- 1 Research Institute for Drug Discovery, School of Pharmacy, Kumamoto University, Kumamoto, Japan
- 2 Department of Microbiology, Institute of Health Biosciences, The University of Tokushima Graduate School, Tokushima, Japan
- 3 Department of Bioorganic Medicinal Chemistry, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan
Both human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) encode a unique set of accessory proteins that enhance viral replication in the host. Two similar accessory proteins, Vpx and Vpr, are encoded by HIV-2. In contrast, HIV-1 encodes Vpr but not Vpx. Recent studies have indicated that Vpx counteracts a particular host restriction factor, thereby facilitating reverse transcription in myeloid cells such as monocyte-derived macrophages and monocyte-derived dendritic cells. This mechanism of counteraction is similar to that of the accessory proteins Vif and Vpu which antagonize other host factors. In 2011, the protein SAMHD1 was identified as the restriction factor counteracted by Vpx. Studies have since revealed that SAMHD1 degrades deoxynucleoside triphosphates (dNTPs), which are components of viral genomic cDNA, in order to deprive viruses of dNTPs. Although interactions between SAMHD1 and Vpx continue to be a major research focus, Vpx has also been shown to have an apparent ability to enhance nuclear import of the viral genome in T lymphocytes. This review summarizes the current knowledge regarding SAMHD1-dependent and -independent functions of Vpx, and discusses possible reasons why HIV-2 encodes both Vpx and Vpr, unlike HIV-1.
Introduction
Human and simian immunodeficiency viruses (HIV/SIVs) carry a unique set of accessory proteins, Vif, Vpx, Vpr, Vpu, and Nef, which enhance viral replication in the host. Of these accessory proteins, Vpx is unique to HIV-2-type viruses, defined in this paper as the HIV/SIVs carrying both Vpr and Vpx, such as HIV-2, SIVsmm (Sooty mangabey), and SIVmac (Rhesus monkey) (Fujita et al., 2010). Vpr and Vpx are small proteins of approximately 100 amino acids and similar sequence (approximately 20–25% similarity). Both Vpr and Vpx are predicted to have a similar structure consisting of three major helices (Khamsri et al., 2006). In contrast, while HIV-1 carries Vpr, it does not carry Vpx. The answer to the question why HIV-2 viruses encode these two similar proteins while HIV-1 carries only one remains elusive, and must await the determination of their functional details.
Extensive research over the past decade has revealed that lentiviruses carry genes for accessory proteins that overcome host antiviral factors. The first such accessory protein identified was Vif, which inactivates APOBEC3 proteins, cellular cytidine deaminases that restrict the replication of retroviruses by hypermutating viral cDNA and/or inhibiting reverse transcription (Sheehy et al., 2002; Goila-Gaur and Strebel, 2008; Kitamura et al., 2011). Vif reduces the amount of APOBEC3 through proteasome-mediated degradation and other degradation-independent mechanisms. The second major finding in this area was that the viral protein Vpu counteracts host BST-2/tetherin, which normally blocks the release of virions by directly tethering viral particles to the membranes of infected cells (Neil et al., 2008; Van Damme et al., 2008; Arias et al., 2011). The mechanism through which Vpu antagonizes the function of BST-2/tetherin may be proteasome/lysosome degradation or relocalization from the cell surface.
Recently, it was reported that the viral accessory protein Vpx inhibits the host restriction factor SAMHD1 in monocyte-derived macrophages (MDMs) and monocyte-derived dendritic cells (MDDCs) (Hrecka et al., 2011; Laguette et al., 2011), stimulating interest in SAMHD1 and Vpx. In addition to inhibiting SAMHD1 in MDMs and MDDCs, Vpx is also capable of enhancing viral replication in T lymphocytes (Guyader et al., 1989; Kappes et al., 1991; Yu et al., 1991; Akari et al., 1992; Gibbs et al., 1994; Kawamura et al., 1994; Tokunaga et al., 1997; Ueno et al., 2003; Doi et al., 2011). In this review, we summarize current research into SAMHD1-dependent and -independent functions of Vpx and discuss the virological significance of this protein.
SAMHD1-Dependent Functions of Vpx
Several studies have shown that while wild-type HIV-2-type viruses grow well in MDMs, growth of these Vpx-deletion mutants is completely suppressed, demonstrating that Vpx is essential for viral replication in MDMs (Ueno et al., 2003; Fujita et al., 2008a). It is known that Vpx is packaged in virions and functions in the target cell. Independent work in our laboratory and that of another group revealed that Vpx is critical for reverse transcription of the viral RNA genome in MDMs (Fujita et al., 2008a; Srivastava et al., 2008), correcting the long-held misconception that Vpx contributes to nuclear import of the viral genome but does not play a role in reverse transcription. Furthermore, Vpx was shown to induce proteasome-degradation of an unknown restriction factor to facilitate reverse transcription of the viral genome. It was demonstrated that degradation of the unknown factor involves formation of a Cul4-DDB1-DCAF1 E3 ligase complex (Sharova et al., 2008; Bergamaschi et al., 2009; Kaushik et al., 2009). Considerable effort was subsequently directed toward identification of the unknown factor, and in 2011 SAMHD1 was identified as the MDM host factor from co-immunoprecipitation studies of Vpx expressed in THP-1 cells and in 293T cells (Hrecka et al., 2011; Laguette et al., 2011). SAMHD1 has a tandem sterile alpha motif (SAM) and HD domain with potential phosphohydrolase activity. The SAMHD1 protein was initially identified from MDDCs as a homolog of mouse interferon-γ-induced protein (Li et al., 2000), and is upregulated in response to viral infection (Prehaud et al., 2005; Hartman et al., 2007; Zhao et al., 2008). Furthermore, SAMHD1 is believed to be involved in regulating cellular intrinsic antiviral responses (Rice et al., 2009).
