- Department of Microbiology, School of Pharmacy, Aichi Gakuin University, Nagoya, Japan
The emergence of multidrug-resistant Pseudomonas aeruginosa has become a serious problem in medical settings. P. aeruginosa clinical isolate PA7 is resistant to fluoroquinolones, aminoglycosides, and most β-lactams but not imipenem. In this study, enhanced efflux-mediated fluoroquinolone resistance of PA7 was shown to reflect increased expression of two resistance nodulation cell division (RND) -type multidrug efflux operons, mexEF-oprN and mexXY-oprA. Such a clinical isolate has rarely been reported because MexEF-OprN-overproducing mutants often increase susceptibility to aminoglycosides apparently owing to impairment of the MexXY system. A mutant of PA7 lacking three RND-type multidrug efflux operons (mexAB-oprM, mexEF-oprN, and mexXY-oprA) was susceptible to all anti-pseudomonas agents we tested, supporting an idea that these RND-type multidrug efflux transporters are molecular targets to overcome multidrug resistance in P. aeruginosa. mexEF-oprN-upregulation in P. aeruginosa PA7 was shown due to a MexS variant harboring the Valine-155 amino acid residue. This is the first genetic evidence shown that a MexS variant causes mexEF-oprN-upregulation in P. aeruginosa clinical isolates.
Introduction
Pseudomonas aeruginosa is a metabolically versatile bacterium that can cause a wide range of severe opportunistic infections in patients with serious underlying medical conditions (Gellatly and Hancock, 2013). Infections caused by P. aeruginosa often are hard to treat; inappropriate chemotherapy readily selects multidrug-resistant P. aeruginosa against which very few agents are effective (Morita et al., 2014; Poole, 2014). This so-called “antibiotic resistance crisis” has been compounded by the lagging in antibiotic discovery and development programs occurred in recent years, and is jeopardizing the essential role played by antibiotics in current medical practices (Rossolini et al., 2014). To combat this organism, it is very useful to understand its antimicrobial resistance mechanisms (see reviews such as Breidenstein et al., 2011; Poole, 2011; Morita et al., 2015).
The complete genome sequence of the multiresistant P. aeruginosa PA7 has been determined (Roy et al., 2010). The sequence of this strain, which exhibits resistance to fluoroquinolones (FQs), aminoglycosides, and various β-lactams but is susceptible to carbapenems (imipenem), includes typical FQ-resistance mutations in gyrA (Thr83Ile) and parC (Ser87Leu) (Roy et al., 2010). PA7 has additionally acquired a mutated aacA4 gene, the product of which (AAC(6′)-II) endows the strain with aminoglycoside (gentamicin and tobramycin) resistance (Roy et al., 2010). This clinical isolate is amenable to the construction of gene knock-out mutations, thereby facilitating the analysis of molecular mechanisms of multidrug antimicrobial resistance in this isolate. Previously we showed that the effect of the modifying enzyme is enhanced by the MexXY resistance nodulation cell division (RND) -type multidrug efflux system, especially in the presence of divalent cations, providing high-level aminoglycoside resistance in P. aeruginosa (Morita et al., 2012b). This observation emphasizes the importance of the MexXY multidrug efflux system for aminoglycoside resistance in multidrug-resistant P. aeruginosa clinical isolates (Morita et al., 2012a).
Four members of the RND family of multidrug efflux systems, MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY have been implicated in FQ resistance in P. aeruginosa clinical isolates (Poole, 2011). As in other organisms, these RND family drug efflux transporters in P. aeruginosa operates as three-component pumps with an RND cytoplasmic membrane (CM) protein (MexB, MexD, MexF, and MexY) linked to an outer membrane channel-forming protein (OprM, OprJ, and OprN) by a CM-tethered periplasmic protein (MexA, MexC, MexE, and MexX) (Poole, 2013). Intriguingly PA7 possesses the mexXY-oprA multidrug efflux operon of which oprA is missing in most P. aeruginosa strains (Roy et al., 2010). We showed that the MexXY can utilize either the OprA or OprM as an outer membrane component of the tripartite efflux pump (Morita et al., 2012b). Multidrug-resistant P. aeruginosa clinical isolates, including carbapenem-resistant P. aeruginosa, often have been reported to be MexXY and/or MexAB-OprM overproducers (e.g., Hocquet et al., 2007; Cabot et al., 2011; Fuste et al., 2013; Khuntayaporn et al., 2013; Vatcheva-Dobrevska et al., 2013). FQ efflux is mediated by the MexAB-OprM system constitutively produced at moderate levels in wild-type P. aeruginosa PAO1 (Morita et al., 2001; Poole, 2013). Expression of mexAB-oprM is controlled directly or indirectly by three repressors encoded by the mexR, nalC and nalD genes, with inactivating mutations in any of these resulting in increased mexAB-oprM expression (Poole, 2011, 2013). In addition, nfxB and nfxC mutants (which overproduce components of the MexCD-OprJ and MexEF-OprN systems, respectively) are well-known as efflux-mediated FQ-resistant mutants of laboratory and clinical isolates of P. aeruginosa, although nfxB mutations appear to be rare in clinical settings (Poole, 2011). nfxC mutants also show increased resistance to carbapenems such as imipenem, not because MexEF-OprN accommodates these agents but because of a coordinated, MexT-dependent reduction of OprD production in such mutants (Poole, 2011, 2013). Hyperexpression of mexEF-orpN (and reduction in oprD expression) is also seen in laboratory isolates disrupted in the mexS gene, which encodes a putative oxidoreductase (a.k.a qrh) of unknown function (Sobel et al., 2005; Lamarche and Deziel, 2011). There are, however, no reports (to our knowledge) with experimental evidence of mexEF-oprN-expressing clinical isolates harboring mutations in mexS (Poole, 2013), although some studies suggested possible mutation of mexS in nfxC-type clinical isolates (e.g., Llanes et al., 2011; Fournier et al., 2013). The incidence of MexXY overproducers among clinical isolates has shown to be linked to the use of ciprofloxacin (Hocquet et al., 2008) and MexXY overproducers have been often reported to be one of major phenotypes in multidrug resistant clinical isolates (Morita et al., 2012a; Poole, 2013). However, MexXY-expressing FQ-resistance or its contribution to FQ-resistance in clinical settings has not well been studied compared to the other three pumps. In this study, we used reverse genetics to investigate the role of efflux-mediated FQ resistance in multidrug-resistant P. aeruginosa clinical isolate PA7.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
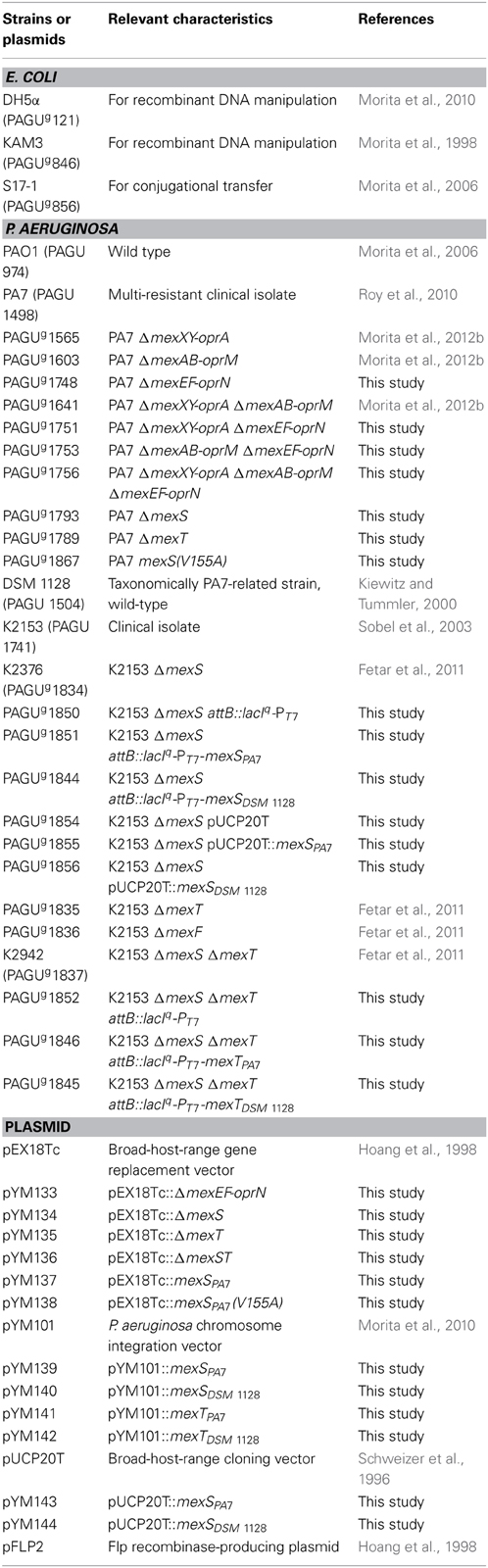
The bacterial strains and plasmids used in this study are listed in Table 1. Bacterial cells were grown in Luria (L) broth and on L agar (1.5%) under aerobic conditions at 37°C as previously described, unless otherwise indicated, with antibiotics as necessary (Morita et al., 2010). Bacterial growth was quantified by measuring the optical density at 600 nm on an Ultrospec 2100 Pro Spectrophotometer (GE Healthcare Corp., Tokyo, Japan), unless otherwise indicated. The plasmids pEX18Tc (Hoang et al., 1998), pYM101 (Morita et al., 2010), and their derivatives were maintained and selected using medium supplemented with 2.5-10 μg tetracycline ml−1 for E. coli or 50–150 μg tetracycline ml−1 for P. aeruginosa. The plasmids pUCP20T (Schweizer et al., 1996) and pFLP2 (Hoang et al., 1998) and their derivatives were maintained and selected using medium supplemented with 100 μg ampicillin ml−1 for E. coli or 200 μg carbenicillin ml−1 for P. aeruginosa.
Molecular Biology Techniques
Plasmid DNA isolation from E. coli, DNA purification, measuring DNA concentration, DNA digestion with restriction enzymes, DNA dephosphorylation, DNA ligation, isolation of chromosomal DNA from P. aeruginosa, PCR conditions, nucleotide sequencing, competent cell preparation from E. coli, transformation of E. coli, and transfer of plasmids into P. aeruginosa via conjugation were performed as described previously (Morita et al., 2010), unless otherwise indicated. DNA sequences and amino acid sequences were analyzed through 2012 to 2013 with Pseudomonas Genome Database (Winsor et al., 2011), Basic Local Alignment Search Tool (BLAST) (Boratyn et al., 2013), SWISS-MODEL (Bordoli et al., 2009), Sorting Tolerant From Intolerant (SIFT) BLink (Kumar et al., 2009), and the software DNASIS Pro (Ver. 2.1; Hitachi, Japan).
Cloning of mexS and mexT from P. aeruginosa
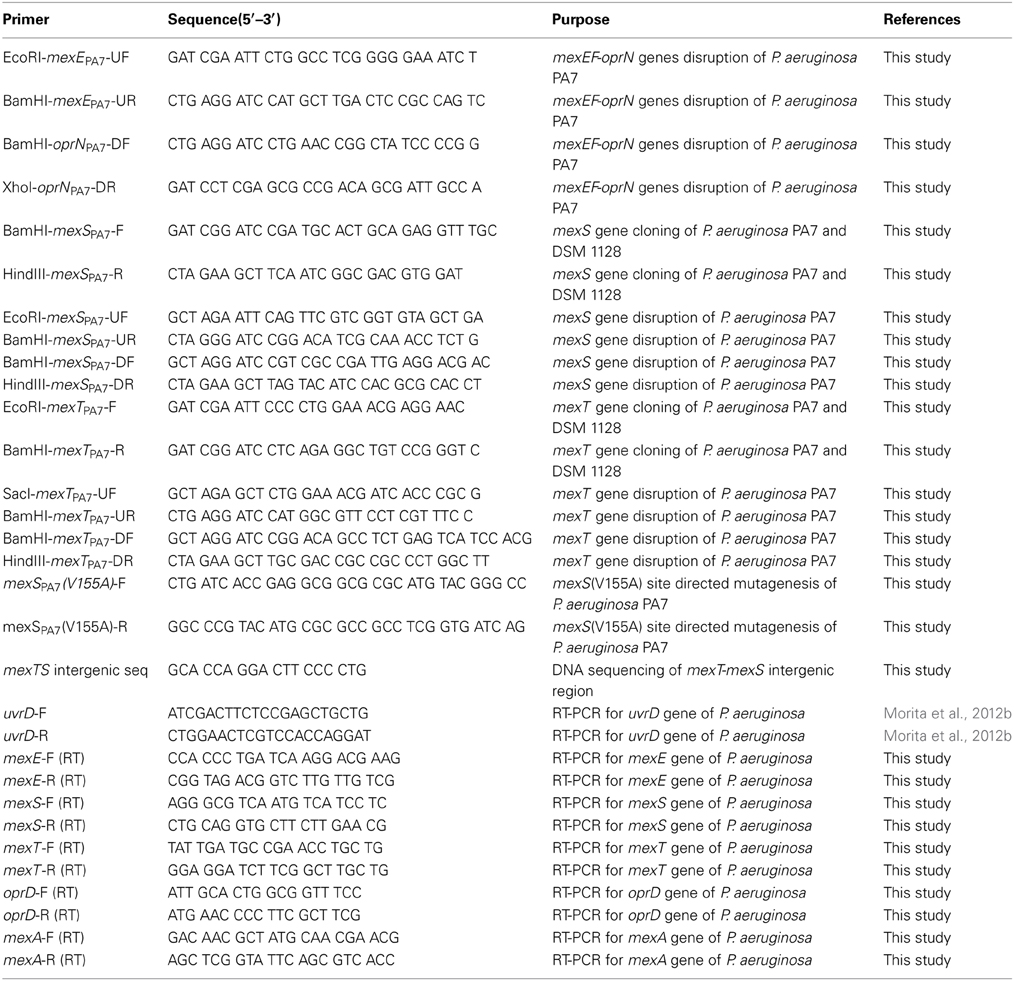
mexS and mexT from P. aeruginosa (without endogenous promoters) were amplified by PCR using the primers listed in Table 2. The purified mexS PCR product, digested with BamHI and HindIII, was cloned into similarly digested, dephosphorylated pUCP20T and pYM101, yielding pUCP20T::mexS and pYM101::mexS. The purified mexT PCR product, digested with EcoRI and BamHI, was cloned into similarly digested, dephosphorylated pUCP20T and pYM101, yielding pUCP20T::mexT and pYM101::mexT.
