- Microbiology Department, Faculty of Biology and Soil Science, St. Petersburg State University, St. Petersburg, Russia
Green cyanobacteria differ from the blue–green cyanobacteria by the possession of a chlorophyll-containing light-harvesting antenna. Three genera of the green cyanobacteria namely Acaryochloris, Prochlorococcus, and Prochloron are unicellular and inhabit marine environments. Prochlorococcus marinus attracts most attention due to its prominent role in marine primary productivity. The fourth genus Prochlorothrix is represented by the filamentous freshwater strains. Unlike the other green cyanobacteria, Prochlorothrix strains are remarkably rare: to date, living isolates have been limited to two European locations. Taking into account fluctuating blooms, morphological resemblance to Planktothrix and Pseudanabaena, and unsuccessful attempts to obtain enrichments of Prochlorothrix, the most successful strategy to search for this cyanobacterium involves PCR with environmental DNA and Prochlorothrix-specific primers. This approach has revealed a broader distribution of Prochlorothrix. Marker genes have been found in at least two additional locations. Despite of the growing evidence for naturally occurring Prochlorothrix, there are only a few cultured strains with one of them (PCC 9006) being claimed to be axenic. In multixenic cultures, Prochlorothrix is accompanied by heterotrophic bacteria indicating a consortium-type association. The genus Prochlorothrix includes two species: P. hollandica and P. scandica based on distinctions in genomic DNA, cell size, temperature optimum, and fatty acid composition of membrane lipids. In this short review the properties of cyanobacteria of the genus Prochlorothrix are described. In addition, the evolutionary scenario for green cyanobacteria is suggested taking into account their possible role in the origin of simple chloroplast.
Introduction
Oxygenic photosynthetic bacteria are represented by blue–green cyanobacteria and green cyanobacteria. This distinction is analogous to that between green chlorosome-containing and purple non-chlorosome-containing anoxygenic photosynthetic bacteria and is based on the supramolecular organization of the light-harvesting complex rather than cell color (certain blue–green cyanobacteria are blue, green, red, violet, or brownish). Blue–green cyanobacteria are characterized by a constantly expressed phycobiliprotein light-harvesting antenna, or phycobilisome, with the exception of inducible mutants and terminally differentiated wild type cells called heterocysts. In contrast, green cyanobacteria have a constitutive chlorophyll-protein light-harvesting antenna instead of the phycobilisome (the former is expressed in some blue–green cyanobacteria in stress conditions). In terms of pigments, green cyanobacteria possess accessory chlorophyll(s), whereas blue–green cyanobacteria have only chl a (Partensky and Garczarek, 2003).
Green cyanobacteria are worth interest primarily for two reasons. First of all, Prochlorococcus marinus dominates pelagic environments for large regions of the world’s oceans. Secondly, the remaining four species (Acaryochloris marina, Prochloron didemni, Prochlorothrix hollandica, and Prochlorothrix scandica) are limited to specific eutrophic habitats, in contrast to ubiquitously distributed P. marinus and blue–green cyanobacteria (Partensky et al., 1999).
Cyanobacteria of the genus Prochlorothrix are unique among green cyanobacteria being filamentous and freshwater, as opposed to the other three marine unicellular species.
In this review, the properties of cyanobacteria of the genus Prochlorothrix are summarized, and the phylogeny of green cyanobacteria is reconsidered in the light of their role in the origin of simple chloroplast.
Scope of Objects
Circumstances of Discovery
Green cyanobacteria were initially discovered by the chromatographic detection of the accessory chl b and/or other accessory chlorophylls (a2, b2, c, etc) in collected samples. This approach lead to the discovery of two unicellular species P. didemni (Lewin, 1981) and A. marina (Miyashita et al., 1996), as well as the filamentous species P. hollandica (Burger-Wiersma et al., 1989) and P. scandica (Pinevich et al., 1999). The third unicellular species, P. marinus, was revealed by flow cytometry in oceanic picoplankton (Chisholm et al., 1988). Later on, pigment-based methods were supplemented with or gave way to PCR detection of the 16S rRNA gene with specific primers (Castiglioni et al., 2004). In order to increase the sensitivity of detection, as well as for a better separation of gene clusters, internal transcribed spacers of 16S–23S-rRNA were also tested (Laloui et al., 2002; Rocap et al., 2002). Screening for green cyanobacteria is also performed using other primers which help amplify genes which code for photosynthetic proteins, e.g., Pcb proteins of chl a/b antenna distinct from CAB proteins in green algae, euglenae, and higher plants (Geiß et al., 2003).
Representatives
The first member of the genus Prochlorothrix, the type species P. hollandica (Figure 1A) was validly described according to the rules of Bacteriological Code (Burger-Wiersma et al., 1989). In the modern taxonomic system, it is ascribed to Subsection III of Phylum X. Cyanobacteria (Oxygenic Photosynthetic Bacteria) or the order “Oscillatoriales” (Castenholz, 2001).
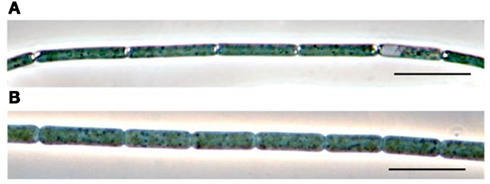
Figure 1. Light micrographs of Prochlorothrix hollandica PCC 9006 (A) and P. scandica NIVA 8/90 (B). Bar, 10 μm.
The second member of the genus Prochlorothrix is Prochlorothrix sp. NIVA 8/90 (Figure 1B). This strain was named P. scandica (Pinevich et al., 1999) legitimized under the rules of Botanical Code (Skulberg, 2008).
Prochlorothrix sp. NIVA 8/90 is considered as separate species primarily based on genomic DNA differences with P. hollandica (Table 1). Noteworthy, the previous demand for 3% dissimilarity of 16S rRNA is now considered to be not obligatory in species delimitation (Wayne et al., 1987). Prochlorothrix hollandica and P. scandica also differ in 16S–23S-ITS, and have one (1,100 bp) and two (1,100 and 620 bp) 16S–23S-ITS, respectively. The larger ITS in Prochlorothrix sp. NIVA 8/90 is 99% identical to the single ITS in P. hollandica. At the same time, the two ITS in Prochlorothrix sp. NIVA 8/90 demonstrate low mutual similarity (Velichko et al., unpublished). In addition, these two cyanobacteria differ by another genetic criterion: the electrophoretic profile of PCR-amplified highly iterated palindromic sequences (HIP-1; Robinson et al., 1995; Smith et al., 1998; Velichko et al., unpublished).
Phenotypic criteria for the species difference between P. hollandica and P. scandica are cell size, temperature optimum (see below), and fatty acid composition of the membrane lipids (Table 2).
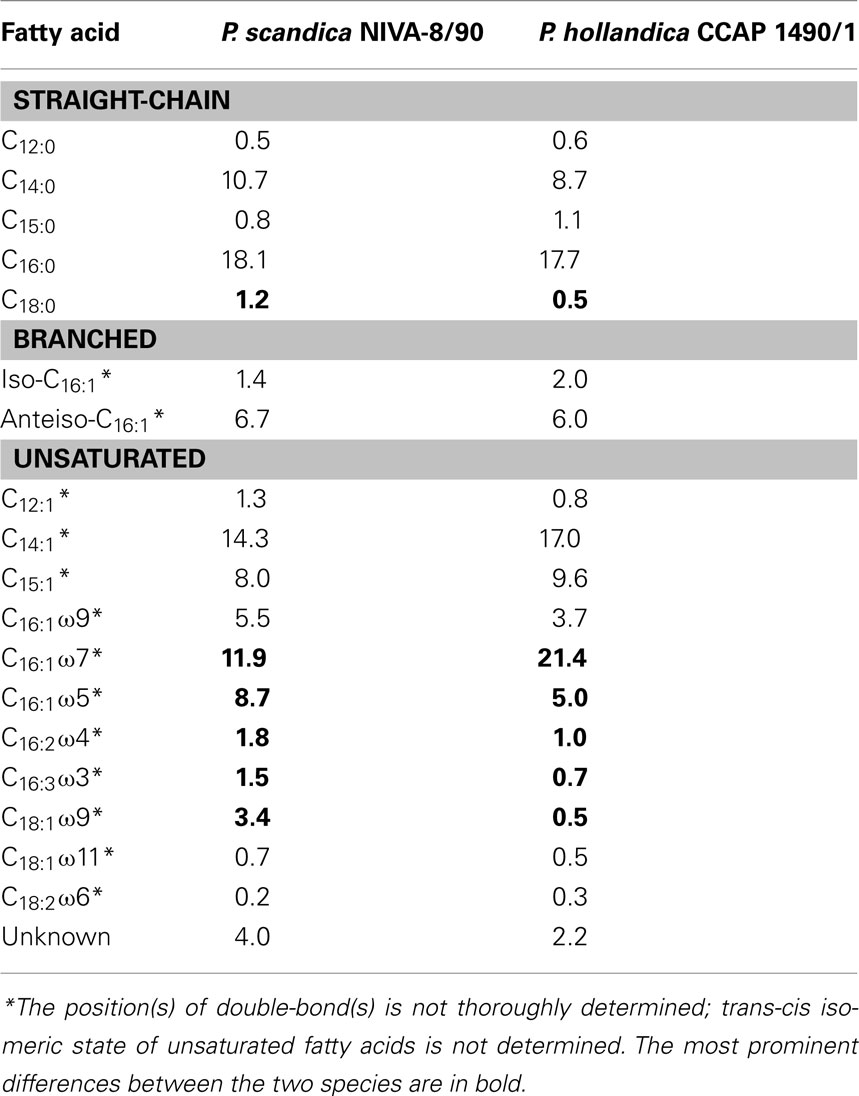
Table 2. Cellular fatty acid composition (%) of P. scandica and P. hollandica grown under identical conditions.
