernández et al. (2023) reported that in four different demosponge species, samples clustered together in non-metric multidimensional scaling (NMDS) space according to species (De Castro-Fernández et al., 2023). Moreover, in an experimental design similar to this current study, the microbiome composition and structure significantly varied across three Fucus algae congeners occupying different heights of the intertidal zone and thus experiencing different levels of abiotic stress (Quigley et al., 2020). Notably, our results provide evidence for phylosymbiosis, in which the microbiomes of more closely related species are more similar to each other. As seen in Figure 4, in multivariate NMDS space the microbiome of T. eiseni is distinct from the other two species T. funebralis and T. gallina, who are more closely related to each other (Hellberg, 1998). Such phylosymbiosis has also been observed when comparing the microbiomes of Chlorostoma (now Tegula) and Littorina intertidal snails (Neu et al., 2019) and of different tropical sponge species (Easson and Thacker, 2014).
Evolutionary codivergence of the microbiome with these Tegula species is one possible explanation for microbiome differentiation, but several other explanations, such as habitat filtering, should also be considered and investigated further (Mazel et al., 2018). Microbial communities are sensitive to environmental parameters such as pH and temperature; potential differences in the gut environment between T. eiseni, T. funebralis, and T. gallina could be contributing to their distinct microbiomes (Neu et al., 2019). Neu et al. also hypothesized that field-collected T. eiseni and T. funebralis could be differentially ingesting microbes through distinct dietary inputs (Neu et al., 2019). Moreover, the cirri microbiome differences in barnacles occupying low vs. high intertidal microhabitats was thought to be due to differential exposure to and/or time underwater (Brown et al., 2020). Comparing the microbiome of T. eiseni, T. funebralis, and T. gallina individuals collected directly from the field to those that have been in a lab common garden environment could help determine how much influence the distinct microhabitats of each species have on their microbiome. Similarly, explicitly manipulating diet and/or time underwater in a laboratory setting could also provide further insight into the mechanisms of microbial differentiation across species.
Microbiome communities do not significantly differ across treatments
We did not observe significant microbiome differentiation between individuals exposed to control vs. heat stress conditions for 5.5 hours. The microbiome of Mytilus coruscus larvae exposed to elevated seawater temperature for 4 hours was also similar to the microbiome of control individuals (Zhu et al., 2024), perhaps indicating that a heat stress period longer than several hours is required to elicit clear differentiation of the microbiome. This lack of differentiation contrasts what has been observed in other intertidal marine mollusks. In Haliotis rufescens red abalone, another type of marine snail, control individuals had a distinct microbial composition compared to heat stressed individuals (Ousley, 2023). Similarly, in the intertidal Sydney rock oyster Saccostrea glomerata exposure to elevated heat wave temperatures significantly changed bacterial community composition (Scanes et al., 2023). Microbial communities also changed in response to elevated temperature in the mussel Mytilus galloprovincialis (Li et al., 2019). One potential cause for the lack of significant differentiation between control and heat stress Tegula microbiomes is high interindividual variability magnified by a small sample size (n = 3 per species per treatment). For example, as shown in Table 1, the abundance of bacterial genera varied widely across individuals of the same species and treatment, especially among T. eiseni control samples with regards to Mycobacterium and among T. gallina heat stress samples with regards to Lutimonas and Polaribacter. Ultimately, the small sample size used in this study limits the statistical power and our ability to reliably detect meaningful biological difference across treatments. Thus, the fact that we did not identify significant differences in the microbiome between control and heat stress treatments could be an artifact of a small sample size.
Enriched bacteria in Tegula eiseni
In T. eiseni, the species with the lowest thermal tolerance, enriched bacteria could be contributing to infection and apoptotic cell death under heat stress conditions. Mycobacterium is much more abundant in T. eiseni under both control and heat stress conditions compared to T. funebralis and T. gallina. This is a pathogenic bacteria that is known to infect other marine mollusks (Davidovich et al., 2020), and enrichment of this taxa could suggest an increased risk of disease in T. eiseni, especially under heat stress conditions. Another taxa enriched in T. eiseni, Halomicronema, is a cyanobacteria that has also been found in marine sponges (Caroppo et al., 2012). Notably, some compounds produced by the Halomicronema genus can trigger apoptotic cell death (Mutalipassi et al., 2019). In other marine mollusks such as the oyster Crassostrea virginica, the density of apoptotic cells increased after exposure to high temperatures of 26 and 30°C (Rahman and Rahman, 2021). Rahman and Rahman (2021) hypothesized that a high level of reactive oxygen species (ROS) could be contributing to this increase in apoptosis (Nash et al., 2019; Nash and Rahman, 2019). Our results suggest components of the microbiome could also be contributing to the induction of apoptosis in heat sensitive marine mollusks, and should be investigated further. The last enriched bacterial genus in T. eiseni, Tunicatomonas, has also been isolated from sea anemones, but there is only one known species, and not much information is available on this taxon (Yoon et al., 2012).
Enriched bacteria in Tegula funebralis
Three bacterial genera were significantly enriched in T. funebralis and were more abundant in heat stress vs. control conditions: Rubritalea, Fluviicola, and Polynucleobacter. Rubritalea belongs to the class Verrucomicrobiae (Yoon et al., 2007) and bacteria of this genus also increase in abundance following high temperature exposure in corals and sponges. For example, in Turbinaria peltata corals repeated heatwaves led to increases of the beneficial bacteria Rubritalea tangerine that facilitates coral health and growth (Zhai et al., 2024). Zhai et al. (2024) hypothesized that such changes in the microbiome could represent adaptive stress responses to improve survival of the coral host under marine heat wave conditions. Moreover, heat wave exposed Crella incrustans sponges produced larvae with significantly more Rubritalea marina (Strano et al., 2023). How this bacterial genus functions during high temperatures, and the potential mechanisms that allow these bacteria to benefit marine invertebrate hosts, remain unknown and require further investigation. Fluviicola, the second genera enriched in T. funebralis, is a gram-negative rod-shaped bacterium in the class Flavobacteriia, family Cryomorphaceae. In this current study Fluviicola was more abundant in heat stress vs. control samples, which contrasts previous results in green-lipped mussels: in Perna canaliculus Fluviicola was more abundant in control vs. heat stress individuals (Ericson et al., 2024). Other studies in marine mollusks found that Fluviicola abundance was high in razor clams exposed to hyposalinity stress (Yang et al., 2024) and that Fluviicola was more abundant in younger vs. older Pacific oyster spat (Zhong et al., 2024). Lastly, bacteria in the genus Polynucleobacter were also significantly more abundant in T. funebralis. Polynucleobacter can be either free-living or symbiotic (Miklós et al., 2023) and are often associated with ciliates (Vannini et al., 2007) and hydra (Fraune et al., 2010; Fraune and Bosch, 2007). In the cold-water adapted hydra Hydra oligactis Polynucleobacter was positively correlated with hydra population size, although the mechanism for this effect requires further research (Miklós et al., 2023). In contrast to the current findings in this study, Polynucleobacter abundance was lower at higher temperatures in H. oligactis (Miklós et al., 2023). Overall, any hypotheses about the potential relationship between Polynucleobacter and T. funebralis should be interpreted with caution because the overall abundance of Polynucleobacter was relatively low (Table 1).