Construction of In-Frame Deletion Mutants from P. aeruginosa
In-frame deletion mutants of mexEF-oprN, mexS, and mexT from P. aeruginosa PA7 and its derivatives were constructed using the previously described sacB-based strategy (Morita et al., 2006, 2010). The plasmids and resulting P. aeruginosa mutants are listed in Table 1, while the primer pairs are listed in Table 2. The selection concentrations of tetracycline for the first homologous recombination event were adjusted to reflect the endogenous tetracycline MICs for the P. aeruginosa strains. These constructs were confirmed by colony PCR.
Construction of the ΦCTX-Based Site-Specific Integrants in P. aeruginosa
For gene complementation experiments in P. aeruginosa, Φ CTX phage-based site-specific integrants were constructed using the integration-proficient, tightly controlled expression vector pYM101 and associated techniques (conjugative transfer, gene replacement at the chromosomal attB site, and curing of the unwanted plasmid backbone from the chromosome via the pFLP2-encoded Flp recombinase) as previously described (Morita et al., 2010).
Site-Directed Mutagenesis of mexSPA7 in the Chromosome of P. aeruginosa PA7
An amino acid substitution (Val155Ala) mutation was introduced into the mexSPA7 gene of the insert in the plasmid pUC20T::mexSPA7. Site-directed mutagenesis within a target plasmid was carried out according to Geiser et al. (2001) and Morita et al. (2009). A 50-μl mixture consisting of 50 ng of plasmid DNA, 0.25 μM of each mutagenic primer pair (Table 2), 0.2 mM each deoxynucleoside triphosphate, 1 mM MgSO4, 2.5 U of KOD Hot Start DNA polymerase -Plus- Ver.2 (TOYOBO Co. Ltd., Osaka, Japan), 1× KOD buffer, and 4.0% (vol/vol) dimethyl sulfoxide was heated to 94°C for 2 min followed by 18 cycles of 0.5 min at 94°C, 1 min at 60°C, and 5 min at 68°C. The resulting DNA products were purified as above, digested overnight with 10 U DpnI (Roche Diagnostics K.K., Tokyo, Japan) to eliminate template plasmid), and used to transform E. coli DH5α. Plasmids were recovered from individual transformants, and the mexS insert was sequenced to identify plasmids bearing the desired mutation. The mexS insert carrying the mutation was digested with BamHI and HindIII and cloned into similarly digested and dephosphorylated pEX18Tc to yield pEX18Tc::mexSPA7(V155A). The mexSPA7(V155A) mutation was gene replaced onto the chromosome of P. aeruginosa PA7 using the previously described sacB-based strategy (Morita et al., 2009, 2012b).
Antibiotic Susceptibility Assay
The susceptibility of P. aeruginosa to antimicrobial agents in cation-adjusted Mueller–Hinton broth was assessed using the 2-fold serial microtiter broth dilution method described previously (Morita et al., 2012b). Minimal inhibitory concentrations (MICs) were defined as the lowest concentration of antibiotic resulting in visible inhibition of growth after 18 h of incubation at 37°C. MICs for the Φ CTX-based site-specific integrants were determined in the presence of the inducer 5 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG). The categorization in susceptible, intermediate, and resistant was performed according to the interpretive standards of the Clinical and Laboratory Standards Institute (CLSI) (Patel et al., 2011).
Antimicrobial Agents
Amikacin, ampicillin, carbenicillin, chloramphenicol, ciprofloxacin, norfloxacin, and tetracycline were purchased from Wako Pure Chemicals Industries, Ltd (Osaka, Japan). Moxifloxacin and levofloxacin were purchased from LKT Laboratories, Inc. (St. Paul, MN, USA). Imipenem/cilastatin was purchased from Sandoz K.K. (Tokyo, Japan).
Real-Time Quantitative Reverse Transcriptase (qRT)-PCR
Overnight cultures of P. aeruginosa strains were diluted 1:100 in 10 ml, incubated with vigorous shaking at 37°C for 3–4 h, and harvested. Total RNA was stabilized with the RNA Protect Bacteria Reagent (Qiagen) and isolated with the RNeasy Mini Kit (Qiagen). The RNA samples were further treated with RQ1 RNase-Free DNase (Promega) and purified by using the RNeasy Mini Kit (Qiagen). Real-time qRT-PCR was performed with primer pairs internal to uvrD, mexE, mexS, mexT, and oprD (Table 2) using the One Step SYBR PrimeScript RT-PCR kit II (TaKaRa) in a Thermal Cycler Dice real-time system (TaKaRa). The transcript levels of the target gene in a given strain were normalized to levels of uvrD and expressed as a ratio (fold change) to that observed in the parental PA7 strain. Gene expression values were calculated based on triplicate experiments.
Nucleotide Sequence Accession Number
The nucleotide sequences of DNA regions containing mexT and mexS in P. aeruginosa DSM 1128 have been deposited in GenBank/EMBL/DDBJ with the accession number AB889539.
