- 1 Institute of Marine and Environmental Technology, University of Maryland, Baltimore, MD, USA
- 2 Department of the Geophysical Sciences, University of Chicago, Chicago, IL, USA
- 3 GE Healthcare, Piscataway, NJ, USA
- 4 Department of Energy, Joint Genome Institute, Walnut Creek, CA, USA
- 5 Bioinformatics, Joint Genome Institute, Los Alamos National Laboratory, Los Alamos, NM, USA
Carbon monoxide (CO), well known as a toxic gas, is increasingly recognized as a key metabolite and signaling molecule. Microbial utilization of CO is quite common, evidenced by the rapid escalation in description of new species of CO-utilizing bacteria and archaea. Carbon monoxide dehydrogenase (CODH), the protein complex that enables anaerobic CO-utilization, has been well-characterized from an increasing number of microorganisms, however the regulation of multiple CO-related gene clusters in single isolates remains unexplored. Many species are extraordinarily resistant to high CO concentrations, thriving under pure CO at more than one atmosphere. We hypothesized that, in strains that can grow exclusively on CO, both carbon acquisition via the CODH/acetyl CoA synthase complex and energy conservation via a CODH-linked hydrogenase must be differentially regulated in response to the availability of CO. The CO-sensing transcriptional activator, CooA is present in most CO-oxidizing bacteria. Here we present a genomic and phylogenetic survey of CODH operons and cooA genes found in CooA-containing bacteria. Two distinct groups of CooA homologs were found: one clade (CooA-1) is found in the majority of CooA-containing bacteria, whereas the other clade (CooA-2) is found only in genomes that encode multiple CODH clusters, suggesting that the CooA-2 might be important for cross-regulation of competing CODH operons. Recombinant CooA-1 and CooA-2 regulators from the prototypical CO-utilizing bacterium Carboxydothermus hydrogenoformans were purified, and promoter binding analyses revealed that CooA-1 specifically regulates the hydrogenase-linked CODH, whereas CooA-2 is able to regulate both the hydrogenase-linked CODH and the CODH/ACS operons. These studies point to the ability of dual CooA homologs to partition CO into divergent CO-utilizing pathways resulting in efficient consumption of a single limiting growth substrate available across a wide range of concentrations.
Introduction
Anaerobic microbial CO-utilization, which was first characterized by Uffen (1976) in a mesophilic α-proteobacterium, Rhodopseudomonas spp., is now recognized as a widespread metabolic capacity (King and Weber, 2007; Techtmann et al., 2009). Carbon monoxide is ubiquitous in geothermal environments and surprisingly common in aerobic microbial ecosystems, albeit in low concentrations (Dunfield and King, 2005; Techtmann et al., 2009). In both aerobic and anaerobic species, optimal conditions for growth span a wide range of CO concentrations. The aerobic utilization of CO via cox genes is extremely common and has been thoroughly studied (Meyer and Schlegel, 1983; Kraut et al., 1989; Hugendieck and Meyer, 1992; Meyer et al., 1993; Schubel et al., 1995; King, 2003a,b; King and Weber, 2007; Wu et al., 2009). The pathways to CO oxidation in aerobes via the cox genes are distinct from the anaerobic CO oxidation mechanisms which use variants of the enzyme CO dehydrogenase encoded by cooS genes (Morton et al., 1991; Ferry, 1995; Ragsdale, 2008). These anaerobes use CO variously as a carbon and/or energy source. Energy conservation is achieved by coupling the oxidation of CO to the reduction of water or various metals. Hydrogenogenic carboxydotrophs utilize the electrons generated from the oxidation of CO for generation of hydrogen via a membrane bound hydrogenase. This physiology is based on the water gas shift reaction CO + H2O → CO2 + H2 (Svetlitchnyi et al., 1991; Kerby et al., 1992, 1995; Sokolova et al., 2004b). Other species shunt the electrons generated from CO oxidation to the reduction of various metals, such as iron or manganese (Slobodkin et al., 1997; Sokolova et al., 2004a).
The extreme thermophile Carboxydothermus hydrogenoformans is the prototype thermophilic hydrogenogenic carboxydotrophic, providing the first genome sequence of a microorganism growing on CO as sole carbon and energy source (Wu et al., 2005). Its genome is highly specialized for CO metabolism, encoding five CO dehydrogenases in different genomic contexts, representing pathways drawing from a common pool of CO (Wu et al., 2005). Coordination of these multiple pathways must be key to the survival of CO-utilizers and is evidently effective in C. hydrogenoformans, which shows extraordinary adaptability, growing from very low CO concentrations up to greater than 1 atm (Svetlitchnyi et al., 1991).
The regulation of anaerobic CO metabolism is usually controlled by the positive gene regulator, CooA (Shelver et al., 1995; Fox et al., 1996; He et al., 1996, 1999; Roberts et al., 2001, 2005; Ibrahim et al., 2007). This heme protein is a member of the CRP family of positive gene regulators, represented by the cAMP binding protein (CAP) that controls hierarchical catabolite utilization in E. coli. Like the CAP protein, CooA binds to promoter sites upstream from the -35 box and activates transcription when bound by the activator ligand, which in the case of CooA is CO (Shelver et al., 1995, 1997). CooA is a dimeric, symmetrical protein containing two heme prosthetic groups that bind CO tightly, replacing the cAMP binding site of CAP. CO-binding induces an allosteric conformational change that results in promoter binding to a specific target sequence resulting in positive gene regulation. In many genomes multiple CODH operons coincide with the occurrence of dual CooA homologs. Here we present a genomic and phylogenetic survey of the number of CODH operons and cooA genes found in CooA-containing bacteria. To further our study of these genomes, the CO-dependent gene regulators were cloned and expressed from the prototype carboxydotrophic C. hydrogenoformans to establish the basis for differential regulation of multiple CODHs in response to CO “feast” and “famine” conditions.
Materials and Methods
Evolutionary Analysis
CooA homologs were identified using criteria similar to those used previously (Youn et al., 2004). Multiple sequence alignment of various cooA genes was performed using the Muscle multiple sequence alignment program (Edgar, 2004a,b). Maximum likelihood trees were constructed using the PhyML 3.0 program (Guindon et al., 2003). Tree topology was verified by bootstrap analysis using 100 iterations.
Bacterial Strains and Plasmids
Carboxydothermus hydrogenoformans Z-2901 was isolated from Stolbovskii Spring on Kunashir Island in the Kuril Islands of Russia as described previously (Svetlitchnyi et al., 1991). C. hydrogenoformans Z-2901 was a gift from Vitali Svetlitchnyi (Bayreuth University, Germany) in 1995, after being serially grown in culture since its original isolation in 1990. C. hydrogenoformans DSMZ was purchased from the DSMZ GmbH (Braunschweig, Germany). C. hydrogenoformans was grown under conditions previously described in Wu et al. (2005). Standard recombinant technique was used and unless otherwise noted, E. coli DH5α, BL21 (DE3), and VJS6737 were cultivated in liquid or solidified (15 g/l Bacto-Agar) Luria Broth. E. coli VJS6737 was a gift from Gary Roberts, University of Wisconsin.
Expression and Purification of CooA-1 and CooA-2
E. coli VJS6737 carrying a CooA-1 expression construct and purified CooA-1 were a gift from Gary Roberts (U. Wisconsin – Madison). CooA-1 was prepared as previously described (Youn et al., 2004; Clark et al., 2006). In summary, E. coli VJS6737 carrying an expression plasmid encoding the cooA-1 gene was cultivated in liquid LB supplemented with appropriate antibiotic and 9.8 mg/l ferric citrate (Sigma F3388). Cultures of the expression strain were grown in stoppered-flasks to an OD600 of 0.4 and expression was induced with 0.5 mM IPTG. Expression was allowed to proceed for 3 h. Cells were harvested by centrifugation at 6000 × g for 10 min at 4°C. The cells were resuspended in Buffer A (25 mM MOPS, pH 7.4, 100 mM NaCl, 5% glycerol). The cells were lysed using a French pressure cell. The lysate was centrifuged at 20,000 × g for 20 min at 4°C. The soluble fraction was collected and loaded onto a Protino Ni-IMAC column (Macherey-Nagel, Bethlehem, PA, USA). The column was washed with 10 column volumes of Buffer A. This was followed by two washes, one with 10 ml of Buffer A plus 10 mM imidazole and another with 10 ml of 20 mM imidazole. The column was then washed with 10 ml each of higher imidazole concentrations (50, 150, and 250 mM). Fractions were screened for the presence of CooA-1 by absorbance at 425 nm and by SDS-PAGE. After the Ni-IMAC step, CooA-1 was precipitated with 50% (NH4)2SO4 and resuspended in 25 mM MOPS, 0.5 M KCl, pH 7.4.
