- 1Department of Biological Sciences, University of Alberta, Edmonton, AB, Canada
- 2Defence Research and Development Canada – Suffield Research Centre, Medicine Hat, AB, Canada
Vaccines developing immune responses toward surface carbohydrates conjugated to proteins are effective in preventing infection and death by bacterial pathogens. Traditional production of these vaccines utilizes complex synthetic chemistry to acquire and conjugate the glycan to a protein. However, glycoproteins produced by bacterial protein glycosylation systems are significantly easier to produce, and could possible be used as vaccine candidates. In this work, we functionally expressed the Burkholderia pseudomallei O polysaccharide (OPS II), the Campylobacter jejuni oligosaccharyltransferase (OTase), and a suitable glycoprotein (AcrA) in a designer E. coli strain with a higher efficiency for production of glycoconjugates. We were able to produce and purify the OPS II-AcrA glycoconjugate, and MS analysis confirmed correct glycan was produced and attached. We observed the attachment of the O-acetylated deoxyhexose directly to the acceptor protein, which expands the range of substrates utilized by the OTase PglB. Injection of the glycoprotein into mice generated an IgG immune response against B. pseudomallei, and this response was partially protective against an intranasal challenge. Our experiments show that bacterial engineered glycoconjugates can be utilized as vaccine candidates against B. pseudomallei. Additionally, our new E. coli strain SDB1 is more efficient in glycoprotein production, and could have additional applications in the future.
Introduction
Burkholderia pseudomallei, a Gram-negative saprophyte, is the causative agent for melioidosis and is endemic in Southeast Asia and Northern Australia (Cheng and Currie, 2005). It is highly resistant to harsh environmental pressures, and it is classified as a potential class B bioterrorism weapon due to its high infectivity when aerosolized (Silva and Dow, 2013). Several virulence factors have been identified, including multiple Type III and VI secretion systems, toxins, capsular polysaccharide, and lipopolysaccharide (LPS; Nandi and Tan, 2013). Two different LPS structures named O-polysaccharide (OPS) I and II are present in B. pseudomallei, and OPS II was shown to be required for serum resistance and virulence (Knirel et al., 1992; Perry et al., 1995; DeShazer et al., 1998). B. pseudomallei has an intrinsically high resistance to several different classes of antibiotics, which increases the potential danger of this organism. Due to the increasing prevalence of new resistance genes, and the increasing number of cases, new alternatives to treat and prevent melioidosis are required.
Immunization is one of the best available tools against infection, and it is significantly more cost effective than treatment after disease has occurred. Three main classes of vaccines are commercially produced. Live attenuated bacteria that have been shown to be highly effective as vaccine candidates however, drawbacks such as safety, reactogenicity, stability, and manufacturing remain problematic (Galen and Curtiss, 2013). Whole-cell-killed bacterial vaccines are easy to commercially manufacture, but have problems with stability, long-term protection, and present biosafety risks in the case of class III pathogens. Purified surface carbohydrates have been utilized as a vaccine candidate, but typically only produce short-term protection and are not effective in children or mature individuals (Lockhart, 2003). Traditional conjugate vaccines, where bacterial surface polysaccharides are chemically conjugated to a carrier protein, have been demonstrated to be highly effective. The best example is the Haemophilus influenzae type b conjugate vaccine, which has nearly eliminated infections in vast parts of the world (Pollard et al., 2009). However, manufacturing these conjugate vaccines requires complex synthetic chemistry for the attachment of the glycan to protein carriers. Additionally, the polysaccharides are either obtained from the target pathogen, which constitutes a major health hazard, or by laborious, chemical synthesis. Often, bacterial polysaccharides are too complex to be synthesized efficiently, making this process economically prohibitive. Finally, chemical attachment of the sugar to the carrier protein can result in large and heterogeneous conjugates, modifying the native structure, and thus decreasing the protective nature of the vaccine. Both live attenuated and killed bacterial vaccines have been tested against B. pseudomallei, but provide little to no protection against disease and mortality in murine virulence models (Peacock et al., 2012). Additionally, since B. pseudomallei requires class III biosafety facilities, manufacturing glycoconjugates containing glycans from its native host is challenging and possibly hazardous. Recently, it has been demonstrated a protein chemically conjugated with the B. pseudomallei OPS and CPS was able to increase survival against B. pseudomallei infection (Scott et al., 2014). Additionally, protection has been demonstrated using B. thailandensis E555 as a live vaccine due to homology of CPS structures (Scott et al., 2013).
A novel method of synthesizing conjugate vaccines is through the exploitation of the protein glycosylation machineries of bacteria (Iwashkiw et al., 2012; Cuccui et al., 2013; Wetter et al., 2013). The cornerstone of bacterial glycosylation is the oligosaccharyltransferase (OTase) enzymes, which covalently attach glycan structures to either asparagine (N-linked) or serine/threonine (O-linked) residues (Nothaft and Szymanski, 2010). OTases have high substrate promiscuity, and thus can transfer a wide range of glycan structures to acceptor proteins, in a process called OTase-dependent glycosylation. The best characterized N-glycosylation system in bacteria is from Campylobacter jejuni (Nothaft et al., 2010). Briefly, a unique initiating glycosyltransferase attaches a nucleotide-activated monosaccharide-1P to the lipid carrier undecaprenyl phosphate (Und-P) in the cytoplasmic face of the inner membrane. Subsequently, a series of other glycosyltransferases attach additional monosaccharides to first residue, and when completed, the lipid-linked oligosaccharide (LLO) is translocated to the periplasmic face of the inner membrane by a flippase. Finally, PglB (N-OTase) covalently attaches the glycan to asparagine residues with the sequon D/E-X-N-Y-S/T (X,Y ≠P; Kowarik et al., 2006). Earlier studies demonstrated PglB has relaxed glycan specificity, allowing the transfer of a vast array of glycans, including O antigens, to acceptor proteins (Feldman et al., 2005). It was later shown that O-OTases share this feature (Faridmoayer et al., 2008). Thus, bacterial glycosylation systems can be exploited to synthesize novel glycoconjugates for vaccination and diagnostic purposes as previously demonstrated (Ihssen et al., 2010; Iwashkiw et al., 2012; Cuccui et al., 2013; Wetter et al., 2013; Wacker et al., 2014). Glycoconjugates produced by this method are significantly less expensive, less challenging to produce, and produce less hazardous waste than conventional chemical methods.