In contrast to the other three bacterial genera that were significantly more abundant in T. funebralis, the genus Shewanella was more abundant in control vs. heat stress samples. These results match previous papers that have identified bacteria in this genus that are adapted to low temperatures (Kloska et al., 2020; Zhao et al., 2010). In general, Shewanella are gram-negative, aerobic and facultatively anaerobic γ-Proteobacteria (Garrity and Holt, 2001; Gauthier et al., 1995; MacDonell and Colwell, 1985; Satomi et al., 2003; Venkateswaran et al., 1999). Shewanella have been isolated from a variety of marine invertebrates including hydrocorals, the benthic marine “peanut worm” (Ivanova et al., 2004), the sea urchin Sterechinus neumayeri (González-Aravena et al., 2016), and the soft coral Alcyonium antarcticum (Webster and Bourne, 2007). Notably, exopolysaccharides (EPS) produced by Shewanella have been isolated from Antarctic sponges (Caruso et al., 2018). These EPS were more abundant at low temperatures, and they are thought to serve protective functions under low temperature stress conditions (Lo Giudice and Rizzo, 2022). Although this current study did not expose any individuals to low temperatures, the fact that Shewanella was significantly more abundant in T. funebralis, the only Tegula species examined in this study whose geographic range extends up to Vancouver Island on the western coast of North America, suggests that this bacterium could be contributing to T. funebralis’ ability to withstand the lower temperatures indicative of the northern parts of its range. For example, sites occupied by T. funebralis in northern California can reach absolute minimum temperatures of 8.8°C, which is roughly 5-6°C colder than the absolute minimum temperatures in southern California, where T. eiseni and T. gallina reside (Gleason and Burton, 2016b).
Enriched bacteria in Tegula gallina
Lutimonas was found in relatively high abundance in T. gallina under both control and heat stress conditions. These results contrast a previous study that observed a decrease in Lutimonas under heat stress conditions in Pacific white shrimp Litopenaeus vannamei (Duan et al., 2021). Lutimonas is a nitrifying bacteria that degrades ammonium (Fu et al., 2009). In other marine mollusks such as the ark shell Scapharca subcrenata (Jiang et al., 2020) and in Daphnia (N. Nash et al., 2022), elevated temperatures increase ammonia excretion rates. High levels of ammonia are known to have an array of negative consequences for aquatic invertebrates (Zhang et al., 2023); thus, the relatively high abundance of Lutimonas in T. gallina, even under heat stress conditions, could suggest that the microbiome helps regulate ammonia levels to prevent toxic overaccumulation.
Our finding that the genus Polaribacter is enriched in T. gallina, including under heat stress conditions, contrasts a previous study in another marine mollusk Mytilus galloprovincialis that found Polaribacter was a dominant genus under control conditions, but its abundance decreased when water temperature was increased (Li et al., 2019). However, in other marine invertebrates, including the mussel Mytilus coruscus (Li et al., 2019) and the common yellow sponge M. acerate (De Castro-Fernández et al., 2023), abundance of Polaribacter was also high under heat stress conditions. Polaribacter has also been detected in seawater (Fukui et al., 2013; Yoon et al., 2006) and in diatom phytoplankton blooms (Xing et al., 2015). Overall, not much is known about this genus, and further research must be done before hypotheses regarding the ability of this genus to increase heat tolerance of T. gallina can be formed.
The third and final bacterial genus that was enriched in T. gallina is Pelagicoccus. Pelagicoccus, a member of the family Puniceicoccaceae, contains four different species (Feng et al., 2021; Yoon et al., 2007). These species have previously been isolated from sea grass (Yoon et al., 2007), seawater (Yoon et al., 2007), and marine sediment (Feng et al., 2021). Based on the genomic analysis performed by Feng et al., it is thought that all bacteria in this genus produce exopolysaccharides (EPS) to better cope with extreme stress conditions, including high temperatures, low nutrients, drought, salinity stress, and antimicrobial agents (Liu et al., 2013; Nichols et al., 2005; Wagh et al., 2022; Wang et al., 2019). For example, EPS are produced in hydrothermal vent communities exposed to extreme high temperatures (Nichols et al., 2005). Importantly, there is evidence in other species that EPS produced by bacteria have positive effects on their host: EPS produced by plant-associated plant-growth promoting rhizobacteria reduce the negative effects of heat shock and growth in plant hosts such as tomatoes (Morcillo and Manzanera, 2021). It is possible the Pelagicoccus observed in this study could similarly be contributing to host thermal tolerance in T. gallina, although current evidence is merely correlational and it is not currently known if the relatively low abundance of Pelagicoccus would be sufficient to produce beneficial effects. Ultimately, more direct functional experiments are needed to confirm whether this particular bacterium plays a role in T. gallina’s high thermal tolerance.
Study design limitations
As with any study, there are limitations that should be considered. Most notably, we did not collect or sequence environmental samples from the field or the lab to characterize the surrounding microbial habitat (e.g., seawater and/or artificial seawater in the aquarium system) of the Tegula snails. Although we did surface sterilize each sample by thoroughly rinsing tissue samples with ethanol before extracting DNA (Brown et al., 2020; Neu et al., 2019), we nevertheless cannot determine whether the Tegula microbiome communities we characterized are subsets of the surrounding bacterial community in the artificial seawater and/or the algal diet of the snails. In particular, because Tegula snails were housed in non-sterile artificial seawater during the three-week common garden period, it is possible this water was an environmental source of bacteria that contributed to the Tegula microbiomes analyzed in this study. We do, however, note that a similar study conducted in the Gleason Lab’s same aquarium system found that the fecal microbiome of juvenile red abalone Haliotis rufescens was significantly different from the microbiome of the surrounding aquarium water (Ousley, 2023). Regarding the potential effects of feeding snails dried green algae sheets, a previous study of Haliotis rufescens red abalone marine snails fed red vs. brown vs. green algae found significant differences in the microbiomes of snails fed the distinct diets (Guo, 2017). Thus, the choice of diet in our current study could have affected the microbiome composition of the Tegula snails. Specifically, Guo (2017) found that bacteria in the Mollicutes and Alphaproteobacteria classes and in the Mycoplasma genus were more abundant in H. rufescens fed green algae diets compared to red and brown algae diets. Thus, the abundance of these bacteria may also be enriched in our current study that used a green algae diet.
