- 1 Department of Biomolecular Engineering, University of California, Santa Cruz, CA, USA
- 2 Janelia Farm Research Campus, Howard Hughes Medical Institute, Ashburn, VA, USA
A great diversity of small, non-coding RNA (ncRNA) molecules with roles in gene regulation and RNA processing have been intensely studied in eukaryotic and bacterial model organisms, yet our knowledge of possible parallel roles for small RNAs (sRNA) in archaea is limited. We employed RNA-seq to identify novel sRNA across multiple species of the hyperthermophilic genus Pyrobaculum, known for unusual RNA gene characteristics. By comparing transcriptional data collected in parallel among four species, we were able to identify conserved RNA genes fitting into known and novel families. Among our findings, we highlight three novel cis-antisense sRNAs encoded opposite to key regulatory (ferric uptake regulator), metabolic (triose-phosphate isomerase), and core transcriptional apparatus genes (transcription factor B). We also found a large increase in the number of conserved C/D box sRNA genes over what had been previously recognized; many of these genes are encoded antisense to protein coding genes. The conserved opposition to orthologous genes across the Pyrobaculum genus suggests similarities to other cis-antisense regulatory systems. Furthermore, the genus-specific nature of these sRNAs indicates they are relatively recent, stable adaptations.
Introduction
Archaeal species are known to encode a plethora of small RNA (sRNA) molecules. These sRNAs have a multitude of functions including suppression of messenger RNA (mRNA; Straub et al., 2009), targeting modifications to ribosomal (rRNA) or transfer RNA (tRNA; Omer et al., 2000; Bernick et al., 2012), specifying targets of the CRISPR immune defense system (Barrangou et al., 2007; Hale et al., 2008; Hale et al., 2009), cis-antisense regulation of transposase mRNA (Tang et al., 2002; Tang et al., 2005; Jager et al., 2009; Wurtzel et al., 2010), and encoding short proteins less than 30 amino acids in length (Jager et al., 2009).
Only a few previous studies have described sRNA genes in the phylum Crenarchaeota. In the Sulfolobus genus, C/D box and H/ACA-box guide sRNAs have been studied, including 18 guide sRNAs in Sulfolobus acidocaldarius (Omer et al., 2000), nine in S. solfataricus (Zago et al., 2005), and corresponding homologs detected computationally in S. tokodaii (Zago et al., 2005). These sRNAs form two distinct classes of guide RNAs: C/D box sRNAs which guide 2′-O-methylation of ribose, and H/ACA-box guide RNAs which direct isomerization of uridine to pseudouridine. Eukaryotes also share these two classes of guide RNAs with the same functions, but these homologs are dubbed small nucleolar RNAs (snoRNAs) because of their cellular localization. Recently, we employed high-throughput sequencing to identify ten conserved, novel families of H/ACA-like sRNA within the genus Pyrobaculum (Bernick et al., 2012).
Sulfolobus solfataricus has been further characterized using high-throughput sequencing (Wurtzel et al., 2010), revealing 18 CRISPR-associated sRNAs, 13 C/D box sRNAs, 28 cis-antisense encoded transposon-associated sRNAs, and 185 sRNA genes encoded antisense to other, non-transposon protein coding genes. It is unclear how many of the latter antisense transcripts are the result of transcriptional noise, overlapping but non-interacting gene products, or biologically relevant products of functional ncRNA genes. The diversity of sRNA genes is just beginning to be studied in depth in other members of the Crenarchaeota.
Genes that produce sRNA antisense to mRNA are known in all three domains of life and many of these sRNA have provided interesting examples of novel regulation. Within bacteria, antisense sRNAs are known and well-studied (Repoila et al., 2003; Aiba, 2007; Vogel, 2009). For example, utilization and uptake of iron in Escherichia coli is modulated by the sRNA RyhB that acts in concert with the ferric uptake regulator (Fur) protein (Masse et al., 2007). The sRNA is coded in trans to its regulatory targets, and the Sm-like protein Hfq is required for its function. In Pseudomonas aeruginosa, an analogous regulatory mechanism exists with the PrrF regulatory RNA (Wilderman et al., 2004).
In this study, we adapted techniques pioneered by researchers studying microRNA in eukaryotes (Lau et al., 2001; Henderson et al., 2006; Lu et al., 2006), to execute parallel high-throughput pyrosequencing of sRNAs across four Pyrobaculum species. This comparative transcriptomic approach enabled us to identify novel conserved sRNA transcripts among four related hyperthermophiles (Pyrobaculum aerophilum, P. arsenaticum, P. calidifontis, and P. islandicum). We provide an overview of the distribution of sRNAs across species, and focus on two major classes: the highly abundant C/D box sRNAs, and sRNAs antisense to three biologically important protein coding genes. We augment our transcriptional analyses further with comparative genomics utilizing two additional Pyrobaculum species with sequenced genomes: P. neutrophilum (recently renamed from Thermoproteus neutrophilus) and P. oguniense (NCBI GenBank accession NC_016885.1).
Materials and Methods
Culture Conditions
Pyrobaculum aerophilum cells were grown anaerobically in media containing 0.5 g/L yeast extract, 1× DSM390 salts, 10 g/L NaCl, 1× DSM 141 trace elements, 0.5 mg/L Fe(SO4)2(NH4)2, pH 6.5, with 10 mM NaNO3. P. islandicum and P. arsenaticum cells were grown anaerobically in media containing 10 g/L tryptone, 2 g/L yeast extract, 1× DSM390 salts, 1× DSM88 trace elements, and 20 mM Na2S2O3. P. calidifontis cells were grown aerobically in 1 L flasks using 500 mL media containing 10 g/L tryptone, 2 g/L yeast extract, 1× DSM88 trace metals, 15 mM Na2S2O3, pH 6.8, loosely capped with moderate shaking at 125 rpm. Anaerobic cultures were grown in 2 L flasks with 1 L media, prepared under nitrogen with resazurin as a redox indicator at 0.5 mg/L; 0.25 mM Na2S was added as a reductant. All cultures were grown at 95°C to late log or stationary phase, monitored at OD600.
The 10× DSM390 salts are comprised of (per liter ddH2O) 1.3 g (NH4)2SO4, 2.8 g KH2PO4, 2.5 g MgSO4·7H2O. The 100× DSM88 trace metal solution is comprised (per liter 0.12 N HCl), 0.9 mM MnCl2, 4.7 mM Na2B4O7, 76 μM ZnSO4, 25 μM CuCl2, 12.4 μM NaMoO4, 18 μM VOSO4, 6 μM CoSO4. The 100× DSM141 trace metal solution is comprised of 7.85 mM Nitrolotriacetic acid, 12.2 mM MgSO4, 2.96 mM MnSO4, 17.1 mM NaCl, 0.36 mM FeSO4, 0.63 mM CoSO4, 0.68 mM CaCl2, 0.63 mM ZnSO4, 40 μM CuSO4, 42 μM KAl(SO4)2, 0.16 mM H3BO3, 41 μM Na2MoO4, 0.1 mM NiCl2, 1.14 μM Na2SeO3.
CDNA Library Preparation
Two preparations were constructed for each of P. aerophilum, P. islandicum, P. arsenaticum, and P. calidifontis cultures, yielding a total of eight cDNA libraries. The following protocol was used for each preparation.
Total RNA was extracted from exponential or stationary cultures; 100 μg of each preparation was loaded onto a 15% polyacrylamide gel, and size selected in the range 15–70 nt. The gel was post-stained with SYBR Gold and the tRNA band was used as the upper exclusion point. The lower exclusion point was set at 75% of the region between xylene cyanol (XC) and bromophenol blue (BP) loading dye bands (Ambion protocol). Samples were eluted, EtOH precipitated, and 3′ linker (5′-adenylated, 3′ ddC) was added as described by Lau et al., 2001; IDTDNA, Linker 1). A second gel purification was performed as above, excising the gel fragment above the XC dye band to remove excess 3′ linker. The recovered linked RNAs were reverse transcribed (RT) using Superscript III (Invitrogen) with a DNA primer complementary to Linker 1. Following RT, Exonuclease I (EXO1, Thermo) was added to the RT reaction mixture, and incubated for 30 min to remove excess primer. We utilized standard alkaline lysis treatment with NaOH-EDTA at 80°C for 15 min to remove any residual RNA, as well as to inactivate the reverse transcriptase and the EXO1 ssDNA nuclease. Neutralization and small fragment removal was performed with water-saturated G50 columns (Ambion NucAway). The recovered single stranded cDNA was dried to near completion using a Servo SpeedVac, followed by a second 5′-adenylated linker addition (IDTDNA – Linker 2) to the cDNA using T4 RNA ligase (Ambion).