CooA-2 was purified according to a similar protocol with the following changes. After the Ni-IMAC step, CooA-2 containing fractions were pooled and loaded onto a Bio-Scale Mini Macro-Prep High S column (BioRad, Hercules, CA, USA) equilibrated with Buffer A. CooA-2 was eluted via a linear gradient from 100 to 800 mM NaCl over five column volumes at a flow rate of 1 ml/min. CooA-2 eluted around 300 mM NaCl. Presence and purity of CooA-2 were determined by SDS-PAGE. Protein concentration was determined using the Bradford assay (Bradford, 1976) using a standard curve constructed from various concentrations of BSA.
Visible spectra for both CooA-1 and CooA-2 were recorded in a Beckman DU60 spectrophotometer. CooA spectra were obtained in 25 mM MOPS, 100 mM NaCl, pH 7.4 under oxic conditions, reduced conditions (N2 sparged buffer with 5 mM DTT), and reduced conditions with 1 atm CO.
Promoter Fragments for Use in DNA-Binding Assays
The promoter upstream of the CODH-linked hydrogenase (henceforth referred to as pcooChyd) and the promoter upstream of the CODH/ACS (to be referred to as pacs) were examined for specific CooA binding. We identified probable regions pcooChyd and pacs by examining the sequences 500 bp upstream of the start codon of the first gene in both the CODH-linked hydrogenase operon (cooChyd) and the ACS operon (acs) from the genome sequence of C. hydrogenoformans. These sequences were run through the bprom program (http://softberry.com) to predict the putative -10 and -35 sites. From this prediction, primers were designed to create a product that started at the putative -150 site and stretched to start codon. The fragments were the following sizes: pcooChyd was 217 bp and the pacs was 381 bp. These fragments were used in all of the DNA-binding experiments described below.
DNA-Binding Assays Using Surface Plasmon Resonance
Surface plasmon resonance (SPR) experiments were carried out on the Biacore T100 at 25°C. 5′-biotinylated promoter regions were PCR amplified. The purified biotin labeled DNA was loaded onto a streptavidin sensor chip (Biacore Sensor Chip SA). The cooChyd promoter was immobilized on flow cell 2 until the response reached 300 response units (RU). RU is the unit used to measure the amount of ligand bound to the target molecule attached to the sensor chip. Typically one RU is equivalent to 1 pg of ligand bound per mm2 of the sensor chip surface. The acs promoter was immobilized on flow cell 4 until the response reached 500 RU. Flow cells 1 and 3 and any free streptavidin on flow cells 2 and 4 were blocked with free biotin. Flow cells 1 and 3 were used as controls to subtract out any non-specific binding.
All of the experiments were done using buffer HBS-P (10 mM HEPES, pH 7.4, 0.15 M NaCl, 10 mM MgCl2, 0.05% P20; Biacore) supplemented with 5% glycerol and 5 mM DTT. Kinetics tests were performed with either CooA-1 or CooA-2 at concentrations ranging from 8 to 200 nM. These CooA solutions were prepared in anaerobic CO-saturated HBS-P supplemented with 5% glycerol and 5 mM DTT. The kinetic measurements were performed at a flow rate of 30 μl/min with 120-s injections. Dissociation data were collected for 300 s.
DNA-Binding Assay Using Electrophoretic Mobility Shift Assays
The two promoter fragments were end-labeled as follows: pcooChyd (15 pmol) and pacs (12 pmol); 5 μl [γ32-P] ATP (10 mCi/ml; Perkin Elmer); 2 μl 10× polynucleotide kinase buffer (500 mM tris–HCl, pH 7.6, 100 mM MgCl2 and 100 μM β-mercaptoethanol); 2 μl T4 polynucleotide kinase (10 U) were incubated at 37°C for 30 min, followed by 15 min at 70°C. End-labeled fragments were purified from unincorporated radionucleotide using the Qiaquick Nucleotide Purification Kit (Qiagen) according to the manufacturers instructions. Fragments were eluted from the filters into 100 μl of water. The efficiency of labeling and recovery was about 20%, determined through scintillation counting. Scintillation counting revealed that the pacs fragment was 126,000 cpm/μl and the pcooChyd fragment was 120,000 cpm/μl.
For electrophoretic gel shift analysis, 0.3 μl of BSA (5 mg/ml), 0.15 μl MgCl2 (0.5 M), 11.5 μl HBS-P (10 mM HEPES, pH 7.4, 0.15 M NaCl, 10 mM MgCl2, 0.005% P20; Biacore) supplemented with 5% glycerol and 5 mM DTT, and 0.5 μg of sonicated salmon sperm DNA were mixed with 1 μl of radiolabeled promoter fragment. To this reaction 1 μl of a CooA-1 or CooA-2 protein solution was added and allowed to bind at 25°C for 30 min. Various concentrations of CooA were used (as described in the legends of Figure 4). One microliter of loading dye was added to each reaction and the reactions were run on an 8% polyacrylamide gel at 150 V for 1.5 h in TBE. The gel was dried on a vacuum blotter at 60°C for 2 h and subsequently exposed to a phosphor screen overnight. The screens were imaged on a Typhoon scanner (GE healthcare).
CO-Binding Assays
CO-binding was determined by examining the changes in the visible absorbance spectrum for CooA between 400 and 600 nm (Beckman DU60). Absorbance changes in the Soret wavelength region were used to determine fractional saturation (Y). A stoppered glass cuvette was filled with a 2.2 μM CooA-1 or CooA-2 solution in anaerobic buffer (25 mM MOPS, 100 mM NaCl, pH 7.4). The buffer was made anaerobic by repeatedly evacuating and sparging the buffer with argon. Two stock solutions were prepared, both with 5 mM DTT. One of the stock solutions was purged with argon, and the other was purged with CO. The pressure in the bottles was equilibrated to 1 atm. The concentration of the dissolved CO in the CO-purged bottle was calculated to be 1000 μM. This was determined by using Henry’s law with a Henry’s law constant appropriate for 25°C of 9.9 × 10−4 mol CO/liter/atm CO (Lide and Frederikse, 1995) and a headspace CO partial pressure of 1 atm as described previously (Puranik et al., 2004). Using a Gastight Hamilton syringe, a 2.2 μM solution of anaerobic CooA was titrated with the 1000 μM CO solution. After each addition of CO the cuvette was gently mixed and the visible spectrum was recorded. The CO solution was added until there was no change in the visible spectrum of CooA. The maximum saturation of CooA was determined by sparging the CooA solution with CO for 5 min then recording the visible spectrum.
Because CO binds tightly to CooA, the concentration of CO was corrected for the bound fraction as follows: [CO]free = [CO]total − (Y × [CooA]total). Fractional saturation (Y) was determined experimentally by using the change in absorbance of the Soret peak relative to the no-CO spectrum divided by the maximum change in absorbance (Y = ΔA/ΔAmax). Fractional saturation was graphed versus [CO]free. The data was fitted to the Hill equation using Kaleidagraph (Synergy Software).
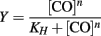
KH is the Hill equilibrium constant. In our experiments it was the disassociation constant, proportional to the affinity of CO-binding to CooA. Here we report P50, which is defined as the concentration of CO needed to occupy half of the CO-binding sites in CooA. n is the Hill coefficient, which describes the cooperativity of the binding of CO by CooA. An n = 1 is indicative of a binding reaction that is non-cooperative, whereas an n = 2 for a dimer is a binding reaction that is fully cooperative.
