- Department of Microbiology, School of Pharmacy, Aichi Gakuin University, Nagoya, Japan
Anti-pseudomonas aminoglycosides, such as amikacin and tobramycin, are used in the treatment of Pseudomonas aeruginosa infections. However, their use is linked to the development of resistance. During the last decade, the MexXY multidrug efflux system has been comprehensively studied, and numerous reports of laboratory and clinical isolates have been published. This system has been increasingly recognized as one of the primary determinants of aminoglycoside resistance in P. aeruginosa. In P. aeruginosa cystic fibrosis isolates, upregulation of the pump is considered the most common mechanism of aminoglycoside resistance. Non-fermentative Gram-negative pathogens possessing very close MexXY orthologs such as Achromobacter xylosoxidans and various Burkholderia species (e.g., Burkholderia pseudomallei and B. cepacia complexes), but not B. gladioli, are intrinsically resistant to aminoglycosides. Here, we summarize the properties (e.g., discovery, mechanism, gene expression, clinical significance) of the P. aeruginosa MexXY pump and other aminoglycoside efflux pumps such as AcrD of Escherichia coli, AmrAB-OprA of B. pseudomallei, and AdeABC of Acinetobacter baumannii. MexXY inducibility of the PA5471 gene product, which is dependent on ribosome inhibition or oxidative stress, is noteworthy. Moreover, the discovery of the cognate outer membrane component (OprA) of MexXY in the multidrug-resistant clinical isolate PA7, serotype O12 deserves special attention.
Introduction
Pseudomonas aeruginosa has been recognized as an increasingly important and worrisome species in health care-associated infections (Poole, 2011). This bacterium possesses intrinsic resistance to many antimicrobials because of the low permeability of its outer membrane barrier and the presence of multidrug efflux transporters (Nikaido, 1994; Hancock, 1998). Although fluoroquinolones (e.g., ciprofloxacin), broad-spectrum β-lactams (e.g., imipenem), and anti-pseudomonas aminoglycosides (e.g., amikacin) are often available for treatment, P. aeruginosa readily acquires resistance to these anti-pseudomonas agents via chromosomal mutations and lateral gene transfer (Poole, 2011). The emergence and spread of multidrug-, extensive drug-, and pandrug-resistant P. aeruginosa infections is of great concern as very few agents are effective against these strains (Fischbach and Walsh, 2009; Poole, 2011). The problem of increasing antimicrobial resistance is even more threatening when considering the very limited number of new antimicrobial agents in development (Fischbach and Walsh, 2009). In particular, the prospects of finding new antibiotics for Gram-negative pathogens are poor because of the low permeability of their outer membrane barriers and the presence of multidrug efflux transporters (Fischbach and Walsh, 2009). To combat these bacteria efficiently, it is necessary to understand the molecular basis of the efflux mechanisms involved in limiting the intracellular (or periplasmic) concentration of many antibiotics (Nikaido and Pages, 2012).
The most clinically relevant multidrug efflux systems in Gram-negative bacteria are those of the resistance-nodulation-cell division (RND) family (Poole, 2004, 2012; Piddock, 2006; Lister et al., 2009). P. aeruginosa expresses several RND-type multidrug efflux systems, of which MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY are significant determinants of multidrug resistance in laboratory and clinical isolates (Poole, 2004, 2012; Piddock, 2006; Lister et al., 2009). These pumps are three-component systems comprising antiporters of the RND family driven by proton motive force (MexB, MexD, MexF, and MexY), outer membrane factors (OMF; OprM, OprJ, and OprN), and periplasmic membrane fusion proteins (MFP; MexA, MexC, MexF, and MexX; Piddock, 2006; Lister et al., 2009). These pumps probably function in a similar manner with AcrAB-TolC, which is the best-studied RND-type multidrug pump of Escherichia coli (Nakashima et al., 2011; Nikaido, 2011).
Among them, the MexXY system is intriguing in that it is a significant determinant of aminoglycoside resistance only in P. aeruginosa, with numerous reports of clinical isolates during the last decade (Nikaido and Pages, 2012; Poole, 2012). Relatively few bacterial drug efflux systems are known to accommodate aminoglycosides (Poole, 2012). In addition to MexXY, AmrAB-OprA of B. pseudomallei is noteworthy for its contribution to this organism’s intrinsic aminoglycoside resistance, while AdeABC of Acinetobacter baumannii is implicated in acquired aminoglycoside resistance (Poole, 2012). The upregulation of the MexXY pump is considered the most common mechanism of resistance (Armstrong and Miller, 2010) and appears to be the major determinant of aminoglycoside resistance in cystic fibrosis (CF) lung isolates of P. aeruginosa (Poole, 2011). Here, we summarize the properties of these pumps and discuss how to combat efflux-mediated aminoglycoside resistance.
PRE-MexXY Discovery Era: Rnd Multidrug Efflux Pumps as Determinants of Resistant to a Wide Range of Antimicrobials, But not Aminoglycosides
In 1993, the first RND-type multidrug efflux system of P. aeruginosa, MexAB-OprM (OprM was called OprK at that time), was discovered at approximately the same time as the AcrAB (AcrB was called AcrE at that time) system of E. coli (Poole et al., 1993b). It was the first genetic evidence that an efflux operon was involved in multiple antibiotic resistance in P. aeruginosa (Poole et al., 1993b). The following year, the efflux activity of tetracycline, chloramphenicol, norfloxacin, and benzylpenicillin was shown using antibiotic accumulation assays in intact cells, which was the first biochemical evidence of the role of efflux in intrinsic multidrug resistance in P. aeruginosa (Li et al., 1994a,b; Li et al., 1995). Taken together, the MexAB-OprM system was shown to contribute to the intrinsic resistance of P. aeruginosa to a wide range of antimicrobial compounds including fluoroquinolones, tetracycline, chloramphenicol, and β-lactams such as carbenicillin (Poole et al., 1993b; Li et al., 1995). Its homologs (MexCD-OprJ and MexEF-OprN) were then discovered as determinants of nfxB and nfxC fluoroquinolone-resistant (e.g., norfloxacin) mutants from P. aeruginosa (Poole et al., 1996; Kohler et al., 1997). Incidentally, these two pumps are not expressed in normal laboratory growth conditions, but are induced under some conditions in wild-type P. aeruginosa (e.g., Morita et al., 2003; Fetar et al., 2011). An unidentified efflux system that requires OprM was shown to contribute to resistance to quinolones and cephalosporins, such as cefpirome, erythromycin, and tetracycline, but not β-lactams, such as cefoperazone and carbenicillin, in the P. aeruginosa PAO1 background (Zhao et al., 1998). More details on these three pumps can be found in recent reviews (e.g., Li and Nikaido, 2004, 2009; Lister et al., 2009).
In those days, RND multidrug efflux systems such as MexAB-OprM and AcrAB-TolC, which can handle a wide variety of drugs that appear to contain hydrophobic domains of significant sizes (Nikaido, 1996), were considered to be similar to the P-glycoprotein multidrug efflux pump of mammalian cells, which extrudes not only basic compounds but also neutral and weakly acidic compounds (Nikaido, 1994). However, there was no evidence for the efflux of aminoglycosides, which are very hydrophilic compounds, among the antibiotics used to treat P. aeruginosa infections (Li et al., 1994a; Nikaido, 1996).
MexXY System of P. aeruginosa was Identified by three Different Groups
The MexXY system was discovered in 1999 in Japan as the fourth RND-type multidrug efflux system of P. aeruginosa PAO1 (Mine et al., 1999). This system was functionally expressed and conferred resistance to fluoroquinolones, tetracycline, erythromycin, etc. in the E. coli KAM3 mutant (Morita et al., 1998) lacking the acrB gene, which is an RND transporter component of the major multidrug efflux pump (AcrAB-TolC) in E. coli. Interestingly, unlike the other three already known systems, no open-reading frame encoding the outer membrane component, such as OprM, was found in the region downstream from the mexY gene (Mine et al., 1999). However, this system was found to function cooperatively with OprM of P. aeruginosa and TolC of E. coli (Mine et al., 1999).
Nine months after the discovery described above, a French group showed that MexXY was involved in the natural resistance of P. aeruginosa PAO1 to aminoglycosides as well as tetracycline and erythromycin (Aires et al., 1999). Although the overexpression of MexXY increased the level of resistance to fluoroquinolones in P. aeruginosa PAO1 cells, disruption of mexXY from PAO1 had no detectable effect on susceptibility to these agents (Aires et al., 1999). mexZ, which is located upstream of but transcribed separately from mexXY, was identified (Aires et al., 1999). Its product, MexZ, contains a helix-turn-helix motif, which is characteristic of DNA-binding proteins, at its N-terminus, similar to the repressors of RND-type multidrug efflux genes (e.g., AcrR, a repressor of acrAB in E. coli), supporting the notion that mexZ negatively controls the expression of the operon (Aires et al., 1999).
The following month (10 months after the first discovery), a group in the USA showed that MexXY, which they called AmrAB, was an aminoglycoside impermeability factor in spontaneous aminoglycoside-resistant mutants of the impermeability phenotype from P. aeruginosa PAO1 (Westbrock-Wadman et al., 1999). Interestingly, a dramatic decrease in the amount of OprM was observed in the mutants compared to wild-type PAO1, indicating that OprM is unlikely to be the outer membrane component associated with this efflux system in the mutants (Westbrock-Wadman et al., 1999). In addition, MexXY was shown to be upregulated in clinical P. aeruginosa isolates displaying aminoglycoside impermeability, suggesting that the pump is a clinically relevant mechanism of aminoglycoside resistance in P. aeruginosa (Westbrock-Wadman et al., 1999).
The following year, the complete genomic sequence of P. aeruginosa strain PAO1 (PAO1-UW) was published in Nature (Stover et al., 2000). Although the locus IDs PA2019-18 of the PAO1-UW genome sequence correspond to the mexXY genes, the nucleotide sequences of PA2019-18 were not identical with those of previously published mexXY (Aires et al., 1999; Mine et al., 1999) findings. This is probably because the DNA sequencing technology at that time was unable to analyze GC-rich bacteria such as P. aeruginosa (66–67% GC content; Winsor et al., 2011). Therefore, we analyzed MexXY using the nucleotide sequences from the PAO1-UW complete genome (Winsor et al., 2011) because we live in the post-genome era.
Structure and Function of MexY
The RND components of RND-type tripartite multidrug efflux pumps determine substrate specificity (e.g., Srikumar et al., 1997; Eda et al., 2003); therefore, we focused on the structure and function of MexY rather than MexX or OprM. Very recently, the crystal structure of the RND-type multidrug efflux pump AcrB of E. coli revealed the presence of two discrete, high-volume multisite binding pockets that contribute to the remarkably broad substrate recognition of AcrB and its homologs (Nakashima et al., 2011). Although we basically assume that MexY pumps out antimicrobials in a similar manner as AcrB, it will be intriguing to uncover the molecular basis of how MexY accommodates aminoglycosides because they are strongly hydrophilic molecules that are completely different from the relatively hydrophobic compounds (e.g., minocycline, doxorubicin, rifampicin, and erythromycin) used as substrates of AcrB (Nakashima et al., 2011).
