- 1Institute for Microbiology, Department of Infectious Diseases, University of Veterinary Medicine Hannover, Hannover, Germany
- 2Genome Analytics, Helmholtz Centre for Infection Research, Braunschweig, Germany
- 3Molecular Immunology, Helmholtz Centre for Infection Research, Braunschweig, Germany
The ferric uptake regulator A (FurA) is known to be involved in iron homeostasis and stress response in many bacteria. In mycobacteria the precise role of FurA is still unclear. In the presented study, we addressed the functional role of FurA in the ruminant pathogen Mycobacterium avium ssp. paratuberculosis (MAP) by construction of a furA deletion strain (MAPΔfurA). RNA deep sequencing revealed that the FurA regulon consists of repressed and activated genes associated to stress response or intracellular survival. Not a single gene related to metal homeostasis was affected by furA deletion. A decisive role of FurA during intracellular survival in macrophages was shown by significantly enhanced survival of MAPΔfurA compared to the wildtype, indicating that a principal task of mycobacterial FurA is oxidative stress response regulation in macrophages. This resistance was not associated with altered survival of mice after long term infection with MAP. Our results demonstrate for the first time, that mycobacterial FurA is not involved in the regulation of iron homeostasis. However, they provide strong evidence that FurA contributes to intracellular survival as an oxidative stress sensing regulator.
Introduction
For efficient infection and colonization, pathogenic bacteria need to adapt their metabolism to the host environment and to combat innate antimicrobial host defense mechanisms such as iron limitation by iron sequestering molecules such as ferritin, transferrin and lactoferrin, and the production of toxic reactive oxygen (ROS) and nitrogen species (RNS).
Reactive oxygen and nitrogen species comprise highly reactive molecules such as the superoxide anion (O2-), hydroxyl radical (OH-), and peroxynitrite (ONOO-) as well as more stable oxidants like hydrogen peroxide (H2O2) and nitric oxide (NO). These products are generated during auto oxidative metabolic processes and play important roles in prokaryotic and eukaryotic homeostasis and cell signaling (Yoshikawa and Naito, 2000; Devasagayam et al., 2004; Gough and Cotter, 2011). However, when enriched in infected phagocytes by the activity of phagosomal myeloperoxidases and NO-oxidases, they represent effective antimicrobial measures. Accordingly, intracellular persisting mycobacteria have developed host defense escape mechanisms and the regulation of these is often linked to the expression of virulence factors.
Like other pathogenic mycobacteria, Mycobacterium avium ssp. paratuberculosis (MAP), the causative agent of Johne’s disease, a chronic, non-curable granulomatous inflammation of the ruminant intestine (Harris and Barletta, 2001; Lombard, 2011), is able to persist and multiply intracellularly in the phagosome of macrophages (Kuehnel et al., 2001). Specifically, MAP adapts to the intestinal environment by adjusting its metabolism to the host environment and responding to antimicrobial defense mechanisms indicated by enhanced expression of KatG and SodA (Weigoldt et al., 2011, 2013). Hence, MAP is clearly able to withstand the host innate defense response, but its enabling mechanisms are still unclear.
In general, oxidative stress response in bacteria is regulated by the major transcription factors OxyR and PerR (Zheng et al., 1999; Herbig and Helmann, 2001). OxyR belongs to the family of LysR-regulators and is not only found in Gram negative bacteria but also in some Gram positive bacteria (Oh et al., 2007). OxyR is a transcriptional activator under oxidizing conditions acting through direct interaction with the RNA polymerase α subunit (Kullik et al., 1995; Tao et al., 1995). PerR is a member of the ferric uptake regulator (FUR) family of metallo-regulators and functions as a peroxide-responsive repressor (Herbig and Helmann, 2001). PerR is found mostly in Gram positive bacteria where it seems to substitute for OxyR (Mongkolsuk and Helmann, 2002). Nevertheless, in some bacterial species PerR is present together with OxyR (van Vliet et al., 1999; Hahn et al., 2002; Tseng et al., 2003). To prevent damage by ROS, OxyR, and/or PerR regulate, amongst others, the expression of a catalase-peroxidase (kat) and an alkyl hydroperoxide reductase (ahp) in many bacteria (Christman et al., 1989; Storz and Altuvia, 1994; Bsat et al., 1996, 1998).
Iron plays a particular role in bacterial metabolism. On the one hand, iron is an essential structural and catalytic cofactor for many metabolic enzymes. On the other hand, iron excess in the cell is harmful due to the spontaneous generation of ROS by the Fenton reaction. Therefore, the bacterial intracellular iron homeostasis must be tightly regulated and often is closely linked to the response to oxidative stresses (Cornelis et al., 2011). In Mycobacterium tuberculosis (Mtb), three genes encoding putative iron-dependent regulatory proteins have been described (Cole et al., 1998): IdeR (Schmitt et al., 1995) and SirR (Hill et al., 1998), both belonging to the diptheria-toxin repressor (DtxR) family, as well as ferric uptake regulator A (FurA), a member of the FUR family (Lee and Helmann, 2007; Fillat, 2014).
IdeR is the global iron-dependent regulator in mycobacteria. As an iron-loaded homodimer, it controls the expression of about 50 genes in Mtb which are mostly involved in iron homeostasis, and in addition a number of virulence associated factors (Rodriguez et al., 2002). The ideR gene is essential in pathogenic mycobacteria like Mtb (Rodriguez et al., 2002) or MAP (Janagama et al., 2009), but not in saprophytic species such as M. smegmatis (Dussurget et al., 1996). Using a conditional ideR mutant, Pandey and Rodriguez (2014) recently demonstrated that IdeR is a factor important for the survival of Mtb in macrophages as well as for the infection of mice. SirR was originally identified in Staphylococcus epidermidis with a sequence identity to Mtb IdeR of 33% (Hill et al., 1998). The biological role of SirR homologs in pathogenic mycobacteria is still unclear.
The FUR family of transcriptional metallo-regulator proteins is widespread in bacteria and archaea (Fillat, 2014). The best characterized member of the FUR family, the Escherichia coli Fur protein, employs Fe2+ as co-repressor and acts as a global regulator, influencing the expression of more than 100 genes involved in iron homeostasis, intermediary metabolism, and oxidative stress response (McHugh et al., 2003). The classical mechanism of Fur regulation includes Fur-binding of Fe2+, formation of a metal-containing dimer which subsequently binds to a specific Fur-box found in the promoter sequences of the corresponding target genes (Bagg and Neilands, 1987). Recent studies indicated that the regulatory mechanisms governed by Fur exceed this classical repressor model. Thus, Fur homologs in different bacterial species can also act as metal-dependent positive regulators or even repress gene expression in the absence of the iron cofactor [apo-Fur repression; see review by Carpenter et al. (2009)]. In addition, a metal-independent activation of genes by apo-Fur was recently described (Butcher et al., 2012).
In all mycobacterial species sequenced so far furA is located upstream of katG. Furthermore, for some mycobacteria species, co-expression of furA and katG indicated an involvement of FurA in mycobacterial oxidative stress response. Indeed, it was proposed that, in mycobacteria, FurA could functionally replace the peroxide repressor PerR (Lucarelli et al., 2008). Nevertheless, the precise regulatory role of FurA and the regulation of stress response in mycobacteria are still not completely clear. One explanation for this might be the absence of a functional OxyR regulator in some mycobacterial species like those of the M. tuberculosis complex and M. smegmatis. In these species, it was found that FurA is involved in the oxidative stress response by regulating katG (Pym et al., 2001; Zahrt et al., 2001) but there was no contribution of FurA to ahpC expression. In contrast, many other mycobacterial species, including the pathogens M. avium and M. marinum, harbor a functional oxyR gene (Deretic et al., 1995; Sherman et al., 1995). In these species it was shown that ahpC but not katG is under control of OxyR (Dhandayuthapani et al., 1997; Pagan-Ramos et al., 1998). However, the regulation of katG in these species has not been addressed. Overall, these data suggest a more complex regulatory network of oxidative stress response regulation in mycobacteria.
In the present work we aimed to analyze the relevance of mycobacterial FurA in iron homeostasis and stress response. We characterized the FurA regulon in MAP by construction and analysis of an isogenic MAPΔfurA mutant. By this, we were able to clarify the influence of FurAMAP in iron metabolism, stress response, and intracellular survival.
Materials and Methods
Chemicals, Bacterial Strains, and Growth Conditions
All chemicals were purchased from Sigma–Aldrich (Munich, Germany) if not stated otherwise. Bacterial strains, oligonucleotides, plasmids, and phages are listed in Tables S1 and S2. E. coli strains were cultivated at 37°C in Luria-Bertani (LB) broth or on LB-agar supplemented with 100 μg/ml ampicillin or 100 μg/ml hygromycin, as required.
Mycobacterium smegmatis mc2 155 and transformants were grown in LB media or on Middlebrook phage (MBP) agar (Supplementary Methods) supplemented with 100 μg/ml hygromycin if necessary. Liquid cultures were incubated at 37°C in a shaking incubator at 100 rpm. MAP DSM44135 wildtype (wt), MAPΔfurA, and the complemented strain MAPΔfurAC were grown in DifcoTM Middlebrook 7H9 broth or on solid Middlebrook 7H10 agar (Beckton Dickinson, Franklin Lakes, NJ, USA) supplemented with mycobactin J (2 mg/l, Allied Monitor, Fayette, MO, USA), 2.5% glycerol and 10% OADC (0.06% oleic acid, 5% albumin, 2% dextrose, 0.085% NaCl, 0.003% catalase; MB-complete broth) and 50 μg/ml hygromycin and/or 50 μg/ml kanamycin, as required. Agar plates were sealed in plastic bags after inoculation and incubated at 37°C for 8–10 weeks. Liquid cultures were grown in Duran® laboratory glass bottles on a magnetic stirrer at 100 rpm and 37°C until they reached an OD600 of 1.0 and used for further experiments.
For growth kinetics, a starter culture was grown to an OD600 of 1.0 in MB-complete broth. Bacteria were harvested by centrifugation at 3,800 ×g for 15 min at 4°C, resuspended in MB-complete broth, singularized with glass beads (Ø 3 mm) by vortexing and inoculated into fresh medium to obtain an initial OD600 of 0.2. The OD600 of cultures was measured twice a week and bacteria were grown until they entered the stationary phase.
The response of MAPwt and MAPΔfurA to oxidative stress was examined upon exposure of bacteria to hydrogen peroxide (H2O2). Cells were grown in MB-complete broth and harvested at an OD600 of 1.0 as described above. The bacterial pellet was washed twice with PBS and singularized cells were transferred to 50 ml Middlebrook 7H9 broth supplemented with 2 mg/l mycobactin, 2.5% glycerol, and 10% ADC lacking catalase (5% albumin, 2% dextrose, 0.085% NaCl; MB-cat broth) to obtain an OD600 of 1.0. H2O2 was added to a final concentration of 10 mM and cultures were exposed for 2 h at 37°C with gentle shaking (100 rpm) in Erlenmeyer flasks. Following, bacteria were harvested and immediately used for RNA-extraction.
Construction of a MapΔfurA Strain and Complementation of the Mutant Strain
Construction of a furA (map1669c) deletion mutant of MAPwt was performed by specialized transduction according to Park et al. (2008) with minor modifications. A detailed protocol is given in the Supplementary Methods file.
