- 1State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
- 2University of Chinese Academy of Sciences, Beijing, China
DNA replication initiation, which starts at specific chromosomal site (known as replication origins), is the key regulatory stage of chromosome replication. Archaea, the third domain of life, use a single or multiple origin(s) to initiate replication of their circular chromosomes. The basic structure of replication origins is conserved among archaea, typically including an AT-rich unwinding region flanked by several conserved repeats (origin recognition box, ORB) that are located adjacent to a replication initiator gene. Both the ORB sequence and the adjacent initiator gene are considerably diverse among different replication origins, while in silico and genetic analyses have indicated the specificity between the initiator genes and their cognate origins. These replicator–initiator pairings are reminiscent of the oriC-dnaA system in bacteria, and a model for the negative regulation of origin activity by a downstream cluster of ORB elements has been recently proposed in haloarchaea. Moreover, comparative genomic analyses have revealed that the mosaics of replicator-initiator pairings in archaeal chromosomes originated from the integration of extrachromosomal elements. This review summarizes the research progress in understanding of archaeal replication origins with particular focus on the utilization, control and evolution of multiple replication origins in haloarchaea.
Introduction
DNA replication is a fundamental cellular process that is functionally conserved across all three domains of life (bacteria, archaea, and eukaryote). The precise regulation of DNA replication ensures the accurate duplication of genomic information, and replication initiation is the first and most important stage of this regulation. The first model of DNA replication initiation was proposed for Escherichia coli in 1963, postulating that a trans-acting factor binds to a cis-acting site which triggers replication initiation (Jacob et al., 1963). In the subsequent 50 years, this “replicon model” has been demonstrated to be essentially true in all organisms, and the cis-acting site is now known as the replication origin. Bacterial chromosomes are typically replicated from a single origin, whereas the replication of eukaryotic chromosomes initiates from a number of discrete origins (Leonard and Mechali, 2013). DNA replication origins have been well-defined in bacteria and unicellular eukaryotes, and relative topics are covered in a number of excellent reviews (Messer, 2002; Mott and Berger, 2007; Zakrzewska-Czerwinska et al., 2007; Mechali, 2010; Aparicio, 2013). In contrast, focus on DNA replication origins in archaea, the third domain of life, commenced only approximately a decade ago. DNA replication origins have been mapped primarily for a few representatives of archaeal species distributed in the three main phyla, Euryarchaeota, Crenarchaeota, and Thaumarchaeota (Myllykallio et al., 2000; Lundgren et al., 2004; Robinson et al., 2004; Grainge et al., 2006; Norais et al., 2007; Majernik and Chong, 2008; Coker et al., 2009; Pelve et al., 2012, 2013; Wu et al., 2012, 2014). In addition, more detailed characterization has been revealed in several model systems, such as Pyrococcus species (Myllykallio et al., 2000; Matsunaga et al., 2001, 2003), Sulfolobus species (Lundgren et al., 2004; Robinson et al., 2004; Duggin et al., 2008; Samson et al., 2013), Haloferax volcanii (Norais et al., 2007; Hawkins et al., 2013) and Haloarcula hispanica (Wu et al., 2012, 2014). It is now known that archaea use a single or multiple origin(s) to replicate their circular chromosomes (Kelman and Kelman, 2004; Robinson and Bell, 2005; Hyrien et al., 2013). The multiple origins together with their adjacent initiator genes in certain archaeal chromosomes may have arisen from the capture of extrachromosomal elements and appear to be mosaics of distinct replicator–initiator pairings (Robinson and Bell, 2007; Wu et al., 2012).
This replicator–initiator system consists of an origin region and an initiator gene (the cdc6 gene in most cases and whiP in the oriC3 of Sulfolobus species). The origin region usually has a high content of adenine and thymine residues (AT-rich) flanked by several conserved repeated motifs known as origin recognition boxes (ORBs). The initiator protein Cdc6 (also denoted Orc or Orc1/Cdc6 in other papers) shows homology to both Orc1 and Cdc6 of eukaryotes and therefore is considered to be involved in both specific recognition of the origin region and loading of the minichromosome maintenance helicase (MCM; Robinson and Bell, 2005). Despite the conservation of the replicator-initiator structure, archaeal replication origins exhibit considerable diversity in terms of both the ORB elements within different origins and their adjacent initiator genes. The specificity of the initiator genes and their cognate origins was recently established by means of in silico and genetic analyses in Sulfolobus species (Samson et al., 2013) and Haloarcula hispanica (Wu et al., 2012, 2014). The cis organization of the replication origin and the initiator gene (replicator–initiator) is reminiscent of the oriC-dnaA system in bacteria. Recently, we revealed that bacterial-like control mechanisms may be used by different replication origins in haloarchaea, and a model has been proposed for the negative regulation of oriC2 by a downstream cluster of ORB elements in Haloarcula hispanica (Wu et al., 2014).
The goal of this review is to present an overview of progress made over the past decade in our understanding of DNA replication origins of archaeal genomes, including the identification (mapping), characterization and evolution of multiple replication origins on the chromosomes. We focus on the utilization and control of multiple replication origins in haloarchaea, as well as comparisons of replication origins from different archaeal species to draw the generality and evolution of multiple replication origins in archaea.
