- Section for Eukaryotic Biotechnology, Department of Systems Biology, Technical University of Denmark, Kongens Lyngby, Denmark
Fungal natural products comprise a wide range of compounds. Some are medically attractive as drugs and drug leads, some are used as food additives, while others are harmful mycotoxins. In recent years the genome sequence of several fungi has become available providing genetic information of a large number of putative biosynthetic pathways. However, compound discovery is difficult as the genes required for the production of the compounds often are silent or barely expressed under laboratory conditions. Furthermore, the lack of available tools for genetic manipulation of most fungal species hinders pathway discovery. Heterologous expression of the biosynthetic pathway in model systems or cell factories facilitates product discovery, elucidation, and production. This review summarizes the recent strategies for heterologous expression of fungal biosynthetic pathways in Aspergilli.
Introduction
Filamentous fungi produce a plethora of secondary metabolites, SMs, like polyketides (PK), terpenes, and non-ribosomal peptides (NRP). Several fungal SMs dramatically impact human life either because they are harmful mycotoxins, like carcinogenic aflatoxin (Eaton and Gallagher, 1994) and fumonisin (Voss and Riley, 2013), or because they are used to efficiently combat human disease, e.g., penicillin and lovastatin (Campbell and Vederas, 2010). Importantly, analyses of fully sequenced fungi show that the number of SMs known to be produced by these fungi is too low to account for the number of genes and gene clusters that potentially may lead to production of SMs (Szewczyk et al., 2008). This strongly suggests that the chemical diversity of the metabolomes produced by filamentous fungi is much larger than what is currently known, and it is therefore very likely that new harmful mycotoxins and new blockbuster drugs await discovery.
The rapid accumulation of fully sequenced genomes has accelerated the discovery of novel SMs dramatically. However, this sequence resource cannot be directly translated into chemical structures of new compounds despite that genes and gene clusters are often readily identified by bioinformatics tools (Khaldi et al., 2010; Andersen et al., 2013; Blin et al., 2013). For example, the exact structures of products released by fungal type I polyketide synthases are difficult to predict due to the iterative use of the different catalytic domains in these enzymes. Similarly, subsequent decorations performed by tailoring enzymes encoded by other genes in the cluster toward formation of the mature end product(s) are complex and not easy to predict. Another challenge is that many SMs are not readily produced under laboratory conditions although several approaches have been successfully employed to activate silent clusters (for reviews, see Brakhage and Schroeckh, 2011; Chiang et al., 2011; Klejnstrup et al., 2012; Wiemann and Keller, 2014; Yaegashi et al., 2014). To link novel SMs to genes, and to map novel biosynthetic pathways, extensive genetic manipulations of the strains are typically required. Since, most new gene clusters uncovered by sequencing projects will be situated in fungi with no available genetic tools, this type of analysis may not be straight forward. Moreover, it may be difficult to purify sufficient amounts of a desired compound from these fungi to allow for thorough characterization of its bioactivity. An alternative approach is to transfer genes and gene clusters to hosts with strong genetic toolboxes thereby facilitating product discovery, production, and characterization. Here we review SM production in Aspergilli based cell factories. Considerations and strategies concerning central steps toward fungal SM production are presented for inspiration: choice of host, how to produce the first intermediate in the pathway, and how to establish the remaining part of the pathway.
Host Choice for Heterologous Expression of Fungal Secondary Metabolites
Heterologous expression of SM genes has mainly been performed in baker’s yeast Saccharomyces cerevisiae (Tsunematsu et al., 2013) and in the filamentous fungi Aspergillus oryzae and A. nidulans. Each of these model organisms offers specific advantages. For S. cerevisiae a superior genetic toolbox for strain construction has been developed and novel genes can easily be engineered into a wealth of single- and multi-copy expression plasmids or into chromosomes. For example, gene targeting and fusion of DNA fragments by homologous recombination (HR) is highly efficient in S. cerevisiae. Moreover, S. cerevisiae contains an insignificant endogenous secondary metabolism (Siddiqui et al., 2012). This fact simplifies the analysis of strains equipped with new pathways as they are not complicated by the presence of a multitude of other SMs; and the risk of undesirable side reactions due to cross chemistry between the novel and endogenous pathways is minimized. However, lack of secondary metabolism also means that yeast is not naturally geared for SM production and may contain limiting amounts of, or even lack, relevant building blocks (Kealey et al., 1998; Mutka et al., 2006). Moreover, localization of relevant enzymes for aflatoxin production into specialized vesicles in A. parasiticus indicate that fungi may possess specialized compartments for SM production, which yeast may not contain (Roze et al., 2011); and as introns are few in S. cerevisiae (Spingola et al., 1999) and differ from those in filamentous fungi (Kupfer et al., 2004), mRNA splicing could be problematic. For these reasons filamentous fungi may often be more appropriate for heterologous SM production. A. oryzae is often used for this purpose because it possesses a limited endogenous secondary metabolism and A. nidulans because a strong genetic toolbox has been developed for this fungus (for review see, Meyer, 2008; Meyer et al., 2011). Importantly, the recent development of efficient tools for gene targeting in filamentous fungi, including strains where random integration is minimized due to mutation of genes required for non-homologous end-joining (Ninomiya et al., 2004; Nayak et al., 2006; Takahashi et al., 2006), has further stimulated the use of these organisms as hosts for SM pathway reconstitution experiments.
