- Department of Molecular Microbiology, School of Biotechnology, Madurai Kamaraj University, Madurai, India
Pathogenic microorganisms have evolved sophisticated mechanisms to inactivate antibiotics and rendered an urgent need for new antibiotics that would target the emerging multidrug-resistance (Butler et al., 2013). Consequently, search for novel sources of potent antibiotics is desperately needed to develop potent drugs. Microbial resources have made an incredible contribution to the antibiotic drug discovery and development process over the last seven decades (Demain and Sanchez, 2009). In particular, actinomycetes are the most important source of bioactive natural compounds with a long track record of producing novel molecules, including several commercially important drugs like Streptomycin, Gentamicin, Vancomycin, Clindamycin, Erythromycin, Amphotericin, Rifampicin, and Tetracycline. More novel molecules with potential therapeutic applications are still on the row to be discovered from these actinomycetes (Baltz, 2007).
Microbial assemblages and their species distributions in specific environments are mostly determined by their specific environmental conditions, which may translate into novel chemistry (Genilloud et al., 2011). The actinomycetes that produce antibiotics are abundant in soil. In recent decades, isolation and exploitation of actinomycetes for novel compounds from conventional environments have led to rediscovery of known compounds (Walsh, 2003; Tulp and Bohlin, 2005). However, diverse actinomycetes from poorly studied unusual environments promises a raise in the prospect of discovering novel compounds with potential activities that can be developed as a resource for drug discovery (Peraud et al., 2009; Jose et al., 2011; Poulsen et al., 2011; Carr et al., 2012; Subramani and Aalbersberg, 2012; Hamedi et al., 2013; Jose et al., 2013; Yuan et al., 2014). In this respect, current actinomycetes isolation programs are reoriented toward largely unexplored, unusual and extreme environments like hypersaline marine environments, extreme inland saline zones, volcanic zones, hyperarids and glaciers (Hamedi et al., 2013; Jose and Jebakumar, 2013b).
The extreme habitats are characterized by chemical or physical conditions that differ significantly from those found in environments that support more abundant and varied life forms (Zhao et al., 2011). Hypersaline environments are typical extreme habitats, in which the high salt concentration, alkalinity, and low oxygen concentrations are environmental factors that may limit their biodiversity (Ventosa, 2006). There are several reports on inhabitance of microorganisms including actinomycetes in diverse hypersaline environments such as salt lakes (Swan et al., 2010; Guan et al., 2011; Phillips et al., 2012), solar salterns (Baati et al., 2008; Sabet et al., 2009), salt mines (Chen et al., 2007) and brine wells (Xiang et al., 2008). However, these hypersaline environments remain largely unexplored as a source of novel actinomycetes in many tropical and subtropical regions throughout the world. In a recent review, Hamedi et al. (2013) described that actinomycetes form a stable, metabolically active and persistent population in various marine hypersaline ecosystem and it is accepted that halophilic actinomycetes will provide a valuable resource for novel products of industrial interest, including antimicrobial, cytotoxic, neurotoxic, antimitotic, antiviral and antineoplastic activities. Series of bioactive metabolites like Pyrostatins, Salinosporamides, Abyssomicins, Trioxacarcin A, Gutingi- mycin, Sporolides, Marinomycins, Himal- omycins, Diazepinomicin, Helquinoline, Lajollamycin, Tetrodotoxin, Mecherchar- mycins, Cyanosporasides, Erythronolides, and Ammosamide D have been reported from this physiological group of actinomycetes (Hamedi et al., 2013). In continued efforts to discover novel natural products from new extremophilic actinomycetes, Zhao et al. (2011) discovered new linear polyketides, actinopolysporins A, B, and C, as well as the known antitneoplastic antibiotic tubercidin from the halophilic actinomycete Actinopolyspora erythraea YIM 90600. Tian et al. (2013) reported the isolation and characterization of p-Terphenyls with antifungal, antibacterial, and antioxidant activities from halophilic actinomycete Nocardiopsis gilva YIM 90087. Jang et al. (2013) declared a significant discovery of new antibiotic, anthracimycin, which is produced by a marine-derived actinomycete in saline culture. More recently, an antimicrobial quinoline alkaloid has been isolated from a novel halophilic actinomycete, Nocardiopsis terrae YIM 90022 (Tian et al., 2014).
