- 1 Department of Biopathology and Clinical Microbiology, Medical School, Aeginition Hospital, University of Athens, Athens, Greece
- 2 Department of Statistics and Insurance Science, University of Piraeus, Piraeus, Greece
- 3 Faculty of Human Movement and Quality of Life Sciences, Department of Nursing, University of Peloponnese, Sparta, Greece
- 4 Department of Medical Microbiology, Medical School, University of Athens, Athens, Greece
- 5 Department of Biopathology, Medical School, Evgenidio Hospital, University of Athens, Athens, Greece
Background: Campylobacter spp. are together with Salmonella spp. the leading causes of human bacterial gastroenteritis worldwide. The most commonly isolated species in humans are Campylobacter jejuni and C. coli. The isolation, identification, and antimicrobial resistance of Campylobacter spp. from poultry and raw meat from slaughterhouses, has been investigated for the first time in Greece. During the period from August 2005 to November 2008 a total of 1080 samples were collected: (a) 830 fecal samples from five poultry farms, (b) 150 cecal samples from chicken carcasses in a slaughterhouse, and (c) 100 fecal samples from one pig farm near the region of Attica. The identification of the isolates was performed with conventional (sodium hippurate hydrolysis and commercial identification system (Api CAMPY system, bioMerieux, France), as well as with and molecular methods based on 16S rRNA species specific gene amplification by PCR and subsequent sequence analysis of the PCR products. Results: Sixteen Campylobacter strains were isolated, all collected from the poultry farms. None of the strains was identified as C. jejuni. Antimicrobial susceptibility to six antimicrobials was performed and all the strains were susceptible to ciprofloxacin, amoxicillin–clavulanic acid, and gentamicin. Thirteen out of 14 C. coli were resistant to erythromycin and all C. coli strains were resistant to ampicillin. Conclusion: Our results emphasize the need for a surveillance and monitoring system with respect to the prevalence and antimicrobial resistance of Campylobacter in poultry, as well as for the use of antimicrobials in veterinary medicine in Greece.
Introduction
Campylobacter spp. are together with Salmonella spp. the leading causes of human bacterial gastroenteritis worldwide. The most commonly isolated species in humans are Campylobacter jejuni, C. coli, and C. lari, but other species like C. fetus, seem to be also involved in human disease (Lastovica and Skirrow, 2000). Sporadic human cases have been associated with consumption of undercooked poultry meat, while larger outbreaks are associated with raw milk (Karagiannis et al., 2010; Denis et al., 2011). Contaminated drinking water has been the cause of sporadic cases, as well as of larger outbreaks (Peterson, 2003). Antimicrobial resistance of Campylobacter spp. to fluoroquinolones, which are generally used for the empiric treatment of bacterial gastroenteritis, has increased during the past two decades, mainly as a result of the approval of this group of antimicrobials for the use in food producing animals (Nelson and Harris, 2006; Han et al., 2009). Macrolide-resistant Campylobacter spp. isolated from food, animals, and humans have also been reported (Kang et al., 2000; Belanger and Shryock, 2007; Gallay et al., 2007). In Greece epidemiological data about the prevalence and antimicrobial susceptibility of Campylobacter spp. are restricted to strains from clinical samples belonging mainly to C. jejuni, isolated from children with gastroenteritis (Kafetzis et al., 2001; Chatzipanagiotou et al., 2002, 2003a,b; Maraki et al., 2003; Ioannidis et al., 2006; Papavasileiou et al., 2007).
There is, neither an official surveillance and monitoring system for the presence of Campylobacter in animals, nor for the use of antimicrobials in veterinary medicine. The aim of this study was (1) to evaluate the prevalence of Campylobacter spp. in positive samples in poultry meat, farms, and slaughterhouses; (2) to report for the first time the isolation, identification, and antimicrobial resistance of Campylobacter spp. from poultry in Greece; and (3) to investigate any possible epidemiological association with previous reports referring to isolates from clinical cases.
Materials and Methods
Sample Collection
In the period from August 2005 to November 2008 a total of 1080 samples were collected from poultry farms, pig farms, and slaughterhouses as follows: (a) 830 fecal samples from five poultry farms near the region of Attica, (b) 150 cecal samples from chicken carcasses in a slaughterhouse, and (c) 100 fecal samples from one pig farm near the region of Attica.
