- 1Microbial Interactions Laboratory, Department of Environmental Health Sciences, Public Health Research Center, Arnold School of Public Health, University of South Carolina, Columbia, SC, USA
- 2Department of Chemistry and Biochemistry, JM Palms Center for Graduate Student Research, University of South Carolina, Columbia, SC, USA
The alarming spread of bacterial resistance to traditional antibiotics has warranted the study of alternative antimicrobial agents. Quorum sensing (QS) is a chemical cell-to-cell communication mechanism utilized by bacteria to coordinate group behaviors and establish infections. QS is integral to bacterial survival, and therefore provides a unique target for antimicrobial therapy. In this study, silicon dioxide nanoparticles (Si-NP) were engineered to target the signaling molecules [i.e., acylhomoserine lactones (HSLs)] used for QS in order to halt bacterial communication. Specifically, when Si-NP were surface functionalized with β-cyclodextrin (β-CD), then added to cultures of bacteria (Vibrio fischeri), whose luminous output depends upon HSL-mediated QS, the cell-to-cell communication was dramatically reduced. Reductions in luminescence were further verified by quantitative polymerase chain reaction (qPCR) analyses of luminescence genes. Binding of HSLs to Si-NPs was examined using nuclear magnetic resonance (NMR) spectroscopy. The results indicated that by delivering high concentrations of engineered NPs with associated quenching compounds, the chemical signals were removed from the immediate bacterial environment. In actively-metabolizing cultures, this treatment blocked the ability of bacteria to communicate and regulate QS, effectively silencing and isolating the cells. Si-NPs provide a scaffold and critical stepping-stone for more pointed developments in antimicrobial therapy, especially with regard to QS—a target that will reduce resistance pressures imposed by traditional antibiotics.
Introduction
Excessive use of antibiotics has resulted in widespread bacterial resistance and poses a significant public health threat. Non-cytotoxic methods for controlling infections, such as interference in bacterial communication, are necessary to fight pathogenic infections while limiting the risk of antibiotic resistance (Rasko and Sperandio, 2010; Schuster et al., 2013). Quorum sensing (QS) is a chemical communication scheme used by bacteria to coordinate their activities within a biofilm, and it regulates many translational features that make bacteria dangerous (Davies, 1998; Ng and Bassler, 2009; Hoiby et al., 2010). One of the most significant challenges in the treatment of chronic infections is the effective delivery of drugs to bacterial biofilms. The importance of nanoparticle-based therapies is imperative to the treatment of biofilm infections. Biofilms are surface-associated bacterial communities living in a highly-organized structure at a liquid interface (Costerton et al., 1987), and have been estimated to be responsible for 80% of hospital-acquired infections (Davies, 2003). The protective nature of bacterial biofilms makes them able to physically limit antibiotic penetration (Stewart et al., 2001; Canton and Morosini, 2011), quickly regulate multidrug efflux pumps and stress response genes (Davies, 2003; Hoiby et al., 2010), induce a biofilm-specific phenotype (Allison et al., 2011), and easily trade and enhance antibiotic resistance genes among their bacterial members (Madsen et al., 2012).
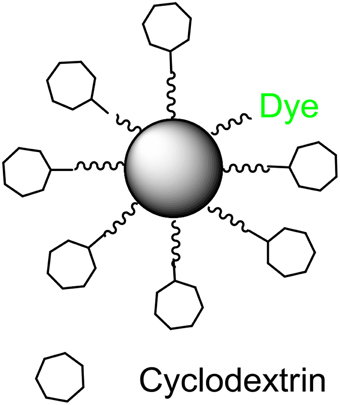
Silicon dioxide nanoparticles (Si-NPs) have a broad range of applications within the biomedical and industrial fields due to their high biocompatibility and controllable particle size (De et al., 2008; Malvindi et al., 2012; Wang and Benicewicz, 2013). Inspired by developments in the field of drug delivery, we have designed Si-NPs (Figure 1) to carry compounds that will quench signal molecules. Removal of signals from the immediate bacterial environment prevents the molecule from reaching its cognate receptor, thus inhibiting the signal/receptor interaction, and interfering with down-stream regulation. We hypothesized that this process will quench signal molecules, “turn off” QS, and silence bacterial communication (schematic seen in Figures 2A,B).
