- 1 Rumen Microbiology, Animal Nutrition and Health Capability Group, Grasslands Research Centre, AgResearch Limited, Palmerston North, New Zealand
- 2 Riddet Institute, Massey University, Palmerston North, New Zealand
Lifestyle adaptation of microbes due to changes in their ecological niches or acquisition of new environments is a major driving force for genetic changes in their respective genomes. Moving into more specialized niches often results in the acquisition of new gene sets via horizontal gene transfer to utilize previously unavailable metabolites, while genetic ballast is shed by gene loss and/or gene inactivation. In some cases, larger genome rearrangements can be observed, such as the incorporation of whole genetic islands, providing a range of new phenotypic capabilities. Until recently these changes could not be comprehensively followed and identified due to the lack of complete microbial genome sequences. The advent of high-throughput DNA sequencing has dramatically changed the scientific landscape and today microbial genomes have become increasingly abundant. Currently, more than 2,900 genomes are published and more than 11,000 genome projects are listed in the Genomes Online Database‡. Although this wealth of information provides many new opportunities to assess microbial functionality, it also creates a new array of challenges when a comparison between multiple microbial genomes is required. Here, functional genome distribution (FGD) is introduced, analyzing the diversity between microbes based on their predicted ORFeome. FGD is therefore a comparative genomics approach, emphasizing the assessments of gene complements. To further facilitate the comparison between two or more genomes, degrees of amino-acid similarities between ORFeomes can be visualized in the Artemis comparison tool, graphically depicting small and large scale genome rearrangements, insertion and deletion events, and levels of similarity between individual open reading frames. FGD provides a new tool for comparative microbial genomics and the interpretation of differences in the genetic makeup of bacteria.
Introduction
Microbial genomes range in size from the smallest microbial genome known to date of Candidatus Tremblaya princeps with just under 139,000 nt (McCutcheon and von Dohlen, 2011) to large genomes with over 13,000,000 nt such as Sorangium cellulosum SO ce56 (Schneiker et al., 2007). With an average gene size of about 1,000 nt, microbial genomes harbor between 140 and 13,000 genes.
Lifestyle adaptation is one of the major driving forces for microbial genome re-arrangement processes such as gene loss and gene acquisition, genome rearrangements, and the movement of whole genetic islands. Until recently, such processes could not be monitored comprehensively and observations were limited to either a few select genes (typing) or the analysis of large scale genome re-arrangement events.
Typing methods such as multi-locus sequence typing (MLST; Chan et al., 2001), are based on the selection of a few widely distributed and conserved house-keeping genes. Today, MLST utilizes whole microbial genome predominantly to identify new select target regions for amplification, rather than analyzing the whole nucleotide sequence (Maiden, 2006).
With the advent of high-throughput DNA sequencing techniques such as automated capillary sequencing, solid-state sequencing1, or pyrosequencing2 or sequencing by synthesis3, access to high-quality draft, and complete bacterial genomes has become feasible and is a commonly used technique. Currently 2,943 complete genomes (including eukaryotic organisms) have been reported, with another 184 archeal and 5,490 bacterial genome projects in progress4. The availability of complete or nearly complete genomes triggered attempts to incorporate selected larger genetic subsets (Makarova et al., 2006; Makarova and Koonin, 2007) or complete genomes (Rohwer and Edwards, 2002; Henz et al., 2005) to infer evolutionary lineages, while less progress has been made in analysis of multiple whole microbial genomes from functional and comparative genomic perspectives. One of the most prominent examples of whole genome comparative analyses is based on Blast Score Ratios (Rasko et al., 2005).
Here, we introduce a new analysis tool, functional genome distribution (FGD). FGD does not attempt to represent the evolutionary path a genome has taken, since different genes will have been acquired by different routes. Instead, FGD investigates the overall levels of similarity between microbial genomes based on amino-acid sequences of the predicted complete ORFeomes. This reflects the impact the evolutionary force has had on genome makeup in the past, resulting in the current level of niche adaptation (Thomson et al., 2003). Thus presence, absence, or modification of individual genes or genetic islands defines the phenotypic potential of a given organism at a given temporal snapshot. The comparison of these ORFeomes to each other ultimately defines the level of similarity of the genomes. This approach then also takes into account important genetic adaptations to specific ecological niches or even to human made environments such as industrial fermentation processes. Such common genotype adaptations might render organisms more similar by FGD analyses than their respective evolutionary heritage would indicate. The presented approach of a FGD is a BLAST-based ORF-position-independent algorithm, implemented in the compACTor software. In the context of FGD analyses the term “functional” is used in the sense of functionality based on sequence and sequence similarity and is not based on annotation classification [i.e., such as implied by COG (Tatusov et al., 2003) or KEGG (Kanehisa, 2002; Kanehisa et al., 2008) databases].
Research in functional genomics (defined as the investigation of gene function by gene inactivation, gene complementation, and in silico analyses) relies heavily on the identification of differences between two or more genomes, identifying differences in the presence or absence of individual genes. The compACTor software also creates all-vs.-all ORFeome distance data files which, in combination with the respective GenBank files of the query microbes and the Artemis comparison tool (ACT; Carver et al., 2005), facilitate visual qualitative comparative in silico analyses of complete and draft phase microbial genomes for the subsequent analysis of changes to gene synteny and operon structures. Furthermore, to identify genes shared and unique between selected clusters a mining tool, FGDfinder, is provided, facilitating rapid identification of relevant gene sets for further in vitro analyses.
Materials and Methods
General Workflow of the compACTor Software
A pool of genomes in GenBank format represents the query space for compACTor (Figure 1). Gene models embedded and subsequently retrieved from the GenBank files of each query genome are parsed and the resulting ORFeome is translated into individual amino-acid sequences. The parsing algorithm considers both gene and CDS features for any given start position, while giving preference to CDS entries. Nucleotide sequences are retrieved from the genome sequence and translated into amino-acid sequence. Gene model errors such as unbalanced number of nucleotides, unrecognized amino-acid codons, or multiple stop-codons are reported but treated as non-critical. The translated ORFeome is then used to build query-specific amino-acid BLAST databases (DBs) using formatdb (Altschul et al., 1990). In subsequent runs previously build BLAST databases can be re-used (Figure 1), reducing the overall runtime of the compactor software. The pooled ORFeomes of all query entries are hashed for subsequent all-vs.-all analyses. Levels of similarities are inferred for each predicted query ORF by an all-vs.-all analysis, submitting sequence-database combinations to a non-filtered gapped BLASTP (Altschul et al., 1997) pipeline. Briefly, the deduced amino-acid sequence for each open reading frame (ORF) of each query entry is compared to all subject databases generated from the entry pool. Individual BLAST results are parsed and collated in ORF-specific ASCII result files. The total number of BLAST analyses performed is a direct function of the number of individual ORFeomes and the pool size.
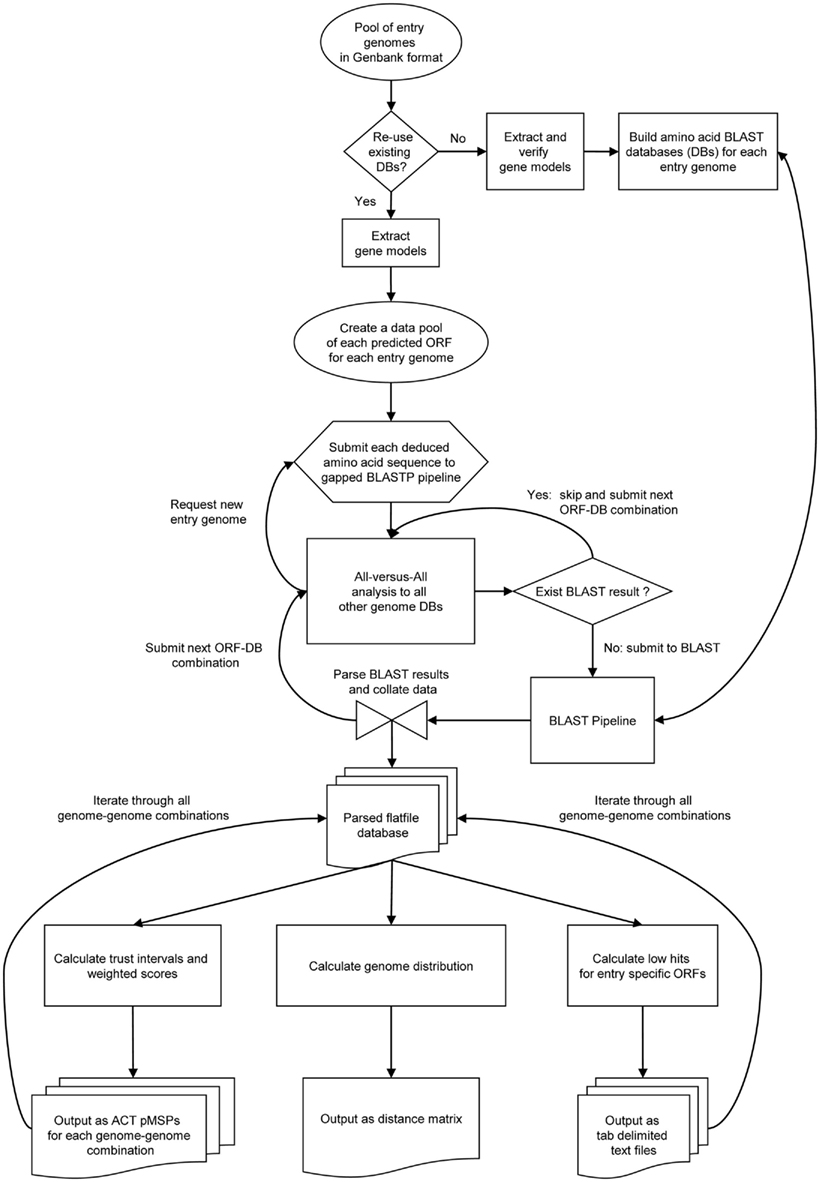
Figure 1. Graphical workflow of the compACTor software. Schematic representation of the algorithm structure. Symbols used are consistent with standard flowchart icons. Abbreviations: DB, database; pMSPs, ORFeome based MSPcrunch comparison format (Sonnhammer and Durbin, 1994) data files. Oval symbol: data pools, diamond symbol: internal decision points, square boxes: internal processes and functions, hourglass symbol: central parsing algorithm, Multidocument symbol: external flatfile databases created by compACTor.