In this work, we demonstrate that the biosynthesis of the B. pseudomallei OPS II can be reconstituted in Escherichia coli (E. coli). Successful generation of the conjugate required the expression of the corresponding B. pseudomallei OPS II genes in an E. coli strain lacking both the waaL ligase and wecA initiating glycosyltransferase (SDB1). This glycoconjugate, when injected into mice, was able to develop a directed IgG immune response toward B. pseudomallei, and provide partial protection against infection in a murine model of melioidosis.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
Escherichia coli strains were grown on LB broth at 37°C. Trimethoprim (100 μg/ml), spectinomycin (80 μg/ml), and ampicillin (100 μg/ml) were added to the media for plasmid selection as needed. The strains and plasmids used are listed in Table 1.
Western Blotting
Glycan expression and glycosylation were analyzed by SDS-PAGE on 10% gels. The gels were electroblotted onto a nitrocellulose membrane via semi-dry membrane transfer and analyzed with antibodies α-His (Santa Cruz Biotechnology) and α-BPs OPSII glycan kindly provided by Dr. Joanne Prior (1:1,000). Membranes were visualized using the Odyssey Infrared Imaging System (Li-Cor Biosciences, USA).
Cloning and Expression of Burkholderia pseudomallei Type II OPS
To obtain the plasmid expressing the type II O-antigen polysaccharide of Burkholderia pseudomallei under an arabinose promoter we used the pCC1FOS-BPF16β_E10 vector kindly provided by Professor R. Titball (University of Exeter), which contains the LPS cluster of B. pseudomallei K96243, coordinates 3191324–3229257. The pCC1FOS-BPF16β_E10 vector was digested with NheI, KpnI, and PciI to get an 8673 bp fragment, corresponding to genes between rmlD and wbiC, and with SnaBI and KpnI to get a 9367 bp fragment that includes the genes between wbiC and wbiI. These two fragments containing the 15 genes required for B. pseudomallei type II OPS expression were inserted into pBAD24 digested with NheI and SmaI. Arabinose-dependent expression of the type II O-antigen was confirmed by Western blot.
Construction of SDB1 waaL and wecA Deficent Strain
Construction of SDB1 strain was done using the P1 transduction protocol adapted from Thomason et al. (2007). The P1 bacteriophage was first grown on the strain (BW25113 rfe::kan.) from the Keio collection library (Baba et al., 2006). This strain has a kanamycin-resistant cassette on the wecA gene as a donor. The resulting phage lysate was used to infect the recipient strain CLM24 (ΔwaaL). Recombinant strains were confirmed by PCR using the oligonucleotides rfe for comp (5′-GCAATGACCAAGACCAATGACG-3′) and rfe rev comp (5′-GCTGCTGCGAGTAATATCCC-3′). The kanamycin cassette was removed using the FLP recombinase expressed from pFLP2.
Production and Purification of Glycosylated AcrA
SDB1 strain transformed with C. jejuni PglB (pMAF10), AcrA (pIH18), and BPs type II O-antigen (pEQ3) was grown overnight at 37°C. Culture was reinnoculated 1/33 into fresh LB media using a culture/flask ratio 1:10. After 2 h at 37°C with shaking at 200 rpm, the cultures were induced with 0.1 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG; Sigma) and 0.2% (w/v) L-(+)-arabinose (MP Biomedicals). To increase the glycosylation yield in SDB1, we also added MnCl2 (4 mM). Five hours after induction at 37°C, arabinose was added again to ensure PglB expression. Cells were harvested by centrifugation after an overnight induction period and the periplasmic extract containing the glycoproteins was extracted using a lysozyme treatment as described previously (Iwashkiw et al., 2012). For purification, the periplasmic fraction was equilibrated with 1/9 vol 10× loading buffer (0.1 M imidazole, 3 M NaCl, 0.2 M Tris-HCl, pH 8.0) and subjected to a Ni2+affinity chromatography as described (Iwashkiw et al., 2012). Purified protein was quantified by Bradford assay (BioRad).
Sugar Quantification of Glycoproteins
The protocol was adapted from Dubois et al. (1956). In a small glass tube was mixed 10 μl of sample, 90 μl of ddH2O, and 100 μl of freshly made 5% phenol in ddH2O. Then 1 ml of concentrated H2SO4 was briskly added into the mixture and immediately vortex for several seconds. An orange color with intensity proportional to concentration began to develop and reached a maximum about 2 h at 30°C. The samples were read against glucose standards at OD500.
Vaccination
BALB/c mice (n = 5 per group, 6-week-old female) were immunized with three doses of purified recombinant bioconjugate vaccine, carrier protein, or gamma-irradiated killed whole cells (3 × 104 Burkholderia pseudomallei K96234), via the intraperitoneal route (i.p.) over 6 weeks. The doses were administered with Imject Alum Adjuvant (Thermo Scientific), not used with whole killed cells or when noted. Sera samples were collected for antibody analysis, prior to immunization 2 weeks after vaccination and boost. The antibody titre of total IgG was analyzed by ELISA. Briefly, wells of microtiter plates were coated (18 h, 4°C) with gamma-irradiated whole Burkholderia pseudomallei cells at a 1/100 dilution in 100 μl of coating buffer (0.05 M Na2CO3, 0.05 M NaHCO3, pH 9.6) and were then blocked with 2% (w/v) BSA in PBS for 2 h at 37°C. Sera samples at a 1/200 dilution in 100 μl of antibody dilution buffer [2% (w/v) BSA, 0.1% (v/v) Tween 20] were incubated for 1 h at 37°C. HRP-conjugated goat anti-mouse IgG at a 1/8000 in antibody dilution buffer was added for 1 h at 37°C and then the reaction was visualized by the addition of 100 μl chromogenic substrate (ultra-TMB) for 5 min. The reaction was stopped with 100 μl H3PO4 and absorbance at 405 nm was measured using ELISA plate reader. Plates were washed five times with washing buffer [0.1% (v/v) Tween 20] after each step.
