- Graduate School of Agriculture, Tokyo University of Agriculture and Technology, Tokyo, Japan
To characterize sulfate-producing microorganisms from the deteriorated sandstones of Angkor monuments in Cambodia, strains of Mycobacterium spp. were isolated from most probable number-positive cultures. All five strains isolated were able to use both elemental sulfur (S0) for chemolithoautotrophic growth and organic substances for chemoorganoheterotrophic growth. Results of phylogenetic and phenotypic analyses indicated that all five isolates were rapid growers of the genus Mycobacterium and were most similar to Mycobacterium cosmeticum and Mycobacterium pallens. Chemolithoautotrophic growth was further examined in the representative strain THI503. When grown in mineral salts medium, strain THI503 oxidized S0 to thiosulfate and sulfate; oxidation was accompanied by a decrease in the pH of the medium from 4.7 to 3.6. The link between sulfur oxidation and energy metabolism was confirmed by an increase in ATP. Fluorescence microscopy of DAPI-stained cells revealed that strain THI503 adheres to and proliferates on the surface of sulfur particles. The flexible metabolic ability of facultative chemolithoautotrophs enables their survival in nutrient-limited sandstone environments.
Introduction
Many stone monuments suffer from various kinds of degradation; biodeterioration caused by algae, cyanobacteria, bacteria, archaea, fungi, and lichens is among the most destructive (Warscheid and Braams, 2000; Scheerer et al., 2009). The ancient structures of Angkor, Cambodia, one of the most famous cultural heritage sites in the world are suffering from serious biodeterioration. For example, the Bayon temple, known for its intricately carved bas-reliefs, are being lost due to exfoliation and the formation of biofilms, which are microbial layers of various colors formed on the stone surface. Exfoliate weathering where sandstone flakes off in large, flat pieces, is also damaging historical cathedrals and churches in Europe (Hirsch et al., 1995).
Sulfur-oxidizing bacteria that excrete sulfuric acid are the dominant group of microorganisms involved in exfoliate weathering of sandstone at the Angkor site (Pochon and Jaton, 1967). However, sulfur-oxidizing bacteria have not been detected on the historic monuments in northern Europe. Therefore, they are considered to play a major role in stone decay only under warm and wet conditions (Warscheid and Braams, 2000), such as in Southeast Asia. The Angkor site has a tropical climate with high humidity. The density of sulfur-oxidizing microorganisms at Angkor, estimated annually using the most probable number (MPN) method, ranged from 101 to 105 MPN g−1 of deteriorated stone sample over a 10-year period. This data indicated their possible involvement of sulfur-oxidizing microorganisms in the deterioration of sandstone in this area (Li et al., 2008).
Sulfur-oxidizing fungal strains, including Fusarium solani strain THIF01, have been isolated from the MPN-positive cultures inoculated with the deteriorated sandstone collected at Angkor monuments. Strain THIF01 exhibited both chemolithoautotrophic sulfur-oxidizing activity and chemoorganoheterotrophic growth using organic substances (Li et al., 2010). Strain THIF01 was able to degrade carbonyl sulfide (COS), the most abundant sulfur compound in the atmosphere, suggesting that ambient COS could provide a source of sulfur for fungi colonizing on stone surfaces. Thus, COS may also be a source of sulfur for sulfur-oxidizing bacteria.
Sulfur-oxidizing ability is a characteristic of many phylogenetically diverse bacteria, including members of the phyla Aquificae, Chlorobi, Proteobacteria, and Firmicutes (Friedrich et al., 2001). Knowledge of this diversity continues to expand. For example, chemolithoautotrophic thiosulfate-oxidizing bacteria that were isolated from rhizosphere soils were identified as belonging to five genera not previously known to be sulfur-oxidizers (Anandham et al., 2008). Furthermore, the well-known soil bacterium Bradyrhizobium japonicum strain USDA110 recently demonstrated the ability to grow chemolithoautotrophically, using thiosulfate as an electron donor (Masuda et al., 2010). Thus, the genes required for sulfur-oxidizing ability may occur in bacteria and fungi that are not currently known to be sulfur-oxidizing microorganisms.
In the present study, five strains of sulfur-oxidizing bacteria of the genus Mycobacterium (phylum Actinobacteria) were isolated from MPN-positive cultures inoculated with samples from Angkor monuments. All five isolates were able to grow both chemolithoautotrophically using elemental sulfur (S0) and chemoorganoheterotrophically using organic substances. Autotrophic growth with hydrogen (Lukins and Foster, 1963; Park and Decicco, 1974) and the ability to use either methanol or carbon monoxide as a sole source of carbon and energy has been reported for several mycobacteria (Park et al., 2003). Structural genes encoding a novel form of ribulose 1,5-bisphosphate carboxylase/oxygenase were identified in Mycobacterium strain JC1 (Park et al., 2009). The role of assimilatory sulfur metabolism in the virulence, antibiotic resistance, and anti-oxidant defense of pathogenic Mycobacterium spp. has been examined (Schelle and Bertozzi, 2006). However, this is the first study to suggest a link between sulfur oxidation, energy production, and chemolithoautotrophic growth in the mycobacteria. The results have important implications for the role of sulfur-oxidizing bacteria in the deterioration of sandstone in Angkor site.
Materials and Methods
Sampling of Deteriorated Sandstone
Sandstone samples were collected from deteriorated walls and near-ground sections of pillars at Angkor Wat, Bayon, and Phnom Krom temples, Angkor, Cambodia during August 2003. Details of the sampling sites and the densities of sulfur-oxidizing microorganisms, enumerated by the MPN method, were previously reported (Li et al., 2008).