Identification (Mapping) of Replication Origins
Similar to bacteria, archaea have simple circular chromosomes (and also several extrachromosomal elements in some archaea); however, many archaea characterized to date harbor multiple replication origins. The approaches developed in bacteria or eukaryotes have been employed to investigate replication origins in archaea, such as GC-skew analysis (Myllykallio et al., 2000), the Z-curve method (Zhang and Zhang, 2003), autonomously replicating sequence (ARS) assay (Berquist and DasSarma, 2003; Norais et al., 2007; Wu et al., 2012), replication initiation point mapping (RIP mapping; Matsunaga et al., 2003), two-dimensional gel analysis (Matsunaga et al., 2001; Robinson et al., 2004), and marker frequency analysis (MFA; Lundgren et al., 2004; Coker et al., 2009; Pelve et al., 2012; Hawkins et al., 2013; Wu et al., 2014). DNA replication origins have been mapped in about a dozen archaeal species.
Single Replication Origin in Pyrococcus Species
The first description of DNA replication origins of archaeal genomes was reported by Myllykallio et al. (2000). These researchers identified a single replication origin (oriC) in Pyrococcus abyssi by means of cumulative skew of GGGT, and the study found that the oriC is flanked with the only cdc6 gene and several eukaryotic-like replication genes (Myllykallio et al., 2000). The origin organization was observed to be highly conserved in two other Pyrococcus species, Pyrococcus horikoshii and Pyrococcus furiosus (Myllykallio et al., 2000). The oriC was then experimentally confirmed via two-dimensional (2D) gel analysis (Matsunaga et al., 2001) and RIP mapping (Matsunaga et al., 2003), and the studies demonstrated that the oriC has several repeated sequences (now named ORBs) and is directly upstream of the cdc6 gene, reminiscent of the oriC-dnaA origin system in bacteria. Furthermore, the specific interaction of the Cdc6 protein with the oriC was detected via chromatin immunoprecipitation assays (Matsunaga et al., 2001). All of these data indicated that the circular chromosome of the Pyrococcus species is bidirectionally replicated from a bacterial mode of replication origin by eukaryotic-type machinery (Figure 1A).
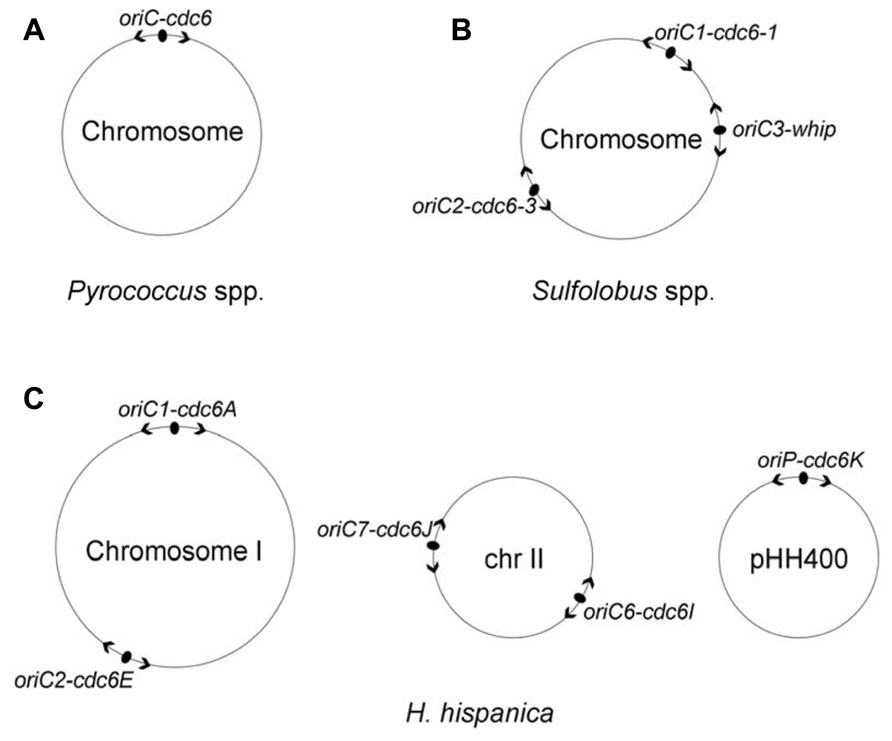
FIGURE 1. Distribution of DNA replication origins in three well-studied archaeal model systems, Pyrococcus species (A), Sulfolobus species (B) and Haloarcula hispanica (C). Origins are indicated with filled ovals, and arrowheads designate bidirectional replication. Replicator-initiator indicates that each origin is specified by its proximally encoded initiator. Both Pyrococcus species and Sulfolobus species contain a single chromosome; the chromosome of Pyrococcus species is replicated from a single origin (Myllykallio et al., 2000), whereas the chromosome of Sulfolobus species is replicated from three origins in near synchrony (Duggin et al., 2008). The Haloarcula hispanica genome consists of a main chromosome and two extrachromosomal elements with five active replication origins: oriC1-cdc6A and oriC2-cdc6E in the main chromosome I, oriC6-cdc6I and oriC7-cdc6J in the minichromosome II, and oriP-cdc6K in the megaplasmid pHH400 (Wu et al., 2012).
