- 1 School of Science, Monash University, Bandar Sunway, Selangor, Malaysia
- 2 School of Biomedical Science, Taylor's University College, Subang Jaya, Selangor, Malaysia
- 3 Department of Microbiology and Immunology, University of Melbourne, Parkville, VIC, Australia
The detection of Legionella pneumophila in environmental and clinical samples is frequently performed by PCR amplification of the mip and/or 16S rRNA genes. Combined with DNA sequencing, these two genetic loci can be used to distinguish different species of Legionella and identify L. pneumophila. However, the recent Legionella genome sequences have opened up hundreds of possibilities for the development of new molecular targets for detection and diagnosis. Ongoing comparative genomics has the potential to fine tune the identification of Legionella species and serogroups by combining specific and general genetic targets. For example, the coincident detection of LPS biosynthesis genes and virulence genes may allow the differentiation of both pathogen and serogroup without the need for nucleotide sequencing. We tested this idea using data derived from a previous genomic subtractive hybridization we performed between L. pneumophila serogroup 1 and L. micdadei. Although not yet formally tested, these targets serve as an example of how comparative genomics has the potential to improve the scope and accuracy of Legionella molecular detection if embraced by laboratories undertaking Legionella surveillance.
Introduction
Bacteria of the genus Legionella are ubiquitous in soil and water environments where they persist and multiply in free living protozoa. Despite the pathogen being adapted for an environmental niche, humans may become infected with Legionella through the inhalation of contaminated aerosols. The ability of Legionella to replicate in environmental protozoa has equipped the bacteria with the capacity to replicate in human alveolar macrophages (Newton et al., 2010). Intracellular replication requires a specialized type IV secretion system termed the defective in organelle trafficking/intracellular multiplication (Dot/Icm) system. Although ancestrally related to DNA conjugation systems, the Dot/Icm system transports multiple effector proteins into the host cell to establish a vacuole that evades lysosome fusion and interacts instead with membranes and vesicles of the secretory pathway (Shin and Roy, 2008; Franco et al., 2009; Isberg et al., 2009; Nora et al., 2009; Newton et al., 2010). The formation of the specialized Legionella containing vacuole (LCV) is critical for pathogen replication and spread to new host cells.
Although several Legionella species have been associated with human infection, Legionella pneumophila is the most common cause of Legionnaire's disease (Fields et al., 2002). In particular, serogroup 1 isolates of L. pneumophila represent the majority of clinical strains and many diagnostic tests are specific for the detection and diagnosis of this serogroup (Yu et al., 2002; Tronel and Hartemann, 2009). The difficulty of culturing of Legionella isolates from clinical and environmental samples has led to the development of rapid molecular tests for the detection of Legionella DNA (Rantakokko-Jalava and Jalava, 2001; Reischl et al., 2002; Templeton et al., 2003; Wilson et al., 2003; Bencini et al., 2007; Wilson et al., 2007). The current gold standard in molecular diagnosis is based on detection of the mip gene specific for L. pneumophila and 16S rRNA for identification of the Legionella genus (Ratcliff et al., 1998; Templeton et al., 2003). The mip gene was one of the first genes associated with the ability of L. pneumophila to replicate in eukaryotic cells and encodes a surface located peptidylprolyl cis/trans isomerase (PPIase) (Cianciotto et al., 1990; Cianciotto and Fields, 1992; Fischer et al., 1992; Wintermeyer et al., 1995). The 24 kDa mip product shares amino acid sequence similarity and is a structural mimic of the mainly eukaryotic family of FK-506 binding proteins, a class of immunophilins (Fischer et al., 1992; Hacker and Fischer, 1993; Riboldi-Tunnicliffe et al., 2001).
The limitation of using mip and 16S rRNA for molecular detection or the 23S–5S rRNA gene spacer region is that without nucleotide sequencing or other post-PCR analysis, these targets cannot distinguish serogroup 1 L. pneumophila from other serogroups and/or cannot detect non-pneumophila species of Legionella (Maurin et al., 2010; Yang et al., 2010). Since other serogroups of L. pneumophila and other species, such as L. longbeachae cause a significant burden of disease in many parts of the world (Yu et al., 2002; Gobin et al., 2009), their diagnosis and detection should be incorporated into any new molecular test. Non-serogroup 1 L. pneumophila and other species are currently likely to be significantly underrepresented given the bias of available tests such as the urine antigen test to the detection of L. pneumophila serogroup 1 (Benin et al., 2002). Therefore, there is significant scope to expand and improve current testing for Legionella.