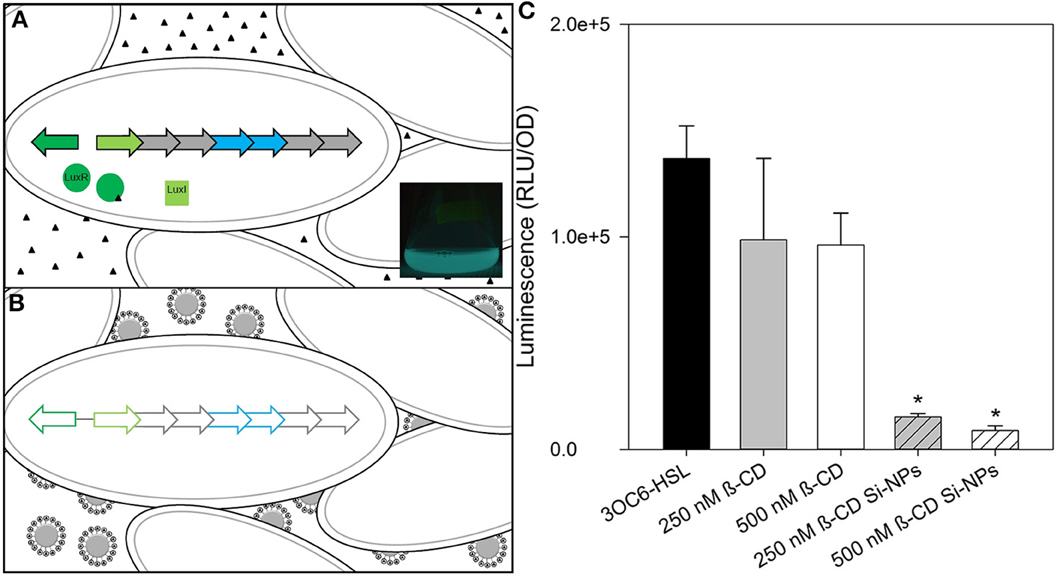
Figure 2. Schematic of nanoparticle-based silencing of bacterial quorum sensing (QS). (A) Diagram of the Gram-negative bacterium Vibrio fischeri lux operon used in luminescence during QS using acyl-homoserine lactones (HSL). Triangles represent 3OC6-HSL. LuxR/3OC6-HSL complex initiates bilateral transcription of lux operon. LuxI produces 3OC6-HSL. Inset: A one-liter V. fischeri culture flask luminescing after treatment with HSLs. (B) Diagram of V. fischeri during nanoparticle treatment. Binding of HSLs by Si-NPs quenches QS and subsequent gene expression in lux. (C) Changes in maximum bioluminescence by V. fischeri during exposure to 2 μ M 3OC6-HSL, with either β-CD or β-CD functionalized Si-NPs. Error bars represent standard error of the mean (n = 3). Asterisks indicate significance (P = 0.05) by ANOVA.
β-cyclodextrin (β-CD) has been shown to non-specifically bind N-acyl-L-homoserine lactone (HSL) molecules (Kato et al., 2006), a common class of signal molecules produced by Gram-negative bacteria. β-CD consists of seven glucopyranose units bound by α-1,4-glycosidic linkages. This molecule takes the shape of a truncated cone with a hydrophilic exterior and a slightly hydrophobic interior. β-CD was chosen as a model compound because the formation of a CD complex can be detected and because the reactivity of a guest molecule in a CD complex is subsequently modified (Szejtli, 1998). In many cases, the CD accelerates the various reactions associated with the guest molecule and modifies the reaction pathway. Herein we present a new approach to disrupting bacterial QS with Si-NPs that have been surface-functionalized with β-CDs.
To test the quorum quenching function of the Si-NPs, the marine bacterium Vibrio fischeri was employed as a model. V. fischeri relies on N-acyl-L-homoserine lactone molecules to trigger coordinated activities such as colonization and bioluminescence in a threshold dependent manner during QS (Kaplan and Greenberg, 1985). Specifically, V. fischeri synthesizes and responds to N-3-oxo-hexanoyl-L-homoserine lactone (3OC6-HSL) (Eberhard et al., 1981; Engebrecht and Silverman, 1984) and N-octanoyl-L-homoserine lactone (C8-HSL) (Engebrecht et al., 1983) via the lux operon, which is also responsible for the proteins that synthesize luminescent luciferase (Stevens et al., 1994). HSLs are also used by pathogenic bacteria such as Actinobacillus, Salmonella, Pseudomonas, and other Vibrio species to regulate their respective QS genes (Defoirdt, 2013; LaSarre and Federle, 2013; Li et al., 2013b).
In this study, the fold change in the luxA and luxR transcription of V. fischeri was quantified to determine the activity of the lux operon during exposure to β-CD. LuxA forms the alpha subunit of luciferase and was used to monitor bacterial luminescence. LuxR is a receptor for both 3OC6-HSL and C8-HSL, and initiates the lux operon, and was used to monitor signal production. In the natural ocean habitat, this QS function allows V. fischeri to establish a symbiotic relationship with the Hawaiian bobtail squid (Euprymna scolopes) and provide counterillumination for the squid (Visick et al., 2000). Environmental conditions are essential for the proper functioning of V. fischeri bioluminescence. A symbiotic culture produces approximately 1000-fold brighter bioluminescence and more 3OC6-HSL than cultured cells at the same density (Boettcher and Ruby, 1990). Therefore, HSLs must be added to the cultures to induce QS and visible bioluminescence in vitro.
In this study we have engineered nanoparticles that weaken the communication network of bacteria, rather than directly target the viability of cells. The functionalized Si-NPs bind small signal molecules as they diffuse between cells, and subsequently block bacterial QS. Using growth and transcription studies, we demonstrate that this novel technology can be used to directly influence bacterial communication and activities regulated by QS, and offer a promising tool that can be further developed to unravel the virulence of pathogenic bacteria. V. fischeri was used in its planktonic form to establish the utility of the functionalized NPs in a simple environment. Also, this bacterium was used because its QS mechanism is well-established in the literature and can be readily detected (by luminescence) in the laboratory. A biofilm model has not yet been employed here because of the inherent complexity found in a biofilm, and the subsequent difficulty in monitoring the QS response.