Three Replication Origins in Sulfolobus Species
The first example of archaeal chromosomes with multiple replication origins was the identification of three replication origins in the single chromosome of Sulfolobus species using 2D gel analysis (Robinson et al., 2004, 2007) and microarray-based MFA (Lundgren et al., 2004), and the results demonstrated that bidirectional replication initiated from three origins in both Sulfolobus acidocaldarius and Sulfolobus solfataricus (oriC1, oriC2, and oriC3; Figure 1B). The oriC1 and oriC2, in each species, are located directly upstream of cdc6-1 and cdc6-3, respectively, which have previously been identified by 2D gel electrophoresis in S. solfataricus (Robinson et al., 2004). The third origin, oriC3, is adjacent to the whiP (Winged-helix initiator protein) gene that is related to the eukaryotic cdt1 gene. An origin comparison between Aeropyrum and Sulfolobus suggested that the oriC3-whiP might have originated from the capture of extrachromosomal elements (Robinson and Bell, 2007). Using synchronized cultures, MFA results indicated that all three origins fire once per cell cycle and are initiated in near synchrony but with a slightly later activation of oriC2 (Lundgren et al., 2004; Duggin et al., 2008). Recently, three replication origins were also mapped in another Sulfolobus species, Sulfolobus islandicus, and a combination of genetic and MF analyses demonstrated that the three origins are specifically dependent on their adjacent initiator genes (two cdc6 genes and one whiP gene; Samson et al., 2013).
Multiple Replication Origins in Haloarchaea
Haloarchaeal genomes are generally composed of multiple genetic elements (chromosome, minichromosome, and megaplasmids) with multiple Cdc6 homologs (Capes et al., 2011), which is suggestive of the occurrence of multiple replication origins. Recently, multiple replication origins were predicted in 15 completely sequenced haloarchaeal genomes by searching for putative ORBs associated with cdc6 genes (Wu et al., 2012), and active replication origins have been experimentally studied in three model systems, Halobacterium sp. NRC-1 (Berquist and DasSarma, 2003; Coker et al., 2009), Haloferax volcanii (Norais et al., 2007; Hawkins et al., 2013) and Haloarcula hispanica (Wu et al., 2012, 2014).
The first prediction of multiple DNA replication origins in haloarchaeal genomes came from Z curve method analysis of the genome of Halobacterium sp. NRC-1, which revealed two cdc6-adjacent replication origins in its chromosome (Zhang and Zhang, 2003). However, only one replication origin was verified to have ARS activity (Berquist and DasSarma, 2003). Whole-genome MFA was employed to map the activation of replication origins in vivo in Halobacterium sp. NRC-1, which demonstrated multiple discrete origin sites in the chromosome, with two being located in the vicinity of cdc6 genes (denoted orc7 and orc10 in the original paper; Coker et al., 2009).
Eleven cdc6 genes are encoded in Haloarcula hispanica, and eight of them have surrounding ORB-like elements. ARS activity assays demonstrated that only five predicted origins, oriC1-cdc6A and oriC2-cdc6E in the main chromosome, oriC6-cdc6I, and oriC7-cdc6J in the minichromosome and oriP-cdc6K in the megaplasmid (pHH400), were able to confer replication ability to a non-replicating plasmid (Figure 1C; Wu et al., 2012). Recently, we combined extensive gene deletion and microarray-based MFA to map the activation of replication origins in vivo in Haloarcula hispanica, demonstrating that the chromosome is bidirectionally replicated from the two initially proven origins, oriC1-cdc6A, and oriC2-cdc6E (Wu et al., 2014). Importantly, our results indicated that one active ori-cdc6 pairing on each replicon, i.e., oriC1-cdc6A or oriC2-cdc6E on the chromosome, oriC6-cdc6I or oriC7-cdc6J on the minichromosome, and oriP-cdc6K on pHH400, is essential for genome replication in Haloarcula hispanica (Wu et al., 2014).
Five replication origins were initially identified in Haloferax volcanii using a combination of bioinformatics and genetic approaches: two within the chromosome and one each within the three megaplasmids pHV1, pHV3, and pHV4 (Norais et al., 2007). Recently, aside from the previously identified origins, a sixth replication origin was mapped in the chromosome via high-throughput sequencing-based MFA (Hawkins et al., 2013). All six replication origins are adjacent to cdc6 genes. Furthermore, four chromosomal replication origins were mapped in the laboratory H26 strain with integration of pHV4 into the chromosome (Hawkins et al., 2013). Surprisingly, the four origins can be deleted simultaneously, and in the absence of these replication origins, the strain even grew 7.5% faster than the wild-type strain (Hawkins et al., 2013). Because the radA gene (the archaeal recA/rad51 homologue) was determined to be essential in the absence of all four origins, the authors proposed that the replication of the origin-less Haloferax volcanii chromosome is dependent on homologous recombination (Hawkins et al., 2013). However, this mode of recombination-dependent replication of the Haloferax volcanii chromosome was not yet observed in other investigated archaea. In contrast, at least one active replication origin has been proven to be essential for chromosome replication in Haloarcula hispanica (Wu et al., 2014), and triple-deletion mutant was not available for the three initiators in the chromosome of S. islandicus (Samson et al., 2013). It would be interesting to investigate how the RadA-dependent replication (if any) efficiently replicates the Haloferax volcanii chromosome, or if there are undetected replication origins functioned in the chromosome lacking the main origins.