Due to the presence of C16:2 and C18:2 fatty acids both species should be categorized as group 5 cyanobacteria (Cohen et al., 1995). In both species, C14:0, C16:0, C14:1, and C16:1 fatty acids are among the major components. The profile of saturated (straight-chain and branched) fatty acids is comparable in the two species, except for a double amount of C18:0 stearic acid in P. scandica. At the same time, cellular content of unsaturated fatty acids is species-specific. The most prominent differences of P. scandica from P. hollandica are a twofold increase (8.7% vs 5.0%) in the amount of the C16:1ω5 isomer palmitoleic acid, sevenfold increase (3.4% vs 0.5%) in the amount of C18:1ω9 oleic acid, and a twofold decrease (11.9% vs 21.4%) in the amount of the C16:1ω7 isomer palmitoleic acid. In addition, the trace amounts of dienoic C16:2ω4 and trienoic C16:3ω3 acids are 1.8% vs 1.0% and 1.5% vs 0.7% in P. scandica and P. hollandica, respectively.
Distribution
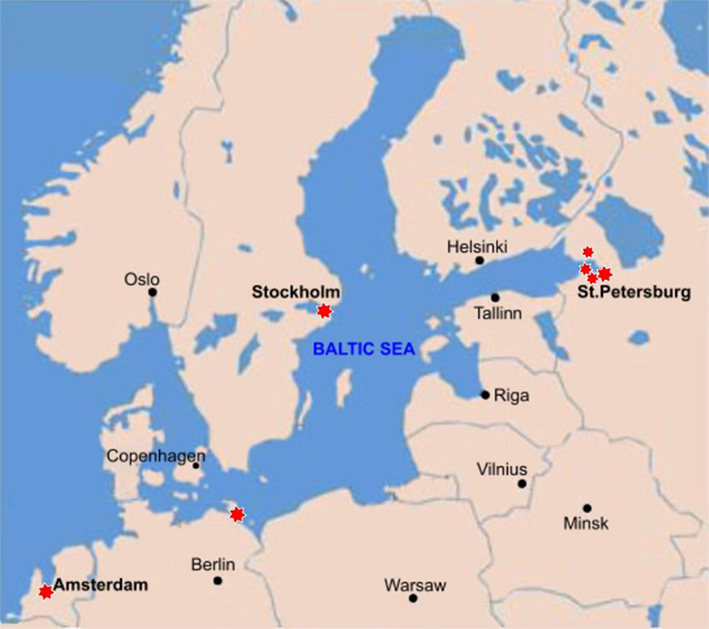
Evidence for the presence of the genus Prochlorothrix has been restricted to eutrophic freshwater or brackish habitats of North Europe (Figure 2). Prochlorothrix hollandica was accidentally encountered during a routine study of cyanobacterial blooms in the Loosdrecht lake system near Amsterdam (The Netherlands; Burger-Wiersma et al., 1989). These polymictic and highly eutrophic (∼60 mg m−3chl a) water pools of average depth ∼2 m originate from a flooded turf pit, with phytoplankton represented mostly by blue–green filamentous cyanobacteria of the group Limnothrix/Pseudanabaena (Zwart et al., 2005). In addition, “Dutch” green cyanobacteria were also found in a shallow eutrophic Lake Tjeukemeer (van Liere et al., 1989). Finally, according to personal communication by J. van der Does (van Liere et al., 1989), the presence of filamentous green cyanobacteria was registered in other shallow eutrophic Dutch lakes by fluorescence microscopy.
The second representative of the genus Prochlorothrix, P. scandica was isolated from Lake Mälaren (1,140 km2, medium depth ∼13 m) near Stockholm, Sweden (Skulberg, 2008).
Based on these discoveries, the representatives of the genus Prochlorothrix are common in eutrophic lakes occupied mainly by cyanobacteria (Lewin et al., 1980; van Liere et al., 1989).
In addition to direct evidence for the presence of these cyanobacteria in eutrophic lakes, they have been indirectly detected by the PCR-amplification of 16S rRNA gene and pcb genes (Geiß et al., 2003). Natural DNA was obtained from water samples of estuary Darss-Zingst located near the South-West coast of the Baltic Sea between the cities Rostock and Stralsund (Germany). This polymictic and eutrophic water body is represented by a chain of lagoons fed by the mouth of Prerow River, separated from the Baltic Sea by the Peninsula Frischland/Darss, and is characterized by opposite gradients of dissolved organic substances and salinity (Geiß et al., 2004). It is reasonable to expect that dynamic combination of these (a)biotic factors favors the development of cyanobacteria from the genus Prochlorothrix.
In summer 2011 Prochlorothrix-like 16S rRNA nucleotide sequences were detected in the brackish waters of the Finnish Bay in the environs of St. Petersburg, Russia (Velichko et al., unpublished data). Also, we came across an environmental Prochlorothrix-like 16S RNA gene sequence revealed during the metagenomic screening of New Orleans area water systems after Hurricanes Katrina and Rita (Amaral-Zettler et al., 2008).
Phylogenetic reconstruction of all available Prochlorothrix 16S rRNA gene sequences demonstrates close clustering of the environmental sequences from different regions of the world within the Prochlorothrix group (Figure 3). In fact, within the Prochlorothrix group sequences of the 16S rRNA gene are 98–99% similar.
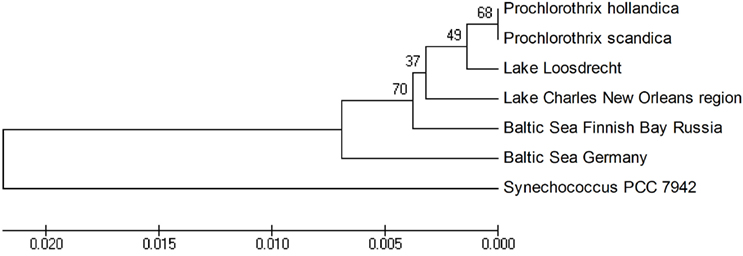
Figure 3. Unrooted dendrogram of a 375-bp fragment of Prochlorothrix 16S rDNA sequences obtained by the UPGMA method using MEGA version 4 (Tamura et al., 2007). Bootstrap values based on 1,000 replications are shown at the nodes.
In seems that the strains of Prochlorothrix detected in brackish coastal waters are merely temporary invaders from the mainland, and they could even sporadically occur in open waters of the Baltic Sea (Geiß et al., 2003). At the same time, there is no indication of their presence in any oligotrophic lake.
Functional Ecology
Filamentous green cyanobacteria never dominate the phytoplankton of small polymictic lakes. Growth rate of natural populations of P. hollandica is usually 30–40% that of Oscillatoria-like blue–green cyanobacteria (0.02–0.14 day−1; see Pel et al., 2004). This is possibly the reason why these blue–green cyanobacteria dominate and even suppress green cyanobacteria in natural environment.
Primary Productivity
The biomass of Prochlorothrix which reaches its maximum during the bloom season can be used as a principal nutrient by the zooplankton. In particular, the number of Euchlanis dilatata (Rotatoria, Nemathelminthes) which is among the main primary suppliers in the Loosdrecht Lakes (Gulati et al., 1993), is increased in late summer and in early spring which coincides with mass development of Prochlorothrix. The intensity of Prochlorothrix biomass consumption by this invertebrate is commeasurable with that of the blue–green cyanobacterium Aphanizomenon flos-aquae (Gulati et al., 1993).
Photoacclimation
Genes pcbABC in P. hollandica constitute an operon (Nikolaitchik and Bullerjahn, 1998; van der Staay et al., 1998). The antenna proteins pcbAB selectively associate with PSII, while the antenna protein PcbC is attached to PSI (Bumba et al., 2005). Photosystems I and II in P. hollandica have the peripheral antennae which produce a 18-meric ring around the PSI trimer and a 14-meric ring around the PSII dimer (Boichenko et al., 2007). Prochlorothrix hollandica is the only green cyanobacterium in which a shift of photosynthetic States 1 ↔ 2 (i.e., preferential light channeling) was demonstrated. However, the details of the structural/functional association between the chl a/b antenna with photosystems remain obscure. According to one of the two conflicting models, in State 1 the main chl a/b antenna (which consists of proteins PcbB and PcbC) transiently associates both PS. In State 2, the minor chl a/b antenna (which consists of PcbA protein) dissociates from the super complex of the main chl a/b antenna and PSII (Post et al., 1992). In another model (van der Staay and Staehelin, 1994), in State 1 the stacked thylakoids are enriched with the super complex of chl a/b antenna and PSII, whereas unstacked thylakoids contain PSI.
Later experimental data suggest a compromise model (Partensky and Garczarek, 2003) based on the spatial redistribution of (de)phosphorylated Pcb proteins.
Temperature
The culture optimum growth of P. hollandica PCC 9006 and P. scandica NIVA-8/90 is 20 and 24°C respectively (Pinevich et al., 1999; Skulberg, 2008). However, in nature they were shown to grow at lower temperatures. Compared to other green cyanobacteria, Prochlorothrix occupies a narrow geographic range characterized by significant seasonal changes in temperature, as well as by the episodic formation of ice cover in winter time. For instance in July 1984 when P. hollandica PCC 9006 was discovered in Lake Loosdrecht the water temperature was 18°C (Burger-Wiersma et al., 1989), whereas in April 1998 it was only 8°C (Zwart et al., 2005). As a rule, mass development of Prochlorothrix in this lake occurs in the end of summer when water temperature is 15–18°C (Post and Bullerjahn, 1994).
Water sampling from Lake Mälaren, where P. scandica NIVA-8/90 was detected was performed in October 1990. During this period, the temperature is 10–13°C throughout the water column due to the active mixing (Skulberg, 2008).
Salinity Response
Cyanobacteria of the genus Prochlorothrix are traditionally considered to inhabit freshwater pools, and they are sensitive even to a small increase in salinity. Prochlorothrix hollandica PCC 9006 demonstrated poor growth in the presence of 25 mM NaCl, and the growth was fully suppressed at 100 mM or after the addition of sea water equivalent (Burger-Wiersma et al., 1989). Our data on the comparative salt resistance in P. hollandica PCC 9006 and P. scandica NIVA-8/90 confirm the inhibitory effect of 50–100 mM NaCl (Pinevich et al., unpublished data). Increased salinity provoked morphological changes in both species: cells were elongated, and the filaments became zigzag. At >200 mM NaCl, the filaments of both species degraded after a week growth.