Can Legionella Genomics Inform Molecular Detection Methods?
The recent L. pneumophila serogroup 1 genome sequences are an invaluable resource for molecular epidemiology and analysis of L. pneumophila genetic diversity. The six available L. pneumophila genome sequences are all serogroup 1 human clinical isolates with worldwide distribution, and include endemic and epidemic strains (McDade et al., 1977; Jepras et al., 1985; Aurell et al., 2003; Nguyen et al., 2006; D'Auria et al., 2010; Schroeder et al., 2010). The core L. pneumophila genome contains many of the factors associated with the ability of the bacteria to replicate in eukaryotic cells but there is also great variability between strains (Cazalet et al., 2008). Comparative analysis of the L. pneumophila genomes has revealed a diverse species where 7–11% of the genes in each L. pneumophila isolate are strain specific (Gomez-Valero et al., 2009). The genome exhibits high plasticity which presumably reflects the ability of the pathogen to acquire new genetic factors that enhance environmental survival and bacterial replication in eukaryotic cells. Some of the diversity occurs among genes encoding Dot/Icm effectors, including those within the same family (Cazalet et al., 2008). Nevertheless, many elements of the L. pneumophila genome are highly conserved and these less variable factors may constitute useful targets for molecular detection and typing. In contrast to L. pneumophila, the L. longbeachae genome appears more highly conserved with few differences between strains and serotypes (Cazalet et al., 2010; Kozak et al., 2010).
One of the most striking features of the L. pneumophila genome is the number and type of genes predicted to encode products that share similarity with eukaryotic proteins (Cazalet et al., 2004; Gomez-Valero et al., 2009; Lomma et al., 2009; Schroeder et al., 2010). For example, L. pneumophila produces two enzymes that belong to the mammalian CD39 family of ecto-nucleoside triphosphate diphosphohydrolases (NTPDases) (Sansom et al., 2007; Galka et al., 2008). NTPDases are associated almost exclusively with eukaryotes and so the L. pneumophila proteins, Lpg1905, and Lpg0971 are likely to contribute to the way the bacteria interact with eukaryotic cells by mimicking eukaryotic NTPDases. Indeed, we showed recently that the bacterial protein is a conserved structural mimic of mammalian NTPDases (Vivian et al., 2010) and that Lpg1905 enhances L. pneumophila intracellular replication (Sansom et al., 2007; Sansom et al., 2008). Virulence genes of L. pneumophila such as the eukaryotic type effectors or even genes of the Dot/Icm type IV secretion system have not yet been used widely as targets for molecular detection. However, since much of the detailed genetic information on L. pneumophila has only been obtained in recent years, the field of molecular diagnostics and detection is perhaps yet to capitalize on the usefulness of this information to inform molecular testing. Given the limited scope of the current PCR based tests, we believe that a knowledge of Legionella genomics could be used to improve rapid molecular detection of Legionella in environmental and clinical samples.
Development of New Generation Molecular Tests: An Example in Progress
Although the identification of Legionella species and serogroup is important for clinical and environmental management as well as epidemiological analysis, few rapid molecular tests can differentiate isolates of L. pneumophila serogroup 1 from other serogroups as well as L. pneumophila from other Legionella species (Tronel and Hartemann, 2009). Prior to determination of the Legionella genome sequences, we identified genes that were specific to L. pneumophila by experimental genomic subtractive hybridization of L. pneumophila with L. micdadei (Newton et al., 2006; Sansom et al., 2007). Two targets emerged as potentially useful for discriminating Legionella species and serogroups based on the fact that one, lpg0774 (lpp0839) (Cazalet et al., 2008), was associated with the serogroup 1 LPS biosynthesis region and another, lpg1905 (lpp1880) encoding one of the L. pneumophila ecto-NTPDases (Sansom et al., 2007; Sansom et al., 2008), was specifically associated with L. pneumophila. We then assessed the suitability of these genes for the detection of L. pneumophila in clinical and environmental samples by designing a multiplex PCR to include lpg0774, lpg1905, and 16S rRNA. This multiplex PCR allowed the simultaneous identification of the genus Legionella, L. pneumophila, and serogroup 1 isolates of L. pneumophila when tested against a culture collection that comprised 36 strains of L. pneumophila of various serogroups and 20 non-pneumophila species (Figure 1 and not shown). The PCR was also effective when tested on a limited number of environmental and clinical samples (Figure 2). To our knowledge this is the first attempt to distinguish L. pneumophila from other Legionella species that does not rely on knowledge of the mip or 16S rRNA nucleotide sequence and that can identify serogroup 1 L. pneumophila without post-PCR analysis.
