- 1 Department of Pathology and Infectious Diseases, Royal Veterinary College, University of London, London, UK
- 2 School of Biology, Newcastle University, Newcastle upon Tyne, UK
The increasing incidence and prevalence of antibiotic resistance in bacteria threatens the “antibiotic miracle.” Conventional antimicrobial drug development has failed to replace the armamentarium needed to combat this problem, and novel solutions are urgently required. Here we review both natural and synthetic RNA silencing and its potential to provide new antibacterials through improved target selection, evaluation, and screening. Furthermore, we focus on synthetic RNA silencers as a novel class of antibacterials and review their unique properties.
Natural RNA Silencing in Bacteria
RNA silencing is widespread in nature. The transcripts involved are often termed non-coding regulatory RNAs or antisense RNA, and there are a range of mechanisms and cellular roles described (Thomason and Storz, 2010). These natural examples of RNA silencing provide important information about how synthetic RNA strategies can be best developed in practice.
RNA-Silencing Mechanisms
In bacteria, there are fascinating examples of trans- and cis-antisense sequences that reversibly repress translation. The regulatory regions within a single mRNA can contain neighboring cis-antisense sequences that form an intra-molecular antisense–sense fold. The folded structure masks or sequesters the ribosome-binding site (RBS), and this prevents initiation. Although, this arrangement is less obvious as a mechanism for antisense control, it was first described as early as 1985, and has now been associated with the regulation of many genes. Also, cis-antisense sequences may arise from transcription from the complementary strand at the same locus, and it is now clear that occur as frequently in simple unicellular bacteria as they do in higher organisms (Georg and Hess, 2011). Many natural antisense sequences are trans-acting regulatory RNAs, where the antisense RNA is transcribed from a distant locus. There may be a bias toward the discovery of such trans-encoded RNAs, as this is the anticipated nature of antisense; however, it seems likely that this is a common mechanism for natural antisense. In prokaryotes, trans-encoded antisense sequences typically work by binding to the start codon region of mRNA. For example, MicF RNA represses the translation of the outer membrane protein gene ompF (Delihas and Forst, 2001), and the OxyS RNA inhibits translation of two target genes, fhlA and rpoS (Altuvia et al., 1998). There are diverse examples of RNA silencing in bacteria (Thomason and Storz, 2010; Georg and Hess, 2011).
Antisense sequences that hybridize to messenger RNA can inhibit target gene expression in several ways; reviewed in-depth in Waters and Storz (2009). Perhaps the simplest mechanism is translation repression. It is easy to envision how this can occur in bacteria, where the ribosome binds to mRNA at the well-characterized RBS (Shine and Dalgarno, 1974). Blocking this initiation process by the positioning of an antisense sequencing at the RBS leads to unmasking of the messenger RNA and decay of the unused transcript. Therefore, RNA-silencing involves a combination of repression of initiation and transcript decay (Figure 1) and natural antisense transcripts have additional confirmed functional roles in transcription termination, co-degradation, transcriptional interference, and enhanced stability of their respective target transcripts. Furthermore, the recently described clustered regularly inter-spaced short palindromic repeats (CRISPR) mechanism provides acquired resistance against bacteriophage and involves RNA-mediated destruction of phage transcripts (Horvath and Barrangou, 2010). Therefore, there are a wide range of natural antisense mechanisms and functional roles, pointing the way to a variety of opportunities for synthetic RNA-silencing tools.
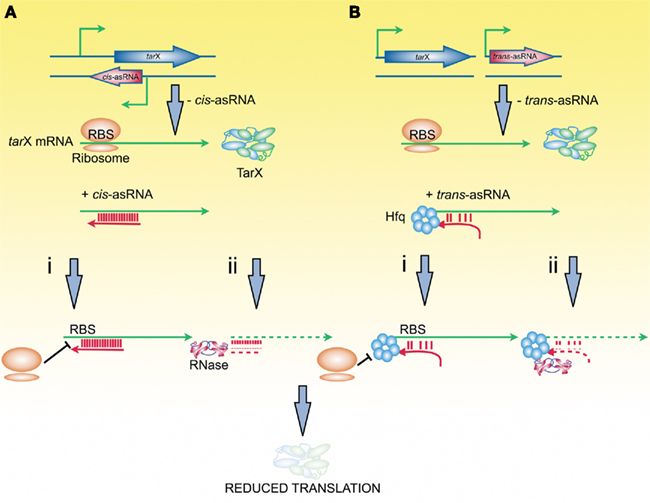
Figure 1. Natural RNA silencing in bacteria. (A) Antisense RNAs (asRNAs) that are cis-encoded share high degrees of complementarity with the target mRNA. (B) asRNAs that are trans-encoded have limited complementarity with the target mRNA and, in some species, require the an RNA chaperone (Hfq) to facilitate binding. In either case, once the asRNA is bound to the target mRNA, translation of the target gene (tarX) is silenced by inhibition of ribosome binding to the target mRNA (i); induced RNase degradation of the asRNA:mRNA hybrid (ii) or a combination of both processes.
Antisense RNAs Encoded in Bacterial Chromosomes and Plasmids
The first natural antisense mechanisms identified were associated with accessory elements in bacteria and viewed as peculiarities within the area of gene expression control. It is now clear that antisense mechanisms are abundant, widespread, and involved in core cellular processes as well as adaptive responses. Therefore, RNA silencing is a common and important form of posttranscriptional gene control.
Structural Features of Short Antisense RNAs and Target Binding
Two typical features of ncRNAs in bacteria are a short length and a stable secondary structure. A folded structure can contribute nuclease resistance and distinct functional domains (Majdalani et al., 1998). Also, the structure itself may aid recognition and binding. Folding may improve target binding through rapid loop–loop interactions. Among bacterial antisense RNAs, comparative sequence analysis revealed the “U-turn” structure, which consists of a short hairpin with a tetra-loop containing the YUNR motif (pyrimidine, uracil, any nucleoside, purine). This is an example of a motif that aids antisense interactions (Franch et al., 1999).
Protein Factors Involved in RNA Silencing
Antisense RNAs have the capacity to work alone as gene regulators through RNA/RNA interactions; however, protein factors may also play a role by binding to antisense and target RNAs. Double-stranded RNA binding domains are well known as factors in RNA processing, and these domains may mediate RNA interactions. Also, a eukaryotic Sm-like sequence motif is present in many antisense RNAs, and the E. coli Sm homolog Hfq recognizes this motif both within the antisense RNA and target mRNA, and Hfq can stabilize an interaction (Møller et al., 2002a). Sm and Sm-like proteins are involved in a number of RNA metabolism/processing steps in eukaryotes, and this role is conserved in bacteria, where Hfq mediates antisense–sense RNA pairing (Figure 1).
Sequence-Selective RNA Silencing
The problem of sequence-specific target recognition must be considered with an appreciation of transcript biology and complexity. Simple sequence uniqueness calculations suggest that gene or sequence-specific inhibitory effects in bacteria require a target sequence of approximately 12 bases (Good et al., 2001), whereas in mammalian cells 15 or more bases are needed due to larger genome sizes. These estimates are supported by the structure and activity of natural and synthetic antisense sequences; natural antisense RNAs typically form 12–30 bp with target mRNA, but binding is often complicated by the presence of mismatches and gapped structures. Designed antisense agents range in size from 6 to 25 residues, and are often near 20 residues for mammalian cell applications. Therefore, antisense agents tend to be just long enough to provide stable binding properties and gene-specific recognition. This is reasonable given that delivery is a major restriction. Natural antisense RNAs are typically longer, but in some cases only a portion of the transcript is involved in target hybridization. The rate-limiting step in RNA/RNA interactions is sequence recognition. Diffusion can be a factor particularly for large RNA molecules; however, RNA/RNA hybridization appears to be inherently constrained, possibly by electrostatic repulsion (Franch et al., 1999). The U-turn motif described above provides one way to reduce such constraints to binding. A prototype example of antisense RNA/mRNA recognition and binding that involves U-turn structures is the copA/copT system involved in plasmid replication control. Known as the “kissing-complex” interaction, hairpin loop heads on both the antisense, and target sequence speed initial binding. Kissing loops provide a meta-stable interaction that can extend to provide the binding stability needed to inhibit the target RNA. Also mismatches between the copA/copT RNAs ease loop extension and formation of the final stable binding complex (Kolb et al., 2000). Therefore, successful antisense interactions and stabilized binding can depend on sequence composition, higher-order structural motifs and in some cases complex RNA dynamics during binding. It may be possible to mimic some aspects of the these fascinating, evolved mechanisms.
