- 1 Department of Cell and Molecular Biology, University of Rhode Island, Kingston, RI, USA
- 2 Institute of Marine Sciences, University of California, Santa Cruz, CA, USA
- 3 School of Marine Sciences, University of Maine, Orono, ME, USA
- 4 Graduate School of Oceanography, University of Rhode Island, Narragansett, RI, USA
Iron (Fe) availability restricts diatom growth and primary production in large areas of the oceans. It is a challenge to assess the bulk Fe nutritional health of natural diatom populations, since species can differ in their physiological and molecular responses to Fe limitation. We assayed expression of selected genes in diatoms from the Thalassiosira genus to assess their potential utility as species-specific molecular markers to indicate Fe status in natural diatom assemblages. In this study, we compared the expression of the photosynthetic genes encoding ferredoxin (a Fe-requiring protein) and flavodoxin (a Fe-free protein) in culture experiments with Fe replete and Fe stressed Thalassiosira pseudonana (CCMP 1335) isolated from coastal waters and Thalassiosira weissflogii (CCMP 1010) isolated from the open ocean. In T. pseudonana, expression of flavodoxin and ferredoxin genes were not sensitive to Fe status but were found to display diel periodicities. In T. weissflogii, expression of flavodoxin was highly responsive to iron levels and was only detectable when cultures were Fe limited. Flavodoxin genes have been duplicated in most diatoms with available genome data and we show that T. pseudonana has lost its copy related to the Fe-responsive copy in T. weissflogii. We also examined the expression of genes for a putative high affinity, copper (Cu)-dependent Fe uptake system in T. pseudonana. Our results indicate that genes encoding putative Cu transporters, a multi-Cu oxidase, and a Fe reductase are not linked to Fe status. The expression of a second putative Fe reductase increased in Fe limited cultures, but this gene was also highly expressed in Fe replete cultures, indicating it may not be a useful marker in the field. Our findings highlight that Fe metabolism may differ among diatoms even within a genus and show a need to validate responses in different species as part of the development pipeline for genetic markers of Fe status in field populations.
Introduction
Diatoms are widespread and abundant primary producers that strongly influence the global cycling of carbon (Nelson et al., 1995). The organic carbon produced by diatoms serves as fuel for the higher trophic levels of marine food webs and dictates the ecological structure of coastal environments. In the open ocean, diatom-bound carbon can sink rapidly out of the photic zone to depths where it is sequestered for long periods of time (Smetacek, 1999). As key players in the ecology and biogeochemistry of the oceans, it is important to understand the variables that regulate diatom growth and the cellular mechanisms employed in attempts to overcome growth restrictions.
It is widely accepted that iron (Fe) availability limits the growth of diatoms in large areas of the oceans (Moore et al., 2004). The physiological response to Fe limitation has been shown to differ among diatom species. For example, diatoms from oceanic environments are able to grow at maximal or near maximal growth rates at low Fe levels that severely restrict the growth of coastal diatoms (e.g., Brand et al., 1983; Sunda et al., 1991). Recent studies have also identified differences at the molecular level. Marchetti et al. (2009) identified an Fe storage protein, ferritin, in several species of pennate diatoms that was absent from centric diatoms. These Fe reserves confer a growth advantage for ferritin-containing diatoms in Fe limiting conditions (Marchetti et al., 2009). This variability in genetic composition and physiological response between diatoms can make it difficult to assess the Fe nutritional status of natural diatom populations.
The photosynthetic proteins ferredoxin and flavodoxin have been used as biomarkers to signal Fe limitation in diatoms. Ferredoxin is an iron–sulfur protein that functions as an electron carrier in the photosynthetic electron transport chain and can also act as an electron donor in other metabolic processes such as nitrogen assimilation and amino acid metabolism. Ferredoxin can be replaced by flavodoxin, an Fe-free protein, a strategy used by cyanobacteria and diatoms to adapt to Fe limitation (reviewed by Zurbriggen et al., 2008). Methods using immunoprobes (LaRoche et al., 1995; McKay et al., 1997, 1999) and high performance liquid chromatography (HPLC; Doucette et al., 1996; Erdner et al., 1999) have shown that induction of flavodoxin and suppression of ferredoxin is a strategy used by a broad range of diatom species to lower cellular Fe demand in Fe limiting conditions.
When applied to natural phytoplankton populations, immunoprobe and HPLC based methods measuring ferredoxin and flavodoxin protein production have yielded complicated results. For example, these methods detected only flavodoxin in diatoms after Fe enrichment in the eastern equatorial Pacific (Erdner and Anderson, 1999) and in the subarctic Pacific Ocean (Suzuki et al., 2009). Ferredoxin was below the limit of detection of both methods, suggesting that protein-based assays are challenging for assessing Fe status in the environment.
In addition, methods targeting the total ferredoxin and flavodoxin pool are unable to distinguish among species in the environment or determine how much an individual species may contribute to the protein pools. They are typically combined with isolation techniques (Erdner and Anderson, 1999) or light microscopy (Hattori-Saito et al., 2010) to provide insight into the diatom community composition and how it changes over time and with Fe additions.
We are developing species-specific molecular probes to understand the dynamics of Fe stress in natural diatom populations. Our efforts are focused on the genus Thalassiosira, as it is a large group with over 100 species that are found distributed throughout the world’s oceans (Round et al., 1990). In this study we targeted two previously reported systems in T. pseudonana, a coastal diatom, that respond to Fe limitation: the flavodoxin and ferredoxin replacement strategy, and genes with homology to members of a putative copper (Cu)-dependent, high affinity Fe uptake system in yeast (Maldonado et al., 2006; Kustka et al., 2007; Thamatrakoln et al., 2011). The Fe uptake proteins have not been functionally characterized in diatoms, but have been well characterized in the yeast, Saccharomyces cerevisiae (Kosman, 2003). In the yeast system, Fe(III) is reduced to biologically available Fe(II) through the activity of plasma membrane-localized iron reductases, followed by re-oxidation and transport into the cell via an Fe oxidase–permease complex. The re-oxidation of Fe(II) is a Cu-dependent reaction because the oxidase protein must be loaded with Cu ions to be functional (Kosman, 2003).
We, along with others (Maldonado et al., 2006; Kustka et al., 2007; Thamatrakoln et al., 2011), have identified genes in Thalassiosira pseudonana (CCMP 1335) encoding proteins with homology to those expressed in the yeast Fe uptake system. In this study, we followed the expression of T. pseudonana Fe uptake system homologs in Fe replete and Fe limited cultures using quantitative reverse transcription-polymerase chain reaction (qRT-PCR) to determine whether members of this system would be a sensitive field-based indicator of Fe status. We also compared ferredoxin and flavodoxin gene expression profiles in response to Fe stress in T. pseudonana and in a related diatom isolated from the open ocean, Thalassiosira weissflogii (CCMP 1010), to discern whether ferredoxin and flavodoxin gene expression could be used as markers to indicate Fe status in field populations.
Materials and Methods
Culture Conditions and Sampling
Thalassiosira pseudonana (CCMP 1335) and Thalassiosira weissflogii (CCMP 1010) isolates were obtained from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton (West Boothbay Harbor, ME, USA). Trace metal limitation experiments were performed using a basal synthetic medium consisting of a modified and combined version of f/2 (Guillard and Hargraves, 1993) and Aquil media (Price et al., 1989). The basal salt medium and individual nutrient stocks were pumped (~2 mL min−1) through an ion-exchange column (Chelex 100, Biorad) containing resin prepared according to Price et al. (1989). Macronutrients were added at initial concentrations of 300 μM 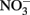 , 15 μM
, 15 μM 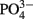 , and 50 μM Si(OH)4. Media batches were sterilized by microwaving in acid-washed Teflon bottles (Keller et al., 1988) and enriched with filter-sterilized (0.2 μm Acrodisc) EDTA-trace metal and vitamin (B12, thiamine, and biotin) solutions. The trace metals zinc (Zn), manganese (Mn), cobalt (Co), selenium (Se), molybdenum (Mo), vanadium (V), nickel (Ni), and chromium (Cr) were added as in previous studies (Maldonado et al., 2002); boric acid (H3BO3) was added at 6.14 × 10−5 M. Metals were buffered with 11.7 μM EDTA and the free ion activities calculated using the chemical equilibrium program MINEQL+ (Schecher and McAvoy, 1998).
, and 50 μM Si(OH)4. Media batches were sterilized by microwaving in acid-washed Teflon bottles (Keller et al., 1988) and enriched with filter-sterilized (0.2 μm Acrodisc) EDTA-trace metal and vitamin (B12, thiamine, and biotin) solutions. The trace metals zinc (Zn), manganese (Mn), cobalt (Co), selenium (Se), molybdenum (Mo), vanadium (V), nickel (Ni), and chromium (Cr) were added as in previous studies (Maldonado et al., 2002); boric acid (H3BO3) was added at 6.14 × 10−5 M. Metals were buffered with 11.7 μM EDTA and the free ion activities calculated using the chemical equilibrium program MINEQL+ (Schecher and McAvoy, 1998).
