- Faculty of Agriculture and Life Science, Hirosaki University, Hirosaki, Japan
Bacteria and virulent bacteriophages are in a prey–predator relationship. Experimental models under simplified conditions with the presence of bacteria and bacteriophages have been used to elucidate the mechanisms that have enabled both prey and predator to coexist over long periods. In experimental coevolution conducted with Escherichia coli and the virulent RNA bacteriophage Qβ in serial transfer, both coexisted for at least for 54 days, during which time they continued to change genetically and phenotypically. By day 16, an E. coli strain partially resistant to Qβ appeared and caused an approximately 104-fold decrease in Qβ amplification. Whole-genome analysis of this strain suggested that a single mutation in TraQ was responsible for the partially resistant phenotype. TraQ interacts with propilin, encoded by the traA gene and a precursor of pilin, which is a component of the F pilus. The present study was performed to elucidate the mechanism underlying the coexistence of E. coli and Qβ by investigating how a mutation in TraQ altered the physiological state of E. coli, and thus the amplification of Qβ. Overexpression of wild-type TraQ in the partially resistant E. coli strain resulted in recovery of both TraA protein content, including propilin and pilin, and Qβ amplification to levels comparable to those observed in the susceptible strain. Intriguingly, overexpression of the mutant TraQ in the partially resistant strains also increased the levels of TraA protein and Qβ amplification, but these increases were smaller than those observed in the wild-type strain or the partially resistant strain expressing wild-type TraQ. The results of this study represent an example of how E. coli can become partially resistant to RNA bacteriophage infection via changes in a protein involved in maturation of a receptor rather than in the receptor itself and of how E. coli can stably coexist with virulent RNA bacteriophages.
Introduction
There have long been ecological and theoretical investigations regarding why predators do not eradicate their prey (Murdoch and Oaten, 1975; Anderson and May, 1978; Alexander, 1981; Berryman, 1992; Abrams, 2000; Briggs and Hoopes, 2004; Pettorelli et al., 2011; Brockhurst and Koskella, 2013). Bacteria and bacteriophages have been used as model systems of prey–predator interactions to elucidate the fundamental mechanism underlying this issue. Due to their short generation time, large population size, and ease of analyzing phenotypic and genomic changes, theoretical and experimental evolutionary research have been conducted extensively using these systems (Campbell, 1961; Levin et al., 1977; Lenski and Levin, 1985; Lenski, 1988; Schrag and Mittler, 1995; Sasaki, 2000; Paterson et al., 2010; Dennehy, 2012; Brockhurst and Koskella, 2013; Bull et al., 2014). The stable coexistence of bacteria and virulent phages has been widely observed in experimental evolution (Horne, 1970; Chao et al., 1977; Levin et al., 1977; Buckling and Rainey, 2002; Lythgoe and Chao, 2003; Kerr et al., 2006; Kashiwagi and Yomo, 2011; Marston et al., 2012), even though virulent phages kill the host bacteria to release progeny phages.
The existence of refuges for sensitive bacteria or the occurrence of endless arms races were suggested to be necessary to explain the coexistence of bacteria and bacteriophages (Chao et al., 1977; Lenski, 1988; Schrag and Mittler, 1995; Brockhurst et al., 2006). The numerical refuge in which density-dependent protection of susceptible cells from over-predation (Chao et al., 1977), spatial refuges such as wall populations on flasks in continuous culture or solid media used in serial passage (Chao et al., 1977; Lenski, 1988; Schrag and Mittler, 1995), and physiological refuges in which cells become transiently resistant or susceptible have been discussed (Lenski, 1988). In most of these previous studies, DNA bacteriophages, such as T2, T5, T7, λ, and Φ2, were used (Bohannan and Lenski, 2000; Buckling and Rainey, 2002; Dennehy, 2012). Although a great deal of knowledge has been accumulated regarding DNA bacteriophages, little is known about RNA bacteriophages in terms of stable coexistence.
In our previous study, Escherichia coli and the lytic RNA bacteriophage Qβ (Qβ) were shown to coevolve for at least 54 days, equivalent to 165 generations, under conditions of serial passage with shaking (Kashiwagi and Yomo, 2011). Qβ is a bacteriophage with a single-stranded RNA genome that specifically infects and lyses E. coli cells to release progeny phages (Van Duin and Tsareva, 2006). Phenotypic and genomic analyses indicated the coevolution of both E. coli and Qβ. In the course of experimental coevolution, partially resistant E. coli appeared in the 54th generation (16th day). Detailed analysis of the partially resistant E. coli is necessary to determine how E. coli and Qβ coexist in this experimental coevolution system. Genetic analysis revealed a single mutation in the traQ gene in the day-16 E. coli population (Kashiwagi and Yomo, 2011). It was reported that TraQ protein is a chaperone for insertion of propilin encoded by the traA gene (Moore et al., 1982; Kathir and Ippen-Ihler, 1991), and propilin was also reported to be unstable in traQ- cells (Maneewannakul et al., 1993). The 13-kDa propilin is processed by peptidase to a 7-kDa pilin and pilin proteins are assembled into filaments (i.e., the F pilus). Qβ adsorbs the F pilus of E. coli at the first step of infection (Van Duin and Tsareva, 2006), and the adsorption rate of Qβ on partially resistant cells estimated by first-order kinetics decreased markedly (Kashiwagi and Yomo, 2011). Therefore, the partially resistant phenotype of E. coli to Qβ infection observed in coevolution may be correlated with F pilus biosynthesis, especially TraA, and we focused on the relationships among mutation in TraQ, TraA content, and Qβ amplification.
Here, we report that a single amino acid change in TraQ was linked to the reduction of TraA content in the E. coli population. In addition, this decrease was recovered by supplying ancestral (wild-type) or mutant-type TraQ from an expression vector, and the ability of Qβ to amplify in the cell also recovered. These results represent one example of how E. coli can become partially resistant to RNA bacteriophage infection, which involves changes in a protein related to the maturation of a receptor, in this case the F pilus, rather than changes to the receptor itself. These results suggest that the mutation in TraQ may cause heterogeneity within the E. coli population, with a small number of cells supporting the phage population and a large number of cells supporting the E. coli population without Qβ infection, even though the E. coli cells were genetically identical.