Mapping of Replication Origins in Other Archaea
DNA replication origins have been well-defined in several bacterial model systems, and have been predicted and/or identified in more than 1300 bacterial genomes (Gao and Zhang, 2007, 2008). Similarly, to understand the general nature of replication origins in archaea, it is necessary to determine and compare replication origins from a broad selection of archaeal species. Fortunately, the genomes of 100s of archaea distributed in different phyla have been sequenced and are publically available, allowing the prediction and mapping of replication origins in these genomes. To date, replication origins have been demonstrated in a dozen archaeal species. Similar to Pyrococcus species, Archaeoglobus fulgidus has been shown to contain a single replication origin (Maisnier-Patin et al., 2002). Two replication origins have been identified in Aeropyrum pernix by using a combination of biochemical and two-dimensional gel electrophoresis (Grainge et al., 2006; Robinson and Bell, 2007). Studies of DNA replication in methanogens have demonstrated that a single origin is responsible for replication initiation of the chromosome of Methanothermobacter thermautotrophicus (Capaldi and Berger, 2004; Majernik and Chong, 2008). Recently, four replication origins were mapped in the single chromosome of Pyrobaculum calidifontis via high-throughput sequencing-based MFA (Pelve et al., 2012). To generate a broader view of modes of origin replication in archaea, Pelve et al. (2013) further completed origin mapping in a thaumarchaeon, revealing a single replication origin in the Nitrosopumilus maritimus chromosome.
Distinct Replicator-Initiator Systems in Archaea
The initiator protein DnaA is highly conserved in bacteria, and bacterial replication origins generally possess conserved sequence elements, DnaA boxes. In contrast, the three replication origins in Sulfolobus species differ from each other. Each of the three origins is specifically recognized by its proximally encoded initiator protein, two distinct Cdc6 proteins and WhiP (Dueber et al., 2011; Samson et al., 2013). In addition, the recognition mechanisms appear to be different, as classic ORB and its shorter version (miniORB) are, respectively, observed in the oriC1 and oriC2 regions, while neither is observed in the oriC3 region (Robinson et al., 2004; Samson et al., 2013).
Haloarchaeal genomes generally contain multiple cdc6 genes and replication origins. Recently, we conducted a comparison of the origin-associated Cdc6 homologs and the corresponding predicted ORB elements. Our results suggested that the replication origins from haloarchaeon are notably diverse in terms of ORB elements and their adjacent cdc6 genes, which could be sorted into distinct families. Based on this phylogenetic analysis, linkage-specificity of Cdc6 homologs and the corresponding ORB elements was proposed, suggestive of their specific interaction (Wu et al., 2012). Very recently, we employed comprehensive genetic studies to investigate the specificity of multiple replication origins and cdc6 genes in Haloarcula hispanica, and our results indicated that each Cdc6 protein specifically recognizes its proximal origin (Wu et al., 2014). Thus, multiple replication origins along with their adjacent cdc6 genes appear to be distinct ori-cdc6 systems. These distinct ori-cdc6 systems in haloarchaeon may have many evolutionary advantages: first, it ensures the compatibility of multiple replication origins, which accounts for the observations that multiple Cdc6 proteins from a haloarchaeal genome are distributed into different families (Wu et al., 2012) and that the oriC2-containing plasmid is incompatible with Haloarcula hispanica (Wu et al., 2014); second, distinct ori-cdc6 pairings help minimize competition among multiple origins for initiators and maintain independent control of replication initiation at different origins. Importantly, as haloarchaeal genomes generally contain multiple replicons, distinct ori-cdc6 origins may be favorable for replicon-specific replication control, similar to the different modes of replication origin adopted by the two chromosomes of Vibrio cholerae (Egan and Waldor, 2003).
To understand the molecular mechanisms involved in the specific recognition of origins by initiators, the structures of two origin-bound Cdc6 proteins from Aeropyrum pernix (Gaudier et al., 2007) and S. solfataricus (Dueber et al., 2007) were crystallized. Both of the two Cdc6 proteins contain an N-terminal AAA+ domain and a C-terminal WH domain. Intriguingly, both of the studies demonstrated that, in addition to the canonical DNA binding WH domain, the AAA+ domains of these two initiators are responsible for recognizing origins (Dueber et al., 2007; Gaudier et al., 2007). Subsequently, biochemical data also demonstrated that both the WH domain and AAA+ domain contribute to the origin-binding specificity of the Cdc6 protein (Dueber et al., 2011).
Control of Replication Initiation at Multiple Origins in Archaea
Multiple mechanisms that regulate replication initiation have been well-characterized in both bacteria and unicellular eukaryotes, and are summarized in a number of excellent reviews (Mott and Berger, 2007; Mechali, 2010; Rajewska et al., 2012; Aparicio, 2013). In contrast, the mechanisms by which archaea regulate replication initiation at multiple origins, either on the same chromosome or from different genetic elements, are far less understood. All of the archaeal replication origins characterized to date are dependent on their adjacent initiator gene (the cdc6 gene in most cases; Samson et al., 2013; Wu et al., 2014), and these distinct ori-cdc6 pairings may contribute to their independent control. In addition, the cis location of the cdc6 gene and the origin is proved to not be required for ARS activity in both Haloferax volcanii and Haloarcula hispanica (Norais et al., 2007; Wu et al., 2014). Therefore, we have proposed that direct linkage of the initiator gene to the origin may facilitate its transcription after replication initiation to sequentially control its cognate origin.
