- 1 Center for Emerging Virus Research, Institute for Virus Research, Kyoto University, Sakyo-ku, Kyoto, Japan
- 2 Laboratory of Viral Pathogenesis, Institute for Virus Research, Kyoto University, Sakyo-ku, Kyoto, Japan
Extensive investigations have identified two cellular proteins in humans that potently inhibit HIV type 1 (HIV-1) replication and are widely accepted as “restriction factors.” APOBEC3G was identified as a restriction factor that diminishes HIV-1 replication by inducing G-to-A hypermutation in the viral genome, while BST2 has been identified as another restriction factor that impairs the release of nascent virions by tethering them on the surface of infected cells. To counter these restriction factors, HIV-1 has equipped itself with its own weapons: viral infectivity factor (Vif) degrades APOBEC3G, while viral protein U (Vpu) antagonizes BST2. These findings have allowed us to further our understanding of virus–host interaction, namely, the interplay between viral factors versus host restriction factors. In the first case, the interplay between APOBEC3G and Vif is clear: vif-deficient HIV-1 is incapable of replicating in APOBEC3G-expressing cells. This insight directly indicates that APOBEC3G is a bona fide restriction factor and has intrinsic immunity against HIV-1, and that Vif is a prerequisite for HIV-1 infection. In other words, the relationship between Vif and APOBEC3G has already “matured,” and Vif has highly evolved to overcome APOBEC3G. On the other hand, although BST2 drastically impairs the release of vpu-deficient HIV-1 virions, it is puzzling that vpu-deficient HIV-1 is still able to replicate in BST2-expressing cells. These insights imply that BST2-mediated anti-HIV-1 activity is vulnerable, and that Vpu is dispensable for HIV-1 infection. If so, why has Vpu acquired the counteracting potential against BST2? Was it necessary or important for HIV-1? Or is the relationship between Vpu and BST2 still “immature”? In this review, we particularly focus on the interplay between Vpu and BST2. We discuss the possibility that Vpu has evolved as a potent antagonist against BST2, and finally, propose a hypothesis that Vpu has evolved as a promoter of human-to-human HIV-1 transmission. Since the first report of acquired immunodeficiency syndrome patients in 1981, HIV-1 has spread explosively worldwide and is currently a pandemic. This review proposes a concept suggesting that the current HIV-1 pandemic may be partly attributed by Vpu.
Introduction
Primate lentiviruses have co-evolved and co-diversified with primate hosts including humans for a myriad of years. During their long history, it is conceivable that hosts have evolved the genes necessary to counteract lentivirus infections, whereas viruses have developed weapons to eliminate these host obstacles. The continuous “cat-and-mouse games” have driven the virus–host co-evolution.
During the last 100 years, human immunodeficiency virus (HIV; particularly HIV type 1, HIV-1), a highly evolved primate lentivirus, has emerged, and has infiltrated the human population (Worobey et al., 2008). HIV-1 was identified as the causative agent of acquired immunodeficiency syndrome (AIDS) in 1983–1984 (Barre-Sinoussi et al., 1983; Gallo et al., 1983; Kitchen et al., 1984), and now, HIV-1 infection is a worldwide pandemic. In 2009, UNAIDS (Joint United Nations Programme on HIV and AIDS) estimated that more than 33 million people worldwide are infected with HIV-1, out of which, more than 2.8 million people are newly infected and approximately 1.8 million people annually die of AIDS (http://www.unaids.org/globalreport/). Since the HIV-1 outbreak has been one of the most urgent issues for mankind, scientists have invested a great deal of effort to reveal the molecular mechanism of HIV-1 replication, which can provide clues to elucidate HIV-1 infection and AIDS.
In the past 10 years, two proteins capable of robustly counteracting HIV-1 infection and encoded in the human genome were identified: apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (APOBEC3G), and bone marrow stromal antigen 2 (BST2, also known as tetherin, CD317, and HM1.24). APOBEC3G and its homologs in humans (APOBEC3A, B, C, DE, F, and H; APOBEC3s) are cellular cytidine deaminases and certain APOBEC3s, particularly APOBEC3G and APOBEC3F, induce G-to-A hypermutation in HIV-1 proviral DNA resulting in diminished HIV-1 replication (Sheehy et al., 2002; Harris and Liddament, 2004). BST2 is a type II transmembrane glycoprotein and tethers nascent HIV-1 particles on the plasma membrane of infected cells resulting in the impairment of virus release (Neil et al., 2008; Van Damme et al., 2008). The identification of these two genes has provided us with a concept of “intrinsic immunity” or “restriction factors” which means that hosts possess innate congenital factors potently capable of restricting viral replication. In parallel, HIV-1 encodes genes that antagonize cellular restriction factors and have given rise to two proteins, also known as accessory proteins, viral infectivity factor (Vif), and viral protein U (Vpu) which counteract the actions of APOBEC3s and BST2, respectively. Taken together, the battle between HIV-1 and humans can be interpreted as the interplay between viral factors (Vif and Vpu) and host factors (APOBEC3s and BST2), which has been a hot topic in the field of HIV-1 virology.
Here, we first provide a brief introduction on the interplay between Vif and APOBEC3s as an excellent example of virus–host interaction. Then, by comparing the Vpu and BST2 relationship with the Vif–APOBEC3s interaction, we summarize and discuss the significance of their interplay from the views of (1) virology, (2) molecular biology, (3) evolutionary biology, and (4) epidemiology.
Virology
Vif Versus APOBEC3s
Certain cell lines (e.g., CEM-SS cells, 293T cells, and HeLa cells) are permissive to vif-deficient HIV-1 replication and are called “permissive cells,” whereas some cell lines (e.g., CEM cells, HUT78 cells, and H9 cells) are not. Two studies using heterokaryons of permissive cells and non-permissive cells suggested that non-permissive cells such as CEM cells express a factor which potently restricts HIV-1 replication and is counteracted by Vif (Madani and Kabat, 1998; Simon et al., 1998). Sheehy et al. (2002) identified APOBEC3G by a subtraction screening assay using CEM cells and CEM-SS cells. Subsequent investigations revealed that certain APOBEC3s including APOBEC3G are incorporated into released virions and mutate viral complementary DNA in newly infected cells, while Vif impedes the incorporation of certain APOBEC3s into progeny virions by degrading these proteins through the ubiquitin–proteasome pathway (Harris and Liddament, 2004; Izumi et al., 2008). Importantly, endogenous APOBEC3s are expressed in primary human CD4-positive T cells and monocyte-derived macrophages (MDMs), which are the primary targets for HIV-1 infection in vivo, and that vif-deficient HIV-1 is not able to replicate in in vitro cultures of these cells. More strikingly, vif-deficient HIV-1 is unable to infect and replicate in a SCID-hu mouse model (Aldrovandi and Zack, 1996) and a humanized mouse model (Sato et al., 2010). These reports indicate that endogenous APOBEC3s are potent restriction factors for HIV-1 infection in vitro and in vivo, and that Vif is absolutely essential for HIV-1 replication.
Vpu Versus BST2
Certain cell lines such as HEK293 cells and Cos-7 cells are able to produce nascent virions of vpu-deficient HIV-1, whereas HeLa cells are not. An experiment using heterokaryons of HeLa cells and Cos-7 cells suggested that HeLa cells exclusively express a factor which potently impairs virus production and is counteracted by Vpu (Varthakavi et al., 2003). Neil et al. (2008) identified BST2 by microarray analyses using HEK293 cells and HeLa cells. The authors revealed that BST2 robustly inhibits the release of budding virions and that Vpu antagonizes the anti-viral action of BST2, leading to the proposal that BST2 tethers nascent virions (Neil et al., 2008). In addition, Van Damme et al. (2008) revealed that BST2 expressed on the surface of HIV-1-producing cells is down-regulated by Vpu.
