- Department of Molecular Virology and Microbiology, Baylor College of Medicine, Houston, TX, USA
Increasing evidence indicates that the host range of primate lentiviruses is in part determined by their ability to counteract innate restriction factors that are effectors of the type 1 interferon (IFN-1) response. For human immunodeficiency virus type 1 (HIV-1), in vitro experiments have shown that its tropism may be narrow and limited to humans and chimpanzees because its replication in other non-human primate species is hindered by factors such as TRIM5α (tripartite motif 5 alpha), APOBEC3G (apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3), and tetherin. Based on these data, it has been hypothesized that primate lentiviruses will infect and replicate in a new species if they are able to counteract and evade suppression by the IFN-1 response. Several studies have tested whether engineering HIV-1 recombinants with minimal amounts of simian immunodeficiency virus sequences would enable replication in CD4+ T cells of non-natural hosts such as Asian macaques and proposed that infection of these macaque species could be used to study transmission and pathogenesis. Indeed, infection of macaques with these viruses revealed that Vif-mediated counteraction of APOBEC3G function is central to cross-species tropism but that other IFN-induced factors may also play important roles in controlling replication. Further studies of these macaque models of infection with HIV-1 derivatives could provide valuable insights into the interaction of lentiviruses and the innate immune response and how lentiviruses adapt and cause disease.
Introduction
Early studies on primate lentiviruses identified key host cell factors required for replication (Hatziioannou and Evans, 2012). More recent investigations have shown that overcoming the suppressive effects of innate restriction factors is also necessary for human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency viruses (SIVs) to replicate in human and non-human primate hosts, respectively. Viral accessory proteins play key roles in antagonizing these inhibitory factors, which are effectors of the type 1 interferon (IFN-1) response (Harris et al., 2012). However, their functional activities are commonly limited to susceptible host species, suggesting that innate immunity may be a significant barrier to transmission of lentiviruses. We, and others, have engineered HIV-1 recombinants with minimal SIV sequences conferring resistance to specific restriction factors and infected macaques to experimentally test this hypothesis. Investigations utilizing these macaque-tropic HIV-1 derivatives may lead to a greater understanding of inter-species transmission of primate lentiviruses as well as the development of a macaque model of HIV-1 infection and disease.
Macaque AIDS Model Development and Species Tropism of Primate Lentiviruses
The development of non-human primate acquired immunodeficiency syndrome (AIDS) models provided initial insights into the species tropism of lentiviruses. In particular, these experiments demonstrated a narrow species tropism for HIV-1. Gibbons and chimpanzees are susceptible to HIV-1 (Gardner and Luciw, 1989; Fultz, 1993). However, due to their endangered status and maintenance cost, they are not reasonable model hosts. On the other hand, Asian macaques, including Macaca mulatta (rhesus macaques, RM) and M. fascicularis (cynomolgus monkeys, CM) and cells from these species appear to be resistant to HIV-1 (Agy et al., 1992; Cowan et al., 2002; Munk et al., 2002), suggesting genetic barriers to infection. In retrospect, these findings are not surprising given that HIV-1 evolved from a novel recombinant SIV infecting chimpanzees (SIVcpz; Gao et al., 1999; Bailes et al., 2003). Uniquely, one species, M. nemestrina (pigtailed macaques, PTM), has been found to be susceptible to transient infection but not disease (Agy et al., 1992, 1997; Gartner et al., 1994), demonstrating that a potent resistance mechanism(s) may indeed control viral replication.
With the absence of a susceptible non-human primate host for HIV-1, a SIV-AIDS macaque model was developed accidently following the discovery that Asian macaques housed with sooty mangabeys at a US primate center had developed AIDS like disease (Gardner, 1996; Apetrei et al., 2005). Although African monkey species harbor SIVs and live with high virus loads without developing disease (Klatt et al., 2012b), SIVs isolated from sooty mangabeys (SM, Cercocebus atys) cause AIDS at varying rates when inoculated into Asian macaques. As a result, SIV infection of macaques has become the most widely used model for studies of AIDS immunopathogenesis and viral fitness (Kimata, 2006; Hatziioannou and Evans, 2012). Quite interestingly, PTMs appear to be more susceptible to infection and disease induced by SIV than RMs, which may be due to a higher level of immune activation and gastrointestinal immune dysfunction (Klatt et al., 2012a; Canary et al., 2013).
Genetic differences in reverse transcriptase and protease of HIV-1 and SIVmac make it difficult to evaluate the efficacy of antiretroviral drugs that target these proteins using the SIVmac-RM model. Evaluating vaccines against HIV-1 is also impossible since cytotoxic T cell epitopes may differ and neutralizing antibodies are not cross-reactive. These shortcomings have been partially addressed by constructing chimeric SIV/HIV-1 viruses (SHIVs) that include certain HIV genes in the SIVmac239 backbone (Shibata et al., 1991; Figure 1).