In accordance with the above reports, we initiated a research program which was designed to discover new natural products from novel actinomycetes originating from previously unexplored hypersaline environments. Saltpan soil samples were collected aseptically from saltpans of coastal solar saltern ponds at Arumuganeri, Tuticorin (about Latitude 8.43 N and Longitude 78.60 E) and inland solar salterns at Sambhar Lake, Jaipur (about Latitude 26.94561 N and Longitude 75.20968 E) in India. Physicochemical characteristics from this study confirmed that this environment is a hypersaline zone. In the course of our program, diverse actinomycetes were isolated by using selective isolation methods which employed stamping, heat and dilution, and succeeded with isolation of both Streptomyces (common actinomycete) and non-Streptomyces including rare actinomycetes Nonomuraea, Actinoalloteichus and Pseudonocardia (Jose and Jebakumar, 2012, 2013a). A total of 83 actinomycetes were isolated and assigned to eight genera (Streptomyces, Micromonospora, Nocardia, Nocardiopsis, Nonomuraea, Saccharopolyspora, Pseudonocardia and Actinoalloteichus) on the basis of their 16S rDNA sequences and it was the first step toward better understanding of actinomycete community from solar saltern ponds in India. Our preliminary biological activity screening and subsequent structural characterisation experiments suggested the occurrence of antimicrobial compounds producing Streptomyces and rare actinomycetes and confirmed that hypersaline solar salterns are reservoirs of antibiotic producing actinomycetes (Jose and Jebakumar, 2013a,b). The range of antimicrobial activities exerted by different actinomycete isolates were summarized in Table 1. In concise, our continuing study contributes in acquaintance of solar saltern associated actinobacteria and further augments the array of actinomycetes available for antibiotic discovery programs.
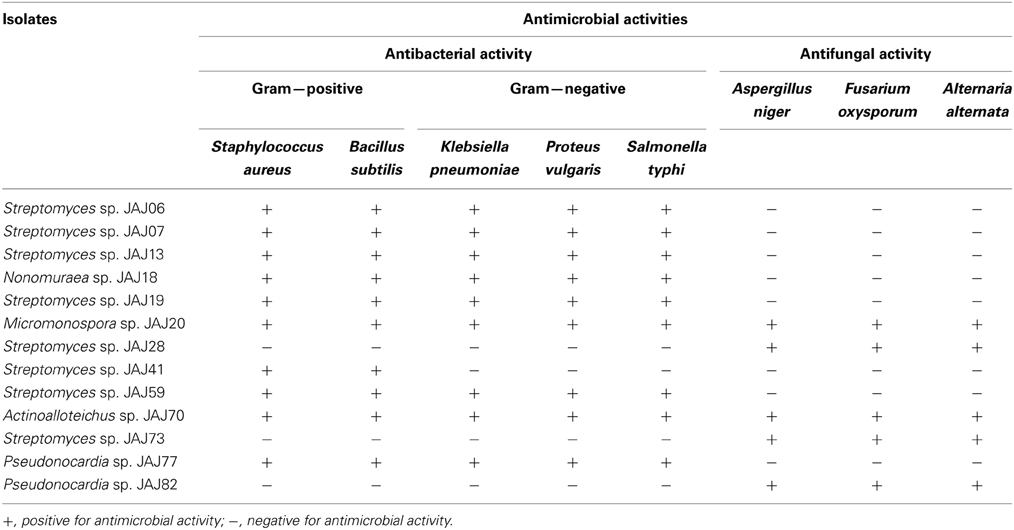
Table 1. Range of antimicrobial activities of the actinomycete isolates obtained from Indian inland (Jose and Jebakumar, 2013a) and coastal (Jose and Jebakumar, 2013b) solar salterns.
-In conclusion, escalating reports on discovery of diverse natural compounds from halophilic and halotolerant actinomycetes which inhabit in hypersaline environments suggested that this physiological group has enormous capacity to produce array of secondary metabolites with disparate activities. Indeed, hypersaline environments warrant significant attention as habitats of novel actinomycetes and unlock new avenues for natural products discovery.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Our research program on actinomycetes is being supported by University Grants Commission (UGC) and Council of Scientific and Industrial Research (CSIR), India.