Why Do Bacteria Use Endogenous RNA Silencing?
RNA structure and function has been described as having high evolvability (Waters and Storz, 2009), and interactions between RNAs can provide subtle and profound changes in biological processes. Bacteria benefit from these features and a range of remarkable RNA-level regulatory processes have evolved. Some of the best understood of these involve antisense repression of translation. An important feature of translation repression is that the mRNA can remain intact during periods of repression, and rapid expression switching may occur in response to cellular and environmental signals. Antibiotic exposure requires a rapid response to ensure cell survival, and it seems reasonable for cells to constitutively transcribe antibiotic resistance genes and then control expression at the translation level. Cis-encoded antisense sequences are within the chloramphenicol and erythromycin resistance genes in E. coli (Moffat et al., 1994). For both the chloramphenicol and erythromycin resistance genes, rapid expression control appears to be provided by antisense sequences that lie within a short open reading frame just upstream of the start codon region. Also, RNA-silencing appears to enable gene control within operons. For example, Spot 42 RNA repression of galK at an internal position within the galETKM operon transcript (Møller et al., 2002b) effectively discoordinating expression of an otherwise coordinated expression system. Finally, RNA silencing is involved in the bacterial CRISPR mechanism to guard against viral infection. Therefore, phenotypes as diverse as antibiotics resistance and phage resistance involve natural RNA silencing and there are clear benefits to the cell in involving RNA-level gene control.
Difficulties in the Practical Application of Synthetic RNA Silencing in Bacteria
There are many potential research and practical applications of RNA silencing. Examples from nature showing how bacteria themselves utilize RNA::RNA interactions to regulate gene expression supports the notion that synthetic RNA silencing can be of practical benefit. This is particularly attractive for species where standard genetics or conventional drug development has yet to provide satisfactory solutions. Nevertheless, despite the promise of the approach and the insight provided by nature, difficulties remain in getting RNA silencing to work well in diverse practical applications.
As described in detail below, there are two general strategies to RNA silencing, where the silencing agent is either expressed as a natural RNA transcript within cells (expressed asRNA) or where the silencing agent is a short non-natural nucleobase polymer that is delivered into bacteria. In both cases it is self evident that the silencing sequence must be sufficiently abundant in cells to sequester the bulk of transcript mRNA to effectively repress translation initiation. To reach such levels, it is necessary to use efficient expression or delivery and stabilizing structures that limit nuclease decay in cells.
In the case of expressed RNA, it is standard practice to use strong and tightly inducible promoters. In addition, evidence for rapid transcript decay in cells has led to effort to stabilize asRNA following synthesis by including stabilizing RNA structures within the asRNA transcript. For example, the transcript termini can be paired by introducing complimentary termini that generate a stem structure that simultaneously protects both the 5′ and 3′ termini, resulting in an accumulation of PTasRNA in cells, providing more effective silencing (Nakashima et al., 2006). As expected, improved silencing correlated with transcript abundance, confirming that a challenge to effective RNA silencing is to achieve sufficient intracellular concentrations of the asRNA sequence (Figure 2).
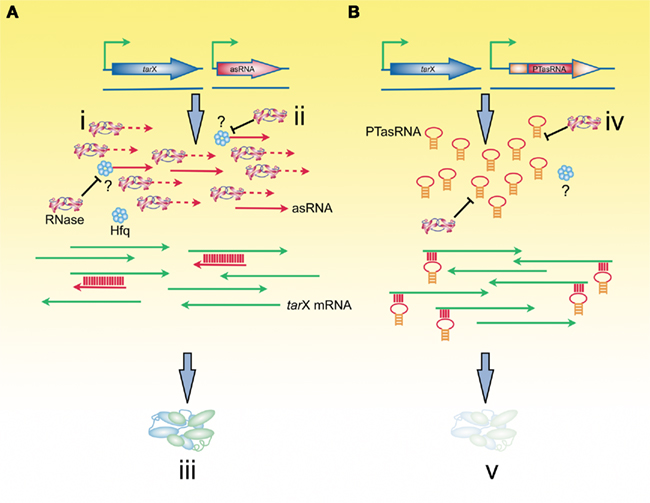
Figure 2. Abundance and stability of antisense RNA is related to the efficacy of gene silencing. (A) Linear asRNAs are subject to degradation by RNases (i) and thus, are not available to form hybrids with their target mRNA. The role of Hfq in trans-encoded asRNAs with a high degree of sequence complementarity with the target mRNA is not elucidated, however, as with natural asRNAs, Hfq may serve to protect asRNAs from degradation (ii). In situations where the abundance and stability of the linear asRNA is low, only partial gene silencing will result (iii). (B) Addition of paired-termini to the asRNA results in asRNAs that have increased abundance, likely as a result of resistance to RNase degradation (iv). The role of Hfq in PTasRNA-mediated gene silencing is unclear. The increased abundance of PTasRNAs results in a higher level of gene silencing (v).
In the case of short non-natural nucleobase polymers, it is standard practice in antisense, and siRNA design to use stabilizing nucleic acid analogs or mimics that resist decay by cellular nucleases. For bacterial applications, we have focused on peptide nucleic acid (PNA; Good et al., 2001), where the sugar phosphate backbone is entirely replaced by a pseudopeptide backbone, apparently providing full protection. Phosphorothioate morpholino oligomers (PMOs) provide similar advantages, involving different chemistry for full backbone replacement (Geller et al., 2003). Indeed, organic chemists provide a variety of analogs and mimics that can potentially act as stable and effective RNA silencers in bacteria. Perhaps the larger challenge is to achieve sufficient intracellular delivery across stringent bacterial cell barriers. Nucleobase oligomers that are long enough for sequence recognition are inherently too large for uptake by simple diffusion. Fortunately, bacterial barriers can be overcome by conjugating the oligomers (PNA or PMO) to cell entry peptides, which are typically cationic (Nikravesh et al., 2007; Mellbye et al., 2009). There are difficulties with developing such peptides as components or drugs, and further developments are needed, but the peptide conjugates that have been described to date provide effective delivery in bacteria grown in culture and as pathogens within animals.
In all applications of RNA silencing it is difficult to design silencers that specifically recognize and inhibit the target mRNA without unintended effects. In the case of bacteria, the situation is somewhat easier, because bacterial genomes are relatively small. Nevertheless, it is important to carefully design and evaluate silencers to ensure gene-specific effects. For expressed RNA, we typically target 100–150 bases surrounding the RBS. For PNA, we typically target a site of 10–15 bases in length, taking care to cover the Shine–Dalgarno sequence or start codon within the RBS region. Our design guidelines have been described previously, along with a description of control experiments that are needed to ensure gene selective silencing (Dryselius et al., 2003a). As controls to ensure gene specificity, we favor phenotype complementation, where the target gene is over expressed to rescue cells from the affects of RNA silencing, thus confirming the mechanism of action (Goh et al., 2009).