Although high sensitivity to salt can be easily explained by the inability to synthesize osmotic protectors, salt adaptation may depend on the local ion climate. In particular, P. hollandica was resistant to 170 mM NaCl when grown in a semi-synthetic medium supplied with water from the estuary Darss-Zingst. Under these conditions, an increase in the cellular content of sucrose was registered (Bergmann et al., 2008). This data are in agreement with the detection of Prochlorothrix-like marker genes in the phytoplankton of the Baltic Sea open waters (Geiß et al., 2003).
Involvement in Symbioses
In contrast to the rest of green cyanobacteria, the strains of Prochlorothrix are easily cultured on liquid mineral media (see below). However, most attempts to obtain pure cultures on solid medium proved unsuccessful. Cultures ceased to grow, when attempts to get rid of heterotrophic contaminants were made by the authors. FISH hybridization with taxon-specific probes EUB338II, EUB338III, ALF968, BET42a, GAM42a, and DELTA495a revealed a vast diversity of heterotrophic bacteria associated with P. hollandica PCC 9006 and P. scandica NIVA-8/90. In case of P. hollandica PCC 9006, a positive signal was obtained with probes specific to alpha-, beta-, and gammaproteobacteria. On the other hand, heterotrophic epiphytes of P. scandica NIVA-8/90 belong to alpha- and betaproteobacteria (Velichko et al., unpublished data).
These data, along with the fact that bacteria-free filaments of P. hollandica PCC 9006 demonstrated negligible growth in liquid culture (Schyns et al., 1997), indicate a mutually favorable consortium-type association between the filamentous green cyanobacteria and their heterotrophic satellites.
Cultivation
Media and Strains
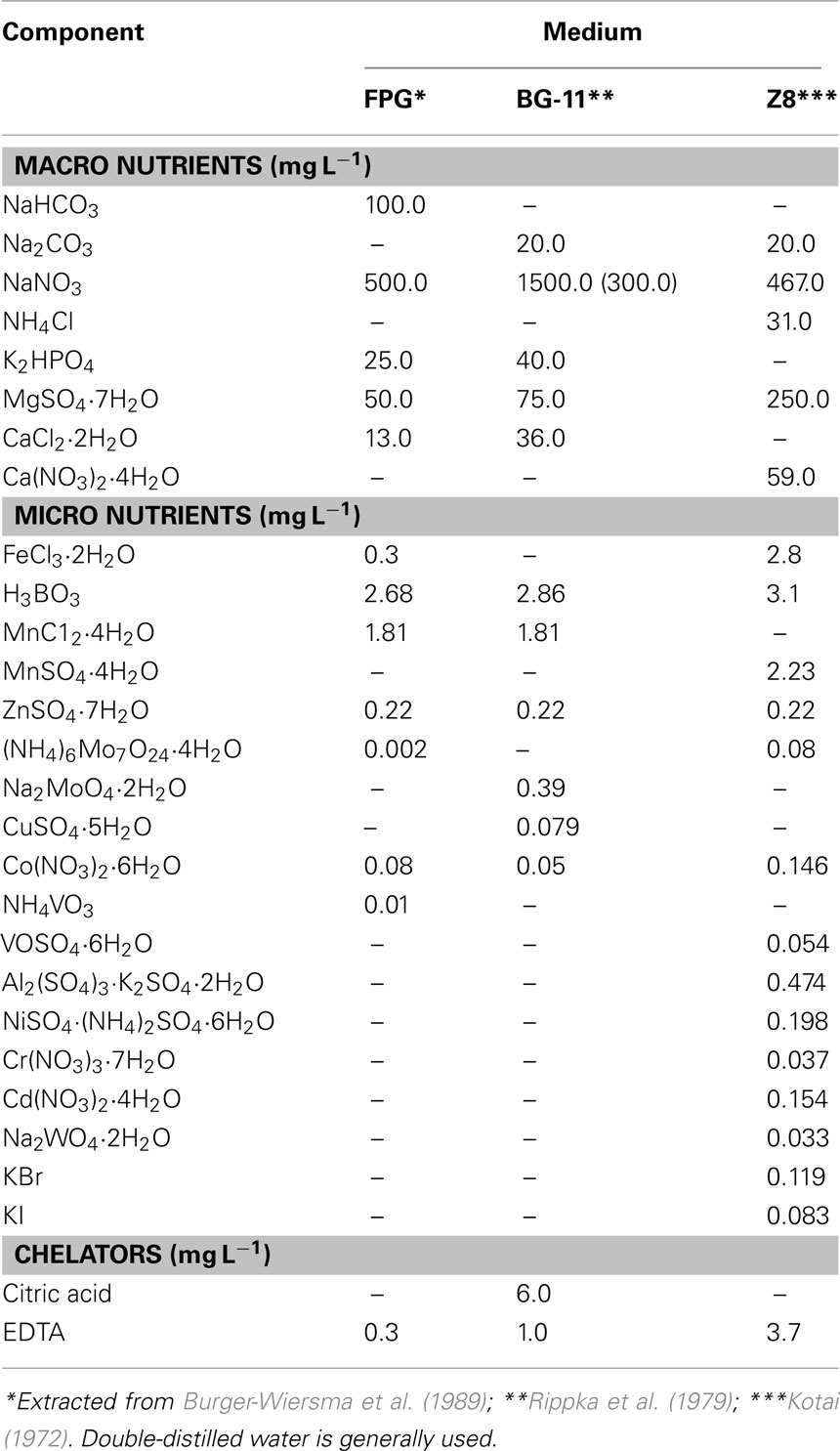
Cyanobacteria of the genus Prochlorothrix are grown in mineral media FPG, BG-11, and Z8 which contain relatively low amounts of inorganic salts and are routinely used for the cultivation of freshwater blue–green cyanobacteria (Table 3). Double-distilled water is necessary. Otherwise, unidentified contaminating chemicals inhibit growth. For slant cultivation, semisolid agar (≤0.5%) is recommended, since ≥1% inhibits growth. In addition to double-distilled water and mineral salts of high quality, purified agar (e.g., Difco) is needed. Cell growth is suppressed by elevated concentrations of NaCl, and the degree of salt resistance depends on ion content of the medium (Burger-Wiersma et al., 1989; Bergmann et al., 2008). Since Prochlorothrix sp. is unable to fix nitrogen even in microaerobic conditions, an addition of combined nitrogen is necessary.
In collections Prochlorothrix sp. is represented by several cultured strains. One of them, isolated in the Netherlands and validly described as P. hollandica (Burger-Wiersma et al., 1989), is maintained under different strain identifiers (CALU 1027, CCAP 1490/1, NIVA-5/89, PCC 9006, and SAG 10.89). Several multixenic strains isolated in Sweden are stored in the Norwegian Institute for Water Research (Oslo, Norway) collection, namely NIVA 8/90, 9/90, and 16/90 (Skulberg, 2008). Strain P. hollandica PCC 9006 (CALU 1027) and strain P. scandica NIVA-8/90 (CALU 1205) are stored at the collection of St. Petersburg University (St. Petersburg, Russia). Various methods of axenization were attempted including streaking, repeated washing, and micromanipulation, as well as differential sensitivity to antibiotics (Burger-Wiersma et al., 1989; Pinevich et al., 1999; Skulberg, 2008). All of them proved unsuccessful because of the loss of viability in both liquid and solid medium. In the latter case, filaments grow not as colonies, but rather as films surrounded by dim layer of colorless heterotrophic bacteria.
Growth Parameters
Batch cultures of P. hollandica PCC 9006 in FPG medium at 40 μE m−2 s−1 and 20°C grew at a maximum rate of 0.02 h−1 (Burger-Wiersma et al., 1989). When the BG-11 medium was used, grow rate varied from 0.007 h−1 (Burger-Wiersma et al., 1989) to 0.026 h−1 (Bergmann et al., 2008). Optimum growth temperature for P. hollandica PCC 9006 is 20°C (Burger-Wiersma et al., 1989; Pinevich et al., 1999), while that for P. scandica is 24°C (Pinevich et al., 1999). Stirring or air bubbling failed to stimulate growth and even resulted in decreased viability due to mechanical damage of the filaments.
Morphology
Immotile, non-branching filaments of Prochlorothrix sp. grow by equal binary fission of intercalary cells and multiply by fortuitous fragmentation. Hormogonia or other types of differentiated structures are never observed.
Filaments of P. hollandica PCC 9006 and P. scandica NIVA-8/90 (Figure 1) are ≤250 μm long. Mean cell size of P. hollandica PCC 9006 is 11.8 ± 0.9 × 1.6 ± 0.1 μm (length vs width, 7.4:1), while that of P. scandica NIVA-8/90 being 7.4 ± 0.7 × 2.1 ± 0.1 μm (3.5:1). In both cases, the width of cell-to-cell junction is 1/8–1/5 of filament diameter. In this respect, the filaments of Prochlorothrix more closely resemble those of Limnothrix than those of Pseudanabaena. In the latter case, the width of cell-to-cell junction is 1/2 of filament diameter (Castenholz, 2001).
Selected Features of Ultrastructure
Thylakoid Stacking
In addition to unstacked thylakoids which are typical of blue–green cyanobacteria, P. hollandica PCC 9006 contains stacked thylakoids vaguely resembling grana thylakoids in chloroplasts (Giddings et al., 1980; Schuster et al., 1984; Miller et al., 1988; Golecki and Jürgens, 1989). In both cases, membrane adhesion is maintained by the PSII proteins and chl a/b antenna proteins (van der Staay and Staehelin, 1994).
Thylakoids in freeze-fractured cells of P. hollandica PCC 9006 were shown to be laterally heterogeneous (van der Staay and Staehelin, 1994). Similarly to chloroplasts, stacked regions are enriched with globular particles which correspond to the super complexes of PSII with chl a/b antenna. By contrast, PSI and ATP synthase are preferentially localized in the unstacked regions (Schuster et al., 1984; van der Staay and Staehelin, 1994).