Intranasal Challenge Model
The murine melioidosis infection model used was carried out under ABSL-3 containment practices. Briefly, female BALB/c mice were challenged via the i.n. route (50 μl) with approximately 2 × 103 CFU (approximately 10–12 LD50) of B. pseudomallei K96243. Mice were weighed prior to inoculation and monitored for 21 days post-infection. Mice were anesthetized, held vertically, and 50 ml of the inoculum was released into the nares for inhalation. Following challenge, the inoculum was back titrated on agar plates to confirm delivered dose. Using this model, control mice died or were euthanized according to predetermined humane end points 3–6 days post-challenge.
Statistics
Survival curves were generated by use of Kaplan–Meier estimators. The survival distributions of each treatment group vs. control protein carrier group were compared by unpaired T test or Mann–Whitney test using GraphPad Prism version 6.0.
Ethics Statement
This study was carried out in accordance with the Canadian animal care guidelines. The protocols were approved by the Animal Care and Use Committees of Defence Research and Development Canada. Mice were anesthetized by intraperitoneal injection of a sodium pentobarbital solution.
Results
Cloning and Expression of the B. pseudomallei k96243 O Antigen Polysaccharide II (OPS II) Locus in E. Coli
Previous work identified a region consisting of 21 potential open reading frames, and further investigation identified a cluster of 15 genes required for the biosynthesis of B. pseudomallei K96243 OPS II (Figure 1A; DeShazer et al., 1998). A previous study demonstrated by NMR analysis that the structure of OPS II is a polymer of a disaccharide repeating structure composed of -3-)-β-D-glucopyranose-(1–3)-α-L-6-deoxy-talopyranose-(1-, with variable O-methyl and O-acetyl modifications (Figure 1B; Perry et al., 1995). In order to recombinantly express the B. pseudomallei OPS II in E. coli, the 15 essential genes were subcloned (genes rmlB to wbiI) from the plasmid pCC1FOS-BPF16β_E10 by restriction digest into the arabinose inducible expression vector pBAD24, generating pEQ3 (Figure 1A). Expression of the B. pseudomallei OPS II in E. coli CLM37 was visualized by Western blot as a typical ladder of immunoreactive bands, confirming the production of the carbohydrate structure (Figure 1C).
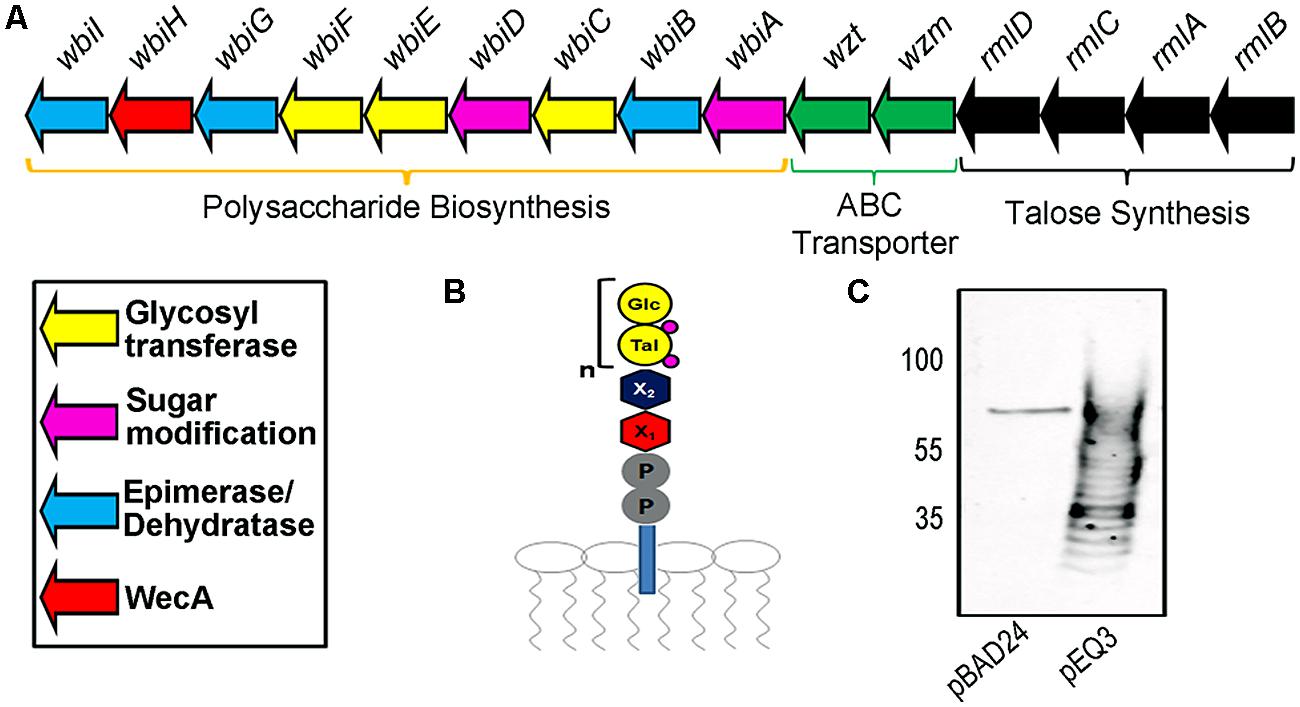
FIGURE 1. Description of Burkholderia pseudomallei O-polysaccharide II (OPSII) cluster. (A) Representation of the required 15 genes for the synthesis of OPS II and predicted functions. (B) Expected glycan intermediate structure attached to Undecaprenyl pyrophosphate. (C) Western blot of 10 μl of whole-cell extracts utilizing an anti-Glycan antibody reveals expression of OPS II (subcloned in pEQ3) in E. coli produces a polymer in whole-cell extracts.