Synthetic RNA Silencing for Gene Function Studies and Antimicrobial Discovery
The use of synthetic RNA silencing has applications in the discovery of novel antimicrobial targets, determination of the stringency of requirement for those targets, the development of highly sensitized antimicrobial screens, mode of action studies, and in the growing field of polypharmacology.
Targets
In the post-genomic era, the process of discovering novel antimicrobial compounds can be broadly described as: (i) identification of novel essential genes, (ii) development of novel, preferably whole-cell, screens, and (iii) validation of the mode of action (MOA) of antibacterial compounds (Thomson et al., 2004). RNA silencing is applicable to each of these areas, and its use has resulted in the discovery of potent and novel antimicrobial compounds (see below). The construction of artificial antisense RNA silencers, to aid in the identification of novel antimicrobial targets, can follow random or targeted design strategies. The random approach is exemplified by the studies of Ji et al. (2001) and Forsyth et al. (2002). In both studies, genomic DNA of Staphylococcus aureus was shotgun fragmented (200–800 bp), blunt-ended, and cloned into an inducible expression vector. Transformation of S. aureus with the fragment library followed by phenotype comparison, on inducing and non-inducing plates, revealed clones that had growth defective or lethal phenotypes. Sequencing of these clones revealed 70% to contain genome fragments in the antisense orientation (Forsyth et al., 2002). An analysis of the binding sites of the randomly generated RNA silencers showed them to be distributed across the length of their target genes. Interestingly, in contrast to natural trans-encoded antisense RNAs (see above), the authors noted that there was a notable absence of RNA silencers that mapped to the N-terminal of the gene. They suggested that there may be preferential target sites for antisense inhibitory activity or that RNA silencers targeting the N-terminus of the gene were deleterious to the growth of E. coli (used to amplify the shotgun library prior to S. aureus transformation). The principal aim of both of the above studies was the facile, unbiased identification of essential genes; a prerequisite for the development of novel antimicrobial compounds. The adoption of a similar approach in Gram-negative bacteria has yet to be published. The reason for this is not clear, but may include different RNA chaperone requirements for trans-acting antisense RNAs (Waters and Storz, 2009); in S. aureus Hfq is not required for functional regulatory RNAs and lacks the C-terminal domain that binds to A-rich domains of the target mRNA (Lorenz et al., 2010). Furthermore, a lower abundance of the expressed antisense in Gram-negative bacteria could be explain the different efficacies in Gram-negative and positive species: S. aureus lacks an RNAse E homolog, which may explain the efficacy of expressed antisense silencers in this species. (Xu et al., 2010) noted that use of an E. coli RNAse E mutant resulted in a significant improvement in the frequency of isolation of clones expressing antisense RNA that were complementary to essential genes. The use of PTasRNA (see above) may also enable shotgun-based approaches to be employed in the identification of essential targets in Gram-negative species. It remains to be determined whether Hfq is required for RNA silencing with PTasRNAs (Figure 3).
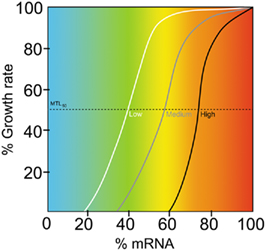
Figure 3. Measuring the degree of gene essentiality. The relationship between the reduction of growth rate and the reduction in mRNA (antisense-induced gene silencing) is a measure of the degree of essentiality, or stringency, of an essential gene (Goh et al., 2009). The minimum transcript level for a 50% reduction in growth (MTL50, dotted line) can be used to compare genes. In this hypothetical example three genes are compared and show low (white line), medium (gray), and high (black) stringencies of requirement.
Target Stringency
The fact that a gene is essential is not necessarily a predictor of the drugability of its cognate protein, i.e., inhibition of the essential protein may not lead to arrested growth or cell death. Thus, the identification of targets that are stringently required (those that cause significant growth defects when subject to comparatively low degrees of inhibition) is highly desirable. In order to assess such target stringency at the protein level, specific inhibitors, with known inhibition kinetics would be required for all essential proteins and is not currently feasible. However, a quantitative measure of the stringency of requirement of essential genes can be used for predictive purposes: Goh et al. (2009) hypothesized that essential genes differ in requirement stringencies, and that those differences could be revealed by measuring the relationship between the decrease in a specific mRNA and bacterial growth rate decline (Figure 3). In their study they selected four essential genes in E. coli (acpP, fabI, ftsZ, and murA) and applied both expressed (PTasRNAs) and oligo-nucleobase (PNAs) RNA silencers to selectively titrate down gene expression and measure growth responses. The authors used a minimum transcript level for a 50% reduction in growth rate (MTL50) to compare the four genes and showed that the hierarchy of stringency had the order acpP > ftsZ > fabI > murA. The results indicated that to maintain 50% viability E. coli requires >70% of the acpP transcripts relative to a normal cell, whereas only 40% of the murA transcripts are required. Interestingly, MurA is the target of a clinically used antimicrobial (fosfomycin), suggesting that AcpP may be an interesting target for antimicrobial discovery; FtsZ has been the subject of a number of antibacterial discovery programs (Lock and Harry, 2008). The assessment of the stringency of requirement may prevent the selection of poor targets early in the discovery phase of antimicrobial development.
Screening
Novel antimicrobial compounds are assessed for their ability to inhibit growth (bacteriostatic) and kill (bacteriocidal) bacteria in vitro. Currently, there is a lack of clinical data to support the preferential development of either class of compounds, as clinical outcomes are often overlooked in evaluation (Pankey and Sabath, 2004). However, bacteriocidal properties are often sought in antimicrobial development. In particular, bacteriocidal agents are preferred in the treatment of slow growing organisms such as Mycobacterium tuberculosis, where the majority of viable anti-tubercular agents are bactericidal. (Kaur et al., 2009) showed that conditional RNA silencing of an essential gene target at relatively high cell number, with subsequent monitoring of cell number over time, can be applied to determine whether the inhibition of the cognate essential protein is likely to result in bacteriostasis or bacteriocide (Figure 4). In their study, mycobacterial genes that encode proteins that are the targets of standard anti-tubercular drugs (inhA, rpoB, and gyrA) were shown to be bactericidal when subject to RNA silencing, whereas silencing of ftsZ was bacteriostatic.
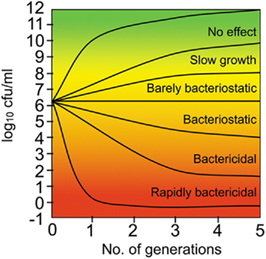
Figure 4. Antisense prediction of bacteriostatic and bactericidal targets. Antisense-induced silencing of essential genes and subsequent monitoring of survival kinetics can be used to delineate targets that will likely produce cidal, static, or no effect upon inhibition of the cognate protein (adapted from Kaur et al., 2009).