A 2 μL volume of this reaction was amplified by PCR (20 μL reaction, 16 cycles). This was followed by a second amplification (20 μL reaction, 16 cycles) using 2 μL from the first amplification as template using Roche 454-specific hybrid adapters based on the method described by Hannon1. A four-base barcode was included in the 5′ hybrid primer. The final reaction was cleaned using the Zymo clean kit following the manufacturer’s protocol.
Sequencing and Read Mapping
Sequencing was performed using a Roche/454 GS FLX sequencer, and the GS emPCR Kit II (Roche). Sequencing reads described in this work are provided online via the UCSC Archaeal Genome Browser2 (Chan et al., 2012).
Reads that included barcodes and sequencing linkers were selected from the raw sequencing data and used to identify reads from each of the eight pooled cDNA libraries. Reads were further consolidated, combining identical sequences with associated counts for viewing with the Archaeal Genome Browser. Reads were mapped to the appropriate genome [P. aerophilum (NC_003364.1); P. arsenaticum (NC_009376.1); P. calidifontis (NC_009073.1); P. islandicum (NC_008701.1); P. oguniense (NC_016885.1); P. neutrophilum (T. neutrophilus: NC_010525.1)] using BLAT (Kent, 2002), requiring a minimum of 90% identity (-minIdentity), a maximal gap of 3 (-maxIntron) and a minimum score (matches minus mismatches) of 16 (-minScore) using alignment parameters for this size range (-tileSize = 8 -stepSize = 4). Reads that mapped equally well to multiple positions in the genome were excluded from this study. The remaining, uniquely mapped reads were formatted and visualized as BED tracks within the UCSC Archaeal Genome Browser.
Of the 216,538 raw sequencing reads obtained, those that had readable barcodes and could be uniquely mapped to their respective genomes were: 39,294 in P. calidifontis, 30,827 in P. aerophilum, 31,206 in P. arsenaticum, and 42,951 in P. islandicum.
Northern Analysis
Northern blots were prepared using ULTRAhyb-Oligo (Ambion) following the manufacturer protocol3 using Hybond-N+ (GE life sciences) membranes to transfer 10 μg/lane denatured total RNA (45 min, 50°C with glyoxyl loading buffer – Ambion). Size separation was conducted using 23 cm × 25 cm gels (1% agarose) in BPTE running buffer (30 mM bis-Tris, 10 mM PIPES, 1 mM EDTA, pH 6.5). The following DNA oligomers (Integrated DNA technologies) were used as probes: TFBiiSense (CCTCCTCTGGAAAGCCCCTCAAGCTCCGA), TFBiiAnti (TCGGAGCTTGAGGGGCTTTCCAGAGGAGG), PAEsR53sense (GACCCCGATCGCCGAAAAATGACGAGTGGT).
Computational Prediction of Orthologous Gene Clusters
Computational prediction of orthologous groups was established by computing reciprocal best BLASTP (Altschul et al., 1990; RBB) protein coding gene-pairs among pairs of four Pyrobaculum species. When at least three RBB gene-pairs select the same inter-species gene set (for example A pairs with B, B pairs with C, and C pairs with A), the cluster was considered an orthologous gene cluster.
Computational Prediction of C/D Box SRNA Homolog Families
C/D box sRNA homolog families were constructed from computational predictions with core C/D box features that were supported by transcripts from one or more of the four Pyrobaculum species (data from this study). Six Pyrobaculum genomes were searched for orthologs using these sRNA candidates as queries to BLASTN (Camacho et al., 2009). The highest scoring candidates were manually curated, then grouped into homologous C/D box sRNA families by multiple alignment.
Results
Small RNA Populations
We prepared eight barcoded sequencing libraries using sRNA fractions (size range 16–70 nt) from anaerobic cultures of P. aerophilum, P. arsenaticum, P. islandicum, and an aerobic culture of P. calidifontis. These libraries were prepared using a 5′-independent ligation strategy (Pak and Fire, 2007) which preserves RNA strand orientation, captures both the 5′ and 3′ ends of the sRNA, and does not impose a bias for molecule selection based on 5′-phosphorylation state. Pyrosequencing, followed by selection of uniquely mapped sequence reads, allowed detection of reads associated with both known and novel genomic features (Figure 1), including:
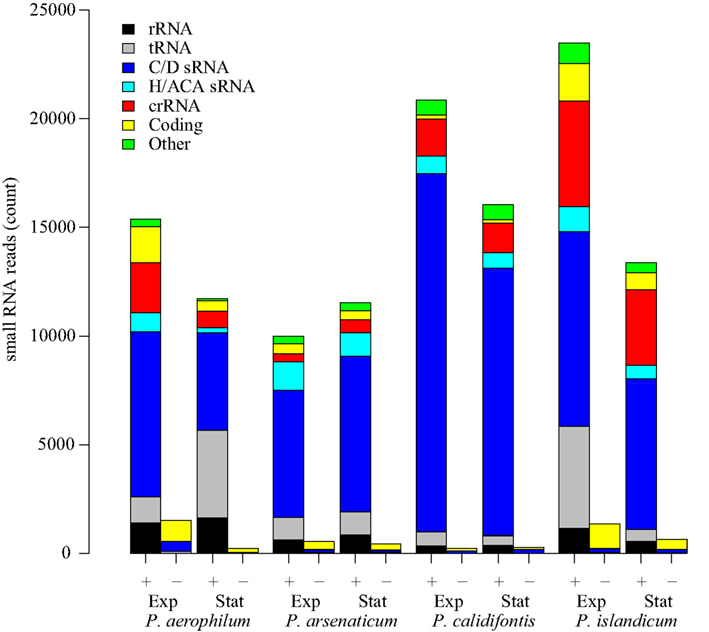
Figure 1. Small RNA transcript abundance in four species of Pyrobaculum. Sense oriented reads (+) and antisense-oriented reads (−) shown in barplots for each species. Samples of each species were taken at both exponential (Exp) and stationary phases (Stat). RNA classifications were made based on mapping to genes coding for C/D box sRNA (C/D sRNA), H/ACA-like sRNA, CRISPR arrays (crRNA), fragments of coding regions (coding), ribosomal RNA (rRNA), and transfer RNA (tRNA).
(i) snoRNA-like guide RNAs, including known and novel C/D box sRNA and a new class of H/ACA-like sRNA (Bernick et al., 2012),
(ii) RNA sequences encoded cis-antisense (asRNA) to known protein coding genes,
(iii) RNA sequences derived from CRISPR arrays, thought to guide the CRISPR-mediated immune response,
(iv) unclassified novel sRNA, and
(v) degradation products of larger RNA including ribosomal RNA, messenger RNA and transfer RNA.
Most antisense-oriented sequencing reads are associated with coding regions (Figure 1) in each of the species and growth phases examined. Antisense-oriented reads are frequently the result of convergent expression of a protein coding gene and a snoRNA-like guide RNA (Tables A1– A4 in Appendix). We find, in some cases, that sequencing reads that appear to be antisense to snoRNA-like RNAs appear to be fragments of novel 3′ untranslated regions (3′ UTRs) of a convergently expressed protein coding region. These antisense-oriented sRNA reads are counted as antisense to the associated snoRNA-like sRNA. We made use of this transcriptional pattern to find novel C/D box sRNA and H/ACA-like sRNA; in these cases, highly abundant antisense reads to coding transcripts often proved to be a hallmark of novel C/D box and H/ACA-like sRNA (Tables A2 and A4 in Appendix). In a few remaining cases, we found novel cis-encoded antisense reads that were not derived from known classes of sRNA. We note that the proportion of reads belonging to each type of classified RNA is relatively stable across species and conditions (Figure 1), with the exception of two conditions in which tRNA fragments were enriched (P. aerophilum stationary phase, P. islandicum exponential phase). We are further investigating these differences, however the purpose and design of the sequencing portion of this study was aimed at qualitative discovery of novel sRNAs.