Results
Contribution of RND Multidrug Efflux Systems to FQ Resistance in P. aeruginosa PA7
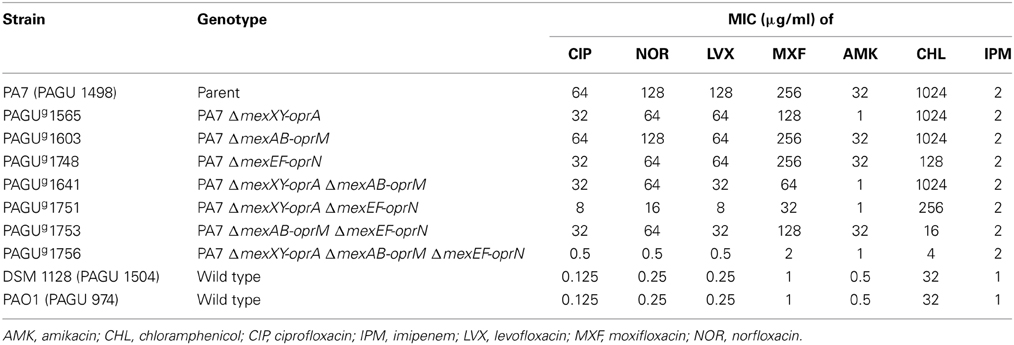
FQ resistance in P. aeruginosa PA7 was compared with the two FQ-susceptible strains, PAO1 (the standard laboratory strain) and DSM 1128 (a strain taxonomically related to PA7) (Morita et al., 2012b). The MICs of the FQs (ciprofloxacin, levofloxacin, moxifloxacin, and norfloxacin) for P. aeruginosa PA7, PAO1, and DSM 1128 are shown in Table 3. P. aeruginosa PA7 exhibited increased resistance (ca. 256-fold) to these FQs compared to P. aeruginosa PAO1 and DSM 1128. P. aeruginosa DSM 1128 showed the same level of resistance to the FQs as P. aeruginosa PAO1. Based on the interpretive standards of the CLSI (Patel et al., 2011), P. aeruginosa PA7 was considered highly resistant to these FQs. This observation is not surprising, because P. aeruginosa PA7 possesses typical target mutations, one in gyrA (Thr83Ile) and one in parC (Ser87Leu), well-known to be associated with FQ resistance (Lomovskaya et al., 1999). However, target mutations alone are not sufficient to explain high-level FQ resistance in P. aeruginosa; the additional effect of up-regulation of one of the four RND efflux pumps (MexXY-OprA, MexAB-OprM, MexCD-OprJ, or MexEF-OprN) is necessary (Lomovskaya et al., 1999; Bruchmann et al., 2013).
Previously we showed that the ΔmexXY-oprA ΔmexAB-oprM double mutant of PA7 was slightly more susceptible (ca. 2- to 4-fold) to ciprofloxacin, while retaining high-level resistance (MIC = 32 μg/ml) to this antibiotic when compared to its parental strain (Morita et al., 2012b). That observation was inconsistent with data, derived via similar genetic analyses, in which a PAO1-derived ΔoprM gyrA parC triple mutant (i.e., a strain deleted for oprM and carrying the target mutations) exhibited increased susceptibility (64-fold) to levofloxacin compared to an isogenic MexAB-OprM overexpressing (nalB) mutant, while exhibiting an MIC (0.5 μg/ml) of levofloxacin similar to that of the wild-type PAO1 (0.25 μg/ml) (Lomovskaya et al., 1999). We noted that deletion of oprM in PAO1 was expected to inactivate any efflux pumps that require OprM as an outer membrane factor (e.g., MexAB-OprM, MexXY-OprM, MexVW-OprM) (Morita et al., 2001; Li et al., 2003). We therefore hypothesized that overexpression of other RND pumps such as MexCD-OprJ and MexEF-OprN was compensating for the effects of the mexAB-oprM mexXY-oprA double deficiency in PA7. As a first step in testing our hypothesis, we examined the primary sequences of the efflux system-encoding genes of PA7. Sequence analysis revealed the presence of mexCD-oprJ (PSPA7_0540- 0539- 0538) and mexEF-oprN (PSPA7_2745-2744-2743) homologs in P. aeruginosa PA7 (Roy et al., 2010), with predicted amino acid lengths (in PA7) and amino acid sequence identity and similarity [respectively; via BLAST of PA7 vs. PAO1-UW (Winsor et al., 2011)] as follows: MexC (387 aa; 88 and 95%), MexD (1043 aa; 95 and 98%,), OprJ (475 aa; 89 and 93%); MexE (414 aa; 99 and 99%), MexF (1062 aa; 99 and 99%), and OprN (472 aa; 97 and 99%). As a second step in testing our hypothesis, we examined the regulation of these genes in PA7. qRT-PCR showed that expression of mexE was increased (ca. 22-fold) in PA7 compared to the wild-type strain DSM 1128. This observation indicated that FQ resistance in P. aeruginosa can be mediated by overexpression of the mexEF-oprN operon, in addition to the previously reported overexpression of mexXY-oprA due to mutation of the local repressor gene (mexZ) (Morita et al., 2012b).
We also observed that a ΔmexXY-oprA ΔmexAB-oprM ΔmexEF-oprN triple mutant derived from PA7 exhibited increased susceptibility to FQs (32–128-fold) compared to an isogenic ΔmexXY-oprA ΔmexAB-oprM double mutant, while exhibiting only mild elevation of FQ MIC (0.5–2 μg/ml) compared to wild-type PAO1 and DSM 1128 (0.125–1 μg/ml) (Table 3). Moreover, the triple mutant was much more sensitive (512-fold) to chloramphenicol than the double mutant, a further indicator of MexEF-OprN-mediated resistance (Sobel et al., 2005) (Table 3). We excluded a role for overexpression of mexCD-oprJ in PA7, as judged by the susceptibilities of the mutant (ΔmexXY-oprA ΔmexAB-oprM ΔmexEF-oprN construct) to not only FQs but also to tetracycline and chloramphenicol, which are substrates of MexCD-OprJ (Poole et al., 1996).
To clarify the roles of MexXY-OprA, MexAB-OprM and MexEF-OprN multidrug efflux pumps in the FQ resistance of P. aeruginosa PA7, we constructed a series of deletion mutants of the three mex operons and examined their drug susceptibility profiles (Table 3). We observed that the ΔmexXY-oprA ΔmexEF-oprN double deletion mutant was more susceptible (4-fold) than the ΔmexAB-oprM ΔmexXY-oprA double mutant and the ΔmexAB-oprM ΔmexEF-oprN double mutant (Table 3). These data suggested that the MexXY-OprA and MexEF-OprN systems make similar contributions to FQ resistance in P. aeruginosa PA7, each having an effect larger than that of MexAB-OprM. The apparent modest MexAB-OprM contribution to resistance of FQs and chloramphenicol in PA7 [PA7 ΔmexXY-oprA ΔmexEF-oprN vs. PA7 ΔmexXY-oprA ΔmexAB-oprM ΔmexEF-oprN (Table 3)] is typically observed in wild type P. aeruginosa such as PAO1 already shown in the previous results (e.g., PAO1 ΔmexXY vs. PAO1 ΔmexXY ΔmexAB-oprM) (e.g., Morita et al., 2001), implying that mexAB-oprM is expressed at moderate levels in PA7. We concluded that high-level FQ resistance in P. aerguinosa PA7 was due to overproduction of MexXY-OprA and MexEF-OprN, in addition to the typical target mutations.
