- 1 Max Planck Institute for Informatics, Saarbrücken, Germany
- 2 Department of Viral Infections, Research Institute for Microbial Diseases, Osaka University, Suita, Osaka, Japan
Human immunodeficiency virus type 2 (HIV-2) and simian immunodeficiency virus isolated from a macaque monkey (SIVmac) are assumed to have originated from simian immunodeficiency virus isolated from sooty mangabey (SIVsm). Despite their close similarity in genome structure, HIV-2 and SIVmac show different sensitivities to TRIM5α, a host restriction factor against retroviruses. The replication of HIV-2 strains is potently restricted by rhesus (Rh) monkey TRIM5α, while that of SIVmac strain 239 (SIVmac239) is not. Viral capsid protein is the determinant of this differential sensitivity to TRIM5α, as the HIV-2 mutant carrying SIVmac239 capsid protein evaded Rh TRIM5α-mediated restriction. However, the molecular determinants of this restriction mechanism are unknown. Electrostatic potential on the protein-binding site is one of the properties regulating protein–protein interactions. In this study, we investigated the electrostatic potential on the interaction surface of capsid protein of HIV-2 strain GH123 and SIVmac239. Although HIV-2 GH123 and SIVmac239 capsid proteins share more than 87% amino acid identity, we observed a large difference between the two molecules with the HIV-2 GH123 molecule having predominantly positive and SIVmac239 predominantly negative electrostatic potential on the surface of the loop between α-helices 4 and 5 (L4/5). As L4/5 is one of the major determinants of Rh TRIM5α sensitivity of these viruses, the present results suggest that the binding site of the Rh TRIM5α may show complementarity to the HIV-2 GH123 capsid surface charge distribution.
Introduction
The host range of human immunodeficiency virus type 1 (HIV-1) is narrow, limited to humans and chimpanzees (Gao et al., 1999). HIV-1 fails to replicate in activated CD4-positive T lymphocytes from Old World monkeys (OWM), such as rhesus (Rh; Shibata et al., 1995; Himathongkham and Luciw, 1996) and cynomolgus (CM) monkeys (Akari et al., 1996, 1999). On the other hand, simian immunodeficiency virus (SIV) isolated from sooty mangabey (SIVsm) and SIV isolated from African green monkey (SIVagm) replicate well in their natural hosts (VandeWoude and Apetrei, 2006). SIV isolated from a macaque monkey (SIVmac) evolved from SIVsm in captive macaques, and replicates efficiently in Rh (Shibata et al., 1995; Himathongkham and Luciw, 1996) and CM (Akari et al., 1996, 1999) monkeys. Human immunodeficiency virus type 2 (HIV-2) is assumed to have originated from SIVsm as the result of zoonotic events involving monkeys and humans (Hahn et al., 2000). Previous studies have shown that HIV-2 strains vary widely in their ability to grow in cells of OWM (Castro et al., 1990, 1991; Locher et al., 1998, 2003; Fujita et al., 2003).
TRIM5α was identified as an anti-HIV-1 host restriction factor in Rh monkey cells (Stremlau et al., 2004). TRIM5 proteins are members of the tripartite motif family containing RING, B-box, and coiled-coil domains. The α isoform of TRIM5 has an additional C-terminal PRYSPRY domain (Reymond et al., 2001). TRIM5α recognizes the multimerized capsid (viral core) of an incoming virus by its PRYSPRY domain and causes degradation of the core (Sebastian and Luban, 2005; Stremlau et al., 2006). In CM monkey, TRIM5α has also been shown to restrict HIV-1 infection (Nakayama et al., 2005).
