- 1Institute of Microbiology and Biotechnology, University of Latvia, Riga, Latvia
- 2Department of Computer Systems, Latvia University of Agriculture, Jelgava, Latvia
- 3SIA TIBIT, Jelgava, Latvia
- 4Department of Biological and Medical Sciences, Oxford Brookes University, Oxford, UK
Mathematical modeling of metabolism is essential for rational metabolic engineering. The present work focuses on several types of modeling approach to quantitative understanding of central metabolic network and energetics in the bioethanol-producing bacterium Zymomonas mobilis. Combined use of Flux Balance, Elementary Flux Mode, and thermodynamic analysis of its central metabolism, together with dynamic modeling of the core catabolic pathways, can help to design novel substrate and product pathways by systematically analyzing the solution space for metabolic engineering, and yields insights into the function of metabolic network, hardly achievable without applying modeling tools.
Introduction
Zymomonas mobilis, a member of the family of Sphingomonadaceae, is an unusual facultatively anaerobic Gram-negative bacterium, which has a very efficient homoethanol fermentation pathway. High ethanol yields, outstanding ethanol productivity (exceeding by 3–5 fold that of yeast; see Rogers et al., 1982), and tolerance to high ethanol and sugar concentrations, keep Z. mobilis in the focus of biotechnological research over four decades. The complete genome sequence of Z. mobilis ZM4, consisting of a single circular chromosome of 2,056,416 bp, was reported by Seo et al. (2005), followed by the genomes of several other strains (Kouvelis et al., 2009; Pappas et al., 2011; Desiniotis et al., 2012). Its small genome size, together with high specific rate of sugar catabolism via the Entner–Doudoroff (ED) pathway, and a relatively simple central metabolic network, make Z. mobilis a promising candidate for metabolic engineering (Sprenger, 1996; Rogers et al., 2007). Currently, recombinant Z. mobilis capable of fermenting pentose sugars is regarded as a potential alternative to yeast and recombinant Escherichia coli for ethanol biofuel synthesis from agricultural and forestry waste (Dien et al., 2003; Panesar et al., 2006; Rogers et al., 2007; Lau et al., 2010).
In spite of the seeming simplicity of its metabolism, Z. mobilis is a bacterium with an interesting physiology (Kalnenieks, 2006), posing researchers some long-standing challenges. Its extremely rapid glucose catabolism, far exceeding the biosynthetic demands of the cell, and the presence of an active respiratory chain with a low apparent P/O ratio (Bringer et al., 1984; Strohdeicher et al., 1990; Kalnenieks et al., 1993) are major manifestations of its so-called uncoupled growth. There are serious gaps in our understanding of the mechanistic basis of uncoupled growth, and in particular, the reason for the low degree of coupling in the respiratory chain of Z. mobilis.
Mathematical modeling and in silico simulations are the most powerful tools of systems biology for understanding of complex metabolic phenomena, and often lead to novel, counterintuitive conclusions. A quantitative picture of physiology and metabolism is a key for rational, model-driven metabolic engineering. Some of the different metabolic modeling approaches that can support the design of novel metabolic engineering strategies are summarized in Figure 1 (for reviews see: Schuster et al., 2000; Liu et al., 2010; Santos et al., 2011; Schellenberger et al., 2011; Rohwer, 2012). Compared with qualitative, pathway-oriented approaches, computational network analyses can enforce strict mass, energy and redox balancing and give an overall stoichiometric equation for predicted conversions (c.f. de Figueiredo et al., 2009). Here we outline recent advances and perspectives from applying such systems biology approaches to the physiology of Z. mobilis. We discuss some recent results gained by stoichiometric and kinetic modeling of its central metabolism, and their potential application to the design of novel substrate pathways, synthesis of novel products, and to the study of the uncoupled growth phenomenon per se.