References
Baati, H., Guermazi, S., Amdouni, R., Gharsallah, N., Sghir, A., and Ammar, E. (2008). Prokaryotic diversity of a Tunisian multipond solar saltern. Extremophiles 4, 505–518. doi: 10.1007/s00792-008-0154-x
Butler, M. S., Blaskovich, M. A., and Cooper, M. A. (2013). Antibiotics in the clinical pipeline in 2013. J. Antibiot. 66, 571–591. doi: 10.1038/ja.2013.86
Carr, G., Poulsen, M., Klassen, J. L., Hou, Y., Wyche, T. P., Bugni, T. S., et al. (2012). Microtermolides A and B from Termite-sssociated Streptomyces sp. and structural revision of vinylamycin. Org. Lett. 14, 2822–2825. doi: 10.1021/ol301043p
Chen, Y. G., Cui, X. L., Pukall, R., Li, H. M., Yang, Y. L., Xu, L. H., et al. (2007). Salinicoccus kunmingensis sp. nov., a moderately halophilic bacterium isolated from a salt mine in Yunnan, south-west China. Int. J. Syst. Evol. Microbiol. 57, 2327–2332. doi: 10.1099/ijs.0.64783-0
Demain, A. L., and Sanchez, S. (2009). Microbial drug discovery: 80 years of progress. J. Antibiot. 62, 5–16. doi: 10.1038/ja.2008.16
Genilloud, O., González, I., Salazar, O., Martín, J., Tormo, J. R., and Vicente, F. (2011). Current approaches to exploit actinomycetes as a source of novel natural products. J. Ind. Microbiol. Biotechnol. 38, 375–389. doi: 10.1007/s10295-010-0882-7
Guan, T. W., Wu, N., Xia, Z. F., Ruan, J. S., and Zhang, X. P. (2011). Saccharopolyspora lacisalsi sp. nov., a novel halophilic actinomycete isolated from a salt lake in Xinjiang, China. Extremophiles 15, 373–378. doi: 10.1007/s00792-011-0369-0
Hamedi, J., Mohammadipanah, F., and Ventosa, A. (2013). Systematic and biotechnological aspects of halophilic and halotolerant actinomycetes. Extremophiles 17, 1–13. doi: 10.1007/s00792-012-0493-5
Jang, K. H., Nam, S.-J., Locke, J. B., Kauffman, C. A., Beatty, D. S., Paul, L. A., et al. (2013). Anthracimycin, a potent Anthrax antibiotic from a marine-derived actinomycete. Angew. Chem. Int. Ed. Engl. 52, 7822–7824. doi: 10.1002/anie.201302749
Jose, P. A., and Jebakumar, S. R. D. (2012). Phylogenetic diversity of actinomycetes cultured from coastal multipond solar saltern in Tuticorin, India. Aquat. Biosyst. 8:23. doi: 10.1186/2046-9063-8-23
Jose, P. A., and Jebakumar, S. R. D. (2013a). Phylogenetic appraisal of antagonistic, slow growing actinomycetes isolated from hypersaline inland solar salterns at Sambhar salt Lake, India. Front. Microbiol. 4:190. doi: 10.3389/fmicb.2013.00190
Jose, P. A., and Jebakumar, S. R. D. (2013b). Diverse actinomycetes from Indian coastal solar salterns - a resource for antimicrobial screening. J. Pure Appl. Microbiol. 7, 2569–2575.