Membranes of stacked thylakoids in P. scandica NIVA-8/90 are covered with particles (viridosomes) which are 10.0 ± 2.3 nm diameter and are located with the periodicity of 20.0 ± 5.6 nm. These are suggested to correspond to the epimembrane domain of chl a/b antenna (Pinevich et al., 1999). The analogous particles (xanthosomes) are present in both the chromophyte algae (e.g., Bacillariophyceae and Phaeophyceae) and Dinozoa (Katoh et al., 1993; Green and Durnford, 1996).
Thylakoid stacking in Prochlorothrix may help redistribute excitation energy between the adjoining thylakoids (Dekker and Boekema, 2005). Alternatively, it may be a functionless result of segregation of PSII centers in thylakoid membranes (Bullerjahn and Post, 1993; see Functional Ecology and Photoacclimation).
Thylakoid Centers
An intriguing detail of ultrastructure in P. hollandica PCC 9006 and P. scandica NIVA-8/90, also rarely observed in blue–green cyanobacteria, is the thylakoid center. The latter corresponds to a ∼200-nm electron dense zone between cell membrane and two to four radiating thylakoids (Pinevich et al., 1996). These metastable structures possibly channel the molecules of chl-ide aGG (the precursor of chl a) from the sites of synthesis in cell membrane to the functional sites in thylakoids (Nierzwicki-Bauer et al., 1983; Bryant, 1986; Hinterstoisser et al., 1993).
Gas Vesicles
Similarly to many planktonic filamentous blue–green cyanobacteria, P. hollandica PCC 9006 contains honeycomb-like aggregates of gas vesicles. An individual vesicle is a cylinder of 50 nm diameter with conical ends. Its 125–265 nm length mirrors different stages of self-assembly (Golecki and Jürgens, 1989; Pinevich et al., 1999).
Carboxysomes
Similarly to blue–green cyanobacteria, the carboxysomes of P. hollandica PCC 9006 are enveloped in a non-unit membrane. However, the carboxysomes of P. scandica NIVA-8/90 lack this shell resembling the pyrenoid of green algae and higher plants (McKay and Gibbs, 1990; Morden and Golden, 1991; Pinevich et al., 1996). According to the immune electron microscopy data, all RuBisCO of P. hollandica PCC 9006 is associated with carboxysomes. Whereas, the RuBisCO in P. scandica NIVA-8/90 exists in both the immobilized and soluble form (Pinevich et al., 1996). Carboxysomes in Prochlorothrix were shown to contact thylakoids (Pinevich et al., 1999), which is not the case in blue–green cyanobacteria. This association may promote phosphorylation of ribulose-5-phosphate, as well as the activity of carbonic anhydrase (Pinevich et al., 1999).
Genetics
Genome size of Prochlorothrix is ∼5.5 Mb (Post and Bullerjahn, 1994; Schyns et al., 1997) which is within the range of most cyanobacteria. The G + C Mol% of genomic DNA is 51–53 (Herdman, 1981; Coleman and Lewin, 1983; Burger-Wiersma et al., 1989; Dufresne et al., 2003).
Genome Sequencing
Genome sequencing of P. hollandica PCC 9006 is currently performed by the authors. Automatic annotation using The RAST Server (Rapid Annotations using Subsystems Technology; Aziz et al., 2008) was performed on the draft sequence, and the manual curation of automatically annotated genes is being done. We are focusing primarily on photosynthetic genes. Phylogenetic affiliation for two of them, namely psbO and psaF, are given below (see Phylogeny).
Energy Assimilation
In contrast to unicellular green cyanobacteria, Prochlorothrix has not been tested for dark chemotrophy; the only studied mode of energy assimilation in this case is phototrophy.
Photosynthetic Pigments
Prochlorothrix hollandica PCC 9006 contains accessory chl b (Burger-Wiersma et al., 1989). It is suggested to be synthesized from chl b2 in one stage using 4-vinyl-reductase (Helfrich et al., 1999). The alternative way involves chl a oxygenase which was the enzyme also found in Prochloron didemni and higher plants (Tanaka et al., 1998; Tomitani et al., 1999; Nagata et al., 2005).
Among the carotenoids in P. hollandica PCC 9006 are β-carotene and zeaxanthin (van der Staay et al., 1992). The content of β-carotene in P. hollandica PCC 9006 is four times that of zeaxanthin. The former preferentially associates with PSI, while the latter complexes with 56/58 kDa proteins integrated in the cell membrane (Omata et al., 1985; Engle et al., 1991).
Light-Harvesting Antenna
In contrast to blue–green cyanobacteria, P. hollandica PCC 9006 and P. scandica NIVA 8/90 possess the light-harvesting antenna containing both chl a and chl b (Bryant, 1986; Burger-Wiersma and Post, 1989; Hess et al., 1996; Marquardt et al., 1998). Although it was originally thought that Pcb proteins of this antenna share the origin with CAB proteins in chloroplasts, in fact they have no similarity (La Roche et al., 1996; van der Staay et al., 1998). In particular, Pcb proteins of P. hollandica PCC 9006 do not react with antibodies raised against CAB proteins of green algae and higher plants (Hiller and Larkum, 1985; Bullerjahn et al., 1990; van der Staay and Staehelin, 1994). At the same time, Pcb proteins of P. hollandica PCC 9006 are similar to PsbB and PsbC (CP43) proteins of PSII core antenna. In addition, they share similarity with IsiA (CP43′) protein of PSI stress antenna which is formed in blue–green cyanobacteria under iron starvation (Bullerjahn et al., 1987, 1990; van der Staay and Staehelin, 1994; Nikolaitchik and Bullerjahn, 1998; van der Staay et al., 1998; Partensky et al., 1999). Hexadomain proteins Pcb/Cp43/IsiA vary in the length of the loop exposed at the thylakoid exoplasmic surface between helices V and VI (Herbstová et al., 2010).
Genes pcbABC of P. hollandica PCC 9006 form an operon (Nikolaitchik and Bullerjahn, 1998; van der Staay et al., 1998). The genes pcbA and pcbB genes belong to the evolutionary lineage of IsiA and code for polypeptides of 32 and 33 kDa. The third gene, pcbC, belongs to a separate lineage and codes for a polypeptide of 38 kDa (van der Staay and Staehelin, 1994; van der Staay et al., 1998; Garczarek et al., 2000).
Pcb proteins aggregate in multimeric super complexes (Bibby et al., 2001, 2003; Chen et al., 2002, 2005; Partensky and Garczarek, 2003) which surround the oligomers of PSI and PSII (Bibby et al., 2003; Boichenko et al., 2007). PcbA/PcbB proteins build up the mobile antenna of both photosystems, while the PcbC proteins associate with a portion of antenna tightly bound to PSI (Herbstová et al., 2010).
Photosystems
PSI polypeptides of P. hollandica PCC 9006 are the same as in blue–green cyanobacteria (Schuster et al., 1984; Bullerjahn et al., 1987; van der Staay et al., 1998; Garczarek et al., 2000). They include major PsaA/PsaB proteins (66 kDa) and 6 minor proteins PsaC–F, I–M (20 kDa; Burger-Wiersma and Post, 1989; van der Staay et al., 1993). PSII proteins of P. hollandica PCC 9006 include CP43/CP47, D1/D2, PsbH, and PsbO, as well as low molecular weight polypeptides PsbJ and PsbK (Greer, 1991; Greer and Golden, 1992; Mor et al., 1993; Post and Bullerjahn, 1994). Interestingly, psbA gene has a 3′-terminal 21-nucleotide deletion; the same deletion is present in the chloroplast ortholog (Morden and Golden, 1991).
The oxygen evolving complex (OEC) in P. hollandica PCC 9006 lacks proteins PsbP (17 kDa) and PsbQ (23 kDa) typical of chloroplasts (Mor et al., 1993). The interaction of OEC with Cl− and Ca2+ is provided by the hydrophobic Mn-stabilizing protein PsbO (37 kDa) different from the more amphiphilic analog in blue–green cyanobacteria (26.5 kDa; Mor et al., 1993; Barber and Kühlbrandt, 1999).
Facultative Anoxygenic Photosynthesis
Among green cyanobacteria, P. hollandica PCC 9006 is uniquely capable of photosynthesis with sulfide as electron donor and, respectively, without dioxygen production (Post and Arieli, 1997). OEC of this cyanobacterium retains activity at ≤1 mM sulfide. High resistance to sulfide stress is provided by the PsbO protein which protects the catalytic site of OEC (Mor et al., 1993; Post and Arieli, 1997). The transition to anoxygenic photosynthesis occurs at >1 mM sulfide when this substance inhibits both the biogenesis and activity of PSII, especially that of OEC.
A relative resistance to sulfide allows P. hollandica dominate the phytoplankton of water bodies with sulfide accumulating in warm seasons (Post and Arieli, 1997). Given the trophic state of shallow waters harboring Prochlorothrix, sulfide production in the anoxic sediments may mix with surface waters. Thus, sulfide resistance may be an essential adaptation in this habitat.
Substrate Assimilation and Enzymology
Carbon Assimilation
Data on the ability to use organic substrates (photomixotrophy or photoheterotrophy, in the light; chemoheterotrophy, in darkness) in Prochlorothrix is absent. Similarly to most blue–green cyanobacteria, Prochlorothrix demonstrates photoautoptrophy based on Calvin cycle with RuBisCO as key enzyme. Prochlorothrix hollandica PCC 9006 contains IB-subform of RuBisCO which clusters together with RuBisCO of blue–green β-cyanobacteria from the so-called “green” phylogenetic branch of the enzyme (Morden and Golden, 1991; Shimada et al., 1995; Chen et al., 2004; Badger et al., 2006).
RuBisCO of P. hollandica PCC 9006 is encoded by the dicistronic operon rbcLS which consists of genes for the large (L, 57 kDa) and small (S, 13 kDa) subunits, respectively. The holoenzyme (560 kDa) belongs to the L8S8-type (Hawthornthwaite et al., 1990; Morden and Golden, 1991). Enzymatic activity is maximum at 30°C and pH 8; Mg2+ is used as a cofactor (Hawthornthwaite et al., 1990).