Results
Co-occurrence of CooA-2 with the CODH/ACS
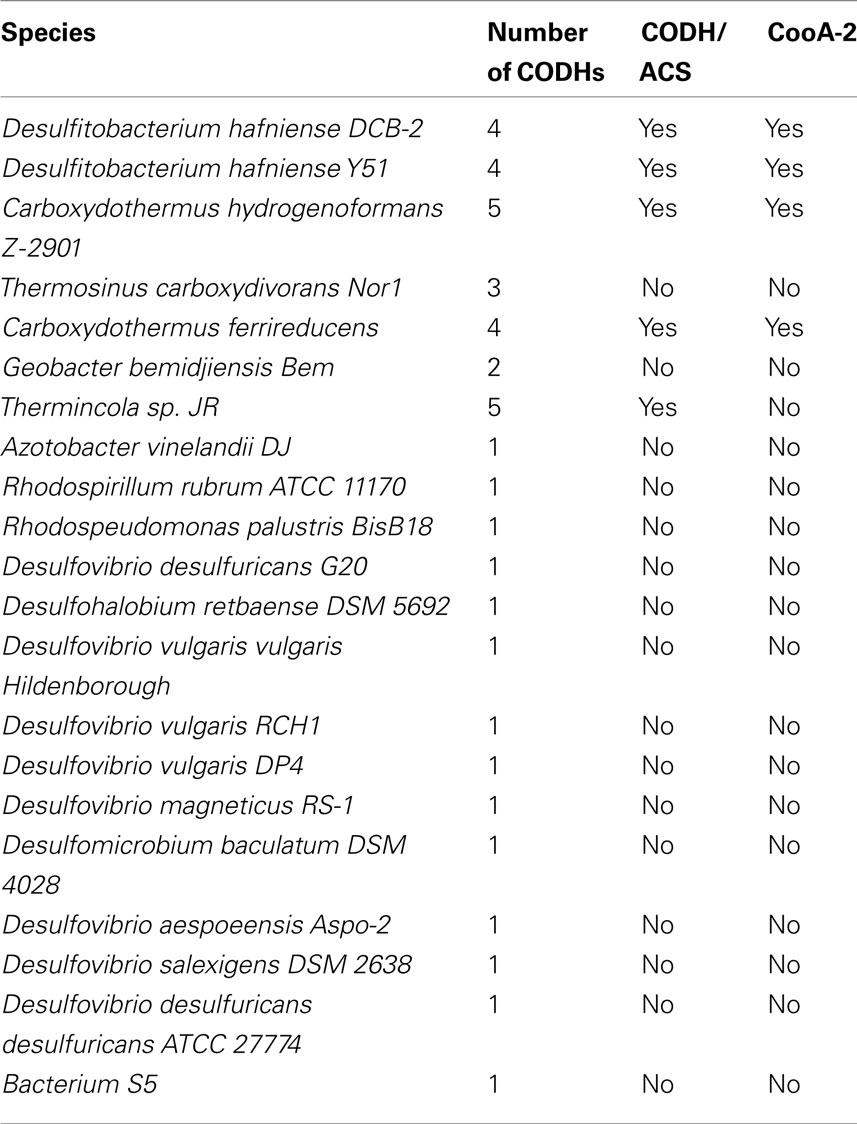
Phylogenetic analysis of the CooA regulators using a maximum likelihood algorithm revealed the presence of two distinct clades of CooAs (CooA-1 and CooA-2 in Figure 1). The larger clade with 21 members contains the well-characterized CooA-1 from Rhodospirillum rubrum and CooAs from diverse species spanning divergent bacterial phyla. In general, the CooA-1 homologs are distributed in subclades that follow evolutionary divisions defined by 16S rDNA phylogeny (i.e., CooAs from Firmicute bacteria cluster with other Firmicute CooAs). The CooA-2 clade contains four homologs and is deeply branching relative to all CooA-1 protein sequences. The CooA-2 genes found to date are all found in the genomes of Firmicute bacteria that also encode multiple CODH operons.
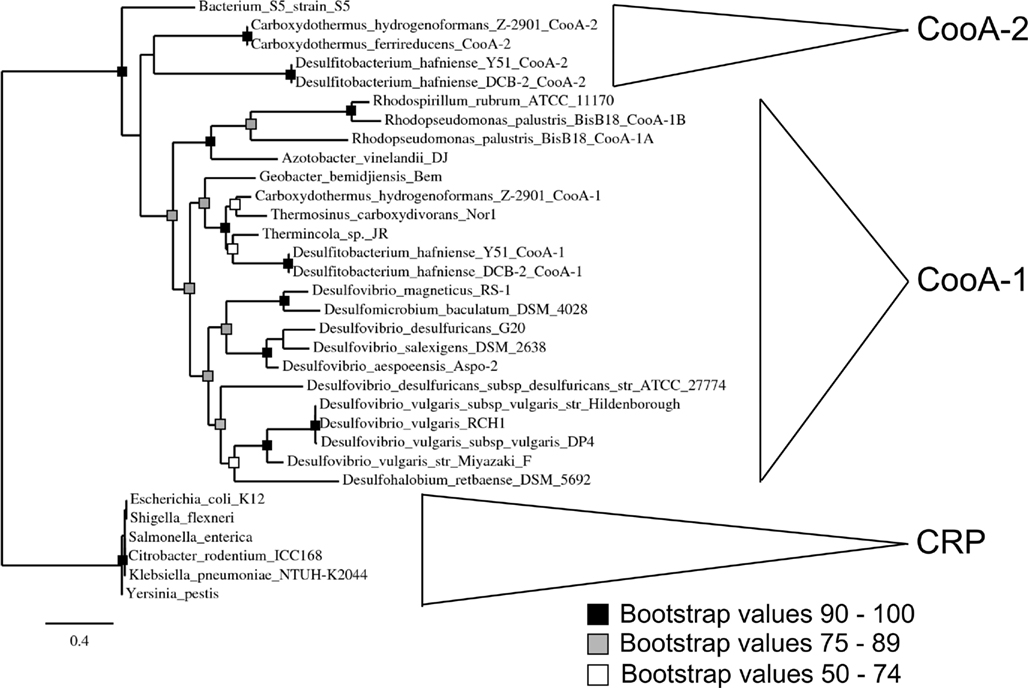
Figure 1. Phylogenetic analysis of CooA showing a maximum likelihood tree of CooA and CRP sequences from various organisms. Major phylogenetic divisions are indicated on the tree. The topology of the tree was confirmed with bootstrap analysis with 100 iterations. See text for details on multiple sequence alignment and tree construction.
Table 1 details the copy number of CODHs contained in the genomes of cooA encoding bacteria. With the exception of Thermincola sp. JR, the genomes that encode CooA-2 also encode CODH/ACS clusters. However, the CODH/ACS cluster may also occur in species without CooA, e.g., Moorella thermoacetica and all the homoacetogens. C. hydrogenoformans was chosen as a model system to explore the role of CooA-2 in regulating the CODH/ACS since it contains all the genetic elements in the CO “toolkit.”
Identification of Key Frameshift Mutations
Further evidence supporting a role for CooA-2 in regulating the CODH/ACS was uncovered upon analysis of the C. hydrogenoformans genome. The strain of C. hydrogenoformans that was used for sequencing was C. hydrogenoformans Z-2901. This strain was grown serially in our laboratory over the course of several years. The standard growth medium was supplemented with pyruvate in order to promote high growth yields. Manual annotation of the genome identified multiple frameshifts (Wu et al., 2005), many of which were in genes associated with proper Coenzyme A processing. Two mutations were detected in genes related to carbon monoxide oxidation. One frameshift was found in the catalytic subunit of the CODH/ACS operon (cooS-III; Wu et al., 2005). The cooS-III gene was amplified and re-sequenced from strain Z-2901, confirming that this frameshift was not a sequencing error. Additionally cooS-III was sequenced from the type strain of C. hydrogenoformans, which was deposited in the DSMZ culture collection shortly after isolation by Svetlitchnyi et al. (1991). This strain is referred to as C. hydrogenoformans DSMZ 6008, and maintained under non-growing conditions since 1991. The strain thus archived contained an intact cooS-III gene.
Upon further analysis of the cooA genes reported by Wu et al. (2005), it became evident that the cooA-2 annotated in the published genome also contained a frameshift mutation, which was absent in the DSMZ strain. It appears likely that serially culturing C. hydrogenoformans under lab conditions (high CO and pyruvate) resulted in mutations in the cooS-III and cooA-2 genes. Concerted mutation of these genes under the same conditions suggested a possible role for CooA-2 in regulating carbon fixation via the CODH/ACS. This led to the expression and characterization of the CooA proteins in this present study.
Expression and Characterization of CooA-2
E. coli overexpressing wild type CooA-2 acquired a red color similar to the color of CooA-1 expressing E. coli as described in Youn et al. (2004) due to the accumulation of the heme containing protein (Figure 2D). The purified CooA-2 exhibited typical heme spectral features similar to CooA-1 with a strong Soret peak at 425 nm (Figures 2A,B). Upon addition of CO the Soret peak increased in height and was blue-shifted to 421 nm. This response is characteristic of other CO-binding heme proteins, such as hemoglobin, and has been demonstrated in all of the characterized CooA homologs (Youn et al., 2004). As an indicator of CooA regulator activity, an E. coli reporter strain was constructed with genomic integration of a lacZ gene under the control of a CooA-responsive promoter (R. rubrum cooF promoter). This strain has been used to identify functional CooAs from various organisms (Youn et al., 2004). As Youn et al. (2004) have previously shown, CooA-1 is able to activate lacZ expression in a CO-responsive manner. CooA-2, like other CooAs, is able to activate expression of LacZ in response to CO, confirming that it is a CO-dependent transcriptional activator (Figure 2C).