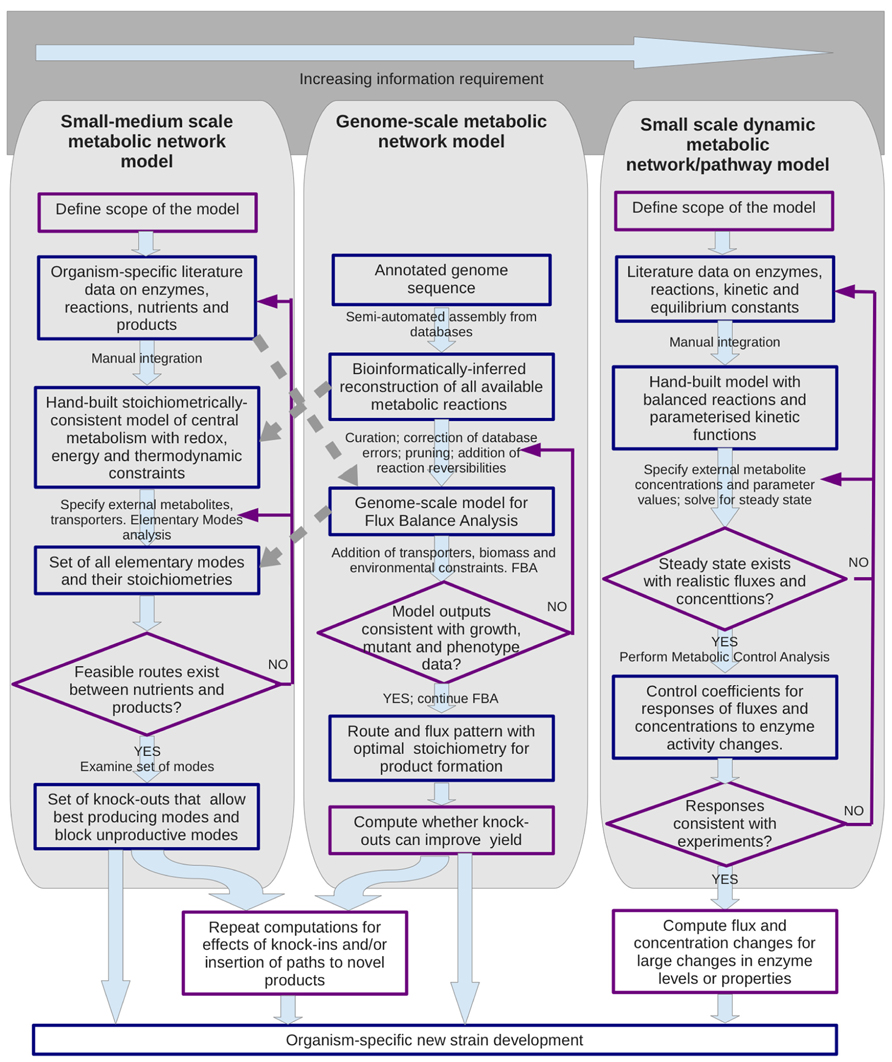
FIGURE 1. Metabolic modeling support for metabolic engineering strategies. Three main types of modeling approach are shown that have been used to analyze metabolic networks and to design alterations. The methodologies for the two structural modeling approaches shown on the left and center can have some cross-talk as indicated by the broken arrows. Blue boxes indicate entities such as data sets and models; purple outlines indicate procedures.
Reconstruction of Z. mobilis Central Metabolic Network
Two medium-scale (Tsantili et al., 2007; Pentjuss et al., 2013) and two genome-scale stoichiometric reconstructions of Z. mobilis (Lee et al., 2010; Widiastuti et al., 2010) have been reported so far, representing instances of the left and center panels of Figure 1 respectively. These reconstructions were based on the available genome annotation (Seo et al., 2005; Kouvelis et al., 2009) and provided an overall picture of Z. mobilis metabolism. The recent reconstruction made by Pentjuss et al. (2013) was focussed solely on the reactions of central metabolism and for the first time for Z. mobilis provided simulation-ready model files. That decreased the scale, yet allowed an improvement in the accuracy of reconstruction, by combining the genome-derived information with the preexisting biochemical evidence on Z. mobilis, available mostly for the reactions of catabolism and central metabolism.
Notably, several key reactions of central metabolism, common for the majority of the chemoheterotrophic, facultatively anaerobic bacteria, are absent in Z. mobilis. The Embden–Meyerhof–Parnas (EMP) glycolytic pathway is not operating in this bacterium. Absence of the EMP pathway has been confirmed by [1-13C]glucose experiments (Fuhrer et al., 2005), and furthermore, the gene for phosphofructokinase is lacking in the genome (Seo et al., 2005). Z. mobilis is the only known microorganism that uses the ED pathway anaerobically in place of the EMP glycolysis. Since the EMP pathway produces two ATP per glucose while the ED produces only one, it might seem that Z. mobilis suffers from ATP deficiency. However, it has been recently shown by means of thermodynamic analysis that, for a given glycolytic flux, the ED pathway requires significantly less enzymatic protein than the EMP pathway (Flamholz et al., 2013). On the other hand, the amount of the ED pathway enzymes in Z. mobilis cell is reported to be very high, reaching 50% of the cell’s soluble protein (Algar and Scopes, 1985; An et al., 1991). The high level of expression of the pathway together with its inherent speed, therefore, makes ATP production by the Z. mobilis ED pathway very rapid and, in fact, excessive for the needs of cell. Energy dissipation in order to regenerate ADP is thus essential for its balanced operation (Kalnenieks, 2006).