Isolation of Sulfur-oxidizing Bacteria
Sulfur-oxidizing bacteria were isolated from MPN-positive cultures, grown in a liquid WS5 medium (3.0 g L−1 KH2PO4, 0.2 g L−1 (NH4)2SO4, 0.5 g L−1 MgSO4 7H2O, 0.01 g L−1 FeSO4 7H2O, 0.25 g L−1 CaCl2 2H2O, 10 g L−1 S0, pH 5.0; Li et al., 2008). Oxidation of S0 was monitored by measuring the decrease in pH of the culture medium, due to the production of sulfuric acid, with a pH meter (Twin pH, Type B-212, Horiba, Kyoto, Japan). Sulfur-oxidizing bacteria from the most dilute MPN-positive cultures were isolated on agar plates of WS5 medium (Li et al., 2010) by serial streaking, and the colonies were then transferred to agar plates of PYG medium [2.0 g L−1 Polypepton (Nihon Pharmaceutical, Tokyo, Japan), 1.0 g L−1 Bacto yeast extract (Difco Laboratories, Detroit, MI, USA), 0.5 g L−1 glucose, pH 7.2] to which 15 g L−1 of Bacto agar (Difco Laboratories) was supplemented.
Morphological and Physiological Characterization
Morphological characteristics of the cultured bacteria were observed after Gram-staining. Catalase and oxidase activities were determined for all isolates. Catalase activity was tested in the presence of 3% hydrogen peroxide and oxidase activity was determined by a Pourmedia oxidase test (Eiken Chemical, Tokyo, Japan). To observe growth of strain THI503, cells were grown for 9 day in liquid medium of WS5 at 30°C with reciprocal shaking at 120 rpm, in the dark. The cells were stained with 4,6-diamidino-2-phenylindole (DAPI), and microphotographs were taken by single z-stack steps, using a BZ-8000 fluorescence microscope (KEYENCE, Osaka, Japan).
Phylogenetic Identification
For phylogenetic analysis, isolates were cultured for 3 day in liquid PYG medium at 30°C with reciprocal shaking at 120 rpm, and genomic DNA was isolated from the harvested cells according to the procedure described by Saito and Miura (1963). The DNA concentration was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The 16S rRNA gene was amplified with primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1525R (5′-AAAGGAGGTGATCCAGCC-3′; Lane, 1991), and then sequenced using a 3130x genetic analyzer (Applied Biosystems, CA, USA) as previously described (Li et al., 2010). Sequence data were compiled with the GENETYX-MAC/ATSQ program version 4.2.1 (GENETYX Corporation, Tokyo, Japan) and compared with sequences in the GenBank database using BLAST version 2.2.24 (Altschul et al., 1990). The data were then analyzed using the Clustal W Program version 1.83 (Thompson et al., 1994) with Kimura’s (1980)distance method. A multiple sequence alignment was performed, and bootstrapping analysis with MEGA version 4 (Tamura et al., 2007) was used to generate a neighbor-joining phylogenetic tree (Felsenstein, 1985), which was viewed with MEGA. The 16S rRNA gene sequences obtained have been deposited in the DNA Data Bank of Japan (DDBJ) under the following accession numbers: AB605019 for strain THI501, AB605020 for strain THI502, AB605021 for strain THI503, AB605022 for strain THI504, and AB605023 for strain THI505.
Oxidation of S0 by Strain THI503
Oxidation of S0 was examined by measuring concentrations of thiosulfate and sulfate ions, and the decrease of medium pH in sulfate-free WS5 (WS5-S) medium (Li et al., 2010). The sulfate salts in WS5 medium were replaced with chloride salts, and the production of sulfuric acid was measured. Strain THI503 was pre-incubated for 7 day in three 50-mL conical polypropylene tubes, each containing 15 mL of WS5-S medium, with reciprocal shaking at 120 rpm, at 30°C in the dark. The contents of the three tubes were then combined, and three 225-mL polypropylene conical tubes containing 50 mL of WS5-S medium were each inoculated with 5 mL of the pre-incubation culture. The tubes were incubated at 30°C with reciprocal shaking at 120 rpm in the dark. Medium that was not inoculated, and medium with strain THI503 but no S0, served as negative controls. pH and concentrations of thiosulfate and sulfate ions in the medium were measured periodically. Thiosulfate and sulfate concentrations were measured using an ion chromatograph (Model 861 Advanced Compact IC, Metrohm, Switzerland) as previously reported (Li et al., 2010). All experiments were done in triplicate.
Extraction and Measurement of ATP
To evaluate bacterial growth, ATP was measured by the bioluminescence method (Stanley and Williams, 1969). Strain THI503 was cultured as described in section Oxidation of S0 by strain THI503 except for using WS5 medium. ATP was extracted as reported (Rakotonirainy et al., 2003) and assayed using ATP assay mix (FLAAM, Sigma-Aldrich, MO, USA) and a LUMI-COUNTER 700 (Microtec, Chiba, Japan) at room temperature. ATP disodium salt hydrate (FLAAS, Sigma-Aldrich, MO, USA) was used to prepare the standard curve.
Degradation of COS by Strain THI503
Degradation of COS by strain THI503 was measured according to the procedure described by Kato et al. (2008). Strain THI503 was pre-incubated for 3 day in a test tube containing 10 mL of PYG medium, with reciprocal shaking at 120 rpm at 30°C in the dark. The pre-culture was then added to a 500-mL Sakaguchi flask containing 100 mL of PYG medium and incubated for 24 h. Cells were collected by centrifugation at 5,000 × g for 15 min, and then rinsed three times with autoclaved 25 mM phosphate buffer (PB, pH 7.0). The aqueous phase was discarded, and fresh PB was added to prepare the cell suspension. Volumes of 10 mL were dispensed into Pyrex test tubes (180 mm × 18 mm), which were sealed with butyl rubber caps. We then added 11.5 μL 10.5% COS gas with N2 as the balance gas (Nissan Tanaka, Saitama, Japan) to headspace to make a final COS mixing ratio of 25 parts per million by volume (ppmv). As a control, 10 mL of sterile PB was dispensed into test tubes. A gas-tight microsyringe was used to obtain 50 μL of headspace gas, which was injected into a gas chromatograph (GC-14B, Shimadzu, Kyoto, Japan) with a flame photometric detector and a glass column packed with Porapak QS (50-80 mesh, Waters Associates, MA, USA) as described previously (Katayama et al., 1992). Organic carbon was measured by using a wet oxidation-non-dispersive infrared gas analyzer (Seto and Tange, 1980).