C/D Box SRNA Account for the Largest Fraction of Reads in All Species Tested
In each of the eight small transcriptomes studied (four species sampled at exponential and stationary phase), C/D box sRNA accounted for the largest fraction of reads (Figure 1). A previous study (Fitz-Gibbon et al., 2002) has provided computational evidence for 65 C/D box sRNA candidates encoded in the genome of P. aerophilum. We now find an additional 23 C/D box sRNA candidates in that genome, representing a 35% increase in family size. By using transcriptional support from the four examined genomes (this study), combined with comparative genomic evidence that includes P. oguniense and P. neutrophilum, we find at least 74 C/D box sRNA in each Pyrobaculum spp. (Table 1). Of those genes, 70 appear to be conserved among all six genomes examined (Figure 2).

Table 1. C/D box sRNA genes in each Pyrobaculum species based on transcriptional evidence or inferred by homology (P. oguniens e and P. neutrophilum).
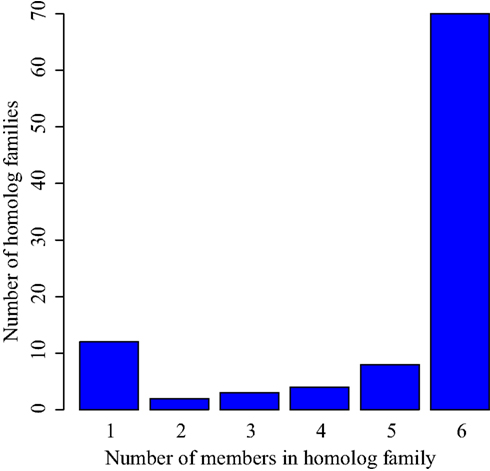
Figure 2. Conservation of C/D box sRNA genes among six Pyrobaculum genomes. C/D box sRNA genes were organized by homolog family based on the location targeted by the encoded guide regions. These homolog families were then compared among the six studied species to verify conservation. While each individual species encodes more than 74 C/D box sRNA genes, 70 of those are conserved among each of the six studied Pyrobaculum spp. (group 6).
Convergently Oriented ncRNA are Frequently Found at the 3′ Terminus of Protein Coding Genes
It has been noted previously that in the genomes of S. acidocaldarius and S. solfataricus, C/D box sRNA genes occasionally exhibit antisense overlap to the 3′ end of protein encoding genes (Dennis et al., 2001). In the Pyrobaculum clade, we find numerous instances of a convergently oriented C/D box or H/ACA-like guide RNA gene that partially overlap, by a few nucleotides, the 3′ end of a protein-coding gene (Tables A2 and A4 in Appendix).
To find conserved, novel cis-encoded antisense RNA, we ranked conserved transcript abundance that overlapped orthologous protein coding genes. Among the top 34 predicted ortholog groups of genes with well-annotated function and conserved 3′ antisense transcription (Table A2 in Appendix), 28 are convergent with C/D box sRNA and three are convergent with H/ACA-like sRNA. Among the top 19 predicted ortholog groups of unknown function with 3′ antisense transcription (Table A4 in Appendix), 11 are convergent with C/D box sRNA, four are convergent H/ACA-like sRNA, and one is adjacent to a tRNA. Together, 87% of conserved, cis-antisense encoded sRNA are snoRNA-like guides, while only 2.6% are tRNA. In P. aerophilum, C/D box sRNA genes are nearly twice as abundant (88 compared to 46) as tRNA genes, but the sRNA genes are over 40-fold more likely to have a conserved overlap with the orthologous protein coding region. This may be an indication that these C/D box sRNA play a regulatory role with respect to the associated protein coding genes.
A notable example of a convergent ncRNA occurs at the 3′ terminus of the electron transport flavoprotein (etf) operon, where a C/D box sRNA, PAEsR53, overlaps the terminal gene (PAE0721 in P. aerophilum) in this four-gene operon. Like other operons within the Pyrobaculum genus, multiple promoters appear to drive expression of the etf operon (Figure 3). For this operon, an upstream promoter generates a 3400-nt-long full length etfDH-ferredoxin-etfB-etfA transcript. Two predicted internal promoters appear to generate respectively, the ferredoxin-etfB-etfA ∼2250 nt transcript, and the etfA-only 1040 nt transcript.
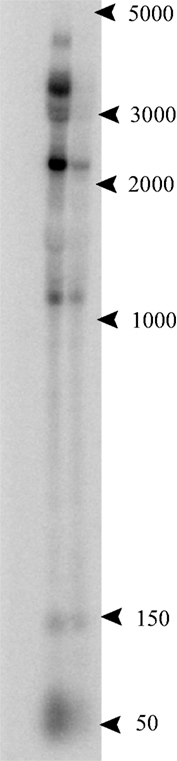
Figure 3. Northern analysis of the 3′ UTR of the electron transport flavoprotein (etf) operon. P. aerophilum total RNA, exponential phase (left lane) and stationary phase (right lane). The probe was designed to anneal beyond the stop codon of the terminal gene in the etf operon, in the region of the convergently oriented PAEsR53 C/D box sRNA. Multiple bands at 3400, 2250, and 1040 nt are consistent with the etf operon and suboperon transcripts. The band near 50 nt, consistent with the RNA sequencing data, shows an apparent antisense transcript to PAEsR53 (sense relative to the 3′ UTR of the etf operon).
The P. aerophilum sRNA sequencing data revealed a strong abundance of sequences mapping to PAEsR53, as well as sequences of the same general size and location, mapping to the opposite strand (the UTR of the etf operon). Northern hybridization was performed to determine the origin of these “anti-PAEsR53” reads. Figure 3 shows that these reads likely originate from the overlapping 3′ UTR of the etf operon, suggesting a possible interaction of the C/D box machinery with the etf mRNA. Predicted orthologs of this C/D box sRNA (PAEsR53) are syntenic with etfA in all Pyrobaculum species studied, overlapping the 3′ end of etfA orthologs by ∼12 bases. The overlap positions the D box guide sequence of PAEsR53 over the etfA stop codon in all Pyrobaculum species. If the guide RNA interacts through complementarity with the etfA mRNA, it could enable a 2′-O-methyl modification of the central “A” nucleotide within the conserved TAA stop codon in all four species.
The Transcription Initiation Factor B Genes, tfb1 and tfb2
The genomes of Pyrobaculum species contain a pair of paralogous genes that encode alternate forms of transcription initiation factor B (TFB). This factor is required for the initiation of basal level transcription at archaeal promoters (Santangelo et al., 2007).
In every sequenced Pyrobaculum species, TFB1 (PAE1645 and orthologs) contains a short N-terminal extension (22 amino acids in P. aerophilum) that is not present in the TFB2 proteins (PAE3329 and orthologs). Sequencing data reveals the presence of an abundant sRNA (asR1) encoded on the antisense strand that overlaps the 5′ end of tfb1 (Figure 4A) in all four Pyrobaculum species examined (Table A1 in Appendix). Tfb1 also appears to have two promoters separated by 17–18 nt, such that the upstream promoter (Pu) is positioned to drive expression of full length tfb1, while the downstream promoter (Pd) generates transcripts that would lack a start codon near the start of the transcript.