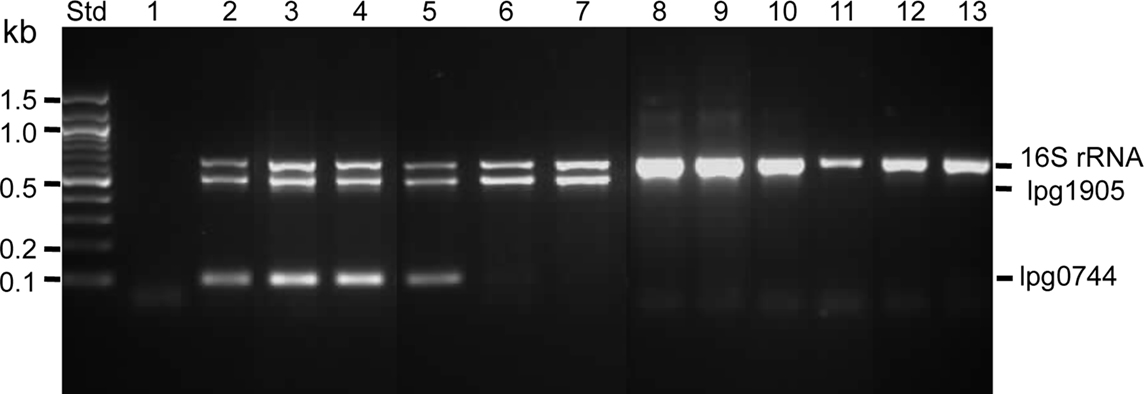
Figure 1. Representative gel showing simultaneous detection of lpg1905, lpg0774, and 16SrRNA by multiplex PCR. Std, 100 base pair marker (Promega); Lane 1, no DNA control; Lane 2, L. pneumophila serogroup 1 strain 02/41; Lane 3, L. pneumophila serogroup 1 strain B6; Lane 4, L. pneumophila serogroup 1 strain CS1; Lane 5, L. pneumophila serogroup 1 strain 02/41; Lane 6, L. pneumophila serogroup 2–14 strain C11(1); Lane 7, L. pneumophila serogroup 2–14 strain C4(1); Lane 8, L. gormanii strain C9; Lane 9, L. anisa strain L041; Lane 10, L. gormanii 03/69; 11, L. longbeachae A4C5; 12, L. longbeachae ATCC33462; 13, L. longbeachae Atlanta 5. For lpg0774 (Gene Bank: AY688227) the upstream primer started at base 46: 5′-TGCTAACAACCACTATCCCAAA-3′ and downstream primer started at base 202: 5′-GTTTCAATAAAAGCGTGCTCCT-3′. The upstream primer of lpg1905 (Gene Bank: NC_002942) started at base 328: 5′-TTGCCTAAAACTCACCACAGAA-3′ and downstream primer started at base 857: 5′-5′ATGCCGCCCAAAATATACC-3′. The 16S rRNA primers included in the triplex PCR to identify the genus Legionella have been described previously (Miyamoto et al., 1997). Triplex PCR was performed using 20 ng of template DNA in a 25 μL PCR reaction mix containing 1 × Green GoTaq® Flexi Buffer (Promega), 2 mM MgCl2, 200 μM dNTP, 0.5 μM of each primer and 1 U GoTaq® DNA polymerase. The optimized triplex PCR condition was performed in MyCycler™ (BIORAD) at initial denaturation of 95°C for 4 min followed by 35 cycles of 95°C for 1 min, 57.5°C for 1 min, and 72°C for 1 min with a final extension at 72°C for 5 min. The amplified products were then analyzed by DNA gel electrophoresis.