Results
Isolation and Phylogenetic Analysis of Sulfur-Oxidizing Bacteria
Bacterial colonies on WS5 agar plates were around 0.1 mm in diameter, circular, entire, convex, and transparent. Five strains – THI501, THI502, THI503, THI504, and THI505 – were isolated from the deteriorated stone samples from the Angkor Wat, Bayon, and Phnom Krom temples (Table 1). On PYG agar medium, all isolates formed colonies within 3 day at 30°C in the dark that were approximately 1 mm diameter, circular, smooth, entire, and convex. Color of the colonies of strains THI501, THI504, and THI505 were pale yellow, and that of strains THI502 and THI503 were creamy white. All isolates were catalase positive and oxidase negative. Microscopic observation revealed that all isolates were Gram-positive rods and acid-fast positive.
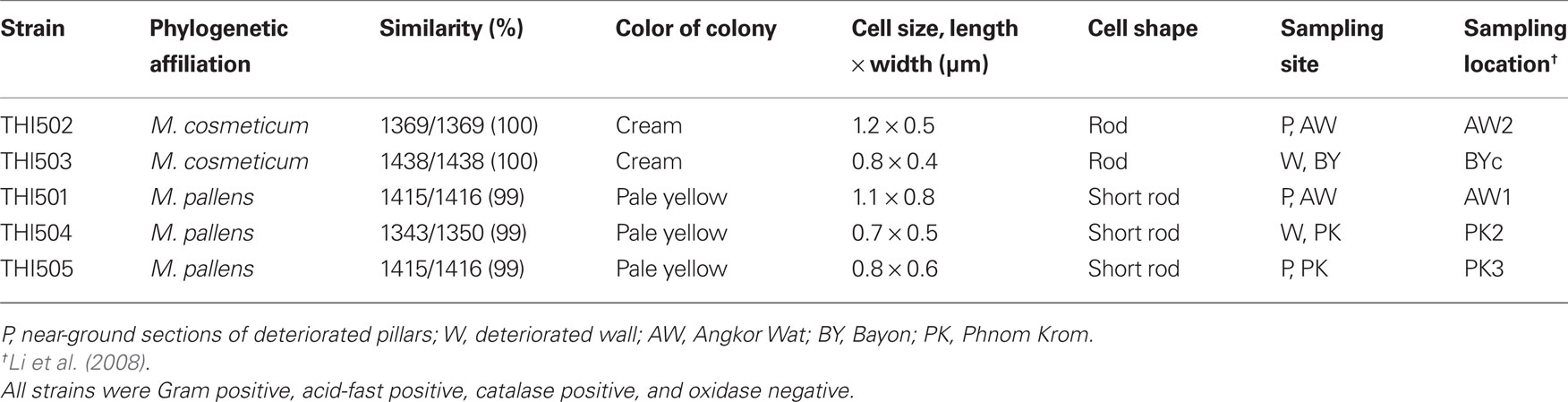
Table 1. Phylogenetic affiliation by 16S rRNA gene sequences and some phenotypic characteristics of the five sulfur-oxidizing isolates.
Phylogenetic analysis of 16S rRNA gene sequences showed that all isolates belonged to the rapid grower group of the genus Mycobacterium (Figure 1). Strains THI501, THI504, and THI505 were most closely related to Mycobacterium pallens (DQ370008; Hennessee et al., 2009), and strains THI502 and THI503 to Mycobacterium cosmeticum (AY449728; Cooksey et al., 2004). Because all isolates grew in both the mineral salts medium supplemented with S0 and the PYG medium, they were considered to be facultative chemolithoautotrophic bacteria.
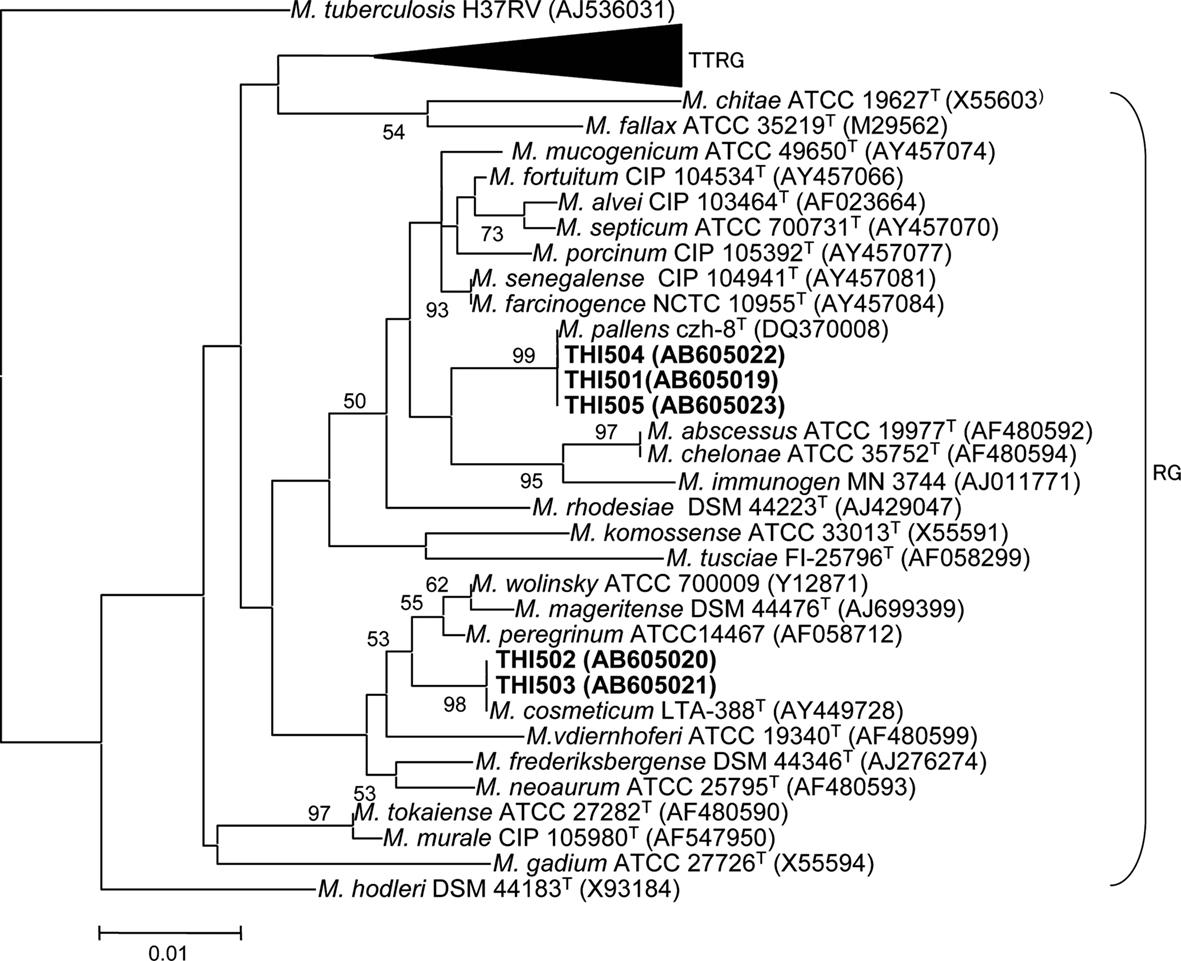
Figure 1. Phylogenetic relationship between the five sulfur-oxidizing isolates and other rapid growing Mycobacterium species based on a comparison of their 16S rRNA gene sequences. A slow-growing species, M. tuberculosis, was included as the outgroup. GenBank accession numbers are shown in parentheses. Numbers at the branch nodes are bootstrap values per 1000 trials; only values greater than 50% are shown. The scale bar corresponds to 1% estimated sequence divergence. TTRG, thermotolerant rapid growers; RG, rapid growers.