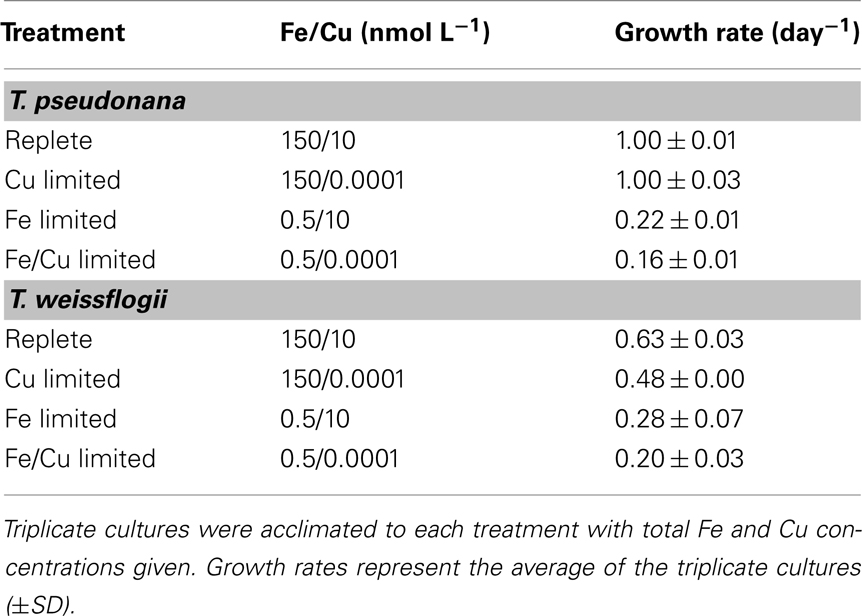
Iron stock solutions were added separately to achieve the desired conditions. Cultures of T. pseudonana and T. weissflogii grown under replete conditions received 150 nmol L−1 total Fe and Fe limited cultures either received 0.5 nmol L−1 total Fe or did not receive any additional Fe, depending on the experiment being conducted. In some experiments, Cu limitation was also explored. In these experiments, Cu was added separately with cultures grown under replete conditions receiving 10 nmol L−1 total Cu and cultures grown under low Cu conditions receiving 0.1 pmol L−1 total Cu. All bottles and apparatus were acid cleaned and sample manipulations were conducted within a laminar-flow hood (HEPA, class 100) using trace metal clean techniques (Maldonado et al., 2002; Wells et al., 2005).
To investigate the effects of Fe limitation, T. pseudonana and T. weissflogii were grown under two experimental conditions: cultures were acclimated to Fe deficient conditions as well as subjected to rapid Fe stress. For the acclimation experiments, triplicate cultures were grown at 13.5°C on a 12:12-h light:dark cycle and maintained at exponential growth by successive additions of culture media. There were four acclimation conditions: replete, low Cu, low Fe, and low Fe/Cu. Cultures were considered acclimated and harvested for molecular analysis when the growth rate differed by <10% upon consecutive media additions. Growth rates were calculated from daily in vivo chlorophyll a fluorescence measurements (Turner fluorometer). Cell counts were done on samples collected at the time of harvesting; in T. pseudonana, cell abundance was determined using a flow cytometer (Cytopeia) and in T. weissflogii cell concentration was measured in one culture replicate from each experimental treatment using a nannoplankton counting chamber.
To rapidly stress T. pseudonana and T. weissflogii for Fe, cells were transferred from replete media into media with replete Fe levels or media with no added Fe and grown as semi-continuous batch cultures at 13.5°C under continuous light. Samples were collected daily with 200 mL (T. pseudonana) or 300 mL (T. weissflogii) of culture volume harvested and an equal volume of fresh media added; samples were also collected for cell counts (Cytopeia flow cytometer) and chlorophyll a fluorescence (Turner) measurements. The semi-continuous batch cultures were maintained for 11 days for T. pseudonana and 15 days for T. weissflogii; the duration of the experiments represents the time when a notable difference in cell abundance was observed between Fe replete and Fe limited cultures. T. weissflogii cultures did not reach steady state growth until day four, so work presented here represents the 9-days of steady state growth.
Trace metal clean techniques were used to filter cells onto 47 mm, 2 μm or 5 μm polycarbonate or polyester filters for both of the Fe limitation experiments described. The filters were flash frozen in liquid N2 and stored at −80°C until RNA extractions were conducted.
A second semi-continuous batch experiment designed to monitor gene expression changes associated with a light:dark cycle was performed with T. pseudonana. Triplicate cultures were grown in f/2 media (Guillard, 1975) at 13.5°C at 150 μE m−2 s−1 on a 12:12-h light:dark cycle for 48 h after which the light regime shifted to continuous light for another 48 h. Samples were collected every 3 h by gentle filtration onto 2 μm polyester filters and flash frozen; at each time point 150 mL of culture was removed for genetic analysis and 150 mL of fresh f/2 media was added. To ensure cell numbers were not drifting from the expected concentration, cell counts were taken every 12 h and fluorescence measurements every 24 h.
RNA Extraction and CDNA Synthesis
Total RNA was extracted using the RNeasy Midi Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol, with the following exceptions: cells were lysed using 0.5 mm zirconia/silica beads (BioSpec) mixed with the lysis buffer and vortexed until the solution looked homogenous 2–3 min, the lysis solution was then put over Qiashredder columns (Qiagen) to remove any large plant material that could clog the spin columns. To aid in the removal of DNA, two DNase digestions were performed. First, Qiagen’s RNase-free DNase Set (an on-column treatment) was used according to the manufacturer’s instructions. The RNA was then quantified in triplicate using the Mx3005 or Mx4000 Multiplex Quantitative PCR System (Stratagene, La Jolla, CA, USA) and the Quant-iTRiboGreen RNA Assay Kit (Invitrogen, Carlsbad, CA, USA). Following quantification, all RNA samples were diluted to 31.25 ng/μL; this allowed us to add equal volumes (8 μL) to the cDNA synthesis reactions. After this dilution, a second DNA removal step was conducted using the Turbo DNA-free kit (Ambion, Austin, TX, USA) according to the manufacturer’s protocol.
cDNA was synthesized from 250 ng of DNase-treated RNA using Invitrogen’s SuperScript III First-Strand Synthesis System for RT-PCR. An oligo (dT) primer was used to generate cDNA for measuring expression of nuclear-encoded genes. T. pseudonana (5′-CCAGCACCCATTTGATCATCATC-3′) and T. weissflogii (5′-TACAAAACCAGCACCCATTT-3′) gene specific primers were used to generate ferredoxin cDNA as it is a plastid-encoded gene. The cDNA synthesis reactions were conducted in triplicate for each sample from the acclimation experiments as well as the T. pseudonana semi-continuous batch rapid Fe stress experiment. Because the greatest variance observed in these experiments came from the biological replicates, it was determined that cDNA syntheses could be carried out in single replicates for the T. weissflogii semi-continuous batch rapid Fe stress experiment as well as the T. pseudonana semi-continuous batch diel experiment (experiments with the largest sample number) without affecting the results. Control reactions lacking reverse transcriptase were also performed on each sample.
Quantitative Real-Time PCR
Multiplexed qRT-PCR was performed using the Mx3005 qPCR System and the Brilliant II qPCR Master Mix (Stratagene). Multiplex qPCR allows for the simultaneous amplification of multiple genes (reference and target gene) as the probes included in the reaction are labeled with different fluorophores (Table 1 and 2). Primers and probes, designed using Primer Express software (Applied Biosystems, Carlsbad, CA, USA), were used at final concentrations of 200 and 100 nM, respectively. To each reaction, 2 μL of cDNA or plasmid standards were added to 25 μL qPCR reactions containing the master mix and reference dye and set up following the manufacturer’s protocol. The qRT-PCR cycling conditions were: 1 cycle at 95°C for 10 min, followed by 40 cycles of 95°C for 30 s and 60°C for 30 s. Amplification efficiencies for the reference and target genes were determined by analyzing serial dilutions of selected cDNA samples. When plotting Ct (cycle threshold) versus log of cDNA input, the slope of the line can be used to calculate qPCR efficiencies with the formula: E = 10(−1/slope). The average efficiency for all genes was 1.92 (±0.05).