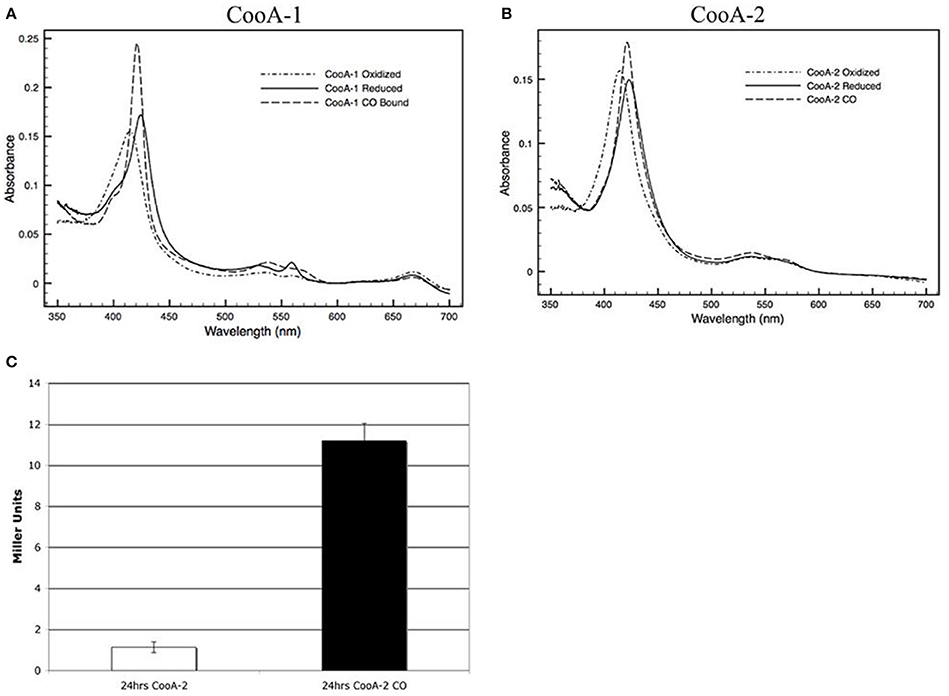
Figure 2. Visible spectra of a 2.2 μM (A) CooA-1 and (B) CooA-2. Dotted lines indicate the spectrum of oxidized CooA. Solid line indicates reduced CooA. Dashed line indicates reduced CooA under 1 atm of CO. (C) β-galactosidase activity of CooA-2 expressing E. coli DH5α with a lacZ under the control of the R. rubrum cooF promoter. Experiments with CO were grown with 2% CO in the headspace of the culture.
Identification of Differential Promoter Binding
At first glance, the occurrence of dual CO-sensing transcriptional activators in one species appears to be a functional redundancy. To further define their physiological roles the CooA proteins were expressed, purified and tested as follows. SPR was used to measure promoter affinity and kinetic constants. The SPR binding curves for CooA-1 binding to the cooChyd promoter exhibited a binding curve typically associated with specific high-affinity binding of a protein with DNA. CooA-1 accumulated on the DNA during the injection and then slowly disassociated after the injection was stopped (Figure 3B). However the binding of CooA-1 to the acs promoter demonstrated a binding curve typical of non-specific low-affinity binding. In this case the overall RU were much lower than with CooA-1 binding to the hydrogenase promoter and the signal dropped instantaneously upon termination of the injection (Figure 3A). The binding of CooA-2 to both promoters tested is typical of specific high-affinity binding (Figures 3C,D). Classical electrophoretic mobility shift assays (EMSA; Figure 4) confirmed the affinity order and lack of binding of CooA-1 to acs promoter. Affinity constants were determined by fitting the SPR binding data to a two-state binding model (Figure 3). The use of this model was supported by the results of a linked-reactions test. CooA-1 bound with extremely low-affinity to pacs, and did not fit a binding model in a meaningful way typical of the non-specific binding of regulator proteins to DNA sequences unrelated to their cognate target motifs.
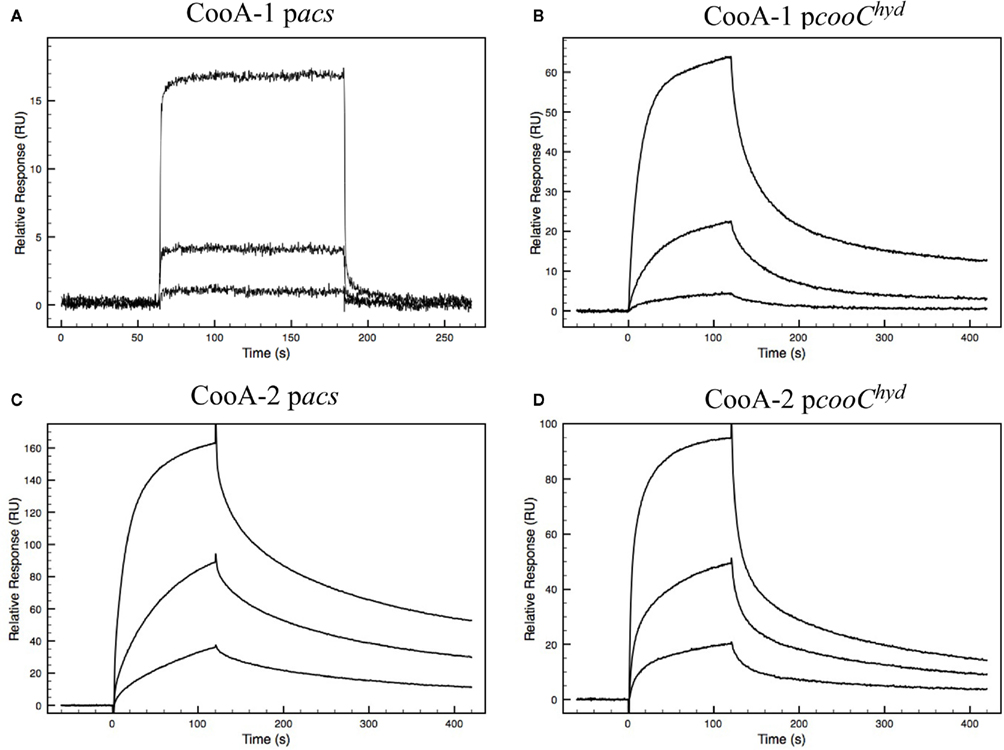
Figure 3. Biacore sensograms representing the binding of either CooA-1 or CooA-2 to either pacs or pcooChyd promoters fixed to the surface of the sensor chip. Varying concentrations of protein were injected. Each injection is represented by a different curve. The protein concentrations used were 8, 40, and 200 nM for CooA-1 and 12, 40, and 200 nM for CooA-2. (A) CooA-1 binding to the pacs promoter fragment. (B) CooA-1 binding to the pcooChyd promoter fragment. (C) CooA-2 binding to the pacs promoter fragment. (D) CooA-2 binding to the pcooChyd promoter fragment.
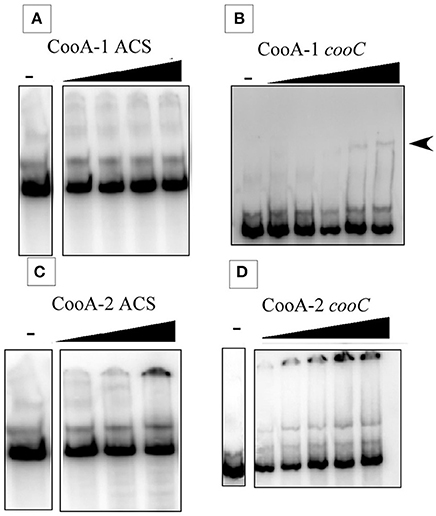
Figure 4. Electrophoretic mobility shift assays at 25°C. Gel shift results showing the binding of CooA-1 or CooA-2 to either pacs or the pcooChyd promters from C. hydrogenoformans. Arrows indicate formation of the CooA-promoter complex. (A) CooA-1 binding to pacs (Protein added: 3, 6, 12, 18 μg/ml). (B) CooA-1 binding to pcooChyd (Protein added: 3, 6, 9, 12, 15 μg/ml). (C) CooA-2 binding to pacs (Protein added: 3, 6, 9 μg/ml). (D) CooA-2 binding to pcooChyd (Protein added: 3, 6, 9, 12, 15 μg/ml).
The fitted SPR data confirms that CooA-1 binds to the hydrogenase promoter with a KD of 82.8 nM. CooA-2 bound to both of these promoters with higher affinity (pcooChyd: KD = 30.7 nM; pacs: KD = 16.8 nM; Figure 3; Table 2). These constants resemble the semi-quantitative data obtained in the EMSA experiment, but with greater precision and control of CO concentration (Figure 4). Table 2 describes both the kinetic and affinity data for CooA-1 and CooA-2 binding.