Once a target has been selected for antibacterial discovery, RNA silencing can be applied to for the development of antibacterial screens. In theory, a strain expressing antisense RNA, or exposed to a nucleobase RNA silencer, targeting a specific mRNA, will have a lower cellular pool of the cognate protein and thus will become sensitized to inhibitors of that protein. RNA-silencing based antimicrobial screens require that the expression of only the target(s) of interest be knocked down; an RNA silencer may target a single gene, or multiple genes via binding site similarities or their presence in operons downstream of the target gene (Dryselius et al., 2006). The specificity of RNA silencing has been demonstrated by a number of methods including: Western blot analysis of the target protein in induced and non-induced strains (Ji et al., 2004); reversible knockdown of auxotrophy (Kaur et al., 2009); complementation of the target gene (Dryselius et al., 2003b; Goh et al., 2009); silencing of reporter genes (Nekhotiaeva et al., 2004; Dryselius et al., 2006); and qRT-PCR (Dryselius et al., 2006; Goh et al., 2009; Kaur et al., 2009). In addition, the utility of expressed antisense RNA silencers in drug discovery was proven by the fact that an S. aureus strain expressing an antisense RNA to the gene fabF was 12-fold more sensitive to cerulenin (a specific inhibitor of FabF) when RNA silencing was induced. In contrast, that same strain was not significantly sensitized to other antibiotics (Forsyth et al., 2002). Specificity has also been demonstrated in E. coli where sensitization to triclosan (FabI inhibitor) but not trimethoprim or rifampicin, was observed upon induction of antisense fabI RNA (Nakashima et al., 2006). Furthermore, combination treatment of both E. coli and S. aureus with PNAs and protein-level inhibitors only resulted in antimicrobial synergy when both compounds were directed to the same target (Dryselius et al., 2005), indicating that nucleobase RNA silencers have similar target specificity. The use of RNA silencing for antimicrobial screening has a particular advantage when screening natural product extracts; bioactive secondary metabolites are often produced in very small quantities (Olano et al., 2008). Thus, the increased sensitization of the RNA-silencing based screens enables detection of compounds that would be missed in standard whole-cell antimicrobial screens, or other target-directed whole-cell screens, such as those where promoters of genes that are specifically upregulated in response to antibacterial compounds with specific modes of action are fused to reporter genes (Hutter et al., 2004). The most convincing argument for the employment of RNA silencing in antibacterial screening has come from the discovery of inhibitors of the fatty acid biosynthesis pathway. Merck Research Laboratories used S. aureus strains expression antisense RNA to fabF to screen 250,000 natural product extracts for FabF/FabH inhibitors (fabF and fabH are organized in an operon). All known natural product FabF and FabH inhibitors (cerulenin, thiolactomycin, thiotetramycin, and Tü3010) were discovered using this approach, in addition Phomallenic acids, new inhibitors of FabF were discovered (Young et al., 2006). This discovery was followed shortly after by the discovery of platensimycin a potent selective inhibitor of FabF and platencin a similarly potent dual FabF/H inhibitor (Wang et al., 2007). The fact that previous antimicrobial screening of the same extract library had failed to identify these compounds, a novel class of antimicrobials, is a convincing vindication of the use of RNA silencing in antimicrobial discovery. Use of RNA-silencing based screens in Merck Research Laboratories has also been demonstrated effective in the isolation of inhibitors of rpsD (small ribosomal protein S4; Singh et al., 2008; Zhang et al., 2009).
Mode of Action
The identification of a MOA is an important step in the development of antibacterial compounds, not least to avoid rediscovery of known molecules, but to deselect compounds that kill through non-selective mechanisms. Furthermore, the elucidation of a MOA can be used to determine if chemical modifications of a lead structure result in compounds that retain the MOA of the parent (Haydon et al., 2008) The fact that RNA-silencing based antimicrobial screens are specifically sensitized for inhibition of a known target, can be exploited to identify the MOA for known antimicrobial compounds. A recent example was the proof that berberine, a plant alkaloid used as an anti-infective in traditional medicine, was an inhibitor of bacterial cell division (Boberek et al., 2010). Domadia et al. (2008) had reported that berberine was a likely inhibitor of FtsZ; data supporting this hypothesis included NMR spectroscopy, FtsZ polymerization and GTPase assays, and the observation of cell filamentation in E. coli cells treated with berberine. However, cell filamentation in E. coli can result from induction of the SOS response (SulA binds to FtsZ to prevent cell division). Boberek et al. (2010) used RNA silencing in both wild-type and sulA deletion strains of E. coli and showed that berberine was indeed an inhibitor of cell division. While, RNA-silencing studies of this nature are proven in the discovery of antimicrobial compounds and their MOA, they have the drawback that only one target is screened at a time. In response to this limitation, arrays of strains can be used to provide comprehensive target coverage combined with accurate and facile determination of MOA (Xu et al., 2010). For antimicrobial discovery, the general approach of using a population of cells composed of strains that have an individual deficiency for a specific protein was first demonstrated in fungal species, where deletion of one allele in a diploid strain results in haploid insufficiency (HI) that can result in lower fitness relative to the parent strain (Giaever et al., 2002). Strains that are HI for the gene target of an antifungal are thus hypersensitized to inhibition. (Xu et al., 2007) used a population of HI Candida albicans strains that were genetically barcoded so that population shifts resulting from antifungal treatment could be observed using microarray analysis; HI strains conferring both hypersensitivity and resistance to the test antifungal could be simultaneously identified. Such a fitness test was applied in the discovery of the parnafungins, a novel class of antifungal that inhibit mRNA polyadenylation (Bills et al., 2009). The use of an equivalent fitness tests in S. aureus has recently been described, where the use of RNA silencing was used to create the conditional alleles of essential genes – termed the S. aureus TargetArray (Xu et al., 2010). In the study, 236 RNA-silencing clones were produced to knockdown expression of cognate targets. Pools of the strains were subject to fitness tests in the presence and absence of antibacterial compounds, with strain responses measured by multiplex PCR. The TargetArray methodology has been has been validated by the elucidation of the MOA of protein synthesis and peptidoglycan inhibitors (Donald et al., 2009; Huber et al., 2009).
Polypharmacology
Polypharmacology describes the phenomenon that a single compound can bind specifically to two or more targets. As noted above, antibacterial discovery in the post-genomic era has focused on a model of development from single targets. However, this model has failed to live-up to its initial promise. Reasons for this failure are multifaceted, but include the observation that, for single-target inhibitors, individual point mutations in the target gene may be sufficient to overcome inhibition (Yeh et al., 2009). Furthermore, identification of targets based on the deletion of individual genes, and assessment of in vitro viability, may overlook targets that are essential in vivo. Network analysis predictions have highlighted the importance of perturbing multiple proteins in order to overcome the limitations of single-target inhibitors (Hopkins, 2008). The polypharmacological approach to antimicrobial development is validated by the fact that most of the targets of successful (i.e., those in clinical use) antimicrobials are rarely encoded by a single gene (Brötz-Oesterhelt and Brunner, 2008). Thus, the development of screens that can identify combinations of targets that can be inhibited by one compound are required to identify novel antimicrobials with low resistance rates. Sensitized antimicrobial screens are well suited for the development of screens for polypharmacological antimicrobials, as bacteria are known to use single antisense RNAs to regulate multiple genes (see above). Furthermore, multiple antisense RNAs can be expressed in a single host (Nakashima and Tamura, 2009). Synthetic antisense methods could be applied to the identification of combinations of known essential genes that display synergistic properties when simultaneously silenced (dual essential), or combinations of genes that are not essential when individually silenced, but are essential in combination (synthetic lethals). A systematic approach to constructing antisense strains for all of the combinations of essential genes in a bacterium would be an insuperable task (In E. coli >83,000 strains for two-pair constructs alone). However, the process could be rationalized and focused by an understanding of the antimicrobial interaction network (Figure 5). Such analysis has revealed that antimicrobial drugs can be separated into classes such that any two will interact purely synergistically or purely antagonistically (Yeh et al., 2009). Thus antisense strains could be constructed that silence combinations of genes in classes known to have synergistic interactions. A random approach for the identification of synthetic lethals could be achieved by using the shotgun cloning method in strains carrying deletions of non-essential genes, for example, the Keio collection of single-gene knockouts in E. coli (Baba et al., 2006). Cloning of the shotgun fragments into a mixture of the Keio strains, selection of clones that show growth defects when the antisense is induced, and sequencing of clones, could identify those that carry antisense fragments to non-essential genes that are synthetically lethal in combination with the individual gene knockout carried by the host strain. Oligo-nucleobase RNA silencers have promise in the area of polypharmacology, either as tools to identify and evaluate targets (as above) or as polypharmacological antibacterials themselves. As with expressed RNA silencers, oligo-nucleobase silencers can be applied in combination. To our knowledge, a study specifically demonstrating antimicrobial synergy using multiple PNAs has yet to be undertaken, but it would seem logical to use the drug interaction network to identify novel gene targets that would likely yield antimicrobial synergy. In our labs we have observed that closely related species show different sensitivity to the same antimicrobial PNA, preliminary bioinformatic studies have indicated that this may be due to silencing of more than one essential gene; evidence of polypharmacology (unpublished). This is to be expected as the target site for PNAs is constrained (see above), and offers a means to specifically design single PNAs that silence multiple essential genes. Thus, synthetic gene silencing applied singularly, or in combination with chemical inhibition approaches, represents a powerful tool in the search for polypharmacological antimicrobial targets and drugs.