Results
Si-NPs Carry β-Cyclodextrin
The unique properties of surface-functionalized Si-NPs make them ideal for targeting the multiple avenues of bacterial defenses and weakening an infection. Surface functionalization plays a critical role in tailoring the properties of Si-NPs via well-developed surface chemistry (Li et al., 2013a, 2014). For this study, the synthesized β-CD functionalized Si-NPs were purified via dialysis to remove un-reacted (i.e., free) β-CD molecules, as confirmed by 1H NMR (Supplementary Figure 1). Thermogravimetric analysis (TGA) confirmed that the monolayer of β-CD accounted for 2.78% of the total weight of particles and indicated a surface graft density of 0.27 and 0.11 groups/nm2 for the 15 and 50 nm nanoparticles, respectively (Supplementary Figure 2).
β-Cyclodextrin Binds HSLs in Solution
NMR spectroscopy was used to determine binding strengths between β-CD and HSLs. NMR diffusion experiments demonstrated that both C8-HSL (produced by V. fischeri) and C6-HSL (produced by Pseudomonas aeruginosa) could bind to β-CD in a 1:1 ratio; however, there was no evidence that 3OC6-HSL (also produced by V. fischeri) formed a complex with β-CD. Further calculations revealed the binding and dissociation constants of the experimental complexes. The binding between the HSLs and β-CD accounted for only 17% of C6-HSL and 35% of C8-HSL at any one time in the 1:1 solution (Supplementary Tables 1, 2). Despite this relatively low binding, we proceeded to quantify the quenching ability of β-CD in vitro. Using growth and luminescence studies we observed the influence of β-CD on V. fischeri bioluminescence.
Nanoparticles Enhance Ability of β-Cyclodextrin to Quench HSLs and Dim Bioluminescence
V. fischeri cultures grown in the presence of 2 μ M 3OC6-HSL exhibited strong bioluminescence and normal growth. When treated with concentrations of free β-CD ranging from 250 nM to 7 mM, the growth study showed that exposure to 2 mM β-CD resulted in the most significant decrease in bioluminescence (Supplementary Figure 3). A preliminary analyses of β-CD functionalized Si-NPs with 2 μ M 3OC6-HSL demonstrated that the β-CD moietie was significantly more-effective at dimming bioluminescence of V. fischeri when functionalized to a Si-NP than it was as a free-compound (Figure 2C). Statistical significance for all experiments was determined by a repeated measures analysis of variance test followed by an aposteriori pairwise muliple comparision procedure (SigmaPlot, Systat Software, San Jose, CA). Because 2 μ M 3OC6-HSL likely exceeds the expected HSL production of symbiotic V. fischeri cultures [which is unknown, but luminescence is induced in the nanomolar range (Lupp and Ruby, 2005)], and does not induce the lux operon like C8-HSL, we altered the HSL conditions to more-closely mimic the natural environment.
Growth experiments were repeated with V. fischeri cultured with 125 nM 3OC6-HSL and 0.25 nM C8-HSL. Optical density and bioluminescence were monitored throughout growth to determine the impact of free and functionalized β-CD. All cultures were grown in defined minimal medium (Studer et al., 2008), and therefore were not in a rich nutrient environment. Additional growth studies indicated that the bacteria did not thrive on β-CD or the various Si-NPs. Also, when all carbon sources in the defined minimal medium were replaced with β-CD, no growth occurred. Cultures grown without the addition of HSLs produced negligible levels of bioluminescence. Therefore, β-CD, with or without NPs, did not influence the growth or relative health of the cells.
V. fischeri cultures grown in the presence of 125 nM 3OC6-HSL and 0.25 nM C8-HSL produced measureable bioluminescence; although, bioluminescence in general was lower compared to the cultures treated with 2 μ M 3OC6-HSL (as indicated by the different scales in Figures 2C, 3). Our study found that at environmentally-relevant levels of HSLs, bioluminescence was significantly reduced in vitro by β-CD (P = 0.05). By functionalizing β-CD to 50 nm NPs, 133 nM β-CD was able to produce the same result as a 2x higher concentration of free 250 nM β-CD (Figure 3). We found that 15 nm Si-NPs with or without β-CD decreased bioluminescence to the same extent, and thus may not have a significant role in dimming luminescence.
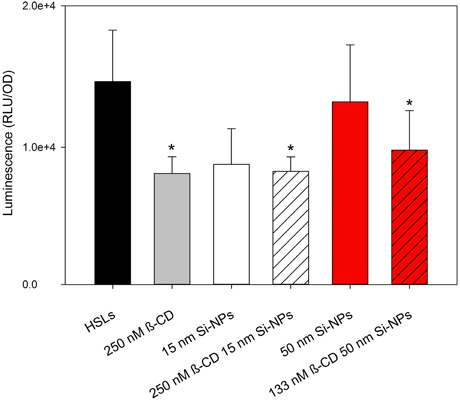
Figure 3. Mean relative maximum bioluminescence per OD (600 nm) of V. fischeri during exposure to 125 nM 3OC6-HSL and 0.25 nM C8-HSL treated with 250 nM β-CD, bare 15 nm Si-NPs, 155 nM β-CD functionalized 15 nm Si-NPs, bare 50 nm Si-NPs, or 133 nM β-CD functionalized 50 nm Si-NPs. Error bars represent standard error of the mean (n = 3). Asterisks indicate significance (P = 0.05).