All samples were collected with a sterile swab from fresh feces immediately dropped to the ground. The sampling places covered geographically the location of Sterea Ellada, which is situated in Central Greece and includes the Department of Attica, the most populous Department in Greece, and also the Departments of Viotia, Evia, Evritania, Fthiotida, and Fokida. Thirteen percent of the total Greek meat production is concentrated in this location and five per cent in Attica.
At these locations, 61 poultry farms, 52 pig farms, and approximately 9 slaughterhouses are situated, most of them located in the Department of Attica and Viotia (40 poultry farms, 29 pig farms, and 4 slaughterhouses).
In this study, the method of two-stage cluster sampling was used. Under the well known cluster sampling scheme the population is divided in representative clusters, then a sample of these clusters is randomly chosen and all the units contained in the chosen clusters are deemed as the sample. The units within a cluster should ideally be as heterogeneous as possible, but there should be homogeneity between cluster means. Here we assume that there is homogeneity between all the farms as well as all the slaughterhouses since there are EU laws that impose a minimum level of neatness.
Nevertheless, in our case the cluster sampling is not adequate, since the units that are contained in each cluster are thousands, thus we choose the method of two-stage cluster sampling. There are many ways to modify cluster sampling for more complex sampling situations. However, the most common modification is to take a sample of secondary units from within sampled primary units (clusters) instead of inspecting every secondary unit within each primary unit (cluster).
Specifically, let us suppose that a simple random sample of n primary units is selected from a population of N primary units. Then a simple random sample of secondary units of sizes m1, m2, …, mn are selected from within these n primary units. The total sample of 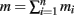 secondary units is called a two-stage cluster sample. In two-stage cluster sampling, we select mi units from the Mi secondary units (mi ≤ Mi) in primary unit i.
secondary units is called a two-stage cluster sample. In two-stage cluster sampling, we select mi units from the Mi secondary units (mi ≤ Mi) in primary unit i.
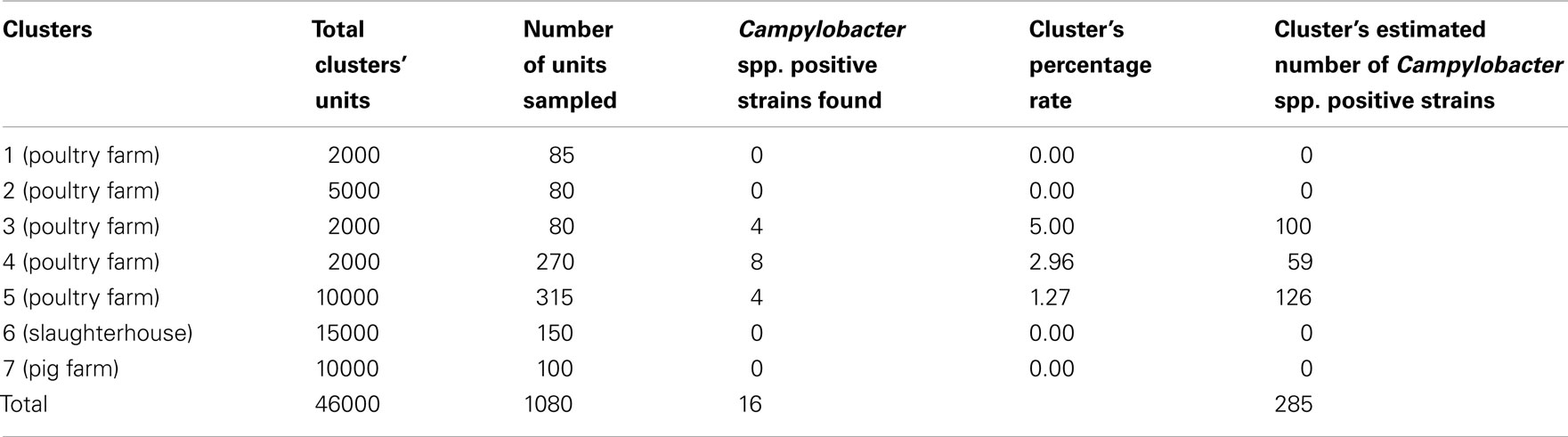
Specifically in our case, at the first stage, we selected seven clusters (each farm and slaughterhouse is deemed as a cluster), which represent about the 5.75% of the total number of the farms and slaughterhouses in the area under study (122 units in total population). The seven clusters (five poultry farms, one pig farm, and chicken carcasses) have a total animal population equal to 46000 animals (Table 1). At the second stage, from each one of the seven clusters, random samples were selected. The distribution of the samples taken from each cluster as well as the each cluster’s estimated number of Campylobacter spp. positive strains are given in Table 1 (an unbiased estimator of the unit total is 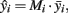 where
where 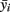 is cluster’s percentage rate).
is cluster’s percentage rate).