We previously evaluated the sensitivity of HIV-2 and SIVmac to Rh and CM TRIM5αs, and found that HIV-2 strain GH123 carrying P at position 120 of the capsid protein (CA) was potently restricted by CM TRIM5α, while the HIV-2 GH123 mutant in which P was replaced with Q was resistant to CM TRIM5α (Song et al., 2007). In contrast, Rh TRIM5α potently restricted the replication of both viruses (Kono et al., 2008). Three amino acid residues, TFP, at positions 339–341 in the PRYSPRY domain of Rh TRIM5α were necessary for restricting HIV-2 strains that were resistant to CM TRIM5α (Kono et al., 2008). Although SIVmac239 CA possesses Q at position 118 corresponding to position 120 of GH123, SIVmac239 was resistant to both of CM and Rh TRIM5αs (Kono et al., 2008, 2010). Therefore, we attempted to identify the viral determinant of SIVmac239 underlying evasion from Rh TRIM5α-mediated restriction, and found that multiple regions including the N-terminal loop, a loop between α-helices 4 and 5 (L4/5), and a loop between α-helices 6 and 7 (L6/7) in the N-terminal half of SIVmac239 CA are necessary for complete evasion of Rh TRIM5α restriction (Kono et al., 2010).
Apart from the sequence and structural characteristics regulating protein–protein interaction, the electrostatic potential at the binding site is an important factor allowing molecular interactions. The electrostatic potential on the protein surface is generated through redistribution of electrons according to local electrical fields. It is defined as the potential energy of a proton at a particular location near a molecule. Negative electrostatic potential results in attraction of the proton by the concentrated electron density. Positive electrostatic potential results in repulsion of the proton by the atomic nuclei in regions where low electron density exists and nuclear charge is incompletely shielded. Electrostatic effects were shown to be a major factor in determining the nature and strength of the interactions between protein surfaces (Dong and Zhou, 2002; Kortemme and Baker, 2002). A complementary charge on the binding site of both proteins may result in an attractive force allowing binding to occur.
In the present study, we analyzed the electrostatic potentials of the surface regions of the CA loop. We analyzed two CA variants, HIV-2 GH123 and SIVmac239 CAs, showing opposite restriction phenotypes. We first modeled the 3-D structures of the proteins by homology modeling and next calculated the electrostatic potentials in the regions of interest based on Adaptive Poisson–Boltzmann Solver and non-local electrostatic method. We found a large difference in the electrostatic potentials of the loop surface between the HIV-2 GH123 and SIVmac239 CAs, potentially responsible for the differential TRIM5α sensitivity of these two viruses.
Materials and Methods
Modeling
The structure of the N-terminal domain of the HIV-1 CA (PDB number 1GWP; Tang et al., 2002) was used as a template for building the corresponding domain models of HIV-2 GH123 and SIVmac239 CAs. The models were built using Modeller 9v4 (Eswar et al., 2007) and visualized with PyMOL (http://www.pymol.org).
Calculation of Electrostatic Potentials
As the initial step preceding electrostatic potential modeling, we added missing hydrogen atoms and estimated the ionization (protonation) of the molecules. We used H++ server (Gordon et al., 2005) http://biophysics.cs.vt.edu/H++, which adds protons to the input structure according to the calculated ionization states at the specified pH of the solvent. The H++ method models molecules as a low dielectric medium εin in a solvent with a high dielectric constant εout. It additionally allows the user to define the salt concentration of the medium and its pH. We used the most biologically relevant parameters of human cells: pH = 7.2, salinity 1%, molecule dielectric εin = 10, and medium dielectric εin = 80. The dielectric parameters were chosen according to the suggestions of the authors of the H++ method as appropriate for modeling protonation of surface residues. We also inspected electrostatic potential profiles resulting from several other parameter combinations. Other parameter regimes did not produce markedly different electrostatic potentials in the region of interest. Therefore, we chose the initial parameters as the most relevant for biological settings.
We next applied two methods of electrostatic potential calculation: Adaptive Poisson–Boltzmann Solver (APBS; Baker et al., 2001) and non-local electrostatic method (Hildebrandt et al., 2007). In both methods, electrostatic properties are described by the Poisson–Boltzmann equation, a second-order non-linear partial differential equation. APBS method solves the equation using finite element techniques based on parameter discretization and iterative parallel refinement of the equation solution. The non-local electrostatic method allows inclusion of the structure of water molecules in the calculation and describes the system as a continuum. This method captures the effects of the dipole polarization of water molecules and the effects of the surrounding hydrogen bond network, and is therefore a more accurate model of the electrostatic potential estimations close to the molecule-solvent interface.