The TCA cycle is truncated, and consists of two branches, leading to 2-oxoglutarate and fumarate as the end products (Bringer-Meyer and Sahm, 1989). The genes for the 2-oxoglutarate dehydrogenase complex and malate dehydrogenase are absent (Seo et al., 2005), and accordingly, 13C-labeling patterns of 2-oxoglutarate and oxaloacetate do not support cyclic function of this pathway in Z. mobilis (de Graaf et al., 1999). Also, the pentose phosphate pathway is incomplete: transaldolase activity is lacking (Feldmann et al., 1992; de Graaf et al., 1999). The activity of 6-phosphogluconate dehydrogenase, the first reaction of the oxidative part of the pentose phosphate pathway, was reported to be very low (Feldmann et al., 1992). Subsequently, the corresponding gene (gnd) could not be identified in the sequenced genomes.
The aerobic redox cofactor balance and the function of electron transport chain represent yet another part of Z. mobilis metabolism that differs from that typically found in other bacteria. Z. mobilis is one of the few known bacteria in which both NADH and NADPH can serve as electron donors for the respiratory type II NADH dehydrogenase (ZMO1113; Bringer et al., 1984; Strohdeicher et al., 1990; Kalnenieks et al., 2008). Because of the truncated Krebs cycle, the ED pathway is the only source of reducing equivalents in catabolism, and therefore the electron transport chain competes for the limited NADH with the highly active alcohol dehydrogenases (Kalnenieks et al., 2006). Withdrawal of NADH from the alcohol dehydrogenase reaction would cause accumulation of acetaldehyde, which inhibits growth of aerobic Z. mobilis culture (Wecker and Zall, 1987). Nevertheless, this bacterium possesses a respiratory chain with high rates of oxygen consumption. The apparent P/O ratio of its respiratory chain is low (Bringer et al., 1984; Kalnenieks et al., 1993) though the mechanistic basis for that is not clear. However, for metabolic engineering purposes, an active, yet energetically inefficient electron transport has advantages for the needs of redox balancing during synthesis of novel products via metabolic pathways for which regeneration of NAD(P)+ is essential, whereas the aerobic increase of biomass yield is unwanted.
Quest for Novel Substrates and Products: Stoichiometric and Thermodynamic Analysis
Much of the metabolic engineering in Z. mobilis has been devoted to broadening of its substrate spectrum and expanding its product range beyond bioethanol with a particular focus on the pathway of pentose sugar utilization for synthesis of bioethanol (Sprenger, 1996; Rogers et al., 2007). Advanced pentose-assimilating strains of Z. mobilis have been developed during the last couple of decades that can, in several respects, compete with the analogous recombinant strains of E. coli and S. cerevisiae (Lau et al., 2010). We were interested to explore the biotechnological potential of the low-efficiency respiratory chain of this bacterium for expanding its substrate and product spectrum.
Based on a medium-scale reconstruction of central metabolism (Pentjuss et al., 2013), stoichiometric modeling was used to search the whole solution space of the model, finding maximum product yields and the byproduct spectra with glucose, xylose, or glycerol as the carbon substrates for respiring cultures (Figure 1, left hand side). This was done by Flux Balance Analysis approach, using the COBRA Toolbox (Schellenberger et al., 2011). The stoichiometric analysis suggested several metabolic engineering strategies for obtaining products, such as glycerate, succinate, and glutamate that would use the electron transport chain to oxidize the excess NAD(P)H, generated during synthesis of these metabolites. Oxidation of the excess NAD(P)H would also be needed for synthesis of ethanol from glycerol.
It is essential, however, to complement the stoichiometric analysis with estimation of the thermodynamic feasibility of the underlying reactions. Glycerol utilization can serve as an example. Being a cheap, renewable carbon source, a byproduct of biodiesel technology, glycerol represents an attractive alternative substrate for Z. mobilis metabolic engineering. It is not expected to have serious growth-inhibitory effects, and also, little genetic engineering seems to be needed to make it consumable by Z. mobilis, and to channel it into the rapid ED pathway. Conversion of glycerol to ethanol by Z. mobilis would require expression of a heterologous transmembrane glycerol transporter and a glycerol kinase. Its genome contains genes for the two subsequent conversion steps, glycerolphosphate dehydrogenase and triose phosphate isomerase, leading to the ED intermediate glyceraldehyde-3-phosphate although, their overexpression might be needed. The further reactions from the glyceraldehyde-3-phosphate to ethanol represent a part of Z. mobilis natural ethanologenic pathway, and should be both rapid and redox-balanced. The extra NAD(P)H, generated by the glycerolphosphate dehydrogenase reaction could be oxidized by the respiratory chain. If succinate is the desired product (Pentjuss et al., 2013), the extra reducing equivalents could be used for reduction of fumarate by the respiratory fumarate reductase.