The collated flatfile database is the basis for all subsequent calculations such as the generation of ACT (Carver et al., 2005) pMSP-datafiles, the prediction of putative strain-specific genes, and the generation of FGD trees. It is also possible to add new query entries to existing analyses. The compACTor software tests for the presence of respective ORF-database BLAST results and omits the BLAST pipeline where possible. This significantly reduces runtime requirements and facilitates future expansions of existing comparative analyses.
Generation of ACT Comparison Data Files (pMSP)
Based on the all-vs.-all principle, all genome–genome pair combinations are analyzed. A bi-directional BLASTP approach was implemented for the creation of ORF-specific ACT comparison files. Genome pairs analyzed in respective comparison files are reflected within the filename which features both filenames in the order query-to-subject. Each predicted ORF of the query genome was allowed a maximum of 20 similarity hits to ORFs present in the respective subject genome. ACT allows two quality parameters, namely “Score cutoff” and “Percent ID cutoff” in its comparison file format. These two cutoffs determine the stringency of sequence similarities, and were replaced in the compACTor software output by the alternative parameters of “e-value” (“Percent ID”) and “weighted score” (“Score”). The parameter “e-value” is derived from individual e-values which are grouped into empirically determined trust level ranges (Table a1 in Appendix). Similarly, for the second parameter, a weighted score is calculated, placing additional emphasis on alignment quality and length (Eq. 1). Briefly, the initial alignment-centric local BLAST score is reflecting the maximum level of sequence similarity between a given ORF pair (ς, Eq. 1). To account for changes outside the local alignment and to further assess the alignment quality, the score is then subjected to three penalty blocks, assessing the quality of amino-acid similarity [(P − Id/2) + Id/λ, Eq. 1], the size of gaps (1 − G/A, Eq. 1), and the length of the BLAST alignment in relation to the query ORF length (A − G/λ, Eq. 1). These penalty parameters take into account differences in amino-acids sequence alignments which are not or insufficiently covered by the regular BLAST score. This process effectively reduces the level of similarity between both ORFs when the BLAST alignment covers only partial ORF sequences and/or the alignments show significant levels of insertions or deletions and results in a weighted BLAST score ψ.
The weighted score is a sub-parameter of the distance calculation between two genomes (see Eq. 2) and allows for stringency shifting in ACT while maintaining continuity to the FGD algorithm.
Weighted BLAST score
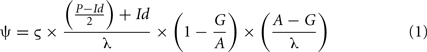
with: ψ = weighted score, ς = BLAST score between query sequence and respective BLAST hit, λ = length of deduced amino-acid in query sequence, P = identified positives in BLAST alignment, Id = identified identities in BLAST alignment, A = length of BLAST alignment, G = number of gaps in BLAST alignment
Artemis comparison tool compatible comparison files (pMSP) created with the compACTor software adhere to the MSPcrunch (Sonnhammer and Durbin, 1994) format. An example dataset of a two-way genome-to-genome comparison is provided in the Appendix.
Functional Genome Distribution
In prokaryotes, genome plasticity (re-arrangement of genomic regions between species), the presence of metabolic islands (clusters of genes with defined and specific metabolic functions), gene acquisition via horizontal gene transfer (HGT), and varied distribution of mobile genetic elements are well described and contribute significantly to phenotypic differences (Desiere et al., 2001; Makarova et al., 2006; Berger et al., 2007; Nicolas et al., 2007). Also, adaptation to new ecological niches and the resulting genetic drift can lead to rapid acquisition or the shedding of genetic elements without changes in selected sequence or gene subsets used in typing analyses (Makarova et al., 2006). Organisms, virtually indistinguishable based on selected individual gene (set) evolutionary analyses, may be markedly different in their complete genetic blueprint. To reflect this widespread diversity, a new algorithm, FGD, was empirically developed to distinguish organisms in the multi-dimensional genome space. Here, the focus distinctly lies on a snapshot picture of a current genome and how the genotype relates to other organisms. From a functional genomics perspective, the presence, absence, or modification of a gene determines a potential metabolic capability, while the respective position with the genome may influence the respective levels of gene expression (Sousa et al., 1997). Therefore, a position independent approach was embraced that investigates the phenotypic potential (absence/presence/similarity of genes) at the cost of loci dependent modulations of gene expression.
Each predicted ORF of a given query genome is analyzed via gapped BLASTP to a subject organism specific BLAST database. Result parameters of the best BLAST hit are applied to the following equations (Eqs 2 and 3).
Distance between two genomes

with: δ = final similarity score between two genomes, ψ = weighted BLAST score as described in Eq.1, λ = length of deduced amino-acid query sequence, TORFquery = total number of ORFs in query genome, n = current query ORF, and ρORF = ORF number ratio as described in Eq.3.
ORFeome ratio between query and subject genome
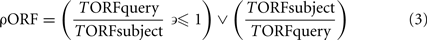
with: ρORF = ORF number ratio between query and subject genome, always ≤ 1, TORFquery = total number of ORFs in query genome, TORFsubject = total number of ORFs in subject genome.
Motivation of the FGD Algorithm
The FGD algorithm initially investigates the level of similarity for each predicted ORF in a query genome to a subject ORF and is based on the BLAST score. This score describes the overall quality of the best local alignment found for a given query–subject sequence alignment. However, the score alone does not necessarily reflect the overall level of similarity between two sequences outside the local alignment. Also, from a functional genomics perspective, insertion, and deletion events in individual amino-acid sequences may change the properties of the gene product, thus decreasing the level of similarity. Furthermore, the relation of the local alignment to the overall sequence is an important factor. While stretches of highly conserved sequence contribute to a higher BLAST score, the presence of unique sequence outside the local alignment is likely to contribute to changing the properties of respective gene products. Therefore, the maximum level of similarity as described by the BLAST score will be decreased by the FGD algorithm if insertion/deletion events and imbalanced alignment/deduced sequence ratios are detected.
The FGD analysis then summarizes individual similarity scores determined for each ORF in the ORFeome. The presence of unique strain-specific genes and differences in the overall number of predicted ORFs in respective microbial genome pairs contribute to different genotypes and consequently decrease the level of overall functional similarity between genome pairs.
The initial alignment BLAST score is subjected to quality assessments as described above for Eq. 1. In Eq. 2 the resulting weighted score is normalized by the number of deduced amino-acids of the query ORF, resulting in a score per amino-acid [ψ/λ(n)]. Individual similarity scores are then are summed up over the query ORFeome and normalized over total number of predicted ORFs, resulting in a weighted score per ORF. Assessing differences in ORFeome sizes by calculating the ratio between query and subject ORFeomes (Eq. 3), the similarity score is adjusted to result in a final genome similarity score (δGquery → Gsubject).
Genome similarity scores for each query–subject combination are entered into a distance matrix. A symmetrical distance matrix, based on the geometrical means of each genome–genome pair combination is calculated according to Eq.4.
Symmetrical distance matrix
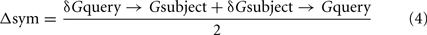
with: Δsym = final score for symmetrical distance matrix between a given genome–genome pair and δGquery → Gsubject = similarity score between a given genome–genome combination.
Distance matrices of larger datasets become increasingly complex to read directly. Therefore a traditional tree representation was used as a visual aid to the underlying data structure. The resulting symmetrical distance matrix can be imported into software packages such as Mega4 (Tamura et al., 2007). Genome clusters are approximated using the Unweighted Pair Group Method with Arithmetic mean (UPGMA) algorithm (Sneath and Sokal, 1962). Approximated branch lengths, representing the level of similarity between genomes are depicted by distance units (du). Although similar in appearance to phylogenetic trees, the resulting functional distribution tree (FDT) depicts a different concept, clustering organisms by their respective ORFeome similarities rather than by their inferred evolutionary line.