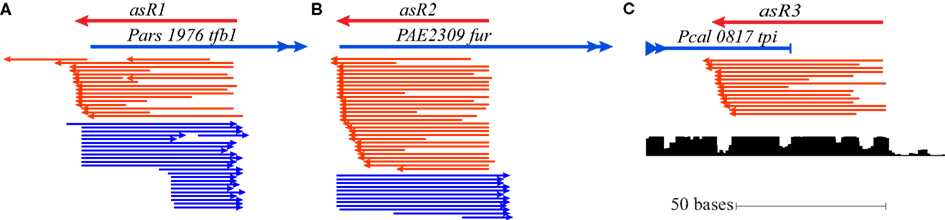
Figure 4. Cis-antisense transcription occurring at three conserved loci in Pyrobaculum spp. The antisense sRNA genes (indicated as asR1, asR2, and asR3) are defined over the region of antisense-oriented sequencing reads (shown in red) with the associated gene (sense orientation) shown in blue. (A) Sequencing reads map to the 5′ terminus of transcription initiation factor B in both sense and antisense orientations (Pars_1976 is shown from Pyrobaculum arsenaticum). Sense oriented transcripts (blue) are present in two populations, consistent with the two conserved promoters, Pu and Pd. (B) The antisense sRNA (asR2) gene is defined over the region of antisense-oriented reads that map to the 5′ terminus of the ferric uptake regulator gene (fur; PAE2309 from Pyrobaculum aerophilum is shown). (C) Antisense-oriented reads map to the 3′ end of the triose-phosphate isomerase (tpi) locus (Pcal_0817 from Pyrobaculum calidifontis). Sequence conservation [lower graphic (C)] extends beyond the tpi stop codon (UCSC Archaeal Browser; Chan et al., 2012).
In P. aerophilum, asR1 sRNA is about 59 nt in length (Table 2; Figure 4), with a well-defined 5′ end that overlaps the extension region of the tfb1 gene. The 3′ end of asR1 is located just upstream of the tfb1 translation initiation codon, precisely at the predicted start of transcription consistent with the Pu promoter. Importantly, there is an additional set of asR1 sRNA reads of 41 nt in length, starting at the same 5′ position but terminating early, at the 5′ end of tfb1 transcripts consistent with the alternate Pd promoter. Mirroring the two variants of the antisense asR1 transcript, deep sequencing revealed a large number of short sense strand sequencing reads, consistent with fragments representing the 5′ end of tfb1 transcripts generated by Pu and Pd, spanning 50 and 32 nt in length respectively.
Northern analysis of total RNA from P. aerophilum confirmed the presence of a population of sense oriented transcripts of about 1000 nt in length, consistent with full length mRNA and another transcript population consistent with the sense oriented sRNAs described above (Figure 5A). When the antisense sRNA is probed, a population of short transcripts near 50 nt is detected (Figure 5B). The full length sense transcripts appear to be relatively constant in abundance across growth phase and culture conditions, consistent with data from a prior microarray study using the same RNA samples (Cozen et al., 2009). The correlated abundance of sense and antisense sRNA (Figures 5C–E) suggests that these sense::antisense pairs are associated, potentially as a double-stranded RNA. The elevated abundance of these pairs relative to the mRNA (Figure 5A) suggests that the sRNA pairs are stabilized within a dsRNA complex. The role of asR1 with respect to tfb1 transcripts is unclear, though the modulation of sRNA (both sense and antisense) while tfb1 mRNA remains at constant and low abundance is reminiscent of negative feedback control.
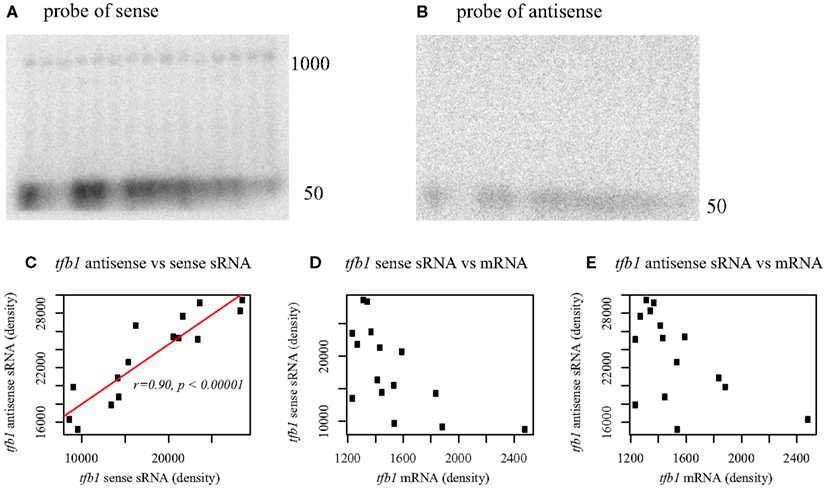
Figure 5. Northern blot analysis of tfb1 from Pyrobaculum aerophilum using probes to the sense strand of tfb (A) and antisense strand (B). Sense transcripts of tfb1 occur at both full length (∼1000 nt) and at 50 nt. Antisense transcripts occur at ∼50 nt. Lanes 1–15 (upper panels, left to right); total RNA across five respiratory growth conditions in three time series. Lanes 1–3 stationary phase, Lanes 4–6 growth with O2, 7–9 growth with NO3, 10–12 growth with As(V), 13–15 Fe(III). Each set of three lanes extracted from a time series for all five respiratory conditions at T = (2.5, 4.5, 7.5 h) with indicated terminal electron acceptor. Sense and antisense sRNA transcript abundance, inferred from band density, is positively correlated across growth conditions (C), while no significant correlation is found between full length tfb1 mRNA and either sRNA population (D,E). Full length tfb1 transcripts [1000 nt] remain nearly constant under all conditions tested (A). Band density established using imagej (http://rsb.info.nih.gov/ij/).
The presence of complementary sense and antisense transcripts has been observed in a previous RNA sequencing study (Tang et al., 2005). Those authors suggested that the presence of an antisense transcript might enhance the stability of the mRNA target. As exemplified with tfb1, the presence of cis-antisense transcripts in our data are often accompanied by the presence of complementary sense strand fragments of similar size. This observation suggests that formation of a dsRNA duplex between the antisense sRNA and the 5′ region of the mRNA target may trigger destabilization of the mRNA; or alternatively, that base pairing between the antisense sRNA and the 5′ end of the nascent mRNA early in elongation may trigger premature transcription termination. For either mechanism, the result appears to be a constant level of tfb1 mRNA under a variety of different culture conditions and growth phases.
The Ferric Uptake Regulator Gene (fur)
In a number of bacteria, the ferric uptake regulator FUR, is a transcriptional regulator of genes encoding proteins involved in iron homeostasis and protection from the toxic effects of iron under aerobic conditions. Some bacteria also encode a FUR-associated sRNA, for example ryhB; its synthesis is negatively regulated by FUR. The ryhB sRNA functions as a negative regulator of genes whose transcription is indirectly activated by FUR. The mechanism of ryhB sRNA negative regulation involves base pairing followed by selective degradation of the targeted mRNA (Andrews et al., 2003).
A homolog of the fur gene is conserved in the genomes of all known Pyrobaculum species. Embedded in each of the associated genes and located about 75 nt downstream from the 5′ start codon is an antisense-oriented, promoter-like sequence. In the two studied facultative aerobes (P. aerophilum and P. calidifontis), we detected a novel 54 nt-long cis-antisense transcript (Table A1 in Appendix), designated as asR2, with precise transcription initiation consistent with the noted antisense promoter-like sequence. The 3′ end of the asR2 transcript (Table 2; Figure 4B) transcript terminates just upstream of the fur translation start codon. Both the asR2 transcript and a complementary RNA fragment apparently derived from the 5′ end of fur mRNA, were present at high levels in anaerobically grown P. aerophilum and at modest levels in P. calidifontis. In the strict anaerobes (P. islandicum, P. arsenaticum), it appears that sequencing depth was insufficient to resolve any antisense-sense pairs under the limited set of growth conditions; however, we note that the predicted promoter for asR2 in the facultative aerobes is equally well-conserved across all Pyrobaculum species.