The long half-life (relative to a 12-h growth experiment) of luciferase likely contributed to the uncertainty in discerning subtle changes in QS over time. To further examine the role of functionalized and free β-CD in V. fischeri QS, we then examined changes in transcription of the lux operon during exposure to β-CD and β-CD functionalized Si-NPs.
Lux Operon Down-Regulated in Presence of β-CD Functionalized Si-NPs
Bioluminescence in V. fischeri is generated by the lux operon and is activated by 3OC6-HSL and C8-HSL. Transcription of the lux operon results in the production of luciferase, the enzyme responsible for bioluminescence. Table 1 summarizes the fold-changes calculated for luxA and luxR from the Ct values generated by qPCR with the Livak method (ΔΔCt) of both HSL-treated cultures.
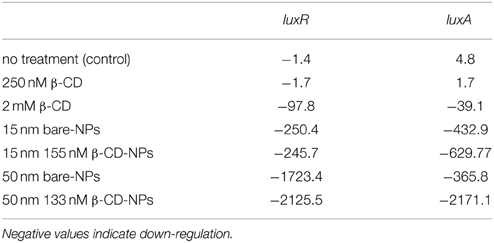
Table 1. Calculated fold change of transcription by V. fischeri after treatment exposures to 125 nM 3OC6- and 0.25 nM C8-HSLs.
The quantity of transcripts produced by untreated cultures (controls) and cultures treated with 250 nM β-CD were not markedly different, indicating that low concentrations of free β-CD are ineffective in down-regulating luxA and luxR at environmental levels of HSLs (data not shown). The addition of much higher (i.e., 2 mM) β-CD significantly reduced the expression of luxA and luxR transcripts, as indicated by the calculated fold changes (P = 0.05). In an attempt to more efficiently deliver β-CD to the bacterial cell, low concentrations of β-CD were applied via Si-NPs. As Table 1 indicates, both the functionalized and non-functionalized 15 nm Si-NPs reduced the expression of luxA and luxR transcripts to a similar extent. The data agree with the growth study, indicating that the influence of 15 nm Si-NPs functionalized with or without 155 nM β-CD is indistinguishable. Conversely, treatment with the larger nanoparticles (i.e., functionalized and non-functionalized 50 nm Si-NPs) resulted in significantly different fold changes (P = 0.05). The 133 nM β-CD, when bound to functionalized 50 nm Si-NPs, resulted in the greatest down-regulation of luxA and luxR transcripts out of all treatments, demonstrating the strong utility of this size of functionalized Si-NPs. The discontinuity of the bioluminescence measurements between the growth and transcription studies was likely influenced by the relatively long half-life of bioluminescence, which contributed to an inability to discern differences using bioluminescence during growth.
Discussion
The bioluminescence and transcription studies indicate that the 15 nm and 50 nm Si-NPs resulted in different levels of QS, although both Si-NPs were synthesized using the same method. Based on the nanoparticle synthesis methods and TGA analyses, we determined that the 15 nm Si-NPs carry 0.06 μmol β-CD/mg Si-NP complex, while the 50 nm Si-NPs carry 0.15 μmol β-CD/mg Si-NP complex, indicating that the surface densities of the two Si-NPs are different. Additionally, the surface area of the 50 nm Si-NPs is more than ten times the surface area of the 15 nm Si-NPs, which may have provided for more space between individual β-CD molecules and allowed for more efficient binding with the HSLs. Given that a 50 nm Si-NP can carry 1.7 times more β-CD than a 15 nm Si-NP, a higher dose of 15 nm Si-NPs was used to treat the samples to achieve comparable levels of β-CD. Three major possibilities exist to explain the difference in results. First, the higher concentration of Si-NPs aggregated in solution, making more of the functional groups unavailable on the smaller Si-NP. Alternatively, the β-CD may have been too dilute among the 15 nm Si-NPs and therefore did not act differently than dissolved β-CD. Lastly, it is possible that the 15 nm Si-NPs behave differently than 50 nm Si-NPs in an aqueous solution (Vertegel et al., 2004; Jiang et al., 2008). It has been shown that the surface modifications of silica affect the dispersion of Si-NPs, and that smaller-sized Si-NPs have unique physicochemical properties and optical absorption characteristics (Rahman et al., 2009). Therefore, smaller nanoparticles with a low surface density may behave differently than larger nanoparticles with a higher surface density. In summary, it is likely that the β-CD dose carried by the 15 nm Si-NPs behaved differently than the 50 nm Si-NPs, and therefore was unable to impact QS more significantly than free β-CD. The larger 50 nm Si-NPs, in contrast, were able to carry a higher dose of β-CD on fewer nanoparticles, and directly influenced QS in V. fischeri.