Heterologous Expression of Polyketide Synthases
The fact that the product(s) released by fungal type I PK synthases (PKSs) cannot easily be predicted from their primary sequence has sparked a major interest in expressing PKS genes in model fungi with the aim of identifying these products. In yeast, two 2 μ based multi-copy plasmids harboring the 6-methylsalicylic acid (6-MSA) synthase gene from Penicillium patulum and the PKS activating PPTase gene from Bacillus subtilis were successfully used to produce 6-MSA (Kealey et al., 1998). Similarly production of green pigment has been achieved in a wAΔ yAΔ (white) A. nidulans (Holm, 2013) via co-expression of the PKS gene wA and laccase gene yA harbored on two AMA1 (Aleksenko and Clutterbuck, 1997) based plasmid. However, if multiple plasmids are needed to form a complex end-product, these vectors may have limited value since sufficient markers may not be available, and since 2 μ and AMA1 plasmids segregate unevenly during mitosis (Albertsen et al., 2011; Holm, 2013; Jensen et al., 2014).
More stable expression has been achieved by integrating PKS genes randomly into the genome of a model filamentous fungus via the non-homologous end-joining pathway. Using this concept, Fujii et al. (1996) successfully linked 6-MSA production to the PKS gene atX from A. terreus by expressing atX host A. nidulans. Considering that foreign SMs may be toxic in the new host, it is advisable to employ an expression strategy that minimizes this risk. For production of 6-MSA and enniatins in A. nidulans and A. niger, this was achieved by fusing the PKS and NRPS genes to the inducible promoters, amyB (Fujii et al., 1996) and Tet-on (Richter et al., 2014), respectively. Over the years, a number of other PKS genes have been linked to products using this strategy in A. nidulans and A. oryzae including the PKS genes for production of 1,3,6,8-tetrahydroxynaphthalene, alternapyrone, and 3-methylorcinaldehyde by (Fujii et al., 1999, 2005; Bailey et al., 2007).
Random integration may trigger unpredictable pleiotropic effects that alter the expression of neighboring genes, hence, complicating subsequent analyses (Verdoes et al., 1995; Palmer and Keller, 2010). Moreover, since multiple copies of the gene often integrate simultaneously into the same site, strains may suffer genetic instability and lose expression over time. Taking advantage of the development of strains and techniques for efficient gene targeting, these problems can be eliminated by inserting genes into a defined locus. This facilitates not only subsequent strain characterization, but also sets the stage for experiments analyzing mutant varieties of the gene where equal expression levels of the alleles are important to fairly judge the impact of individual mutations. Using this approach, Hansen et al. (2011) demonstrated that mpaC from P. brevicompactum encodes a PKS producing 5-methylorsellinic acid. In this case, mpaC was introduced into a defined site, IS1, on chromosome I of A. nidulans, which supports expression of non-toxic genes in a variety of tissues without affecting fitness. Moreover, to simplify the integration of genes into IS1, a set of vectors pre-equipped with targeting sequences, genetic markers, promoters and terminators and a USER-cloning cassette (Nour-Eldin et al., 2006) allowing for seamless ligation free insertion of relevant genes into the vector was developed. Using this technology, ausA, from A. nidulans, and yanA, from A. niger, have been shown to encode PKSs producing 3-,5-dimethyl orsellinic acid and 6-MSA, respectively (Nielsen et al., 2011; Holm et al., 2014). In a variation of this approach, Chiang et al. (2013) used fusion Polymerase chain reaction (PCR) to merge an alcA promoter and PKS genes followed by integration into the wA locus of A. nidulans. Correctly targeted transformants could therefore easily be identified as white colonies. The authors expressed nine non-reducing (NR) PKS genes from A. terrreus in this manner and identified six products. Heterologous production of PKs is complicated by the fact that not all synthases possess a domain providing a product release mechanism (Awakawa et al., 2009; Du and Lou, 2010) and by the fact that some PKSs require a starter unit different from Ac-CoA (Hoffmeister and Keller, 2007). In the study by Chiang et al. (2013), two of the nine NR-PKSs analyzed did not contain such a domain and for one, a product was achieved by co-expressing a gene encoding a thioesterase activity. In addition, two NR-PKS were predicted to employ unusual starter units. For one NR-PKS, production of this starter unit was successfully delivered by co-expressing a gene encoding a highly reducing PKS and the collaborative effort of the two enzymes resulted in production of an intermediate for production of asperfuranone (Chiang et al., 2013).