The Triose-Phosphate Isomerase (tpi) Gene
The tpi gene encodes triose-phosphate isomerase, an enzyme that is central to the modified Embden–Meyerhoff glycolytic pathway in Pyrobaculum species (Reher et al., 2007). We detected a 65-nt-long antisense transcript asR3 (Table A2 in Appendix) that overlaps the 3′ end of the tpi gene (Figure 4C) in all four of the species examined. Upon further examination of the 3′ terminal portion of tpi, we also detected a conserved sequence and associated secondary structure that is present in all sequenced Pyrobaculum spp. (Figure 6), which we term the tpi-element. In P. aerophilum, P. islandicum, and P. calidifontis, the tpi-element includes the stop codon of tpi, while the entire element is encoded immediately downstream of the tpi stop codon in the remaining Pyrobaculum spp.
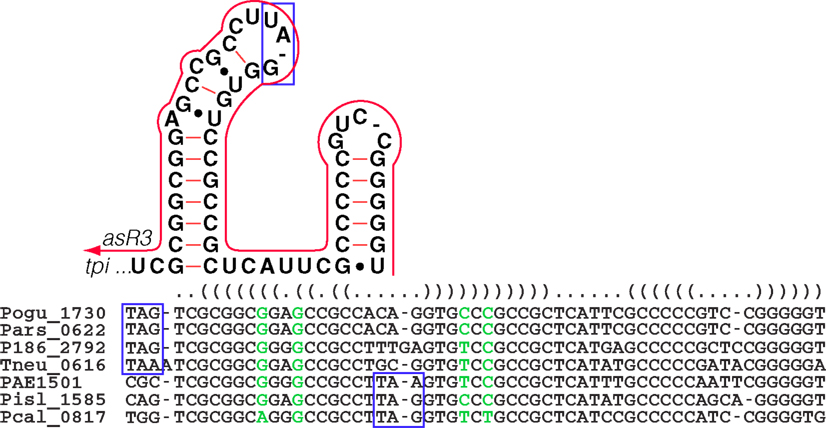
Figure 6. The Pyrobaculum tpi-element and the associated antisense element, asR3. In all sequenced Pyrobaculum species, a highly conserved primary sequence forms a predicted secondary structure element (upper panel) at the 3′ end of the triose-phosphate isomerase gene (tpi). The depicted secondary structure contains the stop codon (blue box) in P. aerophilum (PAE1501), P. islandicum (Pisl_1585), and P. calidifontis (Pcal_0817). In P. oguniense (Pogu_1730), P. arsenaticum (Pars_0622), Pyrobaculum sp.1860 (P186_2792), and P. neutrophilum (Tneu_0616), the stop codon is immediately upstream of the tpi-element. asR3 (red line) is encoded on the opposite strand, and has potential to compete/interfere with the tpi-element secondary structure. The genomic alignment of the 3′ portion of tpi and 3′ UTR is shown with the consensus secondary structure (nested parentheses). Base paired columns with one or more substitutions that maintain secondary structure are highlighted (green).
A dsRNA formed by an interaction of asR3 with the tpi-element could potentially compete against the mRNA intramolecular structure, and thus modulate function of the highly conserved tpi-element. Alternatively, asR3 might itself be the active element of the pair, and in that case, presence of free tpi transcript might act as a repressor of asR3. In this model, asR3 may have other trans targets in the genome and play a more general role in coordination of glycolysis in Pyrobaculum species.
Discussion
Comparative transcriptomics has revealed compelling, conserved cases of novel cis-encoded transcripts that are antisense to core protein coding genes involved in transcription initiation and metabolism. We have considered these most obviously as potential regulators of their opposite strand partners, but they might also have broader regulatory roles.
We found that 28 of the top 34 cases of conserved 3′ antisense expression among orthologous Pyrobaculum proteins of known function coincide with convergent C/D box guide RNAs. This finding suggests that guide directed 2′-O-methylation of the mRNA in the region or downstream of the stop codon might be an unrecognized component of mRNA metabolism and gene regulation. It has been shown that pseudouridine modification of a stop codon can suppress termination of translation (Karijolich and Yu, 2011), but there are currently no studies of the possible implications of 2′-O-methyl modification on mRNA translation or stability. Alternatively, the presence of abundant mRNA fragments at the 3′ end may indicate that a sense-antisense interaction between the C/D box sRNA and mRNA terminus results in truncation of the mRNA by an unknown mechanism, leading to mRNA destabilization and degradation.
The coordinated regulatory program implemented by Fur and its regulatory sRNA ryhB in some bacteria, provides a mechanism that yields both repression of some genes and activation (de-repression) of others. This program balances the needs of iron storage and utilization while protecting from iron-induced toxicity under oxic conditions. These dual roles are mediated by the inverse expression patterns of Fur and ryhB. Fur negatively regulates ryhB, which negatively regulates downstream genes. This circuit allows Fur to derepress (activate) those downstream genes. In published studies, active transcription in one direction can negatively regulate expression of the cis-encoded antisense partner (Lapidot and Pilpel, 2006), thus creating exclusive access to the shared genomic region. Likewise in Pyrobaculum, the cis-antisense transcription observed may yield the same type of inverse expression pattern. In this view, if the cis-antisense gene product is capable of repressing transcription or translation of targets in trans, then positive expression of Pyrobaculum fur, tfb1, or tpi may act through their corresponding antisense partner to activate (derepress) additional members of the associated regulon. Identification and verification of targets in trans is difficult in species that are not amenable to genetic manipulation such as Pyrobaculum, although future studies will explore computationally predicted targets.
The presence of asR1, a cis-encoded antisense RNA found within tfb1 but not tfb2 is of special interest when we consider these possible models of action for the cis-encoded antisense RNA. A specific TFB and TATA binding protein (TBP) pair in the archaeal halophile Halobacterium sp. NRC-1 has been shown to activate transcription under heat shock conditions (Coker and DasSarma, 2007). The observations that there are two instances of tfb in all Pyrobaculum genomes, and that only one harbors an antisense gene, suggest that tfb1 might be essential only under particular conditions and/or initiate transcription for a subset of Pyrobaculum genes. Under this model, tfb1 transcription might be held at low levels by the presence of asR1 and possibly a dsRNA-binding complex. Under the alternative view, the cis-encoded asR1 might facilitate activation of a trans-encoded regulon via de-repression. In the former view, the mechanism(s) that regulate sRNA transcription, stability, and mRNA interaction are central, while in the latter model, the sRNA is a downstream effector molecule of the independently regulated top-strand mRNA partner. In either case, resolving the molecular details of the sRNA’s interaction with tfb1 are needed to better understand this potential high-level mechanism for broad gene regulation in Pyrobaculum.
The tpi-element and its associated antisense partner, asR3, may provide a novel regulatory circuit acting from the 3′ UTR of tpi. The structure of the tpi-element (Figures 4C and 6) contains the stop codon in some species while in other species the conserved structure is just downstream of the tpi stop codon. Possibilities for the function of the tpi-element include early transcription termination or translation termination. In either case, the tpi-element could be acting as a novel 3′ UTR riboswitch by binding a small molecule, or alternatively may be involved in a protein-binding event. Just as described above, the cis-antisense element asR3, encoded opposite the tpi-element, may act as a repressor of tpi-element function, or may have a role in trans with other genes in the tpi regulon.
In this study, we have described 74 or more expressed C/D box sRNA in each of four transcriptomes, most of which are conserved among multiple Pyrobaculum species. We have shown evidence that an unexpectedly large number of these sRNA overlap protein coding genes. Three novel sRNAs asR1, asR2, and asR3 overlap genes involved in core transcription, iron regulation and core metabolism. Sequencing data have revealed the presence of sRNA originating from both strands, and these transcripts can be supported by promoter analysis, and verified by northern analyses. By contrast, less than 1% of transcripts mapped to CRISPR arrays show any evidence of dual strand transcripts (Figure 1). We suggest that the presence of dual-stranded transcript reads is an indication of an interaction of an sRNA with a convergently oriented mRNA, potentially mediated by one or more unknown dsRNA-binding complexes.
