- 1Marine Biological Laboratory, Josephine Bay Paul Center for Comparative Molecular Biology and Evolution, Woods Hole, MA, USA
- 2Department of Geological Sciences, Brown University, Providence, RI, USA
Extremely acidic (pH < 3) and extremely alkaline (pH > 9) environments support a diversity of single-cell and to a lesser extent, multicellular eukaryotic life. This study compared alpha and beta diversity in eukaryotic communities from seven diverse aquatic environments with pH values ranging from 2 to 11 using massively-parallel pyrotag sequencing targeting the V9 hypervariable region of the 18S ribosomal RNA (rRNA) gene. A total of 946 operational taxonomic units (OTUs) were recovered at a 6% cut-off level (94% similarity) across the sampled environments. Hierarchical clustering of the samples segregated the communities into acidic and alkaline groups. Similarity percentage (SIMPER) analysis followed by indicator OTU analysis (IOA) and non-metric multidimensional scaling (NMDS) were used to determine which characteristic groups of eukaryotic taxa typify acidic or alkaline extremes and the extent to which pH explains eukaryotic community structure in these environments. Spain's Rio Tinto yielded the fewest observed OTUs while Nebraska Sandhills alkaline lakes yielded the most. Distinct OTUs, including metazoan OTUs, numerically dominated pH extreme sites. Indicator OTUs included the diatom Pinnularia and unidentified opisthokonts (Fungi and Filasterea) in the extremely acidic environments, and the ciliate Frontonia across the extremely alkaline sites. Inferred from NMDS, pH explained only a modest fraction of the variation across the datasets, indicating that other factors influence the underlying community structure in these environments. The findings from this study suggest that the ability for eukaryotes to adapt to pH extremes over a broad range of values may be rare, but further study of taxa that can broadly adapt across diverse acidic and alkaline environments, respectively present good models for understanding adaptation and should be targeted for future investigations.
Introduction
Life thrives at many extremes, but how eukaryotes adapt and diversify in these environments remains underexplored. Extreme pH environments are typically described as those possessing a pH less than 3 and greater than 9 (Gross and Robbins, 2000; Horikoshi, 2004). Both acidic and alkaline pH extreme environments often include other abiotic extremes as well (Rothschild and Mancinelli, 2001). Eukaryotes that thrive at acidic extremes are often subjected to not only orders of magnitude differential concentrations of hydrogen ions outside their cells, but also high concentrations of toxic metals (Whitton, 1970), low nutrient levels (Brake and Hasiotis, 2010), and/or extreme temperatures (Brock, 1978). Likewise extreme temperatures and salinities often characterize extreme alkaline environments (Horikoshi, 2004).
Both extremely acidic and extremely alkaline environments can be attributed to natural and anthropogenic sources. Classic examples of acidic environments include acid rock drainage (ARD) or acid mine drainage (AMD) systems often involving current or past mining activities, hydrothermal vent fluids, and terrestrial geothermal environments. Alkaline environments include soda lakes, hot springs, hydrothermal vent systems, as well as environments shaped by industries such as papermaking and textiles. In general, acidophilic eukaryotic diversity is more extensively studied, probably due to a larger number of accessible environments characterized by these extremes and relevance to biotechnological applications. Examples include ARD (Amaral Zettler et al., 2002) and AMD environments (Baker et al., 2003), acidic geothermal springs (Aguilera et al., 2010) and acidic lakes (Brown and Wolfe, 2006). Several well-known pH extreme environments such as alkaline Mono Lake in California, have been extensively examined for their bacterial and archaeal diversity but remain underexplored with respect to microbial eukaryotic diversity (Hollibaugh et al., 2001; Humayoun et al., 2003). Others such as acidic Nymph Creek in Wyoming have been the focus of targeted (Sheehan et al., 2003; Ferris et al., 2005) microbial eukaryotic work but have never been surveyed for overall eukaryotic diversity.
Despite the wealth of studies that focus on individual extreme environments, comparisons across multiple extremes are rare. Studies of eukaryotes in extremely acidic environments have revealed representatives from multiple evolutionary lineages, suggesting that the ability to adapt to pH extremes may be widespread (Amaral Zettler et al., 2002; Costas et al., 2007; Lopez-Rodas et al., 2008, 2011). This raises the question of whether there are cosmopolitan eukaryotic taxa that have adapted to a wide range of pH extremes. Which environmental parameters are most influential in shaping eukaryotic microbial diversity patterns at pH extremes also remains underexplored.
Investigations of eukaryotes at pH extremes have a long history (Brock, 1978), but an obstacle to unveiling the extent of eukaryotic diversity at pH extremes is that many of these environments have low evenness with a small number of species that make up biofilm communities dominating a given site. A lack of consistent taxonomic coverage in biodiversity inventories has also hindered comprehensive morphology-based surveys in general. It was not until the advent of molecular approaches employing general eukaryotic primers that the extent of eukaryotic diversity at pH extremes was more fully revealed. The application of next-generation DNA sequencing has helped to unveil the rarer forms of eukaryotes that exist behind the backdrop of more abundant forms (Stoeck et al., 2010). Yet, despite efforts to catch up with the bacterial and archaeal efforts, investigations of eukaryotic diversity at pH extremes and extreme environments in general still lag behind those of other domains. A major challenge to quantifying eukaryotic diversity is PCR-bias associated with the sometimes extreme length variation in ribosomal RNA (rRNA) genes. Another challenge (seldom acknowledged) is accurately representing the relative abundance of a given eukaryotic species or phylotype in the environment, because microbial eukaryotes can have copy numbers of their rRNA genes that differ by 4 orders of magnitude. This confounds our ability to apply abundance-based metrics in quantifying eukaryotic diversity in natural environments. Techniques such as fluorescence in situ hybridization (FISH) offer a solution to this accounting problem but are not appropriate for biodiversity studies where the goal is to discover novel taxa. Inconsistent taxonomic assignments for reference sequences in public databases further challenge comparative molecular descriptive studies. Despite these obstacles, we have learned much about the phylogenetic diversity of microbial eukaryotes using culture-independent approaches.
This study compared eukaryotic communities from naturally occurring and anthropogenic sites with pH values from 2 to 11 in diverse aquatic environments ranging from ARDs of the Rio Tinto, Spain to the alkaline hydrothermal vent fields of Lost City using massively-parallel pyrotag sequencing targeting the V9 hypervariable region of the 18S rRNA gene. The aim of this study was to determine what groups of eukaryotic taxa typify extremely acidic or extremely alkaline environments and how cosmopolitan these groups are at pH extremes. This was tested using a combination of hierarchical clustering, indicator species analysis (ISA), non-metric multidimensional scaling (NMDS), and oligotyping analyses. The advent of next-generation sequencing and its application to biodiversity studies in extreme environments is broadening our understanding of the limits to life on our planet and will aid in determining which groups of taxa are best suited to become model organisms to increase our understanding of adaptation to life's extremes.