Using the Haloarcula hispanica model system, we suggested that some bacterial-like mechanisms may be employed at different replication origins in haloarchaea (Wu et al., 2014). A G-rich inverted-repeat directly inside each ORB element of Haloarcula hispanica oriC1 was shown to be a replication enhancer that stimulated origin activation at oriC1. Because of the repeat’s close location to ORB elements, we proposed that the G-rich inverted-repeat enhances the binding of initiator or regulatory factors at oriC1, similar to many repeated sequences in bacteria that are binding sites for initiation proteins or regulatory factors, playing a crucial role in the control of replication initiation (Rajewska et al., 2012). In addition, a model has been proposed, and partly tested, for the negative regulation of oriC2 by a downstream cluster of Cdc6 binding elements (ORBs), likely via Cdc6E titration, similar to the negative control of replication initiation via a datA locus exhibiting DnaA-titrating activity in E. coli (Kitagawa et al., 1998). More interestingly, many additional predicted replication origins have the oriC2-like structure, suggesting that this strategy of negative replication origin control is used generally by haloarchaea.
Despite the bacterial-like structure of archaeal replication origins, archaea use eukaryotic-type replication machinery (Robinson and Bell, 2005), indicating that archaea may adopt eukaryotic-like mechanisms to control replication proteins and thus replication initiation. Interestingly, genome-wide transcription mapping indicated that serine–threonine protein kinases show cyclic induction in Sulfolobus species, indicating that regulatory factors similar to eukaryotic cyclin-dependent kinase (CDK) complexes may be present in archaea (Lundgren and Bernander, 2007). Recently, an ATP-ADP binary switch model for Cdc6-mediated replication control was proposed in S. islandicus, postulating that binding of ATP remodels Cdc6 conformation for efficient MCM recruitment, and subsequent ATP hydrolysis renders Cdc6 incapable of further recruiting MCM (Samson et al., 2013). In addition, as almost all replication origins are dependent on Cdc6 proteins, conformational changes of Cdc6 proteins may play important roles in coordinating replication initiation at different origins within a cell.
Evolution of Multiple Replication Origins in Archaea
Although considerable diversity of replication origins has been observed in haloarchaea, comparison analysis revealed a conserved replication origin, oriC1, which is positioned in the main chromosome of all analyzed haloarchaeal genomes (Coker et al., 2009; Wu et al., 2012). Both the ORBs within oriC1 and the oriC1-associated Cdc6 homologs are highly conserved. In addition, gene order analysis found that genes around oriC1 are highly syntenic among haloarchaea (Figure 2; Capes et al., 2011). Notably, other studies (Robinson et al., 2004; Coker et al., 2009) and our results indicated that the oriC1 replication origin is broadly conserved in archaea, in terms of both function and structure, which strongly suggested that the ancestral chromosome was dependent on oriC1. Variations were observed in oriC1 homologs from different archaeal phyla, which may contribute to the adaptability of archaea to different extreme environments. For example, an extended halophile-specific “G-string” element has been identified at the end of each ORB in haloarchaea, and these “G-string” elements have been proven to be essential for autonomous replication based on the oriC1 in Haloarcula hispanica (Wu et al., 2014).
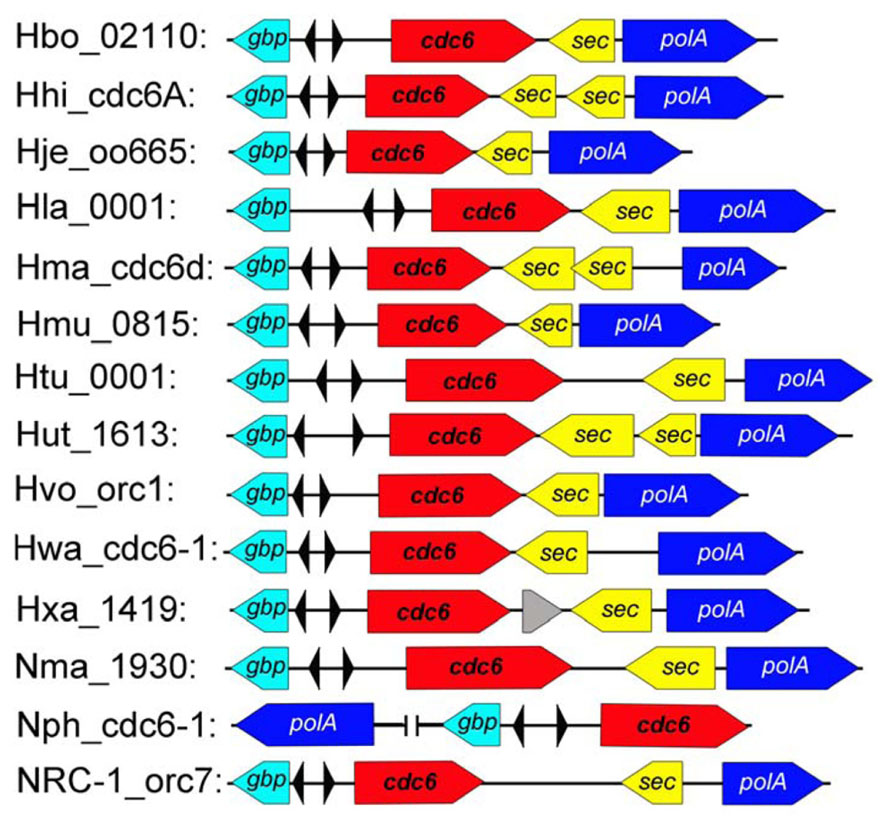
FIGURE 2. The conserved oriC1 origin of replication in sequenced haloarchaeal genomes. The oriC1 context region was mapped as shown in the sequenced haloarchaea. The colored boxed arrows represent different genes as follows: GTP-binding protein (gbp, teal), initiator protein (cdc6, red), signal sequence peptidase (sec, yellow) and DNA-directed DNA polymerase (polA, blue). The inverted ORB elements are indicated by small triangles.