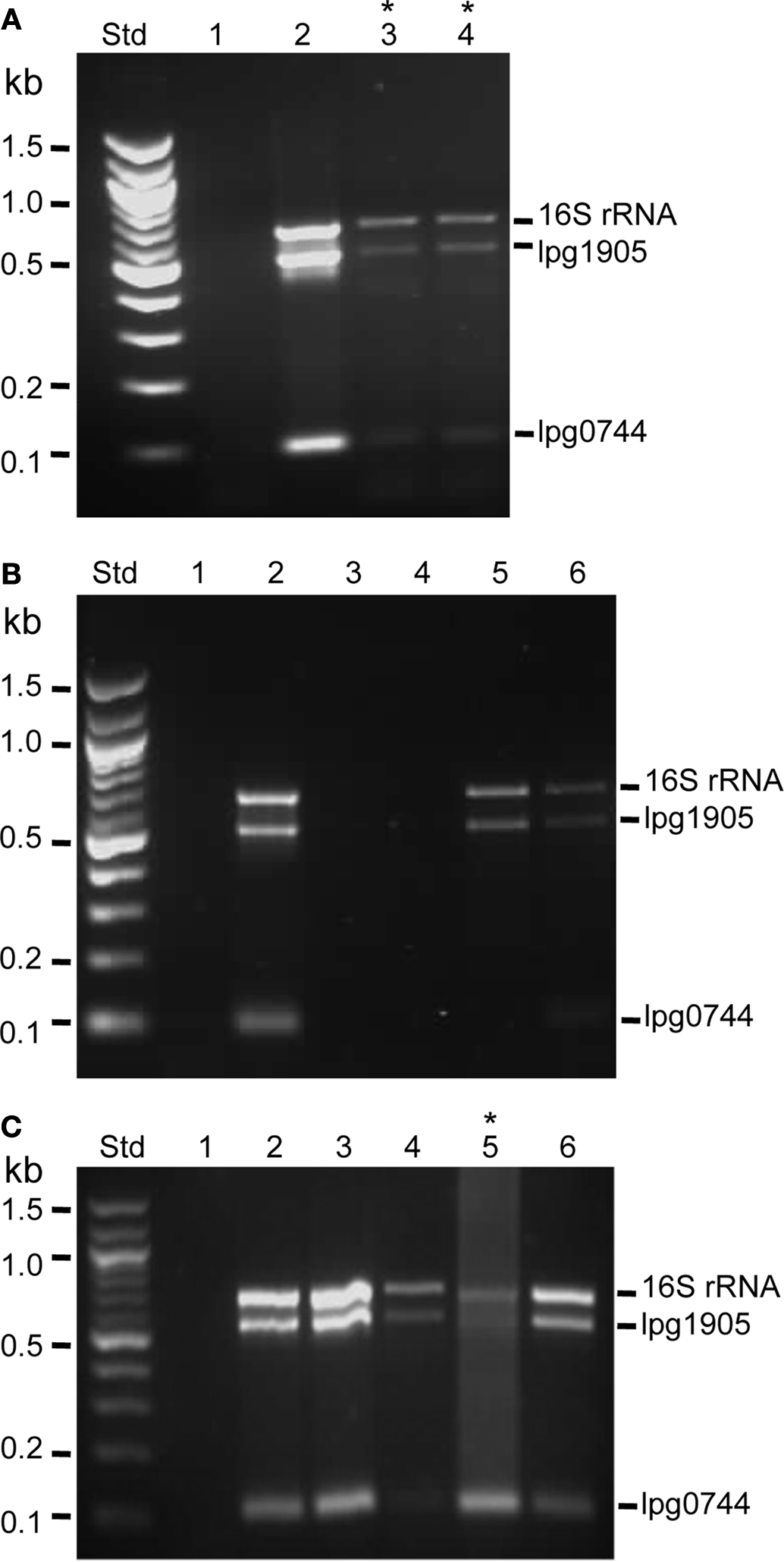
Figure 2. Detection of lpg1905, lpg0774, and 16SrRNA in environmental and clinical samples by multiplex PCR. (A) Detection of Legionella spp. in cooling towers at an office building. Std, 100 base pair marker (Promega); Lane 1, negative control (no DNA); Lane 2, L. pneumophila serogroup 1 strain 02/41; Lane 3, Cooling tower 1; Lane 4, Cooling tower 2. (B) Detection of Legionella spp. in cooling tower and shower head water samples collected from a hotel. Std, 100 base pair marker (Promega); Lane 1, negative control (no DNA); Lane 2, L. pneumophila serogroup 1 strain 02/41; Lane 3, Cooling tower surface water; Lane 4, Cooling tower sediment; Lane 5, Shower head 915; Lane 6, Shower head 1617. (C) Detection of Legionella spp. in patient samples. Std, 100 base pair marker (Promega); Lane 1, negative control (no DNA); Lane 2, L. pneumophila serogroup 1 strain 02/41; Lane 3, Sputum spiked with L. pneumophila serogroup 1 strain 02/41; Lane 4, Patient 1 (male, 22 years old), sputum sample; Lane 5, Patient 2 (male, 70 years old), bronchial wash; Lane 6, Patient 3 (male, 56 years old), sputum sample. In the above examples, 500 ml of cooling tower water was collected by immersing a sterilized 1000 ml bottle approximately 10 cm below water surface. To collect a showerhead sample, hot water was turned on for 5 min prior to collection of 50 ml of sample. Water samples were pressure filtered through a 0.45 μm cellulose nitrate membrane (Millipore), and eluted with 5 ml of sterile phosphate buffered saline (PBS pH 7.2). Following centrifugation (4,000 rpm, 20 min) the resulting sediment was resuspended in 2 ml sterile distilled water. Total DNA was extracted from 200 μl of the sediment suspension using the QIAamp DNA Mini Kit (Qaigen, Germany). A further 1 ml of sediment suspension was treated with 9 ml of HCl–KCl pH 2.2 (0.1 M Tris HCl, 0.1 M KCl) for 20 min then cultured onto BCYE agar containing GVPC selective supplement (Oxoid). 100 ml of the non-acid treated sediment suspension was also diluted 10-fold and cultured on BCYE-GVPC media. Total DNA was extracted from patient sputum and bronchial washes using QIAamp DNA blood mini kit (Qiagen). To liquefy viscose and sticky sputum samples, 40 μl of freshly prepared sputasol (0.75% [wt/vol]) (Oxoid) was added into 200 μl of sputum sample and the mixture was incubated at 37°C for 30 min (Bencini et al., 2007). The remaining 200 μl sputum or bronchial wash was cultured on BCYE media containing BMPA selective supplement (Oxoid). * indicates samples that were also positive for L. pneumophila by bacteriological culture.
What is the Scope for Future Molecular Detection of Legionella?