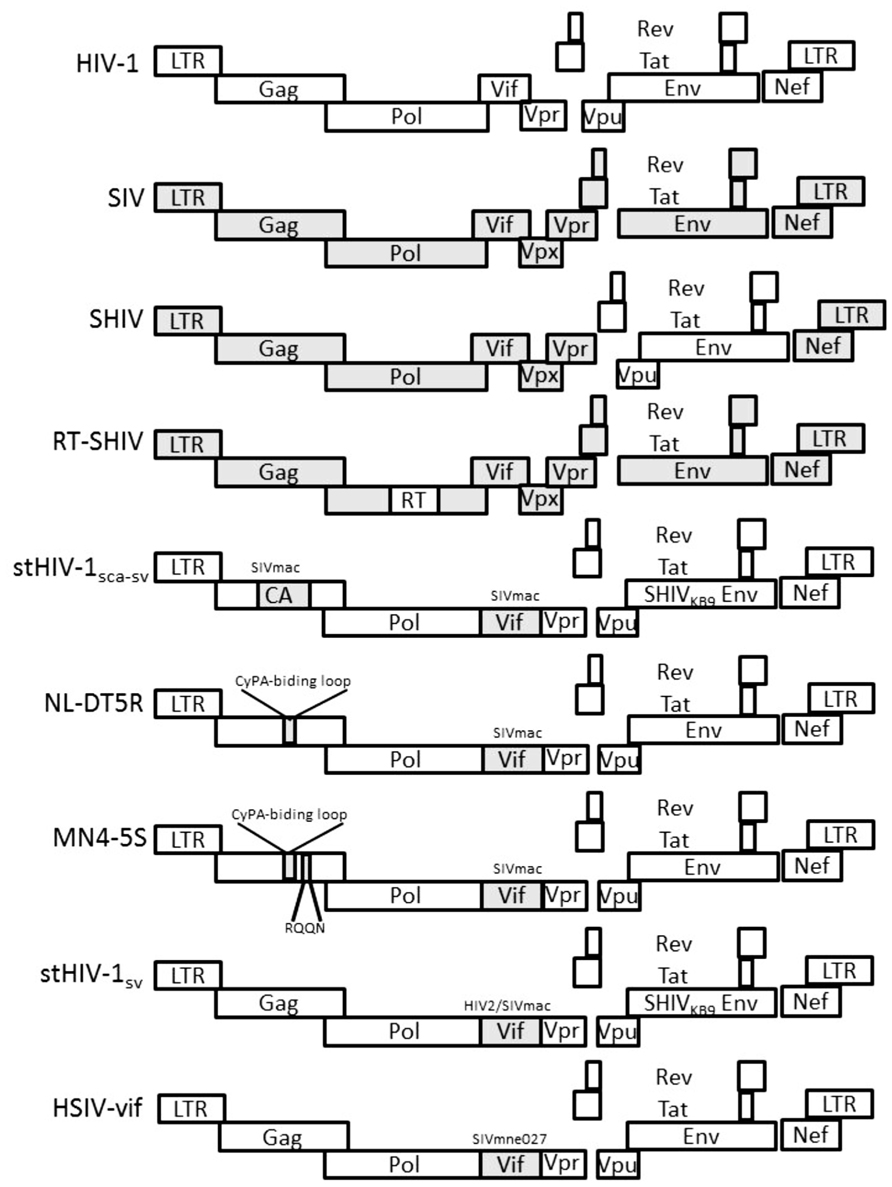
FIGURE 1. Genetic organization of HIV-1, SIV, and HIV-1/SIV chimeric proviruses. HIV-1 sequences are in white. SIV sequences are shaded gray.
Aside from the obvious utility for translational studies, the development of SHIVs revealed important clues about the functional activity of HIV-1 proteins in macaques. SIVmac based chimeras that include HIV-1 gene substitutions in env, tat, and rev (Env-SHIV) or nef (Nef-SHIV) are pathogenic in macaques (Li et al., 1995; Luciw et al., 1995; Reimann et al., 1996; Sinclair et al., 1997; Alexander et al., 1999). Chimeras with HIV-1 rt substitutions (RT-SHIVs) also persistently replicate in macaque hosts (Uberla et al., 1995; Ambrose et al., 2007). While not required, vpu of HIV-1 enhances the pathogenicity of Env-SHIV (Stephens et al., 2002). Thus, a significant amount of HIV-1 sequences can functionally replace SIV sequences, but determinants within gag–pol and vif of SIV appear necessary for infection of Asian macaques.
Innate Restriction Factors of Primate Lentiviruses
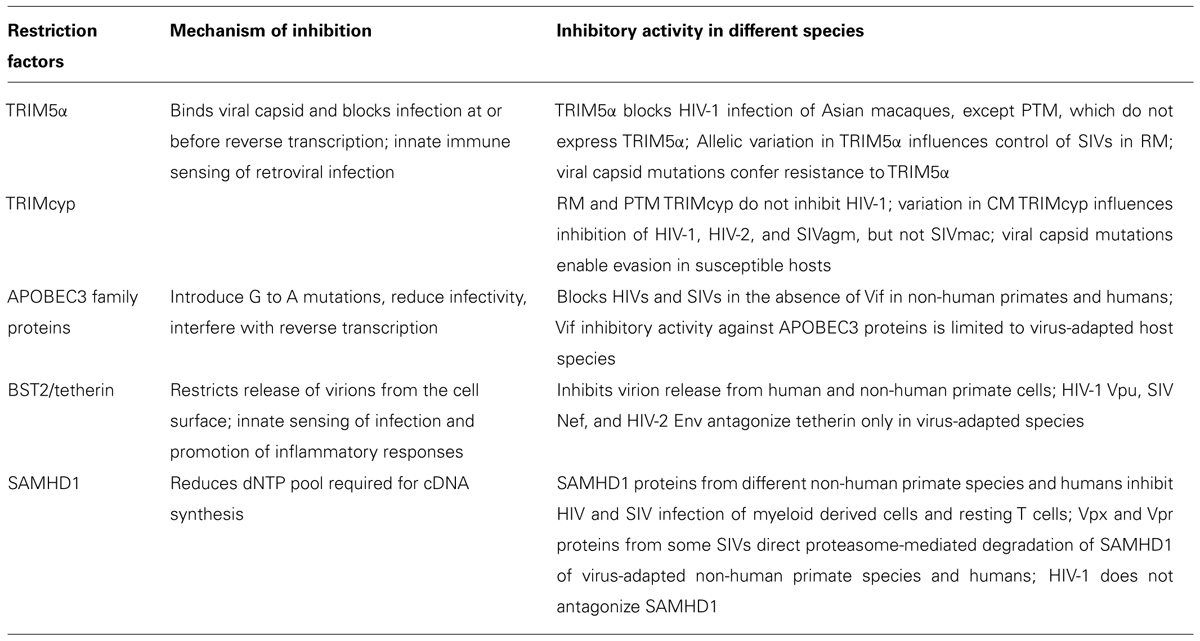
Several cellular restriction factors have been identified that can limit replication of primate lentiviruses in different species, but whose activities are specifically inhibited or evaded (Table 1). These include apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3 (APOBEC3) proteins, tripartite motif 5 alpha (TRIM5α) and related TRIM5–cyclophilin A fusion proteins (TRIMcyp), tetherin/BST2/CD317, and sterile alpha motif (SAM) domain and HD domain-containing protein 1 (SAMHD1; Thippeshappa et al., 2012). All are regulated by IFN-1, suggesting that innate immunity plays a critical role in preventing infection and that viral adaptations that antagonize or escape the effects of the factors may be required for successful transmission of lentiviruses.