Sulfur-Oxidizing Ability of Strain THI503
Strain THI503 was selected for further analysis because it originated in a deteriorated sandstone wall in the Bayon temple where sulfur-oxidizing microbes were found every year through the observation (Li et al., 2008). Epifluorescence microscopy of DAPI-stained cells showed that strain THI503 cultivated in WS5 liquid medium grew on the surface S0 particles (Figures 2A,B). Planktonic cells of strain THI503 were much less abundant than attached cells (data not shown). Growth of cells adhering to solid surfaces, particularly to insoluble substrates, such as S0, have been reported for Thiobacillus thiooxidans (now Acidithiobacillus thiooxidans; Schaeffer et al., 1963), Ferrobacillus ferrooxidans and Thiobacillus ferrooxidans (now Acidithiobacillus ferrooxidans; Dugan and Lundgren, 1964; McGoran et al., 1969), and Sulfolobus acidocaldarius (Brock et al., 1972; Shivvers and Brock, 1973).
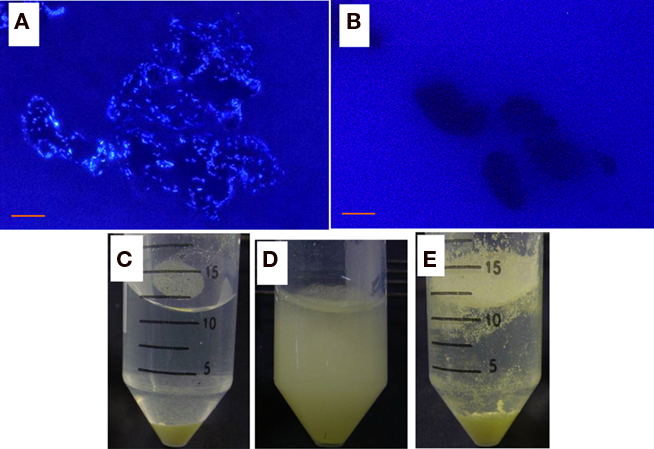
Figure 2. Growth of strain THI503 with S0. (A,B) Fluorescence microphotographs of WS5 liquid medium culture with S0 after 9 day. (A) with strain THI503 and (B) without strain THI503. Bar indicates 10 μm. (C,D,E) S0 in WS5 media after 7 day of incubation. (C,D) With or (E) without strain THI503. Cultures were allowed to stand for 5 min (C), followed by brief shaking (D). In the absence of strain THI503, some S0 remained floating on the surface of WS5 medium (E).
S0 is hydrophobic. When incubated with strain THI503 for 7 day, however, the sulfur became “wet” (Figure 2C) and easily diffused in the WS5 medium (Figure 2D), indicating that being covered with bacterial cells reduced its hydrophobicity. S0 in the non-inoculated tube remained floating on the medium, due to hydrophobicity (Figure 2E). Adherence of strain THI503 cells may promote solubilization of S0 and help the bacteria acquire nutrients (Knickerbocker et al., 2000). When strain THI503 was incubated in mineral salts medium with S0 as the sole energy source, the pH of the medium decreased from 4.7 to 3.6, and 1.1 ± 0.3 mol thiosulfate and 0.1 ± 0.1 mol sulfate accumulated in 50 mL culture after 22 day (Figure 3).
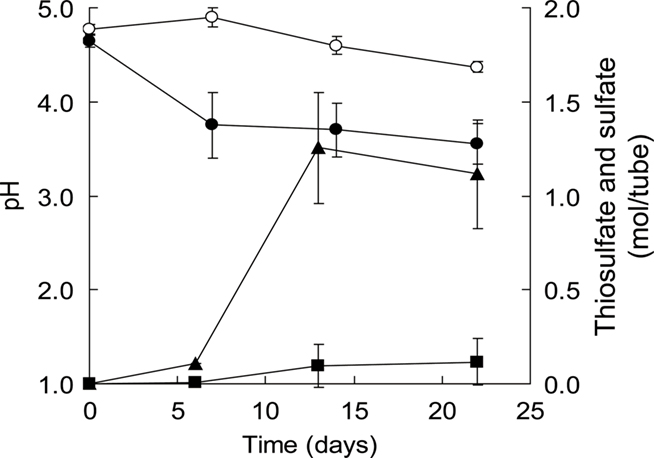
Figure 3. Changes in the medium pH and thiosulfate and sulfate ions during growth of strain THI503 in WS5-S medium with S0 as the sole energy source. Cells were grown in a 225-mL conical tube containing 50 mL culture. Error bars indicates SD, n = 3. Symbols: circles, pH; triangles, thiosulfate; square, sulfate, filled symbols, +THI503 + S0; empty symbols, −THI503 + S0.