Future RNA-seq studies employing deeper sequencing technologies, alternative growth conditions, and other archaeal species will likely uncover many more cases of candidate regulatory antisense RNA. This work suggests multiple new research directions and will require complementary methodologies to better understand the complexity of sRNA function in Archaea. Given the conserved patterns of cis-antisense RNA transcripts now apparent, we anticipate rapid progress from follow-up studies that will demonstrate new modes of gene regulation homologous or analogous to those found in bacteria and eukaryotes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to members of the Joint Genome Institute for making 454 sequencing possible (P. Richardson and J. Bristow for providing resources, and E. Lindquist and N. Zvenigorodsky for sample preparation and analysis). We thank Aaron Cozen for his generous procedural guidance and for the use of RNA blots used in the study. This work was supported by National Science Foundation Grant EF-082277055 (Todd M. Lowe and David L. Bernick); the Graduate Research and Education in Adaptive Bio-Technology (GREAT) Training Program sponsored by the University of California Bio-technology Research and Education Program (David L. Bernick); and by the National Science Foundation while Patrick P. Dennis was working at the Foundation. The opinions, findings, and conclusion expressed in this publications are ours and do not necessarily reflect the views of the National Science Foundation.
Author Contributions
David L. Bernick designed and performed the experimental and computational analyses, and wrote the manuscript. Lauren M. Lui analyzed the C/D box sRNA sequencing data. Patrick P. Dennis provided assistance with the manuscript, collaborative review, and structure determination of C/D box sRNA. Todd M. Lowe provided scientific direction, contributed to interpretation of results, and edited the manuscript.
Footnotes
References
Aiba, H. (2007). Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 10, 134–139.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Andrews, S. C., Robinson, A. K., and Rodriguez-Quinones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237.
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., Romero, D. A., and Horvath, P. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712.
Bernick, D. L., Dennis, P. P., Hochsmann, M., and Lowe, T. M. (2012). Discovery of Pyrobaculum small RNA families with atypical pseudouridine guide RNA features. RNA 18, 402–411.
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., and Madden, T. L. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10, 421.
Chan, P. P., Holmes, A. D., Smith, A. M., Tran, D., and Lowe, T. M. (2012). The UCSC Archaeal Genome Browser: 2012 update. Nucleic Acids Res. 40, D646–D652.
Coker, J. A., and DasSarma, S. (2007). Genetic and transcriptomic analysis of transcription factor genes in the model halophilic archaeon: coordinate action of TbpD and TfbA. BMC Genet. 8, 61.
Cozen, A. E., Weirauch, M. T., Pollard, K. S., Bernick, D. L., Stuart, J. M., and Lowe, T. M. (2009). Transcriptional map of respiratory versatility in the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. J. Bacteriol. 191, 782–794.
Dennis, P. P., Omer, A., and Lowe, T. (2001). A guided tour: small RNA function in archaea. Mol. Microbiol. 40, 509–519.
Fitz-Gibbon, S. T., Ladner, H., Kim, U. J., Stetter, K. O., Simon, M. I., and Miller, J. H. (2002). Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl. Acad. Sci. U.S.A. 99, 984–989.
Hale, C., Kleppe, K., Terns, R. M., and Terns, M. P. (2008). Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA 14, 2572–2579.
Hale, C. R., Zhao, P., Olson, S., Duff, M. O., Graveley, B. R., Wells, L., Terns, R. M., and Terns, M. P. (2009). RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139, 945–956.
Henderson, I. R., Zhang, X., Lu, C., Johnson, L., Meyers, B. C., Green, P. J., and Jacobsen, S. E. (2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38, 721–725.
Jager, D., Sharma, C. M., Thomsen, J., Ehlers, C., Vogel, J., and Schmitz, R. A. (2009). Deep sequencing analysis of the Methanosarcina mazei Go1 transcriptome in response to nitrogen availability. Proc. Natl. Acad. Sci. U.S.A. 106, 21878–21882.
Karijolich, J., and Yu, Y. T. (2011). Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474, 395–398.
Lapidot, M., and Pilpel, Y. (2006). Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep. 7, 1216–1222.
Lau, N. C., Lim, L. P., Weinstein, E. G., and Bartel, D. P. (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862.
Lu, C., Kulkarni, K., Souret, F. F., Muthuvalliappan, R., Tej, S. S., Poethig, R. S., Henderson, I. R., Jacobsen, S. E., Wang, W., Green, P. J., and Meyers, B. C. (2006). MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 16, 1276–1288.
Masse, E., Salvail, H., Desnoyers, G., and Arguin, M. (2007). Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 10, 140–145.
Omer, A. D., Lowe, T. M., Russell, A. G., Ebhardt, H., Eddy, S. R., and Dennis, P. P. (2000). Homologs of small nucleolar RNAs in Archaea. Science 288, 517–522.
Pak, J., and Fire, A. (2007). Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315, 241–244.
Reher, M., Gebhard, S., and Schonheit, P. (2007). Glyceraldehyde-3-phosphate ferredoxin oxidoreductase (GAPOR) and nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN), key enzymes of the respective modified Embden-Meyerhof pathways in the hyperthermophilic crenarchaeota Pyrobaculum aerophilum and Aeropyrum pernix. FEMS Microbiol. Lett. 273, 196–205.
Repoila, F., Majdalani, N., and Gottesman, S. (2003). Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol. Microbiol. 48, 855–861.
Santangelo, T. J., Cubonova, L., James, C. L., and Reeve, J. N. (2007). TFB1 or TFB2 is sufficient for Thermococcus kodakaraensis viability and for basal transcription in vitro. J. Mol. Biol. 367, 344–357.
Straub, J., Brenneis, M., Jellen-Ritter, A., Heyer, R., Soppa, J., and Marchfelder, A. (2009). Small RNAs in haloarchaea: identification, differential expression and biological function. RNA Biol. 6, 281–292.
Tang, T. H., Bachellerie, J. P., Rozhdestvensky, T., Bortolin, M. L., Huber, H., Drungowski, M., Elge, T., Brosius, J., and Huttenhofer, A. (2002). Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. U.S.A. 99, 7536–7541.
Tang, T. H., Polacek, N., Zywicki, M., Huber, H., Brugger, K., Garrett, R., Bachellerie, J. P., and Huttenhofer, A. (2005). Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 55, 469–481.
Vogel, J. (2009). A rough guide to the non-coding RNA world of Salmonella. Mol. Microbiol. 71, 1–11.
Wilderman, P. J., Sowa, N. A., Fitzgerald, D. J., Fitzgerald, P. C., Gottesman, S., Ochsner, U. A., and Vasil, M. L. (2004). Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. U.S.A. 101, 9792–9797.
Wurtzel, O., Sapra, R., Chen, F., Zhu, Y., Simmons, B. A., and Sorek, R. (2010). A single-base resolution map of an archaeal transcriptome. Genome Res. 20, 133–141.
Zago, M. A., Dennis, P. P., and Omer, A. D. (2005). The expanding world of small RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 55, 1812–1828.