Transfer of Gene Clusters to Heterologous Hosts
Reconstitution of most SM pathways depends on the expression of multiple genes since the SM scaffold delivered by the synthase is further decorated by tailoring enzymes. Moreover, genes providing transcriptions factors, transporters and/or a resistance mechanism may also be required. Construction of strains for heterologous end-product production is therefore a major challenge as it requires not only transfer, but also activation, of large gene clusters. Two principles are generally employed for constructing DNA fragments that allow transfer of gene clusters into another fungal host. Firstly, DNA fragments harboring entire, or a large part of, gene clusters have been identified in cosmid/fosmid libraries and transferred into vectors with a selectable fungal marker (Figure 1A). Secondly, PCR fragments covering the gene cluster have been stitched together using a variety of methods including USER Fusion, Gateway cloning and yeast recombination to create suitable transformation vectors (Figure 1B). When gene clusters have been transformed into the host, activation has been achieved by three different methods. Firstly, in cases where the native gene cluster harbors a TF gene, it has been possible to activate the genes in the cluster by equipping the TF gene with a constitutive or inducible promoter known to work in the host. Secondly, in gene clusters without a TF gene, activation has been achieved either by overexpressing the global regulator LaeA or by individually swapping cluster gene promoters for constitutive or inducible promoters. Like for integration of PKS genes, and for the same reasons, integration strategies based on random or directed integration have been used (Figure 1C). In many cases these strategies have been combined and successful examples are provided below.
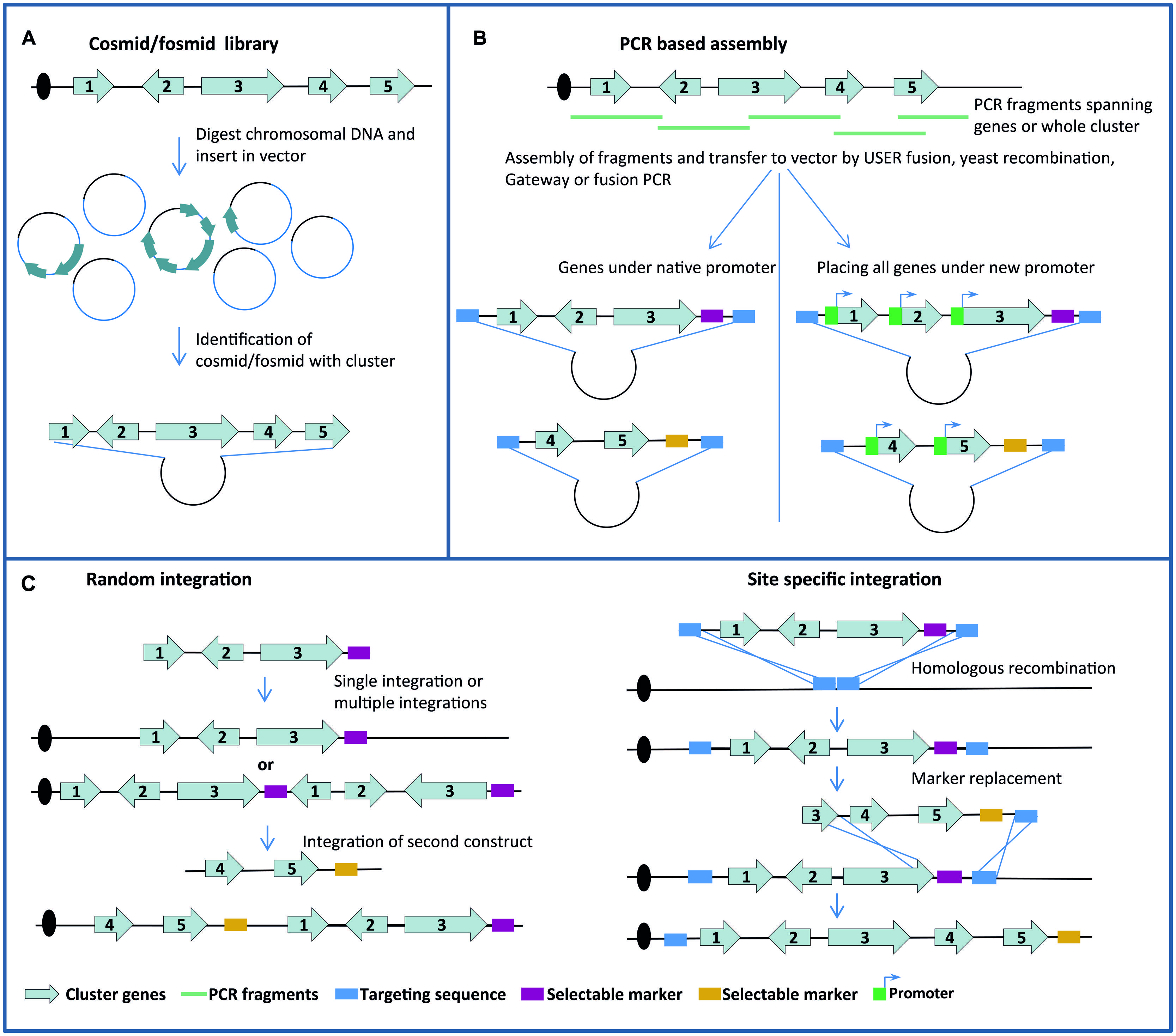
FIGURE 1. Overview of principles employed for constructing DNA fragments (A,B) and for integration in host genomes (C).