Methods
Site Characteristics
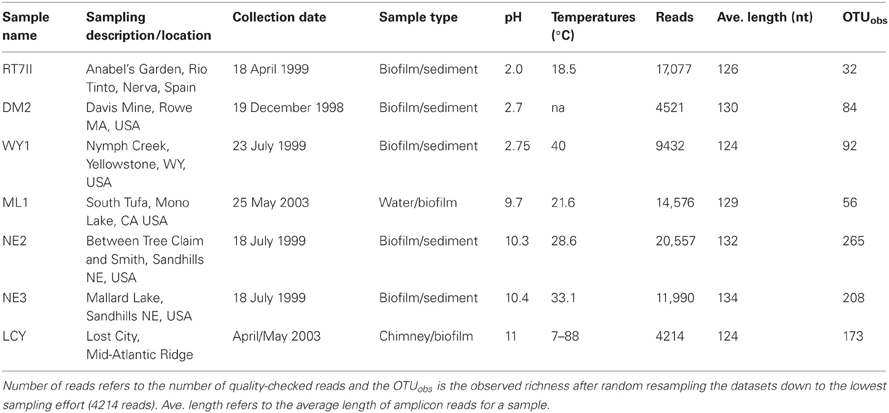
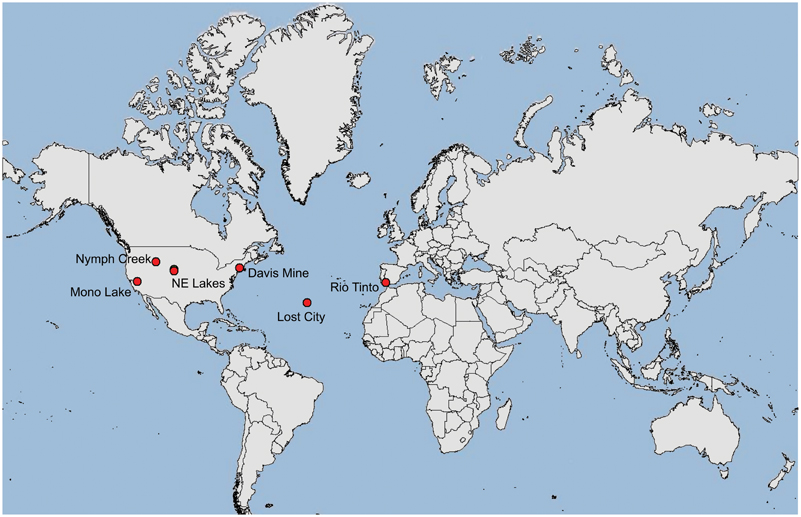
The pH extreme environments examined in this study ranged from extremely acidic (pH 2) to extremely alkaline (pH 11) (Figures 1A–G, Table 1). The extreme acidic sites included Nymph Creek, YNP, WY, Davis Mine in Rowe, MA, and Anabel's Garden in the Rio Tinto in southwestern Spain, while our extreme alkaline sites included two Sandhills region lakes in Nebraska, Mono Lake in California, and the Lost City Hydrothermal Vent on the Mid-Atlantic Ridge in the Atlantic Ocean. Nymph Creek (Figure 1A) is located near the Norris Geyser basin in YNP, WY and is characterized as a ferrous iron (2.3 mg/L) and arsenite-rich (0.086 mg/L) shallow thermal spring with temperatures ranging from 38°C to 52°C (Nordstrom et al., 2005). Its most salient feature is the green microbial mat that blankets the bottom of the creek composed of red and green algal species (Brock, 1978). Davis Mine (Figure 1B) in Rowe, MA, is the former site of the Davis Sulphur Ore Company that operated during the late 1800's to provide a commercial source of sulfuric acid (McCarthy, 1977). The site now contains AMD streams of high iron content. Anabel's Garden (Figure 1G) in the Rio Tinto, Spain is the site of an extensively studied ARD area that is situated on the Iberian Pyritic belt, the largest metal sulfide surface deposit in the world. Abundant and distinct biofilms of Euglena and diatoms typify this site fed by a small stream and groundwater derived from a lake in an abandoned mining pit. Alkaline lakes in the Sandhills region of NE are located in the Crescent Lake National Wildlife Refuge of Garden County (Figures 1C,F). Two of these lakes were included in this study and are typical of alkaline lakes in this region. While no direct physico-chemical measurements other than pH accompanied the sampling of the alkaline lakes, Sandhills lakes in general are classified as sodium and potassium-bicarbonate-carbonate-hydroxide varieties where sodium, potassium, calcium, and magnesium are the most numerous cations found (McCarraher, 1977). Mono Lake (Figure 1D) in California served as an alkaline, hypersaline site characterized by dominance of carbonate, sulfate and sodium ions. Mono Lake contains elevated dissolved organic carbon (DOC) and high bacterial concentrations (Hollibaugh et al., 2001). The final alkaline site, Lost City (Figure 1E) is an ancient hydrothermal vent system rich in calcium but low in heavy metals with abiogenic sources of hydrogen, as well as methane and hydrocarbons (Proskurowski et al., 2008). Table 1 lists the origin and associated contextual data for all samples and Figure 2 shows the geographic locations of each site.

Figure 1. pH extreme sites examined in this study. (A) Nymph Creek, Yellowstone National Park (YNP), WY, USA. (B) Davis Mine, Rowe, MA, USA. (C) Sandhills Lake Between Tree Claim and Smith, Crescent Lake Wildlife Refuge, NE, USA. (D) South Tufa at Mono Lake, CA, USA. (E) Lost City Hydrothermal Vent Field, Mid-Atlantic Ridge (photo credit, IFE, URI—IAO, UW, Lost City science party, and NOAA). (F) Mallard Lake, Crescent Lake Wildlife Refuge, NE, USA. (G) Anabel's Garden, Rio Tinto, Nerva, Spain. See text for details of site descriptions.
DNA Extraction
Genomic DNA extraction methods varied and followed protocols optimized for a given extreme environment. Sediment samples were collected aseptically using a 50-ml centrifuge tube and immediately frozen in a liquid nitrogen dry shipper, then transferred to a −80°C freezer upon return to the laboratory. The Mono Lake water sample containing resuspended biofilm material (500 ml) was filtered onto a 0.2 μm Sterivex filter (Millipore Corp., Beverley, MA) and extracted using the Puregene Tissue kit (Qiagen, Valencia, CA) and methods. Biofilm/sediment extractions (1 g each) followed a modified phenol-CTAB extraction procedure (Hugo et al., 1992). The biofilm/sediment sample from the Rio Tinto (RT7II) was extracted as described in Amaral-Zettler et al. (2002) and Lost City chimney samples were extracted according to protocols in Brazelton et al. (2010). The Lost City sample was a physical pool of four separate samples so represents a range of temperatures as indicated in Table 1.
Pyrosequencing and Data Analysis
Pyrosequencing of all samples followed protocols established in Amaral-Zettler et al. (2009) and applied barcoded primers to achieve multiplexing of samples on the Genome Sequencer FLX Platform (Roche, Basel, Switzerland) housed in the Marine Biological Laboratory Keck Sequencing Facility and used the GS-LR70 long-read sequencing kit (Roche). The tag recovery and read length for all samples varied and is summarized in Table 1. Sequences were trimmed and screened for quality after Huse et al. (2007). To assign taxonomy to the remaining quality-controlled tags, we used the global alignment for sequence taxonomy (GAST) algorithm (Huse et al., 2008) with version 111 of the Silva-ARB database (Pruesse et al., 2007) that has recently been updated with improved eukaryotic taxonomic assignments. V9 amplicon reads were grouped into operational taxonomic units (OTUs) using SLP-PWAN (Huse et al., 2010) at 6% cluster widths. The open source investigation/study/assay (Sansone et al., 2012) metadata-tracking framework was used to curate the datasets and format them for submission to the NCBI sequence read archive (SRA) database under the SRA number SRA059386. All data are MIMARKS compliant (Yilmaz et al., 2011).
Tools available in R (www.r-project.org) helped to calculate Venn diagrams constructed from the full dataset that were visualized using the Venn Diagram Plotter (http://ncrr.pnl.gov/ or http://www.sysbio.org/resources/staff/). For the purposes of drawing the Venn diagrams alone, the OTU data for the Sandhills lakes were pooled so as to represent the overlapping communities in a three-way Venn diagram. All multivariate analyses were performed on a randomly resampled data matrix to standardize for sampling effort (4214 reads). The matrix was subsequently transformed into incidence (presence/absence) data. Hierarchical clustering and similarity profile (SIMPROF) permutation tests were implemented in Primer E (Clarke and Warwich, 2001) and used Jaccard distances. Similarity percentage (SIMPER) analysis, also implemented in Primer E, identified the OTUs that contributed the most to the separation between the acidic and alkaline groups. NMDS (Mather, 1976) was implemented in the PC-ORD software (Peck, 2010). Replicate stress tests allowed for selection of the optimal dimensionality for the NMDS. The final NMDS solution employed a Jaccard distance measure, used the local computer time to generate a random start seed, and included 250 runs with real data and 250 runs with randomized data. A 3-dimension final solution with 24 iterations was deemed optimal with a P value of 0.0199 using a Monte Carlo test. The final stress and final instability were 0 and 0.00053, respectively.
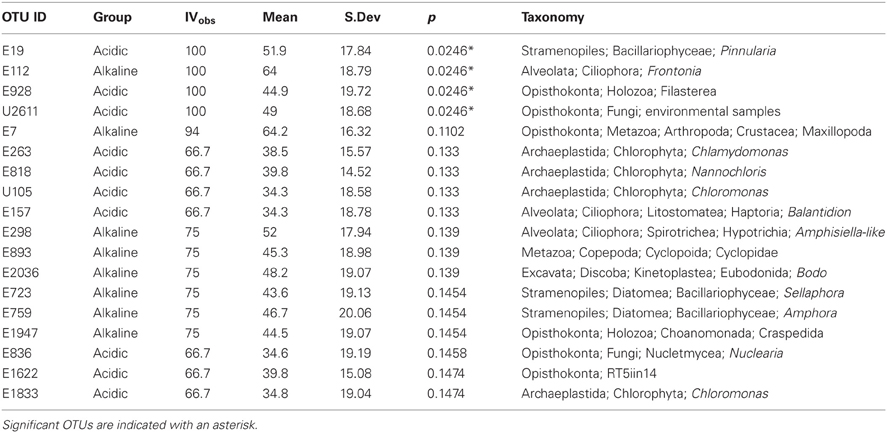
ISA (Peck, 2010) helped to test the hypothesis that different extremophile OTUs define extremely acidic and extremely alkaline environments based on constancy and abundance in a given group. Since the V9 region of 18S rRNA genes is not typically able to differentiate between OTUs at the species level it is more appropriate to refer to this analysis as indicator OTU analysis (IOA). Two groups were chosen on the basis of pH as determined by the hierarchical clustering results: samples falling into the extreme acidic (0–3) pH group and those falling into the extreme alkaline (9–11) pH group. This method can involve either abundance or presence/absence matrices. Randomly resampled (4214 reads for each sample) abundance and binary matrices were employed for this analysis as appropriate. IOA was performed using the Dufrêne and Legendre (1997) method to obtain an “indicator value” (IV), the product of relative abundance and constancy that ranges from 0 to 100. A second method employed the Tichý and Chytrý approach (2006) wherein IV values are reported as a perfect negative indication (−1) or a perfect positive indication (1). Random reassignment of the samples to groups followed by iteration of the IV calculation provided a test of the statistical significance of the IV values.