Additional limitations of study design include the fact that this study used a relatively small number of samples (n = 3 per species per treatment). As noted above, this small sample size could have amplified the high degree of interindividual variability and contributed to the lack of significant variation detected between control and heat stress microbiome samples. Although previous similar microbiome studies in marine snails have used similar sample sizes (Y.-J. Zhu et al., 2021), we do acknowledge that our results may not be representative of each Tegula species’ full range of biological responses to high temperature conditions. Similarly, this work investigates the microbiome from a single field site in San Diego, California. Neu et al., 2019 found that for T. funebralis, Bray-Curtis dissimilarities significantly distinguish snails from north vs. south of Point Conception along the California coast (although the southern site only had a sample size of two). Thus, our findings likely cannot be generalized to T. funebralis individuals from further north in California. Previous studies using RNA sequencing and ddRAD sequencing have identified gene expression and gene sequence differences across geographically distinct T. funebralis populations in northern and southern California that experience unique climates and differ in thermal tolerance (Gleason and Burton, 2015, 2016a). To determine if there are also microbiome differences within a single Tegula species across different geographic regions, future work could sample and subsequently compare the microbiome of T. eiseni and T. gallina from multiple additional sites in southern California (e.g., Abalone Cove in Los Angeles County, Aliso Beach in Orange County, La Jolla in San Diego County), and T. funebralis from northern (e.g., Slide Ranch in Marin County, Pigeon Point and Pescadero in San Mateo County) and southern (e.g., Abalone Cove in Los Angeles County, Aliso Beach in Orange County, La Jolla in San Diego County) California sites, population size permitting (Sato, 2001). Lastly, this study only examines the bacterial component of the microbiome. Any fungal (Chin et al., 2020; Grice, 2015), archaeal, and viral components of the holobiont that could also be contributing to thermal tolerance or host-microbiome interactions are thus not captured in our current dataset.
Future research
At this point it is unknown if the bacterial species we observed in the T. eiseni, T. funebralis, and T. gallina microbiomes are transient (i.e., driven by external factors or stochastic processes) or resident (i.e., explicitly selected for by the host). Resident bacterial species that are a stable, persistent part of the microbiome are more likely to contribute to host thermal tolerance, and thus distinguishing resident vs. transient bacteria in each Tegula host remains an important area of future research. To distinguish these different factions of the Tegula microbiome, further research could follow the approach of Unzueta-Martínez et al. (2022) to differentiate transient vs. resident bacteria in the Eastern oyster Crassostrea virginica. To identify resident bacteria, we could 1) identify any microbiome differences in Tegula species collected directly from multiple different field sites, and 2) determine if these differences persist after several weeks of common garden acclimation in an identical environment. Alternatively, we could determine if certain bacteria persist in each Tegula host in sterile seawater that doesn’t contribute any environmental bacteria (Lokmer et al., 2016, 2016). To identify transient bacteria, we could again collect individuals from multiple different field sites and identify bacterial differences in Tegula individuals held in a common garden environment vs. those sampled directly from the same field site (Unzueta-Martínez et al., 2022).
In addition, to date, we have only characterized the microbiome following a short 5.5-hour heat stress representative of a single low tide period in the field. No assessment has yet been performed of the recovery period following this heat stress; in other words, it is not yet known if the microbial community reverts to the same “baseline” composition after return to non-stressful control conditions. Current marine invertebrate research directly addressing this question is limited, although results in another marine mollusk the green-lipped mussel Perna canaliculus indicate that the microbiome of recovered heat stress samples is more similar to control samples than to sustained heat stress samples (Ericson et al., 2024). Overall, based on the sampling design of this current study, we cannot make any conclusions about the holobiont’s long-term response to heat stress in Tegula species. Thus, more research is needed examining the microbiome at various time points after heat stress to fully understand how the microbiome of Tegula individuals affects thermal tolerance, especially for these intertidal species that regularly experience multiple stressful low tides in a single day.
Conclusion
This study demonstrates that the microbiome of Tegula species is distinct and suggests that components of the microbiome could be contributing to the differential thermal tolerance of T. eiseni, T. funebralis, and T. gallina intertidal snails. Specifically, the pathogenic bacteria Mycobacterium is significantly enriched in the thermally sensitive T. eiseni, and the nitrifying bacteria Lutimonas and the exopolysaccharide-producing bacteria Pelagicoccus are significantly enriched in the thermally tolerant T. gallina. Overall, our results provide further insight into how the microbiome differs across host congeners living in uniquely stressful microhabitats and illustrate the additional information gained when considering non-genetic mechanisms of thermal tolerance.
Data availability statement
The data presented in this study are deposited in the Sequence Read Archive (SRA), accession number PRJNA1309766.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
BA: Writing – review & editing, Investigation, Formal analysis. MB: Investigation, Formal analysis, Writing – review & editing. HC: Investigation, Formal analysis, Writing – review & editing. BG: Writing – review & editing, Investigation, Formal analysis. M-FK: Formal analysis, Writing – review & editing, Investigation. JG: Investigation, Formal analysis, Writing – review & editing. AS: Investigation, Writing – review & editing, Formal analysis. BN: Resources, Funding acquisition, Writing – review & editing. RE: Methodology, Resources, Funding acquisition, Writing – review & editing, Investigation. LG: Conceptualization, Investigation, Writing – review & editing, Supervision, Formal analysis, Writing – original draft, Methodology, Data curation, Visualization.
Funding
The authors declare financial support was received for the research and/or publication of this article. This study was supported by a National Science Foundation IUSE grant, NSF DUE-1821657 awarded to BN and RAE. Write up of this manuscript was supported by a Fall 2024 Sabbatical Leave to LUG.
Acknowledgments
We thank Caity Fox, Instructional Support Technician II at CSU Sacramento, for preparing reagents for sample processing and DNA extractions and Karen Barnard-Kubow at the JMU CGEMS for technical assistance with library preparation and DNA sequencing. We also acknowledge two reviewers whose comments greatly improved the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1645537/full#supplementary-material
Supplementary Table 1 | Lists of bacteria found to be common or unique across the three Tegula species under control conditions, as shown in the Venn diagram in Figure 6A. Bacteria representing each region of the Venn diagram are listed in separate tabs (TE, T. eiseni; TF, T. funebralis; TG, T. gallina). Each bacterium is listed across a single row, with the different taxonomic classifications (e.g. kingdom, phylum, class, order, family, genus) shown in separate columns.