The recent advances in Legionella genomics offer the possibility to rethink the targets currently used for molecular detection and diagnostics. Analysis of the nucleotide sequence of the LPS biosynthesis regions for various serogroups could be adapted to the type of multiplexing described above and used to detect less common serogroups that have been associated with infection such as serogroup 3 (Chien et al., 2010). Additionally, with new knowledge of the genomes of other Legionella pathogens, such as L. longbeachae and L. micdadei, simple tests could be developed to identify multiple Legionella species using species-specific genetic targets. Targets drawn from comparative genomics will need to be validated against a large and diverse Legionella culture collection to ensure that they are as sensitive and accurate as possible. For example, although our own work suggested that lpg0774 was exclusively associated with serogroup 1 strains of L. pneumophila, a recent genome screen by microarray suggested that lpg0774 was present in 1 of 66 non-serogroup 1 strains of L. pneumophila (Cazalet et al., 2008). In addition lpg0774 was present in the majority but not all serogroup 1 L. pneumophila (128 of 150 isolates) (Cazalet et al., 2008). The multi-genome analysis performed by Cazalet et al. (2008) did suggest however that other LPS biosynthesis genes, lpg0766 (lpp0831), lpg0772 (lpp0837/wzm), and lpg0773 (lpp0873/wzt) may be useful markers of serogroup 1 strains of L. pneumophila as they were present in all serogroup 1 strains examined (150 isolates) and no non-serogroup 1 strains (66 isolates). Thus multiplex PCR using targets identified from comparative genomics, possibly combined with a recently described PCR based typing scheme that discriminates between monoclonal antibody subgroups of serogroup 1 strains (Thurmer et al., 2009), could significantly enhance our ability to detect and identify species and subgroups of Legionella rapidly and accurately. With an ever increasing knowledge of genomics and gene variation in Legionella, it is timely to update detection procedures to provide more precise and discriminatory testing for Legionella in clinical and environmental samples.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants awarded to Elizabeth L. Hartland from the Australian National Health and Medical Research Council (NHMRC) and Stacey F. Y. Yong from the Ministry of Science, Technology and Innovation, Malaysia. Elizabeth L. Hartland is supported by an Australian Research Council Future Fellowship. Hayley J. Newton and Fiona M. Sansom are supported by NHMRC training fellowships.
References
Aurell, H., Etienne, J., Forey, F., Reyrolle, M., Girardo, P., Farge, P., Decludt, B., Campese, C., Vandenesch, F., and Jarraud, S. (2003). Legionella pneumophila serogroup 1 strain Paris: endemic distribution throughout France. J. Clin. Microbiol. 41, 3320–3322.