Multiple replication origins along with their adjacent cdc6 genes appear to be mosaics of distinct replicator–initiator systems. A comparison between Aeropyrum and Sulfolobus origins suggested that the capture of extrachromosomal elements accounts for replicon evolution (Robinson and Bell, 2007). In particular, it has been proposed that the three replication origins of the Sulfolobus species arose by the integration of extrachromosomal elements into a single-origin ancestral chromosome (oriC1-cdc6-1), and the acquisition of oriC3-whiP occurred prior to the integration of oriC2-cdc6-3 (Samson et al., 2013). Similarly, genomic context analyses of ori-cdc6 systems in haloarchaea revealed that 40% of predicted replication origins were observed with transposases or integrases nearby, indicative of the translocation of a subset of replication origins among haloarchaea. In addition, comparative analyses of the selected replication origins suggested that different evolutionary mechanisms, including ancestral conservation and coupled acquisition and deletion events, may account for the current mosaics of multiple replication origins in the haloarchaeal genomes. Importantly, a comparative genomic analysis of two Haloarcula species, Haloarcula hispanica and Haloarcula marismortui, revealed that the species-specific origins are located in extremely variable regions, suggesting that these novel origins were recently acquired, via either integration into the chromosome or rearrangement of extrachromosomal elements (Wu et al., 2012). Further work may focus on comparisons of replication origins from closely related species to reveal the dynamics of origin evolution and whether origin evolution alters the mode of genomic replication.
Perspectives
To date, the number of archaea with mapped replication origins is still limited, which to some extent has affected us to get a panoramic view of the generality and evolution of replication origins in archaea. In addition to the mapping of replication origins, the development of prediction algorithms for replication origins in archaeal genomes and the construction of databases with these predicted origins (Gao et al., 2013) will be useful for comparing replication origins from a broader range of archaeal species. Fortunately, the rapid increase in the number of complete archaeal genomic sequences that are publically available will promote our studies of archaeal replication origins.
In addition, the control and coordination of replication initiation at multiple origins in archaea is far less understood. The multireplicon structure of haloarchaeal genomes allows for precise control and coordination of replication initiation at multiple origins. As the chromosome and extrachromosomal elements within a haloarchaeon are generally different sizes and have different copy numbers (Breuert et al., 2006; Liu et al., 2013), it will be interesting to reveal whether they initiate synchronously and how they maintain different copy numbers, as well as what roles multiple replication origins play in governing polyploidy in haloarchaea. In addition, the coordination of multiple origins may play important roles in maintaining the multireplicon structure of haloarchaeal genomes. As most replication origins are dependent on Cdc6 proteins in haloarchaea (excluding the origins of small plasmids), we propose that the coordination of replication initiation at different origins may be obtained by conformational changes of Cdc6 proteins via an ATP-ADP binary switch, which has recently been proposed for chromosome replication in S. islandicus (Samson et al., 2013). Thus, more exhaustive work should be taken into account to uncover the control and coordination of the replication initiation from multiple origins, either on the same chromosome or from different genetic elements, in haloarchaeal multireplicon genomes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was partially supported by grants from the National Natural Science Foundation of China (30925001, 31100893, 31271334).