For complementation of the mutant strain the furA gene, including 88 bp of the upstream UTR, was amplified by PCR using primers oMAP-furA-K1/oMAP-furA-K2. The PCR product was digested with XbaI and HindIII and ligated into the corresponding sites of the mycobacterial integrative vector pMV306 (Stover et al., 1991), carrying a kanamycin resistance. The resulting plasmid pMAP-furA1101 was transformed into electro-competent MAPΔfurA cells and plated on MB-complete agar with 50 μg/ml hygromycin and kanamycin. Successful deletion and integration of furA was confirmed by PCR.
Cell Culture of Macrophages and Survival Assay of Intracellular Mycobacteria
The intracellular survival of MAPwt, MAPΔfurA, and MAPΔfurAC was determined in the mouse macrophage cell line J774A.1 (Ralph et al., 1975). Macrophage cells were seeded in tissue culture dishes and were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), 1% glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin at 37°C and 8% CO2. Twenty-four hours prior to infection, medium was changed to antibiotic-free DMEM. For infection experiments, MAP cells were grown to an OD600 of 1.0, harvested, singularized and stored in MB-complete broth containing 10% glycerol at -80°C until further usage. For infection, MAP cells were thawed on ice, diluted in antibiotic-free DMEM to an OD600 of 0.15 and incubated with the macrophages (2.0 × 106 cells per cell culture dish) as described earlier (Kuehnel et al., 2001), resulting in a MOI of 10. The mean values of the initial inocula (five experiments) of all strains (MAPwt, MAPΔfurA, MAPΔfurAC) are shown in the Supplementary data files. At indicated time points, infected macrophages were washed twice with PBS and scraped off the plates in 1 ml PBS containing 0.1% SDS. J774A.1 cells were disrupted by passage through a 24-gauge needle. The homogenates as well as the infection inocula were 10-fold serial diluted in PBS and 25 μl of the 10-4 and 10-5 dilutions were plated on supplemented MB-complete agar plates in duplicate. Colony forming units (CFU) were determined after incubation for up to 10 weeks at 37°C and normalized to the initial inoculum.
Determination of Reactive Oxygen Species in Macrophage Infection Experiments
Induction of ROS production by macrophages upon infection with MAPwt, MAPΔfurA, and MAPΔfurAC was determined by use of CellRox® Deep Red (Life Technologies), according to the manufacturer’s protocol. In brief, macrophages were cultured and infected as described above and incubated for 2 h at 37°C/8% CO2 or treated with the chemical menadione as a positive control at a final concentration of 100 μM (Malorni et al., 1993). CellRox® Deep Red was added to a final concentration of 2.5 μM 30 min before harvesting and detection. Signals were measured at 660 nm by flow cytometry (Guava® easyCyte 8HT, Millipore, Billerica, MA, USA) and shown as mean fluorescence intensity (MFI) of four independent experiments.
Immunofluorescence and Confocal Microscopy
Macrophages were seeded on coverslips and infected as described above. After 30 min and 2 h of infection, cells were fixed with 3% formaldehyde in PBS for 10 min, washed twice with PBS and treated with blocking buffer (PBS/1% BSA/10% FCS) for 20 min at room temperature. For staining of extracellular mycobacteria, coverslips were incubated with a 1:100 dilution of a polyclonal rabbit anti-MAP-HBHA serum (Supplementary Methods) in blocking buffer at room temperature in a humid chamber for 45 min. After washing with PBS, coverslips were incubated with goat anti-rabbit IgG coupled to Alexa Fluor® 488 (Life Technologies, Darmstadt, Germany) for 30 min and subsequently washed with PBS. To label intracellular mycobacteria, cells were permeabilized with Triton X-100 (0.1%) for 5 min at room temperature, washed in PBS, followed by incubation with a 1:100 dilution of polyclonal rabbit anti-MAP-HBHA serum in blocking buffer for 45 min at room temperature. After washing, coverslips were treated with goat anti-rabbit IgG coupled to Alexa Fluor® 568 (Life Technologies, Darmstadt, Germany) for 30 min. Coverslips were washed three times in PBS and, after one brief washing step with ddH2O, samples were mounted using ProLong® Gold with DAPI (Life Technologies, Darmstadt, Germany). Mounted samples were examined using a TCS SP5 confocal laser scanning microscope equipped with a 63×/1.4–0.6 NA HCX PL APO objective (Leica, Wetzlar, Germany). Image stacks were acquired using 1 Airy unit pinhole diameter in sequential imaging mode to avoid bleed through. Image stacks were deconvolved using Huygens® Essential (Scientific Volume Imaging, Hilversum, The Netherlands), maximum intensity projections were calculated for display purposes and adjusted for brightness and contrast using ImageJ/Fiji (Schindelin et al., 2012). After this labeling procedure, extracellular mycobacteria appear yellow, whereas intracellular mycobacteria are stained red.
Animal Experiment
Mouse infection experiments were approved by the Lower Saxony Federal State Office for Consumer Protection and Food Safety, Germany (reference number 08/1504). In each group, 9 female C57BL/6 mice aged 8 weeks (Charles River, Erkrath, Germany) were challenged intraperitoneally with an infection dose of 1 × 108 bacteria of MAPwt and MAPΔfurA in 200 μl Dulbecco’s Phosphate-Buffered Saline (DPBS, Life Technologies GmbH, Darmstadt, Germany). DPBS was used as negative control. Mice were sacrificed after 4 weeks, and liver and spleen were weighed. Bacteria were quantified by plating of homogenized tissue on MB-complete agar plates. Histology was performed in the Mouse Pathology Platform at HZI Braunschweig as recently described (Meißner et al., 2014; Suwandi et al., 2014).
Extraction of Nucleic Acids and Quantitative Real-Time PCR (qRT-PCR)
Genomic DNA was prepared as previously described (Stratmann et al., 2004). Plasmids were prepared using NucleoBond® AX kit (Macherey Nagel GmbH, Düren, Germany) according to the manufacturer’s protocol. Total RNA from bacteria was isolated according to Rustad et al. (2009) with minor modifications. In brief, bacterial pellets were resuspended in TRIzol® reagent, mechanically disrupted in a FastPrep® instrument (ThermoSavant, Carlsbad, CA, USA) and RNA was separated by chloroform, chloroform-isoamylalcohol (49:1 v/v) extraction, and precipitation with 2-propanol. Total RNA was treated twice with 50 U DNase I (Roche, Mannheim, Germany) according to the manufacturer’s protocol. Integrity and quality of RNA was determined by gel electrophoresis and spectrophotometric analysis using an Epoch instrument (Biotek, Bad Friedrichshall, Germany) at 260 nm.
For cDNA synthesis, 4 μg of DNA-depleted RNA were diluted with RNase free water up to a volume of 20 μl and incubated for 10 min at 70°C with 0.4 μg random primers (Promega, Madison, WI, USA). After 5 min cooling on ice, samples were split in two and mixed with 5x RT buffer, 10 mM dNTP’s, and either 200 U MMLV-superscript transcriptase (Promega) or buffer without transcriptase as a negative control. The mix was incubated for 1 h at 42°C, following incubation for 5 min at 85°C. Samples were diluted with 90 μl ddH2O and stored at -20°C for further analysis. 2.5 μl of each cDNA sample was mixed with 0.4 μM primer and 10 μl SYBR-Green Mix (Qiagen, Düsseldorf, Germany) in a total volume of 20 μl. Quantitative real-time PCR (qRT-PCR) experiments were performed using a Mx3005P qPCR system (Agilent Technologies, Santa Clara, CA, USA) with a thermal cycling profile as follows: segment 1, 20 min at 95°C, 1 cycle; segment 2, 45 s at 95°C, 1 min at 58°C, 1 min at 72°C, 45 cycles; segment 3, 1 min 95°C, 30 s 55°C, 30 s 95°C, 1 cycle. Each sample was analyzed in duplicate. Ct values were shown either as absolute data or normalized to the housekeeping gene gap and expressed as fold change to the untreated control (ΔΔct). All oligonucleotide primer pairs (Table S1) were tested for efficacy with a serial dilution of genomic DNA.
RNA Deep Sequencing and Analysis
RNA deep sequencing was used to determine the regulon of FurA in MAP. Sequencing libraries of MAPwt and MAPΔfurA RNA, of three independent samples each, were prepared and sequenced using 36 bp single-ends sequencing on a Genome Analyzer IIx (Illumina, San Diego, CA, USA) or 50 bp single-ends sequencing on a HiSeq2500 instrument (Illumina), respectively. In brief, libraries of 300 bp were prepared according the manufacturer’s instructions “Preparing Samples for Sequencing of mRNA” (Illumina) and “Protocol for ScriptSeqTM v2 RNA-Seq Library Preparation” (Epicentre Biotechnologies, Madison, WI, USA). Quality of the libraries was validated using an Agilent Bioanalyzer (Agilent Technologies) following the manufacturer’s instruction. Cluster generation was performed using the Illumina cluster station; sequencing on the Genome Analyzer IIx or HiSeq2500 followed a standard protocol. The fluorescent images were processed to sequences and transformed to FastQ format using the Genome Analyzer Pipeline Analysis software 1.8.2 (Illumina). The sequence output was controlled for general quality features using the fastq-mcf tool of ea-utils (Aronesty, 2013) and was mapped against the genome sequence of the reference strain MAP-K10 (AE016958) using BWA version 0.7.5 (Li and Durbin, 2009) and SAMtools (Li et al., 2009) for storing nucleotide sequence alignments. Strain MAP-K10 was used as a reference strain as the genome of MAP DSM44135 has not been sequenced yet. Raw data sets are available in the European Nucleotide Archive Repository1. For data analyses, reads of the 50 bp sequencing were clipped to 36 bp reads. Following, all sequences were computed with Rockhopper tool (McClure et al., 2013). Genes with a q-value ≤0.01 were considered as significantly differentially expressed and genes with raw counts in all replicates were included for further analysis (Table S3). The fold change was calculated as the expression value of MAPwt/MAPΔfurA.
Bioinformatics and Statistics
Differentially expressed genes identified by Rockhopper analysis were further processed with Blast2Go tool (Conesa et al., 2005) and NCBI blastx2 as well as tuberculosis database3 to investigate functions of putative proteins. Additionally, the putative proteins were clustered into orthologous groups (COG), based on the genome of MAP-K104. Data are expressed as mean ± SEM, and statistical analyses were performed using GraphPad Prism 5.03 (GraphPad, San Diego, CA, USA). Depending on the experiment, either the non-parametric t-test (Mann–Whitney) or one-way ANOVA test (Kruskal–Wallis) were used. A p-value of <0.05 between samples and controls was considered as statistically significant.
Prediction of putative FurA binding sites in MAP (NCBI accession No. NC_002944) was performed by FIMO analysis (Bailey et al., 2009). Initially, we generated a consensus sequence for FurA binding by comparison of sequences located -70/+30 bp up-/downstream of predicted FurA translation start sites in Mtb (NC_000962, Rv1909c), M. bovis BCG pasteur (NC_008769, BCG_1948c), M. avium ssp. avium 104 (NC_008595, MAV_2752), MAP (NC_002944, MAP_1669c), and M. smegmatis (NC_008596, MSMEG_6383) with MEME Suite (Bailey et al., 2009). The resulting binding motif T[CT]TTGACT[CG][AG]TTCCA[GA]A[AT]AA[GT][GT][GC][ACG][GC][TG]CAT[AT] was subsequently submitted to FIMO analysis. Nucleotides in brackets are variable, single nucleotides are conserved.