The R package routines gplots and heatmap.2 were used to generate the heatmap summary of all OTUs that were encountered with a frequency of greater than 1% in a given site. An oligotyping analysis (Eren et al., 2011) was performed on the two OTUs identified to the genus level identified using IOA. Oligotyping involved retrieving all reads that fell into the E19 (assigned the genus Pinnularia) and E112 (assigned to the genus Frontonia) 6% OTUs clusters, respectively. Sequence reads were then aligned in MUSCLE (Edgar, 2004) to obtain a multiple sequence alignment. Shannon entropy analysis was performed on the aligned OTU reads to quantify nucleotide uncertainty along the columns of aligned sequences and identify highly variable nucleotide positions. Oligotyping then decomposed the data to identify meaningful groups within the samples examined. The power in this technique is that it can identify evolutionary units missed by distance-based approaches commonly employed in microbial community analyses.
Results and Discussion
Alpha Diversity
Alpha diversity, the number of OTUs (OTU richness) within a given site, is a metric often employed in biodiversity studies to contrast communities from different environments. It is generally accepted that harsh environments experiencing one or more stressors tend to harbor fewer species (Frontier, 1985). This study is the first to compare and contrast eukaryotic diversity in extreme environments spanning 9 orders of magnitude in hydrogen ion concentration. A total of 946 OTUs were recovered at a 6% cut-off level (94% similarity) across seven sampled pH extreme environments. Clustering at this conservative cut-off helped to compensate for possible intraspecific heterogeneities in rRNA gene copies (Amaral Zettler et al., 1998), and the SLP-PWAN approach with 2% single linkage preclustering followed by 6% average neighbor clustering further minimized OTU inflation that can occur from pyrosequenced-generated homopolymers (Huse et al., 2010; Quince et al., 2011). Only observed and not estimated richness values were calculated on these datasets because replicated samples were not available to perform incidence-based diversity estimation better suited to taxa with highly variable marker gene copy number. Although sequenced amplicon read recoveries varied from 4214 to 20,557, resampling to normalize for sampling effort revealed richness values (recorded as observed OTUs) wherein the alkaline samples (e.g., Between Tree-Claim and Smith) were the most OTU-rich while acidic ones (e.g., Anabel's Garden, Rio Tinto) were the least (Table 1).
Within acidic sites, the Rio Tinto yielded the fewest observed numbers of OTUs (32) while Nymph Creek revealed the largest (92). In contrast, alkaline sites were much more OTU-rich, on average yielding several times greater OTU richness than their acidic counterparts. The exception was Mono Lake, where only 56 OTUs were recovered after normalizing for sampling effort. If one associates decreased richness with greater stress in a given environment, then these results support the hypothesis of overall lower diversity at increased environmental extremes. For example, Mono Lake possesses high pH, high levels of certain toxic metals (e.g., arsenic) and is hypersaline. Similarly, in the Rio Tinto, a combination of low pH and high toxic metal concentrations both likely contribute to habitat filtering excluding the existence of a more phylogenetically diverse assemblage of organisms (Amaral-Zettler et al., 2011). Like the Rio Tinto and Mono Lake, where both extreme pH and high metals prevail, the Lost City site extremes are multi-faceted and include high pH, temperature and pressure extremes. Surprisingly, Lost City still yielded relatively higher observed OTU-richness despite the added extremes encountered. One possible explanation may have been that the comparatively reduced metal concentrations characteristic of the Lost City geochemistry (Brazelton et al., 2010) reduces the effects of habitat filtering in this environment. Another important consideration in contrasting richness patterns using any PCR-based approach is the contribution of DNA derived from allochthonous sources (e.g., surrounding seawater) and environmental DNA from inactive or dead cells (Pawlowski et al., 2011). It is well-known that DNA is more chemically stable under alkaline than acidic conditions and thus one might expect that extremely acidic environments would be less likely to contain large amounts of persistent DNA from inactive organisms or foreign sources (Bernhardt and Tate, 2012) while alkaline environments might reflect the opposite trend. Work by Lopez-Garcia et al. (2003) on acidic hydrothermal vents suggested only autochthonous communities were detected based on an absence of photosynthetic taxa. The application of next-generation sequencing to environmental biodiversity surveys, however, is a much more sensitive technique and thus data resulting from next-generation approaches need to be interpreted with caution even in environments seemingly inhospitable to DNA preservation outside the cell.
Beta Diversity and Community Composition
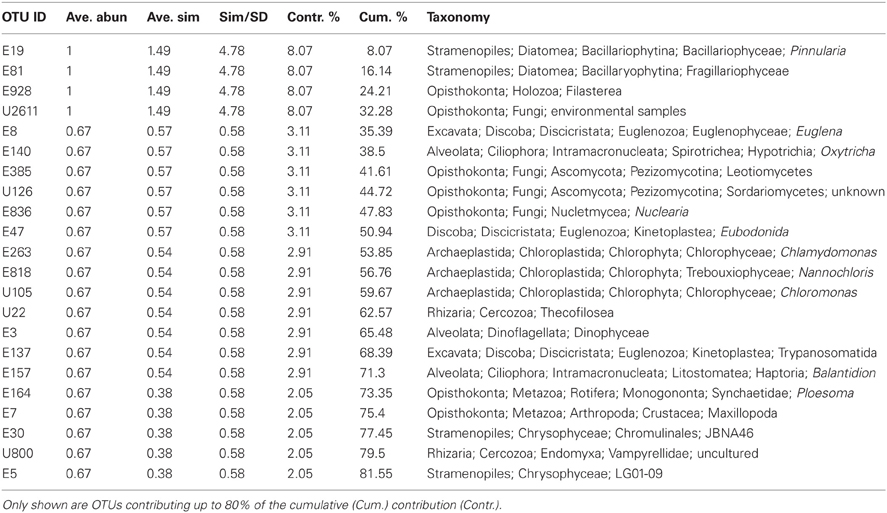
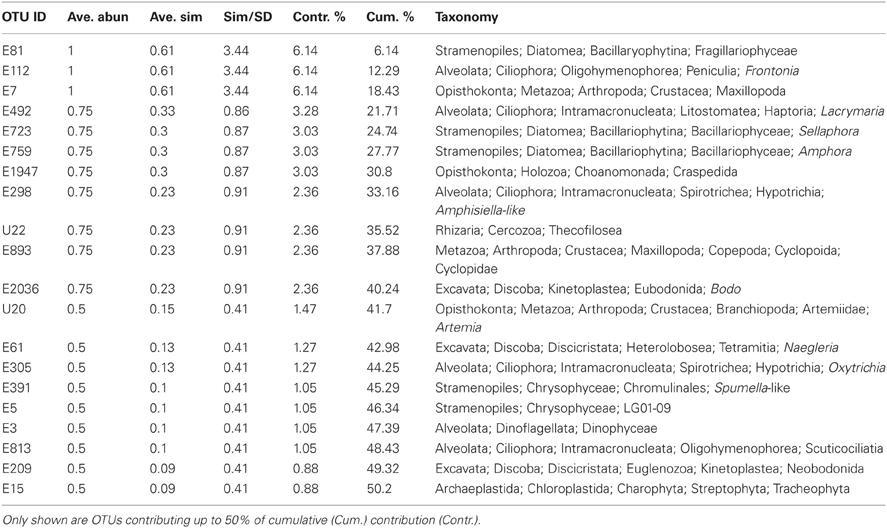
Hierarchical clustering of the sites separated acidic from alkaline communities with greatest support for the clustering of the three acidic sites and the two Nebraskan Sandhills Lake sites (Figure 3). Despite the segregation of the acidic sites from alkaline sites, overall similarities between sites were low. All sites had large proportions of unique OTUs but shared OTUs with other sites as well (Figure 4). For example, Rio Tinto's acidic Anabel's Garden contained ~45% unique OTUs and shared close to a third of its OTUs with either Davis Mine or Nymph Creek. Alkaline sites had proportionally fewer shared OTUs with ~76% to ~94% unique OTUs (when OTUs from the Sandhills Lake sites were pooled). A SIMPER analysis highlighted the shared OTUs within acidic and alkaline sites, respectively (Tables A1 and A2). The average similarity between the acidic sites was 18.48% while that for the alkaline sites was 9.92%. The most cosmopolitan OTU was a diatom that occurred across all seven sites. This OTU (E81) was assigned to the diatom class Fragillariophyceae with an exact match over the entire V9 region to several different diatom genera including three different species of Diatoma, and a single species each of Staurosira, Tabellaria, and Asterionella. Although the level of resolution attainable using the eukaryotic V9 hypervariable region does not readily differentiate between species (and in this example, genera), some authors argue that, with a few exceptions, most diatom genera are cosmopolitan (Vanormelingen et al., 2008). It is noteworthy that members of the genus Diatoma in particular have been reported from alkaline (Reavie and Smol, 2001) and acidic (Denicola, 2000) environments. What is less certain is whether a given species is capable of surviving extremes captured by the sites examined in this study (but see, Costas et al., 2007). However, at least some algal species such as Picocystis sp. are known to be capable of existing at pH values ranging from 4 to 12 (Roesler et al., 2002) despite a preference for alkaline pH, indicating that the ability to adapt to pH over broad ranges does exist in some evolutionary lineages.