We used two different surface approximations: solvent-accessible surface (SAS) of two different sizes. SAS is the surface of a molecule that is accessible to a solvent. It is estimated using a “rolling ball” approach (Shrake and Rupley, 1973) in which a sphere of solvent of a particular radius is used to probe the surface of the molecule, the surface is then described by the center of the probing sphere. We used the approximate radius of a water molecule of 1.4 Å and an additional 3 Å to determine how the electrostatic potential changes with distance from the molecule.
Results
The 3-D Structural Models of HIV-2 GH123 and SIVmac239 CA N-Terminal Domains
Previously, we evaluated the sensitivity of HIV-2 GH123 and SIVmac239 to Rh and CM TRIM5αs, and found that HIV-2 GH123 was sensitive to CM and Rh TRIM5αs (Song et al., 2007; Kono et al., 2008; Figure 1A). In contrast, SIVmac239 was resistant to CM and Rh TRIM5αs (Kono et al., 2008, 2010; Figure 1A). CA is the determinant for this differential sensitivity to TRIM5α between HIV-2 GH123 and SIVmac239, as the HIV-2 GH123 mutant carrying SIVmac239 CA (HIV-2 GH/SCA) was also resistant to CM and Rh TRIM5αs (Figure 1A; Kono et al., 2010). Despite this marked difference in TRIM5α sensitivity between HIV-2 GH123 and SIVmac239, CA of these two viruses share more than 87% amino acid identity (Figure 1B). Therefore, we compared the structural properties of HIV-2 GH123 CA with those of SIVmac239.

Figure 1. (A)Sensitivities of HIV-2 GH123, HIV-2 GH123 mutant carrying SIVmac239 capsid protein (HIV-2 GH/SCA), and SIVmac239 to cynomolgus (CM) and rhesus (Rh) monkey TRIM5α. The replication of HIV-2 GH123 was potently restricted by CM and Rh TRIM5α (sensitive), while that of SIVmac239 and the HIV-2 GH123 mutant carrying SIVmac239 capsid was not (resistant). (B) Alignment of amino acid sequences of HIV-2 GH123 and SIVmac239 capsid proteins. Positions of the N-terminal loop (N-terminal), a loop between α-helices 4 and 5 (L4/5), and a loop between α-helices 6 and 7 (L6/7) are indicated above the amino acid sequences.
We first constructed 3-D models of HIV-2 GH123 and SIVmac239 CA N-terminal domains by homology modeling. In the constructed models, HIV-2 GH123 and SIVmac239 CA N-terminal domains showed the most striking differences in shape of surface exposed loops (Figure 2). SIVmac239 CA is characterized by a more contracted shape as compared to the expanded loop structure of HIV-2 GH123. To confirm that this shape difference is not due to modeling noise, we remodeled both proteins using each one as a template for the other. The remodeled structures showed similar shape differences (data not shown), suggesting that the real structures differ.
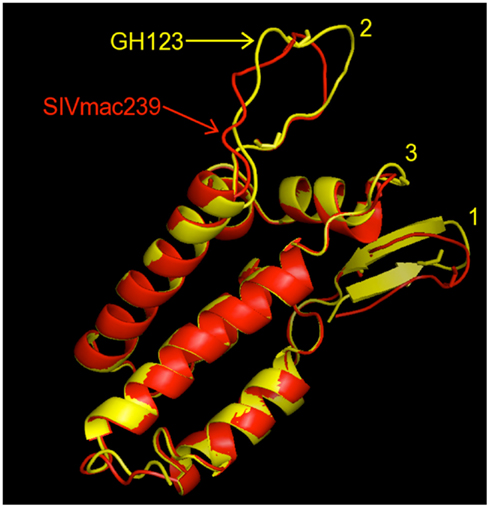
Figure 2. Superposition of modeled structures of the N-terminal domains of HIV-2 GH123 (GH123, yellow) and SIVmac239 (red) capsid proteins. The three loops containing sites important for the TRIM5α interaction are numbered as follows: (1) N-terminal loop, (2) loop between α-helices 4 and 5 (L4/5), (3) loop between α-helices 6 and 7 (L6/7).