Bencini, M. A., van den Brule, A. J., Claas, E. C., Hermans, M. H., Melchers, W. J., Noordhoek, G. T., Salimans, M. M., Schirm, J., Vink, C., van der Zee, A., and Jansen, R. (2007). Multicenter comparison of molecular methods for detection of Legionella spp. in sputum samples. J. Clin. Microbiol. 45, 3390–3392.
Benin, A. L., Benson, R. F., Arnold, K. E., Fiore, A. E., Cook, P. G., Williams, L. K., Fields, B., and Besser, R. E. (2002). An outbreak of travel-associated Legionnaires disease and Pontiac fever: the need for enhanced surveillance of travel-associated legionellosis in the United States. J. Infect. Dis. 185, 237–243.
Cazalet, C., Gomez-Valero, L., Rusniok, C., Lomma, M., Dervins-Ravault, D., Newton, H. J., Sansom, F. M., Jarraud, S., Zidane, N., Ma, L., Bouchier, C., Etienne, J., Hartland, E. L., and Buchrieser, C. (2010). Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires’ disease. PLoS Genet. 6, e1000851.
Cazalet, C., Jarraud, S., Ghavi-Helm, Y., Kunst, F., Glaser, P., Etienne, J., and Buchrieser, C. (2008). Multigenome analysis identifies a worldwide distributed epidemic Legionella pneumophila clone that emerged within a highly diverse species. Genome Res. 18, 431–441.
Cazalet, C., Rusniok, C., Bruggemann, H., Zidane, N., Magnier, A., Ma, L., Tichit, M., Jarraud, S., Bouchier, C., Vandenesch, F., Kunst, F., Etienne, J., Glaser, P., and Buchrieser, C. (2004). Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36, 1165–1173.
Chien, S. T., Hsueh, J. C., Lin, H. H., Shih, H. Y., Lee, T. M., Ben, R. J., Chou, S. T., Fong, C. M., Lin, Y. E., Tseng, L. R., and Chiang, C. S. (2010). Epidemiological investigation of a case of nosocomial Legionnaires’ disease in Taiwan: implications for routine environmental surveillance. Clin. Microbiol. Infect. 16, 761–763.
Cianciotto, N. P., Bangsborg, J. M., Eisenstein, B. I., and Engleberg, N. C. (1990). Identification of mip-like genes in the genus Legionella. Infect. Immun. 58, 2912–2918.
Cianciotto, N. P., and Fields, B. S. (1992). Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. U.S.A. 89, 5188–5191.
D'Auria, G., Jimenez-Hernandez, N., Peris-Bondia, F., Moya, A., and Latorre, A. (2010). Legionella pneumophila pangenome reveals strain-specific virulence factors. BMC Genomics 11, 181.
Fields, B. S., Benson, R. F., and Besser, R. E. (2002). Legionella and Legionnaires’ disease: 25 years of investigation. Clin. Microbiol. Rev. 15, 506–526.
Fischer, G., Bang, H., Ludwig, B., Mann, K., and Hacker, J. (1992). Mip protein of Legionella pneumophila exhibits peptidyl-prolyl-cis/trans isomerase (PPlase) activity. Mol. Microbiol. 6, 1375–1383.
Franco, I. S., Shuman, H. A., and Charpentier, X. (2009). The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell. Microbiol. 11, 1435–1443.
Galka, F., Wai, S. N., Kusch, H., Engelmann, S., Hecker, M., Schmeck, B., Hippenstiel, S., Uhlin, B. E., and Steinert, M. (2008). Proteomic characterization of the whole secretome of Legionella pneumophila and functional analysis of outer membrane vesicles. Infect. Immun. 76, 1825–1836.
Gobin, I., Newton, P. R., Hartland, E. L., and Newton, H. J. (2009). Infections caused by non-pneumophila species of Legionella. Rev. Med. Microbiol. 20, 1–11.
Gomez-Valero, L., Rusniok, C., and Buchrieser, C. (2009). Legionella pneumophila: population genetics, phylogeny and genomics. Infect. Genet. Evol. 9, 727–739.
Hacker, J., and Fischer, G. (1993). Immunophilins: structure-function relationship and possible role in microbial pathogenicity. Mol. Microbiol. 10, 445–456.
Isberg, R. R., O'Connor, T. J., and Heidtman, M. (2009). The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24.
Jepras, R. I., Fitzgeorge, R. B., and Baskerville, A. (1985). A comparison of virulence of two strains of Legionella pneumophila based on experimental aerosol infection of guinea-pigs. J. Hyg. (Lond.) 95, 29–38.