ATP Measurement
Estimating the growth of strain THI503 in mineral salts medium with S0 after DAPI staining and epifluorescence microscopy was difficult, because solid particles of S0 impeded microscopic observation and the cells adhered so tightly to the surface of S0 that detaching them was problematic. Therefore, an increase in ATP was used to confirm growth in the mineral salts medium, assuming that the energy was produced through the oxidation of S0. ATP reached a maximum of 300 pmol in 50 mL culture after 12 day (Figure 4). Control culture without added sulfur reached only around half that ATP concentration; the increased ATP probably derived from ∼1 g L−1 S0 transferred from the pre-culture medium. By comparing to the reported data on the content of ATP in Mycobacterium bovis (1.014 fg per a colony-forming unit, CFU; Prioli et al., 1985), bacterial density in WS5-S medium was calculated to be 1.5 × 107 CFU tube−1 at the maximum of 300 pmol ATP (1.5 × 107 fg).
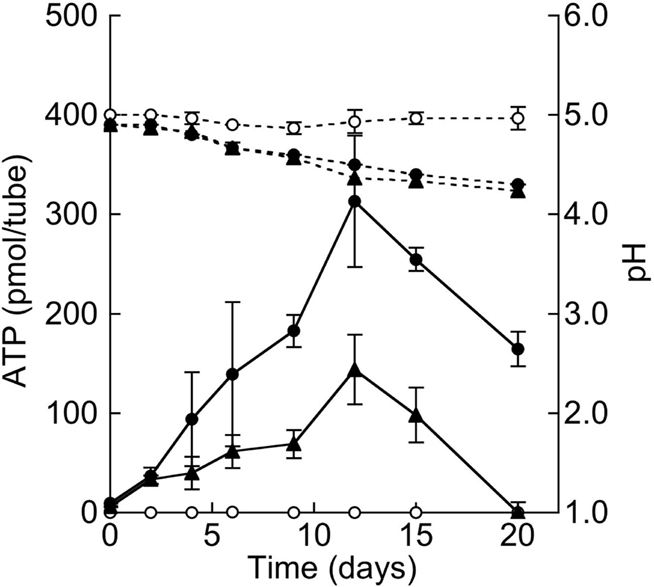
Figure 4. Growth of strain THI503 in WS5 medium with S0 as the sole energy source. Growth of strain THI503 was assessed based on ATP of the culture in a 225-mL conical tube containing 50 mL culture. Error bars indicate SD, n = 3. Symbols: solid line, ATP, dashed line, medium pH, filled circles, +THI503 + 10 g S0 L−1; triangles, +THI503 – S0 (final concentration of S0 was 1 g L−1 due to the carry-over from pre-culture); empty circles, −THI503 + 10 g S0 L−1.
Degradation of COS by Strain THI503
Degradation of COS by strain THI503 was analyzed to know of the possibility whether COS is one of the sources of sulfur for sulfuric acid in the deteriorated sandstone. COS in the headspace of the liquid cultures with no bacterial inoculation decreased from 24 ppmv (39 nmol) to 18 ppmv (30 nmol) during the 6.5-h incubation (Figure 5). The decrease indicated that COS gas dissolved into the liquid medium, because the solubility of COS in water is 1.15 g kg–1 (19 nmol mL–1; Yaws, 1999). In contrast, test tubes inoculated with strain THI503 showed a decrease in COS in the headspace from 25 ppmv (40 nmol) to 1 ppmv (2 nmol) in 6.5 h. Thus, COS degraded by strain THI503 was 29 nmol in 6.5 h. At the same time, total organic carbon increased to 1.86 ± 0.03 mg C in the inoculated tubes, while only 0.24 ± 0.04 mg C was found in the non-inoculated tube. Total organic carbon from strain THI503 was estimated to be 1.62 mg C. By comparing to the reported data on the carbon content of cell in Pseudomonas putida (1.1 × 10−13 to 3.1 × 10−13 g of carbon cell−1, Bratbak, 1985), bacterial cell in the test tube was calculated as 4.8 × 109 to 1.4 × 1010 cells and estimated COS degraded per cell per hour was estimated as 3.3 × 10−10 to 9.2 × 10−10 nmol COS cell−1 h−1. Figure 5 showed that 25 ppmv COS was degraded by strain THI503, however, at ambient atmospheric levels (500 pptv; Torres et al., 1980; Bandy et al., 1992; Chin and Davis, 1995), COS was not degraded by strain THI503 under experimental condition used (data not shown).
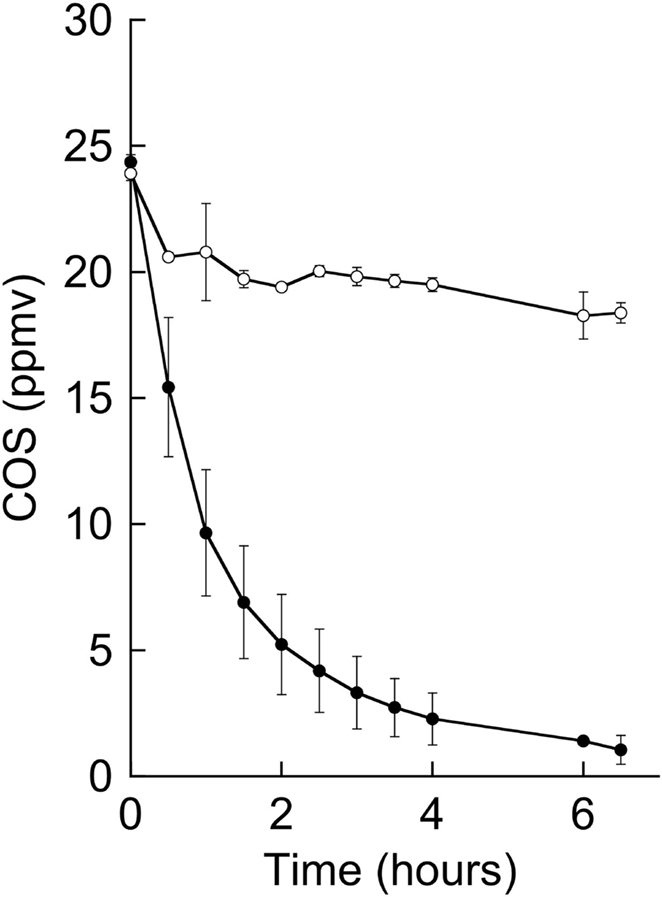
Figure 5. Degradation of 25 ppmv of COS by strain THI503. Cells were suspended in PB. Error bars indicate SD, n = 3. Symbols: Filled circles, +THI 503; empty circles, −THI503.