Identification of Putative Strain-Specific Genes
A key interest in comparative functional genomics is the identification of strain and cluster specific and shared gene sets, identifying the genetic differences, and similarities between selected microbial genomes. These gene sets can then be used to investigate potentially unique metabolic capabilities of microbes and may reveal specific genome adaptation to lifestyle changes. Cluster specific genes or genes shared between clusters are identified using the flatfile BLAST database created by compACTor. Query ORFs exhibiting a minimum e-value threshold with respect to a given subject genome are identified and stored if they fulfill the stringency restriction. Stringency between selected clusters can be set by selecting the number of allowable mismatches. A mismatch is defined as an ORF in the query cluster that has at least one ORF–ORF pairing above the minimum e-value threshold to the subject cluster. Objects implemented in the output are: name of the query GenBank file, respective ORF identifier, the name of the subject GenBank file, annotation of the subject ORF, and the corresponding e-value, query ORF length, score, query ORF start and stop positions, and gene, locus_tag, product, and note annotation. Data for each query ORFeome – subject genome combination are collated and summarized in a tab-delimited ASCII file. The individual data files can easily be imported into spreadsheet programs such as MS Excel or OpenOffice Calc and then be further re-grouped and analyzed. For example, grouping by query ORF number quickly identifies the degree to which a respective ORF is unique among the pool of genome entries.
Implementation
The compACTor and FGDfinder software has been realized as PERL scripts (ActivePerl version 5.8.8.822), using the official NCBI BLAST distribution5. The software is freely available for non-commercial use (commercial use requires a license).
The algorithms and BLAST utilities used in compACTor render the application CPU intensive. compACTor is multi-threaded and support multiple CPUs/cores to reduce runtime in a near linear fashion. For the analysis depicted in Figure 2, 39 genomes, ranging from ∼1.7 to ∼6.6 Mbp, were analyzed. A total number of 113,397 ORFs resulted in ∼4,300,000 individual BLAST queries. Artemis Comparison Files, the prediction of putative strain-specific ORFs and the phylogenetic tree were calculated using a collated and parsed BLAST flatfile database of >11 Gb. 1,560 Artemis comparison files were generated, reflecting all possible genome–genome combinations (a datapool of >2.5 Gb).
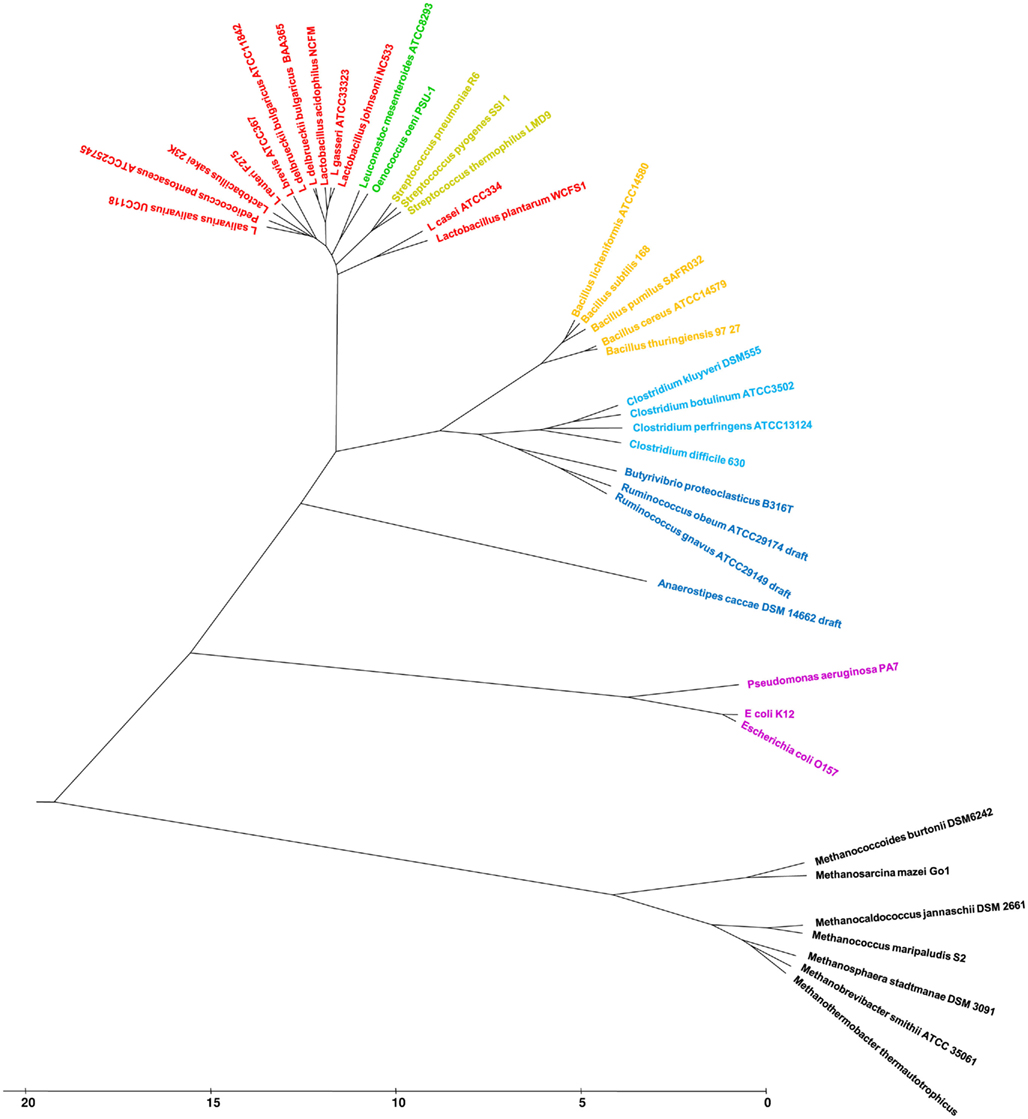
Figure 2. Functional genome distribution of 39 taxa. Publicly available complete genomes were downloaded in GenBank format from the NCBI genome database. Publicly available draft phase genomes were downloaded in FASTA format, concatenated using a universal spacer-stop-spacer sequence and automatically annotated using GAMOLA (Altermann and Klaenhammer, 2003). The in-house draft phase genome of Butyrivibrio proteoclasticus was assembled into an artificial genome and annotated using GAMOLA (publication in preparation). Predicted ORFeomes of all genomes were subjected to an FGD analysis and the resulting distance matrix was imported into MEGA4. The functional distribution was visualized using the UPGMA method (Sneath and Sokal, 1962). The optimal tree with the sum of branch length = 133.1 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the functional distances used to infer the distribution tree.
Known Limitations
Known limitations of the algorithm at this time are the lack of integrated support for extrachromosomal genetic elements such as plasmids and additional chromosomes. A workaround exists by concatenating DNA sequences of all genetic information present in the respective organism and then performing an automated annotation using pipelines such as GAMOLA (Altermann and Klaenhammer, 2003). The implemented GenBank parser currently ignores joined features. In prokaryotes, joined features such as introns [e.g., the mobilization protein MobA E1 (genome position 1344187.0.1345428, 1347858.0.1348370) in Lactococcus lactis subsp. cremoris MG1363 (GI:124491690)] are very rare and usually do not interfere with the analyses. Also, identical genomes still incur a distance to each other, resulting in a small branching when visualizing the dissimilarity matrix. A normalization algorithm will be implemented in the next release to address this issue. compACTor and FGDfinder are actively developed and future releases will resolve those issues.
Results and Discussion
Evaluation of Functional Distribution Trees
To assess the FGD concept 35 completed and four draft phase genomes from different phyla, including several archeal genomes, were selected (Table a2 in Appendix). The purpose of this diverse taxonomy is to investigate how FGD places individual genomes into clusters and how similar these genomes are to each other on a functional level. The majority of the genomes selected are members of the family Lactobacillaceae in the order Lactobacillales (12 genomes). To investigate the discriminatory power of the FGD algorithm, the range was expanded and five genomes of the family Bacillaceae (order Bacillales), three genomes of the family Streptococcaceae (order Lactobacillales) and two genomes of the family Leuconostocaceae (order Lactobacillales) were added. Eight more distantly related genomes of the order Clostridiales (class Clostridia) were chosen to broaden the taxonomic selection to different classes. All of these genomes are members of the phylum Firmicutes. Three representatives of the class Gammaproteobacteria, (phylum Proteobacteria) were included to investigate inter-phylum relationships. As a final outlier, six genomes of archeal Euryarchaeota were included in the analysis.
Figure 2 represents the functional distribution of the selected taxa within an FDT. To test the influence of early, incomplete draft phase genomes on functional placement, the draft phase sequence of Anaerostipes caccae DSM14662 (Schwiertz et al., 2002) which consisted of ∼1.69 Mbp at the time of analysis was included. FGD penalized the missing genetic information and set A. caccae apart from the Clostridiales as a separate cluster. When the updated genome sequence of A. caccae encompassing ∼3.6 Mbp was included instead, A. caccae shifted its position and clustered with Ruminococcus obeum and Ruminococcus gnavus (branch length 1.99 du), while showing a deeper branching to Butyrivibrio proteoclasticus (branch length 3.13 du; data not shown). This clearly highlights the necessity of obtaining high-coverage genome sequence data for FGD analyses. However, with high-throughput genome sequencing techniques currently available, initial draft phase genomes usually encompass 85–95% of the genome, thus allowing an initial representative functional placement.