The pathway from glycerol to glyceraldehyde-3-phosphate via phosphorylation and following oxidation and isomerization steps is presented in biochemistry textbooks as the pathway of glycerol catabolism after breakdown of triacylglycerols in higher animals and humans (see e.g., Lehninger Principles of Biochemistry, 6th edition, Fig. 17-4). Though feasible from the stoichiometric point of view, it reveals problems when subjected to thermodynamic analysis. The equilibrium of the glycerolphosphate dehydrogenase reaction appears to be shifted very much toward formation of glycerol phosphate. In silico kinetic simulations of glycerol uptake for a putative engineered Z. mobilis demonstrate a dramatic accumulation of glycerol-3-phospate, reaching concentrations of several molar even at a high rate of NAD(P)H withdrawal by the respiratory chain (Rutkis et al., unpublished). Apparently, while the estimated overall stoichiometry of aerobic glycerol conversions is correct, thermodynamic analysis suggests the need to search for alternative reaction sequences to avoid excessive intracellular accumulation of metabolites.
Aerobic Elementary Flux Modes of the Pentose Phosphate Pathway
A metabolic network can function according to many different pathway options. Elementary flux mode (EFM) analysis has emerged as a systems biological tool that dissects a metabolic network into its basic building blocks, the EFMs (Schuster et al., 2000, 2002). All metabolic capabilities in steady states represent a weighted average of the EFMs, which are the minimal sets of enzymes that can each generate a valid steady state. The EFM approach has proved to be efficient for designing sets of knock-out mutations in order to minimize unwanted metabolic functionality in the producer strains. For example, in engineered E. coli, EMF-based mutation analysis helped to eliminate catabolite repression and to increase carbon flux toward the target product ethanol (Trinh et al., 2008).
By decomposing a network of highly interconnected reactions, the EFM analysis may reveal unexpected flux options. Recently we applied EFM analysis to the interaction between the ED, pentose phosphate pathway and respiratory chain in an engineered Z. mobilis, which expresses heterologous gnd and enzymes for pentose conversion, using the metabolic modeling package ScrumPy (Poolman, 2006). We were interested in the EFMs that such non-growing engineered Z. mobilis might employ for aerobic catabolism of glucose and xylose. Analysis revealed several EFMs in respiring cells (Figure 2) that have considerable interest for study of aerobic energy-coupling in this bacterium. With both monosaccharides, knocking out edd (encoding 6-phosphogluconate dehydratase), and overexpressing heterologous gnd (encoding 6-phosphogluconate dehydrogenase), would lead to generation of additional NAD(P)H and CO2 in the pentose phosphate pathway, while lowering the ethanol yield. Yet, most importantly, decrease of the ethanol yield would not be accompanied by accumulation of acetaldehyde and acetoin.
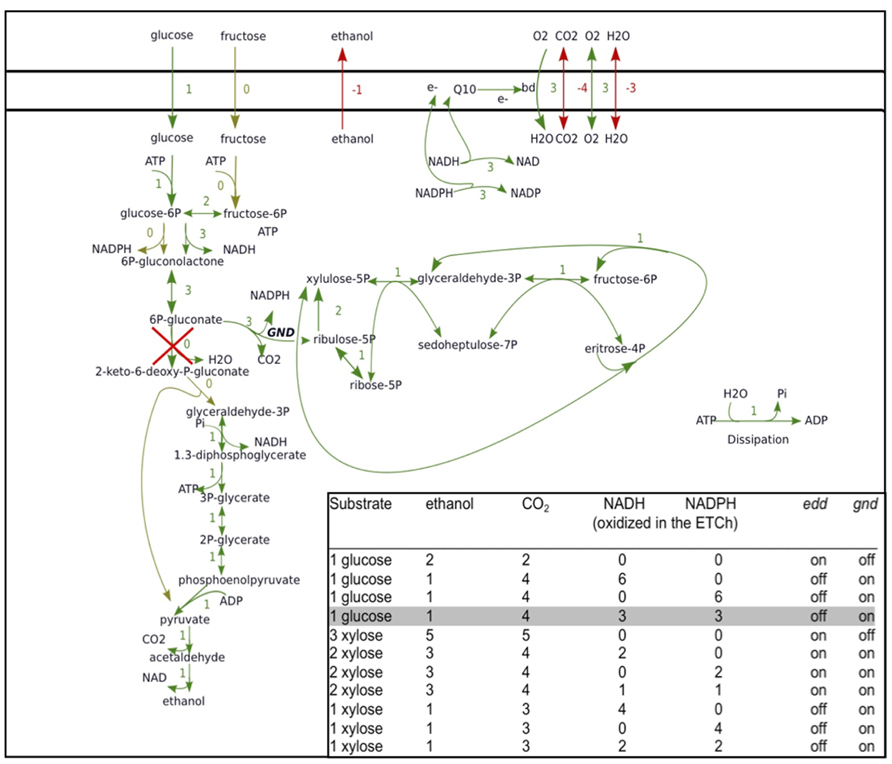
FIGURE 2. Elementary flux modes of aerobic glucose and xylose catabolism for a strain with engineered pentose phosphate pathway enzymes. Elementary flux mode for catabolism of glucose in cells with knocked-out edd and overexpressed heterologous gnd via the Entner–Doudoroff and pentose phosphate pathway, involving both NADH- and NADPH-oxidizing activity of the respiratory chain, is shown. ScrumPy modeling software EFM drawing algorithm (Pentjuss et al., unpublished) was used for visualization. Inset: complete list of elementary flux modes of glucose and xylose catabolism in Z. mobilis, involving the Entner–Doudoroff pathway, pentose phosphate pathway and the respiratory chain, with ethanol and carbon dioxide as the sole products. The explicitly shown elementary flux mode is shaded in gray.