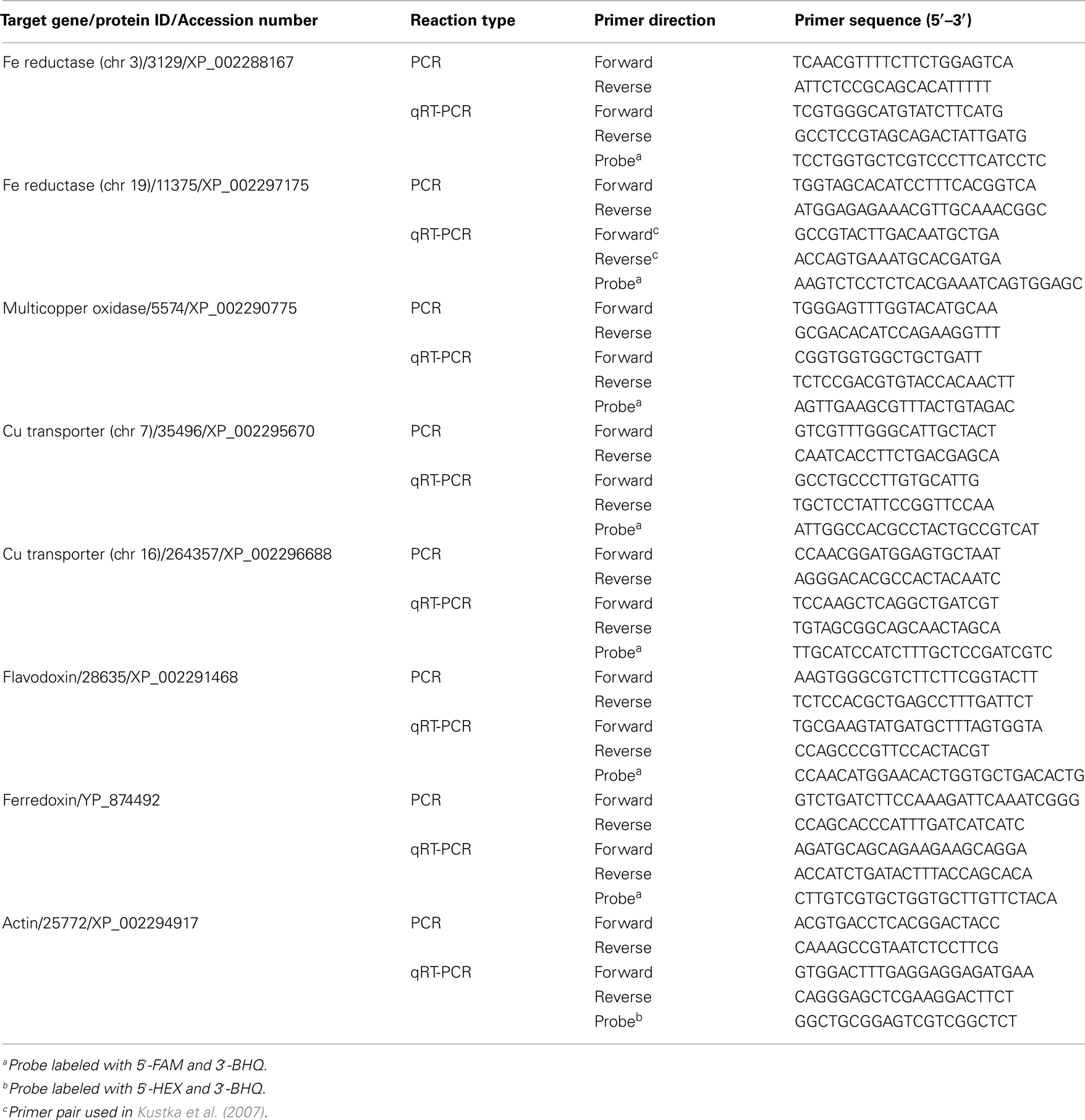
Table 1. Genes and primers used to amplify targets for plasmid standards (PCR) and measure transcript abundance (qRT-PCR) in T. pseudonana.
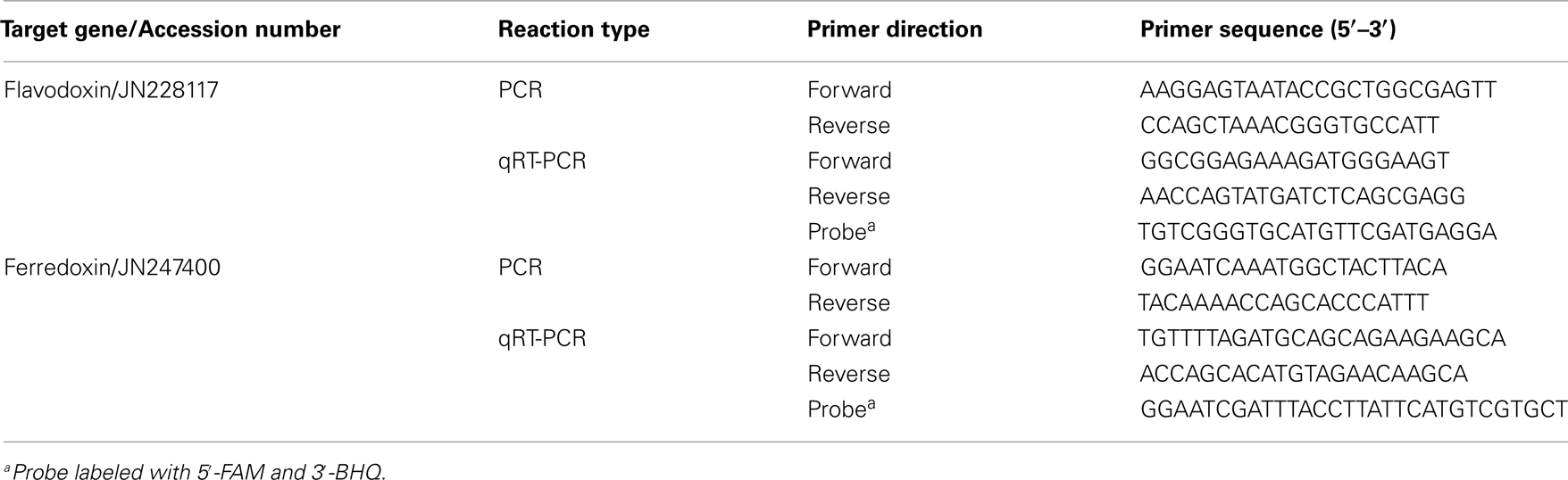
Table 2. Genes and primers used to amplify targets for plasmid standards (PCR) and measure transcript abundance (qRT-PCR) in T. weissflogii.
Actin was used as a reference gene and was included in the qRT-PCR reactions for nuclear-encoded genes. Actin was selected as a reference gene because its expression was found to be constitutive and any changes in its expression were random and not correlated with experimental treatment. Standards made from plasmid dilutions for each gene to be amplified (reference and target gene) were run in triplicate. Controls were also included from the cDNA reactions lacking reverse transcriptase as well as no-cDNA template controls.
To generate the qRT-PCR plasmid standards, genes of interest were isolated from the study organisms using a PCR amplification strategy. First, T. pseudonana genomic DNA was extracted from a culture grown in f/2 media using the DNeasy Plant Mini Kit (Qiagen). Five microliters of diluted DNA was added to PCR reactions consisting of 0.5 μM of each primer, 25 μL of BIO-X-ACT Short Mix (Bioline, Randolph, MA, USA), and 15 μL of AccuGENE Molecular Biology Grade Water (Cambrex Bio Science Rockland, Inc., Rockland, ME, USA). Reactions were performed in an Eppendorf Mastercycler ep thermocycler (Eppendorf North America, Westbury, NY, USA) using the following parameters: a denaturation step at 94°C for 3 min, followed by 30 extension cycles (94°C 30 s, melting for 30 s, 72°C 1 min), a final 5 min extension step at 72°C and then an incubation at 4°C until analyzed. Primers used for target gene isolation in T. pseudonana are listed in Table 1 (PCR).
Degenerate primers were used to amplify target genes from T. weissflogii cDNA as its genome has not been sequenced. The PCR reaction setup was the same as described for T. pseudonana, except the number of extension cycles was increased to 35 cycles. PCR products were sequenced and putative functions were investigated by BLAST analysis against other ferredoxin and flavodoxin sequences. Accession numbers for these genes are indicated in Table 2. T. weissflogii gene specific primers were then designed and used for subsequent steps (Table 2).
After amplification, the PCR products were loaded on to a 1% agarose (wt/vol) TAE gel. Bands were purified using either the QIAquick Gel Extraction Kit or the QIAEX II Gel Extraction Kit according to the manufacturer’s protocol (Qiagen).
The purified products were cloned into pGEM T vector (Promega, Madison, WI, USA), transformed in either JM 109 (Promega) or α-Select Silver Efficiency (Bioline) competent cells and identified by blue-white screening. The plasmids were purified using the QIAprep Spin Miniprep kit (Qiagen) and subsequently sequenced on either the Beckman Colter CEQ 8000 Genetic Analysis System (Beckman Colter, Inc., Fullerton, CA, USA) or the Applied Biosystems 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Purified plasmids were linearized using the restriction endonuclease, SpeI (New England Biolabs), and quantified using the Quant-iTPicoGreen DNA Assay Kit (Molecular Probes, Invitrogen) and the Mx3005 (Stratagene).
The quantified plasmids carrying the target inserts were then used to generate standards in triplicate for qRT-PCR. Serial dilutions of the plasmids, ranging from <10 to >106 copies per reaction, were used to quantify transcript abundance of the target genes in experimental samples.
Statistical Analyses
Analysis of variance (ANOVA) tests were conducted using SigmaStat version 3.5 to determine statistically significant differences among samples.