Subsequent analysis of complete genomes included in the FDT revealed the deepest branching (20.6 du) for euryarcheal genomes. The selected methanogens form two distinct genome clusters (node at 5.3 du), separating Methanococcoides burtonii and Methanosarcina mazei (Deppenmeier et al., 2002) from the remaining four taxa. Initial comparisons of habitat, growth temperature, and GC content did not indicate a consistently shared denominator between the two groups. Further analyses will be necessary to determine the imminent functional similarities indicated by the FGD approach.
Interestingly, Clostridiaceae, Bacillaceae, and a subcluster comprised of Rumincocci and B. proteoclasticus B316 formed a new functional node within the FDT (branch depth from node to Lactobacillales was 7.8 du), combining the taxonomic families into one genome cluster. Although the genomes of members of Clostridiaceae and Bacillaceae are still placed into distinct functional groups (internal cluster branch depth was 4 du) and no taxon shuffling was observed between both sub-clusters, it appears that lifestyle adaptation has led to similar genome content, potentially indicative of a high level of HGT between both families or from one family to the other.
A survey of the nine ORFeomes of the Bacillus and Clostridium clusters revealed 154 ORFs that are highly conserved in both groups (e-value threshold 1e-100). As expected, most ORFs could be assigned to central house-keeping functions such as DNA synthesis and repair (21%), tRNA genes and related processes (12%), central metabolism (35%), transcription and translation (6%), cellular processes (9%), and molecule transport (8%). However, besides these central functions, a significant number of ORFs related to sporulation were present and highly conserved in both clusters (8%). Based on the algorithms used, it is reasonable to hypothesize that the presence of these highly conserved sporulation genes may be one of the key drivers for the observed clustering of bacilli and true clostridia.
When compared to the Ruminococcus subcluster a similar conserved gene set was found with the notable absence of most sporulation genes (e-value threshold 1e-60). This is in agreement with the observed non-sporulating phenotype. It is noteworthy, that two conserved genes involved in sporulation (stage V sporulation protein D, spoVD, and Sporulation initiation inhibitor protein, soj) were identified in the Ruminococcus and Butyrivibrio genomes. This may indicate an ongoing genetic loss in response to adaptation to a new environment (rumen) where sporulation is no longer offering an advantage in fitness. The other identified shared genes are likely to be present in most of the other microbial genomes analyzed, and thus would contribute to higher-level genome clustering.
In addition, the ORFeomes were analyzed for predicted genes which are conserved in one genome cluster but not in the other – and vice versa (threshold conserved: 1e-100; threshold unique: 1e-10). Overall, 84 ORFs were identified to be group specific. Seventy of these were found only in genomes assigned to the Bacillus cluster and 14 in genomes in the Clostridium group. Remarkably, genes involved in heme and cytochrome biogenesis (hemE, hemH, hemY, resB, and resC), cytochrome reduction (gcrB, gcrC, cypD), and cytochrome oxidation (qoxB, cydA, ctaB, ctaD, cydA) were identified, indicating a Bacillus-specific electron transport chain. It is thus tempting to speculate that, functionally, Bacilli are aerobic Clostridia, having acquired the capability of oxidative phosphorylation. Furthermore, a subset of the propionate metabolism pathways identified to be Bacillus-specific. This subset is involved in the conversion of propanoyl-CoA to succinate and succinyl-CoA (prpD, prpB, pccB, sucD, sucC) and might present an additional energy conversion option for Bacilli which is absent in Clostridia.
Only a few ORFs were identified to be Clostridium cluster specific. Among those a cobyric acid synthase cobQ was identified to be Clostridium specific. CobQ is part of the porphyrin metabolic pathway, involved in converting cobyrinic acid into coenzyme vitamin B12. Notably, a central branching point in this pathway leads to the synthesis of hemes and cytochromes found in Bacilli (see above). Interestingly, CobQ is also absent in the Ruminococcus subcluster, providing further support for the proposed ongoing adaptation to the new rumen environment. In contrast, the Ruminococcus subcluster acquired a number of membrane and sugar utilization (e.g., beta-glucosidases bgl3A, bgl3B, and bgl3D and an L-fucose isomerase) which may aid in the adhesion to and degradation of plant fibers in the rumen (threshold conserved: 1e-60; threshold unique: 1e-10; Kelly et al., 2010).
While the function of these genes has been well described in the past, they deliver the proof-of-concept that FGD analyses are able to identify gene sets involved in lifestyle adaptation processes. Because the initial similarity analysis does not rely on existing gene annotation, uncharacterized ORFs (e.g., genes annotated as “conserved hypothetical”) can be identified as potential targets to contributing to respective phenotypes. This is particular important for poorly annotated microbial genomes with a high level of conserved hypothetical ORFs.
In summary, results obtained from the test dataset provide strong support for the usefulness of FGD analyses, by illustrating the ability of the method to draw together groups into common nodes based on shared core (shared by all genomes in a specific cluster) and lifestyle elements, yet distinguishing them into distinct sub-clusters based on relevant genotypic differences and lifestyle adaptation processes. Importantly, FGD subsequently allows identifying gene sets likely to be responsible for the observed clustering, providing meaningful new target selections for functional genomics analyses independent of other means of classification or prior annotation.
Furthermore, distinct placements in genome clustering of Leuconostoc mesenteroides and Oenococcus oeni (Ze-Ze et al., 2000; Makarova et al., 2006; Leuconostocaceae) were observed in the FDT. Both genera can be found epiphytically on fruits, fruit mashes, and vegetables and are used in industrial and food fermentation processes. Interestingly, Functional Distribution placed L. casei and L. plantarum into a separate cluster, branching deeply within the Bacilli (Figure A1 in Appendix). Both genomes are significantly larger than the average Lactobacillus genome of 1.8–2.0 Mbp with 2.8 and 3.3 Mbp, respectively. The significantly increased genome size likely reflects a more generalized lifestyle, capable of thriving in a variety of habitats such as raw and fermented dairy products, plants, and the intestinal and reproductive tracts of animals and humans. In contrast, the smaller genomes of other lactobacilli often reflect the more specialized lifestyle to one habitat such as the human or animal gastrointestinal system or specific fermentation processes.
These results may indicate that lifestyle adaptation can lead to a similar genetic makeup of taxa defined as being distinctively different by heritage based phylogeny.