Culture, Isolation, and Identification of Bacteria
Campylobacter isolation and bacteriological identification
The swabs were immediately inoculated into 10 mL transport Campylobacter selective enrichment broth (Preston broth, Oxoid, CM0067 + SR0084 + SR117E, Basingstoke, UK) and refrigerated at 5°C for a maximum of 2 h after sampling and before dispatch to the laboratory.
The samples in Preston broth were incubated for 48 h at 42°C and were then subcultured onto Campylobacter selective Skirrow agar (manufacturer Oxoid, CM169 + SR0069, Basingstoke UK). The agar plates were further incubated at 42°C under microaerophilic conditions (GENbox microaer, bioMerieux, France).
All isolates were stored in brain heart infusion broth with 50% glycerol at −80°C for any further investigation.
The suspected colonies, showing typical morphology and positive oxidase and catalase reaction were microscopically examined after Gram-stain and typical isolates were primarily identified through sodium hippurate hydrolysis as well as by means of a commercial identification system (Api CAMPY system, bioMerieux, France).
Campylobacter sequence identification
All isolates were further identified by a molecular method based on 16S rRNA species specific gene amplification by PCR and subsequent sequence analysis of the PCR products.
DNA extraction and PCR. Bacterial DNA was extracted using the commercially available InstaGene matrix reagent (6% Polystyrene divinylbenzene iminodiacetate in Aqua bides; BioRad Laboratories, CA, USA). For the PCR reaction JumpStart REDTaq ReadyMix solution (Sigma Laboratories, St. Louis, USA) was used. The following oligonucleotide primer pairs were applied corresponding to three 16S rRNA gene fragments, run in separate reactions (Gorkiewicz et al., 2003): (1) Ps5/1 (5′-TATGGAGAGTTTGATCCTGG-3′) and Ps3/1 (5′ GTTAAGCTGTTAGATTTCAC-3′), (2) Ps5/2 (5′-AGCGTTACTCGGAATCACTG-3′) and Ps3/2 (5′-ACAGCCGTGCAGCACCTGTC-3′), (3) Ps5/3 (5′-AACCTTACCTGGGCTTGATA-3′) and Ps3/3 (5′-AAGGAGGTGATCCAGCCGCA-3′).
Additional oligonucleotide primers were applied to amplify the variable 16S rRNA regions (Vc regions) in order to facilitate detection of sequence variation: The primer pair Vc5/6-F (5′-AAAGCGTGGGGAGCAAACAG-3′) and Vc5/6-R (5′-ACTTAACCCAACATCTCACG-3′) was used for a 334-bp DNA fragment and the primer pair Vc1/2-F (5′-AGAGTTTGATCCTGGCTCAG-3′) andVc1/2R (5′-TGATCATCCTCTCAGACCAG-3′) was used to amplify a 330-bp DNA fragment (Gorkiewicz et al., 2003).