Electrostatic Potentials of HIV-2 GH123 and SIVmac239 CA N-Terminal Domains
Figure 3 shows the distributions of calculated electrostatic potentials of HIV-2 GH123 and SIVmac239 CA N-terminal domains. We observed strong differences between the two molecules on the surface of the loops with the GH123 molecule having predominantly positive and SIVmac239 predominantly negative electrostatic potential on this part of the surface (Figure 3).
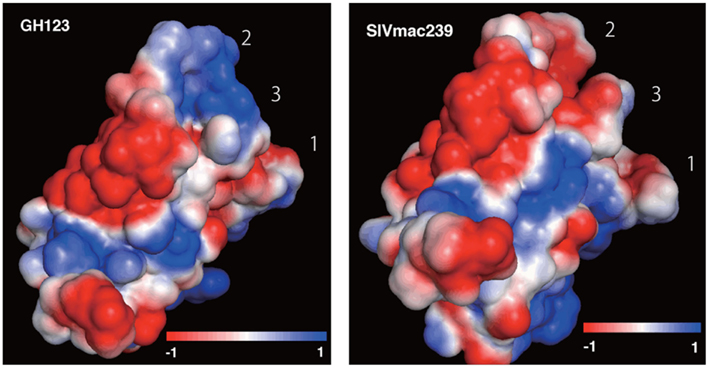
Figure 3. Electrostatic potential on the surface of HIV-2 GH123 (GH123) and SIVmac239 capsid protein N-terminal domains. Structures are positioned as in Figure 2 with the loops directed toward the upper right of the image. Electrostatic potential was calculated and visualized using the APBS plugin in PyMOL. The three loops containing sites important for the TRIM5α interaction are numbered as follows: (1) N-terminal loop, (2) loop between α-helices 4 and 5 (L4/5), (3) loop between α-helices 6 and 7 (L6/7).
To quantify this observation and obtain further insight into the specific region where the electrostatic potential differences are strong, we extracted the electrostatic potential values on the surfaces of the two molecules. From the electrostatic potential values estimated in a grid covering the entire space around the molecules, we extracted grid points neighboring the points of triangulation of each surface type. We grouped these electrostatic potential values according to the atoms of the closest loop residues. This comparison of grouped electrostatic potential values of corresponding residues in the two analyzed molecules allowed us to quantitatively confirm the differences in electrostatic potential in the region of interest and to point to specific residues around which the differences were stronger. The strongest difference in electrostatic potential between HIV-2 GH123 and SIVmac239 CAs was observed on the surface of L4/5, with HIV-2 GH123 and SIVmac239 showing positive and negative electrostatic potential, respectively. Eight of nine residues in this loop showed significant differences in mean electrostatic potential and clear separation of the electrostatic potential values on the grid neighboring to the loop residues by both local ABPS and non-local electrostatic methods (Table 1).