The APOBEC3 (A3) proteins belong to a seven-member family of cytidine deaminases (Jarmuz et al., 2002). A3G was identified as a Vif-targeted inhibitory factor of HIV-1 during a screen for cellular factors that blocked post-entry steps of infection prior to integration (Sheehy et al., 2002). In the absence of Vif, it interferes with viral replication by incorporating into the virion and disrupting reverse transcription or causing accumulation of deleterious G to A mutations (Mangeat et al., 2003; Zhang et al., 2003; Bishop et al., 2008). Hypermutated viral genomes may be degraded or produce non-functional truncated or misfolded viral proteins that are processed and serve as antigens for cellular immune responses (Casartelli et al., 2010).
In virus producing cells, Vif binds A3G and links it to an E3 ubiquitin ligase complex, thereby redirecting it for degradation by the proteasome (Conticello et al., 2003) and preventing its incorporation into assembling virions. Interestingly, Vif function appears to be species-specific. For example, the HIV-1 Vif antagonizes the human A3G protein but not A3G of other non-human primate species. By contrast, the Vif protein of SIVagm antagonizes African green monkey (AGM) A3G but not human A3G (Mariani et al., 2003). These findings suggest that Vif-mediated inhibition of the A3G proteins is likely essential for transmission of a virus to a new host species.
Of the innate restriction factors, only TRIM5α was initially discovered as an inhibitory factor of HIV-1 in Old World Monkeys (OWMs; Stremlau et al., 2006; Grutter and Luban, 2012). TRIM5α blocks a post-entry stage of HIV-1 replication through an interaction with the capsid protein. It belongs to the tripartite family of proteins, and contains a RING finger, B-box2, and coiled coil domain, which are responsible for E3 ubiquitin ligase activity and higher order self-association. It also has a B30.2/SPRY domain that detects the incoming viral capsid proteins, linking the viral core to an ubiquitin-proteasome-dependent pathway. This disrupts the preintegration complex, thereby blocking reverse transcription. However, in cases where the proteasome pathway is inhibited, nuclear entry of viral DNA is impaired. Recent studies also establish TRIM5α as an innate immune sensor of the retroviral capsid (Pertel et al., 2011). Sequence variation in B30.2/SPRY of TRIM5α and amino acid variations in the viral capsid are responsible for species-specific restriction and evasion, respectively (Nakayama et al., 2005; Sawyer et al., 2005). Additionally, allelic variation in TRIM5 influences transmission and modulates disease progression in SIV-infected RM (Kirmaier et al., 2010; Lim et al., 2010b; Reynolds et al., 2011). Interestingly, PTMs do not express a TRIM5α isoform, partially explaining their unique susceptibility to HIV-1 (Brennan et al., 2007).
Novel TRIMcyp also interfere with post-entry steps in HIV/SIV infection. First identified in New World Owl Monkeys (Sayah et al., 2004), the fusion protein appears to have arisen via line-mediated retrotransposition of the cyclophilin A gene into the TRIM5 locus. Subsequent studies have also identified TRIMcyp fusion proteins in RM, CM, and PTM that evolved independently (Brennan et al., 2008; Newman et al., 2008; Virgen et al., 2008; Wilson et al., 2008; Dietrich et al., 2011). Allelic variation in the cyclophilin A domain of the macaque TRIMcyp proteins affects recognition and inhibition of HIV-1 and 2 and SIVagm but not SIVmac. Interestingly, RMs and CMs are polymorphic for TRIM5α alleles and TRIMcyp, although geographically distinct CM populations show different frequencies of TRIMcyp. PTMs, on the other hand, are homozygous for TRIMcyp, again demonstrating a unique genotype for PTMs in comparison to other Asian macaques (Brennan et al., 2008; Newman et al., 2008; Kuang et al., 2009; Dietrich et al., 2011; Saito et al., 2012).
Tetherin or BST2 is interferon inducible type II membrane protein that interferes with the release of HIV-1 progeny virions from the surface of infected human T cells and also functions as an innate immune sensor of viral infection to promote inflammatory responses (Neil et al., 2008; Van Damme et al., 2008; Galao et al., 2012). Initially, it was discovered that the HIV-1 protein Vpu inhibits tetherin and is required for the efficient release of progeny virions (Neil et al., 2008; Van Damme et al., 2008). Subsequent studies have shown that primate lentiviruses that do not encode Vpu evolved other strategies to antagonize tetherin. For example, HIV-2 and SIV use Env- and Nef-dependent mechanisms to counteract the restrictive effect of tetherin (Jia et al., 2009; Le Tortorec and Neil, 2009), respectively. Additionally, the effects of the viral antagonists are specific for the host species in which they evolved. The HIV-1 Vpu evolved to overcome the activity of human tetherin, but it is ineffective against tetherin from chimpanzees, RM, AGM, and mustached monkeys (Jia et al., 2009; Sauter et al., 2009; Lim et al., 2010a; Yang et al., 2010). Despite the close relatedness of HIV-1 and SIVcpz, Vpu of SIVcpz does not antagonize chimpanzee tetherin. Instead it uses Nef to downregulate chimpanzee tetherin expression like other SIVs, which also exhibits species-specific activity (Jia et al., 2009; Zhang et al., 2009).