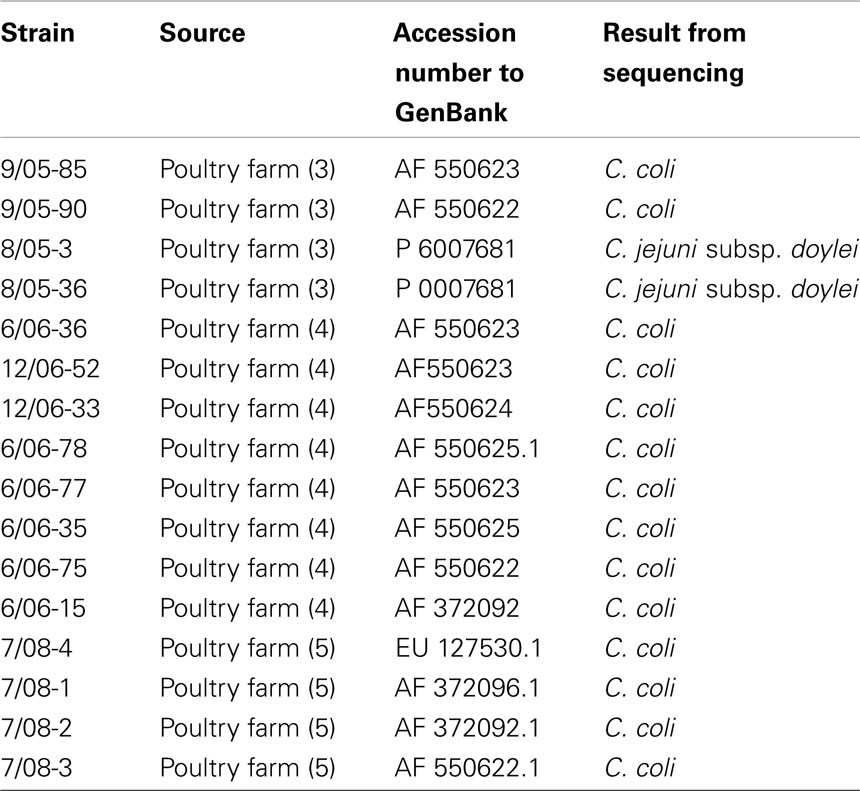
Campylobacter. 16S rRNA sequencing identification The 16S rRNA sequencing of the PCR products was performed by Macrogen Incorporation (908 World Meridian Center #60-24 Gasan-dong, Geumcheon-Gu Seoul, Republic of Korea) and the analyzed 16S rRNA gene fragments were submitted to the GenBank (using nucleotide BLAST) and compared with sequences already accessed from other studies.
For the quality assessment of the method the following reference strains were used: C. jejuni 0:37 NCO 12539, C. jejuni 0:38 NCO 12540, C. jejuni 0:41 NCO 12542, C. coli NCO 11366, C. lari NCO 11352.
Campylobacter Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed by using the standard E-test method (AB Biodisk, Sweden). The antibiotics included were nalidixic acid, ciprofloxacin, ampicillin, amoxicillin–clavulanic acid, gentamicin, and erythromycin. The bacterial inoculum was adjusted to 0.5 McFarland standard turbidity and Mueller–Hinton agar supplemented with 5% lysed sheep blood were used. Plates were incubated at 37°C for 48 h under microaerophilic conditions. Results were interpreted according to the Clinical Laboratory Standards Institute (CLSI) guidelines (CLSI document M45-A and M100-S17; Clinical and Laboratory Standards Institute, 2006, 2007) using the following resistance breakpoints of C. jejuni/coli: ampicillin >16 mg/L, ciprofloxacin >4 mg/L, gentamicin >16 mg/L, erythromycin >32 mg/L, nalidixic acid >32 mg/L, and amoxicillin–clavulanic acid >16 mg/L).
The following reference strains were used as controls: C. jejuni 0:37 NCO 12539, C. jejuni 0:38 NCO 12540, C. jejuni 0:41 NCO 12542, C. coli NCO 11366, C. lari NCO 11352.
Statistical Analysis
For the analysis the statistical package SPSS (version 18) was used. The chi-square test was the statistical method used (as a distribution goodness of fit test).
The chi-square test is used when we are interested as to how well a model actually reflects empirical data. Specifically, when we are interested in how “close” the observed values are to those which would be expected under a theoretical model. In our case, we used it in order to analyze multi-category percentages (e.g., test the null hypothesis that the percentage of Campylobacter strains isolated in poultry farms was equal to the percentages of Campylobacter strains isolated in farm pigs and chicken carcasses).
Details about the chi-square test (as a distribution goodness of fit test) are given in Armitage et al. (2002). Finally, the significance of the test was set equal to 0.05.
Results
Campylobacter Isolation
From the total of 1080 collected samples 16 Campylobacter strains were isolated: (a) 16 out of 830 fecal samples (1.93%) from the five poultry farms, (b) none out of the 150 cecal samples from chicken carcasses, and (c) none out of the 100 fecal samples from the pig farm (Table 2). According to the statistical analysis the percentage of Campylobacter strains isolated in poultry farms was significantly different (p-value <0.05%) from the percentage of Campylobacter strains isolated from the farm pigs and chicken carcasses.