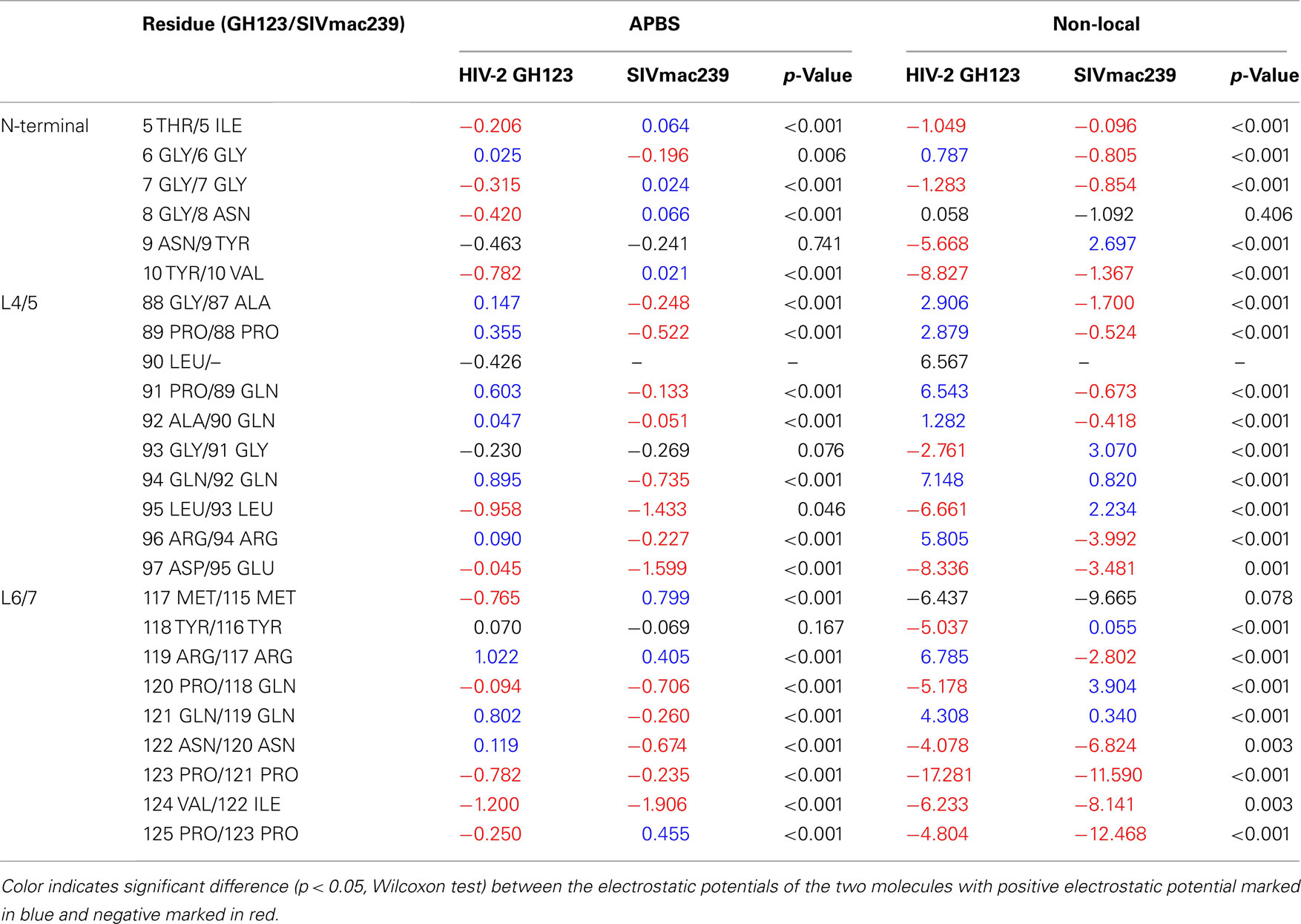
Table 1. Mean electrostatic potential on the surface surrounding residues of the N-terminal loop (N-terminal), the loop between α-helices 4 and 5 (L4/5), and the loop between α-helices 6 and 7 (L6/7) of HIV-2 GH123 and SIVmac239 CAs calculated using the local Adaptive Poisson–Boltzmann Solver (APBS) and non-local electrostatic methods.
Residues in L6/7 showed weak but similar electrostatic potential differences to those of L4/5 by the local ABPS method, but these differences were not confirmed by the non-local electrostatic method (Table 1). The N-terminal loop showed the opposite pattern, with HIV-2 GH123 and SIVmac239 having negative and positive electrostatic potentials, respectively, according to the local APBS method (Table 1). However, the differences were smaller and were not confirmed by the non-local electrostatic method (Table 1).
Similar electrostatic potential differences, although spanning a narrower range of values than those described above, were observed on the SAS of the 3 Å probe radius (data not shown). These observations reflect the electrostatic potential decrease with distance from the molecule surface.