Materials and Methods
Strains, Culture Media, and Plasmid DNA
Escherichia coli Anc(C), the partially Qβ infection-resistant mutant strain, M54(C) (Kashiwagi and Yomo, 2011), and DH1 ΔleuB::(gfpuv5-Kmr) (hereafter called LKG; Kishimoto et al., 2010) were used to characterize the effects of S21P mutation in TraQ protein and a control F- strain. E. coli A/λ (Watanabe et al., 1979) was used as the host strain for titer assay. LB medium (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl; Nakalai Tesque, Kyoto, Japan) was used.
To construct TraQ with a Strep-tag II sequence (Schmidt and Skerra, 2007) at the C-terminus of the ancestral-type TraQ (TraQAnctag) and mutant-type TraQ (TraQS21Ptag), the traQ gene was amplified by PCR with Anc(C) and M54(C) genome as the template, the primers traQ_XbaI and traQ_strep_HindIII, and Phusion® High-Fidelity DNA polymerase (New England Biolabs, Ipswich, MA, USA). The oligonucleotide DNAs used in this study are listed in Table 1. The traQAnctag and traQS21Ptag genes with XbaI and HindIII sites at both ends were subcloned into the XbaI/HindIII sites of pASK-IBA3plus (IBA Biologics GmbH, Goettingen, Germany). The resulting plasmid DNAs were designated as pASK-traQAnctag and pASK-traQS21Ptag, respectively.
DNA Sequencing of the traQ Gene of Ancestral and Partially Resistant E. coli
To determine the traQ gene sequneces of Anc(C) and M54(C), the traQ region in 10 colonies each of Anc(C) and M54(C) was amplified by PCR with the primers F_4f_2 and F_4r_2 and Phusion® High-Fidelity DNA polymerase (New England BioLabs), and PCR products were directly sequenced by the dideoxynucleotide chain termination sequencing method (Sanger et al., 1977).
Estimation of Qβ Amplification
Anc(C)/pASK-traQAnctag, Anc(C)/pASK-traQS21Ptag, Anc(C)/pASK-IBA3plus, M54(C)/pASK-traQAnctag, M54(C)/pASK-traQS21Ptag, and M54(C)/pASK-IBA3plus were cultured in 5 mL of LB with 100 μg/mL ampicillin overnight and 50 μL of the culture was inoculated into 5 mL of LB with 100 μg/mL ampicillin for approximately 2 h. Aliquots of 1 mL of the culture were transferred into 4 mL of LB medium with 100 μg/mL ampicillin and 100 nM doxycycline-HCl (Dox) and cultured for a further 2 h. Qβ was added to infect the bacterial cells and the free phage was separated immediately or 4 h after infection by centrifugation at 13400 × g for 1 min. The free phage in the supernatant was diluted, the number of plaque forming units per milliliter was determined (PFU/mL), and the amplification ratio was calculated as x = N4/N0, where N4 and N0 represent 4 h after and initial (0 h) free phage density, respectively. The titer assay was conducted according to the standard method described previously (Carlson, 2005).
Western Blotting Analysis
A polyclonal antibody to TraA protein raised against the keyhole limpet hemocyanin-conjugated peptide (CDLMASGNTTVKATFGKDSS) was obtained from Sigma-Aldrich Japan (Tokyo, Japan). Cell preculture was conducted as described in the Section “Estimation of Qβ Amplification.” For induction with 100 nM Dox, Dox was added to the 2-h culture and cells were cultured for a further 5.75 h. Without Dox induction, the cells were cultured for 7.25 h in LB medium. Proteins from the cells obtained from 0.1 mL of culture with OD600 = 2.0 were subjected to SDS-PAGE using Any kDTM Mini-PROTEAN® TGXTM precast gels (Bio-Rad Laboratories, Hercules, CA, USA), and the TraA protein was determined by Western blotting analysis with anti-TraA antibody and HRP-conjugated goat anti-rabbit polyclonal antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) as the primary and secondary antibodies, respectively, diluted with Can Get Signal® immunoreaction enhancer solution (Toyobo Co. Ltd., Osaka, Japan). TraQ with a Strep-tag II at the C-terminus was detected using Precision ProteinTM Strep-Tactin-HRP conjugate (Bio-Rad Laboratories). The signals were detected with Chemi-Lumi One L (Nakalai Tesque).
Northern Hybridization Analysis
To compare the traA mRNA contents by Northern hybridization, Anc(C), M54(C), and LKG with pASK-IBA3plus were cultured in LB medium and total RNA was extracted from the cells in logarithmic phase using the SV Total RNA isolation system (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Total RNA from cells obtained from 0.16 mL of culture with OD600 = 0.27 was subjected to Northern hybridization using a digoxigenin-labeled single-stranded DNA probe, i.e., traA1 for traA and 16SrRNA_1 for 16SrRNA, as an indicator of the amount of total RNA used (Moran et al., 1995). The signals were detected with CDP-Star (GE Healthcare UK Ltd., Little Chalfont, UK). DynaMarker® Prestain Marker for RNA High (BioDynamics Laboratory Inc., Tokyo, Japan) was used to obtain the standard curve for RNA length.