The identification of SAMHD1 as a target of Vpx was not sufficient to explain all the related phenomena, suggesting the involvement of another factor (Hrecka et al., 2011; Planelles, 2011). However, based on reports indicating that SAMHD1 is a deoxynucleoside triphosphate (dNTP) triphosphohydrolase (Goldstone et al., 2011; Powell et al., 2011), it was hypothesized that SAMHD1 degrades dNTPs (which are small molecule components of viral genomic cDNA) in order to deprive viruses of dNTPs by keeping their concentration low. Lahouassa et al. (2012) recently demonstrated the validity of this hypothesis (Figures 1 and 2). Thus, the additional factor targeted by Vpx appears to be dNTPs. Although dNTPs are utilized for reverse transcription in the cytosol, they are small enough to freely diffuse through nuclear pores in and out of the nucleus. Since SAMHD1 is a nuclear protein (Rice et al., 2009), it is most likely that the concentration of dNTPs in the cytosol is controlled by SAMHD1 in the nucleus. In fact, it was suggested that Vpx-mediated degradation of SAMHD1 is initiated in the nucleus (Brandariz-Nuñez et al., 2012; Figure 2).
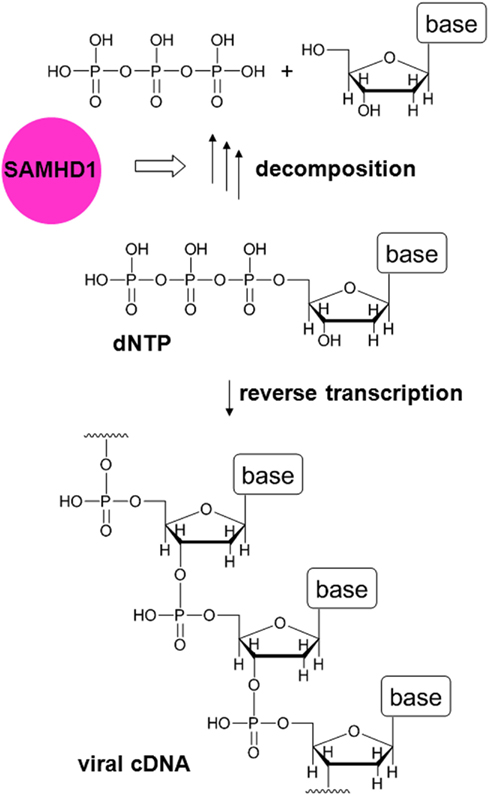
Figure 1. Function of SAMHD1 in MDMs. In reverse transcription, viral cDNA is synthesized from dNTPs. The host restriction factor SAMHD1 is a dNTP triphosphohydrolase that degrades dNTPs. Thus, in the presence of SAMHD1, the concentration of dNTPs becomes low and reverse transcription is inhibited.
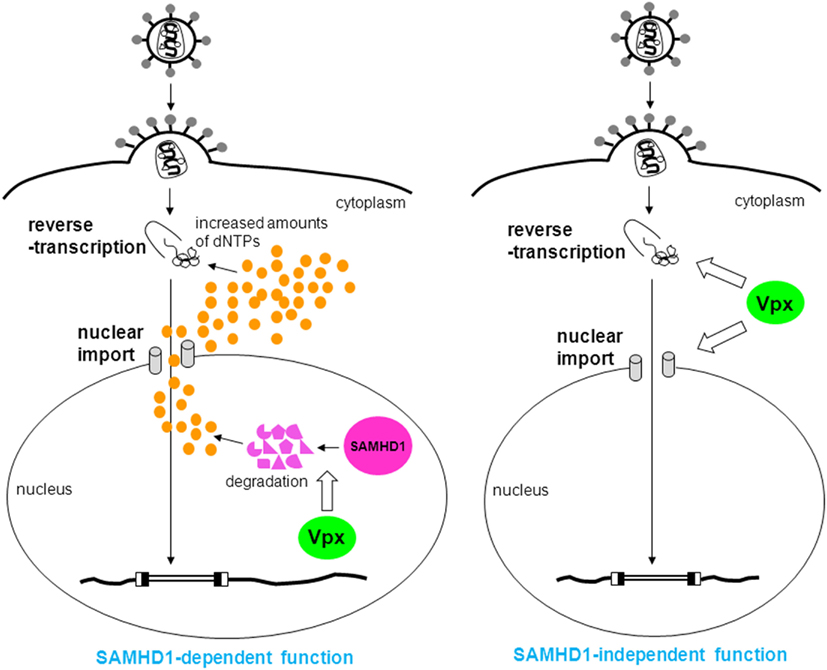
Figure 2. SAMHD1-dependent and -independent functions of Vpx. In the former function, Vpx degrades SAMHD1 to increase amounts of dNTPs (orange circles), thus, reverse transcription proceeds (left). In the latter function, Vpx enhances reverse transcription and nuclear import by an unknown mechanism (right).