Appendix
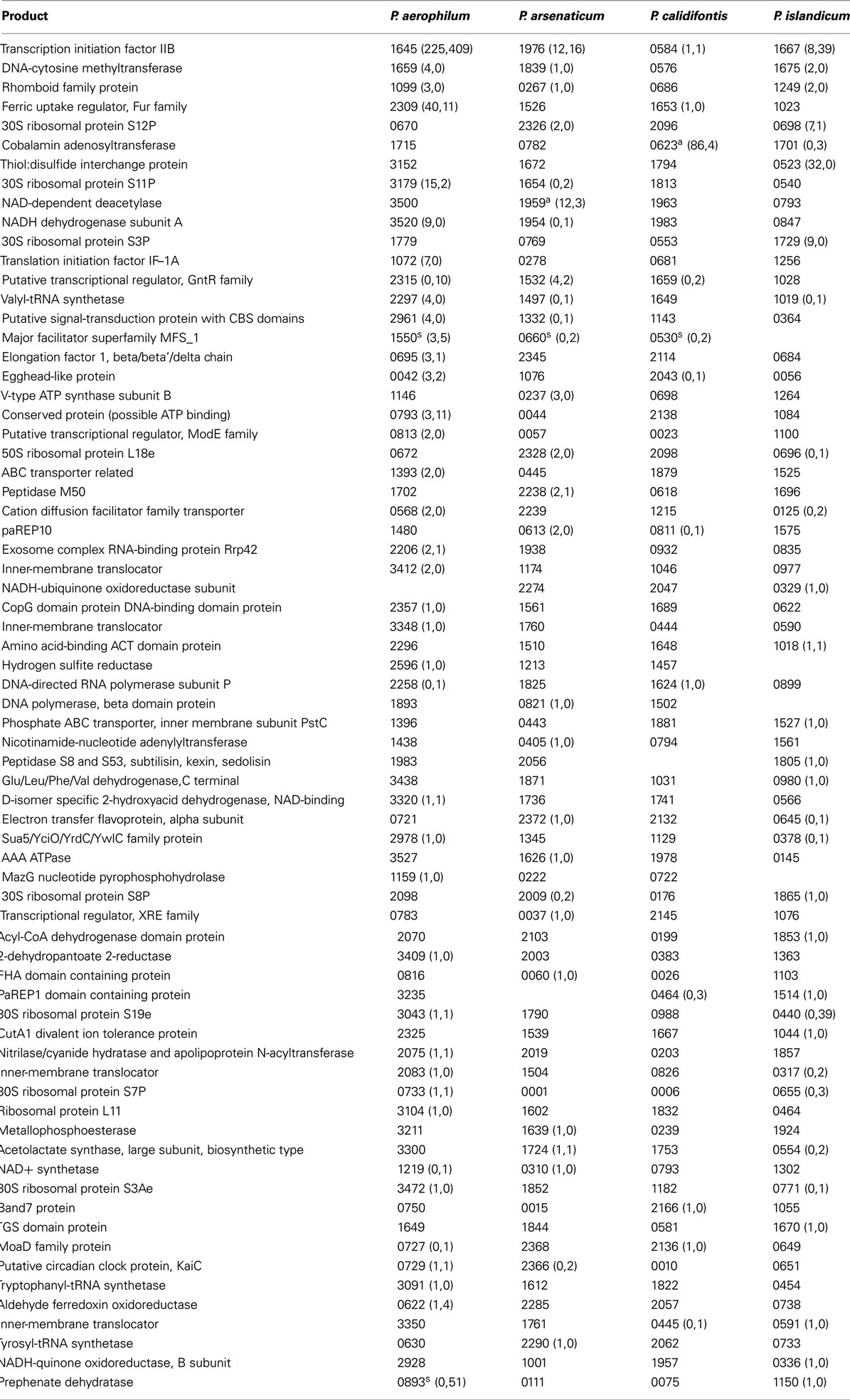
Table A1. Orthologous genes with 5’ sequencing reads. Orthologous groups are shown in each row where the locus tag number (e.g., 1645 for gene PAE1645) is followed by counts of (antisense, sense) reads. Groups are ranked by the total number of reads found within groupings formed by the number of species in a group with antisense sequencing reads. Read counts are accumulated by considering the largest region covered by at least one read in an overlapping region along a given strand, and assigning the read count to that region. Footnoted gene IDs have associated snoRNA-like sRNA (C/D box or H/ACA-like) – a, antisense oriented; s, sense oriented.
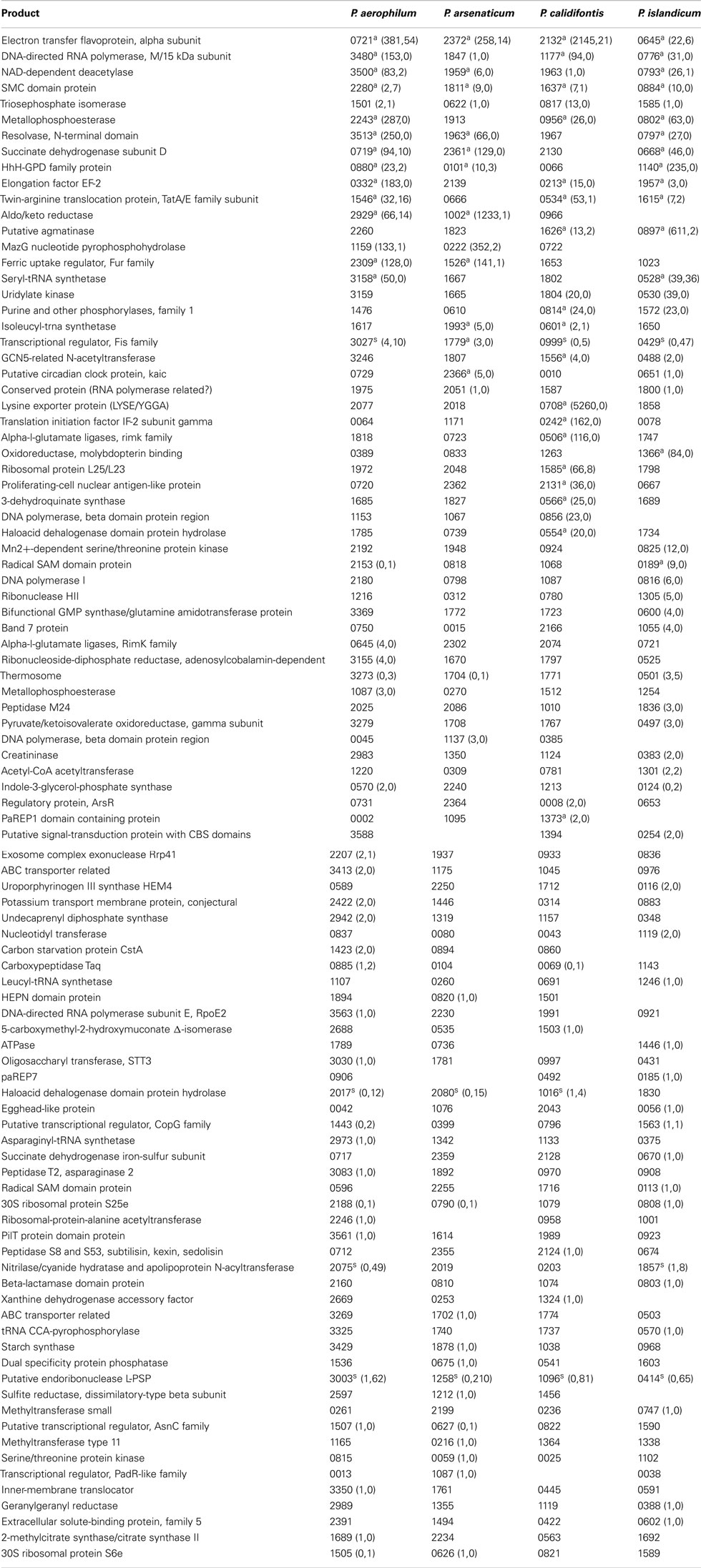
Table A2. Orthologous genes with 3’ sequencing reads. Orthologous groups, read counts, and footnotes displayed are as described in Table A1.