SAMHD1 is a restriction factor that inhibits HIV-1 infection of myeloid cells (Hrecka et al., 2011; Laguette et al., 2011). Although its exact biological function is unclear, mutations in SAMHD1 can result in Aicardi Goutieres syndrome whose symptoms mimic that of a viral infection (Rice et al., 2009). Vpx protein from either HIV-2 or the SIVsm lineage inhibit human SAMHD1, resulting in its degradation through the proteasome. It has been noted that Vpx expression or SAMHD1 depletion increases the amount of dNTP’s in macrophages, which suggests that SAMHD1 decreases the dNTP pool required for viral cDNA synthesis (Lahouassa et al., 2012). Structural studies also indicate that SAMHD1 is a dNTP triphosphate triphosphohydrolase (Goldstone et al., 2011). Interestingly, SAMHD1 only restricts infection of HIV-1 in non-dividing cells such as macrophages and resting T cells but not activated proliferating T cells (Baldauf et al., 2012; Descours et al., 2012). New data also indicate that phosphorylation may regulate SAMHD1’s restriction activity (Cribier et al., 2013; White et al., 2013).
Like the other restriction factors, Vpx appears to antagonize SAMHD1 in a species-specific manner since human and gibbon SAMHD1 can be degraded by Vpx proteins from HIV-2rod, SIVmac, and SIVsm but not by Vpx from SIVrcm and SIVmnd2. However, Vpx proteins from different SIV and HIV-2 strains can induce degradation of RM and SMSAMHD1 (Laguette et al., 2012; Lim et al., 2012). Interestingly, some SIVs inhibit SAMHD1 of their natural hosts via Vpr. Thus, targeting SAMHD1 appears critical for replication and persistence of SIVs in OWMs. It is therefore interesting that HIV-1 does not have a mechanism to antagonize SAMHD1 in human cells. One hypothesis is that this may help the virus avoid immune sensing.
Other innate restriction factors such as interferon inducible transmembrane proteins (IFITM), and 2′,3′-cyclic-nucleotide 3′-phosphodiesterase (Lu et al., 2011; Wilson et al., 2012) have been shown to interfere with early and late stages of the viral life cycle, respectively. However, whether these factors have species-specific activity against primate lentiviruses is unknown.
Engineering Macaque-Tropic HIV-1 Derivatives
The species-specific effects of innate restriction factors and requirement for particular SIV sequences for replication competent SHIV chimeras suggested that engineering macaque-tropic recombinant viruses consisting of mainly HIV-1 sequences may be possible as long as the virus can evade or antagonize key host restriction factors (Figure 1). Hatziioannou et al. (2006) generated the initial HIV-1 chimera with minimal SIV sequences that could replicate in RM peripheral blood mononuclear cells (PBMCs; stHIV-1sca-sv). The virus included ca and vif substitutions from SIVmac in order to escape restriction by RM TRIM5α and A3G, respectively. In other studies, a macaque-tropic HIV-1 derivative with the SIV vif gene and a short 21 base pair segment corresponding to the HIV-1 cyclophilin A binding loop from SIV was constructed (NL-DT5R; Kamada et al., 2006; Igarashi et al., 2007). The virus showed increased infectivity in both CM and PTM T cells. However, only after passaging in a CM T cell line was the virus able to replicate efficiently in CD8+ cell-depleted PBMCs from either PTM or RM. While these HIV-1 derivatives infected PTM, they were rapidly controlled and did not cause disease. Additional studies selected gag variants better able to escape restriction by CM TRIMcyp (e.g., MN4-5S), but replication only modestly improved in CMs (Kuroishi et al., 2009; Saito et al., 2011).
Because of the absence of a post-entry block to HIV-1 infection and potential for more rapid AIDS progression, PTMs were hypothesized to be the most susceptible to macaque-tropic HIV-1 derivatives. Indeed, substituting vif in HIV-1 with alleles from SIVmne (HSIV-vif) or SIVmac or HIV-2 (stHIV-1) is sufficient for HIV-1 to replicate in PTM CD4+ T cells (Hatziioannou et al., 2009; Thippeshappa et al., 2011). Infection of PTMs with mtHIV-1 resulted in acute infection and viremia that was controlled within 25 weeks post-infection. Interestingly, replication of HSIV-vif in PTMs extended for over 90 weeks post-infection, although plasma viral loads were low. Moreover, one animal demonstrated a steep drop in CD4+ T cell counts, persistent but low viremia, and opportunistic infections after three years of infection (unpublished observations). It will be important to reisolate variants from this animal and examine the genetic and phenotypic changes that have occurred during infection. Since the different variants of HIV-1 and SIV used in these studies seem to make a difference in persistence and disease, other variants should be considered for future in vivo infection experiments.
What accounts for virological control in the PTMs remains unclear. There is suggestion from CD8+ cell-depletion studies that cellular immune responses may be limiting replication of the macaque-tropic HIV-1 clones (Hatziioannou et al., 2009). Additionally, the IFN-1 response might restrict viral replication. IFNs are upregulated during HIV-1 and SIV infections (Neil and Bieniasz, 2009; Thippeshappa et al., 2012). Thus, these viruses must be able to overcome the induction of restrictive interferon-stimulated genes (ISGs) in order to replicate to high levels and cause disease. Indeed, new studies demonstrate that the prototype macaque-tropic HIV-1 derivatives are inhibited by IFNα in PTM cells. By contrast, pathogenic SIVmne and SIVmac clones are highly resistant to IFNα-induced inhibition (Bitzegeio et al., 2013; Thippeshappa et al., 2013). Interestingly, suppression of replication of the HIV-1 derivatives by IFNα may not be due to the induction of known restriction factors such as tetherin, TRIM5α, TRIMcyp, A3G, or SAMHD1, indicating that other ISGs may be responsible for potently blocking replication of macaque-tropic HIV-1 in PTMs. Furthermore, IFNα resistance may be acquired by mutations in env, enabling escape from an early block in replication (Thippeshappa et al., 2013). Infection of PTMs with this variant could provide insight into whether evasion of IFNα is critical for viral replication in the host.