Nitrogen Assimilation
There are no data on the ability of P. hollandica PCC 9006 for aerobic nitrogen fixation. Initial attempts to induce nitrogenase under the conditions of nitrogen starvation proved unsuccessful (Burger-Wiersma et al., 1989). Furthermore, there are no nif genes sequences in the partially reconstructed genome of this cyanobacterium (Pinevich et al., unpublished data).
At the same time, P. hollandica PCC 9006 assimilates both ammonium and nitrate. Maximum growth rate in culture conditions is observed at 0.75 mM NH4+, whereas growth is inhibited at 3 mM. Growth is also absent if 1.5 mM urea substitutes for nitrate, indicating lack of the urease enzyme (Burger-Wiersma et al., 1989).
Phosphorus Assimilation
According to field experiments, the most extensive growth of P. hollandica PCC 9006 takes place in those parts of water bodies where phytoplankton is limited by phosphorus deficiency (Burger-Wiersma et al., 1989). Prochlorothrix hollandica PCC 9006 is characterized by higher maximum rate of phosphorus uptake and by lower saturation constant for this substrate as compared to other filamentous cyanobacteria, in particular Planktothrix sp. (Ducobu et al., 2002; Dignum et al., 2005).
Special enzymology: RNAse P
This enzyme performs the maturation of 5′-end of tRNA and is ubiquitous in prokaryotes including plastids and mitochondria. Prochlorothrix hollandica PCC 9006 contains a rare type of this RNAse characterized by long domain P15/16, as well as by a GGU motif which binds 3′-CCA of pre-tRNA and is absent from the ribozymes of other cyanobacteria (Fingerhut and Schön, 1998).
Phylogeny
Tracing phylogeny of green cyanobacteria raises the questions regarding: (i) relations within the green cyanobacteria group; (ii) relations between green and blue–green cyanobacteria; (iii) relations between green cyanobacteria and simple plastids; (iv) the ancestor of green cyanobacteria; and (v) the origin of chlorophyll light-harvesting antenna.
Here, two phylogenies are involved. One of them is the general evolution of cyanobacteria reconstructed by the comparative analysis of the major informational (in particular, 16S rNNA gene and rpoS) and photosynthetic genes (in particular, psbA and petE). Secondly, it is the evolution of green cyanobacteria traced by the analysis of the pcb genes coding for the chlorophyll light-harvesting antenna.
16S rRNA Gene
According to this evolutionary criterion, green cyanobacteria are polyphyletic (Urbach et al., 1992). Paradoxically, P. hollandica PCC 9006 clusters close to the unicellular strain Synechococcus PCC 6301 (Pinevich et al., 2010). Detailed dendrograms showing the distribution of green cyanobacteria between separate clades of cyanobacterial tree, as well as mutual subordination of strains from the same genus, are presented elsewhere (Turner, 1997; Moore et al., 1998; Partensky et al., 1999; Miyashita et al., 2003; Partensky and Garczarek, 2003; Rocap et al., 2003; de los Rios et al., 2007; Münchhoff et al., 2007; Scanlan et al., 2009).
RpoC Gene
Another informational gene which helps reconstruct the evolutionary history of cyanobacteria is rpoC1 coding for a subunit of the DNA dependent RNA polymerase. Here, again, green cyanobacteria were shown to be polyphyletic, and P. hollandica PCC 9006 groups with Synechococcus PCC 6301 (Palenik and Haselkorn, 1992).
The phylogenies of the major photosynthesis genes [psaF, coding for the conservative PsaF protein, a 21-kDa three-domain component of the reaction center complex; psbA, coding for the 32 kDa core PSII protein; psbO, coding for the Mn-stabilizing protein PsbO; and petE, coding for the copper-containing soluble protein plastocyanin (∼11 kDa), which donates electrons to PSI] repeat that of the 16S rRNA gene with green cyanobacteria being polyphyletic and P. hollandica PCC 9006 clustering with Synechococcus PCC 6301 (Figures 4 and 5; Pinevich et al., 2010).
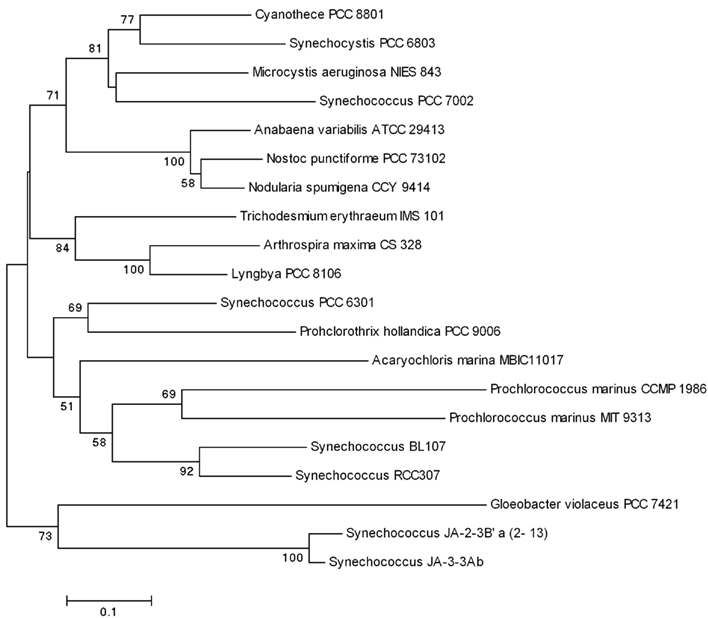
Figure 4. Phylogenetic relationship of the PsaF protein constructed by neighbor-joining method using MEGA version 4 (Tamura et al., 2007). Selected bootstrap values (greater than 50%) based on 1,000 replications are shown at nodes.
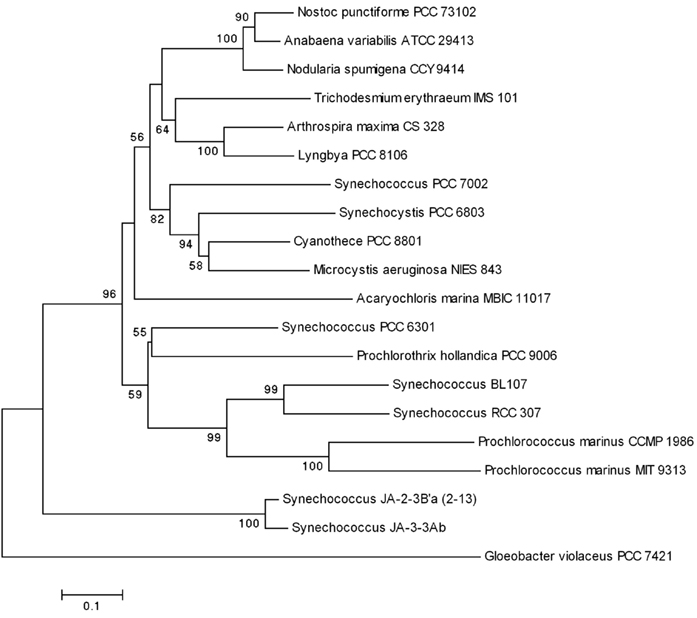
Figure 5. Phylogenetic relationship of PsbO protein constructed by neighbor-joining method using MEGA version 4 (Tamura et al., 2007). Selected bootstrap values (greater than 50%) based on 1,000 replications are shown at nodes.
Pcb Genes
The pcbABC genes of P. hollandica PCC 9006 which code for the antenna proteins of family Pcb/IsiA/CP43 cluster together with their counterparts in unicellular green cyanobacteria from the genera Acaryochloris and Prochloron (Pinevich et al., 2010). The highly conserved proteins of Pcb/IsiA/CP43 family are thought to be inherited from a common ancestor of cyanobacteria. They might evolve according to two scenarios.
According to the first scenario, these proteins may have appeared in Synechococcus-like ancestor of P. marinus which inhabited oceanic systems poor in iron (Ting et al., 2002). In these stress conditions, they constitutively expressed both the IsiA (CP43′) protein and the phycobilisome. Among the descendants of this hypothetical cyanobacterium were phycoerythrin-rich strains of Synechococcus, as well as phycobiliprotein free or phycobiliprotein-poor (phycobilisome free) strains of P. marinus.
By the second scenario, pcb and isiA genes evolved independently by duplication and subsequent specialization of the copied psbC-like gene in a protocyanobacterium (Chen et al., 2005). This scenario is more plausible since it explains the existence of a set of chl-binding proteins in blue–green and green cyanobacteria.
Both scenarios assume a transient coexistence of the chl-based antenna with the phycobiliprotein-based antenna (Shimada et al., 2003).
Genes Coding for Enzymes of the Chromophore Biosynthesis
The evolution of these genes is distinct from that of the pcb genes. In fact, presence of the conservative His residues in Pcb proteins is only a prerequisite of chl binding. In other words, the formation of chl-containing antennae, in particular the chl a/b antenna, needs biosynthetic enzymes for the respective chromatophores. Since the ancestor of Pcb proteins is thought to be chl a-binding protein (see above), the incorporation of chl b, a2, b2, and d in antennae of different green cyanobacteria suggests the formation of these pigments as byproducts in the metabolic pathway of chl a. In fact, monovinyl chls are synthesized from divinyl precursors by reductases, whereas chl b and chl d are formed by oxygenases (Beale, 2008).
The cao genes coding for chl a oxygenase in P. hollandica PCC 9006 and P. didemni are highly similar (Tomitani et al., 1999). Correspondingly, these green cyanobacteria did not acquire this gene independently, but rather the ability to synthesize chl b was a trait of common ancestor of cyanobactetria.
The evolutionary loss of phycobilins resulted in the appearance of green cyanobacteria like P. hollandica PCC 9006 and P. didemni, while the loss of chl b resulted in the lineage of blue–green cyanobacteria, which allows to consider both these green cyanobacteria as a model for the protocyanobacterium (Tomitani et al., 1999).
Relation of Green Cyanobacteria to the Origin of the Simple Chloroplast
Although cyanobacterial origin of the simple chloroplast is beyond doubt (Morden et al., 1992; McFadden, 1996), the specific nature of ancestry is debatable.
The Prochloron-type green cyanobacteria were rejected as a candidate, since Pcb proteins are dissimilar with the members of CAB family (La Roche et al., 1996; van der Staay et al., 1998). This conclusion prematurely deprived green cyanobacteria of any role in the origin of the simple chloroplast.