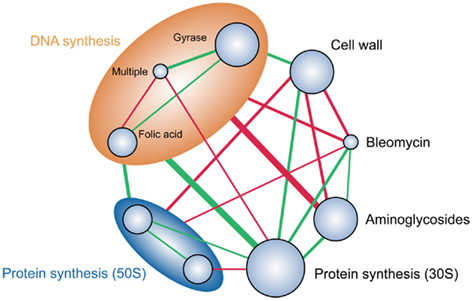
Figure 5. Functional classification of antimicrobial drugs by their pairwise interactions. Adapted form (Yeh et al., 2006). Synergistic (red) and antagonistic (green) interactions are highlighted. Circle size of the drug classes represents the number of compounds examined and the line thickness the number of interactions. The larger ellipses show higher-level functional classification of DNA synthesis inhibitors (orange) and protein synthesis inhibitors of the 50S ribosome subunit. Selection of gene targets from classes known to have synergistic inhibition properties could aid in the selection of genes for the design of antisense-based screens for polypharmacological targets and inhibitors.
Antisense Antibacterials
Synthetic RNA silencers, such as PNAs and PMOs, are not only valuable research tools in the discovery of novel antibacterial compounds, but are themselves a novel class of antibacterial; use of both the above compounds as antibacterials is well documented (Good and Nielsen, 1998; Good et al., 2001; Eriksson et al., 2002; Dryselius et al., 2003b; Nekhotiaeva et al., 2004; Tan et al., 2005; Nikravesh et al., 2007; Mellbye et al., 2009). In this section we focus on the unique properties of these compounds and the likely benefits to antimicrobial development.
Species-Specific Antimicrobial Compounds
Economic and diagnostic pressures have traditionally favored the development of broad-spectrum antibiotics (Casadevall, 2009); these drugs have enabled empirical usage (i.e., without the use of diagnostic identification of the pathogen) and rapid treatment of life-threatening infections. However, the development of resistance to broad-spectrum antibiotics is, in part, a consequence of their indiscriminate MOA. The vast majority of bacteria on and within the human body (the microflora) are non-pathogenic, with many being beneficial to health (Eckburg et al., 2005). Broad-spectrum antibiotics that kill or suppress the growth of these species can have negative outcomes in patient treatment as a result of secondary infection following disruption of the resident microflora (Fowler et al., 2007; Casadevall, 2009) or through the selection of resistance mechanisms in non-target species that are passed, by horizontal transfer, to pathogenic species. Furthermore, the microflora are associated with development and maturation of the immune system; perturbation of this interaction, through the use of broad-spectrum antibiotics, has been implicated in increased risk of diseases such as asthma, eczema, rhinoconjunctivitis, and breast cancer (Foliaki et al., 2009). The degree of microflora perturbation associated with broad-spectrum antimicrobials was highlighted in a recent study by Jakobsson et al. (2010). In their study, the affect of clarithromycin and metronidazole (used in the treatment of Helicobacter pylori) on the microflora was investigated. The authors observed dramatic shifts in the microflora of treated individuals, compared with the relatively stable populations of untreated controls. A striking finding from this study was the observation that, in some cases, the microflora perturbation remained for up to 4 years post-treatment associated with an increased level of the macrolide resistance gene erm(B). Narrow-spectrum (genus- or species-specific) antimicrobial agents, that are only active against the causative pathogen could potentially overcome some of the drawbacks of broad-spectrum antibiotic use, by ameliorating the impact on non-target microflora. Theoretically, RNA-silencing agents such as PNAs and PMOs have the greatest potential to have a tailored spectrum of activity, as the degeneracy of the genetic code ensures that there is greater variation at the genetic, rather than phenotypic level. We have exploited the bactericidal and sequence-specific properties of peptide-PNA conjugants to design species-specific antimicrobial compounds that are capable of discriminating between closely related bacteria such as E. coli, S. enterica, and K. pneumoniae in mixed culture (unpublished). Such selectivity was not possible with single or mixed application of known antibiotics. Furthermore, bioinformatic comparison of E. coli and S. enterica revealed >100 target sites for potential species-discriminating PNAs. We predict that it is unlikely that a similar number of species-discriminating phenotypic targets could be identified for these closely related species. The development and clinical implementation of species-specific antimicrobials alongside immunotherapy approaches aimed at improving host responses, is predicted to be the Third Age of antimicrobial therapy (Casadevall, 2006, 2009; Then and Sahl, 2010) the unique MOA of bactericidal RNA-silencing compounds make them good candidates for the development of narrow-spectrum antibiotics. A further benefit of the MOA of RNA silencers, is their potential to overcome antibacterial resistance (see below).
Delivery of RNA Silencers at Sites of Infection
Efforts to develop RNA silencers as antimicrobials can be criticized on the basis that nucleobase oligomers are considered to be inherently too large to achieve efficient cell uptake and in vivo distribution. However, peptide-PNA and peptide-PMO oligomer-based antimicrobials perform well in cell kill assays in vitro and in vivo. Also, there is renewed interest from clinicians in the efficacy of larger molecular weight antimicrobials (Falagas and Michalopoulos, 2006). Nevertheless, restricted cell uptake of oligomer-based antibiotics is an obvious difficulty. On the other hand, whereas conventional antibiotics are rapidly expelled from bacteria by efflux pumps (George and Hall, 2002), PNAs are retained for several hours, and not removed by efflux pumps. This leads to accumulation in cells and a long post-antibiotic effect (Nikravesh et al., 2007). Such properties help to explain the potent bactericidal effects observed in vitro and in vivo. Whereas delivery of antisense agents into bacteria remains a significant challenge, once delivered, the molecule is surprisingly well retained. These accumulation and retention properties may prove particularly appropriate in new strategies for treating persistent, dormant, or drug resistant infections. Therefore, if drug delivery can be achieved at the site of infection, these antibiotics appear more likely to be retained within pathogens and at the site of infection. Strategies to improve delivery include altered oligomer chemistry, conjugation to carrier molecules, and encapsulation within nanoparticles. To date, most progress has been made by conjugation to carrier peptides; however, each of these areas is complex and researchers are only beginning to explore the various opportunities available to enhance uptake.
Can We Silence Antimicrobial Resistance in Clinical Applications?