Phylogenetic Analysis
The flavodoxin protein sequence from T. pseudonana was used for homology-based searches to identify gene copies from the sequenced diatom genomes, Phaeodactylum tricornutum and Fragilariopsis cylindrus, as well as from the recently sequenced pelagophyte, Aureococcus anophagefferens (http://genome.jgi-psf.org/Auran1/Auran1.home.html). Gene copies were also isolated from preliminary genomes (unpublished) of P. multiseries (JN226577, JN226578) and Thalassiosira oceanica (JN226574, JN226575). CLUSTAL W (Thompson et al., 1994) was used to align the translated fragments (58 amino acids in length). The resulting alignment was used to generate an arbitrarily rooted, maximum-likelihood tree with bootstrap values calculated from 1000 replicates in PhyML (Guindon and Gascuel, 2003) in the Geneious software package (Drummond et al., 2010). Bayesian analysis was also performed using MrBayes (Huelsenbeck and Ronquist, 2001) in the Geneious software package. The Bayesian search was run using the Poisson model during 1,100,000 generations, with a burn-in of 250,000, and every 200th tree was saved.
Results
Growth Response to Fe Limitation in T. pseudonana
Thalassiosira pseudonana was acclimated to low Fe and Cu conditions, as well as a low Fe/Cu condition, through the successive addition of the experimental media. Growth rates were highest in cells grown in replete media and similar growth rates were obtained in the low Cu media (Table 3). A significant drop in growth rate was seen in cultures acclimated to low Fe and low Fe/Cu conditions (p < 0.001, one-way ANOVA; Table 3).
Gene Expression Analysis
The expression levels of all target genes were normalized to beta-actin with the exception of ferredoxin and flavodoxin. Actin was determined to be a suitable reference gene since its transcript copy number per cell did not significantly change with growth rate (R2 = 0.07; data not shown) or experimental treatment (data not shown). To compare gene expression levels of ferredoxin and flavodoxin, transcript abundances of those two genes were normalized to cell number since ferredoxin is a plastid-encoded gene and subject to different cellular regulatory processes than nucleus-encoded genes. We assayed the expression of several plastid-encoded genes (including DNA-directed RNA polymerase beta chain, accession number YP_874523 and 50S ribosomal protein L12, accession number YP_874519) for their use as reference genes; however their expression was found to change with experimental treatment and could not be used to normalize ferredoxin abundance (data not shown).
Regulation of Putative Fe Uptake Genes in T. pseudonana
Five genes representing key components of the putative inducible Fe uptake system were selected for expression analysis in T. pseudonana. These include genes encoding two Fe reductases (located on chromosomes 3 and 19), a multicopper oxidase, as well as two Cu transporters (located on chromosomes 7 and 16) that putatively function to supply the multicopper oxidase with Cu ions (Table 1). Gene expression of the multicopper oxidase in cells acclimated to low Fe and low Fe/Cu conditions decreased approximately 2-fold and 2.5-fold, respectively, when compared to expression of cells grown under replete conditions (p < 0.005, one-way ANOVA; Figure 1A). The gene encoding Fe reductase (chr 19) was the most highly expressed gene in all treatment conditions and was found to be significantly induced in cells acclimated to low Fe, increasing approximately 1.8-fold (p = 0.002, one-way ANOVA; Figure 1B). A significant increase in Fe reductase (chr 19) expression was also seen in Fe/Cu co-limited cells; expression increased 1.6-fold when compared to the replete cells (p = 0.013, one-way ANOVA; Figure 1B). No significant differences were detected when the relative transcript levels of these genes were compared between low Cu and replete-grown cells. There was also no significant difference in the relative transcript abundances of the Cu transporters or the gene encoding Fe reductase (chr 3) between cultures grown in replete media and trace metal limiting media (Figures 1C,D).
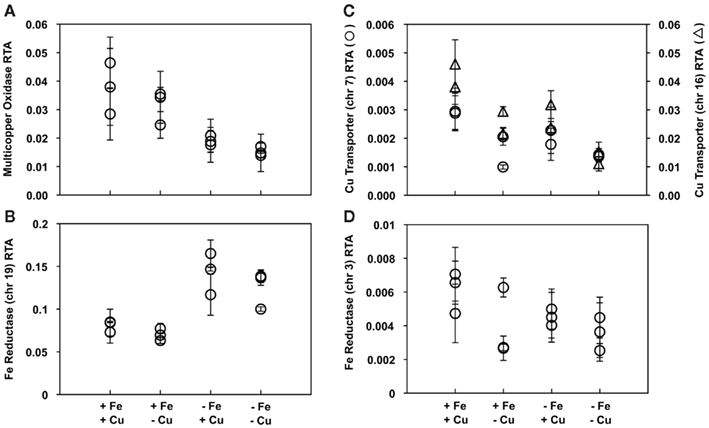
Figure 1. Expression patterns of genes in the putative Fe uptake system in triplicate T. pseudonana cultures acclimated to different treatments. Relative transcript abundance (RTA) of genes encoding (A) a multicopper oxidase, (B) Fe reductase located on chr 19, (C) two Cu transporters located on chr 7 (circles) and chr 16 (triangles), and (D) Fe reductase located on chr 3, all normalized to expression levels of actin. Results from culture replicates are shown; each symbol within a treatment indicates a replicate. Error bars represent the SD of triplicate relative transcript abundance determinations (separate cDNA syntheses) for each culture replicate.
Transcript Abundances of Ferredoxin and Flavodoxin in T. pseudonana
Ferredoxin and flavodoxin transcripts were detected in all experimental treatments with no significant changes in expression regardless of the cells’ Fe nutritional status (Figures 2A,B, respectively). Ferredoxin, the iron-containing protein, was expressed at levels several orders of magnitude higher than flavodoxin, the iron-free protein, in all experiments.
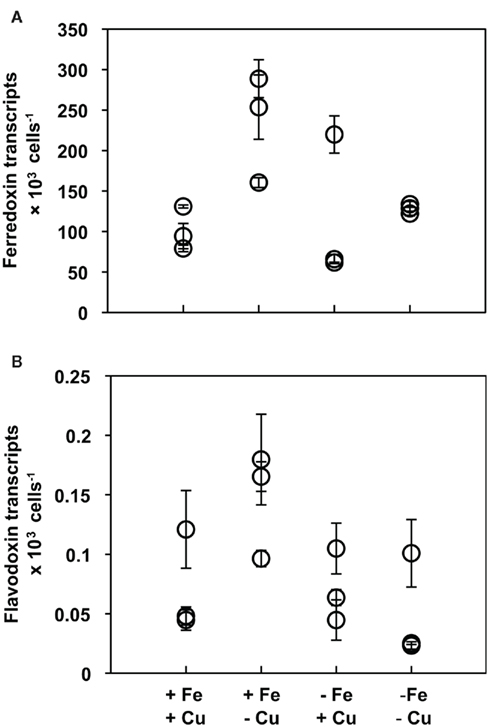
Figure 2. (A) Ferredoxin and (B) flavodoxin transcript abundances normalized to cell number in T. pseudonana cultures acclimated to different culture treatments. Results from culture replicates are shown; each symbol within a treatment indicates a replicate. Error bars represent the SD of triplicate relative transcript abundance determinations (separate cDNA syntheses) for each culture replicate.
Ferredoxin and flavodoxin gene expression was also measured in cells collected daily from Fe limited semi-continuous batch cultures. Unlike the acclimation experiment where cells were exposed to chronic Fe stress, in this experiment T. pseudonana was subjected to rapid Fe deficiency as cells were transferred from replete media into media without any added Fe and sampled daily for 11 days. Cell abundances of the replete cultures and cultures with no added Fe were similar for the first 7 days of the experiment, after which cell numbers in the Fe-minus cultures dropped relative to the replete cultures (Figure 3A). Time points selected for genetic analysis were strategically chosen as they represent cells before experiencing Fe stress (days 1 and 3), at the early onset of stress (day 7), and at the end of the experiment when growth is dramatically restricted (day 11; Figure 3A, indicated with asterisks).
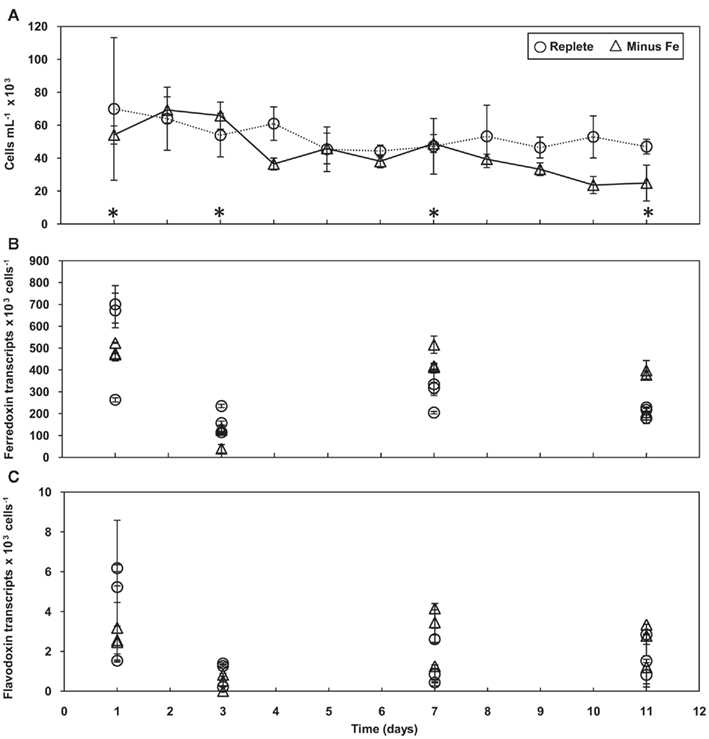
Figure 3. Growth and gene expression patterns of T. pseudonana cells grown in Fe replete and deplete media. (A) Time course of average cell concentration with error bars representing the SD of the biological replicates. Asterisks indicate days analyzed for gene expression. Transcript copies of (B) ferredoxin and (C) flavodoxin normalized to cell number. Results from each culture replicate are shown. Error bars represent the SD of triplicate relative transcript abundance determinations (separate cDNA syntheses) for each culture replicate.