Previous studies have reported the use of QS antagonists and inhibitors to successfully interfere with bacterial QS (Dong et al., 2007; Hong et al., 2012; Stacy et al., 2012; Welsh et al., 2015). QS antagonists are often plant- and algal-based compounds that bind to LuxR-type receptors and prevent the complex from initating QS. Alternatively, QS inhibitors include both natural and synthetic compounds that either degrade (i.e., enzymes) or inhibit HSLs. Many propose that these anti-virulence strategies will decrease selective pressure; however, the risk of antibiotic resistance cannot be eliminated given the rapid rate of bacterial revolution (Rasko and Sperandio, 2010). Our nanoparticle-based QS inhibitors also reduce the risk of antibiotic resistance by delivering high concentrations of the quenching compound without excessive loss of the compound into the microbial environment. This method allows us to specficially target harmful bacteria while reducing side effects often incurred by broad-spectrum antibiotics. By delivering a highly concentrated compound to a localized environment, the bacteria are treated quickly and have less time to respond and adapt. Our study represents one of the first attempts to deliver QS quenching compounds to bacteria via functionalized nanoparticles. Other studies have used antimicrobial nanoparticles, such as silver and titanium dioxide, to kill bacteria or eliminate an infection. However, bacteria have evolved multiple resistance mechanisms against these nanoparticles (Rizzello and Pompa, 2014). Our study is significant because it utilizes the size and capacity of nanoparticles and the strength and relative safety of a QS quenching compound to target and weaken bacteria. This technology could be potentially redesigned to carry highly specific compounds in an effort to further reduce selective pressure and eliminate harmful side effects.
In this study, we have demonstrated that functionalized silica nanoparticles can be used to enhance the role of quenching agents in vitro. Specifically, we demonstrated that β-CD is able to bind HSLs and down-regulate bacterial QS genes. We have found that the quenching ability of β-CD was much greater on functionalized 50 nm Si-NPs, and provides a strong model for future designs. Here, with the advent of nanotechnology, we have demonstrated a unique ability (and tool) to specifically target bacterial communication, which may be used to penetrate biofilms and eliminate infections.
Materials and Methods
Diffusion Study
NMR diffusion measurements were performed on a Varian Mercury/VX 400 spectrometer operating at 400.273 MHz (1H). Pulsed-field gradient spin echoes with varying gradient intensity were collected with the Doneshot (Pelta et al., 2002) pulse sequence that was included with VNMRJ 2.2D software. The standard Varian Performa I pulsed field gradient amplifier and 5 mm broadband probe were capable of producing a maximum of 20 gauss/cm field gradients. Spectra were taken at ambient temperature with bipolar gradient pulses of 4 ms and a diffusion delay of 100 ms. All data processing was done with the Varian DOSY software package.
Bacteria and Growth Media
V. fischeri JB10 is a derivative of the ES114 strain that contains a chromosomal gfp reporter within the lux operon, resulting in luxI-gfp-CDABEG (Bose et al., 2007). The strain was prepared by and obtained from Professor Eric Stabb (Univ. Georgia, Athens, GA). V. fischeri JB10 was cultured from a glycerol stock and grown in marine broth (Difco 2216) to exponential phase. The cells were then transferred to defined minimal medium (Studer et al., 2008) for an optical density of approximately 0.1 (Abs600nm) for the growth and RNA extraction experiments.
Bioluminescence Response during Exposure to Nanoparticles
V. fischeri JB10 was grown in marine broth to exponential phase and then transferred (0.5%) to a defined minimal medium (Studer et al., 2008). For growth experiments, 96-well plates were incubated at 28°C in a Victor X Multilabel plate reader for 24 h. Optical density was measured at 600 nm and GFP fluorescence was measured with a 485/20 nm excitation filter and a 528/20 nm emission filter. Bioluminescence was also measured. All measurements were conducted every 2 h for 24 h following 10 s of vigorous shaking. The total volume of each well was 200 μ L, which included either 250 nM β-CD, 2 mM β-CD, 15 nm SiO2 nanoparticles (2.75 × 10−3 mg/mL NPs), 15 nm SiO2 nanoparticles functionalized with 155 nM β-CD (2.75 × 10−3 mg/mL NPs), 50 nm SiO2 nanoparticles (8.9 × 10−4 mg/mL NPs), or 50 nm SiO2 nanoparticles functionalized with 133 nM β-CD (8.9 × 10−4 mg/mL NPs). Cultures also included 125 nM 3OC6-HSL, 0.25 nM C8-HSL, or both 125 nM 3OC6-HSL and 0.25 nM C8-HSL. All treatments were added at time zero. A growth experiment was also performed with 600 nm SiO2 particles (data not shown, no cell growth occurred), α-cyclodextrin, 2-hydroxypropyl-β-CD, and methyl-β-CD. Cyclodextrins were obtained from Sigma Aldrich.