Supplementary Table 2 | Lists of bacteria found to be common or unique across the three Tegula species under heat stress conditions, as shown in the Venn diagram in Figure 6B. Bacteria representing each region of the Venn diagram are listed in separate tabs (TE, T. eiseni; TF, T. funebralis; TG, T. gallina). Each bacterium is listed across a single row, with the different taxonomic classifications (e.g. kingdom, phylum, class, order, family, genus) shown in separate columns.
References
Alberdi A., Aizpurua O., Bohmann K., Zepeda-Mendoza M. L., and Gilbert M. T. P. (2016). Do vertebrate gut metagenomes confer rapid ecological adaptation? Trends Ecol. Evol. 31, 689–699. doi: 10.1016/j.tree.2016.06.008, PMID: 27453351
Bayer T., Neave M. J., Alsheikh-Hussain A., Aranda M., Yum L. K., Mincer T., et al. (2013). The microbiome of the red sea coral stylophora pistillata is dominated by tissue-associated endozoicomonas bacteria. Appl. Environ. Microbiol. 79, 4759–4762. doi: 10.1128/AEM.00695-13, PMID: 23709513
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9, PMID: 31341288
Brown B. R. P., Nunez J. C. B., and Rand D. M. (2020). Characterizing the cirri and gut microbiomes of the intertidal barnacle Semibalanus balanoides. Anim. Microbiome 2, 41. doi: 10.1186/s42523-020-00058-0, PMID: 33499976
Brumin M., Kontsedalov S., and Ghanim M. (2011). Rickettsia influences thermotolerance in the whitefly Bemisia tabaci B biotype. Insect Sci. 18, 57–66. doi: 10.1111/j.1744-7917.2010.01396.x
Burke G., Fiehn O., and Moran N. (2010). Effects of facultative symbionts and heat stress on the metabolome of pea aphids. ISME J. 4, 242–252. doi: 10.1038/ismej.2009.114, PMID: 19907504
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., and Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869, PMID: 27214047
Caroppo C., Albertano P., Bruno L., Montinari M., Rizzi M., Vigliotta G., et al. (2012). Identification and characterization of a new Halomicronema species (Cyanobacteria) isolated from the Mediterranean marine sponge Petrosia ficiformis (Porifera). Fottea 12, 315–326. doi: 10.5507/fot.2012.022
Caruso C., Rizzo C., Mangano S., Poli A., Di Donato P., Finore I., et al. (2018). Production and biotechnological potential of extracellular polymeric substances from sponge-associated antarctic bacteria. Appl. Environ. Microbiol. 84, e01624–e01617. doi: 10.1128/AEM.01624-17, PMID: 29180360
Cassol I., Ibañez M., and Bustamante J. P. (2025). Key features and guidelines for the application of microbial alpha diversity metrics. Sci. Rep. 15, 622. doi: 10.1038/s41598-024-77864-y, PMID: 39753610
Chin V. K., Yong V. C., Chong P. P., Amin Nordin S., Basir R., and Abdullah M. (2020). Mycobiome in the gut: A multiperspective review. Mediators Inflammation 2020, 9560684. doi: 10.1155/2020/9560684, PMID: 32322167
Davidovich N., Morick D., and Carella F. (2020). Mycobacteriosis in aquatic invertebrates: A review of its emergence. Microorganisms 8, 1249. doi: 10.3390/microorganisms8081249, PMID: 32824567
de Breuyn M., Ostendarp M., El-Khaled Y. C., Garcias-Bonet N., Carvalho S., Wild C., et al. (2025). Probiotics prevent mortality of thermal-sensitive corals exposed to short-term heat stress. ISME Commun. 5, ycaf039. doi: 10.1093/ismeco/ycaf039, PMID: 40151579
De Castro-Fernández P., Ballesté E., Angulo-Preckler C., Biggs J., Avila C., and García-Aljaro C. (2023). How does heat stress affect sponge microbiomes? Structure and resilience of microbial communities of marine sponges from different habitats. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.1072696
Dorosz J., Castro-Mejia J., Hansen L., Nielsen D., and Skovgaard A. (2016). Different microbiomes associated with the copepods Acartia tonsa and Temora longicornis from the same marine environment. Aquat. Microbial Ecol. 78, 1–9. doi: 10.3354/ame01799
Duan Y., Xiong D., Wang Y., Li H., Dong H., and Zhang J. (2021). Toxic effects of ammonia and thermal stress on the intestinal microbiota and transcriptomic and metabolomic responses of Litopenaeus vannamei. Sci. Total Environ. 754, 141867. doi: 10.1016/j.scitotenv.2020.141867, PMID: 32898779
Dudek M., Adams J., Swain M., Hegarty M., Huws S., and Gallagher J. (2014). Metaphylogenomic and potential functionality of the limpet patella pellucida’s gastrointestinal tract microbiome. Int. J. Mol. Sci. 15, 18819–18839. doi: 10.3390/ijms151018819, PMID: 25334059
Dunbar H. E., Wilson A. C. C., Ferguson N. R., and Moran N. A. (2007). Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PloS Biol. 5, e96. doi: 10.1371/journal.pbio.0050096, PMID: 17425405
Easson C. G. and Thacker R. W. (2014). Phylogenetic signal in the community structure of host-specific microbiomes of tropical marine sponges. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00532, PMID: 25368606
Ericson J. A., Laroche O., Biessy L., Delorme N. J., Pochon X., Thomson-Laing J., et al. (2024). Differential responses of selectively bred mussels (Perna canaliculus) to heat stress—Survival, immunology, gene expression and microbiome diversity. Front. Physiol. 14. doi: 10.3389/fphys.2023.1265879, PMID: 38425477
Faith D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. doi: 10.1016/0006-3207(92)91201-3
Feng X., Gong Y., Ye M.-Q., and Du Z.-J. (2021). Antibiotic Modulation of Capsular Exopolysaccharide in Pelagicoccus enzymogenes sp. Nov. Isolated From Marine Sediment. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.655735
Fontaine S. S. and Kohl K. D. (2023). Ectotherm heat tolerance and the microbiome: Current understanding, future directions and potential applications. J. Exp. Biol. 226, jeb245761. doi: 10.1242/jeb.245761, PMID: 37313684
Fontaine S. S., Mineo P. M., and Kohl K. D. (2022). Experimental manipulation of microbiota reduces host thermal tolerance and fitness under heat stress in a vertebrate ectotherm. Nat. Ecol. Evol. 6. doi: 10.1038/s41559-022-01686-2, PMID: 35256809