In addition to being components of the viral genome, dNTPs are components of the host genome; thus, proliferative CD4+ T cells do not express SAMHD1, and maintain the concentration of dNTPs at an optimal level for cell proliferation (2–4 μM; Lahouassa et al., 2012). In contrast, since MDMs do not proliferate, they do not require high levels of dNTPs, and the low dNTP levels (20–40 nM) resulting from SAMHD1-mediated degradation are therefore not harmful to MDMs (Lahouassa et al., 2012). HIV-2-type viruses carry Vpx for proteasome-mediated degradation of SAMHD1 in order to facilitate replication in MDMs. In contrast, HIV-1 does not require Vpx in order to replicate in MDMs (Fujita et al., 2010) because its reverse transcriptase (RT) is capable of catalyzing viral cDNA synthesis from very low levels of dNTPs (Diamond et al., 2004; Lahouassa et al., 2012). The activity of HIV-2 RT is probably lower [Michaelis constant (Km) of HIV-2 RT is higher] than that of HIV-1 RT, and therefore, to overcome this disadvantage, HIV-2-type viruses may have evolved to carry Vpx.
We previously mapped the functional region of Vpx involved in viral replication in MDMs (Fujita et al., 2008a,b; Figure 3). It is known that the region in major helix 3 containing amino acids Q76 and F80 interacts with DCAF1, a subunit of the Cullin4-based E3 ubiquitin ligase complex (Srivastava et al., 2008). This region could overlap with a region that is critical for virion incorporation (Park and Sodroski, 1995; Jin et al., 2001). Gramberg et al. (2010) suggested that another region, which includes amino acids P9, N12, E15, E16, and T17 in the N-terminal loop, binds to a restriction factor; this region was later confirmed to be a SAMHD1-binding region (Ahn et al., 2012).
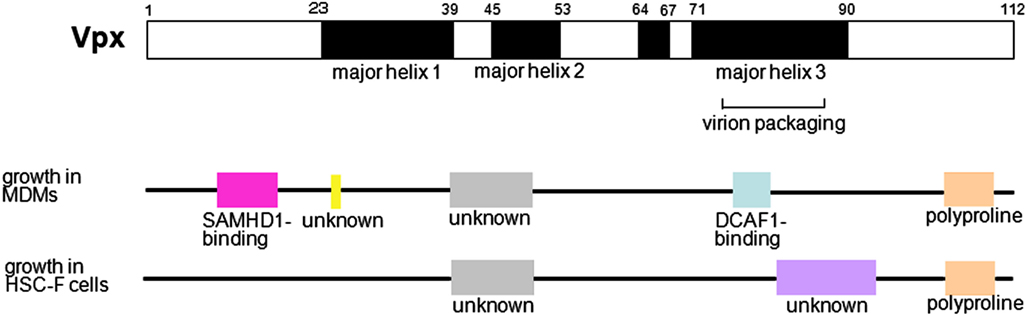
Figure 3. Mapping of the functional region of Vpx. In the upper structure, the positions of predicted helices are shown in black (HIV-2 GL-AN clone; Khamsri et al., 2006). The region important for virion packaging is shown. Middle diagram shows regions critical for replication in MDMs: the SAMHD1-binding region (pink), mechanistically unknown amino acid (yellow), mechanistically unknown region (gray), DCAF1-binding region (light blue), and polyproline motif (pale orange). Lower diagram shows regions involved in replication in HSC-F cells: the mechanistically unknown region (gray), mechanistically unknown and unique region (purple), and polyproline motif (pale orange). The polyproline motif is critical for stable expression of Vpx. All functionally important regions, except those for virion packaging, SAMHD1-binding, and DCAF1-binding, are based on our report (Fujita et al., 2008a,b).
We also identified several other functional regions in Vpx, including a central region located between major helix 1 and major helix 2, and a polyproline motif in a loop in the C-terminus (Fujita et al., 2008a,b). We revealed that the C-terminal polyproline motif is critical for stable expression of Vpx. Although the function of the central region remains unknown, it has been confirmed that this region is not involved in virion incorporation.
Following the identification of SAMHD1, several investigators showed that although some cells such as undifferentiated THP-1 cells express SAMHD1, both the wild-type HIV-2-type virus and its Vpx mutant infect these cells to an equivalent degree (Hrecka et al., 2011; Planelles, 2011). There are several possible explanations for the similar infectivity of wild-type and Vpx mutant viruses: (1) These cells contain large amounts of dNTPs, and thus, even in the presence of SAMHD1 there are sufficient quantities of dNTPs for viral replication, and (2) SAMHD1 does not function in these cells for some as yet unexplained reason. A plausible explanation may be posttranslational modification (phosphorylation, etc.) of the protein. Further study will be required to uncover the molecular basis for this phenomenon.