Determination of 5′- and 3′-Terminal Sequences of traA mRNA
To determine the 5′-terminal sequence of traA mRNA, cDNA was synthesized using total RNA of Anc(C) as the template, SuperScript® III reverse transcriptase (Life Technologies, Carlsbad, CA, USA), and traA_r primer. The 5′-phosphorylated DNA linker 5PpACYC_rev was ligated at the 3′-terminus of the first strand cDNA with T4 RNA ligase 1 (New England Biolabs). PCR was performed using the resultant cDNA as the template, PrimeSTAR® HS DNA polymerase (Takara Bio Inc., Shiga, Japan), and the primers pACYC_rev2 and traA_r2. To determine the 3′-terminal sequence of traA mRNA, universal miRNA cloning linker (5′-rAppCTGTAGGCACCATCAAT-NH2-3′; New England Biolabs) was ligated with the 3′-terminus of total RNA of Anc(C) using T4 RNA ligase 1 (New England Biolabs). The first strand cDNA was synthesized using the primer Linker_r and SuperScript® III reverse transcriptase (Life Technologies), and then purified cDNA was subjected to PCR using the primers Linker_r and traA_f. Three and two bands of PCR products for 5′- and 3′-terminal sequence determination, respectively, were sliced from the gel and subcloned using a Zero Blunt® TOPO® PCR Cloning Kit for Sequencing (Life Technologies). Three to six clones were randomly picked and sequenced by the dideoxynucleotide chain termination sequencing method.
Statistical Analysis
Amplification ratios were compared by one-way ANOVA with the post hoc Tukey’s test (Zar, 2010). In all analyses, values of log10 (N4/N0) of each strain were used for statistical analysis. The Studentized range, q, is shown in the text. In all analyses, P < 0.01 was taken to indicate statistical significance.
Results
Recovery of Qβ Amplification in Resistant E. coli by Supplying TraQ
We first analyzed the traQ gene sequences from 10 single colonies derived from the coevolved E. coli population to confirm that the majority harbored the T61C mutation. We picked 10 single colonies from each of Anc(C) and M54(C) populations, which were the initial and day-16 E. coli populations in the coevolution experiment (Kashiwagi and Yomo, 2011). All 10 colonies of Anc(C) had T and all 10 colonies of M54(C) had C at position 61, and this T61C mutation resulted in S21P in the TraQ protein.
We analyzed the amplification ratio of Qβ on Anc(C) and M54(C) that harbored only the vector (pASK-IBA3plus) to determine the extent of reduction in the amplification ratio of Qβ on M54(C). Anc(C) and M54(C) expressed inherent ancestral-type TraQ protein and mutant-type TraQ protein from the F plasmid, respectively. The amplification ratio of Qβ was calculated as described in the Section “Materials and Methods.” The amplification ratios of Qβ on Anc(C)/pASK-IBA3plus and M54(C)/pASK-IBA3plus were 3.5 × 104 and 1.8, respectively (Figure 1). Although the amplification ratio on M54(C) was much lower than that on Anc(C), Qβ could undergo amplification on M54(C), indicating that M54(C) had a partially rather than fully resistant phenotype.
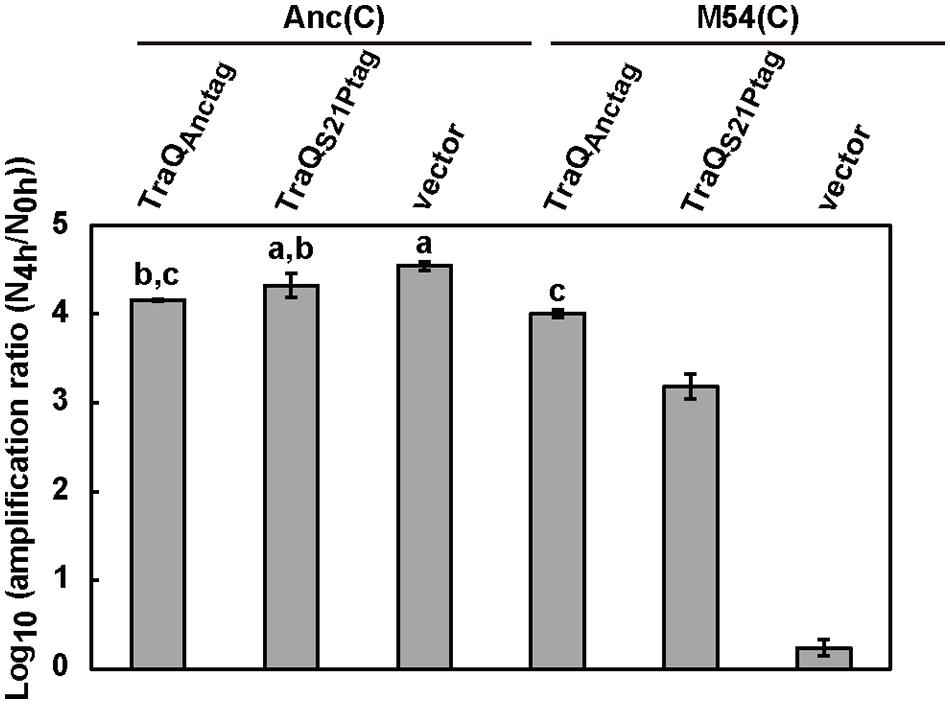
FIGURE 1. Amplification ratio of Qβ on each strain. The amplification ratios of Qβ were measured as the increase of free phage after 4 h of infection on Anc(C) and M54(C) harboring three different plasmids, i.e., pASK-traQAnctag, pASK-traQS21Ptag, and vector (pASK-IBA3plus). Data are expressed as averages ±SD (n = 3). Bars with identical letters are not significantly different from each other (Tukey’s test, P < 0.01).