Kozak, N. A., Buss, M., Lucas, C. E., Frace, M., Govil, D., Travis, T., Olsen-Rasmussen, M., Benson, R. F., and Fields, B. S. (2010). Virulence factors encoded by Legionella longbeachae identified on the basis of the genome sequence analysis of clinical isolate D-4968. J. Bacteriol. 192, 1030–1044.
Lomma, M., Gomez-Valero, L., Rusniok, C., and Buchrieser, C. (2009). Legionella pneumophila – host interactions: insights gained from comparative genomics and cell biology. Genome Dyn. 6, 170–186.
Maurin, M., Hammer, L., Gestin, B., Timsit, J. F., Rogeaux, O., Delavena, F., Tous, J., Epaulard, O., Brion, J. P., and Croize, J. (2010). Quantitative real-time PCR tests for diagnostic and prognostic purposes in cases of legionellosis. Clin. Microbiol. Infect. 16, 379–384.
McDade, J. E., Shepard, C. C., Fraser, D. W., Tsai, T. R., Redus, M. A., and Dowdle, W. R. (1977). Legionnaires’ disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 297, 1197–1203.
Miyamoto, H., Yamamoto, H., Arima, K., Fujii, J., Maruta, K., Izu, K., Shiomori, T., and Yoshida, S. (1997). Development of a new seminested PCR method for detection of Legionella species and its application to surveillance of legionellae in hospital cooling tower water. Appl. Environ. Microbiol. 63, 2489–2494.
Newton, H. J., Ang, D. K., van Driel, I. R., and Hartland, E. L. (2010). Molecular pathogenesis of infections caused by Legionella pneumophila. Clin. Microbiol. Rev. 23, 274–298.
Newton, H. J., Sansom, F. M., Bennett-Wood, V., and Hartland, E. L. (2006). Identification of Legionella pneumophila-specific genes by genomic subtractive hybridization with Legionella micdadei and identification of lpnE, a gene required for efficient host cell entry. Infect. Immun. 74, 1683–1691.
Nguyen, T. M., Ilef, D., Jarraud, S., Rouil, L., Campese, C., Che, D., Haeghebaert, S., Ganiayre, F., Marcel, F., Etienne, J., and Desenclos, J. C. (2006). A community-wide outbreak of legionnaires disease linked to industrial cooling towers– how far can contaminated aerosols spread? J. Infect. Dis. 193, 102–111.
Nora, T., Lomma, M., Gomez-Valero, L., and Buchrieser, C. (2009). Molecular mimicry: an important virulence strategy employed by Legionella pneumophila to subvert host functions. Future Microbiol. 4, 691–701.
Rantakokko-Jalava, K., and Jalava, J. (2001). Development of conventional and real-time PCR assays for detection of Legionella DNA in respiratory specimens. J. Clin. Microbiol. 39, 2904–2910.
Ratcliff, R. M., Lanser, J. A., Manning, P. A., and Heuzenroeder, M. W. (1998). Sequence-based classification scheme for the genus Legionella targeting the mip gene. J. Clin. Microbiol. 36, 1560–1567.
Reischl, U., Linde, H. J., Lehn, N., Landt, O., Barratt, K., and Wellinghausen, N. (2002). Direct detection and differentiation of Legionella spp. and Legionella pneumophila in clinical specimens by dual-color real-time PCR and melting curve analysis. J. Clin. Microbiol. 40, 3814–3817.
Riboldi-Tunnicliffe, A., Konig, B., Jessen, S., Weiss, M. S., Rahfeld, J., Hacker, J., Fischer, G., and Hilgenfeld, R. (2001). Crystal structure of Mip, a prolylisomerase from Legionella pneumophila. Nat. Struct. Biol. 8, 779–783.
Sansom, F. M., Newton, H. J., Crikis, S., Cianciotto, N. P., Cowan, P. J., d'Apice, A. J., and Hartland, E. L. (2007). A bacterial ecto-triphosphate diphosphohydrolase similar to human CD39 is essential for intracellular multiplication of Legionella pneumophila. Cell. Microbiol. 9, 1922–1935.
Sansom, F. M., Riedmaier, P., Newton, H. J., Dunstone, M. A., Muller, C. E., Stephan, H., Byres, E., Beddoe, T., Rossjohn, J., Cowan, P. J., d'Apice, A. J., Robson, S. C., and Hartland, E. L. (2008). Enzymatic properties of an ecto-nucleoside triphosphate diphosphohydrolase from Legionella pneumophila: substrate specificity and requirement for virulence. J. Biol. Chem. 283, 12909–12918.