Ferredoxin expression levels were detected at extremely high levels throughout the duration of the time course experiment in the control cultures (Figure 3B). Similarly, high copies of ferredoxin per cell were detected in the cultures with no added Fe; these high levels were maintained throughout the experiment, even on day 11 when cell numbers decreased with Fe limitation. With the exception of day 3, there were no significant changes in ferredoxin expression over time in either the control or minus Fe cultures. On day 3, ferredoxin levels were significantly lower when compared to expression levels on the other days (p < 0.008, one-way ANOVA). Flavodoxin was expressed at levels 100-fold less than ferredoxin, but transcript abundances were similar between cultures grown in replete and Fe-minus media throughout the experiment with no significant changes over time (Figure 3C).
Flavodoxin Gene Expression Displays Diel Periodicities in T. pseudonana
Results from a previous Fe limitation experiment (unpublished) suggested flavodoxin expression in T. pseudonana coordinated with a diel cycle. To investigate this further, gene expression was evaluated in semi-continuous batch cultures of T. pseudonana grown on a light:dark cycle. Triplicate cultures were grown in nutrient replete media under a 12:12 h light:dark cycle and subsampled every 3 h with an equal volume of media added back. The dilution kept the cells at exponential growth for the duration of the experiment and maintained the cell concentration around 1.25 × 106 cells mL−1. The experiment lasted for 96 h; during the final 48 h of the experiment the light regime was shifted to continuous light, a method commonly used to determine if genes are under circadian control.
The effect of the light cycle on ferredoxin and flavodoxin gene expression was examined every 3 h in two culture replicates (Figures 4A,B). Expression of both genes was found to be regulated over the diel cycle, but with opposite trends. Ferredoxin levels reached their maximum during the light cycle and their minimum at the onset of the dark period with an average reduction in transcript abundance of ~7-fold (Figure 4A). As in the previous experiments (Figures 2 and 3), ferredoxin was expressed at levels 100-times greater than flavodoxin. Flavodoxin transcript abundance was greatest at the onset of the dark period, decreased by ~20-fold over the course of the night, and increased at the end of the light cycle (Figure 4B). These oscillating ferredoxin and flavodoxin expression patterns were similar over two light:dark cycles, but were eliminated when shifted into continuous light indicating expression of these genes was not under circadian control (Figures 4A,B).
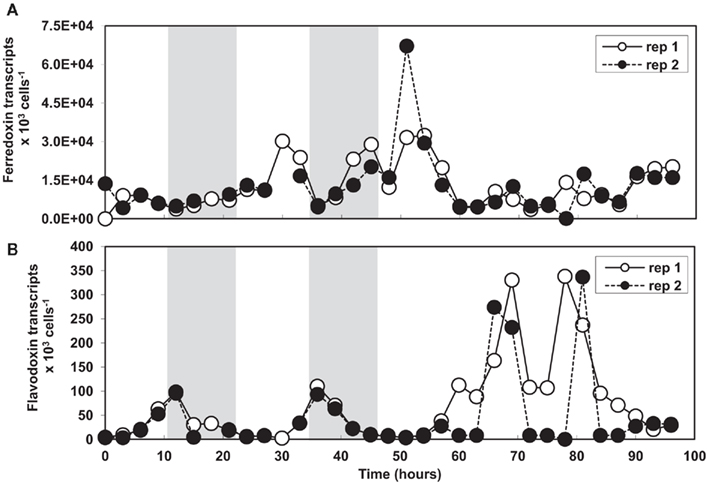
Figure 4. (A) Ferredoxin and (B) flavodoxin transcript abundances normalized to cell number in T. pseudonana semi-continuous batch cultures grown in replete media under a 12:12 h light:dark cycle and shifted into continuous light. Results from culture replicate 1 are shown in white and replicate 2 results are shown in black. Data points not connected in replicate 2 represent missing time points. Shaded boxes indicate the dark period.
Growth Response to Fe Limitation in T. weissflogii
The molecular response to Fe limitation was also studied in an oceanic strain of T. weissflogii, isolated from the North Atlantic. T. weissflogii cells were grown under the same conditions as those previously described for T. pseudonana. Cultures were acclimated to low Cu, low Fe and low Fe/Cu conditions as well as subjected to rapid Fe stress in a semi-continuous batch, time course experiment. T. weissflogii cultures acclimated to trace metal limiting conditions had reduced growth rates when compared to cells acclimated to nutrient replete conditions with the greatest drop seen in cultures chronically stressed for both Fe and Cu (Table 3).
Ferredoxin and Flavodoxin Gene Expression in T. weissflogii
Ferredoxin was expressed at high levels in T. weissflogii cells acclimated to both replete and trace metal limiting conditions (Figure 5). There was no significant difference in the amount of normalized ferredoxin between replete and trace metal limited T. weissflogii cultures. Flavodoxin, however, was undetectable in cells acclimated to replete and low Cu conditions but was significantly induced in cultures acclimated to low Fe media (p = 0.017, one-way ANOVA; Figure 5). Flavodoxin transcripts relative to cell number increased from essentially zero in cultures acclimated to replete growth conditions to levels averaging 2.5 × 103 transcripts per cell in the low Fe cultures. Flavodoxin was also induced in cells co-limited by low Fe and Cu to levels averaging 2.75 × 102 transcripts per cell; approximately 10-fold less when stressed for Fe alone.
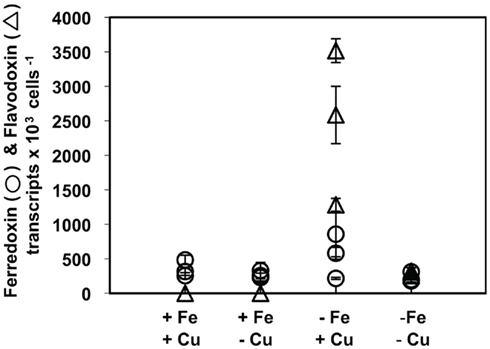
Figure 5. Ferredoxin (circles) and flavodoxin (triangles) transcript abundances normalized to cell number in T. weissflogii cultures acclimated to different culture treatments. Results from culture replicates are shown; each symbol within a treatment indicates a replicate. Error bars represent the SD of triplicate relative transcript abundance determinations (separate cDNA syntheses) for each culture replicate.
To investigate the molecular response to rapid Fe limitation, semi-continuous batch cultures started from T. weissflogii cells grown in nutrient replete media were transferred into media lacking added Fe. Once steady state growth was reached, samples were collected every day for cell counts, relative fluorescence readings, and genetic analysis. When comparing control and Fe deficient culture treatments, cell abundances were similar for the first 7 days of the experiment after which the Fe deficient cultures decreased (Figure 6A).
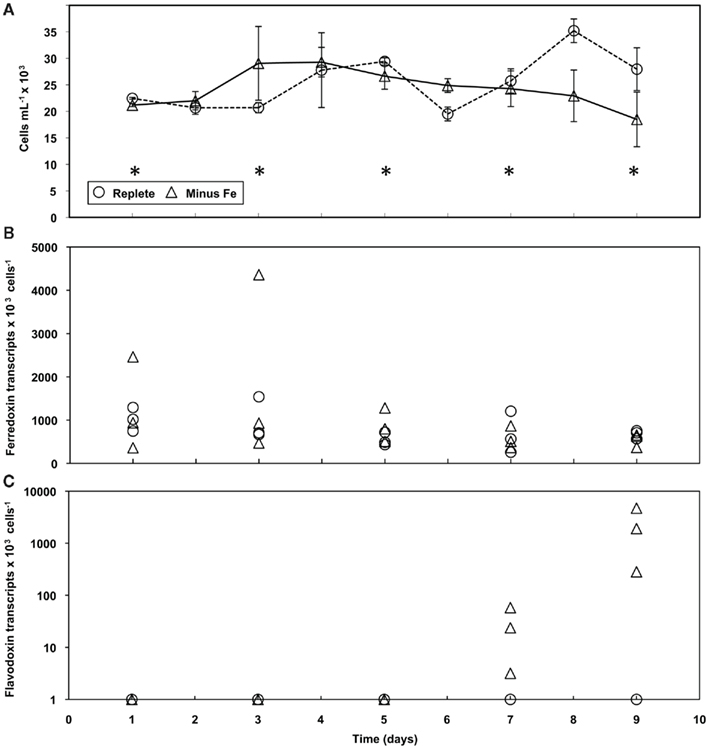
Figure 6. Growth and gene expression patterns of T. weissflogii cells grown in Fe replete and deplete media. (A) Time course of average cell concentration with error bars representing the SD of the biological replicates. Asterisks indicate days analyzed for gene expression. Transcript copies of (B) ferredoxin and (C) flavodoxin normalized to cell number (note the change in scale). Results from each culture replicate are shown.