Quantitative PCR and Transcript Analysis: lux Transcription during Exposure to Nanoparticles
V. fischeri JB10 was grown in marine broth to exponential phase and then transferred (1%) to a defined minimal medium (Studer et al., 2008). Cultures were grown at 28°C and 200 rpm in 250 mL shake flasks with a working volume of 50 mL for 8 h. Treatments were added at 8 h and exposure/growth continued for 4 h. At 12 h of total growth, 2 mL of cells were harvested by centrifugation at 2000 × g (5 min). Cells were treated with 2 mg/mL lysozyme in TE buffer for 10 minutes and then homogenized with a needle and syringe. RNA was extracted with PureLink RNA Mini Kit (Ambion) and on-column DNA digestion was performed with PureLink PCR Micro Kit (Invitrogen). qPCR was performed on a BioRad CFX96 Real Time System with a C1000 Thermal Cycler. Data was analyzed by the Livak method (ΔΔCt) and fold changes were determined by using 2−ΔΔCt.
Primers were designed for luxR and luxA with NCBI's Primer-BLAST. Primer sequences for luxA were ATCCCCATCTTCGTGAACGG and ACAGAACATGGCCACGACAT. Primer sequences for luxR were CGTGGGCGAGTGAAGGAAAA and TGGCGCCAGTTAAAATTGCT. Primer sequences for the 16S rRNA gene were GTTTGATCATGGCTCAGATTG and CTACCTTGTTACGACTTCACC (Hoffmann et al., 2010). The 16S rRNA gene was used as the reference gene. All primers were ordered from Integrated DNA Technologies (Coralville, Iowa). Genomic DNA from V. fischeri JB10 was amplified with the primers and sequenced by Selah Genomics (Columbia, SC) to check the accuracy of the primers.
Reagents for Nanoparticle Synthesis
All chemicals were obtained from Fisher or Acros and used as received unless otherwise specified. 3-aminopropyldimethyl-ethoxysilane was obtained from Gelest, Inc. (Morrisville, PA).
Nanoparticle Characterization
The nanoparticles were characterized according to methods establish by Wang and Benicewicz (2013) and Wang et al. (2014). Infrared spectra were determined with a BioRad Excalibur FTS3000 spectrometer. UV-vis spectra were measured with a Perkin-Elmer Lambda 4C UV-vis spectrophotometer. Infrared spectra were recorded with a Perkin-Elmer Spectrum 100 spectrometer. TGA was conducted using a SDT Q600 TGA system (TA Instruments) with a temperature ramping from 25 to 900°C at a rate of 10°C/min under nitrogen.
Preparation of Monolayer Dye-Labeled β-CD Functionalized Silica Nanoparticles
A method has recently been developed for controlling the density of surface carboxylic acid functional groups coated onto the synthesized silicon dioxide Si-NPs (Cash et al., 2012; Wang and Benicewicz, 2013). Using this method, we coated a monolayer of β-CD onto the fluorescent Si-NPs via a coupling reaction between the β-CD hydroxyl groups and the carboxylic acids on the monolayer of dye-labeled Si-NPs. The carboxylic acid coated Si-NPs were prepared based on a ring-opening reaction between succinic anhydride and amino-functionalized silica Si-NPs with surface graft densities ranging from 0.10 to 0.65 groups/nm2. Thus, the graft density of β-CD functionalized Si-NPs were tailored by varying the feed ratio between bare Si-NPs and amino-silane compound. Specifically, a dimethylformamide (DMF) solution of β-CD (70.56 mg, 62.16 μmol), N,N′-Dicyclohexylcarbodiimide (DCC, 10.3 mg, 49.73 μmol) and 4-Dimethylaminopyridine (DMAP, 0.5063 mg, 4.144 μmol) were added to a 15 mL DMF solution of dye-labeled carboxylic acid-functionalized silica nanoparticles (graft density: 0.24 groups/nm2, 0.7281 g). The reaction was stirred at room temperature overnight. The reaction solution was then poured into 200 mL ethyl ether followed by centrifugation at 3000 rpm for 5 min. The recovered particles were then redispersed in 20 mL of ethanol and subjected to dialysis process to further remove impurities. The dye-labeled β-CD coated silica nanoparticles were finally dissolved in ethanol/water mixture solvents for further use. The preparation of dye-labeled monolayer carboxylic acid functionalized silica nanoparticles was based on our previous work (Cash et al., 2012).
Statistical Analysis
Statistical significance for all experiments was determined by a repeated measures analysis of variance test followed by an aposteriori pairwise muliple comparision procedure (SigmaPlot, Systat Software, San Jose, CA).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the US National Science Foundation (BME-1032579) and the NanoCenter at the University of South Carolina. Dr. BCB also acknowledges support from the SC SmartState Program.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2015.00189/abstract
References
Allison, K. R., Brynildsen, M. P., and Collins, J. J. (2011). Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473, 216–220. doi: 10.1038/nature10069
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Boettcher, K. J., and Ruby, E. G. (1990). Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172, 3701–3706.