Discussion
In the present study, we constructed 3-D models of HIV-2 GH123 and SIVmac239 CA N-terminal domains by homology modeling and analyzed the electrostatic potential distributions on the SASs of these molecules. We observed a large difference between the HIV-2 GH123 and SIVmac239 CA N-terminal domains, with the HIV-2 GH123 molecule having predominantly positive and SIVmac239 predominantly negative electrostatic potential on the surface of L4/5. This result may be relevant to the previous findings that CA L4/5 was one of the major determinants for the differential sensitivity to Rh TRIM5α between HIV-2 and SIVmac239 (Ylinen et al., 2005; Kono et al., 2010).
Precise calculation of the interaction electrostatics is challenging due to the large surfaces involved and the large structural changes that can occur upon binding. Here, our quantitative approach based on two different methods for calculation of electrostatic potential indicated negative electrostatic potential on the surface of the resistant CA variant SIVmac239 and positive electrostatic potential of the non-resistant HIV-2 GH123 variant. The presence of positive electrostatic potential on the surface of L4/5 may therefore be a prerequisite for the interactions with Rh TRIM5α. This loop is the most outward pointing part of the CA protein. Complementarity to the HIV-2 GH123 surface charge distribution at the binding site of the host protein may be necessary for binding. Therefore, similar studies of TRIM5α surface electrostatic potentials could help to point to the specific site of this interaction, although the 3-D structural analysis of TRIM5α PRYSPRY domain is required for this goal.
It was recently reported that a recombinant TRIM5α protein carrying TRIM21 RING domain (TRIM5-21R) assembled to form 2-D paracrystalline hexagonal arrays in vitro (Ganser-Pornillos et al., 2011). This assembly requires RING and B-box 2 domains, and the hexagonal lattices of HIV-1 CA that mimic the surface of core act as template for stabilization of TRIM5-21R arrays in a PRYSPRY-dependent manner (Ganser-Pornillos et al., 2011). As the interaction between individual CA monomers and TRIM5α is very weak, CA recognition by TRIM5α is thought to be a synergistic combination of direct binding interactions with the PRYSPRY domain, higher-order assembly of TRIM5α, template-based assembly, and lattice complementarity. Therefore, the electrostatic potential might be the crucial determinant of this binding allowing TRIM5α for recognition of a broader range of CA sequence variants.
In addition to L4/5, our previous study revealed that the N-terminal loop and L6/7 in the N-terminal half of SIVmac239 CA are also necessary for complete evasion of Rh TRIM5α restriction (Kono et al., 2010). Electrostatic potentials of these 2 loops did not show large differences between HIV-2 GH123 and SIVmac239. Therefore, it is possible that a certain interaction other than the electrostatic interaction would be involved in binding of Rh TRIM5α PRYSPRY domain with the N-terminal loop and L6/7 of HIV-2 GH123.
On sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, SIVmac239 CA is known to migrate at a molecular weight of 27 kDa, while HIV-2 GH123 CA migrates at a molecular weight of 25 kDa (Kono et al., 2010). However, the number of amino acid residues in SIVmac239 CA is smaller than that in HIV-2 GH123 CA (Figure 1B), and the molecular weight of SIVmac239 CA is therefore smaller than that of HIV-2 GH123. We reported previously that the amino acid sequences of L4/5 determined this differential electrophoretic mobility of CAs (Kono et al., 2010). The difference seems to be attributable to the presence of non-polar P and A residues at positions 91 and 92, respectively, in L4/5 of HIV-2 GH123 CA, where two more hydrophilic Q residues are located in SIVmac239 CA L4/5 (Figure 1; Table 1). In addition, HIV-2 GH123 CA L4/5 has a hydrophobic L insertion at position 90 (Figure 1; Table 1). Therefore, L4/5 of HIV-2 GH123 CA is more hydrophobic and would attract larger numbers of SDS molecules than that of SIVmac239 leading to accelerated electrophoretic speed of the CA. It is therefore possible that hydrophobic interactions between Rh TRIM5α and viral CAs would also be involved in determining the anti-viral specificity of TRIM5α in addition to the electrostatic interactions discussed above. Further biochemical studies of TRIM5α and viral CAs are necessary to address this question.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Thomas Lengauer for his support and Ms. Noriko Teramoto for her help. This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology, and the Ministry of Health, Labour and Welfare, Japan.
