- 1Food Safety and Technology Research Centre, Hong Kong Polytechnic University – Shenzhen Research Institute, Shenzhen, China
- 2State Key Laboratory of Chirosciences, Department of Applied Biology and Chemical Technology, The Hong Kong Polytechnic University, Kowloon, China
- 3Department of Microbiology, The Prince of Wales Hospital – The Chinese University of Hong Kong, Shatin, China
Emergence of multidrug-resistant Salmonella typhimurium strains, especially the ACSSuT and nalidixic acid R types, has significantly compromised the effectiveness of current strategies to control Salmonella infections, resulting in increased morbidity and mortality. Clinical S. typhimurium isolates recovered in Hong Kong during the period of 2005–2011 were increasingly resistant to ciprofloxacin (CIP) and antibiotics of the ACSSuT group. Our data revealed that oqxAB and aac(6′)Ib-cr were encoded on plasmids of various sizes and the presence of these two elements together with a single gyrA mutation in S. typhimurium were sufficient to mediate resistance to CIP. Acquisition of the oqxAB and aac(6′)Ib-cr encoding plasmids by S. typhimurium caused a fourfold increase in CIP minimal inhibitory concentration. Furthermore, the presence of oqxAB and aac(6′)Ib-cr in Salmonella dramatically increased the mutation prevention concentration of CIP which may due to mutational changes in the drug target genes. In conclusion, possession of oqxAB and aac(6′)Ib-cr encoding plasmid facilitate the selection of CIP resistant S. typhimurium, thereby causing a remarkable increase of CIP resistance among clinical Salmonella strains in Hong Kong.
Introduction
Non-typhoidal Salmonella are among the principal bacterial pathogens implicated in food-borne gastroenteritis worldwide (Gomez et al., 1997). Antimicrobial agents are not usually required for treatment in salmonellosis but can be lifesaving in cases of severe or systemic infections, as well as treatment for elderly and immunocompromised patients (Hohmann, 2001). Multidrug resistance in Salmonella has been documented since 1980, a representative class of resistant organisms being the ACSSuT resistance type of Salmonella typhimurium DT104, which originated in the UK and spread rapidly to the US and other parts of the world (Centers for Disease Control and Prevention, 1997; Glynn et al., 1998; Markogiannakis et al., 2000). Hence fluoroquinolones and the extended-spectrum cephalosporins have become the drugs of choice for treatment of acute gastroenteritis caused by Salmonella and other enteric pathogens. Resistance toward quinolone and fluoroquinolone antimicrobials is mainly due to target mutations in quinolone resistance determining region (QRDR) of DNA gyrases (gyrA and gyrB) and Type IV topoisomerases (parC and parE), which subsequently prevent drugs from binding (Hawkey, 2003). Although high level fluoroquinolone-resistant Salmonella are known to be associated with specific serotypes of Salmonella and have been reported in scattered regions around the world, their prevalence remains low. This is probably due to the fact that the process of selection of double gyrA and single parC mutations is not very efficient. Nevertheless, several lines of evidence have suggested that emergence of multidrug resistant non-typhoidal Salmonella strains has significant impact on the effectiveness of current strategies, including reduced efficacy of early empirical treatment to control and manage diseases associated with food-borne infections. These include reduced efficacy of early empirical treatment as well as limited choice of treatment.
Plasmid mediated quinolone resistance (PMQR) genes such as qnrA, qnrB, qnrC, qnrD, qnrS, qepA, and aac(6′)Ib-cr have been increasingly reported in bacterial pathogens. The qnr type PMQR genes bind to DNA gyrase and topoisomerase to block the action of fluoroquinolones resulting in reduced susceptibility to fluoroquinolones (Tran et al., 2005). QepA encodes a MFS-type efflux pump which is able to excrete quinolone into extracelluar space (Yamane et al., 2007). AAC(6′)Ib-cr acetylates ciprofloxacin (CIP) and norfloxacin (Robicsek et al., 2006). It is postulated that these PMQR genes are able to contribute to the development of quinolone resistance in these organisms (Cattoir and Nordmann, 2009). Recently, a novel transmissible resistance-nodulation-division (RND) efflux pump OqxAB, which mediated resistance to olaquindox, chloramphenicol, nalidixic acid, and elevated minimal inhibitory concentrations (MICs) of other antimicrobial reagents including ampicillin, gentamicin, and CIP (MIC between 0.06 and 0.25 μg/ml), has been identified (Hansen et al., 2007). OqxAB was encoded on an IncXI plasmid, pOLA52, harbored by a swine Escherichia coli isolate (Hansen et al., 2004). More recently, OqxAB was reported to be prevalent in organisms isolated from pork and pig farms in China (Zhao et al., 2010; Liu et al., 2011; Wong and Chen, 2012), as well as from human food (18). On the other hand, the oqxAB gene has not been found in clinical isolates until recently, when it became detectable in clinical strains of E. coli, Salmonella, and Klebsiella pneumoniae (Kim et al., 2009; Park et al., 2012; Ruiz et al., 2012; Yuan et al., 2012). The functional and clinical significance of this and other PMQR genes such as qnrA, B, D, and S, which have also become prevalent in clinical Salmonella isolates (Cavaco et al., 2009; Ferrari et al., 2011; Kim et al., 2013), remains unclear.