To analyze whether ancestral-type TraQ expression in M54(C) could compensate for Qβ amplification in this strain, ancestral-type TraQ with the Strep-tag II sequence at the C-terminus was expressed from the PtetA promoter by doxycycline (Dox) induction. We designated ancestral and mutant-type TraQ with Strep-tag II sequence at the C-terminus expressed from the expression vector as TraQAnctag and TraQS21Ptag to allow them to be distinguished from the inherent TraQAnc and TraQS21P derived from the F plasmid, respectively. First, to examine whether TraQAnctag overexpression altered the amplification ratio of Qβ on Anc(C), we compared the amplification ratios of Qβ on Anc(C)/pASK-traQAnctag and Anc(C)/pASK-IBA3plus under conditions of Dox induction. The amplification ratio of Qβ on Anc(C) overexpressing TraQAnctag was 1.4 × 104, which was lower than that on Anc(C) harboring only the vector, 3.5 × 104, suggesting that overexpression of TraQ may be slightly deleterious for Qβ amplification (one-way ANOVA F5,12 = 920, P < 0.01; post hoc Tukey’s test q = 7.3, P < 0.01; Figure 1). Second, the amplification ratio of Qβ on M54(C) overexpressing TraQAnctag was 1.0 × 104, which was greater than the value of 1.8 on M54(C) carrying only the vector (one-way ANOVA F5,12 = 920, P < 0.01; post hoc Tukey’s test q = 70.5, P < 0.01), and the amplification ratio of M54(C) overexpressing TraQAnctag was comparable to that of Anc(C)/pASK-TraQAnctag (one-way ANOVA F5,12 = 920, P < 0.01; post hoc Tukey’s test q = 2.76, P = 0.42; Figure 1). These results showed that the amplification ratio of Qβ on M54(C) was recovered by supplying TraQAnctag. Intriguingly, supplying mutant-type TraQ, TraQS21Ptag, to M54(C) also rescued the amplification of Qβ on this strain. When TraQS21Ptag was overexpressed by Dox induction in M54(C), the amplification ratio of Qβ on the strain was 1.6 × 103, which was greater than that of M54(C) with the vector alone (one-way ANOVA F5,12 = 920, P < 0.01; post hoc Tukey’s test q = 55.1, P < 0.01), but was lower than that of M54(C) overexpressing TraQAnctag (one-way ANOVA F5,12 = 920, P < 0.01; post hoc Tukey’s test q = 15.4, P < 0.01; Figure 1). These results indicated that overexpression of mutant-type TraQ in M54(C) could partially, but not completely, compensate for the decrease in amplification of Qβ on the partially resistant cells.
Recovery of Qβ Amplification Linked to an Increase in TraA
We analyzed the TraA content by Western blotting to investigate the links between the mutation in TraQ and TraA content. Proteins derived from the same cell numbers calculated by the values of OD600 were subjected to SDS-PAGE. When we compared the levels of TraA produced by Anc(C) and M54(C) with vector (pASK-IBA3plus) and Anc(C) without the vector, the signal strength of TraA of Anc(C) with vector was almost the same as that of Anc(C) without the vector independent of Dox induction (Figure 2), but the TraA content of M54(C) with vector was extremely low independent of Dox induction (Figure 2). As no signal was detected in the lane for the F- control strain, LKG, M54(C) had not entirely lost TraA. These results showed that M54(C) had markedly decreased propilin and/or pilin content.
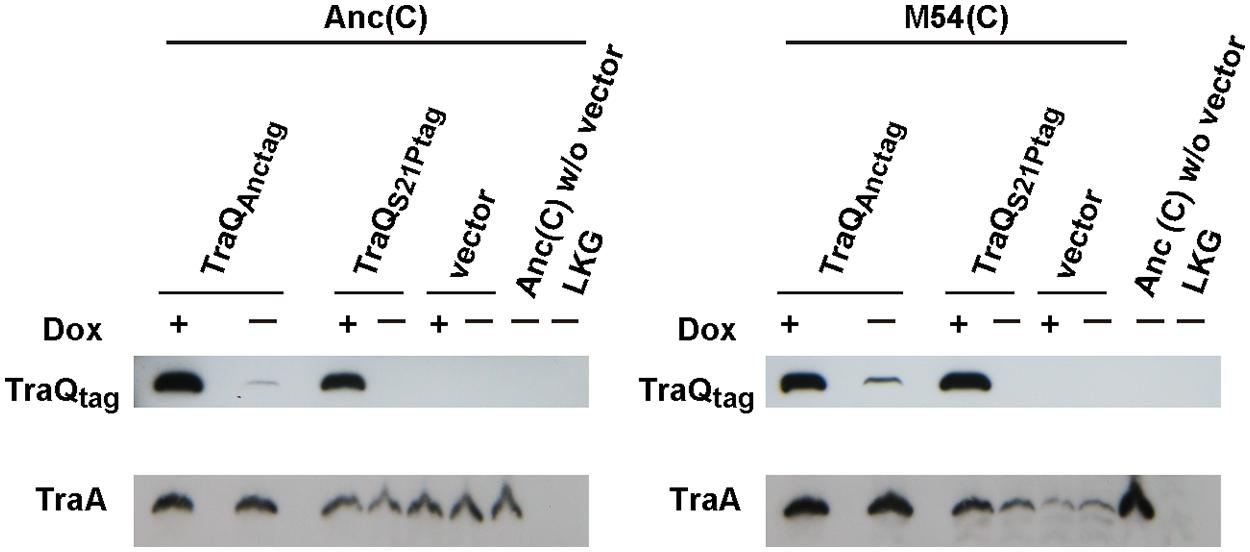
FIGURE 2. TraA expression analysis. The TraA and TraQ with Strep-tag II sequence contents of Anc(C) and M54(C) harboring three different plasmids, i.e., pASK-traQAnctag, pASK-traQS21Ptag, and vector (pASK-IBA3plus), were analyzed. Anc(C) without vector and LKG were used as control strains that expressed and did not express TraA, respectively. (+) and (-) represent with and without 100 nM Dox, respectively. Top and bottom represent Strep-Tactin detection for TraQ with Strep-tag II sequence at the C-terminus and anti-TraA detection for TraA.