Thus, a simple EFM analysis suggests how to modify Z. mobilis aerobic metabolism so that its electron transport chain would receive more reducing equivalents without accumulation of inhibitory byproducts. Strains with such metabolic modifications might be very useful for study of the mechanisms underlying the uncoupled mode of oxidative phosphorylation in this bacterium.
Kinetic Modeling of the Entner–Doudoroff Pathway
Despite the diverse studies of Z. mobilis physiology and genetics, little has been done so far to combine the accumulated knowledge in a form of kinetic model of central metabolism that would be comparable to the existing models for E. coli and yeast, and could be used to develop efficient metabolic engineering strategies (c.f. Figure 1, right-hand panel). A kinetic model reported by Altintas et al. (2006) focussed mainly on the interaction between the heterologous enzymes of pentose phosphate pathway and the native Z. mobilis ED glycolysis. Providing predictions for optimization of expression levels of the heterologous genes, this study contributed to strategies for maximizing xylose conversion to ethanol. However, the authors assumed constant intracellular concentrations of all adenylate cofactors. Since the ED pathway itself is a major player in ANP and NAD(P)(H) turnover, this might lead to erroneous conclusions on the pathway kinetics and restrict the range of model application. The recent kinetic model by Rutkis et al. (2013): (i) treated the cofactor levels as variables, making the interplay between adenylate cofactor levels and the pathway kinetics explicit, and (ii) introduced equilibrium constants in the kinetic equations to account for the reversibility of reactions more correctly. Metabolic control analysis (MCA) carried out with the model pointed to the ATP turnover as a major bottleneck, showing that the ATP consumption (dissipation) exerts a high level of control over glycolytic flux under various conditions (Rutkis et al., 2013).
Indeed, experimental studies of the ED pathway flux have shown that moderate overexpression of the ED pathway and alcohol dehydrogenase genes do not affect the glycolytic flux (Arfman et al., 1992; Snoep et al., 1995). Larger increases of the expression levels even caused a decrease in flux, exerting also a negative impact on Z. mobilis growth rate (Snoep et al., 1995). This clearly indicated that glycolytic flux in Z. mobilis must be controlled at some point(s) outside the ED pathway itself. The negative effects of overexpression apparently did not result from intrinsically negative flux control coefficients of the ED enzymes, but were attributable to the protein burden effect (Snoep et al., 1995), whereby overexpression of an enzyme with a small flux control coefficient caused reductions in the expression of other enzymes that have a greater influence on the flux. These results together with MCA studies on the kinetic model suggested that, due to the negligible flux control coefficients for the majority of reactions, single enzymes of the ED pathway should not be considered as prime targets for overexpression to increase the glycolytic flux in Z. mobilis (Rutkis et al., 2013). The calculated effects of several glycolytic enzyme (gap, pgk, pgm) and both alcohol dehydrogenase isoenzyme (adhA and adhB) overexpression, in accordance with previous experimental observations, predicted little or no increase of glycolytic flux (Arfman et al., 1992; Snoep et al., 1995). The somewhat higher flux control coefficient for the pyruvate decarboxylase (pdc) reaction suggested that overexpression of this enzyme by more than 3-fold, might lead to an increase of glycolytic flux of almost 23% (Rutkis et al., 2013). However, quite the opposite was observed experimentally: approximately 10-fold increase of pdc was shown to slow down glycolysis by up to 25%, thereby implying that the protein burden might be a serious side effect of catabolic enzyme overexpression in Z. mobilis. Usually effects of protein burden are of minor importance in optimization of catabolic fluxes, due to relatively low concentrations of the enzymes in catabolic routes. This is not the case for Z. mobilis catabolism, however, since over 50% of the cell protein already is engaged in the function of the ED pathway (Algar and Scopes, 1985). Fortunately, flux control coefficient estimations still indicate a certain solution space for flux improvement: simultanous overexpression of pdc, eno, pgm within the 3-fold range of initial enzyme activities (wich most probably would be below the putative protein burden threshold), has the potential to increase the glycolytic flux by up to 25% (to reach 6.6 g glucose, g dry wt-1h-1; Rutkis et al. (2013).