Cosmids harboring the entire penicillin biosynthetic pathway from P. chrysogenum were introduced to Neurospora crassa and A. niger, resulting in the production of penicillin (Smith et al., 1990). Similary, cosmids harboring the citrinin biosynthetic pathway from Monascus purpureus and the monacolin K gene cluster from Monascus pilosus were individually integrated into random positions in the genome of A. oryzae. In the case of citrinin, the transformant directly produced citrinin, but in small amounts. However, as the cluster contains a TF gene, additional copies of the activator gene (ctnA) controlled by the A. nidulans trpC promoter were subsequently introduced in the strain to boost production. Impressively, this resulted in a 400 fold increase of citrinin production (Sakai et al., 2008). The monacolin K gene cluster does not contain a TF gene. In this case, the cluster was activated by overexpressing the gene encoding the global activator LaeA (Sakai et al., 2012). A limitation of this strategy may be difficulties in isolating cosmids containing a fragment that harbors the entire gene cluster, especially if clusters are large. For example, the reconstruction of the terrequinone A gene cluster in A. oryzae was based on a fosmid containing an incomplete gene cluster. The remaining part of the cluster was subsequently obtained by PCR, cloned into a vector and transformed into the A. oryzae strain harboring the partial terrequinone A gene cluster (Sakai et al., 2012).
Several PCR based strategies have been used for transferring gene clusters from the natural producer to a model fungus. For clusters harboring a TF gene, PCR fragments covering the entire gene cluster have been amplified, fused, and inserted via a single cloning step into vectors predestined for site specific integration in the genome of the host by HR. Multiple PCR fragments can be orderly assembled by different strategies. For example, PCR fragments of the geodin and neosartoricin B clusters were physically linked by E. coli based USER fusion and by yeast based HR, respectively (Nielsen et al., 2013; Yin et al., 2013). Importantly, in both cases the promoter controlling expression of the TF gene was swapped for a strong constitutive promoter during the cluster re-assembly process. Large inserts (>15 kb) may not be propagated stably in a cloning vector and large clusters need to be subdivided into smaller fragment cassettes, which together represent the entire cluster. Multiple subsequent integrations depend on marker recycling, which can be achieved by using pyrG as a selectable/counterselectable marker. A faster method employs a two marker system for cluster transfer (Nielsen et al., 2013). During one transformation cycle, one of the markers is used to select for integration of the first cluster cassette and the other marker for the next cassette. By ensuring that integration of one cassette eliminates the marker contained by the preceding cassette, numerous cluster cassettes can be integrated sequentially by alternating the use of the two markers. Advantageously, when the gene clusters is inserted in a controlled manner it can be subjected to further genetic dissection to clarify the biochemical pathway toward end product. With the geodin cluster this was exploited to demonstrate that gedL encodes a halogenase using sulochrin as substrate (Nielsen et al., 2013).
Polymerase chain reaction based reconstruction of clusters that do not contain an activating TF gene requires more elaborate genetic engineering as all cluster genes need to be equipped with new promoters and terminators. In one strategy, cluster ORFs were inserted either individually or in pairs into expression cassettes in plasmids carrying different selection markers. Using this approach several small gene clusters containing four to five genes have been, fully or partially, reconstituted by randomly introducing the genes into the genome of A. oryzae. Several SMs have been achieved by this method including tennelin, pyripyropene, aphidicolin, terretonin, and andrastin A (Heneghan et al., 2010; Itoh et al., 2010; Fujii et al., 2011; Matsuda et al., 2012,2013). Construction of larger clusters in A. oryzae has been limited by the number of available markers. To bypass this problem, Tagami et al. (2013, 2014) used the high co-transformation frequency with A. oryzae to integrate two vectors in one round of transformation using selection for only one marker. This allowed for reconstituting clusters with six and seven genes for production of paxilline and aflatrem, respectively (Tagami et al., 2013, 2014). Addressing the same problem, Gateway cloning was used to construct expression vectors containing up to four genes (Pahirulzaman et al., 2012; Lazarus et al., 2014). Utilizing this approach Wasil et al. (2013) expressed different combinations of the synthase and tailoring genes from the aspyridone pathway from A. nidulans in A. oryzae. An alternative approach to save markers is to generate synthetic polycistronic genes where all genes in the construct are under the control of a single promoter and where all ORFs are separated by a sequence encoding the viral 2A peptide that results in co-translational cleavage, hence, resulting in the formation of independent enzymes (Kim et al., 2011). Using this concept Unkles et al. (2014) reconstituted the penicillin gene cluster from P. chrysogenum as a single three ORF polystronic gene by yeast mediated HR. Random genomic integration of this construct resulted in penicillin production in A. nidulans.
A strategy for gene cluster activation based on promoter/terminator swapping has also been implemented in gene cluster transfer methods where genes are inserted into defined integration sites (Hansen et al., 2012; Mikkelsen et al., 2012; Chiang et al., 2013). Specifically, expression plasmids containing one to two cluster genes were constructed by USER cloning or by fusion PCR and integrated into the expression sites in S. cerevisiae and A. nidulans to allow for production of the pigment precursor rubrofusarin in yeast (Rugbjerg et al., 2013) and for partial and fully reconstitution of the pathways for mycophenolic acid and asperfuranone production, respectively, in A. nidulans (Hansen et al., 2012; Chiang et al., 2013).