Generally speaking, the function of a transporter (e.g., substrate specificity and energy coupling) should be determined by its time course efflux assay and evaluated using kinetic constants (e.g., Yerushalmi et al., 1995; Edgar and Bibi, 1997; Mine et al., 1998; Morita et al., 2000). However, it is difficult to conduct such an assessment in a small bacteriology laboratory. The reconstitution of proteoliposomes revealed that AcrB, AcrD, and MdtBC of E. coli were H+/drug antiporters (Zgurskaya and Nikaido, 1999; Aires and Nikaido, 2005; Kim et al., 2010), and we assume that MexY pumps out antimicrobials coupled with the same energy. In addition, five charged and polar amino acid residues that are involved in the proton translocation pathway are conserved between MexY and AcrB of E. coli (Takatsuka and Nikaido, 2006). Unfortunately, the purification, reconstitution, and characterization of the MexXY pump remain to be established, and the energy coupling and substrate specificity of MexXY has not been examined through its efflux activity. On the other hand, the MexXY-mediated energy-dependent efflux activity of ethidium (Mine et al., 1999), aminoglycosides (Aires et al., 1999; Vogne et al., 2004), tetracycline (Aires et al., 1999), Ala-Nap (MC-005,556) (Mao et al., 2001), and fluorescein-di-β-D-galactopyranoside (Matsumoto et al., 2011) has been measured in whole cells.
It is conventional to use differences in minimum inhibitory concentrations (MICs) between bacterial cells with and without a multidrug efflux transporter to estimate substrate specificity (e.g., Nishino and Yamaguchi, 2001; Nishino et al., 2003). Although the comparison of MICs can sometimes be a misleading indicator of pump function (e.g., Nagano and Nikaido, 2009), it can still indicate the possible clinical relevance of a pump (e.g., Poole, 2004, 2012; Piddock, 2006). Mutant strains lacking major multidrug efflux pump(s) have been used to determine substrate specificity (e.g., Nishino and Yamaguchi, 2001; Morita et al., 2001b). The substrate specificity of MexXY-OprM was determined using a mutant from PAO1 that overproduced MexXY-OprM, but not MexAB (and AmpC in the case of β-lactams), and were compared with a mutant lacking MexXY/MexAB-OprM (and AmpC in the case of β-lactams; Masuda et al., 2000b). MexXY-OprM-mediated resistance was then observed for quinolones, macrolides, tetracyclines, aminoglycosides, chloramphenicol, lincomycin, and most β-lactams, but not for novobiocin, polymyxin B, and some β-lactams (carbenicillin, sulbenicillin, cefsulodin, ceftazidime, oxacephem, imipenem, and aztreonam) among a wide variety of antimicrobial agents (Masuda et al., 2000b). In conclusion, MexXY-OprM is a multidrug efflux transporter whose specificity is extraordinary broad, but different compared with MexAB-OprM, MexCD-OprJ, MexEF-OprN, and other RND efflux transporters in P. aeruginosa. In addition, MexXY-OprM was the only pump to mediate aminoglycoside resistance and was thus considered to recognize aminoglycosides as substrates (Masuda et al., 2000b).
Basic local alignment search tool (BLAST) analysis showed that MexY was highly conserved in P. aeruginosa strains: more than 99% (99%) identity (positive) for most strains and 97% (98%) identity (positive) for PA7 (Morita et al., 2012). There was no functional difference between the MexYs of PAO1 and PA7 when they were expressed in either E. coli or P. aeruginosa (Morita et al., 2012). MexY was more similar [70–73% (83–86%) identity (positive)] to orthologs of B. pseudomallei and various B. cepacia complexes than other RND pumps of P. aeruginosa and other Pseudomonas species(Morita et al., 2012). These Burkholderia species, except for B. mallei, are intrinsically resistant to aminoglycosides (e.g., Kenny et al., 1999; Thibault et al., 2004; Vermis et al., 2003; Jassem et al., 2011). B. gladioli is also known to be involved in human infections (Segonds et al., 2009); however, no MexY (AmrB) ortholog exists in B. gladioli BSR3 (Seo et al., 2011), consistent with the fact that all isolates tested were susceptible to aminoglycosides (Segonds et al., 2009). Interestingly, the most similar functional ortholog to MexY exists in Achromobacter xylosoxidans and has a 74% identity (86% positive); this pump was named AxyY in strain AXX-A (Bador et al., 2012). A. xylosoxidans is also an opportunistic human pathogen capable of causing a wide range of infections (Glupczynski et al., 1988; Bador et al., 2011). Most A. xylosoxidans clinical isolates were resistant to the tested aminoglycosides, including amikacin (Ngeow and Puthucheary, 1985; Glupczynski et al., 1988). The AxyY pump contributes to aminoglycoside resistance in a similar manner to MexY and AmrBs (Bador et al., 2012).
COBALT analysis is a multiple sequence alignment tool for finding a collection of pairwise constraints. Such constraints are derived from data of the conserved domain database, protein motif database, and sequence similarity of RND pumps (Papadopoulos and Agarwala, 2007), including all pumps from P. aeruginosa PAO1-UW and E. coli K12 (MG1655). The exception is heavy metal efflux pumps, which are characterized by their relationships. Therefore, we focused on the four branches containing the four Mex pumps in P. aeruginosa (Figure 1). The MexY branch is located next to the MexD branch and includes the AmrBs of Burkholderia species (e.g., Mima and Schweizer, 2010) and AxyY of A. xylosoxidans (Bador et al., 2012). The MexD branch includes the AdeB pump of A. baumannii (Magnet et al., 2001), MtrD of Neisseria gonorrhoeae (Hagman et al., 1997),and BdeB of Bradyrhizobium japonicum (Lindemann et al., 2010). The SmeZ pump of S. maltophilia, which can mediate aminoglycoside resistance (Crossman et al., 2008), also belongs to the MexD branch. Many pumps in the MexY/MexD branches can mediate aminoglycoside resistance (e.g., Magnet et al., 2001; Crossman et al., 2008; Lindemann et al., 2010; Mima and Schweizer, 2010), which hints at the structure-function relationship of pumps involved in aminoglycoside resistance. MexB is located in the branch that contains the AcrB/D/F and MdtF pumps of E. coli (Nishino and Yamaguchi, 2001; Nishino et al., 2003), AdeJ of A. baumannii (Damier-Piolle et al., 2008), BpeB of B. pseudomallei (Mima and Schweizer, 2010), AxyB of A. xylosoxidans (Bador et al., 2011), and VmeB of Vibrio parahaemolyticus (Matsuo et al., 2007). Among them, some pumps (e.g., AcrD and MexB) were reported to be involved in aminoglycoside resistance under some conditions (Li et al., 2003; Aires and Nikaido, 2005). The MexF branch includes AdeG of A. baumannii (Coyne et al., 2010) and MdsB of Salmonella enterica (Nishino et al., 2006).
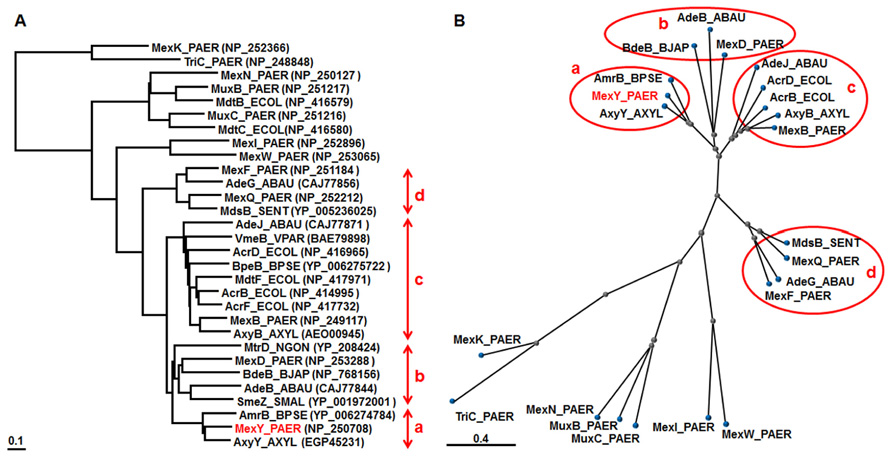
FIGURE 1. Phylogenetic trees for representative RND transporters. According to the COBALT program, the trees were constructed using the Fast evolution method and rendered with (A) Rectangle and (B) Radical. Protein names are abbreviated; for example, “MexY_PAER” stands for “MexY of P. aeruginosa.”Accession numbers are shown in parentheses. The four branches shown in red are: (a) MexY, (b) MexD, (c) MexB, and (d) MexF.
Substrate specificity is determined almost entirely by the periplasmic domain (i.e., two large extramembrane loops that largely protrude toward the periplasmic space) of MexY (and MexB; Eda et al., 2003). MexYF1018L (the F1018L mutation is located in TMS-12 of MexY base on the structure of AcrB; Murakami et al., 2002), enhanced the function of MexY, presumably by increasing the efflux of aminoglycosides, cefepime, and fluoroquinolones, which was the first example of an improved efflux pump in vivo (Vettoretti et al., 2009b).
Gene Expression of the MexXY System
MexXY was shown to be induced by sub-inhibitory concentrations of tetracycline, erythromycin, aminoglycosides, tigecycline, and LMB415 (a peptide deformylase inhibitor), but not ofloxacin in P. aeruginosa PAO1 (Masuda et al., 2000a; Dean et al., 2003; Caughlan et al., 2009). Moreover, ofloxacin and cefpirome were also shown to be inducers, but only in a PAO1 mutant lacking MexAB (and AmpC in the case of cefpirome; Masuda et al., 2000a,b). MexZ was shown to bind an inverted repeat region located in the mexZ-mexX intergenic region directly as a homodimer, which encompasses the putative mexXY promoter, but the inducers failed to alter the MexZ-operator interactions (Matsuo et al., 2004). The crystal structure of MexZ has since been solved (Alguel et al., 2010). Induction of the MexXY efflux pump in P. aeruginosa PAO1 was shown to be dependent on drug–ribosome interactions (Jeannot et al., 2005), and the pump remained inducible, but to a lesser degree, by ribosomal inhibitors, even in the mexZ mutant (Jeannot et al., 2005). These data demonstrate the physiological interactions between MexXY and the ribosome and are suggestive of an alternative function for MexXY beyond the efflux of antibiotics (Jeannot et al., 2005). Microarray analysis showed that mexXY were the most highly upregulated genes in P. aeruginosa PAO1 after 4 h of interaction with primary normal human airway epithelial cells (Frisk et al., 2004) and in response to sub-inhibitory concentrations of tobramycin under normal aerobic conditions, but not under lethal aerobic conditions or anaerobic conditions (Kindrachuk et al., 2011).