References
Aparicio, O. M. (2013). Location, location, location: it’s all in the timing for replication origins. Genes Dev. 27, 117–128. doi: 10.1101/gad.209999.112
Berquist, B. R., and DasSarma, S. (2003). An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. J. Bacteriol. 185, 5959–5966. doi: 10.1128/JB.185.20.5959-5966.2003
Breuert, S., Allers, T., Spohn, G., and Soppa, J. (2006). Regulated polyploidy in halophilic archaea. PLoS ONE 1:e92. doi: 10.1371/journal.pone.0000092
Capaldi, S. A., and Berger, J. M. (2004). Biochemical characterization of Cdc6/Orc1 binding to the replication origin of the euryarchaeon Methanothermobacter thermoautotrophicus. Nucleic Acids Res. 32, 4821–4832. doi: 10.1093/nar/gkh819
Capes, M. D., Coker, J. A., Gessler, R., Grinblat-Huse, V., Dassarma, S. L., Jacob, C. G., et al. (2011). The information transfer system of halophilic archaea. Plasmid 65, 77–101. doi: 10.1016/j.plasmid.2010.11.005
Coker, J. A., Dassarma, P., Capes, M., Wallace, T., Mcgarrity, K., Gessler, R., et al. (2009). Multiple replication origins of Halobacterium sp. strain NRC-1: properties of the conserved orc7-dependent oriC1. J. Bacteriol. 191, 5253–5261. doi: 10.1128/JB.00210-09
Dueber, E. C., Costa, A., Corn, J. E., Bell, S. D., and Berger, J. M. (2011). Molecular determinants of origin discrimination by Orc1 initiators in archaea. Nucleic Acids Res. 39, 3621–3631. doi: 10.1093/nar/gkq1308
Dueber, E. L., Corn, J. E., Bell, S. D., and Berger, J. M. (2007). Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science 317, 1210–1213. doi: 10.1126/science.1143690
Duggin, I. G., Mccallum, S. A., and Bell, S. D. (2008). Chromosome replication dynamics in the archaeon Sulfolobus acidocaldarius. Proc. Natl. Acad. Sci. U.S.A. 105, 16737–16742. doi: 10.1073/pnas.0806414105
Egan, E. S., and Waldor, M. K. (2003). Distinct replication requirements for the two Vibrio cholerae chromosomes. Cell 114, 521–530. doi: 10.1016/S0092-8674(03)00611-1
Gao, F., Luo, H., and Zhang, C. T. (2013). DoriC 5.0: an updated database of oriC regions in both bacterial and archaeal genomes. Nucleic Acids Res. 41, D90–D93. doi: 10.1093/nar/gks990
Gao, F., and Zhang, C. T. (2007). DoriC: a database of oriC regions in bacterial genomes. Bioinformatics 23, 1866–1867. doi: 10.1093/bioinformatics/btm255
Gao, F., and Zhang, C. T. (2008). Ori-Finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinformatics 9:79. doi: 10.1186/1471-2105-9-79
Gaudier, M., Schuwirth, B. S., Westcott, S. L., and Wigley, D. B. (2007). Structural basis of DNA replication origin recognition by an ORC protein. Science 317, 1213–1216. doi: 10.1126/science.1143664
Grainge, I., Gaudier, M., Schuwirth, B. S., Westcott, S. L., Sandall, J., Atanassova, N., et al. (2006). Biochemical analysis of a DNA replication origin in the archaeon Aeropyrum pernix. J. Mol. Biol. 363, 355–369. doi: 10.1016/j.jmb.2006.07.076
Hawkins, M., Malla, S., Blythe, M. J., Nieduszynski, C. A., and Allers, T. (2013). Accelerated growth in the absence of DNA replication origins. Nature 503, 544–547. doi: 10.1038/nature12650
Hyrien, O., Rappailles, A., Guilbaud, G., Baker, A., Chen, C. L., Goldar, A., et al. (2013). From simple bacterial and archaeal replicons to replication N/U-domains. J. Mol. Biol. 425, 4673–4689. doi: 10.1016/j.jmb.2013.09.021
Jacob, F., Cuzin, F., and Brenner, S. (1963). On Regulation of DNA Replication in Bacteria. Cold. Spring Harb. Symp. Quant. Biol. 28, 329–348. doi: 10.1101/SQB.1963.028.01.048
Kelman, L. M., and Kelman, Z. (2004). Multiple origins of replication in archaea. Trends Microbiol. 12, 399–401. doi: 10.1016/j.tim.2004.07.001
Kitagawa, R., Ozaki, T., Moriya, S., and Ogawa, T. (1998). Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 12, 3032–3043. doi: 10.1101/gad.12.19.3032
Leonard, A. C., and Mechali, M. (2013). DNA replication origins. Cold Spring Harb. Perspect. Biol. 5:a010116. doi: 10.1101/cshperspect.a010116
Liu, X., Miao, D., Zhang, F., Wu, Z., Liu, J., and Xiang, H. (2013). Characterization of the minimal replicon of pHM300 and independent copy number control of major and minor chromosomes of Haloferax mediterranei. FEMS Microbiol. Lett. 339, 66–74. doi: 10.1111/1574-6968.12052
Lundgren, M., Andersson, A., Chen, L., Nilsson, P., and Bernander, R. (2004). Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc. Natl. Acad. Sci. U.S.A. 101, 7046–7051. doi: 10.1073/pnas.0400656101
Lundgren, M., and Bernander, R. (2007). Genome-wide transcription map of an archaeal cell cycle. Proc. Natl. Acad. Sci. U.S.A. 104, 2939–2944. doi: 10.1073/pnas.0611333104
Maisnier-Patin, S., Malandrin, L., Birkeland, N. K., and Bernander, R. (2002). Chromosome replication patterns in the hyperthermophilic euryarchaea Archaeoglobus fulgidus and Methanocaldococcus (Methanococcus) jannaschii. Mol. Microbiol. 45, 1443–1450. doi: 10.1046/j.1365-2958.2002.03111.x
Majernik, A. I., and Chong, J. P. (2008). A conserved mechanism for replication origin recognition and binding in archaea. Biochem. J. 409, 511–518. doi: 10.1042/BJ20070213
Matsunaga, F., Forterre, P., Ishino, Y., and Myllykallio, H. (2001). In vivo interactions of archaeal Cdc6/Orc1 and minichromosome maintenance proteins with the replication origin. Proc. Natl. Acad. Sci. U.S.A. 98, 11152–11157. doi: 10.1073/pnas.191387498
Matsunaga, F., Norais, C., Forterre, P., and Myllykallio, H. (2003). Identification of short “eukaryotic” Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Rep. 4, 154–158. doi: 10.1038/sj.embor.embor732
Mechali, M. (2010). Eukaryotic DNA replication origins: many choices for appropriate answers. Nat. Rev. Mol. Cell Biol. 11, 728–738. doi: 10.1038/nrm2976
Messer, W. (2002). The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 26, 355–374.