Around the time SAMHD1 was identified, it was reported that another host restriction factor, APOBEC3A, inhibits HIV-1 infection of MDMs, and that APOBEC3A is degraded by Vpx (Berger et al., 2010, 2011). In addition, APOBEC3A reportedly decreases the amount of viral cDNA synthesized during reverse transcription. Presumably, degradation of SAMHD1 alone is not sufficient to enable reverse transcription to proceed smoothly, and therefore degradation of APOBEC3A is also required for viral replication in MDMs, suggesting that Vpx functions to counteract the antiviral effects of both APOBEC3A and SAMHD1. A comparative study between APOBEC3A and SAMHD1 must be performed in order to establish each protein’s contribution to restricting lentivirus infection in myeloid cells.
SAMHD1-Independent Functions of Vpx
Prior to the time that Vpx was found to act on reverse transcription (Fujita et al., 2008a; Srivastava et al., 2008), it was thought that Vpx is critical for nuclear import of the viral genome (Fletcher et al., 1996; Pancio et al., 2000), based on the results of non-quantitative polymerase chain reaction (PCR) studies. This notion was supported by the tendency of Vpx to localize in the nucleus when Vpx is transduced to a cell solely (Pancio et al., 2000; Mahalingam et al., 2001). It has been well established that Vpx is critical for reverse transcription in MDMs, but this does not preclude participation of Vpx in nuclear import in these cells. We previously identified several Vpx mutants that are defective in both reverse transcription and nuclear import (Fujita et al., 2010) in MDMs, which suggests that Vpx also enhances nuclear import in these cells. We hypothesize that this function of Vpx is SAMHD1-independent, since it is plausible that there is no connection between the amount of dNTPs and nuclear import. Further investigations are underway in order to determine if this is indeed the case.
In T cells, such as peripheral blood lymphocytes (PBLs), peripheral blood mononuclear cells (PBMCs), and cultured simian cell lines immortalized by Herpesvirus saimiri such as HSC-F and M1.3S cells, HIV-2-type viruses grow well, but Vpx-deletion mutants exhibit defective replication (Guyader et al., 1989; Kappes et al., 1991; Yu et al., 1991; Akari et al., 1992; Gibbs et al., 1994; Kawamura et al., 1994; Tokunaga et al., 1997; Ueno et al., 2003; Doi et al., 2011). These results indicate that Vpx is also important for viral replication in T cells. Dispensability of Vpx for the infection of T cells has been believed by some researchers (Bergamaschi et al., 2009; Belshan et al., 2012), but this belief was probably based on the results of infectious experiments using high-titer virus. Our research showed that Vpx enhances nuclear import of the viral genome in HSC-F cells, and that the smaller effect of Vpx on reverse transcription was also observed (Ueno et al., 2003; Fujita et al., 2008a; Figure 2). We mapped the region of Vpx involved in viral replication in HSC-F cells (Fujita et al., 2008b; Figure 3) and found that as is the case in MDMs, the central region and the C-terminal polyproline motif are critical for replication. There is also a unique functional region spanning from major helix 3 to the C-terminal loop, but how this region influences infectivity is unclear. The apparent SAMHD1- and DCAF-1-binding regions are not necessary for viral replication in HSC-F cells, in contrast to MDMs. Furthermore, in HSC-F and M1.3S cells, expression of SAMHD1 was below the detectable level (Nomaguchi, M. and Adachi, A., in preparation). Thus, in these cells, Vpx enhances reverse transcription and nuclear import of the viral genome through an unknown SAMHD1-independent mechanism. Not only cultured cell lines, but also primary T cells are considered to have SAMHD1-independent functions, since SAMHD1- and DCAF-1-binding regions are dispensable for viral replication in PBLs (Fujita, M. and Adachi, A., unpublished data).
It has been reported that Vpx is important for SIV infection in monkeys, and the predominantly infected cells are the intraepithelial T lymphocytes rather than myeloid cells such as macrophages (Hirsch et al., 1998; Belshan et al., 2012). The Vpx in T cells is considered to play a significant role in infection by HIV-2-group viruses in vivo. Thus, we strongly suggest that SAMHD1-independent functions of Vpx are also important, although almost all the recent Vpx research has focused on SAMHD1-dependent functions.
Why Do HIV-2 Viruses have Two Similar Proteins?
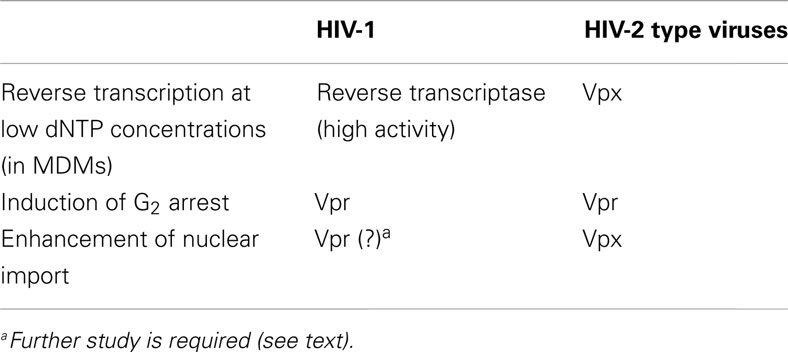
Lim et al. (2012) recently revealed that Vpr, a Vpx-related protein found in HIV-1 and HIV-2-type viruses, is not involved in degradation of SAMHD1. Instead, the Vpr carried by HIV-1 and HIV-2 arrests cells in the G2 phase of the cell cycle, a function not associated with Vpx (Fletcher et al., 1996; Stivahtis et al., 1997; Fujita et al., 2010; Table 1). This G2 arrest is known to be induced via formation of a Cul4-DDB1-DCAF1 E3 ligase complex that includes Vpr as an adaptor. Formation of the complex is followed by proteasomal degradation of an unknown cellular target. This pathway is similar to that involving Vpx, which also functions as an adaptor for the Cul4-DDB1-DCAF1 E3 ligase complex to facilitate proteasomal degradation of SAMHD1 (Ahn et al., 2012). Although the virological significance of the Vpr-mediated G2 arrest has not been determined, this function is likely to be important since it is broadly conserved among HIV/SIV. Since the activity of HIV-2 RT is lower than that of the enzyme found in HIV-1-type viruses, HIV-2 may require SAMHD1 degradation in order to increase the concentration of dNTPs, in addition to induction of G2 arrest.