The antibiotic inducibility of the MexXY multidrug efflux system of P. aeruginosa was shown to be involved in the modulation of MexZ activity by the antibiotic-inducible PA5471 gene product (Morita et al., 2006; Table 1). PA5471 encodes a predicted product of 43.5 kDa, which was identified as a hypothetical protein conserved between bacteria and archaea, and is a representative of the uncharacterized protein family UPF0027 in the Pfam protein families database (Morita et al., 2006) or the PRK09588 cluster in ProtClustDB (NCBI Protein Clusters Database; Klimke et al., 2009). Recently, it was demonstrated that RctB of E. coli, which is related to members of this family, is a novel RNA ligase and functions as a bona fide RNA repair protein in vivo (Tanaka and Shuman, 2011). PA5471 is found upstream of and in a possible operon with an open-reading frame dubbed PA5470; RT-PCR confirmed both the drug inducibility of PA5470 and its expression from a polycistronic message that also contains PA5471 (Morita et al., 2006). PA5470 is predicted to encode a peptide chain release factor of 22.3 kDa (Morita et al., 2006). A homolog of PA5471 from E. coli K12, ykfJ (b0235), which was, however, C-terminally truncated (approximately 1 kb; Baranov et al., 2006), was also shown to be inducible by 4-azaleucine, which is known to interfere with translation, and it too is linked to a putative peptide release factor gene (Morita et al., 2006). P. aeruginosa senses antibiotic-mediated ribosomal disruption and links it to PA5471 gene expression by monitoring the translation of a 13-amino-acid-leader peptide region (PA5471.1) found ~250 bp upstream of the PA5471 coding sequence on PA5471 mRNA (Morita et al., 2009). The antimicrobial-inducible PA5471 gene product has been shown to interact with the repressor MexZ and interfere with its DNA binding activity in vitro (Yamamoto et al., 2009), and this finding contributed to elucidating the molecular mechanisms of the MexXY induction. However, PA5471 is not sufficient for MexXY recruitment in response to antibiotic exposure, and additional antibiotic-dependent effects are needed in P. aeruginosa (Morita et al., 2009). Exposure to reactive oxygen species (ROS; e.g., peroxide) induces the expression of the PA5471 gene, leading to MexXY-dependent aminoglycoside resistance (Fraud and Poole, 2011). Moreover, long-term (8-day) exposure of P. aeruginosa to peroxide (mimicking chronic in vivo ROS exposure) increased the frequency of PA5471- and mexXY-dependentaminoglycoside resistance (Fraud and Poole, 2011). Recently, reduced (approximately twofold) expression of the rplU-rpmA operon (encoding the 50S ribosomal proteins L21 and L27) was shown to promote mexXY expression via the PA5471 gene in pan-aminoglycoside resistant mutants from PAO1 and a CF clinical isolate (Lau et al., 2012). Such expression was in the form of ribosomal protein mutations that influence mexXY expression, including rplY (encoding ribosomal protein L25; El’Garch et al., 2007) and rplA (encoding ribosomal protein L1; Westbrock-Wadman et al., 1999). Transcriptome profiling revealed that significantly increased expression was observed for the mexXY and PA5471 genes in both the PA2572 and PA2573 mutants compared with the wild-type PAO1 strain during exponential growth in Luria–Bertani media (McLaughlin et al., 2012). PA2572 encodes a putative response regulator of a two-component system required for full virulence to Galleria mellonella (Wax moth) and PA2573 also encodes an ophan chemotaxis sensor which seems to function in part through signal transduction involving PA2572 (McLaughlin et al., 2012).
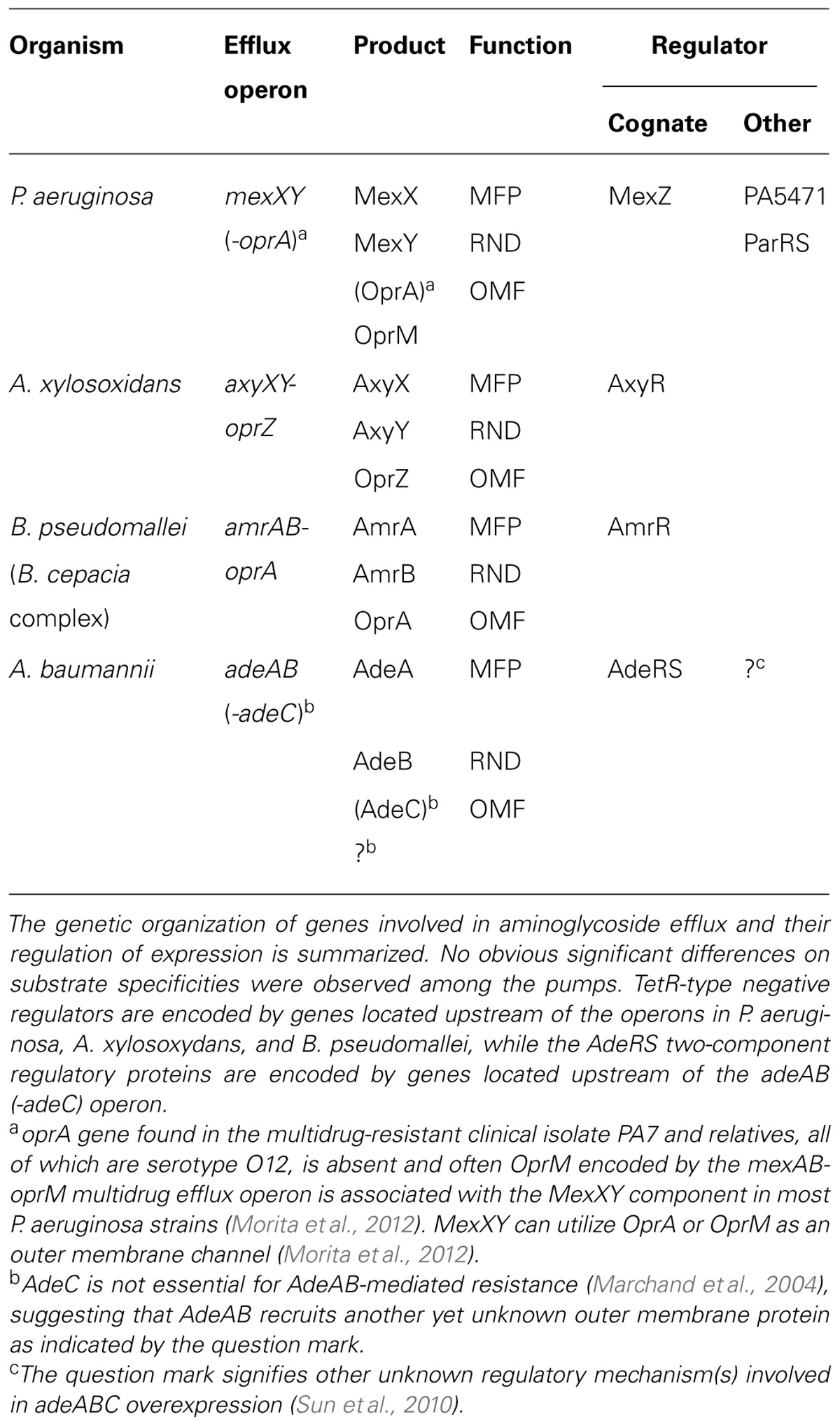
TABLE 1. Genetic organization of aminoglycoside efflux operons of clinical significance and their regulators in non-fermentative Gram-negative pathogens.
A recent study identified a gene, parR, encoding the response regulator of a two-component system, ParRS, which promotes either induced or constitutive mexXY upregulation, thereby activating the MexXY efflux system as well as OprD porin loss and lipopolysaccharide modification in a MexZ-independent manner (Muller et al., 2011). Overexpression of PaeIII, a small non-coding RNA between PA3505 and PA3536 in the genome of P. aeruginosa PAO1, in the stationary phase increased the expression of the mexXY and mexZ genes as well as type III secretion genes, while reducing the expression of genes for arginine metabolism (Goldberg et al., 2008).
MexXY System as an Antimicrobial Resistance Determinant in P. aeruginosa
Pseudomonas aeruginosa shows intrinsic resistance against many antimicrobials because of the low permeability of its outer membrane and the presence of efflux systems (Nikaido, 1994; Hancock, 1998). MexXY was shown to be involved in natural resistance to aminoglycosides, tetracycline, tigecycline, erythromycin, and LBM415 in P. aeruginosa PAO1 (Aires et al., 1999; Masuda et al., 2000a; Morita et al., 2001a; Dean et al., 2003; Caughlan et al., 2009). MexXY was also shown to be necessary for the adaptive resistance of P. aeruginosa PAO1 to aminoglycosides (Hocquet et al., 2003). It is of note that MexXY is the only pump of the 12 identified RND systems that mediates aminoglycoside resistance in P. aeruginosa PAO1 (Poole, 2011). The antagonism of aminoglycosides by the divalent cations Mg2+ and Ca2+ is well documented (Medeiros et al., 1971), and culture in cation-adjusted Mueller–Hinton broth is recommended as a susceptibility test to ensure acceptable results when P. aeruginosa isolates are tested (Barry et al., 1992). MexXY was shown to be required for the antagonism of aminoglycosides by divalent cations in P. aeruginosa PAO1 (Mao et al., 2001). Although Phe-Arg-β-naphthylamide (PAβN, MC-207,110) is known as a non-specific inhibitor against RND-type multidrug efflux pumps (Lomovskaya et al., 2001), this inhibitor, as observed for divalent cations, antagonized the activity of aminoglycosides (amikacin and netilmicin) in a MexXY-dependent manner, even though it also inhibited MexXY-dependent fluoroquinolone (levofloxacin) resistance (Mao et al., 2001). Conversely, PAβN inhibited MexXY-mediated aminoglycoside (gentamicin) resistance (Mesaros et al., 2007). The reason for the discrepancy between these two results remains unknown. Increased susceptibility to aminoglycosides in nfxB mutants, which upregulate mexCD-oprJ expression, was correlated with increased resistance to fluoroquinolones and some β-lactams, such as cefepime, concomitant with a higher susceptibility to aminoglycosides and some β-lactams, such as ticarcillin, aztreonam, and imipenem. This was shown to be partly due to the impaired activity of MexXY-OprM because of major changes in cell physiology, but not the expression/production of mexY/MexY and oprM/OprM (Jeannot et al., 2008; Mulet et al., 2011). The increased susceptibility to aminoglycosides in MexEF-OprN-overproducing nfxC mutants was also observed, apparently owing to impairment of the MexXY system (Sobel et al., 2005). mexXY expression (and so MexXY-mediated resistance) was independent of the AmgRS two-component system in which mutations enhanced aminoglycoside action to control an adaptive response to membrane stress (Lee et al., 2009).