Generation of an E. Coli Strain Optimized for OPS II Production and Protein Glycosylation
We attempted to generate a N-linked glycoprotein with the OPS II by exploiting the C. jejuni N-glycosylation system as previously described (Ihssen et al., 2010; Iwashkiw et al., 2012; Cuccui et al., 2013; Wetter et al., 2013). In earlier work, N-glycosylated AcrA was synthesized in E. coli by co-expression of C. jejuni PglB and AcrA with an appropriate carbohydrate structure. We therefore expressed PglB (pMAF10), AcrA (pIH18), and the B. pseudomallei OPS II antigen (pEQ3) in both a traditional expression (EPI300) and wecA- (CLM37) strains and tested for glycosylation by Western blot. We were unable to detect any evidence of glycosylation of purified AcrA (data not shown).
One issue with exploiting O antigens using protein glycosylation may be the precursor can also be used by the WaaL ligase in E. coli, thus siphoning off the substrate, and decreasing the glycosylation efficiency (Figure 2). Additionally, E. coli strains express the initiating glycosyl transferase (wecA), which transfers a GlcNAc onto the undecaprenyl-diphosphate (Und-PP) carrier. This would interfere with the synthesis of the glycan of interest onto the same lipid if the first sugar in the structure is not a GlcNAc, as in the case of the OPS II. We hypothesized that deletion of WaaL and WecA would result in an increased efficiency of protein glycosylation (Figure 2C).
FIGURE 2. Hypothesis for the lack of glycosylation due to glycan competition and utilization, resulting in the development of the E. coli strain SDB1. (A) When expressing pEQ3 in a WT E. coli background, OPS II is produced at a lower quantity than the native O-antigen, and both structures are preferentially transferred by WaaL to Lipid A, instead of to the acceptor protein by PglB (OTase). (B) If wecA is deleted, the native O-antigen cannot be produced, allowing for a greater production of OPS II. However, OPS II would be still selectively transferred by WaaL over PglB. (C) If both wecA and waaL are deleted, the OPS II structure would be highly produced, and only available to PglB for synthesis of glycoproteins.
We therefore constructed the E. coli wecA- waaL- mutant strain SDB1. Using the KEIO strain collection (Baba et al., 2006), the wecA mutation was transduced into CLM24, an E. coli waaL mutant (Feldman et al., 2005), creating SDB1. To functionally confirm the double mutation, we analyzed the LPS produced by SDB1 transformed with plasmid pJHCV32 (Figure 3). The plasmid pJHCV32 drives the constitutive expression of the E. coli O7 antigen, but relies on the chromosomal copy of the glycosyltransferase WecA (Valvano and Crosa, 1989). Expression in trans of either WecA or WaaL individually in this background did not result in the production of a smooth LPS containing the O7 antigen (lanes 1–8). However, when both genes were co-expressed in SDB1, we observed the characteristic polymerization of O antigen previously observed for the E. coli O7 LPS (lanes 9 and 10), confirming the creation of the double mutation.
FIGURE 3. Confirmation of a wecA- waaL- double mutant (SDB1) in E. coli W3110. To verify the double mutant, LPS extractions (20 μl per sample) were analyzed by silverstain for the ability to produce exogenously expressed E. coli O7 LPS (pJHCV32). No LPS was observed when either WecA or WaaL was expressed in trans (lanes 1–8). However, when both were coexpressed, a laddering pattern was observed, confirming complementation of the double mutant (lanes 9 and 10).
In Vivo Synthesis and Purification of an N-Linked Glycoconjugate with the Burkholderia OPSII Glycan
To create an N-linked glycoconjugate, we transformed the E. coli strain SDB1 with the pEQ3 (OPS II), pMAF10 (PglB), and pIH18 (AcrA). Cultures of the transformed strain were grown and induced as required, and AcrA was purified from periplasmic extracts by Ni2+ affinity chromatography. To determine if AcrA was glycosylated, we analyzed the purified protein by either Western blot with antibodies specific to either AcrA or OPS II, or Coomassie stain, and when visualized together, we observed an overlap of the signal, suggesting glycosylation of AcrA with the OPSII glycan (Figure 4).
FIGURE 4. Analysis of purified AcrA by Western Blot. SDB1 transformed with pMAF10 (pglB), pIH18 (acrA), and pEQ3 (OPS II) were grown (+/-) induction of OPS II, harvested, and AcrA was purified by Ni2+-NTA affinity chromatography. Purified glycoconjugate was separated on 10% SDS-PAGE and analyzed by western blot (5 μl) or Coomassie stain (15 μl). A high-molecular-weight ladder is co-detected by both α-His and α-Glycan antibodies, and overlap of the two signals strongly suggests glycosylation of AcrA. Staining of the purified protein is observed by Coomassie stain.
To confirm AcrA was glycosylated with the B. pseudomallei OPS II carbohydrate, we employed mass spectrometry (MS) techniques. The purified glycoprotein was tryptically digested in-solution, and the resulting peptides were examined by LC-ESI-Q-TOF MS and MS/MS. Manual analysis of the MS data (MassLynx; Waters Corporation) revealed a peak with an m/z 1152.063+, and further inspection of this peak by MS/MS revealed a glycopeptide that corresponded to the previously identified second glycosylation site of AcrA (AVFDNNNSTLLPGAFATITSEGFIQK; m/z 2754.1) with the addition of an m/z 700.2 modification (Figure 5). De novo peak annotation identified the modification to be a tetramer of 188–162–188–162. The mass of 188 Da is consistent with an O-acetyl deoxyhexose residue, and the 162 Da is consistent with a hexose residue. We were also able to identify in the low-molecular region both an individual O-acetyl deoxyhexose (189.0 Da), and a subunit of the dimer with a mass of 351.1 Da. Our MS characterization of the B. pseudomallei OPS II glycan is consistent with the previously published data identifying it to be a polymer of dimers of O-acetylated deoxytalose and glucose (Perry et al., 1995). These data combined with the immunoreactivity of our glycoconjugate to the B. pseudomallei OPS II antibody confirm that we were able to synthesize and glycosylate AcrA with the correct glycan structure in E. coli SDB1.