Summary and Conclusions
The engineering of macaque-tropic HIV-1 derivatives has shed light on the significance of counteracting or escaping restriction factors of the innate immune response for cross-species transmission. Macaque models have provided experimental in vivo systems to demonstrate the importance of Vif-mediated antagonism of A3 proteins and evasion of TRIM5 isoforms. Indeed, in the absence of inhibitory TRIM5α or TRIMcyp alleles in the PTM, Vif-mediated inhibition of A3G is necessary and sufficient for transmission and persistence of HIV-1 in PTMs. However, the SIV Vif is not sufficient for robust replication of macaque-tropic HIV-1 chimeras in PTMs because these viruses fail to adequately overcome the IFNα-induced antiviral state. Additional adaptations like those we have identified in an env sequence may be necessary for HIV-1 to replicate to high levels in the PTM or other macaque hosts. What other restriction factors might play a role in controlling HIV-1 replication in OWMs like Asian Macaques is unclear, but the IFNα resistance mutations may help identify new mechanisms of escape. Finally, it is curious that lentiviruses of OWMs target SAMHD1 for degradation via Vpx or Vpr, and that Vpx enhances transmission and pathogenesis of SIV in PTMs (Hirsch et al., 1998; Belshan et al., 2012), but HIV-1 did not evolve a mechanism to inhibit this protein in humans. Macaque-tropic HIV-1 derivatives provide a way to test whether antagonizing the activity of SAMHD1 is necessary for replication in OWM species.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Support provided by NIH grant AI099007 to Jason T. Kimata.
References
Agy, M. B., Frumkin, L. R., Corey, L., Coombs, R. W., Wolinsky, S. M., Koehler, J., et al. (1992). Infection of Macaca nemestrina by human immunodeficiency virus type-1. Science 257, 103–106.doi: 10.1126/science.1621083
Agy, M. B., Schmidt, A., Florey, M. J., Kennedy, B. J., Schaefer, G., Katze, M. G., et al. (1997). Serial in vivo passage of HIV-1 infection in Macaca nemestrina. Virology 238, 336–343.doi: 10.1006/viro.1997.8832
Alexander, L., Du, Z., Howe, A. Y., Czajak, S., and Desrosiers, R. C. (1999). Induction of AIDS in rhesus monkeys by a recombinant simian immunodeficiency virus expressing nef of human immunodeficiency virus type 1. J. Virol. 73, 5814–5825.
Ambrose, Z., Palmer, S., Boltz, V. F., Kearney, M., Larsen, K., Polacino, P., et al. (2007). Suppression of viremia and evolution of human immunodeficiency virus type 1 drug resistance in a macaque model for antiretroviral therapy. J. Virol. 81, 12145–12155.doi: 10.1128/JVI.01301-07
Apetrei, C., Kaur, A., Lerche, N. W., Metzger, M., Pandrea, I., Hardcastle, J., et al. (2005). Molecular epidemiology of simian immunodeficiency virus SIVsm in U.S. primate centers unravels the origin of SIVmac and SIVstm. J. Virol. 79, 8991–9005.doi: 10.1128/JVI.79.14.8991-9005.2005
Bailes, E., Gao, F., Bibollet-Ruche, F., Courgnaud, V., Peeters, M., Marx, P. A., et al. (2003). Hybrid origin of SIV in chimpanzees. Science 300, 1713.doi: 10.1126/science.1080657
Baldauf, H. M., Pan, X., Erikson, E., Schmidt, S., Daddacha, W., Burggraf, M., et al. (2012). SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 18, 1682–1687.doi: 10.1038/nm.2964
Belshan, M., Kimata, J. T., Brown, C., Cheng, X., McCulley, A., Larsen, A., et al. (2012). Vpx is critical for SIVmne infection of pigtail macaques. Retrovirology 9, 32.doi: 10.1186/1742-4690-9-32
Bishop, K. N., Verma, M., Kim, E. Y., Wolinsky, S. M., and Malim, M. H. (2008). APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 4:e1000231. doi: 10.1371/journal.ppat.1000231
Bitzegeio, J., Sampias, M., Bieniasz, P. D., Hatziioannou, T., and Sampias, M. (2013). Adaptation to the interferon-induced antiviral state by human and simian immunodeficiency viruses. J. Virol. 87, 3549–3560.doi: 10.1128/JVI.03219-12
Brennan, G., Kozyrev, Y., and Hu, S. L. (2008). TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc. Natl. Acad. Sci. U.S.A. 105, 3569–3574.doi: 10.1073/pnas.0709511105
Brennan, G., Kozyrev, Y., and Kodama, T. (2007). Novel TRIM5 isoforms expressed by Macaca nemestrina. J. Virol. 81, 12210–12217.doi: 10.1128/JVI.02499-06
Canary, L. A., Vinton, C. L., Morcock, D. R., Pierce, J. B., Estes, J. D., Brenchley, J., et al. (2013). Rate of AIDS progression is associated with gastrointestinal dysfunction in simian immunodeficiency virus-infected pigtail macaques. J. Immunol. 190, 2959–2965.doi: 10.4049/jimmunol.1202319
Casartelli, N., Guivel-Benhassine, F., Bouziat, R., Brandler, S., Schwartz, O., and Moris, A. (2010). The antiviral factor APOBEC3G improves CTL recognition of cultured HIV-infected T cells. J. Exp. Med. 207, 39–49.doi: 10.1084/jem.20091933