Limits of the FGD Resolution
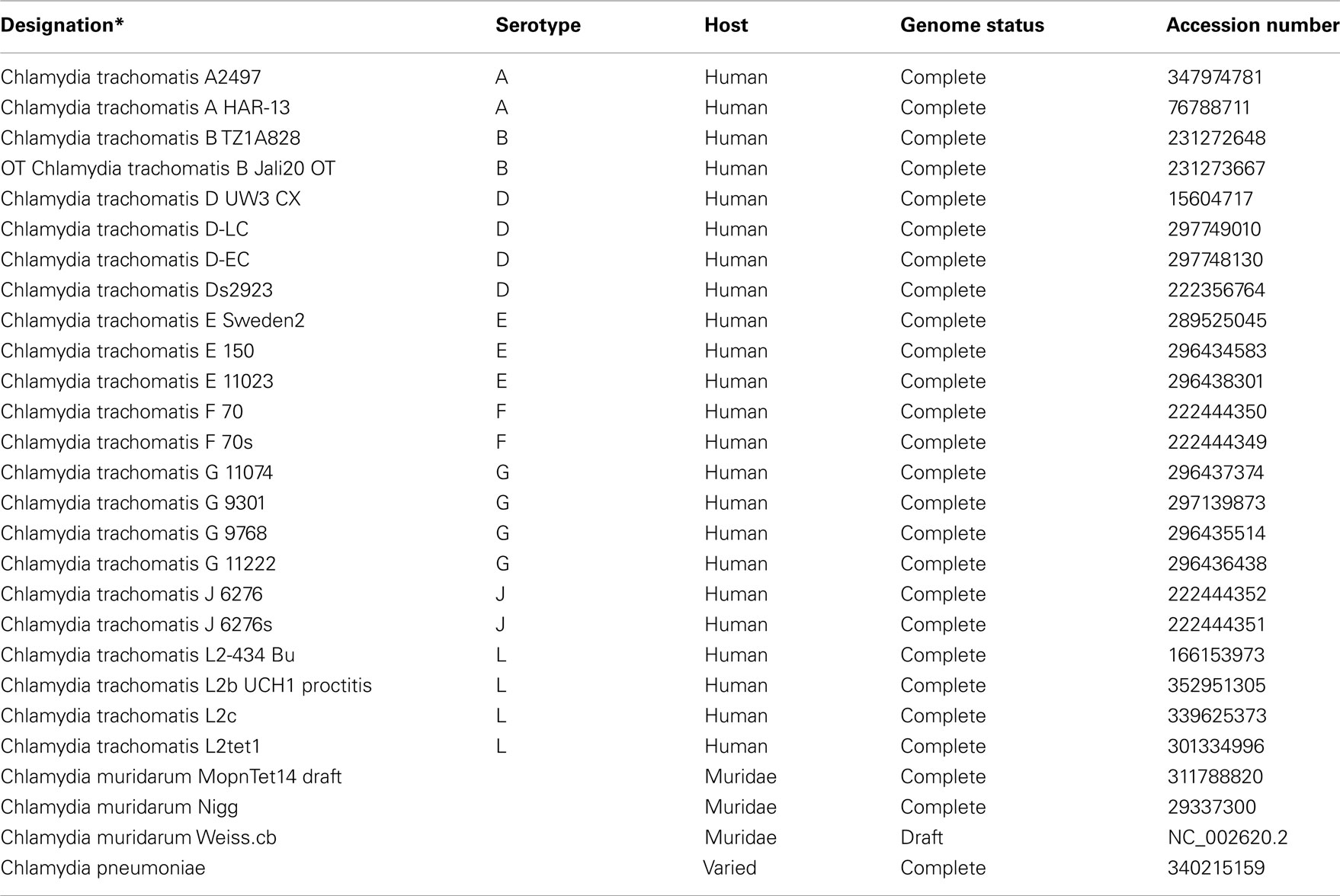
In the previous example microbial genomes from a wider range of species were investigated. To determine if the algorithm can discriminate strains from the same species, 23 Chlamydia trachomatis genomes (host: human), three Chlamydia muridarum genomes (host: members of the family Muridae), and one Chlamydia pneumoniae genome (host: varied; see Table a3 in Appendix) were subjected to an FGD analysis. It is interesting to note that until 1999 C. muridarum, infecting only members of the family Muridae, was designated as C. trachomatis (Everett et al., 1999). C. pneumoniae, which can infect a wide variety of different hosts and causes atypical pneumonia, clusters away from both C. trachomatis and C. muridarum genomes, indicating a different – and possible more flexible – genome makeup (Figure A2 in Appendix). In contrast to the other Chlamydia genomes, C. pneumoniae harbors an additional ∼200 kb of genetic information, and a more detailed analysis will be necessary to determine whether gene loss or gene acquisition is the major driving force. Similarly, the C. muridarum group is clearly forming its own cluster, albeit indicating a higher-level of similarity to C. trachomatis than to C. pneumoniae. On this high level, FGD can clearly resolve genomic differences and support observed host specificities (varied hosts – Muridae – human). Within the C. trachomatis cluster, three distinct sub-clusters could be identified with little reshuffling observed. Cluster 1 comprises serotypes E, F, G, and J (and C. trachomatis Ds2923), Cluster 2 harbors serotypes A, B, D, and L (and C. trachomatis E Sweden2), and Cluster 3 groups two serotype L and one serotype A (Figure A2 in Appendix). The observed positioning of the serovar D strain C. trachomatis Ds2923 into Cluster 1 supports the pairwise alignment of several chlamydial isolates (Jeffrey et al., 2010) which identified the least number of nucleotide substitutions between Ds/2926 and E/11023. In contrast, different major groups were identified in this nucleotide based analysis; two major clades (D/G/J and E/F; Jeffrey et al., 2010) are contrasted by three clusters (E/F/G/J, A/B/D/L, and L/A).
Overall, the FGD analysis was able to resolve strains from the same species to a similar level and with similar results as other whole genome comparative approaches. However, one of the limitations seen in the analysis of very similar genomes from the same species was the difficulty in identifying cluster specific gene sets based on e-value using FGDfinder [only three hypothetical ORFs were found to be cluster specific to Cluster 3 (see Figure A2 in Appendix, threshold conserved: 1e-20; threshold unique: 1e-10)]. In its current version FGDfinder uses calculated e-values to determine respective conserved or cluster specific gene sets. A future version of the software will incorporate the FGD scoring algorithm, increasing the power of resolution when very similar ORFeome sets are compared.
Topographical Stability of Functional Distribution Trees
The overall stability of inferred trees and respective genome clusters was tested by a Jackknife analysis (James and McCulloch, 1990). Individual observations (genome entries) from the calculated distance matrix were removed sequentially and resulting UPGMA-based FDTs were approximated. This was done for each genome in the dataset, resulting in 39 subset trees. Respective topologies were assessed individually. To investigate the impact of whole genome clusters on tree robustness, a Jackknife analysis was performed defining observed genome clusters as individual observations. Again, FDTs were approximated and evaluated for each resulting subset (data not shown). In summary, tree topology remained stable and only a swapping of neighboring branches was observed while in no instance shuffling was found for individual genome entries between genome clusters in either Jackknife analysis. This strongly supports the overall stability and discriminatory power of the FGD analysis.
Similarly, a Jackknife analysis was performed for the narrow strain FGD analysis (Figure A2 in Appendix). Individual entries, individual clusters, and complete serotypes were removed and the resulting tree topology investigated. Removal of individual entries and complete clusters did not change the topology of the FGD tree. Only minor reshuffling was observed within a respective cluster when complete serotypes were removed from the analyses (e.g., removal of serotype L caused a repositioning of C. trachomatis D UW3 CX into the serotype D subcluster within Cluster 2.
As expected, the removal of individual entries reduces discriminatory power, resulting in grouping together previously more separated genome clusters, without any entry-reshuffling.
Comparative Analysis Using the Artemis Comparison Tool
Comparison of the degree of genome similarity between two or more genomes relies on analyses of presence/absence of genes and their respective syntenies in an operon or genome-wide context. In combination with ORFeome based (ORF-to-ORF comparison on amino-acid level, a maximum of 20 similarity hits per ORF is permitted) MSPcrunch (Sonnhammer and Durbin, 1994) comparison format data files (pMSPs) created by the compACTor software and annotated GenBank files, the ACT (Carver et al., 2005) provides an excellent visualization platform for mobility through the entire genome. Figure 3 illustrates the differences and similarities found between closely related strains such as between members of the acidophilus-complex of the Lactobacilli or between subspecies as well as between less similar genomes of more distantly related microbes. For example, small scale [inversion of a specific gene locus between both L. delbrueckii ssp. bulgaricus genomes (Makarova et al., 2006; van de Guchte et al., 2006)] and large scale (double inversion ∼700 and ∼150 kb, respectively) of the terminus of DNA replication between L. gasseri (Azcarate-Peril et al., 2008) and L. johnsonii (Pridmore et al., 2004) genome inversions, deletion, and insertion events [between L. acidophilus (Altermann et al., 2005) and L. delbrueckii ssp. bulgaricus], localized gene synteny (between L. delbrueckii ssp. bulgaricus and L. brevis), general presence/absence of individual genes and larger synteny trends (between L. casei and L. plantarum) can be immediately identified.
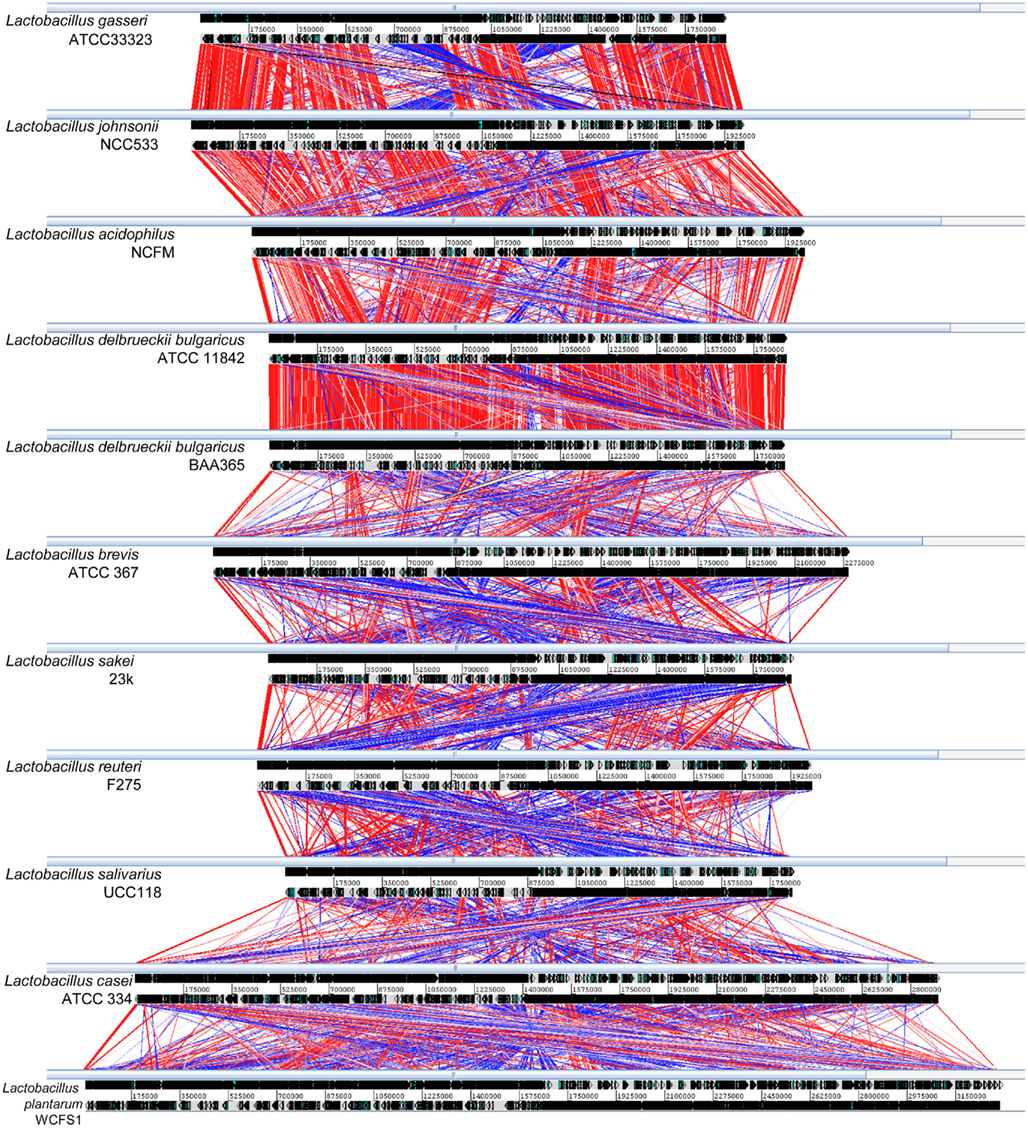
Figure 3. ORFeome based comparative ACT visualization of 11 Lactobacillus genomes. Based on the distribution observed in Figure 2, 11 Lactobacillus genomes and their ORFeome similarities were visualized in ACT using pMSP-datafiles. Respective genome designations are indicated on the left hand side of each genome line. Genomes are shown in full and drawn to scale. Genomic nucleotide sequences are represented by gray lines indicating sense and anti-sense strands and position markers are shown in between. Predicted ORFs are shown on each strand in their respective orientation as arrowed boxes. Direct amino-acid similarity between individual ORFs of neighboring genomes are shown as red lines, inverted similarities are indicated by blue lines. Color shadings indicate the level of similarity, the more saturated a similarity line the more conserved are two ORF-pairs. A trust level value of 40 was employed as display threshold to visualize similarity hits below an e-value of 1e-60.