Multidrug resistant P. aeruginosa clinical isolates have often been reported to be MexXY overproducers (e.g., Llanes et al., 2004, 2006; Wolter et al., 2004; Deplano et al., 2005; Henrichfreise et al., 2007; Hocquet et al., 2007; Maniati et al., 2007; Vettoretti et al., 2009a; Beaudoin et al., 2010; Xavier et al., 2010; Fehlberg et al., 2012; Pasca et al., 2012). Time series analysis (January 1999 to January 2005) revealed a significant relationship between antibiotic use (aminoglycosides, fluoroquinolones, and cefepime, but not carbapenems) and the incidence of MexXY-overproducing P. aeruginosa in a French hospital (Hocquet et al., 2008). MexXY (n = 39) and MexAB (n = 31) were the most frequently overproduced pumps in 85 non-CF P. aeruginosa strains with low-level ciprofloxacin resistance (MICs ranging from 0.25 to 2 µg/mL, which are still susceptible or intermediate according to the CLSI breakpoints; Llanes et al., 2011). A large proportion of the strains were MexXY overproducers in genotypically distinct P. aeruginosa clinical isolates that were less susceptible to cefepime than to ceftazidime, and these were identified in Europe (Hocquet et al., 2006; Pena et al., 2009; Campo Esquisabel et al., 2011) and the USA (Laohavaleeson et al., 2008). In contrast, both cefepime and ceftazidime are potent β-lactam antibiotics with similar MICs (1–2 µg/mL) for wild-type P. aeruginosa strains. Moreover, ceftobiprole, similar to cefepime, selected MexXY overproducers in clinical studies (Baum et al., 2009). Actually, a single step MexXY overproducer was selected in vitro by cefepime and ceftobiprole, but not ceftazidime (Queenan et al., 2010). MexXY contributed very significantly to the development of high-level (100–1000 µg/mL MIC) aminoglycoside resistance via a combination of aminoglycoside-modifying enzymes (AMEs) in multidrug resistant P. aeruginosa non-CF clinical isolates (Morita et al., 2012). However, AMEs are common determinants of aminoglycoside resistance in P. aeruginosa,except for CF isolates (Poole, 2011). In clinical CF isolates, MexXY has been primarily implicated in pan-aminoglycoside resistance (e.g., Sobel et al., 2003; Vogne et al., 2004; Islam et al., 2004, 2009). MexXY was also shown to be necessary in subpopulations of P. aeruginosa CF isolates that are hypersensitive to ticarcillin (called Tichs; Vettoretti et al., 2009b). mexZ was shown to be one of the most frequently mutated genes during chronic infection by P. aeruginosa in CF patients (Smith et al., 2006; Feliziani et al., 2010). However, a number of studies highlighted the absence of mutations in mexZ or the mexXY promoter region in MexXY-overproducing P. aeruginosa CF isolates (Sobel et al., 2003; Vogne et al., 2004; Islam et al., 2009). To date, three kinds of mutants (agrZ, agrW1, and agrW2) have been recognized as MexXY overproducers as a result of genetic mechanisms: mutants with impaired binding or unbinding of MexZ due to alterations in the mexZ or mexZ-mexX intergenic region (type agrZ); mutants with impaired protein synthesis (type agrW1); and mutants with alterations in parRS (type agrW2; de Bentzmann and Plesiat, 2011). Oxidative stress, a component of the host’s immune system in the CF lung, induced mexXY expression via PA5471 and promoted aminoglycoside resistance (Fraud and Poole, 2011). Under conditions of oxidative stress, P. aeruginosa can develop aminoglycoside resistance, even in the absence of aminoglycosides (Poole, 2012). It is also very plausible that the routine use of aminoglycosides (e.g., tobramycin) might simply select for MexXY-overproducing P. aeruginosa in theCF lung (Smith et al., 2006).
Although it is obvious thatMexXY is one of the determinants of antimicrobial resistance in P. aeruginosa in the clinical setting (Poole, 2011), only a few reports have assessed the in vivo impact of the MexXY system on antibiotic therapy for P. aeruginosa infections (e.g., Martha et al., 2006).
Cognate Outer-Membrane Component OprA of the MexXY Pump is Found in Serotype O12 But is Lost in Others
The mexXY operon lacks a gene coding for the outer membrane protein in P. aeruginosa PAO1 (Mine et al., 1999). OprM is necessary for the function of MexXY and MexAB in P. aeruginosa PAO1 (Aires et al., 1999; Masuda et al., 2000a; Morita et al., 2001a), although overproduced OpmB (PA2525) can function as an outer membrane component of MexXY, MexAB, and MexCD (Murata et al., 2002). Intriguingly, the multidrug resistant taxonomic outlier P. aeruginosa PA7 possesses a unique gene (oprA) downstream of mexXY encoding an outer membrane channel that is absent in most P. aeruginosa strains (Roy et al., 2010). MexXY in this strain utilizes either the OprA or OprM outer membrane channel (Morita et al., 2012; Table 1). While OprM is functional with both MexXY and MexAB, OprA did not associate as strongly with MexAB as it did with MexXY (Morita et al., 2012). We compared the OprA of P. aeruginosa PA7 with the OprM family (Remans et al., 2010) from P. aeruginosa PAO1 as well as TolC of E. coli K12 (Mine et al., 1999), OprA of B. pseudomallei 1026b (Mima and Schweizer, 2010), OprZ of A. xylosoxidans AXX-A (Bador et al., 2012),and AdeC of A. baumannii AYE (Magnet et al., 2001; Figure 2). COBALT analysis showed that OprA of P. aeruginosa PA7 and its close orthologs (OprA of B. pseudomallei 1026b and OprZ of A. xylosoxidans AXX-A) is located close to OprJ and is followed by OprM of the OprM outer membrane family of P. aeruginosa PAO1 (Figure 2).
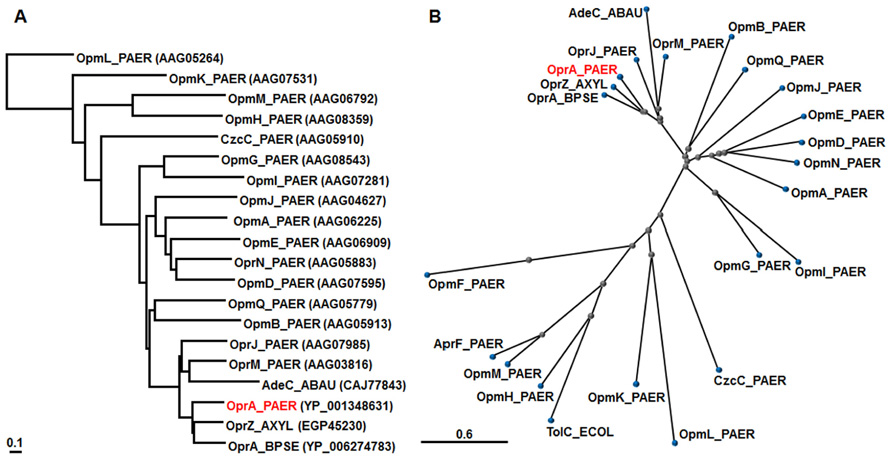
FIGURE 2. Phylogenetic trees for representative OMPs. According to the COBALT program, the trees were constructed using the Fast evolution method and rendered with (A) Rectangle and (B) Radical. Protein names are abbreviated; for example, “OprA_PAER” stands for “OprA of P. aeruginosa.” Accession numbers are shown in parentheses.
Interestingly, a small portion of the oprA gene immediately downstream of the mexY gene in PAO1 was identified, suggesting that non-PA7 P. aeruginosa strains might have possessed, but lost, the intact mexXY-oprA efflux pump locus (Morita et al., 2012; Table 1). Consistent with this, the majority of a panel of serotype strains possessed the truncated oprA, but the serotype O12 isolate had an intact mexXY-oprA locus, similar to PA7 and the related strain DSM 1128 (Morita et al., 2012). O12 is a predominant serotype associated with multidrug resistance to a number of antibiotic classes, including aminoglycosides and β-lactams, although it represents a minor serotype in the environment (Pirnay et al., 2009; Roy et al., 2010). O12 might be more dominant due, in part, to the presence of oprA in hospitals in which antimicrobials promoting MexXY-OprA-mediated multidrug resistance, such as aminoglycosides, were used. P. aeruginosa PA7 isolated before 1984 from a wound infection in Buenos Aires, Argentina (Pirnay et al., 2009; Roy et al., 2010), might also have acquired multidrug resistance via the heavy use of antibiotics, including gentamicin or tobramycin, to treat wounds at that time. Apparently, a slightly increased resistance (two- to fourfold) to amikacin, ciprofloxacin, and cefpirome was shown in the presence and absence of oprA (Morita et al., 2012). Such a small difference might be significant during antibiotic treatment or in the presence of sub-inhibitory concentrations of antibiotics.
AcrD is an Aminoglycoside Efflux Pump that is the Most Similar to MexY Among the rnd Pumps in E. coli K12
AcrD has the highest similarity score at the amino acid level to MexY of the E. coli K12 RND pumps and was shown to be an aminoglycoside efflux pump as judged by MIC determination and the aminoglycoside efflux assay (Rosenberg et al., 2000). However, differences in aminoglycoside resistance (uptake) between the parent strain JC7623 and its acrD-deletion mutant JZM320 was possibly not limited to AcrD function because JZM320 was constructed by inserting the tet gene from pBR322 into acrD (Rosenberg et al., 2000). The increased aminoglycoside uptake might be due to not only AcrD deficiency but also to the production of an aberrant cytoplasmic membrane protein (the product of acrD with the inserted tet) and/or the tetracycline/H+ antiporter itself (Merlin et al., 1989a,b; Wyka and St John, 1990). While disruption of tolC or acrA did not increase the susceptibility of K12 to aminoglycosides (Rosenberg et al., 2000), both of them were necessary for the function of acrD against various antimicrobials; however, no aminoglycosides were used in the study (Hirakawa et al., 2003). We do not rule out the hypothesis of Rosenberg et al. (2000) that AcrD protein can perhaps function without the participation of AcrA and TolC in the case of aminoglycoside efflux.
It is evident that purified AcrD can function as an H+-driven aminoglycoside efflux pump(Aires and Nikaido, 2005). Especially, strong stimulation of proton efflux was observed when aminoglycosides (e.g., streptomycin) were added to the more acidic intra-vesicular space of reconstituted AcrD proteoliposomes containing AcrA and Mg2+(Aires and Nikaido, 2005), indicating that AcrD captures aminoglycosides exclusively from the periplasm in E. coli (Nikaido, 2011). The difference in the MICs of amikacin and gentamicin between a parent strain and its in-frame acrD-deletion mutant or between an acrBD-deletion mutant and its acrD-overexpressing complementation mutant was approximately twofold (Elkins and Nikaido, 2002; Aires and Nikaido, 2005). There was no significant difference in kanamycin resistance in the case of an in-frame deletion (Hirakawa et al., 2003), and a twofold difference was observed in the case of overproduction (Nishino and Yamaguchi, 2001; Nishino et al., 2010). An acrA in-frame deletion mutant also showed an approximately twofold increased susceptibility to aminoglycosides (Aires and Nikaido, 2005). However, similar observations were not seen for AcrB (Nishino and Yamaguchi, 2001; Elkins and Nikaido, 2002; Aires and Nikaido, 2005). A comparison of the entrances of the vestibules, which are found in the central cavities (Murakami et al., 2002) of AcrD (which transports aminoglycosides) and AcrB (which does not) crystal structures, shows that this area in AcrD is in line with many more acidic residues that may attract polycationic substrates (Yu et al., 2003). Treatment with sub-inhibitory concentrations of kanamycin induced adaptive resistance to aminoglycosides, which was dependent on acrD (Sidhu et al., 2012). Aminoglycosides are very hydrophilic and polycationic and assumingly permeate through the porin channel in E. coli, unlike P. aeruginosa (Nikaido and Pages, 2012) in addition to so called “self-promoted” aminoglycoside uptake across the outer membrane of both of E. coli and P. aeruginosa (Hancock et al., 1991; Hancock, 1998). The MICs of aminoglycosides on E. coli, unlike P. aeruginosa,might be poor indicators of aminoglycoside efflux. There are numerous AcrD homologs in other Enterobacteriaceae(Poole, 2004). Although the AcrD of S. enterica serovar Typhimurium ATCC 14028s was studied comprehensively, no significant difference between the AcrDs of E. coli and S. enterica has been observed so far (Nishino et al., 2009; Horiyama et al., 2011; Yamasaki et al., 2011). Interestingly, AcrD pumps mediate resistance to the substrates of MexAB (e.g., carbenicillin, aztreonam, and novobiocin). However, AcrD pumps did not mediate resistance to the substrates of MexXY (e.g., cefpirome, erythromycin, and tetraphenylphosphonium) or shared substrates of both MexAB and MexXY (e.g., fluoroquinolone and tetracycline) when differences of the MICs were compared between a parent strain and its transformant overproducing the pump (Srikumar et al., 1997; Mine et al., 1999; Morita et al., 2001a; Nishino et al., 2003, 2009; Horiyama et al., 2011; Yamasaki et al., 2011). MexAB-OprM was also shown to contribute to aminoglycoside resistance, presumably via active efflux in the low-ionic-strength medium used in this particular study (Li et al., 2003). AcrD and the MdtABC pump were iron-regulated, induced in low-iron conditions, and export the siderophore enterobactin (Bochner et al., 2008), which reminds us that MexAB-OprM was inducible under conditions of iron limitation and compensated for a growth defect in an iron-deficient medium in the presence of the non-metabolizable iron chelator 2,2′-dipyridyl (Poole et al., 1993a,b). AcrD seems to be a functional homolog of MexB rather than MexY, as determined from substrate specificity and physiological function, consistent with the fact that phylogenetic analysis showed that AcrD is closer to MexB than to MexY (Figure 1).