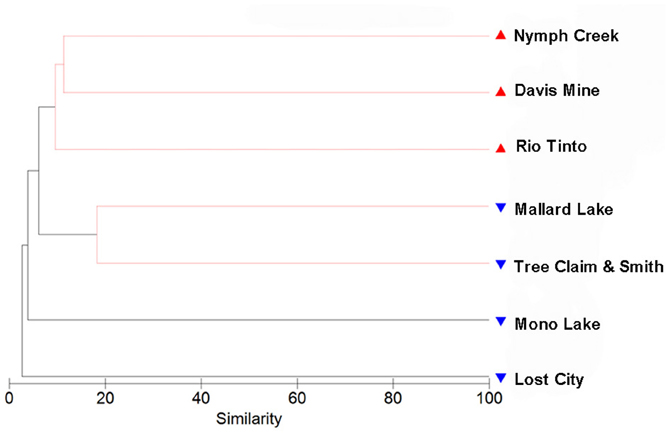
Figure 3. Hierarchical clustering analysis using a resemblance matrix composed of Jaccard derived similarities between samples. Acidic samples are indicated with red symbols while alkaline samples are indicated with blue symbols. A similarity profile (SIMPROF) permutation test highlights in red clusters that show significant internal structure.
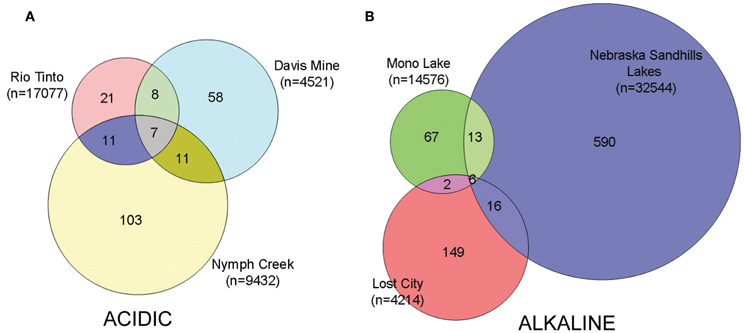
Figure 4. Venn diagrams indicating the degree of overlap between (A) acidic OTUs and (B) alkaline OTUs. Total number of amplicon reads per sample is indicated in parentheses under the sample names. Nebraska Sandhills samples were pooled only for the purposes of drawing the Venn diagram. Non-resampled data sets were used to generate these diagrams.
Apart from the afore-mentioned diatom E81 OTU, a total of 5 additional OTUs were shared across all alkaline sites while 6 were shared across acidic ones. Among the shared alkaline OTUs were ciliates assigned to members of the genera Frontonia and Lacrymaria, a cercozoan OTU assigned to the genus Protaspa, a maxillopod metazoan OTU and a tracheophyte OTU. Shared acidic OTUs included those assigned to the diatom genus Pinnularia, a chrysophyte OTU, and opisthokont OTUs including fungi, a holozoan and a maxillopod. Figure 5 illustrates the OTUs found in the seven sites at a frequency greater than 1%. Some of these OTUs were found exclusively at a given site and were the dominant OTU at that site. Examples included an unidentified opisthokont OTU from Lost City with a top BLAST hit of only 90% similarity to a gammarid amphipod and the unicellular chlorophyte Picocystis from Mono Lake. Picocystis is well-documented in the Mono Lake ecosystem where it accounts for up to 50% of the primary production there at certain times of the year (Roesler et al., 2002).
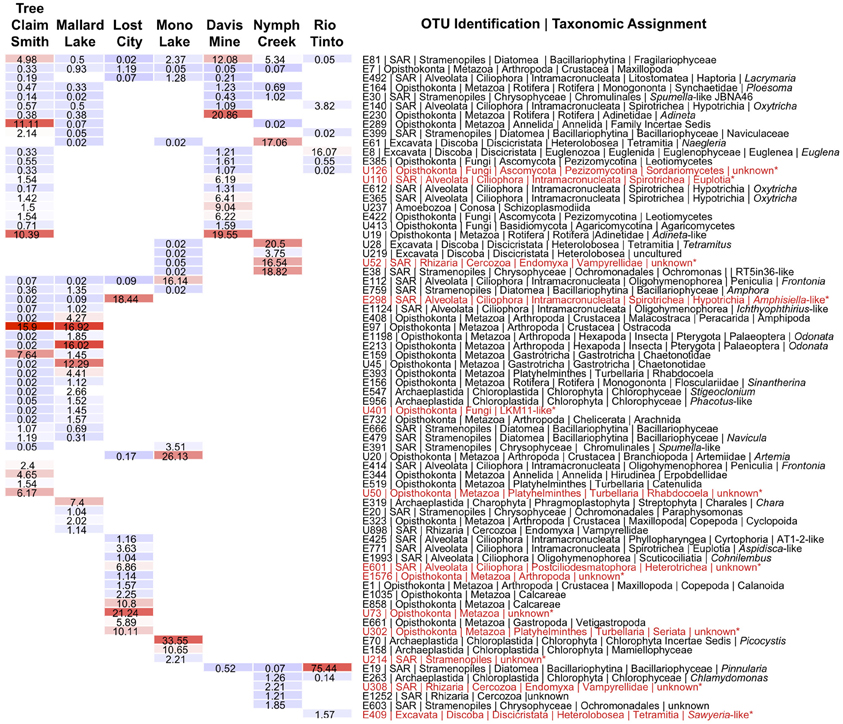
Figure 5. Heatmap of OTUs occurring at a frequency of more than 1% at a given site. Alkaline sites are the four on the left and acidic sites the three on the right and are arranged in order of decreasing OTU richness from left to right. OTUs that occur at both alkaline and acidic sites are arranged at the top of the table from most to least cosmopolitan, while OTUs occurring at only alkaline or acidic sites are arranged below. Hot colors indicate higher percentages while cooler colors indicate lower percentages, with actual values superimposed over colors. Highlighted in red and with an asterisk are OTUs that had either GAST or BLAST distances of greater than 9% to any known sequence in GenBank. Data shown are normalized by randomly resampling data to the lowest sampling effort. The depth of taxonomic resolution available in the V9 region varied by taxon and is displayed to the right.
The heatmap (Figure 5) also highlights the relative dominance of the diatom OTU E19 assigned to the genus Pinnularia at the Rio Tinto site, which unlike the E81 diatom OTU, could be definitively assigned to this genus. Pinnularia is a known acidophilic diatom genus that includes several species that have been reported from both AMD environments (Luis et al., 2012) and hot springs in Yellowstone National Park (Denicola, 2000). Previous work in the Rio Tinto using a polyphasic approach combining molecular and morphological approaches has also shown Pinnularia to be the dominant diatom at this site (Souza-Egipsy et al., 2008).