Obviously, another option would be to raise ATP dissipation. That could be done by overexpression of the H+-dependent F0F1-ATPase, a major ATP-dissipating activity. Reyes and Scopes (1991) have estimated the F0F1-ATPase contribution being over 20% of the total intracellular ATP turnover. It should be noted, however, that overexpression of ATP-dissipating reaction(s) might disturb the intracellular ATP homeostasis, with successive suspension of glycolysis (by slowing down the first reaction of the ED pathway, phosphorylation of glucose). Co-response analysis indicates (Rutkis et al., 2013) that, at the highest glycolytic flux considered (4.6 g/g/h), the cellular capacity to maintain the ATP homeostasis is close to its limit, since even 1% further increase of glycolytic flux due to rise of ATP dissipation would be associated with a 4% decrease in ATP concentration.
Conclusion
Although Z. mobilis metabolism has been subject to extensive research, and genome sequence data for several strains are now also available, it is only quite recently that modeling of its central metabolic network has started to gain momentum. These latest results of modeling Z. mobilis illustrate the relevance of combined stoichiometric, thermodynamic and kinetic analysis of central metabolism at different scales for microorganisms producing biorenewables. Concerted application of structural and dynamic modeling will help to identify targets for future metabolic engineering in a systematic manner, and provide novel insights into the biotechnological potential of this bacterium.
Author Contributions
All authors have equally contributed to the manuscript and have accepted the final version to be published.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Latvian ESF projects 2009/027/1DP/1.1.1.2.0/ 09/APIA/VIAA/128 and 2009/0138/1DP/1.1.2.1.2/09/IPIA/VIAA/004, and by the Latvian Council of Science project 536/2012.
References
Algar, E. M., and Scopes, R. K. (1985). Studies on cell-free metabolism: ethanol production by extracts of Zymomonas mobilis. J. Biotechnol. 2, 275–287.
Altintas, M. M., Eddy, C. K., Zhang, M., McMillan, J. D., and Kompala, D. S. (2006). Kinetic modeling to optimize pentose fermentation in Zymomonas mobilis. Biotechnol. Bioeng. 94, 273–295. doi: 10.1002/bit.20843
An, H., Scopes, R. K., Rodriguez, M., Keshav, K. F., and Ingram, L. O. (1991). Gel electrophoretic analysis of Zymomonas mobilis glycolytic and fermentative enzymes: identification of alcohol dehydrogenase II as a stress protein. J. Bacteriol. 173, 5975–5982.
Arfman, N., Worell, V., and Ingram, L. O. (1992). Use of the tac promoter and lacIq for the controlled expression of Zymomonas mobilis fermentative genes in Escherichia coli and Zymomonas mobilis. J. Bacteriol. 174, 7370–7378.
Bringer-Meyer, S., and Sahm, H. (1989). Junctions of catabolic and anabolic pathways in Zymomonas mobilis: phosphoenolpyruvate carboxylase and malic enzyme. Appl. Microbiol. Biotechnol. 31, 529–536.
Bringer, S., Finn, R. K., and Sahm, H. (1984). Effect of oxygen on the metabolism of Zymomonas mobilis. Arch. Microbiol. 139, 376–381.
de Figueiredo, L. F., Schuster, S., Kaleta, C., and Fell, D. A. (2009). Can sugars be produced from fatty acids? a test case for pathway analysis tools. Bioinformatics 25, 152–158. doi: 10.1093/bioinformatics/btn621
de Graaf, A. A., Striegel, K., Wittig, R. M., Laufer, B., Schmitz, G., Wiechert, W., et al. (1999). Metabolic state of Zymomonas mobilis in glucose-, fructose-, and xylose-fed continuous cultures as analysed by 13C- and 31P-NMR spectroscopy. Arch. Microbiol. 171, 371–385.
Desiniotis, A., Kouvelis, V. N., Davenport, K., Bruce, D., Detter, C., Tapia, R., et al. (2012). Complete genome sequence of the ethanol-producing Zymomonas mobilis subsp. mobilis centrotype ATCC 29191. J. Bacteriol. 194, 5966–5967. doi: 10.1128/JB.01398–1312
Dien, B. S., Cotta, M. A., and Jeffries, T. W. (2003). Bacteria engineered for fuel ethanol production: current status. Appl. Microbiol. Biotechnol. 63, 258–266. doi: 10.1007/s00253-003-1444-y
Feldmann, S., Sahm, H., and Sprenger, G. (1992). Pentose metabolism in Zymomonas mobilis wild-type and recombinant strains. Appl. Microbiol. Biotechnol. 38, 354–361.