We have previously characterized 239 human clinical S. typhimurium isolates recovered from hospital patients in Hong Kong during the period of 2005–2011 for their susceptibilities to fluoroquinolones and other antibiotics and the prevalence of PMQR genes. Two PMQR genes, oqxAB and aac(6′)Ib-cr, were found to exhibit close relationship with fluoroquinolone resistance in S. typhimurium. Approximately 44% of the oqxAB-positive S. typhimurium were resistant to CIP and around 89% of oqxAB, aac(6′)Ib-cr-positive isolates were resistant to CIP, while only 11% of the oqxAB-negative isolates were resistant to CIP (Wong et al., 2013). In the current study, we want to investigate the direct association of oqxAB/aac(6′)Ib-cr with the development of fluoroquinolone resistance in S. typhimurium. We confirm that oqxAB and aac(6′)Ib-cr can mediate rapid development of fluoroquinolone resistance in S. typhimurium and could be a contributive factors accounting for the increasing prevalence of fluoroquinolone-resistant Salmonella in Hong Kong in recent years.
Materials and Methods
Bacterial Strains
239 S. typhimurium previously described clinical isolates were included in this study (Wong et al., 2013).
PCR and Target Gene Mutation Screening in S. typhimurium
The QRDRs of gyrA and parC were amplified by PCR as previously described (Chen et al., 2007), followed by determination of their nucleotide sequences and comparison to the wild-type S. typhimurium LT2 strain to identify target gene mutations in the test strains. The gyrA and parC sequences of four Salmonella isolates, S08-52, S10-9, S05-23, and S05-30, were submitted to GenBank with the accession numbers for gyrA, KM504240, KM504241, KM504242, and KM504243 and parC, KM513651, KM513652, KM513653, and KM513654. The association of Insertion sequence IS26 with oqxAB was determined by PCR using primers IS26-F(5′GCTGTTACGACGGGAGGAG) and oqx-R (5′ GGAGACGAGGTTGGTATGGA).
Conjugation Experiments
A conjugative experiment was carried out as previously described (Jacoby et al., 2003) using sodium azide-resistant E. coli J53 strain as recipient. Briefly, overnight culture of donor and recipient strains were mixed and collected on a filter, which was subjected to overnight incubation on a blood agar plate. The mixture was then spread on double selective blood agar plates containing olaquindox (128 μg/ml) and sodium azide (100 μg/ml).
S1-PFGE and Hybridization
S1-PFGE was conducted to determine the size of large plasmids. Briefly, agarose-embedded DNA was digested with S1 nuclease (New England BioLab) at 37°C for 1 h. The restriction fragments were separated by electrophoresis in 0.5 Tris-borate-EDTA buffer at 14°C for 18 h using a Chef Mapper electrophoresis system (Bio-Rad, Hercules, CA, USA) with pulse times of 2.16 to 63.8 s. Phage Lambda PFGE ladder (New England BioLab) was used as DNA size marker. The gels were stained with GelRed, and DNA bands were visualized with UV transillumination (Bio-Rad). Chromosomal and plasmid DNA of S. typhimurium strains were transferred and cross-linked onto nylon membrane and hybridized with a DIG-labeled oqxAB probe using DIG High Prime DNA Labeling and Detection Starter Kit I (Roche) following manufacturer’s instructions to determine the localization of oqxAB and aac(6′)-Ib-cr genes in S. typhimurium genetic materials.
oqxAB Cloning, Plasmid Transformation, and Plasmid Curing
Cloning of oqxAB into pTrcHisB (Life Technologies) vector was done by PCR using primers pTrc-oqxAB-F (5′TTACTACTCGAGAATGAGCCTGCAAAAAAC) and pTrcoqxAB-R (5′AGGATCGAATTCCTAGGCGGGCAGATCCTC). pTrc-oqxAB was transformed into S. typhimurium LT2. Plasmids from clinical strains were extracted by Qiagen Mini-prep kit, electroporated into S. typhimurium LT2 and a nalidixic acid and CIP susceptible S. typhimurium clinical strain 11–28, and selected on plates containing 32 μg/ml olaquindox. Plasmid curing was performed on clinical S. typhimurium strain 10–63 as previously described with slight modification (Sato et al., 2013). The strain was grown in 3 ml LB at 43°C for 2 weeks and selected on plates containing 0, 8, 16, 32 μg/ml olaquindox.
Mutation Prevention Concentration (MPC)
Mutation prevention concentration (MPC) of oqxAB, aac(6′)Ib-cr positive, and negative strains was determined as described previously (Gebru et al., 2011, 2012). Briefly, MPC was determined by spreading 1 × 109 cells on LB agar plates containing a range of concentration of CIP: 0, 0.05, 0.1, 0.25, 0.5, 1, 2, 4, 8, 16, 32 μg/ml. Plates containing CIP were incubated for up to 72 h, whereas CIP-free plates were incubated for 24 h. Viable counts on each plate were recorded. MPC was defined as the lowest antibiotic concentration at which no colonies were observed. For each strain, MPC was determined on the basis of the results of at least three independent experiments.