The ability of BST2 to restrict the release of nascent virions is not limited to HIV-1. BST2 can impair the release of various enveloped viruses belonging to Retroviridae [HIV type 2 (HIV-2), simian immunodeficiency viruses (SIVs), equine infectious anemia virus (EIAV), feline immunodeficiency virus (FIV), prototype foamy virus (PFV), Mason-Pfizer monkey virus (MPMV), human T-cell leukemia virus type 1 (HTLV-1), Rous sarcoma virus (RSV), and murine leukemia virus (MLV; Douglas et al., 2009; Jia et al., 2009; Jouvenet et al., 2009; Le Tortorec and Neil, 2009; Sauter et al., 2009; Zhang et al., 2009; Yang et al., 2010], Filoviridae [Ebola virus (EBOV) and Marburg virus; Jouvenet et al., 2009; Kaletsky et al., 2009; Sakuma et al., 2009], Arenaviridae (Lassa fever virus; Kaletsky et al., 2009; Sakuma et al., 2009), Herpesviridae (Kaposi’s sarcoma-associated herpesvirus, KSHV; Mansouri et al., 2009; Pardieu et al., 2010), Rhabdoviridae (vesicular stomatitis virus; Weidner et al., 2010), Orthomyxoviridae (influenza A virus; Watanabe et al., 2011), and Paramyxoviridae (Nipah virus; Radoshitzky et al., 2010). On the other hand, the suppression of virus release can be elicited by not only human BST2 but also its orthologs in several mammalians including chimpanzees (McNatt et al., 2009; Sauter et al., 2009), gorillas (Sauter et al., 2009; Lim et al., 2010), rhesus macaques (McNatt et al., 2009), cats (Dietrich et al., 2011; Fukuma et al., 2011), and mice (McNatt et al., 2009; Kobayashi et al., 2011). Since ectopically expressed human BST2 impairs HIV-1 release from cells derived from a broad range of species including potoroo and quail, human BST2 can function without any co-factors exclusively expressed in primates (Sato et al., 2009).
Like HIV-1 Vpu, certain viruses possess their own counterparts to antagonize BST2. For example, HIV-2 envelope glycoprotein (Env), which has been known as the enhancer of virus release (Bour and Strebel, 1996; Bour et al., 1996), counteracts human BST2 (Douglas et al., 2009; Jia et al., 2009; Le Tortorec and Neil, 2009; Hauser et al., 2010). On the other hand, certain SIVs from chimpanzees (SIVcpz), sooty mangabeys (SIVsmm), and gorillas (SIVgor) impair simian BST2 by their commonly shared accessory protein, negative factor (Nef; Douglas et al., 2009; Jia et al., 2009; Sauter et al., 2009; Zhang et al., 2009; Lim et al., 2010). EBOV glycoprotein (GP; Kaletsky et al., 2009) and KSHV K5/MIR2 protein (Mansouri et al., 2009; Pardieu et al., 2010) also possess the potential to counteract human BST2. Given these findings, it is reasonable to assume that the ability of BST2 to impair the release of a broad spectrum of viruses plays a crucial role in host immunity, and that various viruses have evolved a way to counteract the anti-viral action of BST2.
Despite the fact that endogenously and ectopically expressed BST2 proteins are prominent in restricting the release of a broad range of viruses, its capacity to control virus replication remains questionable. In the case of HIV-1, vpu-deficient virus is able to replicate in in vitro cultures of BST2-expressing cells such as primary human CD4-positive T cells (Schubert et al., 1995, 1996, 1999; Neil et al., 2007) and MDMs (Schubert et al., 1995, 1999; Theodore et al., 1996; Neil et al., 2007) and in vivo in a humanized mouse model (Sato et al., in press). Furthermore, SIVagm [an SIV from African green monkey (AGM)] does not encode any accessory factors to antagonize AGM BST2 (Lim and Emerman, 2009), yet naturally infects AGM. Moreover, although human BST2 can impair the release of influenza A virus and EBOV, it does not suppress the spread of these viruses in in vitro cell cultures (Radoshitzky et al., 2010; Watanabe et al., 2011). These findings imply a limitation in BST2’s anti-viral function.
Why is BST2 unable to “restrict” the replication of viruses despite its robust ability to impair the release of virions? One possible explanation is the effect of BST2 on cell-to-cell transmission of viruses. Viruses are usually propagated by at least two modes: cell-free virus-mediated transmission and cell-to-cell transmission (Sattentau, 2008; Martin and Sattentau, 2009). In fact, one study has clearly shown that cell-to-cell HIV-1 transmission overcomes BST2-mediated restriction (Jolly et al., 2010), although it remains controversial (Casartelli et al., 2010; Kuhl et al., 2010). In addition, it has been suggested that Vpu is not required for efficient cell-to-cell spread of HIV-1 in the culture of Jurkat cells (Gummuluru et al., 2000), which express endogenous BST2. Interestingly, feline BST2 can impair the release of FIV but enhances the spread of FIV (Dietrich et al., 2011). Moreover, the entry of human cytomegalovirus is promoted by human BST2 (Viswanathan et al., 2011). Therefore, it is conceivable that BST2 does not diminish the cell-to-cell spread of viruses even if the cell-free virus-mediated infection of these viruses is severely restricted.
Molecular Biology
BST2 directly interacts with various anti-BST2 proteins through distinct manners and is subsequently antagonized. For instance, BST2 and HIV-1 Vpu interact through their transmembrane domains (Dube et al., 2010b; Vigan and Neil, 2010; Kobayashi et al., 2011; Skasko et al., 2012). On the other hand, BST2 interacts with HIV-2 Env (Gupta et al., 2009) and EBOV GP (Kaletsky et al., 2009; Lopez et al., 2010) through an extracellular domain, whereas SIVs Nef interacts with the cytoplasmic domain of BST2 (Sauter et al., 2009; Lim et al., 2010). Since KSHV K5/MIR2 is an E3 ubiquitin ligase, this protein ubiquitinates the lysine residues in the cytoplasmic domain of BST2 (Mansouri et al., 2009; Pardieu et al., 2010). The molecular mechanisms for BST2 antagonization by these viral proteins are dependent on each protein, but most likely occur through (i) degradation by the ubiquitin–proteasome pathway and/or lysosome, (ii) sequestration from the functional site, or (iii) enhancement of down-regulation. However, these mechanisms remain controversial. For more details on this topic, please see the following review papers available elsewhere (Douglas et al., 2010; Dube et al., 2010a; Evans et al., 2010; Arias et al., 2011; Martin-Serrano and Neil, 2011).
Given the role of BST2 in restricting virus release, is BST2 physiologically tethering something other than enveloped viruses? Human BST2 was first cloned as one of the candidates for the surface molecules involved in pre-B cell development (Ishikawa et al., 1995). It is known that BST2 is localized in lipid rafts on the plasma membrane of the apical side and is internalized by clathrin (Kupzig et al., 2003; Rollason et al., 2007, 2009). BST2 provides a physical link between lipid rafts and the apical actin network, which is crucial for the maintenance of microvilli in polarized epithelial cells (Rollason et al., 2009). However, it is unlikely that BST2 plays a role as a natural “tethering” factor in our cells. If this is the case, it is important to understand what BST2 is doing in addition to its anti-viral function.