Results
Characterization of a MapΔfura Deletion Mutant
Ferric uptake regulator A (FurA) has been described as a global regulator in E. coli and was proposed to be involved in oxidative stress response and regulation of iron homeostasis (Zheng et al., 1999). In mycobacteria, iron homeostasis is maintained by IdeR, and FurA is assumed to take part in general stress response due to its genomic co-localization and its (partly) co-transcription with katG. However, the regulatory role of FurA has not yet been fully understood.
To gain insight into the role of FurA in MAP, we generated a furA (map1669c) deletion mutant MAPΔfurA by specialized transduction. In addition, a complemented strain was constructed by introducing the integrative plasmid pFurA-MAP1101, harboring the furA gene under control of its own promoter, into the ΔfurA mutant strain, resulting in strain MAPΔfurAC. Deletion of the furA gene in MAPΔfurA and complementation of MAPΔfurAC were confirmed by PCR (Figure 1A). Expression of furA and the adjacent gene katG were analyzed by qRT-PCR. furA transcripts were detected in MAP wildtype (MAPwt) and MAPΔfurAC but not in MAPΔfurA. The expression levels of katG were similar in all strains (Figure 1B), indicating that the genetic manipulation had no polar effect on the expression of downstream located genes. Next we compared growth of MAPwt, MAPΔfurA, and MAPΔfurAC in MB-complete broth. As shown in Figure 1C, all strains exhibited similar growth kinetics. Growth of MAPΔfurA and MAPΔfurAC was slightly reduced in comparison to MAPwt, and both strains reached stationary growth phase at slightly lower OD600 than MAPwt. These effects might be due to the antibiotic supplements in the growth media.
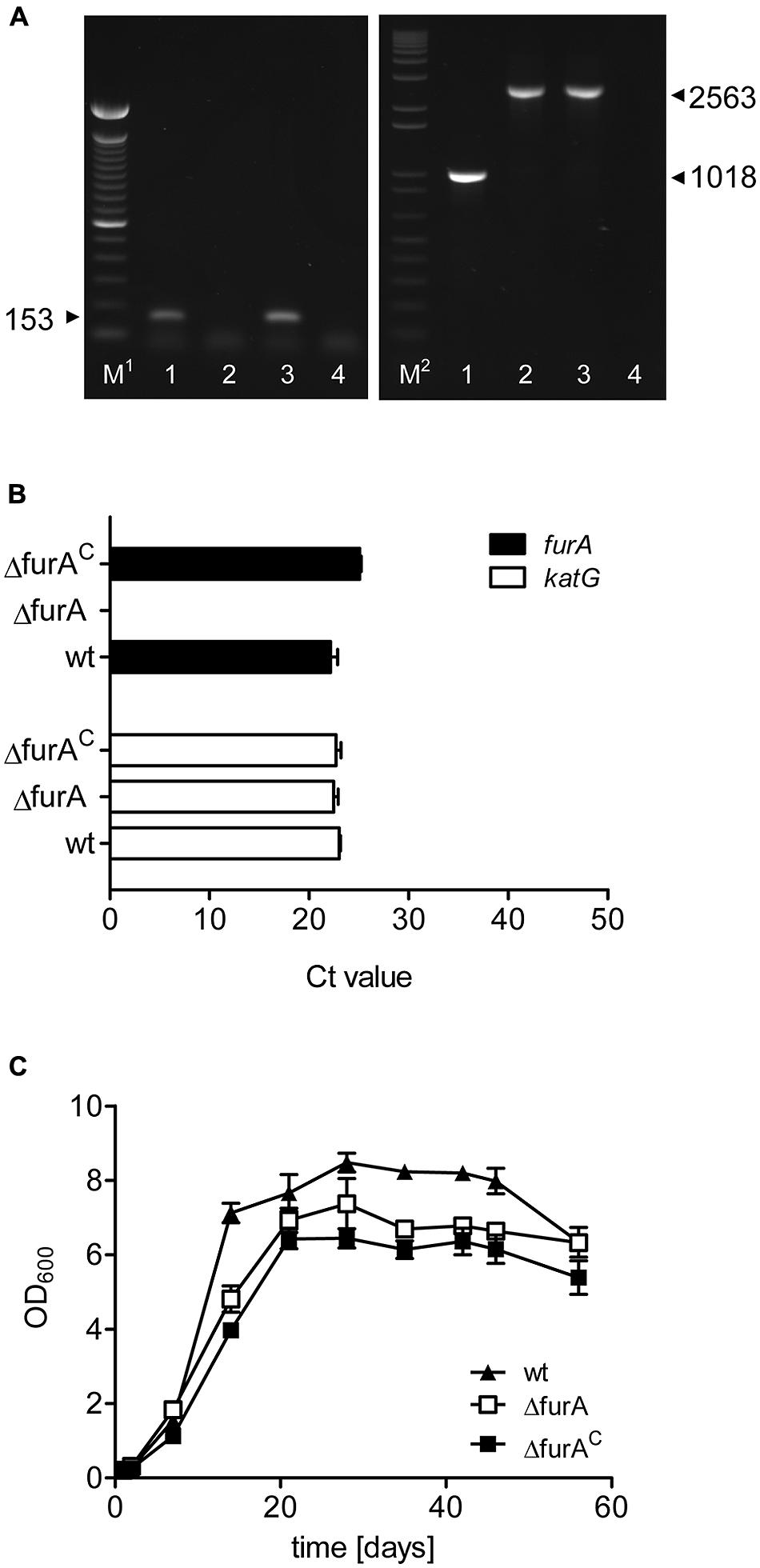
FIGURE 1. Characterization of a MAPΔfurA mutant. (A) Confirmation of deletion and complementation of furA. Chromosomal DNA was prepared from MAPwt (lanes 1), MAPΔfurA (lanes 2), and MAPΔfurAC (lanes 3) grown in MB-complete broth to an OD600 of 1.0 and was subsequently tested in PCR for the presence of furA (left panel) with primers oRTfurA1 fw/rev and hygromycin (right panel) with primers oTKfurAKatGfw/oRTlppsrev. A negative control with water only is shown in lanes 4. M1 = 100 bp DNA ladder (Invitrogen), M2 = 1 kb plus DNA ladder. Samples were run on a 1.5% agarose gel. (B) Total RNA was extracted from all strains grown to an OD600 of 1.0 in MB-complete broth. Expression of furA (black bars) and the adjacent gene katG (white bars) was analyzed by qRT-PCR. Results are expressed as mean Ct values of three independent experiments. The level of the housekeeping gene gap for all samples was 19.27 ± 0.4060. (C) Growth of MAPwt (filled triangles), MAPΔfurA (open rectangle), and the complemented strain MAPΔfurAC (filled rectangle) was monitored for 60 days by measurement of the optical density (OD600) in MB-complete broth. Each growth experiment was conducted in triplicate for each strain. Shown are representative results of one out of two independent experiments.
Determination of the FurA Regulon of Map
To analyze the regulatory role of FurA in MAP, we prepared RNA from MAPwt and MAPΔfurA grown to an OD600 of 1.0 in MB-complete broth and performed RNA deep sequencing. Gene expression profiles of three independent replicates of each strain were analyzed with the Rockhopper analysis tool (McClure et al., 2013), and genes with a q-value of ≤0.01 were considered as significantly differentially expressed. Differences in gene expression were calculated as fold change of expression values calculated by Rockhopper analysis (MAPΔfurA vs. MAPwt).
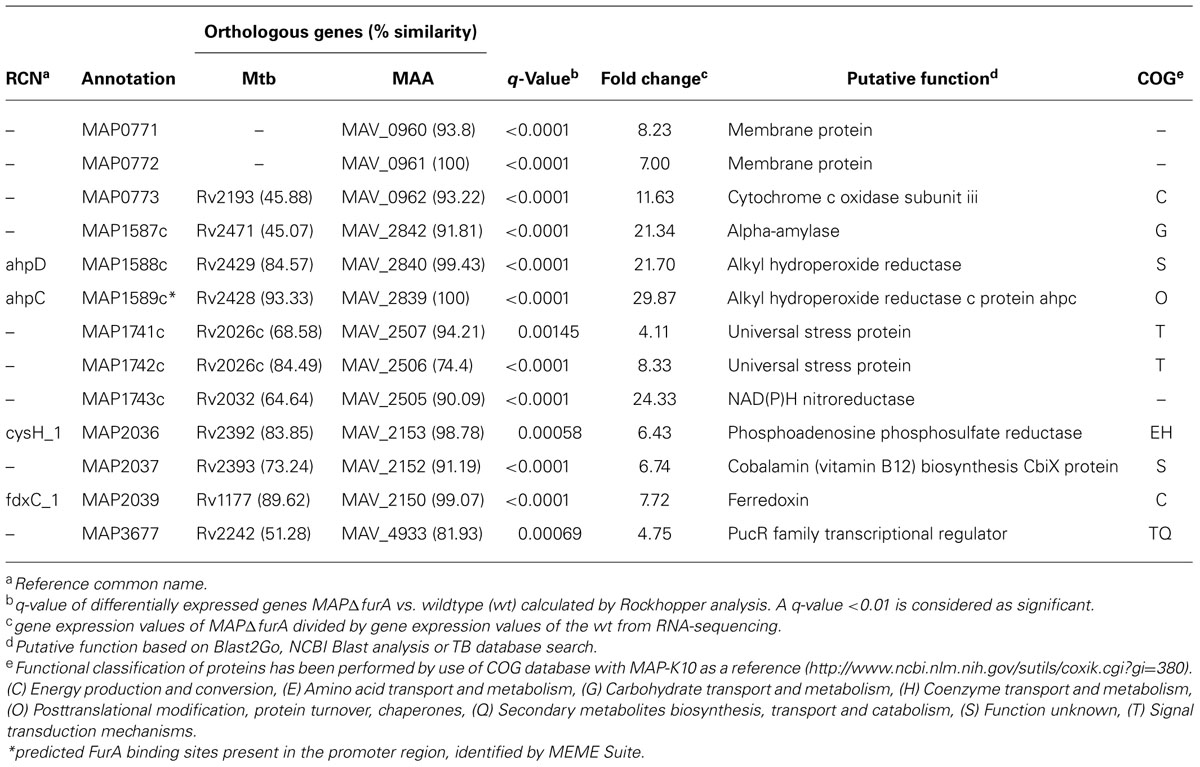
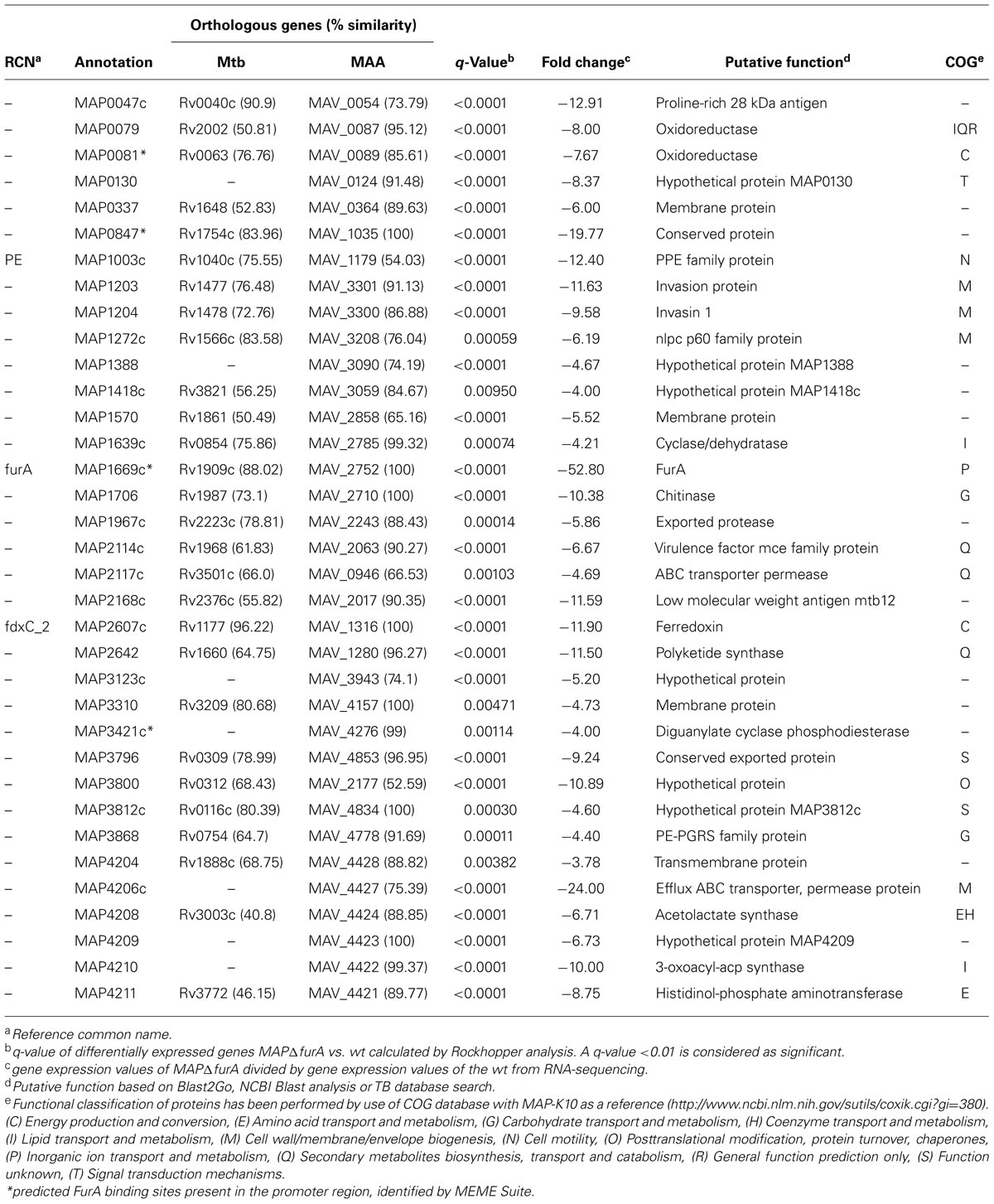
In total, we identified 48 genes, which were differentially expressed in the mutant strain. Compared to the wt strain, 13 genes were higher (Table 1) and 35 lower (Table 2) expressed in MAPΔfurA. Clustering of putative proteins encoded by the differentially expressed genes into orthologous groups (COG) revealed that almost 40% of the differentially expressed genes were assigned to metabolism, 20.75% to cellular processes and signaling, 9.43% belong to the group of poorly characterized proteins, and 30.19% were not assigned to any COG (Tables 1 and 2). No genes related to iron homeostasis, such as siderophores (mycobactin) and siderophore uptake systems (Neilands, 1992; Escolar et al., 1998; McHugh et al., 2003) or iron storage proteins bfr, fntA (Abdul-Tehrani et al., 1999; Wildermann et al., 2004), were affected by the furA deletion in MAP.