Perspectives
The rapid development of molecular tools for cluster transfer and re-engineering in heterologous hosts is now at a stage where high-throughput experiments can be performed, and we therefore predict that novel SMs, genes, pathways and enzymes routinely will be discovered using this approach. For now most efforts have been proof of principle cases analyzing genes and gene clusters from genetically well-characterized organisms, but the next wave of breakthroughs will likely concern SMs originating from genetically exotic fungi. In addition, the natural reservoir of SMs will likely expand dramatically as synthetic biology based approaches using bio-bricks of promoters, terminators and SM genes are combined in intelligent or in random ways in model fungi to deliver compounds that nature never invented. Together, we envision that heterologous production will serve as a major driver for SM discovery and development delivering compounds that can be used in the food and pharma industries. Accordingly, physiologically well-characterized fungal cell factories should preferentially be employed as platforms for novel SMs discovery and development. These fungi display superior fermentation properties and extensive metabolic engineering toolboxes, hence, shortening the way toward large scale production.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Rasmus Frandsen and Jakob Nielsen for helpful discussions; and the Danish Council for Technology and Production Sciences for financial support via grant 09-064967.
References
Albertsen, L., Chen, Y., Bach, L. S., Rattleff, S., Maury, J., Brix, S.,et al. (2011). Diversion of flux toward sesquiterpene production in Saccharomyces cerevisiae by fusion of host and heterologous enzymes. Appl. Environ. Microbiol. 77, 1033–1040. doi: 10.1128/AEM.01361-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Aleksenko, A., and Clutterbuck, A. J. (1997). Autonomous plasmid replication in Aspergillus nidulans: AMA1 and MATE elements. Fungal Genet. Biol. 21, 373–387. doi: 10.1006/fgbi.1997.0980
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Andersen, M. R., Nielsen, J. B., Klitgaard, A., Petersen, L. M., Zachariasen, M., Hansen, T. J.,et al. (2013). Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc. Natl. Acad. Sci. U.S.A. 110, E99–E107. doi: 10.1073/pnas.1205532110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Awakawa, T., Yokota, K., Funa, N., Doi, F., Mori, N., Watanabe, H.,et al. (2009). Physically discrete beta-lactamase-type thioesterase catalyzes product release in atrochrysone synthesis by iterative type I polyketide synthase. Chem. Biol. 16, 613–623. doi: 10.1016/j.chembiol.2009.04.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bailey, A. M., Cox, R. J., Harley, K., Lazarus, C. M., Simpson, T. J., and Skellam, E. (2007). Characterisation of 3-methylorcinaldehyde synthase (MOS) in Acremonium strictum: first observation of a reductive release mechanism during polyketide biosynthesis. Chem. Commun. (Camb.) 39, 4053–4055. doi: 10.1039/b708614h
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Blin, K., Medema, M. H., Kazempour, D., Fischbach, M. A., Breitling, R., Takano, E.,et al. (2013). antiSMASH 2.0–a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 41, W204–W212. doi: 10.1093/nar/gkt449
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brakhage, A. A., and Schroeckh, V. (2011). Fungal secondary metabolites - strategies to activate silent gene clusters. Fungal Genet. Biol. 48, 15–22. doi: 10.1016/j.fgb.2010.04.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Campbell, C. D., and Vederas, J. C. (2010). Biosynthesis of lovastatin and related metabolites formed by fungal iterative PKS enzymes. Biopolymers 93, 755–763. doi: 10.1002/bip.21428
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chiang, Y.-M., Chang, S.-L., Oakley, B. R., and Wang, C. C. C. (2011). Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr. Opin. Chem. Biol. 15, 137–143. doi: 10.1016/j.cbpa.2010.10.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chiang, Y.-M., Oakley, C. E., Ahuja, M., Entwistle, R., Schultz, A., Chang, S.-L.,et al. (2013). An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans. J. Am. Chem. Soc. 135, 7720–7731. doi: 10.1021/ja401945a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Du, L., and Lou, L. (2010). PKS and NRPS release mechanisms. Nat. Prod. Rep. 27, 255–278. doi: 10.1039/b912037h
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eaton, D. L., and Gallagher, E. P. (1994). Mechanisms of aflatoxin carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 34, 135–172. doi: 10.1146/annurev.pa.34.040194.001031
Fujii, I., Mori, Y., Watanabe, A., Kubo, Y., Tsuji, G., and Ebizuka, Y. (1999). Heterologous expression and product identification of Colletotrichum lagenarium polyketide synthase encoded by the PKS1 gene involved in melanin biosynthesis. Biosci. Biotechnol. Biochem. 63, 1445–1452. doi: 10.1271/bbb.63.1445