Bose, J. L., Kim, U., Bartkowski, W., Gunsalus, R. P., Overley, A. M., Lyell, N. L., et al. (2007). Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol. Microbiol. 65, 538–553. doi: 10.1111/j.1365-2958.2007.05809.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Canton, R., and Morosini, M. I. (2011). Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiol. Rev. 35, 977–991. doi: 10.1111/j.1574-6976.2011.00295.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cash, B. M., Wang, L., and Benicewicz, B. C. (2012). The preparation and characterization of carboxylic acid-coated silica nanoparticles. J. Polym. Sci. A Polym. Chem. 50, 2533–2540. doi: 10.1002/pola.26029
Costerton, J. W., Cheng, K. J., Geesey, G. G., Ladd, T. I., Nickel, J. C., Dasgupta, M., et al. (1987). Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41, 435–464. doi: 10.1146/annurev.mi.41.100187.002251
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Davies, D. (2003). Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2, 114–122. doi: 10.1038/nrd1008
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Davies, D. G. (1998). The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280, 295–298. doi: 10.1126/science.280.5361.295
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
De, M., Ghosh, P. S., and Rotello, V. M. (2008). Applications of nanoparticles in biology. Adv. Mater. 20, 4225–4241. doi: 10.1002/adma.200703183
Defoirdt, T. (2013). Antivirulence therapy for animal production: filling an arsenal with novel weapons for sustainable disease control. PLoS Pathog. 9:e1003603. doi: 10.1371/journal.ppat.1003603
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Derrick, T. S., McCord, E. F., and Larive, C. K. (2002). Analysis of protein/ligand interactions with NMR diffusion measurements: the importance of eliminating the protein background. J. Magn. Reson. 155, 217–225. doi: 10.1006/jmre.2002.2513
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Dong, Y.-H., Wang, L.-H., and Zhang, L.-H. (2007). Quorum-quenching microbial infections: mechanisms and implications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362, 1201–1211. doi: 10.1098/rstb.2007.2045
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Eberhard, A., Burlingame, A. L., Eberhard, C., Kenyon, G. L., Nealson, K. H., and Oppenheimer, N. J. (1981). Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20, 2444–2449. doi: 10.1021/bi00512a013
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Engebrecht, J., Nealson, K., and Silverman, M. (1983). Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32, 773–781. doi: 10.1016/0092-8674(83)90063-6
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Engebrecht, J., and Silverman, M. (1984). Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. U.S.A. 81, 4152–4156. doi: 10.1073/pnas.81.13.4154
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hoffmann, M., Brown, E. W., Feng, P. C., Keys, C. E., Fischer, M., and Monday, S. R. (2010). PCR-based method for targeting 16S-23S rRNA intergenic spacer regions among Vibrio species. BMC Microbiol. 10:90. doi: 10.1186/1471-2180-10-90
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hoiby, N., Bjarnsholt, T., Givskov, M., Molin, S., and Ciofu, O. (2010). Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332. doi: 10.1016/j.ijantimicag.2009.12.011
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Hong, K.-W., Koh, C.-L., Sam, C.-K., Yin, W.-F., and Chan, K.-G. (2012). Quorum quenching revisited – from signal decays to signalling confusion. Sensors 12, 4661–4696. doi: 10.3390/s120404661
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jiang, W., Kim, B. Y., Rutka, J. T., and Chan, W. C. (2008). Nanoparticle-mediated cellular response is size-dependent. Nat. Nanotechnol. 3, 145–150. doi: 10.1038/nnano.2008.30
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kaplan, H. B., and Greenberg, E. P. (1985). Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J. Bacteriol. 163, 1210–1214.
Kato, N., Morohoshi, T., Nozawa, T., Matsumoto, H., and Ikeda, T. (2006). Control of gram-negative bacterial quorum sensing with cyclodextrin immobilized cellulose ether gel. J. Incl. Phenom. Macrocycl. Chem. 56, 55–59. doi: 10.1007/s10847-006-9060-y
LaSarre, B., and Federle, M. J. (2013). Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 77, 73. doi: 10.1128/MMBR.00046-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Li, J., Wang, L., and Benicewicz, B. C. (2013a). Synthesis of Janus nanoparticles via a combination of the reversible click reaction and “grafting to” strategies. Langmuir 29, 11547–11553. doi: 10.1021/la401990d
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Li, L., Sun, L., Song, Y., Wu, X., Zhou, X., Liu, Z., et al. (2013b). Screening of Actinobacillus pleuropneumoniae LuxS inhibitors. Curr. Microbiol. 67, 564–571. doi: 10.1007/s00284-013-0403-9
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Li, Y., Krentz, T. M., Wang, L., Benicewicz, B. C., and Schadler, L. S. (2014). Ligand engineering of polymer nanocomposites: from the simple to the complex. ACS Appl. Mater. Interfaces 6, 6005–6021. doi: 10.1021/am405332a
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Lupp, C., and Ruby, E. G. (2005). Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol. 187, 3620–3629. doi: 10.1128/JB.187.11.3620-3629.2005