However, it should be noted that Pcb proteins cluster with PsbC/IsiA(CP43′) proteins (Bullerjahn et al., 1987, 1990; van der Staay and Staehelin, 1994; Nikolaitchik and Bullerjahn, 1998; van der Staay et al., 1998; Partensky et al., 1999). This finding, as well as data on the relationships between green cyanobacteria and their blue–green relatives (see above) help restore the role of the former as both the ancestor of the chloroplast, as well as the ancestor of oxygenic phototrophic bacteria.
Chloroplast Origins
The ancestor of chloroplasts is hypothesized to possess both the pcb genes, coding for chl-binding antenna proteins, and the pbp genes coding for phycobilisome (Tomitani et al., 1999). This cyanobacterium, engulfed by a non-photosynthetic protist, became a precursor of three lineages of the simple chloroplasts (Figure 6).
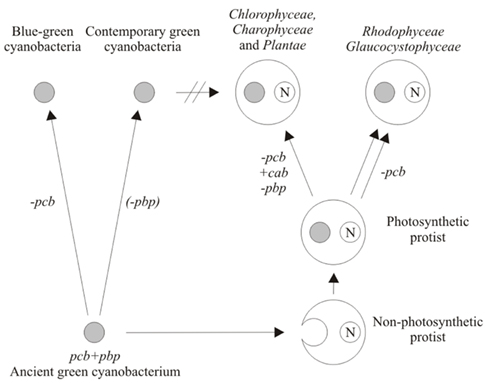
Figure 6. A possible phylogenetic scenario for the origin of green cyanobacteria, blue–green cyanobacteria, and simple plastids. See text for the explanation. N denotes cell nucleus.
One lineage is represented by the chloroplast of Chlorophyceae/Charophyceae/Plantae which lost the ancestral pcb and pbp genes. The loss of “old” chl a/b-binding Pcb proteins was compensated by the acquisition of convergent “new” chl a/b-binding CAB proteins.
Two other lineages are represented by the chloroplasts of Glaucocystophyceae and Rhodophyceae which lost the ancestral pcb genes but retained the ancestral pbp genes.
Cyanobacterial Origins
Cyanobacteria are hypothesized to share the ancestor with chloroplasts, i.e., the primordial cyanobacterium had both the pcb genes, coding for chl-binding antenna proteins, and the pbp genes coding for phycobilisome (Tomitani et al., 1999). This cyanobacterium became a precursor of two lineages of cyanobacteria (Figure 6).
One lineage, resulting from the loss of ancestral pcb genes is represented by blue–green cyanobacteria. The other lineage originating from the complete or partial loss of ancestral pbp genes is represented by green cyanobacteria. Noteworthy, the latter were not chloroplast ancestors.
Interestingly, the genes responsible for chl b synthesis were divergently inherited by green cyanobacteria and chloroplasts.
General Conclusion
The primordial cyanobacterium which possessed both the pcb and pbp genes is by definition a green cyanobacterium Therefore, the ancient green bacteria should be reconsidered as key players of the evolutionary process which resulted in the biodiversity of cyanobacteria and plastids.
Concluding Remarks
Cyanobacteria of the genus Prochlorothrix, in contrast to the other cyanobacteria, are both very rare and largely incapable of axenic growth. They combine trivial features (primarily, oxygenic photosynthesis) with specific, yet obscure aspects of homeostasis and competitive potential. The analysis of their environmental and symbiotic ecology can help better understand this uniqueness. Thereby, the complete annotation of P. hollandica PCC 9006 genome is important, as well as search and isolation of new strains of the genus.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Svetlana Averina and Dr. Olga Gavrilova for fruitful collaboration. Our thanks are due toYaroslav Borisov who participated in the annotation of P. hollandica PCC 9006 genes.
Footnote
† ^ The GenBank accession number for the 16S rRNA gene sequence of Prochlorothrix sp. NIVA-8/90 is HQ316169. The accession number for the sequences of the single 16S–23S-ITS, psbO and PsaF genes in P. hollandica PCC 9006 are JQ739143, JQ739144, and HQ316172 respectively. The accession numbers for the sequences of two 16S–23S-ITS in Prochlorothrix sp. NIVA-8/90 are HQ316170 and HQ316171 respectively. The accession number for the partial 16S rRNA sequence of the uncultured Prochlorothrix of the Finnish Bay is JQ739142.
References
Amaral-Zettler, L. A., Rocca, J. D., Lamontagne, M. G., Dennett, M. R., and Gast, R. J. (2008). Changes in microbial community structure in the wake of Hurricanes Katrina and Rita. Environ. Sci. Technol. 42, 9072–9078.
Aziz, R. K, Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., Formsma, K., Gerdes, S., Glass, E. M., Kubal, M., Meyer, F., Olsen, G. J., Olson, R., Osterman, A. L., Overbeek, R. A., McNeil, L. K., Paarmann, D., Paczian, T., Parrello, B., Pusch, G. D., Reich, C., Stevens, R., Vassieva, O., Vonstein, V., Wilke, A., and Zagnitko, O. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75.
Badger, M. R., Price, G. D., Long, B. M., and Woodger, F. J. (2006). The environmental plasticity and ecological genomics of the cyanobacterial CO2 concentrating mechanism. J. Exp. Bot. 57, 249–265.
Beale, S. I. (2008). Photosynthetic pigments: perplexing persistent prevalence of “superfluous” pigment production. Curr. Biol. 18, 342–343.
Bergmann, I., Geiß-Brunschweiger, U., Hagemann, M., and Schoor, A. (2008). Salinity tolerance of the chlorophyll b-synthesizing cyanobacterium Prochlorothrix hollandica strain SAG 10.89. Microbiol. Ecol. 55, 685–696.
Bibby, T. S., Nield, J., and Barber, J. (2001). Three-dimensional model and characterization of the iron stress-induced CP43’-photosystem I supercomplex isolated from the cyanobacterium Synechocystis PCC 6803. J. Biol. Chem. 276, 43246–43252.
Bibby, T. S., Nield, J., Chen, M., Larkum, A. W., and Barber, J. (2003). Structure of a photosystem II supercomplex isolated from Prochloron didemni retaining its chlorophyll a/b light-harvesting system. Proc. Natl. Acad. Sci. 100, 9050–9054.
Boichenko, V. A., Pinevich, A. V., and Stadnichuk, I. N. (2007). Association of chlorophyll a/b-binding Pcb proteins with photosystems I and II in Prochlorothrix hollandica. Biochim. Biophys. Acta 1767, 801–806.
Bryant, D. A. (1986). The cyanobacterial photosynthetic apparatus: comparison to those of higher plants and photosynthetic bacteria. Can. Bull. Fish. Aquat. Res. 214, 423–500.
Bullerjahn, G. S., Jensen, T. S., Sherman, D. M., and Sherman, L. A. (1990). Immunological characterization of the P proteins from chlorophyll a/b antenna in Prochlorothrix hollandica and Prochloron sp. FEMS Microbiol. Lett. 67, 99–106.
Bullerjahn, G. S., Matthijs, H. C. P., Mur, L. R., and Sherman, L. A. (1987). Chlorophyll-protein composition of the thylakoid membrane from Prochlorothrix hollandica, a prokaryote containing chlorophyll b. Eur. J. Biochem. 168, 295–300.
Bullerjahn, G. S., and Post, A. F. (1993). The prochlorophytes: are they more than just chlorophyll a/b cyanobacteria? CRC Crit. Rev. Microbiol. 19, 43–59.
Bumba, L., Prasil, O., and Vacha, F. (2005). Antenna ring around trimeric photosystem I in chlorophyll b containing cyanobacterium Prochlorothrix hollandica. Biochim. Biophys. Acta 1708, 1–5.
Burger-Wiersma, T., and Post, A. F. (1989). Functional analysis of the photosynthetic apparatus of Prochlorothrix hollandica (Prochlorales), a chlorophyll b containing prokaryote. Plant Physiol. 91, 770–774.
Burger-Wiersma, T., Stal, L. J., and Mur, L. R. (1989). Prochlorothrix hollandicagen. nov., sp. nov., a filamentous oxygenic photoautotrophic procaryote containing chlorophylls a and b: assignment to Prochlovotrichaceae fam. nov. and order Prochlorales Florenzano, Balloni, and Materassi 1986, with emendation of the ordinal description. Int. J. Syst. Bacteriol. 39, 250–257.
Castenholz, R. W. (2001). “Phylum BX Cyanobacteria,” in Bergey’s Manual of Systematic Bacteriology, eds D. R. Boone, R. W. Castenholz, and G. M. Garrity (New York: Springer-Verlag), 473–599.
Castiglioni, B., Rizzi, E., Frosini, A., Sivonen, K., Rajaniemi, P., Rantala, A., Mugnai, M. A., Ventura, S., Wilmotte, A., Boutte, C., Grubisic, S., Balthasart, P., Consolandi, C., Bordoni, R., Mezzelani, A., Battaglia, C., and De Bellis, G. (2004). Development of a universal microarray based on the ligation detection reaction and 16S rRNA gene polymorphism to target diversity of Cyanobacteria. Appl. Environ. Microbiol. 70, 7161–7172.
Chen, F., Wang, K., Kan, J., Suzuki, M. T., and Wommack, K. E. (2004). Phylogenetic diversity of Synechococcus in the Chesapeake Bay revealed by ribulose-1,5-bisphosphate carboxylase-oxygenase (RuBisCO) large subunit gene (rbcL) sequences. Aquat. Microb. Ecol. 36, 153–164.
Chen, M., Hiller, R. G., Howe, C. J., and Larkum, A. W.D. (2005). Unique origin and lateral transfer of prokaryotic chlorophyll-b and chlorophyll-d light-harvesting systems. Mol. Biol. Evol. 22, 21–28.
Chen, M., Quinnell, R. G., and Larkum, A. W. D. (2002). The major light-harvesting pigment protein of Acaryochloris marina. FEBS Lett. 514, 149–152.