Results
PMQRS and Single Gyra Mutation Mediate Development of Fluoroquinolone Resistance in S. typhimurium
To determine if oqxAB alone or the combination of oqxAB and aac(6′)Ib-cr can contribute to fluoroquinolone resistance, the effect of interplay between oqxAB, aac(6′)Ib-cr, and target mutations in mediating fluoroquinolone resistance phenotypes in Salmonella was studied. Among all oqxAB negative S. typhimurium organisms, the vast majority of those which exhibited CIP MIC ≤0.05 μg/ml had no mutation in the gyrA and parC genes. Single amino acid substitution (D87Y or D87N) in GyrA was often detected in strains with CIP MIC between 0.1 and 1 μg/ml. Interestingly, two CIP-resistant isolates (CIP MIC = 1 μg/ml) showed only single mutation at D87Y. Double substitution in GyrA (S83F and D87Y or N) and a single substitution in ParC (S80I) were consistently detected in those with CIP MIC ≥2 μg/ml (Table 1). Among all oqxAB positive S. typhimurium strains, no mutation was detected in gyrA and parC in strains with CIP MIC ≤0.05 μg/ml; single mutation in GyrA (D87Y or D87N), but not in ParC, was detected in strains whose CIP MIC was between 0.25 and 2 μg/ml. Among all isolates which were positive to both oqxAB and aac(6′)Ib-cr, single mutation in GyrA (D87Y or D87N), but not in ParC, was detected in strains whose CIP MIC was between 0.25 and 2 μg/ml, whereas most of the strains from this category exhibited CIP MIC ≥1 μg/ml. Comparative analysis of mutational and drug susceptibility data of oqxAB negative, oqxAB positive, and oqxAB, aac(6′)Ib-cr positive strains showed that similar mutational profiles could result in drastically different CIP MIC, depending on whether the organism harbored the oqxAB or oqxAB, aac(6′)Ib-cr genes. Strikingly, simultaneous presence of a single gyrA mutation and oqxAB, or both oqxAB and aac(6′)Ib-cr genes, was sufficient to produce CIP resistance (CIP MIC = 1 μg/ml); however, double mutations in gyrA plus a single mutation in parC were required to mediate CIP MIC ≥2 μg/ml when oqxAB was absent. Importantly, around 98% of oqxAB positive S. typhimurium strains harbored mutations in the gyrA or parC genes, whereas less than 60% of oqxAB negative S. typhimurium strains had mutations in either or both of these two genes (Data not shown). Taken together, these findings suggest that acquisition of oqxAB or oqxAB, aac(6′)Ib-cr by S. typhimurium could mediate selection of fluoroquinolone resistance in S. typhimurium.
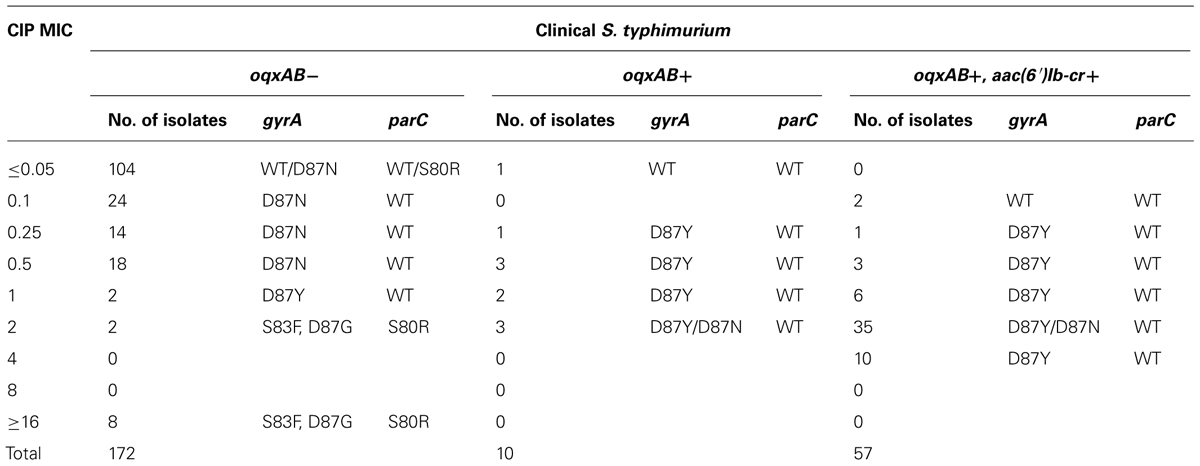
TABLE 1. Presence of target mutations in different level of ciprofloxacin MIC of oqxAB positive and negative Salmonella typhimurium isolates.
Transferability and Genetic Location of oqxAB
Thirty randomly selected oqxAB positive S. typhimurium isolates were subjected to conjugation experiment to determine the transferability of the oqxAB gene that they harbored. Surprisingly, none of the S. typhimurium strains tested was able to transfer this resistance element to E. coli J53 recipient strain through conjugation. S1-PFGE and Southern hybridization were performed on four S. typhimurium isolates and it showed that oqxAB and aac(6′)Ib-cr were concurrently present on plasmids of various sizes in these S. typhimurium isolates, hybridization results of two of which were shown in Figure 1. In all the tested S. typhimurium isolates, the oqxAB gene was found to be flanked by the IS26 fragment in a manner similar to that of the pOLA52 plasmid as previously reported (Norman et al., 2008), suggesting that the oqxAB gene that was becoming prevalent in S. typhimurium could have been derived from the original transferable element located in pOLA52. To test this possibility, we performed PCR screening to determine if pOLA52 specific DNA sequences were prevalent among the test plasmids. To our surprise, however, none of the plasmids that carried oqxAB and aac(6′)Ib-cr contained such sequences of pOLA52 (Data not shown).