Fraune S., Augustin R., Anton-Erxleben F., Wittlieb J., Gelhaus C., Klimovich V. B., et al. (2010). In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc. Natl. Acad. Sci. 107, 18067–18072. doi: 10.1073/pnas.1008573107, PMID: 20921390
Fraune S. and Bosch T. C. G. (2007). Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl. Acad. Sci. 104, 13146–13151. doi: 10.1073/pnas.0703375104, PMID: 17664430
Fu S., Fan H., Liu S., Liu Y., and Liu Z. (2009). A bioaugmentation failure caused by phage infection and weak biofilm formation ability. J. Environ. Sci. 21, 1153–1161. doi: 10.1016/S1001-0742(08)62396-7, PMID: 19862932
Fukui Y., Abe M., Kobayashi M., Saito H., Oikawa H., Yano Y., et al. (2013). Polaribacter porphyrae sp. Nov., isolated from the red alga Porphyra yezoensis, and emended descriptions of the genus Polaribacter and two Polaribacter species. Int. J. Systematic Evolutionary Microbiol. 63, 1665–1672. doi: 10.1099/ijs.0.041434-0, PMID: 22904227
Garrity G. M. and Holt J. G. (2001). “The Road Map to the Manual,” in Bergey’s Manual® of Systematic Bacteriology. Eds. Boone D. R., Castenholz R. W., and Garrity G. M. (New York City, New York: Springer New York), 119–166. doi: 10.1007/978-0-387-21609-6_15
Gauthier G., Gauthier M., and Christen R. (1995). Phylogenetic Analysis of the Genera Alteromonas, Shewanella, and Moritella Using Genes Coding for Small-Subunit rRNA Sequences and Division of the Genus Alteromonas into Two Genera, Alteromonas (Emended) and Pseudoalteromonas gen. Nov., and Proposal of Twelve New Species Combinations. Int. J. Systematic Evolutionary Microbiol. 45, 755–761. doi: 10.1099/00207713-45-4-755, PMID: 7547295
Givens C., Ransom B., Bano N., and Hollibaugh J. (2015). Comparison of the gut microbiomes of 12 bony fish and 3 shark species. Mar. Ecol. Prog. Ser. 518, 209–223. doi: 10.3354/meps11034
Gleason L. U. and Burton R. S. (2013). Phenotypic evidence for local adaptation to heat stress in the marine snail Chlorostoma (formerly Tegula) funebralis. J. Exp. Mar. Biol. Ecol. 448, 360–366. doi: 10.1016/j.jembe.2013.08.008
Gleason L. U. and Burton R. S. (2015). RNA-seq reveals regional differences in transcriptome response to heat stress in the marine snail Chlorostoma funebralis. Mol. Ecol. 24, 610–627. doi: 10.1111/mec.13047, PMID: 25524431
Gleason L. U. and Burton R. S. (2016a). Genomic evidence for ecological divergence against a background of population homogeneity in the marine snail Chlorostoma funebralis. Mol. Ecol. 25, 3557–3573. doi: 10.1111/mec.13703, PMID: 27199218
Gleason L. U. and Burton R. S. (2016b). Regional patterns of thermal stress and constitutive gene expression in the marine snail Chlorostoma funebralis in northern and southern California. Mar. Ecol. Prog. Ser. 556, 143–159. doi: 10.3354/meps11850
González-Aravena M., Urtubia R., Campo K. D., Lavín P., Wong C. M. V. L., Cárdenas C. A., et al. (2016). Antibiotic and metal resistance of cultivable bacteria in the Antarctic sea urchin. Antarctic Sci. 28, 261–268. doi: 10.1017/S0954102016000109
Grice E. A. (2015). The intersection of microbiome and host at the skin interface: Genomic- and metagenomic-based insights. Genome Res. 25, 1514–1520. doi: 10.1101/gr.191320.115, PMID: 26430162
Guo J. (2017). Metabarcoding Analyses of Gut Microbiome Compositions in Red Abalone (Haliotis Rufescens, Swainson 1822) Fed Different Macroalgal Diets. California State University, Monterey Bay. Available online at: https://digitalcommons.csumb.edu/caps_thes_all/229/ (Accessed July 10 2025).
Hector T. E., Hoang K. L., Li J., and King K. C. (2022). Symbiosis and host responses to heating. Trends Ecol. Evol. 37, 611–624. doi: 10.1016/j.tree.2022.03.011, PMID: 35491290
Hellberg M. E. (1998). Sympatric sea shells along the sea’s shore: the geography of speciation in the marine gastropod tegula. Evolution 52, 1311–1324. doi: 10.1111/j.1558-5646.1998.tb02013.x, PMID: 28565375
Hornung B. V. H., Zwittink R. D., and Kuijper E. J. (2019). Issues and current standards of controls in microbiome research. FEMS Microbiol. Ecol. 95, fiz045. doi: 10.1093/femsec/fiz045, PMID: 30997495
IPCC. (2022). Climate Change 2022: Impacts, Adaptation, and Vulnerability. Contribution of Working Group II to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. [Pörtner H.-O., Roberts D. C., Tignor M., Poloczanska E. S., Mintenbeck K., Alegría A., Craig M., Langsdorf S., Löschke S., Möller V., Okem A., and Rama B. (eds.)]. Cambridge, UK and New York, NY, USA: Cambridge University Press. 3056 p. doi: 10.1017/9781009325844
Ivanova E. P., Nedashkovskaya O. I., Sawabe T., Zhukova N. V., Frolova G. M., Nicolau D. V., et al. (2004). Shewanella affinis sp. Nov., isolated from marine invertebrates. Int. J. Systematic Evolutionary Microbiol. 54, 1089–1093. doi: 10.1099/ijs.0.02992-0, PMID: 15280274
Jiang Y., Jiao H., Sun P., Yin F., and Tang B. (2020). Metabolic response of Scapharca subcrenata to heat stress using GC/MS-based metabolomics. PeerJ 8, e8445. doi: 10.7717/peerj.8445, PMID: 32025378
Kers J. G. and Saccenti E. (2022). The power of microbiome studies: some considerations on which alpha and beta metrics to use and how to report results. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.796025, PMID: 35310396
Kloska A., Cech G. M., Sadowska M., Krause K., Szalewska-Pałasz A., and Olszewski P. (2020). Adaptation of the marine bacterium shewanella baltica to low temperature stress. Int. J. Mol. Sci. 21. doi: 10.3390/ijms21124338, PMID: 32570789
Kozich J. J., Westcott S. L., Baxter N. T., Highlander S. K., and Schloss P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13, PMID: 23793624
Li Y.-F., Chen Y.-W., Xu J.-K., Ding W.-Y., Shao A.-Q., Zhu Y.-T., et al. (2019a). Temperature elevation and Vibrio cyclitrophicus infection reduce the diversity of haemolymph microbiome of the mussel Mytilus coruscus. Sci. Rep. 9, 16391. doi: 10.1038/s41598-019-52752-y, PMID: 31704981
Li Y.-F., Xu J.-K., Chen Y.-W., Ding W.-Y., Shao A.-Q., Liang X., et al. (2019b). Characterization of gut microbiome in the mussel mytilus galloprovincialis in response to thermal stress. Front. Physiol. 10. doi: 10.3389/fphys.2019.01086, PMID: 31507449