References
Akari, H., Mori, K., Terao, K., Otani, I., Fukasawa, M., Mukai, R., and Yoshikawa, Y. (1996). In vitro immortalization of old world monkey T lymphocytes with Herpesvirus saimiri: its susceptibility to infection with simian immunodeficiency viruses. Virology 218, 382–388.
Akari, H., Nam, K. H., Mori, K., Otani, I., Shibata, H., Adachi, A., Terao, K., and Yoshikawa, Y. (1999). Effects of SIVmac infection on peripheral blood CD4+CD8+ T lymphocytes in Cynomolgus macaques. Clin. Immunol. 91, 321–329.
Baker, N. A., Sept, D., Joseph, S., Holst, M. J., and McCammon, J. A. (2001). Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041.
Castro, B. A., Barnett, S. W., Evans, L. A., Moreau, J., Odehouri, K., and Levy, J. A. (1990). Biologic heterogeneity of human immunodeficiency virus type 2 (HIV-2) strains. Virology 178, 527–534.
Castro, B. A., Nepomuceno, M., Lerche, N. W., Eichberg, J. W., and Levy, J. A. (1991). Persistent infection of baboons and rhesus monkeys with different strains of HIV-2. Virology 184, 219–226.
Dong, F., and Zhou, H. X. (2002). Electrostatic contributions to T4 lysozyme stability: solvent-exposed charges versus semi-buried salt bridges. Biophys. J. 83, 1341–1347.
Eswar, N., Webb, B., Marti-Renom, M. A., Madhusudhan, M. S., Eramian, D., Shen, M. Y., Pieper, U., and Sali, A. (2007). Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. Chap. 2, Unit 2.9.
Fujita, M., Yoshida, A., Sakurai, A., Tatsuki, J., Ueno, F., Akari, H., and Adachi, A. (2003). Susceptibility of HVS-immortalized lymphocytic HSC-F cells to various strains and mutants of HIV/SIV. Int. J. Mol. Med. 11, 641–644.
Ganser-Pornillos, B. K., Chandrasekaran, V., Pornillos, O., Sodroski, J. G., Sundquist, W. I., and Yeager, M. (2011). Hexagonal assembly of a restricting TRIM5alpha protein. Proc. Natl. Acad. Sci. U.S.A. 108, 534–539.
Gao, F., Bailes, E., Robertson, D. L., Chen, Y., Rodenburg, C. M., Michael, S. F., Cummins, L. B., Arthur, L. O., Peeters, M., Shaw, G. M., Sharp, P. M., and Hahn, B. H. (1999). Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397, 436–441.
Gordon, J. C., Myers, J. B., Folta, T., Shoja, V., Heath, L. S., and Onufriev, A. (2005). H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 33, W368–W371.
Hahn, B. H., Shaw, G. M., De Cock, K. M., and Sharp, P. M. (2000). AIDS as a zoonosis: scientific and public health implications. Science 287, 607–614.
Hildebrandt, A., Blossey, R., Rjasanow, S., Kohlbacher, O., and Lenhof, H. P. (2007). Electrostatic potentials of proteins in water: a structured continuum approach. Bioinformatics 23, e99–e103.
Himathongkham, S., and Luciw, P. A. (1996). Restriction of HIV-1 (subtype B) replication at the entry step in rhesus macaque cells. Virology 219, 485–488.
Kono, K., Song, H., Shingai, Y., Shioda, T., and Nakayama, E. E. (2008). Comparison of anti-viral activity of rhesus monkey and cynomolgus monkey TRIM5alphas against human immunodeficiency virus type 2 infection. Virology 373, 447–456.
Kono, K., Song, H., Yokoyama, M., Sato, H., Shioda, T., and Nakayama, E. E. (2010). Multiple sites in the N-terminal half of simian immunodeficiency virus capsid protein contribute to evasion from rhesus monkey TRIM5alpha-mediated restriction. Retrovirology 7, 72.