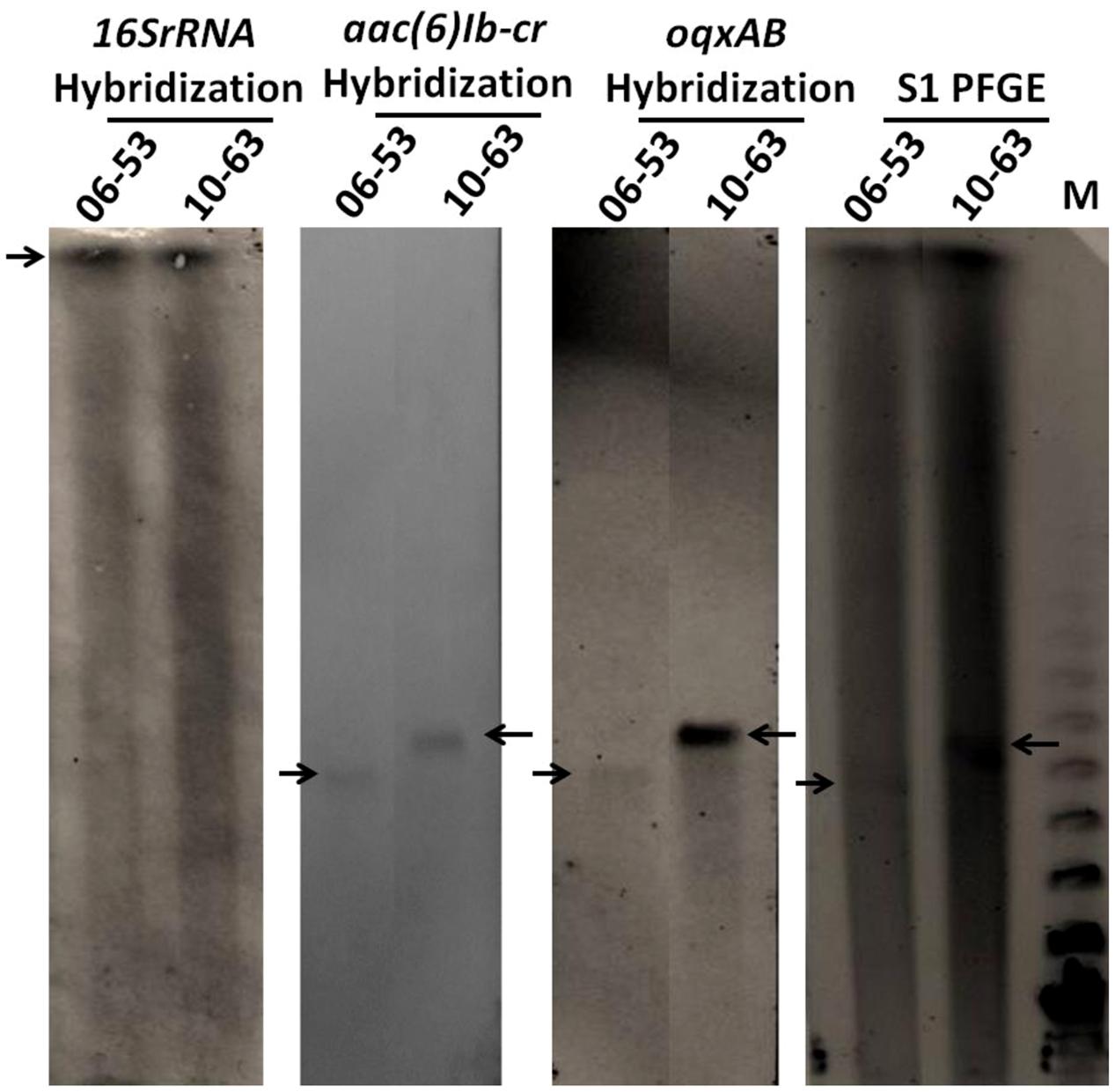
FIGURE 1. S1-PFGE and southern hybridization of 16SrRNA, oqxA, and aac(6′)Ib-cr on two oqxAB-positive isolates. Arrows indicated chromosomal DNA or plasmids harboring oqxAB and aac(6′)Ib-cr. 06–53 and 10–63 are oqxAB-positive S. typhimurium clinical isolates; M, Lambda PFGE marker.