When exposed to rapid Fe stress, ferredoxin expression in T. weissflogii was detected at similar levels for both treatments with no significant differences throughout the duration of the experiment (Figure 6B). As was seen in the acclimation experiment, flavodoxin expression in T. weissflogii was below detectable limits in the control cultures throughout the entirety of the time course experiment (Figure 6C). In cultures lacking added Fe, expression was detected on day 7, with transcript levels averaging 28 per cell, but flavodoxin was significantly induced by day 9 (Figure 6C). Flavodoxin transcripts per cell reached levels as high as 4.7 × 103 in one culture replicate; transcript levels were not as high in the other Fe deficient cultures, but were significantly higher when compared to earlier time points (p < 0.05, one-way ANOVA).
Phylogenetic Analysis of Flavodoxin Genes in Diatoms
Diatoms can contain more than one copy of the flavodoxin gene in their nuclear genomes. Two copies were identified in the complete genomes of Fragilariopsis cylindrus and Phaeodactylum tricornutum and the recently sequenced pelagophyte, Aureococcus anophagefferens. Two copies were also identified in the draft genomes of Pseudo-nitzschia multiseries and Thalassiosira oceanica. Phylogenetic comparison of these flavodoxin genes shows that they group into two distinct clades with high bootstrap support (Figure 7). Clade I contains the T. pseudonana flavodoxin gene copy. The T. weissflogii Fe-responsive flavodoxin gene copy is found in clade II (Figure 7), suggesting that the two different flavodoxin copies have different functions. It should be noted that it is unknown whether T. weissflogii contains a second copy of flavodoxin as its genome has not been sequenced and attempts at isolating an additional copy with degenerate PCR techniques were unsuccessful. The two A. anophagefferens flavodoxin genes also group into separate clades (Figure 7). The presence of two copies in the evolutionarily “younger” pelagophyte (Brown and Sorhannus, 2010) suggests that flavodoxin underwent an ancient gene duplication that preceded the divergence of diatoms and pelagophytes, and that T. pseudonana has lost a copy of flavodoxin.
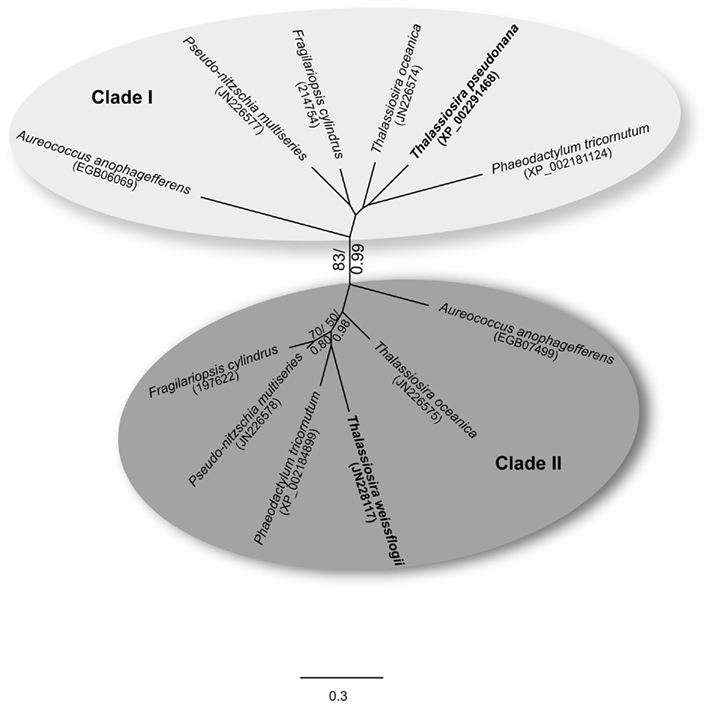
Figure 7. Maximum-likelihood tree (1000 replicates) depicting the relationships of flavodoxin protein sequences from six diatoms and the pelagophyte, Aureococcus anophagefferens. T. pseudonana and T. weissflogii are highlighted in bold; clades I and II are indicated. GenBank accession numbers are shown in parentheses for T. pseudonana, T. weissflogii, T. oceanica, Pseudo-nitzschia multiseries, Phaeodactylum tricornutum, and A. anophagefferens; JGI protein ID numbers are provided for Fragilariopsis cylindrus. Bootstrap proportion values greater than 50 are indicated at the branch points. Included at these branch points are posterior probability values generated from a Bayesian analysis (bootstrap proportion/posterior probability). Scale bar indicates substitutions per site.
Discussion
The aim of this study was to characterize genes that respond to Fe limitation and to identify potential gene-based molecular markers that could be used to detect Fe status and Fe limitation in Thalassiosira species. Gene-based markers provide a means to identify species-specific responses to changing environmental conditions and the extent of their response. An ideal field-based gene target would have a robust transcriptional response in diatoms whose growth is limited by Fe availability. In this study, we analyzed the response of candidate genes that encode components of a putative inducible high affinity Fe uptake system as well as genes encoding ferredoxin and flavodoxin, proteins that have been shown to respond to iron status in a variety of diatom species. The response of these genes was analyzed in the model diatom T. pseudonana, as its complete genome facilitates gene identification and it is a member of a genus with many species that live in different ocean environments.
Fe Limitation Induces Expression of a Putative Fe Reductase in T. pseudonana
Chronically stressing T. pseudonana for Fe or Fe and Cu together resulted in a significant decrease in growth rate, but did not cause an increase in expression of genes putatively involved in the Fe uptake system with the exception of the iron reductase gene copy on chromosome 19 (Figure 1). These results suggest the T. pseudonana putative Fe uptake system is constitutively expressed and may function to acquire Fe in all cells regardless of the cells’ Fe nutritional status. As the only induced gene, Fe reductase (chr 19), which was also the most highly expressed, may be important in supplying the constitutively expressed uptake system with reduced Fe under Fe limiting conditions.
Upregulation of the putative Fe reductase (chr 19) in Fe limited T. pseudonana as measured by qRT-PCR has been seen in previous studies (Kustka et al., 2007; Thamatrakoln et al., 2011) and should be considered a potential Fe status marker. However, this gene is also highly expressed at basal levels in acclimated cells (Figure 1) so it may be difficult to detect induction over background in wild populations of T. pseudonana. More work is needed to determine if this response is seen throughout the genus or is specific to T. pseudonana. Attempts to isolate this gene from T. weissflogii using degenerate PCR techniques were unsuccessful and thus await investigation until more sequenced diatom genomes or transcriptomes become available.
Ferredoxin and Flavodoxin Gene Expression are Constitutive in Fe Limited T. pseudonana
Expression of ferredoxin and flavodoxin was also measured in Fe limited cultures of T. pseudonana. These genes were selected as potential molecular markers as they have been shown to be useful indicators of Fe stress when measuring protein abundances in diatoms. Studies have shown that diatoms, such as P. tricornutum (LaRoche et al., 1995; McKay et al., 1997), T. weissflogii (CCMP 1336; Doucette et al., 1996; Erdner et al., 1999) and T. weissflogii (Gru.) Fryxell et Hasle (clone T-VIC; McKay et al., 1997, 1999) respond to Fe stress by increasing flavodoxin protein abundances. Coupled to the increase in flavodoxin was a decrease in ferredoxin protein levels, thus lowering the cells’ Fe demand (Doucette et al., 1996; Erdner et al., 1999; McKay et al., 1999).
In this study, transcript abundances of ferredoxin and flavodoxin in Fe limited T. pseudonana cells were expected to simulate protein abundance patterns seen in the above-mentioned studies. However, the ferredoxin and flavodoxin transcriptional response was not dependent on iron levels in T. pseudonana cultures acclimated to Fe stress nor in cells exposed to rapid Fe limitation (Figures 2 and 3B,C). In both studies, ferredoxin transcripts were present in abundances several orders of magnitude greater than flavodoxin; this likely reflects the use of ferredoxin in many metabolic pathways.
These results could be interpreted to suggest that the T. pseudonana cells in our experiment were not sufficiently Fe stressed to repress ferredoxin and induce flavodoxin. However, it has been shown that flavodoxin proteins were detected in Fe limited T. weissflogii cells before a significant drop in growth rate was observed (McKay et al., 1997; Erdner et al., 1999). In the experiments presented here, Fe limited cultures of T. pseudonana experienced significant reductions in growth. T. pseudonana cultures acclimated to low Fe and low Fe/Cu conditions grew at ~20% of the maximum rate measured in the control and Cu limiting cultures (Table 3). In the rapid Fe starvation time course experiment, cell abundances were similar until day 8 when the growth in the minus Fe cultures decreased by ~26% compared to the control cultures (Figure 3A). The growth rate and cell abundance data show T. pseudonana was sufficiently Fe stressed in our experiments to detect an early onset response, like flavodoxin induction.