Li S., Young T., Archer S., Lee K., Sharma S., and Alfaro A. C. (2022). Mapping the green-lipped mussel (Perna canaliculus) microbiome: A multi-tissue analysis of bacterial and fungal diversity. Curr. Microbiol. 79, 76. doi: 10.1007/s00284-021-02758-5, PMID: 35091849
Liu S.-B., Chen X.-L., He H.-L., Zhang X.-Y., Xie B.-B., Yu Y., et al. (2013). Structure and ecological roles of a novel exopolysaccharide from the arctic sea ice bacterium pseudoalteromonas sp. Strain SM20310. Appl. Environ. Microbiol. 79, 224–230. doi: 10.1128/AEM.01801-12, PMID: 23087043
Lo Giudice A. and Rizzo C. (2022). Bacteria associated with benthic invertebrates from extreme marine environments: promising but underexplored sources of biotechnologically relevant molecules. Mar. Drugs 20. doi: 10.3390/md20100617, PMID: 36286440
Lokmer A., Goedknegt M. A., Thieltges D. W., Fiorentino D., Kuenzel S., Baines J. F., et al. (2016a). Spatial and temporal dynamics of pacific oyster hemolymph microbiota across multiple scales. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01367, PMID: 27630625
Lokmer A., Kuenzel S., Baines J. F., and Wegner K. M. (2016b). The role of tissue-specific microbiota in initial establishment success of Pacific oysters. Environ. Microbiol. 18, 970–987. doi: 10.1111/1462-2920.13163, PMID: 26695476
Lynch J. B. and Hsiao E. Y. (2019). Microbiomes as sources of emergent host phenotypes. Sci. (New York N.Y.) 365, 1405–1409. doi: 10.1126/science.aay0240, PMID: 31604267
MacDonell M. T. and Colwell R. R. (1985). Phylogeny of the vibrionaceae, and recommendation for two new genera, listonella and shewanella. Systematic Appl. Microbiol. 6, 171–182. doi: 10.1016/S0723-2020(85)80051-5
Mazel F., Davis K. M., Loudon A., Kwong W. K., Groussin M., and Parfrey L. W. (2018). Is host filtering the main driver of phylosymbiosis across the tree of life? mSystems 3. doi: 10.1128/msystems.00097-18, PMID: 30417109
Miklós M., Cseri K., Laczkó L., Kardos G., Fraune S., and Tökölyi J. (2023). Environmental bacteria increase population growth of hydra at low temperature. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1294771, PMID: 38088971
Moeller A. H., Ivey K., Cornwall M. B., Herr K., Rede J., Taylor E. N., et al. (2020). The lizard gut microbiome changes with temperature and is associated with heat tolerance. Appl. Environ. Microbiol. 86, e01181–e01120. doi: 10.1128/AEM.01181-20, PMID: 32591376
Moghadam N. N., Thorshauge P. M., Kristensen T. N., de Jonge N., Bahrndorff S., Kjeldal H., et al. (2017). Strong responses of Drosophila melanogaster microbiota to developmental temperature. Fly 12, 1–12. doi: 10.1080/19336934.2017.1394558, PMID: 29095113
Morcillo R. J. L. and Manzanera M. (2021). The effects of plant-associated bacterial exopolysaccharides on plant abiotic stress tolerance. Metabolites 11, 337. doi: 10.3390/metabo11060337, PMID: 34074032
Mutalipassi M., Mazzella V., Romano G., Ruocco N., Costantini M., Glaviano F., et al. (2019). Growth and toxicity of Halomicronema metazoicum (Cyanoprokaryota, Cyanophyta) at different conditions of light, salinity and temperature. Biol. Open 8, bio043604. doi: 10.1242/bio.043604, PMID: 31615766
Nakagawa H., Shiozaki T., Kobatake E., Hosoya T., Moriya T., Sakai F., et al. (2016). Effects and mechanisms of prolongevity induced by Lactobacillus gasseri SBT2055 in Caenorhabditis elegans. Aging Cell 15, 227–236. doi: 10.1111/acel.12431, PMID: 26710940
Nash S., Johnstone J., and Rahman M. S. (2019). Elevated temperature attenuates ovarian functions and induces apoptosis and oxidative stress in the American oyster, Crassostrea virginica: Potential mechanisms and signaling pathways. Cell Stress Chaperones 24, 957–967. doi: 10.1007/s12192-019-01023-w, PMID: 31363994
Nash N., Klymasz-Swartz A. K., Nash M. T., Sachs M., Yoon G. R., and Weihrauch D. (2022). Impact of heatwaves and environmental ammonia on energy metabolism, nitrogen excretion, and mRNA expression of related genes in the indicator model system Daphnia magna. Aquat. Toxicol. 249, 106225. doi: 10.1016/j.aquatox.2022.106225, PMID: 35724523
Nash S. and Rahman M. S. (2019). Short-term heat stress impairs testicular functions in the American oyster, Crassostrea virginica: Molecular mechanisms and induction of oxidative stress and apoptosis in spermatogenic cells. Mol. Reprod. Dev. 86, 1444–1458. doi: 10.1002/mrd.23268, PMID: 31535424
Neu A. T., Allen E. E., and Roy K. (2019). Diversity and composition of intertidal gastropod microbiomes across a major marine biogeographic boundary. Environ. Microbiol. Rep. 11, 434–447. doi: 10.1111/1758-2229.12743, PMID: 30834681
Nichols C. A. M., Guezennec J., and Bowman J. P. (2005). Bacterial exopolysaccharides from extreme marine environments with special consideration of the southern ocean, sea ice, and deep-sea hydrothermal vents: A review. Mar. Biotechnol. 7, 253–271. doi: 10.1007/s10126-004-5118-2, PMID: 16075348
O’Connell L., Gao S., McCorquodale D., Fleisher J., and Lopez J. V. (2018). Fine grained compositional analysis of Port Everglades Inlet microbiome using high throughput DNA sequencing. PeerJ 6, e4671. doi: 10.7717/peerj.4671, PMID: 29761039
Offret C., Paulino S., Gauthier O., Château K., Bidault A., Corporeau C., et al. (2020). The marine intertidal zone shapes oyster and clam digestive bacterial microbiota. FEMS Microbiol. Ecol. 96, fiaa078. doi: 10.1093/femsec/fiaa078, PMID: 32353873
Oksanen J., Simpson G. L., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., et al. (2001). vegan: Community Ecology Package. 2.6–210. doi: 10.32614/CRAN.package.vegan
Ousley E. (2023). Investigating the relationship between the gut microbiome and host thermal tolerance in the economically important red abalone, Haliotis rufescens. California State University, Sacramento. Available online at: https://scholars.csus.edu/esploro/outputs/graduate/Investigating-the-relationship-between-the-gut/99258032962601671 (Accessed July 10, 2025).