Conticello, S. G., Harris, R. S., and Neuberger, M. S. (2003). The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13, 2009–2013.doi: 10.1016/j.cub.2003.10.034
Cowan, S., Hatziioannou, T., Cunningham, T., Muesing, M. A., Gottlinger, H. G., and Bieniasz, P. D. (2002). Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. U.S.A. 99, 11914–11919.doi: 10.1073/pnas.162299499
Cribier, A., Descours, B., Valadão, A. L., Laguette, N., and Benkirane, M. (2013). Phosphorylation of SAMHD1 by Cyclin A2/CDK1 Regulates Its Restriction Activity toward HIV-1. Cell Rep. 3, 1036–1043.doi: 10.1016/j.celrep.2013.03.017
Descours, B., Cribier, A., Chable-Bessia, C., Ayinde, D., Rice, G., Crow, Y., et al. (2012). SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology 9, 87.doi: 10.1186/1742-4690-9-87
Dietrich, E. A., Brennan, G., Ferguson, B., Wiseman, R. W., O’Connor, D., and Hu, S. L. (2011). Variable prevalence and functional diversity of the antiretroviral restriction factor TRIMCyp in Macaca fascicularis. J. Virol. 85, 9956–9963.doi: 10.1128/JVI.00097-11
Fultz, P. N. (1993). Nonhuman primate models for AIDS. Clin. Infect. Dis. 17(Suppl. 1), S230–S235.doi: 10.1093/clinids/17.Supplement_1.S230
Galao, R. P., Le Tortorec, A., Pickering, S., Kueck, T., and Neil, S. J. (2012). Innate sensing of HIV-1 assembly by Tetherin induces NFkappaB-dependent proinflammatory responses. Cell Host Microbe 12, 633–644.doi: 10.1016/j.chom.2012.10.007
Gao, F., Bailes, E., Robertson, D. L., Chen, Y., Rodenburg, C. M., Michael, S. F., et al. (1999). Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397, 436–441.doi: 10.1038/17130
Gardner, M. B. (1996). The history of simian AIDS. J. Med. Primatol. 25, 148–157.doi: 10.1111/j.1600-0684.1996.tb00011.x
Gartner, S., Liu, Y., Polonis, V., Lewis, M. G., Elkins, W. R., et al. (1994). Adaptation of HIV-1 to pigtailed macaques. J. Med. Primatol. 23, 155–163.doi: 10.1111/j.1600-0684.1994.tb00117.x
Goldstone, D. C., Ennis-Adeniran, V., Hedden, J. J., Groom, H. C., Rice, G. I., Christodoulou, E., et al. (2011). HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382.doi: 10.1038/nature10623
Grutter, M. G., and Luban, J. (2012). TRIM5 structure, HIV-1 capsid recognition, and innate immune signaling. Curr. Opin. Virol. 2, 142–150.doi: 10.1016/j.coviro.2012.02.003
Harris, R. S., Hultquist, J. F., and Evans, D. T. (2012). The restriction factors of human immunodeficiency virus. J. Biol. Chem. 287, 40875–40883.doi: 10.1074/jbc.R112.416925
Hatziioannou, T., Ambrose, Z., Chung, N. P. Y., Piatak, M., Yuan, F., Trubey, C. M., et al. (2009). A macaque model of HIV-1 infection. Proc. Natl. Acad. Sci. U.S.A. 106, 4425–4429.doi: 10.1073/pnas.0812587106
Hatziioannou, T., and Evans, D. T. (2012). Animal models for HIV/AIDS research. Nat. Rev. Microbiol. 10, 852–867.doi: 10.1038/nrmicro2911
Hatziioannou, T., Princiotta, M., Piatak, M. Jr., Yuan, F., Zhang, F., Lifson, J. D., et al. (2006). Generation of simian-tropic HIV-1 by restriction factor evasion. Science 314, 95.doi: 10.1126/science.1130994
Hirsch, V. M., Sharkey, M. E., Brown, C. R., Brichacek, B., Goldstein, S., Wakefield, J., et al. (1998). Vpx is required for dissemination and pathogenesis of SIV(SM) PBj: evidence of macrophage-dependent viral amplification. Nat. Med. 4, 1401–1408.doi: 10.1038/3992
Hrecka, K., Hao, C., Gierszewska, M., Swanson, S. K., Kesik-Brodacka, M., Srivastava, S., et al. (2011). Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661.doi: 10.1038/nature10195
Igarashi, T., Iyengar, R., Byrum, R. A., Buckler-White, A., Dewar, R. L., Buckler, C. E., et al. (2007). Human immunodeficiency virus type 1 derivative with 7% simian immunodeficiency virus genetic content is able to establish infections in pig-tailed macaques. J. Virol. 81, 11549–11552.doi: 10.1128/JVI.00960-07
Jarmuz, A., Chester, A., Bayliss, J., Gisbourne, J., Dunham, I., Scott, J., et al. (2002). An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79, 285–296.doi: 10.1006/geno.2002.6718
Jia, B., Serra-Moreno, R., Neidermyer, W., Rahmberg, A., Mackey, J., Fofana, I. B., et al. (2009). Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 5:e1000429. doi: 10.1371/journal.ppat.1000429
Kamada, K., Igarashi, T., Martin, M. A., Khamsri, B., Hatcho, K., Yamashita, T., et al. (2006). Generation of HIV-1 derivatives that productively infect macaque monkey lymphoid cells. Proc. Natl. Acad. Sci. U.S.A. 103, 16959–16964.doi: 10.1073/pnas.0608289103
Kimata, J. T. (2006). HIV-1 fitness and disease progression: insights from the SIV-macaque model. Curr. HIV Res. 4, 65–77.doi: 10.2174/157016206775197628
Kirmaier, A., Wu, F., Newman, R. M., Hall, L. R., Morgan, J. S., O’Connor, S., et al. (2010). TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 8:e1000462. doi: 10.1371/journal.pbio.1000462