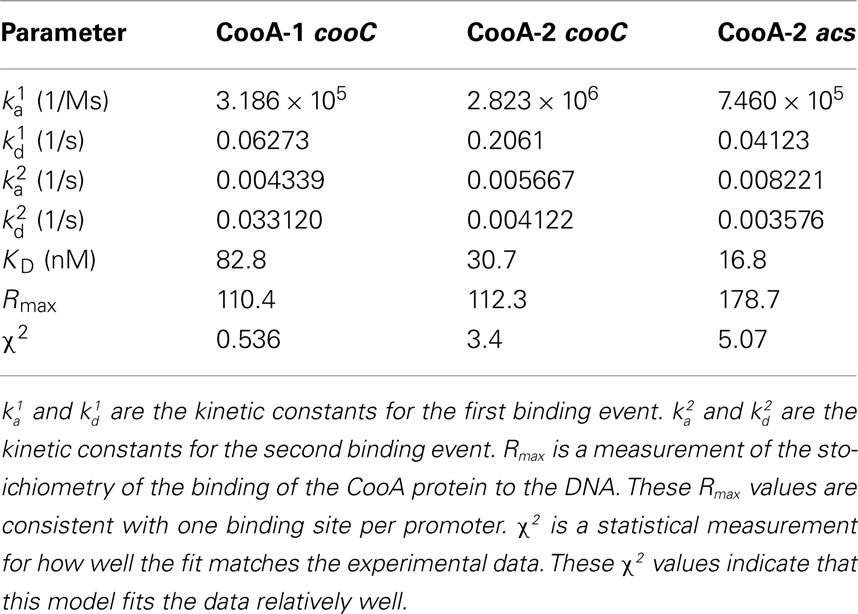
Table 2. Biacore kinetic and affinity data for the binding of CooA-1 and CooA-2 to the cooChyd and the acs promoter regions.
Determination of CO-binding Constants
The variable binding affinities of the CooA proteins for promoters suggested that they might regulate the CODH operons of C. hydrogenoformans differentially in high and low CO concentrations. To detect CO-binding, CooA-1 was titrated with CO at pH 7.4 at 25 and 70°C while monitoring the visible spectrum (Figures 5A,B). Upon addition of CO to CooA-1 the Soret peak increased in amplitude until saturation. By measuring the changes in peak height in relation to [CO], both binding constants and cooperativity were determined. Spectral changes in the Soret peak were used to determine the fractional saturation (Y), which in turn was used to determine P50 as described in Section “Materials and Methods” (Figures 5 and 6). P50 is the concentration of CO at which half of the CO-binding sites are bound by CO. The value of P50 for CooA-1 at 25°C was determined to be 17.4 ± 0.5 μM, whereas at 70°C it was determined to be 5.6 ± 0.2 μM. CooA-2 was titrated with CO at 25 and 55°C (Figures 6A,B). For CooA-2, the P50 at 25°C was 6.2 ± 0.2 μM and at 55°C the P50 is 1.9 ± 0.1 μM (Figures 6C,D). The temperature used for CooA-2 analysis was 55°C because CooA-2 was unstable at 70°C. For comparison the P50 for R. rubrum CooA-1 to CO at 25°C is 2.2 μM (Puranik et al., 2004).
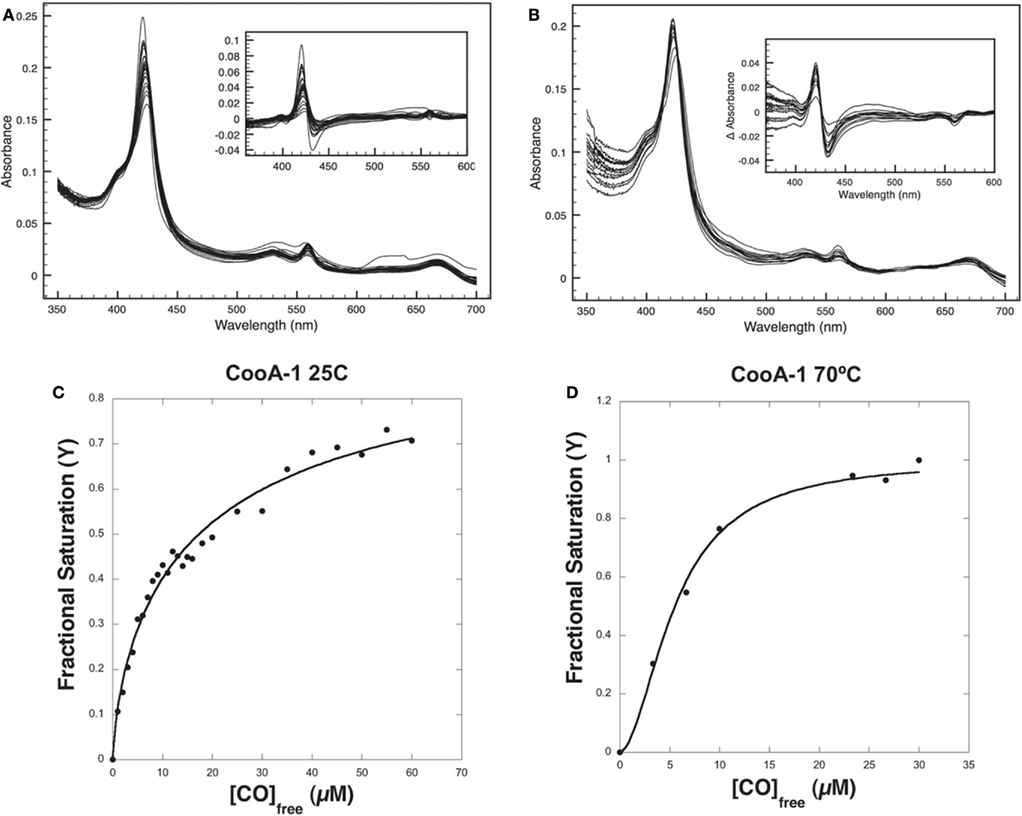
Figure 5. CO-binding assays and determination of P50 for CooA-1. (A) Visible light spectra of a 2.2-μM solution of pure CooA-1 upon successive addition of CO at 25°C were recorded and (B) at 70°C. The inset depicts the difference between each CO-bound curve and the No-CO curve. Fractional Saturation was plotted against [CO] and fit to the Hill equation for (C) CooA-1 at 25°C and (D) CooA-1 at 70°C.
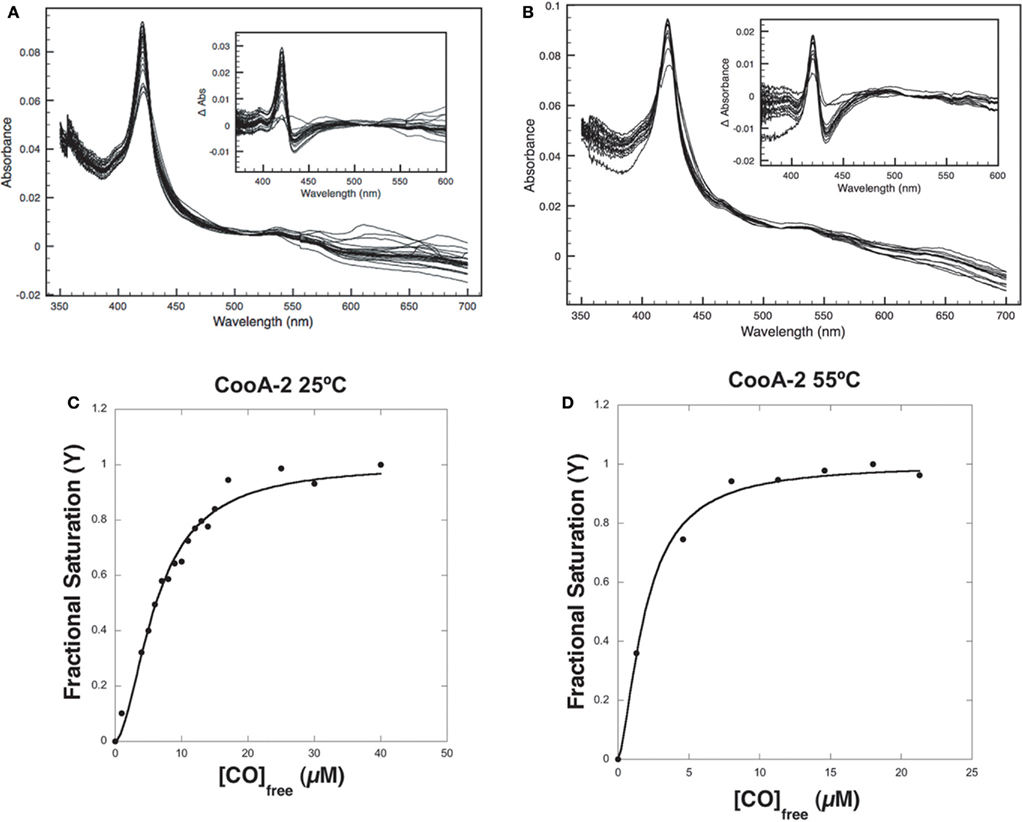
Figure 6. CO-binding assays and determination of P50 for CooA-2. (A) Visible light spectra of a 2.2-μM solution of pure CooA-2 upon successive addition of CO at 25°C were recorded and (B) at 55°C. The inset depicts the difference between each CO-bound curve and the No-CO curve. Fractional Saturation was plotted against [CO] and fit to the Hill equation for (C) CooA-2 at 25°C and (D) CooA-2 at 55°C.