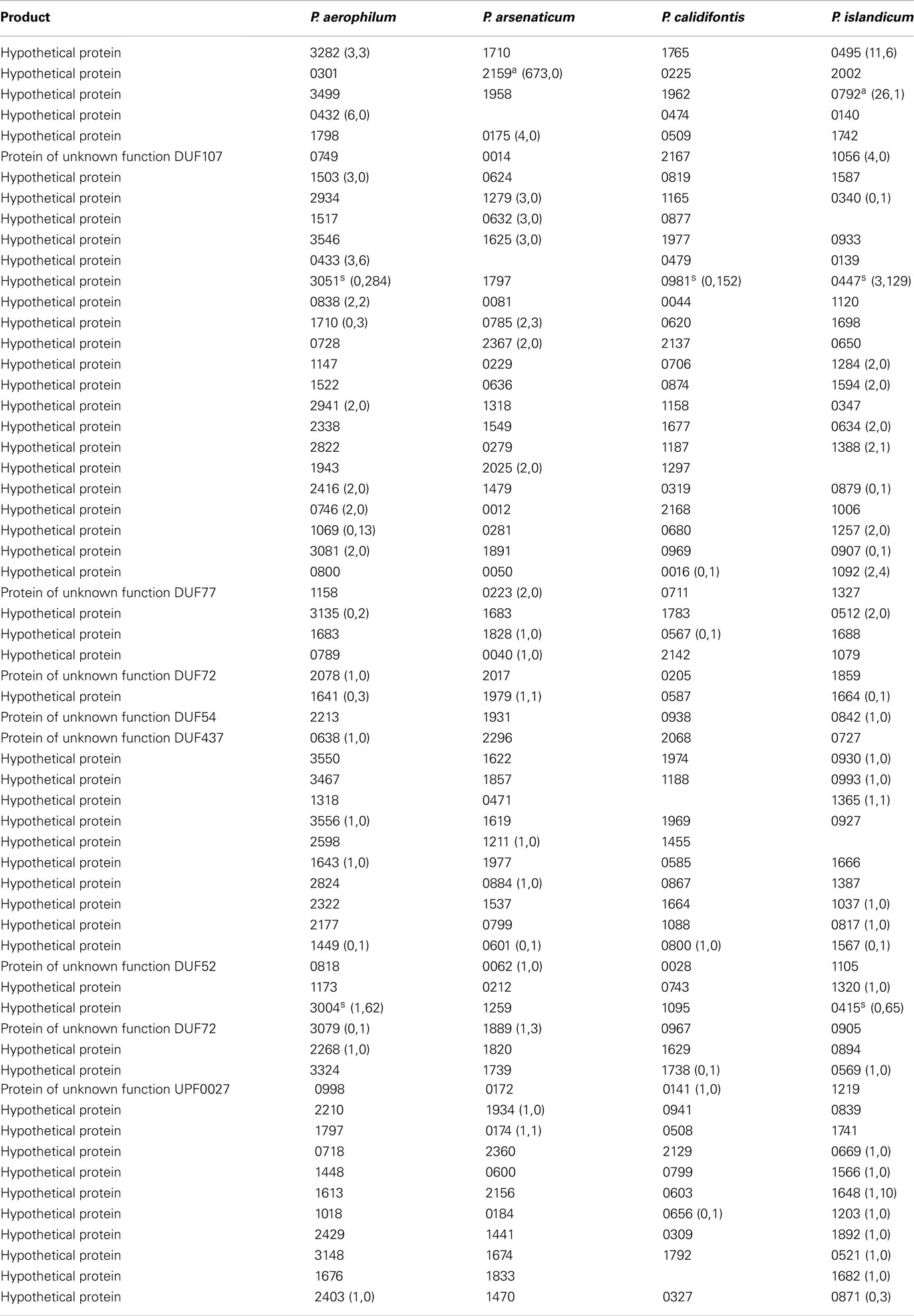
Table A3. Hypothetical genes with 5’ sequencing reads. Orthologous groups, read counts, and footnotes displayed are as described in Table A1.
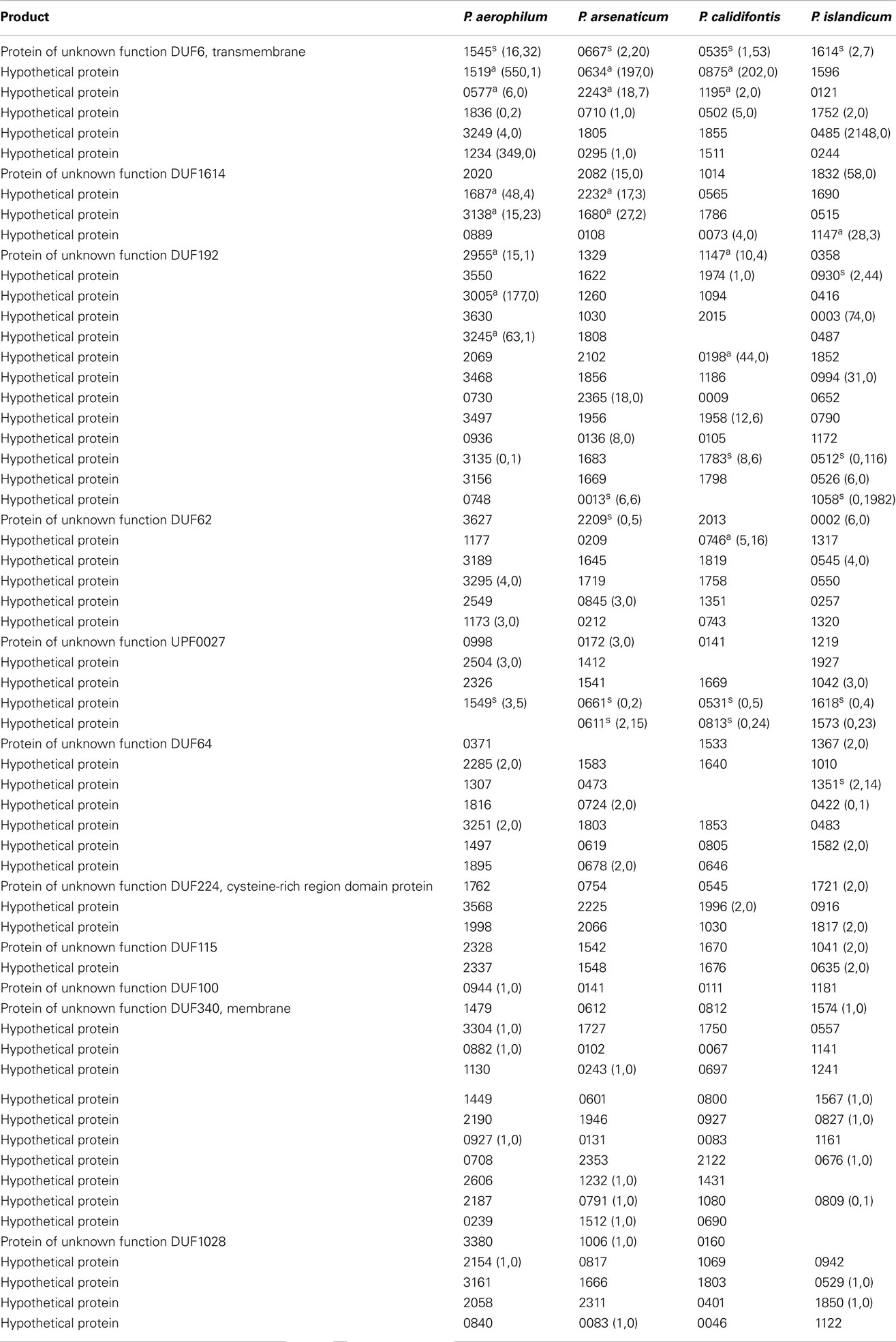
Table A4. Hypothetical genes with 3’ sequencing reads. Orthologous groups, read counts, and footnotes displayed are as described in Table A1.
Keywords: antisense small RNA, archaea, transcriptome sequencing, comparative genomics, gene regulation, C/D box small RNA
Citation: Bernick DL, Dennis PP, Lui LM and Lowe TM (2012) Diversity of antisense and other non-coding RNAs in archaea revealed by comparative small RNA sequencing in four Pyrobaculum species. Front. Microbio. 3:231. doi: 10.3389/fmicb.2012.00231
Received: 28 April 2012; Accepted: 06 June 2012;
Published online: 02 July 2012.
Edited by:
Frank T. Robb, University of California, USAReviewed by:
Mircea Podar, Oak Ridge National Laboratory, USAImke Schroeder, University of California Los Angeles, USA
Matthias Hess, Washington State University, USA
Lanming Chen, Shanghai Ocean University, China
Copyright: © 2012 Bernick, Dennis, Lui and Lowe. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Todd M. Lowe, Department of Biomolecular Engineering, University of California, 1156 High Street, Santa Cruz, CA 95064, USA. e-mail:bG93ZUBzb2UudWNzYy5lZHU=


 Lauren M. Lui1
Lauren M. Lui1