Discussion
Analysis of morphological and physiological characteristics indicated that all five strains isolated as sulfur-oxidizing bacteria were closely related to the genus Mycobacterium. Comparison of 16S rRNA gene sequences supported this conclusion (Figure 1; Table 1). All five strains isolated in the present study developed visible colonies within 7 day on PYG agar media, which is characteristic of rapid-growth species in the genus. Although, both sulfur-oxidizing bacteria and Mycobacterium spp. have been found by means of MPN and DGGE, respectively from corroded concrete sewer pipes (Vincke et al., 2001), no sulfur-oxidizing Mycobacterium had previously been cultured. The present study provides the first evidence of facultative chemolithoautotrophic ability of sulfur oxidation species of the genus Mycobacterium.
All five strains isolated in the present study were found to be facultative sulfur-oxidizing bacteria, able to grow in both mineral salts medium with S0 and chemoorganoheterotrophic PYG medium. Mycobacterium spp. are known to produce sulfated metabolites, such as sulfated glycolipids (Mougous et al., 2002); however, a link between sulfur oxidation and energy metabolism has not been reported. Sulfates were found in most of the samples of the salts and deteriorated sandstones taken from Angkor Wat, Bayon, and Phnom Krom temples. The primary source of sulfur in deteriorated sandstone was assumed to be bat dropping, based on stable isotope analysis (Hosono et al., 2006). Since low concentrations of sulfur compounds can limit survival and growth of sulfur-oxidizing bacteria (Graff and Stubner, 2003; Anandham et al., 2008), the ability to switch between chemolithotrophy and chemoorganotrophy, or to use sulfur compounds by co-oxidation in addition to organic substrates, would be advantageous in nutrient-limited environments such as sandstone surfaces. Strain THI503 degraded COS when it was provided at 25 ppmv, but not at 500 pptv under the experimental conditions employed here. Considering that Mycobacterium spp. isolated from soils (Kato et al., 2008) and sulfur-oxidizing fungi isolated from deteriorated sandstones in Angkor site (Li et al., 2010) degraded COS at both high (30 ppmv) and ambient (500 pptv) levels, it is unclear whether strain THI503 can use COS as a sulfur source under ambient conditions. Experiments to test the utilization of other reduced sulfur compounds such as sulfide, thiosulfate, sulfite, and tetrathionate by these Mycobacterium spp. isolates are in progress.
Although pathogenic species have been reported in the rapid-growth group of Mycobacterium (Brown-Elliott and Wallace, 2002), none of the isolates were closely related to those species. All five isolates fell into either M. pallens or M. cosmeticum by 16S rRNA sequences. M. pallens was isolated from a soil as a bacterium degrading polycyclic aromatic hydrocarbons (Hennessee et al., 2009); M. cosmeticum was isolated from a cosmetic and nail salon (Cooksey et al., 2004).
Strain THI503 achieved a density of around 1.5 × 107 CFU (50 mL culture)−1 in mineral salts medium with S0, with accumulations of 1.1 ± 0.3 mol thiosulfate and 0.1 ± 0.1 mol sulfate. The higher amount of thiosulfate suggests that the sulfate formed was rapidly used for biosynthesis in the sulfate-free medium, WS5-S. Although little is known about the oxidation of sulfur compounds in Gram-positive bacteria, activities of sulfur dioxygenase (SDO) and other relevant enzymes were reported for Sulfobacillus thermosulfidooxidans (Egorova et al., 2004; Rohwerder and Sand, 2007). In mesophilic Gram-negative sulfur-oxidizing bacteria, such as Acidithiobacillus ferrooxidans and Acidithiobacillus thiooxidans, S0 is thought to be oxidized by SDO, resulting in the formation of sulfite: 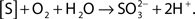 Much of the sulfite is further oxidized to sulfate by the sulfite:acceptor oxidoreductase (SAR). The sulfite can also react abiotically with excess S0 to produce thiosulfate. Another metabolic pathway of S0 found in archaea is the formation of thiosulfate by sulfur oxygenase reductase (SOR):
Much of the sulfite is further oxidized to sulfate by the sulfite:acceptor oxidoreductase (SAR). The sulfite can also react abiotically with excess S0 to produce thiosulfate. Another metabolic pathway of S0 found in archaea is the formation of thiosulfate by sulfur oxygenase reductase (SOR): 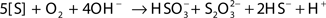 (Chen et al., 2005). SOR activity results in the concomitant formation of sulfite and sulfide; the sulfite can either react abiotically with excess S0 or be oxidized by SAR (Rohwerder and Sand, 2007). Sor genes have been identified in a novel mesophilic Acidithiobacillus sp. strain SM-1, as well as in archaea (Chen et al., 2007; Kletzin, 2008). Based on the higher accumulation of thiosulfate compared to sulfate, strain THI503 probably oxidizes S0 by the SOR pathway. The search for a gene relevant to sulfur oxidation by strain THI503 is in progress.
(Chen et al., 2005). SOR activity results in the concomitant formation of sulfite and sulfide; the sulfite can either react abiotically with excess S0 or be oxidized by SAR (Rohwerder and Sand, 2007). Sor genes have been identified in a novel mesophilic Acidithiobacillus sp. strain SM-1, as well as in archaea (Chen et al., 2007; Kletzin, 2008). Based on the higher accumulation of thiosulfate compared to sulfate, strain THI503 probably oxidizes S0 by the SOR pathway. The search for a gene relevant to sulfur oxidation by strain THI503 is in progress.