Cooperativity of CO-Binding Differs between the Two Regulators
Multisubunit heme proteins may bind to their ligands with cooperative kinetics following the classic model for allosteric intersubunit interactions first described by Hill (1910) for hemoglobin. The method for determining cooperative kinetics is by application of the Hill equation as described in Section “Materials and Methods” (Hill, 1910). Table 3 summarizes the CO-binding data for both CooA-1 and CooA-2. From the Hill equation it was determined that at 25°C CooA-1 had n = 0.73 ± 0.03 indicating non-cooperative binding or negative cooperativity, whereas CooA-2 had an n value of 1.8 ± 0.11 (Figures 5C and 6C) indicating a significantly cooperative process. Cooperativity of CO-binding by CooA-1 was different at a higher temperature corresponding to the normal growth conditions. At 70°C the n value for CooA-1 increased to approximately 1.8 ± 0.2 (Figure 5D). The n value for CO-binding to CooA-2 at 55°C does not change significantly from 25°C with n = 1.6 ± 0.1 (Figure 6D).
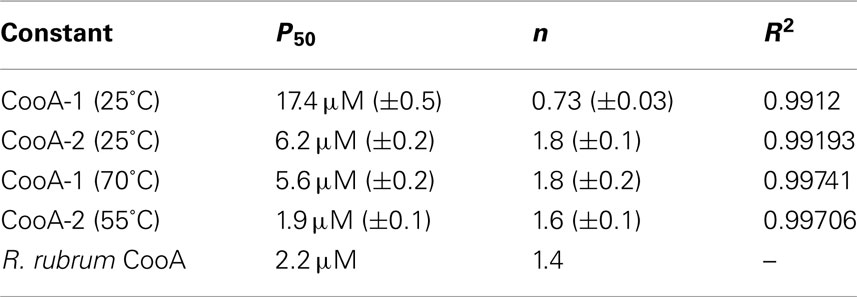
Table 3. Disassociation constants and cooperativity data for CO-binding to CooA-1 or CooA-2 as determined from fitting the changes in fractional saturation to the Hill equation.
Discussion
The ability of an organism to use CO as an electron source for divergent cellular processes may be essential for growth on CO as a sole carbon and energy source. Our work points to a new paradigm for efficient CO-utilization in bacteria. This analysis uses both genomic and biochemical techniques to demonstrate the importance of multiple CO-utilization pathways within C. hydrogenoformans.
Many species that encode a CooA homolog embody a primary physiology that does not include CO, but their possession of cooS and cooA genes imply underlying or backup CO-dependent physiologies. As an example, C. ferrireducens was first isolated as a primary metal reducer growing on fumarate and originally named Thermoterrabacterium ferrireducens (Slobodkin et al., 1997). Its identical 16s rRNA sequence to C. hydrogenoformans led to its renaming as C. ferrireducens (Slobodkin et al., 2006). In collaboration with the Joint Genome Institute, we are currently sequencing the genome of C. ferrireducens. This draft sequence revealed that C. ferrireducens retains four of the five CODH operons found in C. hydrogenoformans and is only lacking the hydrogenase-linked CODH (CODH-I) of C. hydrogenoformans. Additionally C. ferrireducens is missing the cooA-1 but not cooA-2. Furthermore, physiological studies revealed the ability of C. ferrireducens to grow on CO in the presence of an alternative electron acceptor (Henstra and Stams, 2004). The lack of a CODH-I and a CooA-1 in C. ferrireducens further supports the link between the CooA-1 clade of CooAs and the hydrogenase-linked CODH operon. Furthermore, C. hydrogenoformans is an interesting case where CooA-2 is the primary CO-responsive regulator, suggesting perhaps that CooA-2 is able to efficiently regulate the all of the remaining CODH operons.
Monofunctional CODHs are capable of driving many processes from the same substrate, and it is not surprising that they occur in many prokaryotic lineages capable of diverse metabolism. The CODH-linked hydrogenase module is the signature gene cluster predicting hydrogenogenic carboxydotrophy. This complex has been found in bacteria ranging from thermophilic Firmicutes to mesophilic photosynthetic purple non-sulfur bacteria such as R. rubrum, an α-Proteobacterium. The bacterial lineages encoding this complex have recently been joined by archaea in the form of three Thermococcus sp. (Thermococcus sp. AM4, Sokolova et al., 2004b; Thermococcus onnurineus, Lee et al., 2008; Kim et al., 2010; and Thermococcus barophilius, Kim et al., 2010).
Table 1 shows the occurrence of multiple CODH operons in cooA-containing carboxydotrophs. The range of organisms that encode multiple CODH operons had not been fully appreciated until now and includes both bacteria and archaea. This is prevalent in the Peptococcaceae family, including species such as C. hydrogenoformans, Thermincola sp. JR, and the Desulfitobacterium genus. Five species encode either four or five CODH operons and C. hydrogenoformans and Thermincola sp. JR l have five CODH operons apiece (Wu et al., 2005; Byrne-Bailey et al., 2010).
The pathways encoded by these operons constitute different and competing destinations for a single feedstock, CO, and we considered it probable that coordinated regulation of the transcription and activity of the CODHs may have evolved to allow for efficient CO-utilization. The CO-sensing transcriptional activator – CooA – is the most well studied CO-responsive regulator. Therefore, it is important to understand the responses of the CooA dependent regulation to different levels of CO. Interestingly, bacteria encoding multiple CODH operons also encode multiple CooA homologs that fall into two distinct clades (Figure 1). In this context, our original finding that CooA-2 and CooA-1 both regulated the cooChyd promoter linked to a lacZ gene in E. coli (Figure 2C) was an unexpected and seemingly anomalous result.
In an effort to understand the functional difference between members of the two subclasses of CooA, the CooA-1 and CooA-2 proteins from C. hydrogenoformans were expressed, purified, and characterized. Initial work by the Roberts group at the University of Wisconsin characterized CooA in R. rubrum. In these elegant studies, He et al. (1996, 1999) demonstrated that the R. rubrum CooA acted as a positive regulator of the hydrogenase-linked CODH in R. rubrum. Further work by Youn et al. (2004) described the ability of C. hydrogenoformans CooA-1 to activate lacZ expression in a CO-responsive manner in an E. coli reporter strain. Our work expands on this previous work by examining the promoter binding of CooA-1. Here we demonstrated that CooA-1 is a specialized regulator specific for the monofunctional CODH-linked to the CO-tolerant hydrogenase found in CO-dependent hydrogenogens. Based on phylogenetic analysis, the C. hydrogenoformans CooA-1 and the R. rubrum CooA are members of the CooA-1 clade whereas C. hydrogenoformans CooA-2 is a member of the CooA-2 clade. Therefore, the finding that C. hydrogenoformans CooA-1 regulates the CODH/hydrogenase operon like R. rubrum CooA is not surprising. The inability of C. hydrogenoformans CooA-1 to bind to the promoter of the CODH/ACS operon was unexpected.
The CooA-2 from C. hydrogenoformans is a bifunctional regulator and is capable of activating the hydrogenase operon as well as the CODH/ACS operon at low CO partial pressures. CooA-2 was also shown to be able to bind CO cooperatively with a P50 value of 1.9 μM, about three-fold lower than CooA-1 (6.2 μM). These data indicate that the functional difference between the CooA-1 and CooA-2 in C. hydrogenoformans is not only their promoter specificity, but also their threshold concentration for CO activation.