Among the 13 higher expressed genes, 11 were organized in 4 putative operons. The majority of these genes encodes for proteins predicted to be involved in stress response or redox processes. Within this group, the gene cluster map1589c–1587c, encoding for two alkyl hydroperoxide reductases involved in oxidative stress and drug resistance as well as intracellular survival in Mtb (Sherman et al., 1999; Master et al., 2002), and a NAD(P)H nitroreductase (map1743c), which has been shown to be associated with detoxification of nitroaromatic components in Mtb (Purkayastha et al., 2002), exhibited the most significant expression changes (Table 1).
The group of lower expressed genes is comprised of 35 genes. Aside from furA, this group consists of two putative transporters, five genes predicted to be involved in metabolism, three stress response-associated genes and 13 genes were of unknown function. The remaining 11 genes can be associated to virulence which is represented by two putative invasion proteins (map1203/1204), two putative antigens (map0047c/map2168c), one Pro-Pro-Glu protein (PPE; map1003c), one PE-PGRS glycin-rich protein (map3868), one nlpc p60 protein (map1272c), and one mammalian cell entry (mce) family protein (map2114c) as well as three other enzymes (map1706, map1967c, map3421c) for cell entry and persistence. Altogether, map0047c, map0847 (conserved protein), and map4206c (putative efflux ABC transporter) showed the most significant differences in expression compared to the wt (Table 2). In addition, the 5′ UTRs of map1589c (ahpC), map0081 (putative oxidoreductase), map0847, and map3421c (putative diguanylate cyclase phosphodiesterase) featured a predicted FurA binding site (Tables 1 and 2). Overall, these analyses indicated a considerable involvement of MAP FurA in the adaptation of MAP to the host cell environment and general stress response.
To confirm the results of our RNA deep sequencing analyses and to further specify the involvement of FurA in regulation, we analyzed the transcription of ahpC and ahpD (map1589c/1588c, higher expressed in MAPΔfurA), and of map0847 and map0047c (lower expressed in MAPΔfurA) by qRT-PCR. MAPwt, MAPΔfurA, and the complemented strain MAPΔfurAC were grown in MB-complete broth to an OD600 of 1.0, RNA was extracted and analyzed by qRT-PCR. As a control, we included the mbtB gene (map2177c) which has been shown to be regulated iron-dependently by IdeR in Mtb (Gold et al., 2001) and MAP (Janagama et al., 2009). As expected, the expression of mbtB was similar in all strains. In agreement with our RNA sequencing analyses, ahpC, ahpD, map0847, and map0047c were up- and down-regulated in the mutant, respectively. In the complemented mutant MAPΔfurAC expression levels were mostly restored to wt levels (Figure 2). The differences to the wt strain might be due to the slightly lower expression level of furA in MAPΔfurAC compared to MAPwt (Figure 1B). Overall, these results confirm the regulatory influence of FurA on expression of ahpC, ahpD, map0847, and map0047c.
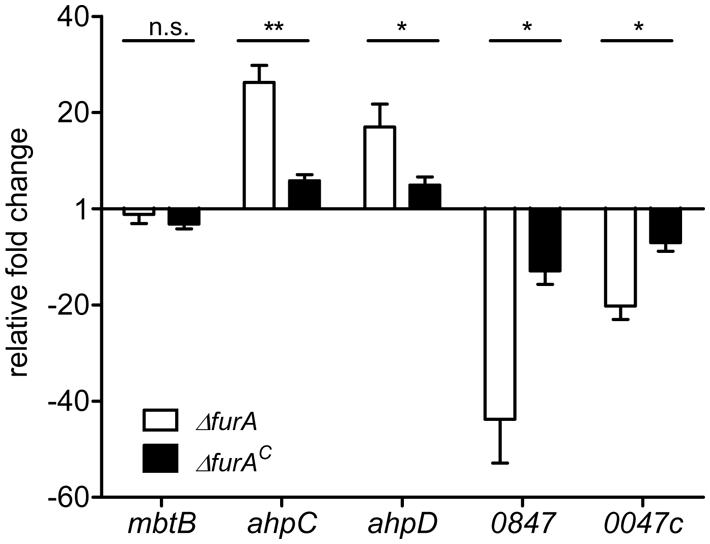
FIGURE 2. Functional complementation of FurA-regulated genes. MAPwt, MAPΔfurA and the complemented strain MAPΔfurAc were grown in MB-complete broth to an OD600 of 1.0. Total RNA was extracted, followed by cDNA synthesis and qRT-PCR analysis. Expression of genes mbtB, ahpC (map1589c), ahpD (map1588c), map0847, and map0047c in MAPΔfurA (white bars) and MAPΔfurAc (black bars) was compared to transcription levels in MAPwt. Three independent experiments were analyzed, qRT-PCR was performed in duplicate for each sample, normalized to the housekeeping gene gap and expressed as fold change compared to the MAPwt (mean ± SEM). Statistical analysis was performed using Mann–Whitney test with *p < 0.05, **p < 0.005.
ahp Expression in Map is Hydrogen Peroxide Dependent
AhpC and AhpD are metal dependent alkyl hydroperoxide reductases and constitute an antioxidant system important in reduction and neutralization of a wide range of organic hydroperoxides and peroxynitrite (Wood et al., 2003). Genes encoding for AhpC/D have been shown to be induced by peroxide stress mediated by OxyR in other mycobacteria (Dhandayuthapani et al., 1997). However, in MAP these genes seem to be regulated by FurA which resembles PerR mediated activation in Bacillus and Campylobacter (van Vliet et al., 1999; Helmann et al., 2003). Thus, we exposed MAPwt and MAPΔfurA cultures without catalase supplement to oxidative stress by addition of 10 mM H2O2 for 2 h and analyzed ahpD expression by qRT-PCR. As a negative control we used the FurB dependent but FurA independent regulated mptA (map3736c) gene (Eckelt et al., 2014).
As shown in Figure 3, expression of ahpD in MAPΔfurA was increased compared to the wt and supports RNA-sequencing data. Upon peroxide treatment, both strains responded with an induction of ahpD expression. However, the relative fold change in MAPΔfurA was considerably higher than in MAPwt compared to the untreated control.
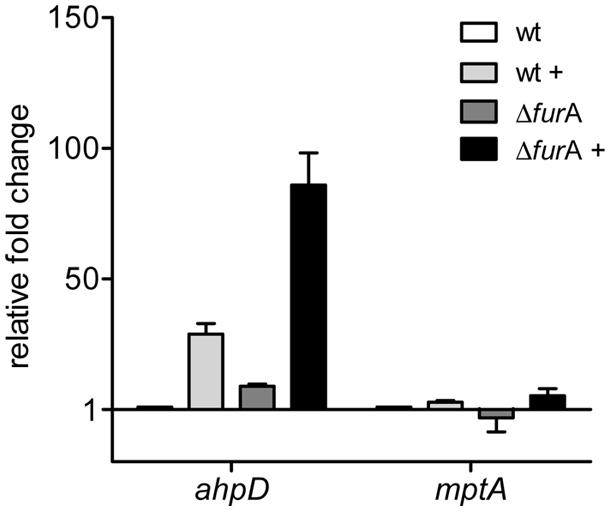
FIGURE 3. Response of the FurA regulated alkyl hydroperoxide reductase system AhpCD to oxidative stress. MAPwt and MAPΔfurA cultures were grown in MB-complete broth, harvested at OD600 of 1.0, washed, transferred to MB-cat broth and incubated for 2 h with or without 10 mM H2O2. RNA was isolated and used for cDNA synthesis. Gene expression analysis by qRT-PCR was performed for the genes ahpD (map1588c) and mptA (map3736c) in untreated (MAPwt, white bars; ΔfurA, dark gray bars) and H2O2 treated samples (MAPwt, light gray bars; ΔfurA, black bars). Results were normalized to the housekeeping gene gap and are expressed as fold change compared to the untreated MAPwt control (mean ± SEM, n = 3).
In contrast mptA expression was similar in all strains at both conditions (standard/oxidative stress).
These data on the one hand side indicate that repression of ahpC/D by FurA can be relieved by peroxide. On the other hand they demonstrate that an additional peroxide sensing regulatory activity, most probably by OxyR or a OxyR-like regulator, co-regulates ahpC/D expression in MAP.