AmrAB-OprA is a Multidrug Efflux System that Mediates Aminoglycoside Resistance in B. pseudomallei
Burkholderia pseudomallei is the etiologic agent of melioidosis, a rare but serious disease endemic to South Asia, Northern Australia, and other parts of the tropics (Mima and Schweizer, 2010). Melioidosis is very difficult to treat because of the intrinsic resistance to many antimicrobial agents including aminoglycosides, macrolides, polymyxins, and some β-lactams (Mima and Schweizer, 2010). AmrAB-OprA was identified as an efflux determinant of resistance to aminoglycosides and macrolides in the B. pseudomallei 1026b clinical isolate (Moore et al., 1999). This pump was actually the first to be demonstrated responsible for the aminoglycoside resistance of RND pumps in Gram-negative bacteria. The gene product of amrR, which is located immediately upstream and divergently transcribed from amrAB-oprA in B. pseudomallei 1026b (Moore et al., 1999), showed strong homology [60% (73%) identity (positive)] to MexZ, which acts as a transcriptional repressor of the mexXY operon of P. aeruginosa PAO1 (Matsuo et al., 2004; Alguel et al., 2010; Table 1).
While the majority of B. pseudomallei clinical isolates exhibit high levels of aminoglycoside and macrolide resistance, rare isolates are susceptible to these antibiotics (Simpson et al., 1999; Trunck et al., 2009). While it is noted that the resistance profile of those isolates matches that of the amrAB-oprA mutants (Simpson et al., 1999), it was shown experimentally that amrAB-oprA was missing in B. pseudomallei 708a, an aminoglycoside- and macrolide-susceptible clinical isolate, and this loss was associated with the deletion of >130 kb of genetic material (Trunck et al., 2009). The expression of amrAB-oprA increased resistance to not only aminoglycosides and macrolides but also fluoroquinolones and tetracyclines in a BpeAB-OprA pump-deficient mutant of 1026b (Mima and Schweizer, 2010). Judging from the substrate specificity and sequence similarity (Mima and Schweizer, 2010), we have no doubt that AmrAB is a functional ortholog of MexXY in B. pseudomallei. BpeAB-OprB of B. pseudomallei also reportedly mediates aminoglycoside resistance in strain KHW (Chan et al., 2004), while this pump did not confer aminoglycoside resistance in 1026b (Mima and Schweizer, 2010). In addition, the BpeB RND transporter was also shown to be closely related to MexB of P. aeruginosa, both functionally and phylogenetically (Mima and Schweizer, 2010), consistent with our phylogenetic analysis (Figure 1).
As described above, AmrB orthologs are conserved among various human pathogens belonging to Burkholderia species, but not B. gladioli. Actually, an AmrAB-OprA ortholog was shown to be a major aminoglycoside resistance contributor in B. cenocepacia, a member of the B. cepacia complex (Hamad et al., 2010).
AdeABC is a Multidrug Efflux System that Mediates Aminoglycoside Resistance in A. baumannii
Acinetobacter baumannii is the most frequently implicated species in nosocomial infections among Acinetobacter spp. (Coyne et al., 2011). AdeABC was identified as an RND-type efflux pump involved in resistance to multiple antimicrobials including aminoglycosides, fluoroquinolones, tetracycline, erythromycin, cefotaxime, trimethoprim, and chloramphenicol in A. baumannii BM4454, a low-level pan-aminoglycoside resistant clinical isolate (Magnet et al., 2001). The polycistronic adeABC transcript was confirmed experimentally to encode AdeA (MFP), AdeB (RND transporter), and AdeC (OMF; Magnet et al., 2001; Marchand et al., 2004). Because the substrate specificity of AdeAB is very similar to that of MexXY, we have no doubt that AdeAB is a functional homolog of MexXY in A. baumannii. AdeC is not essential for AdeAB-mediated resistance (Marchand et al., 2004), indicating that AdeAB recruits another outer membrane protein to form a functional tripartite complex, as observed for the MexXY pump with OprM in P. aeruginosa (Aires et al., 1999; Mine et al., 1999). We do not rule out the possibility that AdeAB is functional with AdeC, as observed for the MexXY pump with its linked outer-membrane channel OprA, which was not essential in P. aeruginosa PA7 (Morita et al., 2012). The adeAB genes are usually present, but the adeC gene was not found in ~40% of clinical isolates (Nemec et al., 2007). Our phylogenetic analysis showed that AdeC is more closely related to OprM and OprJ than to OprA in the OprM outer membrane family of P. aeruginosa (Figure 2).
The adeABC operon is expressed at low levels in natural isolates of A. baumannii due to stringent control by the AdeRS two-component system, which is encoded adjacent to adeABC, but transcribed in the opposite direction (Marchand et al., 2004; Table 1). Mutations (e.g., AdeRPro116Leu, AdeSThr153Met, or AdeSGly30Asp) in AdeRS have been shown to be responsible for the constitutive expression of AdeABC (Marchand et al., 2004), which reminds us that mutations (e.g., ParRMet59Ile) in the ParRS two-component system are responsible for the constitutive expression of MexXY in P. aeruginosa (Muller et al., 2011). Overexpression of the AdeABC system in a tigecycline non-susceptible clinical isolate was due to the transposition of a copy of ISAba1 into adeS (Ruzin et al., 2007). Very recently, a truncated AdeS kinase protein generated by an ISAba1 insertion was shown to be correlated with enhanced adeABC expression in A. baumannii (Sun et al., 2012). Other regulatory mechanism(s) were shown to be involved in adeABC overexpression without any previously known mutation (Sun et al., 2010). Recently the AdeABC ortholog was shown to be a contributor to multiple antimicrobials, including aminoglycosides, in Acinetobacter genomospecies 13TU, a non-A. baumannii species (Roca et al., 2011).
Future Perspectives
MexXY is one of the potential targets for novel anti-pseudomonas agents. Its inhibitor is able to not only potentiate previously used ineffective antimicrobial agents (e.g., aminoglycosides against aminoglycoside-resistant P. aeruginosa and B. cepacia complex), but also to speed up the development of novel anti-pseudomonas agents. Because there are a significant number of potential drug targets encoded by the genome of P. aeruginosa (e.g., products of essential genes; Morita et al., 2010), it is the most promising therapeutic strategy to conquer the impermeability barriers of these bacteria. The efflux inhibitor MP 601384, which has specificity toward aminoglycoside-accommodating RND efflux systems and is not toxic to bacteria, is the only MexXY inhibitor reported so far (Jassem et al., 2011). Uncultured bacteria and plants are predicted to be a significant reservoir of novel antimicrobial agents (Stavri et al., 2007; Piel, 2011). Screening novel antibacterial agents, including a MexXY inhibitor, is currently in progress in our laboratory (e.g., Shiota et al., 2004).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Young Scientists (B) (Kakenhi 23790106) from the Japan Society for the Promotion of Science and a research grant from the Institute of Pharmaceutical Life Sciences, Aichi Gakuin University.
References
Aires, J. R., Kohler, T., Nikaido, H., and Plesiat, P. (1999). Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43, 2624–2628.
Aires, J. R., and Nikaido, H. (2005). Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J. Bacteriol. 187, 1923–1929.
Alguel, Y., Lu, D., Quade, N., Sauter, S., and Zhang, X. (2010). Crystal structure of MexZ, a key repressor responsible for antibiotic resistance in Pseudomonas aeruginosa. J. Struct. Biol. 172, 305–310.
Armstrong, E. S., and Miller, G. H. (2010). Combating evolution with intelligent design: the neoglycoside ACHN-490. Curr. Opin. Microbiol. 13, 565–573.
Bador, J., Amoureux, L., Duez, J. M., Drabowicz, A., Siebor, E., Llanes, C., et al. (2011). First description of an RND-type multidrug efflux pump in Achromobacter xylosoxidans, AxyABM. Antimicrob. Agents Chemother. 55, 4912–4914.
Bador, J., Amoureux, L., Blanc, E. and Neuwirth, C. (2012). The innate aminoglycoside resistance of Achromobacter xylosoxidans is due to AxyXY- OprZ, an RND-type multidrug efflux pump. Antimicrob. Agents Chemother. doi: 10.1128/AAC.01243-12 [Epub ahead of print].
Baranov, P. V., Vestergaard, B., Hamelryck, T., Gesteland, R. F., Nyborg, J., and Atkins, J. F. (2006). Diverse bacterial genomes encode an operon of two genes, one of which is an unusual class-I release factor that potentially recognizes atypical mRNA signals other than normal stop codons. Biol. Direct 1, 28.
Barry, A. L., Reller, L. B., Miller, G. H., Washington, J. A., Schoenknect, F. D., Peterson, L. R., et al. (1992). Revision of standards for adjusting the cation content of Mueller-Hinton broth for testing susceptibility of Pseudomonas aeruginosa to aminoglycosides. J. Clin. Microbiol. 30, 585–589.
Baum, E. Z., Crespo-Carbone, S. M., Morrow, B. J., Davies, T. A., Foleno, B. D., He, W., et al. (2009). Effect of MexXY overexpression on ceftobiprole susceptibility in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53, 2785–2790.
Beaudoin, T., Aaron, S. D., Giesbrecht-Lewis, T., Vandemheen, K., and Mah, T. F. (2010). Characterization of clonal strains of Pseudomonas aeruginosa isolated from cystic fibrosis patients in Ontario, Canada. Can. J. Microbiol. 56, 548–557.
Bochner, B. R., Giovannetti, L., and Viti, C. (2008). Important discoveries from analysing bacterial phenotypes. Mol. Microbiol. 70, 274–280.
Campo Esquisabel, A. B., Rodriguez, M. C., Campo-Sosa, A. O., Rodriguez, C., and Martinez-Martinez, L. (2011). Mechanisms of resistance in clinical isolates of Pseudomonas aeruginosa less susceptible to cefepime than to ceftazidime. Clin. Microbiol. Infect. 17, 1817–1822.