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Madsen, J. S., Burmolle, M., Hansen, L. H., and Sorensen, S. J. (2012). The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 65, 183–195. doi: 10.1111/j.1574-695X.2012.00960.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Malvindi, M. A., Brunetti, V., Vecchio, G., Galeone, A., Cingolani, R., and Pompa, P. P. (2012). SiO2 nanoparticles biocompatibility and their potential for gene delivery and silencing. Nanoscale 4, 486–495. doi: 10.1039/C1NR11269D
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ng, W. L., and Bassler, B. L. (2009). Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43, 197–222. doi: 10.1146/annurev-genet-102108-134304
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Pelta, M. D., Morris, G. A., Stchedroff, M. J., and Hammond, S. J. (2002). A one-shot sequence for high-resolution diffusion-ordered spectroscopy. Magn. Reson. Chem. 40, S147–S152. doi: 10.1002/mrc.1107
Rahman, I. A., Vejayakumaran, P., Sipaut, C. S., Ismail, J., and Chee, C. K. (2009). Size-dependent physicochemical and optical properties of silica nanoparticles. Mater. Chem. Phys. 114, 328–332. doi: 10.1016/j.matchemphys.2008.09.068
Rasko, D. A., and Sperandio, V. (2010). Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 9, 117–128. doi: 10.1038/nrd3013
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Rizzello, L., and Pompa, P. P. (2014). Nanosilver-based antibacterial drugs and devices: mechanisms, methodological drawbacks, and guidelines. Chem. Soc. Rev. 43, 1501. doi: 10.1039/C3CS60218D
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Schuster, M., Sexton, D. J., Diggle, S. P., and Greenberg, E. P. (2013). Acyl-homoserine lactone quorum sensing: from evolution to application. Annu. Rev. Microbiol. 67, 43–63. doi: 10.1146/annurev-micro-092412-155635
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Stacy, D. M., Welsh, M. A., Rather, P. N., and Blackwell, H. E. (2012). Attenuation of quorum sensing in the pathogen Acinetobacter baumannii using non-native N-acyl homoserine lactones. ACS Chem. Biol. 7, 1719–1728. doi: 10.1021/cb300351x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Stevens, A. M., Dolan, K. M., and Greenberg, E. P. (1994). Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc. Natl. Acad. Sci. U.S.A. 91, 12619–12623. doi: 10.1073/pnas.91.26.12619
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Stewart, P., Rayner, J., Roe, F., and Rees, W. (2001). Biofilm penetration and disinfection efficacy of alkaline hypochlorite and chlorosulfamates. J. Appl. Microbiol. 91, 525–532. doi: 10.1046/j.1365-2672.2001.01413.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Studer, S. V., Mandel, M. J., and Ruby, E. G. (2008). AinS quorum sensing regulates the Vibrio fischeri acetate switch. J. Bacteriol. 190, 5915–5923. doi: 10.1128/JB.00148-08
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Szejtli, J. (1998). Introduction and general overview of cyclodextrin chemistry. Chem. Rev. 98, 1743–1754. doi: 10.1021/cr970022c
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vertegel, A. A., Siegel, R. W., and Dordick, J. S. (2004). Silica nanoparticle size influences the structure and enzymatic activity of adsorbed lysozyme. Langmuir 20, 6800–6807. doi: 10.1021/la0497200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Visick, K. L., Foster, J., Doino, J., McFall-Ngai, M., and Ruby, E. G. (2000). Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182, 4578–4586. doi: 10.1128/JB.182.16.4578-4586.2000
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wang, L., and Benicewicz, B. C. (2013). Synthesis and characterization of dye-labeled poly(methacrylic acid) grafted silica nanoparticles. ACS Macro Lett. 2, 173–176. doi: 10.1021/mz3006507
Wang, L., Chen, Y. P., Miller, K. P., Cash, B. M., Jones, S., Glenn, S., et al. (2014). Functionalised nanoparticles complexed with antibiotic efficiently kill MRSA and other bacteria. Chem. Commun. 50, 12030–12033. doi: 10.1039/c4cc04936e
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Welsh, M. A., Eibergen, N. R., Moore, J. D., and Blackwell, H. E. (2015). Small molecule disruption of quorum sensing cross-regulation in Pseudomonas aeruginosa causes major and unexpected alterations to virulence phenotypes. J. Am. Chem. Soc. 137, 1510–1519. doi: 10.1021/ja5110798
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: nanomedicine, nanoparticles, quorum sensing, quorum quenching
Citation: Miller KP, Wang L, Chen Y-P, Pellechia PJ, Benicewicz BC and Decho AW (2015) Engineering nanoparticles to silence bacterial communication. Front. Microbiol. 6:189. doi: 10.3389/fmicb.2015.00189
Received: 12 November 2014; Accepted: 20 February 2015;
Published: 10 March 2015.
Edited by:
Boris Lau, Unversity of Massachusetts Amherst, USAReviewed by:
Robert J. C. McLean, Texas State University, USABinh Thi Thanh Chu, Loyola University Chicago, USA
Copyright © 2015 Miller, Wang, Chen, Pellechia, Benicewicz and Decho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alan W. Decho, Microbial Interactions Laboratory, Department of Environmental Health Sciences, Public Health Research Center, Arnold School of Public Health, University of South Carolina, 921 Assembly Street, Columbia, SC 29208, USAYXdkZWNob0BtYWlsYm94LnNjLmVkdQ==
 Kristen P. Miller
Kristen P. Miller Lei Wang
Lei Wang Yung-Pin Chen1
Yung-Pin Chen1 Alan W. Decho
Alan W. Decho