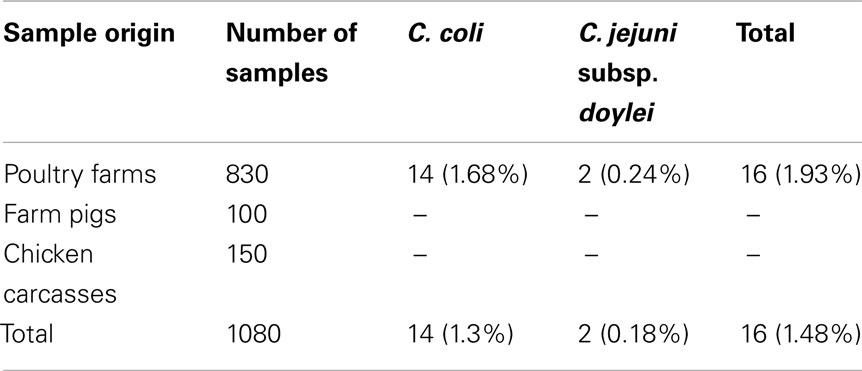
Table 2. 16S rRNA gene sequencing identification of Campylobacte r spp. isolated from poultry and other animal sources.
Campylobacter Bacteriological Identification
All isolated strains were negative for sodium hippurate hydrolysis. Through the commercial identification system (Api CAMPY) only 7 out of the 16 isolated strains were identified: four as C. coli, two as C. lari, and one as Arcobacter cryaerophilus.
Campylobacter Sequencing Identification
The 16S rRNA sequencing analysis identified all the strains as follows: 14 C. coli and 2 C. jejuni subsp. doylei (Table 2). The accession numbers of the nucleotide sequences admitted to GenBank are shown in Table 3. In addition, the statistical analysis revealed that the percentage of C. coli strains isolated in total was significantly different (p-value <0.05%) from the percentage of the isolated C. jejuni subsp. doylei.
Campylobacter Antimicrobial Susceptibility Testing
The results for antimicrobial susceptibility in relation to species and origin are shown in Table 4. All the strains were susceptible to ciprofloxacin and amoxicillin–clavulanic acid, while 13 were resistant to erythromycin, 14 to ampicillin, 2 to nalidixic acid, and 2 to gentamicin.

Table 4. Antimicrobial resistance rates of Campylobacter spp. isolated from poultry and other animal sources in relation to species and sample origin.
Discussion
Our study is the first report ever published from Greece, referring to the isolation, identification, and antimicrobial resistance of Campylobacter spp. from poultry farms. To our knowledge there are only a few published data from Greece upon the infection rate of Campylobacter spp. in human sporadic cases. Kafetzis et al. (2001) isolated Campylobacter spp. in 9% of 294 stool samples of hospitalized children, Maraki et al. (2003) isolated Campylobacter spp. in 4.2% from 7090 human stool samples, and Gousia et al. (2011) reported the prevalence of Campylobacter spp. of raw and processed meat products from retail shops.
In the present study, the isolation rate of positive samples of Campylobacter spp. in the examined poultry farms and in the total of samples was 1.93 and 1.48% respectively, which is low compared to those reported from other countries. In 2009, a total of 24 countries reported data about the presence of Campylobacter in broiler flocks (Scientific Report of European Food Safety Authority, 2011) and the recorded rates of positive samples were variable, ranging from 0 to 100.0%. Low and moderate levels were only observed in Estonia, Finland, Sweden, and Norway and high levels (>50%) were observed in Austria, France, Romania, and Spain. In the present study, we did not isolate any strain of Campylobacter spp. from chicken carcasses and farm pigs and this might be due to good hygiene practices in the slaughterhouse we visited, including daily cleaning and disinfection of the equipment used, existence of sufficient air system ventilation, and use of appropriate clothes (Young et al., 2010). Likewise, we did not isolate any strain of C. jejuni and this could explain the fact that so far no outbreaks due to Campylobacter spp. have been notified in Greece, as all the hitherto reported cases are sporadic and community acquired. The lack of outbreaks can also be explained by the fact that according to the Greek alimentary customs, meat is always well-cooked.
As shown by the results in Table 1, there are 285 estimated positive samples in the 7 primary sampling units (40.71 in each unit on average). Thus, the estimated prevalence rate of Campylobacter spp. positive animals was 0.622%. An upper bound for prevalence rate, with 95% confidence, is approximately 1.226%, while a lower bound is zero. For estimating prevalence rate and confidence intervals, appropriate estimators were used, corresponding to the sampling scheme that was applied (Cochran, 1977; Kish, 1995). In the total of 122 poultry/pig farms and slaughterhouses (which are situated in the examined location of Greece), 4967.2 animals are expected to be found as positive for Campylobacter spp. in the total estimated population of 801715 animals. These figures were estimated using appropriate estimators corresponding to the multistage sampling scheme that was applied (combining and weighting the n independent unbiased estimates 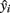 provides us with an unbiased estimate of the population total
provides us with an unbiased estimate of the population total 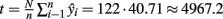 while the prevalence ratio is calculated by the dividing 4967.2 by the estimated total number of units in all the clusters which is 801715).
while the prevalence ratio is calculated by the dividing 4967.2 by the estimated total number of units in all the clusters which is 801715).