Chisholm, S. W., Olson, R. J., Zettler, E. R., Goericke, R., Waterbury, J. B., and Welschmeyer, N. A. (1988). A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature 334, 340–343.
Cohen, Z., Margheri, M., and Tomaselli, L. (1995). Chemotaxonomy of cyanobacteria. Phytochemistry 40, 1155–1158.
Coleman, A. W., and Lewin, R. A. (1983). The disposition of DNA in Prochloron (Prochlorophyta). Phycologia 22, 209–212.
Dekker, J. P., and Boekema, E. J. (2005). Supramolecular organization of thylakoid membrane proteins in green plants. Biochim. Biophys. Acta 1706, 12−–39.
de los Rios, A., Grube, M., Sancho, L. G., and Ascaso, C. (2007). Ultrastructural and genetic characteristics of endolithic cyanobacterial biofilms colonizing Antarctic granite rocks. FEMS Microbiol. Ecol. 59, 386–395.
Dignum, M., Matthijs, H. C. P., Pel, R., Laanbroek, H. J., and Mur, L.R. (2005). “Nutrient limitation of freshwater cyanobacteria,” in Harmful Cyanobacteria, eds J. Huisman, H. C. P. Matthijs, and P. M. Visser (Dordrecht: Kluwer Academic Publishers), 65–86.
Ducobu, H., Huisman, J., Jonker, R. R., and Mur, L. R. (2002). Competition between a prochlorophyte and a cyanobacterium under various phosphorus regimes: comparison with the Dropp model. J. Phycol. 34, 467–476.
Dufresne, A., Salanoubat, M., Parnensky, F., Artiguenave, F., Axmann, I. M., Barbe, V., Duprat, S., Galperin, M. Y., Koonin, E. V., Le Gall, F., Makarova, K. S., Ostrowski, M., Oztas, S., Robert, C., Rogozin, I. B., Scanlan, D. J., Tandeau de Marsac, N., Weissenbach, J., Wincker, P., Wolf, Y. I., and Hess, W. R. (2003). Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl. Acad. Sci. U.S.A. 100, 10020–10025.
Engle, J. M., Burkhart, W., Sherman, D. M., and Bullerjahn, G. S. (1991). Purification and characterization of a surface-associated carotenoid-binding complex from the photosynthetic prokaryote, Prochlorothrix hollandica. Arch. Microbiol. 155, 453–458.
Fingerhut, C., and Schön, A. (1998). Sequence and functional characterization of RNAse P RNA from the chl a/b containing cyanobacterium Prochlorothrix hollandica. FEBS Lett. 428, 161–164.
Garczarek, L., Hess, W. R., Holzendorff, J., van der Staay, G. W. M., and Partensky, F. (2000). Multiplication of antenna genes as a major adaptation to low light in a marine prokaryote. Proc. Natl. Acad. Sci. U.S.A. 97, 4098–4101.
Geiß, U., Bergmann, I., Blank, M., Schumann, R., Hagemann, M., and Schoor, A. (2003). Detection of Prochlorothrix in brackish waters by specific amplifications of pcb genes. Appl. Environ. Microbiol. 69, 6243–6249.
Geiß, U., Selig, U., Schumann, R., Steinbruch, R., Bastrop, R., Hagemann, M., and Schoor, A. (2004). Investigations on cyanobacterial diversity in a shallow estuary (Southern Baltic Sea) including genes relevant to salinity resistance and iron starvation acclimation. Environ. Microbiol. 6, 377–387.
Giddings, T. H, Withers, N. W., and Staehelin, L. A. (1980). Supramolecular structure of stacked and unstacked regions of the photosynthetic membranes of Prochloron sp., a prokaryote. Proc. Natl. Acad. Sci. U.S.A. 77, 352–356.
Golecki, J. R., and Jürgens, U. J. (1989). Ultrastructural studies on the membrane systems and cell inclusions of the filamentous prochlorophyte Prochlorothrix hollandica. Arch. Microbiol. 152, 77–82.
Green, B. R., and Durnford, D. G. (1996). The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 685–714.
Greer, K. L. (1991). Nucleotide sequence of psbB from Prochlorothrix hollandica. Plant Mol. Biol. 17, 915–917.
Greer, K. L., and Golden, S. S. (1992). Conserved relationship between psbH and petBD genes: presence of a shared upstream element in Prochlorothrix hollandica. Plant Mol. Biol. 19, 355–365.
Gulati, R. D., Ejsmont-Karabin, J., and Postema, G. (1993). Feeding in Euchlanis dilatata lucksiana Hauer on filamentous cyanobacteria and a prochlorophyte. Hydrobiologia 255/256, 269–274.
Hawthornthwaite, A. M., Beanland, T. J., Howe, C. J., and Codd, G. A. (1990). The ribulose bisphosphate carboxylase/oxygenase of Prochlorothrix hollandica: purification, subunit structure and partial N-terminal sequence analysis of the large subunit. Z. Naturforsch. 45S, 733–738.
Helfrich, M., Ross, A., and King, G. C. (1999). Identification of 8-vinyl-prochlorophyllide a in phototrophic prokaryotes and algae: chemical and spectroscopic properties. Biochim. Biophys. Acta. 1410, 262–272.
Herbstová, M., Litvin, R., Gardian, Z., Komenda, J., and Vácha, F. (2010). Localization of Pcb antenna complexes in the photosynthetic prokaryote Prochlorothrix hollandica. Biochim. Biophys. Acta 1797, 89–97.
Herdman, M. (1981). Deoxyribonucleic acid base composition and genome size of Prochloron. Arch. Microbiol. 129, 314–316.
Hess, W. R., Parnensky, F., van der Staay, G. W. M., Garcia-Fernandez, J. M., Börner, T., and Vaulot, D. (1996). Coexistence of phycoerythrin and chlorophyll a/b antenna in a marine prokaryote. Proc. Natl. Acad. Sci. U.S.A. 93, 11126–11130.
Hiller, R. G., and Larkum, A. W. D. (1985). The chlorophyll-protein complexes of Prochloron sp. Prochlorophyta). Biochim. Biophys. Acta 806, 107–115.
Hinterstoisser, B., Cichna, M., Kunter, O., and Peschek, G. A. (1993). Cooperation of plasma and thylakoid membranes for the biosynthesis of chlorophyll in cyanobacteria: the role of the thylakoid centers. J. Plant Physiol. 142, 407–413.
Katoh, T., Tanaka, A., and Mimuro, M. (1993). Xanthosomes: supramolecular assemblies of xanthophyll-chlorophyll a/c-protein complexes. Meth. Enzymol. 214, 402–412.
Kotai, J. (1972). Instructions for Preparation of Modified Nutrient Solution Z8 for Algae. Oslo: Norwegian Institute for Water Research, B11–B69.
Laloui, W., Palinska, K., Rippka, R., Partensky, F., De Marsac, N. T., Herdman, M., and Iteman, I. (2002). Genotyping of axenic and non-axenic isolates of the genus Prochlorococcus and the OMGF-“Synechococcus” clade by size, sequence analysis or RFLP of the Internal Transcribed Spacer of the ribosomal operon. Microbiology 148, 453–465.
La Roche, J., van der Staay, G. W. M., Partensky, F., Ducret, A., Aebersold, R., Li, R., Golden, S. S., Hiller, R. G., Wrench, P. M., Larkum, A. W., and Green, B. R. (1996). Independent evolution of the prochlorophyte and green plant chlorophyll a/b light-harvesting proteins. Proc. Natl. Acad. Sci. U.S.A. 93, 15244–15248.
Lewin, R.A. (1981). “The Prochlorophytes,” in The Prokaryotes. A Handbook on Habitats, Isolation, and Identification of Bacteria, eds M. P. Starr, H. Stolp, H. G. Trüper, A. Balows, and H. G. Schlegel (Berlin: Springer-Verlag), 257–266.
Lewin, R. A., Cheng, L., and Lafargue, F. (1980). Prochlorophytes in the Caribbean. Bull. Mar. Sci. 30, 744–745.
Marquardt, J., Hu, Q., Iwasaki, I., Miyashita, H., Kurano, N., Miyachi, S., and Mörschel, E. (1998). “The structure of the biliprotein aggregates of the chl d-containing prokaryote Acaryochloris marina” in Photosynthesis: Mechanism and Effects, ed. G. Garab (Dordrecht: Kluwer Academic Publisher), 217–220.
McFadden, G. I. (1996). Endosymbiosis and evolution of the plant cell. Curr. Opin. Plant Biol. 2, 513–519.
McKay, R. M. L., and Gibbs, S. P. (1990). Composition and function of pyrenoids: cytochemical and immunocytochemical approaches. Can. J. Bot. 69, 1040–1052.
Miller, K. R., Jacob, J. S., Burger-Wiersma, T., and Matthijs, H. C. (1988). Supramolecular structure of the thylakoid membrane of Prochlorothrix hollandica: a chlorophyll b-containing prokaryote. J. Cell Sci. 91, 577–586.
Miyashita, H., Adachi, K., Kurano, N., Ikemoto, H., Chihara, M., and Miyachi, S. (1996). Chlorophyll d as a major pigment. Nature 383, 402.
Miyashita, H., Ikemoto, H., Kurano, N., Miyachi, S., and Chihara, M. (2003). Acaryochloris marina gen. et sp. nov. (Cyanobacteria), an oxygenic photosynthetic prokaryote containing chl d as a major pigment. J. Phycol. 39, 1247–1253.
Moore, L. R., Rocap, G., and Chisholm, S. W. (1998). Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393, 464–467.
Mor, T. S., Post, A. F., and Ohad, I. (1993). The manganese stabilizing protein (MSP) of Prochlorothrix hollandica is a hydrophobic membrane-bound protein. Biochim. Biophys. Acta 1141, 206–212.
Morden, C. W., Delwiche, C. F., Kuhsel, M., and Palmer, J. D. (1992). Gene phylogenies and the endosymbiotic origin of plastids. BioSystems 28, 75–90.
Morden, C. W., and Golden, S. S. (1991). Sequence analysis and phylogenetic reconstruction of the genes encoding the large and small subunits of the ribulose-1,5-bisphosphate carboxylase/oxygenase from the chlorophyll-containing prokaryote Prochlorothrix hollandica. J. Mol. Evol. 32, 379–395.