Human BST2 and murine Bst2 (the ortholog of human BST2 in mouse) are interferon-stimulated genes (ISGs) and their expression is augmented by stimulation with type I interferons (IFNs; Blasius et al., 2006; Neil et al., 2008; Miyagi et al., 2009). However, their expression patterns in cell lineages are different between humans and mice. Human BST2 is expressed in broad lineages of hematopoietic cells (Homann et al., 2011) and certain epithelial cell lines such as HeLa cells (Neil et al., 2008; Van Damme et al., 2008). On the other hand, murine Bst2 is exclusively expressed in plasmacytoid dendritic cells (pDCs), a potent producer of type I IFNs, in naïve mice, and therefore, is used as a specific marker of pDCs (Blasius et al., 2006). Human BST2 expressed on pDCs negatively regulates the production of type I IFNs and inflammatory cytokines by interacting with immunoglobulin-like transcript 7 (ILT7; Cao et al., 2009). Although murine Bst2 is expressed in murine pDCs, a direct ortholog of human ILT7 is absent in mice (Brown et al., 2004). Therefore, the function of BST2 as a negative regulator of type I IFN signaling may have evolved recently (Cao et al., 2009).
Evolutionary Biology
So, why has HIV-1 acquired the weapons to counteract the abilities of APOBEC3s and BST2 in its limited genome space (approximately 10 kb)? From an evolutionary point of view, here we shed light on the interplay of cellular factors and viral factors, illustrated in Figure 1.
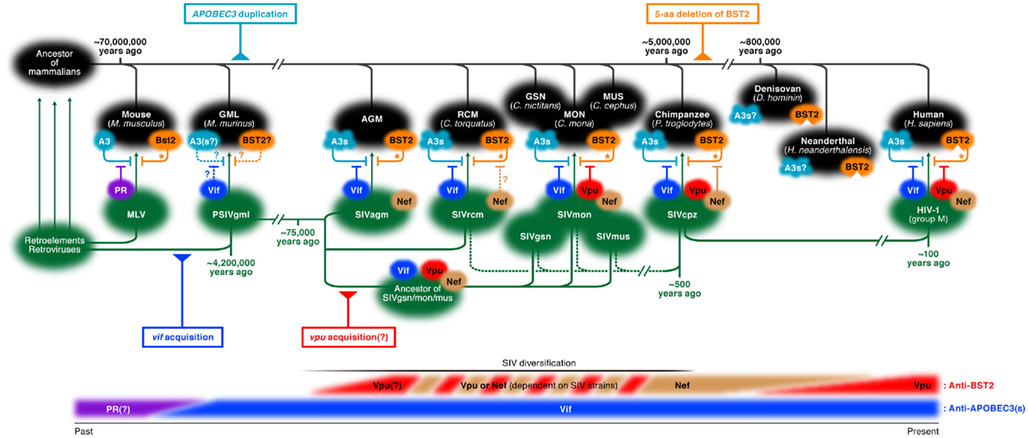
Figure 1. The pedigree chart of the interplay between viral factors (Vif and Vpu) and host factors (APOBEC3s and BST2) during co-evolution and co-divergence of retroviruses, lentiviruses, and hosts. Note that the timescales of hosts (top, shown in black) and viruses (middle, shown in green) are not parallel. (Top) The pedigree chart of hosts. Rodents including mice (Mus musculus) brunched almost 70 million years ago (Eizirik et al., 2001), and gray mouse lemurs (GML; Microcebus murinus) then came into existence. After the divergence of old world monkeys [e.g., African green monkey (AGM), red-capped mangabey (RCM; Cercopithecus torquatus), greater spot-nosed monkey (GSN; Cercopithecus nictitans), mona monkey (MON; Cercopithecus mona), and mustached monkey (MUS; Cercopithecus cephus)], chimpanzees (Pan troglodytes) evolved almost 5 million years ago (Chen and Li, 2001). Two hominids, Neanderthal (Homo neanderthalensis) and Denisovan (Denisova hominin), brunched around 800,000 years ago (Krause et al., 2010; Reich et al., 2010), and then, humans (Homo sapiens) appeared. Regarding host factors, the duplication of APOBEC3 occurred before the divergence of old world monkeys (AGM, RCM, GSN, MON, and MUS; it is unclear whether multiple APOBEC3s are encoded in the genome of GML). On the other hand, after the divergence of chimpanzees, the 5-aa deletion was inserted in the cytoplasmic tail of BST2. Note that BST2/Bst2 is able to impair the release of viruses while it is unable to completely restrict their replication (indicated by asterisks). (Middle) The pedigree chart of retroviruses including lentiviruses. MLV, a retrovirus, counteracts murine Apobec3 (A3) by its protease (PR). Since PSIVgml, a common ancestor of all primate lentiviruses, had already acquired the ancestor of vif [although it is unclear whether the Vif ancestor of PSIVgml is able to antagonize APOBEC3(s) in GML or not], vif would be acquired by lentiviruses at least 4.2 million years ago. All primate lentiviruses including SIVs possess vif and its homologs, and Vifs of all SIVs antagonize certain APOBEC3s (APOBEC3A, B, C, DE, F, G, and H; A3s) of their natural hosts. In contrast to vif, vpu would have been acquired in the lineage of SIVgsn/mon/mus after the divergence of SIVs (e.g., SIVagm, SIVrcm, and the common ancestor of SIVgsn/mon/mus), which had started about 75,000 years ago. Note that SIVagm and SIVrcm do not possess vpu. It has been demonstrated that the other factors of SIVagm (e.g., Nef) are unable to antagonize AGM BST2, while it has been hypothesized that SIVrcm Nef has the potential to counteract RCM BST2. On the other hand, Vpus of SIVgsn, SIVmon, and SIVmus counteract BST2s of their natural hosts. By the evolution and/or the zoonotic swapping between SIVrcm and SIVgsn/mon/mus, SIVcpz has emerged approximately 500 years ago (Wertheim and Worobey, 2009). It is noteworthy that SIVcpz Nef, but not Vpu, is able to counteract chimpanzee BST2. However, HIV-1, particularly HIV-1 group M which infiltrated the human population around 1900s and is the current pandemic virus, antagonizes human BST2 by its Vpu. (Bottom) The pedigree chart of viral factors antagonizing host factors. In the case of the anti-APOBEC3(s) factor, PR might have been the original APOBEC3-antagonizing factor since MLV PR antagonizes murine A3. After the acquisition of vif in the lineage of primate lentiviruses (around 4.2 million years ago by PSIVgml), vif would be turned into a dominant restriction factor that antagonizes APOBEC3s of their natural hosts. On the other hand, in the case of the anti-BST2 factor, certain lineages of primate lentiviruses (e.g., the common ancestor of SIVgsn/mon/mus) would acquire vpu after the initiation of SIV divergence (about 75,000 years ago). It is also of importance that all Vpus of SIVs do not necessarily counteract BST2s of their natural hosts, and that some SIVs (e.g., SIVrcm and SIVcpz) counteract BST2s of their natural hosts by Nef. In the twentieth century, SIVcpz infiltrated the human population as HIV-1, and vpu took over the role of antagonizing BST2 of human.
APOBEC3s and Vif
The orthologs of human (Homo sapiens) APOBEC3s are encoded in the genomes of chimpanzees (Pan troglodytes) and mice (Mus musculus). Although mice have a sole ortholog, Apobec3, which is located on chromosome 15, it is of interest that there are seven paralogs of murine Apobec3 in the genomes of humans and chimpanzees (designated to APOBEC3A, B, C, DE, F, G, and H), which are located on chromosome 22 (Harris and Liddament, 2004). Since gene duplication is strong evidence that the duplicated genes have been exposed to selective pressures, these findings indicate that murine Apobec3 had been subjected to selective pressures and was duplicated during the evolutionary process of rodents and primates. Apobec3/APOBEC3 family proteins have functioned to potently inhibit the replication of retroviruses and retroelements (Harris and Liddament, 2004) suggesting that they have played a role as the selective pressure on Apobec3. On the other hand, although MLV does not encode vif, murine Apobec3 can be degraded by viral protease (Abudu et al., 2006). These findings strongly suggest that the conflict between retroviruses (e.g., the MLV and its ancestors) and their hosts had also taken place in rodents. In the case of monkeys including primates, it has been suggested that primates have been infected by retroviruses for over 30 million years (Belshaw et al., 2004). In addition, it has been suggested that APOBEC3G has been subjected to strong positive selection throughout the history of primate evolution for at least 33 million years (Sawyer et al., 2004) and has rapidly evolved (Zhang and Webb, 2004). Therefore, it is reasonable to consider that the expansion of APOBEC3s in primates was attributed to repeated retrovirus infections and has been further promoted by their antagonists, Vif, and its ancestors.