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fujii, I., Ono, Y., Tada, H., Gomi, K., Ebizuka, Y., and Sankawa, U. (1996). Cloning of the polyketide synthase gene atX from Aspergillus terreus and its identification as the 6-methylsalicylic acid synthase gene by heterologous expression. Mol. Gen. Genet. 253, 1–10. doi: 10.1007/s004380050289
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fujii, I., Yoshida, N., Shimomaki, S., Oikawa, H., and Ebizuka, Y. (2005). An iterative type I polyketide synthase PKSN catalyzes synthesis of the decaketide alternapyrone with regio-specific octa-methylation. Chem. Biol. 12, 1301–1309. doi: 10.1016/j.chembiol.2005.09.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fujii, R., Minami, A., Tsukagoshi, T., Sato, N., Sahara, T., Ohgiya, S.,et al. (2011). Total biosynthesis of diterpene aphidicolin, a specific inhibitor of DNA polymerase α: heterologous expression of four biosynthetic genes in Aspergillus oryzae. Biosci. Biotechnol. Biochem. 75, 1813–1817. doi: 10.1271/bbb.110366
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hansen, B. G., Mnich, E., Nielsen, K. F., Nielsen, J. B., Nielsen, M. T., Mortensen, U. H.,et al. (2012). Involvement of a natural fusion of a cytochrome P450 and a hydrolase in mycophenolic acid biosynthesis. Appl. Environ. Microbiol. 78, 4908–4913. doi: 10.1128/AEM.07955-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hansen, B. G., Salomonsen, B., Nielsen, M. T., Nielsen, J. B., Hansen, N. B., Nielsen, K. F.,et al. (2011). Versatile enzyme expression and characterization system for Aspergillus nidulans, with the Penicillium brevicompactum polyketide synthase gene from the mycophenolic acid gene cluster as a test case. Appl. Environ. Microbiol. 77, 3044–3051. doi: 10.1128/AEM.01768-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heneghan, M. N., Yakasai, A. A., Halo, L. M., Song, Z., Bailey, A. M., Simpson, T. J.,et al. (2010). First heterologous reconstruction of a complete functional fungal biosynthetic multigene cluster. Chembiochem 11, 1508–1512. doi: 10.1002/cbic.201000259
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hoffmeister, D., and Keller, N. P. (2007). Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat. Prod. Rep. 24, 393–416. doi: 10.1039/b603084
Holm, D. K. (2013). Development and Implementation of Novel Genetic Tools for Investigation of Fungal Secondary Metabolism. Ph.D. thesis, Technical University of Denmark, Kongens Lyngby.
Holm, D. K., Petersen, L. M., Klitgaard, A., Knudsen, P. B., Jarczynska, Z. D., Nielsen, K. F.,et al. (2014). Molecular and chemical characterization of the biosynthesis of the 6-MSA-derived meroterpenoid yanuthone D in Aspergillus niger. Chem. Biol. 21, 519–529. doi: 10.1016/j.chembiol.2014.01.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Itoh, T., Tokunaga, K., Matsuda, Y., Fujii, I., Abe, I., Ebizuka, Y.,et al. (2010). Reconstitution of a fungal meroterpenoid biosynthesis reveals the involvement of a novel family of terpene cyclases. Nat. Chem. 2, 858–864. doi: 10.1038/nchem.764
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jensen, N. B., Strucko, T., Kildegaard, K. R., David, F., Maury, J., Mortensen, U. H.,et al. (2014). EasyClone: method for iterative chromosomal integration of multiple genes in Saccharomyces cerevisiae. FEMS Yeast Res. 14, 238–248. doi: 10.1111/1567-1364.12118
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kealey, J. T., Liu, L., Santi, D. V., Betlach, M. C., and Barr, P. J. (1998). Production of a polyketide natural product in nonpolyketide-producing prokaryotic and eukaryotic hosts. Proc. Natl. Acad. Sci. U.S.A. 95, 505–509. doi: 10.1073/pnas.95.2.505
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Khaldi, N., Seifuddin, F. T., Turner, G., Haft, D., Nierman, W. C., Wolfe, K. H.,et al. (2010). SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 47, 736–741. doi: 10.1016/j.fgb.2010.06.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, J. H., Lee, S.-R., Li, L.-H., Park, H.-J., Park, J.-H., Lee, K. Y.,et al. (2011). High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE 6:e18556. doi: 10.1371/journal.pone.0018556
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Klejnstrup, M. L., Frandsen, R. J. N., Holm, D. K., Nielsen, M. T., Mortensen, U. H., Larsen, T. O.,et al. (2012). Genetics of polyketide metabolism in Aspergillus nidulans. Metabolites 30, 100–133. doi: 10.3390/metabo2010100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kupfer, D. M., Drabenstot, S. D., Buchanan, K. L., Lai, H., Zhu, H., Dyer, D. W.,et al. (2004). Introns and splicing elements of five diverse fungi. Eukaryot. Cell 3, 1088–1100. doi: 10.1128/EC.3.5.1088-1100.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lazarus, C. M., Williams, K., and Bailey, A. M. (2014). Reconstructing fungal natural product biosynthetic pathways. Nat. Prod. Rep. 31, 1339–1347. doi: 10.1039/C4NP00084F