FIGURE 5. Mass spectrometry identification of the B. pseudomallei OPS II glycan attached to AcrA. MS analysis of tryptically digested glycosylated AcrA revealed a peak of m/z 1152.063+. MS/MS of this peak showed a previously characterized glycosylation site of AcrA (AVFDNNNSTLLPGAFATITSEGFIQK; 2754.1 Da) with a modification of 700.2 Da. Analysis of the modification revealed a tetrameric glycan, with a structure of 188–162–188–162. This structure is consistent with the previously determined structure of B. pseudomallei OPS II being a polymer of dimers of O-acetylated deoxytalose (188 Da) and glucose (162 Da).
Mice Injected with Purified N-Linked Bioglycoconjugate Developed a Partially Protective IgG Immune Response Toward B. Pseudomallei Whole Cells
To evaluate the potential use of the glycoprotein as conjugate vaccine, the purified AcrA containing OPS II was injected intraperitoneally into mice to measure the immune response compared to whole-cell-killed B. pseudomallei. Groups of five mice were injected with PBS, unglycosylated AcrA as control, glycoconjugate in different quantities, or whole-cell-killed cells. The IgG immune response was tested by ELISA against whole-cell extracts of B. pseudomallei (Figure 6). All test groups were compared to the PBS control, and showed no initial immune response toward B. pseudomallei in the pre-injection sera. The AcrA-injected group had a slight increase in IgG response, but did not increase after additional boosts. A significant increase in IgG response was observed in each of the glycosylated test groups after the primary vaccination, with varying degrees of improvement in immune response after a second and third boost. The best immune response was observed in the mice injected with whole-cell lysates of B. pseudomallei had a significantly stronger immune response as compared to the glycoconjugate sera, but this was expected as whole cells were use as antigen for the ELISA.
FIGURE 6. ELISA of the IgG immune response of immunized mice toward whole-cell extracts of B. pseudomallei. All data were normalized to the PBS-injected control mice, and no response was observed in all groups prior to immunization. A low level of response was observed at a consistent level for the unglycosylated AcrA control, whereas all test groups injected with glycosylated AcrA had a significant IgG response. The results are the median of five mice in each test group.
We next tested the immunized mice for a preliminary evaluation of the efficacy of the glycoconjugate. We employed an intranasal murine melioidosis model against B. pseudomallei infection with a dose of 12× LD50 (Figure 7). Mice vaccinated with only protein carrier died or were euthanized according to predetermined humane end points after 6 days. For the PBS-injected control group, 80% of the mice died or were euthanized after 6 days, and one mouse survived until day 13 of the challenge. All of the mice vaccinated with the glycoconjugate showed a significant increase in survival time as compared to the control protein carrier group. However, contrary to the ELISA results that showed the best IgG immune response in mice vaccinated with 2 μg of glycosylated AcrA, 40% of the mice survived until day 12, with the remaining being sacrificed on day 14. Mice injected with 1 μg of glycoprotein without any adjuvant saw survival until day 18, while one mouse vaccinated with 1 μg survived until day 22. The difference in mean time to death was not statistically significant between the groups receiving the various glycoprotein preparations. In comparing our glycoconjugate to whole-cell-killed bacteria as vaccine candidates, we observed highly similar survival of the mice, with all mice succumbing to infection by day 18 of the challenge. Overall, these results demonstrate that the N-glycoconjugate containing the B. pseudomallei OPS II is capable of providing partial immune protection against a 12 × LD50 dose, and it is comparable to whole-cell-killed bacteria in protection against infection.
FIGURE 7. Murine survival of different test groups to a 12× LD50 i.n. challenge of B. pseudomallei. Mice vaccinated with either PBS or unglycosylated AcrA controls had a significantly lower survival period as compared to the mice injected with OPS II glycosylated AcrA. Interestingly, mice vaccinated with 1 μg glycoconjugate (+/- adjuvant) had a higher survivorship percentage as compared to the 2 μg vaccinated group.
Discussion
Due to a combination of factors including the increasing number of reported cases of B. pseudomallei infections, the risk posed by the bacterium as a potential biological warfare agent, and an absence of an effective vaccine, we explored the possibility that by exploiting the N-linked glycosylation system of C. jejuni, we could produce a glycoconjugate vaccine containing the OPS II of B. pseudomallei. This strategy was only demonstrated in a few cases (Iwashkiw et al., 2012; Cuccui et al., 2013; Wetter et al., 2013). Since B. pseudomallei is a biosafety class III agent we were unable to directly exploit the native OPS II by expressing the N-glycosylation system in the host as previously shown (Iwashkiw et al., 2012). Instead, we utilized previous knowledge of the genetic loci responsible for the biosynthesis of OPS II (DeShazer et al., 1998), and subcloned the key 15 genes into an E. coli expression vector. We observed high levels of expression of the OPS II in E. coli strains by Western blot, and attempted to create the glycoconjugate in a basic expression and wecA- E. coli strains as previously described (Ihssen et al., 2010; Cuccui et al., 2013; Wetter et al., 2013). We did not observe any evidence of glycosylation, and hypothesized that the OPS II was being utilized exclusively to modify the LPS. Therefore, we engineered an E. coli strain (SDB1) lacking both the waaL ligase, and wecA. Construction of the SDB1 strain was confirmed by expressing either or both WecA and WaaL with a plasmid encoding the E. coli O7 LPS cluster that lacks WecA homolog. Analysis of LPS extractions by silverstain showed that the O7 antigen structure was only transferred to lipid A in the presence of both enzymes. Our engineered strain in theory should be able to produce the OPS II-AcrA glycoconjugate with high efficiency due to no other competition for either the undecaprenyl-phosphate lipid carrier by WecA or the OPS II-Und-PP substrate from WaaL.