When was vif acquired by primate lentiviruses? In 2008, a unique “molecular fossil record,” an endogenous lentivirus named gray mouse lemur prosimian immunodeficiency virus (PSIVgml), was identified in the genome of gray mouse lemur (Microcebus murinus), which is a strepsirrhine primate from Madagascar (Gifford et al., 2008). Gifford et al. (2008) demonstrated that PSIVgml is the putative common ancestor of all known primate lentiviruses. Of particular importance, PSIVgml had already acquired the putative ancestor gene of vif. In addition, one study has estimated that gray mouse lemurs were infected with PSIVgml at least 4.2 million years ago (Mya; Gilbert et al., 2009), suggesting that the vif ancestor had already existed in primate lentiviruses during this period. Moreover, it has also been estimated that Carnivora introduced the PSIV ancestor(s) from Africa to Madagascar at least 19 Mya, which would lead to the establishment of lentiviruses in primates (Yoder et al., 2003; Gifford, 2012). Although it is still unknown whether the PSIV ancestor(s) had already possessed a putative vif ancestor, it seems convincing that primates and their ancestors have co-existed and co-evolved with primate lentiviruses encoding vif for more than several million years.
In summary, the interplay between APOBEC3(s) and retroviruses in terms of evolutionary biology, a scenario in which APOBEC3s have co-evolved with retroelements and retroviruses (including lentiviruses) in hosts for over 30 million years, is reasonable. In parallel, Vif has co-evolved as a masterpiece of primate lentiviruses with expertise in counteracting APOBEC3s. In other words, Vif is a consequence of the “Red Queen’s race” with APOBEC3s.
BST2 and Vpu
As in the case of human APOBEC3s, has human BST2 been exposed to selective pressures by its counteracting agents such as HIV-1 Vpu? In sharp contrast to APOBEC3s, BST2 duplication is not observed. BST2 is encoded in the genomes of human and chimpanzee located on chromosome 19, while murine Bst2 is located on chromosome 8. Although it has been suggested that BST2 is a consequence of positive selection (McNatt et al., 2009), others have implied that human BST2 is in an intermediate stage in transition to adaptive evolution (Liu et al., 2010). Therefore, it seems feasible to consider that BST2 has been not undergone extensive selective pressure compared to APOBEC3s. However, it is of note that Nefs of SIVcpz and SIVgor are able to antagonize BST2s of both chimpanzees and gorillas but not that of humans (Lim et al., 2010). By comparing the amino acid (aa) sequences of BST2s in these three species, the absence of a 5-aa motif in the cytoplasmic tail of human BST2, where Nefs of certain SIVs (e.g., SIVcpz and SIVgor) target was found (Sauter et al., 2009; Lim et al., 2010). These findings suggest that the 5-aa deletion made human BST2 resistant to the counteraction by Nefs of certain SIVs. Moreover, the 5-aa deletion in BST2 is detected in two hominids, Neanderthal (Homo neanderthalensis) and Denisovan (Denisova hominin), suggesting that this deletion occurred at least 0.8 Mya (Sauter et al., 2011b). Therefore, it would be possible to assume that the selective pressure by Nefs of SIVs, which can antagonize BST2s of their natural hosts, has resulted in the 5-aa deletion in BST2. However, as mentioned above, other viruses also possess anti-BST2 factors such as EBOV GP (Kaletsky et al., 2009) and KSHV K5/MIR2 (Mansouri et al., 2009; Pardieu et al., 2010). Therefore, a possibility that the resulting 5-aa deletion in BST2 is due to selective pressure by these other factors cannot be excluded.
When was vpu acquired by primate lentiviruses? In contrast to vif, PSIVgml does not encode a putative ancestor of vpu (Gifford et al., 2008), suggesting that vpu participated in the evolutionary history of primate lentiviruses after the period of PSIVgml emergence. In addition, although vif and its homologs has been encoded by a broad range of lentiviruses including caprine arthritis–encephalitis virus in small ruminants, bovine immunodeficiency virus in ruminants, FIVs in carnivores, all SIVs in monkeys, and HIV-1 in humans (Gifford et al., 2008; Gifford, 2012), vpu has only been encoded in certain primate lentiviruses. Out of all primate lentiviruses, vpu has only been encoded by SIVcpz, three SIVs from greater spot-nosed monkeys (SIVgsn), mona monkeys (SIVmon), and mustached monkeys (SIVmus; Courgnaud et al., 2002, 2003), and HIV-1. SIVcpz, the origin of HIV-1, emerged in chimpanzees as a hybrid of SIVgsn/mon/mus and SIVrcm (an SIV from red-capped mangabeys; Bailes et al., 2003), meaning that the common ancestor of vpu was acquired by the ancestral lineage of SIVgsn/mon/mus after the initiation of SIV divergence (Sharp et al., 2005; Gifford et al., 2008). Since primate lentiviruses have co-diversified and co-evolved with hosts along with cross-species swapping during the past millions of years, estimating the evolutionary history of SIVs has been challenging and it seems difficult to pinpoint the period when the lineage of SIVgsn/mon/mus acquired the putative vpu ancestor. However, one study has estimated that the SIV diversification had started about 75,000 years ago (Worobey et al., 2010), which suggests that vpu was acquired by the lineage of SIVgsn/mon/mus after this point. Considering this evidence, it is likely that vpu is much younger than vif in the evolutionary history of primate lentiviruses (Figure 1). More importantly, although Vpus of SIVgsn, SIVmon, and SIVmus are able to antagonize BST2s of their natural hosts, Vpu of SIVcpz (a chimera of SIVgsn/mon/mus and SIVrcm) appears to have transferred its BST2 antagonizing ability to Nef (Kirchhoff, 2010). These series of events resulted in a restoration of BST2 antagonism by Vpu in the only pandemic group of HIV-1 (for more details, see the next section), which emerged during the past 100 years (estimated in 1908–1933; Worobey et al., 2008). Therefore, in terms of co-evolution of lentiviruses and hosts, the interplay between HIV-1 Vpu and BST2 can be considered to be immature and still in the midst of the “Red Queen’s race.”
BST2 and Other Viral Factors
As mentioned above, various families of viruses belonging to Retroviridae (HIV-1 Vpu, HIV-2 Env, and SIV Nef), Filoviridae (EBOV GP), and Herpesviridae (KSHV K5/MIR2) respectively possess their own anti-BST2 factor that can antagonize BST2 through different molecular mechanisms of action. Since these viruses are highly divergent, it should be interpreted that each anti-BST2 factor has independently acquired its BST2 counteracting ability at different periods in time. Therefore, in order to further understand the interplay between BST2 and viruses, it is necessary to focus on the evolutionary history of individual viral anti-BST2 factors. For instance, although EBOV GP is able to counteract human BST2 (Kaletsky et al., 2009), it is still unclear whether GP’s function is a common feature in Filoviridae. Given that EBOV GP counteracts the ortholog(s) of BST2 in its natural host(s), it would be interesting to explore this possibility. In addition, among eight human herpesviruses, it has been suggested that the gene encoding K5/MIR2 is unique in KSHV (Russo et al., 1996; Haque et al., 2000), indicating that the antagonizing potential of human BST2 has been originally acquired by KSHV after the divergence of human herpesviruses. Moreover, since it has been suggested that bovine herpesvirus 4 (BHV4) possesses the homologous gene to K5/MIR2 in KSHV (Russo et al., 1996), it would be interesting to address the antagonizing ability of the protein transcribed by the homologous gene in BHV4 against bovine BST2. Taken together, revealing these insights would provide clues to understand the evolutionary history of viruses and BST2.