Comparison of the FGD Algorithm to Alternative Methods
Although similar algorithms exist to investigate phylogenetic relationships based on whole (or partial) genomes sequences (Snel et al., 1999; Wolf et al., 2001; Henz et al., 2005; Khiripet, 2005; Canchaya et al., 2006; Fuchsman and Rocap, 2006; Berger et al., 2007; Felis and Dellaglio, 2007; Blaiotta et al., 2008), their focus remains mostly to infer a heritage based phylogeny. Furthermore, often only subsets of ORFeomes are chosen for these analyses. These are then analyzed individually (with or without weighting) or as concatenated sequences. Thus an artificial restriction is introduced that may bias the analysis. An example of such a method has been published by Konstantinidis and Tiedje (2005). There, genome information to infer taxonomy of prokaryotes is used, calculating an average amino-acid identity of shared gene subsets. Only few publications investigate the functional relationship of microbial genomes, such as the development of the Blast Score Ratio which analyzes the complete ORFeome but is limited to three genomes at a time (Rasko et al., 2005). Other methods investigating the functional relationship and similarities between gene clusters have been used to address the problem of genomes with different sizes. One example of such a method, GRAST, explores the ongoing genome reductions and rearrangements by identifying clusters of functionally related genes (Toft and Fares, 2006). Subsets of orthologous gene pairs are identified to determine conserved genetic loci. Similar to Blast Score Ratio analyses, the number of input genomes is limited to two at a time. While the output in part shows similarity to FGD analyses (visual representation of a genome plot and the determination of common and non-common genes), the purpose of this method is distinctly different in specifically identifying genome plasticity trends. A combination of genome analysis and visualization tool, GeneComp, has been published earlier (Yang et al., 2003). GeneComp is able to use different BLAST flavors and then visualize the textual output with varying levels of alignment length stringencies. While this solution offers the advantage of providing a combined analysis and visualization package, a number of limitations exist when compared to FGD. Like BSR and GRAST, GeneComp is restricted to a maximum number of three genomes. Furthermore, the algorithm is sequence based, highlighting genome variations such as repeat regions, insertions, deletions, and rearrangements rather than specific similarities to predicted ORFs.
Non-sequence based methods such as MLST use only a relatively small number of conserved genome loci with the primary aim to establish a highly discriminating (microbial) typing system (Chan et al., 2001; Maiden, 2006; Diancourt et al., 2007). Noteworthy, the ability of FGD analyses to identify cluster-conserved gene sets may provide a high-quality starting point for the selection of MLST targets.
Conclusion
16S rRNA and other gene subset analyses mainly focus on the determination of the line of descendants of a given gene or organism (Zhang et al., 2009) or on the identification of protein families (Enright and Ouzounis, 2000; Enright et al., 2002; Kelil et al., 2007). Such phylogenetic studies aim to reconstruct the relationship between organisms and are paramount to analyze the (changing) community structures of complex biological ecosystems. While this type of phylogenetic analysis is well accepted and widely used, it does not reflect the respective comprehensive genotypes. In contrast, FGD provides a different view of microbial similarities to each other. The example data set demonstrated the effects of lifestyle adaptation on genome content. FGD has shown the potential to provide new insights into the relationships between microbes from a comparative genomics perspective. The algorithm has already been used in a variety of analyses ranging from microbial (Goh et al., 2011) and archeal (Leahy et al., 2010) genomes to bacteriophage (Lu et al., 2010) which describe the impact of FGD analyses in more detail within their respective scopes.
Functional genome distribution in combination with the graphical visualization in ACT using ORFeome distance files (pMSPs) and functionally annotated GenBank files offers a powerful tool for comparative genomics that allows comparisons of whole genomes within genome space, encompassing heritage based (vertical transmission), lateral gene transfer (HGT), and lifestyle-driven change (adaptation) in a single analysis. Therefore, rather than attempting to reconstruct the evolution of the core genome with its set of commonly shared genes, FGD allows a representation of whole genome similarity at the functional level.
It will be possible to add further functionality to the algorithm, such as the ability to mask defined or dynamically created gene clusters within groups of organisms, thus identifying potentially important genetic elements independent from otherwise overpowering gene sets, such as central house-keeping or metabolism genes.
Availability
The complete software suite consisting of the compACTor and the FGDfinder software is freely available upon email request for academic use. Commercial use is subject to a license agreement.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Special thanks go to Peter Janssen for helpful and critical discussions during the development of the FGD algorithm, the subsequent interpretation of the data and the critical review of the manuscript. I also thank Sinead Leahy, Graeme Attwood, and Bill Kelly for their scientific contributions to this manuscript. This work was funded by the New Economic Research Fund programs C10 × 0314 and PROJ-13812-LNNR-AGR and supported by the Pastoral Greenhouse Gas Research Consortium (PGgRc).
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Evolutionary_and_Genomic_Microbiology/10.3389/fmicb.2012.00048/abstract
Footnotes
References
Altermann, E., and Klaenhammer, T. R. (2003). GAMOLA: a new local solution for sequence annotation and analyzing draft and finished prokaryotic genomes. OMICS 7, 161–169.
Altermann, E., Russell, W. M., Azcarate-Peril, M. A., Barrangou, R., Buck, B. L., Mcauliffe, O., Souther, N., Dobson, A., Duong, T., Callanan, M., Lick, S., Hamrick, A., Cano, R., and Klaenhammer, T. R. (2005). Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. U.S.A. 102, 3906–3912.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.
Azcarate-Peril, M. A., Altermann, E., Goh, Y. J., Tallon, R., Sanozky-Dawes, R. B., Pfeiler, E. A., O’flaherty, S., Buck, B. L., Dobson, A., Duong, T., Miller, M. J., Barrangou, R., and Klaenhammer, T. R. (2008). Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism. Appl. Environ. Microbiol. 74, 4610–4625.
Berger, B., Pridmore, R. D., Barretto, C., Delmas-Julien, F., Schreiber, K., Arigoni, F., and Brussow, H. (2007). Similarity and differences in the Lactobacillus acidophilus group identified by polyphasic analysis and comparative genomics. J. Bacteriol. 189, 1311–1321.
Blaiotta, G., Fusco, V., Ercolini, D., Aponte, M., Pepe, O., and Villani, F. (2008). Diversity of Lactobacillus strains based on partial HSP60 gene sequences and design of PCR-RFLP assays for species identification and differentiation. Appl. Environ. Microbiol. 74, 208–215.
Canchaya, C., Claesson, M. J., Fitzgerald, G. F., Van Sinderen, D., and O’toole, P. W. (2006). Diversity of the genus Lactobacillus revealed by comparative genomics of five species. Microbiology 152, 3185–3196.
Carver, T. J., Rutherford, K. M., Berriman, M., Rajandream, M. A., Barrell, B. G., and Parkhill, J. (2005). ACT: the Artemis comparison tool. Bioinformatics 21, 3422–3423.
Chan, M.-S., Maiden, M. C. J., and Spratt, B. G. (2001). Database-driven multi locus sequence typing (MLST) of bacterial pathogens. Bioinformatics 17, 1077–1083.