Klatt, N. R., Canary, L. A., Vanderford, T. H., Vinton, C. L., Engram, J. C., Dunham, R. M., et al. (2012a). Dynamics of simian immunodeficiency virus SIVmac239 infection in pigtail macaques. J. Virol. 86, 1203–1213.doi: 10.1128/JVI.06033-11
Klatt, N. R., Silvestri, G., and Hirsch, V. (2012b). Nonpathogenic simian immunodeficiency virus infections. Cold Spring Harb. Perspect. Med. 2, a007153.doi: 10.1101/cshperspect.a007153
Kuang, Y. Q., Tang, X., Liu, F. L., Jiang, X. L., Zhang, Y. P., Gao, G., et al. (2009). Genotyping of TRIM5 locus in northern pig-tailed macaques (Macaca leonina), a primate species susceptible to Human Immunodeficiency Virus type 1 infection. Retrovirology 6, 58.doi: 10.1186/1742-4690-6-58
Kuroishi, A., Saito, A., Shingai, Y., Shioda, T., Nomaguchi, M., Adachi, A., et al. (2009). Modification of a loop sequence between alpha-helices 6 and 7 of virus capsid (CA) protein in a human immunodeficiency virus type 1 (HIV-1) derivative that has simian immunodeficiency virus (SIVmac239) vif and CA alpha-helices 4 and 5 loop improves replication in cynomolgus monkey cells. Retrovirology 6, 70.doi: 10.1186/1742-4690-6-70
Laguette, N., Rahm, N., Sobhian, B., Chable-Bessia, C., Münch, J., Snoeck, J., et al. (2012). Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe 11, 205–217.doi: 10.1016/j.chom.2012.01.007
Laguette, N., Sobhian, B., Casartelli, N., Ringeard, M., Chable-Bessia, C., Ségéral, E., et al. (2011). SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657.doi: 10.1038/nature10117
Lahouassa, H., Daddacha, W., Hofmann, H., Ayinde, D., Logue, E. C., Dragin, L., et al. (2012). SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228.doi: 10.1038/ni.2236
Le Tortorec, A., and Neil, S. J. (2009). Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J. Virol. 83, 11966–11978.doi: 10.1128/JVI.01515-09
Li, J. T., Halloran, M., Lord, C. I., Watson, A., Ranchalis, J., Fung, M., et al. (1995). Persistent infection of macaques with simian-human immunodeficiency viruses. J. Virol. 69, 7061–7067.
Lim, E. S., Fregoso, O. I., McCoy, C. O., Matsen, F. A., Malik, H. S., Emerman, M., et al. (2012). The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11, 194–204.doi: 10.1016/j.chom.2012.01.004
Lim, E. S., Malik, H. S., and Emerman, M. (2010a). Ancient adaptive evolution of tetherin shaped the functions of Vpu and Nef in human immunodeficiency virus and primate lentiviruses. J. Virol. 84, 7124–7134.doi: 10.1128/JVI.00468-10
Lim, S. Y., Rogers, T., Chan, T., Whitney, J. B., Kim, J., Sodroski, J., et al. (2010b). TRIM5alpha modulates immunodeficiency virus control in rhesus monkeys. PLoS Pathog. 6:e1000738. doi: 10.1371/journal.ppat.1000738
Lu, J., Pan, Q., Rong, L., He, W., Liu, S. L., and Liang, C. (2011). The IFITM proteins inhibit HIV-1 infection. J. Virol. 85, 2126–2137.doi: 10.1128/JVI.01531-10
Luciw, P. A., Pratt-Lowe, E., Shaw, K. E., Levy, J. A., and Cheng-Mayer, C. (1995). Persistent infection of rhesus macaques with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV). Proc. Natl. Acad. Sci. U.S.A. 92, 7490–7494.doi: 10.1073/pnas.92.16.7490
Mangeat, B., Turelli, P., Caron, G., Friedli, M., Perrin, L., and Trono, D. (2003). Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103.doi: 10.1038/nature01709
Mariani, R., Chen, D., Schröfelbauer, B., Navarro, F., König, R., Bollman, B., et al. (2003). Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114, 21–31.
Munk, C., Brandt, S. M., Lucero, G., and Landau, N. R. (2002). A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. U.S.A. 99, 13843–13848.doi: 10.1073/pnas.212400099
Nakayama, E. E., Miyoshi, H., Nagai, Y., and Shioda, T. (2005). A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5alpha determines species-specific restriction of simian immunodeficiency virus SIVmac infection. J. Virol. 79, 8870–8877.doi: 10.1128/JVI.79.14.8870-8877.2005
Neil, S., and Bieniasz, P. (2009). Human immunodeficiency virus, restriction factors, and interferon. J. Interferon Cytokine Res. 29, 569–580.doi: 10.1089/jir.2009.0077
Neil, S. J., Zang, T., and Bieniasz, P. D (2008). Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430.doi: 10.1038/nature06553
Newman, R. M., Hall, L., Kirmaier, A., Pozzi, L. A., Pery, E., Farzan, M., et al. (2008). Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 4:e1000003. doi: 10.1371/journal.ppat.1000003
Pertel, T., Hausmann, S., Morger, D., Züger, S., Guerra, J., Lascano, J., et al. (2011). TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472, 361–365.doi: 10.1038/nature09976
Reimann, K. A., Li, J. T., Veazey, R., Halloran, M., Park, I. W., Karlsson, G. B., et al. (1996). A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J. Virol. 70, 6922–6928.