Our working model for the interplay between these regulatory elements is shown in Figure 7. Under “feast” conditions represented by an intracellular concentration of CO > 6 μM, both CooA-1 and CooA-2 are fully activated and hydrogen production and acetyl CoA production are fully induced, corresponding to exponential growth in ad libitum CO. As CO becomes limiting (approaching famine, less than 2 μM CO in the cytoplasm), hydrogen production and energy conservation are limited in favor of acetyl CoA production and C-fixation. This mechanism allows C. hydrogenoformans, and likely the other species encoding CooA-2 homologs, to modulate expression of the CODH operons in ratios appropriate to direct the CO flux for optimal growth and survival across a wide range of CO concentrations. As we have described previously, C. hydrogenoformans can grow under a headspace of 1–2 atm of pure CO, which is lethal to most microbial species (Techtmann et al., 2009). The ability to “convert” excess CO to H2 under toxic CO stress conditions would benefit CO-sensitive neighbor species in the geothermal mat community. C. hydrogenoformans has been the subject of extensive work characterizing its ability to grow synergistically with CO-sensitive sulfate reducing bacteria. In these co-cultures C. hydrogenoformans acts as a safety valve to dispel CO, producing hydrogen which is an essential substrate for sulfate reducers (Parshina et al., 2005). Thus the ability to consume CO and produce H2 is potentially a community asset, which would explain the relatively high counts (104–6 cells/ml) of carboxydotrophic bacteria recorded in hot spring effluent mats (Slepova et al., 2007), as well as the relative ease of isolation of CO oxidizer strains from widespread geothermal sites (Robb et al., 2005; Techtmann et al., 2009). In a recent review we have described how a typical geothermal mat community might use CO as a “currency” metabolite that can be exchanged for other key metabolites such as H2, CH4, and H2S (Techtmann et al., 2009).
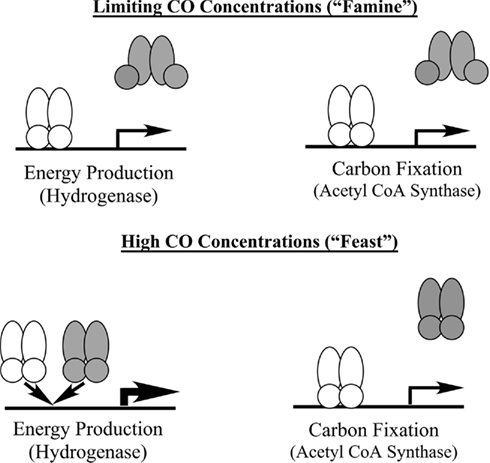
Figure 7. Model of CooA mediated regulation of CO-utilization in C. hydrogenoformans. The shaded symbol represents CooA-1 and the shaded symbol represents CooA-2. When CO levels are high both CooA-1 and CooA-2 are in the CO-bound form. CooA-1 is then able to shunt more CO into energy production through overexpression of the hydrogenase, while CooA-2 continues to shuttle CO toward carbon fixation. Under limiting CO concentrations only CooA-2 is in the CO-bound state and can activate both energy production via balanced expression of the hydrogenase and carbon fixation operons.
This study revealed the concerted laboratory evolution resulting in a null mutation of the CODH/ACS operon and its cognate CooA-2 regulator in response to the relaxation of selective pressure to use CO as a sole carbon source. Paralogous classes of CooA regulators were shown to be capable of regulating different CODH operons in trans. This accentuates the dynamic genome evolution probably taking place in geothermal communities, which represents the majority of CO-utilizing microorganisms isolated to date. Finally, we have characterized the kinetics of CO-binding in the CooA-1 and CooA-2 proteins and can assign them to coarse and fine control ranges that allow the efficient distribution of a single metabolic feedstock into carbon and energy metabolism. So far, little has been published concerning the metabolic crosstalk involving CO/H2 interconversion in microbial mats and biofilms, which may be very significant given the key roles that hydrogen and CO may play as energy sources in geothermal microbial systems (Spear et al., 2005; Shock et al., 2010).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the US National Science Foundation through award numbers EAR 0747394 (Frank T. Robb), EAR 0747412 (Albert S. Colman), and MCB 0605301 (Frank T. Robb and Albert S. Colman). The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
References
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254.
Byrne-Bailey, K. G., Wrighton, K. C., Melnyk, R. A., Agbo, P., Hazen, T. C., and Coates, J. D. (2010). Complete genome sequence of the electricity-producing “Thermincola potens” strain JR. J. Bacteriol. 192, 4078–4079.
Clark, R. W., Lanz, N. D., Lee, A. J., Kerby, R. L., Roberts, G. P., and Burstyn, J. N. (2006). Unexpected NO-dependent DNA binding by the CooA homolog from Carboxydothermus hydrogenoformans. Proc. Natl. Acad. Sci. U.S.A. 103, 891–896.
Dunfield, K. E., and King, G. M. (2005). Analysis of the distribution and diversity in recent Hawaiian volcanic deposits of a putative carbon monoxide dehydrogenase large subunit gene. Environ. Microbiol. 7, 1405–1412.
Edgar, R. C. (2004a). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. doi: 10.1186/1471-2105-5-113
Edgar, R. C. (2004b). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797.
Fox, J. D., Kerby, R. L., Roberts, G. P., and Ludden, P. W. (1996). Characterization of the CO-induced, CO-tolerant hydrogenase from Rhodospirillum rubrum and the gene encoding the large subunit of the enzyme. J. Bacteriol. 178, 1515–1524.
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2003). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321.
He, Y., Gaal, T., Karls, R., Donohue, T. J., Gourse, R. L., and Roberts, G. P. (1999). Transcription activation by CooA, the CO-sensing factor from Rhodospirillum rubrum. The interaction between CooA and the C-terminal domain of the alpha subunit of RNA polymerase. J. Biol. Chem. 274, 10840–10845.
He, Y., Shelver, D., Kerby, R. L., and Roberts, G. P. (1996). Characterization of a CO-responsive transcriptional activator from Rhodospirillum rubrum. J. Biol. Chem. 271, 120–123.
Henstra, A. M., and Stams, A. J. (2004). Novel physiological features of Carboxydothermus hydrogenoformans and Thermoterrabacterium ferrireducens. Appl. Environ. Microbiol. 70, 7236–7240.
Hill, A. V. (1910). The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J. Physiol. 40, iv–vii.
Hugendieck, I., and Meyer, O. (1992). The structural genes encoding CO dehydrogenase subunits (cox L, M and S) in Pseudomonas carboxydovorans OM5 reside on plasmid pHCG3 and are, with the exception of Streptomyces thermoautotrophicus, conserved in carboxydotrophic bacteria. Arch. Microbiol. 157, 301–304.
Ibrahim, M., Kuchinskas, M., Youn, H., Kerby, R. L., Roberts, G. P., Poulos, T. L., and Spiro, T. G. (2007). Mechanism of the CO-sensing heme protein CooA: new insights from the truncated heme domain and UVRR spectroscopy. J. Inorg. Biochem. 101, 1776–1785.
Kerby, R. L., Hong, S. S., Ensign, S. A., Coppoc, L. J., Ludden, P. W., and Roberts, G. P. (1992). Genetic and physiological characterization of the Rhodospirillum rubrum carbon monoxide dehydrogenase system. J. Bacteriol. 174, 5284–5294.
Kerby, R. L., Ludden, P. W., and Roberts, G. P. (1995). Carbon monoxide-dependent growth of Rhodospirillum rubrum. J. Bacteriol. 177, 2241–2244.
Kim, Y. J., Lee, H. S., Kim, E. S., Bae, S. S., Lim, J. K., Matsumi, R., Lebedinsky, A. V., Sokolova, T. G., Kozhevnikova, D. A., Cha, S. S., Kim, S. J., Kwon, K. K., Imanaka, T., Atomi, H., Bonch-Osmolovskaya, E. A., Lee, J. H., and Kang, S. G. (2010). Formate-driven growth coupled with H(2) production. Nature 467, 352–355.
King, G. M. (2003a). Contributions of atmospheric CO and hydrogen uptake to microbial dynamics on recent Hawaiian volcanic deposits. Appl. Environ. Microbiol. 69, 4067–4075.
King, G. M. (2003b). Molecular and culture-based analyses of aerobic carbon monoxide oxidizer diversity. Appl. Environ. Microbiol. 69, 7257–7265.
King, G. M., and Weber, C. F. (2007). Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. Microbiol. 5, 107–118.
Kraut, M., Hugendieck, I., Herwig, S., and Meyer, O. (1989). Homology and distribution of CO dehydrogenase structural genes in carboxydotrophic bacteria. Arch. Microbiol. 152, 335–341.
Lee, H. S., Kang, S. G., Bae, S. S., Lim, J. K., Cho, Y., Kim, Y. J., Jeon, J. H., Cha, S. S., Kwon, K. K., Kim, H. T., Park, C. J., Lee, H. W., Kim, S. I., Chun, J., Colwell, R. R., Kim, S. J., and Lee, J. H. (2008). The complete genome sequence of Thermococcus onnurineus NA1 reveals a mixed heterotrophic and carboxydotrophic metabolism. J. Bacteriol. 190, 7491–7499.
Lide, D. R., and Frederikse, H. P. R. (1995). CRC Handbook of Chemistry and Physics. Boca Raton, FL: CRC Press.