Epidemiology
What is the significance of Vpu’s potential to antagonize BST2? One possible explanation has been provided by Sauter et al. (2009), in which Vpu has contributed to the explosion of HIV-1 infection resulting in a pandemic. HIV-1 has been classified into four lineages; group M (main/major), group O (outlier), group N, and group P. Only HIV-1 group M is pandemic, while the others are endemic in Africa. Within the four groups, Vpus of HIV-1 groups M and O are able to antagonize BST2’s tethering ability, while those of HIV-1 groups N and P are not (Sauter et al., 2009, 2011a; Yang et al., 2011). Although it is well known that HIV-1 Vpu has another ability to rapidly degrade CD4 molecules, the primary receptor of HIV-1, (Willey et al., 1992; Margottin et al., 1998; Schubert et al., 1998), group O Vpu fails to induce the degradation of CD4 (Sauter et al., 2009). Taken together, group M Vpu is the only protein able to antagonize and/or degrade both BST2 and CD4, indicating a clear correlation between the range of virus epidemicity and the degree of Vpu-mediated BST2 and CD4 down-regulation. In other words, these findings suggest that Vpu positively contributes to the efficiency of inter-individual (i.e., human-to-human) spread of HIV-1. HIV-1 is predominantly transmitted sexually, and it has been assumed that HIV-1 transmission can be accomplished by cell-free virions in genital fluids at a low probability (1 in 200–3,000; Gray et al., 2001). Therefore, consistent with a hypothesis proposed by Kirchhoff (2010), it is conceivable that Vpu increases the amount of cell-free virions in genital fluids by antagonizing BST2, which can lead to a rise in the efficiency of human-to-human HIV-1 transmission.
In the field of HIV-1 virology, there is a longstanding unanswered question: why has only group M led to a pandemic – was it just by chance? The hypothesis mentioned above may be the clue to answer this mystery. Moreover, it has been suggested that each HIV-1 group (M, N, O, and P) was independently established through zoonotic transmission of SIVcpz from chimpanzees to the human population (Gao et al., 1999; Heeney et al., 2006). However, it is still unclear whether each HIV-1 group is derived from (1) a single chimpanzee infected with SIVcpz or (2) separate chimpanzees infected with SIVcpz as a respective ancestor of each HIV-1 group (Heeney et al., 2006). Furthermore, as mentioned in the previous section, SIVcpz was derived from a hybrid of SIVgsn/mon/mus and SIVrcm in a chimpanzee (Bailes et al., 2003). Has this recombination event occurred only once in a chimpanzee and the chimpanzee simultaneously transferred the virus to humans as the origin of each HIV-1 group? Or was the hybrid SIV first transmitted to different chimpanzees and then individually transferred from chimpanzees to humans? Since the phenotype of Vpu is clearly different among HIV-1 groups and SIVs, the difference in Vpu’s ability may be an important key to trace the origin of each HIV-1 group.
Future Directions
Here, we briefly described the interplay between Vpu and BST2 from four different aspects: virology, molecular biology, evolutionary biology, and epidemiology. It is particularly noteworthy to consider how Vpu has acquired its anti-BST2 activity during HIV-1 evolution. In addition, various viral anti-BST2 factors including HIV-1 Vpu, HIV-2 Env, SIV Nef, EBOV GP, and KSHV K5/MIR2 have respectively procured individual anti-BST2 activity during viral evolution and diversity since the molecular mechanisms of action to antagonize BST2 are different among these proteins. If so, what was/is the advantage for viruses to acquire this activity? On the other hand, why have human BST2 and its orthologs evolved a function to “tether” viral particles? Finally, it would be difficult to conclude that HIV-1, particularly a group of the pandemic HIV-1, has aimlessly acquired the anti-BST2 activity within its limited genome space (one would expect that artificially inserted unnecessary and/or inadequate portions in HIV-1 genome can be easily deleted after several passages even in cell cultures), and vice versa, it would be also difficult to assume that BST2 has accidentally acquired its activity to tether virions during evolution of hosts. Understanding how these events occurred will be important and interesting beyond the field of science.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Robert Gifford (Aaron Diamond AIDS Research Institute, Rockefeller University, USA), Taichiro Takemura (AIDS Research Center, National Institute of Infectious Diseases, Japan), and Tomoko Kobayashi (Laboratory of Viral Pathogenesis, Institute for Virus Research, Kyoto University, Japan) for helpful suggestions in writing this paper. This work was supported in-part by a Grant-in-Aid for Scientific Research (B21390137 to Yoshio Koyanagi) and a Grant-in-Aid for Young Scientists (B23790500 to Kei Sato) from the Japan Society for the Promotion of Science; the Shimizu Foundation for Immunological Research Grant (to Kei Sato).
References
Abudu, A., Takaori-Kondo, A., Izumi, T., Shirakawa, K., Kobayashi, M., Sasada, A., Fukunaga, K., and Uchi-yama, T. (2006). Murine retrovirus escapes from murine APOBEC3 via two distinct novel mechanisms. Curr. Biol. 16, 1565–1570.
Aldrovandi, G. M., and Zack, J. A. (1996). Replication and pathogenicity of human immunodeficiency virus type 1 accessory gene mutants in SCID-hu mice. J. Virol. 70, 1505–1511.
Arias, J. F., Iwabu, Y., and Tokunaga, K. (2011). Structural basis for the antiviral activity of BST-2/Tetherin and its viral antagonism. Front. Microbiol. 2:250. doi:10.3389/fmicb.2011.00250
Bailes, E., Gao, F., Bibollet-Ruche, F., Courgnaud, V., Peeters, M., Marx, P. A., Hahn, B. H., and Sharp, P. M. (2003). Hybrid origin of SIV in chimpanzees. Science 300, 1713.
Barre-Sinoussi, F., Chermann, J. C., Rey, F., Nugeyre, M. T., Chamaret, S., Gruest, J., Dauguet, C., Axler-Blin, C., Vezinet-Brun, F., Rouzioux, C., Rozenbaum, W., and Montagnier, L. (1983). Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220, 868–871.
Belshaw, R., Pereira, V., Katzourakis, A., Talbot, G., Paces, J., Burt, A., and Tristem, M. (2004). Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. U.S.A. 101, 4894–4899.
Blasius, A. L., Giurisato, E., Cella, M., Schreiber, R. D., Shaw, A. S., and Colonna, M. (2006). Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J. Immunol. 177, 3260–3265.
Bour, S., Schubert, U., Peden, K., and Strebel, K. (1996). The envelope glycoprotein of human immunodeficiency virus type 2 enhances viral particle release: a Vpu-like factor? J. Virol. 70, 820–829.
Bour, S., and Strebel, K. (1996). The human immunodeficiency virus (HIV) type 2 envelope protein is a functional complement to HIV type 1 Vpu that enhances particle release of heterologous retroviruses. J. Virol. 70, 8285–8300.
Brown, D., Trowsdale, J., and Allen, R. (2004). The LILR family: modulators of innate and adaptive immune pathways in health and disease. Tissue Antigens 64, 215–225.