As a general trend, distinct OTUs numerically dominated at different pH extreme sites (Figure 5) regardless of similarity in pH or geographic proximity. Metazoan OTUs were well represented among them. At Davis Mine for example, rotifer OTUs related to the bdelloid genus Adineta encompassed the largest number of reads while in both of the Nebraskan alkaline lakes ostracods were the dominant OTU. In addition to ostracods, additional metazoan OTUs in the Sandhills Lakes included annelid, gastrotrich, amphipod, rotifer, and insect larvae (Odonata). The broad range of metazoa that characterized these sites likely indicates that these particular alkaline environments are more hospitable for multicellular life than the other extreme environments examined in this study. However, some metazoa such as rotifers, are known to thrive at a range of pH extremes including high metal environments (Horvath and Hummon, 1980; Deneke, 2000; Bell, 2012) and this observation was confirmed in this study. While no rotifer OTUs were detected in the sample examined from the Rio Tinto, other sites along this river do contain rotifers (Figure 6A, personal observation) but typically at pH values closer to 3. Metazoan OTUs likewise dominated the Lost City hydrothermal vent site. At Lost City, apart from the potentially novel (metazoan) opisthokont OTU, also common was a calcareous sponge OTU that is likely well-adapted to the calcium-carbonate-rich chemistry of the vent. At other sites protistan OTUs dominated. A dominant ciliate OTU at Lost City was only found at alkaline sites, was not closely related to other sequences recovered in previous molecular surveys of Lost City (López-García et al., 2007), and had an overall low BLAST similarity assignment to sequences in GenBank (91%), indicating it may be quite novel. In contrast, other ciliate OTUs detected in the Lost City site including OTUs assigned to Lost City ciliate clone AT1-2 and the genus Cohnilembus appear to be related to ones detected by López-García and colleagues.
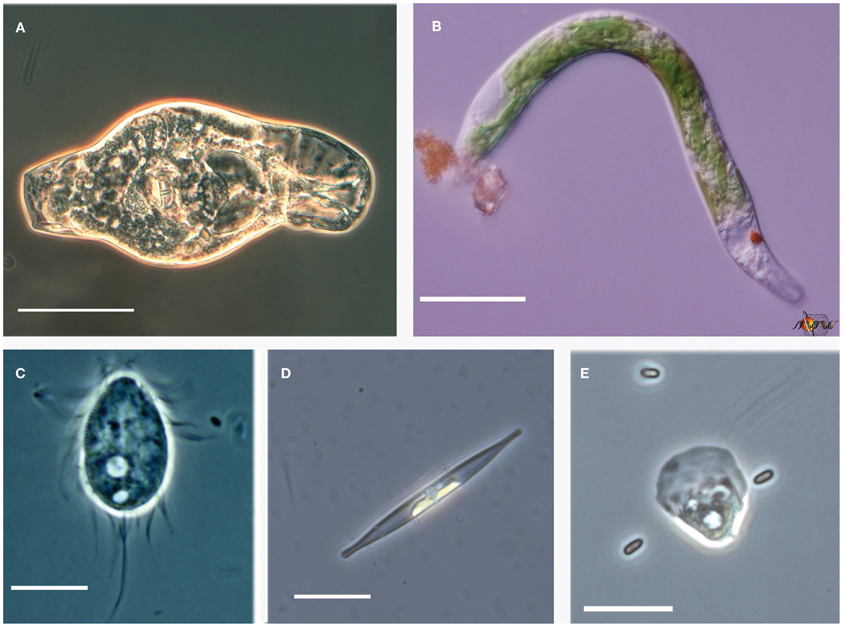
Figure 6. Examples of pH extremophiles. (A) Rotifer from the Rio Tinto (scale bar = 50 μm). (B) Euglena mutabilis from Davis Mine, MA (scale bar = 20 μm). (C) Cyclidium sp. ciliate from Mallard Lake, NE (scale bar = 10 μm). (D) Mono Lake diatom from South Tufa (scale bar = 20 μm). (E) Lobose amoeba with yeast from Rio Tinto (scale bar = 20 μm).
The most abundant OTU at Nymph Creek was 98% identical to the heterolobosean amoeba Tetramitus thermoacidophilus—an amoeboflagellate isolated from acidic hot springs in Italy and Kamchatka (Baumgartner et al., 2009). The Italian site where this amoeba was isolated was similar to Nymph Creek in being covered with Cyanidium-like algae and possessing a pH of 3, but was warmer in temperature at 72°C. The amoeba was described as a benthic thermo-acidophile with optimal growth at low pH and elevated temperatures. This same OTU was rare but also found in Mono Lake so acidity and high temperature may not constrain its distribution. The second most abundant Nymph Creek OTU was a flagellate identical to a clone from Rio Tinto [RT5in36; (Amaral Zettler et al., 2002)], but did not occur at Anabel's Garden examined in this study. Representative sequences from the third most abundant Nymph Creek OTU matched various species of the heterolobosean amoeba Naegleria with 98% similarity including Naegleria. sp. COHH, N. gruberi, N. australiensis, and others. Members of this genus include human pathogenic species reported from other YNP hot springs (Sheehan et al., 2003) and are thus of interest from the human health perspective. The potentially novel vampyrellid OTUs (U308 and U52) may be effective predators on the overlying algal biofilm biomass in Nymph Creek given that many representatives of this group are known to selectively feed on algae as prey (Hess et al., 2012).
The occurrence of similar taxa across seemingly physico-chemically distinct environments calls into the question the relative influence pH imparts on the community structure of eukaryotes living at pH extremes. While most abundant OTUs were predictable, there were exceptions. The presence of the ubiquitous E81 diatom OTU must be interpreted with caution due to the limits of taxonomic resolution afforded by the V9 region for this OTU. Another curious observation, however, was an absence of Cyanidium sequences in the Nymph Creek sample and the low frequency with which Chlorella-like OTUs were detected (<1% frequency) there. These taxa have been reported as dominant in this system, but the absence of Cyanidium in Nymph Creek may corroborate the findings of Ferris et al. (2005) who noted a shift from Cyanidium to Chlorella species in the cooler sections of the Creek. However, the lack of dominance of either Cyanidium to Chlorella is surprising. It may also be possible that extraction methods employed in this study were insufficient to recover larger proportions of Cyanidium or Chlorella-like OTUs from the samples given that these cells are sometimes difficult to lyse. An alternative explanation for the low algal recoveries may be low copy numbers of rRNA genes in these taxa relative to those of the amoebae that dominated the diversity at this site.
Diatom and euglenid OTUs dominated the Rio Tinto site. Both Pinnularia and Euglena mutabilis are “flagship” taxa known to occur in the conspicuous biofilms at this site (Amaral Zettler et al., 2002; Aguilera et al., 2007) and have such distinct morphologies that they allow unambiguous identification (Foissner, 2006) in the absence of molecular ecological approaches. The detection of both the Pinnularia and Euglena OTUs in this study is noteworthy, however, because this same sample was examined during the first descriptions of biofilm diversity in the Rio Tinto (Amaral Zettler et al., 2002) using a clone-library-based approach targeting full-length rRNA genes and did not recover the Euglena OTU. The difference between this study and the previous one was that this study targeted the much shorter and less length variable V9 hypervariable region of the 18S rRNA gene. Length variations rRNA genes are known to cause systematic amplification biases wherein shorter regions are preferentially amplified over longer ones (Huber et al., 2009). The length of the E. mutabilis 18S rRNA gene is ~2500 bp, nearly 700 bp longer than typical eukaryotic rRNA genes and was biased against in earlier work. A similar problem was encountered in a study at the same Rio Tinto Anabel's Garden site sampled during a different season using 3-domain specific primers targeting the V4–V8 region (Amaral-Zettler et al., 2011). In the latter study, only the shorter chloroplast 16S rRNA genes from E. mutabilis were recovered and not the longer nuclear versions. Therefore, for the purposes of biodiversity discovery, there are still advantages to targeting shorter regions such as the V9 that, in addition to being less prone to chimera-formation, are also less prone to amplification length biases.
One of the advantages of deep sequencing is the recovery of both abundant and rare microbial community members (Sogin et al., 2006). The detection of rare taxa also invites the discovery of novel taxa as well. Alongside the more conspicuous Rio Tinto photosynthetic biofilm community members already mentioned were other OTUs that constituted lesser readily observed heterotrophic members of the community including a ciliate OTU assigned to the genus Oxytricha, a Sawyeria-like heterolobosean amoeba OTU, and fungal OTUs related to those previously characterized in the Gadanho et al. (2006) study. The concordance of the most abundant OTUs found in the Rio Tinto with previous studies and the detection of a potentially novel fungal OTU (U126) occurring at another ARD site with greater than 1% representation lend support to the value of the comparative approach taken in this study. Figure 6 provides additional examples of some of the common acidophiles and alkaliphiles observed by microscopy during this study.