Conclusion
The MPN approach used to isolate the bacterial strains reported here indicated that the sulfur-oxidizing microorganisms occurred at 103–104 cells g–1 of deteriorated sandstone in the Angkor monuments. All five isolates were characterized as facultative chemolithoautotrophic sulfur-oxidizing bacteria by growth in both mineral salts medium with S0 and chemoorganoheterotrophic PYG medium. The isolates were most closely related to M. pallens or M. cosmeticum by 16S rRNA gene sequence comparisons. Thus, the present study provides the first evidence of facultative chemolithoautotrophic sulfur oxidation with S0 in the species of the genus Mycobacterium.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank APSARA Authority and Ministry on Environment, Kingdom of Cambodia, for permission to collect stone samples from monuments and transport to Japan, and Plant Protection Station of Japanese Ministry of Agriculture, Forestry, and Fisheries, for permission to import the stone samples. This research was supported by Japanese Government Team for Safeguarding Angkor and by a Grant-in-aid for Scientific Research (No.19251001) from the Ministry of Education, Culture, Sports, and Technology of Japan. Assistance of Hideo Arai, and Ichita Shimoda in sampling is appreciated.
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Anandham, R., Indiragandhi, P., Medhaiyan, M., Ryu, K. Y., Jee, H. J., and Sa, T. M. (2008). Chemolithoautotrophic oxidation of thiosulfate and phylogenetic distribution of sulfur oxidation gene (soxB) in rhizobacteria isolated from crop plants. Res. Microbiol. 159, 579–589.
Bandy, A. R., Thornton, D. C., Scott, D. L., Lalevic, M., Lewin, E. E., and Driedger, A. R. III. (1992). A time series for carbonyl sulfide in the northern hemisphere. J. Atmos. Chem. 14, 527–534.
Bratbak, G. (1985). Bacterial biovolume and biomass estimations. Appl. Environ. Microbiol. 49, 1488–1493.
Brock, T. D., Brock, K. M., Belly, R. T., and Weiss, R. L. (1972). Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Microbiol. 84, 54–68.
Brown-Elliott, B. A., and Wallace, R. J. Jr. (2002). Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15, 716–746.
Chen, Z.-W., Jiang, C.-Y., She, Q., Liu, S.-J., and Zhou, P.-J. (2005). Key role of cysteine residues in catalysis and subcellular localization of sulfur oxygenase-reductase of Acidianus tengchongensis. Appl. Environ. Microbiol. 71, 621–628.
Chen, Z.-W., Liu, Y.-Y., Wu, J.-F., She, Q., Jiang, C.-Y., and Liu, S.-J. (2007). Novel bacterial sulfur oxygenase reductases from bioreactors treating gold-bearing concentrates. Appl. Microbiol. Biotechnol. 74, 688–698.
Chin, M., and Davis, D. D. (1995). A reanalysis of carbonyl sulfide as a source of stratospheric background sulfur aerosol. J. Geophys. Res. 100, 8993–9005.
Cooksey, R. C., de Waard, J. H., Yakrus, M. A., Rivera, I., Chopite, M., Toney, S. R., Morlock, G. P., and Butler, W. R. (2004). Mycobacterium cosmeticum sp. nov., a novel rapidly growing species isolated from a cosmetic infection and from a nail salon. Int. J. Syst. Evol. Microbiol. 54, 2385–2391.
Dugan, P. R., and Lundgren, D. G. (1964). Acid production by Ferrobacillus ferrooxidans and its relation to water pollution. Develop. Ind. Microbiol. 5, 250–257.
Egorova, M. A., Tsaplina, I. A., Zakharchuk, L. M., Bogdanova, T. I., and Krasil’nikova, E. N. (2004). Effect of cultivation conditions on the growth and activities of sulfur metabolism enzymes and carboxylases of Sulfobacillus thermosulfidooxidans subsp. asporogenes strain 41. Appl. Biochem. Microbiol. 40, 381–387.
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791.
Friedrich, C. G., Rother, D., Bardischewsky, F., Quentmeier, A., and Fischer, J. (2001). Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. Environ. Microbiol. 67, 2873–2882.
Graff, A., and Stubner, S. (2003). Isolation and molecular characterization of thiosulfate-oxidizing bacteria from an Italian rice field soil. System. Appl. Microbiol. 26, 445–452.
Hennessee, C. T., Seo, J.-S., Alvarez, A. M., and Li, Q. X. (2009). Polycyclic aromatic hydrocarbon-degrading species isolated from Hawaiian soils: Mycobacterium crocinum sp. nov., Mycobacterium pallens sp. nov., Mycobacterium rutilum sp. nov., Mycobacterium rufum sp. nov. and Mycobacterium aromaticivorans sp. nov. Int. J. Syst. Evol. Microbiol. 59, 378–387.
Hirsch, P., Eckhardt, F. E. W., and Palmer, R. J. Jr. (1995). Methods for the study of rock-inhabiting microorganisms – a mini review. J. Microbiol. Meth. 23, 143–167.
Hosono, T., Uchida, E., Suda, C., Ueno, A., and Nakagawa, T. (2006). Salt weathering of sandstone at the Angkor monuments, Cambodia: identification of the origins of salts using sulfur and strontium isotopes. J. Archaeol. Sci. 33, 1541–1551.
Katayama, Y., Narahara, Y., Inoue, Y., Amano, F., Kanagawa, T., and Kuraishi, H. (1992). A thiocyanate hydrolase of Thiobacillus thioparus. A novel enzyme catalyzing the formation of carbonyl sulfide from thiocyanate. J. Biol. Chem. 267, 9170–9175.
Kato, H., Saito, M., Nagahata, Y., and Katayama, Y. (2008). Degradation of ambient carbonyl sulfide by Mycobacterium spp. in soil. Microbiology 154, 249–255.
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120.
Kletzin, A. (2008). “Oxidation of sulfur and inorganic sulfur compounds in Acidianus ambivalens,” in Microbial Sulfur Metabolism, eds C. Dahl and C. G. Friedrich (Berlin: Springer), 184–201.
Knickerbocker, C., Nordstrom, D. K., and Southam, G. (2000). The role of “blebbing” in overcoming the hydrophobic barrier during biooxidation of elemental sulfur by Thiobacillus thiooxidans. Chem. Geol. 169, 425–433.