Cao, W., Bover, L., Cho, M., Wen, X., Hanabuchi, S., Bao, M., Rosen, D. B., Wang, Y. H., Shaw, J. L., Du, Q., Li, C., Arai, N., Yao, Z., Lanier, L. L., and Liu, Y. J. (2009). Regulation of TLR7/9 responses in plasmacytoid dendritic cells by BST2 and ILT7 receptor interaction. J. Exp. Med. 206, 1603–1614.
Casartelli, N., Sourisseau, M., Feldmann, J., Guivel-Benhassine, F., Mallet, A., Marcelin, A. G., Guatelli, J., and Schwartz, O. (2010). Tetherin restricts productive HIV-1 cell-to-cell transmission. PLoS Pathog. 6, e1000955. doi:10.1371/journal.ppat.1000955
Chen, F. C., and Li, W. H. (2001). Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68, 444–456.
Courgnaud, V., Abela, B., Pourrut, X., Mpoudi-Ngole, E., Loul, S., Delaporte, E., and Peeters, M. (2003). Identification of a new simian immunodeficiency virus lineage with a vpu gene present among different cercopithecus monkeys (C. mona, C. cephus, and C. nictitans) from Cameroon. J. Virol. 77, 12523–12534.
Courgnaud, V., Salemi, M., Pourrut, X., Mpoudi-Ngole, E., Abela, B., Auzel, P., Bibollet-Ruche, F., Hahn, B., Vandamme, A. M., Delaporte, E., and Peeters, M. (2002). Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. J. Virol. 76, 8298–8309.
Dietrich, I., Mcmonagle, E. L., Petit, S. J., Vijayakrishnan, S., Logan, N., Chan, C. N., Towers, G. J., Hosie, M. J., and Willett, B. J. (2011). Feline tetherin efficiently restricts release of feline immunodeficiency virus but not spreading of infection. J. Virol. 85, 5840–5852.
Douglas, J. L., Gustin, J. K., Viswanathan, K., Mansouri, M., Moses, A. V., and Fruh, K. (2010). The great escape: viral strategies to counter BST-2/tetherin. PLoS Pathog. 6, e1000913. doi:10.1371/journal.ppat.1000913
Douglas, J. L., Viswanathan, K., Mccarroll, M. N., Gustin, J. K., Fruh, K., and Moses, A. V. (2009). Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/Tetherin via a {beta}TrCP-dependent mechanism. J. Virol. 83, 7931–7947.
Dube, M., Bego, M. G., Paquay, C., and Cohen, E. A. (2010a). Modulation of HIV-1-host interaction: role of the Vpu accessory protein. Retrovirology 7, 114.
Dube, M., Roy, B. B., Guiot-Guillain, P., Binette, J., Mercier, J., Chiasson, A., and Cohen, E. A. (2010b). Antagonism of tetherin restriction of HIV-1 release by Vpu involves binding and sequestration of the restriction factor in a perinuclear compartment. PLoS Pathog. 6, e1000856. doi:10.1371/journal.ppat.1000856
Eizirik, E., Murphy, W. J., and O’brien, S. J. (2001). Molecular dating and biogeography of the early placental mammal radiation. J. Hered. 92, 212–219.
Evans, D. T., Serra-Moreno, R., Singh, R. K., and Guatelli, J. C. (2010). BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends Microbiol. 18, 388–396.
Fukuma, A., Abe, M., Morikawa, Y., Miyazawa, T., and Yasuda, J. (2011). Cloning and characterization of the antiviral activity of feline Tetherin/BST-2. PLoS ONE 6, e18247. doi:10.1371/journal.pone.0018247
Gallo, R. C., Sarin, P. S., Gelmann, E. P., Robert-Guroff, M., Richardson, E., Kalyanaraman, V. S., Mann, D., Sidhu, G. D., Stahl, R. E., Zolla-Pazner, S., Leibowitch, J., and Popovic, M. (1983). Isolation of human T-cell leukemia virus in acquired immune deficiency syndrome (AIDS). Science 220, 865–867.
Gao, F., Bailes, E., Robertson, D. L., Chen, Y., Rodenburg, C. M., Michael, S. F., Cummins, L. B., Arthur, L. O., Peeters, M., Shaw, G. M., Sharp, P. M., and Hahn, B. H. (1999). Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397, 436–441.
Gifford, R. J. (2012). Viral evolution in deep time: lentiviruses and mammals. Trends Genet. 28, 89–100.
Gifford, R. J., Katzourakis, A., Tristem, M., Pybus, O. G., Winters, M., and Shafer, R. W. (2008). A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc. Natl. Acad. Sci. U.S.A. 105, 20362–20367.
Gilbert, C., Maxfield, D. G., Goodman, S. M., and Feschotte, C. (2009). Parallel germline infiltration of a lentivirus in two Malagasy lemurs. PLoS Genet. 5, e1000425. doi:10.1371/journal.pgen.1000425
Gray, R. H., Wawer, M. J., Brookmeyer, R., Sewankambo, N. K., Serwadda, D., Wabwire-Mangen, F., Lutalo, T., Li, X., Vancott, T., and Quinn, T. C. (2001). Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet 357, 1149–1153.
Gummuluru, S., Kinsey, C. M., and Emerman, M. (2000). An in vitro rapid-turnover assay for human immunodeficiency virus type 1 replication selects for cell-to-cell spread of virus. J. Virol. 74, 10882–10891.
Gupta, R. K., Mlcochova, P., Pelchen-Matthews, A., Petit, S. J., Mattiuzzo, G., Pillay, D., Takeuchi, Y., Marsh, M., and Towers, G. J. (2009). Simian immunodeficiency virus envelope glycoprotein counteracts tetherin/BST-2/CD317 by intracellular sequestration. Proc. Natl. Acad. Sci. U.S.A. 106, 20889–20894.
Haque, M., Chen, J., Ueda, K., Mori, Y., Nakano, K., Hirata, Y., Kanamori, S., Uchiyama, Y., Inagi, R., Okuno, T., and Yamanishi, K. (2000). Identification and analysis of the K5 gene of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 74, 2867–2875.
Harris, R. S., and Liddament, M. T. (2004). Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4, 868–877.
Hauser, H., Lopez, L. A., Yang, S. J., Oldenburg, J. E., Exline, C. M., Guatelli, J. C., and Cannon, P. M. (2010). HIV-1 Vpu and HIV-2 Env counteract BST-2/tetherin by sequestration in a perinuclear compartment. Retrovirology 7, 51.
Heeney, J. L., Dalgleish, A. G., and Weiss, R. A. (2006). Origins of HIV and the evolution of resistance to AIDS. Science 313, 462–466.
Homann, S., Smith, D., Little, S., Richman, D., and Guatelli, J. (2011). Upregulation of BST-2/tetherin by HIV infection in vivo. J. Virol. 85, 10659–10668.
Ishikawa, J., Kaisho, T., Tomizawa, H., Lee, B. O., Kobune, Y., Inazawa, J., Oritani, K., Itoh, M., Ochi, T., and Ishihara, K. (1995). Molecular cloning and chromosomal mapping of a bone marrow stromal cell surface gene, BST2, that may be involved in pre-B-cell growth. Genomics 26, 527–534.
Izumi, T., Shirakawa, K., and Takaori-Kondo, A. (2008). Cytidine deaminases as a weapon against retroviruses and a new target for antiviral therapy. Mini Rev. Med. Chem. 8, 231–238.
Jia, B., Serra-Moreno, R., Neidermyer, W., Rahmberg, A., Mackey, J., Fofana, I. B., Johnson, W. E., Westmoreland, S., and Evans, D. T. (2009). Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 5, e1000429. doi:10.1371/journal.ppat.1000429
Jolly, C., Booth, N. J., and Neil, S. J. (2010). Cell-cell spread of human immunodeficiency virus type 1 overcomes tetherin/BST-2-mediated restriction in T cells. J. Virol. 84, 12185–12199.