Both of these functions, SAMHD1 degradation and G2 arrest, are mediated via the Cul4-DDB1-DCAF1 E3 ligase complex. SIVagm (African Green Monkey) is known to have only one Vpr, which induces both the degradation of SAMHD1 (Lim et al., 2012) and G2 arrest (Planelles et al., 1996; Stivahtis et al., 1997; Zhu et al., 2001). Lim et al. proposed that in the evolution of HIV/SIVs, neofunctionalization of Vpr to degrade SAMHD1 resulted in the rapid evolution of the SAMHD1 protein, which induced the birth of a similar protein, Vpx (subfunctionalization), to maximize its SAMHD1-targeting capability. Here, we propose another reason why HIV-2 viruses have similar proteins, Vpr and Vpx. It is known that the region of HIV-1 Vpr spanning from major helix 3 to the C-terminal loop (which includes a cluster of basic amino acids) is critical for induction of G2 arrest (Di Marzio et al., 1995; Selig et al., 1997; Jacquot et al., 2007). This region corresponds to the mechanistically unknown and unique region of Vpx required for replication in HSC-F cells (Figure 3; Khamsri et al., 2006; Fujita et al., 2008b), but the corresponding region in Vpx does not contain a cluster of basic amino acids in the C-terminal loop. Induction of G2 arrest and enhancement of replication in T cells may be incompatible functions for one protein. The presence of both Vpr and Vpx may facilitate G2 arrest and enhancement of HIV-2 replication in T cells, but a full explanation as to why HIV-2 has two proteins that are so similar will require further study.
Conclusion
Recent findings indicating that Vpx mediates the degradation of SAMHD1 are intriguing, and reveal yet another example of a virus with a means to counteract host defense mechanisms. Table 1 summarizes the roles played by Vpx and Vpr in HIV-1 and HIV-2. HIV-2/SIV Vpx negates the effect of the unique host restriction factor SAMHD1 by inducing its degradation, thereby enabling reverse transcription to occur under conditions of low dNTP concentrations. In addition, Vpx enhances reverse transcription and nuclear import of the viral genome in an SAMHD1-independent manner. We are trying to isolate host factor(s) which concern with SAMHD1-independent function. Different regions of the Vpx protein are involved in mediating SAMHD1-dependent and -independent functions. A number of HIV/SIV accessory proteins have multiple functions, including HIV-1 Vpr (Fujita et al., 2010; Sharifi et al., 2012), Vpu (Nomaguchi et al., 2008; Andrew and Strebel, 2010), and Nef (Foster and Garcia, 2008; Laguette et al., 2010). Importance of those two functions of Vpx in the infected individuals should be revealed in the future.
HIV-1 Vpr has a modest effect on replication in MDMs (Fujita et al., 2010), and there have been reports that Vpr enhances nuclear import of the viral genome in these cells (Tsurutani et al., 2000; Agostini et al., 2002). It is possible that HIV-1 Vpr and HIV-2 Vpx function similarly with respect to nuclear import. However, further study is required to elucidate how these proteins impact nuclear import, since the role of HIV-1 Vpr was demonstrated using non-quantitative PCR, and we could not reproduce this result (Fujita et al., 2010).
Expression of Vpx in MDMs results in the degradation of SAMHD1 and a subsequent increase in the concentration of dNTPs, resulting in an increase in the infectivity of HIV-1. To date, studies of Vpx/SAMHD1 have been mainly restricted to HIV-1, even though Vpx is carried by HIV-2-type viruses. Through future studies involving HIV-2-type viruses, we hope to provide a more complete picture of the roles played by Vpx and Vpr.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research (C) (21590511; to Mikako Fujita) from the Japan Society for the Promotion of Science.
References
Agostini, I., Popov, S., Hao, T., Li, J. H., Dubrovsky, L., Chaika, O., Chaika, N., Lewis, R., and Bukrinsky, M. (2002). Phosphorylation of Vpr regulates HIV type 1 nuclear import and macrophage infection. AIDS Res. Hum. Retroviruses 18, 283–288.
Ahn, J., Hao, C., Yan, J., DeLucia, M., Mehrens, J., Wang, C., Gronenborn, A. M., and Skowronski, J. (2012). HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J. Biol. Chem. 287, 12550–12558.
Akari, H., Sakuragi, J., Takebe, Y., Tomonaga, K., Kawamura, M., Fukasawa, M., Miura, T., Shinjo, T., and Hayami, M. (1992). Biological characterization of human immunodeficiency virus type 1 and type 2 mutants in human peripheral blood mononuclear cells. Arch. Virol. 123, 157–167.