Another possibility, which was not explored in this study, is the potential for post-transcriptional and/or translational level regulation of ferredoxin and flavodoxin in T. pseudonana. This may resolve the differences between this study where transcript abundances were quantified and previous studies where protein levels were measured. However, in T. weissflogii this level of regulation does not seem to occur as flavodoxin gene expression (see discussion below) and upregulation of protein expression (from previous studies, e.g., McKay et al., 1997; Erdner et al., 1999) patterns are similar.
Gene Expression Pattern Suggests Alternate Function for Flavodoxin in T. pseudonana
The expression patterns of ferredoxin and flavodoxin in T. pseudonana suggest that Fe does not regulate these genes; instead their expression was found to respond to diel periodicities. Ferredoxin expression peaked during the light cycle (Figure 4A); its expression pattern was opposite to that of flavodoxin which peaked during the dark cycle (Figure 4B). Regulation of ferredoxin by diel cycles has been documented in higher plants such as pea (Dobres et al., 1987) and wheat (Bringloe et al., 1995), and the green alga, Chlamydomonas reinhardtii (Lemaire et al., 1999). There have not been previous studies in eukaryotic phototrophs assaying flavodoxin in response to diel variations.
Recently Saito et al. (2011) demonstrated that flavodoxin protein production followed diel patterns in the marine diazotrophic cyanobacteria, Crocosphaera watsonii. In this organism, the flavodoxin protein accumulates in the dark concomitant with nitrogen fixation and likely serves as an electron donor for Fe-containing nitrogenase. This allows C. watsonii to reduce its Fe demand during peaks of nitrogen fixation and maintain optimal photosynthesis during the day. Perhaps in T. pseudonana flavodoxin serves a similar function, accumulating at night to help this diatom to meet its high Fe cellular demands, as coastal diatoms have been shown to have higher Fe requirements compared to oceanic isolates (Brand et al., 1983; Brand, 1991; Sunda et al., 1991; Sunda and Huntsman, 1997). Flavodoxin could serve as an electron carrier in plastid-localized processes that occur in the dark, such as fatty acid biosynthesis (Wilhelm et al., 2006); this would allow ferredoxin, the Fe-requiring protein, to fuel photosynthesis and nitrogen assimilation, plastidial processes that occur in the daytime.
Fe Limitation Triggers Flavodoxin Induction in T. weissflogii
In marked contrast to T. pseudonana, flavodoxin expression was undetectable in T. weissflogii (CCMP 1010) cultures grown under replete media and was induced in Fe limiting conditions; a result consistent with the dynamics of flavodoxin protein induction in other isolates of T. weissflogii (McKay et al., 1997; Erdner et al., 1999). When acclimated to trace metal replete and deplete conditions, flavodoxin was detectable only in the low Fe and low Fe/Cu cultures. Expression levels in the Fe limited cells were significantly greater (nine times higher) than in the Fe/Cu co-limited cells (Figure 5). The reduction in flavodoxin expression in the co-limited cultures suggests Cu limitation impedes the cells’ ability to adapt to Fe limitation. This is an interesting response and indicates that expression of the putative high affinity uptake system may be induced by low Fe in T. weissflogii and is Cu-dependent. As previously mentioned, we were unsuccessful in homology-based cloning efforts to isolate T. weissflogii components of the putative high affinity uptake system and further analysis of this system will require genome or transcriptome sequence data.
The daily sampling of the time course study allowed us to capture the window of time where flavodoxin transcription in T. weissflogii was induced in response to rapid Fe starvation. Expression reached detectable levels 7 days after being transferred into media without added Fe, 1 day before a decrease in cell abundance was observed (Figures 6A–C). By day 9, flavodoxin expression had increased on average more than 80-fold when compared to day 7 (Figure 6C). The flavodoxin transcriptional response in T. weissflogii matched the protein response seen by McKay et al. (1997) and Erdner et al. (1999), where flavodoxin protein was detected prior to a major decrease in growth rate.
Repression of ferredoxin expression did not accompany the induction of flavodoxin in Fe stressed T. weissflogii cultures. In this study, ferredoxin was detected at high transcript levels regardless of growth conditions. It is difficult to conclusively determine from this study if ferredoxin expression in this diatom is constitutive and unresponsive to Fe levels as was seen in T. pseudonana, or if it is reduced only under severe Fe stress that exceeds what was experienced by cells in our experiments. However, the latter hypothesis seems probable as flavodoxin, the early-response indicator of Fe stress, was only detected during the last few days of the T. weissflogii time course experiment.
Phylogenetic Analysis Supports Flavodoxins Have Multiple Functions
Phylogenetic analysis of available flavodoxin proteins from diatoms supports the hypothesis that the T. pseudonana gene has a function other than substituting for ferredoxin under Fe limiting conditions (Figure 7). Gene copies encoded by T. pseudonana and T. weissflogii group into separate clades; clade II contains the T. weissflogii flavodoxin gene copy whose expression was regulated by Fe. This suggests members from this clade may be the proteins that respond to Fe deficiency. Clade I contains the T. pseudonana flavodoxin gene copy whose expression did not change with Fe limitation, but rather was regulated by diel periodicities.
One could speculate that diatoms with two gene copies of flavodoxin could have a copy that functions similarly to the one in T. pseudonana and C. watsonii and another copy that is the functional analog of ferredoxin and is induced under Fe limiting conditions. This could be the strategy used by T. weissflogii (CCMP 1010), but may differ among diatoms. For example, Pankowski and McMinn (2009) detected flavodoxin proteins in several coldwater diatom species, despite being grown under Fe replete conditions. The dynamic and complex regulation of flavodoxin among diatoms suggests the species-specific response needs to be investigated before using flavodoxin as a diagnostic marker of Fe stress.
Conclusion
We have shown the expression of a gene encoding a putative Fe reductase may function as a gene marker for Fe status in Thalassiosiroid diatoms. Transcripts accumulated in T. pseudonana when grown under Fe limiting conditions, a response that has been previously documented. It is unknown if this response is specific to T. pseudonana. It was also demonstrated that flavodoxin could be used as a gene-based molecular marker in T. weissflogii. Its expression is specific to Fe stress and appears to be sensitive to the severity of the stress (i.e., not just on/off). This is not true for T. pseudonana where flavodoxin expression is insensitive to Fe status but controlled by a diel cycle. The distinct expression patterns of T. weissflogii and T. pseudonana likely reflect the differences in evolutionary history of the flavodoxin gene copy being analyzed. From this study, we cannot determine if other diatoms are similar to T. pseudonana in that they contain only a single copy of flavodoxin. It is possible that T. pseudonana could have lost the Fe-responsive flavodoxin as it has been maintained for many years in culture. Comparing the transcriptional response to Fe limitation in T. pseudonana to other diatoms would help determine if T. pseudonana is anomalous, but the limited number of sequenced diatom genomes places restrictions on making these comparisons. However, advances are being made on this front. The Marine Microbial Eukaryotic Transcriptome Project (sponsored by the National Center for Genome Resources and the Gordon and Betty Moore Foundation’s Marine Microbiology Initiative) aims to sequence the transcriptomes of nearly 1000 marine microbes. This will vastly improve our ability to compare the molecular response to Fe limitation among species of diatoms as well as aid in the selection of molecular markers that can be used to detect Fe stress in natural populations of diatoms.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge funding from the Chemical and Biological Oceanography Programs at National Science Foundation with grants OCE 0526800 and OCE 0962208 to Bethany D. Jenkins and OCE 0526704 to Mark L. Wells The authors would like to acknowledge instrumentation and sequencing support from NSF-EPSCoR (NSF-0554548 and NSF-1004057) to the University of Rhode Island. The authors would like to thank Drs. Michaela Parker and Virginia Armbrust at the School of Oceanography at University of Washington for sharing data from the Pseudo-nitzschia multiseries genome prior to publication. The authors would also like to thank Dr. Chris Lane at the University of Rhode Island for his help with the phylogenetic analysis. Finally, the authors would like to thank the reviewers, Drs. Coleman, Morgan-Kiss, and Kustka, for improving the manuscript.
References
Brand, L. E. (1991). Minimum iron requirements of marine-phytoplankton and the implications for the biogeochemical control of new production. Limnol. Oceanogr. 36, 1756–1771.
Brand, L. E., Sunda, W. G., and Guillard, R. R. L. (1983). Limitation of marine-phytoplankton reproductive rates by zinc, manganese, and iron. Limnol. Oceanogr. 28, 1182–1198.
Bringloe, D. H., Dyer, T. A., and Gray, J. C. (1995). Developmental, circadian and light regulation of wheat ferredoxin gene-expression. Plant Mol. Biol. 27, 293–306.