Deppenmeier, U., Johann, A., Hartsch, T., Merkl, R., Schmitz, R. A., Martinez-Arias, R., Henne, A., Wiezer, A., Baumer, S., Jacobi, C., Bruggemann, H., Lienard, T., Christmann, A., Bomeke, M., Steckel, S., Bhattacharyya, A., Lykidis, A., Overbeek, R., Klenk, H. P., Gunsalus, R. P., Fritz, H. J., and Gottschalk, G. (2002). The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4, 453–461.
Desiere, F., Mcshan, W. M., Van Sinderen, D., Ferretti, J. J., and Brussow, H. (2001). Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic Streptococci: evolutionary implications for prophage-host interactions. Virology 288, 325–341.
Diancourt, L., Passet, V., Chervaux, C., Garault, P., Smokvina, T., and Brisse, S. (2007). Multilocus sequence typing of Lactobacillus casei (L. paracasei) reveals a clonal population structure with low levels of homologous recombination. Appl. Environ. Microbiol. 73, 6601–6611.
Enright, A. J., and Ouzounis, C. A. (2000). GeneRAGE: a robust algorithm for sequence clustering and domain detection. Bioinformatics 16, 451–457.
Enright, A. J., Van Dongen, S., and Ouzounis, C. A. (2002). An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584.
Everett, K. D., Bush, R. M., and Andersen, A. A. (1999). Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49(Pt 2), 415–440.
Felis, G. E., and Dellaglio, F. (2007). Taxonomy of Lactobacilli and Bifidobacteria. Curr. Issues Intest. Microbiol. 8, 44–61.
Fuchsman, C. A., and Rocap, G. (2006). Whole-genome reciprocal BLAST analysis reveals that planctomycetes do not share an unusually large number of genes with eukarya and archaea. Appl. Environ. Microbiol. 72, 6841–6844.
Goh, Y., Goin, C., O’flaherty, S., Altermann, E., and Hutkins, R. (2011). Specialized adaptation of a lactic acid bacterium to the milk environment: the comparative genomics of Streptococcus thermophilus LMD-9. Microb. Cell Fact. 10, S22.
Henz, S. R., Huson, D. H., Auch, A. F., Nieselt-Struwe, K., and Schuster, S. C. (2005). Whole-genome prokaryotic phylogeny. Bioinformatics 21, 2329–2335.
James, F. C., and McCulloch, C. E. (1990). Multivariate analysis in ecology and systematics: panacea or Pandora’s box? Annu. Rev. Ecol. Syst. 21, 129–166.
Jeffrey, B. M., Suchland, R. J., Quinn, K. L., Davidson, J. R., Stamm, W. E., and Rockey, D. D. (2010). Genome sequencing of recent clinical Chlamydia trachomatis strains identifies loci associated with tissue tropism and regions of apparent recombination. Infect. Immun. 78, 2544–2553.
Kanehisa, M. (2002). The KEGG database. Novartis Found. Symp. 247, 91–101; discussion 101–103, 119–128, 244–152.
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., Katayama, T., Kawashima, S., Okuda, S., Tokimatsu, T., and Yamanishi, Y. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484.
Kelil, A., Wang, S., Brzezinski, R., and Fleury, A. (2007). CLUSS: clustering of protein sequences based on a new similarity measure. BMC Bioinformatics 8, 286.
Kelly, W. J., Leahy, S. C., Altermann, E., Yeoman, C. J., Dunne, J. C., Kong, Z., Pacheco, D. M., Li, D., Noel, S. J., Moon, C. D., Cookson, A. L., and Attwood, G. T. (2010). The glycobiome of the rumen bacterium Butyrivibrio proteoclasticus B316(T) highlights adaptation to a polysaccharide-rich environment. PLoS ONE 5, e11942.
Khiripet, N. (2005). “Bacterial whole genome phylogeny using proteome comparison and optimal reversal distance,” in Computational Systems Bioinformatics Conference, Stanford.
Konstantinidis, K. T., and Tiedje, J. M. (2005). Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187, 6258–6264.
Leahy, S. C., Kelly, W. J., Altermann, E., Ronimus, R. S., Yeoman, C. J., Pacheco, D. M., Li, D., Kong, Z., Mctavish, S., Sang, C., Lambie, S. C., Janssen, P. H., Dey, D., and Attwood, G. T. (2010). The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS ONE 5, e8926.
Lu, Z., Altermann, E., Breidt, F., and Kozyavkin, S. (2010). Sequence analysis of Leuconostoc mesenteroides bacteriophage {Phi}1-A4 isolated from an industrial vegetable fermentation. Appl. Environ. Microbiol. 76, 1955–1966.
Makarova, K., Slesarev, A., Wolf, Y., Sorokin, A., Mirkin, B., Koonin, E., Pavlov, A., Pavlova, N., Karamychev, V., Polouchine, N., Shakhova, V., Grigoriev, I., Lou, Y., Rohksar, D., Lucas, S., Huang, K., Goodstein, D. M., Hawkins, T., Plengvidhya, V., Welker, D., Hughes, J., Goh, Y., Benson, A., Baldwin, K., Lee, J. H., Diaz-Muniz, I., Dosti, B., Smeianov, V., Wechter, W., Barabote, R., Lorca, G., Altermann, E., Barrangou, R., Ganesan, B., Xie, Y., Rawsthorne, H., Tamir, D., Parker, C., Breidt, F., Broadbent, J., Hutkins, R., O’sullivan, D., Steele, J., Unlu, G., Saier, M., Klaenhammer, T., Richardson, P., Kozyavkin, S., Weimer, B., and Mills, D. (2006). Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 15611–15616.
Makarova, K. S., and Koonin, E. V. (2007). Evolutionary genomics of lactic acid bacteria. J. Bacteriol. 189, 1199–1208.
McCutcheon, J. P., and von Dohlen, C. D. (2011). An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr. Biol. 21, 1366–1372.
Nicolas, P., Bessieres, P., Ehrlich, S. D., Maguin, E., and Van De Guchte, M. (2007). Extensive horizontal transfer of core genome genes between two Lactobacillus species found in the gastrointestinal tract. BMC Evol. Biol. 7, 141.
Pridmore, R. D., Berger, B., Desiere, F., Vilanova, D., Barretto, C., Pittet, A. C., Zwahlen, M. C., Rouvet, M., Altermann, E., Barrangou, R., Mollet, B., Mercenier, A., Klaenhammer, T., Arigoni, F., and Schell, M. A. (2004). The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. U.S.A. 101, 2512–2517.
Rasko, D. A., Myers, G. S., and Ravel, J. (2005). Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinformatics 6, 2.
Rohwer, F., and Edwards, R. (2002). The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184, 4529–4535.
Schneiker, S., Perlova, O., Kaiser, O., Gerth, K., Alici, A., Altmeyer, M. O., Bartels, D., Bekel, T., Beyer, S., Bode, E., Bode, H. B., Bolten, C. J., Choudhuri, J. V., Doss, S., Elnakady, Y. A., Frank, B., Gaigalat, L., Goesmann, A., Groeger, C., Gross, F., Jelsbak, L., Kalinowski, J., Kegler, C., Knauber, T., Konietzny, S., Kopp, M., Krause, L., Krug, D., Linke, B., Mahmud, T., Martinez-Arias, R., Mchardy, A. C., Merai, M., Meyer, F., Mormann, S., Munoz-Dorado, J., Perez, J., Pradella, S., Rachid, S., Raddatz, G., Rosenau, F., Ruckert, C., Sasse, F., Scharfe, M., Schuster, S. C., Suen, G., Treuner-Lange, A., Velicer, G. J., Vorholter, F. J., Weissman, K. J., Welch, R. D., Wenzel, S. C., Whitworth, D. E., Wilhelm, S., Wittmann, C., Blocker, H., Puhler, A., and Muller, R. (2007). Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat. Biotechnol. 25, 1281–1289.
Schwiertz, A., Hold, G. L., Duncan, S. H., Gruhl, B., Collins, M. D., Lawson, P. A., Flint, H. J., and Blaut, M. (2002). Anaerostipes caccae gen. nov., sp. nov., a new saccharolytic, acetate-utilising, butyrate-producing bacterium from human faeces. Syst. Appl. Microbiol. 25, 46–51.
Snel, B., Bork, P., and Huynen, M. A. (1999). Genome phylogeny based on gene content. Nat. Genet. 21, 108–110.
Sonnhammer, E. L., and Durbin, R. (1994). A workbench for large-scale sequence homology analysis. Comput. Appl. Biosci. 10, 301–307.
Sousa, C., De Lorenzo, V., and Cebolla, A. (1997). Modulation of gene expression through chromosomal positioning in Escherichia coli. Microbiology 143(Pt 6), 2071–2078.
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0 . Mol. Biol. Evol. 24, 1596–1599.
Tatusov, R., Fedorova, N., Jackson, J., Jacobs, A., Kiryutin, B., Koonin, E., Krylov, D., Mazumder, R., Mekhedov, S., Nikolskaya, A., Rao, B. S., Smirnov, S., Sverdlov, A., Vasudevan, S., Wolf, Y., Yin, J., and Natale, D. (2003). The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4, 41.