In the poultry samples, C. coli was prevailing with 14 out of 16 strains, a relatively high rate compared to other studies from European countries and the USA (Atanassova et al., 2007; Han et al., 2009). This predominance could be specific for the geographical region we visited and might be different in other Greece regions. Besides, the present survey was performed in a specific region near Athens and that could explain a probable clonality of the strains, which might be not representative of the general animal population. In our case another reason could be the protein-based broilers feed, which influences the digestive bacterial flora equilibrium of chickens. Previous reports showed that the ceca of birds that receive plant protein-based feed are less likely to be colonized with C. jejuni, than the ceca of birds that receive other types of feed (Udayamputhoor et al., 2003).
There was a remarkable discrepancy in the identification of Campylobacter strains between biotyping by the commercial Api CAMPY system and the molecular methods. In our study as well as in previous reports, the 16S rRNA gene sequencing was shown to be the most suitable test for the identification of the less common Campylobacter spp. (Al Amri et al., 2007; Caner et al., 2008).
Regarding the antimicrobial susceptibility, there was a remarkably high resistance rate to erythromycin and ampicillin of C. coli isolates from poultry farms (92.85 and 100% respectively). Unfortunately, there is no surveillance and monitoring system for the notification of antibiotic use in veterinary medicine and together with the possibility that antibiotics were administered to animals for growth, the possible use of erythromycin, and ampicillin in poultry can only be an assumption. The potential use of ampicillin and erythromycin in veterinary medicine or for animal growth could increase the prevalence of high-resistant C. coli strains in poultry farms. On the other hand, C. coli isolates are generally more resistant than C. jejuni strains (Gallay et al., 2007). According to a pan–European survey upon the antimicrobial susceptibility of Campylobacter spp. isolated from chickens (de Jong et al., 2009), 60.2% of the isolated C. coli isolates were resistant to ciprofloxacin, 61.4% to nalidixic acid, 12.0% to erythromycin, and 1.2% to gentamicin. Macrolide resistance is based on mechanisms including target modification by point mutation or methylation of 23S rRNA gene, hydrolysis of the drug, and efflux pumps (Belanger and Shryock, 2007). Since no transmission of erythromycin resistance genes through plasmid mechanism has been described so far, either among strains of the same Campylobacter species, or of different Campylobacter species, it seems that no clear association could be assumed, between the use of macrolides in veterinary medicine and the emergence of macrolide-resistant Campylobacter strains in humans.
None of the strains was resistant to ciprofloxacin and amoxicillin–clavulanic acid. Two C. coli isolates from the poultry farms were resistant to nalidixic acid and susceptible to ciprofloxacin, a phenotype which could be considered as a type 2 mutant being previously described (Reina et al., 1994; Ioannidis et al., 2009).
Conclusion
The isolation, identification, and antimicrobial resistance of Campylobacter spp. from poultry farms and slaughterhouses has been investigated and reported for the first time in Greece. The results showed a low prevalence of Campylobacter spp. in a geographical region around Athens, with the predominance of C. coli. There was a remarkably high resistance rate of C. coli isolates from poultry farms to ampicillin and erythromycin, indicating the possible use of erythromycin and ampicillin in poultry. Our results also emphasize the need for a surveillance and monitoring system for the prevalence and antimicrobial resistance of Campylobacter in poultry and other food animals.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by funds from the European Social Founds and National Resources (EPEAEK II, HRAKLEITOS).
References
Al Amri, A., Senok, A. C., Ismaeel, A. Y., Al-Mahmeed, A. E., and Botta, G. A. (2007). Multiplex PCR for direct identification of Campylobacter spp. in human and chicken stools. J. Med. Microbiol. 56(Pt 10), 1350–1355.
Armitage, P., Berry, G., and Matthews, J. N. S. (2002). Statistical Methods in Medical Research, 4th Edn. Oxford: Blackwell Science Ltd.
Atanassova, V., Reich, F., Beckmann, L., and Klein, G. (2007). Prevalence of Campylobacter spp. in turkey meat from a slaughterhouse and in turkey meat retail products. FEMS Immunol. Med. Microbiol. 49, 141–145.