The expression levels of ancestral and mutant-type TraQ with the Strep-tag II sequence at its C-terminus were determined with Strep-Tactin. When TraQAnctag was expressed in M54(C), the signal level for TraA was almost the same as that of Anc(C; Figure 2). In this case, the TraA contents were independent of Dox induction. Next, we supplied TraQS21Ptag to M54(C) with and without Dox induction. The TraA content in M54(C) increased with Dox induction, but the level was lower than that in M54(C) supplied with TraQAnctag (Figure 2). The TraA content of M54(C) also increased without Dox induction, but the strength of the signal was smaller than that under conditions of Dox induction. With supply of TraQS21Ptag, the TraA content was dependent on the expression level of TraQS21Ptag. These observations indicated that a small amount of ancestral-type TraQ in the partially resistant cells was sufficient to recover the TraA content, and a large amount of mutant-type TraQ protein increased the TraA content but the level was lower than that of the ancestral cells. Therefore, these results showed that a single mutation in TraQ resulted in a decrease of TraA content in the partially resistant cells.
traA mRNA Expression Level in M54(C)
To determine whether the decrease of TraA protein content in the partially resistant cells was due to a decrease in traA mRNA content, we analyzed the traA mRNA level by Northern hybridization. No signal was detected in the lane for the F- control strain LKG (Figure 3A), while signals were observed in the lanes for the Anc(C) and M54(C) strains (Figure 3A). The difference in signal strength of traA mRNA between Anc(C) and M54(C) may have been due to differences in the amount of total RNA loaded per lane, which was determined based on the 16SrRNA signal strength, although equal amounts of total RNA were loaded in each lane as calculated from the OD600. Therefore, we assumed that the mRNA levels were the almost the same for Anc(C) and M54(C).
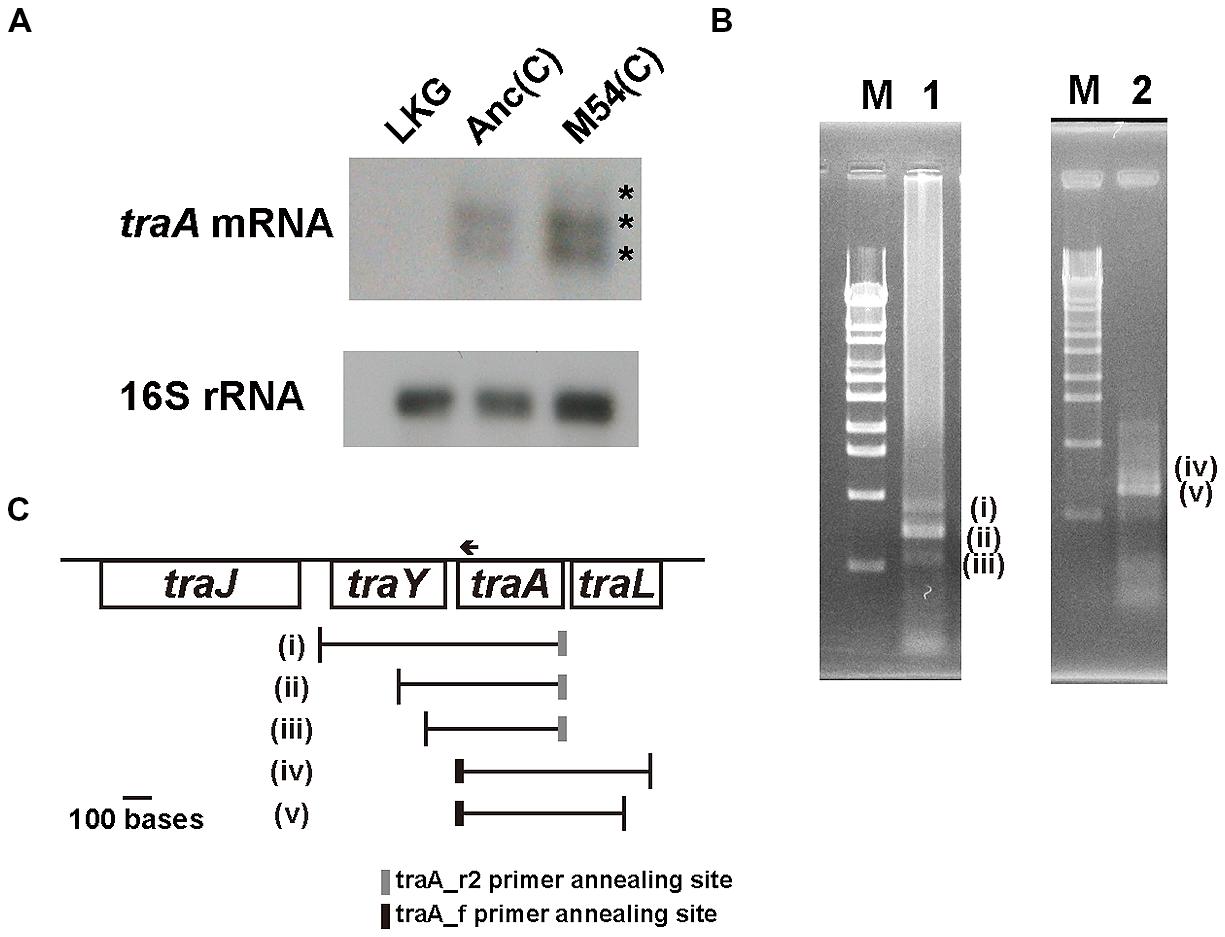
FIGURE 3. mRNA of traA gene analysis. (A) Northern hybridization for traA mRNA and 16S rRNA using the total RNA of LKG/pASK-IBA3plus; left, Anc(C)/pASK-IBA3plus; middle, M54(C)/pASK-IBA3plus; right. The upper and lower figures are X-ray films of traA mRNA and 16S rRNA, respectively. The asterisks (*) indicate the three signals (one weak and two strong). (B) RT-PCR for Anc(C)/pASK-IBA3plus to determine the 5′- and 3′-terminal sequences of traA mRNA. Lambda DNA digested with StyI was used as a molecular size marker; lane M. RT-PCR products for determination of the 5′-terminus. The three bands were designated as (i), (ii), and (iii), respectively; lane 1. RT-PCR products for determination of the 3′-terminus. The two bands were designated as (iv) and (v), respectively; lane 2. (C) Schematic representations of the start and end positions of traA mRNA. The vertical lines represent the positions of 5′- and 3′-terminal sequences of (i)–(v) shown in (B). The gray and black boxes represent the positions of traA_r2 and traA_f primer annealing sites in RT-PCR. The arrow represents the position of the traA1 probe annealing site for Northern hybridization.