A search for taxa indicative of acidic or alkaline extremes is summarized in the IOA analysis (Table A3). These results indicated that Pinnularia (E19) and two different opisthokont OTUs of holozoan (E928) and fungal (U2611) affiliation were significant indicator OTUs of acidic environments. Species within the genus Pinnularia have been described as having the ability to disperse globally with even isolates derived from neutral pH environments being capable of acid-tolerance (Ciniglia et al., 2007). There was only one indicator taxon among the alkaline sites examined, likely because these sites were so geochemically diverse and thus hosted relatively distinct communities. In the IOA analyses, a single OTU (E112) assigned to the ciliate genus Frontonia was identified as a strong candidate for indicating extreme alkaline conditions using both abundance-based and presence/absence-based IOA methods (only Dufrêne and Legendre abundance data are shown). Published reports of Frontonia from saline alkaline environments exist from Kenya (Yasindi et al., 2002) and sequences related to Frontonia were detected in the López-García et al. Lost City study (2007). In the former study, Frontonia was determined to contribute up to 58% of the ciliate production in the lake. Due to their large size and potential to ingest larger prey, these ciliates may serve the role of top predator in extreme environments that don't support a large metazoan population. However, ciliate size does not necessarily correlate with their predatory role. While this study did not attempt to examine indicators at the “species” level, closer examination of the Pinnularia and Frontonia OTUs using an oligotyping approach still revealed one predominant oligotype shared across all the acidic or all the alkaline sites, respectively (Figure A1). For the Pinnularia OTU cluster, two additional oligotypes were detected in the Rio Tinto sample while for the Frontonia OTU cluster, two additional oligotypes were detected with one being shared by Mono Lake and Mallard Lake and the second found only in Mono Lake.
The IOA identified statistically significant pH indicators within acidic and alkaline groups of sites but insights can also be gained from examining shared OTUs across sites. For example, although not the major focus of this study, it has long been recognized that rotifer distributions are linked to pH (Edmondson, 1944) where they have been subsequently used in water quality studies (Horvath and Hummon, 1980; Siegfried et al., 1989). In this study we found that Adineta and Adineta-like OTUs (E230 and U19) occurred at both the acidic Davis Mine, as well as one or more of the alkaline Sandhills Lakes. The occurrence of rotifers over a broad range of pH values is well documented and a study in Sweden reported Adineta vaga with a range of pH from 3 to 10 (Berzins and Pejler, 1987), similar to the pH difference measured at sites in this study. The genus Ploesoma also detected in this study often occurs in humic and acidic waters that may explain its distribution at both acidic and alkaline sites where it may be associated with leaf litter. Other rotifer OTUs such as E156 assigned to the genus Sinantherina were only found in the Sandhills alkaline lakes and members of this genus are known to have species restricted to alkaline waters (Smith, 2001). There is less information on the biogeography of microbial eukaryotes than rotifers with respect to pH environments so it is difficult to say whether a given microbial eukaryotic species possesses broad pH tolerance. While the ability for microbial eukaryotes to adapt to pH extremes over a broad range of values may be rare, this study has revealed that diatoms appear to be good candidate taxa that may be able to span this gamut. Other possible candidates to examine more closely include representative ciliate (e.g., Oxytricha), flagellate (e.g., Spumella), and amoeboid (e.g., Heterolobosean) protists.
The goal of this study was to compare eukaryotic diversity across a range of pH extremes. The distinct sites described here yielded ordinations that separated pH extreme sites but this was only able to explain a modest fraction in the variation of the data (7.1%; Figure 7). Other factors in addition to pH clearly influence the underlying community structure in these environments. Even though pH appears to be a dominant factor structuring bacterial communities in soils (Fierer and Jackson, 2006), other factors such as salinity heavily influences the underlying community structure in aquatic environments (Lozupone and Knight, 2007). In the case of the sites examined in this study that include ranges of salinity and metal ion concentrations, a likely factor shaping community structure is metal ion concentrations, many of which are known to be toxic to microbes and macrobes alike.
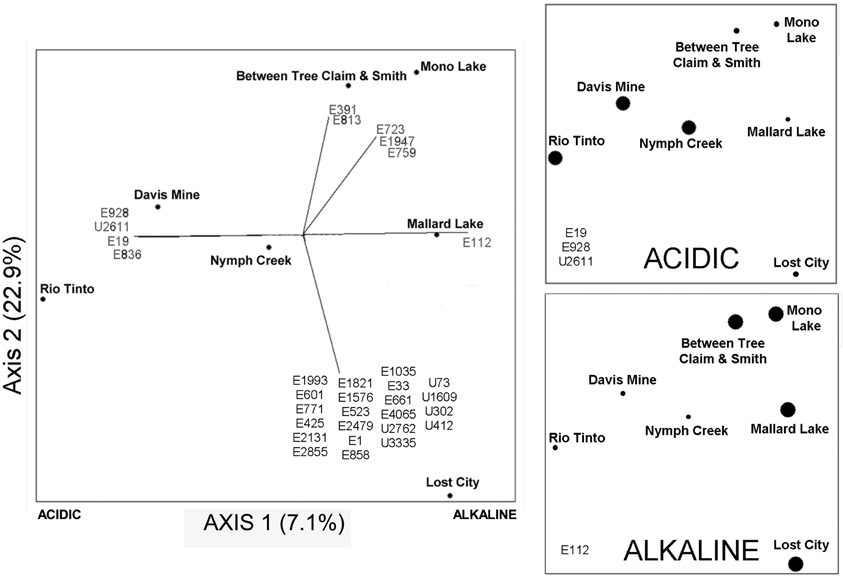
Figure 7. NMDS plot on the left with Jaccard distances showing the proportion of variance contributed by each axis (calculated from the r2 between distance in the ordination space and in the original space). Small circles on the left-hand plot show the location of all samples in the ordination. The two panels on the right show an overlay in the same ordination space of the “indicator” OTUs that are found in all acidic samples, upper panel (Rio Tinto, Davis Mine, and Nymph Creek), and in the lower panel the OTU found in all alkaline samples (Between Tree Claim and Smith, Mallard Lake, Mono Lake, and Lost City). Large circles in the two right-hand panels indicate the presence of numbered indicator OTUs in the lower left-hand corners of these plots, while small circles indicate their absence. See Table A4 for taxonomic assignments of representative OTU numbers shown on all plots.
Conclusions
Comparative molecular ecology studies at pH extremes allow us to identify taxa that may be broadly adapted to both acidic and alkaline pH and thus able to more readily attain cosmopolitan distributions. Many of the OTUs identified in this study were assigned to genera that are capable of widespread passive dispersal via resting or vegetative cysts or active dispersal via animal or human vectors. Active dispersal via ingestion exposes organisms to a broad range of pH within the gut of a given vector and thus tolerance to a wide range of pH values may impart a competitive advantage. Alkaline sites exhibited higher alpha diversity (larger number of observed OTUs) versus acidic sites overall and high metal-containing sites were the least rich within acidic and alkaline groups, respectively. Beta diversity analyses indicated low similarity between and within acidic and alkaline environments with few shared OTUs even with a conservative clustering OTU cut-off of 6%. IOA suggested that the diatom genus Pinnularia and the ciliate genus Frontonia are good indicators of acidic and alkaline conditions, respectively, but the taxonomic resolution achievable via examination of the V9 18S rRNA hypervariable region alone does not allow for discrimination at the species level. Despite this limitation, the V9 region does allow for a relatively unbiased first look comparing eukaryotic diversity in underexplored habitats such as pH extreme environments. Future studies targeting cosmopolitan pH extreme taxa using a battery of additional marker genes (Pawlowski et al., 2012) may provide important insights into the ecology, biogeography, and diversity of eukaryotes at pH extremes.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work has been supported by a US National Institutes of Health INRSA, the National Science Foundation (DEB-0085486), and the NASA Astrobiology Institute (NCC2-1054). The author acknowledges the assistance of the following individuals for sample collection, infrastructural support, or sample donation: Dr. R. Amils (Rio Tinto), Dr. D. Searcy (Davis Mine), Dr. A.-L. Reysenbach (Nymph Creek), Dr. J. Farmer (Mono Lake), Dr. E. Zettler (all terrestrial sites), Drs. J. Baross and W. Brazelton (Lost City Hydrothermal Vent). The author also acknowledges technical support from Abby Laatsch, Nicholas Scalfone and Elizabeth McCliment and SRA submission support from Philippe Rocca-Serra.
References
Aguilera, A., Souza-Egipsy, V., Gonzalez-Toril, E., Rendueles, O., and Amils, R. (2010). Eukaryotic microbial diversity of phototrophic microbial mats in two Icelandic geothermal hot springs. Int. Microbiol. 13, 21–32.
Aguilera, A., Zettler, E., Gomez, F., Amaral-Zettler, L., Rodriguez, N., and Amils, R. (2007). Distribution and seasonal variability in the benthic eukaryotic community of Rio Tinto (SW, Spain), an acidic, high metal extreme environment. Syst. Appl. Microbiol. 30, 531–546.
Amaral Zettler, L. A., Anderson, O. R., and Caron, D. A. (1998). Insights on the diversity within a “species” of Thalassicolla (Spumellarida) based on 16S-like ribosomal RNA gene sequencing. J. Eukaryot. Microbiol. 45, 488–496.