Kortemme, T., and Baker, D. (2002). A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl. Acad. Sci. U.S.A. 99, 14116–14121.
Locher, C. P., Blackbourn, D. J., Herndier, B. G., Reyes-Teran, G., Barnett, S. W., Murthy, K. K., and Levy, J. A. (1998). Transient virus infection and pathogenesis of a new HIV type 2 isolate, UC12, in baboons. AIDS Res. Hum. Retroviruses 14, 79–82.
Locher, C. P., Witt, S. A., Herndier, B. G., Abbey, N. W., Tenner-Racz, K., Racz, P., Kiviat, N. B., Murthy, K. K., Brasky, K., Leland, M., and Levy, J. A. (2003). Increased virus replication and virulence after serial passage of human immunodeficiency virus type 2 in baboons. J. Virol. 77, 77–83.
Nakayama, E. E., Miyoshi, H., Nagai, Y., and Shioda, T. (2005). A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5 alpha determines species-specific restriction of simian immunodeficiency virus SIVmac infection. J. Virol. 79, 8870–8877.
Reymond, A., Meroni, G., Fantozzi, A., Merla, G., Cairo, S., Luzi, L., Riganelli, D., Zanaria, E., Messali, S., Cainarca, S., Guffanti, A., Minucci, S., Pelicci, P. G., and Ballabio, A. (2001). The tripartite motif family identifies cell compartments. EMBO J. 20, 2140–2151.
Sebastian, S., and Luban, J. (2005). TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology 2, 40.
Shibata, R., Sakai, H., Kawamura, M., Tokunaga, K., and Adachi, A. (1995). Early replication block of human immunodeficiency virus type 1 in monkey cells. J. Gen. Virol. 76(Pt 11), 2723–2730.
Shrake, A., and Rupley, J. A. (1973). Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J. Mol. Biol. 79, 351–371.
Song, H., Nakayama, E. E., Yokoyama, M., Sato, H., Levy, J. A., and Shioda, T. (2007). A single amino acid of the human immunodeficiency virus type 2 capsid affects its replication in the presence of cynomolgus monkey and human TRIM5alphas. J. Virol. 81, 7280–7285.
Stremlau, M., Owens, C. M., Perron, M. J., Kiessling, M., Autissier, P., and Sodroski, J. (2004). The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in old world monkeys. Nature 427, 848–853.
Stremlau, M., Perron, M., Lee, M., Li, Y., Song, B., Javanbakht, H., Diaz-Griffero, F., Anderson, D. J., Sundquist, W. I., and Sodroski, J. (2006). Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. U.S.A. 103, 5514–5519.
Tang, C., Ndassa, Y., and Summers, M. F. (2002). Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 9, 537–543.
VandeWoude, S., and Apetrei, C. (2006). Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin. Microbiol. Rev. 19, 728–762.
Keywords: HIV-2, SIVmac, capsid, TRIM5α, electrostatic potential, APBS, SAS
Citation: Bozek K, Nakayama EE, Kono K and Shioda T (2012) Electrostatic potential of human immunodeficiency virus type 2 and rhesus macaque simian immunodeficiency virus capsid proteins. Front. Microbio. 3:206. doi: 10.3389/fmicb.2012.00206
Received: 19 April 2012; Paper pending published: 18 May 2012;
Accepted: 21 May 2012; Published online: 05 June 2012.
Edited by:
Masaru Yokoyama, National Institute of Infectious Diseases, JapanReviewed by:
Masako Nomaguchi, The University of Tokushima Graduate School, JapanMasaru Yokoyama, National Institute of Infectious Diseases, Japan
Copyright: © 2012 Bozek, Nakayama, Kono and Shioda. This is an openaccess article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Tatsuo Shioda, Department of Viral Infections, Research Institute for Microbial Diseases, Osaka University, 3-1, Yamada-oka, Suita, Osaka 565-0871, Japan. e-mail:c2hpb2RhQGJpa2VuLm9zYWthLXUuYWMuanA=


 Ken Kono2
Ken Kono2