Andrew, A., and Strebel, K. (2010). HIV-1 Vpu targets cell surface markers CD4 and BST-2 through distinct mechanisms. Mol. Aspects Med. 31, 407–417.
Arias, J. F., Iwabu, Y., and Tokunaga, K. (2011). Structural basis for the antiviral activity of BST-2/tetherin and its viral antagonism. Front. Microbiol. 2:250. doi:10.3389/fmicb.2011.00250
Belshan, M., Kimata, J. T., Brown, C., Cheng, X., McCulley, A., Larsen, A., Thippeshappa, R., Hodara, V., Giavedoni, L., Hirsch, V., and Ratner, L. (2012). Vpx is critical for SIVmne infection of pigtail macaques. Retrovirology 9, 32.
Bergamaschi, A., Ayinde, D., David, A., Le Rouzic, E., Morel, M., Collin, G., Descamps, D., Damond, F., Brun-Vezinet, F., Nisole, S., Margottin-Goguet, F., Pancino, G., and Transy, C. (2009). The human immunodeficiency virus type 2 Vpx protein usurps the CUL4A-DDB1 DCAF1 ubiquitin ligase to overcome a postentry block in macrophage infection. J. Virol. 83, 4854–4860.
Berger, A., Münk, C., Schweizer, M., Cichutek, K., Schüle, S., and Flory, E. (2010). Interaction of Vpx and apolipoprotein B mRNA-editing catalytic polypeptide 3 family member A (APOBEC3A) correlates with efficient lentivirus infection of monocytes. J. Biol. Chem. 285, 12248–12254.
Berger, G., Durand, S., Fargier, G., Nguyen, X.-N., Cordeil, S., Bouaziz, S., Muriaux, D., Darlix, J.-L., and Cimarelli, A. (2011). APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells. PLoS Pathog. 7, e1002221. doi:10.1371/journal.ppat.1002221
Brandariz-Nuñez, A., Valle-Casuso, J. C., White, T. E., Laguette, N., Benkirane, M., Brojatsch, J., and Diaz-Griffero, F. (2012). Roles of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Retrovirology 9, 49.
Di Marzio, P., Choe, S., Ebright, M., Knoblauch, R., and Landau, N. R. (1995). Mutational analysis of cell cycle arrest, nuclear localization, and virion packaging of human immunodeficiency virus type 1 Vpr. J. Virol. 69, 7909–7916.
Diamond, T. L., Roshal, M., Jamburuthugoda, V. K., Reynolds, H. M., Merriam, A. R., Lee, K. Y., Balakrishnan, M., Bambara, R. A., Planelles, V., Dewhurst, S., and Kim, B. (2004). Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J. Biol. Chem. 279, 51545–51553.
Doi, N., Fujiwara, S., Adachi, A., and Nomaguchi, M. (2011). Rhesus M1.3S cells suitable for biological evaluation of macaque-tropic HIV/SIV clones. Front. Microbiol. 2:115. doi:10.3389/fmicb.2011.00115
Fletcher, T. M. III, Brichacek, B., Sharova, N., Newman, M. A., Stivahtis, G., Sharp, P. M., Emerman, M., Hahn, B. H., and Stevenson, M. (1996). Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIV(SM). EMBO J. 15, 6155–6165.
Fujita, M., Otsuka, M., Miyoshi, M., Khamsri, B., Nomaguchi, M., and Adachi, A. (2008a). Vpx is critical for reverse transcription of the human immunodeficiency virus type 2 genome in macrophages. J. Virol. 82, 7752–7756.
Fujita, M., Otsuka, M., Nomaguchi, M., and Adachi, A. (2008b). Functional region mapping of HIV-2 Vpx protein. Microbes Infect. 10, 1387–1392.
Fujita, M., Otsuka, M., Nomaguchi, M., and Adachi, A. (2010). Multifaceted activity of HIV Vpr/Vpx proteins: the current view of their virological functions. Rev. Med. Virol. 20, 68–76.
Gibbs, J. S., Regier, D. A., and Desrosiers, R. C. (1994). Construction and in vitro properties of SIVmac mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retroviruses 10, 607–616.
Goila-Gaur, R., and Strebel, K. (2008). HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 5, 51.
Goldstone, D. C., Ennis-Adeniran, V., Hedden, J. J., Groom, H. C., Rice, G. I., Christodoulou, E., Walker, P. A., Kelly, G., Haire, L. F., Yap, M. W., de Carvalho, L. P., Stoye, J. P., Crow, Y. J., Taylor, I. A., and Webb, M. (2011). HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382.
Gramberg, T., Sunseri, N., and Landau, N. R. (2010). Evidence for an activation domain at the amino terminus of simian immunodeficiency virus Vpx. J. Virol. 84, 1387–1396.
Guyader, M., Emerman, M., Montagnier, L., and Peden, K. (1989). VPX mutants of HIV-2 are infectious in established cell lines but display a severe defect in peripheral blood lymphocytes. EMBO J. 8, 1169–1175.
Hartman, Z. C., Kiang, A., Everett, R. S., Serra, D., Yang, X. Y., Clay, T. M., and Amalfitano, A. (2007). Adenovirus infection triggers rapid, MyD88-regulated transcriptome response critical to acute-phase and adaptive immune responses in vivo. J. Virol. 81, 1796–1812.