Amaral Zettler, L. A., Gomez, F., Zettler, E., Keenan, B. G., Amils, R., and Sogin, M. L. (2002). Microbiology: eukaryotic diversity in Spain's River of Fire. Nature 417, 137.
Amaral-Zettler, L. A., McCliment, E. A., Ducklow, H. W., and Huse, S. M. (2009). A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE 4:e6372. doi: 10.1371/journal.pone.0006372
Amaral-Zettler, L. A., Zettler, E. R., Theroux, S. M., Palacios, C., Aguilera, A., and Amils, R. (2011). Microbial community structure across the tree of life in the extreme Rio Tinto. ISME J. 5, 42–50.
Baker, B. J., Hugenholtz, P., Dawson, S. C., and Banfield, J. F. (2003). Extremely acidophilic protists from acid mine drainage host Rickettsiales-lineage endosymbionts that have intervening sequences in their 16S rRNA genes. Appl. Environ. Microbiol. 69, 5512–5518.
Baumgartner, M., Eberhardt, S., De Jonckheere, J. F., and Stetter, K. O. (2009). Tetramitus thermacidophilus n. sp., an amoeboflagellate from acidic hot springs. J. Eukaryot. Microbiol. 56, 201–206.
Bell, E. M. (2012). “Alkaline environments,” in Life at Extremes: Environments, Organisms and Strategies for Survival, ed E. M. Bell (Cambridge, MA: CAB International), 554.
Bernhardt, H. S., and Tate, W. P. (2012). Primordial soup or vinaigrette: did the RNA world evolve at acidic pH? Biol. Direct 7:4. doi: 10.1186/1745-6150-7-4
Berzins, B., and Pejler, B. (1987). Rotifer occurrence in relation to pH. Hydrobiologia 147, 107–116.
Brake, S. S., and Hasiotis, S. T. (2010). Eukaryote-dominated biofilms and their significance in acidic environments. Geomicrobiol. J. 27, 534–558.
Brazelton, W. J., Ludwig, K. A., Sogin, M. L., Andreishcheva, E. N., Kelley, D. S., Shen, C. C., et al. (2010). Archaea and bacteria with surprising microdiversity show shifts in dominance over 1, 000-year time scales in hydrothermal chimneys. Proc. Natl. Acad. Sci. U.S.A. 107, 1612–1617.
Brock, T. D. (1978). Thermophilic Micro-Organisms and Life at High Temperatures. New York, NY: Springer-Verlag.
Brown, P. B., and Wolfe, G. V. (2006). Protist genetic diversity in the acidic hydrothermal environments of Lassen Volcanic National Park, USA. J. Eukaryot. Microbiol. 53, 420–431.
Ciniglia, C., Cennamo, P., De Stefano, M., Pinto, G., Caputo, P., and Pollio, A. (2007). Pinnularia obscura Krasske (Bacillariophyceae, Bacillariophyta) from acidic environments: characterization and comparison with other acid-tolerant Pinnularia species. Fund. Appl. Limnol. 170, 29–47.
Clarke, K. R., and Warwich, R. M. (2001). Change in Marine Communities: An Approach to Statistical Analysis and Interpretation. Plymouth: PRIMER-E, Ltd.
Costas, E., Flores-Moya, A., Perdigones, N., Maneiro, E., Blanco, J. L., Garcia, M. E., et al. (2007). How eukaryotic algae can adapt to the Spain's Rio Tinto: a neo-Darwinian proposal for rapid adaptation to an extremely hostile ecosystem. New Phytol. 175, 334–339.
Deneke, R. (2000). Review of rotifers and crustaceans in highly acidic environments. Hydrobiologia 433, 167–172.
Denicola, D. M. (2000). A review of diatoms found in highly acidic environments. Hydrobiologia 433, 111–122.
Dufrêne, M., and Legendre, P. (1997). Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol. Monogr. 67, 345–366.
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Edmondson, W. T. (1944). Ecological studies of the sessile Rotatoria. Part, I. Factors affecting distribution. Ecol. Monogr. 14, 31–66.
Eren, A. M., Zozaya, M., Taylor, C. M., Dowd, S. E., Martin, D. H., and Ferris, M. J. (2011). Exploring the diversity of Gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation. PLoS ONE 6:e26732. doi: 10.1371/journal.pone.0026732
Ferris, M. J., Sheehan, K. B., Kuhl, M., Cooksey, K., Wigglesworth-Cooksey, B., Harvey, R., et al. (2005). Algal species and light microenvironment in a low-pH, geothermal microbial mat community. Appl. Environ. Microbiol. 71, 7164–7171.
Fierer, N., and Jackson, R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 103, 626–631.
Foissner, W. (2006). Biogeography and Dispersal of Micro-Organisms: A Review Emphasizing Protists. Warszawa: Nencki Institute of Experimental Biology.
Frontier, S. (1985). Diversity and structure in aquatic ecosystems. Oceanogr. Mar. Biol. 23, 253–312.
Gadanho, M., Libkind, D., and Sampaio, J. P. (2006). Yeast diversity in the extreme acidic environments of the Iberian Pyrite Belt. Microb. Ecol. 52, 552–563.
Gross, S., and Robbins, E. I. (2000). Acidophilic and acid-tolerant fungi and yeasts. Hydrobiologia 433, 91–109.
Hess, S., Sausen, N., and Melkonian, M. (2012). Shedding light on vampires: the phylogeny of vampyrellid amoebae revisited. PLoS ONE 7:e31165. doi: 10.1371/journal.pone.0031165
Hollibaugh, J. T., Wong, P. S., Bano, N., Pak, S. K., Prager, E. M., and Orrego, C. (2001). Stratification of microbial assemblages in Mono Lake, California, and response to a mixing event. Hydrobiologia 466, 45–60.
Horvath, F. J., and Hummon, W. D. (1980). Influence of mine acid on planktonic rotifers. Ohio J. Sci. 80, 104–107.
Huber, J. A., Morrison, H. G., Huse, S. M., Neal, P. R., Sogin, M. L., and Mark Welch, D. B. (2009). Effect of PCR amplicon size on assessments of clone library microbial diversity and community structure. Environ. Microbiol. 11, 1292–1302.
Hugo, E. R., Stewart, V. J., Gast, R. J., and Byers, T. J. (1992). “Purification of amoeba mtDNA using the UNSET procedure,” in Protocols in Protozoology, eds J. J. Lee and A. T. Soldo (Lawrence: Allen Press Inc.), D-7.1.
Humayoun, S. B., Bano, N., and Hollibaugh, J. T. (2003). Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl. Environ. Microbiol. 69, 1030–1042.
Huse, S. M., Dethlefsen, L., Huber, J. A., Mark Welch, D., Relman, D. A., and Sogin, M. L. (2008). Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4:e1000255. doi: 10.1371/journal.pgen.1000255
Huse, S. M., Huber, J. A., Morrison, H. G., Sogin, M. L., and Mark Welch, D. (2007). Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8:R143. doi: 10.1186/gb-2007-8-7-r143
Huse, S. M., Welch, D. M., Morrison, H. G., and Sogin, M. L. (2010). Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12, 1889–1898.
Lopez-Garcia, P., Philippe, H., Gail, F., and Moreira, D. (2003). Autochthonous eukaryotic diversity in hydrothermal sediment and experimental microcolonizers at the Mid-Atlantic Ridge. Proc. Natl. Acad. Sci. U.S.A. 100, 697–702.
López-García, P., Vereshchaka, A., and Moreira, D. (2007). Eukaryotic diversity associated with carbonates and fluid-seawater interface in Lost City hydrothermal field. Environ. Microbiol. 9, 1462–2920.
Lopez-Rodas, V., Marva, F., Rouco, M., Costas, E., and Flores-Moya, A. (2008). Adaptation of the chlorophycean Dictyosphaerium chlorelloides to stressful acidic, mine metal-rich waters as result of pre-selective mutations. Chemosphere 72, 703–707.
Lopez-Rodas, V., Rouco, M., Sanchez-Fortun, S., Flores-Moya, A., and Costas, E. (2011). Genetic adaptation and acclimation of phytoplankton along a stress gradient in the extreme waters of the Agrio River-Caviahue Lake (Argentina). J. Phycol. 47, 1036–1043.
Lozupone, C. A., and Knight, R. (2007). Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. U.S.A. 104, 11436–11440.
Luis, A. T., Novais, M. H., Van De Vijver, B., Almeida, S. F. P., Ferreira Da Silva, E. A., Hoffman, L., et al. (2012). Pinnularia aljustrelica sp. nov. (Bacillariophyceae), a new diatom species found in the acidic waters in the Aljustrel mining area (Portugal) and further observations on the taxonomy and ecology of P. acidophila Hoffmann et Krammer and P. acoricola Hustedt. Fottea 12, 27–40.