MapΔfurA Exhibits Higher Resistance to Intracellular Killing in Macrophages but FurA is Dispensable During Chronic Infection
It has been shown that ahpC/D are necessary for defense against ROS and RNS in the mycobacterial phagosome (Sherman et al., 1995; Wilson et al., 1998; Master et al., 2002). By this pathogenic mycobacteria are able to survive within macrophages, enabling metabolic adaptation to the phagosomal compartment which is necessary for persistence. The above data showed that FurA is involved in the regulation of MAP response to oxidative stress, as indicated by the regulation of enzymes involved in neutralization of oxygen and nitrogen radicals [ahpC/D, NAD(P)H nitroreductase]. Therefore, we aimed to address the role of FurA for intracellular survival of MAPwt, MAPΔfurA, and MAPΔfurAC in macrophages. First we confirmed that the MAP strains are exposed to ROS after being taken up by the macrophages. For this, J774A.1 macrophages were infected with MAPwt, MAPΔfurA, and MAPΔfurAC. After 1.5 h of infection, cells were treated with the reagent CellROX Deep Red for 30 min. This dye fluoresces upon oxidation and allows the detection of ROS. As positive control we included macrophages treated (+) with menadione (Figure 4B) which is known to induce the generation of ROS in eukaryotic cells (Malorni et al., 1993). As shown in Figures 4A,B, the MFI measured by FACS analysis in macrophages infected with MAPwt, MAPΔfurA and the complemented mutant strain was higher than in untreated (-) macrophages (Figure 4B), indicating the generation of ROS induced by all MAP strains. Next, cells were infected with MAPwt, MAPΔfurA, and MAPΔfurAC as described above and bacterial survival was determined after 2 h, 2 and 7 days by CFU-counting after serial dilution plating. After 2 h and 2 days infection we found significantly higher numbers of viable MAPΔfurA cells compared to MAPwt (Figure 4C). Complementation of MAPΔfurA almost completely restored the phenotype to wt level. Confocal analyses of macrophages infected with the different MAP strains for 30 min or 2 h after inside-outside staining of bacteria showed no differences in mycobacterial uptake by and adhesion to the macrophages (Figure 4D). These findings clearly exclude a better phagocytosis of MAPΔfurA which could account for the initial higher survival of MAPΔfurA in the macrophages. Interestingly, the percentage of viable MAPΔfurA cells decreased over time from ∼60% (of the initial inoculum) at 2 h to ∼40% at day 2 and to ∼18% at day 7. In contrast, no change in survival was visible in the wt and complement strain, indicating that the mutant was also being killed (Figure 4C). Calculating the survival rates at day 2 and 7 relative to the number of bacteria taken up by the macrophages after the 2 h infection period revealed that MAPΔfurA even though being more resistant to initial killing, is much more efficiently killed during the time course of macrophage infection (data not shown). Overall, these data indicate that MAPΔfurA can better resist the initial killing by the macrophage most probably caused by oxidative stress but is susceptible to macrophage defense mechanisms present at later time points of infection or less adapted to the phagosomal life style.
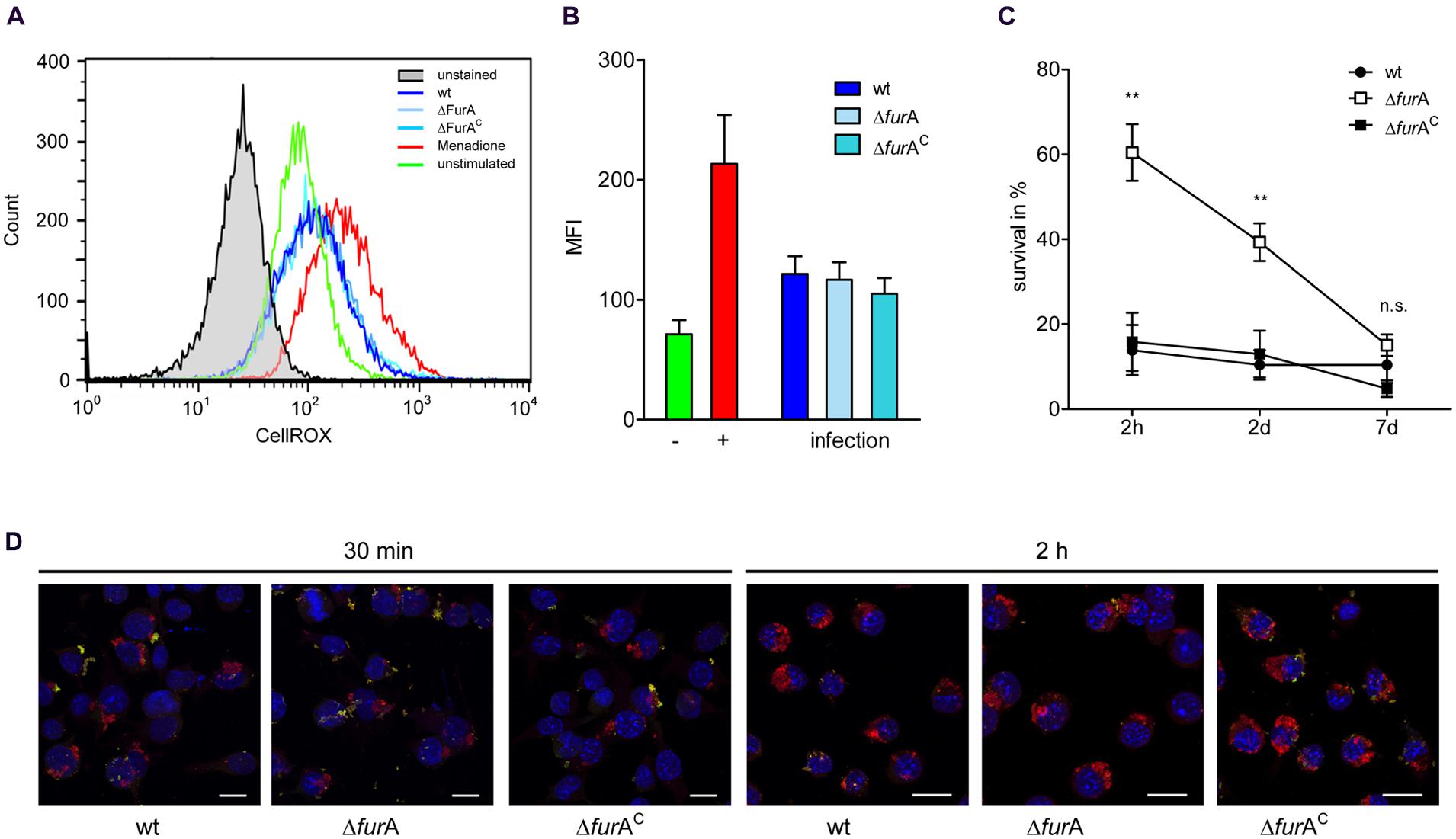
FIGURE 4. Induction of oxidative burst in macrophages and survival of MAP strains in macrophages. J774A.1 macrophages (Mø) were grown in DMEM and infected with an MOI of 10 with MAPwt, MAPΔfurA and the complemented strain MAPΔfurAC grown in MB-complete broth to an OD600 of 1.0. Mø were infected as described above for 1.5 h or treated with 100 μM menadione (+). CellROX Deep Red was then added to a final concentration of 2.5 mM for 30 min. (A) Histograms showing the production of ROS measured by CellROX Deep Red staining of unstimulated cells (green histogram) or infected Mø with MAPwt (dark blue histogram), MAPΔfurA (light blue histogram), and the complemented strain MAPΔfurAC (turquoise histogram). Mø treated with 100 μM menadione were used as a positive control (red histogram). Unstained cells are shown in the gray histogram. One representative experiment out of four is shown. (B) Fluorescence of untreated (-, green bar), menadione treated (+, red bar), and with MAPwt (dark blue bar), MAPΔfurA (light blue bar) or MAPΔfurAC (turquoise bar) infected macrophages was determined by FACS analysis. Shown are the results of four independent experiments (mean fluorescence intensity, MFI ± SEM). (C) Mø were infected as described above and incubated for 2 h, 2 and 7 days. Cells were disrupted and the lysate was diluted prior plating. 25 μl of the dilutions were plated in duplicate on MB-complete agar and incubated for up to 10 weeks. Five independent experiments were conducted, CFU of lysates and of the respective inocula used for macrophage infection were determined, and mycobacterial survival was expressed as percent to the inoculum. Statistical analysis was performed using one-way ANOVA (Kruskal–Wallis) with **p < 0.005. (D) Confocal maximum intensity projections of a double immunofluorescence staining of J774A.1 macrophages infected for 30 min or 2 h with MAPwt, MAPΔfurA, and MAPΔfurAC. Extracellular mycobacteria were labeled with anti-MAP-HBHA and green fluorescent Alexa Fluor® 488, intracellular bacteria with anti-MAP-HBHA and orange fluorescent Alexa Fluor® 568. Nuclei were stained blue with DAPI. Extracellular bacteria appear yellow and intracellular appear red. Bar represents 15 μm.
Hence, we analyzed the role of FurA for survival in the host. For this, C57BL/6 mice were infected intraperitoneally with 1 × 108 bacteria of exponentially grown cultures (OD600 of 1.0) and were sacrificed 4 weeks post infection. As shown in Figure 5, no significant differences in weight of liver and spleen, as well as number of bacteria or granuloma in the liver upon infection with MAPwt and MAPΔfurA were detected. However, as a trend we observed lower CFU numbers in the livers of MAPΔfurA infected cells. Overall, the lack of FurA in MAP did not alter survival rates compared to the wt and suggests that FurA might be dispensable for MAP-infection in the mouse model.