Hirsch, V. M., Sharkey, M. E., Brown, C. R., Brichacek, B., Goldstein, S., Wakefield, J., Byrum, R., Elkins, W. R., Hahn, B. H., Lifson, J. D., and Stevenson, M. (1998). Vpx is required for dissemination and pathogenesis of SIVSMPBj: evidence of macrophage-dependent viral amplification. Nat. Med. 4, 1401–1408.
Hrecka, K., Hao, C., Gierszewska, M., Swanson, S. K., Kesik-Brodacka, M., Srivastava, S., Florens, L., Washburn, M. P., and Skowronski, J. (2011). Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661.
Jacquot, G., Le Rouzic, E., David, A., Mazzolini, J., Bouchet, J., Bouaziz, S., Niedergang, F., Pancino, G., and Benichou, S. (2007). Localization of HIV-1 Vpr to the nuclear envelope: impact on Vpr functions and virus replication in macrophages. Retrovirology 4, 84.
Jin, L., Zhou, Y., and Ratner, L. (2001). HIV type 2 Vpx interaction with Gag and incorporation into virus-like particles. AIDS Res. Hum. Retroviruses 17, 105–111.
Kappes, J. C., Conway, J. A., Lee, S.-W., Shaw, G. M., and Hahn, B. H. (1991). Human immunodeficiency virus type 2 vpx protein augments viral infectivity. Virology 184, 197–209.
Kaushik, R., Zhu, X., Stranska, R., Wu, Y., and Stevenson, M. (2009). A cellular restriction dictates the permissivity of nondividing monocytes/macrophages to lentivirus and gammaretrovirus infection. Cell Host Microbe 6, 68–80.
Kawamura, M., Sakai, H., and Adachi, A. (1994). Human immunodeficiency virus Vpx is required for the early phase of replication in peripheral blood mononuclear cells. Microbiol. Immunol. 38, 871–878.
Khamsri, B., Murao, F., Yoshida, A., Sakurai, A., Uchiyama, T., Shirai, H., Matsuo, Y., Fujita, M., and Adachi, A. (2006). Comparative study on the structure and cytopathogenic activity of HIV Vpr/Vpx proteins. Microbes Infect. 8, 10–15.
Kitamura, S., Ode, H., and Iwatani, Y. (2011). Structural features of antiviral APOBEC3 proteins are linked to their functional activities. Front. Microbiol. 2:258. doi:10.3389/fmicb.2011.00258
Laguette, N., Brégnard, C., Benichou, S., and Basmaciogullari, S. (2010). Human immunodeficiency virus (HIV) type-1, HIV-2 and simian immunodeficiency virus Nef proteins. Mol. Aspects Med. 31, 418–433.
Laguette, N., Sobhian, B., Casartelli, N., Ringeard, M., Chable-Bessia, C., Segeral, E., Yatim, A., Emiliani, S., Schwartz, O., and Benkirane, M. (2011). SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657.
Lahouassa, H., Daddacha, W., Hofmann, H., Ayinde, D., Logue, E. C., Dragin, L., Bloch, N., Maudet, C., Bertrand, M., Gramberg, T., Pancino, G., Priet, S., Canard, B., Laguette, N., Benkirane, M., Transy, C., Landau, N. R., Kim, B., and Margottin-Goguet, F. (2012). SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228.
Li, N., Zhang, W., and Cao, X. (2000). Identification of human homologue of mouse IFN-γ induced protein from human dendritic cells. Immunol. Lett. 74, 221–224.
Lim, E. S., Fregoso, O. I., McCoy, C. O., Matsen, F. A., Malik, H. S., and Emerman, M. (2012). The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11, 194–204.
Mahalingam, S., Van Tine, B., Santiago, M. L., Gao, F., Shaw, G. M., and Hahn, B. H. (2001). Functional analysis of the simian immunodeficiency virus Vpx protein: identification of packaging determinants and a novel nuclear targeting domain. J. Virol. 75, 362–374.
Neil, S. J., Zang, T., and Bieniasz, P. D. (2008). Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430.
Nomaguchi, M., Fujita, M., and Adachi, A. (2008). Role of HIV-1 Vpu protein for virus spread and pathogenesis. Microbes Infect. 10, 960–967.
Pancio, H. A., Vander Heyden, N., and Ratner, L. (2000). The C-terminal proline-rich tail of human immunodeficiency virus type 2 Vpx is necessary for nuclear localization of the viral preintegration complex in nondividing cells. J. Virol. 74, 6162–6167.
Park, I.-W., and Sodroski, J. (1995). Amino acid sequence requirements for the incorporation of the Vpx protein of simian immunodeficiency virus into virion particle. J. Acquir. Immune. Defic. Syndr. Hum. Retrovirol. 10, 506–510.
Planelles, V., Jowett, J. B., Li, Q. X., Xie, Y., Hahn, B., and Chen, I. S. (1996). Vpr-induced cell cycle arrest is conserved among primate lentiviruses. J. Virol. 70, 2516–2524.
Powell, R. D., Holland, P. J., Hollis, T., and Perrino, F. W. (2011). Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286, 43596–43600.