Contribution of oqxAB and aac(6′)Ib-cr to the Elevated Ciprofloxacin Mic in S. typhimurium
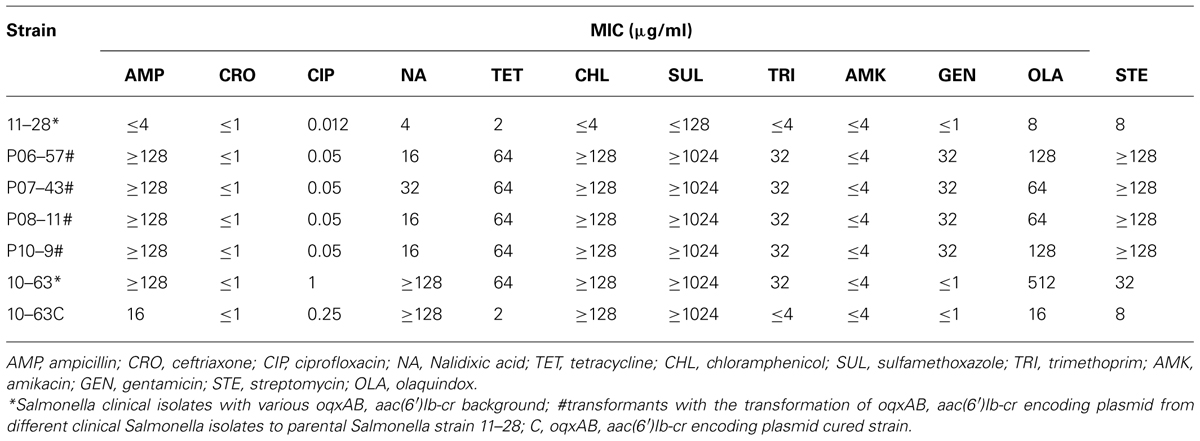
To directly prove the degree of contribution of oqxAB and aac(6′)Ib-cr to the development of fluoroquinolone resistance in S. typhimurium, oqxAB was cloned into a pTrc expression vector and transformed into S. typhimurium LT2 strain. Compared to the original oqxAB negative S. typhimurium LT2 strain, pTrc-oqxAB- carrying S. typhimurium LT2 exhibited a CIP MIC of 0.25 μg/ml, with a 20-fold increase. However, S. typhimurium LT2 carrying pTrc-oqxAB showed much weaker growth than its parental counterpart, which was presumably due to the fitness cost caused by the over-expression of oqxAB in the host strain. To overcome this problem, the plasmids that carried oqxAB and aac(6′)Ib-cr were extracted from different clinical S. typhimurium isolates and electroporated into S. typhimurium LT2 with no success. The plasmids were then electroporated into an oqxAB-negative S. typhimurium strain 11–28. Upon acquisition of such plasmid, the CIP MIC of this S. typhimurium strain increased by approximately fourfold (Table 2). To further prove the contribution of oqxAB and aac(6′)Ib-cr to S. typhimurium fluoroquinolone resistance, the plasmid carrying such genes in a clinical S. typhimurium strain 10–63 was cured and it showed that the curing of the plasmid in 10–63 decreased the CIP MIC by approximately fourfold (Table 2). Taken together, our data had proven that oqxAB and aac(6′)Ib-cr contributed to about four fold increase of CIP MIC in S. typhimurium. The MICs of other antibiotics were also determined for S. typhimurium that acquired oqxAB, aac(6′)Ib-cr encoding plasmids. In addition, it is showed that acquisition of oqxAB and aac(6′)Ib-cr encoding plasmids ensured resistance to ampicillin, chloramphenicol, streptomycin, nalidixic acid, sulfamethoxazole, tetracycline, trimethoprim, and olaquindox in addition to the elevated CIP MIC (Table 2). This is also consistent to our previous finding that the presence of oqxAB in S. typhimurium was associated with the ACSSuT R phenotype. As much as 56% of oqxAB-positive S. typhimurium clinical isolates were resistant to ACSSuT, whereas only 14% of oqxAB-negative isolates were resistant to ACSSuT (Wong et al., 2013).
Contribution of oqxAB and aac(6′)Ib-Cr to Elevated MPC of Fluoroquinolone in S. typhimurium
To validate the hypothesis that oqxAB and aac(6′)Ib-cr contributed to mutation development, MPC of CIP were determined for S. typhimurium with and without oqxAB. As shown in Table 3, oqxAB, aac(6′)Ib-cr positive clinical Salmonella isolates, 06–57, 07–43, and 08–11 exhibited much higher MPC of CIP than the oqxAB, aac(6′)Ib-cr-negative Salmonella strains, 05–41, 07–54, and 10–25 (Table 3). Furthermore, although Salmonella 11–28 strain exhibited MPC for CIP of about 0.1 μg/ml, transformation of plasmids from other clinical Salmonella isolates carrying oqxAB, aac(6′)Ib-cr to Salmonella 11–28 dramatically increased its MPC to 2∼4 μg/ml. On the other hand, Salmonella 10–63 exhibited MPC of 8 μg/ml, yet the curing of the oqxAB, aac(6′)Ib-cr encoding plasmid led to a slightly decreased MPC (4 μg/ml). The minimal effect of curing of oqxAB, aac(6′)Ib-cr encoding plasmid on the MPC of 10–63 may be due to the fact that the long-term starvation stress used to cure the plasmid may have caused stress response to develop in the isolate, thereby indirectly contributing to the elevated MPC for strain.