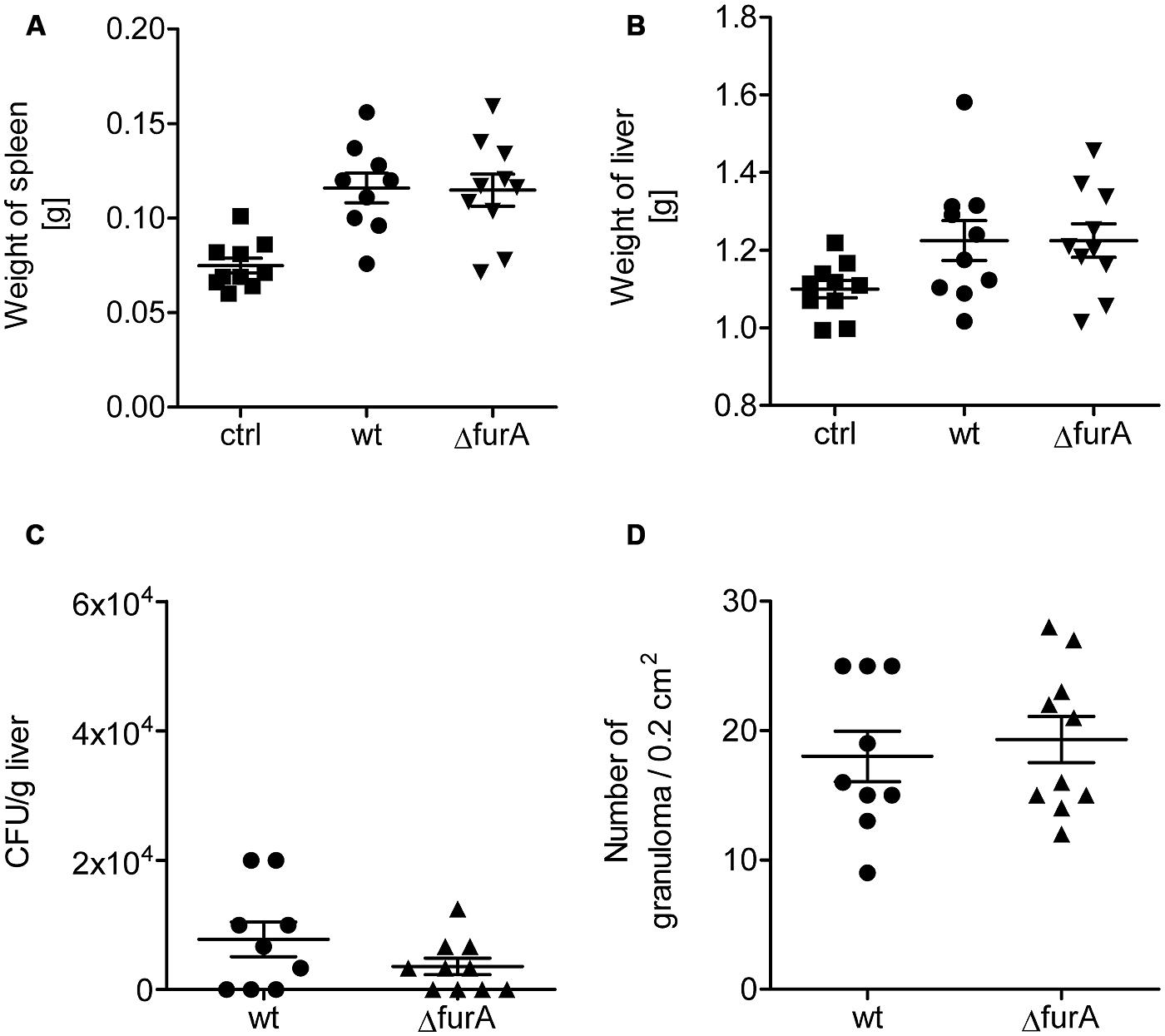
FIGURE 5. Biological fitness of MAPwt and MAPΔfurA in infected mice. Eight weeks old female C57BL/6 mice were infected with MAPwt (circles) and MAPΔfurA (triangle), grown to an OD600 of 1.0, with an infection dose of 1 × 108 cells and sacrificed 4 weeks later. The control group was treated with DPBS buffer (squares). Weight of spleen (A), liver (B), CFU detection of bacteria from liver (C), and the amount of granuloma in the liver (D) were chosen as read-out parameters. The results represent the mean ± SEM of one animal experiment with 9 mice in each group.
Discussion
Bacterial iron homeostasis is tightly regulated and closely linked to the cellular response against cytotoxic effects due to the generation of ROS by free iron ions via the Fenton reaction (Cornelis et al., 2011). Accordingly, FurA was first identified in E. coli as global transcriptional regulator influencing expression of genes associated with iron acquisition and storage, but was later also shown to be involved in the regulation of genes activated by cellular stress (Dubrac and Touati, 2000). In mycobacteria, furA is located immediately upstream of katG encoding a catalase-peroxidase. Both genes are co-induced by peroxide treatment (Master et al., 2001; Milano et al., 2001; Jang et al., 2009). From this and the higher resistance of a furA mutant of the fast growing M. smegmatis to peroxide stress (Zahrt et al., 2001), it was proposed that in mycobacterial species, FurA is involved in the oxidative stress response regulation (Lucarelli et al., 2008). However, the precise role of FurA in pathogenic mycobacteria has not yet been elucidated.
In the present study, we characterized the role of furA in MAP. By RNAseq analyses, we were able to dissect a specific involvement of FurA in the regulation of the MAP response to reactive nitrogen and oxygen radicals, in intracellular survival, and in the expression of predicted virulence-associated genes. Most interestingly, unlike in many other bacterial species, where Fur regulates the expression of iron uptake and storage systems (Bagg and Neilands, 1987; Bsat et al., 1998; Abdul-Tehrani et al., 1999), FurA did not participate in the regulation of iron homeostasis in MAP. The involvement of FurA in modulating stress response and mechanisms for evading host cell defense as well as for invasion and infection is emphasized as 30 of the 48 regulated genes have been shown by works of other groups to be differentially expressed in MAP after infection of macrophages or in vivo infection of cattle (9 M/O, 21 in vivo; Wu et al., 2007; Cossu et al., 2012; Ghosh et al., 2013) and 26 of these genes have been found to be associated to stress response, as they were significantly regulated when MAP was exposed to acidic pH, nitrosamine, hydrogen peroxide, or heat (Wu et al., 2007; Kawaji et al., 2010; Cossu et al., 2012).
Our transcriptomic data also revealed that the FurA regulon includes genes with repressed and with activated transcription. Expression of a defined group of 13 genes encoding proteins of general stress response and enzymes necessary for the generation of reductive activity to detoxify oxygen radicals was higher in MAPΔfurA and is therefore repressed by FurA. For instance map1589c (ahpC) and map1588c (ahpD) were found to be induced in MAP upon oxidative stress (Kawaji et al., 2010) and during macrophage infection (Ghosh et al., 2013). Additionally, we found a peroxide dependent regulation of these genes by FurA in MAP. The alkyl hydroperoxide reductases AhpC and AhpD of the peroxireductase family (Prx) form an antioxidant system that is necessary for the reduction and neutralization of a wide range of organic hydroperoxides and peroxynitrite (Wood et al., 2003) to protect structures particularly sensitive to peroxide-mediated damage, such as lipids and nucleic acids (Halliwell and Gutteridge, 1984). A regulation of ahp genes by the Fur-like regulator PerR has been described for other bacteria, e.g. Bacillus subtilis and Campylobacter jejuni (van Vliet et al., 1999; Helmann et al., 2003). The regulatory mechanism bases on the property of mononuclear iron proteins such as FurA to lose repressive activity during both O2- and H2O2 stress by oxidation of Fe2+ to Fe3+. In addition to the missing activation of iron acquisition systems it supports our hypothesis of the role of FurA as ROS sensing regulator in MAP. CysH has been shown to contribute to protection against oxidative and nitrosative stress in Mtb (Senaratne et al., 2006) and many other FurA-repressed genes encode for enzymes involved in reactions liberating reducing activity in form of H+ ions.
Beyond this, FurA actively contributes to the expression of a subset of 35 genes many of which are suggested to be associated to virulence or intracellular survival. A major group of FurA-activated genes comprises genes encoding invasins (MAP1204/1204), antigens (MAP2168c, Webb et al., 1998), proteins of the Pro-Pro-Glu (PPE) family (MAP1003c, Akhter et al., 2012), PE-PGRS glycine-rich proteins, (MAP3868, Iantomasi et al., 2012), nlpc p60 (MAP1272c, Parthasarathy et al., 2012), and a mce family protein (MAP2114c, Gioffre et al., 2005) as well as several enzymes for cell entry, and it has been demonstrated for other bacteria, that homologs to these genes are important virulence factors (Kulasakara et al., 2006; Frederiksen et al., 2013). Interestingly, also some proteins involved in electron transfer were affected which suggest that FurA contributes to the maintenance of the canonical endogenous redox balance. Finally we found, that katG expression was not affected by furA deletion. Despite the fact, that a co-transcription of both genes and regulation of katG by FurA was suggested for other mycobacteria species (Master et al., 2001; Pym et al., 2001; Zahrt et al., 2001), we could not confirm this for MAP. Hence, katG might be regulated independently of FurA as proposed for Mycobacterium sp. strain JC1 DSM 3803 (Lee et al., 2009) or other bacteria (Italiani et al., 2011).
The particular role of FurA for virulence in mycobacteria has not been shown yet. Like other pathogenic mycobacteria, MAP is able to infect and survive in macrophages. Therefore, MAP must be able to react to the antimicrobial macrophage response such as reactive oxygen radicals. Indeed, our macrophage infection experiments with MAPwt, MAPΔfurA, and MAPΔfurAC showed the induction of ROS formation in the host cell. When monitoring the intracellular fate of MAPwt, MAPΔfurA, and MAPΔfurAC, we observed a significantly higher percentage of viable MAPΔfurA cells compared to the wt and MAPΔfurAC during the initial phase of infection. The higher percentage resulted from higher survival most probably given by an enhanced resistance of MAPΔfurA to oxidative stress mediated by the expression of FurA-repressed genes. Enhanced resistance to oxidative stress was also described for C. jejuni and M. smegmatis: in these species, deletion of perR and furA, respectively, resulted in a hyper resistancy to peroxide stress (van Vliet et al., 1999; Pym et al., 2001). In contrast, the survival rate of MAPΔfurA was considerably lower compared to the wt and MAPΔfurAC, indicated by killing of the mutant over time. This suggests that genes activated by FurA are needed for the adaptation of MAP to the macrophage phagosomal microenvironment.
In the intestinal tissue of the infected host, metabolic adaptation, and oxidative stress defense dominate the metabolism of MAP (Weigoldt et al., 2011, 2013). In contrast to the macrophage infections, the furA deletion seems not to affect MAP survival after 4 weeks intraperitoneal MAP infection of mice. Histological staining did not show significant differences in the number of granuloma and granuloma size as well as composition (data not shown). This suggests that the phenotype of MAPΔfurA is not advantageous when MAP has to counteract a complex immune response or has to metabolically adapt to the host environment. Nevertheless, we cannot exclude that FurA might play a role in a later stage of infection or effects of the FurA deletion might only be notable very early after infection.
Conclusion
In conclusion, in the presented study, we determined for the first time the regulon of FurA in a pathogenic mycobacterial species. Our data provide novel insights into the function of FurA in mycobacteria. They suggest two different physiological roles executed by FurA. Firstly, FurA acts as a repressor for a selective group of genes involved in the response to oxidative stresses. During oxidative stress it might operate as an iron-based sensor of ROS and, therefore, might be functionally closely related to PerR. Secondly, FurA contributes to activation of a second group of genes, many of which are suggested to contribute to MAP long term survival in macrophages. To conclusively investigate the role of FurA in virulence, however, infection experiments in animals developing clinical disease are required.
Author Contributions
Ralph Goethe, Gerald-F. Gerlach, and Jochen Meens designed the experiments; Thorsten Meißner constructed the mutant, Thorsten Meißner and Kristin Laarmann accomplished mutant complementation, Michael Jarek performed RNA sequencing; Thorsten Meißner, Elke Eckelt, and Kristin Laarmann characterized the mutant and the complemented strain; Elke Eckelt performed stress experiments, Andreas Nerlich and Kristin Laarmann performed the macrophage experiments, Ralph Goethe and Siegfried Weiss designed the mouse infection experiments, Thorsten Meißner performed the mouse infections. Thorsten Meißner, Elke Eckelt, Jochen Meens, Ralph Goethe analyzed data; and Elke Eckelt, Ralph Goethe, Gerald-F. Gerlach, and Jochen Meens wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Nina Janze (TiHo) for excellent technical assistance and Abdulhadi Suwandi (HZI) for the help in the mouse infection experiments. This work was supported by a grant from the German Research Foundation (DFG, Ge522/6-1). Ralph Goethe and Gerald-F. Gerlach were additionally supported by the German Federal Ministry of Education and Research (BMBF, ZooMAPII: 01KI1003A, 01KI1003B).
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2015.00016/abstract
Footnotes
- ^http://www.ebi.ac.uk/ena/data/view/PRJEB7916
- ^http://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^http://tbdb.org
- ^http://www.ncbi.nlm.nih.gov/sutils/coxik.cgi?gi=380
References
Abdul-Tehrani, H., Hudson, A. J., Chang, Y.-S., Timms, A. R., Hawkins, C., Williams, J. M.,et al. (1999). Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 181, 1415–1428.