Münchhoff, J., Hirose, E., Maruyama, T., Sunairi, M., Burns, B. P., and Neilan, B. A. (2007). Host specificity and phylogeography of the prochlorophyte Prochloron sp., an obligate symbiont in didemnid ascidians. Environ. Microbiol. 9, 890–899.
Nagata, N., Tanaka, R., Satoh, S., and Tanaka, A. (2005). Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus Species. Plant Cell 17, 233–240.
Nierzwicki-Bauer, S. A., Balkwill, D. L., and Stevens, S. E. (1983). Three-dimensional ultrastructure of a unicellular cyanobacterium. J. Cell Biol. 97, 713–722.
Nikolaitchik, O. A., and Bullerjahn, G. S. (1998). Transcript analysis of the pcbABC genes encoding the antenna apoproteins in the photosynthetic prokaryote Prochlorothrix hollandica. FEMS Microbiol. Lett. 68, 187–194.
Omata, T., Okada, M., and Murata, N. (1985). Separation and partial characterization of membranes from Prochloron sp. Plant Cell Physiol. 26, 579–584.
Palenik, B., and Haselkorn, R. (1992). Multiple evolutionary origins of prochlorophytes, the chlorophyll b-containing prokaryotes. Nature 55, 265–267.
Partensky, F., and Garczarek, L. (2003). “The photosynthetic apparatus of chlorophyll b- and d-containing cyanobacteria,” in Photosynthesis in Algae, eds A. W. Larkum, S. E. Douglas, and J. A. Raven (Dordrecht: Kluwer Academic Publishers), 29–62.
Partensky, F., Hess, W. R., and Vaulot, D. (1999). Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63, 106–127.
Pel, R., Floris, V., Gons, H., and Hoogveld, H. L. (2004). Linking flow cytometric sorting and compound-specific 13C-analysis to determine population-specific isotopic signatures and growth rates in cyanobacteria-dominated lake plankton. J. Phycol. 40, 857–866.
Pinevich, A. V., Averina, S. G., and Velichko, N. V. (2010). Essays in the biology of prochlorophytes (in Russian, with English Table of Contents and Summary). St. Petersburg: Publishing House of St. Petersburg University.
Pinevich, A. V., Matthijs, H. C. P., Gavrilova, O. V., Averina, S. G., and Velichko, N. V. (1996). New ultrastructural aspects of membranes and cell inclusions in Prochlorothrix hollandica (Prochlorales, Cyanobacteria). Microbios 87, 217–225.
Pinevich, A. V., Skulberg, O., Matthijs, H. C. P., Schubert, H., Willen, E., Gavrilova, O. V., and Velichko, N. (1999). Characterization of a novel chlorophyll b-containing Prochlorothrix species (Prochlorophyta) and its photosynthetic apparatus. Microbios 100, 159–174.
Post, A. F., and Arieli, B. (1997). Photosynthesis of Prochlorothrix hollandica under sulfide-rich anoxic conditions. Appl. Environ. Microbiol. 63, 3507–3511.
Post, A. F., and Bullerjahn, G. S. (1994). The photosynthetic machinery in prochlorophytes: structural properties and ecological significance. FEMS Microbiol. Rev. 13, 393–414.
Post, A. F., Gal, A., and Ohad, I. (1992). Characterization of light-activated reversible phosphorylation of a chlorophyll a/b antenna apoprotein in the photosynthetic prokaryote Prochlorothrix hollandica. Biochim. Biophys. Acta 1110, 75–82.
Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M., and Stanier, R. Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Gen. Microbiol. 111, 1–61.
Robinson, N. J., Robinson, J., Gupta, A., Bleasby, A. J., Whitton, B. A., and Morby, A. P. (1995). Singular over-representation of an octameric palindrome, HIP1, in DNA from many cyanobacteria. Nucleic Acids Res. 23, 729–735.
Rocap, G., Dispel, D. L., Waterbury, J. B., and Chisholm, S. W. (2002). Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S–23S ribosomal DNA internal transcribed spacer sequences. Appl. Environ. Microbiol. 68, 1180–1191.
Rocap, G., Larimer, F. W., Lamerdin, J., Malfatti, S., Chain, P., Ahlgren, N. A., Arellano, A., Coleman, M., Hauser, L., Hess, W. R., Johnson, Z. I., Land, M., Lindell, D., Post, A. F., Regala, W., Shah, M., Shaw, S. L., Steglich, C., Sullivan, M. B., Ting, C. S., Tolonen, A., Webb, E. A., Zinser, E. R., and Chisholm, S. W. (2003). Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424, 1042–1047.
Scanlan, D. J., Ostrowski, M., Mazard, S., Dufresne, A., Garczarek, L., Hess, W. R., Post, A. F., Hagemann, M., Paulsen, I., and Partensky, F. (2009). Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 73, 249–299.
Schuster, G., Owens, D. C., Cohen, Y., and Ohad, I. (1984). Thylakoid polypeptide composition and light-independent phosphorylation of the chlorophyll a/b-protein in Prochloron, a prokaryote exhibiting oxygenic photosynthesis. Biochim. Biophys. Acta. 767, 596–605.
Schyns, G., Rippka, R., Namane, A., Campbell, D., Herdman, M., and Houmard, J. (1997). Prochlorothrix hollandica PCC 9006: genomic properties of an axenic representative of the chlorophyll a/b-containing oxyphotobacteria. Res. Microbiol. 148, 345–354.
Shimada, A., Yano, N., Kanai, S., Lewin, R. A., and Maruyama, T. (2003). Molecular phylogenetic relatioship between two symbiotic photo-oxygenic prokaryotes, Prochloron sp. and Synechocystis trididemni. Phycologia 42, 193–197.
Shimada, A. S., Kanai, T., and Maruyama, T. (1995). Partial sequence of ribulose-1,5-bisphosphate carboxylase/oxygenase and the physiology of Prochloron and Prochlorococcus (Prochlorales). J. Mol. Evol. 40, 671–677.
Skulberg, O. (2008). Species of Prochlorothrix: a retrospective view with attention on taxonomy // Algol. Studies 126, 87–101.
Smith, J. K., Parry, J. D., Day, J. G., and Smith, R. J. (1998). A PCR technique based on the Hip1 interspersed repetitive, sequence distinguishes cyanobacterial species and strains. Microbiology 144, 2791–2801.
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0 . Mol. Biol. Evol. 24, 1596–1599.
Tanaka, A., Ito, H., Tanaka, R., Yoshida, K., and Okada, K. (1998). Chlorophyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a. Proc. Natl. Acad. Sci. U.S.A. 95, 12719–12723.
Ting, C. S., Rocap, G., King, J., and Chisholm, S. W. (2002). Cyanobacterial photosynthesis in the oceans: the origins and significance of divergent light-harvesting strategies. Trends Microbiol. 10, 134–142.
Tomitani, A., Okada, K., Miyashita, H., Matthijs, H. C. P., Ohno, T., and Tanaka, A. (1999). Chlorophyll b and phycobilins in the common ancestor of cyanobacteria and chloroplasts. Nature 400, 159–162.
Turner, S. (1997). Molecular systematics of oxygenic photosynthetic bacteria. Plant Syst. Evol. 11, 13–52.
Urbach, E., Robertson, D. L., and Chisholm, S. W. (1992). Multiple evolutionary origins of prochlorophytes within the cyanobacterial radiation. Nature 355, 267–269.
van der Staay, G. W. M., Boekema, E. J., Dekker, J. P., and Matthijs, H. C. P. (1993). Characterization of trimeric photosystem I particles from the prochlorophyte Prochlorothrix hollandica by electron microscopy and image analysis. Biochim. Biophys. Acta 1142, 189–193.
van der Staay, G. W. M., Brouwer, A., Baard, R. L., van Mourikb, F., and Matthij, H. C. P. (1992). Separation of photosystems I and II from the oxychlorobacterium (prochlorophyte) Prochlorothrix hollandica and association of chlorophyll b binding antennae with photosystem II. Biochim. Biophys. Acta 1102, 220–228.
van der Staay, G. W. M., and Staehelin, L. A. (1994). Biochemical characterization of protein composition and protein phosphorylation patterns in stacked and unstacked thylakoid membranes of the prochlorophyte Prochlorothrix hollandica. J. Biol. Chem. 269, 24834–24844.
van der Staay, G. W. M., Yurkova, N., and Green, B. R. (1998). The 38 kDa chlorophyll a/b protein of the rokaryote Prochlorothrix hollandica is encoded by a divergent pcb gene. Plant Mol. Biol. 36, 709–716.
van Liere, L., Breebaart, L., and Dullemont, Y. J. (1989). Determining the relative number of prochlorophytes in lake phytoplankton using epifluorescence microscopy. Br. Phycol. J. 24, 391–394.
Wayne, L. G., Brenner, D. J., Cowell, R. R., Grimont, P. A. D., Kandler, O., Krichevsky, M. I., Moore, L. H., Murray, R. G. E., Stackebrandt, E., Starr, M. P., and Truper, H. G. (1987). Report of the ad hoc committee on reconciliation of approaches to bacterial systematic. Int. J. Syst. Bacteriol. 37, 463–464.
Keywords: cyanobacteria, Prochlorothrix, Prochlorophytes
Citation: Pinevich A, Velichko N and Ivanikova N (2012) Cyanobacteria of the genus Prochlorothrix. Front. Microbio. 3:173. doi: 10.3389/fmicb.2012.00173
Received: 30 January 2012; Accepted: 19 April 2012;
Published online: 21 May 2012.
Edited by:
George S. Bullerjahn, Bowling Green State University, USAReviewed by:
George S. Bullerjahn, Bowling Green State University, USAEdward Hall, United States Geological Survey, USA
Copyright: © 2012 Pinevich, Velichko and Ivanikova. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Alexander Pinevich, Microbiology Department, Faculty of Biology and Soil Science, St. Petersburg State University, 29, Lane 16 Vasilievskii island, St. Petersburg, Russia e-mail:cGluZXZpY2guYUBtYWlsLnJ1
 Alexander Pinevich*
Alexander Pinevich*