Flamholz, A., Noor, E., Bar-Even, A., Liebermeister, W., and Milo, R. (2013). Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc. Natl. Acad. Sci. U.S.A. 110, 10039–10044. doi: 10.1073/pnas.1215283110
Fuhrer, T., Fischer, E., and Sauer, U. (2005). Experimental identification and quantification of glucose metabolism in seven bacterial species. J. Bacteriol. 187, 1581–1590. doi: 10.1128/JB.187.5.1581–1590.2005
Kalnenieks, U. (2006). “Physiology of Zymomonas mobilis: some unanswered questions,” in Advances in Microbial Physiology, Vol. 51, ed. R. K. Poole (London: Academic Press), 73–117. doi: 10.1016/S0065-2911(06)51002-1
Kalnenieks, U., de Graaf, A. A., Bringer-Meyer, S., and Sahm, H. (1993). Oxidative phosphorylation in Zymomonas mobilis. Arch. Microbiol. 160, 74–79.
Kalnenieks, U., Galinina, N., Strazdina, I., Kravale, Z., Pickford, J. L., Rutkis, R., et al. (2008). NADH dehydrogenase deficiency results in low respiration rate and improved aerobic growth of Zymomonas mobilis. Microbiology 154, 989–994. doi: 10.1099/mic.0.2007/012682-0
Kalnenieks, U., Galinina, N., Toma, M. M., Pickford, J. L., Rutkis, R., and Poole, R. K. (2006). Respiratory behaviour of a Zymomonas mobilis adhB::kanr mutant supports the hypothesis of two alcohol dehydrogenase isoenzymes catalysing opposite reactions. FEBS Lett. 580, 5084–5088. doi: 10.1016/j.febslet.2006.08.034
Kouvelis, V. N., Saunders, E., Brettin, T. S., Bruce, D., Detter, C., Han, C., et al. (2009). Complete genome sequence of the ethanol producer Zymomonas mobilis NCIMB 11163. J. Bacteriol. 191, 7140–7141. doi:10.1128/JB.01084-09
Lau, M. W., Gunawan, C., Balan, V., and Dale, B. E. (2010). Comparing the fermentation performance of Escherichia coli KO11, Saccharomyces cerevisiae 424A(LNH-ST) and Zymomonas mobilis AX101 for cellulosic ethanol production. Biotechnol. Biofuels 3, 11. doi: 10.1186/1754-6834-3-11
Lee, K. Y., Park, J. M., Kim, T. Y., Yun, H., and Lee, S. Y. (2010). The genome-scale metabolic network analysis of Zymomonas mobilis ZM4 explains physiological features and suggests ethanol and succinic acid production strategies. Microb. Cell Fact. 9, 94. doi: 10.1186/1475-2859-9-94
Liu, L, Agren, R., Bordel, S., and Nielsen, J. (2010). Use of genome-scale metabolic models for understanding microbial physiology. FEBS Lett. 584, 2556–2564. doi: 10.1016/j.febslet.2010.04.052
Panesar, P. S., Marwaha, S. S., and Kennedy, J. F. (2006). Zymomonas mobilis: an alternative ethanol producer. J. Chem. Technol. Biotechnol. 81, 623–635. doi: 10.1002/jctb.1448
Pappas, K. M., Kouvelis, V. N., Saunders, E., Brettin, T. S., Bruce, D., Detter, C., et al. (2011). Genome sequence of the ethanol-producing Zymomonas mobilis subsp. mobilis lectotype strain ATCC 10988. J. Bacteriol. 193, 5051–5052. doi: 10.1128/JB.05395-11
Pentjuss, A., Odzina, I., Kostromins, A., Fell, D. A., Stalidzans, E., and Kalnenieks, U. (2013). Biotechnological potential of respiring Zymomonas mobilis: a stoichiometric analysis of its central metabolism. J. Biotechnol. 165, 1–10. doi: 10.1016/j.jbiotec.2013.02.014
Poolman, M. G. (2006). ScrumPy: metabolic modelling with python. Syst. Biol. 153, 375–378. doi: 10.1049/ip-syb:20060010
Reyes, L., and Scopes, R. K. (1991). Membrane-associated ATPase from Zymomonas mobilis; purification and characterization. Biochim. Biophys. Acta 1068, 174–178.
Rogers, P., Lee, K., Skotnicki, M., and Tribe, D. (1982). Ethanol production by Zymomonas mobilis. Adv. Biochem. Eng. 23, 37–84.