Jouvenet, N., Neil, S. J., Zhadina, M., Zang, T., Kratovac, Z., Lee, Y., Mcnatt, M., Hatziioannou, T., and Bieniasz, P. D. (2009). Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J. Virol. 83, 1837–1844.
Kaletsky, R. L., Francica, J. R., Agrawal-Gamse, C., and Bates, P. (2009). Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc. Natl. Acad. Sci. U.S.A. 106, 2886–2891.
Kirchhoff, F. (2010). Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8, 55–67.
Kitchen, L. W., Barin, F., Sullivan, J. L., Mclane, M. F., Brettler, D. B., Levine, P. H., and Essex, M. (1984). Aetiology of AIDS – antibodies to human T-cell leukaemia virus (type III) in haemophiliacs. Nature 312, 367–369.
Kobayashi, T., Ode, H., Yoshida, T., Sato, K., Gee, P., Yamamoto, S. P., Ebina, H., Strebel, K., Sato, H., and Koyanagi, Y. (2011). Identification of amino acids in the human tetherin transmembrane domain responsible for HIV-1 Vpu interaction and susceptibility. J. Virol. 85, 932–945.
Krause, J., Fu, Q., Good, J. M., Viola, B., Shunkov, M. V., Derevianko, A. P., and Paabo, S. (2010). The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature 464, 894–897.
Kuhl, B. D., Sloan, R. D., Donahue, D. A., Bar-Magen, T., Liang, C., and Wainberg, M. A. (2010). Tetherin restricts direct cell-to-cell infection of HIV-1. Retrovirology 7, 115.
Kupzig, S., Korolchuk, V., Rollason, R., Sugden, A., Wilde, A., and Banting, G. (2003). Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic 4, 694–709.
Le Tortorec, A., and Neil, S. J. (2009). Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J. Virol. 83, 11966–11978.
Lim, E. S., and Emerman, M. (2009). Simian immunodeficiency virus SIVagm from African green monkeys does not antagonize endogenous levels of African green monkey tetherin/BST-2. J. Virol. 83, 11673–11681.
Lim, E. S., Malik, H. S., and Emerman, M. (2010). Ancient adaptive evolution of tetherin shaped the functions of Vpu and Nef in human immunodeficiency virus and primate lentiviruses. J. Virol. 84, 7124–7134.
Liu, J., Chen, K., Wang, J. H., and Zhang, C. (2010). Molecular evolution of the primate antiviral restriction factor tetherin. PLoS ONE 5, e11904. doi:10.1371/journal.pone.0011904
Lopez, L. A., Yang, S. J., Hauser, H., Exline, C. M., Haworth, K. G., Oldenburg, J., and Cannon, P. M. (2010). Ebola virus glycoprotein counteracts BST-2/Tetherin restriction in a sequence-independent manner that does not require tetherin surface removal. J. Virol. 84, 7243–7255.
Madani, N., and Kabat, D. (1998). An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J. Virol. 72, 10251–10255.
Mansouri, M., Viswanathan, K., Douglas, J. L., Hines, J., Gustin, J., Moses, A. V., and Fruh, K. (2009). Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 83, 9672–9681.
Margottin, F., Bour, S. P., Durand, H., Selig, L., Benichou, S., Richard, V., Thomas, D., Strebel, K., and Benarous, R. (1998). A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1, 565–574.
Martin, N., and Sattentau, Q. (2009). Cell-to-cell HIV-1 spread and its implications for immune evasion. Curr. Opin. HIV AIDS 4, 143–149.
Martin-Serrano, J., and Neil, S. J. (2011). Host factors involved in retroviral budding and release. Nat. Rev. Microbiol. 9, 519–531.
McNatt, M. W., Zang, T., Hatziioannou, T., Bartlett, M., Fofana, I. B., Johnson, W. E., Neil, S. J., and Bieniasz, P. D. (2009). Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 5, e1000300. doi:10.1371/journal.ppat.1000300
Miyagi, E., Andrew, A. J., Kao, S., and Strebel, K. (2009). Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface down-modulation and intracellular depletion. Proc. Natl. Acad. Sci. U.S.A. 106, 2868–2873.
Neil, S. J., Sandrin, V., Sundquist, W. I., and Bieniasz, P. D. (2007). An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe 2, 193–203.
Neil, S. J., Zang, T., and Bieniasz, P. D. (2008). Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430.
Pardieu, C., Vigan, R., Wilson, S. J., Calvi, A., Zang, T., Bieniasz, P., Kellam, P., Towers, G. J., and Neil, S. J. (2010). The RING-CH ligase K5 antagonizes restriction of KSHV and HIV-1 particle release by mediating ubiquitin-dependent endosomal degradation of tetherin. PLoS Pathog. 6, e1000843. doi:10.1371/journal.ppat.1000843
Radoshitzky, S. R., Dong, L., Chi, X., Clester, J. C., Retterer, C., Spurgers, K., Kuhn, J. H., Sandwick, S., Ruthel, G., Kota, K., Boltz, D., Warren, T., Kranzusch, P. J., Whelan, S. P., and Bavari, S. (2010). Infectious Lassa virus, but not filoviruses, is restricted by BST-2/tetherin. J. Virol. 84, 10569–10580.
Reich, D., Green, R. E., Kircher, M., Krause, J., Patterson, N., Durand, E. Y., Viola, B., Briggs, A. W., Stenzel, U., Johnson, P. L., Maricic, T., Good, J. M., Marques-Bonet, T., Alkan, C., Fu, Q., Mallick, S., Li, H., Meyer, M., Eichler, E. E., Stoneking, M., Richards, M., Talamo, S., Shunkov, M. V., Derevianko, A. P., Hublin, J. J., Kelso, J., Slatkin, M., and Paabo, S. (2010). Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060.
Rollason, R., Korolchuk, V., Hamilton, C., Jepson, M., and Banting, G. (2009). A CD317/tetherin-RICH2 complex plays a critical role in the organization of the subapical actin cytoskeleton in polarized epithelial cells. J. Cell Biol. 184, 721–736.
Rollason, R., Korolchuk, V., Hamilton, C., Schu, P., and Banting, G. (2007). Clathrin-mediated endocytosis of a lipid-raft-associated protein is mediated through a dual tyrosine motif. J. Cell. Sci. 120, 3850–3858.
Russo, J. J., Bohenzky, R. A., Chien, M. C., Chen, J., Yan, M., Maddalena, D., Parry, J. P., Peruzzi, D., Edelman, I. S., Chang, Y., and Moore, P. S. (1996). Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. U.S.A. 93, 14862–14867.
Sato, K., Misawa, N., Fukuhara, M., Iwami, S., An, D. S., Ito, M., and Koyanagi, Y. (in press). Vpu augments the initial burst phase of HIV-1 propagation and downregulates BST2 and CD4 in humanized mice. J. Virol. doi: 10.1128/JVI.07062-11
Sakuma, T., Noda, T., Urata, S., Kawaoka, Y., and Yasuda, J. (2009). Inhibition of Lassa and Marburg virus production by tetherin. J. Virol. 83, 2382–2385.
Sato, K., Izumi, T., Misawa, N., Kobayashi, T., Yamashita, Y., Ohmichi, M., Ito, M., Takaori-Kondo, A., and Koyanagi, Y. (2010). Remarkable lethal G-to-A mutations in vif-proficient HIV-1 provirus by individual APOBEC3 proteins in humanized mice. J. Virol. 84, 9546–9556.
Sato, K., Yamamoto, S. P., Misawa, N., Yoshida, T., Miyazawa, T., and Koyanagi, Y. (2009). Comparative study on the effect of human BST-2/Tetherin on HIV-1 release in cells of various species. Retrovirology 6, 53.