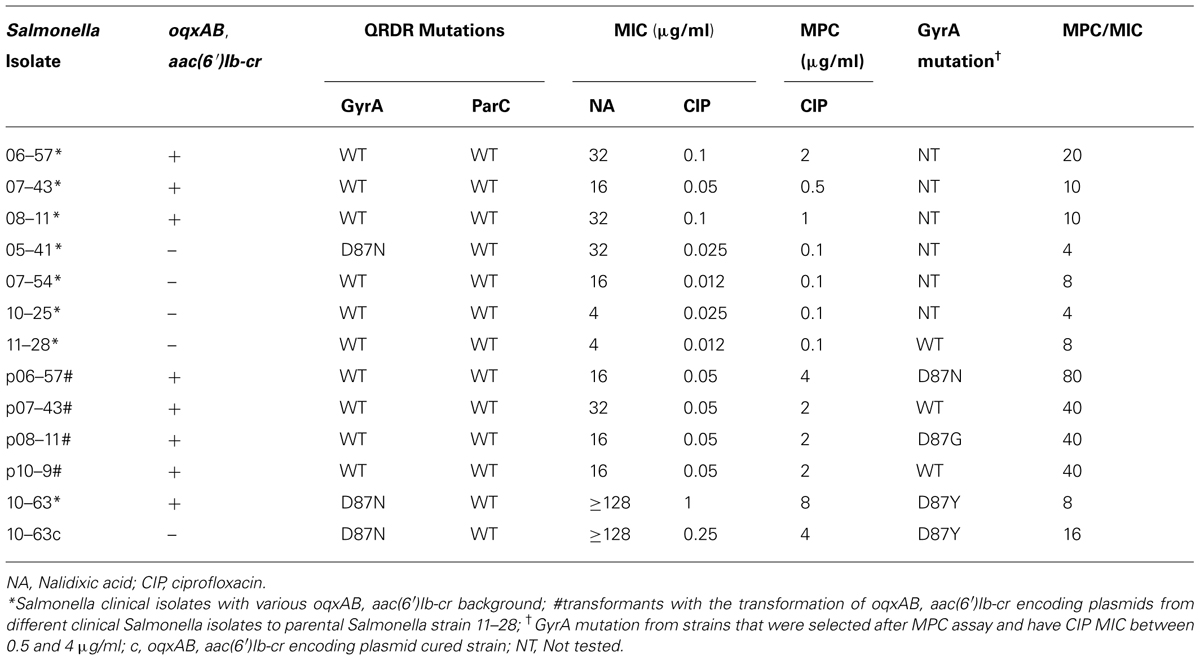
TABLE 3. MICs of nalidixic acid (NA) and ciprofloxacin (CIP) and mutation prevention concentration (MPC) toward CIP of Salmonella isolates with various background of oqxAB and aac(6′)Ib-cr.
Ten to 63 C. It has been shown that long-term starvation stress stimulates the stringent SOS response in bacteria, which is essential in bacteria for acquisition of mutations leading to resistance to some antibiotic drugs (Fung et al., 2010). Most importantly, compared to Salmonella 11–28 alone, which did not develop gyrA mutation in MPC assay, Salmonella 11–28 transformed with oqxAB, aac(6′)Ib-cr encoding plasmids from Salmonella 06–57 and 08–11 developed single mutation in gyrA, which may partly contributed to the increase of CIP MPC (Table 3). It is probably due to that the presence of oqxAB and aac(6′)Ib-cr may enable S. typhimurium to survive under fluoroquinolone stress and facilitate subsequent development of target mutations. Nevertheless, these data confirm that oqxAB, aac(6′)Ib-cr plays a key role in elevated CIP MIC and MPC, and hence resistance to fluoroquinolone in S. typhimurium.
Discussion
An important finding in this work is that the oqxAB and aac(6′)Ib-cr gene products not only directly contribute to the elevated CIP MIC, but also enhance the ability of S. typhimurium to survive in an environment with high dose of CIP, which may in turn facilitate the development of fluoroquinolone resistance. The mechanism of fluoroquinolone resistance in Salmonella has conventionally been attributed to double mutations in gyrA with or without a single parC mutation (Casin et al., 2003; Chu et al., 2005). Unlike E. coli and Campylobacter, double gyrA mutations in Salmonella were rare and presumably difficult to acquire, therefore fluoroquinolone remained an effective treatment of choice for severe Salmonella infections. In this study, we demonstrated that acquisition of the oqxAB or oqxAB, aac(6′)Ib-cr genes in S. typhimurium, could mediate development of resistance to CIP (CIP MIC ≥1 μg/ml). We postulate that the pump activities and enzymatic hydrolysis of fluoroquinolones enable the organisms to withstand antibiotic pressure for a prolonged period, during which mutational changes can occur. Elevation of the antibiotic resistance potential of Salmonella is one way by which oqxAB can help the host strain to successfully launch clinical infection in human, leading to a dramatic increase in the proportion of oqxAB positive strains observable among clinical Salmonella isolates recovered in recent years (Wong et al., 2014). The increased prevalence of oqxAB positive S. typhimurium in clinical isolates also contributes directly to a higher percentage of fluoroquinolone resistance in clinical salmonella strains. In 2011, the proportion of the oqxAB positive S. typhimurium in Hong Kong that were found to be resistant to CIP reached 34% (Data not shown).
The fact that oqxAB could not be found in S. typhimurium until 2006 may be due to its poor ability to replicate in Salmonella initially; this notion is supported by the fact that transformation of oqxAB-borne plasmid to S. typhimurium did not elevate MIC of CIP in these strains and that direct expression of oqxAB into S. typhimurium had a fitness cost in this work (Hansen et al., 2007; Wong et al., 2013). Nevertheless, our data indicate that the oqxAB gene has adapted to co-exist in S. typhimurium. In this study, oqxAB were found to be associated with IS26 but not carried by pOLA52-like plasmids, suggesting oqxAB was excised from pOLA52 and integrated into other plasmids mediated by IS26 transposase. Since no oqxAB encoding plasmid in Salmonella has been sequenced, the mechanism underlying the co-existence of oqxAB and aac(6′)Ib-cr in over 80% of the oqxAB-positive strain is not clear. The quick expansion of oqxAB and aac(6′)Ib-cr positive, CIP-resistant S. typhimurium will pose huge threat to efforts of infection control of Salmonella infections. Urgent actions are required to halt further transmission of the oqxAB positive strains in both environmental and clinical settings. In addition, it remains to be seen if oqxAB has been taken up by other bacterial species and whether it plays a role in the evolution of resistance and virulence traits of various bacterial pathogens. Findings in this work also highlight a need to investigate the impact of oqxAB in a wide range of foodborne and zoonotic pathogens.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Julia Ling for her assistance in the collection of clinical Salmonella isolates in Hong Kong. This work was supported by the Chinese National Key Basic Research and Development (973) Program (2013CB127200) and the Health and Medical Research Fund of the Food and Health Bureau, The Government of Hong Kong (13121412 and 14130402 to Sheng Chen).