Schroeder, G. N., Petty, N. K., Mousnier, A., Harding, C. R., Vogrin, A. J., Wee, B., Fry, N. K., Harrison, T. G., Newton, H. J., Thomson, N. R., Beatson, S., Dougan, G., Hartland, E. L., and Frankel, G. (2010). Legionella pneumophila strain 130b possesses a unique combination of type IV secretion systems and novel Dot/Icm type IV secretion system effector proteins. J. Bacteriol. 192, 6001–6016.
Shin, S., and Roy, C. R. (2008). Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell. Microbiol. 10, 1209–1220.
Templeton, K. E., Scheltinga, S. A., Sillekens, P., Crielaard, J. W., van Dam, A. P., Goossens, H., and Claas, E. C. (2003). Development and clinical evaluation of an internally controlled, single-tube multiplex real-time PCR assay for detection of Legionella pneumophila and other Legionella species. J. Clin. Microbiol. 41, 4016–4021.
Thurmer, A., Helbig, J. H., Jacobs, E., and Luck, P. C. (2009). PCR-based “serotyping” of Legionella pneumophila. J. Med. Microbiol. 58, 588–595.
Tronel, H., and Hartemann, P. (2009). Overview of diagnostic and detection methods for legionellosis and Legionella spp. Lett. Appl. Microbiol. 48, 653–656.
Vivian, J. P., Riedmaier, P., Ge, H., Le Nours, J., Sansom, F. M., Wilce, M. C., Byres, E., Dias, M., Schmidberger, J. W., Cowan, P. J., d'Apice, A. J., Hartland, E. L., Rossjohn, J., and Beddoe, T. (2010). Crystal structure of a Legionella pneumophila ecto-triphosphate diphosphohydrolase, a structural and functional homolog of the eukaryotic NTPDases. Structure 18, 228–238.
Wilson, D. A., Reischl, U., Hall, G. S., and Procop, G. W. (2007). Use of partial 16S rRNA gene sequencing for identification of Legionella pneumophila and non-pneumophila Legionella spp. J. Clin. Microbiol. 45, 257–258.
Wilson, D. A., Yen-Lieberman, B., Reischl, U., Gordon, S. M., and Procop, G. W. (2003). Detection of Legionella pneumophila by real-time PCR for the mip gene. J. Clin. Microbiol. 41, 3327–3330.
Wintermeyer, E., Ludwig, B., Steinert, M., Schmidt, B., Fischer, G., and Hacker, J. (1995). Influence of site specifically altered Mip proteins on intracellular survival of Legionella pneumophila in eukaryotic cells. Infect. Immun. 63, 4576–4583.
Yang, G., Benson, R., Pelish, T., Brown, E., Winchell, J. M., and Fields, B. (2010). Dual detection of Legionella pneumophila and Legionella species by real-time PCR targeting the 23S-5S rRNA gene spacer region. Clin. Microbiol. Infect. 16, 255–261.
Yu, V. L., Plouffe, J. F., Pastoris, M. C., Stout, J. E., Schousboe, M., Widmer, A., Summersgill, J., File, T., Heath, C. M., Paterson, D. L., and Chereshsky, A. (2002). Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J. Infect. Dis. 186, 127–128.
Keywords: Legionella, virulence, genomics, molecular testing
Citation: Yong SFY, Tan SH, Wee J, Tee JJ, Sansom FM, Newton HJ and Hartland EL (2010) Molecular detection of Legionella: moving on from mip. Front. Microbio. 1:123. doi:10.3389/fmicb.2010.00123
Received: 06 September 2010;
Paper pending published: 28 September 2010;
Accepted: 21 October 2010;
Published online: 11 November 2010.
Edited by:
Amal Amer, The Ohio State University, USAReviewed by:
Zhao-Qing Luo, Purdue University, USA;Stephanie M. Seveau, The Ohio State University, USA;
Abul K. Azad, The Ohio State University, USA
Copyright: © 2010 Yong, Tan, Wee, Tee, Sansom, Newton and Hartland. This is an open-access article subject to an exclusive license agreement between the authors and the Frontiers Research Foundation, which permits unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited.
*Correspondence: Elizabeth L. Hartland, Department of Microbiology and Immunology, University of Melbourne, Parkville,VIC 3010, Australia. e-mail:aGFydGxhbmRAdW5pbWVsYi5lZHUuYXU=




 and
and