Akhter, Y., Ehebauer, M. T., Mukhopadhyay, S., and Hasnain, S. E. (2012). The PE/PPE multigene family codes for virulence factors and is a possible source of mycobacterial antigenic variation: perhaps more? Biochemie 94, 110–116. doi: 10.1016/j.biochi.2011.09.026
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Aronesty, E. (2013). Comparison of sequencing utility programs. Open Bioinformatics J. 7, 1–8. doi: 10.2174/1875036201307010001
Bagg, A., and Neilands, J. B. (1987). Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26, 5471–5477. doi: 10.1021/bi00391a039
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L.,et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bsat, N., Chen, L., and Helmann, J. D. (1996). Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes. J. Bacteriol. 178, 6579–6586.
Bsat, N., Herbig, A., Casillas-Martinez, L., Setlow, P., and Helmann, J. D. (1998). Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29, 189–198. doi: 10.1046/j.1365-2958.1998.00921.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Butcher, J., Sarvan, S., Brunzelle, J. S., Couture, J. F., and Stintzi, A. (2012). Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc. Natl. Acad. Sci. U.S.A. 109, 10047–10052. doi: 10.1073/pnas.1118321109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carpenter, B. M., Whitmire, J. M., and Merrell, D. S. (2009). This is not your mother’s repressor: the complex role of fur in pathogenesis. Infect. Immun. 77, 2590–2601. doi: 10.1128/IAI.00116-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Christman, M. F., Storz, G., and Ames, B. N. (1989). OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc. Natl. Acad. Sci. U.S.A. 86, 3484–3488. doi: 10.1073/pnas.86.10.3484
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D.,et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544. doi: 10.1038/31159
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Conesa, A., Gotz, S., Garcia-Gomez, J. M., Terol, J., Talon, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cornelis, P., Wei, Q., Andrews, S. C., and Vinckx, T. (2011). Iron homeostasis and management of oxidative stress response in bacteria. Metallomics 3, 540–549. doi: 10.1039/c1mt00022e
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cossu, A., Sechi, L. A., Zanetti, S., and Rosu, V. (2012). Gene expression profiling of Mycobacterium avium subsp. paratuberculosis in simulated multi-stress conditions and within THP-1 cells reveals a new kind of interactive intramacrophage behaviour. BMC Microbiol. 12:87. doi: 10.1186/1471-2180-12-87
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deretic, V., Philipp, W., Dhandayuthapani, S., Mudd, M. H., Curcic, R., Garbe, T.,et al. (1995). Mycobacterium tuberculosis is a natural mutant with an inactivated oxidative-stress regulatory gene: implications for sensitivity to isoniazid. Mol. Microbiol. 17, 889–900. doi: 10.1111/j.1365-2958.1995.mmi_17050889.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Devasagayam, T. P., Tilak, J. C., Boloor, K. K., Sane, K. S., Ghaskadbi, S. S., and Lele, R. D. (2004). Free radicals, and antioxidants in human health: current status, and future prospects. J. Assoc. Physicians India 52, 794–804.
Dhandayuthapani, S., Mudd, M., and Deretic, V. (1997). Interactions of OxyR with the promoter region of the oxyR and ahpC genes from Mycobacterium leprae and Mycobacterium tuberculosis J. Bacteriol. 179, 2401–2409.
Dubrac, S., and Touati, D. (2000). Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 182, 3802–3808. doi: 10.1128/JB.182.13.3802-3808.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dussurget, O., Rodriguez, M., and Smith, I. (1996). An ideR mutant of Mycobacterium smegmatis has derepressed siderophore production and an altered oxidative-stress response. Mol. Microbiol. 22, 535–544. doi: 10.1046/j.1365-2958.1996.1461511.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eckelt, E., Jarek, M., Frömke, C., Meens, J., and Goethe, R. (2014). Identification of a lineage specific zinc responsive genomic island in Mycobacterium avium ssp. paratuberculosis. BMC Genomics 15:1076. doi: 10.1186/1471-2164-15-1076
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Escolar, L., Perez-Martin, J., and de Lorenzo, V. (1998). Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 283, 537–547. doi: 10.1006/jmbi.1998.2119
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fillat, M. F. (2014). The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch. Biochem. Biophys. 546C, 41–52. doi: 10.1016/j.abb.2014.01.029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Frederiksen, R. F., Paspaliari, D. K., Larsen, T., Storgaard, B. G., Larsen, M. H., Ingmer, H.,et al. (2013). Bacterial chitinases and chitin-binding proteins as virulence factors. Microbiology 159, 833–847. doi: 10.1099/mic.0.051839-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ghosh, P., Wu, C. W., and Talaat, A. M. (2013). Key role for the alternative sigma factor, sigH, in the intracellular life of Mycobacterium avium subsp. paratuberculosis during macrophage stress Infect. Immun. 81, 2242–2257. doi: 10.1128/IAI.01273-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gioffre, A., Infante, E., Aguilar, D., Santangelo, M. P., Klepp, L., Amadio, A.,et al. (2005). Mutation in mce operons attenuates Mycobacterium tuberculosis virulence. Microbes Infect. 7, 325–334. doi: 10.1016/j.micinf.2004.11.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gold, B., Rodriguez, G. M., Marras, S. A., Pentecost, M., and Smith, I. (2001). The Mycobacterium tuberculosis IdeR is a dual functional regulator that controls transcription of genes involved in iron acquisition, iron storage and survival in macrophages. Mol. Microbiol. 42, 851–865. doi: 10.1046/j.1365-2958.2001.02684.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gough, D. R., and Cotter, T. G. (2011). Hydrogen peroxide: a Jekyll and Hyde signalling molecule. Cell Death Dis. 2:e213. doi: 10.1038/cddis.2011.96
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hahn, J. S., Oh, S. Y., and Roe, J. H. (2002). Role of OxyR as a peroxide-sensing positive regulator in Streptomyces coelicolor A3(2). J. Bacteriol. 184, 5214–5222. doi: 10.1128/JB.184.19.5214-5222.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Halliwell, B., and Gutteridge, J. M. (1984). Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 219, 1–14.
Harris, N. B., and Barletta, R. G. (2001). Mycobacterium avium subsp. paratuberculosis in Veterinary Medicine. Clin. Microbiol. Rev. 14, 489–512. doi: 10.1128/CMR.14.3.489-512.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Helmann, J. D., Wu, M. F., Gaballa, A., Kobel, P. A., Morshedi, M. M., Fawcett, P.,et al. (2003). The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J. Bacteriol. 185, 243–253. doi: 10.1128/JB.185.1.243-253.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Herbig, A. F., and Helmann, J. D. (2001). Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 41, 849–859. doi: 10.1046/j.1365-2958.2001.02543.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hill, P. J., Cockayne, A., Landers, P., Morrissey, J. A., Sims, C. M., and Williams, P. (1998). SirR, a novel iron-dependent repressor in Staphylococcus epidermidis. Infect. Immun. 66, 4123–4129.
Iantomasi, R., Sali, M., Cascioferro, A., Palucci, I., Zumbo, A., Soldini, S.,et al. (2012). PE_PGRS30 is required for the full virulence of Mycobacterium tuberculosis. Cell Microbiol. 14, 356–367. doi: 10.1111/j.1462-5822.2011.01721.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Italiani, V. C., da Silva Neto, J. F., Braz, V. S., and Marques, M. V. (2011). Regulation of catalase-peroxidase KatG is OxyR dependent and Fur independent in Caulobacter crescentus. J. Bacteriol. 193, 1734–1744. doi: 10.1128/JB.01339-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Janagama, H. K., Senthilkumar, T. M., Bannantine, J. P., Rodriguez, G. M., Smith, I., Paustian, M. L.,et al. (2009). Identification and functional characterization of the iron-dependent regulator (IdeR) of Mycobacterium avium subsp. paratuberculosis. Microbiology 155, 3683–3690. doi: 10.1099/mic.0.031948-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jang, H. J., Nde, C., Toghrol, F., and Bentley, W. E. (2009). Microarray analysis of Mycobacterium bovis BCG revealed induction of iron acquisition related genes in response to hydrogen peroxide. Environ. Sci. Technol. 43, 9465–9472. doi: 10.1021/es902255q
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kawaji, S., Zhong, L., and Whittington, R. J. (2010). Partial proteome of Mycobacterium avium subsp. paratuberculosis under oxidative and nitrosative stress. Vet. Microbiol. 145, 252–264. doi: 10.1016/j.vetmic.2010.03.025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kuehnel, M. P., Goethe, R., Habermann, A., Mueller, E., Rohde, M., Griffiths, G.,et al. (2001). Characterization of the intracellular survival of Mycobacterium avium ssp. paratuberculosis: phagosomal pH and fusogenicity in J774 macrophages compared with other mycobacteria. Cell. Microbiol. 3, 551–566. doi: 10.1046/j.1462-5822.2001.00139.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kulasakara, H., Lee, V., Brencic, A., Liberati, N., Urbach, J., Miyata, S.,et al. (2006). Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U.S.A. 103, 2839–2844. doi: 10.1073/pnas.0511090103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kullik, I., Toledano, M. B., Tartaglia, L. A., and Storz, G. (1995). Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J. Bacteriol. 177, 1275–1284.
Lee, H.-I., Yoon, J.-H., Nam, J.-S., Kim, Y.-M., and Ro, Y.-T. (2009). Cloning, expression and characterization of the catalase-peroxidase (KatG) gene from a fast-growing Mycobacterium sp. strain JC1 DSM 3803. J. Biochem. 147, 511–522. doi: 10.1093/jb/mvp197
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, J. W., and Helmann, J. D. (2007). Functional specialization within the Fur family of metalloregulators. Biometals 20, 485–499. doi: 10.1007/s10534-006-9070-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N.,et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lombard, J. E. (2011). Epidemiology and economics of paratuberculosis. Vet. Clin. North Am. Food Anim. Pract. 27, 525–535. doi: 10.1016/j.cvfa.2011.07.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lucarelli, D., Vasil, M. L., Meyer-Klaucke, W., and Pohl, E. (2008). The metal-dependent regulators FurA and FurB from Mycobacterium tuberculosis. Int. J. Mol. Sci. 9, 1548–1560. doi: 10.3390/ijms9081548
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Malorni, W., Iosi, F., Santini, M. T., and Testa, U. (1993). Menadione-induced oxidative stress leads to a rapid down-modulation of transferrin receptor recycling. J. Cell Sci. 106(Pt 1), 309–318.
Master, S. S., Springer, B., Sander, P., Boettger, E. C., Deretic, V., and Timmins, G. S. (2002). Oxidative stress response genes in Mycobacterium tuberculosis: role of ahpC in resistance to peroxynitrite and stage-specific survival in macrophages. Microbiology 148, 3139–3144.