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Matsuda, Y., Awakawa, T., and Abe, I. (2013). Reconstituted biosynthesis of fungal meroterpenoid andrastin A. Tetrahedron 69, 8199–8204. doi: 10.1016/j.tet.2013.07.029
Matsuda, Y., Awakawa, T., Itoh, T., Wakimoto, T., Kushiro, T., Fujii, I.,et al. (2012). Terretonin biosynthesis requires methylation as essential step for cyclization. Chembiochem 13, 1738–1741. doi: 10.1002/cbic.201200369
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Meyer, V. (2008). Genetic engineering of filamentous fungi–progress, obstacles and future trends. Biotechnol. Adv. 26, 177–185. doi: 10.1016/j.biotechadv.2007.12.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Meyer, V., Wu, B., and Ram, A. F. J. (2011). Aspergillus as a multi-purpose cell factory: current status and perspectives. Biotechnol. Lett. 33, 469–476. doi: 10.1007/s10529-010-0473-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mikkelsen, M. D., Buron, L. D., Salomonsen, B., Olsen, C. E., Hansen, B. G., Mortensen, U. H.,et al. (2012). Microbial production of indolylglucosinolate through engineering of a multi-gene pathway in a versatile yeast expression platform. Metab. Eng. 14, 104–111. doi: 10.1016/j.ymben.2012.01.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mutka, S. C., Bondi, S. M., Carney, J. R., Da Silva, N. A., and Kealey, J. T. (2006). Metabolic pathway engineering for complex polyketide biosynthesis in Saccharomyces cerevisiae. FEMS Yeast Res. 6, 40–47. doi: 10.1111/j.1567-1356.2005.00001.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nayak, T., Szewczyk, E., Oakley, C. E., Osmani, A., Ukil, L., Murray, S. L.,et al. (2006). A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172, 1557–1566. doi: 10.1534/genetics.105.052563
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nielsen, M. L., Nielsen, J. B., Rank, C., Klejnstrup, M. L., Holm, D. K., Brogaard, K. H.,et al. (2011). A genome-wide polyketide synthase deletion library uncovers novel genetic links to polyketides and meroterpenoids in Aspergillus nidulans. FEMS Microbiol. Lett. 321, 157–166. doi: 10.1111/j.1574-6968.2011.02327.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nielsen, M. T., Nielsen, J. B., Anyaogu, D. C., Anyaogu, D. C., Holm, D. K., Nielsen, K. F.,et al. (2013). Heterologous reconstitution of the intact geodin gene cluster in Aspergillus nidulans through a simple and versatile PCR based approach. PLoS ONE 8:e72871. doi: 10.1371/journal.pone.0072871
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ninomiya, Y., Suzuki, K., Ishii, C., and Inoue, H. (2004). Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. U.S.A. 101, 12248–12253. doi: 10.1073/pnas.0402780101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nour-Eldin, H. H., Hansen, B. G., Nørholm, M. H. H., Jensen, J. K., and Halkier, B. A. (2006). Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 34, e122. doi: 10.1093/nar/gkl635
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pahirulzaman, K. A., Williams, K., and Lazarus, C. M. (2012). A toolkit for heterologous expression of metabolic pathways in Aspergillus oryzae. Methods Enzymol. 517, 241–260. doi: 10.1016/B978-0-12-404634-4.00012-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Palmer, J. M., and Keller, N. P. (2010). Secondary metabolism in fungi: does chromosomal location matter? Curr. Opin. Microbiol. 13, 431–436. doi: 10.1016/j.mib.2010.04.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Richter, L., Wanka, F., Boecker, S., Storm, D., Kurt, T., Vural, Ö.,et al. (2014). Engineering of Aspergillus niger for the production of secondary metabolites. Fungal Biol. Biotechnol. 1, 4. doi: 10.1186/s40694-014-0004-9
Roze, L. V., Chanda, A., and Linz, J. E. (2011). Compartmentalization and molecular traffic in secondary metabolism: a new understanding of established cellular processes. Fungal Genet. Biol. 48, 35–48. doi: 10.1016/j.fgb.2010.05.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rugbjerg, P., Naesby, M., Mortensen, U. H., and Frandsen, R. J. (2013). Reconstruction of the biosynthetic pathway for the core fungal polyketide scaffold rubrofusarin in Saccharomyces cerevisiae. Microb. Cell Fact. 12, 31. doi: 10.1186/1475-2859-12-31
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sakai, K., Kinoshita, H., and Nihira, T. (2012). Heterologous expression system in Aspergillus oryzae for fungal biosynthetic gene clusters of secondary metabolites. Appl. Microbiol. Biotechnol. 93, 2011–2022. doi: 10.1007/s00253-011-3657-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sakai, K., Kinoshita, H., Shimizu, T., and Nihira, T. (2008). Construction of a citrinin gene cluster expression system in heterologous Aspergillus oryzae. J. Biosci. Bioeng. 106, 466–472. doi: 10.1263/jbb.106.466