Molecular Mechanisms of mexEF-oprN-Upregulation in P. aeruginosa PA7
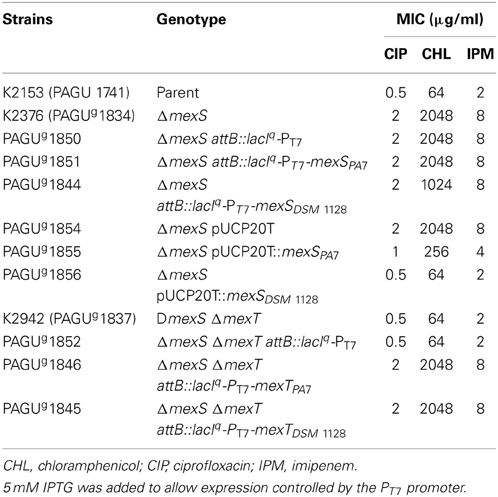
The mexEF-oprN operon is quiescent in wild-type P. aeruginosa cells grown under standard laboratory conditions. In contrast, this operon is expressed in so-called nfxC mutants and in mutants defective in the mexS gene (previously known as qrh), a locus that encodes a putative oxidoreductase of as yet unknown function (Poole, 2013). Expression of mexEF-oprN is regulated by a transcriptional activator, MexT, a LysR family regulator (Kohler et al., 1999; Poole, 2013). mexT occurs upstream of mexEF-oprN and downstream of mexS, the latter gene also positively regulated by MexT (Kohler et al., 1999). Unusually, many so-called wild type strains possess inactive MexT (e.g., 8 bp insertion in mexT present in some PAO1 strains) (Maseda et al., 2000; Poole, 2013). The induction of mexEF-oprN contributes to multidrug resistance, albeit against a rather narrow range of antimicrobials including FQs, trimethoprim, and chloramphenicol (Poole, 2013). The enhanced resistance to imipenem of nfxC mutants or mexS deficient mutants results not from mexEF-oprN expression but the concomitant decrease in the level of outer membrane protein OprD (Poole, 2013), because OprD is an imipenem channel and serves as the primary route of entry of this antibiotic in P. aeruginosa (Trias and Nikaido, 1990). However, P. aeruginosa PA7 was reported to be susceptible to carbapenems, with MICs of 2 and 1 μg/ml for imipenem and meropenem, respectively (Roy et al., 2010). This observation of carbapenem susceptibility is somewhat paradoxical, given that a typical nfxC mutant or a mexS deficient mutant is expected to exhibit carbapenem resistance due to a coordinate, MexT-dependent reduction of OprD levels. We found that P. aeruginosa PA7 exhibited slight but reproducible elevation (2-fold) of resistance to imipenem compared to wild-type P. aeruginosa strains PAO1 and DSM 1128 (Table 4). Deletion of mexS in a PA7 background resulted in slightly increased expression of mexE (2-fold) and increased resistance to imipenem (4-fold) compared to PA7. Thus, PA7 appeared to exhibit susceptibility intermediate between that of wild type and a typical nfxC mutant. Deletion of mexT in a PA7 background resulted in drastically (>200-fold) decreased expression of mexE and mexS compared to PA7 (Table 4). Notably, expression of mexS in PA7 ΔmexT was more than 96-fold reduced compared to the wild-type DSM 1128 and PA7 mexS (V155A). This observation was consistent with a previous report (using β-galacotosidase reporter assays) indicating that mexS (qrh) is constitutively expressed at a moderate level (Kohler et al., 1999; Poole, 2013).
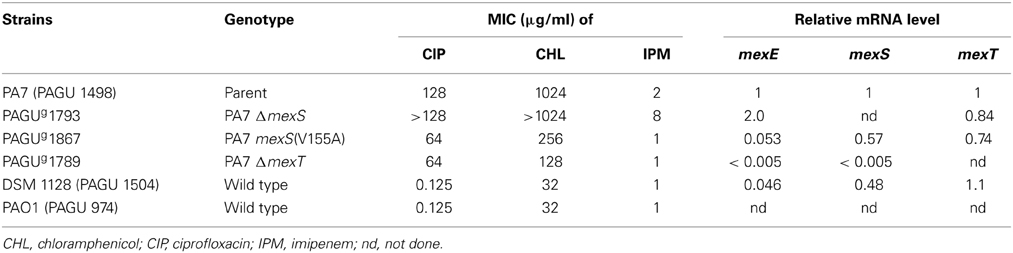
Table 4. Relationship between antimicrobial resistance and mexS-mexT-mediated mexEF-oprN expression in P. aeruginosa.
We therefore determined the nucleotide sequences (ca. 2.4 kb) upstream of the mexEF-oprN operon in P. aeruginosa DSM 1128 (accession no. AB889539). This interval corresponds to the operon's cognate regulatory region, and includes the mexS and mexT loci. Comparison to the corresponding sequences from P. aeruginosa PA7 (data not shown) revealed that MexSPA7 is encoded as an A155V variant, and MexTPA7 as an A256T variant, compared to the DSM 1128 genome. In contrast, alanine-155 of MexS and alanine-256 of MexT are conserved among multiple laboratory and clinical strains, including PAO1, PA14, PAK, and K2153 (Sobel et al., 2005; Jin et al., 2011; Lamarche and Deziel, 2011). To test the function of the mexSPA7 and mexTPA7 loci, we amplified and cloned the individual mexS and mexT genes (without endogenous promoters) from P. aeruginosa PA7 and DSM 1128, and expressed the relevant genes in P. aeruginosa K2153 ΔmexS (Fetar et al., 2011) for a mexS complementation test or in K2153 ΔmexS ΔmexT (Sobel et al., 2005; Fetar et al., 2011) for a mexT complementaton test (Table 5). [As demonstrated by other researchers, P. aeruginosa K2153 and its derivatives are useful for the analysis of mexEF-oprN-dependent antimicrobial resistance as regulated by the MexT activator and MexS function (Sobel et al., 2005; Fetar et al., 2011)]. Interestingly, introduction of mexSDSM 1128 or mexSPA7 into the chromosome of K2153 ΔmexS did not provide complementation of ΔmexS (Table 5). These data contrast previous instances in which we observed attB-site (i.e., single-copy) complementation for several other genes (Morita et al., 2010, 2012b). Presumably, in the present studies, failure to complement reflected insufficient mexS expression from the PT7 promoter in PAGUg1844, given that MexSDSM 1128 is expected to be functional. Introduction of pUCP20T::mexSDSM 1228 into P. aeruginosa K2153 ΔmexS yielded MICs identical to those of the K2153 mexS+ parent; introduction of pUCP20T::mexSPA7 yielded parent-like resistance to imipenem, but only partially restored resistance to ciprofloxacin and chloramphenicol (Table 5). Taken together, these results suggested that MexSPA7 provides reduced function compared to MexSDSM 1228. Additionally it appears that the high levels of mexS expression are required to overcome the nfxC-type antimicrobial resistance.