Master, S., Zahrt, T. C., Song, J., and Deretic, V. (2001). Mapping of Mycobacterium tuberculosis katG promoters and their differential expression in infected macrophages. J. Bacteriol. 183, 4033–4039. doi: 10.1128/JB.183.13.4033-4039.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McClure, R., Balasubramanian, D., Sun, Y., Bobrovskyy, M., Sumby, P., Genco, C. A.,et al. (2013). Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 41:e140. doi: 10.1093/nar/gkt444
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McHugh, J. P., Rodriguez-Quinones, F., Abdul-Tehrani, H., Svistunenko, D. A., Poole, R. K., Cooper, C. E.,et al. (2003). Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278, 29478–29486. doi: 10.1074/jbc.M303381200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Meißner, T., Eckelt, E., Basler, T., Meens, J., Heinzmann, J., Suwandi, A.,et al. (2014). The Mycobacterium avium ssp. paratuberculosis specific mptD gene is required for maintaining of the metabolic homeostasis necessary for full virulence in mouse infections. Front. Cell Infect. Microbiol. 4:110. doi: 10.3389/fcimb.2014.00110
Milano, A., Forti, F., Sala, C., Riccardi, G., and Ghisotti, D. (2001). Transcriptional regulation of furA and katG upon oxidative stress in Mycobacterium smegmatis. J. Bacteriol. 183, 6801–6806. doi: 10.1128/JB.183.23.6801-6806.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mongkolsuk, S., and Helmann, J. D. (2002). Regulation of inducible peroxide stress responses. Mol. Microbiol. 45, 9–15. doi: 10.1046/j.1365-2958.2002.03015.x
Neilands, J. B. (1992). Mechanism and regulation of synthesis of aerobactin in Escherichia coli K12 (pColV-K30). Can. J. Microbiol. 38, 728–733. doi: 10.1139/m92-119
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Oh, S. Y., Shin, J. H., and Roe, J. H. (2007). Dual role of OhrR as a repressor and an activator in response to organic hydroperoxides in Streptomyces coelicolor. J. Bacteriol. 189, 6284–6292. doi: 10.1128/JB.00632-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pagan-Ramos, E., Song, J., McFalone, M., Mudd, M. H., and Deretic, V. (1998). Oxidative stress response and characterization of the oxyR-ahpC and furA-katG loci in Mycobacterium marinum. J. Bacteriol. 180, 4856–4864.
Pandey, R., and Rodriguez, G. M. (2014). IdeR is required for iron homeostasis and virulence in Mycobacterium tuberculosis. Mol. Microbiol. 91, 98–109. doi: 10.1111/mmi.12441
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Park, K. T., Dahl, J. L., Bannantine, J. P., Barletta, R. G., Ahn, J., Allen, A. J.,et al. (2008). Demonstration of allelic exchange in the slow-growing bacterium Mycobacterium avium subsp. paratuberculosis, and generation of mutants with deletions at the pknG, relA, and lsr2 loci. Appl. Environ. Microbiol. 74, 1687–1695. doi: 10.1128/AEM.01208-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Parthasarathy, G., Lun, S., Guo, H., Ammerman, N. C., Geiman, D. E., and Bishai, W. R. (2012). Rv2190c, an NlpC/P60 family protein, is required for full virulence of Mycobacterium tuberculosis. PLoS ONE 7:e43429. doi: 10.1371/journal.pone.0043429
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Purkayastha, A., McCue, L. A., and McDonough, K. A. (2002). Identification of a Mycobacterium tuberculosis putative classical nitroreductase gene whose expression is coregulated with that of the acr gene within macrophages, in standing versus shaking cultures, and under low oxygen conditions. Infect. Immun. 70, 1518–1529. doi: 10.1128/IAI.70.3.1518-1529.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pym, A. S., Domenech, P., Honore, N., Song, J., Deretic, V., and Cole, S. T. (2001). Regulation of catalase-peroxidase (KatG) expression, isoniazid sensitivity and virulence by furA of Mycobacterium tuberculosis. Mol. Microbiol. 40, 879–889. doi: 10.1046/j.1365-2958.2001.02427.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ralph, P., Prichard, J., and Cohn, M. (1975). Reticulum cell sarcoma: an effector cell in antibody-dependent cell-mediated immunity. J. Immunol. 114, 898–905.
Rodriguez, G. M., Voskuil, M. I., Gold, B., Schoolnik, G. K., and Smith, I. (2002). ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70, 3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002
Rustad, T. R., Roberts, D. M., Liao, R. P., and Sherman, D. R. (2009). Isolation of mycobacterial RNA. Methods Mol. Biol. 465, 13–21. doi: 10.1007/978-1-59745-207-6_2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T.,et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schmitt, M. P., Predich, M., Doukhan, L., Smith, I., and Holmes, R. K. (1995). Characterization of an iron-dependent regulatory protein (IdeR) of Mycobacterium tuberculosis as a functional homolog of the diphtheria toxin repressor (DtxR) from Corynebacterium diphtheriae. Infect. Immun. 63, 4284–4289.
Senaratne, R. H., De Silva, A. D., Williams, S. J., Mougous, J. D., Reader, J. R., Zhang, T.,et al. (2006). 5′-Adenosinephosphosulphate reductase (CysH) protects Mycobacterium tuberculosis against free radicals during chronic infection phase in mice. Mol. Microbiol. 59, 1744–1753. doi: 10.1111/j.1365-2958.2006.05075.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sherman, D. R., Mdluli, K., Hickey, M. J., Barry, C. E. III, and Stover, C. K. (1999). AhpC, oxidative stress and drug resistance in Mycobacterium tuberculosis. Biofactors 10, 211–217. doi: 10.1002/biof.5520100219
Sherman, D. R., Sabo, P. J., Hickey, M. J., Arain, T. M., Mahairas, G. G., Yuan, Y.,et al. (1995). Disparate responses to oxidative stress in saprophytic and pathogenic mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 92, 6625–6629. doi: 10.1073/pnas.92.14.6625
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Storz, G., and Altuvia, S. (1994). OxyR regulon. Methods Enzymol. 234, 217–223. doi: 10.1016/0076-6879(94)34088-9
Stover, C. K., de la Cruz, V. F., Fuerst, T. R., Burlein, J. E., Benson, L. A., Bennett, L. T.,et al. (1991). New use of BCG for recombinant vaccines. Nature 351, 456–460. doi: 10.1038/351456a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stratmann, J., Strommenger, B., Goethe, R., Dohmann, K., Gerlach, G. F., Stevenson, K.,et al. (2004). A 38-kilobase pathogenicity island specific for Mycobacterium avium subsp. paratuberculosis encodes cell surface proteins expressed in the host. Infect. Immun. 72, 1265–1274. doi: 10.1128/IAI.72.3.1265-1274.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suwandi, A., Bargen, I., Roy, B., Pils, M. C., Krey, M., Zur Lage, S.,et al. (2014). Experimental colitis is exacerbated by concomitant infection with Mycobacterium avium ssp. paratuberculosis. Inflamm. Bowel Dis. 20, 1962–1971. doi: 10.1097/MIB.0000000000000157
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tao, K., Zou, C., Fujita, N., and Ishihama, A. (1995). Mapping of the OxyR protein contact site in the C-terminal region of RNA polymerase alpha subunit. J. Bacteriol. 177, 6740–6744.
Tseng, H. J., McEwan, A. G., Apicella, M. A., and Jennings, M. P. (2003). OxyR acts as a repressor of catalase expression in Neisseria gonorrhoeae. Infect. Immun. 71, 550–556. doi: 10.1128/IAI.71.1.550-556.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
van Vliet, A. H., Baillon, M. L., Penn, C. W., and Ketley, J. M. (1999). Campylobacter jejuni contains two fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181, 6371–6376.
Webb, J. R., Vedvick, T. S., Alderson, M. R., Guderian, J. A., Jen, S. S., Ovendale, P. J.,et al. (1998). Molecular cloning, expression, and immunogenicity of MTB12, a novel low-molecular-weight antigen secreted by Mycobacterium tuberculosis. Infect. Immun. 66, 4208–4214.
Weigoldt, M., Meens, J., Bange, F. C., Pich, A., Gerlach, G. F., and Goethe, R. (2013). Metabolic adaptation of Mycobacterium avium subsp. paratuberculosis to the gut environment. Microbiology 159, 380–391. doi: 10.1099/mic.0.062737-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Weigoldt, M., Meens, J., Doll, K., Fritsch, I., Mobius, P., Goethe, R.,et al. (2011). Differential proteome analysis of Mycobacterium avium subsp. paratuberculosis grown in vitro and isolated from cases of clinical Johne’s disease. Microbiology 157, 557–565. doi: 10.1099/mic.0.044859-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wildermann, P. J., Sowa, N. A., FitzGerald, D. J., FitzGerald, P. C., Gottesman, S., Ochnser, U. A.,et al. (2004). Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. U.S.A. 101, 9792–9797. doi: 10.1073/pnas.0403423101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wilson, T., deLisle, G. W., Marcinkeviciene, J. A., Blanchard, J. S., and Collins, D. M. (1998). Antisense RNA to ahpC, an oxidative stress defence gene involved in isoniazid resistance, indicates that AhpC of Mycobacterium bovis has virulence properties. Microbiology 144, 2687–2695. doi: 10.1099/00221287-144-10-2687
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wood, Z. A., Schröder, E., Harris, J. R., and Poole, L. B. (2003). Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 28, 32–40. doi: 10.1016/S0968-0004(02)00003-8
Wu, C. W., Schmoller, S. K., Shin, S. J., and Talaat, A. M. (2007). Defining the stressome of Mycobacterium avium subsp. paratuberculosis in vitro and in naturally infected cows. J. Bacteriol. 189, 7877–7886. doi: 10.1128/JB.00780-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yoshikawa, T., and Naito, Y. (2000). The role of neutrophils and inflammation in gastric mucosal injury. Free Radic. Res. 33, 785–794. doi: 10.1080/10715760000301301
Zahrt, T. C., Song, J., Siple, J., and Deretic, V. (2001). Mycobacterial FurA is a negative regulator of catalase-peroxidase gene katG. Mol. Microbiol. 39, 1174–1185. doi: 10.1111/j.1365-2958.2001.02321.x
Keywords: oxidative stress response regulation, furA, mycobacteria, iron, metallo-regulator
Citation: Eckelt E, Meißner T, Meens J, Laarmann K, Nerlich A, Jarek M, Weiss S, Gerlach G-F and Goethe R (2015) FurA contributes to the oxidative stress response regulation of Mycobacterium avium ssp. paratuberculosis. Front. Microbiol. 6:16. doi: 10.3389/fmicb.2015.00016
Received: 12 August 2014; Accepted: 07 January 2015;
Published online: 06 February 2015.
Edited by:
Biswarup Mukhopadhyay, Virginia Polytechnic Institute and State University, USAReviewed by:
Martin I. Voskuil, University of Colorado Denver, USAPatricia Coutinho Dos Santos, Wake Forest University, USA
Adel M. Talaat, University of Wisconsin–Madison, USA
Endang Purwantini, Virginia Polytechnic Institute and State University, USA
Copyright © 2015 Eckelt, Meißner, Meens, Laarmann, Nerlich, Jarek, Weiss, Gerlach and Goethe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ralph Goethe, Institute for Microbiology, Department of Infectious Diseases, University of Veterinary Medicine Hannover, Bischofsholer Damm 15, 30173 Hannover, Germany e-mail:cmFscGguZ29ldGhlQHRpaG8taGFubm92ZXIuZGU=
†Present address: Gerald-F. Gerlach, IVD GmbH, Heisterbergallee 12, 30453 Hannover, Germany
‡These Authors Have Contributed Equally to This Work.
 Elke Eckelt
Elke Eckelt Thorsten Meißner
Thorsten Meißner Jochen Meens
Jochen Meens Kristin Laarmann
Kristin Laarmann Andreas Nerlich1
Andreas Nerlich1 Michael Jarek
Michael Jarek Siegfried Weiss
Siegfried Weiss Gerald-F. Gerlach
Gerald-F. Gerlach Ralph Goethe
Ralph Goethe