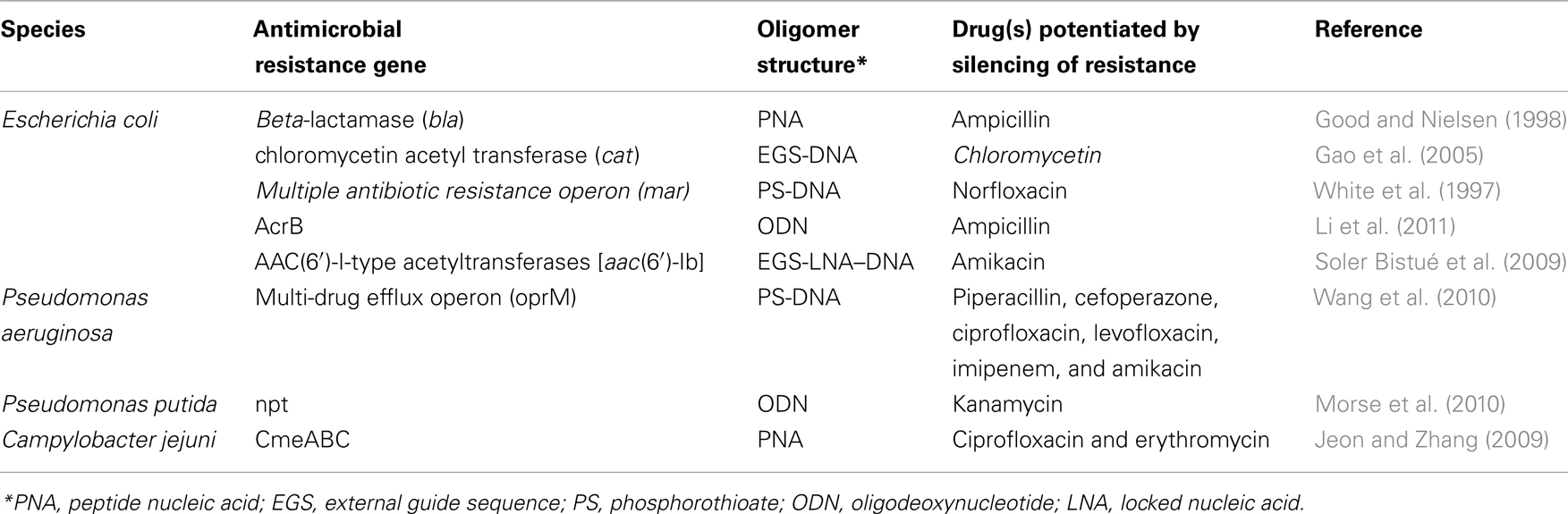
Acquired antimicrobial resistance is typically conferred by expression of one or just a few genes. Therefore, in principle, it should be possible to silence resistance to regain near full potency. Protein-level inhibitors of beta-lactamase enzymes have helped to recover the activity of beta-lactamase antibiotics. Unfortunately, it has proven difficult to extend this idea to other enzymes involved in resistance, which has led to interest in the idea of silencing resistance genes at the mRNA level. Several in vitro studies have demonstrated similar recovery of drug activity by silencing resistance genes at the RNA level (Table 1). Development of the approach for clinical applications would require that the RNA-silencing agents satisfies all requirements as a drug and also works well in combination with the partner antibiotic in vivo.
Conclusion
The requirement for novel antimicrobial compounds and strategies is of upmost importance if we are to continue to reap the benefits of the antibiotic era; improved life expectancy and outcomes. The current dearth of truly novel antimicrobials in the development pipelines requires that we explore multiple research avenues in antimicrobial research. Synthetic RNA-silencing offers several benefits to antimicrobial research, indeed its use has already aided in the discovery of novel antibacterial targets and compounds and the future application of RNA silencing will, no doubt, continue to realize novel antibacterials. Future priorities for expressed RNA silencers include: Determination of target stringency for known essential genes in a number of Gram-negative and positive species, with correlation of data to known antibacterial targets, and implementation of S. aureus TargetArray technology for a broad range of pathogenic bacteria. The latter will likely require development of host strains based on detailed understanding of the fate of expressed RNAs. The potential for oligo-nucleobase RNA silencers such as PNAs and PMOs may require the development of second-generation carrier strategies, possibly involving non-biological solutions. Investigation of PNA antimicrobial efficacy with leaky mutants of E. coli, suggest that very significant improvement could be made upon improved delivery. Furthermore, a range of carrier molecules may aid in target delivery to sights of infection, or to specific species. The development of polypharmacological oligo-nucleobase RNA silencers will also improve antibacterial efficacy. Such improvements could accelerate clinical evaluation of RNA silencers and hopefully deliver a new class of antimicrobial compound with unique design properties.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Altuvia, S., Zhang, A., Argaman, L., Tiwari, A., and Storz, G. (1998). The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 17, 6069–6075.
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., Datsenko, K. A., Tomita, M., Wanner, B. L., and Mori, H. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006–0008.
Bills, G. F., Platas, G., Overy, D. P., Collado, J., Fillola, A., Jiménez, M. R., Martín, J., Del Val, A. G., Vicente, F., Tormo, J. R., Peláez, F., Calati, K., Harris, G., Parish, C., Xu, D., and Roemer, T. (2009). Discovery of the parnafungins, antifungal metabolites that inhibit mRNA polyadenylation, from the Fusarium larvarum complex and other Hypocrealean fungi. Mycologia 101, 449–472.
Boberek, J. M., Stach, J., and Good, L. (2010). Genetic evidence for inhibition of bacterial division protein FtsZ by berberine. PLoS ONE 5, e13745.
Brötz-Oesterhelt, H., and Brunner, N. A. (2008). How many modes of action should an antibiotic have? Curr. Opin. Pharmacol. 8, 564–573.
Casadevall, A. (2009). The case for pathogen-specific therapy. Expert Opin. Pharmacother. 10, 1699–1703.
Delihas, N., and Forst, S. (2001). MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Mol. Biol. 313, 1–12.
Domadia, P. N., Bhunia, A., Sivaraman, J., Swarup, S., and Dasgupta, D. (2008). Berberine targets assembly of Escherichia coli cell division protein FtsZ. Biochemistry 47, 3225–3234.
Donald, R. G. K., Skwish, S., Forsyth, R. A., Anderson, J. W., Zhong, T., Burns, C., Lee, S., Meng, X., Locastro, L., Jarantow, L. W., Martin, J., Lee, S. H., Taylor, I., Robbins, D., Malone, C., Wang, L., Zamudio, C. S., Youngman, P. J., and Phillips, J. W. (2009). A Staphylococcus aureus fitness test platform for mechanism-based profiling of antibacterial compounds. Chem. Biol. 16, 826–836.
Dryselius, R., Aswasti, S. K., Rajarao, G. K., Nielsen, P. E., and Good, L. (2003a). The translation start codon region is sensitive to antisense PNA inhibition in Escherichia coli. Oligonucleotides 13, 427–433.
Dryselius, R., Nekhotiaeva, N., Nielsen, P. E., and Good, L. (2003b). Antibiotic-free bacterial strain selection using antisense peptide nucleic acid. BioTechniques 35, 1060–1064.
Dryselius, R., Nekhotiaeva, N., and Good, L. (2005). Antimicrobial synergy between mRNA- and protein-level inhibitors. J. Antimicrob. Chemother. 56, 97–103.
Dryselius, R., Nikravesh, A., Kulyté, A., Goh, S., and Good, L. (2006). Variable coordination of cotranscribed genes in Escherichia coli following antisense repression. BMC Microbiol. 6, 97.
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., Gill, S. R., Nelson, K. E., and Relman, D. A. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638.
Eriksson, M., Nielsen, P. E., and Good, L. (2002). Cell permeabilization and uptake of antisense peptide-peptide nucleic acid (PNA) into Escherichia coli. J. Biol. Chem. 277, 7144–7147.
Falagas, M. E., and Michalopoulos, A. (2006). Polymyxins: old antibiotics are back. Lancet 367, 633–634.
Foliaki, S., Pearce, N., Björkstén, B., Mallol, J., Montefort, S., Von Mutius, E., International Study of Asthma and Allergies in Childhood Phase III Study Group. (2009). Antibiotic use in infancy and symptoms of asthma, rhinoconjunctivitis, and eczema in children 6 and 7 years old: International Study of Asthma and Allergies in Childhood Phase III. J. Allergy Clin. Immunol. 124, 982–989.