Penn J. L. and Deutsch C. (2022). Avoiding ocean mass extinction from climate warming. Science 376, 524–526. doi: 10.1126/science.abe9039, PMID: 35482875
Pielou E. C. (1966). The measurement of diversity in different types of biological collections. J. Theor. Biol. 13, 131–144. doi: 10.1016/0022-5193(66)90013-0
Porras M. F., Navas C. A., Marden J. H., Mescher M. C., De Moraes C. M., Pincebourde S., et al. (2020). Enhanced heat tolerance of viral-infected aphids leads to niche expansion and reduced interspecific competition. Nat. Commun. 11, 1184. doi: 10.1038/s41467-020-14953-2, PMID: 32132537
Price D. K., West K., Cevallos-Zea M., Cahan S. H., Nunez J. C. B., Longman E. K., et al. (2025). Microbiome composition shapes temperature tolerance in a Hawaiian picture-winged Drosophila (bioRxiv). doi: 10.1101/2025.06.03.657679, PMID: 40501599
Quigley C. T. C., Capistrant-Fossa K. A., Morrison H. G., Johnson L. E., Morozov A., Hertzberg V. S., et al. (2020). Bacterial communities show algal host (Fucus spp.)/zone differentiation across the stress gradient of the intertidal zone. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.563118, PMID: 33072025
Rahman M. S. and Rahman M. S. (2021). Effects of elevated temperature on prooxidant-antioxidant homeostasis and redox status in the American oyster: Signaling pathways of cellular apoptosis during heat stress. Environ. Res. 196, 110428. doi: 10.1016/j.envres.2020.110428, PMID: 33186574
Sato L. M. (2001). Variation in the density, size structure, and reproductive characteristics of intertidal Tegula populations on Southern California rocky shores. California State University, Fullerton. Available online at: https://www.proquest.com/openview/e5c92c5f63a9557b3e0ee4098962d4a8/1?pq-origsite=gscholar&cbl=18750&diss=y (Accessed July 11, 2025).
Satomi M., Oikawa H., and Yano Y. (2003). Shewanella marinintestina sp. Nov., Shewanella schlegeliana sp. Nov. And Shewanella sairae sp. Nov., novel eicosapentaenoic-acid-producing marine bacteria isolated from sea-animal intestines. Int. J. Systematic Evolutionary Microbiol. 53, 491–499. doi: 10.1099/ijs.0.02392-0, PMID: 12710618
Scanes E., Siboni N., Rees B., and Seymour J. R. (2023). Acclimation in intertidal animals reduces potential pathogen load and increases survival following a heatwave. iScience 26. doi: 10.1016/j.isci.2023.106813, PMID: 37213223
Schmitt R. J. (1982). Consequences of dissimilar defenses against predation in a subtidal marine community. Ecology 63, 1588–1601. doi: 10.2307/1938882
Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60. doi: 10.1186/gb-2011-12-6-r60, PMID: 21702898
Strano F., Micaroni V., Thomas T., Woods L., Davy S. K., and Bell J. J. (2023). Marine heatwave conditions drive carryover effects in a temperate sponge microbiome and developmental performance. Proc. R. Soc. B: Biol. Sci. 290, 20222539. doi: 10.1098/rspb.2022.2539, PMID: 37282536
Taur Y. (2024). Ying14/yingtools2. Available online at: https://github.com/ying14/yingtools2 (Accessed August 28, 2024).
Tomanek L. (2002). The heat-shock response: its variation, regulation and ecological importance in intertidal gastropods (genus tegula)1. Integr. Comp. Biol. 42, 797–807. doi: 10.1093/icb/42.4.797, PMID: 21708778
Tomanek L. (2005). Two-dimensional gel analysis of the heat-shock response in marine snails(genus Tegula): Interspecific variation in protein expression and acclimation ability. J. Exp. Biol. 208, 3133–3143. doi: 10.1242/jeb.01748, PMID: 16081611
Tomanek L. and Sanford E. (2003). Heat-shock protein 70 (Hsp70) as a biochemical stress indicator: An experimental field test in two congeneric intertidal gastropods (genus: Tegula). Biol. Bull. 205, 276–284. doi: 10.2307/1543291, PMID: 14672982
Tomanek L. and Somero G. N. (1999). Evolutionary and acclimation-induced variation in the heat-shock responses of congeneric marine snails (genus Tegula) from different thermal habitats: Implications for limits of thermotolerance and biogeography. J. Exp. Biol. 202, 2925–2936. doi: 10.1242/jeb.202.21.2925, PMID: 10518474
Tomanek L. and Somero G. N. (2000). Time course and magnitude of synthesis of heat-shock proteins in congeneric marine snails (Genus tegula) from different tidal heights. Physiol. Biochem. Zoology: PBZ 73, 249–256. doi: 10.1086/316740, PMID: 10801403
Unzueta-Martínez A., Welch H., and Bowen J. L. (2022). Determining the composition of resident and transient members of the oyster microbiome. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.828692, PMID: 35185836
Vandeputte D., Tito R. Y., Vanleeuwen R., Falony G., and Raes J. (2017). Practical considerations for large-scale gut microbiome studies. FEMS Microbiol. Rev. 41, S154–S167. doi: 10.1093/femsre/fux027, PMID: 28830090
Vannini C., Pöckl M., Petroni G., Wu Q. L., Lang E., Stackebrandt E., et al. (2007). Endosymbiosis in statu nascendi: Close phylogenetic relationship between obligately endosymbiotic and obligately free-living Polynucleobacter strains (Betaproteobacteria). Environ. Microbiol. 9, 347–359. doi: 10.1111/j.1462-2920.2006.01144.x, PMID: 17222133
Venkateswaran K., Moser D. P., Dollhopf M. E., Lies D. P., Saffarini D. A., MacGregor B. J., et al. (1999). Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. Nov. Int. J. Systematic Evolutionary Microbiol. 49, 705–724. doi: 10.1099/00207713-49-2-705, PMID: 10319494