Mott, M. L., and Berger, J. M. (2007). DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 5, 343–354. doi: 10.1038/nrmicro1640
Myllykallio, H., Lopez, P., Lopez-Garcia, P., Heilig, R., Saurin, W., Zivanovic, Y., et al. (2000). Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science 288, 2212–2215. doi: 10.1126/science.288.5474.2212
Norais, C., Hawkins, M., Hartman, A. L., Eisen, J. A., Myllykallio, H., and Allers, T. (2007). Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PLoS Genet. 3:e77. doi: 10.1371/journal.pgen.0030077
Pelve, E. A., Lindas, A. C., Knoppel, A., Mira, A., and Bernander, R. (2012). Four chromosome replication origins in the archaeon Pyrobaculum calidifontis. Mol. Microbiol. 85, 986–995. doi: 10.1111/j.1365-2958.2012.08155.x
Pelve, E. A., Martens-Habbena, W., Stahl, D. A., and Bernander, R. (2013). Mapping of active replication origins in vivo in thaum- and euryarchaeal replicons. Mol. Microbiol. 90, 538–550. doi: 10.1111/mmi.12382
Rajewska, M., Wegrzyn, K., and Konieczny, I. (2012). AT-rich region and repeated sequences – the essential elements of replication origins of bacterial replicons. FEMS Microbiol. Rev. 36, 408–434. doi: 10.1111/j.1574-6976.2011.00300.x
Robinson, N. P., and Bell, S. D. (2005). Origins of DNA replication in the three domains of life. FEBS J. 272, 3757–3766. doi: 10.1111/j.1742-4658.2005.04768.x
Robinson, N. P., and Bell, S. D. (2007). Extrachromosomal element capture and the evolution of multiple replication origins in archaeal chromosomes. Proc. Natl. Acad. Sci. U.S.A. 104, 5806–5811. doi: 10.1073/pnas.0700206104
Robinson, N. P., Blood, K. A., McCallum, S. A., Edwards, P. A., and Bell, S. D. (2007). Sister chromatid junctions in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO J. 26, 816–824. doi: 10.1038/sj.emboj.7601529
Robinson, N. P., Dionne, I., Lundgren, M., Marsh, V. L., Bernander, R., and Bell, S. D. (2004). Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell 116, 25–38. doi: 10.1016/S0092-8674(03)01034-1
Samson, R. Y., Xu, Y., Gadelha, C., Stone, T. A., Faqiri, J. N., Li, D., et al. (2013). Specificity and function of archaeal DNA replication initiator proteins. Cell Rep. 3, 485–496. doi: 10.1016/j.celrep.2013.01.002
Wu, Z., Liu, H., Liu, J., Liu, X., and Xiang, H. (2012). Diversity and evolution of multiple orc/cdc6-adjacent replication origins in haloarchaea. BMC Genomics 13:478. doi: 10.1186/1471-2164-13-478
Wu, Z., Liu, J., Yang, H., Liu, H., and Xiang, H. (2014). Multiple replication origins with diverse control mechanisms in Haloarcula hispanica. Nucleic Acids Res. 42, 2282–2294. doi: 10.1093/nar/gkt1214
Zakrzewska-Czerwinska, J., Jakimowicz, D., Zawilak-Pawlik, A., and Messer, W. (2007). Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol. Rev. 31, 378–387. doi: 10.1111/j.1574-6976.2007.00070.x
Keywords: DNA replication origin, origin recognition box, archaea, control, evolution, haloarchaea
Citation: Wu Z, Liu J, Yang H and Xiang H (2014) DNA replication origins in archaea. Front. Microbiol. 5:179. doi: 10.3389/fmicb.2014.00179
Received: 27 February 2014; Accepted: 01 April 2014;
Published online: 29 April 2014.
Edited by:
Feng Gao, Tianjin University, ChinaReviewed by:
Jonathan H. Badger, J. Craig Venter Institute, USAQunxin She, University of Copenhagen, Denmark
Copyright © 2014 Wu, Liu, Yang and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Xiang and Jingfang Liu, State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, No. 1 Beichen West Road, Chaoyang District, Beijing 100101, China e-mail:eGlhbmdoQGltLmFjLmNu;bGl1amZAaW0uYWMuY24=
 Zhenfang Wu
Zhenfang Wu Jingfang Liu
Jingfang Liu Haibo Yang
Haibo Yang Hua Xiang
Hua Xiang