As shown in Figure 3A, three bands were observed on Northern hybridization; one was weak and two were strong. The three bands corresponded to approximately 800, 610, and 460 bases, respectivel y, calculated using the standard curve obtained with the molecular weight markers. As the traA gene is 366 bases in length, the mRNA may contain upstream and/or downstream sequences. To determine the 5′- and 3′-terminal sequences of traA mRNA, RT-PCR was performed as described in the Section “Materials and Methods.” For 5′-terminal analysis of mRNA, we added a linker to the 3′-terminus of the first strand cDNA and conducted PCR. At least three PCR products, two of which were clear and the remaining one was weak, were obtained (Figure 3B, left). For 3′-terminal analysis of mRNA, we added a linker at the 3′-terminus of the mRNA and conducted RT-PCR. At least two PCR products were obtained, one of which was clearly observed and the other was weak (Figure 3B, right). The sequences of these PCR products, designated as (i)–(v), were analyzed and the starting and termination positions of the mRNA including traA were determined (Figure 3C). The starting and terminating positions described below are numbered according to GenBank accession number AP001918.1. Analysis of six clones of (i) showed that mRNAs started from a position upstream of the traY gene; five started at position 67808 and one started at position 67754. Analysis of six clones of (ii) showed that it started from within the traY gene; four started at position 68000 and two started at position 67999. Analysis of five clones of (iii) showed that mRNAs started from within the traY gene; one started at position 68165, two at 68166, one at 68167, and one at 68189 (Figure 3C). Analysis of five clones of (v) showed that mRNAs terminated within the traL gene; four terminated at 68818 and one at 68819. Analysis of three clones of (iv) also showed that the mRNAs terminated within the traL gene; all three terminated at position 68902. These results indicated that the total RNA included mRNAs encoding traA of various lengths with different upstream and downstream sequences. Therefore, all three bands observed on Northern hybridization should contain the traA gene sequence.
Discussion
We reported previously that E. coli and Qβ coexisted in serial passage and both continued to change genetically and phenotypically (Kashiwagi and Yomo, 2011). Here, we characterized partially resistant E. coli obtained in the previous study and demonstrated links among TraQ content, TraA content, and amplification of Qβ.
In this study, overexpression of mutant-type TraQS21Ptag was shown to result in an increase in TraA content and recovery of Qβ amplification. These observations indicated that TraQS21P had not entirely lost its function. The S21P mutation may alter the activity of TraQ, such as changing the binding affinity between TraQ and TraA, and may result in a decrease of TraA content in the M54(C) population and reduce the possibility of Qβ infection by decreasing either the number of cells with F pili or the amount of F pili in each cell in the population, as TraQ protein binds propilin that is a precursor of mature TraA and the first 21 amino acids are important for this binding (Harris et al., 1999). In this study, not only TraQAnctag but also the TraQS21Ptag increased the TraA contents of M54(C) without Dox induction. The copy numbers of pASK-TraQS21Ptag, which has the ColE1 replication origin, and F plasmid in the cell are 15–20 and 1, respectively (Snyder and Champness, 2007). Therefore, introduction of pASK-TraQS21Ptag into the cell increased the copy number of the traQ (T61C) gene and therefore may have increased the concentration of mutant-type TraQ in the cell.
It has been reported that F+ cells in E. coli populations are heterogeneous in the number of F pili per cell and in the length of F pili through the cycles of extension and retraction (Clarke et al., 2008; Silverman and Clarke, 2010). In addition, it is widely accepted that even E. coli with the same genotype show phenotypic diversity due to the stochasticity in living organisms (Elowitz et al., 2002; Kashiwagi et al., 2006; Bressloff, 2014). Therefore, even in Anc(C), the population would be heterogeneous in both number and length of F pili per cell. The single mutation of TraQ would decrease the percentage of cells that could be infected by Qβ in the population, even though the M54(C) population was genetically identical throughout the community. The mutation was introduced into the region involved in binding with propilin (Harris et al., 1999) and protein–protein binding is one of the stochastic processes in a cell because it is a collision reaction and reducing numbers of interacting molecules in a cell would increase the fluctuation in number of bound proteins (Bressloff, 2014). Therefore, there would be at least three types of players in the community: a small proportion of cells supporting the phage population, a large proportion of cells supporting the E. coli population due to escape from Qβ infection, and Qβ itself. This heterogeneity would result in the partially resistant phenotype of the M54(C) population, as we assessed the phenotype based on the amplification ratio of Qβ in the population and not in single infected cells. As we evaluated the TraA and TraQ contents and Qβ amplification of the population and not of single cells, there are at least two plausible explanations for the partial resistance. The first is that every cell had low levels of TraA or F pili, and the second is that only a small portion of cells in the population had sufficient F pili for Qβ adsorption. In both cases, at least two types of cell—a minor population infected by Qβ and a major population that was not infected by Qβ—may emerge from E coli with the identical genotype.
The physiological refuge hypothesis has been reported as one of the mechanisms allowing the coexistence of bacteria and bacteriophages by providing phenotypic heterogeneity in resistance within the bacterial population (Lenski, 1988; Schrag and Mittler, 1995). The results of the present study suggested that the coevolved E. coli in experimental evolution would generate phenotypic heterogeneity with both resistant and susceptible cells, as suggested by the physiological refuge hypothesis.
Many resistance mechanisms of bacteria for DNA bacteriophages have been reported, such as preventing phage adsorption, preventing phage DNA entry, cutting phage nucleic acids, abortive infection, and phase variation (Hancock and Reeves, 1975; Labrie et al., 2010; Bikard and Marraffini, 2012). In preventing phage adsorption, surface receptors of bacteria for phage infection were modified, masked by proteins, or blocked by exopolysaccharide (Labrie et al., 2010). However, there have been few discussions regarding the mechanisms of resistance to RNA bacteriophages. Here, we first reported one of the mechanisms underlying partial resistance of E. coli to the RNA bacteriophage Qβ that would be included in preventing phage adsorption: a decrease in chance of phage adsorption by reducing the receptor contents in the host population by changing a single amino acid on the protein related to production of the mature receptor (F pilus), not the receptor itself. In addition, the results of this study also suggested that the phenotypic fluctuation caused by changing a single amino acid on the protein would facilitate long-term coexistence of both predator (Qβ phage) and prey (E. coli).