Wagh V. S., Said M. S., Bennale J. S., and Dastager S. G. (2022). Isolation and structural characterization of exopolysaccharide from marine Bacillus sp. And its optimization by Microbioreactor. Carbohydr. Polymers 285, 119241. doi: 10.1016/j.carbpol.2022.119241, PMID: 35287863
Wang J., Salem D. R., and Sani R. K. (2019). Extremophilic exopolysaccharides: A review and new perspectives on engineering strategies and applications. Carbohydr. Polymers 205, 8–26. doi: 10.1016/j.carbpol.2018.10.011, PMID: 30446151
Webster N. S. and Bourne D. (2007). Bacterial community structure associated with the Antarctic soft coral, Alcyonium antarcticum. FEMS Microbiol. Ecol. 59, 81–94. doi: 10.1111/j.1574-6941.2006.00195.x, PMID: 17233746
Xie B., Bishop S., Stessman D., Wright D., Spalding M. H., and Halverson L. J. (2013). Chlamydomonas reinhardtii thermal tolerance enhancement mediated by a mutualistic interaction with vitamin B12-producing bacteria. ISME J. 7, 1544–1555. doi: 10.1038/ismej.2013.43, PMID: 23486253
Xing P., Hahnke R. L., Unfried F., Markert S., Huang S., Barbeyron T., et al. (2015). Niches of two polysaccharide-degrading Polaribacter isolates from the North Sea during a spring diatom bloom. ISME J. 9, 1410–1422. doi: 10.1038/ismej.2014.225, PMID: 25478683
Yang Y., Ni J., Niu D., Zheng G., and Li Y. (2024). Physiological response of the razor clam Sinonovacula constricta exposed to hyposalinity stress. Aquaculture Fisheries 9, 663–673. doi: 10.1016/j.aaf.2022.11.002
Yoon J.-H., Kang S.-J., and Oh T.-K. (2006). Polaribacter dokdonensis sp. Nov., isolated from seawater. Int. J. Systematic Evolutionary Microbiol. 56, 1251–1255. doi: 10.1099/ijs.0.63820-0, PMID: 16738100
Yoon J., Matsuo Y., Matsuda S., Adachi K., Kasai H., and Yokota A. (2007a). Rubritalea spongiae sp. Nov. And Rubritalea tangerina sp. Nov., two carotenoid- and squalene-producing marine bacteria of the family Verrucomicrobiaceae within the phylum ‘Verrucomicrobia’, isolated from marine animals. Int. J. Systematic Evolutionary Microbiol. 57, 2337–2343. doi: 10.1099/ijs.0.65243-0, PMID: 17911307
Yoon J., Oku N., Matsuda S., Kasai H., and Yokota A. (2007b). Pelagicoccus croceus sp. Nov., a novel marine member of the family Puniceicoccaceae within the phylum ‘Verrucomicrobia’ isolated from seagrass. Int. J. Systematic Evolutionary Microbiol. 57, 2874–2880. doi: 10.1099/ijs.0.65286-0, PMID: 18048742
Yoon J., Oku N., Park S., Katsuta A., and Kasai H. (2012). Tunicatimonas pelagia gen. Nov., sp. Nov., a novel representative of the family Flammeovirgaceae isolated from a sea anemone by the differential growth screening method. Antonie Van Leeuwenhoek 101, 133–140. doi: 10.1007/s10482-011-9626-6, PMID: 21789597
Yoon J., Yasumoto-Hirose M., Matsuo Y., Nozawa M., Matsuda S., Kasai H., et al. (2007c). Pelagicoccus mobilis gen. Nov., sp. Nov., Pelagicoccus albus sp. Nov. And Pelagicoccus litoralis sp. Nov., three novel members of subdivision 4 within the phylum “Verrucomicrobia”, isolated from seawater by in situ cultivation. Int. J. Systematic Evolutionary Microbiol. 57, 1377–1385. doi: 10.1099/ijs.0.64970-0, PMID: 17625161
Zhai X., Zhang Y., Zhou J., Li H., Wang A., and Liu L. (2024). Physiological and microbiome adaptation of coral Turbinaria peltata in response to marine heatwaves. Ecol. Evol. 14, e10869. doi: 10.1002/ece3.10869, PMID: 38322002
Zhang T.-X., Li M.-R., Liu C., Wang S.-P., and Yan Z.-G. (2023). A review of the toxic effects of ammonia on invertebrates in aquatic environments. Environ. pollut. 336, 122374. doi: 10.1016/j.envpol.2023.122374, PMID: 37634564
Zhao J.-S., Deng Y., Manno D., and Hawari J. (2010). Shewanella spp. Genomic evolution for a cold marine lifestyle and in-situ explosive biodegradation. PloS One 5, e9109. doi: 10.1371/journal.pone.0009109, PMID: 20174598
Zhong K. X., Chan A. M., Collicutt B., Daspe M., Finke J. F., Foss M., et al. (2024). The prokaryotic and eukaryotic microbiome of Pacific oyster spat is shaped by ocean warming but not acidification. Appl. Environ. Microbiol. 90, e00052–e00024. doi: 10.1128/aem.00052-24, PMID: 38466091
Zhu Y.-T., Liang X., Liu T.-T., Power D. M., Li Y.-F., and Yang J.-L. (2024). The mussel larvae microbiome changes in response to a temperature rise. Front. Mar. Sci. 11. doi: 10.3389/fmars.2024.1367608
Zhu Y.-J., Liao M.-L., Ding M.-W., Wang Z.-K., and Dong Y.-W. (2021). Compositional and functional features of the gut microbiota of the intertidal snail Nerita yoldii along China’s coast. J. Mar. Biol. Assoc. United Kingdom 101, 1103–1110. doi: 10.1017/S0025315422000157
Keywords: microbiome, heat stress, 16S sequencing, marine mollusk, Tegula eiseni, Tegula funebralis, Tegula gallina
Citation: Applegate B, Burkhart M, Caddow H, Gover B, Kantola M-F, Gaetos Obenchain J, Smith A, Nash B, Enke RA and Gleason LU (2025) Investigating bacterial contributions to thermal tolerance in three intertidal marine snail Tegula species. Front. Mar. Sci. 12:1645537. doi: 10.3389/fmars.2025.1645537
Received: 11 June 2025; Accepted: 11 August 2025;
Published: 29 August 2025.
Edited by:
Taewoo Ryu, Okinawa Institute of Science and Technology Graduate University, JapanReviewed by:
Ramadoss Dineshram, Council of Scientific and Industrial Research (CSIR), IndiaDang Ha Quyen vu, Nha Trang University, Vietnam
Copyright © 2025 Applegate, Burkhart, Caddow, Gover, Kantola, Gaetos Obenchain, Smith, Nash, Enke and Gleason. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lani U. Gleason, bGFuaS5nbGVhc29uQGNzdXMuZWR1
 Brian Applegate1
Brian Applegate1 Ray A. Enke
Ray A. Enke Lani U. Gleason
Lani U. Gleason