We showed that SDB1 produced the desired glycoconjugate when OPS II (pEQ3), PglB (pMAF10), and AcrA (pIH18) were co-expressed. The generation of the conjugate was shown via Western blot and MS. Interestingly, glycosylation of AcrA required the addition of MnCl2 to the media. Although Mn2+ is required for PglB activity and was observed in the active site of the crystal structure (Lizak et al., 2011), we did not have to add Mn2+ to obtain glycosylated proteins in previous experiments. The direct relationship between its addition and glycosylation efficiency is unclear. The MS examination of the purified glycoconjugate revealed the presence of a tetrameric glycan moiety corresponding to two repeats of O-acetylated deoxyhexose and hexose, in agreement with previously published characterization of the structure (Perry et al., 1995). Interestingly, previous work has shown that the C. jejuni OTase PglB can only transfer glycans with a reducing monomer with an N-acetyl group, whereas the B. pseudomallei OPS II structure has been shown to possess an O-acetyl modification. This finding expands our knowledge on the substrate specificity of PglB. We did not observe any larger glycan structures that were detected by Western blot, but this could be due to limitations of our MS instruments. Most of the Wzy-independent glycans require an adapter composed of two monosaccharides linking the lipid carrier to the polymeric structure (Whitfield, 2006; Greenfield and Whitfield, 2012). The finding that the O-acetylated deoxytalose appears to be directly linked to the protein indicates that a linker glycan (proposed in Figure 1A as X1 and X2) is not present in the OPS II structure. Our experiments demonstrate that in the genetically engineered strain SDB1, we were able to produce a glycoconjugate with the B. pseudomallei OPS II carbohydrate.
We carried out preliminary experiments to determine if our glycoconjugate could be utilized as a vaccine against B. pseudomallei. We injected different quantities of the glycoconjugate into mice. We observed virtually no response toward B. pseudomallei for the mice injected with the protein carrier alone, whereas a significantly stronger response from the mice injected with all groups of the glycoconjugate. Interestingly, an intermediate response was observed in the 1 μg glycoconjugate groups, regardless of the addition of an adjuvant. We then challenged the vaccinated mice with B. pseudomallei K96243, and after 6 days post challenge, none of the carrier control injected mice survived, whereas survival was observed each of the glycoconjugate groups, with the longest survival in the 1 μg group, irrespective of the addition of an adjuvant. Why lower levels of antibody production resulted in better protection remains unknown, although these differences in protection were not statistically significant between groups receiving the glycoprotein. It is possible that the carrier protein, AcrA, due to its high immunogenic nature, acts as an adjuvant itself. This could also possibly explain why both of the 1 μg groups while having a lower detected IgG immune response compared to the 2 μg group have a longer survival period. Our results are consistent with these previous studies with the non-protected mice succumbing to infection after ∼1 week (Nelson et al., 2004; Su et al., 2010; Nieves et al., 2011). Interestingly, our initial studies with the glycoconjugate gave similar protection levels to whole-cell-killed B. pseudomallei. Further optimization of our vaccine candidate is currently undergoing. This includes testing different amounts of conjugate, and replacing the acceptor protein from AcrA of C. jejuni to a B. pseudomallei protein, which may enhance the immunogenicity of the conjugate.
Several other studies have been published attempting to develop a vaccine against B. pseudomallei (reviewed in Silva and Dow, 2013). Many of these vaccine candidates have dealt with either attenuated strains, whole-cell-killed bacteria, or purified proteins directly from B. pseudomallei, which requires class III biosafety facilities, and would lead to higher significant problems for commercialization of the product. Other groups attempted to recombinantly express and purify B. pseudomallei proteins in E. coli, with limited success. However, our work has demonstrated that the OPS II carbohydrate of B pseudomallei can be functionally expressed in E. coli, and be utilized by PglB to create a glycoconjugate that is partially protective against infection. Glycoengineered therapeutics are simple to produce, cost effective, and have been demonstrated to provide long-term protection against several pathogens. Additionally, previous work has used a similar glycoconjugate with the Y. enterocolitica O9 O-antigen as a diagnostic tool (Iwashkiw et al., 2012). Conjugation of the glycoconjugate to magnetic nanobeads has shown significant promise in detection of disease in cattle and human sera samples (Ciocchini et al., 2013, 2014). A similar system may have future potential for diagnosing individuals infected with B. pseudomallei.
In summary, we have demonstrated that the OPS II glycan of B. pseudomallei can be functionally expressed in E. coli. Additionally, this glycan was transferred to the carrier protein AcrA by the OTase PglB, both from C. jejuni, to generate a glycoconjugate. We also described a novel E. coli strain SDB1 which lacks wecA and waaL, resulting in a higher efficiency of glycosylation as compared to previously used strains. Mice injected with this glycoprotein were able to develop a long-term IgG immune response, and showed significantly longer survival when challenged with B. pseudomallei as compared to the naive controls. This new biologically engineered strain may be used for the future creation of commercial bioglycoconjugate therapeutics, and glycoconjugate may have future potential for diagnostic applications or vaccination against B. pseudomallei infections.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the Natural Sciences and Engineering Research Council (NSERC) to Mario F. Feldman and by DRDC ARP funds. Mario F. Feldman is an Alberta Heritage Foundation for Medical Research (AHFMR) Scholar and a Canadian Institutes of Health Research (CIHR) new investigator. Jeremy A. Iwashkiw is an Alberta Innovates Health Solutions (AIHS) scholar. The authors acknowledge the technical assistance of Scott Jager and Melissa Crichton.