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Siddiqui, M. S., Thodey, K., Trenchard, I., and Smolke, C. D. (2012). Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. FEMS Yeast Res. 12, 144–170. doi: 10.1111/j.1567-1364.2011.00774.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Smith, D., Burnham, M., Edwards, J., Earl, A., and Turner, G. (1990). Cloning and heterologous expression of the penicillin biosynthetic gene cluster from Penicillium chrysogenum. Nat. Biotechnol. 8, 39–41. doi: 10.1038/nbt0190-39
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Spingola, M., Grate, L., Haussler, D., and Ares, M. (1999). Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA 5, 221–234. doi: 10.1017/S1355838299981682
Szewczyk, E., Chiang, Y.-M., Oakley, C. E., Davidson, A. D., Wang, C. C. C., and Oakley, B. R. (2008). Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Appl. Environ. Microbiol. 74, 7607–7612. doi: 10.1128/AEM.01743-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tagami, K., Liu, C., Minami, A., Noike, M., Isaka, T., Fueki, S.,et al. (2013). Reconstitution of biosynthetic machinery for indole-diterpene paxilline in Aspergillus oryzae. J. Am. Chem. Soc. 135, 1260–1263. doi: 10.1021/ja3116636
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tagami, K., Minami, A., Fujii, R., Liu, C., Tanaka, M., Gomi, K.,et al. (2014). Rapid reconstitution of biosynthetic machinery for fungal metabolites in Aspergillus oryzae: total biosynthesis of aflatrem. Chembiochem. 15, 2076–2080. doi: 10.1002/cbic.201402195
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Takahashi, T., Masuda, T., and Koyama, Y. (2006). Enhanced gene targeting frequency in ku70 and ku80 disruption mutants of Aspergillus sojae and Aspergillus oryzae. Mol. Genet. Genomics 275, 460–470. doi: 10.1007/s00438-006-0104-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsunematsu, Y., Ishiuchi, K., Hotta, K., and Watanabe, K. (2013). Yeast-based genome mining, production and mechanistic studies of the biosynthesis of fungal polyketide and peptide natural products. Nat. Prod. Rep. 30, 1139–1149. doi: 10.1039/c3np70037b
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Unkles, S. E., Valiante, V., Mattern, D. J., and Brakhage, A. A. (2014). Synthetic biology tools for bioprospecting of natural products in eukaryotes. Chem. Biol. 21, 502–508. doi: 10.1016/j.chembiol.2014.02.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Verdoes, J. C., Punt, P. J., and van den Hondel, C. A. M. J. J. (1995). Molecular genetic strain improvement for the overproduction of fungal proteins by filamentous fungi. Appl. Microbiol. Biotechnol. 43, 195–205. doi: 10.1007/BF00172812
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Voss, K. A., and Riley, R. T. (2013). Fumonisin toxicity and mechanism of action: overview and current perspectives. Food Saf. 1, 2013006–2013006. doi: 10.14252/foodsafetyfscj.2013006
Wasil, Z., Pahirulzaman, K. A. K., Butts, C., Simpson, T. J., Lazarus, C. M., and Cox, R. J. (2013). One pathway, many compounds: heterologous expression of a fungal biosynthetic pathway reveals its intrinsic potential for diversity. Chem. Sci. 4, 3845–3856. doi: 10.1039/c3sc51785c
Wiemann, P., and Keller, N. P. (2014). Strategies for mining fungal natural products. J. Ind. Microbiol. Biotechnol. 41, 301–313. doi: 10.1007/s10295-013-1366-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yaegashi, J., Oakley, B. R., and Wang, C. C. C. (2014). Recent advances in genome mining of secondary metabolite biosynthetic gene clusters and the development of heterologous expression systems in Aspergillus nidulans. J. Ind. Microbiol. Biotechnol. 41, 433–442. doi: 10.1007/s10295-013-1386-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yin, W.-B., Chooi, Y. H., Smith, A. R., Cacho, R. A., Hu, Y., White, T. C.,et al. (2013). Discovery of cryptic polyketide metabolites from dermatophytes using heterologous expression in Aspergillus nidulans. ACS Synth. Biol. 2, 629–634. doi: 10.1021/sb400048b
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: secondary metabolite, Aspergillus, gene clusters, fungi, heterologous expression, polyketide synthase
Citation: Anyaogu DC and Mortensen UH (2015) Heterologous production of fungal secondary metabolites in Aspergilli. Front. Microbiol. 6:77. doi: 10.3389/fmicb.2015.00077
Received: 01 November 2014; Accepted: 21 January 2015;
Published online: 10 February 2015.
Edited by:
Nancy Keller, University of Wisconsin–Madison, USAReviewed by:
Clay Chia Chun Wang, University of Southern California, USAWei Xu, University of California, Los Angeles, USA
Copyright © 2015 Anyaogu and Mortensen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Uffe Hasbro Mortensen, Section for Eukaryotic Biotechnology, Department of Systems Biology, Technical University of Denmark, Soltofts Plads, Building 223, 2800 Kongens Lyngby, Denmark e-mail:dW1AYmlvLmR0dS5kaw==
 Diana Chinyere Anyaogu
Diana Chinyere Anyaogu Uffe Hasbro Mortensen
Uffe Hasbro Mortensen