Author Contributions
AK designed the research. AK, HK, and FST carried out the experiments and analyzed the data. AK wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Tetsuya Yomo (Osaka University) and Dr. Kotaro Mori (Ishihara Sangyo Kaisha, Ltd.) for valuable discussions, and Ms. Hiroko Tohyama for excellent technical assistance. We thank Dr. Takahiro Toba and the members of RNA Research Center (Hirosaki University) for their cooperation. This work was supported in part by MEXT KAKENHI (21770255 and 23570268), Kato Memorial Bioscience Foundation, Hirosaki University Grant for Exploratory Research by Young Scientists, Priority Research Grant for Young Scientists Designated by the President of Hirosaki University, and Hirosaki University Institutional Research Grant for Young Scientists.
References
Abrams, P. A. (2000). The evolution of predator-prey interactions: theory and evidence. Annu. Rev. Ecol. Syst. 31, 79–105. doi: 10.1146/annurev.ecolsys.31.1.79
Alexander, M. (1981). Why microbial predators and parasites do not eliminate their prey and hosts. Annu. Rev. Microbiol. 35, 113–133. doi: 10.1146/annurev.mi.35.100181.000553
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Anderson, R. M., and May, R. M. (1978). Regulation and stability of host-parasite population interactions I. Regulatory processes. J. Anim. Ecol. 47, 219–247. doi: 10.2307/3933
Berryman, A. A. (1992). The origins and evolution of predator-prey theory. Ecology 73, 1530–1535. doi: 10.2307/1940005
Bikard, D., and Marraffini, L. A. (2012). Innate and adaptive immunity in bacteria: mechanisms of programmed genetic variation to fight bacteriophages. Curr. Opin. Immunol. 24, 15–20. doi: 10.1016/j.coi.2011.10.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bohannan, B. J., and Lenski, R. E. (2000). Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol. Lett. 3, 362–377. doi: 10.1046/j.1461-0248.2000.00161.x
Bressloff, P. C. (2014). Stochastic Processes in Cell Biology. Heidelberg: Springer. doi: 10.1007/978-3-319-08488-6
Briggs, C. J., and Hoopes, M. F. (2004). Stabilizing effects in spatial parasitoid-host and predator-prey models: a review. Theor. Popul. Biol. 65, 299–315. doi: 10.1016/j.tpb.2003.11.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brockhurst, M. A., Buckling, A., and Rainey, P. B. (2006). Spatial heterogeneity and the stability of host-parasite coexistence. J. Evol. Biol. 19, 374–379. doi: 10.1111/j.1420-9101.2005.01026.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brockhurst, M. A., and Koskella, B. (2013). Experimental coevolution of species interactions. Trends Ecol. Evol. 28, 367–375. doi: 10.1016/j.tree.2013.02.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Buckling, A., and Rainey, P. B. (2002). Antagonistic coevolution between a bacterium and a bacteriophage. Proc. Biol. Sci. 269, 931–936. doi: 10.1098/rspb.2001.1945
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bull, J. J., Vegge, C. S., Schmerer, M., Chaudhry, W. N., and Levin, B. R. (2014). Phenotypic resistance and the dynamics of bacterial escape from phage control. PLoS ONE 9:e94690. doi: 10.1371/journal.pone.0094690
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Campbell, A. (1961). Conditions for the existence of bacteriophage. Evolution 15, 153–165. doi: 10.2307/2406076
Carlson, K. (2005). “Working with bacteriophages: common techniques and methodological approaches,” in Bacteriophages, Biology and Applications, eds E. Kutter and A. Sulakvelidze (Boca Raton, FL: CRC Press), 437–494.
Chao, L., Levin, B. R., and Stewart, F. M. (1977). A complex community in a simple habitat: an experimental study with bacteria and phage. Ecology 58, 369–378. doi: 10.2307/1935611
Clarke, M., Maddera, L., Harris, R. L., and Silverman, P. M. (2008). F-pili dynamics by live-cell imaging. Proc. Natl. Acad. Sci. U.S.A. 105, 17978–17981. doi: 10.1073/pnas.0806786105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dennehy, J. J. (2012). What can phages tell us about host-pathogen coevolution? Int. J. Evol. Biol. 2012, 12. doi: 10.1155/2012/396165
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Elowitz, M. B., Levine, A. J., Siggia, E. D., and Swain, P. S. (2002). Stochastic gene expression in a single cell. Science 297, 1183–1186. doi: 10.1126/science.1070919
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hancock, R. E., and Reeves, P. (1975). Bacteriophage resistance in Escherichia coli K-12: general pattern of resistance. J. Bacteriol. 121, 983–993.
Harris, R. L., Sholl, K. A., Conrad, M. N., Dresser, M. E., and Silverman, P. M. (1999). Interaction between the F plasmid TraA (F-pilin) and TraQ proteins. Mol. Microbiol. 34, 780–791. doi: 10.1046/j.1365-2958.1999.01640.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Horne, M. T. (1970). Coevolution of Escherichia coli and bacteriophages in chemostat culture. Science 168, 992–993. doi: 10.1126/science.168.3934.992-a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kashiwagi, A., Urabe, I., Kaneko, K., and Yomo, T. (2006). Adaptive response of a gene network to environmental changes by fitness-induced attractor selection. PLoS ONE 1:e49. doi: 10.1371/journal.pone.0000049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kashiwagi, A., and Yomo, T. (2011). Ongoing phenotypic and genomic changes in experimental coevolution of RNA bacteriophage Qβ and Escherichia coli. PLoS Genet. 7:e1002188. doi: 10.1371/journal.pgen.1002188
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kathir, P., and Ippen-Ihler, K. (1991). Construction and characterization of derivatives carrying insertion mutations in F plasmid transfer region genes, trbA, artA, traQ, and trbB. Plasmid 26, 40–54. doi: 10.1016/0147-619X(91)90035-U
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kerr, B., Neuhauser, C., Bohannan, B. J., and Dean, A. M. (2006). Local migration promotes competitive restraint in a host-pathogen ‘tragedy of the commons.’ Nature 442, 75–78. doi: 10.1038/nature04864
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kishimoto, T., Iijima, L., Tatsumi, M., Ono, N., Oyake, A., Hashimoto, T.,et al. (2010). Transition from positive to neutral in mutation fixation along with continuing rising fitness in thermal adaptive evolution. PLoS Genet. 6:e1001164. doi: 10.1371/journal.pgen.1001164
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Labrie, S. J., Samson, J. E., and Moineau, S. (2010). Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8, 317–327. doi: 10.1038/nrmicro2315
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lenski, R. E. (1988). “Dynamics of interactions between bacteria and virulent bacteriophage,” in Advances in Microbial Ecology, ed. K. C. Marshall (New York: Plenum Publishing Corporation), 1–44.