Prehaud, C., Megret, F., Lafage, M., and Lafon, M. (2005). Virus infection switches TLR-3-positive human neurons to become strong producers of beta interferon. J. Virol. 79, 12893–12904.
Rice, G. I., Bond, J., Asipu, A., Brunette, R. L., Manfield, I. W., Carr, I. M., Fuller, J. C., Jackson, R. M., Lamb, T., Briggs, T. A., Ali, M., Gornall, H., Couthard, L. R., Aeby, A., Attard-Montalto, S. P., Bertini, E., Bodemer, C., Brockmann, K., Brueton, L. A., Corry, P. C., Desguerre, I., Fazzi, E., Cazorla, A. G., Gener, B., Hamel, B. C., Heiberg, A., Hunter, M., van der Knaap, M. S., Kumar, R., Lagae, L., Landrieu, P. G., Lourenco, C. M., Marom, D., McDermott, M. F., van der Merwe, W., Orcesi, S., Prendiville, J. S., Rasmussen, M., Shalev, S. A., Soler, D. M., Shinawi, M., Spiegel, R., Tan, T. Y., Vanderver, A., Wakeling, E. L., Wassmer, E., Whittaker, E., Lebon, P., Stetson, D. B., Bonthron, D. T., and Crow, Y. J. (2009). Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41, 829–832.
Selig, L., Benichou, S., Rogel, M. E., Wu, L. I., Vodicka, M. A., Sire, J., Benarous, R., and Emerman, M. (1997). Uracil DNA glycosylase specifically interacts with Vpr of both human immunodeficiency virus type 1 and simian immunodeficiency virus of sooty mangabeys, but binding does not correlate with cell cycle arrest. J. Virol. 71, 4842–4846.
Sharifi, H. J., Furuya, A. M., and de Noronha, C. M. (2012). The role of HIV-1 Vpr in promoting the infection of nondividing cells and in cell cycle arrest. Curr. Opin. HIV AIDS 7, 187–194.
Sharova, N., Wu, Y., Zhu, X., Stanska, R., Kaushik, R., Sharkey, M., and Stevenson, M. (2008). Primate lentiviral Vpx commandeers DDB1 to counteract a macrophage restriction. PLoS Pathog. 4, e1000057. doi:10.1371/journal.ppat.1000057
Sheehy, A. M., Gaddis, N. C., Choi, J. D., and Malim, M. H. (2002). Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650.
Srivastava, S., Swanson, S. K., Manel, N., Florens, L., Washburn, M. P., and Skowronski, J. (2008). Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog. 4, e1000059. doi:10.1371/journal.ppat.1000059
Stivahtis, G. L., Soares, M. A., Vodicka, M. A., Hahn, B. H., and Emerman, M. (1997). Conservation and host specificity of Vpr-mediated cell cycle arrest suggest a fundamental role in primate lentivirus evolution and biology. J. Virol. 71, 4331–4338.
Tokunaga, K., Ishimoto, A., Ikuta, K., and Adachi, A. (1997). Growth ability of auxiliary gene mutants of human immunodeficiency virus types 1 and 2 in unstimulated peripheral blood mononuclear cells. Arch. Virol. 142, 177–181.
Tsurutani, N., Kubo, M., Maeda, Y., Ohashi, T., Yamamoto, N., Kannagi, M., and Masuda, T. (2000). Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J. Virol. 74, 4795–4806.
Ueno, F., Shiota, H., Miyaura, M., Yoshida, A., Sakurai, A., Tatsuki, J., Koyama, A. H., Akari, H., Adachi, A., and Fujita, M. (2003). Vpx and Vpr proteins of HIV-2 up-regulate the viral infectivity by a distinct mechanism in lymphocytic cells. Microbes Infect. 5, 387–395.
Van Damme, N., Goff, D., Katsura, C., Jorgenson, R. L., Mitchell, R., Johnson, M. C., Stephens, E. B., and Guatelli, J. (2008). The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3, 245–252.
Yu, X.-F., Yu, Q.-C., Essex, M., and Lee, T.-H. (1991). The vpx gene of simian immunodeficiency virus facilitates efficient viral replication in fresh lymphocytes and macrophages. J. Virol. 65, 5088–5091.
Zhao, D., Peng, D., Li, L., Zhang, Q., and Zhang, C. (2008). Inhibition of GIP3 expression found in the different display study on respiratory syncytial virus infection. Virol. J. 5, 114.
Keywords: Vpx, HIV-2, SIV, SAMHD1, reverse transcription, dNTP, nuclear import, Vpr
Citation: Fujita M, Nomaguchi M, Adachi A and Otsuka M (2012) SAMHD1-dependent and -independent functions of HIV-2/SIV Vpx protein. Front. Microbio. 3:297. doi: 10.3389/fmicb.2012.00297
Received: 25 April 2012; Paper pending published: 16 May 2012;
Accepted: 25 July 2012; Published online: 10 August 2012.
Edited by:
Atsushi Koito, Kumamoto University, JapanCopyright: © 2012 Fujita, Nomaguchi, Adachi and Otsuka. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Mikako Fujita, Research Institute for Drug Discovery, School of Pharmacy, Kumamoto University, 5-1 Oe-honmachi, Chuo-ku, Kumamoto 862-0973, Japan. e-mail:bWZ1aml0YUBrdW1hbW90by11LmFjLmpw