Slightly increased expression of mexS was observed in P. aeruginosa PA7 compared to P. aeruginosa DSM 1128 (ca. 2-fold), while expression levels of mexT in P. aeruginosa PA7 were similar to those in P. aeruginosa DSM 1128 (c.a. 0.9-fold) (Table 4). Introduction of mexTDSM 1128 or mexTPA7 into the chromosome of K2153 ΔmexS ΔmexT yielded the nfxC phenotype (Table 5), suggesting that the A256T substitution in MexT is not a primary reason for overexpression of mexEF-oprN in P. aeruginosa PA7. Using site-specific mutagenesis, we altered the plasmid-borne mexSPA7 locus to encode a V155A version of the protein and replaced the endogenous PA7 chromosomal locus with the mutated gene. The PA7 mexSPA7(V155A) strain showed decreased mexE expression (0.053-fold compared to that of the PA7 parent), a value similar to that seen in DSM 1128 (0.034-fold compared to that of PA7) (Table 4). The PA7 mexSPA7(V155A) strain also showed decreased MICs for chloramphenicol (0.25-fold) (Table 4). Taken together with the complementation experiments, these data strongly suggested that the A155V substitution in MexSPA7 is the primary reason for increased expression of mexEF-oprN and increased antimicrobial resistance in the PA7 strain.
Discussion
In this study, the multidrug-resistant clinical isolate PA7 was shown to exhibit increased expression of mexEF-oprN as well as mexXY-oprA. Although multidrug-resistant P. aeruginosa clinical isolates have often been reported to be MexXY overproducers (Morita et al., 2012a), clinical strains of P. aeruginosa overproducing MexEF-OprN and MexXY(-OprA) efflux pumps simultaneously have rarely been reported. In fact, PA7 was not a typical nfxC mutant (i.e., a MexEF-OprN overproducer), and instead expressed intermediate levels of mexEF-oprN. We assume that simultaneous overproduction of MexEF-OprN and MexXY(-OprA) impairs P. aeruginosa growth, based on our observation that our PA7 ΔmexS construct (i.e., a simultaneous overproducer of MexEF-OprN and MexXY-OprA compared to the PA7 parent) was unstable even on L agar plates: colonies of the construct exhibited a non-uniform phenotype during growth on plates (data not shown). This observation is consistent with the increased susceptibility to aminoglycosides previously observed in MexEF-OprN-overproducing nfxC mutants, apparently owing to impairment of the MexXY system (Sobel et al., 2005).
Valine-155 of MexSPA7 also was shown as the likely primary reason for increased production of MexEF-OprN in PA7. With the exception of mexSPA7, sequenced P. aeruginosa mexS genes (including those from PAO1, K2153, PA14, PAK, and DSM 1128) encode proteins with an alanine at residue 155 (Sobel et al., 2005; Jin et al., 2011; Lamarche and Deziel, 2011). The Ala155Val substitution is predicted by the SIFT algorithm (Kumar et al., 2009) not to affect the protein's function (data not shown). We hypothesize that MexSPA7 retains function, albeit with decreased activity and/or altered regulation (e.g., allostery), compared with the other MexS orthologs. In fact, there were few differences among the whole structures of MexSPA7, MexSPAO1, and MexSDSM 1128 models developed by using the SWISS-MODEL program (data not shown) (Bordoli et al., 2009). MexS is a member of the cd08268: MDR2 family of the Conserved Domains Database (CDD) of the National Center for Biotechnology Information (NCBI) (Fargier et al., 2012) and alanine-155 corresponds to one of the putative NAD(P) binding sites featured in the MDR2 family.
In addition to antibiotic resistance, an nfxC-type mutation has been linked to reduced levels of homoserine lactone-dependent quorum sensing (QS) -regulated virulence factors, including pyocyanin, elastase, rhamnolipids, and Pseudomonas Quinolone Signal (PQS), and to reduced expression of type-III secretion system (TTSS) effector proteins (Kohler et al., 2001; Linares et al., 2005). QS is a cell-to-cell communication mechanism employing diffusible signal molecules (Jimenez et al., 2012), and the TTSS is a mechanism by which bacterial pathogens can deliver effectors directly into the cytoplasm of eukaryotic host cells (Hauser, 2009). We found that PA7 had reduced level of pyocyanin production, rhamnolipid production, and swarming activity compared to DSM 1128 and PAO1 (data not shown), consistent with the typical nfxC mutant phenotype (Jin et al., 2011). However, PA7 derivatives including PA7 mexSPA7(V155A) also showed almost the same activities of the QS regulated virulence factors with PA7 (data not shown), which suggests that the impaired QS-related phenotype is not derived from the nfxC-like phenotype in PA7. We presume that this lack of correlation reflects the absence from PA7 of TTSS-encoding genes and of the mvfR (pqsR) gene, which is known to encode a LysR-type transcriptional regulator that modulates the expression of multiple QS-regulated virulence factors (Deziel et al., 2005; Roy et al., 2010). These deficiencies might be the source of the mexS mutation and increased mexEF-oprN expression under oxidative stress, sulfide stress, or nitrosative stress (Juhas et al., 2004; Fetar et al., 2011; Fargier et al., 2012).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Tadashi Kumazawa for his contribution. We thank Dr. Keith Poole (Queen's University, Canada) for providing the P. aeruginosa strains. This work was supported in part by a Grant-in-Aid for Young Scientists (B) (Kakenhi 23790106) and a Grant-in-Aid for Scientific Research (C) (Kakenhi 26460080) from the Japan Society for the Promotion of Science, and by a research grant from the Institute of Pharmaceutical Life Sciences, Aichi Gakuin University.