Lenski, R. E., and Levin, B. R. (1985). Constraints on the coevolution of bacteria and virulent phage: a model, some experiments, and predictions for natural communities. Am. Nat. 125, 585–602. doi: 10.1086/284364
Levin, B. R., Stewart, F. M., and Chao, L. (1977). Resource-limited growth, competition, and predation: a model and experimental studies with bacteria and bacteriophage. Am. Nat. 111, 3–24. doi: 10.1086/283134
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lythgoe, K. A., and Chao, L. (2003). Mechanisms of coexistence of a bacteria and a bacteriophage in a spatially homogeneous environment. Ecol. Lett. 6, 326–334. doi: 10.1046/j.1461-0248.2003.00433.x
Maneewannakul, K., Maneewannakul, S., and Ippen-Ihler, K. (1993). Synthesis of F pilin. J. Bacteriol. 175, 1384–1391.
Marston, M. F., Pierciey, F. J. Jr., Shepard, A., Gearin, G., Qi, J., Yandava, C.,et al. (2012). Rapid diversification of coevolving marine Synechococcus and a virus. Proc. Natl. Acad. Sci. U.S.A. 109, 4544–4549. doi: 10.1073/pnas.1120310109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moore, D., Sowa, B. A., and Ippen-Ihler, K. (1982). A new activity in the Ftra operon which is required for F-pilin synthesis. Mol. Gen. Genet. 188, 459–464. doi: 10.1007/BF00330049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moran, M. A., Rutherford, L. T., and Hodson, R. E. (1995). Evidence for indigenous Streptomyces populations in a marine environment determined with a 16S rRNA probe. Appl. Environ. Microbiol. 61, 3695–3700.
Murdoch, W. W., and Oaten, A. (1975). “Predation and population stability,” in Advances in Ecological Research, ed. A. Macfadyen (London: Academic press Inc.), 1–131.
Paterson, S., Vogwill, T., Buckling, A., Benmayor, R., Spiers, A. J., Thomson, N. R.,et al. (2010). Antagonistic coevolution accelerates molecular evolution. Nature 464, 275–278. doi: 10.1038/nature08798
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pettorelli, N., Coulson, T., Durant, S. M., and Gaillard, J. M. (2011). Predation, individual variability and vertebrate population dynamics. Oecologia 167, 305–314. doi: 10.1007/s00442-011-2069-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sanger, F., Nicklen, S., and Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 74, 5463–5467. doi: 10.1073/pnas.74.12.5463
Sasaki, A. (2000). Host-parasite coevolution in a multilocus gene-for-gene system. Proc. Biol. Sci. 267, 2183–2188. doi: 10.1098/rspb.2000.1267
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schmidt, T. G., and Skerra, A. (2007). The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2, 1528–1535. doi: 10.1038/nprot.2007.209
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schrag, S. J., and Mittler, J. E. (1995). Host-parasite coexistence: the role of spatial refuges in stabilizing bacteria-phage interactions. Am. Nat. 148, 348–377. doi: 10.1086/285929
Silverman, P. M., and Clarke, M. B. (2010). New insights into F-pilus structure, dynamics, and function. Integr. Biol. 2, 25–31. doi: 10.1039/B917761B
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Snyder, L., and Champness, W. (2007). “Plasmids,” in Molecular Genetics of Bacteria (Washington, DC: ASM Press), 197–242.
Van Duin, J., and Tsareva, N. (2006). “Single-stranded RNA phages,” in The Bacteriophages, ed. R. Calendar (New York: Oxford University Press), 175–196.
Watanabe, I., Sakurai, T., Furuse, K., and Ando, A. (1979). “Pseudolysogenization” by RNA phage Qβ. Microbiol. Immunol. 23, 1077–1083. doi: 10.1111/j.1348-0421.1979.tb00539.x
Keywords: coevolution, prey–predator, experimental evolution, virulent phage, partial resistance
Citation: Kashiwagi A, Kitamura H and Sano Tsushima F (2015) Characterization of a single mutation in TraQ in a strain of Escherichia coli partially resistant to Qβ infection. Front. Microbiol. 6:124. doi: 10.3389/fmicb.2015.00124
Received: 26 November 2014; Accepted: 02 February 2015;
Published online: 20 February 2015.
Edited by:
Hiroyuki Futamata, Shizuoka University, JapanReviewed by:
Evelien M. Adriaenssens, University of Pretoria, South AfricaHailan Piao, Washington State University in Tri-Cities, USA
Copyright © 2015 Kashiwagi, Kitamura and Sano Tsushima. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akiko Kashiwagi, Faculty of Agriculture and Life Science, Hirosaki University, 3 Bunkyo-cho, Hirosaki, Aomori 036-8561, Japan e-mail:a2FzaGlfYTFAY2MuaGlyb3Nha2ktdS5hYy5qcA==
 Akiko Kashiwagi
Akiko Kashiwagi Hikari Kitamura
Hikari Kitamura Fumie Sano Tsushima
Fumie Sano Tsushima