Reynolds, M. R., Sacha, J. B., Weiler, A. M., Borchardt, G. J., Glidden, C. E., Sheppard, N. C., et al. (2011). The TRIM5{alpha} genotype of rhesus macaques affects acquisition of simian immunodeficiency virus SIVsmE660 infection after repeated limiting-dose intrarectal challenge. J. Virol. 85, 9637–9640.doi: 10.1128/JVI.05074-11
Rice, G. I., Bond, J., Asipu, A., Brunette, R. L., Manfield, I. W., Carr, I. M., et al. (2009). Mutations involved in Aicardi–Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41, 829–832.doi: 10.1038/ng.373
Saito, A., Kono, K., Nomaguchi, M., Yasutomi, Y., Adachi, A., Shioda, T., et al. (2012). Geographical, genetic and functional diversity of antiretroviral host factor TRIMCyp in cynomolgus macaque (Macaca fascicularis). J. Gen. Virol. 93(Pt 3), 594–602.doi: 10.1099/vir.0.038075-0
Saito, A., Nomaguchi, M., Iijima, S., Kuroishi, A., Yoshida, T., Lee, Y. J., et al. (2011). Improved capacity of a monkey-tropic HIV-1 derivative to replicate in cynomolgus monkeys with minimal modifications. Microbes Infect. 13, 58–64.doi: 10.1016/j.micinf.2010.10.001
Sauter, D., Schindler, M., Specht, A., Landford, W. N., Münch, J., Kim, K. A., et al. (2009). Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6, 409–421.doi: 10.1016/j.chom.2009.10.004
Sawyer, S. L., Wu, L. I., Emerman, M., and Malik, H. S. (2005). Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U.S.A. 102, 2832–2837.doi: 10.1073/pnas.0409853102
Sayah, D. M., Sokolskaja, E., Berthoux, L., and Luban, J. (2004). Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430, 569–573.doi: 10.1038/nature02777
Sheehy, A. M., Gaddis, N. C., Choi, J. D., and Malim, M. H. (2002). Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650.doi: 10.1038/nature00939
Shibata, R., Kawamura, M., Sakai, H., Hayami, M., Ishimoto, A., and Adachi, A. (1991). Generation of a chimeric human and simian immunodeficiency virus infectious to monkey peripheral blood mononuclear cells. J. Virol. 65, 3514–3520.
Sinclair, E., Barbosa, P., and Feinberg, M. B. (1997). The nef gene products of both simian and human immunodeficiency viruses enhance virus infectivity and are functionally interchangeable. J. Virol. 71, 3641–3651.
Stephens, E. B., McCormick, C., Pacyniak, E., Griffin, D., Pinson, D. M., Sun, F., et al. (2002). Deletion of the vpu sequences prior to the env in a simian-human immunodeficiency virus results in enhanced Env precursor synthesis but is less pathogenic for pig-tailed macaques. Virology 293, 252–261.doi: 10.1006/viro.2001.1244
Stremlau, M., Perron, M., Lee, M., Li, Y., Song, B., Javanbakht, H., et al. (2006). Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. U.S.A. 103, 5514–5519.doi: 10.1073/pnas.0509996103
Thippeshappa, R., Polacino, P., Yu Kimata, M. T., Siwak, E. B., Anderson, D., Wang, W., et al. (2011). Vif substitution enables persistent infection of pig-tailed macaques by human immunodeficiency virus type 1. J. Virol. 85, 3767–3779.doi: 10.1128/JVI.02438-10
Thippeshappa, R., Ruan, H., and Kimata, J. T. (2012). Breaking barriers to an AIDS model with macaque-tropic HIV-1 derivatives. Biology (Basel) 1, 134–164.doi: 10.3390/biology1020134
Thippeshappa, R., Ruan, H., Wang, W., Zhou, P., and Kimata, J. T. (2013). A variant macaque-tropic human immunodeficiency virus type 1 is resistant to interferon alpha induced restriction in pig-tailed macaque CD4+ T-cells. J. Virol. 87, 6678–6692 doi: 10.1128/JVI.00338-13
Uberla, K., Stahl-Hennig, C., Böttiger, D., Mätz-Rensing, K., Kaup, F. J., Li, J., et al. (1995). Animal model for the therapy of acquired immunodeficiency syndrome with reverse transcriptase inhibitors. Proc. Natl. Acad. Sci. U.S.A. 92, 8210–8214.
Van Damme, N., Goff, D., Katsura, C., Jorgenson, R. L., Mitchell, R., Johnson, M. C., et al. (2008). The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3, 245–252.doi: 10.1016/j.chom.2008.03.001
Virgen, C. A., Kratovac, Z., Bieniasz, P. D., and Hatziioannou, T. (2008). Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc. Natl. Acad. Sci. U.S.A. 105, 3563–3568.doi: 10.1073/pnas.0709258105
White, T. E., Brandariz-Nunez, A., Valle-Casuso, J. C., Amie, S., Anh Nguyen, L., Kim, B., et al. (2013). The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13, 441–451.doi: 10.1016/j.chom.2013.03.005
Wilson, S. J., Schoggins, J. W., Zang, T., Kutluay, S. B., Jouvenet, N., Alim, M. A., et al. (2012). Inhibition of HIV-1 particle assembly by 2′,3′-cyclic-nucleotide 3′-phosphodiesterase. Cell Host Microbe 12, 585–597.doi: 10.1016/j.chom.2012.08.012
Wilson, S. J., Webb, B. L., Ylinen, L. M., Verschoor, E., Heeney, J. L., and Towers, G. J. (2008). Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc. Natl. Acad. Sci. U.S.A. 105, 3557–3562.doi: 10.1073/pnas.0709003105
Yang, S. J., Lopez, L. A., Hauser, H., Exline, C. M., Haworth, K. G., and Cannon, P. M. (2010). Anti-tetherin activities in Vpu-expressing primate lentiviruses. Retrovirology 7, 13.doi: 10.1186/1742-4690-7-13
Zhang, F., Wilson, S. J., Landford, W. C., Virgen, B., Gregory, D., Johnson, M. J., et al. (2009). Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6, 54–67.doi: 10.1016/j.chom.2009.05.008
Keywords: HIV-1, SIV, AIDS, macaque models, tropism, innate restriction
Citation: Misra A, Thippeshappa R and Kimata JT (2013) Macaques as model hosts for studies of HIV-1 infection. Front. Microbiol. 4:176. doi: 10.3389/fmicb.2013.00176
Received: 29 April 2013; Accepted: 11 June 2013;
Published online: 28 June 2013.
Edited by:
Akio Adachi, The University of Tokushima Graduate School, JapanReviewed by:
Francois Villinger, Emory University School of Medicine, USAFrank Kirchhoff, University Clinic Ulm, Germany
Copyright: © 2013 Misra, Thippeshappa and Kimata. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Jason T. Kimata, Department of Molecular Virology and Microbiology, Baylor College of Medicine, One Baylor Plaza, BCM385, Houston, TX 77030, USA e-mail:amtpbWF0YUBiY20uZWR1
 Anisha Misra
Anisha Misra