References
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., and Ma, N. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33. doi: 10.1093/nar/gkt282
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bordoli, L., Kiefer, F., Arnold, K., Benkert, P., Battey, J., and Schwede, T. (2009). Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 4, 1–13. doi: 10.1038/nprot.2008.197
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Breidenstein, E. B., De La Fuente-Nunez, C., and Hancock, R. E. (2011). Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 19, 419–426. doi: 10.1016/j.tim.2011.04.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bruchmann, S., Dotsch, A., Nouri, B., Chaberny, I. F., and Haussler, S. (2013). Quantitative contributions of target alteration and decreased drug accumulation to Pseudomonas aeruginosa fluoroquinolone resistance. Antimicrob. Agents Chemother. 57, 1361–1368. doi: 10.1128/aac.01581-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cabot, G., Ocampo-Sosa, A. A., Tubau, F., Macia, M. D., Rodriguez, C., and Moya, B. (2011). Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa isolates from bloodstream infections: prevalence and impact on resistance in a Spanish multicenter study. Antimicrob. Agents Chemother. 55, 1906–1911. doi: 10.1128/aac.01645-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deziel, E., Gopalan, S., Tampakaki, A. P., Lepine, F., Padfield, K. E., and Saucier, M. (2005). The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol. Microbiol. 55, 998–1014. doi: 10.1111/j.1365-2958.2004.04448.x
Fargier, E., Mac Aogain, M., Mooij, M. J., Woods, D. F., Morrissey, J. P., and Dobson, A. D. (2012). MexT functions as a redox-responsive regulator modulating disulfide stress resistance in Pseudomonas aeruginosa. J. Bacteriol. 194, 3502–3511. doi: 10.1128/jb.06632-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fetar, H., Gilmour, C., Klinoski, R., Daigle, D. M., Dean, C. R., and Poole, K. (2011). mexEF-oprN multidrug efflux operon of Pseudomonas aeruginosa: regulation by the MexT activator in response to nitrosative stress and chloramphenicol. Antimicrob. Agents Chemother. 55, 508–514. doi: 10.1128/aac.00830-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fournier, D., Richardot, C., Muller, E., Robert-Nicoud, M., Llanes, C., and Plesiat, P. (2013). Complexity of resistance mechanisms to imipenem in intensive care unit strains of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 68, 1772–1780. doi: 10.1093/jac/dkt098
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fuste, E., Lopez-Jimenez, L., Segura, C., Gainza, E., Vinuesa, T., and Vinas, M. (2013). Carbapenem-resistance mechanisms of multidrug-resistant Pseudomonas aeruginosa. J. Med. Microbiol. 62, 1317–1325. doi: 10.1099/jmm.0.058354-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geiser, M., Cebe, R., Drewello, D., and Schmitz, R. (2001). Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques 31, 88–92.
Gellatly, S. L., and Hancock, R. E. (2013). Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog. Dis. 67, 159–173. doi: 10.1111/2049-632x.12033
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hauser, A. R. (2009). The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7, 654–665. doi: 10.1038/nrmicro2199
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. J., and Schweizer, H. P. (1998). A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86.
Hocquet, D., Berthelot, P., Roussel-Delvallez, M., Favre, R., Jeannot, K., and Bajolet, O. (2007). Pseudomonas aeruginosa may accumulate drug resistance mechanisms without losing its ability to cause bloodstream infections. Antimicrob. Agents Chemother. 51, 3531–3536. doi: 10.1128/aac.00503-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hocquet, D., Muller, A., Blanc, K., Plesiat, P., Talon, D., Monnet, D. L., et al. (2008). Relationship between antibiotic use and incidence of MexXY-OprM overproducers among clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52, 1173–1175. doi: 10.1128/aac.01212-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jimenez, P. N., Koch, G., Thompson, J. A., Xavier, K. B., Cool, R. H., and Quax, W. J. (2012). The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 76, 46–65. doi: 10.1128/mmbr.05007-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jin, Y., Yang, H., Qiao, M., and Jin, S. (2011). MexT regulates the type III secretion system through MexS and PtrC in Pseudomonas aeruginosa. J. Bacteriol. 193, 399–410. doi: 10.1128/jb.01079-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Juhas, M., Wiehlmann, L., Huber, B., Jordan, D., Lauber, J., and Salunkhe, P. (2004). Global regulation of quorum sensing and virulence by VqsR in Pseudomonas aeruginosa. Microbiology 150, 831–841. doi: 10.1099/mic.0.26906-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Khuntayaporn, P., Montakantikul, P., Santanirand, P., Kiratisin, P., and Chomnawang, M. T. (2013). Molecular investigation of carbapenem resistance among multidrug-resistant Pseudomonas aeruginosa isolated clinically in Thailand. Microbiol. Immunol. 57, 170–178. doi: 10.1111/1348-0421.12021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kiewitz, C., and Tummler, B. (2000). Sequence diversity of Pseudomonas aeruginosa: impact on population structure and genome evolution. J. Bacteriol. 182, 3125–3135. doi: 10.1128/JB.182.11.3125-3135.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kohler, T., Epp, S. F., Curty, L. K., and Pechere, J. C. (1999). Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 181, 6300–6305.
Kohler, T., Van Delden, C., Curty, L. K., Hamzehpour, M. M., and Pechere, J. C. (2001). Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 183, 5213–5222. doi: 10.1128/JB.183.18.5213-5222.2001
Kumar, P., Henikoff, S., and Ng, P. C. (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081. doi: 10.1038/nprot.2009.86
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lamarche, M. G., and Deziel, E. (2011). MexEF-OprN efflux pump exports the Pseudomonas quinolone signal (PQS) precursor HHQ (4-hydroxy-2-heptylquinoline). PLoS ONE 6:e24310. doi: 10.1371/journal.pone.0024310
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, Y., Mima, T., Komori, Y., Morita, Y., Kuroda, T., and Mizushima, T. (2003). A new member of the tripartite multidrug efflux pumps, MexVW-OprM, in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 52, 572–575. doi: 10.1093/jac/dkg390
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Linares, J. F., Lopez, J. A., Camafeita, E., Albar, J. P., Rojo, F., and Martinez, J. L. (2005). Overexpression of the multidrug efflux pumps MexCD-OprJ and MexEF-OprN is associated with a reduction of type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187, 1384–1391. doi: 10.1128/jb.187.4.1384-1391.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Llanes, C., Kohler, T., Patry, I., Dehecq, B., Van Delden, C., and Plesiat, P. (2011). Role of the MexEF-OprN efflux system in low-level resistance of Pseudomonas aeruginosa to ciprofloxacin. Antimicrob. Agents Chemother. 55, 5676–5684. doi: 10.1128/aac.00101-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lomovskaya, O., Lee, A., Hoshino, K., Ishida, H., Mistry, A., and Warren, M. S. (1999). Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43, 1340–1346.