References
Casin, I., Breuil, J., Darchis, J. P., Guelpa, C., and Collatz, E. (2003). Fluoroquinolone resistance linked to GyrA, GyrB, and ParC mutations in Salmonella enterica Typhimurium isolates in humans. Emerg. Infect. Dis. 9, 1455–1457. doi: 10.3201/eid0911.030317
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cattoir, V., and Nordmann, P. (2009). Plasmid-mediated quinolone resistance in gram-negative bacterial species: an update. Curr. Med. Chem. 16, 1028–1046. doi: 10.2174/092986709787581879
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cavaco, L. M., Hasman, H., Xia, S., and Aarestrup, F. M. (2009). qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob. Agents Chemother. 53, 603–608. doi: 10.1128/AAC.00997-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Centers for Disease Control and Prevention. (1997). Multidrug-resistant Salmonella serotype Typhimurium–United States, 1996. MMWR Morb. Mortal. Wkly. Rep. 46, 308–310.
Chen, S., Cui, S., McDermott, P. F., Zhao, S., White, D. G., Paulsen, I.,et al. (2007). Contribution of target gene mutations and efflux to decreased susceptibility of Salmonella enterica serovar typhimurium to fluoroquinolones and other antimicrobials. Antimicrob. Agents Chemother. 51, 535–542. doi: 10.1128/AAC.00600-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chu, C., Su, L. H., Chu, C. H., Baucheron, S., Cloeckaert, A., and Chiu, C. H. (2005). Resistance to fluoroquinolones linked to gyrA and par C mutations and overexpression of acr AB efflux pump in Salmonella enterica serotype Choleraesuis. Microb. Drug Resist. 11, 248–253. doi: 10.1089/mdr.2005.11.248
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ferrari, R., Galiana, A., Cremades, R., Rodriguez, J. C., Magnani, M., Tognim, M. C.,et al. (2011). Plasmid-mediated quinolone resistance by genes qnrA1 and qnrB19 in Salmonella strains isolated in Brazil. J. Infect. Dev. Ctries. 5, 496–498. doi: 10.3855/jidc.1735
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fung, D. K., Chan, E. W., Chin, M. L., and Chan, R. C. (2010). Delineation of a bacterial starvation stress response network which can mediate antibiotic tolerance development. Antimicrob. Agents Chemother. 54, 1082–1093. doi: 10.1128/AAC.01218-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gebru, E., Choi, M. J., Lee, S. J., Damte, D., and Park, S. C. (2011). Mutant-prevention concentration and mechanism of resistance in clinical isolates and enrofloxacin/marbofloxacin-selected mutants of Escherichia coli of canine origin. J. Med. Microbiol. 60, 1512–1522. doi: 10.1099/jmm.0.028654-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gebru, E., Damte, D., Choi, M. J., Lee, S. J., Kim, Y. H., and Park, S. C. (2012). Mutant prevention concentration and phenotypic and molecular basis of fluoroquinolone resistance in clinical isolates and in vitro-selected mutants of Escherichia coli from dogs. Vet. Microbiol. 154, 384–394. doi: 10.1016/j.vetmic.2011.07.033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Glynn, M. K., Bopp, C., Dewitt, W., Dabney, P., Mokhtar, M., and Angulo, F. J. (1998). Emergence of multidrug-resistant Salmonella enterica serotype typhimurium DT104 infections in the United States. N. Engl. J. Med. 338, 1333–1338. doi: 10.1056/NEJM199805073381901
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gomez, T. M., Motarjemi, Y., Miyagawa, S., Kaferstein, F. K., and Stohr, K. (1997). Foodborne salmonellosis. World Health Stat. Q. 50, 81–89.
Hansen, L. H., Jensen, L. B., Sorensen, H. I., and Sorensen, S. J. (2007). Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J. Antimicrob. Chemother. 60, 145–147. doi: 10.1093/jac/dkm167
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hansen, L. H., Johannesen, E., Burmolle, M., Sorensen, A. H., and Sorensen, S. J. (2004). Plasmid-encoded multidrug efflux pump conferring resistance to olaquindox in Escherichia coli. Antimicrob. Agents Chemother. 48, 3332–3337. doi: 10.1128/AAC.48.9.3332-3337.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hawkey, P. M. (2003). Mechanisms of quinolone action and microbial response. J. Antimicrob. Chemother. 51(Suppl. 1), 29–35. doi: 10.1093/jac/dkg207
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hohmann, E. L. (2001). Nontyphoidal salmonellosis. Clin. Infect. Dis. 32, 263–269. doi: 10.1086/318457
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jacoby, G. A., Chow, N., and Waites, K. B. (2003). Prevalence of plasmid-mediated quinolone resistance. Antimicrob. Agents Chemother. 47, 559–562. doi: 10.1128/AAC.47.2.559-562.2003
Kim, H. B., Wang, M., Park, C. H., Kim, E. C., Jacoby, G. A., and Hooper, D. C. (2009). oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 3582–3584. doi: 10.1128/AAC.01574-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, J. H., Cho, J. K., and Kim, K. S. (2013). Prevalence and characterization of plasmid-mediated quinolone resistance genes in Salmonella isolated from poultry in Korea. Avian Pathol. 42, 221–229. doi: 10.1080/03079457.2013.779636
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, B. T., Wang, X. M., Liao, X. P., Sun, J., Zhu, H. Q., Chen, X. Y.,et al. (2011). Plasmid-mediated quinolone resistance determinants oqxAB and aac(6′)-Ib-cr and extended-spectrum beta-lactamase gene blaCTX-M-24 co-located on the same plasmid in one Escherichia coli strain from China. J. Antimicrob. Chemother. 66, 1638–1639. doi: 10.1093/jac/dkr172
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Markogiannakis, A., Tassios, P. T., Lambiri, M., Ward, L. R., Kourea-Kremastinou, J., Legakis, N. J.,et al. (2000). Multiple clones within multidrug-resistant Salmonella enterica serotype Typhimurium phage type DT104. The Greek nontyphoidal Salmonella study group. J. Clin. Microbiol. 38, 1269–1271.