Forsyth, R. A., Haselbeck, R. J., Ohlsen, K. L., Yamamoto, R. T., Xu, H., Trawick, J. D., Wall, D., Wang, L., Brown-Driver, V., Froelich, J. M., C, K. G., King, P., Mccarthy, M., Malone, C., Misiner, B., Robbins, D., Tan, Z., Zhu Zy, Z.-Y., Carr, G., Mosca, D. A., Zamudio, C., Foulkes, J. G., and Zyskind, J. W. (2002). A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43, 1387–1400.
Fowler, S., Webber, A., Cooper, B. S., Phimister, A., Price, K., Carter, Y., Kibbler, C. C., Simpson, A. J. H., and Stone, S. P. (2007). Successful use of feedback to improve antibiotic prescribing and reduce Clostridium difficile infection: a controlled interrupted time series. J. Antimicrob. Chemother. 59, 990–995.
Franch, T., Petersen, M., Wagner, E. G., Jacobsen, J. P., and Gerdes, K. (1999). Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J. Mol. Biol. 294, 1115–1125.
Gao, M.-Y., Xu, C.-R., Chen, R., Liu, S.-G., and Feng, J.-N. (2005). Chloromycetin resistance of clinically isolated E. coli is conversed by using EGS technique to repress the chloromycetin acetyl transferase. World J. Gastroenterol. 11, 7368–7373.
Geller, B. L., Deere, J. D., Stein, D. A., Kroeker, A. D., Moulton, H. M., and Iversen, P. L. (2003). Inhibition of gene expression in Escherichia coli by antisense phosphorodiamidate morpholino oligomers. Antimicrob. Agents Chemother. 47, 3233–3239.
Georg, J., and Hess, W. R. (2011). cis-Antisense RNA, another level of gene regulation in bacteria. Microbiol. Mol. Biol. Rev. 75, 286.
George, A. M., and Hall, R. M. (2002). Efflux of chloramphenicol by the CmlA1 protein. FEMS Microbiol. Lett. 209, 209–213.
Giaever, G., Chu, A., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., Arkin, A., Astromoff, A., El Bakkoury, M., Bangham, R., Benito, R., Brachat, S., Campanaro, S., Curtiss, M., Davis, K., Deutschbauer, A., Entian, K., Flaherty, P., Foury, F., Garfinkel, D., Gerstein, M., Gotte, D., Guldener, U., Hegemann, J., Hempel, S., Herman, Z., Jaramillo, D., Kelly, D., Kelly, S., Kotter, P., Labonte, D., Lamb, D., Lan, N., Liang, H., Liao, H., Liu, L., Luo, C., Lussier, M., Mao, R., Menard, P., Ooi, S., Revuelta, J., Roberts, C., Rose, M., Ross-Macdonald, P., Scherens, B., Schimmack, G., Shafer, B., Shoemaker, D., Sookhai-Mahadeo, S., Storms, R., Strathern, J., Valle, G., Voet, M., Volckaert, G., Wang, C., Ward, T., Wilhelmy, J., Winzeler, E., Yang, Y., Yen, G., Youngman, E., Yu, K., Bussey, H., Boeke, J., Snyder, M., Philippsen, P., Davis, R., and Johnston, M. (2002). Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391.
Goh, S., Boberek, J. M., Nakashima, N., Stach, J., and Good, L. (2009). Concurrent growth rate and transcript analyses reveal essential gene stringency in Escherichia coli. PLoS ONE 4, e6061.
Good, L., Awasthi, S. K., Dryselius, R., Larsson, O., and Nielsen, P. E. (2001). Bactericidal antisense effects of peptide-PNA conjugates. Nat. Biotechnol. 19, 360–364.
Good, L., and Nielsen, P. E. (1998). Inhibition of translation and bacterial growth by peptide nucleic acid targeted to ribosomal RNA. Proc. Natl. Acad. Sci. U.S.A. 95, 2073–2076.
Haydon, D. J., Stokes, N. R., Ure, R., Galbraith, G., Bennett, J. M., Brown, D. R., Baker, P. J., Barynin, V. V., Rice, D. W., Sedelnikova, S. E., Heal, J. R., Sheridan, J. M., Aiwale, S. T., Chauhan, P. K., Srivastava, A., Taneja, A., Collins, I., Errington, J., and Czaplewski, L. G. (2008). An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science 321, 1673–1675.
Hopkins, A. L. (2008). Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 4, 682–690.
Horvath, P., and Barrangou, R. (2010). CRISPR/Cas, the immune system of bacteria and archaea. Science 327, 167–170.
Huber, J., Donald, R. G. K., Lee, S. H., Jarantow, L. W., Salvatore, M. J., Meng, X., Painter, R., Onishi, R. H., Occi, J., Dorso, K., Young, K., Park, Y. W., Skwish, S., Szymonifka, M. J., Waddell, T. S., Miesel, L., Phillips, J. W., and Roemer, T. (2009). Chemical genetic identification of peptidoglycan inhibitors potentiating carbapenem activity against methicillin-resistant Staphylococcus aureus. Chem. Biol. 16, 837–848.
Hutter, B., Fischer, C., Jacobi, A., Schaab, C., and Loferer, H. (2004). Panel of Bacillus subtilis reporter strains indicative of various modes of action. Antimicrob. Agents Chemother. 48, 2588–2594.
Jakobsson, H. E., Jernberg, C., Andersson, A. F., Sjölund-Karlsson, M., Jansson, J. K., and Engstrand, L. (2010). Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE 5, e9836.
Jeon, B., and Zhang, Q. (2009). Sensitization of Campylobacter jejuni to fluoroquinolone and macrolide antibiotics by antisense inhibition of the CmeABC multidrug efflux transporter. J. Antimicrob. Chemother. 63, 946–948.
Ji, Y., Yin, D., Fox, B., Holmes, D. J., Payne, D., and Rosenberg, M. (2004). Validation of antibacterial mechanism of action using regulated antisense RNA expression in Staphylococcus aureus. FEMS Microbiol. Lett. 231, 177–184.
Ji, Y., Zhang, B., Van Horn, S. F., Warren, P., Woodnutt, G., Burnham, M. K., and Rosenberg, M. (2001). Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293, 2266–2269.
Kaur, P., Agarwal, S., and Datta, S. (2009). Delineating bacteriostatic and bactericidal targets in mycobacteria using IPTG inducible antisense expression. PLoS ONE 4, e5923.
Kolb, F. A., Engdahl, H. M., Slagter-Jäger, J. G., Ehresmann, B., Ehresmann, C., Westhof, E., Wagner, E. G., and Romby, P. (2000). Progression of a loop-loop complex to a four-way junction is crucial for the activity of a regulatory antisense RNA. EMBO J. 19, 5905–5915.
Li, B., Yao, Q., Pan, X.-C., Wang, N., Zhang, R., Li, J., Ding, G., Liu, X., Wu, C., Ran, D., Zheng, J., and Zhou, H. (2011). Artesunate enhances the antibacterial effect of {beta}-lactam antibiotics against Escherichia coli by increasing antibiotic accumulation via inhibition of the multidrug efflux pump system AcrAB-TolC. J. Antimicrob. Chemother. 66, 769–777.
Lock, R. L., and Harry, E. J. (2008). Cell-division inhibitors: new insights for future antibiotics. Nat. Rev. Drug Discov. 7, 324–338.
Lorenz, C., Gesell, T., Zimmermann, B., Schoeberl, U., Bilusic, I., Rajkowitsch, L., Waldsich, C., Von Haeseler, A., and Schroeder, R. (2010). Genomic SELEX for Hfq-binding RNAs identifies genomic aptamers predominantly in antisense transcripts. Nucleic Acids Res. 38, 3794–3808.
Majdalani, N., Cunning, C., Sledjeski, D., Elliott, T., and Gottesman, S. (1998). DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. U.S.A. 95, 12462–12467.