References
Alaimo, C., Catrein, I., Morf, L., Marolda, C. L., Callewaert, N., Valvano, M. A.,et al. (2006). Two distinct but interchangeable mechanisms for flipping of lipid-linked oligosaccharides. EMBO J. 25, 967–976. doi: 10.1038/sj.emboj.7601024
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M.,et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 1–11. doi: 10.1038/msb4100050
Cheng, A. C., and Currie, B. J. (2005). Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18, 383–416. doi: 10.1128/CMR.18.2.383-416.2005
Ciocchini, A. E., Rey Serantes, D. A., Melli, L. J., Iwashkiw, J. A., Deodato, B., Wallach, J.,et al. (2013). Development and validation of a novel diagnostic test for human brucellosis using a glyco-engineered antigen coupled to magnetic beads. PLoS Negl. Trop. Dis. 7:e2048. doi: 10.1371/journal.pntd.0002048
Ciocchini, A. E., Serantes, D. A., Melli, L. J., Guidolin, L. S., Iwashkiw, J. A., Elena, S.,et al. (2014). A bacterial engineered glycoprotein as a novel antigen for diagnosis of bovine brucellosis. Vet. Microbiol. doi: 10.1016/j.vetmic.2014.04.014 [Epub ahead of print].
Cuccui, J., Thomas, R. M., Moule, M. G., D’Elia, R. V., Laws, T. R., Mills, D. C.,et al. (2013). Exploitation of bacterial N-linked glycosylation to develop a novel recombinant glycoconjugate vaccine against Francisella tularensis. Open Biol. 3, 130002. doi: 10.1098/rsob.130002
DeShazer, D., Brett, P. J., and Woods, D. E. (1998). The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol. Microbiol. 30, 1081–1100. doi: 10.1046/j.1365-2958.1998.01139.x
Dubois, M., Gilles, K. A., Hamilton, J. K., Rebers, P. A., and Smith, F. (1956). Colorimetric method for the determination of sugars and related substances. Anal. Chem. 28, 350–356. doi: 10.1021/ac60111a017
Dykxhoorn, D. M., St Pierre, R., and Linn, T. (1996). A set of compatible tac promoter expression vectors. Gene 177, 133–136. doi: 10.1016/0378-1119(96)00289-2
Faridmoayer, A., Fentabil, M. A., Haurat, M. F., Yi, W., Woodward, R., Wang, P. G.,et al. (2008). Extreme substrate promiscuity of the Neisseria oligosaccharyl transferase involved in protein O-glycosylation. J. Biol. Chem. 283, 34596–34604. doi: 10.1074/jbc.M807113200
Feldman, M. F., Wacker, M., Hernandez, M., Hitchen, P. G., Marolda, C. L., Kowarik, M.,et al. (2005). Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 102, 3016–3021. doi: 10.1073/pnas.0500044102
Galen, J. E., and Curtiss, R. III. (2013). The delicate balance in genetically engineering live vaccines. Vaccine doi: 10.1016/j.vaccine.2013.12.026 [Epub ahead of print].
Greenfield, L. K., and Whitfield, C. (2012). Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr. Res. 356, 12–24. doi: 10.1016/j.carres.2012.02.027
Guzman, L. M., Belin, D., Carson, M. J., and Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130.
Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. J., and Schweizer, H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86. doi: 10.1016/S0378-1119(98)00130-9
Hug, I., Couturier, M. R., Rooker, M. M., Taylor, D. E., Stein, M., and Feldman, M. F. (2010). Helicobacter pylori lipopolysaccharide is synthesized via a novel pathway with an evolutionary connection to protein N-glycosylation. PLoS Pathog. 6:e1000819. doi: 10.1371/journal.ppat.1000819
Ihssen, J., Kowarik, M., Dilettoso, S., Tanner, C., Wacker, M., and Thony-Meyer, L. (2010). Production of glycoprotein vaccines in Escherichia coli. Microb. Cell Fact. 9, 61. doi: 10.1186/1475-2859-9-61
Iwashkiw, J. A., Fentabil, M. A., Faridmoayer, A., Mills, D. C., Peppler, M., Czibener, C.,et al. (2012). Exploiting the Campylobacter jejuni protein glycosylation system for glycoengineering vaccines and diagnostic tools directed against brucellosis. Microb. Cell Fact. 11, 13. doi: 10.1186/1475-2859-11-13
Kitagawa, M., Ara, T., Arifuzzaman, M., Ioka-Nakamichi, T., Inamoto, E., Toyonaga, H.,et al. (2005). Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12, 291–299. doi: 10.1093/dnares/dsi012
Knirel, Y. A., Paramonov, N. A., Shashkov, A. S., Kochetkov, N. K., Yarullin, R. G., Farber, S. M.,et al. (1992). Structure of the polysaccharide chains of Pseudomonas pseudomallei lipopolysaccharides. Carbohydr. Res. 233, 185–193. doi: 10.1016/S0008-6215(00)90930-3
Kowarik, M., Young, N. M., Numao, S., Schulz, B. L., Hug, I., Callewaert, N.,et al. (2006). Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 25, 1957–1966. doi: 10.1038/sj.emboj.7601087
Lefebre, M. D., and Valvano, M. A. (2002). Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 68, 5956–5964. doi: 10.1128/AEM.68.12.5956-5964.2002
Linton, D., Dorrell, N., Hitchen, P. G., Amber, S., Karlyshev, A. V., Morris, H. R.,et al. (2005). Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol. Microbiol. 55, 1695–1703. doi: 10.1111/j.1365-2958.2005.04519.x
Lizak, C., Gerber, S., Numao, S., Aebi, M., and Locher, K. P. (2011). X-ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–355. doi: 10.1038/nature10151