Rogers, P. L., Jeon, Y. J., Lee, K. J., and Lawford, H. G. (2007). Zymomonas mobilis for fuel ethanol and higher value products. Adv. Biochem. Eng. 108, 263–288. doi: 10.1007/10_2007_060
Rohwer, J. M. (2012). Kinetic modeling of plant metabolic pathways. J. Exp. Bot. 63, 2275–2292. doi:10.1093/jxb/ers080
Rutkis, R., Kalnenieks, U., Stalidzans, E., and Fell, D. A. (2013). Kinetic modeling of Zymomonas mobilis Entner-Doudoroff pathway: insights into control and functionality. Microbiology 159, 2674–2689. doi: 10.1099/mic.0.071340-0
Santos, F., Boele, J., and Teusink, B. (2011). A practical guide to genome-scale metabolic models and their analysis. Methods Enzymol. 500, 509–532. doi: 10.1016/B978-0-12-385118-5.00024-4
Schellenberger, J., Que, R., Fleming, R. M. T., Thiele, I., Orth, J. D., Feist, A. M., et al. (2011). Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat. Protoc. 6, 1290–1307. doi: 10.1038/nprot.2011.308
Schuster, S., Fell, D. A., and Dandekar, T. (2000). A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nat. Biotechnol. 18, 326–332. doi: 10.1038/73786
Schuster, S., Hilgetag, C., Woods, J. H., and Fell, D. A. (2002). Reaction routes in biochemical reaction systems: algebraic properties, validated calculation procedure and example from nucleotide metabolism. J. Math. Biol. 45, 153–181. doi: 10.1007/s002850200143
Seo, J.-S., Chong, H., Park, H. S., Yoon, K.-O., Jung, C., Kim, J. J., et al. (2005). The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat. Biotechnol. 23, 63–68. doi: 10.1038/nbt1045
Snoep, J. L., Yomano, L. P., Westerhoff, H. V., and Ingram, L. O. (1995). Protein burden in Zymomonas mobilis: negative flux and growth control due to overproduction of glycolytic enzymes. Microbiology 141, 2329–2337.
Sprenger, G. A. (1996). Carbohydrate metabolism in Zymomonas mobilis: a catabolic highway with some scenic routes. FEMS Microbiol. Lett. 145, 301–307.
Strohdeicher, M., Neuss, B., Bringer-Meyer, S., and Sahm, H. (1990). Electron trans- port chain of Zymomonas mobilis. Interaction with the membrane-bound glucose dehydrogenase and identification of ubiquinone 10. Arch. Microbiol. 154, 536–543.
Trinh, C. T., Unrean, P., and Srienc, F. (2008). Minimal Escherichia coli cell for the most efficient production of ethanol from hexoses and pentoses. Appl. Environ. Microbiol. 74, 3634–3643. doi: 10.1128/AEM.02708-07
Tsantili, I. C., Karim, M. N., and Klapa, M. I. (2007). Quantifying the metabolic capabilities of engineered Zymomonas mobilis using linear programming analysis. Microb. Cell Fact. 6, 8. doi: 10.1186/1475-2859-6-8
Wecker, M. S. A., and Zall, R. R. (1987). Production of acetaldehyde by Zymomonas mobilis. Appl. Environ. Microbiol. 53, 2815–2820.
Keywords: stoichiometric modeling, elementary flux modes, kinetic modeling, systems biology, metabolic engineering, Entner–Doudoroff pathway, central metabolism, Zymomonas mobilis
Citation: Kalnenieks U, Pentjuss A, Rutkis R, Stalidzans E and Fell DA (2014) Modeling of Zymomonas mobilis central metabolism for novel metabolic engineering strategies. Front. Microbiol. 5:42. doi: 10.3389/fmicb.2014.00042
Received: 16 November 2013; Paper pending published: 29 December 2013;
Accepted: 21 January 2014; Published online: 05 February 2014.
Edited by:
Katherine M. Pappas, University of Athens, GreeceReviewed by:
Jason Warren Cooley, University of Missouri, USAPatrick Hallenbeck, University of Montreal, Canada
Copyright © 2014 Kalnenieks, Pentjuss, Rutkis, Stalidzans and Fell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Uldis Kalnenieks, Institute of Microbiology and Biotechnology, University of Latvia, Kronvalda boulevard 4, Riga, LV-1586, Latvia e-mail:a2FsbmVuQGxhbmV0Lmx2
 Uldis Kalnenieks
Uldis Kalnenieks Agris Pentjuss
Agris Pentjuss Reinis Rutkis
Reinis Rutkis Egils Stalidzans
Egils Stalidzans David A. Fell
David A. Fell