Chan, Y. Y., Tan, T. M., Ong, Y. M., and Chua, K. L. (2004). BpeAB-OprB, a multidrug efflux pump in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 48, 1128–1135.
Caughlan, R. E., Sriram, S., Daigle, D. M., Woods, A. L., Buco, J., Peterson, R. L., et al. (2009). Fmt bypass in Pseudomonas aeruginosa causes induction of MexXY efflux pump expression. Antimicrob. Agents Chemother. 53, 5015–5021.
Coyne, S., Rosenfeld, N., Lambert, T., Courvalin, P., and Perichon, B. (2010). Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 4389–4393.
Coyne, S., Courvalin, P., and Perichon, B. (2011). Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 55, 947–953.
Crossman, L. C., Gould, V. C., Dow, J. M., Vernikos, G. S., Okazaki, A., Sebaihia, M., et al. (2008). The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol. 9, R74.
Damier-Piolle, L., Magnet, S., Bremont, S., Lambert, T., and Courvalin, P. (2008). AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother. 52, 557–562.
de Bentzmann, S., and Plesiat, P. (2011). The Pseudomonas aeruginosa opportunistic pathogen and human infections. Environ. Microbiol. 13, 1655–1665.
Dean, C. R., Visalli, M. A., Projan, S. J., Sum, P. E., and Bradford, P. A. (2003). Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 47, 972–978.
Deplano, A., Denis, O., Poirel, L., Hocquet, D., Nonhoff, C., Byl, B., et al. (2005). Molecular characterization of an epidemic clone of panantibiotic-resistant Pseudomonas aeruginosa. J. Clin. Microbiol. 43, 1198–1204.
Eda, S., Maseda, H., and Nakae, T. (2003). An elegant means of self-protection in gram-negative bacteria by recognizing and extruding xenobiotics from the periplasmic space. J. Biol. Chem. 278, 2085–2088.
Edgar, R., and Bibi, E. (1997). MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J. Bacteriol. 179, 2274–2280.
El’Garch, F., Jeannot, K., Hocquet, D., Llanes-Barakat, C., and Plesiat, P. (2007). Cumulative effects of several nonenzymatic mechanisms on the resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 51, 1016–1021.
Elkins, C. A., and Nikaido, H. (2002). Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominantly by two large periplasmic loops. J. Bacteriol. 184, 6490–6498.
Fehlberg, L. C., Xavier, D. E., Peraro, P. P., Marra, A. R., Edmond, M. B., and Gales, A. C. (2012). Beta-lactam resistance mechanisms in Pseudomonas aeruginosa strains causing bloodstream infections: comparative results between Brazilian and American isolates. Microb. Drug Resist. 18, 402–407.
Feliziani, S., Lujan, A. M., Moyano, A. J., Sola, C., Bocco, J. L., Montanaro, P., et al. (2010). Mucoidy, quorum sensing, mismatch repair and antibiotic resistance in Pseudomonas aeruginosa from cystic fibrosis chronic airways infections. PLoS ONE 5, e12669. doi: 10.1371/journal.pone.0012669
Fetar, H., Gilmour, C., Klinoski, R., Daigle, D. M., Dean, C. R., and Poole, K. (2011). mexEF-oprN multidrug efflux operon of Pseudomonas aeruginosa: regulation by the MexT activator in response to nitrosative stress and chloramphenicol. Antimicrob. Agents Chemother. 55, 508–514.
Fischbach, M. A., and Walsh, C. T. (2009). Antibiotics for emerging pathogens. Science 325, 1089–1093.
Fraud, S., and Poole, K. (2011). Oxidative stress induction of the MexXY multidrug efflux genes and promotion of aminoglycoside resistance development in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55, 1068–1074.
Frisk, A., Schurr, J. R., Wang, G., Bertucci, D. C., Marrero, L., Hwang, S. H., et al. (2004). Transcriptome analysis of Pseudomonas aeruginosa after interaction with human airway epithelial cells. Infect. Immun. 72, 5433–5438.
Glupczynski, Y., Hansen, W., Freney, J., and Yourassowsky, E. (1988). In vitro susceptibility of Alcaligenes denitrificans subsp. xylosoxidans to 24 antimicrobial agents. Antimicrob. Agents Chemother. 32, 276–278.
Goldberg, J. B., Hancock, R. E., Parales, R. E., Loper, J., and Cornelis, P. (2008). Pseudomonas 2007. J. Bacteriol. 190, 2649–2662.
Hagman, K. E., Lucas, C. E., Balthazar, J. T., Snyder, L., Nilles, M., Judd, R. C., et al. (1997). The MtrD protein of Neisseria gonorrhoeae is a member of the resistance/nodulation/division protein family constituting part of an efflux system. Microbiology 143 (Pt 7), 2117–2125.
Hamad, M. A., Skeldon, A. M., and Valvano, M. A. (2010). Construction of aminoglycoside-sensitive Burkholderia cenocepacia strains for use in studies of intracellular bacteria with the gentamicin protection assay. Appl. Environ. Microbiol. 76, 3170–3176.
Hancock, R. E., Farmer, S. W., Li, Z. S., and Poole, K. (1991). Interaction of aminoglycosides with the outer membranes and purified lipopolysaccharide and OmpF porin of Escherichia coli. Antimicrob. Agents Chemother. 35, 1309–1314.
Hancock, R. E. (1998). Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative gram-negative bacteria. Clin. Infect. Dis. 27(Suppl. 1), S93–S99.
Henrichfreise, B., Wiegand, I., Pfister, W., and Wiedemann, B. (2007). Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob. Agents Chemother. 51, 4062–4070.
Hirakawa, H., Nishino, K., Hirata, T., and Yamaguchi, A. (2003). Comprehensive studies of drug resistance mediated by overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Bacteriol. 185, 1851–1856.
Hocquet, D., Vogne, C., El Garch, F., Vejux, A., Gotoh, N., Lee, A., et al. (2003). MexXY-OprM efflux pump is necessary for a adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 47, 1371–1375.
Hocquet, D., Nordmann, P., El Garch, F., Cabanne, L., and Plesiat, P. (2006). Involvement of the MexXY-OprM efflux system in emergence of cefepime resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50, 1347–1351.
Hocquet, D., Berthelot, P., Roussel-Delvallez, M., Favre, R., Jeannot, K., Bajolet, O., et al. (2007). Pseudomonas aeruginosa may accumulate drug resistance mechanisms without losing its ability to cause bloodstream infections. Antimicrob. Agents Chemother. 51, 3531–3536.
Hocquet, D., Muller, A., Blanc, K., Plesiat, P., Talon, D., Monnet, D. L., et al. (2008). Relationship between antibiotic use and incidence of MexXY-OprM overproducers among clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52, 1173–1175.
Horiyama, T., Yamaguchi, A., and Nishino, K. (2011). TolC dependency of multidrug efflux systems in Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 65, 1372–1376.
Islam, S., Jalal, S., and Wretlind, B. (2004). Expression of the MexXY efflux pump in amikacin-resistant isolates of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 10, 877–883.
Islam, S., Oh, H., Jalal, S., Karpati, F., Ciofu, O., Hoiby, N., et al. (2009). Chromosomal mechanisms of aminoglycoside resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Clin. Microbiol. Infect. 15, 60–66.
Jassem, A. N., Zlosnik, J. E., Henry, D. A., Hancock, R. E., Ernst, R. K., and Speert, D. P. (2011). In vitro susceptibility of Burkholderia vietnamiensis to aminoglycosides. Antimicrob. Agents Chemother. 55, 2256–2264.
Jeannot, K., Sobel, M. L., El Garch, F., Poole, K., and Plesiat, P. (2005). Induction of the MexXY efflux pump in Pseudomonas aeruginosa is dependent on drug-ribosome interaction. J. Bacteriol. 187, 5341–5346.
Jeannot, K., Elsen, S., Kohler, T., Attree, I., van Delden, C., and Plesiat, P. (2008). Resistance and virulence of Pseudomonas aeruginosa clinical strains overproducing the MexCD-OprJ efflux pump. Antimicrob. Agents Chemother. 52, 2455–2462.
Kenny, D. J., Russell, P., Rogers, D., Eley, S. M., and Titball, R. W. (1999). In vitro susceptibilities of Burkholderia mallei in comparison to those of other pathogenic Burkholderia spp. Antimicrob. Agents Chemother. 43, 2773–2775.
Kim, H. S., Nagore, D., and Nikaido, H. (2010). Multidrug efflux pump MdtBC of Escherichia coli is active only as a B2C heterotrimer. J. Bacteriol. 192, 1377–1386.
Kindrachuk, K. N., Fernandez, L., Bains, M., and Hancock, R. E. (2011). Involvement of an ATP-dependent protease, PA0779/AsrA, in inducing heat shock in response to tobramycin in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55, 1874–1882.
Klimke, W., Agarwala, R., Badretdin, A., Chetvernin, S., Ciufo, S., Fedorov, B., et al. (2009). The National Center for Biotechnology Information’s Protein Clusters Database. Nucleic Acids Res. 37, D216–D223.
Kohler, T., Michea-Hamzehpour, M., Henze, U., Gotoh, N., Curty, L. K., and Pechere, J. C. (1997). Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23, 345–354.
Laohavaleeson, S., Lolans, K., Quinn, J. P., Kuti, J. L., and Nicolau, D. P. (2008). Expression of the MexXY-OprM efflux system in Pseudomonas aeruginosa with discordant cefepime/ceftazidime susceptibility profiles. Infect. Drug Resist. 1, 51–55.
Lau, C. H., Fraud, S., Jones, M., Peterson, S. N., and Poole, K. (2012). Reduced expression of the rplU-rpmA ribosomal protein operon in mexXY-expressing pan-aminoglycoside-resistant mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 56, 5171–5179.
Lee, S., Hinz, A., Bauerle, E., Angermeyer, A., Juhaszova, K., Kaneko, Y., et al. (2009). Targeting a bacterial stress response to enhance antibiotic action. Proc. Natl. Acad. Sci. U.S.A. 106, 14570–14575.
Li, X. Z., Livermore, D. M., and Nikaido, H. (1994a). Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob. Agents Chemother. 38, 1732–1741.
Li, X. Z., Ma, D., Livermore, D. M., and Nikaido, H. (1994b). Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as a contributing factor to beta-lactam resistance. Antimicrob. Agents Chemother. 38, 1742–1752.
Li, X. Z., Nikaido, H., and Poole, K. (1995). Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39, 1948–1953.
Li, X. Z., Poole, K., and Nikaido, H. (2003). Contributions of MexAB-OprM and an EmrE homolog to intrinsic resistance of Pseudomonas aeruginosa to aminoglycosides and dyes. Antimicrob. Agents Chemother. 47, 27–33.
Li, X. Z., and Nikaido, H. (2009). Efflux-mediated drug resistance in bacteria: an update. Drugs 69, 1555–1623.
Lindemann, A., Koch, M., Pessi, G., Muller, A. J., Balsiger, S., Hennecke, H., et al. (2010). Host-specific symbiotic requirement of BdeAB, a RegR-controlled RND-type efflux system in Bradyrhizobium japonicum. FEMS Microbiol. Lett. 312, 184–191.