Norman, A., Hansen, L. H., She, Q., and Sorensen, S. J. (2008). Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60, 59–74. doi: 10.1016/j.plasmid.2008.03.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Park, K. S., Kim, M. H., Park, T. S., Nam, Y. S., Lee, H. J., and Suh, J. T. (2012). Prevalence of the plasmid-mediated quinolone resistance genes, aac(6′)-Ib-cr, qepA, and oqxAB in clinical isolates of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae in Korea. Ann. Clin. Lab. Sci. 42, 191–197.
Robicsek, A., Strahilevitz, J., Jacoby, G. A., Macielag, M., Abbanat, D., Park, C. H.,et al. (2006). Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12, 83–88. doi: 10.1038/nm1347
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ruiz, E., Saenz, Y., Zarazaga, M., Rocha-Gracia, R., Martinez-Martinez, L., Arlet, G.,et al. (2012). qnr, aac(6′)-Ib-cr and qepA genes in Escherichia coli and Klebsiella spp.: genetic environments and plasmid and chromosomal location. J. Antimicrob. Chemother. 67, 886–897. doi: 10.1093/jac/dkr548
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sato, T., Yokota, S., Uchida, I., Okubo, T., Usui, M., Kusumoto, M.,et al. (2013). Fluoroquinolone resistance mechanisms in an Escherichia coli isolate, HUE1, without quinolone resistance-determining region mutations. Front. Microbiol. 4:125. doi: 10.3389/fmicb.2013.00125
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tran, J. H., Jacoby, G. A., and Hooper, D. C. (2005). Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 49, 118–125. doi: 10.1128/AAC.49.1.118-125.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wong, M. H., and Chen, S. (2012). First detection of oqxAB in Salmonella spp. isolated from food. Antimicrob. Agents Chemother. 57, 658–660. doi: 10.1128/AAC.01144-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wong, M. H., Yan, M., Chan, E. W., Biao, K., and Chen, S. (2014). Emergence of clinical Salmonella Typhimurium with concurrent resistant to ciprofloxacin, ceftriaxone and azithromycin. Antimicrob. Agents Chemother. 58, 3752–3756. doi: 10.1128/AAC.02770-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wong, M. H., Yan, M., Chan, E. W., Liu, L. Z., Kan, B., and Chen, S. (2013). Expansion of Salmonella Typhimurium ST34 clone carrying multiple resistance determinants in China. Antimicrob. Agents Chemother. doi: 10.1128/AAC.01174-13 [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yamane, K., Wachino, J., Suzuki, S., Kimura, K., Shibata, N., Kato, H.,et al. (2007). New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 51, 3354–3360. doi: 10.1128/AAC.00339-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yuan, J., Xu, X., Guo, Q., Zhao, X., Ye, X., Guo, Y.,et al. (2012). Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and Escherichia coli clinical isolates. J. Antimicrob. Chemother. 67, 1655–1659. doi: 10.1093/jac/dks086
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, J., Chen, Z., Chen, S., Deng, Y., Liu, Y., Tian, W.,et al. (2010). Prevalence and dissemination of oqxAB in Escherichia coli isolates from animals, farmworkers, and the environment. Antimicrob. Agents Chemother. 54, 4219–4224. doi: 10.1128/AAC.00139-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: S. typhimurium, ciprofloxacin resistance, ACSSuT R type, oqxAB, aac(6′)Ib-cr
Citation: Wong MH, Chan EW, Liu LZ and Chen S (2014) PMQR genes oqxAB and aac(6′)Ib-cr accelerate the development of fluoroquinolone resistance in Salmonella typhimurium. Front. Microbiol. 5:521. doi: 10.3389/fmicb.2014.00521
Received: 06 August 2014; Accepted: 17 September 2014;
Published online: 02 October 2014.
Edited by:
Attilio Vittorio Vargiu, Universitá di Cagliari, ItalyReviewed by:
Daniela Ceccarelli, University of Maryland, USAEtinosa Igbinosa, University of Benin, Nigeria
Amit Kumar, Kansas State University, USA
Copyright © 2014 Wong, Chan, Liu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheng Chen, State Key Laboratory of Chirosciences, Department of Applied Biology and Chemical Technology, The Hong Kong Polytechnic University, Hung Hom, Kowloon, China e-mail:c2hlbmcuY2hlbkBwb2x5dS5lZHUuaGs=
†Marcus H. Wong and Edward W. Chan have contributed equally to this work.
 Marcus H. Wong
Marcus H. Wong Edward W. Chan3†
Edward W. Chan3† Li Z. Liu
Li Z. Liu Sheng Chen
Sheng Chen