Sattentau, Q. (2008). Avoiding the void: cell-to-cell spread of human viruses. Nat. Rev. Microbiol. 6, 815–826.
Sauter, D., Hue, S., Petit, S. J., Plantier, J. C., Towers, G. J., Kirchhoff, F., and Gupta, R. K. (2011a). HIV-1 Group P is unable to antagonize human tetherin by Vpu, Env or Nef. Retrovirology 8, 103.
Sauter, D., Vogl, M., and Kirchhoff, F. (2011b). Ancient origin of a deletion in human BST2/Tetherin that confers protection against viral zoonoses. Hum. Mutat. 32, 1243–1245.
Sauter, D., Schindler, M., Specht, A., Landford, W. N., Munch, J., Kim, K. A., Votteler, J., Schubert, U., Bibollet-Ruche, F., Keele, B. F., Takehisa, J., Ogando, Y., Ochsenbauer, C., Kappes, J. C., Ayouba, A., Peeters, M., Learn, G. H., Shaw, G., Sharp, P. M., Bieniasz, P., Hahn, B. H., Hatziioannou, T., and Kirchhoff, F. (2009). Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6, 409–421.
Sawyer, S. L., Emerman, M., and Malik, H. S. (2004). Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2, E275. doi:10.1371/journal.pbio.0020275
Schubert, U., Anton, L. C., Bacik, I., Cox, J. H., Bour, S., Bennink, J. R., Orlowski, M., Strebel, K., and Yewdell, J. W. (1998). CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J. Virol. 72, 2280–2288.
Schubert, U., Bour, S., Ferrer-Montiel, A. V., Montal, M., Maldarell, F., and Strebel, K. (1996). The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J. Virol. 70, 809–819.
Schubert, U., Bour, S., Willey, R. L., and Strebel, K. (1999). Regulation of virus release by the macrophage-tropic human immunodeficiency virus type 1 AD8 isolate is redundant and can be controlled by either Vpu or Env. J. Virol. 73, 887–896.
Schubert, U., Clouse, K. A., and Strebel, K. (1995). Augmentation of virus secretion by the human immunodeficiency virus type 1 Vpu protein is cell type independent and occurs in cultured human primary macrophages and lymphocytes. J. Virol. 69, 7699–7711.
Sharp, P. M., Shaw, G. M., and Hahn, B. H. (2005). Simian immunodeficiency virus infection of chimpanzees. J. Virol. 79, 3891–3902.
Sheehy, A. M., Gaddis, N. C., Choi, J. D., and Malim, M. H. (2002). Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650.
Simon, J. H., Gaddis, N. C., Fouchier, R. A., and Malim, M. H. (1998). Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 4, 1397–1400.
Skasko, M., Wang, Y., Tian, Y., Tokarev, A., Munguia, J., Ruiz, A., Stephens, E. B., Opella, S. J., and Guatelli, J. (2012). HIV-1 Vpu protein antagonizes innate restriction factor BST-2 via lipid-embedded helix-helix interactions. J. Biol. Chem. 287, 58–67.
Theodore, T. S., Englund, G., Buckler-White, A., Buckler, C. E., Martin, M. A., and Peden, K. W. (1996). Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res. Hum. Retroviruses 12, 191–194.
Van Damme, N., Goff, D., Katsura, C., Jorgenson, R. L., Mitchell, R., Johnson, M. C., Stephens, E. B., and Guatelli, J. (2008). The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3, 245–252.
Varthakavi, V., Smith, R. M., Bour, S. P., Strebel, K., and Spearman, P. (2003). Viral protein U counteracts a human host cell restriction that inhibits HIV-1 particle production. Proc. Natl. Acad. Sci. U.S.A. 100, 15154–15159.
Vigan, R., and Neil, S. J. (2010). Determinants of tetherin antagonism in the transmembrane domain of the human immunodeficiency virus type 1 Vpu protein. J. Virol. 84, 12958–12970.
Viswanathan, K., Smith, M. S., Malouli, D., Mansouri, M., Nelson, J. A., and Fruh, K. (2011). BST2/Tetherin enhances entry of human cytomegalovirus. PLoS Pathog. 7, e1002332. doi:10.1371/journal.ppat.1002332
Watanabe, R., Leser, G. P., and Lamb, R. A. (2011). Influenza virus is not restricted by tetherin whereas influenza VLP production is restricted by tetherin. Virology 417, 50–56.
Weidner, J. M., Jiang, D., Pan, X. B., Chang, J., Block, T. M., and Guo, J. T. (2010). Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 84, 12646–12657.
Wertheim, J. O., and Worobey, M. (2009). Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLoS Comput. Biol. 5, e1000377. doi:10.1371/journal.pcbi.1000377
Willey, R. L., Maldarelli, F., Martin, M. A., and Strebel, K. (1992). Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J. Virol. 66, 7193–7200.
Worobey, M., Gemmel, M., Teuwen, D. E., Haselkorn, T., Kunstman, K., Bunce, M., Muyembe, J. J., Kabongo, J. M., Kalengayi, R. M., Van Marck, E., Gilbert, M. T., and Wolinsky, S. M. (2008). Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature 455, 661–664.
Worobey, M., Telfer, P., Souquiere, S., Hunter, M., Coleman, C. A., Metzger, M. J., Reed, P., Makuwa, M., Hearn, G., Honarvar, S., Roques, P., Apetrei, C., Kazanji, M., and Marx, P. A. (2010). Island biogeography reveals the deep history of SIV. Science 329, 1487.
Yang, S. J., Lopez, L. A., Exline, C. M., Haworth, K. G., and Cannon, P. M. (2011). Lack of adaptation to human tetherin in HIV-1 group O and P. Retrovirology 8, 78.
Yang, S. J., Lopez, L. A., Hauser, H., Exline, C. M., Haworth, K. G., and Cannon, P. M. (2010). Anti-tetherin activities in Vpu-expressing primate lentiviruses. Retrovirology 7, 13.
Yoder, A. D., Burns, M. M., Zehr, S., Delefosse, T., Veron, G., Goodman, S. M., and Flynn, J. J. (2003). Single origin of Malagasy Carnivora from an African ancestor. Nature 421, 734–737.
Zhang, F., Wilson, S. J., Landford, W. C., Virgen, B., Gregory, D., Johnson, M. C., Munch, J., Kirchhoff, F., Bieniasz, P. D., and Hatziioannou, T. (2009). Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6, 54–67.
Keywords: HIV-1, Vpu, BST2, restriction factor, viral evolution, pandemic
Citation: Sato K, Gee P and Koyanagi Y (2012) Vpu and BST2: Still Not There Yet? Front. Microbio. 3:131. doi: 10.3389/fmicb.2012.00131
Received: 23 January 2012; Paper pending published: 17 February 2012;
Accepted: 20 March 2012; Published online: 09 April 2012.
Edited by:
Atsushi Koito, Kumamoto University, JapanReviewed by:
Akifumi Takaori-Kondo, Kyoto University Graduate School of Medicine, JapanHirofumi Akari, Kyoto University, Japan
Copyright: © 2012 Sato, Gee and Koyanagi. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Kei Sato, Center for Emerging Virus Research, Institute for Virus Research, Kyoto University, 53 Shogoinkawara-cho, Sakyo-ku, Kyoto, Kyoto 606-8507, Japan. e-mail:a3NhdG9AdmlydXMua3lvdG8tdS5hYy5qcA==; Yoshio Koyanagi, Laboratory of Viral Pathogenesis, Institute for Virus Research, Kyoto University, 53 Shogoinkawara-cho, Sakyo-ku, Kyoto, Kyoto 606-8507, Japan. e-mail:eWtveWFuYWdAdmlydXMua3lvdG8tdS5hYy5qcA==

 Peter Gee2
Peter Gee2