Jose, P. A., Santhi, V. S., and Jebakumar, S. R. D. (2011). Phylogenetic−affiliation, antimicrobial potential and PKS gene sequence analysis of moderately halophilic Streptomyces sp. inhabiting an Indian saltpan. J. Basic Microbiol. 51, 348–356. doi: 10.1002/jobm.201000253
Jose, P. A., Sivakala, K. K., and Jebakumar, S. R. D. (2013). Formulation and statistical optimization of culture medium for improved production of antimicrobial compound by Streptomyces sp. JAJ06. Int. J. Microbiol. 2013:526260. doi: 10.1155/2013/526260
Peraud, O., Biggs, J. S., Hughen, R. W., Light, A. R., Concepcion, G. P., Concepcion, G. P., et al. (2009). Microhabitats within venomous cone snails contain diverse Actinobacteria. Appl. Env. Microbiol. 75, 6820–6826. doi: 10.1128/AEM.01238-09
Phillips, K., Zaidan, F., Elizondo, O. R., and Lowe, K. L. (2012). Phenotypic characterization and 16S rDNA identification of culturable non-obligate halophilic bacterial communities from a hypersaline lake, La Sal del Rey, in extreme South Texas (USA). Aquat. Biosyst. 8:5. doi: 10.1186/2046-9063-8-5
Poulsen, M., Oh, D.-C., Clardy, J., and Currie, C. R. (2011). Chemical Analyses of Wasp-Associated Streptomyces bacteria reveal a prolific potential for natural products discovery. PLoS ONE 6:e16763. doi: 10.1371/journal.pone.0016763
Sabet, S., Diallo, L., Hays, L., Jung, W., and Dillon, J. G. (2009). Characterization of halophiles isolated from solar salterns in Baja California, Mexico. Extremophiles 13, 643–656. doi: 10.1007/s00792-009-0247-1
Subramani, R., and Aalbersberg, W. (2012). Marine actinomycetes: an ongoing source of novel bioactive metabolites. Microbiol. Res. 167, 571–580. doi: 10.1016/j.micres.2012.06.005
Swan, B. K., Ehrhardt, C. J., Reifel, K. M., Moreno, L. I., and Valentine, D. L. (2010). Archaeal and bacterial communities respond differently to environmental gradients in anoxic sediments of a California hypersaline lake, the Salton Sea. Appl. Environ. Microbiol. 76, 757–768. doi: 10.1128/AEM.02409-09
Tian, S., Yang, Y., Liu, K., Xiong, Z., Xu, L., and Zhao, L. (2014). Antimicrobial metabolites from a novel halophilic actinomycete Nocardiopsis terrae YIM 90022. Nat. Prod. Res. 28, 344–346. doi: 10.1080/14786419.2013.858341
Tian, S.-Z., Pu, X., Luo, G., Zhao, L.-X., Xu, L.-H., Li, W.-J., et al. (2013). Isolation and characterization of new p-Terphenyls with antifungal, antibacterial, and antioxidant activities from halophilic actinomycete Nocardiopsis gilva YIM 90087. J. Agr. Food Chem. 61, 3006–3012. doi: 10.1021/jf400718w
Tulp, M., and Bohlin, L. (2005). Rediscovery of known natural compounds: nuisance or goldmine? Bioorg. Med. Chem. 13, 5274–5282. doi: 10.1016/j.bmc.2005.05.067
Ventosa, A. (2006). “Unusual microorganisms from unusual habitats: hypersaline environments,” in Prokaryotic Diversity – Mechanisms and Significance (Society for General Microbiology Symposium no. 66), eds N. A. Logan, H. M. Lappin-Scott, and P. C. F. Oyston (Cambridge: Cambridge University Press), 223–253.
Walsh, C. (2003). Where will new antibiotics come from? Nature Rev. Microbiol. 1, 65–70. doi: 10.1038/nrmicro727
Xiang, W., Guo, J., Feng, W., Huang, M., Chen, H., Zhao, J., et al. (2008). Community of extremely halophilic bacteria in historic Dagong Brine Well in southwestern China. World J. Microbiol. Biotechnol. 24, 2297–2305. doi: 10.1007/s11274-008-9744-0
Yuan, M., Yu, Y., Li, H.-R., Dong, N., and Zhang, X.-H. (2014). Phylogenetic diversity and biological activity of actinobacteria isolated from the chukchi shelf marine sediments in the Arctic Ocean. Mar. Drugs 12, 1281–1297. doi: 10.3390/md12031281
Keywords: actinomycetes, hypersaline environments, antibiotic, Streptomyces, drug discovery
Citation: Jose PA and Jebakumar SRD (2014) Unexplored hypersaline habitats are sources of novel actinomycetes. Front. Microbiol. 5:242. doi: 10.3389/fmicb.2014.00242
Received: 07 April 2014; Accepted: 04 May 2014;
Published online: 27 May 2014.
Edited by:
Luis Cláudio Nascimento Da Silva, University of Copenhague, DenmarkReviewed by:
Atte Von Wright, University of Eastern Finland, FinlandDmitri Debabov, NovaBay Pharmaceuticals, USA
Copyright © 2014 Jose and Jebakumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence:anNvbG9tb25fbXJuYUB5YWhvby5jb20=
 Polpass Arul Jose
Polpass Arul Jose Solomon Robinson David Jebakumar
Solomon Robinson David Jebakumar