Meyer, O., Frunzke, K., and Mörsdorf, G. (eds). (1993). Biochemistry of the Aerobic Utilization of Carbon Monoxide. Andover, MA: Intercept, Ltd.
Meyer, O., and Schlegel, H. G. (1983). Biology of aerobic carbon monoxide-oxidizing bacteria. Annu. Rev. Microbiol. 37, 277–310.
Morton, T. A., Runquist, J. A., Ragsdale, S. W., Shanmugasundaram, T., Wood, H. G., and Ljungdahl, L. G. (1991). The primary structure of the subunits of carbon monoxide dehydrogenase/acetyl-CoA synthase from Clostridium thermoaceticum. J. Biol. Chem. 266, 23824–23828.
Parshina, S. N., Kijlstra, S., Henstra, A. M., Sipma, J., Plugge, C. M., and Stams, A. J. (2005). Carbon monoxide conversion by thermophilic sulfate-reducing bacteria in pure culture and in co-culture with Carboxydothermus hydrogenoformans. Appl. Microbiol. Biotechnol. 68, 390–396.
Puranik, M., Nielsen, S. B., Youn, H., Hvitved, A. N., Bourassa, J. L., Case, M. A., Tengroth, C., Balakrishnan, G., Thorsteinsson, M. V., Groves, J. T., Mclendon, G. L., Roberts, G. P., Olson, J. S., and Spiro, T. G. (2004). Dynamics of carbon monoxide binding to CooA. J. Biol. Chem. 279, 21096–21108.
Ragsdale, S. W. (2008). Enzymology of the Wood-Ljungdahl pathway of acetogenesis. Ann. N. Y. Acad. Sci. 1125, 129–136.
Robb, F. T., Gonzalez, J. M., Sokolova, T., Techtmann, S. M., Chernyh, N., Lebedinski, A., Tallon, L. J., Jones, K., Wu, M., and Eisen, J. A. (2005). “Primary energy metabolism in geothermal environments: the role of carbon monoxide,” in Geothermal Biology and Geochemistry in Yellowstone National Park, ed. W. I. A. T. Mcdermott (Bozeman, MT: Montana State University), 163–170.
Roberts, G. P., Kerby, R. L., Youn, H., and Conrad, M. (2005). CooA, a paradigm for gas sensing regulatory proteins. J. Inorg. Biochem. 99, 280–292.
Roberts, G. P., Thorsteinsson, M. V., Kerby, R. L., Lanzilotta, W. N., and Poulos, T. (2001). CooA: a heme-containing regulatory protein that serves as a specific sensor of both carbon monoxide and redox state. Prog. Nucleic Acid Res. Mol. Biol. 67, 35–63.
Schubel, U., Kraut, M., Morsdorf, G., and Meyer, O. (1995). Molecular characterization of the gene cluster coxMSL encoding the molybdenum-containing carbon monoxide dehydrogenase of Oligotropha carboxidovorans. J. Bacteriol. 177, 2197–2203.
Shelver, D., Kerby, R. L., He, Y., and Roberts, G. P. (1995). Carbon monoxide-induced activation of gene expression in Rhodospirillum rubrum requires the product of cooA, a member of the cyclic AMP receptor protein family of transcriptional regulators. J. Bacteriol. 177, 2157–2163.
Shelver, D., Kerby, R. L., He, Y., and Roberts, G. P. (1997). CooA, a CO-sensing transcription factor from Rhodospirillum rubrum, is a CO-binding heme protein. Proc. Natl. Acad. Sci. U.S.A. 94, 11216–11220.
Shock, E. L., Holland, M., Meyer-Dombard, D. R., Amend, J. P., Osburn, G. R., and Fischer, T. P. (2010). Quantifying inorganic sources of geochemical energy in hydrothermal ecosystems, Yellowstone National Park, USA. Geochimica et Cosmochimica Acta 74, 4005–4043.
Slepova, T. V., Rusanov, I. I., Sokolova, T. G., Bonch-Osmolovskaia, E. A., and Pimenov, N. V. (2007). Radioisotopic assays of rates of carbon monoxide conversion by anaerobic thermophilic prokaryotes. Mikrobiologiia 76, 594–601.
Slobodkin, A., Reysenbach, A. L., Strutz, N., Dreier, M., and Wiegel, J. (1997). Thermoterrabacterium ferrireducens gen. nov., sp. nov., a thermophilic anaerobic dissimilatory Fe(III)-reducing bacterium from a continental hot spring. Int. J. Syst. Bacteriol. 47, 541–547.
Slobodkin, A. I., Sokolova, T. G., Lysenko, A. M., and Wiegel, J. (2006). Reclassification of Thermoterrabacterium ferrireducens as Carboxydothermus ferrireducens comb. nov., and emended description of the genus Carboxydothermus. Int. J. Syst. Evol. Microbiol. 56, 2349–2351.
Sokolova, T. G., Gonzalez, J. M., Kostrikina, N. A., Chernyh, N. A., Slepova, T. V., Bonch-Osmolovskaya, E. A., and Robb, F. T. (2004a). Thermosinus carboxydivorans gen. nov., sp. nov., a new anaerobic, thermophilic, carbon-monoxide-oxidizing, hydrogenogenic bacterium from a hot pool of Yellowstone National Park. Int. J. Syst. Evol. Microbiol. 54, 2353–2359.
Sokolova, T. G., Jeanthon, C., Kostrikina, N. A., Chernyh, N. A., Lebedinsky, A. V., Stackebrandt, E., and Bonch-Osmolovskaya, E. A. (2004b). The first evidence of anaerobic CO oxidation coupled with H2 production by a hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Extremophiles 8, 317–323.
Spear, J. R., Walker, J. J., Mccollom, T. M., and Pace, N. R. (2005). Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc. Natl. Acad. Sci. U.S.A. 102, 2555–2560.
Svetlitchnyi, V., Sokolova, T. G., Gerhardt, M., Ringpfeil, M., Kostrikina, N. A., and Ga, Z. (1991). Carboxydothermus hydrogenoformans gen. nov., sp. nov., a CO-utilizing thermophilic anaerobic bacterium from hydrothermal environments of Kunashir Island. Syst. Appl. Microbiol. 14, 254–260.
Techtmann, S. M., Colman, A. S., and Robb, F. T. (2009). “That which does not kill us only makes us stronger”: the role of carbon monoxide in thermophilic microbial consortia. Environ. Microbiol. 11, 1027–1037.
Uffen, R. L. (1976). Anaerobic growth of a Rhodopseudomonas species in the dark with carbon monoxide as sole carbon and energy substrate. Proc. Natl. Acad. Sci. U.S.A. 73, 3298–3302.
Wu, D., Raymond, J., Wu, M., Chatterji, S., Ren, Q., Graham, J. E., Bryant, D. A., Robb, F., Colman, A., Tallon, L. J., Badger, J. H., Madupu, R., Ward, N. L., and Eisen, J. A. (2009). Complete genome sequence of the aerobic CO-oxidizing thermophile Thermomicrobium roseum. PLoS ONE 4, e4207. doi:10.1371/journal.pone.0004207
Wu, M., Ren, Q., Durkin, A. S., Daugherty, S. C., Brinkac, L. M., Dodson, R. J., Madupu, R., Sullivan, S. A., Kolonay, J. F., Haft, D. H., Nelson, W. C., Tallon, L. J., Jones, K. M., Ulrich, L. E., Gonzalez, J. M., Zhulin, I. B., Robb, F. T., and Eisen, J. A. (2005). Life in hot carbon monoxide: the complete genome sequence of Carboxydothermus hydrogenoformans Z-2901. PLoS Genet. 1, e65. doi:10.1371/journal.pgen.0010065
Keywords: carbon monoxide, thermophiles, hydrogenogens, carboxydotrophs, Carboxydothermus hydrogenoformans, carbon monoxide dehydrogenase, CooA
Citation: Techtmann SM, Colman AS, Murphy MB, Schackwitz WS, Goodwin LA and Robb FT (2011) Regulation of multiple carbon monoxide consumption pathways in anaerobic bacteria. Front. Microbio. 2:147. doi: 10.3389/fmicb.2011.00147
Received: 21 March 2011; Accepted: 22 June 2011;
Published online: 11 July 2011.
Edited by:
Ludmila Chistoserdova, University of Washington, USAReviewed by:
Gary M. King, Louisiana State University, USAAlfons Stams, Wageningen University, Netherlands
Copyright: © 2011 Techtmann, Colman, Murphy, Schackwitz, Goodwin and Robb. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Frank T. Robb, Institute of Marine and Environmental Technology, University of Maryland,701 East Pratt Street, Baltimore, MD 21202, USA. e-mail:ZnJvYmJAc29tLnVtYXJ5bGFuZC5lZHU=