Thomson, N., Bentley, S., Holden, M., and Parkhill, J. (2003). Fitting the niche by genomic adaptation. Nat. Rev. Microbiol. 1, 92–93.
Toft, C., and Fares, M. A. (2006). GRAST: a new way of genome reduction analysis using comparative genomics. Bioinformatics 22, 1551–1561.
van de Guchte, M., Penaud, S., Grimaldi, C., Barbe, V., Bryson, K., Nicolas, P., Robert, C., Oztas, S., Mangenot, S., Couloux, A., Loux, V., Dervyn, R., Bossy, R., Bolotin, A., Batto, J. M., Walunas, T., Gibrat, J. F., Bessieres, P., Weissenbach, J., Ehrlich, S. D., and Maguin, E. (2006). The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. U.S.A. 103, 9274–9279.
Wolf, Y., Rogozin, I., Grishin, N., Tatusov, R., and Koonin, E. (2001). Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol. Biol. 1, 8.
Yang, J., Wang, J., Yao, Z.-J., Jin, Q., Shen, Y., and Chen, R. (2003). GenomeComp: a visualization tool for microbial genome comparison. J. Microbiol. Methods 54, 423–426.
Ze-Ze, L., Tenreiro, R., and Paveia, H. (2000). The Oenococcus oeni genome: physical and genetic mapping of strain GM and comparison with the genome of a ‘divergent’ strain, PSU-1. Microbiology 146(Pt 12), 3195–3204.
Zhang, H., Zhong, Y., Hao, B., and Gu, X. (2009). A simple method for phylogenomic inference using the information of gene content of genomes. Gene 441, 163–168.
Appendix
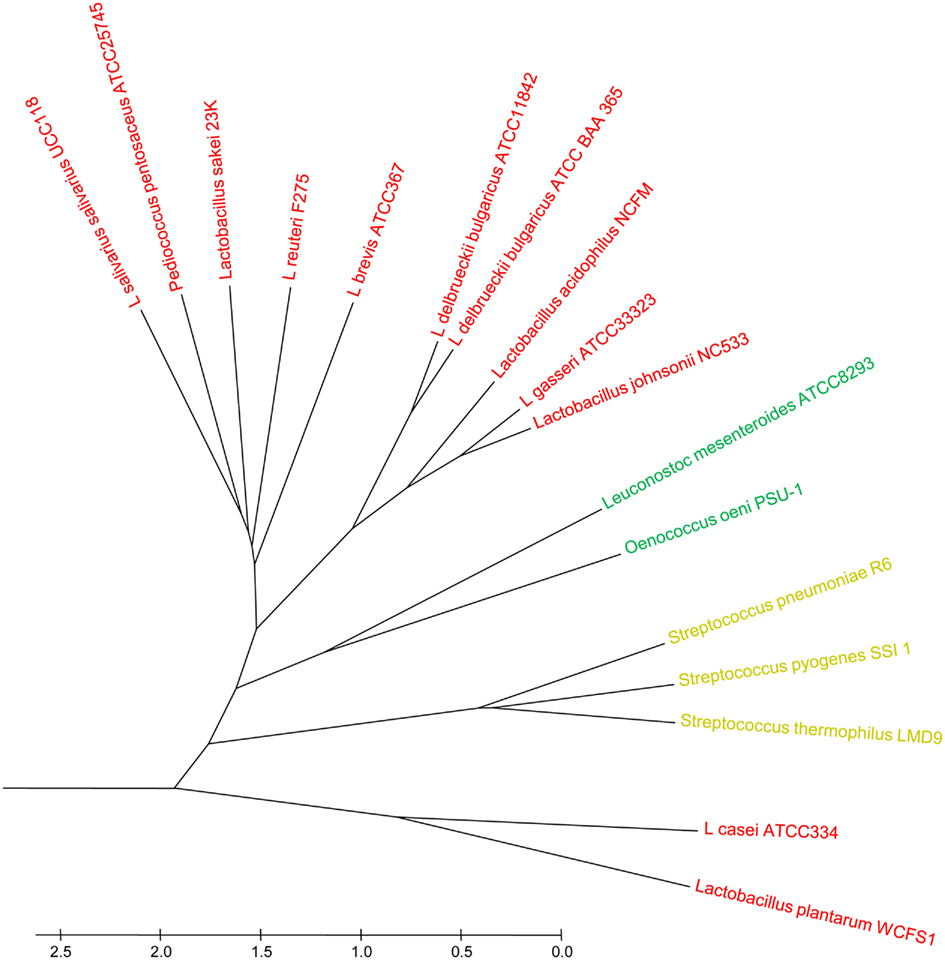
Figure A1. Functional distribution tree of 17 closely related taxa within the class Bacilli. The tree represents a subset of the one shown in Figure 2. Predicted ORFeomes of all genomes were subjected to an FGD analysis and the resulting distance matrix was imported into MEGA4. The functional distribution was visualized using the UPGMA method (32). The tree is drawn to scale, with branch lengths in the same units as those of the functional distances used to infer the distribution tree.
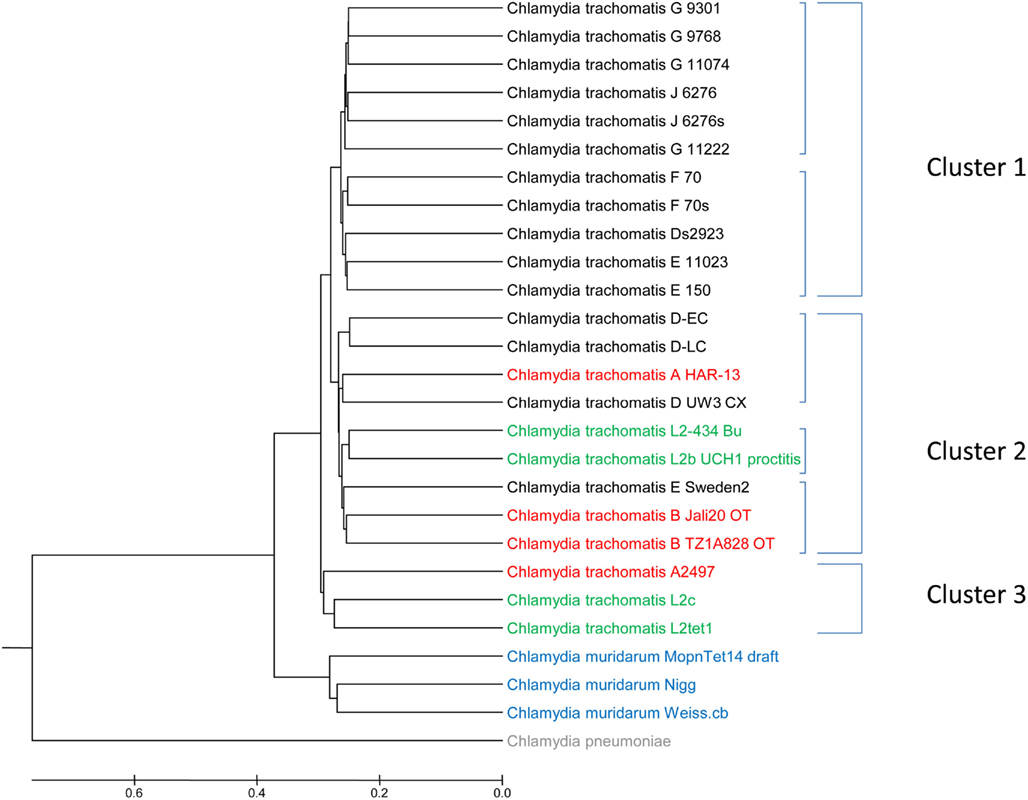
Figure A2. A functional distribution tree comprising of 23 Chlamydia trachomatis genomes (host: human), three C. maridarum genomes (host: members of the family Muridae), and one C. pneumoniae genome were used to investigate genome similarities and the FGD-power-of-resolution on strain level. Entries in red depict Chlamydia trachomatis serotypes A–C (trachoma), entries in black represent serotypes D–K (sexually transmitted pathovars) and entries in green show serotype LGV (L1–L3; lymphogranuloma venereum). Chlamydia muridarum entries are shown in blue and Chalmydia pneumoniae is depicted in gray. Functional clusters and subclusters are indicated by square brackets.
Keywords: functional genomics, genome comparison, lifestyle adaption, horizontal gene transfer, genome plasticity
Citation: Altermann E (2012) Tracing lifestyle adaptation in prokaryotic genomes. Front. Microbio. 3:48. doi: 10.3389/fmicb.2012.00048
Received: 08 November 2011; Accepted: 30 January 2012;
Published online: 21 February 2012.
Edited by:
Jacques Ravel, University of Maryland School of Medicine, USAReviewed by:
David Nelson, Indiana University Bloomington, USABing Ma, The Institute for Genome Sciences, USA
Copyright: © 2012 Altermann. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Eric Altermann, Grasslands Research Centre, AgResearch Limited, Tennent Drive, Private Bag 11008, Palmerston North, New Zealand. e-mail:ZXJpYy5hbHRlcm1hbm5AYWdyZXNlYXJjaC5jby5ueg==