Belanger, A. E., and Shryock, T. R. (2007). Macrolide-resistant Campylobacter: the meat of the matter. J. Antimicrob. Chemother. 60, 715–723.
Caner, V., Cokal, Y., Cetin, C., Sen, A., and Karagenc, N. (2008). The detection of hipO gene by real-time PCR in thermophilic Campylobacter spp. with very weak and negative reaction of hippurate hydrolysis. Antonie Van Leeuwenhoek 94, 527–532.
Chatzipanagiotou, S., Kilidireas, K., Trimis, G., Nicolaou, C., Anagnostouli, M., Athanassaki, C., Giannoulia, A., Legakis, N., and Youroukos, S. (2003a). Campylobacter jejuni O:19 serotype-associated Guillain-Barré syndrome in a child: the first case reported from Greece. Clin. Microbiol. Infect. 9, 69–72.
Chatzipanagiotou, S., Papavasileiou, E., Lakumenta, A., Makri, A., Nicolaou, C., Chantzis, K., Manganas, S., and Legakis, N. (2003b). Heat-stable antigen serotyping of Campylobacter jejuni strains isolated from hospitalized children in Athens, Greece. Eur. J. Epidemiol. 18, 1097–1100.
Chatzipanagiotou, S., Papavasileiou, E., Lakumenta, A., Makri, A., Nicolaou, C., Chantzis, K., Manganas, S., and Legakis, N. I. (2002). Antimicrobial susceptibility patterns of Campylobacter jejuni strains isolated from hospitalized children in Athens, Greece. J. Antimicrob. Chemother. 49, 803–805.
Clinical and Laboratory Standards Institute. (2006). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria: Approved Guideline. CLSI Document M45-A. Wayne: Clinical and Laboratory Standards Institute.
Clinical and Laboratory Standards Institute. (2007). Performance Standards for Antimicrobial Susceptibility Testing; Seventeenth International Supplement. Wayne: Clinical and Laboratory Standards Institute.
de Jong, A., Bywater, R., Butty, P., Deroover, E., Godinho, K., Klein, U., Marion, H., Simjee, S., Smets, K., Thomas, V., Vallé, M., and Wheadon, A. (2009). A pan-European survey of antimicrobial susceptibility towards human-use antimicrobial drugs among zoonotic and commensal enteric bacteria isolated from healthy food-producing animals. J. Antimicrob. Chemother. 63, 733–744.
Denis, M., Tanguy, M., Chidaine, B., Laisney, M. J., Mégraud, F., and Fravalo, P. (2011). Description and sources of contamination by Campylobacter spp. of river water destined for human consumption in Brittany, France. Pathol. Biol. 59, 256–263.
Gallay, A., Prouzet-Mauleon, V., Kempf, I., Lehours, P., Labadi, L., Camou, C., Denis, M., De Valk, H., Desenclos, J. C., and Megraud, F. (2007). Campylobacter antimicrobial drug resistance among humans, broiler chickens, and pigs, France. Emerging Infect. Dis. 13, 259–266.
Gorkiewicz, G., Feierl, G., Schober, C., Dieber, F., Köfer, J., Zechner, R., and Zechner, E. L. (2003). Species-specific identification of campylobacters by partial 16S rRNA gene sequencing. J. Clin. Microbiol. 41, 2537–2546.
Gousia, P., Economou, V., Sakkas, H., Leveidiotou, S., and Papadopoulou, C. (2011). Antimicrobial resistance of major foodborne pathogens from major meat products. Foodborne Pathog. Dis. 8, 27–38.
Han, F., Lestari, S. I., Pu, S., and Ge, B. (2009). Prevalence and antimicrobial resistance among Campylobacter spp. in Louisiana retail chickens after the enrofloxacin ban. Foodborne Pathog. Dis. 6, 163–171.
Ioannidis, A., Nicolaou, C., and Chatzipanagiotou, S. (2009). Correlation between flagellin A (flaA) genotypes and antimicrobial susceptibility patterns of Campylobacter jejuni strains isolated from children with gastroenteritis in Athens, Greece. Mol. Diagn. Ther. 13, 389–395.
Ioannidis, A., Nicolaou, C., Legakis, N. J., Ioannidou, V., Papavasileiou, E., Voyatzi, A., and Chatzipanagiotou, S. (2006). Genotyping of human Campylobacter jejuni isolates in Greece by pulsed-field gel electrophoresis. Mol. Diagn. Ther. 10, 391–396.