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (New York: John Wiley & Sons), 115–175.
Li, X., Arai, H., Shimoda, I., Kuraishi, H., and Katayama, Y. (2008). Enumeration of sulfur-oxidizing microorganisms on deteriorating stone of the Angkor monuments, Cambodia. Microbes Environ. 23, 293–298.
Li, X. S., Sato, T., Ooiwa, Y., Kusumi, A., Gu, J.-D., and Katayama, Y. (2010). Oxidation of elemental sulfur by Fusarium solani strain THIF01 harboring endobacterium Bradyrhizobium sp. Microb. Ecol. 60, 96–104.
Lukins, H. B., and Foster, J. W. (1963). Utilization of hydrocarbons and hydrogen by mycobacteria. Z. Allg. Mikrobiol. 3, 251–264.
Masuda, S., Eda, S., Ikeda, S., Mitsui, H., and Minamisawa, K. (2010). Thiosulfate-dependent chemolithoautotrophic growth of Bradyrhizobium japonicum. Appl. Environ. Microbiol. 76, 2402–2409.
McGoran, C. J. M., Duncan, D. W., and Walden, C. C. (1969). Growth of Thiobacillus ferrooxidans on various substrates. Can. J. Microbiol. 15, 135–138.
Mougous, J. D., Leavell, M. D., Senaratne, R. H., Leigh, C. D., Williams, S. J., Riley, L. W., Leary, J. A., and Bertozzi, C. R. (2002). Discovery of sulfated metabolites in mycobacteria with a genetic and mass spectrometric approach. Proc. Natl. Acad. Sci. U.S.A. 99, 17037–17042.
Park, S. S., and Decicco, B. T. (1974). Autotrophic growth with hydrogen of Mycobacterium gordonae and another scotochromogenic mycobacterium. Int. J. Syst. Bacteriol. 24, 338–345.
Park, S. W., Hwang, E. H., Jang, H. S., Lee, J. H., Kang, B. S., Oh, J. I., and Kim, Y. M. (2009). Presence of duplicate genes encoding a phylogenetically new subgroup of form I ribulose 1,5-bisphosphate carboxylase/oxygenase in Mycobacterium sp. strain JC1 DSM 3803. Res. Microbiol. 160, 159–165.
Park, S. W., Hwang, E. H., Park, H., Kim, J. A., Heo, J., Lee, K. H., Song, T., Kim, E., Ro, Y. T., Kim, S. W., and Kim, Y. M. (2003). Growth of mycobacteria on carbon monoxide and methanol. J. Bacteriol. 185, 142–147.
Pochon, J., and Jaton, C. (1967). The role of microbiological agencies in the deterioration of stone. Chem. Ind. 9, 1587–1589.
Prioli, R. P., Tanna, A., and Brown, I. N. (1985). Rapid methods for counting mycobacteria – comparison of methods for extraction of mycobacterial adenosine triphosphate (ATP) determined by firefly luciferase assay. Tubercle. 66, 99–108.
Rakotonirainy, M. S., Héraud, C., and Lavédrine, B. (2003). Detection of viable fungal spores contaminant on documents and rapid control of the effectiveness of an ethylene oxide disinfection using ATP assay. Luminescence 18, 113–121.
Rohwerder, T., and Sand, W. (2007). Oxidation of inorganic sulfur compounds in acidophilic prokaryotes. Eng. Life. Sci. 7, 301–309.
Saito, H., and Miura, K.-I. (1963). Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72, 619–629.
Schaeffer, W. I., Holbert, P. E., and Umbreit, W. W. (1963). Attachment of Thiobacillus thiooxidans to sulfur crystals. J. Bacteriol. 85, 137–140.
Scheerer, S., Ortega-Morales, O., and Gaylarde, C. (2009). Microbial deterioration of stone monuments – an updated overview. Adv. Appl. Microbiol. 66, 97–139.
Schelle, M. W., and Bertozzi, C. R. (2006). Sulfate metabolism in mycobacteria. ChemBioChem 7, 1516–1524.
Seto, M., and Tange, I. (1980). Rapid and sensitive method for the determination of total organic carbon in soil by potassium persulfate-nondispersive infrared gas analyzer. Jap. J. Soil Sci. Plant Nutr. 51, 27–30 (in Japanese).
Shivvers, D. W., and Brock, T. D. (1973). Oxidation of elemental sulfur by Sulfolobus acidocaldarius. J. Bacteriol. 114, 706–710.
Stanley, P. E., and Williams, S. G. (1969). Use of the liquid scintillation spectrometer for determining adenosine triphosphate by the luciferase enzyme. Anal. Biochem. 29, 381–392.
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599.
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680.
Torres, A. L., Maroulis, P. J., Goldberg, A. B., and Bandy, A. R. (1980). Atmospheric OCS measurements on project Gametag. J. Geophys. Res. 85, 7357–7360.
Vincke, E., Boon, N., and Verstraete, W. (2001). Analysis of the microbial communities on corroded concrete sewer pipes – a case study. Appl. Microbiol. Biotechnol. 57, 776–785.
Keywords: facultative sulfur-oxidizing bacteria, Mycobacterium, biodeterioration, Angkor monuments
Citation: Kusumi A, Li XS and Katayama Y (2011) Mycobacteria isolated from Angkor monument sandstones grow chemolithoautotrophically by oxidizing elemental sulfur. Front. Microbio. 2:104. doi: 10.3389/fmicb.2011.00104
Received: 17 January 2011;
Accepted: 27 April 2011;
Published online: 10 May 2011.
Edited by:
Thomas E. Hanson, University of Delaware, USAReviewed by:
Masaharu Ishii, University of Tokyo, JapanKoji Mori, National Institute of Technology and Evaluation, Japan
Copyright: © 2011 Kusumi, Li and Katayama. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Yoko Katayama, Graduate School of Agriculture, Tokyo University of Agriculture and Technology, 3-5-8 Saiwai-cho, Fuchu-shi, Tokyo 183-8509, Japan. e-mail:a2F0YXlhbWFAY2MudHVhdC5hYy5qcA==