Brown, J. W., and Sorhannus, U. (2010). A molecular genetic timescale for the diversification of autotrophic stramenopiles (Ochrophyta): substantive underestimation of putative fossil ages. PLoS ONE 5, e12759. doi:10.1371/journal.pone.0012759
Dobres, M. S., Elliott, R. C., Watson, J. C., and Thompson, W. F. (1987). A phytochrome regulated pea transcript encodes ferredoxin-i. Plant Mol. Biol. 8, 53–59.
Doucette, G. J., Erdner, D. L., Peleato, M. L., Hartman, J. J., and Anderson, D. M. (1996). Quantitative analysis of iron-stress related proteins in Thalassiosira weissflogii: measurement of flavodoxin and ferredoxin using HPLC. Mar. Ecol. Prog. Ser. 130, 269–276.
Drummond, A. J., Ashton, B., Buxton, S., Cheung, M., Cooper, A., Duran, C., Field, M., Heled, J., Kearse, M., Markowitz, S., Moir, R., Stones-Havas, S., Sturrock, S., Thierer, T., and Wilson, A. (2010). Geneious v5.3. Available at: http://www.geneious.com
Erdner, D. L., and Anderson, D. M. (1999). Ferredoxin and flavodoxin as biochemical indicators of iron limitation during open-ocean iron enrichment. Limnol. Oceanogr. 44, 1609–1615.
Erdner, D. L., Price, N. M., Doucette, G. J., Peleato, M. L., and Anderson, D. M. (1999). Characterization of ferredoxin and flavodoxin as markers of iron limitation in marine phytoplankton. Mar. Ecol. Prog. Ser. 184, 43–53.
Guillard, R. R. L. (1975). “Culture of phytoplankton for feeding marine invertebrates,” in Culture of Marine Invertebrate Animals, eds W. L. Smith and M. H. Chaney (New York: Plenum Press), 26–60.
Guillard, R. R. L., and Hargraves, P. E. (1993). Stichochrysis immobilis is a diatom, not a chyrsophyte. Phycologia 32, 234–236.
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. (Stevenage) 52, 696–704.
Hattori-Saito, A., Nishioka, J., Ono, T., Mckay, R. M. L., and Suzuki, K. (2010). Iron deficiency in micro-sized diatoms in the Oyashio region of the western subarctic Pacific during spring. J. Oceanogr. 66, 105–115.
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755.
Keller, M. D., Bellows, W. K., and Guillard, R. R. L. (1988). Microwave treatment for sterilization of phytoplankton culture media. J. Exp. Mar. Biol. Ecol. 117, 279–283.
Kustka, A. B., Allen, A. E., and Morel, F. M. M. (2007). Sequence analysis and transcriptional regulation of iron acquisition genes in two marine diatoms. J. Phycol. 43, 715–729.
LaRoche, J., Murray, H., Orellana, M., and Newton, J. (1995). Flavodoxin expression as an indicator of iron limitation in marine diatoms. J. Phycol. 31, 520–530.
Lemaire, S. D., Stein, M., Issakidis-Bourguet, E., Keryer, E., Benoit, V., Pineau, B., Gerard-Hirne, C., Miginiac-Maslow, M., and Jacquot, J. P. (1999). The complex regulation of ferredoxin/thioredoxin-related genes by light and the circadian clock. Planta 209, 221–229.
Maldonado, M. T., Allen, A. E., Chong, J. S., Lin, K., Leus, D., Karpenko, N., and Harris, S. L. (2006). Copper-dependent iron transport in coastal and oceanic diatoms. Limnol. Oceanogr. 51, 1729–1743.
Maldonado, M. T., Hughes, M. P., Rue, E. L., and Wells, M. L. (2002). The effect of Fe and Cu on growth and domoic acid production by Pseudo-nitzschia multiseries and Pseudo-nitzschia australis. Limnol. Oceanogr. 47, 515–526.
Marchetti, A., Parker, M. S., Moccia, L. P., Lin, E. O., Arrieta, A. L., Ribalet, F., Murphy, M. E. P., Maldonado, M. T., and Armbrust, E. V. (2009). Ferritin is used for iron storage in bloom-forming marine pennate diatoms. Nature 457, 467–470.
McKay, R. M. L., Geider, R. J., and LaRoche, J. (1997). Physiological and biochemical response of the photosynthetic apparatus of two marine diatoms to Fe stress. Plant Physiol. 114, 615–622.
McKay, R. M. L., La Roche, J., Yakunin, A. F., Durnford, D. G., and Geider, R. J. (1999). Accumulation of ferredoxin and flavodoxin in a marine diatom in response to Fe. J. Phycol. 35, 510–519.
Moore, J. K., Doney, S. C., and Lindsay, K. (2004). Upper ocean ecosystem dynamics and iron cycling in a global three-dimensional model. Global Biogeochem. Cycles 18, 21.
Nelson, D. M., Treguer, P., Brzezinski, M. A., Leynaert, A., and Queguiner, B. (1995). Production and dissolution of biogenic silica in the ocean – revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem. Cycles 9, 359–372.
Pankowski, A., and McMinn, A. (2009). Development of immunoassays for the iron-regulated proteins ferredoxin and flavodoxin in polar microalgae. J. Phycol. 45, 771–783.
Price, N. M., Harrison, G. I., Hering, J. G., Hudson, R. J., Pascale, M. V. N., Palenik, G., and Morel, F. M. M. (1989). Preparation and chemistry of the artificial algal culture medium aquil. Biol. Oceanogr. 6, 443–461.
Round, F. E., Crawford, D. G., and Mann, D. G. (1990). The Diatoms: Biology and Morphology of the Genera. Cambridge: Cambridge University Press, 747.
Saito, M. A., Bertrand, E. M., Dutkiewicz, S., Bulygin, V. V., Moran, D. M., Monteiro, F. M., Follows, M. J., Valois, F. W., and Waterbury, J. B. (2011). Iron conservation by reduction of metalloenzyme inventories in the marine diazotroph Crocosphaera watsonii. Proc. Natl. Acad. Sci. U.S.A. 108, 2184–2189.
Schecher, W. D., and McAvoy, D. C. (1998). MINEQL+: A Chemical Equilibrium Modeling System, Version 4.0 for Windows, User’s Manual, Environmental Research Software.
Sunda, W. G., and Huntsman, S. A. (1997). Interrelated influence of iron, light and cell size on marine phytoplankton growth. Nature 390, 389–392.
Sunda, W. G., Swift, D. G., and Huntsman, S. A. (1991). Low iron requirement for growth in oceanic phytoplankton. Nature 351, 55–57.
Suzuki, K., Saito, H., Isada, T., Hattori-Saito, A., Kiyosawa, H., Nishioka, J., McKay, R. M. L., Kuwata, A., and Tsuda, A. (2009). Community structure and photosynthetic physiology of phytoplankton in the northwest subarctic Pacific during an in situ iron fertilization experiment (seeds-ii). Deep Sea Res. Part 2 Top. Stud. Oceanogr. 56, 2733–2744.
Thamatrakoln, K., Korenovska, O., Niheu, A. K., and Bidle, K. D. (2011). Whole-genome expression analysis reveals a role for death-related genes in stress acclimation of the diatom Thalassiosira pseudonana. Environ. Microbiol. doi: 10.1111/j.1462-2920.2011.02468.x
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). Clustal-w – improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680.
Wells, M. L., Trick, C. G., Cochlan, W. P., Hughes, M. P., and Trainer, V. L. (2005). Domoic acid: the synergy of iron, copper, and the toxicity of diatoms. Limnol. Oceanogr. 50, 1908–1917.
Wilhelm, C., Buchel, C., Fisahn, J., Goss, R., Jakob, T., LaRoche, J., Lavaud, J., Lohr, M., Riebesell, U., Stehfest, K., Valentin, K., and Kroth, P. G. (2006). The regulation of carbon and nutrient assimilation in diatoms is significantly different from green algae. Protist 157, 91–124.
Keywords: diatom, iron, flavodoxin, ferredoxin, gene expression
Citation: Whitney LP, Lins JJ, Hughes MP, Wells ML, Chappell PD and Jenkins BD (2011) Characterization of Putative Iron Responsive Genes as Species-Specific Indicators of Iron Stress in Thalassiosiroid Diatoms. Front. Microbio. 2:234. doi: 10.3389/fmicb.2011.00234
Received: 21 July 2011;
Accepted: 09 November 2011;
Published online: 25 November 2011.
Edited by:
Anton F. Post, MBL, USAReviewed by:
Maureen Coleman, California Institute of Technology, USARachael Marie Morgan-Kiss, Miami University, USA
Adam Kustka, Rutgers University at Newark, USA
Copyright: © 2011 Whitney, Lins, Hughes, Wells, Chappell and Jenkins. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Bethany D. Jenkins, Department of Cell and Molecular Biology, University of Rhode Island, 120 Flagg Road, Kingston, RI 02881, USA. e-mail:YmplbmtpbnNAdXJpLmVkdQ==


 Margaret P. Hughes2
Margaret P. Hughes2