Lister, P. D., Wolter, D. J., and Hanson, N. D. (2009). Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22, 582–610.
Llanes, C., Hocquet, D., Vogne, C., Benali-Baitich, D., Neuwirth, C., and Plesiat, P. (2004). Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 48, 1797–1802.
Llanes, C., Neuwirth, C., El Garch, F., Hocquet, D., and Plesiat, P. (2006). Genetic analysis of a multiresistant strain of Pseudomonas aeruginosa producing PER-1 beta-lactamase. Clin. Microbiol. Infect. 12, 270–278.
Llanes, C., Kohler, T., Patry, I., Dehecq, B., van Delden, C., and Plesiat, P. (2011). Role of the MexEF-OprN efflux system in low-level resistance of Pseudomonas aeruginosa to ciprofloxacin. Antimicrob. Agents Chemother. 55, 5676–5684.
Lomovskaya, O., Warren, M. S., Lee, A., Galazzo, J., Fronko, R., Lee, M., et al. (2001). Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45, 105–116.
Magnet, S., Courvalin, P., and Lambert, T. (2001). Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45, 3375–3380.
Maniati, M., Ikonomidis, A., Mantzana, P., Daponte, A., Maniatis, A. N., and Pournaras, S. (2007). A highly carbapenem-resistant Pseudomonas aeruginosa isolate with a novel blaVIM-4/blaP1b integron overexpresses two efflux pumps and lacks OprD. J. Antimicrob. Chemother. 60, 132–135.
Mao, W., Warren, M. S., Lee, A., Mistry, A., and Lomovskaya, O. (2001). MexXY-OprM efflux pump is required for antagonism of aminoglycosides by divalent cations in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 45, 2001–2007.
Marchand, I., Damier-Piolle, L., Courvalin, P., and Lambert, T. (2004). Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 48, 3298–3304.
Martha, B., Croisier, D., Durand, D., Hocquet, D., Plesiat, P., Piroth, L., et al. (2006). In-vivo impact of the MexXY efflux system on aminoglycoside efficacy in an experimental model of Pseudomonas aeruginosa pneumonia treated with tobramycin. Clin. Microbiol. Infect. 12, 426–432.
Masuda, N., Sakagawa, E., Ohya, S., Gotoh, N., Tsujimoto, H., and Nishino, T. (2000a). Contribution of the MexX-MexY-oprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44, 2242–2246.
Masuda, N., Sakagawa, E., Ohya, S., Gotoh, N., Tsujimoto, H., and Nishino, T. (2000b). Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44, 3322–3327.
Matsumoto, Y., Hayama, K., Sakakihara, S., Nishino, K., Noji, H., Iino, R., et al. (2011). Evaluation of multidrug efflux pump inhibitors by a new method using microfluidic channels. PLoS ONE 6, e18547. doi: 10.1371/journal.pone.0018547
Matsuo, T., Hayashi, K., Morita, Y., Koterasawa, M., Ogawa, W., Mizushima, T., et al.i (2007). VmeAB, an RND-type multidrug efflux transporter in Vibrio parahaemolyticus. Microbiology 153, 4129–4137.
Matsuo, Y., Eda, S., Gotoh, N., Yoshihara, E., and Nakae, T. (2004). MexZ-mediated regulation of mexXY multidrug efflux pump expression in Pseudomonas aeruginosa by binding on the mexZ-mexX intergenic DNA. FEMS Microbiol. Lett. 238, 23–28.
McLaughlin, H. P., Caly, D. L., McCarthy, Y., Ryan, R. P., and Dow, J. M. (2012). An orphan chemotaxis sensor regulates virulence and antibiotic tolerance in the human pathogen Pseudomonas aeruginosa. PLoS ONE 7, e42205. doi: 10.1371/journal.pone.0042205
Medeiros, A. A., O’Brien, T. F., Wacker, W. E., and Yulug, N. F. (1971). Effect of salt concentration on the apparent in-vitro susceptibility of Pseudomonas and other Gram-negative bacilli to gentamicin. J. Infect. Dis. 124(Suppl), S59–S64.
Merlin, T. L., Corvo, D. L., Gill, J. H., and Griffith, J. K. (1989a). Enhanced gentamicin killing of Escherichia coli by tet gene expression. Antimicrob. Agents Chemother. 33, 230–232.
Merlin, T. L., Davis, G. E., Anderson, W. L., Moyzis, R. K., and Griffith, J. K. (1989b). Aminoglycoside uptake increased by tet gene expression. Antimicrob. Agents Chemother. 33, 1549–1552.
Mesaros, N., Glupczynski, Y., Avrain, L., Caceres, N. E., Tulkens, P. M., and Van Bambeke, F. (2007). A combined phenotypic and genotypic method for the detection of Mex efflux pumps in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 59, 378–386.
Mima, T., and Schweizer, H. P. (2010). The BpeAB-OprB efflux pump of Burkholderia pseudomallei 1026b does not play a role in quorum sensing, virulence factor production, or extrusion of aminoglycosides but is a broad-spectrum drug efflux system. Antimicrob. Agents Chemother. 54, 3113–3120.
Mine, T., Morita, Y., Kataoka, A., Mizushima, T., and Tsuchiya, T. (1998). Evidence for chloramphenicol/H+ antiport in Cmr (MdfA) system of Escherichia coli and properties of the antiporter. J. Biochem. 124, 187–193.
Mine, T., Morita, Y., Kataoka, A., Mizushima, T., and Tsuchiya, T. (1999). Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43, 415–417.
Moore, R. A., DeShazer, D., Reckseidler, S., Weissman, A., and Woods, D. E. (1999). Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43, 465–470.
Morita, Y., Kodama, K., Shiota, S., Mine, T., Kataoka, A., Mizushima, T., et al. (1998). NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42, 1778–1782.
Morita, Y., Kataoka, A., Shiota, S., Mizushima, T., and Tsuchiya, T. (2000). NorM of Vibrio parahaemolyticus is an Na(+)-driven multidrug efflux pump. J. Bacteriol. 182, 6694–6697.
Morita, Y., Kimura, N., Mima, T., Mizushima, T., and Tsuchiya, T. (2001a). Roles of MexXY- and MexAB-multidrug efflux pumps in intrinsic multidrug resistance of Pseudomonas aeruginosa PAO1. J. Gen. Appl. Microbiol. 47, 27–32.
Morita, Y., Komori, Y., Mima, T., Kuroda, T., Mizushima, T., and Tsuchiya, T. (2001b). Construction of a series of mutants lacking all of the four major mex operons for multidrug efflux pumps or possessing each one of the operons from Pseudomonas aeruginosa PAO1: MexCD-OprJ is an inducible pump. FEMS Microbiol. Lett. 202, 139–143.
Morita, Y., Murata, T., Mima, T., Shiota, S., Kuroda, T., Mizushima, T., et al. (2003). Induction of mexCD-oprJ operon for a multidrug efflux pump by disinfectants in wild-type Pseudomonas aeruginosa PAO1. J. Antimicrob. Chemother. 51, 991–994.
Morita, Y., Sobel, M. L., and Poole, K. (2006). Antibiotic inducibility of the MexXY multidrug efflux system of Pseudomonas aeruginosa: involvement of the antibiotic-inducible PA5471 gene product. J. Bacteriol. 188, 1847–1855.
Morita, Y., Gilmour, C., Metcalf, D., and Poole, K. (2009). Translational control of the antibiotic inducibility of the PA5471 gene required for mexXY multidrug efflux gene expression in Pseudomonas aeruginosa. J. Bacteriol. 191, 4966–4975.
Morita, Y., Narita, S., Tomida, J., Tokuda, H., and Kawamura, Y. (2010). Application of an inducible system to engineer unmarked conditional mutants of essential genes of Pseudomonas aeruginosa. J. Microbiol. Methods 82, 205–213.
Morita, Y., Tomida, J., and Kawamura, Y. (2012). Primary mechanisms mediating aminoglycoside resistance in the multidrug-resistant Pseudomonas aeruginosa clinical isolate PA7. Microbiology 158, 1071–1083.
Mulet, X., Moya, B., Juan, C., Macia, M. D., Perez, J. L., Blazquez, J., et al. (2011). Antagonistic interactions of Pseudomonas aeruginosa antibiotic resistance mechanisms in planktonic but not biofilm growth. Antimicrob. Agents Chemother. 55, 4560–4568.
Muller, C., Plesiat, P., and Jeannot, K. (2011). A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and beta-lactams in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 55, 1211–1221.
Murakami, S., Nakashima, R., Yamashita, E., and Yamaguchi, A. (2002). Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419, 587–593.
Murata, T., Gotoh, N., and Nishino, T. (2002). Characterization of outer membrane efflux proteins OpmE, OpmD and OpmB of Pseudomonas aeruginosa: molecular cloning and development of specific antisera. FEMS Microbiol. Lett. 217, 57–63.
Nagano, K., and Nikaido, H. (2009). Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 106, 5854–5858.
Nakashima, R., Sakurai, K., Yamasaki, S., Nishino, K., and Yamaguchi, A. (2011). Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 480, 565–569.
Nemec, A., Maixnerova, M., van der Reijden, T. J., van den Broek, P. J., and Dijkshoorn, L. (2007). Relationship between the AdeABC efflux system gene content, netilmicin susceptibility and multidrug resistance in a genotypically diverse collection of Acinetobacter baumannii strains. J. Antimicrob. Chemother. 60, 483–489.
Ngeow, Y. F., and Puthucheary, S. D. (1985). Antibiotic susceptibilities of Achromobacter xylosoxydans. Singapore Med. J. 26, 519–522.
Nikaido, H. (1994). Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264, 382–388.
Nikaido, H. (2011). Structure and mechanism of RND-type multidrug efflux pumps. Adv. Enzymol. Relat. Areas Mol. Biol. 77, 1–60.
Nikaido, H., and Pages, J. M. (2012). Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 36, 340–363.
Nishino, K., and Yamaguchi, A. (2001). Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183, 5803–5812.
Nishino, K., Yamada, J., Hirakawa, H., Hirata, T., and Yamaguchi, A. (2003). Roles of TolC-dependent multidrug transporters of Escherichia coli in resistance to beta-lactams. Antimicrob. Agents Chemother. 47, 3030–3033.
Nishino, K., Latifi, T., and Groisman, E. A. (2006). Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59, 126–141.
Nishino, K., Nikaido, E., and Yamaguchi, A. (2009). Regulation and physiological function of multidrug efflux pumps in Escherichia coli and Salmonella. Biochim. Biophys. Acta 1794, 834–843.
Nishino, K., Yamasaki, S., Hayashi-Nishino, M., and Yamaguchi, A. (2010). Effect of NlpE overproduction on multidrug resistance in Escherichia coli. Antimicrob. Agents Chemother. 54, 2239–2243.
Papadopoulos, J. S., and Agarwala, R. (2007). COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079.
Pasca, M. R., Dalla Valle, C., De Jesus Lopes Ribeiro, A. L., Buroni, S., Papaleo, M. C., Bazzini, S., et al. (2012). Evaluation of fluoroquinolone resistance mechanisms in Pseudomonas aeruginosa multidrug resistance clinical isolates. Microb. Drug Resist. 18, 23–32.