Kafetzis, D. A., Maltezou, H. C., Zafeiropoulou, A., Attilakos, A., Stavrinadis, C., and Foustoukou, M. (2001). Epidemiology, clinical course and impact on hospitalization costs of acute diarrhea among hospitalized children in Athens, Greece. Scand. J. Infect. Dis. 33, 681–685.
Kang, Y. S., Cho, Y. S., Yoon, S. K., Yu, M. A., Kim, C. M., Lee, J. O., and Pyun, Y. R. (2000). Prevalence and antimicrobial resistance of Campylobacter jejuni and Campylobacter coli isolated from raw chicken meat and human stools in Korea. J. Food Prot. 69, 2915–2923.
Karagiannis, I., Sideroglou, T., Gkolfinopoulou, K., Tsouri, A., Lampousaki, D., Velonakis, E. N., Scoulica, E. V., Mellou, K., Panagiotopoulos, T., and Bonovas, S. (2010). A waterborne Campylobacter jejuni outbreak on a Greek island. Epidemiol. Infect. 138, 1726–1734.
Lastovica, A., and Skirrow, M. (2000). “Clinical significance of Campylobacter and related species other than Campylobacter jejuni and C. coli,” in Campylobacter, 2nd Edn, Chap. 5, eds I. Nachamkin and M. J. Blaser (Washington, DC: ASM Press), 89–120.
Maraki, S., Georgiladakism, A., Tselentis, Y., and Samonis, G. (2003). A 5-year study of the bacterial pathogens associated with acute diarrhoea on the island of Crete, Greece, and their resistance to antibiotics. Eur. J. Epidemiol. 18, 85–90.
Nelson, W., and Harris, B. (2006). Flies, fingers, fomites, and food. Campylobacteriosis in New Zealand-food-associated rather than food-borne. N. Z. Med. J. 119, U2128.
Papavasileiou, E., Voyatzi, A., Papavasileiou, K., Makri, A., Andrianopoulou, I., and Chatzipanagiotou, S. (2007). Antimicrobial susceptibilities of Campylobacter jejuni isolates from hospitalized children in Athens, Greece, collected during 2004–2005. Eur. J. Epidemiol. 22, 77–78.
Peterson, M. C. (2003). Campylobacter jejuni enteritis associated with consumption of raw milk. J. Environ. Health 65, 20–21.
Reina, J., Ros, M. J., and Serra, A. (1994). Susceptibilities to 10 antimicrobial agents of 1,220 Campylobacter strains isolated from 1987 to 1993 from feces of pediatric patients. Antimicrob. Agents Chemother. 38, 2917–2920.
Scientific Report of European Food Safety Authority. (2011). The community summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in the European Union in 2009. EFSA J. 9, 2090.
Udayamputhoor, R. S., Hariharan, H., Van Lunen, T. A., Lewis, P. J., Heaney, S., Price, L., and Woodward, D. (2003). Effects of diet formulations containing proteins from different sources on intestinal colonization by Campylobacter jejuni in broiler chickens. Can. J. Vet. Res. 67, 204–212.
Keywords: Campylobacter, animals, sequencing identification, antimicrobial resistance
Citation: Marinou I, Bersimis S, Ioannidis A, Nicolaou C, Mitroussia-Ziouva A, Legakis NJ and Chatzipanagiotou S (2012) Identification and antimicrobial resistance of Campylobacter species isolated from animal sources. Front. Microbio. 3:58. doi: 10.3389/fmicb.2012.00058
Received: 31 October 2011;
Accepted: 03 February 2012;
Published online: 24 February 2012.
Edited by:
Danilo Ercolini, Università degli Studi di Napoli Federico II, ItalyReviewed by:
Sergio Enrique Pasteris, Universidad Nacional de Tucumán, ArgentinaAntonio Valero, University of Cordoba, Spain
Copyright: © 2012 Marinou, Bersimis, Ioannidis, Nicolaou, Mitroussia-Ziouva, Legakis and Chatzipanagiotou. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Stylianos Chatzipanagiotou, Laboratory of Biopathology and Clinical Microbiology, Athens Medical School, Aeginition Hospital, Vasilissis Sophias Avenue 72-74, 115 28 Athens, Greece. e-mail:Y2hhdGxvdWtAaG90bWFpbC5jb20=;c2NoYXR6aUBtZWQudW9hLmdy

 Sotiris Bersimis2
Sotiris Bersimis2