Maseda, H., Saito, K., Nakajima, A., and Nakae, T. (2000). Variation of the mexT gene, a regulator of the MexEF-oprN efflux pump expression in wild-type strains of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 192, 107–112. doi: 10.1111/j.1574-6968.2000.tb09367.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morita, Y., Gilmour, C., Metcalf, D., and Poole, K. (2009). Translational control of the antibiotic inducibility of the PA5471 gene required for mexXY multidrug efflux gene expression in Pseudomonas aeruginosa. J. Bacteriol. 191, 4966–4975. doi: 10.1128/jb.00073-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morita, Y., Kimura, N., Mima, T., Mizushima, T., and Tsuchiya, T. (2001). Roles of MexXY- and MexAB-multidrug efflux pumps in intrinsic multidrug resistance of Pseudomonas aeruginosa PAO1. J. Gen. Appl. Microbiol. 47, 27–32. doi: 10.2323/jgam.47.27
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morita, Y., Kodama, K., Shiota, S., Mine, T., Kataoka, A., and Mizushima, T. (1998). NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42, 1778–1782.
Morita, Y., Narita, S., Tomida, J., Tokuda, H., and Kawamura, Y. (2010). Application of an inducible system to engineer unmarked conditional mutants of essential genes of Pseudomonas aeruginosa. J. Microbiol. Methods 82, 205–213. doi: 10.1016/j.mimet.2010.06.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morita, Y., Sobel, M. L., and Poole, K. (2006). Antibiotic inducibility of the MexXY multidrug efflux system of Pseudomonas aeruginosa: involvement of the antibiotic-inducible PA5471 gene product. J. Bacteriol. 188, 1847–1855. doi: 10.1128/jb.188.5.1847-1855.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morita, Y., Tomida, J., and Kawamura, Y. (2012a). MexXY multidrug efflux system of Pseudomonas aeruginosa. Front. Microbiol. 3:408. doi: 10.3389/fmicb.2012.00408
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morita, Y., Tomida, J., and Kawamura, Y. (2012b). Primary mechanisms mediating aminoglycoside resistance in the multidrug-resistant Pseudomonas aeruginosa clinical isolate PA7. Microbiology 158, 1071–1083. doi: 10.1099/mic.0.054320-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morita, Y., Tomida, J., and Kawamura, Y. (2014). Responses of Pseudomonas aeruginosa to antimicrobials. Front. Microbiol. 4:422. doi: 10.3389/fmicb.2013.00422
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Morita, Y., Tomida, J., and Kawamura, Y. (2015). “Resistance and response to anti-pseudomonas agents and biocides,” in Pseudomonas: New Aspects of Pseudomonas Biology, eds J. Ramos, J. B. Goldberg, and A. Filloux (New York, NY: Springer), 173–187.
Patel, J. B., Tenover, F. C., Turnidge, J. D., and Jorgensen, J. H. (2011). “Susceptibility test methods: dilution and disk diffusion methods,” in Manual of Clinical Microbiology, 10th Edn, eds J. Versalovic, K. C. Carroll, G. Funke, J. H. Jorgensen, M. L. Landry, and D. W. Warnock (Washington, DC: ASM Press), 1122–1143.
Poole, K. (2011). Pseudomonas aeruginosa: resistance to the max. Front. Microbiol. 2:65. doi: 10.3389/fmicb.2011.00065
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Poole, K. (2013). “Pseudomonas aeruginosa efflux pumps,” in Microbial Efflux Pumps: Current Research, eds E. W. Yu, Q. Zhang, and M. H. Brown (Norfolk: Caiser Academic Press), 175–206.
Poole, K. (2014). Stress responses as determinants of antimicrobial resistance in Pseudomonas aeruginosa: multidrug efflux and more. Can. J. Microbiol. 60, 783–791. doi: 10.1139/cjm-2014-0666
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Poole, K., Gotoh, N., Tsujimoto, H., Zhao, Q., Wada, A., and Yamasaki, T. (1996). Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21, 713–724.
Rossolini, G. M., Arena, F., Pecile, P., and Pollini, S. (2014). Update on the antibiotic resistance crisis. Curr. Opin. Pharmacol. 18c, 56–60. doi: 10.1016/j.coph.2014.09.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roy, P. H., Tetu, S. G., Larouche, A., Elbourne, L., Tremblay, S., and Ren, Q. (2010). Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS ONE 5:e8842. doi: 10.1371/journal.pone.0008842
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schweizer, H. P., Klassen, T. R., and Hoang, T. T. (1996). “Improved methods for gene analysis and expression in Pseudomonas,” in Molecular Biology of Pseudomonads, eds T. Nakazawa, D. Haas, and S. Silver (Washington, DC: ASM Press), 229–237.
Sobel, M. L., McKay, G. A., and Poole, K. (2003). Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 47, 3202–3207. doi: 10.1128/AAC.47.10.3202-3207.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sobel, M. L., Neshat, S., and Poole, K. (2005). Mutations in PA2491 (mexS) promote MexT-dependent mexEF-oprN expression and multidrug resistance in a clinical strain of Pseudomonas aeruginosa. J. Bacteriol. 187, 1246–1253. doi: 10.1128/jb.187.4.1246-1253.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Trias, J., and Nikaido, H. (1990). Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J. Biol. Chem. 265, 15680–15684.
Vatcheva-Dobrevska, R., Mulet, X., Ivanov, I., Zamorano, L., Dobreva, E., and Velinov, T. (2013). Molecular epidemiology and multidrug resistance mechanisms of Pseudomonas aeruginosa isolates from Bulgarian hospitals. Microb. Drug Resist. 19, 355–361. doi: 10.1089/mdr.2013.0004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Winsor, G. L., Lam, D. K., Fleming, L., Lo, R., Whiteside, M. D., and Yu, N. Y. (2011). Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39, D596–D600. doi: 10.1093/nar/gkq869
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Pseudomonas aeruginosa, efflux, mexXY-oprA, mexEF-oprN, mexS
Citation: Morita Y, Tomida J and Kawamura Y (2015) Efflux-mediated fluoroquinolone resistance in the multidrug-resistant Pseudomonas aeruginosa clinical isolate PA7: identification of a novel MexS variant involved in upregulation of the mexEF-oprN multidrug efflux operon. Front. Microbiol. 6:8. doi: 10.3389/fmicb.2015.00008
Received: 27 October 2014; Paper pending published: 21 November 2014;
Accepted: 05 January 2015; Published online: 21 January 2015.
Edited by:
Keith Poole, Queen's University, CanadaReviewed by:
Antonio Oliver, Hospital Son Dureta, SpainHerbert P. Schweizer, Colorado State University, USA
Patrick Plesiat, University of Franche-Comté, France
Copyright © 2015 Morita, Tomida and Kawamura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuji Morita, Department of Microbiology, School of Pharmacy, Aichi Gakuin University, 1-100 Kusumoto, Chikusa, Nagoya, Aichi 464-8650, Japan e-mail:eXVqbW9yQGRwYy5hZ3UuYWMuanA=
 Yuji Morita
Yuji Morita Junko Tomida
Junko Tomida