Mather, P. M. (1976). Computational Methods of Multivariate Analysis in Physical Geography. London: Wiley and Sons.
Nordstrom, D. K., Ball, J. W., and McCleskey, R. B. (2005). Ground Water to Surface Water: Chemistry of Thermal Outflows in Yellowstone National Park. Reston, VA: U. S. Geological Survey.
Pawlowski, J., Audic, S., Adl, S., Bass, D., Belbahri, L., Berney, C., et al. (2012). CBOL protist working group: barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLoS Biol. 10:e1001419. doi: 10.1371/journal.pbio.1001419
Pawlowski, J., Christen, R., Lecroq, B., Bachar, D., Shahbazkia, H. R., Amaral-Zettler, L., et al. (2011). Eukaryotic richness in the abyss: insights from pyrotag sequencing. PLoS ONE 6:e18169. doi: 10.1371/journal.pone.0018169
Peck, J. E. (2010). Multivariate Analysis for Community Ecologists: Step-by-Step using PC-ORD. Gleneden Beach, OR: MjM Software Design.
Proskurowski, G., Lilley, M. D., Seewald, J. S., Fruh-Green, G. L., Olson, E. J., Lupton, J. E., et al. (2008). Abiogenic hydrocarbon production at lost city hydrothermal field. Science 319, 604–607.
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196.
Quince, C., Lanzen, A., Davenport, R. J., and Turnbaugh, P. J. (2011). Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12:38. doi: 10.1186/1471-2105-12-38
Reavie, E. D., and Smol, J. P. (2001). Diatom-environmental relationships in 64 alkaline southeastern Ontario (Canada) lakes: a diatom-based model for water quality reconstructions. J. Paleolimnol. 25, 25–42.
Roesler, C. S., Culbertson, C. W., Etheridge, S. M., Goericke, R., Kiene, R. P., Miller, L. G., et al. (2002). Distribution, production, and ecophysiology of Picocystis strain ML in Mono Lake, California. Limnol. Oceanogr. 47, 440–452.
Rothschild, L. J., and Mancinelli, R. L. (2001). Life in extreme environments. Nature 409, 1092–1101.
Sansone, S.-A., Rocca-Serra, P., Field, D., Maguire, E., Taylor, C., Hofmann, O., et al. (2012). Toward interoperable bioscience data. Nat. Genet. 44, 121–126.
Sheehan, K. B., Fagg, J. A., Ferris, M. J., and Henson, J. M. (2003). PCR detection and analysis of the free-living amoeba Naegleria in hot springs in Yellowstone and Grand Teton National Parks. Appl. Environ. Microbiol. 69, 5914–5918.
Siegfried, C. A., Bloomfield, J. A., and Sutherland, J. W. (1989). Planktonic rotifer community structure in Adirondack, New York, U.S.A. lakes in relation to acidity, trophic status and related water quality characteristics. Hydrobiologia 175, 33–48.
Smith, D. G. (2001). Pennak's Freshwater Invertebrates of the United States: Porifera to Crustacea. New York, NY: J. Wiley.
Sogin, M. L., Morrison, H. G., Huber, J. A., Welch, D. M., Huse, S. M., Neal, P. R., et al. (2006). Microbial diversity in the deep sea and the underexplored “Rare Biosphere”. Proc. Natl. Acad. Sci. U.S.A. 103, 12115–12120.
Souza-Egipsy, V., Gonzalez-Toril, E., Zettler, E., Amaral-Zettler, L., Aguilera, A., and Amils, R. (2008). Prokaryotic community structure in algal photosynthetic biofilms from extreme acidic streams in Rio Tinto (Huelva, Spain). Int. Microbiol. 11, 251–260.
Stoeck, T., Bass, D., Nebel, M., Christen, R., Jones, M. D., Breiner, H. W., et al. (2010). Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 19(Suppl. 1), 21–31.
Tichý, L., and Chytrý, M. (2006). Statistical determination of diagnostic species for site groups of unequal size. J. Veg. Sci. 17, 809–818.
Vanormelingen, P., Verleyen, E., and Vyverman, W. (2008). The diversity and distribution of diatoms: from cosmopolitan to narrow endemism. Biodivers. Conserv. 17, 393–405.
Yasindi, A., Lynn, D., and Taylor, W. (2002). Ciliated protozoa in Lake Nakuru, a shallow alkaline-saline lake in Kenya: seasonal variation, potential production and role in the food web Arch. Hydrobiol. 154, 311–325.
Yilmaz, P., Kottmann, R., Field, D., Knight, R., Cole, J. A., Amaral-Zettler, L. A., et al. (2011). The “minimum information about a MARKer gene sequence” (MIMARKS) specification. Nat. Biotechnol. 29, 415–420.
Appendix
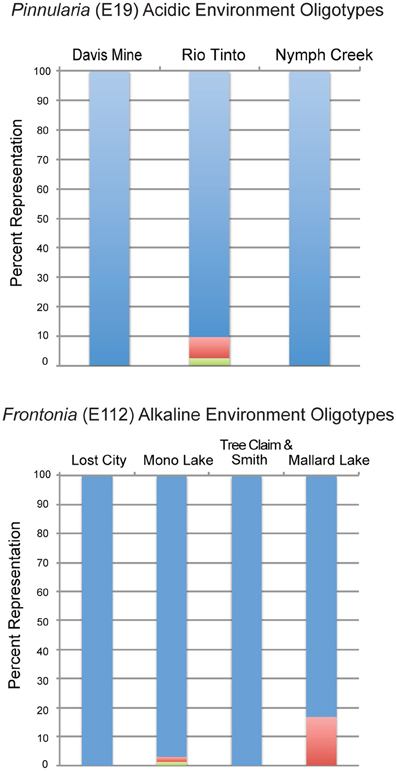
Figure A1. Oligotypes detected in indicator OTUs identified to the genus level. Different colors represent different oligotypes.
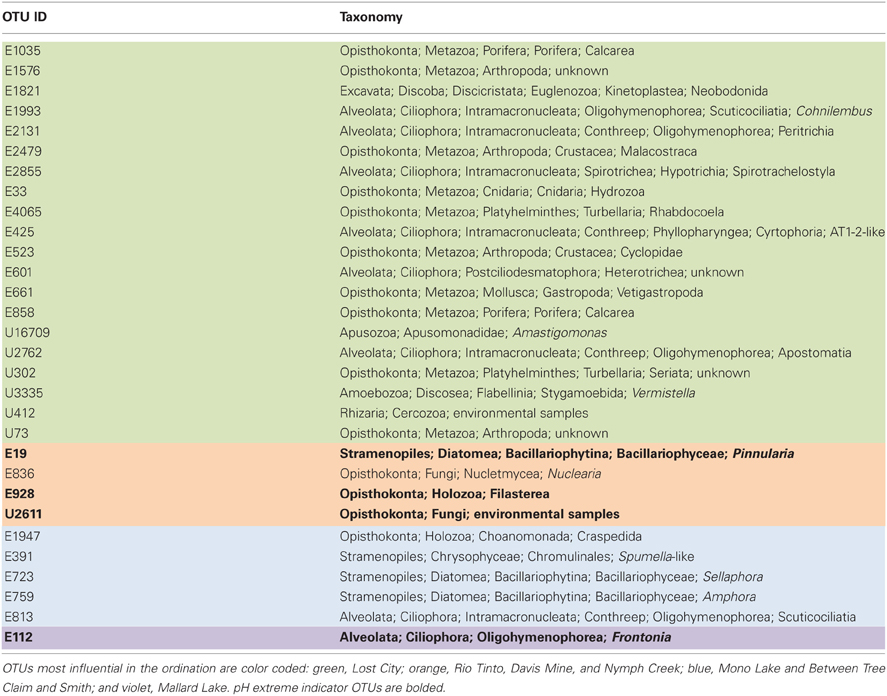
Table A4. Taxonomy for OTUs listed in Figure 7.
Keywords: acidophiles, alkaliphiles, pyrosequencing, protists, V9, indicator OTU analysis
Citation: Amaral-Zettler LA (2013) Eukaryotic diversity at pH extremes. Front. Microbio. 3:441. doi: 10.3389/fmicb.2012.00441
Received: 30 August 2012; Accepted: 21 December 2012;
Published online: 17 January 2013.
Edited by:
Angeles Aguilera, Centro de Astrobiologia (INTA-CSIC), SpainReviewed by:
William D. Orsi, Woods Hole Oceanographic Institution, USAConnie Lovejoy, Université Laval, Canada
Copyright © 2013 Amaral-Zettler. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Linda A. Amaral-Zettler, Marine Biological Laboratory, Josephine Bay Paul Center for Comparative Molecular Biology and Evolution, 7 MBL Street, Woods Hole, MA, USA. e-mail:YW1hcmFsQG1ibC5lZHU=