Lockhart, S. (2003). Conjugate vaccines. Expert Rev. Vaccines 2, 633–648. doi: 10.1586/14760584.2.5.633
Nandi, T., and Tan, P. (2013). Less is more: Burkholderia pseudomallei and chronic melioidosis. MBio 4, e00709–00713. doi: 10.1128/mBio.00709-713
Nelson, M., Prior, J. L., Lever, M. S., Jones, H. E., Atkins, T. P., and Titball, R. W. (2004). Evaluation of lipopolysaccharide and capsular polysaccharide as subunit vaccines against experimental melioidosis. J. Med. Microbiol. 53, 1177–1182. doi: 10.1099/jmm.0.45766-0
Nieves, W., Asakrah, S., Qazi, O., Brown, K. A., Kurtz, J., Aucoin, D. P.,et al. (2011). A naturally derived outer-membrane vesicle vaccine protects against lethal pulmonary Burkholderia pseudomallei infection. Vaccine 29, 8381–8389. doi: 10.1016/j.vaccine.2011.08.058
Nothaft, H., Liu, X., Mcnally, D. J., and Szymanski, C. M. (2010). N-linked protein glycosylation in a bacterial system. Methods Mol. Biol. 600, 227–243. doi: 10.1007/978-1-60761-454-8_16
Nothaft, H., and Szymanski, C. M. (2010). Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8, 765–778. doi: 10.1038/nrmicro2383
Peacock, S. J., Limmathurotsakul, D., Lubell, Y., Koh, G. C., White, L. J., Day, N. P.,et al. (2012). Melioidosis vaccines: a systematic review and appraisal of the potential to exploit biodefense vaccines for public health purposes. PLoS Negl. Trop. Dis. 6:e1488. doi: 10.1371/journal.pntd.0001488
Perry, M. B., Maclean, L. L., Schollaardt, T., Bryan, L. E., and Ho, M. (1995). Structural characterization of the lipopolysaccharide O antigens of Burkholderia pseudomallei. Infect. Immun. 63, 3348–3352.
Pollard, A. J., Perrett, K. P., and Beverley, P. C. (2009). Maintaining protection against invasive bacteria with protein-polysaccharide conjugate vaccines. Nat. Rev. Immunol. 9, 213–220. doi: 10.1038/nri2494
Scott, A. E., Burtnick, M. N., Stokes, M. G., Whelan, A. O., Williamson, E. D., Atkins, T. P.,et al. (2014). Burkholderia pseudomallei capsular polysaccharide conjugates provide protection against acute melioidosis. Infect. Immun. 82, 3206–3213. doi: 10.1128/IAI.01847-14
Scott, A. E., Laws, T. R., D’Elia, R. V., Stokes, M. G., Nandi, T., Williamson, E. D.,et al. (2013). Protection against experimental melioidosis following immunization with live Burkholderia thailandensis expressing a manno-heptose capsule. Clin. Vaccine Immunol. 20, 1041–1047. doi: 10.1128/CVI.00113-13
Silva, E. B., and Dow, S. W. (2013). Development of Burkholderia mallei and pseudomallei vaccines. Front. Cell Infect. Microbiol. 3:10. doi: 10.3389/fcimb.2013.00010
Su, Y. C., Wan, K. L., Mohamed, R., and Nathan, S. (2010). Immunization with the recombinant Burkholderia pseudomallei outer membrane protein Omp85 induces protective immunity in mice. Vaccine 28, 5005–5011. doi: 10.1016/j.vaccine.2010.05.022
Thomason, L. C., Costantino, N., and Court, D. L. (2007). E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. Chap. 1, Unit 1.17. doi: 10.1002/0471142727.mb0117s79
Valvano, M. A., and Crosa, J. H. (1989). Molecular cloning and expression in Escherichia coli K-12 of chromosomal genes determining the O7 lipopolysaccharide antigen of a human invasive strain of E. coli O7:K1. Infect. Immun. 57, 937–943.
Wacker, M., Wang, L., Kowarik, M., Dowd, M., Lipowsky, G., Faridmoayer, A.,et al. (2014). Prevention of Staphylococcus aureus infections by glycoprotein vaccines synthesized in Escherichia coli. J. Infect. Dis. 209, 1551–1561. doi: 10.1093/infdis/jit800
Wetter, M., Kowarik, M., Steffen, M., Carranza, P., Corradin, G., and Wacker, M. (2013). Engineering, conjugation, and immunogenicity assessment of Escherichia coli O121 O antigen for its potential use as a typhoid vaccine component. Glycoconj. J. 30, 511–522. doi: 10.1007/s10719-012-9451-9
Keywords: glycobiology, vaccines, protein glycosylation, microbiology and biotechnology, molecular biology, mass spectrometry
Citation: Garcia-Quintanilla F, Iwashkiw JA, Price NL, Stratilo C and Feldman MF (2014) Production of a recombinant vaccine candidate against Burkholderia pseudomallei exploiting the bacterial N-glycosylation machinery. Front. Microbiol. 5:381. doi: 10.3389/fmicb.2014.00381
Received: 03 June 2014; Paper pending published: 29 June 2014;
Accepted: 08 July 2014; Published online: 29 July 2014.
Edited by:
Ivan Mijakovic, Chalmers University of Technology, SwedenReviewed by:
Brendan Wren, University of London, UKLei Shi, Chalmers University of Technology, Sweden
Copyright © 2014 Garcia-Quintanilla, Iwashkiw, Price, Stratilo and Feldman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mario F. Feldman, Department of Biological Sciences, University of Alberta, CCIS 6-063, 116 Street, 85 Avenue, Edmonton, AB T6G 2R3, Canada e-mail:bWZlbGRtYW5AdWFsYmVydGEuY2E=
†Fatima Garcia-Quintanilla and Jeremy A. Iwashkiw have equally contributed to this work.
 Fatima Garcia-Quintanilla1†
Fatima Garcia-Quintanilla1† Chad Stratilo
Chad Stratilo Mario F. Feldman
Mario F. Feldman