Pena, C., Suarez, C., Tubau, F., Juan, C., Moya, B., Dominguez, M. A., et al. (2009). Nosocomial outbreak of a non-cefepime-susceptible ceftazidime-susceptible Pseudomonas aeruginosa strain overexpressing MexXY-OprM and producing an integron-borne PSE-1 betta-lactamase. J. Clin. Microbiol. 47, 2381–2387.
Piddock, L. J. (2006). Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19, 382–402.
Piel, J. (2011). Approaches to capturing and designing biologically active small molecules produced by uncultured microbes. Annu. Rev. Microbiol. 65, 431–453.
Pirnay, J. P., Bilocq, F., Pot, B., Cornelis, P., Zizi, M., Van Eldere, J., et al. (2009). Pseudomonas aeruginosa population structure revisited. PLoS ONE 4, e7740. doi: 10.1371/journal.pone.0007740
Poole, K., Heinrichs, D. E., and Neshat, S. (1993a). Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol. Microbiol. 10, 529–544.
Poole, K., Krebes, K., McNally, C., and Neshat, S. (1993b). Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175, 7363–7372.
Poole, K., Gotoh, N., Tsujimoto, H., Zhao, Q., Wada, A., Yamasaki, T., et al. (1996). Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21, 713–724.
Poole, K. (2004). Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 10, 12–26.
Poole, K. (2011). Pseudomonas aeruginosa: resistance to the max. Front. Microbiol. 2:65. doi: 10.3389/fmicb.2011.00065
Poole, K. (2012). “Efflux-mediated antimicrobial resistance,” in Antibiotic Discovery and Development, Vol. 1, eds T. J. Dougherty and M. J. Pucci (New York, NY: Springer), 349–395.
Queenan, A. M., Shang, W., Bush, K., and Flamm, R. K. (2010). Differential selection of single-step AmpC or efflux mutants of Pseudomonas aeruginosa by using cefepime, ceftazidime, or ceftobiprole. Antimicrob. Agents Chemother. 54, 4092–4097.
Remans, K., Vercammen, K., Bodilis, J., and Cornelis, P. (2010). Genome-wide analysis and literature-based survey of lipoproteins in Pseudomonas aeruginosa. Microbiology 156, 2597–2607.
Roca, I., Espinal, P., Marti, S., and Vila, J. (2011). First identification and characterization of an AdeABC-like efflux pump in Acinetobacter genomospecies 13TU. Antimicrob. Agents Chemother. 55, 1285–1286.
Rosenberg, E. Y., Ma, D., and Nikaido, H. (2000). AcrD of Escherichia coli is an aminoglycoside efflux pump. J. Bacteriol. 182, 1754–1756.
Roy, P. H., Tetu, S. G., Larouche, A., Elbourne, L., Tremblay, S., Ren, Q., et al. (2010). Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS ONE 5, e8842. doi: 10.1371/journal.pone.0008842
Ruzin, A., Keeney, D., and Bradford, P. A. (2007). AdeABC multidrug efflux pump is associated with decreased susceptibility to tigecycline in Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J. Antimicrob. Chemother. 59, 1001–1004.
Segonds, C., Clavel-Batut, P., Thouverez, M., Grenet, D., Le Coustumier, A., Plesiat, P., et al. (2009). Microbiological and epidemiological features of clinical respiratory isolates of Burkholderia gladioli. J. Clin. Microbiol. 47, 1510–1516.
Seo, Y. S., Lim, J., Choi, B. S., Kim, H., Goo, E., Lee, B., et al. (2011). Complete genome sequence of Burkholderia gladioli BSR3. J. Bacteriol. 193, 3149.
Shiota, S., Shimizu, M., Sugiyama, J., Morita, Y., Mizushima, T., and Tsuchiya, T. (2004). Mechanisms of action of corilagin and tellimagrandin I that remarkably potentiate the activity of beta-lactams against methicillin-resistant Staphylococcus aureus. Microbiol. Immunol. 48, 67–73.
Sidhu, K., Talbot, M., Mil, K. V., and Verstraete, M. (2012). Treatment with sub-inhibitory kanamycin induces adaptive resistance to aminoglycoside antibiotics via the AcrD multidrug efflux pump in Escherichia coli K-12. J. Exp. Microbiol. Immunol. 16, 11–16.
Simpson, A. J., White, N. J., and Wuthiekanun, V. (1999). Aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43, 2332.
Smith, E. E., Buckley, D. G., Wu, Z., Saenphimmachak, C., Hoffman, L. R., D’Argenio, D. A., et al. (2006). Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U.S.A. 103, 8487–8492.
Sobel, M. L., McKay, G. A., and Poole, K. (2003). Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 47, 3202–3207.
Sobel, M. L., Neshat, S., and Poole, K. (2005). Mutations in PA2491 (mexS) promote MexT-dependent mexEF-oprN expression and multidrug resistance in a clinical strain of Pseudomonas aeruginosa. J. Bacteriol. 187, 1246–1253.
Srikumar, R., Li, X. Z., and Poole, K. (1997). Inner membrane efflux components are responsible for beta-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J. Bacteriol. 179, 7875–7881.
Stavri, M., Piddock, L. J., and Gibbons, S. (2007). Bacterial efflux pump inhibitors from natural sources. J. Antimicrob. Chemother. 59, 1247–1260.
Stover, C. K., Pham, X. Q., Erwin, A. L. Mizoguchi, S, D., Warrener, P., Hickey, M. J., et al. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964.
Sun, J. R., Chan, M. C., Chang, T. Y., Wang, W. Y., and Chiueh, T. S. (2010). Overexpression of the adeB gene in clinical isolates of tigecycline-nonsusceptible Acinetobacter baumannii without insertion mutations in adeRS. Antimicrob. Agents Chemother. 54, 4934–4938.
Sun, J. R., Perng, C. L., Chan, M. C., Morita, Y., Lin, J. C., Su, C. M., et al. (2012). A truncated AdeS kinase protein generated by ISAba1 insertion correlates with tigecycline resistance in Acinetobacter baumannii. PLoS ONE . doi: 10.1371/journal.pone.0049534
Takatsuka, Y., and Nikaido, H. (2006). Threonine-978 in the transmembrane segment of the multidrug efflux pump AcrB of Escherichia coli is crucial for drug transport as a probable component of the proton relay network. J. Bacteriol. 188, 7284–7289.
Tanaka, N., and Shuman, S. (2011). RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J. Biol. Chem. 286, 7727–7731.
Thibault, F. M., Hernandez, E., Vidal, D. R., Girardet, M., and Cavallo, J. D. (2004). Antibiotic susceptibility of 65 isolates of Burkholderia pseudomallei and Burkholderia mallei to 35 antimicrobial agents. J. Antimicrob. Chemother. 54, 1134–1138.
Trunck, L. A., Propst, K. L., Wuthiekanun, V., Tuanyok A., Beckstrom-Sternberg, S. M., Beckstrom-Sternberg, J. S., et al. (2009). Molecular basis of rare aminoglycoside susceptibility and pathogenesis of Burkholderia pseudomallei clinical isolates from Thailand. PLoS Negl. Trop. Dis. 3, e519. doi: 10.1371/journal.pntd.0000519
Vermis, K., Vandamme, P. A., and Nelis, H. J. (2003). Burkholderia cepacia complex genomovars: utilization of carbon sources, susceptibility to antimicrobial agents and growth on selective media. J. Appl. Microbiol. 95, 1191–1199.
Vettoretti, L., Floret, N., Hocquet, D., Dehecq, B., Plesiat, P., Talon, D., et al. (2009a). Emergence of extensive-drug-resistant Pseudomonas aeruginosa in a French university hospital. Eur. J. Clin. Microbiol. Infect. Dis. 28, 1217–1222.
Vettoretti, L., Plesiat, P., Muller, C., El Garch, F., Phan, G., Attree, I., et al. (2009b). Efflux unbalance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 53, 1987–1997.
Vogne, C., Aires, J. R., Bailly, C., Hocquet, D., and Plesiat, P. (2004). Role of the multidrug efflux system MexXY in the emergence of moderate resistance to aminoglycosides among Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob. Agents Chemother. 48, 1676–1680.
Westbrock-Wadman, S., Sherman, D. R., Hickey, M. J., Coulter, S. N., Zhu, Y. Q., Warrener, P., et al. (1999). Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43, 2975–2983.
Winsor, G. L., Lam, D. K., Fleming, L., Lo, R., Whiteside, M. D., Yu, N. Y., et al. (2011). Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39, D596-600.
Wolter, D. J., Smith-Moland, E., Goering, R. V., Hanson, N. D., and Lister, P. D. (2004). Multidrug resistance associated with mexXY expression in clinical isolates of Pseudomonas aeruginosa from a Texas hospital. Diagn. Microbiol. Infect. Dis. 50, 43–50.
Wyka, M. A., and St John, A. C. (1990). Effects of production of abnormal proteins on the rate of killing of Escherichia coli by streptomycin. Antimicrob. Agents Chemother. 34, 534–538.
Xavier, D. E., Picao, R. C., Girardello, R., Fehlberg, L. C., and Gales, A. C. (2010). Efflux pumps expression and its association with porin down-regulation and beta-lactamase production among Pseudomonas aeruginosa causing bloodstream infections in Brazil. BMC Microbiol. 10, 217. doi: 10.1186/1471-2180-10-217
Yamamoto, M., Ueda, A., Kudo, M., Matsuo, Y., Fukushima, J., Nakae, T., et al. (2009). Role of MexZ and PA5471 in transcriptional regulation of mexXY in Pseudomonas aeruginosa. Microbiology 155, 3312–3321.
Yamasaki, S., Nagasawa, S., Hayashi-Nishino, M., Yamaguchi, A., and Nishino, K. (2011). AcrA dependency of the AcrD efflux pump in Salmonella enterica serovar Typhimurium. J. Antibiot. (Tokyo) 64, 433–437.
Yerushalmi, H., Lebendiker, M., and Schuldiner, S. (1995). EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J. Biol. Chem. 270, 6856–6863.
Yu, E. W., Aires, J. R., and Nikaido, H. (2003). AcrB multidrug efflux pump of Escherichia coli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J. Bacteriol. 185, 5657–5664.
Zgurskaya, H. I., and Nikaido, H. (1999). Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 96, 7190–7195.
Keywords: aminoglycoside resistance, Pseudomonas aeruginosa, efflux, MexXY, PA5471, OprA
Citation: Morita Y, Tomida J and Kawamura Y (2012) MexXY multidrug efflux system of Pseudomonas aeruginosa. Front. Microbio. 3:408. doi: 10.3389/fmicb.2012.00408
Received: 11 October 2012; Accepted: 13 November 2012;
Published online: 28 November 2012.
Edited by:
Kunihiko Nishino, Institute of Scientific and Industrial Research, Osaka University, JapanReviewed by:
Herbert P. Schweizer, Colorado State University, USAAixin Yan, The University of Hong Kong, Hong Kong
Copyright: © 2012 Morita, Tomida and Kawamura. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Yuji Morita, Department of Microbiology, School of Pharmacy, Aichi Gakuin University, 1-100 Kusumoto, Chikusa, Nagoya, Aichi 464-8650, Japan. e-mail:eXVqbW9yQGRwYy5hZ3UuYWMuanA=

 Junko Tomida and Yoshiaki Kawamura
Junko Tomida and Yoshiaki Kawamura