Mellbye, B. L., Puckett, S. E., Tilley, L. D., Iversen, P. L., and Geller, B. L. (2009). Variations in amino acid composition of antisense peptide-phosphorodiamidate morpholino oligomer affect potency against Escherichia coli in vitro and in vivo. Antimicrob. Agents Chemother. 53, 525–530.
Moffat, J. G., Tate, W. P., and Lovett, P. S. (1994). The leader peptides of attenuation-regulated chloramphenicol resistance genes inhibit translational termination. J. Bacteriol. 176, 7115–7117.
Møller, T., Franch, T., Højrup, P., Keene, D. R., Bächinger, H. P., Brennan, R. G., and Valentin-Hansen, P. (2002a). Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell 9, 23–30.
Møller, T., Franch, T., Udesen, C., Gerdes, K., and Valentin-Hansen, P. (2002b). Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 16, 1696–1706.
Morse, T. O., Morey, S. J., and Gunsch, C. K. (2010). Microbial inactivation of Pseudomonas putida and Pichia pastoris using gene silencing. Environ. Sci. Technol. 44, 3293–3297.
Nakashima, N., and Tamura, T. (2009). Conditional gene silencing of multiple genes with antisense RNAs and generation of a mutator strain of Escherichia coli. Nucleic Acids Res. 37, e103.
Nakashima, N., Tamura, T., and Good, L. (2006). Paired termini stabilize antisense RNAs and enhance conditional gene silencing in Escherichia coli. Nucleic Acids Res. 34, e138.
Nekhotiaeva, N., Awasthi, S. K., Nielsen, P. E., and Good, L. (2004). Inhibition of Staphylococcus aureus gene expression and growth using antisense peptide nucleic acids. Mol. Ther. 10, 652–659.
Nikravesh, A., Dryselius, R., Faridani, O. R., Goh, S., Sadeghizadeh, M., Behmanesh, M., Ganyu, A., Klok, E. J., Zain, R., and Good, L. (2007). Antisense PNA accumulates in Escherichia coli and mediates a long post-antibiotic effect. Mol. Ther. 15, 1537–1542.
Olano, C., Lombó, F., Méndez, C., and Salas, J. A. (2008). Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab. Eng. 10, 281–292.
Pankey, G. A., and Sabath, L. D. (2004). Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of Gram-positive bacterial infections. Clin. Infect. Dis. 38, 864–870.
Shine, J., and Dalgarno, L. (1974). The 3’-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U.S.A. 71, 1342–1346.
Singh, S., Zink, D., Herath, K., Salazar, O., and Genilloud, O. (2008). Discovery and antibacterial activity of lucensimycin C from Streptomyces lucensis. Tetrahedron Lett. 49, 2616–2619.
Soler Bistué, A. J. C., Martín, F. A., Vozza, N., Ha, H., Joaquín, J. C., Zorreguieta, A., and Tolmasky, M. E. (2009). Inhibition of aac(6’)-Ib-mediated amikacin resistance by nuclease-resistant external guide sequences in bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 13230–13235.
Tan, X. -X., Actor, J. K., and Chen, Y. (2005). Peptide nucleic acid antisense oligomer as a therapeutic strategy against bacterial infection: proof of principle using mouse intraperitoneal infection. Antimicrob. Agents Chemother. 49, 3203–3207.
Then, R. L., and Sahl, H.-G. (2010). Anti-infective strategies of the future: is there room for species-specific antibacterial agents? Curr. Pharm. Des. 16, 555–566.
Thomason, M. K., and Storz, G. (2010). Bacterial antisense RNAs: how many are there, and what are they doing? *. Annu. Rev. Genet. 44, 167–188.
Thomson, C. J., Power, E., Ruebsamen-Waigmann, H., and Labischinski, H. (2004). Antibacterial research and development in the 21(st) Century – an industry perspective of the challenges. Curr. Opin. Microbiol. 7, 445–450.
Wang, H., Meng, J., Jia, M., Ma, X., He, G., Yu, J., Wang, R., Bai, H., Hou, Z., and Luo, X. (2010). oprM as a new target for reversion of multidrug resistance in Pseudomonas aeruginosa by antisense phosphorothioate oligodeoxynucleotides. FEMS Immunol. Med. Microbiol. 60, 275–282.
Wang, J., Kodali, S., Lee, S. H., Galgoci, A., Painter, R., Dorso, K., Racine, F., Motyl, M., Hernandez, L., Tinney, E., Colletti, S. L., Herath, K., Cummings, R., Salazar, O., González, I., Basilio, A., Vicente, F., Genilloud, O., Pelaez, F., Jayasuriya, H., Young, K., Cully, D. F., and Singh, S. B. (2007). Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc. Natl. Acad. Sci. U.S.A. 104, 7612–7616.
White, D. G., Maneewannakul, K., Von Hofe, E., Zillman, M., Eisenberg, W., Field, A. K., and Levy, S. B. (1997). Inhibition of the multiple antibiotic resistance (mar) operon in Escherichia coli by antisense DNA analogs. Antimicrob. Agents Chemother. 41, 2699–2704.
Xu, D., Jiang, B., Ketela, T., Lemieux, S., Veillette, K., Martel, N., Davison, J., Sillaots, S., Trosok, S., Bachewich, C., Bussey, H., Youngman, P., and Roemer, T. (2007). Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog. 3, e92.
Xu, H., Trawick, J. D., Haselbeck, R. J., Forsyth, R. A., Yamamoto, R. T., Archer, R., Patterson, J., Allen, M., Froelich, J. M., Taylor, I., Nakaji, D., Maile, R., Kedar, G. C., Pilcher, M., Brown-Driver, V., Mccarthy, M., Files, A., Robbins, D., King, P., Sillaots, S., Malone, C., Zamudio, C. S., Roemer, T., Wang, L., Youngman, P. J., and Wall, D. (2010). Staphylococcus aureus TargetArray: comprehensive differential essential gene expression as a mechanistic tool to profile antibacterials. Antimicrob. Agents Chemother. 54, 3659–3670.
Yeh, P., Tschumi, A., and Kishony, R. (2006). Functional classification of drugs by properties of their pairwise interactions. Nat. Genet. 38, 489–494.
Yeh, P. J., Hegreness, M. J., Aiden, A. P., and Kishony, R. (2009). Drug interactions and the evolution of antibiotic resistance. Nat. Rev. Microbiol. 7, 460–466.
Young, K., Jayasuriya, H., Ondeyka, J., and Herath, K. (2006). Discovery of FabH/FabF inhibitors from natural products. Antimicrob. Agents Chemother. 50, 512–526.
Zhang, C., Ondeyka, J. G., Zink, D. L., Basilio, A., Vicente, F., Salazar, O., Genilloud, O., Dorso, K., Motyl, M., Byrne, K., and Singh, S. B. (2009). Discovery of okilactomycin and congeners from Streptomyces scabrisporus by antisense differential sensitivity assay targeting ribosomal protein S4. J. Antibiot. 62, 55.
Keywords: antisense, resistance, antimicrobial, gene silencing, peptide nucleic acid, drug discovery
Citation: Good L and Stach JEM (2011) Synthetic RNA silencing in bacteria – antimicrobial discovery and resistance breaking. Front. Microbio. 2:185. doi: 10.3389/fmicb.2011.00185
Received: 13 June 2011; Accepted: 20 August 2011;
Published online: 12 September 2011.
Edited by:
Tzi Bun Ng, The Chinese University of Hong Kong, ChinaReviewed by:
Fiona Walsh, Agroscope Changins Wädenswil, SwitzerlandPaul Alan Hoskisson, University of Strathclyde, UK
Copyright: © 2011 Good and Stach. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: James E. M. Stach, School of Biology, Newcastle University, Newcastle upon Tyne NE1 7RU, UK. e-mail:amVtLnN0YWNoQG5jbC5hYy51aw==
 Liam Good1
Liam Good1